Abstract
Aberrant activation of the PI3K-mTOR signaling pathway occurs in >80% of head and neck squamous cell carcinomas (HNSCC), and overreliance on this signaling circuit may in turn represent a cancer-specific vulnerability that can be exploited therapeutically. mTOR inhibitors (mTORi) promote tumor regression in genetically defined and chemically induced HNSCC animal models, and encouraging results have been recently reported. However, the mTOR-regulated targets contributing to the clinical response have not yet been identified. Here, we focused on EIF4E-BP1 (4E-BP1), a direct target of mTOR that serves as key effector for protein synthesis. A systematic analysis of genomic alterations in the PIK3CA-mTOR pathway in HNSCC revealed that 4E-BP1 is rarely mutated, but at least one 4E-BP1 gene copy is lost in over 35% of the HNSCC patients, correlating with decreased 4E-BP1 protein expression. 4E-BP1 gene copy number loss correlated with poor disease-free and overall survival. Aligned with a tumor suppressive role, 4e-bp1/2-knock out mice formed larger and more lesions in models of HNSCC carcinogenesis. mTORi treatment or conditional expression of a mutant 4E-BP1 that cannot be phosphorylated by mTOR was sufficient to disrupt the translation initiation complex and prevent tumor growth. Furthermore, CRISPR/Cas9 targeted 4E-BP1 HNSCC cells resulted in reduced sensitivity to mTORi in vitro and in vivo. Overall, these findings indicate that in HNSCC, mTOR persistently restrains 4E-BP1 via phosphorylation and that mTORi can restore the tumor-suppressive function of 4E-BP1. Our findings also support 4E-BP1 expression and phosphorylation status as a mechanistic biomarker of mTORi sensitivity in HNSCC patients.
Keywords: mTOR, signal transduction, 4E-BP1, INK128, HNSCC
Introduction:
Each year approximate 600,000 new cases of head and neck squamous cell carcinoma (HNSCC), including cancers the oral cavity, oropharynx, larynx, and hypopharynx are diagnosed worldwide, resulting in 300,000 deaths, 13,000 of which occur in the United States alone (1). The main risk factors for HNSCC include tobacco and alcohol use and high risk human papillomavirus (HPV) infection (2). The incidence of HNSCC is rising with the increasing incidence of HPV+ oropharyngeal cancer (3). HNSCC has a poor five-year survival rate, approximately 63% (1), which emphasizes the urgent need to develop new effective options to prevent and treat this malignancy.
Early studies by our team revealed that aberrant activation of the PI3K-mTOR signaling network is one of the most frequent molecular alterations in HNSCC (4–6). Indeed, this study of a large collection of HNSCC tissues and multiple HNSCC mouse models have provided evidence that PI3K-mTOR activation is an early and necessary step for HNSCC development. The underlying molecular mechanisms resulting in pathway over activity have been recently elucidated by Nex-Gen Sequencing approaches as part of the Cancer Genome Atlas (TCGA) Network (7–10). This large deep sequencing initiative has revealed numerous mutations, copy number variations, and altered DNA methylation profiles in individual HNSCC lesions, thus providing an in-depth genomic characterization of HNSCC (7–10). Of interest, the PI3K-mTOR circuitry is among the most commonly altered signaling mechanisms. In particular, multiple genetic and epigenetic changes, including frequent PIK3CA mutations and gene copy number gain, and PTEN gene copy number loss and mutations, converge to sustain persistent aberrant PI3K-mTOR pathway activation in HNSCC (reviewed in (10,11). In turn, the overreliance on this pathway for HNSCC progression and metastasis may represent a vulnerability that can be exploited therapeutically for HNSCC treatment.
In this regard, mTOR inhibition is quite effective in promoting the regression of tumor lesions in multiple HNSCC xenografts, as well as in chemically-induced and genetically-defined HNSCC mouse models (4,6,12,13). These findings provided the rationale for launching a Phase IIb clinical trial targeting mTOR with its allosteric inhibitor, rapamycin, in HNSCC patients in the neoadjuvant setting(14). This trial (NCT01195922), which was recently completed, achieved objective clinical responses (≥30% tumor volume reduction, including a complete pathological response) in 25% of the patients, in spite of a short duration of the trial (3 weeks)(14). However, given the extraordinary complexity of the mTOR network, we still do not know which of the mTOR-regulated targets contributes to the clinical response. This prevents identifying genetic alterations that can have predictive value regarding the sensitivity or resistance to mTOR inhibitors in spite of encouraging clinical results in unselected HNSCC patients.
While conducting an in depth PI3K-mTOR-pathway specific analysis of genetic alterations in HNSCC, we found that a high percentage of lesions exhibit loss of at least one copy of EIF4E-BP1. This gene encodes a translational repressor, 4E-BP1, which blocks the translation of a subset of growth promoting genes (reviewed in (15,16). Specifically, cap-dependent translation initiation is activated by binding of mRNA to the eukaryotic initiation factor complex, eIF4F, which is comprised of several subunit proteins: eIF4A, eIF4E, and eIF4G (15,16). eIF4E physically binds to the m7G cap structure at the 5’ end of the mRNA, and eIF4G functions as a scaffold by interacting with eIF4E, eIF4A and eIF3 (15,16). eIF4E-eIF4G association is essential for cap-dependent translation initiation (15,16). 4E-BP1 (eIF4E binding protein 1) and its related 4E-BP2 and 4E-BP3 proteins, displace eIF4G by binding to eIF4E, thereby preventing translation initiation (15,16). This interaction of 4E-BPs with eIF4E is disrupted upon phosphorylation by mTOR in its mTORC1 complex (17,18). This finding, and the emerging role of protein translation control in cancer growth (15,16,19), prompted us to explore whether 4E-BP1 contributes to HNSCC progression and its therapeutic response to mTOR inhibition. We provide evidence that EIF4E-BP1 acts as tumor suppressor gene in HNSCC, and that the therapeutic response to mTOR blockade is dependent, at least in part, on the ability to reactivate 4E-BP1 translation repressive function. We also provide evidence that 4E-BP1 protein levels and status of phosphorylation may represent mechanistic biomarkers predicting sensitivity to mTORi in HNSCC.
Materials and Methods
Cell lines and tissue culture
Human head and neck cancer cell lines Cal33 and HN12 were developed as part of the NIDCR Oral and Pharyngeal Cancer Branch cell collection and have been described previously (4,20). All cell lines underwent DNA authentication by multiplex STR profiling (Genetica DNA Laboratories, Inc. Burlington, NC) prior to the described experiments to ensure consistency in cell identity. No presence of mycoplasma were found according to Mycoplasma Detection Kit-QuickTest from Biomake (Houston, TX, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum supplemented with antibiotic/ antimycotic solution at 37 ºC in the presence of 5% CO2.
DNA constructs and lentivirus
Cal33 and HN12 cells stably expressing the reverse tetracycline-controlled transactivator fused to VP16 (rtTA) were generated by infection with pLESIP rtTA lentivirus. A phosphorylation-defective mutant of 4E-BP1 (T37A, T46A, S65A, and T70A, 4E-BP1 M) was engineered using appropriate oligonucleotides and the QuikChange II method, and cloned into a tetracycline-inducible lentiviral vector tagged with GFP (pLTI-GFP-4E-BP1 mut)(21,22). An empty GFP vector was used as a control. After lentivirus infection, 4E-BP1 mut (fused to GFP) was expressed in cells by adding doxycycline to the medium, and GFP+ cells were sorted by FACS.
CRISPR/CAS9
Lenti-CRISPR-v2 plasmid was purchased from Addgene (Cambridge, MA, USA). A single guide RNA (sgRNA) to facilitate genome editing was designed according to ZhangLab protocol (23). The sgRNAs of 4E-BP1 are as following, FWD: 5’CACCGCACCACCCGGCGAGTGGCG3’; REV: 5’AAACCGCCACTCGCCGGG-TGGTGC3’.
Immunoblot analysis
Cells were lysed in lysis buffer supplemented with protease phosphatase inhibitors, and Western blot assays were performed as described (24). See Supplemental Materials for additional details.
7-methyl GTP pull down and immunoprecipitation (IP) assay
Cells lysates were incubated with γ-Aminophenyl-m7GTP (C10-spacer)-Agarose (catalog number AC-155L) from Jena Bioscience (Jena, Germany) or incubated with Protein A Agarose (catalog number 16–125) from EMD Millipore (Billerica, MA, USA), conjugated with elF4G antibody. Beads were washed three times with lysis buffer. Proteins were released with SDS–polyacrylamide gel electrophoresis loading buffer and analyzed by western blot analysis using the antibodies listed above.
In vivo mouse experiments and analysis
All the animal studies using HNSCC tumor xenografts and oral carcinogenesis studies in wild type and 4e-bp1/2 double KO mice were approved by the Institutional Animal Care and Use Committee (IACUC) of University of California, San Diego, with protocol ASP # S15195. See Supplemental Materials for additional details.
Immunohistochemistry.
Immunohistochemical analysis of pS6, 4E-BP1, p4E-BP1, Cleaved Caspase-3, and Ki67 were performed following our previously reported procedures (24). Staining for 4E-BP1 and p4E-BP1 in human HNSCC was performed on tissue arrays (US Biomax OR601a). See Supplemental Materials for additional details.
RNA isolation from RNA-binding proteins (RBPs), polysome analysis, and qPCR.
To isolate RNA from ribonucleoprotein elF4G, we used protocols as described previously (25). To analyze polysome profiling, we used protocols as described previously (26,27). For qPCR, see Supplemental Materials for additional details and Supplemental Table 1 for the list of oligonucleotides.
The Cancer Genome Atlas analysis
Data regarding the copy number of 4E-BP1, analysis of disease-free or overall survival of a HNSCC cohort, and analysis DNA methylation-mediated gene silencing were all extracted from the cBio Portal for Cancer Genomics (http://www.cbioportal.org/public-portal/). See Supplemental Materials for additional details.
Statistical Analysis
Data analyses, variation estimation and validation of test assumptions were performed with GraphPad Prism version 7 for Windows (GraphPad Software, San Diego, CA). See Supplemental Materials for additional details. The asterisks of figures denote statistical significance (non-significant or ns, P>0.05; *P<0.05; **P<0.01; and ***P<0.001). All the data are reported as mean ± standard error of the mean (S.E.M.).
Results:
Loss of 4E-BP1 expression in HNSCC: Poor patient prognosis and enhanced carcinogenesis in 4e-bp1/2 knockout mice.
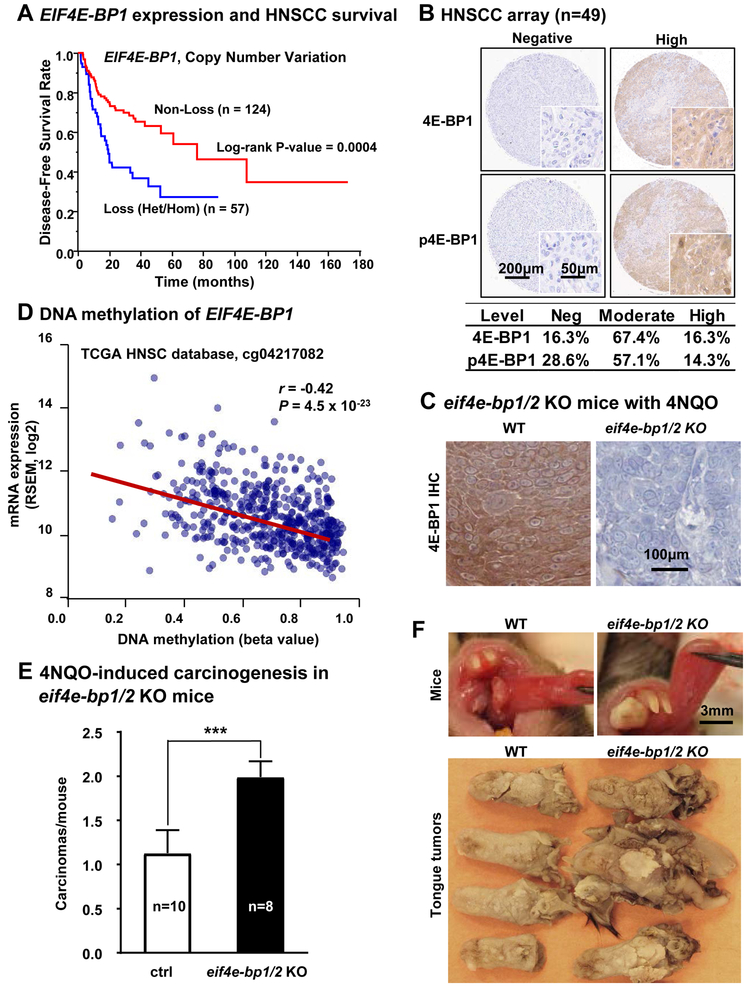
While analyzing genetic alterations in the PIK3CA-mTOR pathway in HNSCC, we observed that although the 4E-BP1 gene (EIF4E-BP1) is rarely mutated, at least one gene copy is lost in over 35% of HNSCC patients (34.9% heterozygous and 1.4% homozygous, respectively) (Figure S1A). To begin exploring the importance of 4E-BP1 loss in the progression of HNSCC, we initially analyzed the correlation between genomic alterations and disease-free (Figure 1A, p = 0.0004, n=181) and overall (Figure S1B, n=511, p=0.0061) survival of HNSCC patients from The Cancer Genome Atlas (TCGA). In both analyses, 4E-BP1 gene loss was a strong predictor of poor prognosis.
Figure 1: EIF4EBP1 is a candidate HNSCC tumor suppressor gene.
(A) The TCGA (The Cancer Genome Atlas) database was used to determine the relationship between EIF4EBP1 copy number variation (CNV) and disease-free survival (DFS). Data of CNV predicted by GISTIC algorism were available for 181 HNSCC patients in DFS (Log-rank test; p = 0.0004). Non-loss includes normal, copy gain and amplification. Loss includes heterozygous deletion and homozygous deletions. (B) Top, representative cores of HNSCC lesions stained with total 4E-BP1 (4E-BP1) and phospho-4E-BP1 (p4E-BP1, Thr37/46) using a HNSCC tissue microarray. Bottom, the intensity of staining was scored as previously described, (n=49) (6) and divided into negative, moderate and high expressed groups. (C) Representative immunohistochemical analysis of 4E-BP1 in WT and eif4ebp1/2 KO mice, respectively. (D) Significant negative correlation between DNA methylation and gene expression in the first intron of EIF4EBP1, suggestive of DNA methylation-mediated gene silencing (n=566, r = −0.42, p = 4.56 × 10−23). (E) 4NQO-induced carcinogenesis in eif4e-bp1/2 KO mice. Numbers of squamous cell carcinomas at the end of 4NQO-carcinogen treatment (mean ± SEM, n of WT mice = 10, n of 4e-bp1/2 KO mice= 8). (F) Top, representative pictures of live mice tongue in WT and eif4e-bp1/2 KO mice on week 26 of 4NQO treatment. Bottom, representative pictures of tongue lesions in WT and eif4e-bp1/2 KO mice on week 26 (time point when mice were sacrificed) of 4NQO treatment. These tumors in eif4e-bp1/2 KO mice were larger than those in wild type C57Bl/6 mice.
At the protein level, we evaluated the expression of 4E-BP1 and phospho-4E-BP1 (p4E-BP1, 4E-BP1 phosphorylated in Thr37/46) in HNSCC tissue microarray (TMA, n=49) sections by immunohistochemistry (IHC) (Figure 1B). In the TMA, we observed 4E-BP1 expression loss in 16% (n=8) of the HNSCC cases, with 67% (n=33) of the cases displaying moderate expression. As controls, no immune staining for 4E-BP1 was observed in eif4e-bp1/2 knock out (KO) mice (Figure 1C), and in HNSCC cells in which 4E-BP1 was genome edited (see below), but rescued by reexpression of the corresponding gene (Figure S1C). Analysis of the HNSCC TCGA cohort showed that DNA copy number of 4E-BP1 was significantly correlated with both mRNA (n=511, P<0.0001) and protein (n=351, P<0.0001) expression of 4E-BP1 (Figure S1D). While homozygous deletion of 4E-BP1 can occur in 1.4% of HNSCC cases (above), reduced expression could also result from loss of one gene copy and gene or promoter methylation of the remaining allele. Indeed, we observed a strong correlation between gene expression and DNA methylation in the gene body of 4E-BP1 (n=566, P <0.001) (Figure 1D, figure S2A). We also examined the status of phosphorylation of 4E-BP1, p4E-BP1 (Thr37/46) by IHC, and found that 71% (n=35) of the cases in the HNSCC tissue microarray sections were positive (n=49), while all cases that lack detectable 4E-BP1 protein expression were also negative for p4E-BP1, thus serving also as an internal control. As additional controls, p4E-BP1 immunodetection was lost in CRISPR/Cas9 targeted 4E-BP1 HNSCC cells and rescued by re-expression of wild type 4E-BP1 but not of a mutant that cannot be phosphorylated in the corresponding residues (Figure S1C and see below), and its immunodetection was abolished by mTOR inhibition (see below). This suggests that in most HNSCC cases in which 4E-BP1 is expressed, this translational inhibitor is persistently phosphorylated, thus repressing its function (15,16).
To investigate whether 4E-BP1 loss contributes to HNSCC progression, we examined the impact of 4E-BP1/2 (eif4e-bp1/2) gene deletion (28) on cancer development in the oral-specific 4NQO-carcinogenesis model (13). These mice lack 4E-BP1 immune reactivity (Figure 1C). 4E-BP1/2 KO mice do not develop tumors spontaneously (28,29), and as expected, these mice did not exhibit spontaneous HNSCC lesions. However, we found that they are highly susceptible to chemically-induced (4NQO) oral carcinogenesis. These KO mice had nearly double the number of squamous carcinomas in the tongue, and these tumors were larger than those in wild type C57Bl/6 mice (Figure 1E-F and figure S2B). Collectively, these findings and the analysis of the HNSCC oncogenome revealing that a high percentage of lesions exhibit loss of 4E-BP1 function, either by gene copy loss, gene methylation, or persistent phosphorylation, suggest that 4E-BP1 may represent a potential tumor suppressor in HNSCC.
De-phosphorylation of 4E-BP1 disrupts translation initiating complexes, and is sufficient to prevent HNSCC tumor growth.
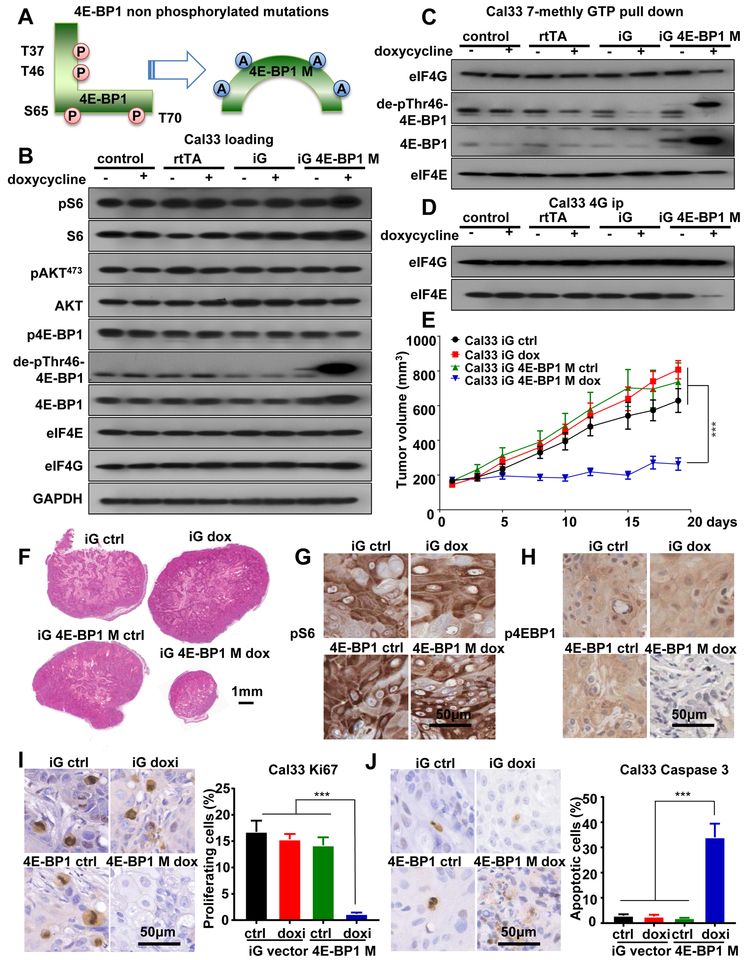
mTOR regulates ribosomal biogenesis and protein synthesis through at least two eukaryotic translation regulators, 4E-BP1 and p70S6K (p70-S6 kinase); the latter phosphorylates ribosomal protein S6 (S6) and several initiation factors, including eIF4B (17,30). To confirm the contribution of 4E-BP1 regulated ribosomal biogenesis and protein synthesis in this process, we took advantage of a 4E-BP1 mutant protein engineered previously (15,16), in which the residues T37, T46, S65 and T70 were all mutated to alanine (termed 4E-BP1 M) (Figure 2A). This mutant cannot be regulated by mTOR due to lack of phosphorylated residues, thus mimicking the accumulation of the hypophosphorylated form of 4E-BP1 (termed de-phospho-Thr46–4E-BP1) induced by mTOR inhibition and its consequent effect on cap-dependent translation (31). We conditionally expressed 4E-BP1 M in representative HNSCC cells, HN12 and Cal33, both of which exhibit elevated mTOR activity, as most SCC cell lines (4,20). These cells harbor wild type and PIK3CA mutant, respectively (20). Cells expressing DNA vector or rtTA (reverse tetracycline-controlled transactivator) alone served as controls. An antibody that detects 4E-BP1 only when dephosphorylated at Thr46 was used to recognize de-phospho-Thr46–4E-BP1. Expression of 4E-BP1 M increased de-phospho-Thr46–4E-BP1 levels, but did not affect mTOR signaling as it did not reduce phospho-S6 (pS6) and phospho-AKT (pAKTS473) levels, the latter a typical target of the mTORC2 complex (32) (Figure 2B). To explore whether expression of 4E-BP1 M in HNSCC can disrupt the protein-protein interactions between eIF4E and eIF4G, both key elements of translation initiation complex, we used m7GTP pulldown and eIF4G immunoprecipitation experiments (scheme in Figure S3A-B). This study revealed that expression of 4E-BP1 M in Cal33 and HN12 HNSCC cells did not change the expression of eIF4E and eIF4G (Figure 2B), but disrupted their association (Figure 2C-D, figure S3C, S4A-D). Remarkably, inducible expression of 4E-BP1 M halted HNSCC tumor growth (Figure 2E-F, Figure S4E-F). Of importance, this mutant did not affect S6 phosphorylation (Figure 2G-H), suggesting that in HNSCC disruption of the 4E-BP1 signaling branch may be sufficient to cause tumor reduction, in spite of a pS6 accumulation. Moreover, IHC for Ki67 and cleaved Caspase 3 showed that, 4E-BP1 M also caused a reduction of cell proliferation and increased cell apoptosis in HNSCC tumors (Figures 2I-J), similar to mTOR inhibition (see below).
Figure 2: Reduced growth and apoptotic effect of HNSCC cells engineered to express a mutant form of 4E-BP1 lacking mTOR phosphorylation sites.
(A) Scheme of 4E-BP1 M. The amino acids T37, T46, S65 and T70 of 4E-BP1 were mutated into alanine (A). This 4E-BP1 M remain non-phosphorylated when expressed. (B). Western blot analysis of signaling events in HNSCC expressing 4E-BP1 M. Wild-type Cal33 cells (control), cells expressing rtTA (rtTA), cells infected with rtTA and inducible GFP fusion empty lentiviral virus (iG), or cells infected with rtTA and inducible GFP fusion 4E-BP1 M lentiviral virus (iG 4E-BP1) were turned on by doxycycline for 2 days, and lysates were analyzed as indicated. (C) 7mGTP pull down and (D) eIF4G co-IP analyzing the regulation of complex formation by 4E-BP1. Cells (same as panel B) were treated as described above, and analyzed as indicated. (E) Cal33 cells expressing empty vector (iG) or 4E-BP1 M (iG 4E-BP1 M) were transplanted into athymic nude mice, and when they reached approximately 200 mm3, mice were fed with either regular food (ctrl) or doxycycline food (dox) to turn on 4E-BP1 M expression. *** P < .001 when comparing the 4E-BP1 M group with empty vector groups or 4E-BP1 control group (regular food) mice (n = 10 per group). (F) Representative histological tissue sections from each treatment group in panel E. Scale bars represent 1 mm. (G and (H) Representative immunohistochemical analysis of pS6 and p4E-BP1 in tumors from panel E. (I) and (J) Representative immunohistochemical analysis (left) and quantification (right) of Ki67 and cleaved-Caspase 3 in tumors from panel E. Data are represented as mean ± SEM, n= 3 in each group.
INK128, an mTOR kinase inhibitor display anti-tumor effect in HNSCC and disrupts the translation initiation eIF4F complex through 4E-BP1
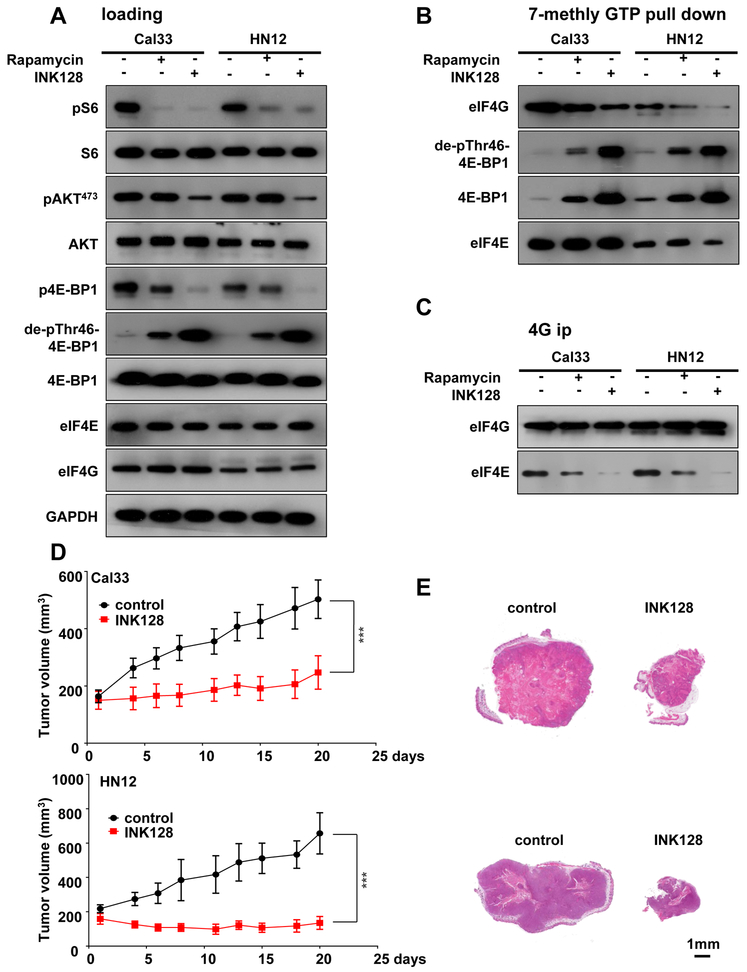
As 4E-BP1 acts downstream of mTOR, we asked whether mTOR inhibition is sufficient to influence its tumor suppressive effect in HNSCC. For these studies, we used rapamycin (sirolimus), a first generation of mTOR allosteric inhibitor (33), and INK128 (also known as MLN-0128 and TAK-228), which is representative of a second generation of small molecule active-site mTOR kinase inhibitors (34,35). As expected, both rapamycin and INK128 decreased pS6 levels. Interestingly, both drugs decreased p4E-BP1 and increased de-phospho-Thr46–4E-BP1 levels, with INK128 exerting a stronger effect. INK128 also decreased pAKTS47 3 levels, a typical target of the mTORC2 complex (32), while rapamycin did not (Figure 3A). Although ERK was reported in some studies to inhibit 4E-BP1 function (36), in our study, inhibition of ERK with trametinib, a MEK1/2 inhibitor, had limited effect on the levels of phosphorylated 4E-BP1, as determined with use of p4E-BP1 and dephospho-Thr46–4E-BP1 antibodies. Rapamycin and LY294002, which inhibit of mTORC1 and PI3K respectively, the latter acting upstream of mTOR, served as additional controls (Figure S5A). Immunoblotting, m7GTP pulldown and eIF4G co-immune precipitation (IP) experiments revealed that mTOR inhibition promotes the association of 4E-BP1, and specifically its dephosphorylated form, with eIF4E, and disrupts the association between eIF4E and eIF4G (Figure 3B-C, S5B). Aligned with the results of 4E-BP1 dephosphorylation (above), INK128 showed higher effect than rapamycin. This provided a strong rationale for the preclinical evaluation of the efficacy of INK128 in HNSCC. Indeed, we observed that INK128 displays potent anti-tumor effects, achieving statistically significant differences in tumor burden as early as three days after treatment initiation (n=10, p<0.001) (Figure 3D-E).
Figure 3: Anti-tumor effect of mTOR kinase inhibition with INK128: Disruption of 4E-BP1 protein complexes.
(A) Western blot analysis of signaling events in HNSCC cells treated by mTOR inhibitors. Cal33 and HN12 cells were treated by Rapamycin (20nM) or INK128 (20nM) for 1 hour, and lysates were analyzed as indicated. (B) 7mGTP pull down and (C) eIF4G co-IP to analyze the regulation of translational initiation complex formation by mTOR inhibition. Cells (similar to panel A) were treated as described above, and analyzed as indicated. (D) Cal33 (top) and HN12 (bottom) were transplanted into athymic nude mice, and, when they reached approximately 200 mm3, mice were treated with vehicle diluent or INK128 for approximately 20 days, as indicated. (E) Representative histological sections from each treatment group in panel D. Scale bars represent 1 mm. (***P < .001 when compared with the control-treated group, n = 10 per group).
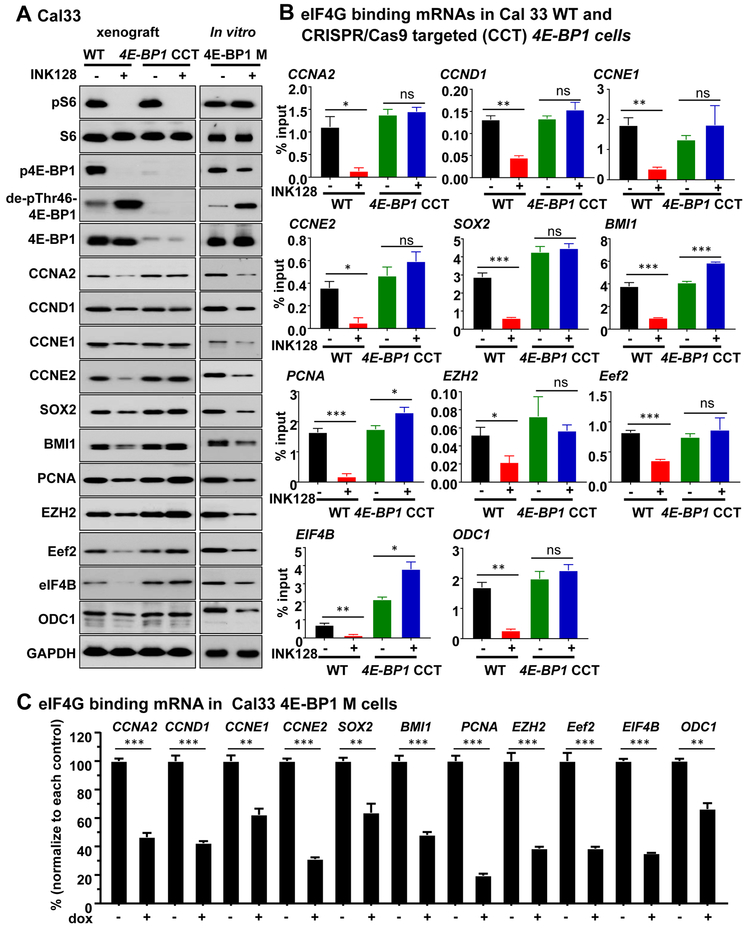
CRISPR/Cas9 targeting of 4E-BP1 in HNSCC reduces the sensitivity to mTOR inhibition, and reveals a role for the mTOR-4E-BP1 axis in the regulation of mRNA translation of proliferative genes
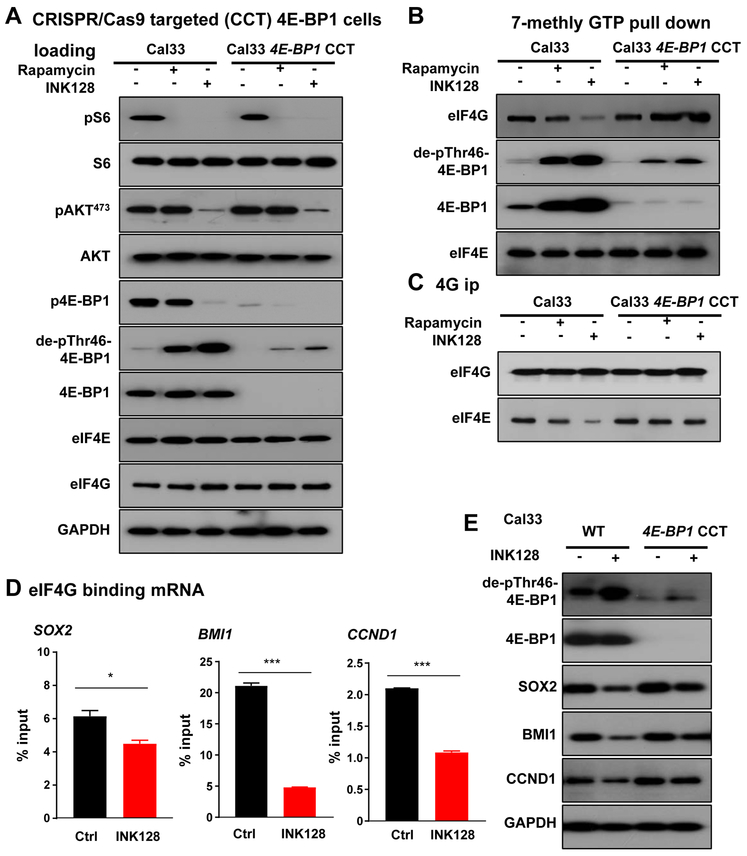
To further explore the role of 4E-BP1 in the antitumor activity of mTOR inhibition, we targeted 4E-BP1 in Cal33 and HN12 HNSCC cell lines using the CRISPR/Cas9 system. Nearly complete 4E-BP1 expression suppression was achieved (Figure 4A). In these CRISPR/Cas9 targeted 4E-BP1 cells (4E-BP1 CCT), mTOR inhibition remained sensitive to reduce pS6 levels. As expected, mTOR inhibition in cells lacking 4E-BP1 did not result in changes in p4E-BP1 and total 4E-BP1 levels, with slight immunoreactivity remaining likely revealing the presence of few cells retaining residual 4E-BP1, or due to cross reactivity with 4E-BP2 (Figure 4A and S4A). In light of these results, disruption of the eIF4E- eIF4G complex by mTOR inhibition was almost completely reversed in 4E-BP1 CCT cells compared to parental cells (Figure 4B-C, S5C and S6A-D). We next investigated the molecular mechanism by which 4E-BP1 may mediate cancer progression in HNSCC. We isolated mRNAs associated with eIF4G, the component of the eIF4F cap binding complex that binds mRNAs (15,16). Initially we used a targeted approach, based on prior findings that SOX2 (37) and cyclin D1 (CCND1) (38) can be regulated by eIF4E/4E-BP1 dependent translation, and observed decreased association of these well-known HNSCC proliferative (20) genes, with eIF4G (Figure 4D-E). This prompted us to also explore whether BMI1, a key HNSCC cancer proliferative and stemness gene (39), is also regulated by 4E-BP1. Indeed, BMI1 mRNA association with eIF4G was significantly reduced by mTORi treatment, which was reflected by reduced expression levels. Of importance, in 4E-BP1 CCT cells, INK128 was less effective compared to its parental cells (Figure 4E), supporting the conclusion that translation of these HNSCC growth promoting genes is dependent on 4E-BP1 expression and phosphorylation status.
Figure 4: CRISPR-CAS9 targeted 4E-BP1 HNSCC cells results in insensitivity to mTORi in vitro, and reveals a role for 4E-BP1 in the regulation of the expression of proliferative molecules.
(A) CRISPR/Cas9 targeted 4E-BP1 cells (4E-BP1 CCT) was achieved. Western blot analysis of signaling events in HNSCC cells treated by mTOR inhibitors. Cal33 and Cal33 4E-BP1 CCT cells were treated by Rapamycin (20nM) or INK128 (20nM) for 1 hour, and lysates were analyzed as indicated. (B) 7mGTP pull down and (C) eIF4G co-IP analyses of the regulation of translation initiation complex formation by mTOR inhibition. Cells (similar to panel A) were treated as described above, and analyzed as indicated. (D) Cal33 cells were treated with INK128 for 2 days, the lysates were subjected to eIF4G co-IP and associated RNAs analyzed (bound RNA). The same treated lysates were used to isolate total RNA. RNAs were followed by qPCR to assess the 4G-binding levels of regulated genes. Data are mean ±SEM % of total input (ns P>.05, *P<.05, **P<.01, ***P < .001 when compared with the control-treated group, n = 3 per group). (E) Cells were treated by INK128 for 2 days, lysates were analyzed as indicated.
To further investigate the impact in translational control caused by mTOR inhibition in HNSCC, polysome-bound mRNAs were isolated by sucrose gradient fractionation after INK128 treatment of Cal33 HNSCC cells, and subjected to RNA sequencing. The complete list of genes whose polysome association was significantly altered by mTOR inhibition is shown in Supplemental Table 2. We compared this gene list with those whose mRNA translation has been previously reported to be regulated by mTORC1 in mouse embryonic fibroblasts (MEFs) and prostate cancer (40,41), and developed an extened list of shared genes of interest based on their relevance to HNSCC. These included CCNA2, CCNE1, CCNE2, BMI1, PCNA, EZH2, Eef2, eIF4B and ODC1, and validated them in 4E-BP1 CCT tumor samples and 4E-BP1 M expressing cells. Firstly, the protein expression of these selected molecules was dependent on 4E-BP1 expression and phosphorylation status in HNSCC (Figure 5A). Moreover, their mRNA association with eIF4G was significantly reduced by INK128 treatment as well as in 4E-BP1 M cells, and the mTORi induced reduction was rescued in 4E-BP1 CCT tumors (Figure 5B-C), thus supporting their translational control by mTOR through 4E-BP1.
Figure 5: Representative 4E-BP1 regulated molecules.
(A) Western blot analysis of signaling and 4E-BP1 regulated molecules in vitro and in vivo in 4E-BP1 control and 4E-BP1 CCT HNSCC xenografts (left), and HNSCC cells expressing the 4E-BP1 M (right). Left, cells were transplanted into athymic nude mice, and when they reached approximately 200 mm3, mice were treated with vehicle diluent or INK128 for 5 days, as indicated. Right, cells infected with rtTA and inducible GFP fusion 4E-BP1 M lentiviral virus (iG 4E-BP1 M) were turned on by doxycycline for 5 days. Lysate were analyzed as indicated. (B) The lysates of 4E-BP1 WT and 4E-BP1 CCT HNSCC xenografts (same as panel A) were subjected to eIF4G co-IP and associated RNAs analyzed (bound RNA). The same treated lysates were used to isolate total RNA. RNAs were followed by qPCR to assess the eIF4G-binding levels of regulated genes. (C) The lysates of 4E-BP1 M (same as panel A) were subjected to eIF4G co-IP and associated RNAs analyzed (bound RNA). The same treated lysates were used to isolate total RNA. RNAs were followed by qPCR to assess the eIF4G-binding levels of regulated genes. For each gene, data were normalized to its control group. Data are mean ±SEM % of total input (ns P>0.05, *P<0.05, **P<0.01, ***P < 0.001 when compared with the control-treated group, n = 3 per group).
mTOR inhibition resistance in vivo in CRISPR/Cas9 targeted 4E-BP1 HNSCC cells
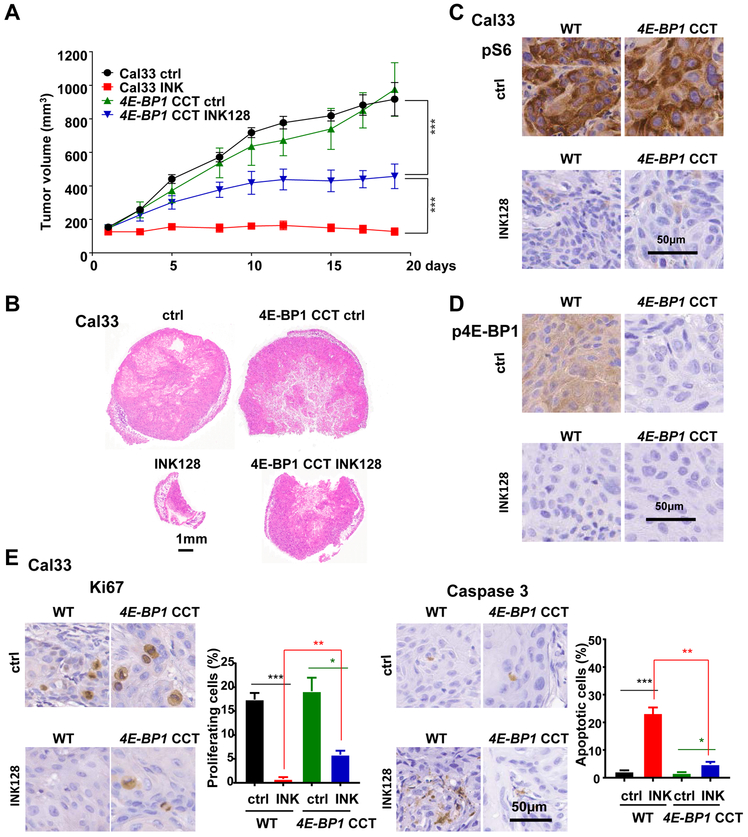
The reduced sensitivity of 4E-BP1 CCT tumors to mTOR inhibition was also recapitulated in a xenograft model. INK128 had reduced anti-tumor activity in 4E-BP1 CCT tumors (Figure 6A-B). Of note, the partial response of 4E-BP1 CCT tumors to INK128 may be due to other effects of mTORC1/2, such as AKT inhibition (Figure 4A). Consistent with our in vitro data (Figure 4A), mTOR inhibition reduced pS6 in 4E-BP1 parental and 4E-BP1 CCT xenografts (Figure 6C-D). While p4E-BP1 was reduced in parental cells, there was no p4E-BP1 immunoreactivity in CCT cells already under basal conditions. Consistent with the reduced anti-tumor effect of INK128 in 4E-BP1 CCT cells, this mTORi reduced Ki67 in CCT cells less effectively than in parental cells (Figure 6E left panels). Moreover, HNSCC treated with INK128 failed to accumulate cleaved Caspase3 in 4E-BP1 CCT cells, in contrast to its pro-apoptotic activity in wild type cells (Figure 6E right panels).
Figure 6: Reduced sensitivity to mTOR inhibition in CRISPR/Cas9 targeted 4E-BP1 HNSCC cells in vivo.
(A) Cal33 control and 4E-BP1 CCT cells were transplanted into athymic nude mice and treated by INK128. (Data are the mean ±SEM of the tumor volume; ***P < .001 when compared with the control-treated group, n = 10 per group). (B) Representative histological sections from each treatment group in panel A. Scale bars represent 1 mm. (C-D) Representative immunohistochemical analysis of pS6 (C) and p4E-BP1(D) in tumors from panel A. (E) Representative immunohistochemical analysis and quantification of Ki67 (left) and cleaved-Caspase 3 to determine the percentage of apoptotic cells (right) in tumors from panel A.
Discussion:
The elucidation of the genomic landscape of most solid tumors, including HNSCC, has provided a unique opportunity to identify new precision therapeutic options to prevent and treat cancer. Most of the cancer driver and tumor suppressive alterations identified to date occur in a discrete number of genes whose protein products are organized in pathways and networks. For example, in HNSCC many distinct alterations, ranging from EGFR and FGFR1 mutations and overexpression, HRAS mutations, PIK3CA mutations and gene amplification, as well as few mutations and copy number variations in AKT1, PTEN, and TSC1/TSC2 converge in the activation of the PI3K-mTOR signaling circuitry (11). This prompted the exploration of the clinical benefit of PI3Ki and mTORi in this malignancy (reviewed in (11). However, we still do not know the molecular events mediating the therapeutic response of these PI3K/mTORi and their mechanisms of sensitivity and resistance, which may prevent the selection of the patients that may benefit the most from these novel anticancer agents. Here we provide evidence that 4E-BP1, a direct mTOR substrate, acts as a HNSCC tumor suppressor, and that while 4E-BP1 dephosphorylation mediates the therapeutic response to mTORi, its gene copy loss or reduced expression render HNSCC lesions resistant to mTOR inhibition.
Indeed, we observed that one third of HNSCC lesions lose at least one allele of 4E-BP1, exhibiting lower progression free and overall survival, and that eif4e-bp1/2 double KO mice are highly susceptible to chemically-induced oral carcinogenesis. We also engineered a mutant 4E-BP1 that cannot be phosphorylated by mTOR resembling de-phospho-Thr46–4E-BP1, and generated HNSCC cells expressing this mutant in a tetracycline-inducible fashion. Expression of 4E-BP1 M was sufficient to disrupt the function of the translation initiation complex, and resulted in tumor regression in HNSCC xenograft models. Certainly, overexpression of this mutant in the conext of a wild type 4E-BP1 may have additional effects to those caused by dephosporylation of endogenous 4E-BP1, a possibility that warrants further investigation. In this regard, as a complementary approach we observed that mTORi could not affect the translation initiation complex in 4E-BP1 CCT HNSCC cells, and that this limited the tumor suppressive activity of mTORi. Taken together, our findings suggest that endogenous 4E-BP1 may exert tumor suppressive activity by restraining the expression of oncogenic translational programs in HNSCC, and that cancer progression requires bypassing the growth inhibitory properties of 4E-BP1 by gene loss and methylation, or via persistent phosphorylation by mTOR. In the latter case, which involves nearly 70% of all HNSCC cases, the latent tumor suppressive activity of 4E-BP1 can in turn be reactivated by mTOR inhibition, thus contributing to the anti-tumor activity of mTORi in this cancer type.
Our findings support the idea that in HNSCC mTORi causes 4E-BP1 dephosphorylation and consequently suppresses cancer by inhibiting translation initiation. Indeed, both the mTORC1 inhibitor rapamycin and mTORC1/2 inhibitor INK128 induced the accumulation of de-phospho-Thr46–4E-BP1. Interestingly, the same dose of INK128 showed relatively higher activity in promoting 4E-BP1 dephosphorylation and disruption of eIF4E-eIF4G association, suggesting a stronger inhibition of translation initiation. The reason for this difference might be that rapamycin and its analogues (rapalogs), which represent the first generation of mTOR inhibitors, block primarily mTOR in its complex 1 (mTORC1) indirectly by binding to FKBP12; while INK128, which represents a second generation of mTOR inhibitors, blocks both mTORC1 and mTORC2 by inhibiting the mTOR kinase directly (34,35), hence displaying a stronger activity (reviewed in (32). Specifically, a catalytic cleft exists within FKBP12-rapamycin-binding (FRB) domain of mTOR, enabling limited access to 4E-BP1 as a substrate, while ATP-competitive inhibitors, such as INK128, can bind deeper inside the catalytic cleft abolishing the ability to phosphorylate 4E-BP1 (42,43).
Inhibition of translation initiation by directly targeting the eIF4E-eIF4G association represents an attractive therapeutic option. 4EGI-1, a small molecular inhibitor that was developed to displace eIF4G from eIF4E has displayed antitumor effects (44). However, the affinity of 4EGI-1 for the eIF4G/eIF4E complex is lower than 4E-BP1 for eIF4E (45), indicating that this approach may require further optimization to achieve its full potential. Certainly, mTOR inhibition may represent a readily available, effective, and clinically relevant choice to disrupt the eIF4G/eIF4E complex by unleashing the latent 4E-BP1 tumor suppressive function in cancers expressing it. Regarding the latter, 4E-BP1 CCT cells showed reduced sensitivity in vitro and in vivo to mTORi, suggesting that 4E-BP1 expression is required for the effectiveness of mTORi. In this case, the assessment of 4E-BP1 expression and status of phosphorylation may help select patients most likely to respond to mTORi. HNSCCs in which 4E-BP1 protein is lost may not respond fully to these agents, and they may instead be suitable candidates for future clinically relevant direct eIF4G/eIF4E complex inhibitors.
Accumulation of de-phospho-Thr46–4E-BP1, either engineered (4E-BP1 M) or when induced by INK128 administration, reduced cell proliferation and caused apoptosis in HNSCC xenografts, aligned with a proposed role for 4E-BP1 in the control of apoptosis in breast cancer, glioma, lymphomas, and other cancers (46–48). Recent high resolution transcriptome-scale ribosome profiling studies revealed the impact of mTORi on mRNA translational efficiency (40,41). Specifically, most direct mTOR target mRNAs possess a pyrimidine-rich translational element (PRTE) and/or a 5’ terminal oligopyrimidine track (TOP) within their 5’ untranslated regions (UTR) (40,41). These include Eef2 and eIF4B, two genes harboring PRTE and TOP 5’ sequences whose mRNA translation and eiF4G association were highly repressed by mTOR blockade in HNSCC, and recued by 4E-BP1 CCT. However, prolonged mTOR blockade can also result in reduced translation of multiple mRNAs that may not harbor these targeting sequences (40,41). Indeed, our ribosomal profiling revealed that many mRNAs are regulated by mTOR in HNSCC, including multiple drivers of HNSCC proliferation and likely cancer cell stemness, all of which were resistant to the reduction of their association to eiF4G and protein expression in 4E-BP1 CCT cells and tumors. These findings raise the possibility that multiple molecules often associated with cancer stem cells (or cancer-initiating cells) and tumor growth may be under mTOR/4E-BP1 translational control. Together, these findings support a key role for the mTOR/4E-BP1 axis in HNSCC growth, survival, and CSC characteristics, which can be exploited therapeutically for HNSCC treatment.
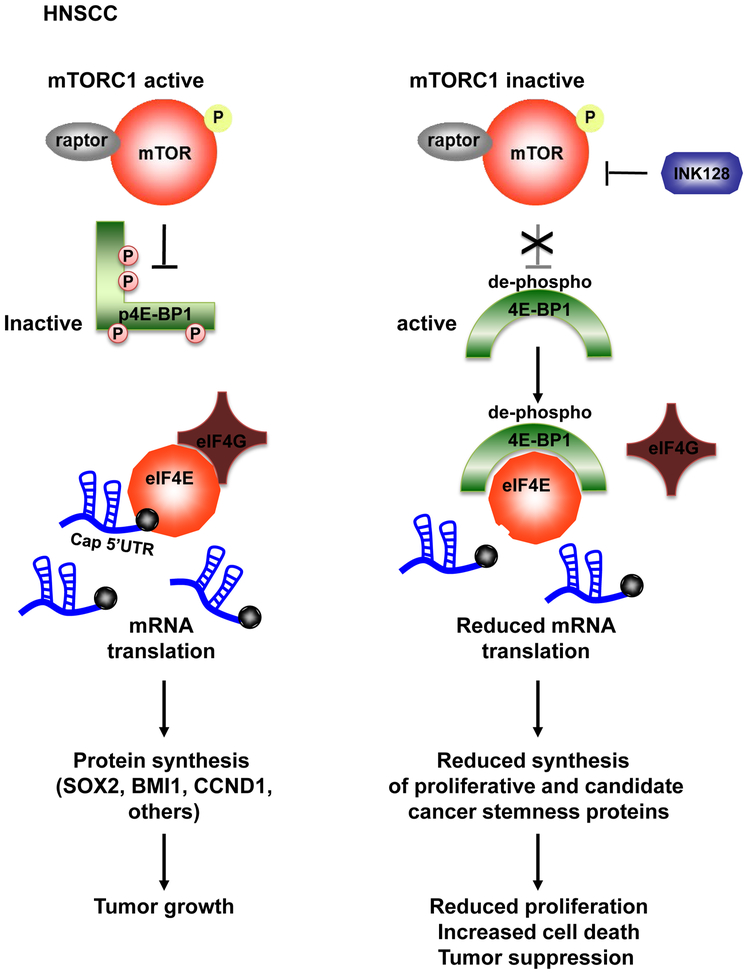
Overall, deep sequencing approaches are now making it possible to identify precision medicine strategies for cancer treatment. In this regard, our findings support a tumor suppressive role of 4E-BP1 in HNSCC, and that its expression and phosphorylation status are associated with prognosis and the clinical response of mTORi in HNSCC. In addition, these findings suggest that direct mTOR kinase blockers will be even more efficacious in the clinic than its allosteric inhibitors, such as rapalogs, as they are more potent in promoting the accumulation of de-phospho-Thr46–4E-BP1, thereby restoring its endogenous growth suppressive and pro-apoptotic function. Taken together, we can conclude that mTORi may unleash 4E-BP1’s tumor suppressive activity, by dephosphorylating 4E-BP1 thus reducing the translation of growth promoting, survival, and candidate cancer stemnes genes, and that in turn 4E-BP1 expression and phosphorylation may serve as a prognostic biomarker for mTORi activity in HNSCC patients (Figure 7)
Figure 7: Schematic representation of the mechanism by which mTOR inhibition acts in HNSCC through 4E-BP1.
See discussion for details.
Supplementary Material
Significance.
Findings suggest that EIF4E-BP1 acts as a tumor suppressor in HNSCC and that 4E-BP1 dephosphorylation mediates the therapeutic response to mTORi, providing a mechanistic biomarker for future precision oncology trials.
Acknowledgements
This project was supported by grants from National Institute of Dental and Craniofacial Research (NIH/NIDCR, 1R01DE026644, 1R01DE026870), National Natural Science Foundation of China (81602376, 81520108009, 81621062), and 111 Project of MOE (B14038), China. We thank Dr. Robert A.J. Signer, Kosuke Yamaguchi and Yusuke Goto for insightful suggestions.
Footnotes
Competing financial interests
On behalf of all other authors, the corresponding author states that there is no conflict of interest
Disclaimers: The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians; 2018;68:7–30 [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9–22 [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011;29:4294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amornphimoltham P, Patel V, Sodhi A, Nikitakis NG, Sauk JJ, Sausville EA, et al. Mammalian target of rapamycin, a molecular target in squamous cell carcinomas of the head and neck. Cancer research 2005;65:9953–61 [DOI] [PubMed] [Google Scholar]
- 5.Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clinical cancer research : an official journal of the American Association for Cancer Research 2007;13:4964–73 [DOI] [PubMed] [Google Scholar]
- 6.Molinolo AA, Marsh C, El Dinali M, Gangane N, Jennison K, Hewitt S, et al. mTOR as a molecular target in HPV-associated oral and cervical squamous carcinomas. Clinical Cancer Research 2012;18:2558–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science 2011;333:1154–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov 2013;3:770–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov 2013;3:761–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iglesias-Bartolome R, Martin D, Gutkind JS. Exploiting the head and neck cancer oncogenome: widespread PI3K-mTOR pathway alterations and novel molecular targets. Cancer Discov 2013;3:722–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun ZJ, Zhang L, Hall B, Bian Y, Gutkind JS, Kulkarni AB. Chemopreventive and chemotherapeutic actions of mTOR inhibitor in genetically defined head and neck squamous cell carcinoma mouse model. Clinical cancer research : an official journal of the American Association for Cancer Research 2012;18:5304–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czerninski R, Amornphimoltham P, Patel V, Molinolo AA, Gutkind JS. Targeting mammalian target of rapamycin by rapamycin prevents tumor progression in an oral-specific chemical carcinogenesis model. Cancer prevention research (Philadelphia, Pa) 2009;2:27–36 [DOI] [PubMed] [Google Scholar]
- 14.Day TA, Shirai K, O’Brien PE, Matheus MG, Godwin K, Sood AJ, et al. Inhibition of mTOR Signaling and Clinical Activity of Rapamycin in Head and Neck Cancer in a Window of Opportunity Trial. Clinical cancer research : an official journal of the American Association for Cancer Research 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009;136:731–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 2014;83:779–812 [DOI] [PubMed] [Google Scholar]
- 17.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & Development 2004;18:1926–45 [DOI] [PubMed] [Google Scholar]
- 18.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010;328:1172–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer 2010;10:254–66 [DOI] [PubMed] [Google Scholar]
- 20.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget 2014;5:8906–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell 2010;18:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llanos S, Garcia-Pedrero JM, Morgado-Palacin L, Rodrigo JP, Serrano M. Stabilization of p21 by mTORC1/4E-BP1 predicts clinical outcome of head and neck cancers. Nat Commun 2016;7:10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Martin D, Molinolo AA, Patel V, Iglesias-Bartolome R, Degese MS, et al. mTOR co-targeting in cetuximab resistance in head and neck cancers harboring PIK3CA and RAS mutations. J Natl Cancer Inst 2014;106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 2006;1:302–7 [DOI] [PubMed] [Google Scholar]
- 26.Galban S, Fan J, Martindale JL, Cheadle C, Hoffman B, Woods MP, et al. von Hippel-Lindau protein-mediated repression of tumor necrosis factor alpha translation revealed through use of cDNA arrays. Molecular and cellular biology 2003;23:2316–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahba A, Rath BH, Bisht K, Camphausen K, Tofilon PJ. Polysome Profiling Links Translational Control to the Radioresponse of Glioblastoma Stem-like Cells. Cancer research 2016;76:3078–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 2007;117:387–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nature medicine 2001;7:1128–32 [DOI] [PubMed] [Google Scholar]
- 30.Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 2004;23:1761–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin D, Nguyen Q, Molinolo A, Gutkind JS. Accumulation of dephosphorylated 4EBP after mTOR inhibition with rapamycin is sufficient to disrupt paracrine transformation by the KSHV vGPCR oncogene. Oncogene 2014;33:2405–12 [DOI] [PubMed] [Google Scholar]
- 32.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell 2007;12:9–22 [DOI] [PubMed] [Google Scholar]
- 34.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest 2011;121:1231–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janes MR, Vu C, Mallya S, Shieh MP, Limon JJ, Li LS, et al. Efficacy of the investigational mTOR kinase inhibitor MLN0128/INK128 in models of B-cell acute lymphoblastic leukemia. Leukemia 2013;27:586–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolli-Derkinderen M, Machavoine F, Baraban JM, Grolleau A, Beretta L, Dy M. ERK and p38 inhibit the expression of 4E-BP1 repressor of translation through induction of Egr-1. J Biol Chem 2003;278:18859–67 [DOI] [PubMed] [Google Scholar]
- 37.Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, et al. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 2014;14:606–16 [DOI] [PubMed] [Google Scholar]
- 38.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene 2008;27:1106–13 [DOI] [PubMed] [Google Scholar]
- 39.Chen D, Wu M, Li Y, Chang I, Yuan Q, Ekimyan-Salvo M, et al. Targeting BMI1(+) Cancer Stem Cells Overcomes Chemoresistance and Inhibits Metastases in Squamous Cell Carcinoma. Cell Stem Cell 2017;20:621–34 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012;485:109–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012;485:55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell 2010;38:768–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature 2013;497:217–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007;128:257–67 [DOI] [PubMed] [Google Scholar]
- 45.Peter D, Igreja C, Weber R, Wohlbold L, Weiler C, Ebertsch L, et al. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol Cell 2015;57:1074–87 [DOI] [PubMed] [Google Scholar]
- 46.Li S, Sonenberg N, Gingras AC, Peterson M, Avdulov S, Polunovsky VA, et al. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Molecular and cellular biology 2002;22:2853–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi C, Zhang X, Lu T, Zhang X, Wang X, Meng B, et al. Inhibition of 4EBP phosphorylation mediates the cytotoxic effect of mechanistic target of rapamycin kinase inhibitors in aggressive B-cell lymphomas. Haematologica 2017;102:755–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yellen P, Saqcena M, Salloum D, Feng J, Preda A, Xu L, et al. High-dose rapamycin induces apoptosis in human cancer cells by dissociating mTOR complex 1 and suppressing phosphorylation of 4E-BP1. Cell Cycle 2011;10:3948–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.