Abstract
Aims: Precise regulation of cellular protein degradation is essential for maintaining protein and redox homeostasis. The ubiquitin proteasome system (UPS) represents one of the major degradation machineries, and UPS disturbances are strongly associated with neurodegeneration. We have previously shown that the transcription factor TCF11/Nrf1 induces antioxidant response element-mediated upregulation of UPS components in response to proteotoxic stress. Knockout of TCF11/Nrf1 is embryonically lethal, and therefore, the present investigation describes the role of oxidative stress in regulating TCF11/Nrf1-dependent proteasome expression in a model system relevant to Parkinson's disease.
Results: Using the human dopaminergic neuroblastoma cell line SH-SY5Y and mouse nigrostriatal organotypic slice cultures, gene and protein expression analysis and functional assays revealed oxidative stress is induced by the proteasome inhibitor epoxomicin or the mitochondrial complex I inhibitor rotenone and promotes the upregulation of proteasome expression and function mediated by TCF11/Nrf1 activation. In addition, we show that these stress conditions induce the unfolded protein response. TCF11/Nrf1, thus, has a cytoprotective function in response to oxidative and proteotoxic stress.
Innovation and Conclusion: We here demonstrate that adaption of the proteasome system in response to oxidative stress is dependent on TCF11/Nrf1 in this model system. We conclude that TCF11/Nrf1, therefore, plays a vital role in maintaining redox and protein homeostasis. This work provides a vital insight into the molecular mechanisms of neurodegeneration due to oxidative stress by rotenone, and further studies investigating the role of TCF11/Nrf1 in the human condition would be of considerable interest. Antioxid. Redox Signal. 25, 870–885.
Keywords: : neurodegeneration, oxidative stress, proteasome, rotenone, transcription factor TCF11/Nrf1, ubiquitin
Introduction
Intracellular protein degradation is a tightly regulated process, essential for protein homeostasis or proteostasis. The ubiquitin–proteasome system (UPS) is one of the major protein degradation machineries in eukaryotic cells, important for protein quality control and the regulation of processes such as cell cycle progression and differentiation, in addition to stress and immune responses (14). Proteins targeted for degradation by the UPS are labeled with ubiquitin and degraded by the 26S proteasome. The 26S proteasome consists of the 20S core complex and the 19S regulatory particle. The 20S complex is made up of four stacked heteroheptameric rings, the two outer alpha rings and the two inner beta rings harboring the proteolytic active sites β1, β2, and β5. The 19S particle performs substrate binding, deubiquitylation, unfolding, and translocation through the 20S pore, an ATP-dependent process (6, 21).
To adjust the UPS to changing environmental conditions, the degradation system is subordinated to precise regulatory mechanisms influencing proteasomal gene expression and composition. Besides upregulation of the standard 26S proteasome, under certain stressful conditions, the additional incorporation of interferon-induced catalytic immunosubunits or interaction with the PA28 regulator enables further rapid adaption of the proteasome system to inflammation (45, 57, 64). We and others have previously identified the transcription factor TCF11/Nrf1 as the main regulator of UPS-related gene expression. In response to proteasome inhibition, TCF11/Nrf1 induces the transcription of the 26S proteasome subunit genes resulting in newly synthesized proteasome complexes and increased proteolytic activity (56, 61).
Innovation.
The relationship between protein and redox homeostasis is currently poorly understood. We here uncovered a TCF11/Nrf1-dependent early cellular response mechanism, which enables the cell to adjust proteasomal protein degradation to contend oxidative damage and, thereby, promotes cell viability. Given that neurodegenerative disorders are known to involve disturbances of protein and redox homeostasis, these data indicate TCF11/Nrf1-dependent regulation may be an important therapeutic target.
TCF11/Nrf1 is encoded by the NFE2L1 gene and two different isoforms, namely Nrf1 (742 amino acids) and TCF11 (772 amino acids) are expressed in humans (10, 24, 40). TCF11/Nrf1, like Nrf2 and Nrf3, belongs to the Cap'n'Collar family of basic leucine zipper (CNC-bZIP) transcription factors (63). Nrf's (nuclear factor-erythroid 2-like) bind as heterodimers to antioxidant response elements (ARE) and thereby regulate the transcription of antioxidative and cytoprotective genes (25, 26, 66). Although Nrf2 is believed to be the main regulator of the antioxidative response, TCF11/Nrf1 also transactivates some detoxifying and antioxidant genes. The interplay between these transcription factors is, however, not fully understood and appears to be dependent on the type of stimulation and target gene (12, 30, 48, 66). The proteasome subunit and other UPS-related genes are under control of a promoter harboring ARE regions and are thus regulated by Nrf2 or TCF11/Nrf1 (32, 53, 61). In response to proteasome inhibition, however, the upregulation of the UPS depends on TCF11/Nrf1 rather than Nrf2 (61).
While the mechanism of oxidative stress-induced nuclear translocation of Nrf2 has been widely investigated (9), the mechanism of activation and nuclear translocation of TCF11/Nrf1 is poorly understood. Under nonstressed conditions, TCF11/Nrf1 resides into the endoplasmic reticulum (ER) membrane and is targeted by the E3 ubiquitin ligase HRD1 and p97/VCP-dependent ER-associated protein degradation (61). The turnover of nuclear TCF11/Nrf1 is mediated by the SCF (Skp1-Cullin1-F-box protein) E3 ubiquitin ligase containing the F-box protein Fbw7 or β-TrCP (4, 5, 65). Upon proteasome inhibition, TCF11/Nrf1 is retrotranslocated depending on p97/VCP and transferred into the nucleus. During the nuclear translocation process, TCF11/Nrf1 is known to be proteolytically processed, although the identity of the responsible protease is still under discussion (54, 58, 61).
The loss of the transcription factor in mice is lethal in the embryonic stage (11), and other TCF11/Nrf1 siRNA and knockout studies emphasize the importance of this transcription factor in maintaining cellular redox as well as protein homeostasis. The absence of Nrf1 sensitizes the cells to oxidative stress (33), and TCF11/Nrf1 deficiency also prohibits proteasome upregulation and aggravates cell death during proteasome inhibition (56, 61). UPS activity is critical for preventing the accumulation of oxidant-damaged and misfolded proteins (19, 31), and impairment of the UPS is associated with neurodegenerative disorders like Alzheimer's and Parkinson's diseases (15, 43). The specific loss of TCF11/Nrf1 in the mouse brain results in dysregulation of proteasome gene expression, accumulation of polyubiquitylated proteins, and neurodegeneration (29, 34).
Importantly, recent data demonstrate that chronic proteotoxic stress by impairment of proteasome function in patients with proteasome mutations causes autoinflammation (6, 7).
We here studied the adaption of the standard proteasome in the human dopaminergic neuroblastoma cell line, SH-SY5Y, upon exposure to the proteasome inhibitor epoxomicin or the mitochondrial complex I inhibitor rotenone. Treatment with rotenone results in the selective degeneration of dopaminergic neurons in the substantia nigra, and it is widely used in animal and cell culture models for Parkinson's disease (47). Rotenone causes mitochondrial dysfunction, leading to the production of reactive oxygen species (ROS), and UPS deficiency, resulting in the accumulation of ubiquitylated proteins (3, 36, 39, 68, 69). Researchers have now shown that Parkinson's disease in humans is associated with increased levels of oxidant-damaged proteins (1, 16), accumulation of ubiquitylated protein aggregates, increased unfolded protein response activation markers (23), and indicators of neuroinflammation, including activated glial cells (18, 42).
We here demonstrate that rotenone leads to a rapid induction of TCF11/Nrf1-dependent proteasome expression and further investigate the mechanism behind and the effects of TCF11/Nrf1 induction by inhibition of the proteasome. The current data show that depletion of the transcription factor with siRNA results in severe proteotoxic stress and enhances the sensitivity of SH-SY5Y neuronal cells to rotenone, indicating that TCF11/Nrf1 protects the cells against cytotoxic stress via upregulation of the UPS.
Results
Proteasome inhibition leads to oxidative stress and TCF11/Nrf1-dependent upregulation of proteasome subunit expression
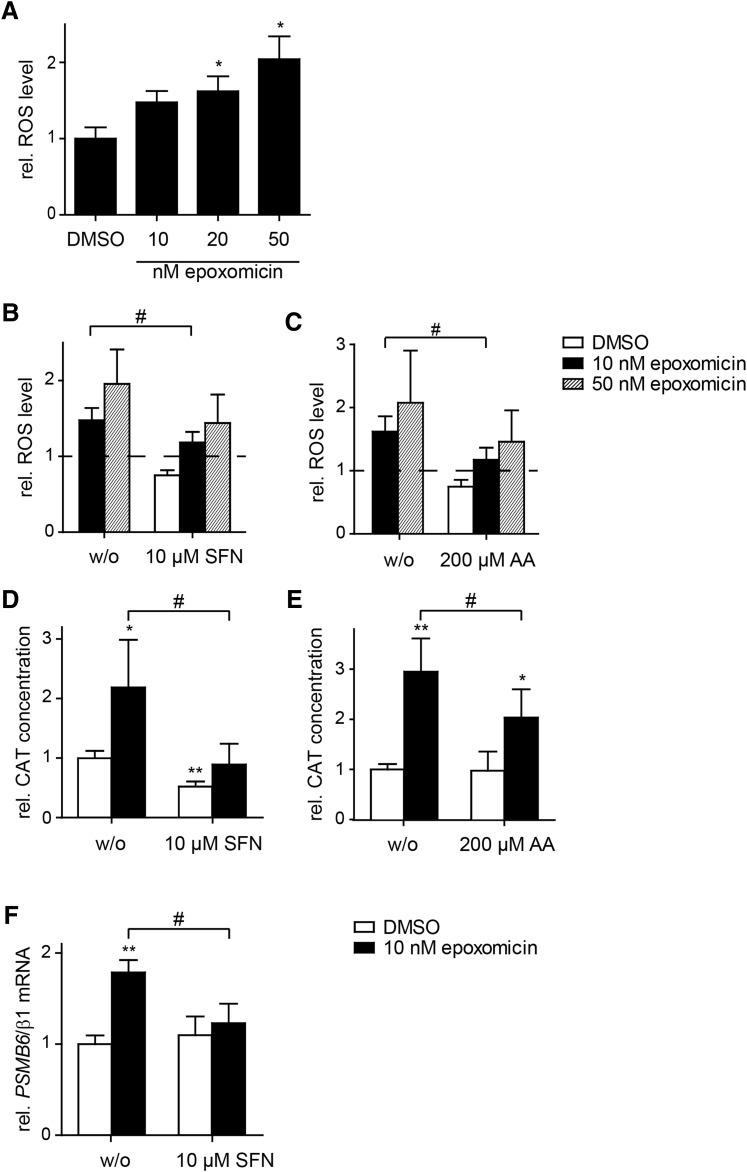
Previous studies have shown adaption of the UPS in response to proteasome inhibition (45, 61). To investigate the role of TCF11/Nrf1 in proteotoxic and oxidative stress, we induced its activation in SH-SY5Y cells by incubation with the proteasome inhibitor epoxomicin. Epoxomicin induced intracellular ROS production in SH-SY5Y cells in a concentration-dependent manner and led to accumulation of oxidant-damaged proteins, detected by staining of protein carbonyl groups (Fig. 1A and Supplementary Fig. S1C; Supplementary Data are available online at www.liebertpub.com/ars). Levels of polyubiquitylated proteins were increased by epoxomicin treatment, proving inhibition of protein clearance (Supplementary Fig. S1A).
FIG. 1.
Oxidative stress promotes epoxomicin-induced proteasome subunit expression. (A) Cultured SH-SY5Y cells were exposed to the indicated concentrations of epoxomicin or 0.1% DMSO for 18 h. Relative ROS level was measured by flow cytometry using the DCFH-DA dye. The mean values of the relative ROS level ± SD from four independent experiments are shown. (B, C) Cultured SH-SY5Y cells were cotreated with 10 nM epoxomicin or 0.1% DMSO and 10 μM SFN or 200 μM AA for 20 h. Relative ROS level was measured by flow cytometry using the DCFH-DA dye. The mean values of the relative ROS level ± SD from five (B) or six (C) independent experiments are shown. (D, E) SH-SY5Y cells were transfected with PSMB6/β1 promoter containing CAT reporter gene construct and cotransfected with pcDNA3.1-lacZ. For the last 20 h of transfection, cells were exposed to 10 nM epoxomicin or 0.1% DMSO and 10 μM SFN or 200 μM AA. The CAT concentration of cell lysates was determined by ELISA and normalized to β-Galactosidase activity. Shown are the mean values of the relative CAT concentration ± SD from six replicates of three independent experiments. (F) Cultured SH-SY5Y cells were cotreated with 10 nM epoxomicin or 0.1% DMSO and 10 μM SFN for 20 h. The relative mRNA level of PSMB6/β1 was analyzed by quantitative real-time PCR. Shown are the mean values ± SD from three independent experiments (*/#p < 0.05; **p < 0.005). AA, ascorbic acid; CAT, chloramphenicol acetyltransferase; DCFH-DA, dichlorodihydrofluorescein diacetate; DMSO, dimethyl sulfoxide; PCR, polymerase chain reaction; ROS, reactive oxygen species; SD, standard deviation; SFN, sulforaphane.
Furthermore, proteasome inhibition resulted in a concentration-dependent decrease in SH-SY5Y cell viability (Supplementary Fig. S1B). Similarly, acute exposure to the proteasome inhibitor MG-132 led to apoptosis shown by a rise in caspase-3/7 activity, thus verifying that proteasome inhibition leads to cytotoxicity and cell death (Supplementary Fig. S3A). All further experiments were performed with nonlethal concentrations of proteasome inhibitor.
Epoxomicin treatment increased the whole-cell TCF11/Nrf1 protein level in SH-SY5Y cells and other cell types as measured by immunoblotting, suggesting a general mechanism (Supplementary Fig. S2A). We observed a slower migrating larger form along with the faster migrating protein, which is most likely to be the deglycosylated and proteolytically processed active form of TCF11/Nrf1 (54, 61, 70). The TCF11/Nrf1 protein was more strongly increased in the undifferentiated compared to the differentiated SH-SY5Y cells. The undifferentiated cell model was preferred, because mainly standard proteasome subunits were induced. In the differentiated cell model also immunoproteasome subunits are induced by retinoic acid (RA; Supplementary Fig. S2A), which contradicts in vivo models indicating that immunoproteasome subunits are only expressed in glia cells and not in neurons (49). This supports the use of the undifferentiated model to study the mechanisms and effects of TCF11/Nrf1-mediated proteasome induction (Supplementary Fig. S2D, E).
In addition to the total rise in TCF11/Nrf1 protein level, subcellular fractionation separating cytosolic, membrane, nuclear, and chromatin-bound proteins revealed an increase of nuclear and chromatin-bound TCF11/Nrf1 (Supplementary Fig. S2B), demonstrating the translocation of the transcription factor. Transfer of TCF11/Nrf1 into the nucleus after epoxomicin treatment was also shown by the colocalization of TCF11/Nrf1 with the nuclear staining (Supplementary Fig. S2C).
We next investigated the proteasome subunit regulation by using a reporter gene assay and quantitative real-time polymerase chain reaction (PCR). Cells expressing a chloramphenicol acetyltransferase (CAT) reporter construct under control of the −1500 bp region of the PSMB6/β1 promoter showed an epoxomicin-dependent significant increase in CAT expression, indicating PSMB6/β1 upregulation (Supplementary Fig. S2D). Furthermore, we observed induction of PSMA2/α2 and PSMC4/Rpt3 mRNA in response to proteasome inhibition in SH-SY5Y cells (Supplementary Fig. S2E). The epoxomicin-induced rise in the proteasome subunit mRNA level in SH-SY5Y cells was significantly attenuated by TCF11/Nrf1 depletion using siRNA, implying that the epoxomicin-induced upregulation of the proteasome subunit expression is TCF11/Nrf1 dependent (Supplementary Fig. S2E). The efficiency of TCF11/Nrf1 depletion was verified by quantitative real-time PCR showing a reduction of 70% (Fig. S2F).
In addition, we determined activation of TCF11/Nrf1 in primary keratinocytes from healthy subjects and patients with proteasome mutations (Supplementary Fig. S2G), who suffer from chronic impairment of proteolytic function (6, 7). Moreover, TCF11/Nrf1 binds specifically to proteasome promoters exemplified by the PSMB6 gene as shown by chromatin immunoprecipitation (ChIP). The specificity of the TCF11/Nrf1 antibody in immunoprecipitation was proven by immunoblot (Supplementary Fig. S2H).
The epoxomicin-induced rise in ROS levels was diminished by cotreatment of the SH-SY5Y cells with the antioxidants sulforaphane (SFN) or ascorbic acid (AA; Fig. 1B, C), and the antioxidants attenuated the CAT reporter induction (Fig. 1D, E). A reduction of the increase in PSMB6/β1 mRNA level in response to proteasome inhibition was, additionally, observed in SFN-treated cells (Fig. 1F). These results show that ROS formation is required for epoxomicin-induced upregulation of the proteasome system.
Rotenone induces oxidative stress and upregulation of the proteasome system
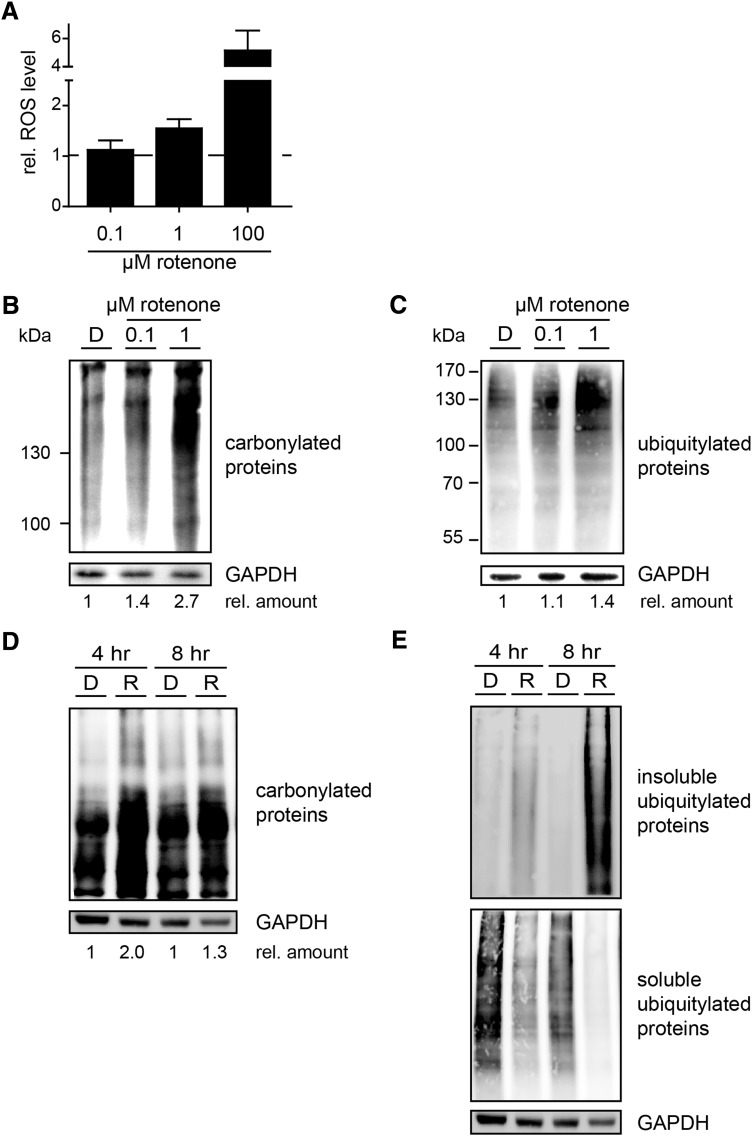
We next investigated how the mitochondrial inhibitor rotenone affects the proteasome system in SH-SY5Y cells. Rotenone treatment led to a concentration-dependent (0.1–100 μM) increase in intracellular ROS production (Fig. 2A). As expected, carbonylated proteins, indicating oxidant damage, were accumulated in cells treated with 100 nM rotenone and further increased after exposure to 1 μM of the complex I inhibitor (Fig. 2B). An accumulation of polyubiquitylated proteins was observed via immunoblotting against ubiquitin (Fig. 2C). These results were also confirmed in an ex vivo model using nigrostriatal organotypic slice cultures (NOSCs) of mouse brains treated with 100 nM rotenone for 4 and 8 h (Fig. 2D, E). Of note, rotenone elevated the levels of insoluble and detergent-resistant ubiquitin conjugates, indicating more protein damage and aggregation (Fig. 2E).
FIG. 2.
Rotenone exposure leads to oxidative and proteotoxic stress. (A, B) Cultured SH-SY5Y cells were exposed for 6 h to the indicated concentrations of rotenone. (A) The relative intracellular ROS level was measured by flow cytometry using the DCFH-DA dye. The mean of the relative ROS level ± SD from three independent experiments is shown. (B) Levels of carbonylated proteins and GAPDH in cell lysates were analyzed by immunoblotting. Carbonylated proteins were marked using DNPH and detected using an antibody against DNP. The numbers indicate the relative increase in carbonyl proteins (rotenone/DMSO). (C) Cultured SH-SY5Y cells were exposed to the indicated concentrations of rotenone or 0.1% DMSO for 24 h. Polyubiquitylated proteins and GAPDH were analyzed by immunoblotting. The numbers indicate the relative increase in ubiquitin conjugates (rotenone/DMSO). (D) NOSCs of mouse brains were treated with 100 nM rotenone for 4 and 8 h, and extracted proteins were stained for carbonyls indicating an increase in oxidant-damaged proteins in rotenone-treated NOSCs. The numbers indicate the relative increase in carbonyl proteins (rel. amount). (E) NOSCs treated as in (D) were fractionated into detergent soluble and detergent-resistant fraction by urea and stained for ubiquitin. Rotenone elevated the levels of insoluble and detergent-resistant ubiquitin conjugates, indicating more protein damage and aggregation. DNP, 2,4-dinitrophenol; DNPH, 2,4-dinitrophenylhydrazine; NOSC, nigrostriatal organotypic slice culture.
In addition, we here confirmed that rotenone induces cell death in a concentration-dependent manner in this SH-SY5Y model by determining caspase-3/7 activity and cell viability (Supplementary Fig. S3A, B). Furthermore, rotenone exposure activated the unfolded protein response, shown by increased as well as spliced XBP-1 mRNA and the phosphorylation of eIF2α (Supplementary Fig. S3C, D). From these experiments, it becomes evident that the metabolic function (XTT-test) is already slightly affected by low rotenone concentrations of 1 nM (Supplementary Fig. S3A), whereas the activation of apoptosis by caspase-3/7 activity is not increased at 100 nM rotenone treatment for 24 h (Supplementary Fig. S3B). Thus, we used the nontoxic dose of 100 nM rotenone for shorter time points further on.
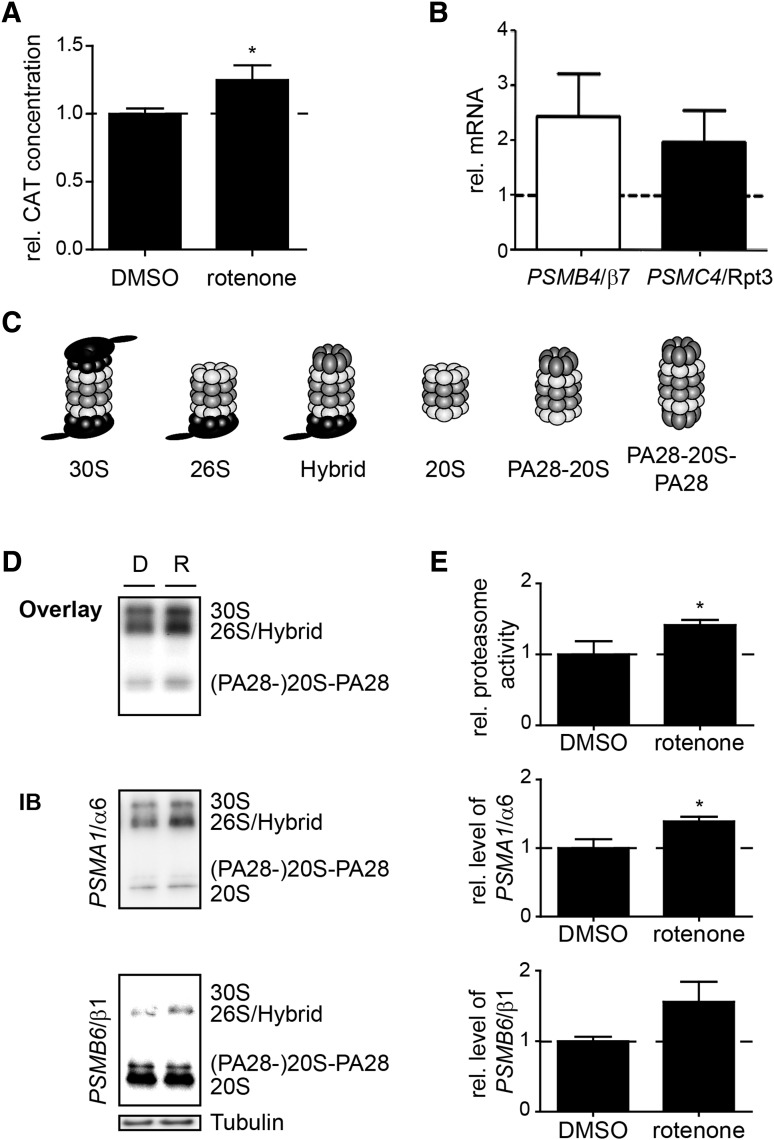
Exposure to rotenone was shown to affect UPS component expression and function. Cells transfected with the CAT reporter construct showed a significant increase in CAT expression after exposure to rotenone, indicating PSMB6/β1 upregulation (Fig. 3A). Our NOSCs model treated with 100 nM rotenone for 8 h showed upregulation of proteasome genes exemplified by PSMB4/β7 (20S subunit) and PSMC4/Rpt3 (19S subunit) (Fig. 3B). The upregulation of PSMB6/β1 in SH-SY5Y cells could be counteracted by the antioxidant AA (Supplementary Fig. S3E). We then investigated proteasome function after rotenone exposure. The activity of the 20S, 26S/hybrid, and 30S complexes (for illustration, see Fig. 3C) in native polyacrylamide gel electrophoresis (PAGE) substrate overlay experiments was increased, which correlates with the total amount of the complexes (Fig. 3D, E). The presence of PA28-containing complexes was proven by immunoblotting against PSME2/PA28β (Supplementary Fig. S4).
FIG. 3.
Short rotenone exposure increases the amount of proteasome complexes. (A) SH-SY5Y cells were transfected with the PSMB6/β1 promoter containing the CAT reporter gene construct and cotransfected with pcDNA3.1-lacZ. The next day, cells were exposed to 100 nM rotenone or 0.1% DMSO for 4 h. The CAT concentration of cell lysates was determined by ELISA and corrected to β-galactosidase activity. Shown are the mean values of the relative CAT concentration normalized to DMSO control ± SD from six replicates of three independent experiments. (B) The relative mRNA levels of PSMC4/Rpt3 and PSMB4/β7 from NOSCs treated with 100 nM rotenone for 8 h were analyzed by quantitative real-time PCR. Shown are the mean values normalized to the DMSO control (set to 1) ± SD from pooled four NOSCs from at least six mouse pups. (C) Schematic illustration of the proteasome complexes relevant to (D). (D) Cultured SH-SY5Y cells were exposed to 100 nM rotenone or 0.1% DMSO for 8 h. Once proteasome complexes were separated in native gels, the activity was analyzed by an overlay with the fluorogenic Suc-LLVY-AMC substrate. The levels of PSMA1/α6 and PSMB6/β1 were analyzed by immunoblotting. Acting as a loading control, the native lysates were separated by SDS-PAGE and immunoblotted against β-tubulin. (E) Densitometrical analysis of substrate overlays relative proteasome activity and the corresponding PSMA1/α6 and PSMB6/β1 IB. Shown are the mean values ± SD from three (activity and α6 IB) or two (β1 IB) independent experiments (*p < 0.5). IB, immunoblots; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Rotenone induces nuclear translocation of TCF11/Nrf1
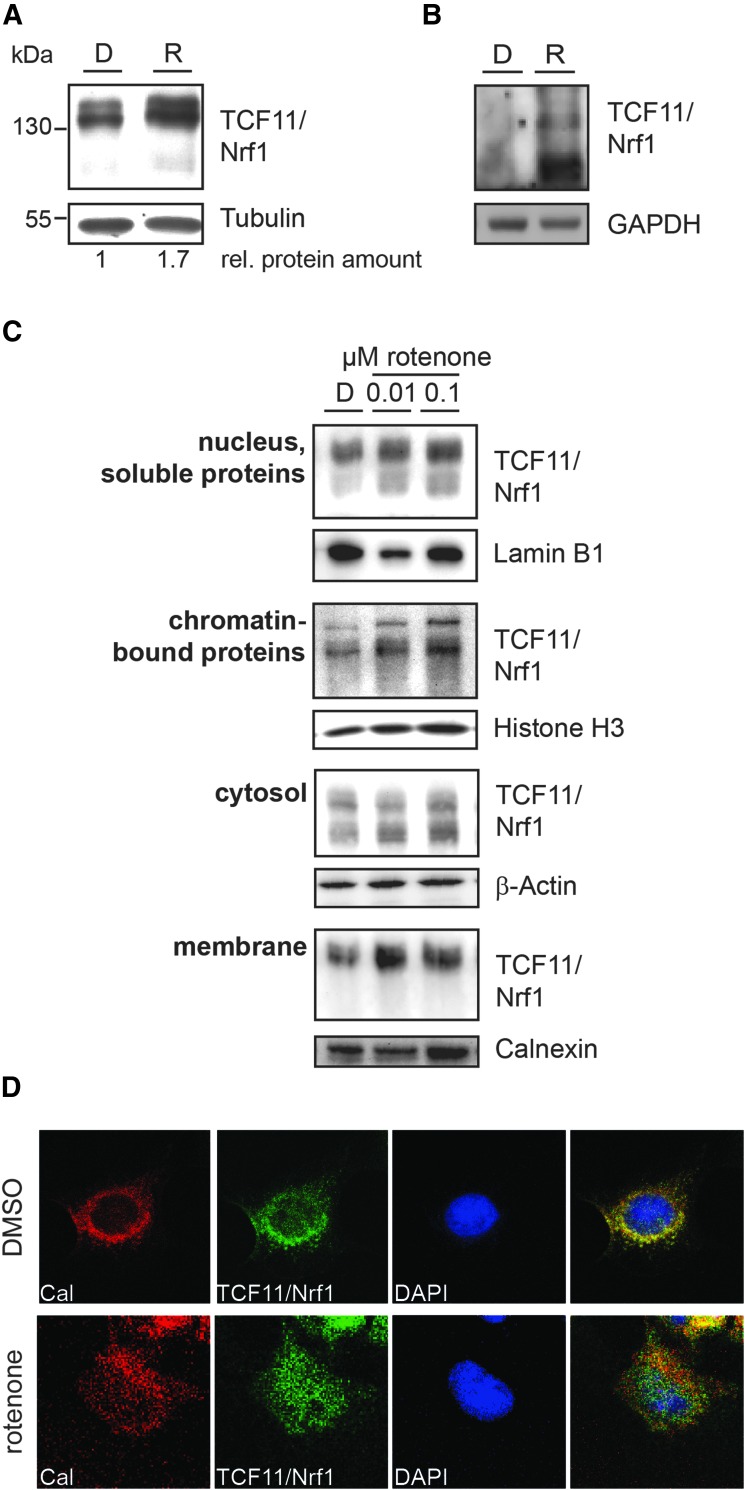
We next explored whether TCF11/Nrf1 is translocated into the nucleus in response to rotenone-induced oxidative stress in SH-SY5Y cells. In a time course experiment, immunoblotting against TCF11/Nrf1 revealed an increase in the total TCF11/Nrf1 level after rotenone treatment (Fig. 4A). Furthermore, a significant rotenone-induced activation of Nrf1 revealed the NOSC model (Fig. 4B).
FIG. 4.
Rotenone increases TCF11/Nrf1 level and promotes its nuclear translocation. (A) Cultured SH-SY5Y cells were exposed to 100 nM rotenone (R) or 0.1% DMSO (D) for 4 h. Levels of TCF11/Nrf1 and β-tubulin in cell lysates were analyzed by immunoblotting. The numbers indicate the relative increase in activated TCF11/Nrf1 (rotenone/DMSO). (B) NOSCs were treated with 100 nM rotenone for 8 h, and extracted proteins were stained for activated TCF11/Nrf1. (C) Cultured SH-SY5Y cells were exposed to rotenone or 0.1% DMSO (D) for 3 h and afterward separated into cytosolic, membrane, soluble nuclear, and chromatin-bound fractions. Levels of TCF11/Nrf1 and compartment controls Lamin B1, Histon H3, Calnexin, and β-actin were analyzed by immunoblotting. (D) For immunocytochemistry, cells were incubated with 100 nM rotenone or 0.1% DMSO for 2 h and stained for the ER marker calnexin (red) and TCF11/Nrf1 (green). Nuclei are stained using DAPI (blue). ER, endoplasmic reticulum; Nrf, nuclear factor-erythroid-2-related factor.
Subcellular fractionation separating cytosolic, membrane, nuclear, and chromatin-bound proteins showed that rotenone induced an increase in the TCF11/Nrf1 protein level in all fractions, with an increase observed in the nuclear and chromatin-bound fractions (Fig. 4C), demonstrating the translocation of the transcription factor into the nucleus. The pattern for the subcellular distribution after rotenone treatment was comparable to that observed in cells exposed to epoxomicin (Supplementary Fig. S2B and Fig. 4C).
The nuclear translocation of TCF11/Nrf1 was verified using immunofluorescence analysis with ER staining for control and nuclear staining upon rotenone exposure (Fig. 4D). A similar pattern was seen after treatment with epoxomicin (Supplementary Fig. S2C) and also after treatment with the mitochondrial toxin arsenite (Supplementary Fig. S5).
Rotenone-induced proteasome expression and synthesis are dependent on TCF11/Nrf1
The increase of nuclear TCF11/Nrf1 as well as the upregulation of the proteasome complex in rotenone-treated SH-SY5Y cells indicate that, under these conditions, TCF11/Nrf1 is transcriptionally active and induces proteasome expression.
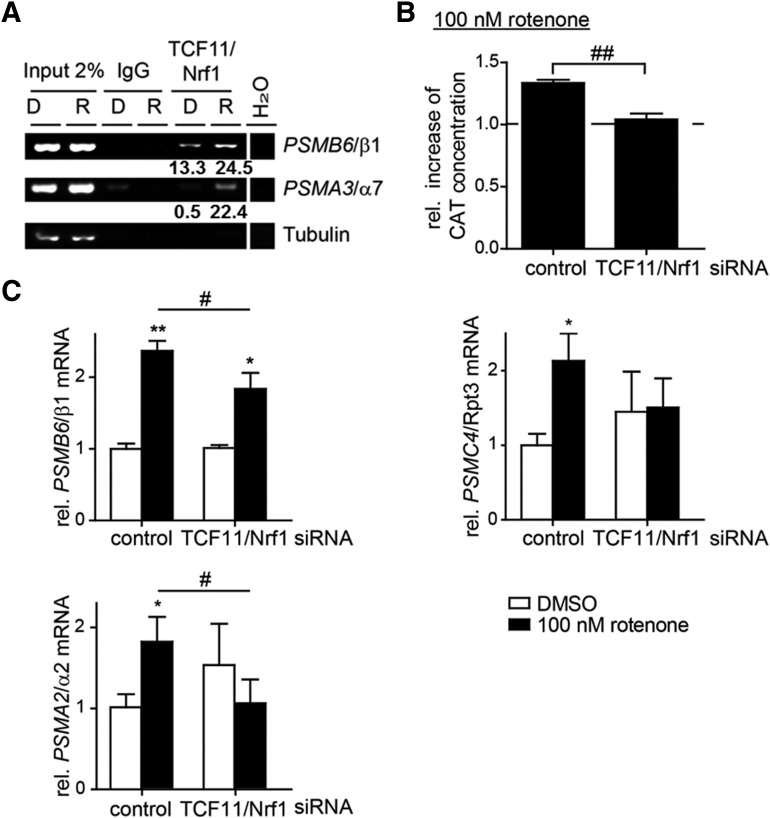
To confirm this hypothesis, we assessed binding of TCF11/Nrf1 to the ARE region of the PSMB6/β1 and PSMA3/α7 promoter by ChIP. The intensity of the coimmunoprecipitated promoter DNA was increased after rotenone exposure in comparison to the dimethyl sulfoxide (DMSO) control, implying increased TCF11/Nrf1 binding to these ARE regions (Fig. 5A). To investigate whether the induction of the proteasome subunit expression depends on TCF11/Nrf1, we depleted TCF11/Nrf1 by siRNA. Indeed, the increase of the CAT reporter after rotenone exposure dropped to control levels in TCF11/Nrf1-depleted cells (Fig. 5B), indicating PSMB6/β1 upregulation is mostly dependent on TCF11/Nrf1. Furthermore, the upregulation of the PSMB6/β1, PSMA2/α2, and PSMC4/Rpt3 mRNA level in control siRNA-transfected cells (Fig. 5C) induced by rotenone treatment was reduced in TCF11/Nrf1-depleted cells.
FIG. 5.
Rotenone-induced proteasome subunit transcription depends on TCF11/Nrf1. (A) Cultured SH-SY5Y cells were exposed to 100 nM rotenone for 3 h. Transcription factor DNA complexes were chromatin immunoprecipitated by an antibody against TCF11/Nrf1 or an unspecific IgG antibody used as the control. The binding of the transcription factors to the PSMB6/β1 or PSMA3/α7 promoter was analyzed by PCR. Primers amplifying a promoter region of tubulin were used for the negative control. Densitometric evaluation of this experiment is shown as the ratio between TCF11/Nrf1 precipitates to the mock (IgG) signal (TCF11/Nrf1/IgG) as numbers below the lanes. (B) Cultured SH-SY5Y cells were transfected with the PSMB6/β1 promoter containing the CAT reporter gene construct and cotransfected with pcDNA3.1-lacZ. The next day, cells were exposed to rotenone for 4 h. The CAT concentration of cell lysates was determined by ELISA and normalized to β-galactosidase activity. Shown are the mean values of the fold increase in relative CAT concentration corrected to the DMSO control ± SD from six replicates of three independent experiments. (C) For the last 16–18 h of a 48-h TCF11/Nrf1 knockdown, SH-SY5Y cells were exposed to 100 nM rotenone. The relative mRNA levels of PSMC4/Rpt3, PSMA2/α2, PSMB6/β1, and TCF11/Nrf1 were analyzed by quantitative real-time PCR. Shown are the mean values normalized to the DMSO control ± SD from three independent experiments (rotenone/DMSO) (*/#p < 0.05; **/##p < 0.005).
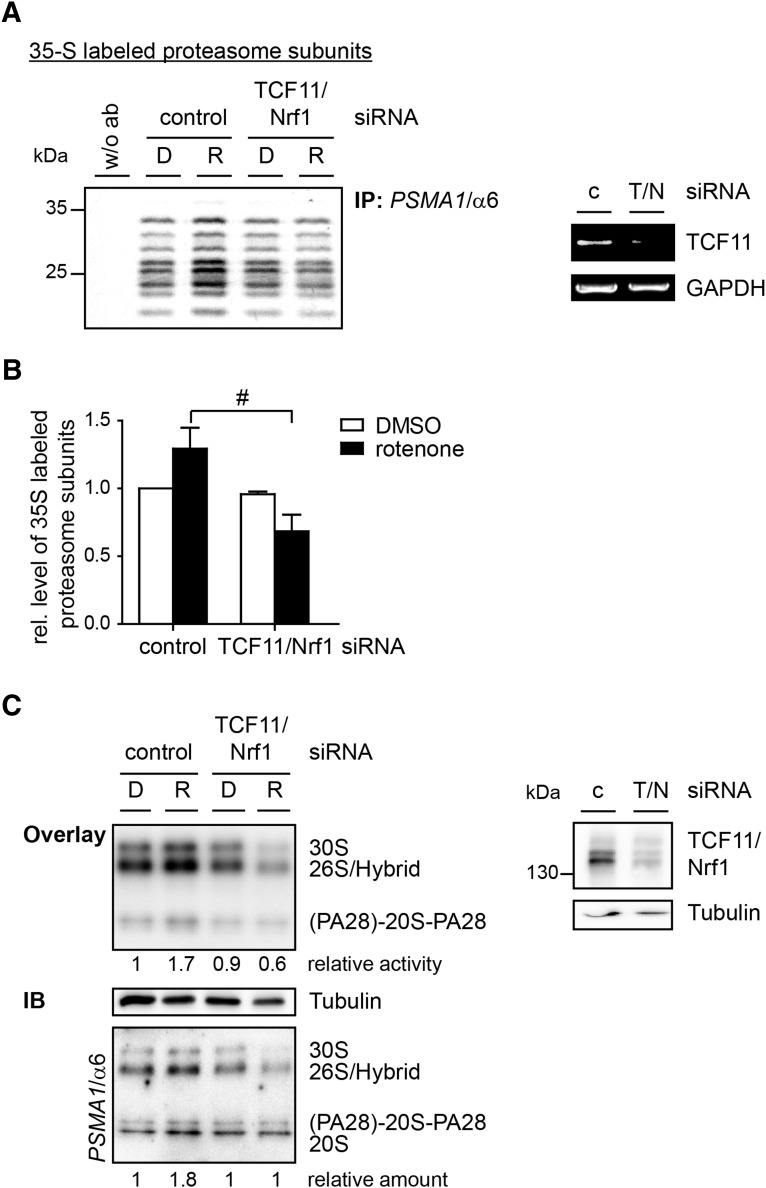
We next examined the effect of TCF11/Nrf1 on formation of new proteasome complexes using TCF11/Nrf1 siRNA-transfected cells. Newly synthesized proteins were metabolically labeled and the proteasome complexes then immunoprecipitated. The amount of newly synthesized proteasome complexes increased in control cells exposed to rotenone, and this effect was abolished by TCF11/Nrf1 (Fig. 6A, B).
FIG. 6.
Rotenone induced proteasome expression depends on TCF11/Nrf1. (A, B) TCF11/Nrf1-deficient SH-SY5Y cells or control cells were exposed to 100 nM rotenone (R) or 0.1% DMSO (D) for 20 h. The cultured cells were metabolically labeled with [35S] methionine/[35S] cysteine. The 20S proteasome was then immunoprecipitated with an antibody against PSMA1/α6 and separated by SDS-PAGE (A). The knockdown efficiency was analyzed by PCR (right). (B) The mean values of the relative amount of labeled proteasome subunit relative to the input signal ± SD from the densitometric analysis from three independent experiments are plotted. (C) Cultured SH-SY5Y cells were exposed to 100 nM rotenone (R) or 0.1% DMSO (D) for 8 h. Once proteasome complexes were separated in native gels, activity was analyzed by an overlay with the fluorogenic Suc-LLVY-AMC substrate. The level of PSMA1/α6 was analyzed by immunoblotting. The loading was proofed by SDS-PAGE and immunoblotting against β-tubulin. For a knockdown control, RIPA lysates were separated by SDS-PAGE and analyzed by immunoblotting (right) (#p < 0.05).
To verify that the increased expression of proteasome subunits also results in a higher amount of active proteasome complexes, we separated the complexes in native gels and performed an overlay with a fluorogenic substrate. We observed a rise in the proteasome activity in rotenone-treated control cells, which was not apparent in TCF11/Nrf1-depleted cells (Fig. 6C). These alterations in activity correlate with the alteration in the amount of proteasome complexes, shown by immunoblotting against PSMA1/α6. The knockdown efficiency was here checked by immunoblotting against TCF11/Nrf1 using tubulin as a loading control.
Taken together, these findings demonstrate that rotenone induces proteasome expression in a TCF11/Nrf1-dependent manner, resulting in newly assembled and active proteasome complexes.
TCF11/Nrf1 prevents cell death following rotenone exposure
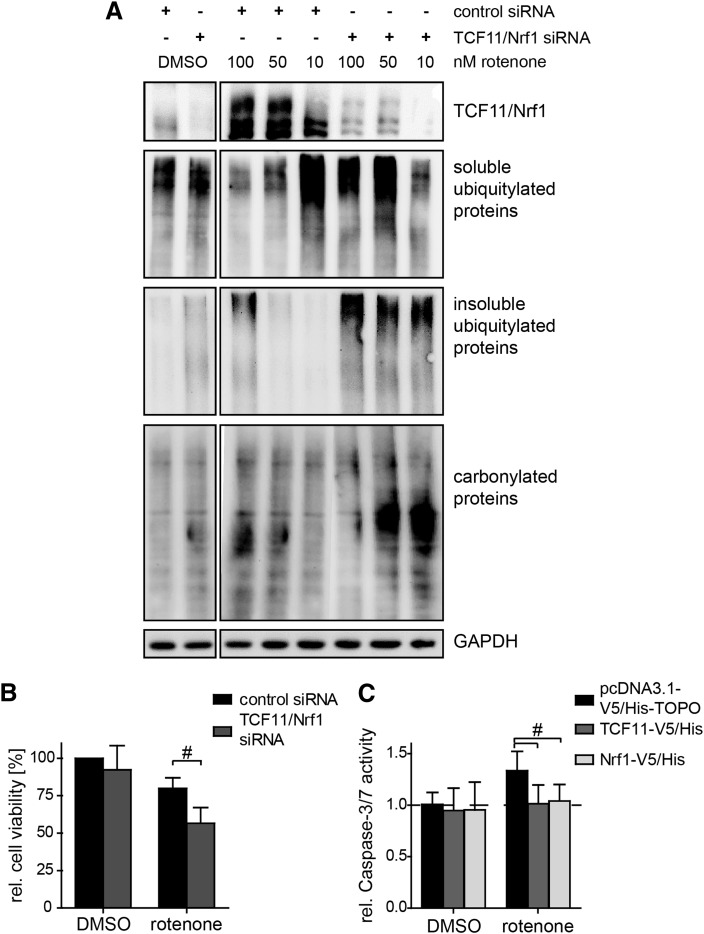
We finally showed that TCF11/Nrf1 impacts the extent of protein damage and survival of cells after rotenone exposure. Increasing rotenone concentrations trigger the formation of protein carbonyls along with accumulation of ubiquitin conjugates. In TCF11/Nrf1-depleted cells, however, the increase in protein carbonyls was more pronounced and the major proportion of ubiquitin conjugates was present in the insoluble detergent-resistant fraction of the protein indicating highest levels of protein damage (Fig. 7A). Moreover, the staining for the insoluble fraction of ubiquitin conjugates was even increased in untreated cells depleted for TCF11/Nrf1, which indicated that TCF11/Nrf1 is also important for basal transcription of proteasome subunits.
FIG. 7.
TCF11/Nrf1 diminishes rotenone-induced proteotoxic stress and cell death. (A) SH-SY5Y cells were treated with the rotenone concentrations indicated for 6 h and depleted or not for TCF11/Nrf1 by siRNA. Proteins were extracted by RIPA and were fractionated into detergent-soluble and detergent-resistant fraction by urea and stained for TCF11/Nrf1, GAPDH (loading control), ubiquitin conjugates, and carbonyls. TCF11/Nrf1 depletion cells promote the accumulation of ubiquitin conjugates in the detergent-resistant fraction. (B) For the final 20 h of a 48 h TCF11/Nrf1 depletion, SH-SY5Y cells were exposed to 1 nM rotenone and cell viability analyzed by XTT assay. Shown are the mean values of the relative cell viability ± SD from three independent experiments. (C) Cultured SH-SY5Y cells were transfected with TCF11-V5/His, Nrf1-V5/His, or pcDNA3.1-V5/His-TOPO and then exposed to 10 μM rotenone for 24 h. The caspase-3/7 activity was then measured. Shown are the mean values of the relative activity ± SD from nine replicates of three independent experiments. The overexpression was proofed by immunoblotting against the V5 tag, TCF11/Nrf1, and β-tubulin (#p < 0.05).
Rotenone exposure decreased cell viability as measured using the XTT assay, and the decline was significantly more pronounced in TCF11/Nrf1-depleted cells, indicating that TCF11/Nrf1 positively affects cell viability (Fig. 7B). In addition, we analyzed caspase-3/7 activity in TCF11 and Nrf1-overexpressing cells incubated with rotenone (Fig. 7C). In cells transfected with the empty control vector (pcDNA3.1-V5/His-TOPO), rotenone induced caspase-3/7 activity, representing apoptosis. The overexpression of TCF11 and Nrf1 prevented this increase in caspase activity, implying that the transcription factor prevents cell death due to apoptosis.
Discussion
The UPS is constantly being adapted to match changing proteolytic requirements. Disturbance of the UPS is associated with neurodegenerative disorders like Alzheimer's and Parkinson's disease, in which misfolded ubiquitylated proteins accumulate intra- or extracellularly (15, 30, 31, 43). Protein accumulation may result from impairments in ubiquitylation or substrate delivery to the proteasome, or the apparent age-related decrease in proteasome activity (41). We here demonstrate that 26S proteasome expression is induced in response to inhibition of the mitochondrial complex I by rotenone or proteasome inhibition in a TCF11/Nrf1-dependent manner and that TCF11/Nrf1 thereby plays a cytoprotective role.
The present data confirm and extend previous findings identifying TCF11/Nrf1 as the main regulator of the UPS in response to proteotoxic and oxidative stress (61). Proteasome inhibition leads to oxidative stress shown by an increase in the intracellular ROS level and the accumulation of oxidative damaged proteins. Recently, Kirstein et al. demonstrated, using a redox sensor, that the redox state shifts to more oxidizing conditions in the cytosol in response to proteotoxic stress (27). Oxidative stress is required for the epoxomicin-induced activation of TCF11/Nrf1. Beside proteasome inhibition, rotenone exposure leads to the upregulation and nuclear translocation of TCF11/Nrf1 resulting in increased transcription, translation, and, finally, assembly of active proteasome complexes. TCF11/Nrf1 transactivates proteasome expression via binding to the ARE promoter regions in response to rotenone-induced oxidative stress, and depletion of the transcription factor partly abolishes the induction of proteasome subunit expression. In murine C2C12 cells, acute exposure to rotenone has been shown to suppress the nuclear translocation of TCF11/Nrf1 (17), but in human keratinocytes, TCF11/Nrf1 contributes to the cellular response to arsenite, which also induces oxidative stress by regulating cytoprotective genes like GCLC or GCLM, encoding for the glutamate cysteine ligase (71). These data may therefore signal differences relating to species or cell type. In the SH-SY5Y model used here, TCF11/Nrf1 is necessary for the upregulation of proteasome subunits in response to cellular stress, and levels of active, high-molecular, ATP- and ubiquitin-dependent 19S-containing proteasome complexes were increased. Newly synthesized proteins are very vulnerable to oxidation, and oxidant-damaged proteins induced by interferon exposure were reported to be partly polyubiquitinated, implying a demand on 26S/30S activity (44, 57). Previous work has shown that H2O2 exposure leads to an upregulation of the proteasome system, especially of the interferon-inducible immunoproteasome and the PA28 regulator, and that depletion of the proteasome subunits or PA28 regulator sensitizes the cells to H2O2-induced cell death. These data from Pickering et al. highlight the importance of the proteasome system in the cellular response to oxidative stress, even though upregulation of the 19S regulatory particle was not observed (51, 52). Rotenone quickly induces intracellular ROS formation and oxidative stress, but rotenone treatment is clearly not directly comparable to pure application of H2O2. The current data do not exclude the role of inducible components of the proteasome system; indeed, we believe that the ATP-and ubiquitin-independent PA28 regulator is additionally induced and binds to the 20S core complex in response to rotenone exposure (Supplementary Fig. S4). We argue that there is a TCF11/Nrf1-dependent upregulation of the 26S/30S proteasome concurrent to the upregulation of inducible proteasome subunits upon rotenone-induced stress.
Alongside the impairment of redox homeostasis, several studies have shown disturbances of protein homeostasis in response to rotenone. Protein aggregates, specifically those containing α-synuclein and ubiquitin, were observed in vivo as well as in cell culture (3, 35, 60), and here, rotenone treatment led to an accumulation of ubiquitylated proteins indicating impairment of the UPS. Previous work reported that rotenone diminishes proteasome activity, although possibly after a period of increased proteasome activity preceding a collapse (46, 59). We propose the effect of rotenone is strongly dependent on concentration and exposure time. We suggest that the cell first tries to compensate for the impairment of redox and protein homeostasis by an upregulation of the proteasome system, assumedly dependent on the maintenance of ATP/ADP levels. However, intracellular oxidative stress will lead to protein aggregation and cell death if homeostatic mechanisms prove insufficient. This is supported by an in vivo study showing that, after rotenone injection, brain areas showing lesions have a decreased proteasome activity in contrast with areas without lesions, assumed to be in a lesser state of degeneration, where the activity is increased (2).
Oxidative and proteotoxic stress interfere with ER homeostasis and activate the unfolded protein response, which, in concert with UPS dysregulation, creates a vicious cycle leading to protein aggregation, inflammation, and cell death, characteristic of neurodegenerative disorders as well as proteasome-associated autoinflammatory syndromes (PRAAS) (6, 7). In keratinocytes of these patients, TCF11/Nrf1 is permanently activated most likely to compensate for the impaired proteasome function. Intracellular oxidative stress, proteotoxic stress, ER stress, and finally cell death are all outcomes of mitochondrial inhibition, and the mechanism leading to rotenone-induced cell death has been widely discussed. Both apoptotic mechanisms, involving mitochondrial- and ER-dependent caspases, and the necrosis-associated caspase-1-mediated pathway have been indicated (20, 28, 50). TCF11/Nrf1 clearly plays an important cytoprotective role. In this study, we confirmed that depletion of TCF11/Nrf1 sensitizes cells to rotenone exposure and overexpression of the transcription factor prevents cell apoptosis. We argue that TCF11/Nrf1-dependent induction of ARE-driven target genes, here the proteasome subunit genes, is part of the antioxidative cellular stress response. This is supported by studies showing that overexpression of standard proteasome subunits or the PA28 regulator attenuates oxidative stress and promotes cell viability in response to oxidants (13, 38). The failure of these mechanisms leads to the breakdown of the UPS and finally degeneration (29, 34).
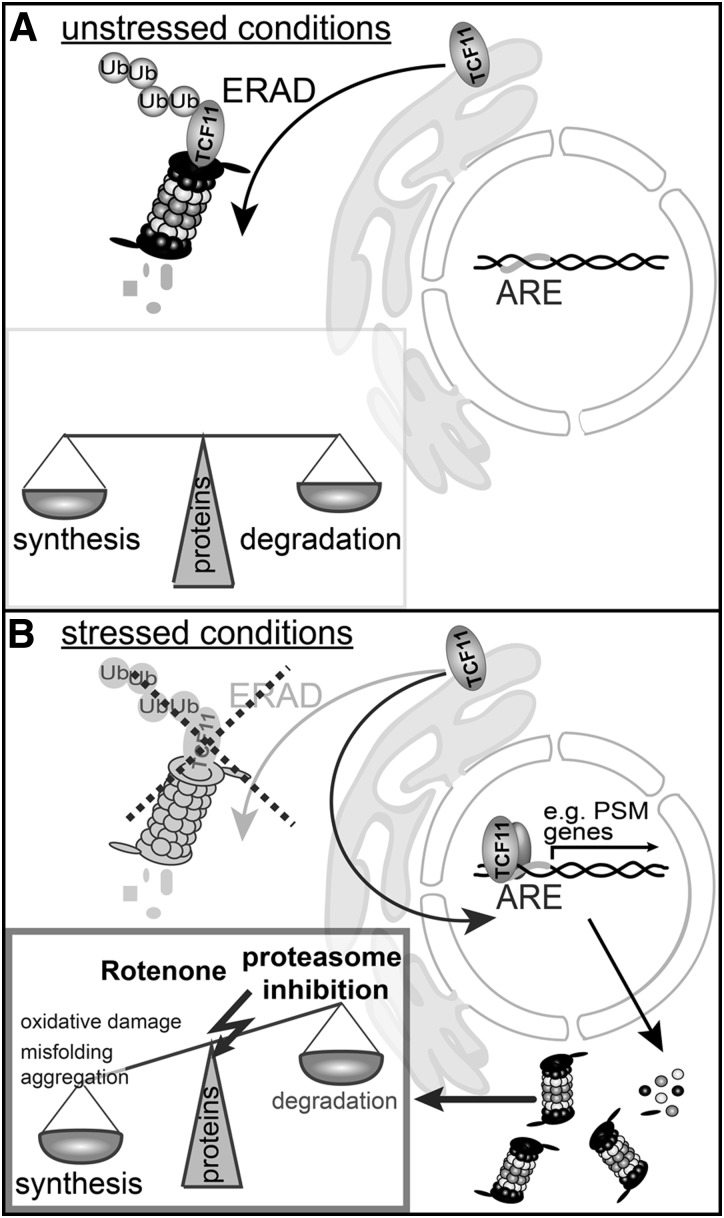
In summary, our investigation shows that the ER membrane protein TCF11/Nrf1 is an essential component of the cellular stress response mechanism (illustrated in Fig. 8). Proteasomal as well as mitochondrial inhibition leads to disruption of redox and protein homeostasis by ROS formation, oxidative protein damage, or accumulation of misfolded proteins (Fig. 8B). In response to cytotoxic stress, TCF11/Nrf1 is retrotranslocated and transferred to the nucleus where it induces proteasome subunit expression via binding to the ARE region of the relevant promoter. The following increase of newly synthesized, active proteasome complexes enables the cell to balance disturbed protein homeostasis, and TCF11/Nrf1 thus promotes cell viability under stressed conditions. This work provides a vital insight into the cellular mechanisms of neurodegeneration in rotenone-based models of Parkinson's disease, and further studies investigating the role of TCF11/Nrf1 in the human condition would be of considerable interest.
FIG. 8.
Proposed TCF11/Nrf1-dependent cellular adaptive mechanism. (A) Under unstressed conditions, protein synthesis and protein degradation/recycling are balanced, maintaining protein homeostasis. TCF11/Nrf1 resides in the ER membrane and is degraded by the ERAD pathway. (B) Cytotoxic stress leads to increased levels of oxidative damaged or misfolded proteins and therefore requires adaptation of the protein homeostasis for efficient clearance. To effectuate the increase in the proteolytic requirement, TCF11/Nrf1 is retrotranslocated and transferred to the nucleus. There, it binds to the ARE region of the promoter of proteasome subunit genes, thereby inducing their expression. The increased expression of the proteasome subunits finally results in an upregulation of newly synthesized active proteasome complexes, increasing the degradation capacity of the cell. ARE, antioxidant response elements.
Materials and Methods
Cell culture
Cells of the established SH-SY5Y (available from ATCC, catalog number CRL-2266) human neuroblastoma cell line, derived from a 4-year-old female, were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS), 2 mM L-glutamine (Biochrom), 100 U/ml penicillin, and 100 μg/ml streptomycin (Pan-Biotech) at 37°C and 5% CO2 in a humidified atmosphere. For differentiation, SH-SY5Y cells were incubated in culture media containing 10 μM RA (Sigma-Aldrich) for at least 7 days. The media were changed every second day. Rotenone (Sigma-Aldrich), expoxomicin, MG132 (Calbiochem), and SFN (Sigma-Aldrich) were dissolved in DMSO, and AA (Sigma-Aldrich) was prepared in culture media. Rotenone and AA solutions were freshly prepared for each experiment.
Nigrostriatal organotypic slice culture
NOSC was prepared from postnatal 3–6-day-old mice pups (C57BL/6J) according to the regulations of animal care and protection. Pups were anesthetized by isoflurane and sacrificed by decapitation. Brains were rapidly dissected and separated in hemispheres. The cut side was glued to the metal block of the vibratome (Leica VT1200S), which was then placed in cold Hank's balanced salt solution (HBSS; Gibco) supplemented with 1% penicillin/streptomycin (P/S; Pan Biotech), 0.6% glucose (Fluka), and 20 mM HEPES (Sigma-Aldrich), purged with Carbogen for dissection in sagittal slices of 300 μm. Cortex, hippocampus, and cerebellum were removed using a razor blade to obtain NOSC. Slices were collected and placed onto membrane inserts (Millicell, nitrocellulose 0.45 μm; Merck Millipore) in six-well plates containing 1 ml of minimum essential medium Eagle (Gibco) supplemented with 25% horse serum (Gibco), 20.7% HBSS, 1% P/S, 0.6% glucose, and 2% B27 (Gibco). Two slices per insert were cultivated at 35°C and 5% CO2 for 8 days before starting treatments (100 nM Rotenone or solvent control: 0.1% DMSO). Medium was replaced every other day, and slice cultures were observed regularly using light microscopy.
Plasmids, DNA, and siRNA transfection
The plasmids TCF11-V5, Nrf1-V5, and the −1500 PSMB6/β1 CAT reporter construct were generated as previously described (61). pcDNA3.1-V5/His-TOPO and pcDNA3.1-Zeo/lacZ were purchased from Invitrogen and pCAT3basic from Promega.
Cells were transfected with plasmids using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. SMARTpool siRNA was purchased from Dharmacon and was transfected using siRNA X-treme gene reagent (Roche) according to the manufacturer's instructions. The siRNA was prediluted to the working concentration of 0.8 μM in Opti-MEM.
CAT reporter gene assay
SH-SY5Y cells were seeded on 24-well plates (250 × 104 cells/cm2) and the next day transfected with 0.9 μg CAT construct and 0.1 μg pcDNA3.1-LacZ as the internal control. The media were changed 6 h post-transfection and at least 18 h later; the CAT concentration was determined using the CAT-ELISA kit (Roche) according to the manufacturer's instructions and normalized to the β-galactosidase activity, measured using chlorophenol red-β-D-galactopyranoside (Roche). The activity buffer (1.2 mg/ml CPRG, 100 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) was added to the lysates at a ration 7.5:1. The color change was measured following incubation at 37°C for circa 1 h using a Synergy HT Multi-Detection microplate reader (BioTek) at 570 nm.
Chromatin immunoprecipitation
ChIP was performed as previously described (61). Equal loads of protein were included in each precipitation. Protein G sepharose was saturated with 1 mg/ml BSA and 0.5 mg/ml sonicated salmon sperm DNA before use. The following primer sets were used to amplify ARE regions in the promoters: PSMB6/β1 forward GCTTCGCTCCCAGAAGGCC and reverse GAAACTTCTCGGCTTTCCCAGTC, and PSMA3/α7 forward CCACAATACTCCTCGTCCTGG and reverse CAAAGGGACGCGTAGGGATTC (BioTeZ). As previously described, α-tubulin served as the control (62). A constant input signal was taken to indicate that the same amount of total DNA was used in the preparation, and the specificity of immunoprecipitation was tested using an unspecific IgG antibody (Cell Signaling).
Reverse transcription-PCR and real-time PCR
RNA isolation and cDNA synthesis were performed using the High Pure RNA Isolation kit and the Transcriptor High Fidelity cDNA Synthesis kit (Roche) according to the manufacturer's instructions.
For reverse transcription-PCR, the primers against TCF11 were as follows; forward CTAGTGGATGGAGAGACTGG and reverse GCACTGCTTCTGTTATGCTGG. The GAPDH primers sequences were forward CAGCAGTGAGGGTCTCTCTC and reverse CAGACACCATGGGGAAGGTG. Real-time PCR using TaqMan gene expression assays (Applied Biosystems) for PSMB6/β1, PSMA2/α2, PSMC4/Rpt3, and HPRT1 were performed using a Rotor-Gene RG-3000 (Corbett Research).
Subcellular fractionation and cell lysis
Cells were fractionated using a subcellular protein fractionation kit (Thermo Scientific). RIPA lysis and nondenaturating TSDG lysis were executed as previously described (61). Protein concentration was determined by a BCA Protein Assay (Thermo Scientific).
Substrate overlay and immunoblotting
Equal amounts of protein were separated either by sodium dodecyl sulfate (SDS)-PAGE or native PAGE and further processed by substrate overlay or immunoblotting as previously described (61). Detection was performed with antibodies against calnexin (BD Bioscience), eIF2α, histone H3, P-eIF2α (Ser-51), PSME2/PA28β, TCF11/Nrf1 (D5B10) (Cell Signaling), β-tubulin (Covance), ubiquitin (Dako), lamin B1, V5 epitope tag (Invitrogen), PSMA1/α6, PSMB6/β1 (laboratory stock), β-actin (C4), GAPDH (FL-335) and TCF11/Nrf1 (H285) (Santa Cruz), and 2,4-dinitrophenol (DNP; Sigma-Aldrich). Densitometrical analysis was performed using ImageJ Software (U.S. National Institutes of Health).
Oxyblot
Oxidizing agents such as the hydroxyl radical directly modify the amino acid side chains of proteins, resulting in a diverse array of altered amino acids. The generation of free carbonyls is one of the most widespread of these modifications and is considered specific for oxidative damage (37). In this study, carbonyl groups were modified with 2,4-dinitrophenylhydrazine (DNPH) and carbonyl derivates analyzed by immunoblotting against DNP. Equal loads of RIPA lysates were denaturated using 6% SDS before carbonyl groups were modified in the presence of 5 mM DNPH for 30 min in the dark. Following the addition of a 0.375-fold volume of neutralization solution (2 M Tris, 30% glycerol) and 1 mM DTT, samples were analyzed by immunoblotting.
Metabolic [35S] labeling
SH-SY5Y cells were seeded on a six-well plate (210 × 104 cells/cm2) and transfected either with control siRNA or siRNA against TCF11/Nrf1 for 2 days. During the last 16–18 h, the cells were treated with rotenone or DMSO. The cells were then fasted for 1.5 h in methionine and cysteine-free culture media, and afterward pulsed with 100 μCi [35S] methionine/[35S] cysteine mixture (TRAN35S-LABEL™; MP Biomedicals) for 3 h. Cells were washed four times with phosphate-buffered saline, lysed (50 mM Tris pH 7.5, 10% glycerine, 50 mM NaCl, 0.5% NP-40) and processed as previously described (22). The proteasome complexes were immunoprecipitated using an antibody against PSMA1/α6 (MCP20). The intensity of the proteasome subunits was determined using ImageJ.
Detection of intracellular ROS
Intracellular ROS were detected by the cell permeable dichlorodihydrofluorescein diacetate (DCFH-DA) dye, which can be deacetylated by intracellular esterases and afterward oxidized to the highly fluorescent 2′,7′-dichlorodihydrofluorescein (DCF) (67). SH-SY5Y cells were seeded on a 24-well plate (250 × 104 cells/cm2) and treated accordingly with rotenone, epoxomicin, or DMSO the next day. Cells were washed with HBSS (Gibco) then cultured in 10 μM DCFH-DA (Sigma-Aldrich)-containing, FCS-free culture media for 30 min. Cells were then washed twice with HBSS, trypsinated, and resuspended in culture media. The fluorescence of the intracellular oxidized product DCF was measured by flow cytometry using a FACSCalibur™ cytometer (BD Bioscience) 6 h after rotenone treatment.
Microscopy
Immunostaining and confocal fluorescence microscopy was performed as previously described (61).
Cell viability and apoptosis
Cell apoptosis was determined using an Apo-ONE® Homogeneous caspase-3/7 assay (Promega), and cell viability was measured using a XTT assay (AppliChem) according to the manufacturer's instructions.
Statistical analysis
All values are presented as mean ± standard deviation of the indicated number of independent experiments. Statistical significance was evaluated using a two tailed, unpaired Student's t-test.
Supplementary Material
Abbreviations Used
- AA
ascorbic acid
- ARE
antioxidant response elements
- bZIP
basic leucine zipper
- CAT
chloramphenicol acetyltransferase
- ChIP
chromatin immunoprecipitation
- DCF
2′,7′-dichlorodihydrofluorescein
- DCFH-DA
dichlorodihydrofluorescein diacetate
- DMSO
dimethyl sulfoxide
- DNP
2,4-dinitrophenol
- DNPH
2,4-dinitrophenylhydrazine
- ER
endoplasmic reticulum
- FCS
fetal calf serum
- HBSS
Hank's balanced salt solution
- NOSC
nigrostriatal organotypic slice culture
- Nrf
nuclear factor-erythroid-2-related factor
- PCR
polymerase chain reaction
- PRAAS
proteasome-associated autoinflammatory syndromes
- RA
retinoic acid
- ROS
reactive oxygen species
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SFN
sulforaphane
- UPS
ubiquitin proteasome system
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (KO 4144/1-1; KR 1915/5-1; SFB740 B3; SFBTRR 43 B7). Elke Bürger (Charité) is acknowledged for her excellent technical assistance and Yin Liu (NIH, Bethesda) for his support with cells from PRAAS patients.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Alam ZI, Jenner A, Daniel SE, Lees AJ, Cairns N, Marsden CD, Jenner P, and Halliwell B. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J Neurochem 69: 1196–1203, 1997 [DOI] [PubMed] [Google Scholar]
- 2.Betarbet R, Canet-Aviles RM, Sherer TB, Mastroberardino PG, McLendon C, Kim JH, Lund S, Na HM, Taylor G, Bence NF, Kopito R, Seo BB, Yagi T, Yagi A, Klinefelter G, Cookson MR, and Greenamyre JT. Intersecting pathways to neurodegeneration in Parkinson's disease: effects of the pesticide rotenone on DJ-1, alpha-synuclein, and the ubiquitin-proteasome system. Neurobiol Dis 22: 404–420, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, and Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci 3: 1301–1306, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Biswas M, Kwong EK, Park E, Nagra P, and Chan JY. Glycogen synthase kinase 3 regulates expression of nuclear factor-erythroid-2 related transcription factor-1 (Nrf1) and inhibits pro-survival function of Nrf1. Exp Cell Res 319: 1922–1931, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas M, Phan D, Watanabe M, and Chan JY. The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J Biol Chem 286: 39282–39289, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm A. and Kruger E. Dysfunction in protein clearance by the proteasome: impact on autoinflammatory diseases. Semin Immunopathol 37: 323–333, 2015 [DOI] [PubMed] [Google Scholar]
- 7.Brehm A, Liu Y, Sheikh A, Marrero B, Omoyinmi E, Zhou Q, Montealegre G, Biancotto A, Reinhardt A, Almeida de Jesus A, Pelletier M, Tsai WL, Remmers EF, Kardava L, Hill S, Kim H, Lachmann HJ, Megarbane A, Chae JJ, Brady J, Castillo RD, Brown D, Casano AV, Gao L, Chapelle D, Huang Y, Stone D, Chen Y, Sotzny F, Lee CC, Kastner DL, Torrelo A, Zlotogorski A, Moir S, Gadina M, McCoy P, Wesley R, Rother K, Hildebrand PW, Brogan P, Kruger E, Aksentijevich I, and Goldbach-Mansky R. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest 125: 4196–4211, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. This reference has been deleted.
- 9.Bryan HK, Olayanju A, Goldring CE, and Park BK. The Nrf2 cell defence pathway: keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85: 705–717, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Chan JY, Han XL, and Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci U S A 90: 11371–11375, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, and Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J 17: 1779–1787, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chepelev NL, Zhang H, Liu H, McBride S, Seal AJ, Morgan TE, Finch CE, Willmore WG, Davies KJ, and Forman HJ. Competition of nuclear factor-erythroid 2 factors related transcription factor isoforms, Nrf1 and Nrf2, in antioxidant enzyme induction. Redox Biol 1: 183–189, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chondrogianni N, Tzavelas C, Pemberton AJ, Nezis IP, Rivett AJ, and Gonos ES. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J Biol Chem 280: 11840–11850, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Bioorg Med Chem 21: 3400–3410, 2013 [DOI] [PubMed] [Google Scholar]
- 15.Ciechanover A. and Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40: 427–446, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Floor E. and Wetzel MG. Increased protein oxidation in human substantia nigra pars compacta in comparison with basal ganglia and prefrontal cortex measured with an improved dinitrophenylhydrazine assay. J Neurochem 70: 268–275, 1998 [DOI] [PubMed] [Google Scholar]
- 17.Furuya N, Ikeda S, Sato S, Soma S, Ezaki J, Oliva Trejo JA, Takeda-Ezaki M, Fujimura T, Arikawa-Hirasawa E, Tada N, Komatsu M, Tanaka K, Kominami E, Hattori N, and Ueno T. PARK2/Parkin-mediated mitochondrial clearance contributes to proteasome activation during slow-twitch muscle atrophy via NFE2L1 nuclear translocation. Autophagy 10: 631–641, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhard A, Pavese N, Hotton G, Turkheimer F, Es M, Hammers A, Eggert K, Oertel W, Banati RB, and Brooks DJ. In vivo imaging of microglial activation with [11C](R)-PK11195 PET in idiopathic Parkinson's disease. Neurobiol Dis 21: 404–412, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Goswami P, Gupta S, Biswas J, Joshi N, Swarnkar S, Nath C, and Singh S. Endoplasmic reticulum stress plays a key role in rotenone-induced apoptotic death of neurons. Mol Neurobiol 53: 285–298, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, and Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386: 463–471, 1997 [DOI] [PubMed] [Google Scholar]
- 22.Heink S, Ludwig D, Kloetzel PM, and Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci U S A 102: 9241–9246, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM, and Scheper W. Activation of the unfolded protein response in Parkinson's disease. Biochem Biophys Res Commun 354: 707–711, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Husberg C, Murphy P, Bjorgo E, Kalland KH, and Kolsto AB. Cellular localisation and nuclear export of the human bZIP transcription factor TCF11. Biochim Biophys Acta 1640: 143–151, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322, 1997 [DOI] [PubMed] [Google Scholar]
- 26.Johnsen O, Skammelsrud N, Luna L, Nishizawa M, Prydz H, and Kolsto AB. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res 24: 4289–4297, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirstein J, Morito D, Kakihana T, Sugihara M, Minnen A, Hipp MS, Nussbaum-Krammer C, Kasturi P, Hartl FU, Nagata K, and Morimoto RI. Proteotoxic stress and ageing triggers the loss of redox homeostasis across cellular compartments. EMBO J 34: 2334–2349, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitamura Y, Inden M, Miyamura A, Kakimura J, Taniguchi T, and Shimohama S. Possible involvement of both mitochondria- and endoplasmic reticulum-dependent caspase pathways in rotenone-induced apoptosis in human neuroblastoma SH-SY5Y cells. Neurosci Lett 333: 25–28, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi A, Tsukide T, Miyasaka T, Morita T, Mizoroki T, Saito Y, Ihara Y, Takashima A, Noguchi N, Fukamizu A, Hirotsu Y, Ohtsuji M, Katsuoka F, and Yamamoto M. Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Genes Cells 16: 692–703, 2011 [DOI] [PubMed] [Google Scholar]
- 30.Koch A, Steffen J, and Kruger E. TCF11 at the crossroads of oxidative stress and the ubiquitin proteasome system. Cell Cycle 10: 1200–1207, 2011 [DOI] [PubMed] [Google Scholar]
- 31.Kriegenburg F, Poulsen EG, Koch A, Kruger E, and Hartmann-Petersen R. Redox control of the ubiquitin-proteasome system: from molecular mechanisms to functional significance. Antioxid Redox Signal 15: 2265–2299, 2011 [DOI] [PubMed] [Google Scholar]
- 32.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, and Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol 23: 8786–8794, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwong M, Kan YW, and Chan JY. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. J Biol Chem 274: 37491–37498, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Lee CS, Lee C, Hu T, Nguyen JM, Zhang J, Martin MV, Vawter MP, Huang EJ, and Chan JY. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc Natl Acad Sci U S A 108: 8408–8413, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HJ, Shin SY, Choi C, Lee YH, and Lee SJ. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J Biol Chem 277: 5411–5417, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Lev N, Melamed E, and Offen D. Proteasomal inhibition hypersensitizes differentiated neuroblastoma cells to oxidative damage. Neurosci Lett 399: 27–32, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Levine RL, Williams JA, Stadtman ER, and Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol 233: 346–357, 1994 [DOI] [PubMed] [Google Scholar]
- 38.Li J, Powell SR, and Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. FASEB J 25: 883–893, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, and Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem 278: 8516–8525, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Luna L, Skammelsrud N, Johnsen O, Abel KJ, Weber BL, Prydz H, and Kolsto AB. Structural organization and mapping of the human TCF11 gene. Genomics 27: 237–244, 1995 [DOI] [PubMed] [Google Scholar]
- 41.Mattson MP. and Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci 7: 278–294, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGeer PL, Itagaki S, Boyes BE, and McGeer EG. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson's and Alzheimer's disease brains. Neurology 38: 1285–1291, 1988 [DOI] [PubMed] [Google Scholar]
- 43.McKinnon C. and Tabrizi SJ. The ubiquitin-proteasome system in neurodegeneration. Antioxid Redox Signal 21: 2302–2321, 2014 [DOI] [PubMed] [Google Scholar]
- 44.Medicherla B. and Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol 182: 663–673, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, and Kruger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J Biol Chem 278: 21517–21525, 2003 [DOI] [PubMed] [Google Scholar]
- 46.Naoi M, Maruyama W, Shamoto-Nagai M, Yi H, Akao Y, and Tanaka M. Oxidative stress in mitochondria: decision to survival and death of neurons in neurodegenerative disorders. Mol Neurobiol 31: 81–93, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Norazit A, Meedeniya AC, Nguyen MN, and Mackay-Sim A. Progressive loss of dopaminergic neurons induced by unilateral rotenone infusion into the medial forebrain bundle. Brain Res 1360: 119–129, 2010 [DOI] [PubMed] [Google Scholar]
- 48.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, and Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem 283: 33554–33562, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orre M, Kamphuis W, Dooves S, Kooijman L, Chan ET, Kirk CJ, Dimayuga Smith V, Koot S, Mamber C, Jansen AH, Ovaa H, and Hol EM. Reactive glia show increased immunoproteasome activity in Alzheimer's disease. Brain 136: 1415–1431, 2013 [DOI] [PubMed] [Google Scholar]
- 50.Pei W, Liou AK, and Chen J. Two caspase-mediated apoptotic pathways induced by rotenone toxicity in cortical neuronal cells. FASEB J 17: 520–522, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Pickering AM. and Davies KJ. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch Biochem Biophys 523: 181–190, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, and Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J 432: 585–594, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pickering AM, Staab TA, Tower J, Sieburth D, and Davies KJ. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster. J Exp Biol 216: 543–553, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radhakrishnan SK, den Besten W, and Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife 3: e01856, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. This reference has been deleted.
- 56.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, and Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38: 17–28, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, and Kruger E. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142: 613–624, 2010 [DOI] [PubMed] [Google Scholar]
- 58.Sha Z. and Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol 24: 1573–1583, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shamoto-Nagai M, Maruyama W, Kato Y, Isobe K, Tanaka M, Naoi M, and Osawa T. An inhibitor of mitochondrial complex I, rotenone, inactivates proteasome by oxidative modification and induces aggregation of oxidized proteins in SH-SY5Y cells. J Neurosci Res 74: 589–597, 2003 [DOI] [PubMed] [Google Scholar]
- 60.Sherer TB, Betarbet R, Stout AK, Lund S, Baptista M, Panov AV, Cookson MR, and Greenamyre JT. An in vitro model of Parkinson's disease: linking mitochondrial impairment to altered alpha-synuclein metabolism and oxidative damage. J Neurosci 22: 7006–7015, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steffen J, Seeger M, Koch A, and Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell 40: 147–158, 2010 [DOI] [PubMed] [Google Scholar]
- 62.Sun Z, Zhang S, Chan JY, and Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol 27: 6334–6349, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sykiotis GP. and Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal 3: re3, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, and Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem 275: 14336–14345, 2000 [DOI] [PubMed] [Google Scholar]
- 65.Tsuchiya Y, Morita T, Kim M, Iemura S, Natsume T, Yamamoto M, and Kobayashi A. Dual regulation of the transcriptional activity of Nrf1 by beta-TrCP- and Hrd1-dependent degradation mechanisms. Mol Cell Biol 31: 4500–4512, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venugopal R. and Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A 93: 14960–14965, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H. and Joseph JA. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic Biol Med 27: 612–616, 1999 [DOI] [PubMed] [Google Scholar]
- 68.Xiong N, Long X, Xiong J, Jia M, Chen C, Huang J, Ghoorah D, Kong X, Lin Z, and Wang T. Mitochondrial complex I inhibitor rotenone-induced toxicity and its potential mechanisms in Parkinson's disease models. Crit Rev Toxicol 42: 613–632, 2012 [DOI] [PubMed] [Google Scholar]
- 69.Zeevalk GD. and Bernard LP. Energy status, ubiquitin proteasomal function, and oxidative stress during chronic and acute complex I inhibition with rotenone in mesencephalic cultures. Antioxid Redox Signal 7: 662–672, 2005 [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Lucocq JM, Yamamoto M, and Hayes JD. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem J 408: 161–172, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao R, Hou Y, Xue P, Woods CG, Fu J, Feng B, Guan D, Sun G, Chan JY, Waalkes MP, Andersen ME, and Pi J. Long isoforms of NRF1 contribute to arsenic-induced antioxidant response in human keratinocytes. Environ Health Perspect 119: 56–62, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.