Abstract
MicroRNAs (miRNAs) are important posttranscriptional regulators of gene expression. However, comprehensive expression profiles of miRNAs during mammalian spermatogenesis are lacking. Herein, we sequenced small RNAs in highly purified mouse spermatogenic cells at different stages. We found that a family of X-linked miRNAs named spermatogenesis-related miRNAs (spermiRs) is predominantly expressed in the early meiotic phases and has a conserved testis-specific high expression pattern in different mammals. We identified one spermiR homolog in opossum; this homolog might originate from THER1, a retrotransposon that is active in marsupials but extinct in current placental mammals. SpermiRs have expanded rapidly with mammalian evolution and are diverged into two clades, spermiR-L and spermiR-R, which are likely to have been generated at least in part by tandem duplication mediated by flanking retrotransposable elements. Notably, despite having undergone highly frequent lineage-specific duplication events, the sequences encoding all spermiR family members are strictly located between two protein-coding genes, Slitrk2 and Fmr1. Moreover, spermiR-Ls and spermiR-Rs have evolved different expression patterns during spermatogenesis in different mammals. Intriguingly, the seed sequences of spermiRs, which are critical for the recognition of target genes, are highly divergent within and among mammals, whereas spermiR target genes largely overlap. When miR-741, the most highly expressed spermiR, is knocked out in cultured mouse spermatogonial stem cells (SSCs), another spermiR, miR-465a-5p, is dramatically upregulated and becomes the most abundant miRNA. Notably, miR-741−/− SSCs grow normally, and the genome-wide expression levels of mRNAs remain unchanged. All these observations indicate functional compensation between spermiR family members and strong coevolution between spermiRs and their targets.
Keywords: X-linked miRNAs, THER1, spermatogenesis, evolution, meiosis, transposable element, MSCI
Introduction
In mammals, male germ cell differentiation starts from primordial germ cells (PGCs) and ends with mature fertile spermatozoa. Male gametes are produced in the seminiferous tubules of the testes, and spermatogenesis mainly involves three phases: self-renewal and proliferation of spermatogonial stem cells (SSCs), meiosis of spermatocytes, and spermiogenesis. PGCs, which emerge at embryonic day 7.5 (E7.5), reach the genital ridges at E11.5, enter mitotic arrest at ∼E13.5 and initiate differentiation to SSCs on postnatal day 2 (P2) by mitotic proliferation. Following mitotic division, the first meiotic division commences at P10. With the initiation of the first wave of spermatogenesis, male germ cells enter the preleptotene/leptotene, zygotene, pachytene, and diplotene stages at P10, P12, P14, and P17, respectively (Bellve et al. 1977). Thereafter, two subsequent fast divisions (MI and MII) occur, with round spermatids first appearing at ∼P20. Round spermatids then go through a complex differentiation program called spermiogenesis, during which they undergo dramatic morphological changes to form sperm-specific structures such as the acrosome and flagellum, as well as nuclear shaping and chromatin compaction (O’Donnell 2014). Spermatozoa are released into the lumen of seminiferous tubules and continue their journey along the convoluted epididymal duct, where they mature and acquire the capacity for full motility (Kotaja 2014; Yadav and Kotaja 2014).
The precise spatial and temporal regulation of gene expression is fundamentally important for the normal progression of spermatogenesis (Kotaja 2014). In recent decades, accumulating evidence has shown that microRNAs (miRNAs) are important posttranscriptional regulators of gene expression and are involved in numerous biological processes (Bartel 2009; Kim and Kim 2012). Both Dicer and Drosha are essential genes for male fertility (Bernstein et al. 2003; Wu et al. 2012; Yadav and Kotaja 2014). The transient inhibition of miR-21 in cultured SSC increases the number of cells undergoing apoptosis and reduces SSC potency (Niu et al. 2011). The miR-34/449 family is highly expressed in late spermatocytes and round spermatids (Bouhallier et al. 2010; Romero et al. 2011; Liang et al. 2012), and the depletion of the entire miR-34/449 family causes male infertility (Song et al. 2014). These results indicate that miRNAs are involved in the orchestrated and stage-specific control of gene expression during spermatogenesis (Yadav and Kotaja 2014). P-element-induced wimpy testis-interacting RNAs (piRNAs) are another type of 25–32-nt small RNA present in germ line cells, and they have important functions in silencing mobile elements and regulating gene expression during spermatogenesis (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Watanabe et al. 2006). Several genome-wide analyses of small RNAs have been performed using either purified late- or postmeiotic spermatogenic cells or whole testes from mice of different ages (Yu et al. 2005; Ro et al. 2007; Yan et al. 2007; Smorag et al. 2012). However, a comprehensive profiling of small RNAs throughout the entire course of spermatogenesis, especially during the early meiotic phases, is lacking.
In the present study, we isolated spermatogenic cells with a high purity at different stages, including SSCs; spermatocytes at the preleptotene, leptotene, zygotene, and pachytene stages; haploid round spermatids; and spermatozoa. The small RNAs in these cells demonstrated a highly dynamic expression pattern. We investigated an X-linked miRNA family that is predominantly expressed in the early meiotic phases and provided insights into the evolution and potential functions of this miRNA family in mammalian spermatogenesis.
Results
Purification of Male Germ Cells in Different Phases during Mouse Spermatogenesis
SSCs are significantly enriched in the Thy1 (CD90)-positive and C-kit-negative cell fraction of mouse testis (Kubota et al. 2004, 2003). PGCs start to differentiate into SSCs at P2, and the first meiosis is initiated in a synchronous manner at ∼P10. Because SSCs constitute <0.02% of the total germ cells in the testes of adult mice, it is very difficult to isolate a large amount of SSCs with a high purity. Instead, we used the testes of mice at P5 to collect Thy1-positive (Thy1+) and C-kit-negative (C-kit-) cells as SSC-enriched populations by magnetic-activated cell sorting, an efficient method for enriching rare cells (supplementary information, Supplementary Material online). The high purity of the collected cells was verified, as ∼90% were positive for Lin28, a defined marker of undifferentiated spermatogonia (Zheng et al. 2009) (supplementary fig. S1A and B, Supplementary Material online). The purified primary SSCs were used for the further profiling of small RNAs.
Meiosis stages can be distinguished on the basis of differences in the relationships of the homologous chromosome pairs and their degrees of condensation. We took advantage of the highly synchronized cell differentiation during the first wave of spermatogenesis and separated different types of spermatogenic cells using the testes of prepubertal mice at different ages by fluorescence-activated cell sorting, as previously reported (Bellve et al. 1977), a method that efficiently prevented contamination by later-stage spermatogenic cells (supplementary information, Supplementary Material online). Preleptotene and leptotene spermatocytes were isolated from the testes of mice at P10; zygotene spermatocytes were isolated from the testes of mice at P13; and pachytene spermatocytes and round spermatids were isolated from the testes of mice at P25 (supplementary fig. S1C, Supplementary Material online). The purity of the isolated spermatocytes in each stage and the haploid round spermatids was >90%, as verified by chromatin spread and immunofluorescence staining assays (supplementary fig. S1D and E, Supplementary Material online). Spermatozoa were obtained from the adult mouse epididymis and collected by the swim-up method (supplementary fig. S1D, Supplementary Material online), as previously reported (Mortimer 1991).
Temporal Expression Patterns of Small RNAs in Mouse Spermatogenic Cells
We sequenced the small RNAs in purified SSCs; spermatocytes in the preleptotene, leptotene, zygotene, and pachytene stages; haploid round spermatids; and spermatozoa. Known and novel small RNAs were identified following the procedure in the Materials and Methods section. miRNAs and piRNAs were the two major types of small RNAs present in these cell stages, and their overall abundance was dynamically regulated during spermatogenesis (fig. 1A and B;supplementary Data S1, Supplementary Material online). miRNAs accounted for 19% and piRNAs accounted for 47% of the total small RNAs in the SSCs. The miRNA abundance strongly increased at the onset of meiosis, and miRNAs represented over 60% of the total small RNAs in the preleptotene stages. In contrast, piRNAs decreased to 8% of the total small RNAs in this stage. As the meiotic process progressed, piRNA expression started to increase dramatically; piRNAs represented 76% of the total small RNAs in the pachytene stage and 80% in round spermatids, whereas the abundance of miRNAs decreased to <5% in these stages. In addition to the change in the predominant small RNAs from miRNAs to piRNAs at the onset of the transition between early and late meiosis, the peak of piRNA length shifted from 27 nt for prepachytene piRNAs to 30 nt for pachytene piRNAs (fig. 1B), which was consistent with the findings of previous studies using whole mouse testes at different ages (Aravin et al. 2006; Li et al. 2013). In spermatozoa, the abundance of piRNAs decreased to 12% and that of miRNAs increased to 40% of the total small RNAs. Meanwhile, the tRNA-derived small RNAs constituted as much as 40% of the total small RNAs in spermatozoa, implying that these small RNAs probably have important functions in spermatozoa maturation and early embryonic development, as reported previously (Sharma et al. 2016).
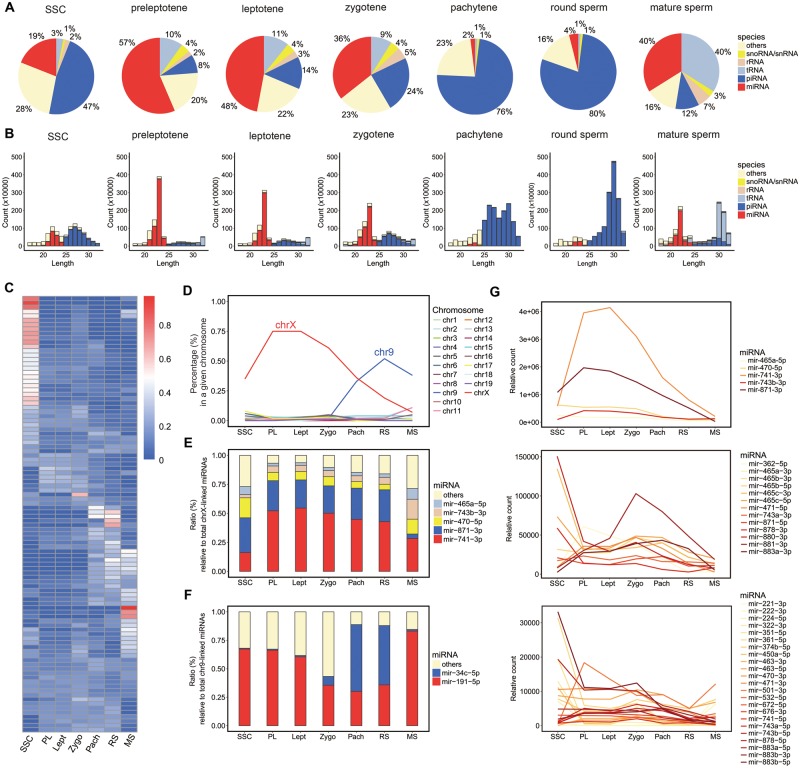
Fig. 1.
Profiling small RNAs in purified mouse spermatogenic cells at different stages of spermatogenesis. (A) Composition of small RNA categories in spermatogenic cells at each stage. (B) Length distribution in each small RNA category. The relative count represents the mapped reads per 10 million total reads. (C) Heat map of the expression levels of the top 100 miRNAs in seven stages during mouse spermatogenesis. The cells within the matrix display the ratio of each miRNA in one stage relative to the total expression level in the seven stages. SSC; preleptotene (PL); leptotene (Lept); zygotene (Zygo); pachytene (Pach); round spermatids (RS); mature sperm (MS). (D) Relative expression levels of miRNAs derived from each chromosome. (E) Expression level of the five most abundant chrX-derived miRNAs relative to the total chrX-derived miRNA abundance. (F) Expression levels of miR-34c and miR-191 relative to the total chr9-derived miRNA abundance. (G) Expression pattern of X-linked miRNAs during mouse spermatogenesis. To clearly display the expression pattern of each miRNA, the miRNAs were divided into three groups according to expression level. The relative count of each miRNA was normalized to the total miRNA abundance.
Using the clustered expression patterns of individual miRNAs, the entire process of mouse spermatogenesis could be classified into four developmental stages: SSCs, early meiotic stages (including the preleptotene, leptotene, and zygotene stages), late meiotic stages (including the pachytene and round spermatid stages), and spermatozoa (fig. 1C and supplementary Data S2, Supplementary Material online). Dozens of the top 100 miRNAs were expressed at their highest levels in SSCs, with expression significantly decreasing after the initiation of the meiotic process (fig. 1C). In contrast, the expression of a few miRNAs, most of which were X linked, increased when the meiotic process started and remained high until the late meiotic phases. Intriguingly, when the expression of miRNAs on each chromosome was combined, miRNAs on the X chromosome were more abundant than those on other chromosomes in the SSCs, and these miRNAs became predominant in the early meiotic stages, accounting for up to 75% of all miRNAs in leptotene spermatocytes (fig. 1D). Among the X-derived miRNAs, miR-741-3p, miR-871-3p, miR-470-5p, miR-743a-3p, and miR-465a-5p constituted up to 95% of the leptotene spermatocytes (fig. 1E). Accompanied by a decrease in X-derived miRNAs in the late meiotic stages, the expression levels of some miRNAs significantly increased. For example, the expression of chromosome 9 (chr9)-derived miR-34c and miR-191 increased sharply, and these miRNAs became the predominant miRNAs in round spermatids (fig. 1D and F). In spermatozoa, miR-34c expression was downregulated to a low level, and miR-191 became the most abundant miRNA. The observation that each spermatogenic stage was marked by a few stage-specifically highly expressed miRNAs indicates the important functions of these miRNAs in the temporal regulation of gene expression during spermatogenesis.
During mammalian spermatogenesis, X and Y chromosomes are compartmentalized into a perinuclear subdomain called the XY body (Handel 2004). Transcriptome analyses demonstrated that almost all protein-coding genes on the X chromosome were subjected to meiotic sex chromosome inactivation (MSCI) when the XY body formed in the late meiotic phases in mice (Handel 2004; Turner 2007; Cloutier and Turner 2010). In contrast to a previous study, which proposed that X-linked miRNAs escaped silencing during MSCI (Song et al. 2009), our deep sequencing results using purified spermatocytes at different stages and round spermatids demonstrated that the total amount of X-derived miRNAs was significantly decreased after the meiotic process entered the pachytene stage and continuously decreased to a low level in round spermatids (fig. 1D). Notably, not only was this expression decrease observed in several of the most abundant miRNAs but also almost all X-linked miRNAs were downregulated (fig. 1G). These results support the possibility that the expression of X-derived miRNAs is repressed in MSCI, similar to that of the protein-coding genes, although we cannot rule out the possibility that these X-linked miRNA precursors are continuously transcribed without being processed into their mature forms.
Identification and Evolution of an X-Linked miRNA Family Located between Slitrk2 and Fmr1
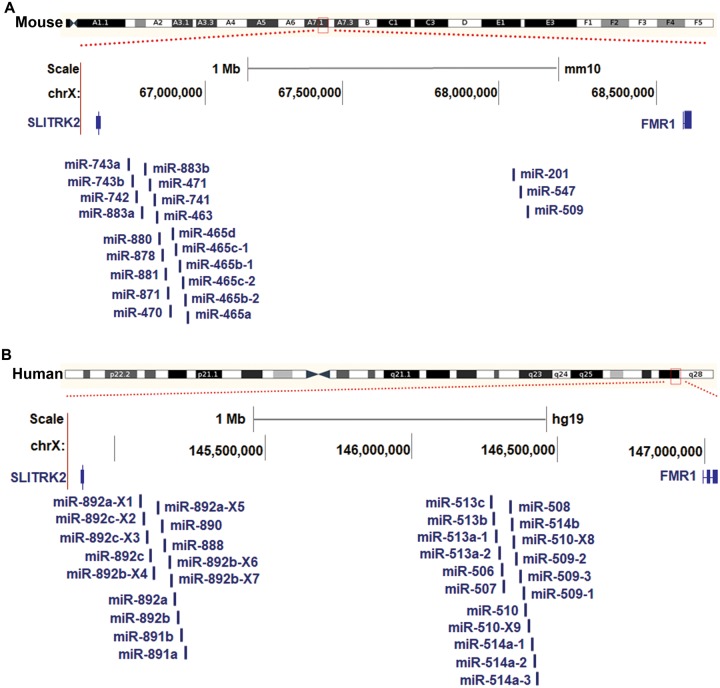
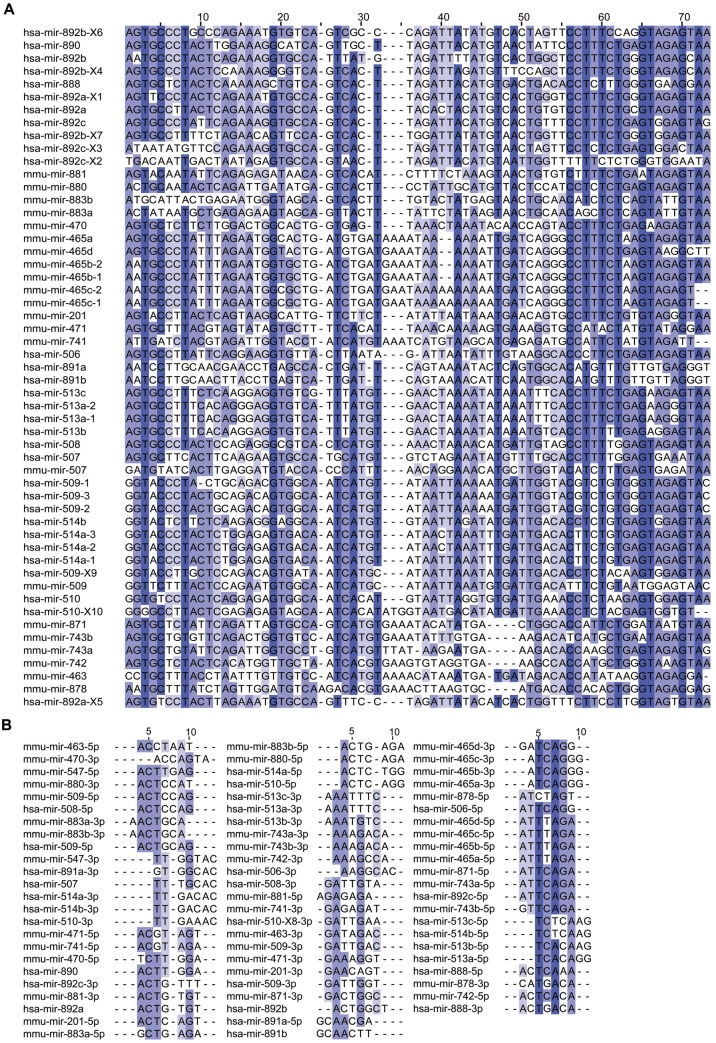
Analyses of the genomic locations of those X-linked miRNAs highly expressed in spermatogenesis revealed the intriguing finding that all were adjacent to each other within a small region between two conserved protein-coding genes, Slitrk2 and Fmr1, in the mouse genome (Rosenbloom et al. 2015) (University of California Santa Cruz [UCSC] genome version mm10) (fig. 2A). Among the 22 miRNA genes encoded in this region, 19 were located proximal to Slitrk2, and the other 3 were located proximal to Fmr1. Interestingly, we also found 31 miRNAs encoded in a corresponding region between Slitrk2 and Fmr1 in the human genome (fig. 2B) (Rosenbloom et al. 2015) (UCSC genome version hg19). In humans, 14 of these miRNA genes were located proximal to Slitrk2, and the other 17 were located near Fmr1. Intriguingly, all of these miRNAs in mice and humans showed high similarity in their precursor sequences (fig. 3A), suggesting that they may have expanded from a common ancestor. As they were preferentially expressed at the highest level during mouse spermatogenesis, this family of X-linked miRNAs was named the spermatogenesis-related miRNA (spermiR) family. Furthermore, the spermiRs located near Slitrk2 were named spermiR-Ls, and those located near Fmr1 were named spermiR-Rs. The sequence of ribonucleotides at positions 2–8 from the miRNA 5′ end, known as the seed sequence, is important for miRNA-target recognition. Highly frequent nucleotide variations, including substitutions, insertions, and deletions, were observed in both the spermiR precursors and the seed sequences (fig. 3A and B), indicating the rapid evolution of this miRNA family. Notably, some insertions and deletions in the nonseed region of spermiR precursors could also alter the Drosha/Dicer cleavage position during miRNA biogenesis, resulting in a shift of the seed region sequence, which is common in spermiRs.
Fig. 2.
Genomic distribution of spermiRs in mice and humans. (A–B) Schematic representation of the spermiR distribution in the mouse (A) and human (B) genomes. The newly identified spermiRs are marked by the addition of X1–9 to the end of the corresponding miRNA name. The relative distance between the spermiRs is not proportional to their physical distance in the mouse or human genome.
Fig. 3.
Sequence divergence of spermiR precursors and seed regions. (A–B) Multiple sequence alignment of spermiR precursors (A) and spermiR seed regions (B) in humans and mice. The darkness of the color represents the degree of sequence similarity.
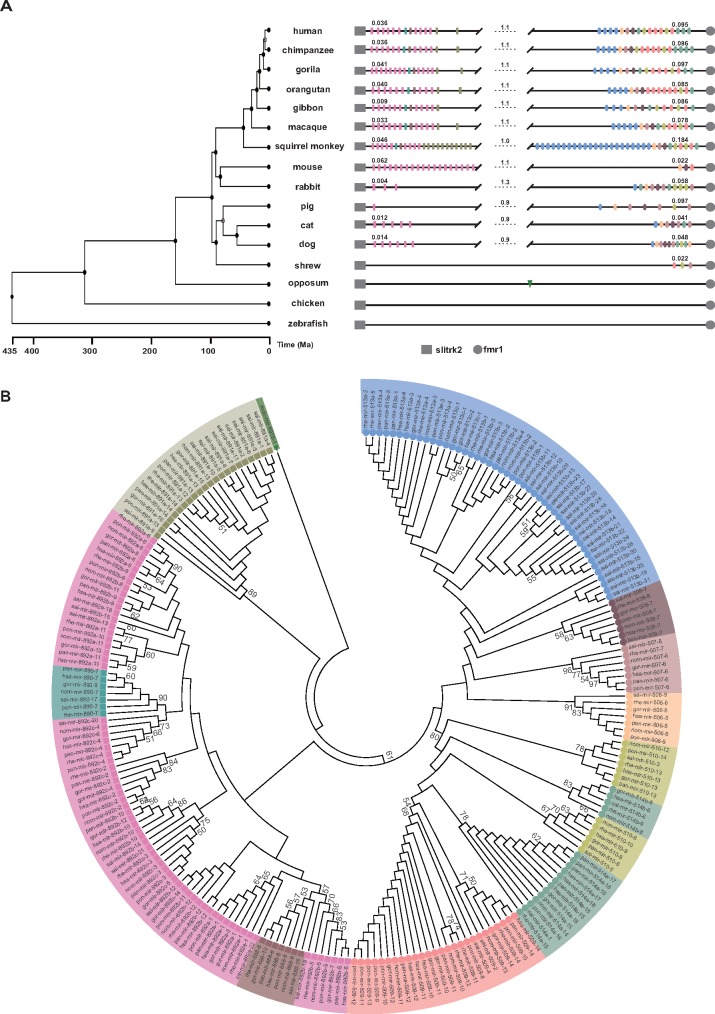
When we used Blast (Altschul et al. 1990) and Exonerate (Slater and Birney 2005) to search for spermiR homologs in the genomes of representative species other than mice and humans, we identified spermiR family members in all the placental mammals and a marsupial (Monodelphis domestica) but not in the monotreme (Ornithorhynchus anatinus) or any other nonmammalian vertebrate species (fig. 4A). In addition to 19 spermiR-Ls and 3 spermiR-Rs in mice, we identified 6 spermiR-Ls and 11 spermiR-Rs in dogs, 5 spermiR-Ls and 7 spermiR-Rs in cats, 1 spermiR-L and 7 spermiR-Rs in pigs, and 3 spermiR-Ls and 11 spermiR-Rs in rabbits (fig. 4A and supplementary Data S3 and S4, Supplementary Material online). Only three spermiR-Rs, but no spermiR-L, were discovered in common shrews, implying that either spermiR-L has not emerged in this species or their sequences have diverged too much to be recognized. Interestingly, compared with the highly divergent copy numbers of the two spermiR clades in nonprimate species, both have undergone more frequent duplication in lower primates and maintained comparable copy numbers for ∼30 million years (My) without any major structural change in higher primate species (fig. 4A and supplementary Data S3 and S4, Supplementary Material online). These observations suggest that both spermiR-Ls and spermiR-Rs may have evolved through lineage-specific duplication events. Notably, none of the spermiRs were identified outside the genomic locus between Slitrk2 and Fmr1 in any of the representative mammals we investigated.
Fig. 4.
Evolutionary history of spermiRs. (A) Schematic representation of spermiRs in the genomes of representative species. The branch length on the phylogenetic tree reflects the divergence times of different species in millions of years ago (Ma) (Hedges et al. 2015). The rectangles represent spermiR-Ls, and the ellipses represent spermiR-Rs. The pink rectangles represent the miR-892 subfamily; cadet-blue, miR-890; deep khaki, miR-891; and deep salmon, miR-888. The sky-blue ellipses represent the miR-513 subfamily; light orange, miR-506; light rosybrown, miR-507; deep rosybrown, miR-508; light sea-green, miR-514; deep yellow, miR-510; and light salmon, miR-509. The green triangle represents the spermiR homolog in opossum. The physical length of the spermiR-L and spermiR-R clusters or the region between them in each species (in Mb) is indicated by the number above the lines. The genomic coordinate of each spermiR in the different species is provided in supplementary Data S4, Supplementary Material online. (B) Neighbor-joining tree of the spermiR precursor sequences (200 bp) in the representative primate species using the spermiR homolog in opossum as the outgroup. The colored shapes represent different spermiR subfamilies or outgroups, as described in (A). Only the bootstrap value ≥50 is shown.
With the opossum spermiR as the outgroup, phylogenetic analyses for all spermiR precursors in the representative primates revealed that except for the miR-891 subfamily, which clustered more closely with the outgroup, spermiR-Ls and spermiR-Rs clustered together (fig. 4B). Intriguingly, although the miR-891 subfamily clustered separately from other spermiR-Ls and appeared to be more closely related to the original spermiR, members of this subfamily were not identified in the nonprimate mammals (fig. 4A), probably because the corresponding homologs were extinct during the evolution of the nonprimates. We also built a phylogenetic tree for all spermiRs in the representative nonprimate mammals and found that the members of the spermiR-Ls clustered more closely to the outgroup than did the spermiR-Rs, implying the possibility that the spermiR-L clade might have emerged relatively earlier than the spermiR-R clade in nonprimate mammals (supplementary fig. S2A, Supplementary Material online). A previous study showed that the phylogenetic relationships between spermiR-Rs correlated with their physical distances in humans (Zhang et al. 2007). However, our analysis of all spermiRs in humans showed that the correlation between phylogenetic proximity and physical distances in spermiR-Ls was significantly lower than that in spermiR-Rs (supplementary fig. S2B, Supplementary Material online).
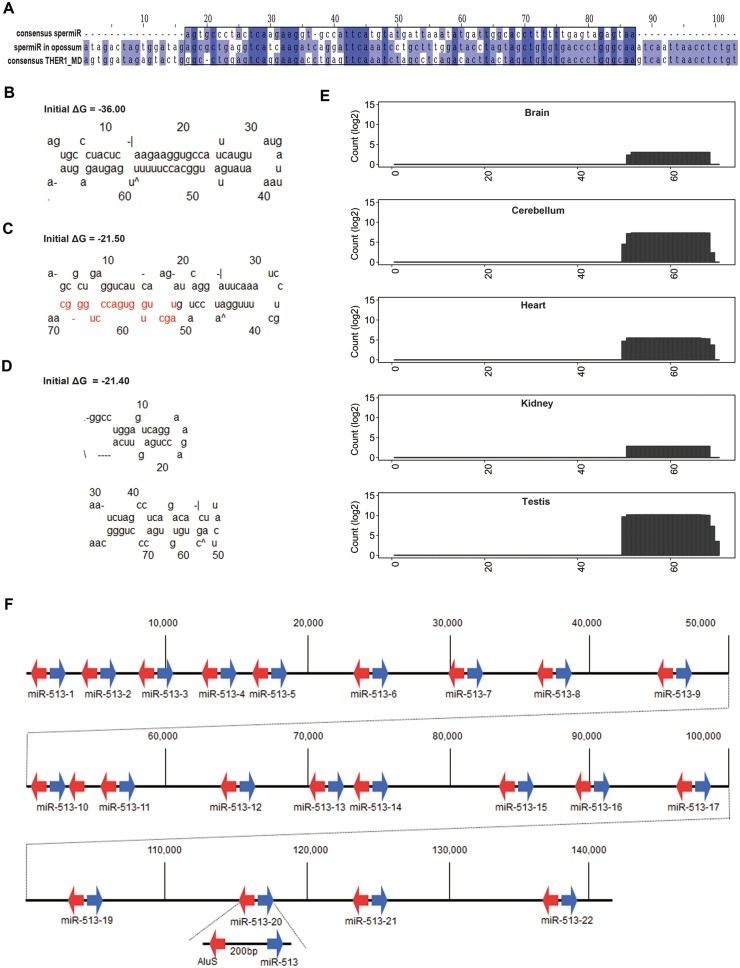
In the opossum genome, 34 Blast hits on sequences similar to spermiRs from representative placental mammalian were found, all of which were further characterized as the remnants of a transposable element, THER1, by both RepeatMasker (Smit and Riggs 1996) and CENSORS (Jurka et al. 1996). THER1 was an active retrotransposon in the opossum genome, but it became extinct during mammalian evolution, and the remnants have been named mammalian-wide interspersed repeats (Gilbert and Labuda 1999). Compared with the consensus sequence of THER1, all of these 34 similar hits accumulated many mutations, including a hit located between Slitrk2 and Fmr1 (fig. 5A). In contrast to the THER1 consensus sequence, the sequence of the THER1-related hit located between Slitrk2 and Fmr1 could be folded into a classical stem-loop structure, although the pre-miRNA-like structure appeared to be less stable than the spermiR consensus sequence from representative placental mammalian (fig. 5B–D). Analyzing the previous small RNA sequencing data from different opossum tissues (Meunier et al. 2013), we identified a 19-nt small RNA derived from the 3′ arm of the stem-loop structure (fig. 5C); this small RNA was expressed at a significantly higher level in the testes than in the other tissues (fig. 5E), although the expression was relatively low compared with that of other abundant miRNAs, indicating that this spermiR may be neofunctionalized in opossum testes. Newborn miRNAs were often expressed at low levels because of weaker promoters for primary transcription and/or less efficient biogenesis, which might prevent potential detrimental effects from targeting many genes improperly (Liu et al. 2008). Intriguingly, the density of X-linked miRNAs in opossum was much higher than that in birds and placental mammals (supplementary fig. S3A, Supplementary Material online), indicating a high birth rate of new miRNAs on the opossum X chromosome.
Fig. 5.
Retrotransposon-related evolution of spermiRs. (A) Multiple sequence alignment of the consensus sequence of spermiR precursors in representative placental mammals, the THER1-derived spermiR homolog in opossum, and the THER1_MD consensus sequence in opposum. The darkness of the color represents the degree of similarity. (B–D) Predicted secondary structure of the consensus sequence of spermiR precursors (B), the precursor of the spermiR homolog in opossum (C), and the THER1_MD consensus sequence in opossum (D). The sequence of spermiR in opossum is marked in red in (C). (E) Expression level of spermiRs in different tissues of opossum. The height of the bar in each position represents the cumulative read count that mapped to the spermiR precursor sequence. The count was normalized to the total count mapped to the genome. (F) Physical map of 21 miR-513 subfamily genes and the flanking AluS fragments in the squirrel monkey genome. A repeat unit composed of a miR-513 precursor, a 200-bp constant intervening sequence and a 150-bp AluS fragment is shown below.
Transposable elements have been reported to be the sequence resources for miRNA birth (Smalheiser and Torvik 2005), as well as to facilitate the expansion of miRNAs (Zhang et al. 2008; Lehnert et al. 2009). In our study, the miR-513 subfamily from the spermiR-R clade was shown to experience frequent expansion in lower primates (fig. 4A and supplementary Data S3 and S4, Supplementary Material online). Of the 22 sequential copies of miR-513 genes found in squirrel monkeys, 21 were flanked with a constant AluS remnant. Further sequence analysis identified that the duplication unit was composed of a 150-nt sequence derived from the 3′ end of AluS, a miR-513 precursor, and a constant intervening sequence of ∼200 bp (fig. 5F). Phylogenetic analysis showed that all miR-513 family members clustered closer than other spermiRs (supplementary fig. S3B, Supplementary Material online), indicating potential amplification by tandem duplication. In primates, Alu-mediated rearrangement events are known to be a common cause for local deletions and duplications associated with genome structural changes and genetic diseases (Smith et al. 1996; Deininger and Batzer 1999). Alu elements can be categorized into three major subfamilies, which were active in primates at different divergence times: AluJ (65–40 million years ago (Ma)), AluS (45–25 Ma), and AluY (30 Ma to present) (Batzer and Deininger 2002; Price et al. 2004). The most abundant Alu subfamily is AluS (accounting for 57% of Alu elements), which underwent a major activity burst after the split of New World monkeys and prosimians (Bailey et al. 2003). A previous study investigated the evolution of the miR-513 subfamily in representative primate species and found that the duplications of miR-513 genes appeared to be independent in Platyrrhini (New World monkeys) and Catarrhini (Old World monkeys, apes and humans) after their divergence (Sun et al. 2013). Here, we propose that the miR-513 subfamily in the squirrel monkey, a representative New World monkey, may be descended from a single common ancestor and amplified via a series of AluS-related local duplication events. In addition, the physical structure of the spermiR-L clade is almost identical in the mouse and rat genomes, except there are six sequential copies of the miR-465 gene in mice but only one copy in rats. Multiple sequence alignment identified six repeat cassettes in the miR-465 locus of the mouse genome, each of which is ∼1,000 bp in length and consists of an Lx5c-derived sequence, followed by a simple (TAAA)n repeat, a miR-465 gene, and an Lx7-derived sequence (supplementary fig. S3C, Supplementary Material online). Moreover, the precursor sequences of the miR-465 subfamily clustered together in the phylogenetic tree of all spermiRs in mice (supplementary fig. S3D, Supplementary Material online), suggesting that Lx, a retrotransposable element in the LINE family that flanks the miR-465 locus, may have mediated the copy number divergence in the rodent genome by local duplication events.
Conserved Testes-Specific Expression Pattern of SpermiRs in Mammals
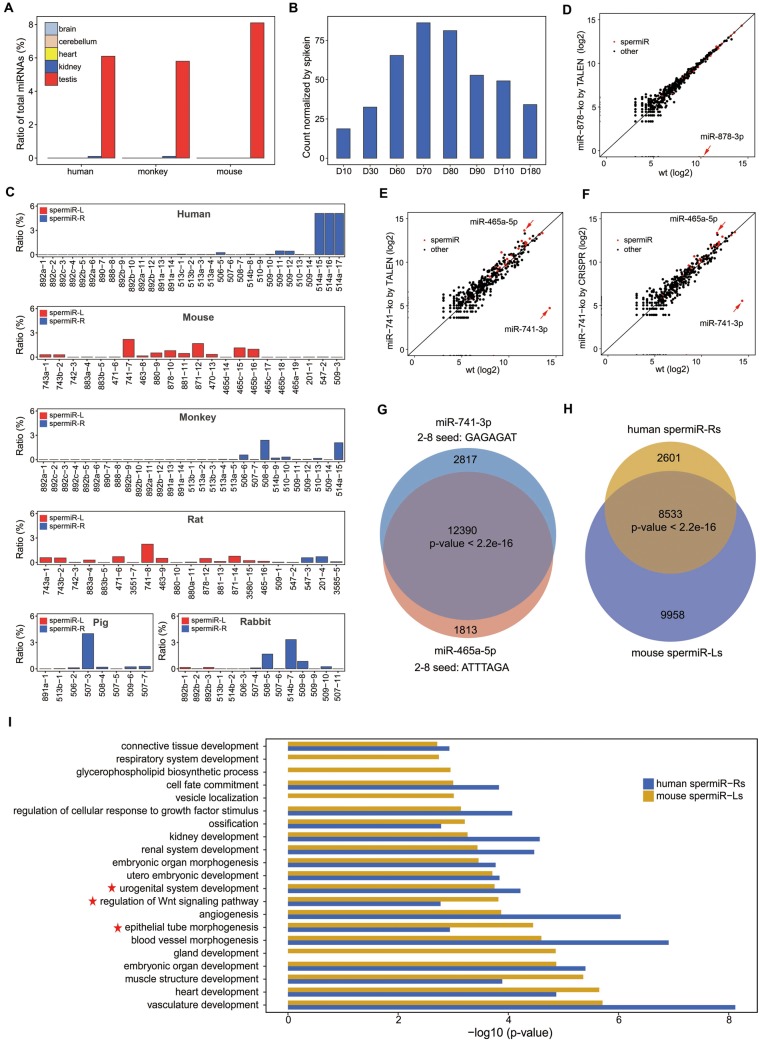
Multiple cell types are present in adult testes, of which early meiotic spermatocytes account for only a small proportion (Bellve et al. 1977). Analysis of previous small RNA sequencing data in tissues from different species revealed that spermiRs were specifically highly expressed in adult testes, accounting for 6.1%, 5.8%, and 8.1% of all miRNAs in humans, monkeys, and mice, respectively (Meunier et al. 2013) (fig. 6A). SpermiRs also accounted for 82%, 93%, and 99% of the X-linked miRNAs in the adult testes of these species, respectively. SpermiRs presented comparable expression levels in the adult testes of humans, monkeys, and mice, indicating that their predominant expression in the early meiotic phases is likely conserved in mammals. This speculation was further confirmed by our sequencing results of small RNAs in rabbit testes on postnatal days 10–180. The highest expression of spermiRs was detected on postnatal days 60, 70, and 80 (fig. 6B), on which the meiotic stage of rabbit spermatogenesis had begun and the early spermatocytes presented at a higher density in the seminiferous tubules than in the other stages (supplementary fig. S4, Supplementary Material online). Consistent with the observations using purified spermatocytes in mice, the overall abundance of spermiRs started to decrease after postnatal day 80, possibly due to the increased late meiotic spermatocytes in the rabbit testes during development.
Fig. 6.
Conserved testis-specific expression pattern of spermiRs and functional compensation between spermiRs with divergent seed sequences. (A) Expression of all spermiRs relative to that of total miRNA in the different tissues from humans, monkeys, and mice. (B) Expression levels of spermiRs in the testes of rabbits at different ages. D10, 30, 60, 70, 80, 90, 110, and 180 represent postnatal days 10–180, as indicated. The count of spermiR is normalized to that of exogenous spike-ins. (C) Proportion of each spermiR relative to total miRNA in the adult testes of humans, mice, monkeys, rats, pigs, and rabbits. The order of the bars represents the physical order of the spermiRs in the genome of different species. (D–F) Expression levels of the top 200 most highly expressed miRNAs in wild-type (Wt) and miR-878 knockout SSCs (D) and in Wt and miR-741 knockout SSCs by TALEN (E) and by CRISPR (F). The expression level of each miRNA was normalized to the total miRNA abundance. In (D–F), miR-878-3p, miR-741-3p, and miR-465-5p are indicated by a red arrow. (F) Venn diagram showing the number of genes predicted to be targeted by miR-741-3p and miR-465-5p. (G) Venn diagram showing the number of genes predicted to be targeted by the top five most highly expressed spermiR-Ls in mice and the top five most highly expressed spermiR-Rs in humans. In (F) and (G), Welch’s two-sample t-test was used for statistical analysis. (H) Bar plot showing the GO term categories of the genes predicted to be targeted by the five most highly expressed spermiR-Ls in mice and spermiR-Rs in humans. The P values of the top 20 GO term categories with the highest fold enrichment are plotted. GO terms related to cell differentiation, epithelial tube morphogenesis, and male reproductive-related pathways are indicated with red five-pointed stars.
We proceeded to investigate the expression patterns of individual spermiRs during spermatogenesis in different species. Unlike spermiR-Ls, which were highly expressed in mouse and rat testes, spermiR-Rs contributed to the high expression of spermiRs mainly in the testes of humans, monkeys, pigs, and rabbits (fig. 6C and supplementary Data S5, Supplementary Material online). These results indicate that a lineage-specific expression pattern evolved between the two spermiR clades in mammalian spermatogenesis. Furthermore, we calculated the transcript abundance of each spermiR precursor by analyzing RNA sequencing data from the testes of different species and found that the lineage-specific expression patterns of the two spermiR clades were mainly regulated at the transcriptional level (supplementary fig. S5, Supplementary Material online), but we could not rule out the possibility that the processing efficiency of miRNA precursors also contributed to this pattern.
Functional Compensation of Different SpermiRs with Divergent Seed Sequences
In mice, spermiR-Ls were the major contributors to the predominant expression of spermiRs in the early meiotic stages. In contrast, three spermiR-Rs were expressed at a very low level throughout spermatogenesis (fig. 6C and supplementary fig. S6A and supplementary Data S1, Supplementary Material online). The spermiR-Ls could be mainly divided into two subgroups according to their expression patterns during spermatogenesis. One subgroup was expressed at the highest level in the early meiotic stages and showed significantly decreased expression in the late stages. The other subgroup was expressed at the highest level in SSCs and was downregulated when spermatogenesis entered the meiotic process (supplementary fig. S6A, Supplementary Material online). A previous ChIP-Seq study of RNA polymerase II indicated that spermiR-Ls in mice were likely derived from one primary transcript and that posttranscriptional regulation might be involved in the production of individual spermiR-Ls (Royo et al. 2015).
miR-741 was the most highly expressed miRNA in the early meiotic phases, accounting for >40% of all miRNAs in the preleptotene/leptotene spermatocytes. We validated the miR-741 expression level by quantitative reverse transcription-polymerase chain reaction using miR-21, a housekeeping miRNA expressed at a high level throughout mouse spermatogenesis, as an internal control (supplementary fig. S6B, Supplementary Material online). Secondary structure analyses indicated that the precursor sequence of miR-741 could be folded into a typical pre-miRNA-like stem-loop (supplementary fig. S6C, Supplementary Material online). Moreover, we cloned the precursors and flanking sequences of the three most highly expressed miRNAs (miR-741, miR-871, and miR-470) into plasmids with a Cytomegalovirus (CMV) promoter and expressed them in HEK293 cells (supplementary fig. S6D, Supplementary Material online). The reporter assays demonstrated that all three expressed spermiR-Ls could repress the cotransfected luciferase reporter bearing the corresponding partially complementary target sequences (supplementary fig. S6D and E, Supplementary Material online), indicating that these spermiR-Ls can be processed and function as typical miRNAs in mammalian cells.
Isolated SSCs can grow in vitro under defined culture conditions and restore fertility after being transplanted into the seminiferous tubules of infertile recipient mice. Moreover, cultured SSCs express high levels of spermiRs, accounting for ∼45% of all miRNAs, making these cells an ideal model to study the function of spermiRs in vitro. To investigate the function of individual spermiRs, we generated miR-741 knockout SSC lines using CRISPR/Cas9. To rule out potential off-target effects by CRISPR/Cas9, we also generated miR-741−/− SSCs using transcription activator-like effector nucleases (TALENs). TALENs have unique mechanisms for DNA target recognition and cleavage, making it unlikely to have the same off-target effects in the genome as CRISPR/Cas9 does. We generated miR-878 (a spermiR-L expressed at a low level in SSCs) knockout SSCs as an additional negative control. miR-741−/− SSCs, as well as the miR-878−/− SSCs, grew normally. When miR-741−/− SSCs were transplanted into the seminiferous tubules of infertile recipient mice, fertility was restored (data not shown). We sequenced the small RNAs in wild-type, miR-741−/−, and miR-878−/− SSCs. Intriguingly, a member of the spermiR-L family, miR-465a-5p, showed strong upregulation (>8-fold) and was the most abundant miRNA in miR-741−/− SSCs, regardless of knockout mediated by CRISPR/Cas9 or TALEN (fig. 6E and F). In contrast, no obvious change in the miRNA profile from that of wild-type SSCs was observed in miR-878−/− SSCs (fig. 6D).
We further examined the mRNA expression levels in miR-741−/− SSCs using RNA-seq. Surprisingly, compared with the control profile, the gene expression profile of miR-741−/− SSCs did not significantly change (supplementary fig. S6F and G, Supplementary Material online), implying that the upregulation of miR-465a-5p may compensate for the loss of miR-741 function in SSCs. Remarkably, despite the distinct seed sequences of miR-741-3p and miR-465a-5p, >80% of the genes predicted to be targeted by miR-741-3p were also potential targets of miR-465a-5p (fig. 6G), suggesting that the compensatory effect of miR-465a-5p for miR-741-3p function was probably mediated by the targeting of different sites in the same genes. We further compared the predicted target genes of the top five spermiR-Ls and spermiR-Rs expressed at the highest levels in the testes of mice and humans, respectively, and found that most of the genes predicted to be targeted by spermiR-Rs in humans were also targeted by spermiR-Ls in mice (fig. 6H). We conducted functional enrichment analyses of the genes targeted by these two groups of spermiRs and found that similar functional categories were overrepresented (fig. 6I), which included functions related to differentiation, epithelial tube morphogenesis, and regenerative system-related processes. These results suggest that despite the rapid evolution of spermiR sequences, the functions of the spermiR family may be highly conserved by the targeting of a similar group of genes with spermatogenesis-related functions in mammals.
Discussion
Functional Compensation of SpermiRs in Cultured SSCs
In this study, we isolated mouse male germ cells with high purity at different stages and identified an X-linked miRNA family, the spermiR family, which is collectively predominantly expressed in the early meiotic phases and then undergoes a rapid decrease in expression during the late and postmeiotic processes (fig. 1D and E). Along with the decrease in spermiR expression, the miR-34/449 family, mainly miR-34c, starts to be upregulated in the pachytene stage and constitutes nearly 30% of the total expressed miRNAs in haploid round spermatids (fig. 1D and F). Emerging evidence supports the functional importance of redundant, polycistronic miRNAs in normal development and stress responses (Olive et al. 2015). miR-34a, miR-34b/c, and miR-449a/b/c belong to the same miRNA family and share the same seed sequences but are located on different chromosomes. In a previous study, the depletion of individual miR-34/449 family members resulted in no obvious phenotype in mouse fertility. However, the simultaneous depletion of all six miR-34/449 family members resulted in a severe infertility phenotype (Song et al. 2014). A recent study reported that miR-449 was induced in miR-34b/34c-deficient mice, whereas miR-34b and miR-34c induction was observed in miR-449-deficient mice, suggesting compensatory expression among miR-34/449 family members (Wu et al. 2014). However, these reported miRNAs have the same seed sequences and functional compensation between miRNAs with different seed sequences has not been reported. In this study, we show that when miR-741 is knocked out in cultured SSCs, the expression of miR-465a-5p (another spermiR family member), but not other miRNAs, is dramatically upregulated and replaces miR-741-3p as the most abundant miRNA (fig. 6E and F). Although miR-741-3p and miR-465a-5p have distinct seed sequences (fig. 3B), their predicted target genes are highly overlapped (fig. 6G). Transcriptome analyses revealed that the mRNA levels remained virtually unchanged after the deletion of miR-741-3p in SSCs (supplementary fig. S6F and G, Supplementary Material online), supporting the possibility that miR-465a-5p expression might be upregulated to compensate for the function of miR-741-3p by regulating a similar group genes. However, these evidences for function compensation between spermiRs, including highly overlapping of the predicted target genes between miR-741-3p and miR-465a-5p and the unchanged transcriptome upon miR-741 deletion are indirect. It is possible that the miR-465a-5p upregulation upon miR-741 deletion in the cultured SSCs is merely a coincidence caused by certain unknown mechanism. If so, miR-741/miR-465a double knockout might have similar phenotype in spermatogenesis as miR-741 knockout after the SSCs being transplanted into infertile recipient mice, which warrants future investigations. Further experiments to identify the miRNA-target sites by ultraviolet crosslinking-immunoprecipitation and high-throughput sequencing (CLIP-seq) of Argonaute protein in the wild-type and spermiR-knockout SSCs, and verify the spermiR target site individually by dual-luciferase reporter assay may provide direct evidences for validating the hypothesis whether miR-741-3p and miR-465a-5p target a similar group of genes despite of the difference in their seed sequences.
The precise mechanism that mediates the induction of other miRNAs upon the depletion of one or several miRNAs remains elusive. Besides transcriptional regulation in miRNA expression, accumulating evidence has demonstrated that both the tertiary structure of miRNA precursors and trans-acting RNA binding proteins could impact miRNA biogenesis and/or miRNA turnover, resulting in cell type- and context-dependent differential miRNA expression (Olive et al. 2015). Notably, miR-465a-5p displays an expression pattern similar to that of miR-741-3p during spermatogenesis (supplementary fig. S6A, Supplementary Material online). The deletion of miR-741 might turn on a feedback response to change the processing efficiency and/or turnover of miR-465a-5p, resulting in the upregulation of miR-465a-5p; this interesting mechanism warrants further investigation. The predominant production of spermiRs in early meiosis and the functional redundancy among the family members, as well as the coevolution of spermiRs with their target genes, suggests the functional importance of spermiRs. Therefore, it is reasonable to expect that the deletion of an entire spermiR family but not individual one might produce discernible phenotypes in spermatogenesis.
Amplification and Divergence of SpermiRs in Mammals
The rapid evolution of the miR-513-514 and miR-892-891 clusters has previously been investigated in primates respectively (Bentwich et al. 2005; Zhang et al. 2007; Li et al. 2010). Our studies showed that these two miRNA clusters belong to the same miRNA family, which originated from a common ancestor and underwent fast amplification and rapid divergence. We optimized the parameters for homology detection, which helped us identify more spermiR family members than were identified in previous studies (Bentwich et al. 2005; Zhang et al. 2007; Li et al. 2010) and establish a more comprehensive evolutionary picture of this miRNA family in eutherians and marsupials (opossum) beyond primates. Although the expression of many spermiRs has been detected in the testes of some representative species, we could not rule out the possibility that some of the putative spermiRs might be pseudogenes.
In this study, we identified one spermiR homolog located between Slitrk2 and Fmr1 on the X chromosome of opossum (marsupial), which appears to be derived from a mutated THER1 (figs. 4A and5A). Previous studies proposed that sex chromosomes emerged from autosomes in the common ancestor of eutherians and marsupials (Potrzebowski et al. 2008; Veyrunes et al. 2008). Intriguingly, we found that the density of miRNA genes on the X chromosome of opossum is much higher than that on the X chromosome of birds and placental mammals (supplementary fig. S3A, Supplementary Material online), suggesting the rapid birth of new X-linked miRNAs around the time of emergence of the X chromosome. After divergence from the common eutherian ancestor, different mammals underwent frequent lineage-specific spermiR expansion, and two clades were formed, namely, spermiR-L (the miR-743a-465 cluster in rodents or the miR-892-891 cluster in other mammals) and spermiR-R (the miR-513-514 cluster) (fig. 4A), which might have been due at least in part to tandem duplication mediated by flanking retrotransposable elements (fig. 5F and supplementary fig. S3B–D, Supplementary Material online). A previous study compared the relative density of genes on individual chromosomes across 450 My and found that two bursts of protein-coding gene expansion occurred on the X chromosome (Zhang et al. 2010). One of these bursts occurred around the time of the divergence of eutherian mammals and marsupials, and the duplication of genomic loci was the major origin of the new genes. Our analyses show that spermiRs undergo rapid duplication in eutherian mammals but not in marsupials, indicating that similar events might have driven the amplification of spermiRs as protein-coding genes during mammalian evolution.
It has been reported that selective pressure to compensate for the silencing of the sex chromosome during MSCI results in the duplication of many X-linked genes as functional retrogenes on autosomes and that these retrogenes are biased toward expression in the testes, especially in the meiotic and postmeiotic phases of spermatogenesis (Vinckenbosch et al. 2006; McLysaght 2008; Potrzebowski et al. 2008). In addition, more than half of human miRNA clusters have corresponding paralogous clusters in distant genomic loci or on different chromosomes (Yu et al. 2006). Interestingly, even though spermiRs have undergone extensive amplification in placental mammals and are the predominant miRNA family on the X chromosome in primates and rodents, no spermiRs have moved outside the relatively small genomic region between the genes Slitrk2 and Fmr1 in the representative species that we investigated (fig. 4A), possibly due to some unknown selective pressure and/or structural barriers in the adjacent chromosome regions that constrain the movement of spermiRs. Whether small RNAs, such as miRNAs or piRNAs, escape the X chromosome as protein-coding genes has not yet been investigated. We believe that this question is interesting and warrants further investigation.
The X chromosome may experience stronger positive selection than autosomes because recessive beneficial mutations will be more visible to selection on the X chromosome, where they will spend less time being masked by a dominant, less-beneficial allele—a proposal known as the faster-X hypothesis (Kayserili et al. 2012; Coolon et al. 2015). Sex-linked protein-coding genes have been shown to have higher rates of adaptive sequence evolution than autosome-linked protein-coding genes in fruit flies, birds, and mammals, supporting the faster-X hypothesis (Mank et al. 2007; Hvilsom et al. 2012; Kayserili et al. 2012). The testis-specific high expression pattern (fig. 6A), as well as the divergence in the precursor and seed sequences within and among mammalian species (fig. 2), makes spermiRs an excellent example fitting the faster-X hypothesis. Variations in miRNA genes and/or their target repertoires have been proposed to be key drivers for phenotypic differences between species (Liu et al. 2008; Rosa and Brivanlou 2009; Zheng et al. 2011). Multiple members with diverse expression patterns, extensive sequence polymorphisms in the seed region, and effective functional compensatory mechanisms among different members might have made spermiRs more flexible at generating a complex influence on the reproductive system across different lineages during evolution.
Are X-Linked miRNAs Silenced during MSCI?
Whether X-linked miRNA genes are silenced in a similar manner as protein-coding genes during MSCI remains controversial (Song et al. 2009; Royo et al. 2015; Sosa et al. 2015). Our small RNA sequencing results using purified male germ cells at different stages of mouse spermatogenesis showed that X-linked miRNAs account for over 75% of all miRNAs in the leptotene stage, whereas their expression starts to be downregulated in zygotene spermatocytes and is continuously decreased to a very low level in round spermatids (fig. 1D). This decrease is not caused by a few highly abundant miRNAs. Instead, almost all X-linked miRNAs are downregulated in the late meiotic phases (fig. 1G). A recent study using RNA FISH to detect individual primary miRNA transcripts showed that the transcription of X-linked miRNA genes was active in spermatogonia but was silenced in cells in the pachytene stage (Royo et al. 2015). The discrepancy between our results and those of a previous study (Song et al. 2009) might be explained by the fact that miRNAs have a longer half-life than their primary transcripts (Gantier et al. 2011); X-linked miRNAs, mainly spermiRs, are produced at extremely high levels in the early stages of meiosis and even a small portion of those remaining might still be readily detected even though the pri-miRNAs are no longer transcribed during MSCI.
Materials and Methods
Small RNA Library Preparation and Categorization
Total cellular RNA was extracted using TRIzol reagent (Takara). Approximately 1 µg of total cellular RNA was used to construct a small RNA library according to the Illumina protocol. Equal amounts of six exogenous synthetic spike-in mixes were added to each RNA sample from rabbit testis tissue at different stages before small RNA library construction. High-throughput RNA sequencing was performed using Hi-Seq 2000 with 50 or 150 run cycles (Genergy Biotechnology). The raw fastq data were preprocessed by FASTX_Toolkit. After quality filtering, the 3′ adapters were clipped from the sequencing reads. Read sequence that was unable to match the 3′ adapter or with a length shorter than 17 bp after clipping of the 3′ adapter was discarded. The redundant sequences were collapsed as useful reads for further analysis. The useful reads were mapped to the genome by Bowtie (Langmead et al. 2009) (Bowtie options –best –strata –v 0 –k 100). The reads were aligned sequentially to known miRNA, tRNA, rRNA, snoRNA, and snRNA sequences by Bowtie without any mismatch allowed. The remaining 25–32-nt sequences were used to identify piRNAs according to the previously described method (Jung et al. 2014) with slight modifications. The clustering parameters were determined to be MinReads = 4 and Eps = 2,500 bp by running a series of k-dist analyses with different Eps and MinReads values. All of the candidate clusters that satisfied these parameters were considered piRNA clusters, and the remaining 25–32-nt sequences located in these clusters were defined as piRNAs without any further filtering. Only the reads that exactly matched the 5′ start site of annotated miRNAs and matched the 3′ ends with at most 2-nt deletions and/or 2-nt additional sequences derived from pre-miRNAs were counted in the abundance of miRNAs. The counts of the individual miRNAs were normalized to that of the total miRNAs or spike-ins. The sequences of the six spike-ins are provided in the supplementary information, Supplementary Material online.
RNA Sequencing and Data Analysis
Approximately 1 µg of total RNA was used to construct a cDNA library using the NEBNext Ultra Directional RNA Library Prep Kit (NEB) according to the manufacturer’s protocol. The cDNA libraries were sequenced by Hi-Seq 2000 with 2× 150 run cycles. The raw fastq data were mapped to the genome by STAR with the default parameters. The gene expression (in FPKM) was calculated by RSEM. The gene annotation was downloaded from GENCODE (version M2). The processed RNA sequencing data for human adult testes were downloaded from the ENCODE in bigWig format (https://www.encodeproject.org/experiments/ENCSR344MQK/; last accessed November 15, 2018). The processed RNA-seq data for rabbit, monkey, rat, and pig adult testes were downloaded from Ensembl (http://ftp.ensembl.org/pub/release-83/bamcov/; last accessed July 10, 2017) in bigWig format. The transcript abundance of each spermiR precursor was calculated as the total reads mapped to the location of the spermiR precursor with a sequence extension of 200-bp upstream and downstream.
Annotation Sources and Genome Assemblies
Genome sequences for all surveyed species were downloaded from the UCSC Genome Browser (Rosenbloom et al. 2015). The following genome assemblies were used in this study: human, hg19; chimpanzee, panTro4; gorilla, gorGor3; gibbon, omLeu3; orangutan, ponAbe2; monkey, rheMac3; squirrel monkey, saiBai1; mouse, mm10; rat, rn6; pig, susScr3; rabbit, oryCun2; dog, canFam3; cat, felCat5; common shrew, sorAra2; opossum, monDom5; chicken, galGal4; and zebrafish, danRer7. Known RNA annotations were retrieved from the following databases: miRNA and miRBase (Kozomara and Griffiths-Jones 2011) (version 18.0); tRNAs, Genomic tRNA Database; rRNAs, 18S and 28S from NCBI GenBank and 5S and 5.8S from Ensembl; and snoRNAs and snRNAs from Ensembl.
Genome-Wide Identification of miRNA Homologs
We searched for spermiR homologs with Blast (Altschul et al. 1990), which was optimized to detect locally similar sequences, and Exonerate (Slater and Birney 2005), which was optimized to detect globally similar regions. The known spermiRs from four species (human, monkey, mouse, and rat) were used as queries to search for homologs in the genomes of other species. We determined the Blast parameter E-value ≤ 10−2 and the Exonerate parameter –percent ≥ 50% by mapping known spermiRs across these four species. After precursor sequences were determined in one species, the identified spermiRs were accumulated as queries for the next ortholog search in other species. After searching the orthologs in one species, the identified orthologs were also accumulated as queries to search for paralogs in this species. The precursor sequence of each identified spermiR was manually inspected by multiple alignments, named according to the most similar known spermiR from humans, monkeys, mice, and rats and assigned a number in order of physical location on the genome. Data for all of the identified spermiR names, precursor sequences, mature sequences, genomic loci, predicted secondary structures, and annotation status by miRBase version 18 are provided in supplementary Data S4, Supplementary Material online.
Sequence Analysis and Phylogenetic Tree
Slitrk2-Fmr1 syntenic regions were identified using UCSC whole-genome chained sequence alignments (Rosenbloom et al. 2015). Multiple sequence alignments were performed using ClustalW (Thompson et al. 1994). RNA secondary structures were predicted by RNAfold (Martinez 1990) and mfold (Zuker 2003) with the default settings. Phylogenetic trees were constructed using the neighbor-joining method in MEGA5 (Tamura et al. 2011) with 1,000 bootstrap replications. A Kimura two-parameter model was used to calculate the distance between spermiR precursors. RepeatMasker (Smit and Riggs 1996) and CENSORS (Jurka et al. 1996) were used to identify repeat elements.
miRNA-Target Prediction and Regulatory Function Analysis
miRNA-target prediction was performed using Probability of Interaction by Target Accessibility (Kertesz et al. 2007), which can identify nonconserved target sites. The target gene enrichment analysis of the five most highly expressed spermiR-Ls in mice or spermiR-Rs in humans was based on the biological process Gene ontology (GO) terms (Barrell et al. 2009). Welch’s two-sample t-test was performed for statistical analysis.
Data Access
All data used to obtain the conclusions are presented in the article and the supplementary information, Supplementary Material online. The deep sequencing data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE101933. Novel miRNA genes have been submitted to miRBase (http://www.mirbase.org/). Other data may be requested from the authors.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Zhi Lu for direction on the analysis of the small RNA deep sequencing data. We thank Chen Chu from Dr Yonglian Zhang’s lab for providing the segmental epididymis of adult mice. We thank Dr Renfu Shang for performing the reporter assays. We thank Guangqin Wang for implementing the HE staining of rabbit testis tissue at different stages of spermatogenesis. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19040102 to L.W.), the Ministry of Science and Technology of China (2017YFA0504400 and 2014CB943100 to L.W.), the CAS-Shanghai Science Research Center (CAS-SSRC-YJ-2015-01 to L.W.), the National Natural Science Foundation of China (91440107 and 31470781 to L.W.), the Shanghai Municipal Commission for Science and Technology (12JC1409400 to L.W.), and the State Key Laboratory of Molecular Biology.
Author Contributions
F.Z., Y.Z., and L.W. conceived and designed the study. F.Z. performed the computational analysis. Y.Z. and X.L. isolated and characterized the male germ cells from mice. B.X. and H.Z. constructed the cDNA libraries of the small RNAs. F.Z., Y.Z., J.Y., H.L., and L.W. wrote the manuscript.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442(7099):203–207. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE.. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 73(4):823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R.. 2009. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 37(Database):D396–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL.. 2002. Alu repeats and human genomic diversity. Nat Rev Genet. 3(5):370–379. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M.. 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 74(1):68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 37(7):766–770. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ.. 2003. Dicer is essential for mouse development. Nat Genet. 35(3):215–217. [DOI] [PubMed] [Google Scholar]
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP.. 2010. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16(4):720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier JM, Turner JM.. 2010. Meiotic sex chromosome inactivation. Curr Biol. 20(22):R962–R963. [DOI] [PubMed] [Google Scholar]
- Coolon JD, Stevenson KR, McManus CJ, Yang B, Graveley BR, Wittkopp PJ.. 2015. Molecular mechanisms and evolutionary processes contributing to accelerated divergence of gene expression on the Drosophila X chromosome. Mol Biol Evol. 32(10):2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA.. 1999. Alu repeats and human disease. Mol Genet Metab. 67(3):183–193. [DOI] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39(13):5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Labuda D.. 1999. CORE-SINEs: eukaryotic short interspersed retroposing elements with common sequence motifs. Proc Natl Acad Sci U S A. 96(6):2869–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA.. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442(7099):199–202. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H.. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20(13):1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA. 2004. The XY body: a specialized meiotic chromatin domain. Exp Cell Res. 296(1):57–63. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32(4):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C, Qian Y, Bataillon T, Li Y, Mailund T, Salle B, Carlsen F, Li R, Zheng H, Jiang T, et al. 2012. Extensive X-linked adaptive evolution in central chimpanzees. Proc Natl Acad Sci U S A. 109(6):2054–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I, Park JC, Kim S.. 2014. piClust: a density based piRNA clustering algorithm. Comput Biol Chem. 50:60–67. [DOI] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P.. 1996. CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 20(1):119–121. [DOI] [PubMed] [Google Scholar]
- Kayserili MA, Gerrard DT, Tomancak P, Kalinka AT.. 2012. An excess of gene expression divergence on the X chromosome in Drosophila embryos: implications for the faster-X hypothesis. PLoS Genet. 8(12):e1003200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E.. 2007. The role of site accessibility in microRNA target recognition. Nat Genet. 39(10):1278–1284. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim VN.. 2012. MicroRNA factory: RISC assembly from precursor microRNAs. Mol Cell 46(4):384–386. [DOI] [PubMed] [Google Scholar]
- Kotaja N. 2014. MicroRNAs and spermatogenesis. Fertil Steril. 101(6):1552–1562. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S.. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39(Database):D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL.. 2003. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A. 100(11):6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL.. 2004. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol Reprod. 71(3):722–731. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S, Van Loo P, Thilakarathne PJ, Marynen P, Verbeke G, Schuit FC.. 2009. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS One 4(2):e4456.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Dong D, Zhang Z.. 2010. Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol. 27(3):671–683. [DOI] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50(1):67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, Cui S.. 2012. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One 7(3):e33861.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC.. 2008. The evolution and functional diversification of animal microRNA genes. Cell Res. 18(10):985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H.. 2007. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 17(5):618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez HM. 1990. Detecting pseudoknots and other local base-pairing structures in RNA sequences. Methods Enzymol. 183:306–317. [DOI] [PubMed] [Google Scholar]
- McLysaght A. 2008. Evolutionary steps of sex chromosomes are reflected in retrogenes. Trends Genet. 24(10):478–481. [DOI] [PubMed] [Google Scholar]
- Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H.. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res. 23(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer D. 1991. Sperm preparation techniques and iatrogenic failures of in-vitro fertilization. Hum Reprod. 6(2):173–176. [DOI] [PubMed] [Google Scholar]
- Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, Brinster RL.. 2011. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 108(31):12740–12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L. 2014. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis 4(2):e979623.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Minella AC, He L.. 2015. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci Signal. 8(368):re2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H.. 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 6(4):e80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Eskin E, Pevzner PA.. 2004. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 14(11):2245–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W.. 2007. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 311(2):592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Y, Meikar O, Papaioannou MD, Conne B, Grey C, Weier M, Pralong F, De Massy B, Kaessmann H, Vassalli JD, et al. 2011. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One 6(10):e25241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH.. 2009. MicroRNAs in early vertebrate development. Cell Cycle 8(21):3513–3520. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43(D1):D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Seitz H, ElInati E, Peters AH, Stadler MB, Turner JM.. 2015. Silencing of X-linked microRNAs by meiotic sex chromosome inactivation. PLoS Genet. 11(10):e1005461.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351(6271):391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6(1):31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI.. 2005. Mammalian microRNAs derived from genomic repeats. Trends Genet. 21(6):322–326. [DOI] [PubMed] [Google Scholar]
- Smit AF, Riggs AD.. 1996. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A. 93(4):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TM, Lee MK, Szabo CI, Jerome N, McEuen M, Taylor M, Hood L, King MC.. 1996. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 6(11):1029–1049. [DOI] [PubMed] [Google Scholar]
- Smorag L, Zheng Y, Nolte J, Zechner U, Engel W, Pantakani DV.. 2012. MicroRNA signature in various cell types of mouse spermatogenesis: evidence for stage-specifically expressed miRNA-221, -203 and -34b-5p mediated spermatogenesis regulation. Biol Cell 104(11):677–692. [DOI] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W.. 2009. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet. 41(4):488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, et al. 2014. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510(7503):115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa E, Flores L, Yan W, McCarrey JR.. 2015. Escape of X-linked miRNA genes from meiotic sex chromosome inactivation. Development 142(21):3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhang Y, Zhang R, Qi X, Su B.. 2013. Functional divergence of the rapidly evolving miR-513 subfamily in primates. BMC Evol Biol. 13(1):255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ.. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. 2007. Meiotic sex chromosome inactivation. Development 134(10):1823–1831. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, et al. 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18(6):965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H.. 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 103(9):3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H.. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20(13):1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W.. 2014. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A. 111(28):E2851–E2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, Griswold MD, Namekawa SH, Royo H, Turner JM, et al. 2012. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem. 287(30):25173–25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RP, Kotaja N.. 2014. Small RNAs in spermatogenesis. Mol Cell Endocrinol. 382(1):498–508. [DOI] [PubMed] [Google Scholar]
- Yan N, Lu Y, Sun H, Tao D, Zhang S, Liu W, Ma Y.. 2007. A microarray for microRNA profiling in mouse testis tissues. Reproduction 134(1):73–79. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW.. 2006. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 349(1):59–68. [DOI] [PubMed] [Google Scholar]
- Yu Z, Raabe T, Hecht NB.. 2005. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 73(3):427–433. [DOI] [PubMed] [Google Scholar]
- Zhang R, Peng Y, Wang W, Su B.. 2007. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 17(5):612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang YQ, Su B.. 2008. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol. 25(7):1493–1502. [DOI] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M.. 2010. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 8: e1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Ravi A, Gould GM, Burge CB, Sharp PA.. 2011. Genome-wide impact of a recently expanded microRNA cluster in mouse. Proc Natl Acad Sci U S A. 108(38):15804–15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ.. 2009. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol. 9(1):38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31(13):3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.