Abstract
Purpose:
Uveal melanoma (UM) is the most prevalent and lethal intraocular malignancy in adults. Here, we examined the importance of hypoxia in UM growth and tested the anti-tumor effects of arylsulfonamide 64B, an inhibitor of the hypoxia-induced factor (HIF) pathway in animal models of UM and investigated the related mechanisms.
Experimental Design:
UM cells were implanted in the uvea of mice eyes and mice systemically treated with 64B. Drug effect on primary eye tumor growth, circulating tumor cells, metastasis formation in liver and survival were examined. 64B effects on UM cell growth, invasion and hypoxia-induced expression of C-X-C chemokine receptor type 4 (CXCR4) and mesenchymal-epithelial transition factor (c-Met) were measured. Luciferase reporter assays, chromatin immunoprecipitation, co-immunoprecipitation and cellular thermal shift assays were used to determine how 64B interferes with the HIF transcriptional complex.
Results:
Systemic administration of 64B had potent anti-tumor effects against UM in several orthotopic mouse models, suppressing UM growth in the eye (~70% reduction) and spontaneous liver metastasis (~50% reduction), and extending mice survival (p < 0.001) while being well tolerated. 64B inhibited hypoxia-induced expression of CXCR4 and c-Met, two key drivers of tumor invasion and metastasis. 64B disrupted HIF-1 complex by interfering with HIF-1α binding to p300/CBP co-factors, reducing p300 recruitment to the MET and CXCR4 gene promoters. 64B could thermo-stabilize p300, supporting direct 64B binding to p300.
Conclusions:
Our pre-clinical efficacy studies support the further optimization of the 64B chemical scaffold towards a clinical candidate for the treatment of UM.
Keywords: HIF, uveal melanoma, metastasis, CXCR4, c-Met, arylsulfonamide, experimental therapeutics, drug discovery
Introduction
Uveal melanoma (UM) accounts for ~70% of all adult primary eye cancers, and has a mean annual incidence of ~4.3 per million in adults (1). For the last 30 years, UM patient mortality has remained stagnant with a 5-year death rate of ~50% (2). Standard of care for primary eye tumor control includes surgical enucleation combined with plaque radiotherapy, and patients suffer permanent vision loss and cosmetic defects. About 40% of UM patients develop metastasis within ten years of diagnosis (3), and liver is the main organ colonized by circulating UM cells. Patient survival rate drops to less than one year after onset of liver metastasis despite active treatments, such as Dacarbazine or Temozolomide (1, 2), and liver failure due to metastatic burden is the major cause of death. Hence, new treatment options are direly needed to improve overall survival. Recent genomic analysis has classified UM into subgroups with different metastatic risk (4), and specific markers in the primary eye tumor can be used to stratify patients (4, 5), suggesting that high-risk patients could benefit from neoadjuvant metastasis prevention treatments. Developing new anti-metastatic therapeutics that can antagonize primary UM growth in eye, its dissemination into the bloodstream, seeding into other organs, and growth from micro- to macro- metastases, is an important therapeutic goal.
Hypoxia inducible factor (HIF), the master transcriptional regulator of hypoxia-mediated gene responses, plays a key role in tumorigenesis and metastasis (6, 7). Growing tumors experience limited oxygen supply due to inadequate vascular development, which leads to hypoxia-mediated genome reprogramming mediated by HIF. Recent genomic studies have revealed that hypoxia signaling is a major transcriptional signature of specific subsets of human UM (4). Active HIF is composed of a heterodimer of an oxygen-regulated alpha (HIF-1α or 2α) subunit and a beta subunit (HIF-1β) and recruits its co-activators p300/CBP to form an active transcription factor complex that transactivates target genes by binding to hypoxia response elements (HRE) in their promoters (8). HIF activation in tumors leads to the expression of over 100 target genes that mediate critical biological functions such as metabolic adaptation to anaerobic growth, angiogenesis, invasion and metastasis (7, 9). Previous studies have suggested hypoxia is involved in ocular melanoma cell adhesion, migration/invasion and angiogenesis (10, 11), but the role of hypoxia and HIF-1 in in vivo UM growth and metastasis remains to be established.
Two important cell surface mediators of hypoxia-induced migration, invasion and metastasis are tyrosine kinase receptor c-Met (12) and C-X-C chemokine receptor type 4 (CXCR4) (13). Binding of c-Met through its ligand hepatocyte growth factor (HGF) activates multiple oncogenic intracellular signaling pathways, driving cancer cell proliferation, survival, motility, invasion, and metastatic ability (12). Elevated c-Met expression in patient’s tumors correlates with unfavorable outcome, including in uveal melanoma (14). CXCR4 is the receptor for stromal cell-derived factor 1 (SDF-1/CXCL12), and can increase cancer cell migration and tumor metastasis (15). HGF and SDF-1 are produced by liver hepatocytes and may act as homing factors for circulating UM cells (16). Prior studies have evidenced the importance of CXCR4 and c-Met in UM growth and metastasis (14, 17), but targeting their hypoxia-induced activation in UM has not been realized.
Our laboratory is focused on the development of novel therapeutics based on the HIF pathway (18–22). We have previously developed a series of natural product-like small molecule HIF inhibitors (23–28), and the lead compound exhibited potent anti-tumor activity against several cancers with minimal side effects (29, 30). Structural optimization of the chemical scaffold subsequently yielded arylsulfonamide 64B, a more potent lead compound in the HRE reporter assay (27). The current study aimed to test the anti-cancer efficacy and toxicity of systemic delivery of 64B in orthotopic uveal melanoma mouse models and the related mechanisms.
Methods
Arylsulfonamide 64B synthesis and formulation
The discovery and synthesis of N-cyclobutyl-N-((2,2-dimethyl-2H-pyrano[3,2-b]pyridin-6-yl)methyl)-3,4-dimethoxybenzenesulfonamide (abbreviated as 64B) was previously described (27). For cell culture experiments a stock solution of 64B was dissolved in DMSO (10mM) and final concentration of solvent in culture medium was kept to 0.5% DMSO. For mice experiments, a large-scale crystalline batch of 64B was synthesized, and its purity was verified by mass spectrometry to be >95% (Supplementary Fig.S1 A, B). For mice delivery, crystalline 64B was suspended in a 1:1 cremophor EL/ethanol (1:1 volume) formulation by heating at 90oC and extensive vortexing. Before intraperitoneal (i.p.) injection, the mix was diluted 1:5 with PBS (preheated to 37oC) for a total injection volume of about 500 μl adjusted to mice body weight.
Cell Culture
Human 92.1 (31) and Mel290 (32) uveal melanoma cells authenticated by STR (Emory Genomics core facility) (Supplementary Fig. S1C), as well as B16LS9 mouse melanoma cells (33) were cultured in RPMI medium supplemented with 10% FBS, sodium pyruvate, and non-essential amino acids as previously described. We thank Dr. Bruce Ksander (Schepens Eye Institute, Boston, MA) for providing Mel290 cells, Dr. Jerry Niederkorn, (Dept. of Ophthalmology, UT Southwestern, Dallas, TX) for 92.1 cells, and Dr. Dario Rusciano (Friedrich Miescher Institute, Basel, Switzerland) for providing B16LS9 cells. Media for culture of human fibroblasts were as follows: HFF-1 (DMEM + 15% FBS), HDF [McCoy’s 5A; Gibco BRL, Cat #: 16600–082), and IMR-90 (MEM + 10% FBS). For experiments involving hypoxia (1% O2), cells were first pre-treated with 64B (2 μM) or vehicle control (0.5% DMSO) for 1 h. under normoxia (21% O2) and then transferred to a hypoxia (1% O2) incubator (Thermo Forma).
Sulforhodamine B (SRB) assay
SRB cytotoxicity assays were performed in 96 well plates with 4×103 cells/well for 48–72 hrs. as described (34). The inhibitory effects of 64B on cell growth were calculated by using the equation: % growth inhibition = (1 - At / Ac) x 100, where At and Ac represent the absorbance in treated and control cultures, respectively.
RNA extraction and qRT-PCR
Total RNA was extracted using Trizol Reagent (Thermo Fisher Scientific) and 1st strand cDNA was synthesized with ProtoScript First Strand cDNA Synthesis Kit (NEB). Amplifications were performed in 7500 Fast Real-Time PCR System (Applied Biosystems) with gene-specific primers (C-MET: forward: AGCGTCAACAGAGGGACCT, Reverse: GCAGTGAACCTCCGACTGTATG; CXCR4: forward: ACTACACCGAGGAAATGGGCT, Reverse: CCCACAATGCCAGTTAAGAAGA). β-actin was used as internal control, data were analyzed with 7500 Software, v2.3, and are presented as expression relative to normoxic control.
Western blot analysis
Cells were harvested by scraping in RIPA buffer supplemented with protease and phosphatase inhibitors (Thermo Scientific). Cell lysates were kept on ice for 30 min, centrifuged for 15 min at 13,500 x g in a microfuge at 4° C, and supernatants separated by sodium dodecyl sulfate poly-acrylamide gel electrophoresis (SDS-PAGE). Proteins in gel were transferred to nitrocellulose membranes (Bio-Rad; Cat: 1620112) and probed with anti-HIF-1α (BD Transduction Laboratories™; Cat: 610959), HIF-1β (BD Transduction Laboratories™; Cat: 611078), p300 (LS Bio; LS-B6081), CBP (Santa Cruz; SC-48343), c-Met (Abcam; ab10728), CXCR4 (Abcam; ab124824), and β-actin (Cell Signaling Technology; D6A8) antibodies (all used at 1:1000 dilution) as described (29).
Hypoxia/HIF transcription reporter assays
Mel290-V6R cells were generated by stably co-transfecting Mel290 cells with pBIGL-V6R and pcDNA3 (expresses Neomycin resistance gene), followed by G418 (200 μg/ml) selection. pBIGL-V6R is a bi-directional HIF-1 reporter construct in which the firefly luciferase and LacZ reporter genes are under the control of 6 head to tail copies of the VEGF hypoxia-responsive element (HRE) coupled to a mini-CMV promoter (20). Luciferase reporter assays were performed as described (20).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were conducted with the EZChIP Assay Kit (Cell Signaling Technology). A total of 4 × 107 cells was pretreated with 1% DMSO (control) or 2 μM 64B for 1 hour, transferred to hypoxia for 24 hours, and then fixed in 1% formaldehyde at room temperature for 20 minutes. Isolated nuclei were lysed, followed by chromatin digestion with micrococcal nuclease. Chromatin fragments were immunoprecipitated with anti-p300 polyclonal antibodies (Abcam) or rabbit IgG as a control. After reversal of cross-linking and DNA purification, DNA from input (1:20 dilution) or immunoprecipitated samples was analyzed with PCR, and products were separated by 2% agarose gel electrophoresis. Primers for the following genes were used.
CXCR4: 5’-TCGTGCCAAAGCTTGTCC-3’ (sense) and 5’-GCGGTAACCAATTCGCGAATAGTGC-3’ (antisense); c-Met: 5’-AGCGTCAACAGAGGGACCT-3’ (sense) and 5’-GCAGTGAACCTCCGACTGTATG-3’ (antisense); and DAPK1: 5’-CATTTCAGCTCAACAATAGCCAATA-3’ (sense) and 5’- GGAATAGAGAATGGCAGGAAGG-3’ (antisense)
Orthotopic uveal melanoma mouse models
Female nu/nu mice (10–15 mice/group) were used under IACUC approval and procedures adhered to the NIH Guide for the Care and Use of Laboratory Animals (See figure legends for mice strains used in the different experiments). Mice were identified by individual tattoos (35). On day 0, melanoma cells were inoculated into the suprachoroidal space of the right murine eye using a trans-scleral technique as previously described (36). 64B was administered i.p. at 120 mg/kg for 5 days/week starting on day 1 after tumor inoculation and animal weight was monitored weekly. Tumor-bearing eyes were enucleated on the 7th (B16LS9) or 9th day (92.1) after inoculation, and livers with metastases were collected at the time of sacrifice 4 (B16LS9) to 8 weeks (92.1) after inoculation. Number and size of liver metastases were evaluated as described (36). For Kaplan-Meier survival experiments, mice underwent enucleation on day 7 or 9, and then were left alive until reaching IACUC criteria for termination (usually >20% of body weight loss). Eyes, livers, and other tissues were harvested for pathological examination. For some experiments, mice were injected i.p. with pimodinazole (60 mg/kg; Hypoxyprobe ™, Cat No: 06172016), a hypoxia marker 1 h before sacrifice.
Systemic mouse metastasis assay
Male outbred nu/nu mice (5 mice/group; 10 weeks old; Jackson labs) were injected in the tail vein with 4 × 106 92.1 or Mel290 cells. Four days after systemic injection, mice were treated daily with 64B (60 mg/kg i.p. twice per day) or vehicle for 45 days, at which time they were terminated, livers sectioned and total number of metastases/section counted. Blood collected by cardiac puncture was collected and evaluated for blood cell counts using an automatic hematology analyzer (HemaTrue Veterinary Hematology Analyzer, Heska).
Immunohistochemistry (IHC)
Formalin-fixed tumors were cut into 4μm sections and processed for IHC using the UltraVision AEC Detection System (Thermo Scientific, TL-015-HAJ) with the following antibodies: c-Met (Abcam; ab10728), CXCR4 (Abcam; ab124824), Ki67 (Abcam; ab92742), Pimo (Hypoxyprobe ™), HIF-1α (BD Transduction Laboratories™; Cat: 610959), HIF-1β (BD Transduction Laboratories™; Cat: 611078) overnight at 4° C. Secondary antibodies conjugated with Alex 488, Alex 594, Alex 647 (Life Technologies™) were used to visualize the primary antibody’s staining. Images were acquired using Olympus Confocal Laser Scanning Biological Microscope FV1000 equipped with four lasers ranging from 405 to 635 nm. Images were processed with ImageJ software. The quantification of the positive cells was processed by using CellProfiler software (37).
Cellular thermal shift assay
64B drug target engagement was monitored by using the cellular thermal shift assay (38). In brief, cell lysates were collected, diluted and divided into two aliquots, treated with drug or diluent as control. After incubation for 30 min at room temperature, respective lysates were divided into smaller aliquots and heated individually at different temperatures (thermal cycler, Applied Biosystems/Life Technologies™). Lysates were centrifuged to eliminate temperature destabilized proteins, and supernatants analyzed by SDS-PAGE followed by Western blot analysis. A shift in temperature stability reflects drug-protein binding.
Microarray analysis
Two microarray datasets were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), including (GSE27831(39) and GSE22138(40)). These datasets contained gene expression data derived from the Affymetrix U133_plus2 platform. For microarray analysis, expression and raw expression data (CEL files) were summarized and normalized using the Robust Multi-array Average algorithm (http://www.bioconductor.org/packages/2.0/bioc/html/affy.html) from the Bioconductor library for the R statistical programming system. TIBCO Spotfire software package (TIBCO Software, Palo Alto, CA) was used for visualization of microarray data. Hierarchical clustering was performed using Ward’s method.
Outcome analysis
Gene expression and survival data of UM patients were obtained from the Gene Expression Omnibus (GEO) database, SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) and Kaplan-Meier Plotter (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). Survival curves were constructed by Kaplan-Meier analysis and compared with the log-rank test using SPSS Statistics 18 software (IBM, Chicago, IL, http://www.ibm.com).
Network analysis
STRING 10.5 (www.string-db.org) (41) was used for network analysis. STRING database contains known and predicted protein–protein interactions. Proteins considered to be interacting proteins were based on the original criteria in STRING 10.5. Highly interacting proteins were defined as having over 4 interactions..
Statistical analysis
Differences between two groups were analyzed using two-sided unpaired Student’s t-test. Differences between three or more groups were analyzed using one-way ANOVA. P values < 0.05 were considered statistically significant. Statistical analysis was performed with Graphpad Prism 5 software (San Diego, CA).
Results
Increased expression of c-Met and CXCR4 correlates with poor prognosis and liver metastasis in uveal melanoma patients
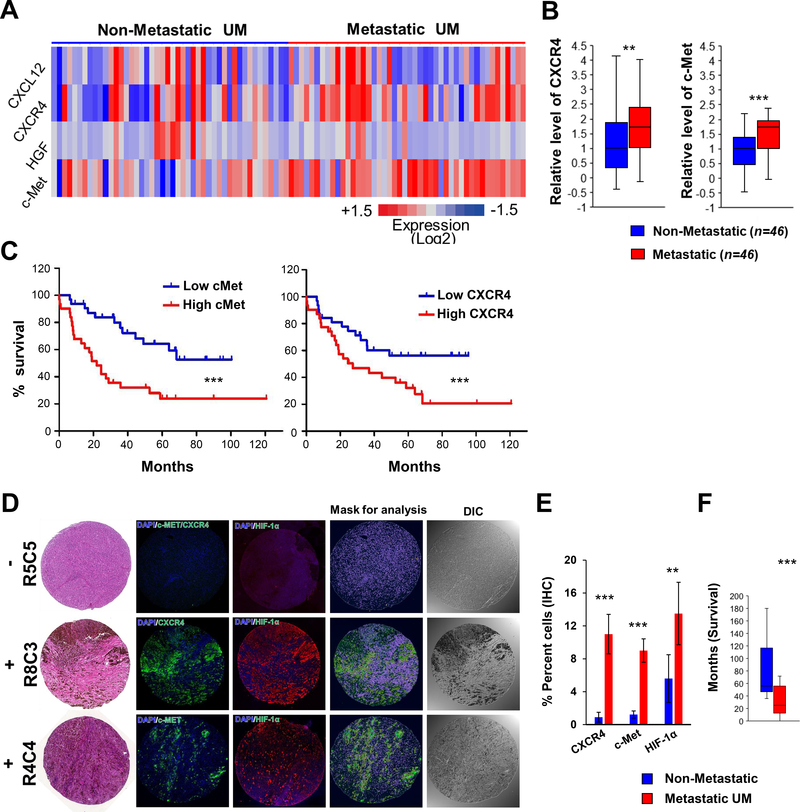
To identify hypoxia-induced genes whose expression might underlie metastasis and UM patient survival, we performed comparative gene expression analysis on primary melanoma samples of patients who had or not developed metastasis using two publicly available databases (GSE27831(39) and GSE22138(40)). 168 genes were found up-regulated in common in the metastatic groups, and pathway analysis revealed that 32 of them were highly inter-connected. Outcome analysis using an independent dataset (TCGA Uveal melanoma (4)) evidenced that gene expression of 22 of these was correlated with poor outcome, two of which (CXCR4 and MET) are hypoxia-inducible (Supplementary Fig. S2A, B and Supplementary Table 1). Kaplan-Meier analyses on two datasets (GSE22138 and TCGA) showed that elevated mRNA expression of CXCR4 and MET is strongly correlated with patient survival (Fig. 1C and Supplementary Fig. S2C). In contrast, expression levels of genes encoding HGF and CXCL12, the respective ligands for c-Met and CXCR4 and other main hypoxia response genes showed no difference between groups (Fig. 1A and Supplementary Fig. S2D). Unbiased hierarchical clustering further showed that most patients with low expression of both MET and CXCR4 mRNAs did not metastasize, while those with elevated MET metastasize (Supplementary Fig. S2E). Immunohistochemistry on a UM tissue array confirmed expression levels of CXCR4, c-Met and HIF-1α were significantly higher in samples collected from metastatic (n=17) as compared to non-metastatic UM patients (n=14) (Fig. 1D and E; Supplementary Fig. S3 and Supplementary Table 2). Survival analysis of matching patients evidenced that those with metastasis had worse outcome (Fig. 1F and Supplementary Table 1). Altogether, these results show that c-Met and CXCR4 expression are useful markers for prediction of UM metastatic progression and prognostic for patient outcome. They also suggest that both c-Met and CXCR4 may play an important role in the metastatic process in UM.
Figure 1.
Elevated expression of CXCR4 and MET correlates with poor prognosis and metastasis in uveal melanoma patients. A, Heatmap comparison of CXCR4, CXCL12 and MET, HGF mRNA expression in human uveal melanoma patients with or without metastasis (n = 46/group). B, Quantification of (A). C, Kaplan-Meier survival curves for UM patients (GSE22138) with high (n = 35) or low (n = 28) MET (Left) and CXCR4 (Right) mRNA levels. D, Representative immunofluorescence staining of CXCR4, c-Met and HIF-1α expression in ocular tumor tissue array from UM patients with or without metastasis. E, Quantification of percent positive tumor cells in (D). F, Survival time for metastatic patients and follow-up time for non-metastatic patients used for tissue array. **, p < 0.01; ***, p < 0.001;
c-Met/CXCR4 are hypoxia/HIF-1 target genes in uveal melanoma cells and their expression is inhibited by arylsulfonamide 64B, a novel HIF inhibitor
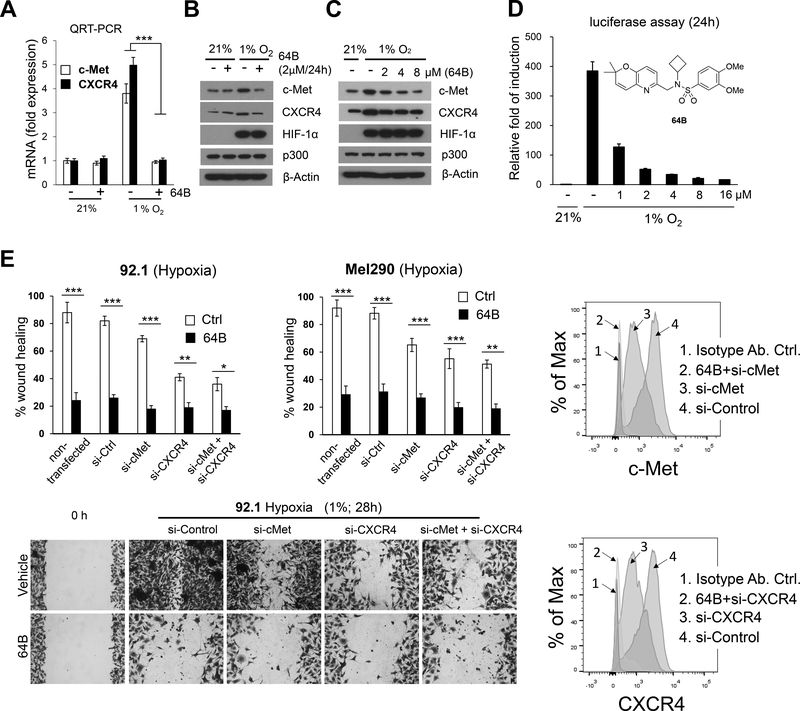
To examine whether tumor hypoxia could contribute to upregulated c-Met and CXCR4 expression in UM, we exposed cultured UM cells to 1% hypoxia. Hypoxia led to HIF-1 stabilization and increased expression of both c-Met and CXCR4 mRNA and protein, responses which were inhibited by HIF pathway inhibitor 64B (Fig. 2A-C and Supplementary Fig. S4A). 64B dose-dependently inhibited hypoxia-induced transcription of a luciferase reporter gene under the control of a promoter driven by hypoxia-response elements in UM cells (Fig. 2D). This effect was not due to cytotoxicity or direct inhibition of luciferase activity as 64B had no effect on cell viability of Mel290 cells under the culture conditions used in this assay, and did not affect luciferase expression driven by a constitutive CMV promoter (data not shown). 64B was not cytotoxic to normal cells either as tested in three human fibroblasts cell cultures (Supplementary Fig. S4B).
Figure 2.
64B blocks hypoxia-induced expression of c-Met and CXCR4 and UM cell migration. A, Quantification of mRNA expression level of CXCR4 and MET by QRT-PCR in Mel290-V6R UM cells under normoxia (21% O2) or hypoxia (1% O2) with or without treatment with 64B (2 μmol/L) for 24 h. B, C, Western blot analysis for indicated proteins from Mel290-V6R cells cultured under normoxia or hypoxia with or without 64B treatment (at indicated doses) for 24 hours. D, Dose-response inhibition of hypoxia-inducible gene expression with 64B in Mel290-V6R cells stably expressing an HRE-driven luciferase reporter gene. n = 18 independent experiments carried out in triplicate. E, Tumor cell scratch wound assay. Percent wound closure was measured in 64B-treated versus control from 92.1 (28 h) and Mel290 (16 h) cells grown under hypoxia (top). Representative pictures of wound closure in 92.1 UM cells after 28 h. of hypoxia (bottom). Cells were pre-treated for 48 h with control (Ctrl.), cMet or CXCR4 siRNAs for 48h. with or without treatment with 64B (2 μmol/L; 1h. pre-treatment), and silencing of cMet and CXCR4 cell surface expression was verified by FACS (right). Representative data of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001;
To further test whether 64B-mediated inhibition of c-Met/CXCR4 expression had any effect on UM cell motility, a prerequisite for metastasis, we performed scratch wound closure assays. 64B caused a significant decrease in hypoxia-induced wound closure in the assay (Fig. 2E), while have little effect under normoxic conditions (Supplementary Fig.S4C). Treatment of the cells with siRNA for CXCR4 and c-Met also inhibited cell migration under hypoxia, supporting their role in this process (Fig. 2E). Combination with 64B yielded slightly further inhibition, likely because the siRNA effect was incomplete (Fig. 2E), or due to additional 64B-inhibited factors. These results show that hypoxia enhances UM cell migration at least in part through c-Met and CXCR4 expression and that this response can be potently antagonized by HIF transcription inhibitor 64B.
64B blocks HIF transcriptional activity by disrupting the interaction between HIF-1α and p300, independently of Factor inhibiting HIF-1 (FIH-1)
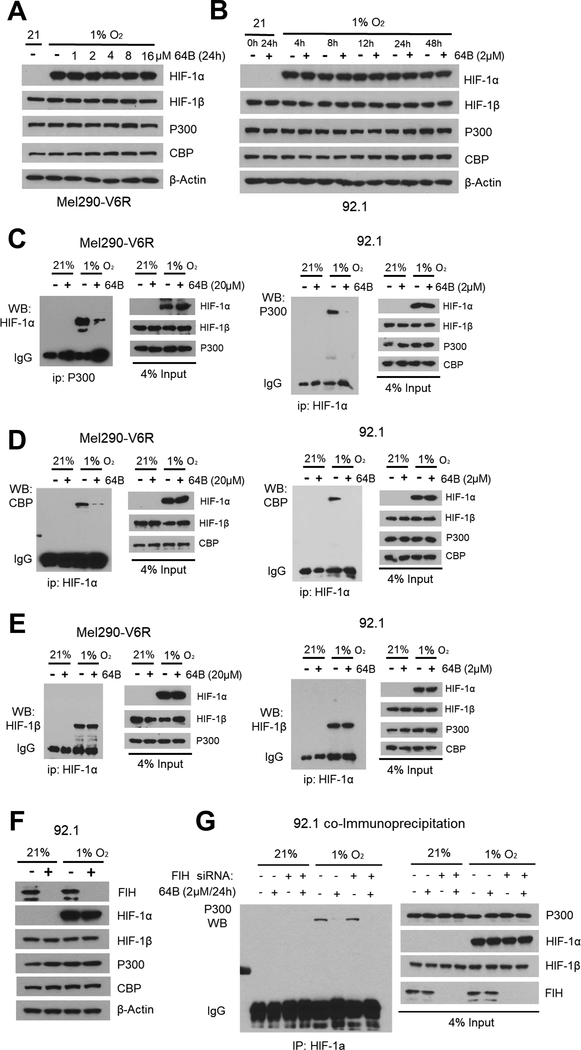
We next aimed to define the mechanism underlying compound 64B inhibition of HIF transcriptional activity in UM cells. We first tested if it alters hypoxia-activated HIF-1α protein stability. A 24h dose response experiment showed that 64B at concentrations up to 16 μM did not disrupt hypoxia-mediated stabilization of HIF-1α (Fig. 3A). A time course experiment showed that HIF-1α is stabilized by hypoxia as early as 4h, remains stable up to 48 h, and is not affected by 64B (Fig. 3B).
Figure 3.
64B prevents the binding of p300 and CBP to HIF-1α. A, Western blot of indicated protein expression after dose-response of 64B in Mel290-V6R cells after 24 h of hypoxia. B, Western blot of indicated protein expression after time-course of hypoxia +/− 64B (2 μmol/L) treatment. C, D, E, Co-immunoprecipitation (co-IP) of cell extracts from Mel290-V6R and 92.1 UM cells grown for 24 h. under normoxia or hypoxia +/− 64B (2 μmol/L) treatment. Note reduction in binding between HIF-1α and p300/CBP cofactors, but not HIF-1α. Western blot analysis of an aliquot (4%) of the cell extracts was used as a control for input protein (input). F, Western blot analysis of indicated protein expression in 92.1 cells under normoxia and hypoxia +/− FIH sliencing. G, Effect of siRNA-mediated silencing of FIH, on 64B’s ability to inhibit p300 - HIF-1α interaction in hypoxic 92.1 cells as measured by co-IP.
Under hypoxia, HIF-1α heterodimerizes with HIF-1β and forms a transcriptional complex with p300/CBP cofactors in the nucleus. Co-immunoprecipitation (co-IP) experiments showed 64B could disrupt the interaction between HIF-1α and both p300 and CBP paralogs, but not with HIF-1β (Fig. 3C-E). Levels of hypoxia-induced HIF-1α, and constitutive expression of p300, and CBP were unaffected by 64B (Fig. 3C-E; see input fraction).
The interaction of HIF with p300/CBP co-factors is regulated by the oxygen-dependent hydroxylation of asparagine 803 by Factor Inhibiting HIF-1 (FIH) on the C-terminus of HIF-1α (42). To probe for FIH role in 64B action, we silenced it by siRNA. FIH knockdown did not abrogate 64B disruption of HIF-1α/p300 interaction, suggesting that 64B does not act through FIH (Fig. 3F and G). Altogether, these results show that 64B can disrupt the recruitment of p300/CBP co-factors to the HIF transcription complex in a FIH-independent manner.
64B can bind to p300 and directly interferes with the interaction between HIF-1α and p300
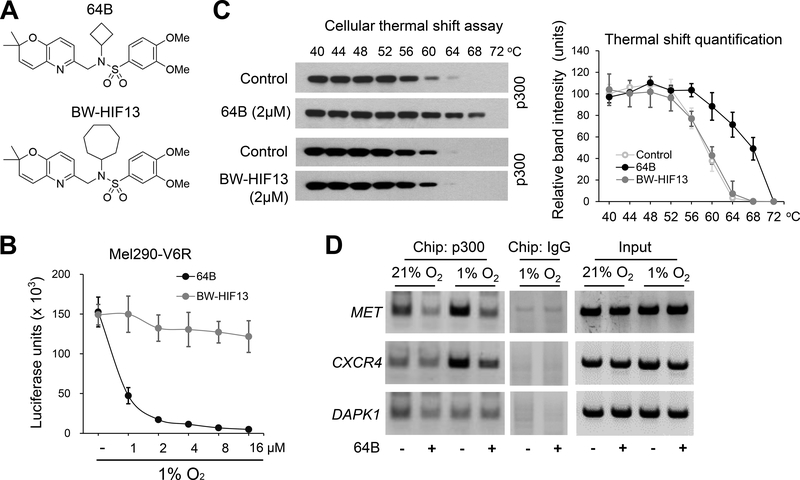
To further examine whether 64B disrupted HIF-1α/p300 interaction through direct binding of p300, we utilized the cellular thermal shift assay (38). As a negative control, we used BW-HIF13, a structural analog of 64B in which the cyclobutane is replaced with a benzene ring (Fig. 4A) that does not inhibit hypoxia-activated luciferase gene expression (Fig. 4B), Cell extracts of UM cells were prepared and then treated with 64B (2 μM) or BW-HIF13 in vitro for 30 min. Thereafter, aliquots (20 μg protein each) were prepared and each tube was heated to a different temperature, then centrifuged to eliminate any heat-denatured precipitated proteins, and the supernatants examined by Western blot for the protein of interest. In this assay, protein binding to a small molecule of interest will thermo-stabilize the protein and shift its denaturation temperature. This assay revealed that 64B could shift the thermal stability of p300 by 8°C (Fig. 4C), supporting a direct interaction between 64B and p300. In contrast, BW-HIF-13 did not elicit any shift (Fig. 4C), indicating 64B structural specificity to bind p300.
Figure 4.
64B directly binds to p300 and interferes with p300 recruitment to HIF-1 activated MET and CXCR4 genes. A, Chemical formula of 64B and inactive analog BW-HIF13. B, Dose-response of 64B and BW-HIF13 on hypoxia-induced luciferase expression in Mel290-V6R cells with a stably integrated HRE-luciferase reporter. (n = 3). C, Cellular thermal shift assays on 92.1 cell extracts treated with 64B or BW-HIF-13 (2 μM for 2 hours). Western blot (Left) of soluble fraction of individually heated cell extract aliquots (44–72°C) show 64B (but not BW-HIF-13) extends the thermal stability of p300. Quantification of p300 bands (Right). (n = 3). D, Chromatin immunoprecipitation (Chip) analysis shows hypoxia-induced binding of p300 on the CXCR4 and MET promoter regions is inhibited by 64B (2 μmol/L) in 92.1 cells. P300 recruitment to control gene DAPK1 was not affected. Data are representative of three independent experiments.
These findings suggested that 64B might prevent CXCR4 and MET gene activation under hypoxia by preventing p300 co-factor recruitment to the HIF transcriptional complex on the gene promoters in the nucleus. To test this, we used chromatin immunoprecipitation (ChIP), and found that 64B reduced p300 binding to both genes under hypoxia (Fig. 4D). In contrast, 64B did not prevent recruitment of p300 to the DAPK1 gene, which is activated by transcription factor C/EBPβ, independently of hypoxia/HIF. P300 binds to HIF-1 through its CH1 domain (43), while it binds to C/EBPβ via its CH3 domain (44). These findings suggest that 64B may exhibit CH1 domain specificity in its ability to interfere with p300/CBP transcription factor interactions.
64B inhibits primary tumor growth, liver metastasis formation, and prolongs survival in orthotopic mice models of uveal melanoma
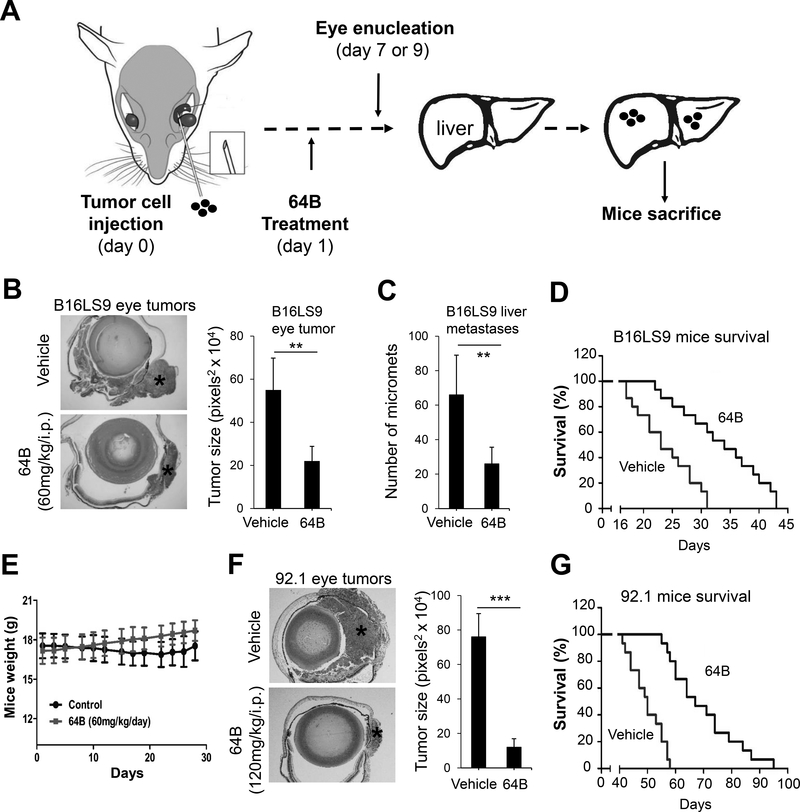
Next examined the in vivo effects of 64B on primary and metastatic uveal melanoma growth. To establish a treatment dose, we carried out a pilot acute toxicity study and with a single i.p. injection of 64B at 100, 200 and 400mg/kg. This did not result in any mice death, significant loss of mice weight or other abnormal behavioral changes (data not shown). Based on this, we selected a dose of 120 mg/kg for testing in two complementary orthotopic uveal melanoma models in mice. Both models involve the injection of melanoma cells in the sub-uveal area of the mouse eye where they form melanomas (Fig. 5A), which can spontaneously metastasize to the liver, mimicking human uveal melanoma disease process. The first is an immunocompetent model where mouse skin melanoma cells (B16LS9) are injected in syngeneic C57BL/6 mice. The second uses human uveal melanoma cells (92.1) injected in immunocompromised athymic nude mice. After eye inoculation, mice are treated i.p. with 64B or vehicle control 5 days/week. After primary tumor growth, tumor-bearing eyes are removed, fixed and stained with H&E to evaluate tumor burden. Mice are continually treated with 64B till they are sacrificed to observe metastasis development in the liver. In the B16LS9 model, 64B treatment strongly suppressed eye tumor formation with a ~60% decrease in tumor size 7 days post-inoculation (Fig. 5B). At day 28, mice were terminated and the formation of liver micro-metastases quantified in liver sections. Mice treated with 64B showed a ~50% reduction in metastatic foci (Fig. 5C). To further examine whether 64B could prolong mice survival, we conducted a separate experiment where after enucleation on day 7 mice were kept till they reached IACUC criteria for termination. 64B significantly extended mice lifespan by ~30% (Fig. 5D). Moreover, treatment of 64B (60mg/kg/day) in mice accompanied by slightly body weight gain (Fig. 5E). Similarly, in the 92.1 model we observed ~90% decrease of eye tumor size 7 days post-inoculation (Fig. 5F). 64B significantly extended mice lifespan (Fig. 5G). Altogether, these findings demonstrate that 64B reduces primary and metastatic growth of melanoma in the eye and prolongs mice life span.
Figure 5.
64B inhibits primary eye tumor growth, liver metastasis, and prolongs survival in UM mouse models. A, Schematic showing timeline and procedure for the animal experiments. B, C, D, E, B16LS9 mouse skin melanoma cells (2 × 106/ eye) were injected in the sub-uveal area of C57BL/6 mice eyes where they form melanomas in the uvea. One day after eye inoculation (day 1), mice (8/group) were treated with either vehicle control or 60mg/kg/i.p of 64B 5 days per week. On day 7, tumor-bearing eyes were enucleated, fixed and stained to evaluate tumor burden (B). Representative H&E sections are shown (Left) and quantification of eye tumor (*) size was performed in 4 randomly selected mice/group (Right). Mice were sacrificed on day 28 and number of liver metastases counted (8 mice/group) (C). In a repeat experiment with identical design, mice were kept under treatment after eye enucleation and terminated when reaching IACUC criteria to measure survival by Kaplan-Meier analysis (15 mice/group) (D). E. Weight of mice used in (D). F, G 92.1 human uveal melanoma cells (2 × 106/ mouse) were injected in the suprachoroidal space of 3 ½ month old female nu/nu mice (Envigo Corp; Strain: Hsd: Athymic Nude-Foxn1nu) (15 mice/group). On day 1, mice were treated with either vehicle control or 120mg/kg/i.p of 64B 5 days per week. On day 7, eyes were removed and evaluated for tumor burden. Representative H&E stained eyes are shown (left) and tumor size (*) quantified in 6 mice per group (F). Survival rate of mice (15 mice/group) (G).
64B reduces circulating UM cells, antagonizes HIF-1 induced activation of c-Met and CXCR4 expression, and inhibits the metastasis of uveal melanoma cells to the liver.
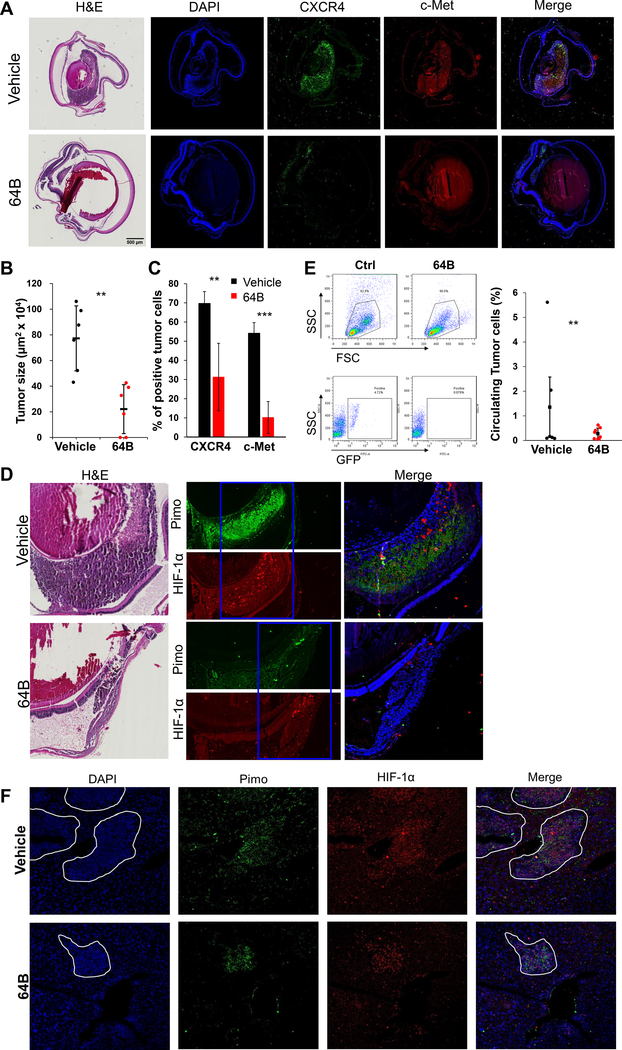
To test the efficacy of 64B in inhibition of circulating tumor cells and liver metastasis formation, we conducted another independent experiment with the 92.1 model (Supplementary Fig. S5A-D). GFP labeled 92.1 cells were used for cell tracking, and mice were injected with pimonidazole, a hypoxia marker before enucleation or sacrifice. Eyes were enucleated on day 9, and sections showed reduced tumor size in 64B treatment group (Fig. 6A and B). Immunofluorescence analysis of eye tumor sections showed CXCR4 and c-Met expression was strongly reduced in 64B treated mice compared to vehicle controls (Fig. 6A and C; Supplementary Fig. S5E). Staining for pimonidazole revealed that vehicle control primary tumors were hypoxic, while control tumors had little evidence for hypoxia, likely due to their smaller size (Fig. 6D). Flow cytometry of blood collected on day 9 showed that mice treated with 64B exhibited reduced amounts of circulating tumor cells (Fig. 6E and F). Mice were sacrificed on day 56, and livers from mice treated with 64B had a reduced metastatic burden and showed the number of micrometastases was also dramatically decreased in the 64B treatment group (Supplementary Fig. S5F). Immunofluorescence staining of Pimo and HIF-1α in liver sections evidenced that micro-metastases were hypoxic in both control and 64B (Fig. 6G). Prolonged systemic treatment with 64B was well tolerated as mice did not lose weight (Supplementary Fig. S5A), and examination of blood cell populations at day 56 did not evidence any alteration (Supplementary Fig. S5D).
Figure 6.
64B reduced UM tumor growth, circulating tumor cells and metastasis and inhibited c-Met and CXCR4 expression in hypoxic/HIF-1α expressing tumors in eye and liver in a uveal melanoma mouse model. 92.1-GFP UM cells (2× 106/ eye) were injected in six-week-old female nu/nu mice [JAX Lab; Strains: Outbred athymic nude; J:NU (007850)] (10 mice/group). On day 1, mice received either vehicle control or 120mg/kg/i.p of 64B 5 days per week. Eyes were enucleated on day 9. A, Representative H&E pictures (left) and immunofluorescence analysis for CXCR4 and c-Met (right) in tumor-bearing eyes. B, Quantification of tumor size on 6 representative mice/group). C, Quantification of CXCR4 and cMet positive cells from (A). D, HIF-1α and Pimo immunofluorescence staining in representative control and 64B-treated tumor-bearing eyes (5 mice/group). E, Representative flow cytometry analysis (left) and quantification (right) of circulating tumor cells (GFP-labeled) in mice blood collected on day 9 (6 mice/group). F, Representative pictures of HIF-1α and Pimo immunofluorescence staining in livers from vehicle and 64B-treated mice (7 mice/group). * p < 0.05; ** p < 0.01; ***, p < 0.001.
Finally, we sought to determine whether the reduced metastatic load in 64B-treated mice was primarily due to a reduction in primary eye tumor burden, or was also related to tumor cell survival in blood circulation and colonization of liver. We directly injected tumor cells (92.1 and Mel290) in the tail vein and counted liver metastasis formation five weeks post-injection. We observed ~10-fold reduction in liver metastases in 64B-treated mice versus vehicle-treated controls (Supplementary Fig. 6A,B). The treatment was well tolerated, with no weight loss (Supplementary Fig. 6C).
Discussion
Developing new systemic treatments for patients with uveal melanomas is a pressing medical need. Ideally, such treatments will reduce primary tumor growth and block metastasis formation and progression, which is the main cause of death. The HIF transcription factor pathway is an attractive cancer target as HIF upregulates a wide range of tumor survival and metastasis related genes (9), including CXCR4 and MET (45, 46). Here, we showed that UM, like other solid cancers exhibits extended areas of hypoxia, and tumor cells in those areas had strong HIF-1α, c-Met and CXCR4 expression. To test whether targeting hypoxia/HIF in UM has promise for therapeutic treatment, we tested small molecule 64B, a novel arylsulfonamide targeting the HIF pathway we previously discovered (27), and found strong anti-tumor effects on primary melanoma growth in the eye and formation of distant liver metastases in two animal models.
Our laboratory is interested in the development of novel therapeutic approaches to target the HIF pathway (18–22, 28, 29). Previously, we described KCN1 (N-((2,2-dimethyl-2H-chromen-6-yl) methyl)-3,4-dimethoxy-N-phenylbenzenesulfonamide), the initial lead compound of a screen for HIF inhibitors using a HIF-responsive reporter assay and a combinatorial library of 10,000 natural product-like compounds (29). Arylsulfonamide 64B is an analog of KCN1, but is about twice as potent in inhibiting HRE-driven reporters (IC50 ~ 0.3μM), is more soluble, and is well tolerated. 64B has desirable pharmaceutical properties: it is small (MW: 444.54), chemically stable, and sufficiently lipophilic to ensure good cell membrane penetration and is compatible for oral delivery. These data suggest that 64B is well suited for the treatment of ocular melanoma and its liver metastases.
Our findings evidence that 64B potently inhibits UM tumor growth, invasion, extravasation into the circulation and metastasis, important hallmarks of cancer. To investigate the molecular determinants of the pleiotropic anti-tumor activities of 64B in UM melanoma, we further studied its mechanism of action. We found that 64B disrupts the hypoxia inducible factor (HIF) transcription complex by interfering with HIF-1α recruitment of p300/CBP co-factors on hypoxia-responsive elements on target genes, which is essential for their activation under hypoxia. Using co-immunoprecipitation experiments we evidenced that 64B blocks HIF-1α association with co-factors p300 and CBP at low micromolar concentrations. This was not mediated by indirect activation of FIH, an asparagyl hydroxylase that modifies the C-terminus of HIF-1α and prevents p300/CBP binding (25, 26). Cellular thermal shift assays showed that 64B thermo-stabilizes p300, supporting direct 64B binding to p300, thereby preventing HIF-1 binding. We found reduced binding of p300 to the MET and CXCR4 gene promoters, explaining 64B inhibition of hypoxia-induced CXCR4 and MET transcription and c-Met and CXCR4-dependent UM cell migration.
In contrast, 64B had no effect on p300 recruitment to the DAPK1 gene, which depends on the C/EBPβ transcription factor for activation and is not hypoxia-HIF activated. The p300/CBP proteins remodel chromatin structure through their histone acetyltransferase activity and serve as co-activators by recruiting the RNA polymerase II transcription complex to gene promoters. They are large multi-domain proteins and each domain mediates binding to specific transcription factors. The CH1 domain interacts with HIF-1 (43), while the CH3 domain mediates C/EBPβ binding (44). Further genome-wide studies are necessary to confirm that 64B displays specificity in blocking only CH1-dependent p300/CBP transcription factor interactions, and whether there might be even further sub-domain specificity.
The importance of c-Met and CXCR4 has long been recognized in multiple cancers, both are important regulators of cell survival and metastasis (13, 47). CXCL12 (SDF-1) binding to CXCR4 elicits signaling that facilitates cancer cell survival, proliferation, chemotaxis, invasion and adhesion. Blockade of CXCR4 signaling inhibits chemotaxis in skin melanomas, a pre-requisite for metastasis (48). c-Met is a receptor tyrosine kinase that, after binding with its ligand, hepatocyte growth factor (HGF), activates a wide range of different cellular signaling pathways, including those involved in proliferation, motility, migration and invasion (47). CXCR4 and c-Met promote uveal melanoma cell proliferation, migration, invasion and metastasis (14, 47, 49–51). Here we uncovered that the oncogenic signaling from both tyrosine kinase receptors can be simultaneously inhibited through upstream targeting of the HIF-1 pathway, which is activated in hypoxic UM tissue and upregulates their expression.
In summary, our findings demonstrate that arylsulfonamide 64B has strong anti-cancer activity in syngeneic and xenogeneic orthotopic UM mouse models. 64B is a potent inhibitor of the hypoxia-induced CXCR4 and c-Met-mediated signaling axis that controls in vitro tumor cell motility. 64B has potent anti-tumor activity at the primary melanoma growth site in the eye, reduces circulating tumor cells, and strongly inhibits spontaneous formation and expansion of metastases in the liver with minimal toxicity. These pre-clinical data provide the rationale for further development of the 64B chemical scaffold towards a clinical candidate for inhibiting primary tumor growth or as a metastasis prevention therapy in uveal melanoma patients at high risk for metastasis.
Supplementary Material
Translational relevance.
There is a pressing need for new therapies to treat ocular melanoma and prevent its spread and growth to distant organs such as liver. Here we tested the anti-tumor efficacy of a novel small chemical molecule targeting the HIF survival pathway in mouse models of uveal melanoma and showed reduction of primary tumor growth and metastasis formation in the liver. Our translational study supports the further translation of this chemical scaffold towards a novel treatment option for patients suffering from ocular melanoma in the clinic.
Acknowledgements
We thank Zhengjia Chen (Department of Biostatistics & Bioinformatics, Emory Rollins School of Public Health) for providing statistical consultation. We appreciate the helpful advice and assistance of all members of the Laboratory of Molecular Neuro-Oncology. We thank the NCI-60 Development Therapeutics Program for performing the acute cytotoxicity study. This work was supported in part by the NIH (grants R01 CA116804, R01 CA176001, R01 CA180805, R24 EY017045, P30 EY06360 and P30 CA138292), a Fight For Sight Postdoctoral Award, the Emory Melanoma Prevention and Research Discovery Fund, a Winship Cancer Institute pilot grant, a Central South University Lieying pilot grant, the V Foundation, the Max Cure Foundation, the Samuel Waxman Cancer Research Foundation, the Alan B. Slifka Foundation, an unrestricted grant from Research to Prevent Blindness, Inc., and a grant from the National Natural Science Foundation of China (#81502544 to S.Y.).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby-marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
Erwin G. Van Meir and Binghe Wang have ownership interest as co-inventors on patents jointly owned by Emory University and Georgia State University. The other authors disclosed no potential conflicts of interest.
References
- 1.Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD. 2017. Uveal melanoma: epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol 11:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luke JJ, Triozzi PL, McKenna KC, Van Meir EG, Gershenwald JE, Bastian BC, Gutkind JS, Bowcock AM, Streicher HZ, Patel PM, Sato T, Sossman JA, Sznol M, Welch J, Thurin M, Selig S, Flaherty KT, Carvajal RD. 2015. Biology of advanced uveal melanoma and next steps for clinical therapeutics. Pigment Cell Melanoma Res 28:135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolandjian NA, Wei C, Patel SP, Richard JL, Dett T, Papadopoulos NE, Bedikian AY. 2013. Delayed systemic recurrence of uveal melanoma. Am J Clin Oncol 36:443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, Lichtenberg TM, Kucherlapati M, Kimes PK, Tang M, Penson A, Babur O, Akbani R, Bristow CA, Hoadley KA, Iype L, Chang MT, Network TR, Cherniack AD, Benz C, Mills GB, Verhaak RGW, Griewank KG, Felau I, Zenklusen JC, Gershenwald JE, Schoenfield L, Lazar AJ, Abdel-Rahman MH, Roman-Roman S, Stern MH, Cebulla CM, Williams MD, Jager MJ, Coupland SE, Esmaeli B, Kandoth C, Woodman SE. 2017. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 32:204–220 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onken MD, Worley LA, Davila RM, Char DH, Harbour JW. 2006. Prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. J Mol Diagn 8:567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giaccia A, Siim BG, Johnson RS. 2003. HIF-1 as a target for drug development. Nat Rev Drug Discov 2:803–11. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol 15:551–78. [DOI] [PubMed] [Google Scholar]
- 8.Burroughs SK, Kaluz S, Wang D, Wang K, Van Meir EG, Wang B. 2013. Hypoxia inducible factor pathway inhibitors as anticancer therapeutics. Future Med Chem 5:553–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankin EB, Giaccia AJ. 2016. Hypoxic control of metastasis. Science 352:175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asnaghi L, Lin MH, Lim KS, Lim KJ, Tripathy A, Wendeborn M, Merbs SL, Handa JT, Sodhi A, Bar EE, Eberhart CG. 2014. Hypoxia promotes uveal melanoma invasion through enhanced Notch and MAPK activation. PLoS One 9:e105372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.el Filali M, Missotten GS, Maat W, Ly LV, Luyten GP, van der Velden PA, Jager MJ. 2010. Regulation of VEGF-A in uveal melanoma. Invest Ophthalmol Vis Sci 51:2329–37. [DOI] [PubMed] [Google Scholar]
- 12.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. 2012. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 12:89–103. [DOI] [PubMed] [Google Scholar]
- 13.Balkwill F 2004. Cancer and the chemokine network. Nat Rev Cancer 4:540–50. [DOI] [PubMed] [Google Scholar]
- 14.Gardner FP, Serie DJ, Salomao DR, Wu KJ, Markovic SN, Pulido JS, Joseph RW. 2014. c-MET expression in primary and liver metastases in uveal melanoma. Melanoma Res 24:617–20. [DOI] [PubMed] [Google Scholar]
- 15.Kollmar O, Rupertus K, Scheuer C, Junker B, Tilton B, Schilling MK, Menger MD. 2007. Stromal cell-derived factor-1 promotes cell migration and tumor growth of colorectal metastasis. Neoplasia 9:862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liepelt A, Tacke F. 2016. Stromal cell-derived factor-1 (SDF-1) as a target in liver diseases. Am J Physiol Gastrointest Liver Physiol 311:G203–9. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Yang W, Chen PW, Alizadeh H, Niederkorn JY. 2009. Inhibition of chemokine receptor expression on uveal melanomas by CXCR4 siRNA and its effect on uveal melanoma liver metastases. Invest Ophthalmol Vis Sci 50:5522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang SH, Cho HT, Devi S, Zhang Z, Escuin D, Liang Z, Mao H, Brat DJ, Olson JJ, Simons JW, Lavallee TM, Giannakakou P, Van Meir EG, Shim H. 2006. Antitumor effect of 2-methoxyestradiol in a rat orthotopic brain tumor model. Cancer Res 66:11991–7. [DOI] [PubMed] [Google Scholar]
- 19.Post DE, Devi NS, Li Z, Brat DJ, Kaur B, Nicholson A, Olson JJ, Zhang Z, Van Meir EG. 2004. Cancer therapy with a replicating oncolytic adenovirus targeting the hypoxic microenvironment of tumors. Clin Cancer Res 10:8603–12. [DOI] [PubMed] [Google Scholar]
- 20.Post DE, Van Meir EG. 2001. Generation of bidirectional hypoxia/HIF-responsive expression vectors to target gene expression to hypoxic cells. Gene Ther 8:1801–7. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, Cao Z, Li F, Post DE, Van Meir EG, Zhong H, Wood WC. 2004. Tumor-specific gene expression using the survivin promoter is further increased by hypoxia. Gene Ther 11:1215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Peng G, Hicks JM, Weiss HL, Van Meir EG, Brenner MK, Yotnda P. 2008. Engineering human tumor-specific cytotoxic T cells to function in a hypoxic environment. Mol Ther 16:599–606. [DOI] [PubMed] [Google Scholar]
- 23.Tan C, de Noronha RG, Devi NS, Jabbar AA, Kaluz S, Liu Y, Mooring SR, Nicolaou KC, Wang B, Van Meir EG. 2011. Sulfonamides as a new scaffold for hypoxia inducible factor pathway inhibitors. Bioorg Med Chem Lett 21:5528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Q, Yin S, Kaluz S, Ni N, Devi NS, Mun J, Wang D, Damera K, Chen W, Burroughs S, Mooring SR, Goodman MM, Van Meir EG, Wang B, Snyder JP. 2012. Binding Model for the Interaction of Anticancer Arylsulfonamides with the p300 Transcription Cofactor. ACS Med Chem Lett 3:620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson J, De Los Santos Z, Devi N, Van Meir E, Zingales S, Wang B. 2017. Examining the structure-activity relationship of benzopyran-based inhibitors of the hypoxia inducible factor-1 pathway. Bioorg Med Chem Lett 27:1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson JH, De Los Santos Z, Devi SN, Kaluz S, Van Meir EG, Zingales SK, Wang B. 2017. Design and synthesis of benzopyran-based inhibitors of the hypoxia-inducible factor-1 pathway with improved water solubility. J Enzyme Inhib Med Chem 32:992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mooring SR, Jin H, Devi NS, Jabbar AA, Kaluz S, Liu Y, Van Meir EG, Wang B. 2011. Design and synthesis of novel small-molecule inhibitors of the hypoxia inducible factor pathway. J Med Chem 54:8471–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narita T, Yin S, Gelin CF, Moreno CS, Yepes M, Nicolaou KC, Van Meir EG. 2009. Identification of a novel small molecule HIF-1alpha translation inhibitor. Clin Cancer Res 15:6128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin S, Kaluz S, Devi NS, Jabbar AA, de Noronha RG, Mun J, Zhang Z, Boreddy PR, Wang W, Wang Z, Abbruscato T, Chen Z, Olson JJ, Zhang R, Goodman MM, Nicolaou KC, Van Meir EG. 2012. Arylsulfonamide KCN1 inhibits in vivo glioma growth and interferes with HIF signaling by disrupting HIF-1alpha interaction with cofactors p300/CBP. Clin Cancer Res 18:6623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Ao L, Rayburn ER, Xu H, Zhang X, Zhang X, Nag SA, Wu X, Wang MH, Wang H, Van Meir EG, Zhang R. 2012. KCN1, a novel synthetic sulfonamide anticancer agent: in vitro and in vivo anti-pancreatic cancer activities and preclinical pharmacology. PLoS One 7:e44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Waard-Siebinga I, Blom DJ, Griffioen M, Schrier PI, Hoogendoorn E, Beverstock G, Danen EH, Jager MJ. 1995. Establishment and characterization of an uveal-melanoma cell line. Int J Cancer 62:155–61. [DOI] [PubMed] [Google Scholar]
- 32.Verbik DJ, Murray TG, Tran JM, Ksander BR. 1997. Melanomas that develop within the eye inhibit lymphocyte proliferation. Int J Cancer 73:470–8. [DOI] [PubMed] [Google Scholar]
- 33.Elia G, Ren Y, Lorenzoni P, Zarnegar R, Burger MM, Rusciano D. 2001. Mechanisms regulating c-met overexpression in liver-metastatic B16-LS9 melanoma cells. J Cell Biochem 81:477–87. [DOI] [PubMed] [Google Scholar]
- 34.Vichai V, Kirtikara K. 2006. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 1:1112–6. [DOI] [PubMed] [Google Scholar]
- 35.Van Meir EG. 1997. Identification of nude mice in tumorigenicity assays. Int J Cancer 71:310. [DOI] [PubMed] [Google Scholar]
- 36.Dithmar S, Rusciano D, Grossniklaus HE. 2000. A new technique for implantation of tissue culture melanoma cells in a murine model of metastatic ocular melanoma. Melanoma Res 10:2–8. [PubMed] [Google Scholar]
- 37.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, Martinez Molina D. 2014. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9:2100–22. [DOI] [PubMed] [Google Scholar]
- 39.Gangemi R, Mirisola V, Barisione G, Fabbi M, Brizzolara A, Lanza F, Mosci C, Salvi S, Gualco M, Truini M, Angelini G, Boccardo S, Cilli M, Airoldi I, Queirolo P, Jager MJ, Daga A, Pfeffer U, Ferrini S. 2012. Mda-9/syntenin is expressed in uveal melanoma and correlates with metastatic progression. PLoS One 7:e29989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurent C, Valet F, Planque N, Silveri L, Maacha S, Anezo O, Hupe P, Plancher C, Reyes C, Albaud B, Rapinat A, Gentien D, Couturier J, Sastre-Garau X, Desjardins L, Thiery JP, Roman-Roman S, Asselain B, Barillot E, Piperno-Neumann S, Saule S. 2011. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res 71:666–74. [DOI] [PubMed] [Google Scholar]
- 41.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, Haddad GG, Poellinger L, Peet DJ, Powell F, Johnson RS. 2010. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab 11:364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. 2002. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A 99:5367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. 2008. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol Cell Biol 28:2528–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. 2003. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 3:347–61. [DOI] [PubMed] [Google Scholar]
- 46.Schioppa T, Uranchimeg B, Saccani A, Biswas SK, Doni A, Rapisarda A, Bernasconi S, Saccani S, Nebuloni M, Vago L, Mantovani A, Melillo G, Sica A. 2003. Regulation of the chemokine receptor CXCR4 by hypoxia. J Exp Med 198:1391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otsuka T, Takayama H, Sharp R, Celli G, LaRochelle WJ, Bottaro DP, Ellmore N, Vieira W, Owens JW, Anver M, Merlino G. 1998. c-Met autocrine activation induces development of malignant melanoma and acquisition of the metastatic phenotype. Cancer Res 58:5157–67. [PubMed] [Google Scholar]
- 48.O’Boyle G, Swidenbank I, Marshall H, Barker CE, Armstrong J, White SA, Fricker SP, Plummer R, Wright M, Lovat PE. 2013. Inhibition of CXCR4-CXCL12 chemotaxis in melanoma by AMD11070. Br J Cancer 108:1634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bi J, Li P, Li C, He J, Wang Y, Zhang H, Fan X, Jia R, Ge S. 2016. The SDF-1/CXCR4 chemokine axis in uveal melanoma cell proliferation and migration. Tumour Biol 37:4175–82. [DOI] [PubMed] [Google Scholar]
- 50.Chattopadhyay C, Grimm EA, Woodman SE. 2014. Simultaneous inhibition of the HGF/MET and Erk1/2 pathways affect uveal melanoma cell growth and migration. PLoS One 9:e83957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Bosch T, Koopmans AE, Vaarwater J, van den Berg M, de Klein A, Verdijk RM. 2013. Chemokine receptor CCR7 expression predicts poor outcome in uveal melanoma and relates to liver metastasis whereas expression of CXCR4 is not of clinical relevance. Invest Ophthalmol Vis Sci 54:7354–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.