Abstract
Inhibition of factor Xa (fXa) by antithrombin (AT) enabled by heparin or heparan sulfate is critical for controlling blood coagulation. AT activation by heparin had been investigated extensively, while heparin interaction with the trapped AT/fXa intermediates received relatively little attention. We use native electrospray ionization mass spectrometry to study the role of heparin chains of varying length (hexa-, octa-, deca- and eicosa-saccharides; dp6, dp8, dp10 and dp20) in AT/fXa complexes assembly. Despite being critical promoters of the AT/Xa binding, shorter heparin chains are excluded from the final products (trapped intermediates). However, replacement of short heparin segments with dp20 gives rise to a prominent ionic signal of ternary complexes. These species are also observed when the trapped intermediate is initially prepared in the presence of a short oligoheparin (dp6), followed by addition of a longer heparin chain (dp20), indicating that heparin binding to AT/fXa complexes takes place after the inhibition event. The importance of the heparin chain length for its ability to associate with the trapped intermediate suggests that the binding likely occurs in a bidentate fashion (where two distinct segments of oligoheparin make contacts with the protein components, while the part of the chain separating these two segments is extended into solution to minimize electrostatic repulsion). This model is corroborated by both molecular dynamics simulations with an explicit solvent and ion mobility measurements in the gas phase. The observed post-inhibition heparin binding to the trapped AT/fXa intermediates hints at the likely role played by heparan sulfate in their catabolism.
Introduction
A century following its discovery,1 heparin (as well as polysaccharides derived from2 and inspired by it3) remains the most effective and commonly used anti-coagulation agent despite growing availability of direct inhibitors of proteases initiating blood clotting.4 The inhibition of these proteases (such as thrombin and factor Xa, fXa) is triggered by the interaction of heparin or heparan sulfate with antithrombin (AT), one of the most important elements in regulating the entire blood coagulation cascade in vertebrates5, 6 (structurally related polysaccharides, such as chondroitin sulfate, also possess the ability to modulate coagulation via interaction with AT7, 8). Binding of a heparin chain to an AT molecule results in an allosteric conformational change, exposing a reactive center loop (RCL), which upon insertion into the catalytic site of the protease forms a Michaelis-like complex.9 This, in turn, triggers a series of events (common to all trypsin-like serpins10) whereby the catalytic serine residue of fXa reacts with the arginine residue of the serpin’s RCL, leading to the formation of an O-acyl intermediate. Although this cleaves the RCL chain, AT remains covalently bound to fXa via arginine’s carbonyl at the cleavage site. Cleavage of the RCL chain allows AT to assume a lower-energy conformation by undergoing a large-scale transition where the cleaved RCL is inserted (as an additional strand) in the middle of a five-strand β-sheet, moving the covalently attached fXa molecule to the side opposite to that where the initial contact had been made.9 This results in a significant deformation of the fXa active site structure (which remains covalently bound to RCL), preventing it from completing the final (hydrolytic de-acylation) step in proteolysis, and trapping the acyl intermediate.9
While AT acts as a suicide inhibitor (i.e., undergoes irreversible structural changes), the heparin chain activating this serpin is not “consumed” and can act as a catalyst (it is released from the trapped acyl intermediate and can participate in another reaction5). However, even though the large conformational change within AT upon fXa inhibition is believed to result in a significant reduction of heparin affinity, the glycosaminoglycan/protein interactions may still occur. In fact, both latent and cleaved forms of AT (which bear significant structural resemblance to the AT molecule trapped within the fXa/AT acyl intermediate – a trait shared by nearly all serpins11) are capable of binding both the pentasaccharide molecule (a strong minimal-length AT binder3) and longer heparin chains, although the affinity is reduced at least 30-fold compared to native AT.12 Despite the relatively low affinity, the interactions of the cleaved and latent forms of AT with heparin and heparan sulfate are physiologically relevant and had been shown to play a role in a variety of processes ranging from angiogenesis13 to inflammation.14
Despite the well-documented involvement of heparin (and its next-of-kin heparan sulfate) in mediation of physiological effects exerted by the latent and cleaved forms of AT, the possibility (as well as potential implications) of these glycosaminoglycans’ interactions with AT within the trapped serpin/protease intermediate had been largely overlooked. This might be a result of a common perception of the AT/fXa trapped intermediate as the endpoint of the inhibition process, which is inert and destined for degradation. However, even though the proteolytic activity of fXa within the trapped intermediate is greatly suppressed, it is not completely eliminated. In fact, the serpin/protease traps are kinetic, and ultimately they do break down to release a cleaved (inactive) serpin and a fully active protease.15 The de-acylation rate in the trapped complex is low (minutes to months depending on a particular serpin/protease pair15), but can be accelerated by certain co-factors (e.g., Ca2+ ions in the case of AT16). Clearly, there must be an effective clearance mechanism in place to remove the trapped AT/fXa intermediates from circulation prior to their breakdown and release of the active protease.
The search for “serpin receptors”17 responsible for the clearance of kinetically trapped (but thermodynamically unstable) protease/inhibitor complexes initially focused on cell-surface receptors that may recognize the unique conformation of such complexes,18 and eventually led to the identification of the low density lipoprotein receptor-related protein (LRP)19 as the receptor assisting in internalization of the complexes followed by their routing to lysosomes for degradation.20, 21 Intriguingly, heparan sulfate has been shown to be intimately involved in the LRP-mediated degradation pathway for at least two serpin/protease trapped intermediates,22, 23 one of which is the AT-inhibited thrombin,23 a protease that is highly homologous to fXa. However, the AT/thrombin complex also contains an accessory protein vitronectin (acting as an opsonin promoting the clearance of trapped thrombin molecules), which has significant affinity to heparan sulfate,24 and it remains unclear whether heparan sulfate can bind to (and assist in internalization of) the AT/fXa complexes without the help of accessory proteins. In fact, it is still not known whether the trapped AT/fXa complexes remain bound to or can re-bind the polyanionic glycosaminoglycan chains, such as heparin or heparan sulfate.
We set out to investigate the possibility of interactions between the trapped AT/fXa intermediate and heparin oligomers using native electrospray ionization mass spectrometry (ESI MS). This technique had been enjoying growing acceptance within the biochemistry and biophysics communities in the past decade as a reliable tool to study non-covalent associations of biopolymers, including proteins, nucleic acids and polysaccharides.25 Until recently, the use of native MS in the studies of protein/heparin interactions has been limited to relatively short heparin oligomers26–30 due to the extreme structural heterogeneity exhibited by heparin and other heparin-like glycosaminoglycans, which makes interpretation of the ESI MS data challenging. However, recent advances in the ESI MS analysis of highly heterogeneous biopolymers, such as the introduction of the limited charge reduction in the gas phase31 have resulted in a significant expansion of the range of glycosaminoglycans for which meaningful ESI MS analyses can be carried out, including protein interaction with intact unfractionated heparin.32
The approach adopted in this work relies on the ability of ESI MS to deduce both composition and stoichiometry of covalent and non-covalent complexes from their masses. If the assignment becomes ambiguous due to significant structural heterogeneity exhibited by heparin oligomers, ESI MS measurements are aided by limited charge reduction of ions in the gas phase. We observe that despite being critical promoters of the AT/Xa binding, shorter heparin chains (up to at least a decasaccharide) are not incorporated in the trapped intermediates (only binary AT/fXa complexes are detected by native ESI MS). However, formation of ternary complexes (AT/fXa/heparin) is observed when longer heparin chains (eicosaccharides) are either used to activate AT or added to the protein solution after the trapped intermediate had been formed. The observation that only relatively long heparin chains are capable of binding to the trapped intermediate suggests that heparin interaction with either of the two proteins in the complex alone is too weak, and the binding must occur in a bidentate fashion, where a single heparin chain makes contacts with both AT and fXa. The occurrence of the post-inhibition binding of long glycosaminoglycan chains to the AT/fXa trapped intermediate suggests that heparan sulfate is likely to play a role in catabolizing these trapped intermediates by presenting them to cell-surface receptors that mediate their internalization via endocytosis. Facilitation of the endosomal uptake (followed by degradation in the lysosomal compartments) is important for maintaining the fidelity of the blood coagulation cascade, as accumulation of a large pool of the AT/fXa trapped intermediates (where the proteolytic step is greatly decelerated, but not completely stopped) will inevitably lead to release of active fXa molecules back to circulation.
Materials and Methods
Materials.
Human Factor Xa (fXa) was purchased from Hematologic Technologies, Inc. (Essex Junction, VT). Factor Xa contains two subunits: heavy chain (contains the catalytic domain) and light chain (contains the N-terminal gamma-carboxyglutamic acid-rich (Gla)-domain and two EGF-like domains). The α-glycoform of native human antithrombin (AT) and latent AT were generously provided by CSL Behring, Inc. (Marburg, Germany). All protein samples were desalted in 150 mM ammonium acetate (pH 7.0) by ultrafiltration with a 10 kDa MW cutoff filter devices (Millipore, Billerica, MA). The protein solution concentrations were verified by measuring UV-VIS absorbance (using molar absorptivity of 37275 M−1 cm−1 and 48370 M−1 cm−1 at 280 nm for AT and fXa, respectively). Heparin oligomers of fixed length (dp6, dp8, dp10 and dp20), prepared by partial heparin digestion using bacterial heparinase followed by high resolution gel filtration, were generously donated by Dr. John Gallagher (Iduron, Manchester, UK). All other chemicals and solvents used in this work were of analytical grade or higher. Formation of AT/fXa trapped intermediates was carried out by incubating AT with fixed-length heparin oligomer at 7.5 or 15 μM for 5 min. followed by addition of fXa and incubating this mixture for 10 min. (the final molar ratio: [AT]/[fXa] = 1.2) in 150 mM ammonium acetate (pH 7.0). All incubation steps were performed at 23 °C.
Size Exclusion Chromatography.
All size exclusion chromatography (SEC) measurements were carried out using an HP1100 (Agilent, Santa Clara, CA) liquid chromatograph equipped with a TSKgel G2000xl (TOSOH, Tokyo, Japan) column. Ammonium acetate (150 mM, pH 6.9) was used as a mobile phase. All separations were carried out at a flow rate of 0.3 mL/min. UV absorption at 232 nm was used for detection.
Mass Spectrometry.
Native ESI MS measurements were carried out with a QStar-XL (ABI/Sciex, Toronto, Canada) hybrid quadrupole/time-of-flight mass spectrometer in the nanospray mode. Glass nanospray capillaries (New Objective, Woburn, MA, USA) of 2 μm id were used in this work. The stability of non-covalent protein complexes in the gas phase was maximized using the collisional cooling in the ESI interface region and the following setting for the ion optics: DP, 140; FP, 280–300; and DP2, 25. Characterization of the ternary AT/fXa/dp20 complex formed via binding of dp20 to a pre-formed trapped intermediate was carried out by adding dp20 (15 μM) to the AT/fXa (molar ratio: [AT]/[fXa] = 1.5) complex formed in the presence of dp6 (30 μM) in 150 mM ammonium acetate (pH 7.0) followed by MS analysis with a Synapt HD (Waters, Milford, MA) hybrid quadrupole/time-of-flight mass spectrometer equipped with a tri-wave ion mobility analyzer. Limited charge reduction of multiply charged ions in the gas phase was initiated by isolating the ions in the m/z window 4871–4921 followed by reactions with 1,4-dicyanobenzene radical anions prior to acquisition of the product ion mass and mobility spectra. The trap wave height was set at 0.1 V, and trap wave velocity was 300 m/s. The IMS wave height and wave velocity were set at 33V and 300 m/s, respectively. The transfer wave height was 3 V, and the transfer wave velocity was 85 m/s.
Molecular Modeling.
All computational procedures were carried out using applications (Prime, Maestro, Epik, Desmond) from the Schrödinger software package (v. 2016–1, Schrodinger, LLC, New York, NY, USA). The model of the covalently trapped AT/fXa O-acyl intermediate was prepared using a homology model of human AT (Homology Modeling Workflow within Prime) templated by a cleaved form of bovine AT III (PDB id: 1ATT; 89% sequence identity) and apo form of fXa (PDB id: 1C5M). All-atom models of both proteins with optimized protonation assignments were produced using Protein Preparation Wizard (Maestro, Epik). The model of human AT was furnished with the energy-optimized 57SPEKK61 loop, absent in the template (Prime’s Refine Loops tool), and was truncated by a single residue from the C-terminal end of the cleaved site to reveal the carbonyl of Arg426 as the O-acylation partner. The fXa structure was also truncated to the β-fXa form by removing unstructured C-terminal residues removed via autoproteolysis between Arg245 and Gly246. The elastase/serpin trapped intermediate (PDB id: 2D26) was used as a template for both relative orientation of the two proteins and conformational preferences of the regions involved in the formation of the covalent intermediate (i.e., 424GR425 acyl fragment of AT, unstructured in the 1ATT template, and catalytic Ser195 of fXa). The covalent complex was then energy-minimized using OPLS3 force field and VGSB solvation model (Prime). The dp20 model was created by removing four saccharide units from the reducing end of dp24 (PDB id: 3IRJ, model 1). The non-reducing end of the resulting oligosaccharide was placed in a close contact with the heparin-binding site of fXa, as revealed by the ternary AT/thrombin/SR123781 complex (PDB id: 1TB6) featuring uncleaved (Michaelis) complex between a protease, its cognate serpin and the synthetic 16-mer heparin mimetic. Thus, the sulfate groups of the terminal saccharides at the non-reducing end of the dp20 model were manually superimposed with sulfates of SR123781 in contact with a positive patch on the surface of thrombin, which is partially conserved in fXa (R93, K236, K240). Minimization of the resulting complex provided a starting structure for a subsequent simulated annealing experiment. The 1.2-ns simulation was set up using a neutralized system (with 16 Na+ ions placed in a close proximity to the sulfate groups of the eight intervening saccharides) with explicit water and 150 mM NaCl ions (Desmond). The following program was used: 200-ps transition from 0 to 300K, 100-ps transition to 400K followed by 200-ps Molecular Dynamics (MD) simulation at 400K, 500-ps annealing to 300K and 200-ps MD simulation at 300K. The final frame of the simulation was minimized using the OPLS force field to provide a model for the ternary AT/fXa/dp20 complex.
Results and Discussion
Short heparin oligomers are effective triggers of fXa inhibition by AT, but do not bind to the trapped AT/fXa intermediate.
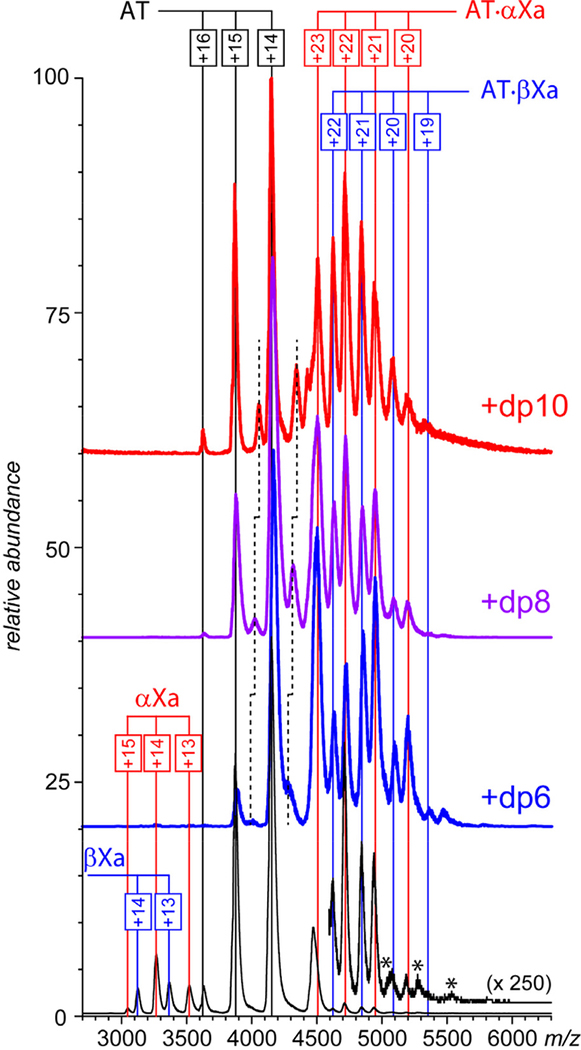
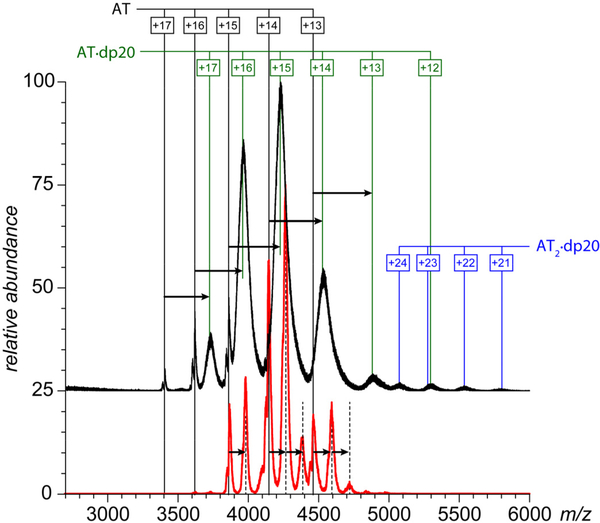
AT is typically represented in circulation by two glycoforms (termed α and β), the latter being less abundant, but more potent vis-à-vis fXa inhibition.33 In order to decrease the heterogeneity within the populations of protein complexes and avoid ambiguity in mass assignment, all experiments were carried out with the α-form of the protein. The fXa sample used in this work comprised two isoforms (α-fXa and β-fXa). The β-fXa isoform is formed as a result of autoproteolysis of α-fXa,34 and both proteins have equal activity in coagulation.35 Both proteins display abundant and well-defined ion peaks in ESI mass spectra (data not shown), which remain unchanged upon their mixing. The only novel feature observed in the mass spectrum of the AT/fXa mixture (with AT being present at a 20% molar excess) in the absence of heparin oligomers is the low-abundance signal in the high m/z region (> 4500 u) corresponding to ions whose masses are consistent with the AT·fXa complex formation (Figure 1, bottom). The intensity of these ion peaks is two orders of magnitude below those of free proteins, suggesting that the complexation process is very anemic.
Figure 1.
ESI mass spectra of mixtures of AT (1.8 μM) and fXa (1.5 μM) incubated in the absence of hepain oligomers (bottom) and in the presence of ca. 15 μM short-chain oligomers (as indicated on each panel) in 150 mM ammonium acetate (pH 7.0). Note that the detectable fXa signal (m/z region 3,000 – 3,700) can be observed only in the absence of heparin oligomers.
The appearance of the mass spectra of the AT/fXa mixture changes dramatically upon introduction of short heparin oligomers (dp6, dp8 or dp10) to the protein solution (at the AT/fXa/dpX molar ratio of 1.2 : 1.0 : 10). Abundant ionic signal corresponding to both AT·αfXa and AT·βfXa heterodimers is now observed in the high m/z region of the mass spectra (Figure 1), and the signal of both isoforms of fXa is completely eliminated (suggesting near-complete consumption of this protein). A signal of monomeric AT is still present in all mass spectra due to its molar excess (20%) in the original solution, along with the lower abundance AT·dpX complexes (indicated by dashed lines in Figure 1). The observed mass increases of the AT·dpX complexes formed in the presence of different heparin oligomers correspond to the differences in their average molecular weight (ca. 674 Da mass increments in the dp6-dp8-dp10 series). However, no such mass differences are observed for the AT·fXa complexes, and their measured masses are consistent with those calculated by summing the masses of the protein components (AT and αfXa, or AT and βfXa). This is fully consistent with the commonly accepted view of heparin serving as a catalyst in fXa inhibition by AT, and not being a part of the trapped intermediate.5
While the short heparin segments actively promote the interaction of AT with fXa very effectively (as manifested by a complete elimination of the fXa signal from the mass spectra), they fail to do so when the native AT is substituted with its latent form. The mass spectrum of the latent AT/fXa mixture incubated in the presence of dp10 shows no signs of either AT·αfXa or AT·βfXa ions, even though some binding of the heparin oligomer to latent AT is evident (Figure 2). This observation is fully consistent with the notion of latent AT being insensitive to activation with heparin and unable to interact with fXa, as it is already in the thermodynamically stable (inactive) form.11 The observed binding of heparin to the latent AT (which was also reported and studied previously12) fails to activate this form of the serpin, as the RCL segment has been already converted from a loop to a β-strand.11
Figure 2.
ESI mass spectra of mixtures of AT (1.8 μM) and fXa (1.5 μM) incubated in the presence of 0.02 mg/mL (ca. 15 μM) heparin decasaccharides (dp10). The red and blue traces show the results of experiment where active and latent forms of AT were used, respectively.
Longer heparin oligomers trigger fXa inhibition, and appear to remain bound to the trapped AT/fXa intermediate.
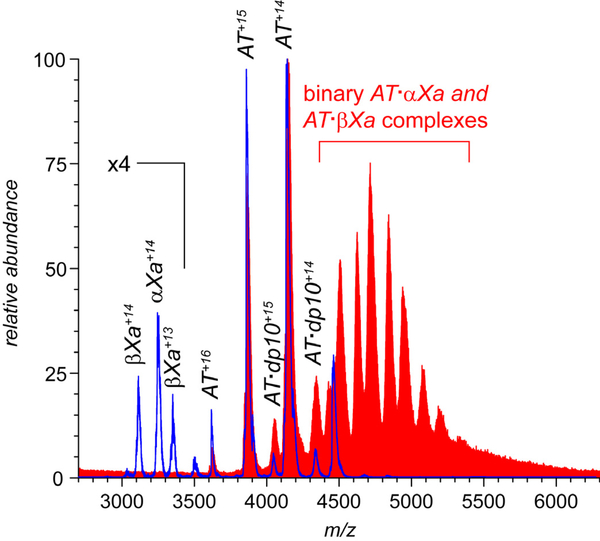
All MS data sets presented in Figures 1 and 2 are fully consistent with the commonly accepted view of heparin’s role in promoting the inhibition of fXa by AT: (i) the AT/fXa interaction in the absence of heparin is rather anemic leading only to negligible complex formation; (ii) heparin is incapable of inducing the interaction between AT and fXa when the former is already in the conformationally relaxed (thermodynamically favorable) latent state; and (iii) heparin acts as a catalyst in promoting the interaction between fXa and native (conformationally strained) AT leading to the AT·fXa heterodimer formation, but not being a part of this complex. However, introduction of a longer heparin oligomer (dp20) to the AT/fXa mixture (at the AT/fXa/dp20 molar ratio of 1.2 : 1.0 : 5.0) leads to a surprising result: while dp20 clearly promotes the AT/fXa interaction (as did shorter heparin oligomers), the resulting complexes are ternary, i.e. they incorporate the heparin chain in addition to the serpin and the protease (Figure 3). While the ionic peaks representing the macromolecular complexes in Figure 3 appear to be broad, the mass ranges derived from the spectra are in agreement with the heterogeneity of the dp20 chains (estimated to be 5,400±195 Da). A more definitive proof that the mass/charge assignments for the observed ionic species are done correctly is obtained by using limited charge reduction, a recently introduced experimental technique in which interpretation of MS data for highly heterogeneous macromolecules is aided by ion chemistry (electron capture or electron transfer reactions) and/or ion mobility measurements in the gas phase31, 32 (an example of such analysis is shown later).
Figure 3.
ESI mass spectra of mixtures of AT (1.8 μM) and fXa (1.5 μM) incubated in the presence of ca. 7.5 μM dp20 (top trace) and dp10 (bottom) in 150 mM ammonium acetate (pH 7.0).
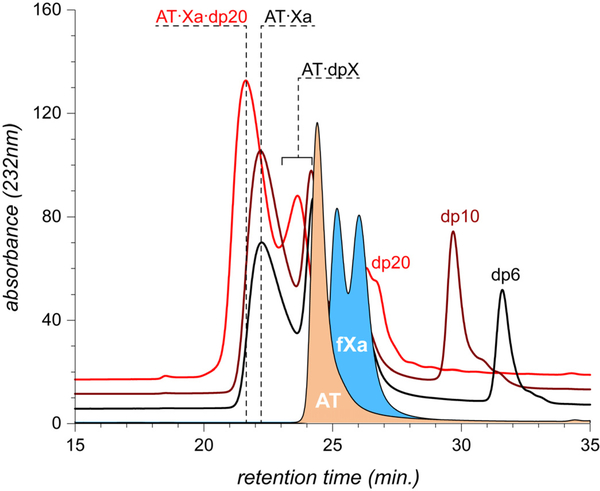
While mass spectrometry provides a clear indication that the composition of the AT·fXa complexes generated in the presence of the dp20 is different from those generated in the presence of shorter heparin chains, it may be argued that the results of native MS measurements do not always reflect the composition of the non-covalent assemblies in solution due to the possibility of their dissociation in the gas phase. The gas-phase instability of non-covalent macromolecular complexes is typically correlated with the role played by the hydrophobic interactions in stabilizing such assemblies in solution (elimination of the solvent reduces the hydrophobic interactions to weak van der Waals forces, which are frequently insufficient for maintaining the integrity of the non-covalent assemblies in the solvent-free environment36). Since hydrophobic forces are not likely to play a significant role in formation of the protein/heparin complexes, and the interaction is driven primarily by the electrostatic interactions, it seems unlikely that the removal of the solvent may compromise the stability of these macromolecular associations. Nevertheless, in order to completely exclude the possibility of the short heparin chains (e.g., dp6) being bound to the trapped AT·fXa intermediate in solution, but dissociating from it in the gas phase, these macromolecular assemblies were characterized by size exclusion chromatography (Figure 4). Formation of the trapped AT·fXa intermediate manifests itself via the appearance of an abundant early-eluting peak in all chromatograms where a heparin oligomer (dp6, dp10 or dp20) was present alongside AT and fXa. Importantly, the elution times of the trapped intermediate formed in the presence of dp6 and dp10 are identical, while the presence of dp20 in the AT/fXa mixture shortens the elution time of the complex by nearly a minute, consistent with the notion of dp20 being an integral part of the complex, unlike shorter heparinoids used in this study. MS analysis of the high molecular weight SEC fraction does not provide any signs of the presence of the shorter heparin chains, while dp20 is clearly seen as the part of the macromolecular assembly (see Supplementary Information for more detail).
Figure 4.
SEC chromatograms of AT/fXa mixture incubated in the presence of dp6 (black trace), dp10 (brown) and dp20 (red). In all cases protein concentrations were kept at 17 μM (fXa) and 22 μM (AT), while concentrations of the heparin oligomers were adjusted to 170 μM. The traces at the bottom of the diagram correspond to AT and fXa, respectively.
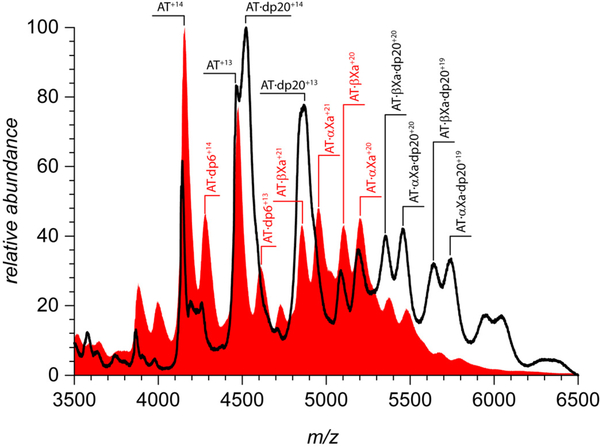
We also note that dp20 appears to be a more effective AT binder compared to shorter heparin oligomers: the most abundant ionic species in the mass spectrum of the AT/fXa/dp20 mixture shown in Figure 3 correspond to the AT/heparin oligomer complexes. While AT·dpX complexes are clearly present in the mass spectra shown in Figure 1 as well, their relative abundance is significantly lower. The same trend is seen in the mass spectra of AT/dp20 and AT/dp10 mixtures: even though in both cases AT/heparin oligomer complexes are predominant ionic species, longer chain lengths clearly favor the interaction with the protein (as becomes evident upon comparison of IAT/IAT·dpX ratios for dp10 and dp20 in Figure 5). The enhanced ability of longer heparin chains to interact with AT had been previously demonstrated using calorimetry,37, 38 and is likely to be related to the greater degree of structural diversity (sulfation and N-acetylation patterns) displayed by longer glycosaminoglycan chains compared to their shorter fragments.39 This would be true regardless of whether the protein/heparin (or protein/heparan sulfate) interactions are viewed through the prism of specific structurally defined “recognition elements” (such as the pentasaccharide sequence40) or less specific “distributed” characteristics (such as the sulfation patterns).41 Given the stronger AT affinity of longer heparin chains, one might be tempted to argue that they would be more likely to remain attached to the protein throughout the entire activation and fXa inhibition process. This, however, contradicts the observations by Olson and co-workers12 that longer heparin chains have in fact lower affinity to the cleaved and latent forms of AT (which are structurally similar to AT conformation within the trapped AT·fXa intermediate). Therefore, a more plausible explanation for the presence of the abundant signal of the ternary complexes AT·αfXa·dp20 and AT·βfXa·dp20 in the mass spectra of AT/fXa/dp20 mixture (Figure 5) would invoke the notion of the heparin oligomer dissociating from AT upon its activation and re-binding the AT·fXa complex following completion of the inhibition process.
Figure 5.
ESI mass spectra of 1.8 μM AT acquired in the presence of ca. 7.5 μM dp20 (top trace) and dp10 (bottom) in 150 mM ammonium acetate (pH 7.0). Ionic signals corresponding to AT/heparin oligomer complexes are prominent in each mass spectrum.
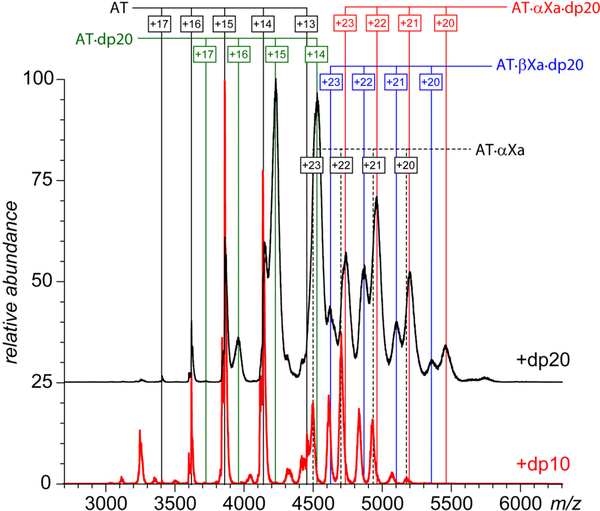
In order to check the feasibility of this scenario, the AT·fXa trapped intermediate was initially formed by incubating the AT/fXa mixture in the presence of short heparin oligomers (dp6) followed by addition of longer heparin chains (dp20). The resultant mass spectrum (black trace in Figure 6) provides unequivocal evidence for the formation of the ternary complexes (both AT·αfXa·dp20 and AT·βfXa·dp20), even though only binary AT·fXa complexes were present in the mass spectra (alongside unconsumed AT and AT·dp6 adducts) prior to the dp20 addition (at the AT/fXa/dpX molar ratio of 1.5 : 1.0 : 10). This clearly suggests that the ternary serpin/protease/heparin complexes (such as those observed in Figure 3) are formed during the final stage of a multi-step process,9 which includes (i) initial AT/heparin contact leading to AT activation; (ii) interaction of the activated AT with fXa leading to RCL cleavage and formation of the AT-fXa covalent bond; (iii) large scale conformational transition resulting in RCL insertion as a strand into a β-sheet leading to relocation of the covalently bound fXa to the opposite site of AT molecule and dissociation of the heparin chain; (iv) binding of another heparin to the kinetically trapped AT·fXa complex. In most of the current literature it is generally assumed that the trapped AT·fXa intermediate remains heparin-free, a supposition that can be linked to the original work by Craig et al., who did not observe a detectable difference in fluorescence signal for this complex in the presence and in the absence of heparin.42 It must be noted, however, that the fluorescence measurements may not reveal the presence of the binding partner if no tryptophan residues are affected by it. Furthermore, most of the original fluorescence measurements were carried out in high-salt solutions (300 mM), which typically results in effective screening of electrostatic interactions and, therefore, disfavors heparin interactions with its client proteins.43 On the contrary, our measurements were carried out at the physiological ionic strength, which should provide a relevant environment for electrostatically driven protein-heparin interactions.
Figure 6.
ESI mass spectra of mixtures of AT and fXa (4.5 and 3.0 μM, respectively) incubated in the presence of dp6 (30 μM) followed by the isolation of the trapped intermediate and addition of dp20 to a final concentration of 15 μM (black trace). The red trace shows a control mass spectrum acquired without post-isolation addition of the long heparin chains.
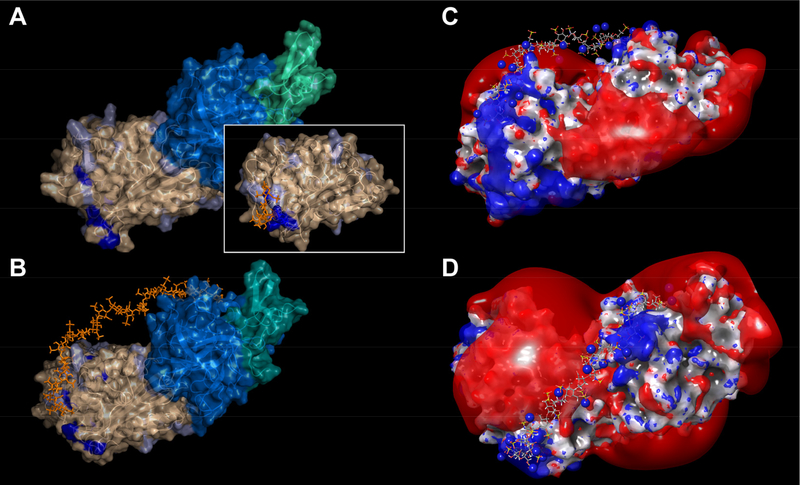
The fact that short heparin chains fail to re-bind to AT·fXa following completion of the inhibition process (Figure 1) suggests that the positive patches existing on the surfaces of the overall negatively charged AT (pI values ranging from 4.8 to 5.344, 45) and fXa (calculated pI value is 5.7) are insufficient to provide the requisite local attraction for the short polyanions on the background of strong repulsion due to the total negative charge on both protein components. Since the crystal structure of the trapped AT·fXa complex remains unavailable, we generated a structural model of this complex using a homology model of human AT, templated by a structure of a cleaved bovine AT38 and an apo-form of fXa,46 truncated to βfXa to reduce conformational complexity of the unstructured C-terminus. The initial fXa structure (PDB id: 1C5M) was carefully chosen from a wide pool of available fXa structures to feature the flexible 144–155 loop (missing in the available O-acyl complex crystal structures) in an conformational state compatible with the serpin structure brought into proximity in the AT·fXa complex. The two protein components were then linked covalently, using the existing crystal structure of a trapped intermediate for another serpin/protease pair: alpha1-proteinase inhibitor (α1P) and porcine pancreatic elastase (PPE).47 These two proteins are sufficiently homologous to fXa and AT (32% and 40% identity, respectively), allowing their covalent complex to be used as a template for modeling the structure of the AT·fXa trapped intermediate. Minimization of the complex upon covalent linkage has resulted in the perturbation of the catalytic triad residue of fXa in a manner consistent with the α1P·PPE structure.45 The energy-minimized model of the resulting O-acyl complex is presented in Figure 7A. We note that the contiguous positive patch accommodating the pentasaccharide on the surface of the free AT (shown as an inset in Figure 7A) loses its cohesion upon formation of the trapped intermediate (dark blue color in Figure 7A). While the basic residues appear to be spread out across the surface of both proteins in this model, the overall charge is negative, and calculations of the electrostatic potential reveal the extended segments of the protein surface generating strong negative potentials. Nevertheless, positively charged patches are also evident, with one such patch on the surface of AT corresponding to the cluster of amino acids, some of which participate in forming a pentasaccharide-binding site on the surface of native AT.48, 49 Likewise, several amino acid residues involved in a heparin-binding exosite on the surface of native fXa50, 51 also appear to cluster together in the homology model of the AT·fXa trapped intermediate, and generate positive potential locally.
Figure 7.
A model of a trapped AT/fXa intermediate prepared using a homology model of AT (PDB id: 1ATT) a conformationally compatible structure of fXa (PDB id: 1C5M) and an O-acyl serpin/elastase complex (PDB id: 2D26) as structural templates (A). The AT surface is colored in wheat, and fXa heavy and light chains are colored in marine blue and teal, respectively. The amino acid residues participating in the heparin pentasaccharide binding in free AT are colored in dark blue. A structure of free AT, rendered in the same fashion, is shown as an inset. Model of the ternary AT·fXa·dp20 complex produced by docking the dp20 chain (prepared by eliminating four saccharide units from a dp24 chain, PDB id 3IRG) to the AT/fXa intermediate and subsequent optimization by a simulated annealing sequence (B). The dp20 chain is shown in orange, and pale blue color on the AT surface identifies the positions of all basic residues beyond the initial pentasaccharide binding domain (which are shown in dark blue). Two representative orthogonal views of the ternary complex AT·fXa·dp20 generated by MD simulations showing the final position of the heparin chain with respect to the kT/2e isoelectric surfaces of the trapped intermediate (C and D).
Both of these positive patches may interact with polyanionic heparin chains, but these attractive forces will be mitigated to a significant extent by the repulsion from the negatively charged regions of the protein complex. Out of the four oligoheparins tested in this work, only the dp20 chain has sufficient length to (i) create contacts with the positive patches on the surfaces of both AT and fXa and (ii) create a kink to minimize the repulsion form the extensive negatively charged region(s) separating the positive patches. Heparin (as well as heparan sulfate) chains are known to exhibit a significant degree of flexibility in solution,52 and it can be readily envisioned that such flexibility of the chain would allow a sufficiently long heparin oligomer to bind the trapped intermediate in a bidentate fashion. This can be accomplished by making contacts with the positive patches on the surfaces of both AT and fXa, and extending the rest of the chain into solution to minimize the repulsion from the negatively charged segment of the complex, which could be partially compensated by associated counterions.
To test this supposition, we used MD simulation to evaluate the interaction between the trapped intermediate and a representative dp20 chain. Although it might be tempting to consider a comprehensive docking study of a heparin oligomer with the AT·fXa trapped intermediate, the amount of computational resources that would be required for completion of such a study makes it impractical (the only simulation work carried out to date focused on AT activation by heparin within the context of formation of the initial Michaelis-type complex, considering binary interactions within the native AT/heparin53, 54 and the native fXa/heparin55 pairs). To limit the conformational space in our MD simulations, the non-reducing end of dp20 was initially placed in contact with the positive patch on the surface of fXa in an arrangement reminiscent of that found in a ternary complex of uncleaved AT, thrombin and a synthetic heparinoid,56 involving conserved basic sites in thrombin and fXa. In addition, the backbone charge density in the middle segment of dp20 (saccharide residues 7 through 14) was reduced by placing sixteen Na+ counterions in the vicinity of sulfate groups to minimize unfavorable electrostatic interactions with the overall negatively charged protein surface during the simulation experiment.
Following several cycles of annealing/MD simulation, it was observed that while the non-reducing end of dp20 remained bound to the positive patch on the fXa surface, the other end of the chain was accommodated on the AT surface by prominent patch of basic residues, which become accessible to the reducing end of dp20 through conformational reorganization of unstructured C-terminus of cleaved AT. Although the length of the chain was not sufficient for making a contact with the majority of basic residues that form pentasaccharide-binding site on the free AT surface, a significant re-arrangement of basic residues on the AT surface allowed them to make multiple contacts with the sulfate groups on the dp20 chain. In the final energy-minimized structure of the ternary complex (Figure 7B) saccharides 1 through 5 are involved in favorable electrostatic interactions with the basic residues on the AT surface, while saccharides 15 through 20 still interact with the corresponding positive patch on the fXa surface. Saccharides 6 through 14, thus, do not make stable contacts with either of the two proteins; instead, this segment arches away from the surface of the complex to minimize the electrostatic repulsion between the negative charges on the protein surface and the sulfate groups within that segment of dp20 (Figure 7C & D). Although this simulation experiment is not exhaustive, and other conformations of the ternary complex may exist, it certainly demonstrates the feasibility of the bidentate mode of dp20 binding to the trapped intermediate.
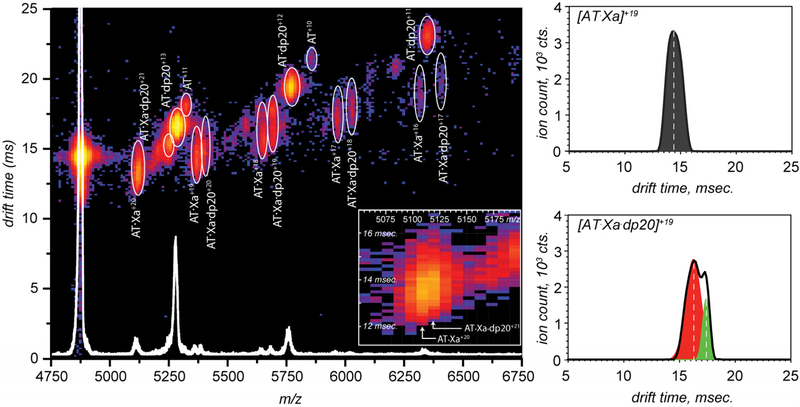
Additional evidence for the existence of the bidentate mode of binding of the long heparin chain (dp20) to the AT·fXa trapped intermediate was provided by analyzing ion mobility profiles of the ternary complexes. Since the ionic signals of ternary complexes observed in the mass spectra of AT/fXa/dp20 mixtures (Figures 3 and 6) have significant overlap with those of AT·dp20 and other ionic species, the mobility profiles of the AT·fXa·dp20 ions are likely to contain contributions from other species. In order to minimize the interference from ionic species having similar m/z values in the crowded mass spectrum of the pre-formed AT·fXa incubated in the presence of dp20 (Figure 6), a less heterogeneous ionic population was obtained by limited charge reduction31 of precursor ions within the 4871–4921 m/z window. The resulting mass spectrum shows distinct contributions from several species, corresponding to free AT, as well as AT·dp20, AT·fXa, and AT·fXa·dp20 complexes at different charge states (Figure 8). Representative mobility profiles for the binary and ternary AT·fXa complexes are also shown in Figure 8.
Figure 8.
Drift time distributions for ions representing binary (AT·fXa) and ternary AT·fXa·dp20 complexes produced by limited charge reduction of ionic species within the m/z window 4871–4921 u in the ESI mass spectrum of mixtures of AT and fXa (4.5 and 3.0 μM, respectively) incubated in the presence of dp6 (30 μM) followed by isolation of the trapped intermediate and addition of dp20 to a final concentration of 15 μM.
Since the mobility of an ion is determined by its mass, total electric charge, and physical dimensions (collisional cross-section), the AT·fXa·dp20 ions are expected to have slightly lower mobility (and longer drift time) compared to the AT·fXa ions of the same charge even if their collisional cross-sections are identical (e.g., in the case of the glycosaminoglycan chain being tightly wrapped around the protein). Therefore, the difference between the average drift times of these two ions observed in our measurements (Figure 8) does not necessarily prove the bidentate character of the heparin chain interaction with the protein complex. However, careful examination of the mobility profiles of these two ions reveals significant conformational heterogeneity within the ternary complex. Indeed, while the drift time distribution of [AT·fXa]+19 ions is nearly-symmetrical, the mobility profile of the [AT·fXa·dp20]+19 ions has a convoluted shape and appears to be a sum of at least two distinct components with Gaussian shapes (shown in red and green Figure 8). These stark differences in mobility profiles were also observed for the [AT·fXa]+20 and [AT·fXa·dp20]+20 pair of charge-reduced species, even though the binary complex at this charge state was barely resolved from the [AT·fXa·dp20]+21 ion (see inset in Figure 8). Importantly, the bimodal shape of the drift time distributions is not observed for the ionic species representing AT·dp20 complexes. This provides a clear indication that the bimodal shapes of the mobility profiles of [AT·fXa·dp20]+20 and [AT·fXa·dp20]+21 ions do not reflect the heterogeneous nature the dp20 chains (variation in the extent of sulfation), but arise due to the conformational heterogeneity (multiple ternary structures in the gas phase).
The observed conformational heterogeneity of the ternary complex in the gas phase is significant, as it supports the bidentate model of the heparin oligomer interaction with the AT·fXa trapped intermediate. Indeed, transition of a protein molecule from solution to the gas phase during the positive ion-ESI process necessarily gives rise to polycationic species even if the molecular species in solution carry a net negative charge.57, 58 Therefore, the electrostatic repulsion between the heparin chain and the large negatively charged patch on the surface of the AT·fXa complex that is expected to exist in solution (vide supra) will have all but disappeared upon transition of the complex to the gas phase. This should result in a dramatic increase of the heparin chain flexibility, allowing it to assume a variety of different conformations. This is in contrast to the expected behavior of the binary protein complex, where the majority of the hydrogen bonds stabilizing the conformation are expected to survive the transition to the solvent-free environment, keeping the higher order structure largely intact (although some alterations are inevitable in the absence of the solvent59). Therefore, the binary complex is expected to remain conformationally homogeneous in the gas phase, while the ternary complex is expected to display much greater heterogeneity and populate a range of conformers in the gas phase, which agrees with the results of ion mobility measurements shown in Figure 8.
The observed binding of longer heparin chains to the trapped AT·fXa intermediate raises an intriguing question of whether this interaction has physiological relevance. It seems conceivable that binding of a heparin (or heparan sulfate) chain to the trapped intermediate may provide additional stabilization to the “crushed” conformation of the protease, which is trapped kinetically, while being disfavored thermodynamically. Given sufficient time, the proteolysis will be completed, resulting in degradation of AT and release of fully functional fXa. Therefore, binding of the glycosaminoglycan chain to the distorted conformation of the protease may provide additional stabilization and further decelerate the final step of the proteolytic reaction. The occurrence of the post-inhibition binding of long glycosaminoglycan chains to the AT/fXa trapped intermediates may also hint at another role that is likely played by heparan sulfate in fXa inhibition, namely facilitation of catabolism of the trapped AT·fXa intermediates by presenting them to the cell-surface receptors that mediate their internalization followed by lysosomal degradation. Heparan sulfate chains of the proteoglycans present in the extracellular matrix are known to facilitate cell-surface receptor recognition by a variety of signaling proteins,60–62 and even have been suggested to act in some cases as autonomous endocytosis receptors.63
Identification of LRP as a receptor assisting in internalization of a number of serpin/protease trapped intermediates20, 21 is significant, as this receptor is involved in a variety of other tasks via recognition of a large number of ligands.19 Promiscuity inevitably results in modest affinities for individual ligands, which frequently requires involvement of facilitators of the recognition process. It seems plausible that heparan sulfate may play such a role not only via effective retention of the trapped AT/fXa intermediates in the extracellular matrix, but also their presentation to the receptor. The ability of sufficiently long glycosaminoglycan chains to associate with the trapped AT/fXa intermediates suggests that the effective clearance of these complexes from circulation may be carried out by scavenging receptors even without the help of vitronectin-like opsonins. Efficient removal of the kinetically trapped fXa from circulation is obviously very important for maintaining the fidelity of the blood coagulation cascade, and could be another aspect of hemostasis (in addition to AT activation) where heparin-like glycosaminoglycans play an important role.
Conclusions
Heparin and its next-of-kin heparan sulfate are key players in the fXa inhibition by AT, a critical element in the blood coagulation cascade. Much effort had been directed in the past two decades towards understanding the mechanisms of AT activation by these glycosaminoglycans, while relatively little attention has been paid to their possible interactions with the trapped AT/fXa complex. Native ESI MS provides a unique opportunity to study association of the heparin chains with these proteins before and after the inhibition event. While the assembly of the AT/fXa complex is anemic in the absence of heparin oligomers, it increases dramatically in the presence of the glycosaminoglycans, completely eliminating vacant fXa molecules from solution. Intriguingly, the shorter heparin oligomers (dp6, dp8 and dp10) are not incorporated in the serpin/protease complexes (despite being critical promoters of the AT/Xa binding). However, replacement of the relatively short heparin chains with a longer one (dp20) gives rise to a prominent ionic signal of ternary complexes in ESI MS. These ternary complexes reflect interactions that occur after the inhibition event (in which heparin acts simply as a catalyst, and is not a part of the “final product”), as their formation is also observed when the trapped AT/fXa intermediate is initially prepared by incubating the AT/fXa mixture in the presence of a short heparin oligomer (dp6), followed by addition of a longer heparin oligomer (dp20). The critical importance of the heparin chain length for its ability to associate with the trapped AT/fXa intermediate suggests that the binding is likely to occur in a bidentate fashion (where two distinct segments of the heparin chain make contacts with the protein components, while the part of the chain separating these two segments is extended into solution to minimize the unfavorable electrostatic interactions). A homology model of the AT/fXa trapped intermediate suggests that physical dimensions of dp20 molecules are sufficient for such a scenario to be implemented, while other heparin oligomers tested in this work (dp6, dp8 and dp10) are not sufficiently long. The bidentate model of heparin interaction with the AT/fXa complex is also corroborated by the results of ion mobility measurements in the gas phase, which reveal anomalous flexibility within the complex-bound dp20 in the gas phase. The observed post-inhibition binding of long heparin chains to the AT/fXa trapped intermediate hints at the possible role played by heparan sulfate in stabilizing and catabolizing these intermediates by presenting them to the cell-surface receptors that mediate their uptake via endocytosis. Facilitation of the endosomal uptake (followed by degradation in the lysosomal compartments) is important for maintaining the fidelity of the blood coagulation cascade, as accumulation of a large pool of the AT/fXa trapped intermediates (where the proteolytic step is greatly decelerated, but not completely stopped) will inevitably lead to release of active fXa molecules back to circulation.
Supplementary Material
Acknowledgements
This work was supported in part by a grant from the National Institutes of Health R01 GM112666. All MS measurements were carried out in the Mass Spectrometry Core facility at UMass-Amherst. The authors wish to thank Prof. Anne Gershenson (Department of Biochemistry and Molecular Biology, UMass-Amherst) for helpful discussions.
Footnotes
Conflicts of Interest
The authors declare no competing financial interest.
References
- [1].Howell WH, and Holt E (1918) Two new factors in blood coagulation ‐ heparin and pro‐antithrombin, Am. J. Physiol 47, 328–341. [Google Scholar]
- [2].Messmore HL (1986) Clinical efficacy of heparin fractions: issues and answers, Crit. Rev. Clin. Lab. Sci 23, 77–94. [DOI] [PubMed] [Google Scholar]
- [3].Petitou M, and van Boeckel CAA (2004) A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next?, Angew. Chem. Int. Ed. Engl 43, 3118–3133. [DOI] [PubMed] [Google Scholar]
- [4].Di Nisio M, Middeldorp S, and Büller HR (2005) Direct Thrombin Inhibitors, N. Engl. J. Med 353, 1028–1040. [DOI] [PubMed] [Google Scholar]
- [5].Versteeg HH, Heemskerk JWM, Levi M, and Reitsma PH (2013) New Fundamentals in Hemostasis, Physiol. Rev 93, 327–358. [DOI] [PubMed] [Google Scholar]
- [6].Mann KG (1999) Biochemistry and physiology of blood coagulation, Thromb. Haemost. 82, 165–174. [PubMed] [Google Scholar]
- [7].Berteau O, and Mulloy B (2003) Sulfated fucans, fresh perspectives: structures, functions, and biological properties of sulfated fucans and an overview of enzymes active toward this class of polysaccharide, Glycobiology 13, 29R–40R. [DOI] [PubMed] [Google Scholar]
- [8].Mourao PA, and Pereira MS (1999) Searching for alternatives to heparin: sulfated fucans from marine invertebrates, Trends Cardiovasc. Med 9, 225–232. [DOI] [PubMed] [Google Scholar]
- [9].Olson ST, Richard B, Izaguirre G, Schedin‐Weiss S, and Gettins PGW (2010) Molecular mechanisms of antithrombin–heparin regulation of blood clotting proteinases. A paradigm for understanding proteinase regulation by serpin family protein proteinase inhibitors, Biochimie 92, 1587–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gettins PGW (2012) Mechanisms of Serpin Inhibition, In Molecular and Cellular Aspects of the Serpinopathies and Disorders in Serpin Activity, pp 67–100, WORLD SCIENTIFIC. [Google Scholar]
- [11].Pearce MC, Pike RN, Lesk AM, and Bottomley SP (2012) Serpin Conformations, In Molecular and Cellular Aspects of the Serpinopathies and Disorders in Serpin Activity, pp 35–66, WORLD SCIENTIFIC. [Google Scholar]
- [12].Schedin‐Weiss S, Richard B, Hjelm R, and Olson ST (2008) Antiangiogenic forms of antithrombin specifically bind to the anticoagulant heparin sequence, Biochemistry 47, 13610–13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang W, Swanson R, Xiong Y, Richard B, and Olson ST (2006) Antiangiogenic antithrombin blocks the heparan sulfate‐dependent binding of proangiogenic growth factors to their endothelial cell receptors: evidence for differential binding of antiangiogenic and anticoagulant forms of antithrombin to proangiogenic heparan sulfate domains, J. Biol. Chem 281, 37302–37310. [DOI] [PubMed] [Google Scholar]
- [14].Wang J, Wang Y, Wang J, Gao J, Tong C, Manithody C, Li J, and Rezaie AR (2013) Antithrombin is protective against myocardial ischemia and reperfusion injury, J. Thromb. Haemost 11, 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Plotnick MI, Samakur M, Wang ZM, Liu X, Rubin H, Schechter NM, and Selwood T (2002) Heterogeneity in Serpin−Protease Complexes As Demonstrated by Differences in the Mechanism of Complex Breakdown, Biochemistry 41, 334–342. [DOI] [PubMed] [Google Scholar]
- [16].Calugaru SV, Swanson R, and Olson ST (2001) The pH Dependence of Serpin‐Proteinase Complex Dissociation Reveals a Mechanism of Complex Stabilization Involving Inactive and Active Conformational States of the Proteinase Which Are Perturbable by Calcium, J. Biol. Chem 276, 32446–32455. [DOI] [PubMed] [Google Scholar]
- [17].Pizzo SV (1989) Serpin receptor 1: a hepatic receptor that mediates the clearance of antithrombin III‐proteinase complexes, Am. J. Med 87, 10s–14s. [DOI] [PubMed] [Google Scholar]
- [18].Enghild JJ, Valnickova Z, Thogersen IB, and Pizzo SV (1994) Complexes between serpins and inactive proteinases are not thermodynamically stable but are recognized by serpin receptors, J. Biol. Chem 269, 20159–20166. [PubMed] [Google Scholar]
- [19].Pieper‐Fürst U, and Lammert F (2013) Low‐density lipoprotein receptors in liver: Old acquaintances and a newcomer, Biochim. Biophys. Acta 1831, 1191–1198. [DOI] [PubMed] [Google Scholar]
- [20].Wells MJ, Sheffield WP, and Blajchman MA (1999) The clearance of thrombin‐antithrombin and related serpin‐enzyme complexes from the circulation: role of various hepatocyte receptors, Thromb. Haemost 81, 325–337. [PubMed] [Google Scholar]
- [21].Kounnas MZ, Church FC, Argraves WS, and Strickland DK (1996) Cellular internalization and degradation of antithrombin III‐thrombin, heparin cofactor II‐thrombin, and alpha 1‐antitrypsin-trypsin complexes is mediated by the low density lipoprotein receptor‐related protein, J. Biol. Chem 271, 6523–6529. [DOI] [PubMed] [Google Scholar]
- [22].Ho G, Broze GJ Jr., and Schwartz AL (1997) Role of heparan sulfate proteoglycans in the uptake and degradation of tissue factor pathway inhibitor‐coagulation factor Xa complexes, J. Biol. Chem 272, 16838–16844. [DOI] [PubMed] [Google Scholar]
- [23].Wells MJ, and Blajchman MA (1998) In vivo clearance of ternary complexes of vitronectin‐thrombin‐antithrombin is mediated by hepatic heparan sulfate proteoglycans, J. Biol. Chem 273, 23440–23447. [DOI] [PubMed] [Google Scholar]
- [24].Wilkins‐Port CE, and McKeown‐Longo PJ (1996) Heparan sulfate proteoglycans function in the binding and degradation of vitronectin by fibroblast monolayers, Biochem. Cell Biol 74, 887–897. [DOI] [PubMed] [Google Scholar]
- [25].Kaltashov IA, Bobst CE, and Abzalimov RR (2013) Mass spectrometry‐based methods to study protein architecture and dynamics, Protein Sci. 22, 530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Abzalimov RR, Dubin PL, and Kaltashov IA (2007) Glycosaminoglycans as naturally occurring combinatorial libraries: Developing a mass spectrometry‐based strategy for characterization of anti‐thrombin interaction with low molecular weight heparin and heparin oligomers, Anal. Chem 79, 6055–6063. [DOI] [PubMed] [Google Scholar]
- [27].Harmer NJ, Ilag LL, Mulloy B, Pellegrini L, Robinson CV, and Blundell TL (2004) Towards a resolution of the stoichiometry of the fibroblast growth factor (FGF)‐FGF receptor‐heparin complex, J. Mol. Biol 339, 821–834. [DOI] [PubMed] [Google Scholar]
- [28].Crown SE, Yu Y, Sweeney MD, Leary JA, and Handel TM (2006) Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding, J. Biol. Chem 281, 25438–25446. [DOI] [PubMed] [Google Scholar]
- [29].Fermas S, Gonnet F, Varenne A, Gareil P, and Daniel R (2007) Frontal analysis capillary electrophoresis hyphenated to electrospray ionization mass spectrometry for the characterization of the antithrombin/heparin pentasaccharide complex, Anal. Chem 79, 4987–4993. [DOI] [PubMed] [Google Scholar]
- [30].Minsky BB, Dubin PL, and Kaltashov IA (2017) Electrostatic Forces as Dominant Interactions Between Proteins and Polyanions: an ESI MS Study of Fibroblast Growth Factor Binding to Heparin Oligomers, J. A. Soc. Mass Spectrom 28, 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Abzalimov RR, and Kaltashov IA (2010) Electrospray ionization mass spectrometry of highly heterogeneous protein systems: Protein ion charge state assignment via incomplete charge reduction, Anal. Chem 82, 7523–7526. [DOI] [PubMed] [Google Scholar]
- [32].Zhao Y, Abzalimov RR, and Kaltashov IA (2016) Interactions of Intact Unfractionated Heparin with Its Client Proteins Can Be Probed Directly Using Native Electrospray Ionization Mass Spectrometry, Anal. Chem 88, 1711–1718. [DOI] [PubMed] [Google Scholar]
- [33].McCoy AJ, Pei XY, Skinner R, Abrahams JP, and Carrell RW (2003) Structure of beta‐antithrombin and the effect of glycosylation on antithrombin’s heparin affinity and activity, J. Mol. Biol 326, 823–833. [DOI] [PubMed] [Google Scholar]
- [34].Pryzdial ELG, and Kessler GE (1996) Autoproteolysis or Plasmin‐mediated Cleavage of Factor Xaα Exposes a Plasminogen Binding Site and Inhibits Coagulation, J. Biol. Chem 271, 16614–16620. [DOI] [PubMed] [Google Scholar]
- [35].Fujikawa K, Titani K, and Davie EW (1975) Activation of bovine factor X (Stuart factor): conversion of factor Xaalpha to factor Xabeta, Proceedings of the National Academy of Sciences 72, 3359–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kaltashov IA, and Eyles SJ (2002) Studies of biomolecular conformations and conformational dynamics by mass spectrometry, Mass Spectrom. Rev 21, 37–71. [DOI] [PubMed] [Google Scholar]
- [37].DeLauder S, Schwarz FP, Williams JC Sr., and Atha DH (1992) Thermodynamic analysis of heparin binding to human antithrombin, Biochim. Biophys. Acta 1159, 141–149. [DOI] [PubMed] [Google Scholar]
- [38].Mourey L, Samama JP, Delarue M, Petitou M, Choay J, and Moras D (1993) Crystal structure of cleaved bovine antithrombin III at 3.2 A resolution, J. Mol. Biol 232, 223–241. [DOI] [PubMed] [Google Scholar]
- [39].Lin P‐H, Sinha U, and Betz A (2001) Antithrombin binding of low molecular weight heparins and inhibition of factor Xa, Biochimica et Biophysica Acta (BBA) ‐ General Subjects 1526, 105–113. [DOI] [PubMed] [Google Scholar]
- [40].Desai UR, Petitou M, Bjork I, and Olson ST (1998) Mechanism of heparin activation of antithrombin. Role of individual residues of the pentasaccharide activating sequence in the recognition of native and activated states of antithrombin, J. Biol. Chem 273, 7478–7487. [DOI] [PubMed] [Google Scholar]
- [41].Kreuger J, Spillmann D, Li J. p., and Lindahl U (2006) Interactions between heparan sulfate and proteins: the concept of specificity, J. Cell Biol 174, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Craig PA, Olson ST, and Shore JD (1989) Transient kinetics of heparin‐catalyzed protease inactivation by antithrombin III. Characterization of assembly, product formation, and heparin dissociation steps in the factor Xa reaction, J. Biol. Chem 264, 5452–5461. [PubMed] [Google Scholar]
- [43].Kayitmazer AB, Seeman D, Minsky BB, Dubin PL, and Xu YS (2013) Protein‐polyelectrolyte interactions, Soft Matter 9, 2553–2583. [Google Scholar]
- [44].Kera Y, and Yamasawa K (1982) ISOELECTRIC‐FOCUSING PATTERN OF HUMAN ANTITHROMBIN‐III (AT‐III) ‐ EFFECTS OF HEPARIN OR THROMBIN ON A MICROHETEROGENEOUS FORM OF AT‐III, Electrophoresis 3, 157–161. [Google Scholar]
- [45].Kremser L, Bruckner A, Heger A, Grunert T, Buchacher A, Josic D, Allmaier M, and Rizzi A (2003) Characterization of antithrombin III from human plasma by two‐dimensional gel electrophoresis and capillary electrophoretic methods, Electrophoresis 24, 4282–4290. [DOI] [PubMed] [Google Scholar]
- [46].Katz BA, Mackman R, Luong C, Radika K, Martelli A, Sprengeler PA, Wang J, Chan H, and Wong L (2000) Structural basis for selectivity of a small molecule, S1‐binding, submicromolar inhibitor of urokinase‐type plasminogen activator, Chem. Biol 7, 299–312. [DOI] [PubMed] [Google Scholar]
- [47].Dementiev A, Dobo J, and Gettins PG (2006) Active site distortion is sufficient for proteinase inhibition by serpins: structure of the covalent complex of alpha1‐proteinase inhibitor with porcine pancreatic elastase, J. Biol. Chem 281, 3452–3457. [DOI] [PubMed] [Google Scholar]
- [48].Huntington JA, McCoy A, Belzar KJ, Pei XY, Gettins PG, and Carrell RW (2000) The conformational activation of antithrombin. A 2.85‐A structure of a fluorescein derivative reveals an electrostatic link between the hinge and heparin binding regions, The Journal of biological chemistry 275, 15377–15383. [DOI] [PubMed] [Google Scholar]
- [49].Johnson DJ, Li W, Adams TE, and Huntington JA (2006) Antithrombin–S195A factor Xa‐heparin structure reveals the allosteric mechanism of antithrombin activation, EMBO J. 25, 2029–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rezaie AR (2000) Identification of basic residues in the heparin‐binding exosite of factor Xa critical for heparin and factor Va binding, J. Biol. Chem 275, 3320–3327. [DOI] [PubMed] [Google Scholar]
- [51].Rezaie AR (2000) Heparin‐Binding Exosite of Factor Xa, Trends Cardiovasc. Med 10, 333–338. [DOI] [PubMed] [Google Scholar]
- [52].Khan S, Fung KW, Rodriguez E, Patel R, Gor J, Mulloy B, and Perkins SJ (2013) The Solution Structure of Heparan Sulfate Differs from That of Heparin: IMPLICATIONS FOR FUNCTION, Journal of Biological Chemistry 288, 27737–27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Verli H, and Guimaraes JA (2005) Insights into the induced fit mechanism in antithrombin‐heparin interaction using molecular dynamics simulations, J. Mol. Graph. Model 24, 203–212. [DOI] [PubMed] [Google Scholar]
- [54].Sarkar A, Yu W, Desai UR, MacKerell AD, and Mosier PD (2016) Estimating glycosaminoglycan‐protein interaction affinity: water dominates the specific antithrombin-heparin interaction, Glycobiology 26, 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pol‐Fachin L, and Verli H (2014) Structural glycobiology of heparin dynamics on the exosite 2 of coagulation cascade proteases: Implications for glycosaminoglycans antithrombotic activity, Glycobiology 24, 97–105. [DOI] [PubMed] [Google Scholar]
- [56].Li W, Johnson DJ, Esmon CT, and Huntington JA (2004) Structure of the antithrombin‐thrombin‐heparin ternary complex reveals the antithrombotic mechanism of heparin, Nat. Struct. Mol. Biol 11, 857–862. [DOI] [PubMed] [Google Scholar]
- [57].Kaltashov IA, and Abzalimov RR (2008) Do ionic charges in ESI MS provide useful information on macromolecular structure?, J. Am. Soc. Mass Spectrom 19, 1239–1246. [DOI] [PubMed] [Google Scholar]
- [58].Frimpong AK, Abzalimov RR, Eyles SJ, and Kaltashov IA (2007) Gas‐phase interference‐free analysis of protein ion charge‐state distributions: detection of small‐scale conformational transitions accompanying pepsin inactivation, Anal. Chem 79, 4154–4161. [DOI] [PubMed] [Google Scholar]
- [59].Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, and Heck AJR (2010) Ion mobility mass spectrometry of proteins and protein assemblies, Chem. Soc. Rev 39, 1633–1655. [DOI] [PubMed] [Google Scholar]
- [60].Sarrazin S, Lamanna WC, and Esko JD (2011) Heparan sulfate proteoglycans, Cold Spring Harb. Perspec.t Biol 3, a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Park PW, Reizes O, and Bernfield M (2000) Cell surface heparan sulfate proteoglycans: selective regulators of ligand‐receptor encounters, J. Biol. Chem 275, 29923–29926. [DOI] [PubMed] [Google Scholar]
- [62].Gallagher J (2015) Fell‐Muir Lecture: Heparan sulphate and the art of cell regulation: a polymer chain conducts the protein orchestra, Int. J. Exp. Pathol 96, 203–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Christianson HC, and Belting M (2014) Heparan sulfate proteoglycan as a cell‐surface endocytosis receptor, Matrix Biol. 35, 51–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.