Proteins of the L-type amino acid transporter (LAT) subfamily take up amino acids from the environment for use in the cell. Bartoccioni et al. show that the bacterial amino acid exchanger BasC is functionally similar to the human LAT Asc1, making BasC a useful model for this class of transporters.
Abstract
Many key cell processes require prior cell uptake of amino acids from the environment, which is facilitated by cell membrane amino acid transporters such as those of the L-type amino acid transporter (LAT) subfamily. Alterations in LAT subfamily amino acid transport are associated with several human diseases, including cancer, aminoacidurias, and neurodegenerative conditions. Therefore, from the perspective of human health, there is considerable interest in obtaining structural information about these transporter proteins. We recently solved the crystal structure of the first LAT transporter, the bacterial alanine-serine-cysteine exchanger of Carnobacterium sp AT7 (BasC). Here, we provide a complete functional characterization of detergent-purified, liposome-reconstituted BasC transporter to allow the extension of the structural insights into mechanistic understanding. BasC is a sodium- and proton-independent small neutral amino acid exchanger whose substrate and inhibitor selectivity are almost identical to those previously described for the human LAT subfamily member Asc-1. Additionally, we show that, like its human counterparts, this transporter has apparent affinity asymmetry for the intra- and extracellular substrate binding sites—a key feature in the physiological role played by these proteins. BasC is an excellent paradigm of human LAT transporters and will contribute to our understanding of the molecular mechanisms underlying substrate recognition and translocation at both sides of the plasma membrane.
Introduction
Amino acid availability regulates cellular physiology. The transfer of amino acids across the plasma membrane is mediated by specific amino acid transporters. Heteromeric amino acid transporters (HATs) comprise two subunits, a polytopic membrane protein (the light subunit; SLC7 family) and a disulfide-linked N-glycosylated type II membrane glycoprotein (the heavy subunit; SLC3 family; Fotiadis et al., 2013). The light subunits of HATs belong to the L-type amino acid transporter (LAT) subfamily, which is part of the large amino acids, polyamines, and organic cations (APC) family of transporters, and are the catalytic component of the transporter (Reig et al., 2002). In contrast, the heavy subunit appears to be essential only for trafficking to the plasma membrane. Two heavy subunits (4F2hc and rBAT) and eight light subunits have been identified in humans (Fernández et al., 2006).
Several human pathologies highlight the physiological roles of HATs. Two transporters of this family are responsible for inherited aminoacidurias, and mutations in either of the two genes coding for the subunits of system b(0,+) (rBAT and b(0,+)AT) lead to cystinuria (MIM 220100; Calonge et al., 1994; Feliubadaló et al., 1999), while mutations in y+LAT1 (a 4F2hc-associated system y+L) result in lysinuric protein intolerance (LPI; MIM222700; Torrents et al., 1998; Borsani et al., 1999). Additionally, mutations in LAT1 and LAT2 are associated with autism and age-related hearing loss, respectively (Tărlungeanu et al., 2016; Espino Guarch et al., 2018), and Asc-1/CD98hc is a druggable target in schizophrenia as it is the major D-serine transporter in brain, which acts as a coagonist of NMDA glutamate receptors (Sakimura et al., 2016). Moreover, xCT and LAT1 are overexpressed in many human tumors, thereby suggesting that amino acid transporters are essential for tumor cell survival and progression (del Amo et al., 2008; Lo et al., 2008; Savaskan and Eyüpoglu, 2010). In this regard, two different anticancer therapies involving both xCT and LAT1 have been proposed. On the one hand, these transporters mediate the uptake of several amino acid–derived anticancer drugs (del Amo et al., 2008; Lo et al., 2008; Savaskan and Eyüpoglu, 2010). This observation thus suggests that these proteins are involved in the cellular internalization of these antineoplasic drugs. On the other hand, a novel strategy based on the inhibition of xCT and LAT1 activities has been described (Chung et al., 2005; del Amo et al., 2008; Lo et al., 2008; Savaskan and Eyüpoglu, 2010), thus reducing tumor proliferation and progression. Targeting amino acid transporters in cancer is conceptually novel since specific inhibitors are scarce. In this regard, the development of drugs with greater activity against these transporters is expected to challenge the scene.
The atomic structures of plasma membrane transporter proteins are the most powerful tools for the design of more specific and active therapeutic molecules and for understanding substrate binding and translocation mechanisms. Considerable homology with human amino acid transporters may provide crucial insights into intelligent drug design for the treatment of several cancers and neuronal diseases, as well as knowledge of the effect of pathological mutations on transporter protein activity (Morth et al., 2009; Wu et al., 2014). To date, the atomic structure of a LAT transporter has not been published. The closest available crystal structures correspond to those of remote bacterial homologues (17–22% sequence identity [SI]), namely the amino acid exchangers AdiC and GadC, and the proton-coupled amino acid transporters ApcT and GkApcT, which belong to the APC family of transporters (Fang et al., 2009; Gao et al., 2009, 2010; Shaffer et al., 2009; Kowalczyk et al., 2011; Ma et al., 2012; Ilgü et al., 2016; Jungnickel et al., 2018). Nevertheless, the generation of robust and reliable structural models of LAT transporters based on these distant crystallized homologues is precluded by low amino acid SI with human LAT (hLAT) transporters. In this regard, an hLAT1 model based on a mixed sequence alignment of AdiC and ApcT with hLAT1 has been reported as a tool to identify structurally novel molecules of pharmacological interest (Geier et al., 2013).
Close prokaryotic homologues have emerged as excellent structural and functional paradigms of their human counterparts (Penmatsa et al., 2013). In this regard, the serine/threonine exchanger (SteT) was shown to be a good functional homologue of hLAT (Reig et al., 2007). However, despite significant efforts that have led to an increase in the crystalizability of the protein, no crystal structure was obtained (Rodríguez-Banqueri et al., 2016). We recently solved the crystal structure of the first transporter of the LAT subfamily (bacterial asc; BasC; unpublished data). Here we report the complete functional characterization of BasC, showing extremely similar functional features to those of hLAT, thus making it a suitable protein for further studies on LAT subfamily members.
Materials and methods
Identification and sequence analysis of prokaryotic LAT transporters
To identify prokaryotic homologues of LAT subfamily transporters, initial sequence analysis was performed using the BLAST algorithm (default settings; Altschul et al., 1990) to compare a prototypical hLAT transporter sequence (particularly LAT1) against a nonredundant protein sequence database. Seven bacterial sequences encoding for putative LAT transporters sharing between 25% and 30% SI with LAT1 were selected for protein expression and purification. Multi-sequence alignment was done with the PSI/TM-Coffee server (Floden et al., 2016) using the members of the APA (2.A.3.2), CAT (2.A.3.3), ABT (2.A.3.6), GGA (2.A.3.7), and LAT (2.A.3.8) subfamilies of the APC family (www.tcdb.org; Saier et al., 2016), and a phylogenetic tree was drawn by iTOL tool (Letunic and Bork, 2016).
Cloning of prokaryotic LAT transporters
Ecolized complementary DNAs encoding the seven selected putative amino acid transporter proteins were obtained from GenScript and subcloned into the EcoRI and PstI sites of a modified PTTQ18 expression vector. A variant of a GFP, lacking the start codon, was added in frame to the C terminus of the transporter sequence. Additionally, the sequence of HRV-3C protease was inserted between the transporter protein and GFP sequences. Finally, four additional histidines were added to the preexistent hexahistidine tag in the PTTQ18 vector. Single point mutations were introduced using the QuikChange site-directed mutagenesis kit (Agilent). All DNA constructs were verified by sequencing. The A20C mutant was generated on a cys-less mutant (C427A) background to finally generate the double A20C-C427A mutant (referred to as A20C mutant) that has been used to study the sidedness of the substrate interaction of BasC.
Protein expression of prokaryotic LAT transporters in Escherichia coli
Expression experiments were performed with freshly transformed cultures of E. coli strain BL21 Star (DE3). Initially, the expression of all seven putative LAT transporters was studied in 50-ml samples cultured in Luria-Bertani medium containing 50 µg/ml ampicillin. When the absorbance measured at 600 nm (A600) had reached 0.5, protein expression was induced at different isopropyl-β-D-thiogalactopyranoside (Roche) concentrations (0.1 mM, 0.5 mM, or 1 mM), incubation times, 4 h or overnight, and temperatures (25°C, 30°C, or 37°C). To estimate protein expression, GFP fluorescence was measured and protein quantity calculated as previously reported (Drew et al., 2005). The putative LAT transporter of Carnobacterium sp AT7 (BasC hereafter) was identified as a stable protein species suitable for crystallographic studies by application of fluorescence-detection size exclusion chromatography (Drew et al., 2005). BasC was overexpressed in E. coli BL21 Star (DE3) cells grown in Luria-Bertani media as a C-terminal fusion with GFP. Expression was induced by 0.1 mM isopropyl-β-D-thiogalactopyranoside at 37°C for 22 h. Cells were harvested at 5,000×g for 15 min and stored at −80°C until use.
Preparation of E. coli membranes
Cell pellets were thawed and resuspended in 20 mM Tris-Base, 150 mM NaCl, pH 7.4. Cells were pelleted again at 5,000×g for 15 min, resuspended in lysis buffer (20 mM Tris-Base, 350 mM NaCl, pH 7.4, 1 mM Pefabloc, and complete mini protease inhibitor cocktail (Roche), and broken using a Cell Disruptor (20,000 psi, four cycles; Constant Systems). Cell debris was removed by centrifugation (1 h at 15,000×g, 4°C), and the supernatant was subjected to ultracentrifugation (2 h at 200,000×g, 4°C). The membrane pellet was resuspended in 20 mM Tris-Base, 150 mM NaCl, pH 7.4, and 10% glycerol at a protein concentration of between 8 and 12 mg/ml, frozen in liquid nitrogen, and stored at −80°C until use.
Purification of prokaryotic LAT transporters
All subsequent steps were performed at 4°C. Membranes at 3 mg/ml protein concentration were solubilized using 1% (wt/vol) n-dodecyl-β-D-maltopyranoside (DDM; Affymetrix) for 1 h in purification buffer (20 mM Tris-Base, 150 mM NaCl, pH 7.4, 0.05% DDM, and 10% glycerol). After ultracentrifugation (2 h, 200,000×g, 4°C), the soluble fraction was incubated for 3 h at 4°C with equilibrated Ni2+–nitrilotriacetic acid (NTA) Superflow beads (QIAGEN) with washing buffer (20 mM Tris-Base, 150 mM NaCl, pH 7.4, 0.05% DDM, 10% glycerol, and 20 mM imidazole). Protein-bound beads were washed three times with 10 column volumes of washing buffer before elution with 15 ml of elution buffer (washing buffer supplemented with 350 mM imidazole). The purified protein was concentrated by centrifugation in an Amicon Ultra (100,000 D molecular weight cut-off; Millipore) at 3,220xg until reaching the desired protein concentration. To assess BasC quality and purity after Ni2+-NTA purification, size exclusion chromatography (SEC) of GFP-fused BasC was performed in a Superdex 200 10/300 GL (GE Healthcare) in SEC buffer (20 mM Tris-Base, 150 mM NaCl, pH 7.4, and 0.05% DDM). AdiC was purified for functional studies as previously described (Kowalczyk et al., 2011).
Reconstitution into proteoliposomes (PLs)
BasC and AdiC proteins were reconstituted in E. coli polar lipids (Sigma-Aldrich), as previously described (Reig et al., 2007; Kowalczyk et al., 2011). Lipid was dried under N2 and suspended in reconstitution buffer (20 mM Tris-Base and 150 mM NaCl, pH 7.4). The suspension was sonicated to clarity, and purified BasC protein was added to the desired protein/lipid ratio (wt/wt; 1:100). To destabilize the liposomes, 1.25% β-D-octylglucoside was added and incubated in ice with occasional agitation for 5 min. DDM and β-D-octylglucoside were removed by dialysis for 40 h at 4°C against 100 volumes of dialysis buffer. PL suspensions were used or frozen in liquid N2 and stored at −80°C until use.
Amino acid transport assays in PLs
For transport measurements, PLs were prepared as previously described (Tsai et al., 2012). For uptake experiments, the desired intracellular amino acid concentration was added to the PL suspension, which was then subjected to three freeze–thaw cycles. The extraliposomal amino acid content was then removed by ultracentrifugation. Amino acid uptake assays were initiated after mixing 10 µl of cold PLs with 180 µl of transport buffer (20 mM Tris-Base and 150 mM NaCl, pH 7.4), 0.5–1 µCi/data point of radiolabeled L-amino acid (PerkinElmer), and unlabeled amino acid to the desired concentration, and this mixture was then incubated at room temperature for the indicated periods of time. Efflux measurements were performed by filling the liposomes by three freeze–thaw cycles with transport buffer plus 10 µM L-serine and 0.5 µCi/data point of [3H]L-serine. The release of [3H]L-serine was measured by adding 180 µl of transport buffer with or without 4 mM cold amino acid. Transport experiments were stopped by the addition of 2 ml of ice-cold stop buffer (5 mM L-serine in transport buffer) and filtration through 0.45-µm pore size membrane filters (Sartorius Stedim Biotech). Filters were then washed two times with 2 ml of stop buffer and dried, and the trapped radioactivity was counted. Transport measured in PLs containing no amino acid was subtracted from each data point in order to calculate the net exchange. Transport measurements are normalized to the BasC protein concentration measured for each proteoliposome batch. Saturation kinetics were evaluated by nonlinear regression analysis, and the kinetic parameters derived from this method were confirmed by linear regression analysis of the derived Eadie–Hofstee plots. Data are expressed as the mean ± SEM of three experiments performed on different days and on different batches of protein and PLs.
Use of cysteine-modifying reagents
MTSES or MTSEA (Anatrace) were freshly prepared by dissolving the reagent in ice-cold water just before use. Liposomes were then centrifuged and resuspended in fresh buffer (20 mM Tris-Base and 150 mM NaCl, pH 7.4) and treated with 1–5 mM MTSES or MTSEA for 15–60 min (as indicated) before [3H]L-serine uptake or efflux measurements were performed. In labeling experiments, liposomes with BasC A20C mutant were treated with HRV-3C protease (2 h at 4°C), adding 1 mM DTT and 0.5 mM EDTA to the buffer. Then, 1 mM Cy5-maleimide was added (ON at 4°) after 1–5 mM MTSES or MTSEA treatment for 60 min. In-gel fluorescence was detected by Odyssey Fc (Bonsai Advanced Technologies) at 600 nm (GFP) and 700 nm (Cy5). N-His–tagged HRV-3C protease was obtained from the Institute for Research in Biomedicine Barcelona Protein Expression Core Facility.
Online supplemental material
Fig. S1 shows BasC sequence alignment with human LATs. Additionally, mutated residues associated with autism (LAT1), age-related hearing loss (LAT2), cystinuria (b(0+)AT), and LPI (y+LAT1) are also highlighted. Fig. S2 shows that cysless BasC L-serine/L-alanine exchange activity in PLs is not affected by cysteine-modifying reagents MTSES and MTSEA.
Results
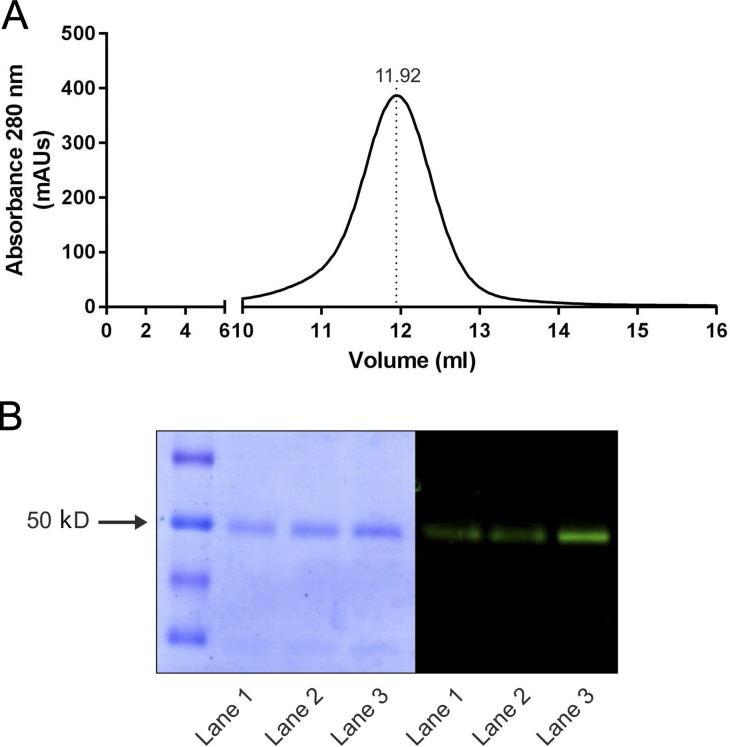
To identify a suitable prokaryotic candidate for the structure–function analysis of LAT transporters, BLAST analysis on hLAT1 sequence retrieved a number of candidates with SIs ranging from 25 to 30%. The seven bacterial putative amino acid transporters with higher SI were expressed as a GFP-fused version, thus analyzing protein expression, detergent solubilization efficiency, and stability by means of GFP fluorescence (data not shown). As a result, a putative amino acid transporter (BasC hereafter) from Carnobacterium sp AT7 was selected due to its high yield of purified protein (up to 10 mg/liter of bacterial culture), high stability, and monodisperse behavior in DDM, as shown by SEC (Fig. 1 A).
Figure 1.
BasC purification and reconstitution in PLs. (A) SEC of DDM-solubilized Ni2+-NTA purified BasC-GFP protein. mAUs, milli absorbance units. (B) Coomassie blue–stained gel and in-gel fluorescence of purified BasC. Purification of His-tagged BasC-GFP by nickel affinity chromatography with or without a further SEC step. Coomassie blue–stained (left) and in-gel fluorescence (right) SDS-PAGE (12% polyacrylamide gel) of the different purification steps is shown: Ni2+-NTA purified BasC-GFP (lane 1), BasC reconstituted in PLs (lane 2), and SEC-purified BasC (lane 3). BasC-GFP migrates as a prominent band at ∼50 kD.
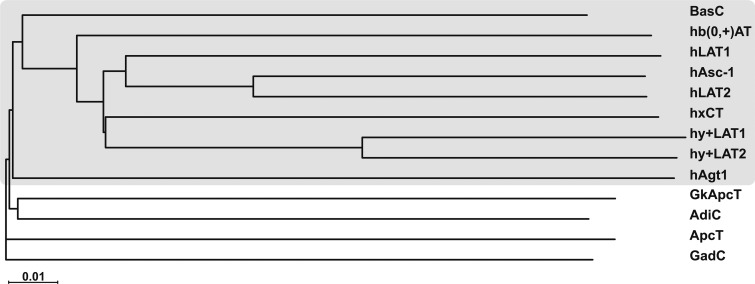
In a wide phylogenetic study of the APC superfamily of transporters, sequence comparisons and the constructed phylogenetic tree indicated that BasC clustered with all of the members of the LAT subfamily (Fig. 2). BasC contains 434 amino acid residues that can be aligned in their entirety with almost the full length of the sequences of hLAT transporters, in contrast to the crystallized distant hLAT homologues AdiC, ApcT, and GadC (Fang et al., 2009; Gao et al., 2009, 2010; Shaffer et al., 2009; Kowalczyk et al., 2011; Ma et al., 2012; Ilgü et al., 2016; Fig. 2 and Fig. S1) that have been the structural paradigms of the hLAT transporter subfamily until now. Very recently, the atomic structure of a prokaryotic homologue of the cationic amino acid transporters subfamily belonging to the SLC7 family has been solved (Jungnickel et al., 2018). This new transporter, named GkApcT, has been shown to be an excellent model of eukaryotic cationic amino acid transporters, but a distant one to the LAT transporters both in sequence and function (Jungnickel et al., 2018). Indeed, BasC presents 26–28% of SI to the core of human LATS (LAT1, LAT2, y+LAT1, y+LAT2, Asc-1, b0,+AT, and xCT), whereas GkApcT shows 18–22% SI to these human LATs. Additionally, mapping of the pathogenic mutations found in various LAT transporters, such as LAT1 in autism, y+LAT1 in LPI, b(0+)AT in cystinuria, and LAT2 in age-related hearing loss (Font-Llitjós et al., 2005; Sperandeo et al., 2008; Tărlungeanu et al., 2016; Espino Guarch et al., 2018), showed that 62.8% of these residues are conserved or semi-conserved in BasC (Fig. S1), compared with 41.0%, 46.1%, 51.3%, and 53.1% in GadC, ApcT, AdiC, and GkApcT, respectively. The main differences between BasC and hLATs are shorter N and C termini in the former and the absence of the cysteine residue in the loop between TM3 and TM4 that is involved in the disulfide bridge between the light and the heavy subunits in metazoan LAT transporters.
Figure 2.
Phylogenetic relationships of BasC with other prokaryotic and eukaryotic members of the APC family. The neighbor-joining tree shows the phylogenetic relationships of BasC with human members of the LAT subfamily, within the APC family, and the crystallized prokaryotic members of the APC family. The tree was performed using all the alignable member sequences within the APC family present in the Transporter Classification Database (www.tcdb.org).
To assess BasC transport characteristics, C-terminally GFP-His10–tagged BasC (BasC-GFP) was overexpressed in E. coli, and DDM-solubilized protein was purified with a single immobilized metal affinity chromatography step. Purified BasC-GFP ran as a single and monodisperse peak in size exclusion chromatography, its purity and identity being confirmed by Coomassie blue–stained gels and in gel fluorescence (Fig. 1, A and B). Purified BasC-GFP was reconstituted in the presence of E. coli lipids to form PLs (BasC-GFP-PLs) at a protein/lipid ratio of 1:100. SDS-PAGE analysis of the resulting PLs revealed a single band at the same molecular weight as the protein purified in detergent solution (Fig. 1 B), thereby confirming incorporation of the protein.
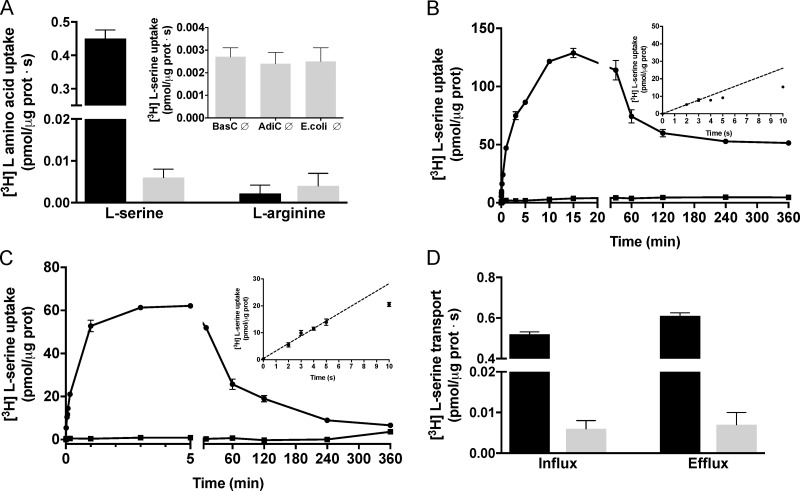
Because BasC was an orphan transporter, to identify BasC transport activity, the influx of 10 µM [3H]L-serine (SteT substrate) or [3H]L-arginine (AdiC substrate) was then measured. In the presence of [3H]L-serine, but not [3H]L-arginine, BasC-GFP-PLs showed higher transport in PLs filled with a mixture of 10 representative cold amino acids (at 1 mM each) than in the empty ones (i.e., trans-stimulation; Fig. 3 A). Additionally, 10 µM [3H]L-serine accumulation inside empty BasC-GFP-PLs was identical to that observed for both empty AdiC-PLs and empty transporter-free liposomes (Fig. 3 A, inset). 10 µM [3H]L-serine transport was then examined over time in BasC-GFP-PLs containing 4 mM L-serine or no amino acid. BasC showed accumulation of [3H]L-serine over the equilibrium levels, as indicated by the overshoot observed in L-serine–containing BasC-GFP-PLs, which was not observed in the empty BasC-GFP-PLs (Fig. 3 B).
Figure 3.
Functional characterization of BasC reconstituted into E. coli liposomes. (A) 10 µM [3H]L-serine (1 µCi/µl) or [3H]L-arginine (1 µCi/µl) influx (pmol/µg protein · s) into BasC-GFP-PLs containing a mixture of 10 representative amino acids (L-Arg, L-Orn, Gly, L-Pro, L-Ala, L-Leu, L-Met, L-Phe, L-Tyr, and L-Glu at 1 mM each; black bars) or no amino acid (gray bars). Inset: 10 µM [3H]L-serine (1 µCi/µl) uptake (pmol/µg protein · s) in empty BasC-PLs, AdiC-PLs or E. coli polar lipid liposomes. Data (mean ± SEM) are from three experiments with three replicates per condition. (B) Time course (0–360 min) of 10 µM [3H]L-serine influx (pmol/µg protein) into BasC-GFP-PLs containing 4 mM L-serine (closed circles) or no amino acid (closed squares). Inset: Time course (0–10 s) of 10 µM [3H]L-serine/4 mM L-serine exchange (pmol/µg protein). Data correspond to a representative experiment, performed using three replicates. A second independent experiment gave similar results. (C) Time course (0–360 min) of 10 µM [3H]L-serine influx (pmol/µg protein) into BasC-GFP-PLs containing 4 mM L-alanine (closed circles) or no amino acid (closed squares). Inset: Time course (0–10 s) of 10 µM [3H]L-serine/4 mM L-alanine exchange (pmol/µg protein). Data correspond to a representative experiment, performed using three replicates. A second independent experiment gave similar results. (D) Comparison of 10 µM [3H]L-serine (1 µCi/data point) influx (left) and 10 µM [3H]L-serine (1 µCi/data point) efflux (right) in BasC-GFP-PLs, expressed in pmol/µg protein · s. Efflux measurements were performed by filling the liposomes by three freeze–thaw cycles with transport buffer plus 10 µM L-serine and 1 µCi/data point of [3H]L-serine. The release of [3H]L-serine was measured by adding 180 µl of transport buffer to the BasC-GFP-PL suspension with or without 4 mM cold amino acid. Efflux of radiolabeled L-serine was dramatically stimulated by the presence of L-serine in the external medium. Data (mean ± SEM) are from three experiments with three replicates per condition.
Nevertheless, BasC was a fast [3H]L-serine/L-serine exchanger, the uptake being barely linear below 5 s (Fig. 3 B, inset). Due to this experimental drawback, we decided to analyze [3H]L-serine uptake over time in BasC-PLs containing 4 mM of individual amino acids. Thus, 10 µM [3H]L-serine/4 mM L-alanine exchange was demonstrated to be linear until 5 s (Fig. 3 C, inset). As expected, 10 µM [3H]L-serine in exchange with 4 mM L-alanine showed an overshoot, not observed in empty PLs (Fig. 3 C). The peak of the overshoot (62 pmol L-serine/µg protein) represents ∼5 molecules of substrate per the estimated number of binding sites of BasC in the assay. This corresponds to an accumulation of ∼100 µM L-serine inside BasC-PLs (internal volume: 0.62 µl/µg protein), which represents ∼10-fold versus the 10 µM [3H]L-serine in the medium and 12-fold versus equilibrium conditions (360 min). The uptake of radiolabeled L-serine in BasC-GFP-PLs filled with 4 mM L-alanine at 4 s was considered ideal working conditions for further transporter characterization. Similarly to the influx of 10 µM radiolabeled L-serine in exchange with 4 mM L-alanine, efflux of 10 µM radiolabeled L-serine from preloaded BasC-GFP-PLs was dramatically trans-stimulated by the presence of L-serine in the external medium, thereby demonstrating that BasC is a bidirectional obligate exchanger (Fig. 3 D).
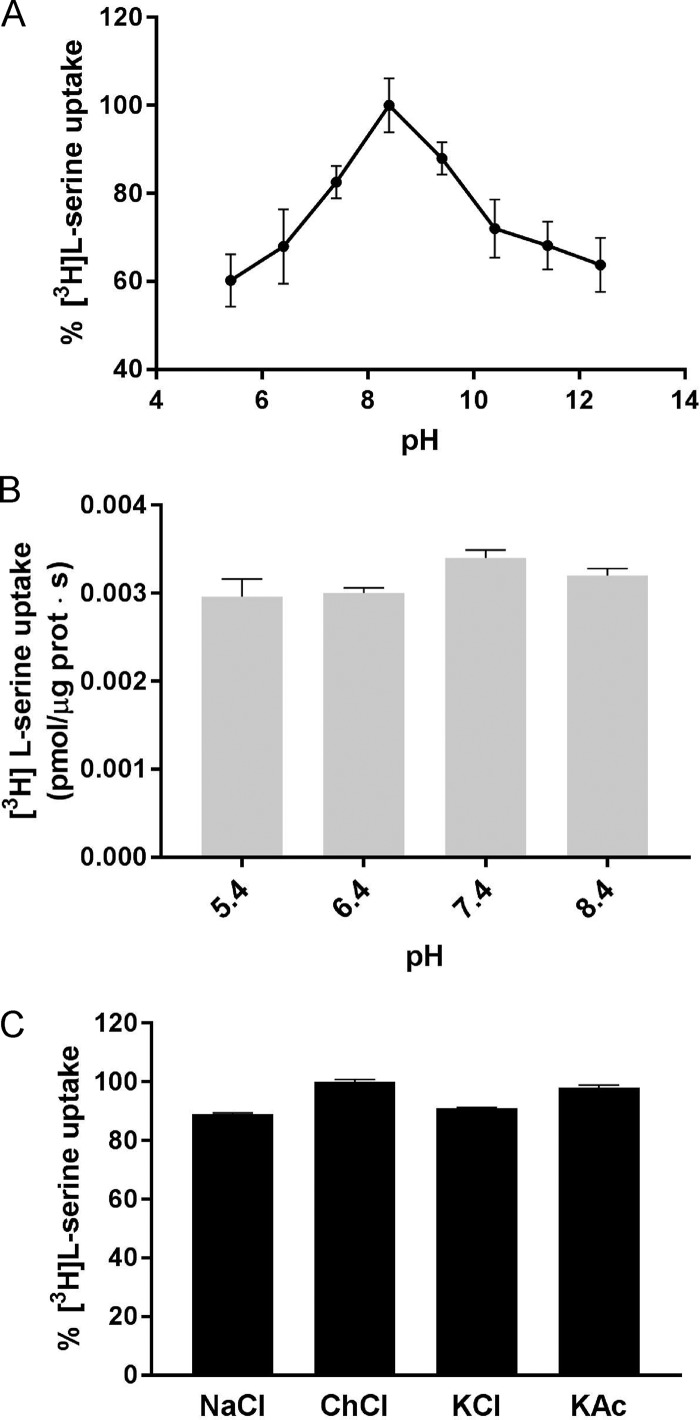
Given that several APC transporters are exchangers that are active over a range of pHs, we explored various external proton concentrations at a fixed internal pH value of 7.4. BasC-GFP–mediated 10 µM [3H]L-serine/L-alanine exchange was optimal between pH 7.0 and 9.4, being strongly reduced at both acidic and highly basic pHs (Fig. 4 A). We observed [3H]L-serine/L-alanine exchange at all the external proton concentrations assayed, this exchange being dependent exclusively on the presence of amino acid substrate inside PLs. No 10 µM [3H]L-serine uptake was observed at any external pH value in empty PLs (Fig. 4 B). Moreover, BasC exchange activity was independent of Na+, K+, and Cl− in the external medium (Fig. 4 C).
Figure 4.
Effect of external pH and salts on BasC activity. (A) 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-GFP-PLs containing 4 mM L-alanine at different pHs. Internal pH was maintained at 7.4 while external pH was changed (5.4–11.4). Transport was expressed as the percentage of [3H]L-serine transport, considering 100% the highest [3H]L-serine uptake value obtained. Data are from three experiments with three replicates per condition. (B) 10 µM [3H]L-serine (1 µCi/µl) influx (pmol/µg protein · s) into BasC-GFP-PLs containing no amino acid and at different pHs. Internal pH was maintained at 7.4 while external pH was changed (5.4–8.4). We observed [3H]L-serine exchange in all the assayed external proton concentrations, this exchange being dependent solely on the internal amino acid concentration. No significant increase in [3H]L-serine uptake was observed at any external pH value in empty PLs. Data (mean± SEM) are from three experiments with three replicates per condition. (C) Effect of ions on BasC activity. Uptake of 10 µM [3H]L-serine (1 µCi/µl) into BasC-PLs containing 4 mM L-alanine was performed in uptake buffer (20 mM Tris-Base and 150 mM NaCl, pH 7.4). NaCl was then replaced by 150 mM of either choline chloride (ChoCl), potassium chloride (KCl), or potassium acetate (KAc), and radiolabeled serine uptake was measured. Transport was expressed as the percentage of transport in BasC-GFP-PLs containing 4 mM L-alanine in uptake buffer with NaCl. Data are from three experiments with three replicates per condition.
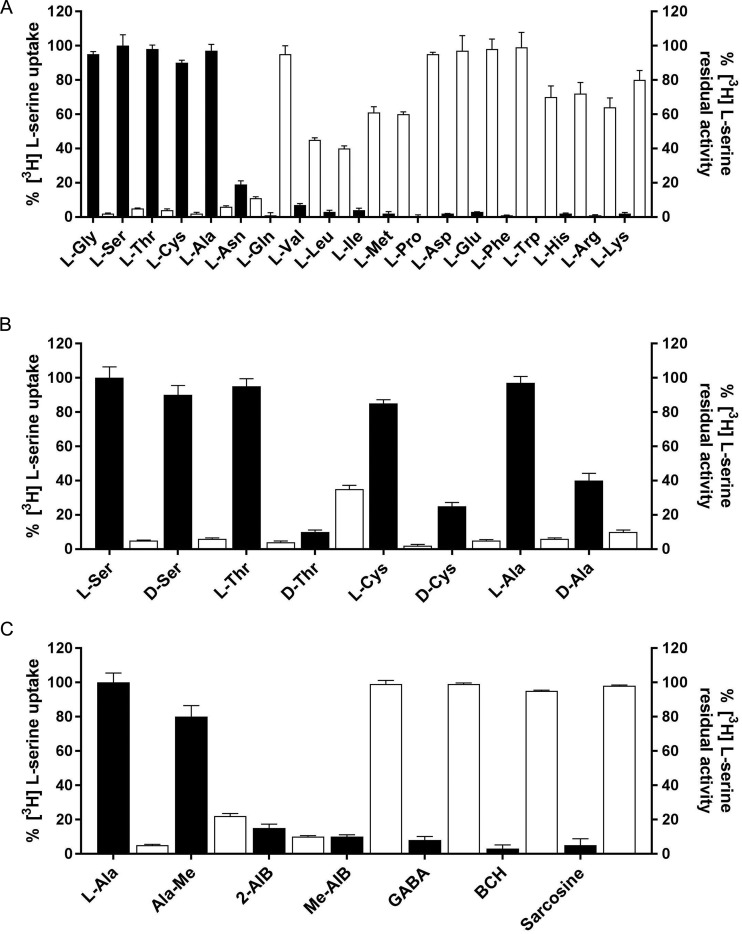
The substrate selectivity of BasC was studied initially using a competition assay, in which the exchange of 10 µM [3H]L-serine with 4 mM L-alanine inside BasC-GFP-PLs was measured in the presence of 4 mM nonlabeled amino acids and derivatives in the external medium. [3H]L-serine uptake was strongly inhibited by small neutral amino acids (glycine, L-alanine, L-serine, L-threonine, and L-cysteine) and L-asparagine, while acidic, aromatic (except tryptophan) as well as proline and glutamine, did not inhibit [3H]L-serine uptake (Fig. 5 A). Consistent with these results, high uptake levels of [3H]L-serine were observed in BasC-GFP-PLs containing 4 mM small neutral amino acids (and significant for those containing L-asparagine), whereas negligible levels of uptake were detected for the rest of the amino acids assayed (Fig. 5 A). Valine, leucine, isoleucine, methionine, tryptophan, and basic L–amino acids had significant inhibitory effects on BasC-mediated [3H]L-serine uptake. Surprisingly, no [3H]L-serine uptake was observed in BasC-GFP-PLs containing 4 mM of these amino acids (Fig. 5 A). In this regard, exchange of either 10 µM or 100 µM [3H]L-isoleucine in 4 mM L-alanine containing BasC PLs was negligible (data not shown). On the other hand, 4 mM D-serine, D-threonine, D-cysteine, and D-alanine exerted strong inhibitory effects on [3H]L-serine/L-alanine exchange, but only D-serine trans-stimulated this uptake at a similar rate as that achieved by L-serine (Fig. 5 B). Additionally, experiments in which 10 µM [3H]L-serine/L-alanine exchange was measured in the presence of 4 mM of compounds structurally related to alanine revealed that alanine methyl ester and 2-aminoisobutyric acid (AIB) strongly inhibited serine uptake, while N-methylalanine, methyl AIB, sarcosine, and γ-aminobutyric acid (GABA) did not inhibit BasC-mediated [3H]L-serine/L-alanine exchange (Fig. 5 C). In agreement, 4 mM alanine methyl ester and 4 mM 2-AIB trans-stimulated 10 µM [3H]L-serine uptake, although at different rates. Additionally, [3H]L-serine/L-alanine exchange was unaffected by system L-specific inhibitor 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH) (Fig. 5 C). Given that the substrate and inhibitor selectivity highly resembled that of the human and mouse (Fukasawa et al., 2000; Brown et al., 2014) alanine-serine-cysteine exchanger (Asc-1), we proposed that this new prokaryotic LAT be named BasC (bacterial alanine-serine-cysteine exchanger).
Figure 5.
Substrate and inhibitor specificity of BasC transport activity. (A) BasC substrate specificity. 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-GFP-PLs containing 4 mM of the indicated individual amino acids (black bars). Transport was expressed as the percentage of the transport in BasC-PLs containing 4 mM L-alanine. 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-GFP-PLs measured in the presence of 5 mM of the indicated amino acids in the external medium (white bars). Inhibition was expressed as the percentage of the transport in BasC-GFP-PLs containing 4 mM L-alanine with no cis-inhibitors. Data are from three experiments with three replicates per condition. (B) BasC stereoselectivity. 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-PLs containing 4 mM of the indicated individual L– or D–amino acids (black bars). Transport was expressed as the percentage of the transport in BasC-GFP-PLs containing 4 mM L-alanine. 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-GFP-PLs measured in the presence of 4 mM of the indicated L– or D–amino acids in the external medium (white bars). Inhibition was expressed as the percentage of transport in BasC-GFP-PLs containing 4 mM L-alanine with no cis-inhibitors. Data are from three experiments with three replicates per condition. (C) BasC amino acid derivative specificity. 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-GFP-PLs containing 4 mM of the indicated individual amino acid derivatives (black bars). Transport was expressed as the percentage of transport in BasC-GFP-PLs containing 4 mM L-alanine. 10 µM [3H]L-serine (1 µCi/µl) influx into BasC-GFP-PLs measured in the presence of 4 mM of the indicated amino acid derivatives in the external medium (white bars). Inhibition was expressed as the percentage of transport in BasC-GFP-PLs containing 4 mM L-alanine with no cis-inhibitors. Data are from three experiments with three replicates per condition.
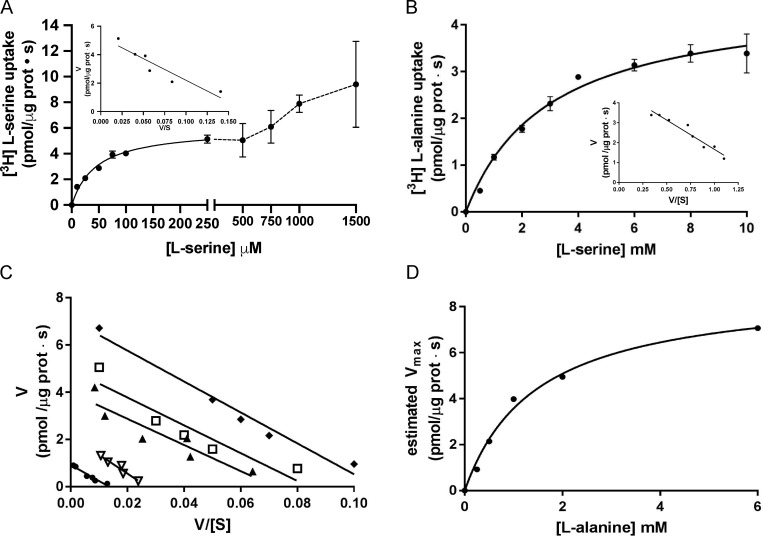
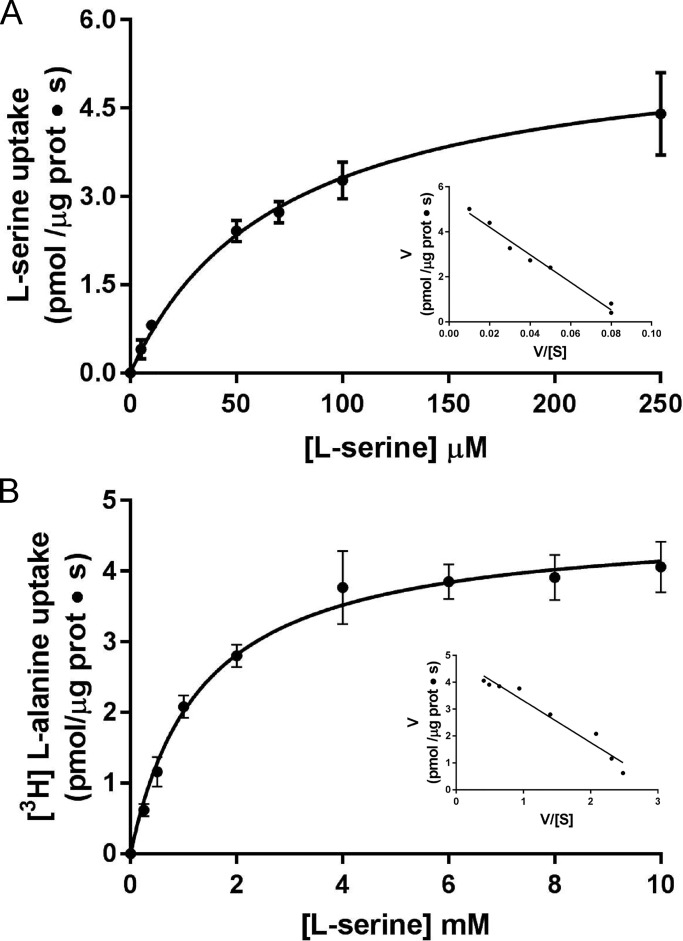
The kinetics of [3H]L-serine/L-alanine or [3H]L-alanine/L-serine exchange in BasC-GFP-PLs were examined by varying L-serine concentrations outside and inside the PLs, respectively. Radiolabeled [3H]L-serine (up to 1.5 mM)/4 mM L-alanine exchange was saturable but showed a complex kinetics, suggesting more than one component of transport (Fig. 6 A). A good fit to one-component Michaelis–Menten kinetics was obtained when the range of [3H]L-serine concentrations was set up to 250 µM in exchange with 4 mM L-alanine inside BasC-GFP-PLs. In these conditions, the estimated Km values of L-serine measured from outside were 45 ± 5 µM with a Vmax value of 6.0 ± 0.2 pmol [3H]L-serine/µg BasC protein · s (Fig. 6 A). In contrast to the kinetics from outside PLs, the kinetics varying the L-serine concentration inside the liposomes (up to 10 mM) in exchange with 10 µM [3H]L-alanine in the external medium showed a good fit to one-component Michaelis–Menten kinetics with a Km of 2.5 ± 0.4 mM from inside the BasC-PLs (Fig. 6 B). Similar values for the external and internal Km and Vmax were obtained with non-GFP-tagged BasC reconstituted in PLs (Fig. 7).
Figure 6.
Kinetic characterization of [3H]L-serine uptake in BasC PLs. (A) Determination of extraliposomal kinetic parameters. Michaelis–Menten plot of the transporter-mediated uptake of [3H]L-serine (1 µCi/µl, 4 s pmol/µg protein · s) in BasC-GFP-PLs containing 4 mM L-alanine, varying extracellular L-serine concentrations (0–1,500 µM). Mediated transport (exchange) was calculated as [3H]L-serine uptake in PLs containing 4 mM L-alanine minus empty PLs. Data correspond to a representative experiment, performed using three replicates. Inset: Eadie–Hofstee plot of the kinetics covering only L-serine concentrations between 0–250 µM (i.e., the high apparent affinity component). Km and Vmax values were 45 ± 5 µM and 6.0 ± 0.2 pmol [3H]L-serine/µg prot · s, respectively. V/[S]: pmol [3H]L-serine/µg prot · s / substrate concentration. (B) Determination of intraliposomal kinetic parameters. Michaelis–Menten plot of the transporter-mediated uptake of [3H]L-alanine (10 µM, 1 µCi/µl, pmol/µg protein · s) in BasC-GFP-PLs containing 0.5–10 mM cold L-serine. Mediated transport was calculated as [3H]L-alanine uptake in L-serine–containing PLs minus empty PLs. Data correspond to a representative experiment, performed using three replicates. Inset: Eadie–Hofstee plot. Km value was 2.5 ± 0.4 mM. In A and B, three independent experiments were performed, giving similar results. (C) Extraliposomal kinetics as in A at different intraliposomal concentrations of L-alanine (0.2, 0.5, 1.0, 2.0, and 6.0 mM). Eadie–Hofstee plots from a representative experiment with the mean values from three replicates. (D) Michaelis–Menten plot of the estimated Vmax values at the different intraliposomal L-alanine concentrations from the kinetic series shown in C.
Figure 7.
Extraliposomal and intraliposomal kinetics of BasC without GFP. (A) Determination of extraliposomal kinetic parameters. Michaelis–Menten plot of the transporter-mediated uptake of [3H]L-serine (1 µCi, 4 s) in BasC-PLs containing 4 mM L-alanine, varying extracellular L-serine concentrations (0–250 µM). Mediated transport was calculated as [3H]L-serine uptake in PLs containing 4 mM L-alanine minus empty PLs. Inset: Eadie–Hofstee plot. Km and Vmax values were 71.7 ± 4.3 µM and 5.7 ± 0.1 pmol [3H]L-serine/µg prot · s, respectively. (B) Determination of intraliposomal kinetic parameters. Michaelis–Menten plot of the transporter-mediated uptake of [3H]L-alanine (10 µM, 1 µCi, 4 s) in BasC-PLs containing 0.5–10 mM cold L-serine. Mediated transport was calculated as [3H]L-alanine uptake in L-serine–containing PLs minus empty PLs. In A and B, data correspond to a representative experiment, performed using three replicates. Inset: Eadie–Hofstee plot. Km value was 1.3 ± 0.1 mM. Data (mean ± SEM) correspond to triplicates from representative experiments (A and B).
As expected for a “ping-pong” mechanism of exchange, the Km values for L-serine estimated from outside the liposomes, using the conditions indicated above, were constant, independently of the concentration of L-alanine inside the liposomes (Fig. 6 C). Moreover, the Km for L-alanine inside the liposomes estimated with the Vmax values of this seriated experiment revealed again a Km in the mM range (1.5 ± 0.2 mM; Fig. 6 D). In all, these experiments suggest that BasC reconstituted in liposomes shows two components of transport with different apparent affinities.
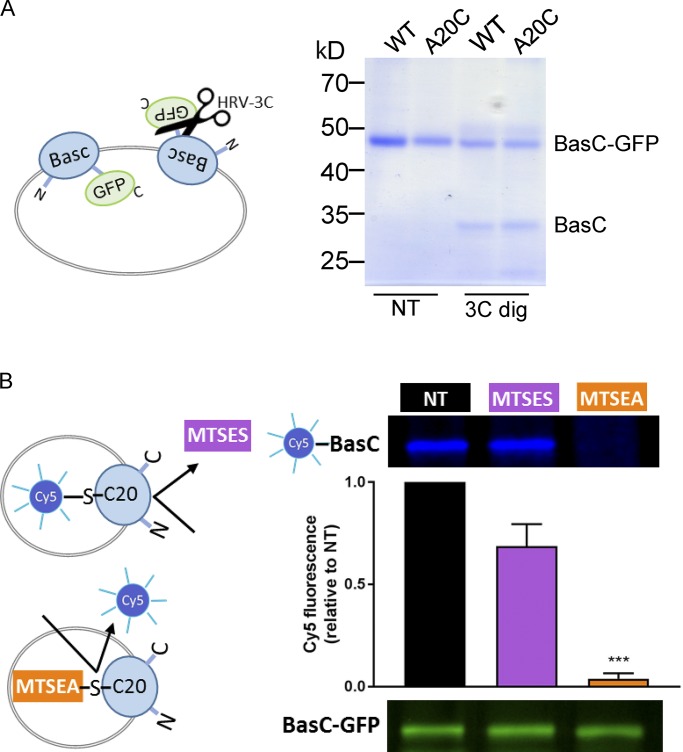
We next address whether these two components correspond to different apparent affinities for the substrate at the extracellular and intracellular sides of BasC. Indeed, BasC-GFP is inserted randomly into the PLs as the arginine/agmatine exchanger AdiC (Tsai et al., 2012). This was evidenced by the partial cleavage of GFP by HRV-3C protease from outside the PLs. Because GFP is fused to the C terminus (intracellular side), the cleaved BasC corresponds to the inside-out inserted transporters (Fig. 8 A). To address whether the apparent high-affinity component would correspond to the extra- or intracellular sides of the transporter, we used a strategy similar to that previously reported to study the sidedness of substrate interaction in AdiC (Tsai et al., 2012). We used BasC-GFP A20C mutant in an otherwise BasC cysless background (BasC-A20C-GFP). This mutant is homologous to the mutant S26C in AdiC, in which residue Cys 26 is only accessible from the extracellular side. Similarly, Cys 20 in BasC is also only accessible from the extracellular side. Thus, BasC-A20C-GFP PLs were treated with HRV-3C protease to cleave GFP from the inside-out oriented transporter molecules as in Fig. 8 A. Then, the sample was pretreated with 1 mM MTSES (nonpermeable sulfhydryl-reagent) or with 5 mM MTSEA (permeable sulfhydryl-reagent; Fig. 8 B) to block Cys20 residue. Then, the permeable Cy5-maleimide reagent was added to covalently bind the fluorophore to nonblocked cysteine residues. Pre-treatment with MTSES did not block Cy5 labeling of the inside-out oriented BasC-A20C molecules (i.e., BasC with GFP cleaved out), indicating that MTSES reached residue A20C only from the extracellular face of BasC (Fig. 8 B). As expected, the permeable reagent MTSEA almost completely abolished Cy5 labeling of Cys 20 (Fig. 8 B). Thus, Cys 20 in inside-out inserted BasC molecules is not accessible from outside the PLs.
Figure 8.
Vectorial modification of BasC A20C mutant by MTSES from the extracellular side. (A) Random incorporation of the BasC-GFP protein in PLs. BasC-GFP-PLs for wild-type and A20C mutant were treated with HRV-3C protease (3C dig) for 2 h at 4°C. The protease site is accessible in the inside-out BasC-GFP–inserted molecules, resulting in GFP cleavage. Protease-treated PLs were then mixed with loading sample buffer and analyzed in a 10% polyacrylamide gel. As a result, approximately half of the wild-type and mutant proteins were inserted right-side-out when reconstituted in E. coli polar lipid liposomes. (B) MTSEA but not MTSES pretreatment of BasC-A20C-GFP-PLs protected inside-out molecules from Cy5-maleimide dyeing. BasC-A20C-GFP-PLs were digested with HRV-3C protease for 2 h at 4°C to remove GFP from the inside-out BasC-A20C molecules. Then, either MTSES (1 mM; 15 min) or MTSEA (5 mM; 30 min) pretreatment was performed before BasC-A20C labeling with the membrane-permeable reagent Cy5-maleimide (1 mM; overnight). Finally, samples were mixed with loading sample buffer and analyzed in a 10% polyacrylamide gel. In-gel fluorescence at 600 nm (GFP) and 700 nm (Cy5) is shown. The graph represents quantification of Cy5 fluorescence in BasC-A20C normalized by GFP fluorescence in BasC-A20C-GFP and relative to aliquots nontreated with MTSES or MTSES in each experiment (NT). Data (mean ± SEM) are from four independent experiments. Student’s t test for paired data was used for statistical comparison between NT and MTSES (no significant differences) or MTSEA (***, P ≤ 0.001) treatments.
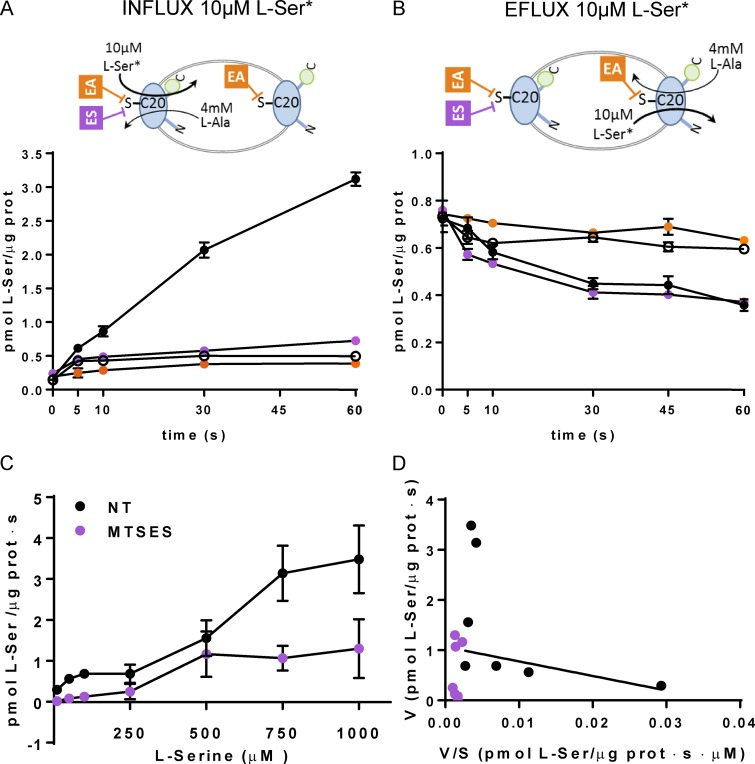
Next we address the sidedness of the substrate apparent affinities of BasC taking advantage of the vectorial inactivation by MTSES of the BasC-A20C-GFP activity. Fortunately, this mutant retained significant transport activity (∼20% of BasC-GFP; 0.35 ± 0.08 and 1.84 ± 0.15 pmol [3H[L-serine/µg protein · s in the mutant and wild-type BasC for 10 µM [3H]L-serine/4 mM L-alanine exchange, respectively). Treatment of PLs of BasC-A20C-GFP with 1 mM MTSES (impermeable) fully inhibited the influx of 10 µM [3H]L-serine, but not its efflux, in exchange with 4 mM L-alanine in the trans-side (Fig. 9, A and B). In contrast, treatment of BasC-A20C-GFP PLs with 5 mM MTSEA (permeable) resulted in complete inhibition of both 10 µM [3H]L-serine influx and efflux in exchange with 4 mM L-alanine (Fig. 9, A and B). As expected, 10 µM [3H]L-serine/4 mM L-alanine exchange in PLs reconstituted with cysless BasC-GFP was not affected by either MTSES or MTSEA treatment (Fig. S2). These results indicate that the side of BasC-A20C-GFP showing detectable transport of 10 µM [3H]L-serine (i.e., the apparent high-affinity side) is accessible to the nonpermeant reagent (MTSES) and therefore corresponds to the extracellular side of BasC.
Figure 9.
Sidedness of the transport activity of BasC in PLs. (A and B) Effect of cysteine-modifying reagents MTSES (ES) and MTSEA (EA) on L-serine transport activity. Time course (0–60 s) influx of 10 µM [3H]L-serine (0.5 µCi/data point) in empty (empty circles) or filled with 4 mM L-alanine (filled circles) BasC-A20C-GFP-PLs (A) and time course (0–60 s) efflux of 10 µM [3H]L-serine (0.5 µCi/data point) filled BasC-A20C-GFP-PLs against medium without amino acid (empty circles) or with 4 mM alanine and nontreated (filled circles), treated with 1 mM MTSES (ES) for 15 min (magenta circles), or treated with 5 mM MTSEA (EA) for 30 min (orange circles; B). 10 µM [3H]L-serine influx and efflux occurs via right-side-out– and inside-out–oriented BasC in PLs, respectively. MTSES only reaches Cys 20 (C20) in right-side-out–oriented BasC. MTSEA reaches C20 independently of the orientation of BasC insertion in PLs. Influx was determined by the incorporation of [3H]L-serine into PLs, whereas efflux was determined by the decay of [3H]L-serine remaining in PLs. MTSES or MTSEA treatment abolished L-serine influx, while only treatment with MTSEA, but not MTSES, completely inhibited L-serine efflux. Data (mean ± SEM) correspond to quadruplicates of a representative experiment. Two additional independent experiments gave similar results. (C and D) Kinetics of [3H]L-serine/4 mM L-alanine exchange in BasC-A20C-GFP-PLs treated (magenta circles) and nontreated (black circles) with 1 mM MTSES for 15 min. (D) Eadie–Hofstee plot of the kinetics shown in C. Nontreated PLs (black circles) showed a complex kinetics with low (line) and high Km components. MTSES treatment abolished the low Km (i.e., apparent high-affinity) component. Data (mean ± SEM) correspond to quadruplicates of a representative experiment. Another independent experiment gave similar results.
To finally support this asymmetry of the substrate interaction of BasC, the external kinetics of the [3H]L-serine (up to 1 mM; outside)/4 mM L-alanine (inside) exchange by BasC-A20C-GFP was performed before and after treatment with 1 mM MTSES (Fig. 9, C and D). As expected, nontreated BasC-A20C-GFP PLs showed a complex extracellular kinetics for [3H]L-serine/4 mM L-alanine exchange with a component of apparent high affinity (Km for L-serine: 35 ± 2.8 µM) and another of apparent low affinity. In contrast, MTSES-treated PLs did not show the extracellular apparent high-affinity component. In these conditions and due to the low activity of A20C mutant, the low-affinity Km could not be determined because of the low signal/background ratio at high L-serine concentrations. In all, these results support that BasC has asymmetric interaction with substrates, being the extracellular face of apparent high affinity (Km in the µM range) and the intracellular face of apparent low affinity (Km in the mM range) for substrates. Indeed, this is similar to human LATs (Meier et al., 2002).
Discussion
HATs are gaining relevance as their essential physiological role joins the increasing number of mutations associated with several diseases (Feliubadaló et al., 1999; Sperandeo et al., 2008; Tărlungeanu et al., 2016; Espino Guarch et al., 2018). Furthermore, in recent years it has been shown that the pharmacological inhibition of some of these transporters provides an effective strategy for the treatment of some types of cancer (Chung et al., 2005; del Amo et al., 2008; Lo et al., 2008; Savaskan and Eyüpoglu, 2010). However, knowledge of the effects of deleterious mutations on the structure or function of these transporters, as well as the generation of specific target drugs, has been greatly hindered by the lack of structural information. In this regard, only structures of distant homologues both in sequence (18–22% SI) and function have been solved (Fang et al., 2009; Gao et al., 2009; Kowalczyk et al., 2011; Ma et al., 2012; Ilgü et al., 2016; Jungnickel et al., 2018).
Some years ago, we cloned, expressed, and purified SteT, the L-serine/L-threonine antiporter of Bacillus subtilis, the first prokaryotic member of LATs to be characterized (Reig et al., 2007). For this reason, SteT emerged as a good bacterial paradigm of LATs. Nevertheless, although a great deal of effort in recent years has been channeled into improving SteT stability (Rodríguez-Banqueri et al., 2016), crystallization assays have failed to render results, thus fueling the search for a new prokaryotic paradigm suitable for atomic structure determination.
The crystal structure of BasC (unpublished data) is the only high-resolution structural information available for the LAT subfamily of transporters. The present study reports on the functional reconstitution and characterization of BasC to identify those features shared with other LAT members: (1) phylogenetic analysis clustered the BasC amino acid sequence with those of LAT family members within the APC superfamily; (2) BasC showed obligate amino acid exchange activity, which is characteristic of the catalytic subunits of HATs (Fotiadis et al., 2013); (3) kinetic and biochemical studies support an asymmetric binding site in terms of apparent affinity, as reported for several human LATs (Torras-Llort et al., 2001; Meier et al., 2002); and (4) both substrate and inhibitor selectivities in BasC are nearly identical to the eukaryotic LAT subfamily member Asc-1 (Fukasawa et al., 2000).
The phylogenetic tree of the APC family placed BasC within the LAT subfamily, and sequence alignments of BasC with the core members of the human LATs revealed amino acid identities ranging from 26 to 28% (Fig. 2 and Fig. S1). Additionally, nearly 63% of pathogenic mutations present in LAT transporters are conserved or semi-conserved in BasC (Fig. S1). This higher SI, compared with the distant homologues AdiC, ApcT, GadC, and GkApcT, together with its nearly complete sequence alignment with hLATs, makes BasC an excellent template for homology modeling of human transporters. Reliable models of hLAT transporters would help to unravel the molecular defects underlying pathogenic mutations, as well as identify specific molecules that regulate LAT activity. In this regard, molecular docking for ligand discovery in structural models for LAT-1 based on atomic structures of distantly related prokaryotic homologues (AdiC and GadC) has produced promising results (Geier et al., 2013).
BasC is a protein from Carnobacterium sp. AT7, a piezophilic strain isolated from the Aleutian trench (Lauro et al., 2007). Protein adaptation to high pressure is conferred, in part, by weaker intramolecular interactions that favor higher catalytic efficiencies and greater molecular flexibility (Somero, 1992; Brindley et al., 2008). In this regard, BasC is an extremely fast and efficient amino acid exchanger. Additionally, the lack of intramolecular disulfide bridges, due to the presence of only one cysteine in the whole BasC amino acid sequence (Cys 427), is consistent with these adaptations to high-pressure environments. This particular characteristic of BasC makes it an excellent model for biochemical studies involving single cysteine substitutions. In this regard, the single cysteine mutant A20C-C427A (mutant A20C) used in the present study to address the sidedness of the substrate’s apparent affinity is a clear example (Fig. 8 and Fig. 9).
Analysis of BasC in PLs by SDS-PAGE, after HRV-3C protease treatment, indicated that the protein is integrated into PLs in two orientations (Fig. 8). Distinct substrate affinities can be expected for the two orientations (almost two orders of higher apparent affinity for the external than the cytoplasmic side), as demonstrated for the reconstituted catalytic subunit (b(0+)AT) of system b(0+) (Torras-Llort et al., 2001) and for other HATs expressed in oocytes (Meier et al., 2002). In agreement, fine-tuning the experimental conditions both outside and inside BasC-PLs allowed us to calculate two distinct apparent Km values of 45 ± 5 µM and 2.5 ± 0.4 mM for L-serine, respectively (Fig. 6). Nevertheless, whether these different affinities corresponded to the extra- or intracellular sides of the transporter remained unclear. To address this issue, using a strategy similar to that of Tsai et al. (2012) developed for the AdiC transporter and adapted to BasC, we identified that the high-affinity binding site corresponded to the extracellular site and the low-affinity one to the intracellular site (Figs. 8 and 9). Interestingly, our kinetic analysis showed that the substrate extracellular Km was independent of the concentration of substrate in the cytosolic medium (Fig. 6), suggesting a ping-pong mechanism compatible with the alternative access model of transport (Kowalczyk et al., 2011). This is compatible with a binary complex (one substrate molecule per transporter molecule) that induces transport, as revealed by the available structures of APC transporters (Kowalczyk et al., 2011; Wang et al., 2012; Jungnickel et al., 2018).
This asymmetry in the substrate apparent affinity is a key feature of mammalian LAT transporters (Torras-Llort et al., 2001; Meier et al., 2002) that allows them to fulfill their physiological role in maintaining intracellular amino acid pools in the mM range while taking up extracellular amino acids at µM concentrations. The molecular bases underlying this asymmetry in the substrate apparent affinity of BasC and LATs is at present unknown. Different substrate binding affinities at both faces of the transporter and/or impact of different steps of the translocation mechanisms might be at the basis of this asymmetry.
Among the members of the LAT family, BasC showed the highest substrate and inhibitor selectivity similarity to Asc-1 (Fig. 5), thereby suggesting a very similar substrate binding site design in both cases. In this regard, the extracellular binding site of Asc1 accepts, although with lower affinity, larger neutral amino acids (e.g., leucine, isoleucine, and valine), while the intracellular site seems to be strictly designed to recognize small neutral amino acids (Fukasawa et al., 2000). In contrast, BasC intra- and extracellular binding sites seem to be restricted to small neutral amino acids (and asparagine) as uptake of larger neutral amino acids is negligible. Asc-1, like BasC, is a Na+-independent amino acid transporter that is noninhibitable by 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid, and it has a distinctive property among LATs, namely the capacity to mediate D-serine uptake (Fukasawa et al., 2000; Fig. 5, A and B). D-serine is present in the mammalian brain, where Asc-1 is strongly expressed (Ehmsen et al., 2016) and has been proposed to be an endogenous modulator of N-methyl-D-aspartate–type glutamate receptors. Thus, further knowledge of this specific characteristic shared by Asc-1 and BasC would help elucidate pathological situations associated with altered D-serine levels in the brain (Sason et al., 2017). In this regard, human Asc-1 is considered a potential target to treat schizophrenia and cognitive deficits (Fujita et al., 2016; Sakimura et al., 2016).
Little is known about the structure of the light subunits of HATs (LAT subfamily). The elucidation of their structure is limited mainly by the low HAT expression in native cells and tissues and the difficulty to overexpress these proteins in heterologous systems. The elucidation of the atomic structure of a prokaryotic transporter with high SI to eukaryotic light subunits of HATs emerges as an excellent alternative to unravel the molecular mechanisms underlying HATs. To this end, we have identified, cloned, overexpressed, and purified a novel prokaryotic Na+-independent neutral amino acid transporter, BasC, which exhibits the functional properties of the LAT subfamily. BasC is also the first member of this LAT subfamily to be crystallized, and analysis of its atomic structure would help elucidate the amino acid recognition and translocation mechanisms of LATs, the study of pathological mutations associated with LAT transporters, and the generation of reliable models of the hLAT transporters that would contribute to the intelligent design of more specific and potent drugs.
Supplementary Material
Acknowledgments
We thank Nick Berrow (Core Facility of Protein Expression, Institute for Research in Biomedicine, Barcelona Institute of Science and Technology, Barcelona, Spain) for providing us with HRV-3C protease.
E. Errasti-Murugarren has been supported by a Sara Borrell contract and P. Bartoccioni and J. Fort by Spanish Biomedical Research Center in Rare Diseases contracts. This work has been supported by the Spanish Ministry of Science and Innovation (SAF2015-64869-R-FEDER), Fundació la Marató TV3 (20132330) and Generalitat de Catalunya (SGR2009-1355).
The authors declare no competing financial interests.
Author contributions: P. Bartoccioni designed and performed experiments. J. Fort designed and performed experiments and wrote the manuscript. E. Errasti-Murugarren designed the strategy and experiments, performed experiments, and wrote the manuscript. M. Palacín designed the strategy and experiments and wrote the manuscript.
Merritt C. Maduke served as editor.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., and Lipman D.J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Borsani G., Bassi M.T., Sperandeo M.P., De Grandi A., Buoninconti A., Riboni M., Manzoni M., Incerti B., Pepe A., Andria G., et al. . 1999. SLC7A7, encoding a putative permease-related protein, is mutated in patients with lysinuric protein intolerance. Nat. Genet. 21:297–301. 10.1038/6815 [DOI] [PubMed] [Google Scholar]
- Brindley A.A., Pickersgill R.W., Partridge J.C., Dunstan D.J., Hunt D.M., and Warren M.J.. 2008. Enzyme sequence and its relationship to hyperbaric stability of artificial and natural fish lactate dehydrogenases. PLoS One. 3:e2042 10.1371/journal.pone.0002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.M., Hunihan L., Prack M.M., Harden D.G., Bronson J., Dzierba C.D., Gentles R.G., Hendricson A., Krause R., Macor J.E., and Westphal R.S.. 2014. In vitro Characterization of a small molecule inhibitor of the alanine serine cysteine transporter -1 (SLC7A10). J. Neurochem. 129:275–283. 10.1111/jnc.12618 [DOI] [PubMed] [Google Scholar]
- Calonge M.J., Gasparini P., Chillarón J., Chillón M., Gallucci M., Rousaud F., Zelante L., Testar X., Dallapiccola B., Di Silverio F., et al. . 1994. Cystinuria caused by mutations in rBAT, a gene involved in the transport of cystine. Nat. Genet. 6:420–425. 10.1038/ng0494-420 [DOI] [PubMed] [Google Scholar]
- Chung W.J., Lyons S.A., Nelson G.M., Hamza H., Gladson C.L., Gillespie G.Y., and Sontheimer H.. 2005. Inhibition of cystine uptake disrupts the growth of primary brain tumors. J. Neurosci. 25:7101–7110. 10.1523/JNEUROSCI.5258-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Amo E.M., Urtti A., and Yliperttula M.. 2008. Pharmacokinetic role of L-type amino acid transporters LAT1 and LAT2. Eur. J. Pharm. Sci. 35:161–174. 10.1016/j.ejps.2008.06.015 [DOI] [PubMed] [Google Scholar]
- Drew D., Slotbloom D.J., Friso G., Reda T., Genevaux P., Rapp M., Meindl-Beinker N.M., Lambert W., Lerch M., Daley D.O., et al. . 2005. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci. 14:2011–2017. 10.1110/ps.051466205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen J.T., Liu Y., Wang Y., Paladugu N., Johnson A.E., Rothstein J.D., du Lac S., Mattson M.P., and Höke A.. 2016. The astrocytic transporter SLC7A10 (Asc-1) mediates glycinergic inhibition of spinal cord motor neurons. Sci. Rep. 6:35592 10.1038/srep35592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino Guarch M., Font-Llitjós M., Murillo-Cuesta S., Errasti-Murugarren E., Celaya A.M., Girotto G., Vuckovic D., Mezzavilla M., Vilches C., Bodoy S., et al. . 2018. Mutations in L-type amino acid transporter-2 support SLC7A8 as a novel gene involved in age-related hearing loss. eLife. 7:e31511 10.7554/eLife.31511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Jayaram H., Shane T., Kolmakova-Partensky L., Wu F., Williams C., Xiong Y., and Miller C.. 2009. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 460:1040–1043. 10.1038/nature08201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliubadaló L., Font M., Purroy J., Rousaud F., Estivill X., Nunes V., Golomb E., Centola M., Aksentijevich I., Kreiss Y.; International Cystinuria Consortium, et al. . 1999. Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat. Genet. 23:52–57. 10.1038/12652 [DOI] [PubMed] [Google Scholar]
- Fernández E., Jiménez-Vidal M., Calvo M., Zorzano A., Tebar F., Palacín M., and Chillarón J.. 2006. The structural and functional units of heteromeric amino acid transporters. The heavy subunit rBAT dictates oligomerization of the heteromeric amino acid transporters. J. Biol. Chem. 281:26552–26561. 10.1074/jbc.M604049200 [DOI] [PubMed] [Google Scholar]
- Floden E.W., Tommaso P.D., Chatzou M., Magis C., Notredame C., and Chang J.M.. 2016. PSI/TM-Coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res. 44(W1):W339–W343. 10.1093/nar/gkw300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font-Llitjós M., Jiménez-Vidal M., Bisceglia L., Di Perna M., de Sanctis L., Rousaud F., Zelante L., Palacín M., and Nunes V.. 2005. New insights into cystinuria: 40 new mutations, genotype-phenotype correlation, and digenic inheritance causing partial phenotype. J. Med. Genet. 42:58–68. 10.1136/jmg.2004.022244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D., Kanai Y., and Palacín M.. 2013. The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34:139–158. 10.1016/j.mam.2012.10.007 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Ishima T., and Hashimoto K.. 2016. Supplementation with D-serine prevents the onset of cognitive deficits in adult offspring after maternal immune activation. Sci. Rep. 6:37261 10.1038/srep37261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y., Segawa H., Kim J.Y., Chairoungdua A., Kim D.K., Matsuo H., Cha S.H., Endou H., and Kanai Y.. 2000. Identification and characterization of a Na(+)-independent neutral amino acid transporter that associates with the 4F2 heavy chain and exhibits substrate selectivity for small neutral D- and L-amino acids. J. Biol. Chem. 275:9690–9698. 10.1074/jbc.275.13.9690 [DOI] [PubMed] [Google Scholar]
- Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., and Shi Y.. 2009. Structure and mechanism of an amino acid antiporter. Science. 324:1565–1568. 10.1126/science.1173654 [DOI] [PubMed] [Google Scholar]
- Gao X., Zhou L., Jiao X., Lu F., Yan C., Zeng X., Wang J., and Shi Y.. 2010. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 463:828–832. 10.1038/nature08741 [DOI] [PubMed] [Google Scholar]
- Geier E.G., Schlessinger A., Fan H., Gable J.E., Irwin J.J., Sali A., and Giacomini K.M.. 2013. Structure-based ligand discovery for the Large-neutral Amino Acid Transporter 1, LAT-1. Proc. Natl. Acad. Sci. USA. 110:5480–5485. 10.1073/pnas.1218165110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgü H., Jeckelmann J.M., Gapsys V., Ucurum Z., de Groot B.L., and Fotiadis D.. 2016. Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC. Proc. Natl. Acad. Sci. USA. 113:10358–10363. 10.1073/pnas.1605442113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel K.E.J., Parker J.L., and Newstead S.. 2018. Structural basis for amino acid transport by the CAT family of SLC7 transporters. Nat. Commun. 9:550 10.1038/s41467-018-03066-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk L., Ratera M., Paladino A., Bartoccioni P., Errasti-Murugarren E., Valencia E., Portella G., Bial S., Zorzano A., Fita I., et al. . 2011. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc. Natl. Acad. Sci. USA. 108:3935–3940. 10.1073/pnas.1018081108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro F.M., Chastain R.A., Blankenship L.E., Yayanos A.A., and Bartlett D.H.. 2007. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Environ. Microbiol. 73:838–845. 10.1128/AEM.01726-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., and Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M., Ling V., Wang Y.Z., and Gout P.W.. 2008. The xc- cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br. J. Cancer. 99:464–472. 10.1038/sj.bjc.6604485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Lu P., Yan C., Fan C., Yin P., Wang J., and Shi Y.. 2012. Structure and mechanism of a glutamate-GABA antiporter. Nature. 483:632–636. 10.1038/nature10917 [DOI] [PubMed] [Google Scholar]
- Meier C., Ristic Z., Klauser S., and Verrey F.. 2002. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 21:580–589. 10.1093/emboj/21.4.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morth J.P., Poulsen H., Toustrup-Jensen M.S., Schack V.R., Egebjerg J., Andersen J.P., Vilsen B., and Nissen P.. 2009. The structure of the Na+,K+-ATPase and mapping of isoform differences and disease-related mutations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:217–227. 10.1098/rstb.2008.0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A., Wang K.H., and Gouaux E.. 2013. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 503:85–90. 10.1038/nature12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig N., Chillarón J., Bartoccioni P., Fernández E., Bendahan A., Zorzano A., Kanner B., Palacín M., and Bertran J.. 2002. The light subunit of system b(o,+) is fully functional in the absence of the heavy subunit. EMBO J. 21:4906–4914. 10.1093/emboj/cdf500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig N., del Rio C., Casagrande F., Ratera M., Gelpí J.L., Torrents D., Henderson P.J., Xie H., Baldwin S.A., Zorzano A., et al. . 2007. Functional and structural characterization of the first prokaryotic member of the L-amino acid transporter (LAT) family: a model for APC transporters. J. Biol. Chem. 282:13270–13281. 10.1074/jbc.M610695200 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Banqueri A., Errasti-Murugarren E., Bartoccioni P., Kowalczyk L., Perálvarez-Marín A., Palacín M., and Vázquez-Ibar J.L.. 2016. Stabilization of a prokaryotic LAT transporter by random mutagenesis. J. Gen. Physiol. 147:353–368. 10.1085/jgp.201511510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, et al. . 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic acids research. D372–D379. 10.1093/nar/gkv1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura K., Nakao K., Yoshikawa M., Suzuki M., and Kimura H.. 2016. A novel Na(+) -Independent alanine-serine-cysteine transporter 1 inhibitor inhibits both influx and efflux of D-Serine. J. Neurosci. Res. 94:888–895. 10.1002/jnr.23772 [DOI] [PubMed] [Google Scholar]
- Sason H., et al. . 2017. Asc-1 Transporter Regulation of Synaptic Activity via the Tonic Release of d-Serine in the Forebrain. Cereb. Cortex. 27:1573–1587. [DOI] [PubMed] [Google Scholar]
- Savaskan N.E., and Eyüpoglu I.Y.. 2010. XCT modulation in gliomas: Relevance to energy metabolism and tumor microenvironment normalization. Ann. Anat. 192:309–313. 10.1016/j.aanat.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Shaffer P.L., Goehring A., Shankaranarayanan A., and Gouaux E.. 2009. Structure and mechanism of a Na+-independent amino acid transporter. Science. 325:1010–1014. 10.1126/science.1176088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero G.N. 1992. Adaptations to high hydrostatic pressure. Annu. Rev. Physiol. 54:557–577. 10.1146/annurev.ph.54.030192.003013 [DOI] [PubMed] [Google Scholar]
- Sperandeo M.P., Andria G., and Sebastio G.. 2008. Lysinuric protein intolerance: update and extended mutation analysis of the SLC7A7 gene. Hum. Mutat. 29:14–21. 10.1002/humu.20589 [DOI] [PubMed] [Google Scholar]
- Tărlungeanu D.C., Deliu E., Dotter C.P., Kara M., Janiesch P.C., Scalise M., Galluccio M., Tesulov M., Morelli E., Sonmez F.M., et al. . 2016. Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell. 167:1481–1494.e18. 10.1016/j.cell.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torras-Llort M., Torrents D., Soriano-García J.F., Gelpí J.L., Estévez R., Ferrer R., Palacín M., and Moretó M.. 2001. Sequential amino acid exchange across b(0,+)-like system in chicken brush border jejunum. J. Membr. Biol. 180:213–220. 10.1007/s002320010072 [DOI] [PubMed] [Google Scholar]
- Torrents D., Estévez R., Pineda M., Fernández E., Lloberas J., Shi Y.B., Zorzano A., and Palacín M.. 1998. Identification and characterization of a membrane protein (y+L amino acid transporter-1) that associates with 4F2hc to encode the amino acid transport activity y+L. A candidate gene for lysinuric protein intolerance. J. Biol. Chem. 273:32437–32445. 10.1074/jbc.273.49.32437 [DOI] [PubMed] [Google Scholar]
- Tsai M.-F., Fang Y., and Miller C.. 2012. Sided functions of an arginine-agmatine antiporter oriented in liposomes. Biochemistry. 51:1577–1585. 10.1021/bi201897t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Elferich J., and Gouaux E.. 2012. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat. Struct. Mol. Biol. 19:212–219. 10.1038/nsmb.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Guang W., Chen X., and Hong A.. 2014. Homology modeling and molecular docking of human pituitary adenylate cyclase‑activating polypeptide I receptor. Mol. Med. Rep. 10:1691–1696. 10.3892/mmr.2014.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.