Abstract
Cyclic dinucleotides (CDNs) are important second messenger molecules in prokaryotes and eukaryotes. Within host cells, cytosolic CDNs are detected by STING and alert the host by activating innate immunity characterized by type I interferon (IFN) responses. Extracellular bacteria and dying cells can release CDNs, but sensing of extracellular CDNs (eCDNs) by mammalian cells remains elusive. Here, we report that endocytosis facilitates internalization of eCDNs. The DNA sensor cGAS facilitates sensing of endocytosed CDNs, their perinuclear accumulation, and subsequent STING‐dependent release of type I IFN. Internalized CDNs bind cGAS directly, leading to its dimerization, and the formation of a cGAS/STING complex, which may activate downstream signaling. Thus, eCDNs comprise microbe‐ and danger‐associated molecular patterns that contribute to host–microbe crosstalk during health and disease.
Keywords: cyclic dinucleotides, cyclic guanosine monophosphate–adenosine monophosphate synthase, endocytosis, pathogen‐associated molecular pattern
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Signal Transduction
Introduction
Recognition of conserved microbial molecules termed microbe‐associated molecular patterns (MAMPs), through germline‐encoded pattern‐recognition receptors (PRRs), initiates innate immune responses and shapes adaptive immunity 1. Cyclic dinucleotides (CDNs) of prokaryotic and eukaryotic origin represent intracellular microbial cues or alarmins 2, 3, 4, 5, 6, which alert the host by inducing type I interferons (IFNs) 2, 5, 6, 7. CDNs encompass bacterial (c‐di‐AMP, c‐di‐GMP, and canonical cGAMP, including 2′2′‐cGAMP and 3′3′‐cGAMP) and mammalian secondary messengers (noncanonical 2′3′‐cGAMP). The nucleotidyl transferase cyclic GMP‐AMP synthase (cGAS) generates mammalian CDNs upon recognition of cytosolic DNA 6. To date, the endoplasmic reticulum (ER)‐resident adaptor protein stimulator of interferon genes (STING) and ER adaptor protein (ERAdP) as well as the mouse oxidoreductase RECON and the cytosolic DNA receptor DDX41 were identified as unique sensors for CDNs 3, 8, 9, 10. Binding of CDNs to STING leads to activation of the TANK‐binding kinase 1 (TBK1)/IFN regulatory transcription factor 3 (IRF3) axis for type I IFN induction 11. Of note, most investigations have used purified CDNs co‐delivered with permeabilizing agents 3, 5, 6 or employed liposome transfection 4 for STING activation. However, in mammalian species, the vast majority of CDNs are likely generated by commensal bacteria thus representing extracellular cues, which must find their way into host cells to induce STING activation 12. Although STING appears critical for CDN‐induced immune modulation 13, mechanistic insights into how extracellular CDNs (eCDNs) activate innate immune responses within host cells are missing. External CDNs, such as c‐di‐GMP 14, 15, and more recently 2′3′‐cGAMP 13, have been exploited as adjuvants, and such approaches suggest alternative receptors for extracellular CDNs 16. Here, we show that clathrin‐dependent endocytosis facilitates the internalization of eCDNs. Internalized CDNs bind cGAS directly, leading to its dimerization and promoting the formation of cGAS/STING complexes. cGAS thus serves as a scaffolding protein and nucleates the formation of perinuclear signalosomes encompassing eCDNs/cGAS/STING which enable STING activation. We conclude that eCDNs comprise microbe‐ and danger‐associated molecular patterns engaged by cGAS to initiate STING activation and type I IFN responses.
Results
eCDNs trigger innate immune responses
To investigate host cell responses to eCDNs, we stimulated macrophages, namely the human monocytic cell line THP‐1, human PBMC‐derived monocytes, murine macrophage cell line RAW264.7, and murine bone marrow‐derived macrophages (mBMDMs) with eCDNs. We employed 2′3′‐cGAMP as a proxy CDN and measured IFNβ transcript abundance. Induction of IFNB1 mRNA was observed in all phagocytes regardless of the origin (Fig 1A). mBMDMs responded to various eCDNs of prokaryotic (c‐di‐AMP, c‐di‐GMP, 2′2′‐cGAMP, 3′3′‐cGAMP) and eukaryotic (2′3′‐cGAMP) origin by upregulating Ifnb1 (Fig 1B) and interleukin (IL) 6 (Il6) transcripts (Appendix Fig S1A). Induction of type I IFN by eCDNs was further validated by the enzyme‐linked immunosorbent assay (ELISA) of IFNβ release into the supernatants (Fig 1C and D). We then stimulated mBMDMs with extracellular 2′3′‐cGAMP (ecGAMP) solution pretreated with snake venom phosphodiesterase (SVPDE) which cleaves cGAMP but leaves potential trace amounts of contaminants intact 17, 18. Our results demonstrated that SVPDE completely blocked the effect of ecGAMP on the induction of IFNβ (Appendix Fig S1B), indicating that cGAMP itself, but not the contamination of DNA, manganese, or endotoxin, was responsible for macrophage responses. Human macrophage‐like cell line THP‐1 showed similar responses to eCDNs (Fig 1E and F, and Appendix Fig S1C). Moreover, CD14+ monocytes from healthy donors produced IFNβ (Fig 1G and H) and IL6 (Appendix Fig S1D) after eCDN stimulation. Collectively, eCDNs induced innate immune responses in monocytes and macrophages of mouse and man.
Figure 1. eCDNs trigger innate immune responses.
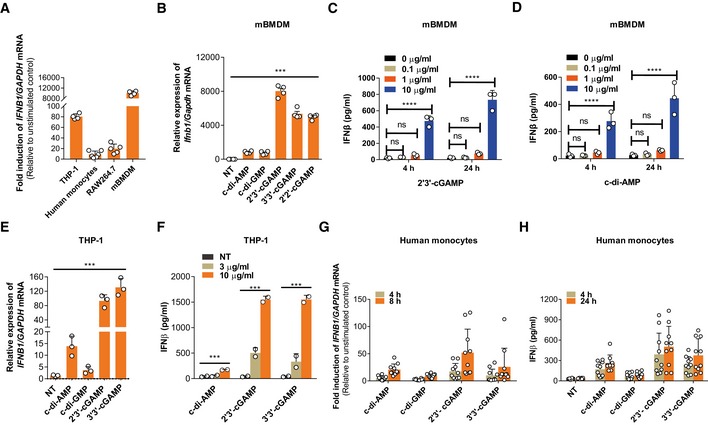
-
AqRT–PCR detection of the fold induction of IFNB1 mRNA relative to unstimulated condition in different cell types. Cells were stimulated with ecGAMP (5 μg/ml) for 4 h.
-
BqRT–PCR detection of Ifnb1 mRNA abundance in mBMDMs treated with different eCDNs (5 μg/ml) for 4 h.
-
C, DELISA detection of IFNβ release by mBMDMs treated for 4 h or 24 h with extracellular 2′3′‐cGAMP (C) or c‐di‐AMP (D) at indicated concentrations.
-
EqRT–PCR detection of IFNB1 mRNA in THP‐1 cells stimulated with indicated eCDNs (5 μg/ml) for 4 h.
-
FELISA detection of IFNβ in supernatants of THP‐1 cells stimulated with indicated eCDNs at indicated concentrations for 4 h.
-
GqRT–PCR detection of the fold induction of IFNB1 mRNA relative to unstimulated condition in human CD14+ monocytes derived from PBMC stimulated with indicated eCDNs (5 μg/ml) for 4 and 8 h. Each symbol represents one individual donor.
-
HELISA detection of IFNβ in supernatants of human CD14+ monocytes derived from PBMC stimulated with indicated eCDNs (5 μg/ml) for 4 h. Each symbol represents result from one individual donor.
eCDNs are less potent than iCDNs in inducing innate immune responses
Next, we employed digitonin, a commonly used detergent for cytosolic delivery of ligands 6, to compare macrophage responses to eCDNs and intracellular CDNs (iCDNs). Cytosolic delivery of CDNs such as cGAMP and c‐di‐AMP strongly induced IFNβ in a dose‐dependent manner and is much more prone to type I IFN induction in THP‐1 cells (Fig 2A–D). Next, we stimulated mBMDMs with eCDNs and iCDNs at different concentrations. iCDNs were consistently more potent than eCDNs in inducing type I IFN responses (Fig 2E–H). To determine whether responses to eCDNs were due to different internalization rates, we stimulated THP‐1 cells with fluorescein isothiocyanate (FITC)‐labeled‐2′3′‐cGAMP. Digitonin did not affect uptake of eCDNs at late time points (4 h; Appendix Fig S2A and B), indicating that uptake per se was not the determining factor for the differential cell response to eCDNs versus iCDNs.
Figure 2. eCDNs are less potent than iCDNs in inducing innate immune responses.
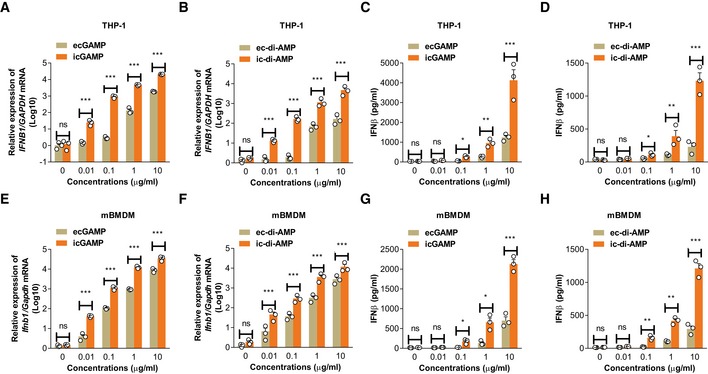
-
A, BqRT–PCR detection of IFNB1 mRNA abundance in THP‐1 cells treated with ecGAMP and icGAMP (A) or ec‐di‐AMP and ic‐di‐AMP (B) at indicated concentrations for 4 h.
-
C, DELISA detection of IFNβ release from THP‐1 cells stimulated with ecGAMP and icGAMP (C) or ec‐di‐AMP and ic‐di‐AMP (D) at indicated concentrations for 4 h.
-
E, FqRT–PCR detection of Ifnb1 mRNA abundance in mBMDMs treated with ecGAMP and icGAMP (E) or ec‐di‐AMP and ic‐di‐AMP (F) at indicated concentrations for 4 h.
-
G, HELISA detection of IFNβ release from mBMDMs stimulated with ecGAMP and icGAMP (G) or ec‐di‐AMP and ic‐di‐AMP (H) at indicated concentrations for 4 h.
eCDNs require endocytosis to activate type I IFN
To understand the mechanisms directing uptake of eCDNs by host cells, we used inhibitors that block endocytic pathways. Dynasore and chlorpromazine (CPZ), which inhibit clathrin‐dependent endocytosis by targeting dynamin and adaptor complex 2 (AP2), respectively 19, consistently inhibited internalization of eCDNs in both THP‐1 cells and HEK293T cells as assessed by flow cytometry (Fig 3A and Appendix Fig S2C). In contrast, incubation with dimethylamiloride (DMA), an inhibitor of pinocytosis, inhibited ecGAMP uptake in THP‐1 cells, but not in HEK293T cells (Fig 3A and Appendix Fig S2C). Polyinosinic acid or mannan, specific inhibitors of cellular entry via scavenger or mannose receptors 20, respectively, did not affect ecGAMP uptake in either cell type (Fig 3A and Appendix Fig S2C). Interestingly, dynasore almost completely blocked the induction of Ifnb1 and Il6 expression in macrophages in response to ecGAMP in both THP‐1 cells (Fig 3B and C) and mBMDMs (Fig 3D and E), indicating that endocytosis plays a major role in eCDN‐induced innate immune activation. However, dynasore treatment dramatically reduced expression of Ifnb1 and Il6 (Fig 3B–E) while leaving uptake of FITC‐icGAMP unchanged (Appendix Fig S2D), indicating that dynasore abrogates macrophage responses to iCDNs in an endocytosis‐independent manner. To further clarify the role of endocytosis in sensing of eCDNs, we assessed compartmentalization of eCDNs and observed that eCDNs colocalized with the early endosome antigen 1 (EEA1), a marker for early endosomes (Fig 3F), and with the lysosome‐associated membrane protein 2 (LAMP2), a late endosome/lysosome marker (Fig 3G). Application of bafilomycin A1 (BafA1), an inhibitor of vacuolar‐type H+‐ATPase that interferes with acidification and maturation of early endosomes 21, drastically diminished responses to ecGAMP in both THP‐1 cells (Fig 3H and I) and mBMDM (Fig 3J and K). In contrast, the response to icGAMP remained intact in both types of cells (Fig 3H–K). To exclude involvement of autophagy upon usage of BafA1 22, we employed 3‐methyladenine (3‐MA), an inhibitor of autophagy 23. Exposure to 3‐MA restricted ecGAMP‐induced autophagy (Fig EV1A), whereas changes in IFNB1 and IL6 transcripts were insignificant (Fig EV1B and C). We conclude that endocytosis followed by vesicle maturation, independent of autophagy induction, is important for eCDN‐induced immune activation.
Figure 3. eCDNs require endocytosis to activate type I IFN.
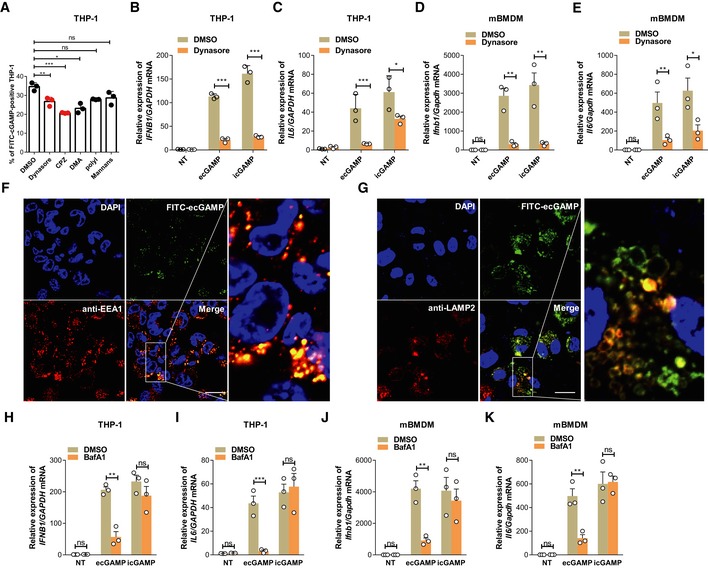
-
AFrequencies of FITC+ THP‐1 cells stimulated with FITC‐ecGAMP for 1 h in the presence of DMSO or indicated inhibitors including dynasore (10 μM), chlorpromazine (CPZ, 10 μM), dimethylamiloride (DMA, 100 μM), polyinosinic acid (Poly I, 50 μg/ml), or mannans from Saccharomyces cerevisiae (mannans, 1 mg/ml).
-
B, CqRT–PCR detection of IFNB1 (B) and IL6 (C) mRNA in THP‐1 cells stimulated with ecGAMP (5 μg/ml) or intracellular 2′3′‐cGAMP (icGAMP) (0.1 μg/ml) for 4 h in the presence of DMSO or dynasore (10 μM).
-
D, EqRT–PCR detection of Ifnb1 (D) and Il6 (E) mRNA in mBMDMs stimulated with ecGAMP (5 μg/ml) or icGAMP (0.1 μg/ml) for 4 h in the presence of DMSO or dynasore (10 μM).
-
F, GImmunostaining for EEA1 (red) (F) and LAMP2 (red) (G) in THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml, green) for 30 min, nucleus in blue (DAPI). Data are representative of three independent experiments. Scale bars, 10 μm.
-
H–KqRT–PCR detection of IFNB1 and IL6 mRNA in THP‐1 cells (H, I) and mBMDMs (J, K) stimulated with ecGAMP (5 μg/ml) and icGAMP (0.1 μg/ml), respectively, for 4 h in the presence of DMSO or bafilomycin A1 (BafA) (1 μM).
Figure EV1. eCDN‐induced type I IFN responses are uncoupled from autophagy.
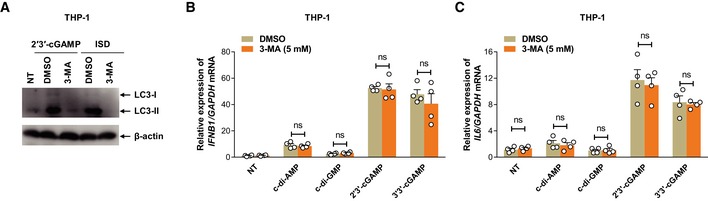
-
AWestern blot detection of indicated proteins in lysates of THP‐1 cells left untreated or treated with ecGAMP or transfected with ISD in the presence of mock or indicated concentrations of 3‐MA (5 mM). Data are representative of three independent experiments.
-
B, CqRT–PCR detection of IFNB1 (B) and IL6 (C) mRNA in THP‐1 cells stimulated with indicated eCDNs (5 μg/ml) for 4 h in the presence of mock or 3‐MA (5 mM). Data are means + SD averaged from four independent experiments performed in technical duplicates, and each symbol represents mean of technical duplicates. Two‐way ANOVA followed by Bonferroni's post hoc test was used for statistical analysis. ns, not significant.
Source data are available online for this figure.
STING is important, but insufficient for eCDN‐induced type I IFN response
Next, we interrogated whether STING is necessary for induction of type I IFN by eCDNs. STING knockdown (KD) THP‐1 cells 24 were impaired in induction of IFNB1 mRNA and release of type I IFN, following treatment with eCDNs or IFN stimulatory DNA (ISD) (dsDNA mimic), but not upon poly(I:C) (RNA mimic) stimulation (Fig 4A and B). Sting knockout (KO) mBMDMs were markedly impaired in induction of Ifnb1 (Fig 4C) and Il6 (Appendix Fig S3A) transcripts as well as in release of IFNβ protein (Fig 4D) irrespective of the CDNs employed. Defective Ifnb1 and Il6 mRNA expression in response to eCDNs was rescued in Sting KO RAW264.7 cells complemented with STING expression (Fig 4E and Appendix Fig S3B). Consistent with these findings, the deficiency of Sting completely blocked phosphorylation of IRF3 in response to ecGAMP or ISD stimulation (Fig 4F). We conclude that STING is indispensable for activation of the TBK1/IRF3/IFN I axis in macrophages downstream of eCDN sensing.
Figure 4. STING is important but not sufficient for eCDN‐induced type I IFN response.
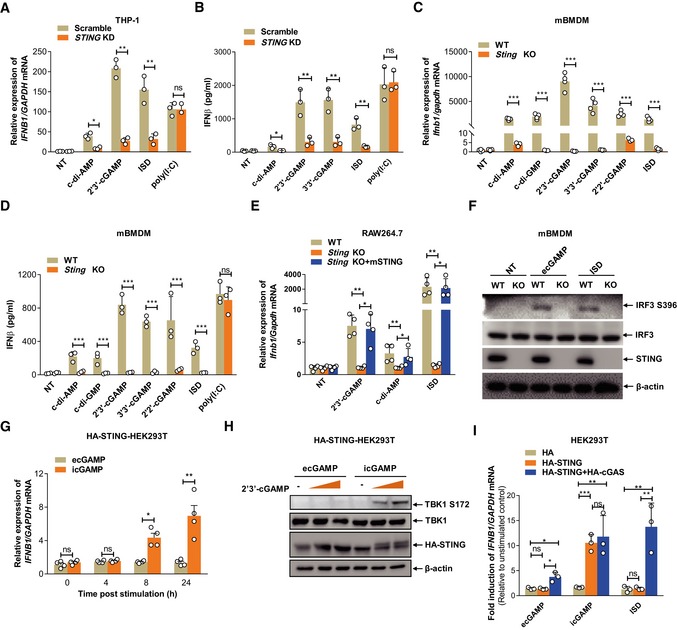
- qRT–PCR detection of IFNB1 mRNA in scrambled (Scramble) or STING shRNA stably transfected (STING KD) THP‐1 cells stimulated with ec‐di‐AMP (5 μg/ml) and ecGAMP (5 μg/ml) or transfected with ISD or poly(I:C).
- ELISA detection of IFNβ protein in supernatants of Scramble or STING KD THP‐1 cells stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD or poly(I:C).
- qRT–PCR detection of Ifnb1 mRNA in WT and Sting −/− (Sting KO) mBMDMs stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD.
- ELISA detection of IFNβ protein secretion in supernatants of mBMDMs stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD or poly (I:C).
- qRT–PCR detection of Ifnb1 mRNA in WT, Sting KO, or Sting KO complemented with mouse STING (Sting KO+mSTING) RAW264.7 cells stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD.
- Western blot detection of indicated proteins in lysates of WT and Sting −/− (KO) mBMDMs stimulated with ecGAMP (5 μg/ml) or transfected with ISD. Data are representative of three independent experiments.
- qRT–PCR detection of IFNB1 mRNA levels in STING stable HEK293T cells (HA‐STING‐HEK293T) stimulated with ecGAMP or icGAMP at 5 μg/ml for indicated times.
- Western blot detection of indicated proteins in HA‐STING‐HEK293T cells stimulated with increasing amounts of ecGAMP and icGAMP at 5 μg/ml for 24 h. Data are representative of three independent experiments.
- qRT–PCR detection of the induction of IFNB1 mRNA in HEK293T cells stably transfected with pcDNA3.1‐HA (HA), HA‐STING, and HA‐STING+HA‐cGAS stimulated with ecGAMP (5 μg/ml) or icGAMP (0.1 μg/ml) or transfected with ISD.
We further generated HEK293T cells stably transfected with HA‐tagged human STING (HA‐STING‐HEK293T). icGAMP, but not ecGAMP, upregulated IFNβ transcription and promoted TBK1 phosphorylation in HA‐STING‐HEK293T cells (Fig 4G and H), while their internalization was comparable at 4 h post‐stimulation (Appendix Fig S3C). We acknowledge the propensity of human adenovirus 5 (hAd5) and simian virus 40 (SV40)‐transformed cell lines, such as HEK293T cells, to restrict type I IFN responses 25 and the lower magnitude of IFNβ induction by eCDN compared to iCDN. Yet, these data suggest that STING per se is not sufficient for detecting eCDNs in HEK293T cells.
cGAS facilitates eCDN detection in macrophages
Surprisingly, unlike HA‐STING‐HEK293T STING cells, HEK293T cells stably expressing both HA‐cGAS and HA‐STING (Fig EV2A) conferred responsiveness to eCDNs and ISD upon HA‐STING‐HEK293T cells without altering that to iCDNs (Fig 4I). This indicates a role for cGAS expression in eCDN sensing by STING. Consistently, CGAS deficiency profoundly reduced IFNB1 expression in THP‐1 cells in response to eCDNs (Fig 5A), while the uptake of eCDNs remained unaffected in CGAS KO THP‐1 cells (Fig EV2B). IFNβ secretion was also significantly reduced in CGAS KO THP‐1 cells upon stimulation with eCDNs or impaired in response to ISD, but was not affected by treatment with poly (I:C) (Fig 5B). In line with our observations in human THP‐1 cells, BMDMs from Cgas‐deficient mice produced significantly less IFNβ both at transcript (Fig 5C) and at protein level (Fig 5D) than those from WT mice. Of note, Ifnb1 transcription was impaired in cGAS KO macrophages treated with ecGAMP, whereas the abundance of IFNB1 transcripts was not affected by icGAMP (Fig 5E and F). In agreement with transcriptional responses, the absence of Cgas remarkably reduced phosphorylation of IRF3 and STING in mBMDMs in response to ecGAMP, but not icGAMP (Fig 5G). As expected, the phosphorylation of IRF3 and STING was completely lost in Cgas‐deficient mBMDMs transfected with ISD (Fig 5G). Defective IFNβ production in response to eCDNs in CGAS KO THP‐1 cells (Fig 5H and I) or RAW264.7 cells (Fig EV2C) was rescued by complementing cGAS expression. Although eCDN‐induced type I IFN responses in HEK293T cells stably expressing both HA‐cGAS and HA‐STING cells in a dose‐dependent manner, the saturated eCDNs were still less potent than iCDNs (Fig EV2D). These results indicate that additional factor(s) other than cGAS are involved in the differential responses to eCDNs and iCDNs. Taken together, cGAS facilitated eCDN sensing in macrophages to activate the STING/TBK1/IRF3 axis leading to innate immune activation.
Figure EV2. cGAS contributes to sensing of eCDNs.
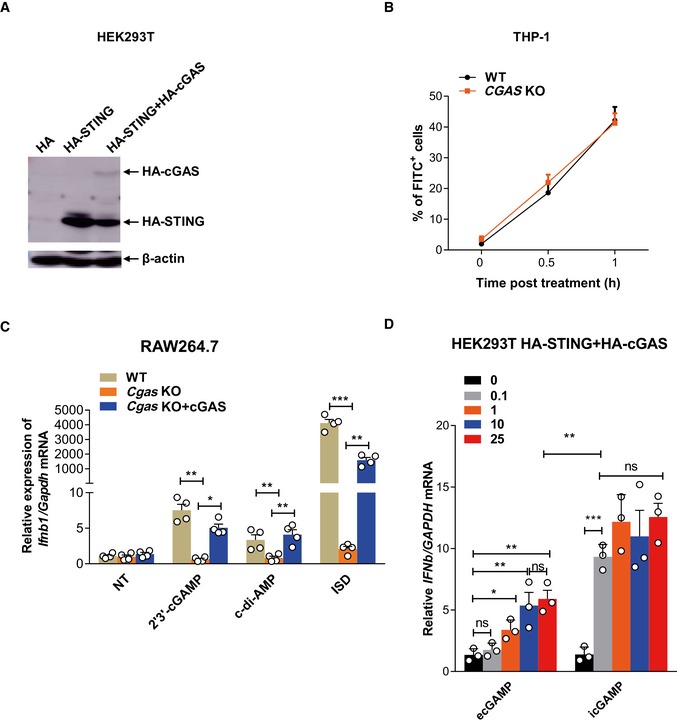
- Western blot detection of indicated proteins in lysates of HEK293T cells stably transfected with pcDNA3.1‐HA (HA), HA‐STING, and HA‐STING + HA‐cGAS. Data are representative of three independent experiments.
- Frequencies of FITC+ WT and CGAS KO THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml) for indicated times.
- qRT–PCR detection of Ifnb1 mRNA in WT, Cgas KO, or Cgas KO complemented with cGAS (Cgas KO+cGAS) RAW264.7 cells stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD.
- qRT–PCR detection of IFNB1 mRNA in HEK293T cells stably transfected with both HA‐STING and HA‐cGAS stimulated with either ecGAMP or icGAMP at the indicated concentrations (μg/ml) for 24 h.
Figure 5. cGAS facilitates eCDN detection in macrophages.
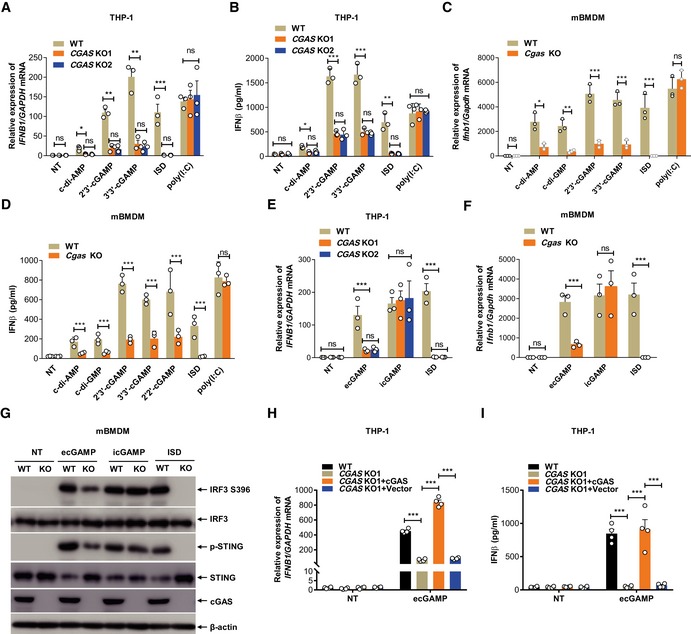
-
AqRT–PCR detection of IFNB1 mRNA in WT and CGAS KO THP‐1 cells stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD or poly (I:C) for 4 h.
-
BELISA detection of IFNβ in supernatants of WT and CGAS KO THP‐1 cells stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD or poly(I:C) for 4 h.
-
CqRT–PCR detection of Ifnb1 mRNA in mBMDMs from WT and Cgas −/− (Cgas KO) mice stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD or poly(I:C) for 4 h.
-
DELISA detection of IFNβ in supernatants of mBMDMs from WT and Cgas KO mice stimulated with indicated eCDNs (5 μg/ml) or transfected with ISD or poly(I:C) for 4 h.
-
EqRT–PCR detection of IFNB1 mRNA in WT and CGAS KO THP‐1 cells stimulated with ecGAMP (5 μg/ml) or icGMAP (0.1 μg/ml) or transfected with ISD.
-
FqRT–PCR detection of Ifnb1 mRNA in mBMDMs from WT and Cgas KO stimulated with ecGAMP (5 μg/ml) or icGMAP (0.1 μg/ml) or transfected with ISD.
-
GWestern blot detection of indicated proteins in lysates of mBMDMs from WT and Cgas KO mice stimulated with ecGAMP (5 μg/ml) or icGAMP (0.1 μg/ml) or transfected with ISD. Data are representative of three independent experiments.
-
H, IqRT–PCR detection of IFNB1 mRNA (H) and ELISA detection of IFNβ in supernatants (I) of WT, CGAS KO, CGAS KO complemented with cGAS (CGAS KO+cGAS) or CGAS KO stably transfected with the empty vector (CGAS KO+Vector) THP‐1 cells stimulated with ecGAMP (5 μg/ml).
CDNs bind cGAS directly leading to its dimerization
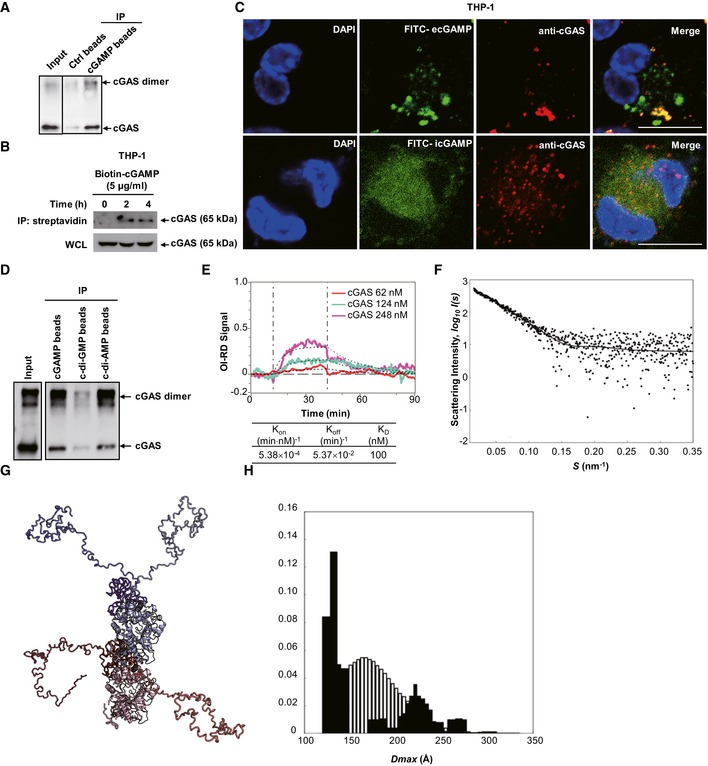
The capacity of cGAS to initiate STING activation by eCDNs prompted us to investigate whether cGAS directly senses endocytosed eCDNs. Immunoprecipitation (IP) assays demonstrated that HA‐tagged human cGAS (HA‐h‐cGAS) coprecipitated with 2′3′‐cGAMP beads in a dose‐dependent manner (Fig EV3A). We also purified cGAS expressed in Escherichia coli (Fig EV3B) and verified its function by measuring production of 2′3′‐cGAMP in the presence of DNA (Fig EV3C). Small‐angle X‐ray scattering (SAXS) and multiangle light scattering (MALS) analysis revealed that purified cGAS is monomeric in solution and can be divided into a flexible N‐terminal domain (residues M1 to A159) that can assume different conformations and a stably folded C‐terminal domain (residues P160 to F522; Fig EV3D–F, and Appendix Table S1). A direct association of purified cGAS with 2′3′‐cGAMP beads was demonstrated (Fig 6A). Interaction of ecGAMP with cGAS was then studied in THP‐1 cells. Stimulation of THP‐1 cells with extracellular biotin‐cGAMP revealed association of cGAS with ecGAMP post‐stimulation (Fig 6B), suggesting that ecGAMP interacts with endogenous cGAS. This was further strengthened by the observed colocalization of cGAS with FITC‐ecGAMP, but not icGAMP, which appeared diffusive in the cytosol (Fig 6C). To evaluate whether cGAS binds to different CDNs, we expressed HA‐h‐cGAS and HA‐tagged mouse cGAS (HA‐m‐cGAS) in HEK293T cells and performed IP with beads coupled to various CDNs. Both human and mouse cGAS were precipitated with all tested CDNs with varied binding affinities (Figs 6D and EV3G). In a similar manner, c‐di‐AMP, c‐di‐GMP, and 2′3′‐cGAMP beads pulled down purified cGAS protein with different binding affinities (Fig 6D). Note that the binding affinities were positively correlated with the magnitude of type I IFN responses to corresponding eCDNs in murine and human macrophage (Fig EV3H). Specific interactions of cGAS with CDNs including 2′3′‐cGAMP, 3′3′‐cGAMP (Fig EV3I), and c‐di‐AMP (Fig EV3J) were observed as coelutants by analytical size exclusion chromatography. Fluorometric binding assays further validated interactions of cGAS with different CDNs (Fig EV3K and L). A label‐free biomolecular interaction assay 26 further demonstrated direct interaction of cGAMP with cGAS (KD = 100 nM; Fig 6E). cGAS purified in the presence of cGAMP formed a head‐to‐tail dimer with a mostly flexible N‐terminus, as revealed by on‐line SAXS coupled to size exclusion chromatography (Fig 6F–H, and Appendix Table S1). Our observation of cGAS in solution is in agreement with the reported crystal structure of the N‐terminal truncated cGAS 27 bound to cGAMP. We conclude that host‐ and pathogen‐derived eCDNs bind cGAS directly, causing its dimerization.
Figure EV3. Purification of cGAS and its binding to CDNs.
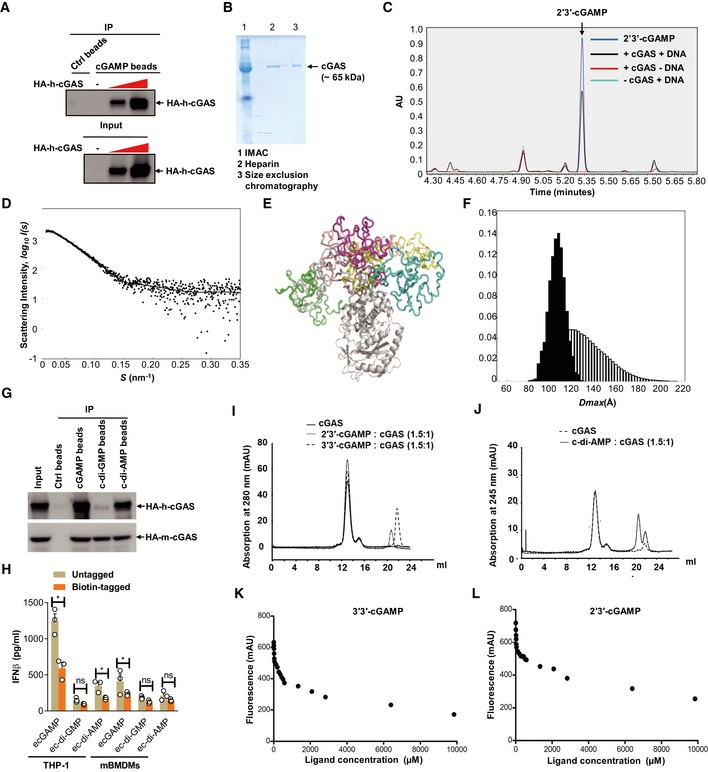
-
ALysates of HEK293T cells transfected with increasing amounts of HA‐cGAS were precipitated with control beads (Ctrl) or 2′3′‐cGAMP beads (cGAMP) and immunoblotted. Data are representative of three independent experiments.
-
BSDS–PAGE gel analysis of purified cGAS protein by indicated methods. IMAC, immobilized metal affinity chromatography. Data are representative of two independent experiments.
-
CEnzyme activity of purified cGAS confirmed by UPLC detection of cGAMP on a Waters BEH Amide Column. Data are representative of three independent experiments.
-
DThe small‐angle X‐ray scattering analysis of the full‐length apo‐cGAS. The EOM fit of the measured SAXS data. The goodness‐of‐the fit χ2 = 1.2.
-
EStructural alignment of the representative structures from cGAS apo EOM analysis.
-
FThe D max distributions (the maximum distance within a particle) derived from the EOM analysis of the measured SAXS profile (pool—the white histogram; the selected structures—the black histogram). The ensemble average of D max is 109.1 Å.
-
GLysates of HEK293T cells transfected with HA‐tagged human cGAS (HA‐h‐cGAS) or mouse cGAS (HA‐m‐cGAS) were precipitated with Ctrl beads or beads coupled with cGAMP, c‐di‐GMP, or c‐di‐AMP followed by immunoblotting.
-
HELISA detection of IFNβ release in the supernatant of THP‐1 cells stimulated with untagged or biotin‐tagged eCDNs (5 μM) including cGAMP, c‐di‐GMP, and c‐di‐AMP for indicated times. Data are means + SD averaged from at least three independent experiments performed with technical triplicates, where each symbol represents the mean of technical triplicates. Two‐way ANOVA followed by Bonferroni's post hoc test was used for statistical analysis, respectively. *P < 0.05; ns, not significant.
-
I, JElution profiles of analytical size exclusion chromatography (Superdex 200, GE Healthcare, 10/300 GL). Absorption profiles at 280 nm for detection of cGAS (solid lines) and cGAMPs (dashed lines) (I) and at 245 nm for c‐di‐AMP (J) are shown. Molecular stoichiometric ratios are indicated. Peak maximum at 13 ml (peak 1) corresponds to dimeric cGAS with a molecular mass of about 120 kDa. Despite the presence of free CDNs (Peaks 2–4), the specific interaction (coelution) of cGAS with CDNs was evident in each case by an increased peak intensity of 36% (2′3′‐cGAMP), 17% (3′3′‐cGAMP), and 2% (c‐di‐AMP), respectively. mAU, milli absorbance units. Data are representative of three independent experiments.
-
K, LFluorometry assay to detect the binding of 3′3′‐cGAMP (K) or 2′3′‐cGAMP (L) with purified cGAS. Data are representative of at least three independent experiments.
Source data are available online for this figure.
Figure 6. CDNs bind cGAS directly leading to its dimerization.
- Purified h‐cGAS protein was precipitated with Ctrl beads or cGAMP beads and then immunoblotted.
- THP‐1 cells were stimulated with biotin‐cGAMP for indicated times, and then, whole‐cell lysates (WCL) were precipitated with streptavidin beads followed by immunoblotting.
- Immunofluorescent staining of cGAS (red) in THP‐1 cells treated with FITC‐ecGAMP (5 μg/ml) (green) or FITC‐icGAMP (0.1 μg/ml) (green) for 4 h, nucleus in blue (DAPI). Scale bar, 10 μm.
- Purified h‐cGAS protein was precipitated with beads coupled with cGAMP, c‐di‐GMP, or c‐di‐AMP followed by immunoblotting.
- Binding curves of surface‐immobilized 2′3′‐cGAMP with His‐cGAS at indicated concentrations. Vertical lines mark the start of association and dissociation phases of the binding events. The dashed lines are global fits to a Langmuir reaction model; global fitting parameters are listed in the table below the plot (n = 4 independent experiment).
- The small‐angle X‐ray scattering analysis of the full‐length cGAS with cGAMP. The EOM fit of the measured SAXS data. The goodness‐of‐the fit χ2 = 1.1.
- Structural alignment of the representative structures from cGAS‐cGAMP EOM analysis. cGAMP subunits are colored red and blue, respectively. Different conformations of the cGAS N‐termini are highlighted in additional shades of red and blue.
- The D max distributions (the maximum distance within a particle) derived from the EOM analysis of the measured SAXS profile (pool—the white histogram, the selected structures—the black histogram). The distribution of the selected structures shows a bimodal behavior with an average value of 166.9 Å.
Of note, the dimerization of cGAS upon recognition of cytosolic DNA is critical for the activation of its enzyme activity and synthesis of 2′3′‐cGAMP which enables activation of STING 6, 28, 29. We determined whether cGAS dimerization after eCDN binding enabled enzymatic generation of 2′3′‐cGAMP. To this end, we reconstituted STING KO RAW264.7 cells with mouse STING R231A mutant (mSTINGR231A; Fig EV4A). These cells could still initiate responses to dsDNA by sensing noncanonical 2′3′‐cGAMP generated by cGAS while lacking responsiveness to canonical CDNs 3. Stimulation with extracellular c‐di‐GMP (ec‐di‐GMP) and ecGAMP as well as ISD failed to induce type I IFN responses in STING KO cells (Fig EV4B and C). mSTINGR231A reconstitution restored the responses to ecGAMP and ISD, but not to ec‐di‐GMP (Fig EV4B and C), arguing against a major role of de novo synthesis or resynthesis of cGAMP by cGAS upon binding of eCDNs in sensing of eCDNs. We further complemented cGAS KO THP‐1 cells with cGAS enzyme‐inactive mutant cGASE225A D227A (Fig EV4D). The impaired type I IFN response of cGAS KO THP‐1 cells was not restored by cGASE225A D227A complementation (Fig EV4E and F), suggesting that the amino acids E225A D227A themselves are critical for the sensing of eCDNs by cGAS independently from its enzyme activity.
Figure EV4. cGAS facilitates eCDN‐induced type I IFN response dispensable of resynthesis of cGAMP by cGAS .
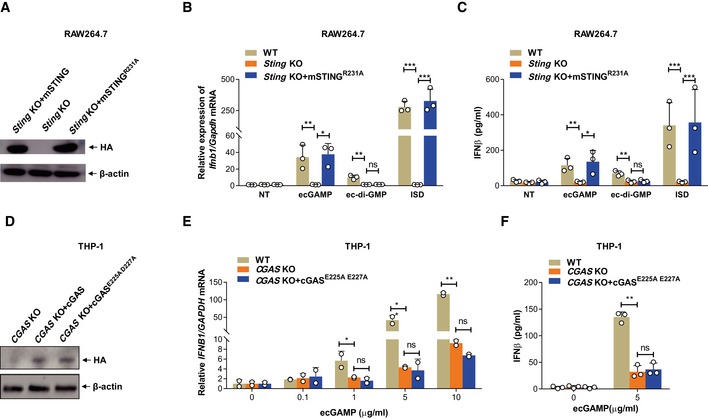
-
AWestern blot detection of indicated proteins in lysates of Sting KO RAW264.7 cells (Sting KO) and Sting KO RAW264.7 cells complemented with WT mouse STING (Sting KO+mSTING) or mSTINGR231A (Sting KO+ mSTINGR231A). Data are representative of three independent experiments.
-
B, CqRT–PCR detection of Ifnb1 mRNA (B) or ELISA detection of IFNβ in the supernatants (C) of WT, Sting KO, and Sting KO+ mSTINGR231A RAW264.7 cells stimulated with ecGAMP (5 μg/ml) or ec‐di‐GMP (5 μg/ml) or transfected with ISD.
-
DWestern blot detection of indicated proteins in lysates of CGAS KO THP‐1 cells (CGAS KO) and CGAS KO THP‐1 cells complemented with WT cGAS (CGAS KO+cGAS) or cGASE225A D227A (CGAS KO+cGASE225A D227A). Data are representative of three independent experiments.
-
E, FqRT–PCR detection of IFNB1 mRNA (D) or ELISA detection of IFNβ in the supernatants (E) of WT, CGAS KO, and cGAS KO+cGASE225A D227A THP‐1 cells stimulated with ecGAMP at indicated concentrations.
eCDNs promote formation of the cGAS/STING complex
To precisely understand how cGAS promotes the sensing of eCDNs, we stimulated THP‐1 cells with extracellular FITC‐cGAMP (FITC‐ecGAMP) and visualized cellular compartmentalization of eCDNs. FITC‐ecGAMP formed perinuclear puncta and significantly colocalized with STING, indicating colocalization of ecGAMP with STING in the perinuclear regions (Fig 7A). CDNs delivered together with permeabilizing agents 3, 5, 6 or by liposome transfection 4 reach the cytosol and activate STING in the ER 30. Activated STING dissociates from the ER exit sites (ERES) and translocates through the Golgi to perinuclear punctate structures, where it can recruit TBK1 and initiate signal transduction 30. Until recently, the ER‐Golgi intermediate compartment (ERGIC) was considered a unique subcellular compartment that serves as platform for the recruitment of TBK1 and IRF3 in the STING signaling cascade 31, 32. We further investigated compartmentalization of FITC‐ecGAMP in different organelles including ER, ERGIC, and Golgi. ecGAMP puncta did not colocalize with any of these subcellular compartment markers (Appendix Fig S4A–C). However, we detected colocalization of the perinuclear 2′3′‐cGAMP puncta with phospho‐TBK1 (Appendix Fig S4D). This observation indicates that ecGAMP forms puncta representing a specialized subcellular compartment which functions as a “platform” for initiation of STING signaling. To determine how cGAS converges signaling to STING downstream of eCDN sensing, we stimulated THP‐1 cells with FITC‐ecGAMP and compared cellular compartmentalization of the ecGAMP in WT and CGAS KO THP‐1 cells. The formation of FITC‐ecGAMP perinuclear puncta was impaired in CGAS KO cells (Fig 7B and C), suggesting that cGAS promoted STING activation by regulating the formation of perinuclear puncta. In an in vitro binding assay, we observed that the GST‐tagged cGAS and His‐tagged STING were immunoprecipitated together with cGAMP agarose (Fig 7D). In addition, a GST pull‐down assay demonstrated direct interaction of cGAS with STING (Fig 7E). Moreover, cGAMP enhanced the interaction of cGAS with STING in a dose‐dependent manner (Fig 7F). These results indicate that cGAMP promotes the formation of a cGAMP/cGAS/STING complex. Stimulation of THP‐1 cells with extracellular biotin‐cGAMP revealed association of cGAS with ecGAMP at an early time point (2 h) post‐treatment. Notably, at a later time point (4 h) post‐stimulation, a complex of ecGAMP with endogenous cGAS and STING was observed (Fig 7G), indicating that interactions of ecGAMP with cGAS preceded the formation of the cGAMP/cGAS/STING complex. icGAMP interacted with STING at an earlier time point (2 h) but did not induce the formation of the cGAMP/cGAS/STING complex (Fig 7G). These observations prompted us to propose that cGAS serves as a scaffolding protein and nucleates the formation of signalosomes including the cGAMP/cGAS/STING complex, a process specifically required for STING activation in response to eCDNs.
Figure 7. CDNs promote formation of cGAS/STING complex.
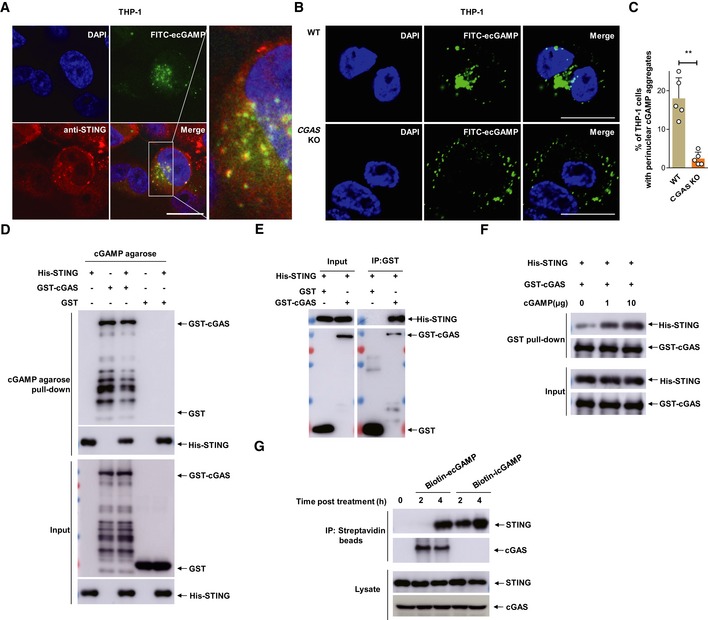
- Immunostaining of STING (anti‐STING, red) in THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml, green) for 2 h, nucleus in blue (DAPI). Data are representative of three independent experiments. Scale bar, 10 μm.
- Cellular localization of ecGAMP in WT and CGAS KO THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml, green) for 2 h, nucleus in blue (DAPI). Data are presentative of five independent experiments. Scale bar, 10 μm.
- Frequency of perinuclear accumulation of FITC‐ecGAMP in WT and CGAS KO THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml) for 2 h. Data are means + SD averaged from five independent experiments, and approximately 100 cells were imaged and counted in each experiment. Each symbol represents the percentage of THP‐1 cells with perinuclear cGAMP aggregates in every independent experiment. Mann–Whitney U‐test was used for statistical analysis. **P < 0.01.
- Western blot detection of the presence of GST‐cGAS and His‐STING in the immunoprecipitates of cGAMP agarose. Purified GST‐cGAS and His‐STING were incubated separately or together with cGAMP agarose. The input and immunoprecipitates were immunoblotted.
- Western blot detection of the presence of His‐STING in the immunoprecipitates of Glutathione Sepharose. Purified GST‐cGAS and His‐STING were incubated together in IP lysis buffer overnight at 4°C.
- Western blot detection of the presence of GST‐cGAS and His‐STING in the immunoprecipitates of Glutathione Sepharose. Purified GST‐cGAS and His‐STING were incubated together in the absence or presence of increasing 2′3′‐cGAMP.
- Western blot detection of the presence of cGAS and STING in the immunoprecipitates of streptavidin beads. THP‐1 cells were stimulated with biotin‐ecGAMP (5 μg/ml) or biotin‐icGAMP (1 μg/ml) for indicated time, and cell lysates were harvested for IP with streptavidin beads.
The puncta formed by CDNs are reminiscent of perinuclear aggresomes that are regulated by dynein 33. Consequently, we examined the role of dynein in eCDN puncta formation. Dynein colocalized with perinuclear ecGAMP puncta (Fig EV5A). The importance of the GTPase was corroborated by the addition of ciliobrevin D, a dynein inhibitor 34, which impaired the formation of perinuclear puncta of ecGAMP (Fig EV5B and C). Moreover, inhibition of dynein by ciliobrevin D significantly reduced IFNB1 mRNA production following ecGAMP, but not icGAMP stimulation (Fig EV5D). Thus, dynein is specifically required for the eCDN‐induced type I IFN response. Moreover, the inhibitory effect of dynein inhibitor on the ecGAMP‐induced type I IFN response was not observed in CGAS KO THP‐1 cells (Fig EV5E). Therefore, dynein‐dependent perinuclear eCDN puncta formation is critical for initiating STING signaling.
Figure EV5. Dynein contributes to cGAS sensing of eCDNs.
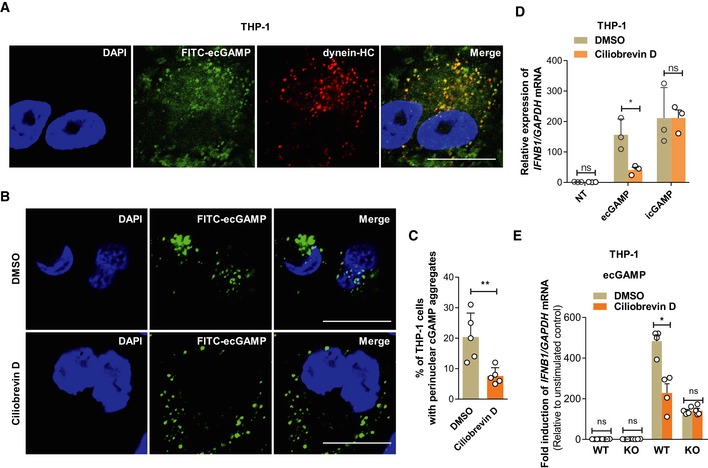
- Immunostaining of dynein heavy chain (HC) (red) in THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml, green) for 2 h, nucleus in blue (DAPI). Data are representative of three independent experiments. Scale bar, 10 μm.
- Cellular localization of ecGAMP in THP‐1 cells stimulated with FITC‐ecGAMP (5 μg/ml, green) for 2 h in the presence of DMSO or dynein inhibitor ciliobrevin D (50 μM), nucleus in blue (DAPI). Data are presentative of five independent experiments. Scale bar, 10 μm.
- Frequency of perinuclear accumulation of 2′3′‐cGAMP in THP‐1 cells stimulated with FITC‐cGAMP (2 μg/ml) for 2 h in the presence of DMSO or dynein inhibitor ciliobrevin D (50 μM). Data are means + SD averaged from five independent experiments, and approximately 100 cells were imaged and counted in each experiment. Each symbol represents the percentage of THP‐1 cells with perinuclear cGAMP aggregates in every independent experiment. Mann–Whitney U‐test was used for statistical analysis. **P < 0.01.
- qRT–PCR detection of IFNB1 mRNA in THP‐1 cells stimulated with ecGAMP (5 μg/ml) or icGAMP (0.1 μg/ml) for 4 h in the presence of DMSO or ciliobrevin D (50 μM).
- qRT–PCR detection of IFNB1 mRNA abundance in WT and cGAS KO THP‐1 cells stimulated with ecGAMP (5 μg/ml) for 4 h in the presence of DMSO or ciliobrevin D (50 μM).
eCDNs promote cGAS‐mediated sensing of DNA virus
Sensing of cytosolic dsDNA and eCDNs converges at the dimerization of cGAS, a biochemical process critical for its enzymatic activation and generation of 2′3′‐cGAMP engaged by STING 6, 28, 29. We therefore interrogated whether eCDN‐induced cGAS dimerization facilitates DNA binding to cGAS and thereby promotes DNA sensing via the canonical cGAS‐cGAMP‐STING pathway. We ascertained the effect of eCDNs on macrophage response to DNA virus infection. Costimulation with eCDNs robustly boosted the production of type I IFN in response to HSV‐1 infection in a dose‐dependent manner (Fig 8A and B) and ecGAMP markedly enhanced HSV‐induced phosphorylation of IRF3 in a synergistic way (Fig 8C). We conclude that eCDNs promote cGAS‐mediated sensing of DNA virus.
Figure 8. eCDNs promote cGAS‐mediated DNA sensing.
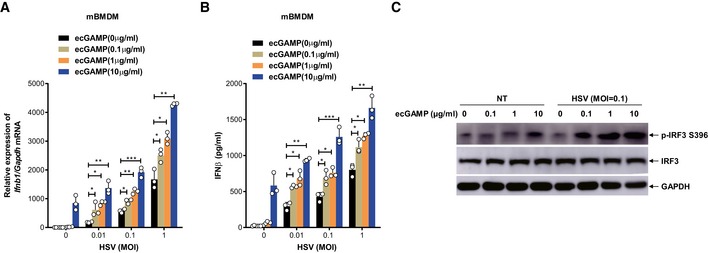
-
A, BqRT–PCR detection of Ifnb1 mRNA (A) or ELISA detection of IFNβ in supernatants (B) of mBMDMs infected with HSV‐1 at indicated MOI together with stimulation with ecGAMP at indicated concentrations for 4 h. Data are means + SD averaged from three independent experiments performed with technical triplicates. Each symbol represents the mean of technical triplicates. Two‐way ANOVA followed by Bonferroni's post hoc test was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001.
-
CWestern blot detection of indicated proteins in lysates of mBMDMs infected with HSV‐1 at indicated MOI together with stimulation with ecGAMP at indicated concentrations for 4 h. Data are representative of three independent experiments.
Source data are available online for this figure.
Discussion
Our data demonstrate that mammalian cells employ unique strategies for differential sensing of eCDNs versus iCDNs and underscore an important role of cGAS in sensing of eCDNs. We conclude that the location of the stimulus determines the type of the intracellular signaling pathway. In contrast to direct binding of iCDNs to STING, eCDNs require clathrin‐dependent endocytosis and binding to cGAS for subsequent STING activation (Appendix Fig S5). This extends the role of cGAS from a generator of the endogenous mammalian signal 2′3′‐cGAMP to a sensor of eCDNs, further supporting the expanding role of cGAS beyond DNA sensing such as inhibition of DNA repair to fuel genome instability 35, 36, 37.
Our data demonstrate that eCDNs critically differ from cGAMP introduced by digitonin permeabilization: eCDNs require cGAS for STING activation, and this may account for differential type I IFN production in response to eCDNs and iCDNs. Although type I IFNs are essential for control of most viral infections, they are often detrimental in bacterial infections 38. Therefore, the elaborate regulation of the magnitude of response to the same type of stimulus but of different origin is critical. This is supported by the finding that R232H variant of human STING and R231A variant of murine STING can confer a selective advantage by impairing responses to canonical bacterial CDNs, while still retaining responsiveness to endogenous noncanonical 2′3′‐cGAMP produced by cGAS in response to viral dsDNA 3, 39. Therefore, we consider it likely that formation of the cGAS/STING complex is specifically involved in sensing of eCDNs.
Understanding the molecular mechanisms orchestrating sensing of eCDNs can form the basis for the development of novel intervention measures since eCDNs are currently exploited as vaccine adjuvants 13, 14, 15 and for cancer therapy 40, 41. Although STING has emerged as a critical receptor for CDN‐induced immunomodulation 13, 16, the mechanisms by which eCDNs precisely activate STING remain elusive. Additional receptors for CDNs have been proposed 16, 42. The ER membrane adaptor ERAdP was recently identified as a direct sensor for c‐di‐AMP 9, 43. Mouse oxidoreductase RECON has been classified as a sensor for some bacterial CDNs which modulate NF‐κB activation independently of STING through which they shape a proinflammatory antibacterial response 10. Here, we determined that the DNA sensor cGAS is involved in the engagement of endocytosed eCDNs, prior to STING activation. This finding can be harnessed for application of eCDNs in cancer therapy since cGAS has been reported to be aberrantly expressed or dysfunctional in tumor cells 40, 41.
CDNs are relevant to homeostasis at mucosal sites, as recently demonstrated by defective intestinal defense mechanisms in the absence of STING 44. By continuously producing CDNs, the gut microbiome can locally activate STING 12. This in turn affects the development of gut‐resident immune effectors such as goblet cells, innate lymphoid cells, and regulatory lymphocytes 44. The role of cGAS in these processes has not been addressed so far. STING, but not cGAS, affects chemically induced intestinal polyp formation 45. However, potent responses downstream of massive cell damage and DNA release may mask fine‐tuning of gut homeostasis by cGAS subsequent to eCDN sensing. The role of cGAS in tailoring the microbiome's composition and the pathogenesis of diseases in the intestine, as well as its role in systemic disorders influenced by the gut microbiome, thus warrants further investigations.
We demonstrate that cGAS is involved in the recognition of endocytosed CDNs. cGAS resides in the cytosol which, in contrast to the ER‐positioned STING, favors its access to vacuolar compartments. In line with our observations, the unrelated cytosolic sensors NOD1 and NOD2 are recruited to endosomes to sense their respective ligands 46. The endocytosed CDNs are presumably membrane bound within endosomes and may be released into the cytosol by an active process, for example, via a transporter or by random endosomal “sterile” damage. Previous work demonstrated that human multidrug transporter P‐glycoprotein (P‐gp) (also named MDR1 or ABCB1) is important for full activation of type I IFN responses against Listeria monocytogenes 47, indicating a possible role of P‐gp in exporting bacteria‐derived c‐di‐AMP from endosome to cytosol. However, whether a CDN transporter on endosomes is required for eCDN sensing warrants further investigation. Of note, the observations that endosome maturation is a prerequisite for eCDN‐induced innate immune activation and delivery of eCDNs to lysosomes raise the possibility that eCDNs enable STING activation after release into the cytosol from permeabilized lysosomes.
Sensing and binding to CDNs resulted in the dimerization of cGAS and generation of complexes characterized by a head‐to‐tail conformation and a mostly flexible N‐terminus. However, such biochemical processes seem insufficient for activation of the enzymatic activity of cGAS and subsequent synthesis of cGAMP. Indeed, reconstitution with mSTINGR231A initiated responses to dsDNA by sensing noncanonical 2′3′‐cGAMP generated by cGAS while lacking responsiveness to canonical CDNs 3. Yet, it did not rescue impaired c‐di‐GMP‐induced type I IFN responses in STING KO cells. The necessity for the catalytic domain of cGAS as indicated by absent immune activation in cGAS catalytic dead cells may reflect a requirement for the enzymatic site for trapping and transporting eCDNs.
Our data demonstrate that costimulation with eCDNs synergistically amplified type I IFN responses in macrophages upon concomitant DNA virus infection, indicating that eCDNs facilitate cGAS‐mediated DNA sensing. Recent studies indicate that the N‐terminus promotes formation of a cGAS‐DNA monomeric complex and enhances the functionality of this molecule 48. Whether conformational changes upon CDN binding enhance the role of the N‐terminus of cGAS in DNA binding and subsequent signaling remains to be evaluated.
The conformational changes in cGAS upon binding to eCDNs promote its interaction with STING, which may be important for the recruitment of the latter to the perinuclear region. Moreover, we observed that eCDNs promoted the formation of perinuclear puncta, which colocalized with cGAS, STING, and TBK1. Hence, it is tempting to propose that cGAS serves as a scaffolding protein which nucleates a complex composed of STING and TBK1 (Appendix Fig S5). The colocalization of endocytosed CDNs with STING at the perinuclear region points to a specialized compartment for STING aggregation/activation. These findings raise the question whether the interaction of cGAS with STING or the formation of the cGAMP/cGAS/STING complex is involved in sensing of dsDNA by cGAS, as well. This is supported by the finding that HEXIM1‐DNA‐PK‐paraspeckle components‐ribonucleoprotein complex (HDP‐RNP) has been established as a key nuclear regulator of DNA sensing through modulating the formation of a signalosome containing both cGAS and STING 49. The accurate mapping of essential amino acid residues critical for the interaction of cGAS with STING, without altering its DNA‐binding capacity or enzyme activity, will help to address this question. In addition, the autophagy‐related features of the perinuclear accumulation of STING 50 raise the question whether and how autophagy modulates the stability of the cGAS/STING/TBK1 signalosome complex and regulates sensing of eCDNs.
CDNs can activate bystander cells by transmission via gap junctions, exosomes, or budding viruses 18, 51, 52. Sensing of eCDNs via clathrin‐dependent endocytosis provides novel insights into the mechanisms underlying bystander cell activation. Damaged or dying infected cells can release host and bacterial CDNs and thereby signal adjacent cells. This strengthens the role of CDNs as alarmins and thereby opens novel avenues for better understanding of intra‐ and inter‐kingdom communication.
Materials and Methods
Reagents and plasmids
FITC‐cGAMP, biotinylated CDNs including c‐di‐AMP, c‐di‐GMP, and 2′3′‐cGAMP as well as beads coupled with c‐di‐AMP, c‐di‐GMP, and 2′3′‐cGAMP were purchased from BIOLOG Life Science Institute. CDNs, including c‐di‐AMP, c‐di‐GMP, 2′3′‐cGAMP, 2′2′‐cGAMP, 3′3′‐cGAMP, poly(I:C), and interferon stimulatory DNA (ISD), were all purchased from InvivoGen. Phosphodiesterase I from Crotalus adamanteus venom, Phorbol 12‐myristate 13‐acetate (PMA), 3‐methyladenine (3‐MA), bafilomycin A1 (BafA1), chlorpromazine (CPZ), dimethylamiloride (DMA), polyinosinic acid (polyI), mannans from Saccharomyces cerevisiae, and 4′,6′‐diamidino‐2‐phenylindole (DAPI) were obtained from Sigma‐Aldrich. Dynasore was purchased from Santa Cruz and ciliobrevin D from Millipore. The following antibodies were used: anti‐cGAS (D3O8O) (mouse specific) (31659), anti‐STING (D1V5L) (Rodent Preferred) (50494), anti‐phospho‐STING (Ser365) (D8F4W) (72971), anti‐TBK1 (3504), anti‐phospho‐TBK1 (Ser172) (5483), anti‐phospho‐IRF3 (Ser396) (29047), anti‐β‐actin (4970), horseradish peroxidase (HRP)‐conjugated anti‐rabbit, or anti‐mouse IgG (all from Cell Signaling); anti‐HA (H6908) and anti‐cGAS (both from Sigma‐Aldrich). The following antibodies were employed: anti‐cGAS (D1D3G, Cell Signaling), anti‐STING (R&D), anti‐phospho‐TBK1 (Ser172) (5483, Cell Signaling), anti‐ERp‐72 (5033, Cell Signaling), anti‐RCAS1 (12290, Cell Signaling), anti‐ERGIC/p58 (Santa Cruz), anti‐EEA1 (BD Biosciences), anti‐LAMP2 (H4B4), and anti‐dynein heavy chain (HC) (Santa Cruz). Plasmids encoding HA‐tagged human cGAS (HA‐cGAS) and human STING (HA‐STING) were purchased from InvivoGen. Mouse STING was purchased from Changsha YouBio Tech (Changsha, China). The corresponding mutated constructs were generated by site‐directed mutagenesis. HA agarose (A2095) used for immunoprecipitation (IP) was purchased from Sigma‐Aldrich.
Mice
Sting −/− mice on C57BL/6 background were kindly provided by Lei Jin (Albany Medical Center, New York, USA) through Bastian Opitz (Charite Medical University, Berlin, Germany). Cgas −/− mice on C57BL/6 background were originally from The Jackson Laboratory and kindly provided by Skip Virgin (Washington University School of Medicine in St. Louis, MO, USA). Sting −/− and Cgas −/− mice were also obtained from The Jackson Laboratory and kept under specific pathogen‐free (SPF) conditions at Tongji University. C57BL/6 mice were purchased from Charles River, Germany, or Shanghai Laboratory Animal Center, CAS, China, and used as WT control. Mice were 6–12 weeks of age for all experiments, matched for age and sex, and kept under specific pathogen‐free (SPF) conditions at the Max Planck Institute for Infection Biology in Berlin, Germany, and at the Tongji University, China. All animal experiments were performed according to institutional guidelines approved by the local ethics committees of the German authorities (Landesamtes für Gesundheit und Soziales Berlin; Landesamtes für Verbraucherschutz und Lebensmittelsicherheit, Animal Application T0087/13, T0157/15) and of Tongji University.
Cells
HEK293T cells (human embryonic kidney epithelial cells, ATCC CRL‐11268) and RAW264.7 cells (mouse macrophage cell line, ATCC TIB‐71) were cultured in DMEM (Gibco) and THP‐1 cells (human monocytic cell line, ATCC TIB‐202) in RPMI‐1640 (Gibco), both supplemented with 10% (v/v) heat‐inactivated fetal bovine serum (Sigma‐Aldrich, F0804), 1 mM sodium pyruvate (Gibco, 11360070), 2 mM l‐glutamine (Gibco, 25030081), 10 mM HEPES buffer (Gibco, 15630080), pH 7.2–7.5, and 50 μM 2‐mercaptoethanol (Gibco, 31350010). Cells were kept at 37°C in 5% CO2. THP‐1 cells were differentiated into macrophages by treatment with 200 nM PMA (Sigma‐Aldrich) for 24 h and then left rested for another 48 h for differentiation followed by subsequent experiments. CGAS KO THP‐1 cells (CGAS KO1) were generously provided by Veit Hornung (Ludwig‐Maximilians‐Universität München, Germany) 53. RAW264.7 cells deficient in Sting and complemented with corresponding genes were a kind gift from Denise M. Monack (Stanford University, Stanford, USA) 54. mBMDMs were obtained from tibial and femoral bones and generated with DMEM containing 20% L929 cell supernatant, 10% FCS, 5% heat‐inactivated horse serum, 1 mM sodium pyruvate, 2 mM l‐glutamine, and 10 mM HEPES buffer. All cells were mycoplasma‐free with regular checks performed by a LookOut Mycoplasma PCR (i.e., polymerase chain reaction) Detection Kit (MP0035, Sigma‐Aldrich).
Human primary monocyte cultures
The buffy coats were purchased from the blood bank of the Shanghai Red Cross. The study encompassed specimens from healthy donors and was approved by the Ethics Committee of the Shanghai Pulmonary Hospital (2018‐fk‐252). Peripheral blood mononuclear cells (PBMCs) were isolated according to their buoyant density using Percoll (Sigma‐Aldrich). Monocytes were purified with CD14+ magnetic microbeads, following a positive selection procedure, as indicated by the vendor (Miltenyi Biotec, DE). Cells were stimulated immediately after isolation.
Generation of stable cell lines
STING KD THP‐1 cells were generated as described previously 24. HEK293T cells stably expressing HA‐STING were generated by transfecting HEK293T cells with pcDNA3.1‐HA‐STING and selected using blasticidin (10 μg/ml). HA‐cGAS was subcloned into pCDH‐CMV‐MCS‐EF1‐Puro vector and transfected in HA‐STING stably expressing HEK293T cells followed by screening with puromycin (10 μg/ml).
LentiCRISPRv2 vectors were used to generate CGAS KO (i.e., knockout) cells. HEK293T cells were transfected by means of Lipofectamine 2000 with pSPAX2, pMD2.G, and LentiCRISPRv2 containing a guide (g)RNA that targeted human CGAS (CACGCAGTTATCAAAGCAG). Lentiviruses were collected 48 h later and were applied to infect THP‐1 cell. Subsequently, selection with puromycin (5 μg/ml) was carried out. Clones derived from single CGAS KO cells were obtained by serial dilutions in a 96‐well plate and were confirmed by Western blot. One confirmed CGAS KO clone was used for further experiment (CGAS KO2). To complement cGAS in CGAS KO THP‐1 cells, the CGAS KO1 cells were electroporated with HA‐cGAS or HA‐cGASE225A D227A subcloned in pcDNA3.1 plasmid (pcDNA3.1‐HA‐cGAS) followed by selection with G418 (400 μg/ml). To complement STING in Sting KO RAW264.7 cells, the KO cells were electroporated with HA‐m‐STING or HA‐m‐STINGR231A subcloned in pcDNA3.1 plasmid followed by selection with G418 (400 μg/ml).
Virus infection
HSV‐1 was collected from supernatants of infected Vero cells 55. Virus titer was determined by standard plaque assay. Cells were infected with HSV‐1 at indicated MOI in the presence of eCDNs at various concentrations for 4 h.
His‐ or GST‐tagged cGAS expression and purification
For purification of His‐tagged cGAS, the cDNA of h‐cGAS was subcloned into pET22b vector and transformed into E. coli BL21 (DE3). The bacteria were grown to optical density at 600 nm (OD600) of 0.5, followed by induction with isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) (0.5 mM) at 18°C O/N. Affinity chromatography was performed using 1 ml HisTrap HP (GE Healthcare; resuspension buffer containing 20 mM Na3PO4 pH 7.4, 500 mM NaCl, 40 mM imidazole, 10% glycerol, 10 mM MgCl2, 1 mM DTT, DNase I, and protease inhibitor (Roche); elution buffer 20 mM Na3PO4, 500 mM NaCl, 400 mM imidazole). Affinity purification was done using HiTrap Heparin HP (GE Healthcare) elution with 20 mM HEPES pH 7.4, 2 M NaCl and size exclusion chromatography (HiLoad Superdex 200 16/60 prep grade, GE Healthcare) with 20 mM HEPES pH 7.4, 500 mM NaCl. For purification of GST‐tagged cGAS, human cGAS cDNA was subcloned into pGEX‐4T‐1 vector and transfected into BL21(DE3) to express the protein. Bacteria grown in LB at OD600 around 0.8 were induced by IPTG (0.1 mM) overnight at 16°C. Recombinant GST‐cGAS was purified from bacterial lysates by GSTrap FF column (GE Healthcare). The concentration of the GST‐cGAS protein was measured by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Endotoxin in the purified protein was < 1,000 EU/mg.
Size exclusion chromatography
Binding of CDNs with cGAS was detected by performing analytical size exclusion chromatography of purified cGAS protein using Superdex 20010/300 (GE Healthcare). Absorption at 280 nm for cGAS, 3′3′‐cGAMP, and 2′3′‐cGAMP and at 245 nm for c‐di‐AMP was monitored. 20 μM cGAS was mixed at a stoichiometric ratio of 1:1.5 with 3′3′‐cGAMP, 2′3′‐cGAMP, or c‐di‐AMP. The area under the peak corresponds to a defined concentration of purified cGAS; loading the same amount of cGAS with CDNs results in a peak whose area was determined.
Fluorometry
Measurements were performed in a fluorescence spectrometer (Perkin Elmer LS55), using a 10‐mm path length quartz cell (Hellma). CDNs including c‐di‐AMP, 2′3′‐cGAMP, and 3′3′‐cGAMP were titrated to purified cGAS (0.85 μM) observing emission from 300 to 400 nm at 1‐nm intervals (excitation 290 nm). Scans were done in triplicates per sample. Excitation and emission slits were set to 5 nm.
cGAS enzyme activity assay
To detect the capability of purified human cGAS for in vitro synthesis of cGAMP, cGAS protein (1 μg) was incubated in a total volume of 20 μl of reaction buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2 with 1 mM ATP and 1 mM GTP) in the presence or absence of 0.3 μg DNA (ISD Naked, InvivoGen). The mixture was then incubated for 1 h at 37°C. The reaction was stopped by the addition of 100 μl chloroform/methanol (2:1, v/v). The aqueous phase was evaporated to dryness and dissolved in 10 μl of 50% acetonitrile, 0.1% formic acid. Detection of cGAMP was performed by UPLC on a Waters BEH Amide Column (1.7 μm, 2.1 × 100 mm) under isocratic conditions with 50% acetonitrile, 0.1% formic acid.
Immunoprecipitation and Western blot
To test the binding of CDNs to cGAS, HEK293T cells were transfected with HA‐cGAS by Lipofectamine 3000 (Invitrogen) and cell lysates were harvested and precipitated with beads coupled with 2′3′‐cGAMP, c‐di‐AMP, and c‐di‐GMP. For direct binding of CDNs with cGAS, recombinant cGAS protein purified as described above was incubated with beads coupled with 2′3′‐cGAMP, c‐di‐AMP, and c‐di‐GMP at 4°C O/N. To test the endogenous binding of 2′3′‐cGAMP with cGAS or STING, THP‐1 cells were stimulated with biotin‐ecGAMP (5 μg/ml) or biotin‐icGAMP (0.1 μg/ml) for indicated times followed by precipitation with Dynabeads MyOne Streptavidin C1 (Thermo Fisher). To test the direct interaction of cGAS with STING, GST or GST‐cGAS protein was incubated with His‐STING protein for 30 min at 4°C followed by incubation with Glutathione Sepharose 4B (GE Healthcare) at 4°C O/N for immunoprecipitation. To test the effect of cGAMP on the interaction of cGAS with STING, GST‐cGAS protein was incubated with His‐STING protein in the presence of cGAMP at indicated concentrations for 30 min at 4°C followed by incubation with Glutathione Sepharose 4B (GE Healthcare) at 4°C O/N for immunoprecipitation. For immunoblotting, cell lysates or precipitates in 1 × SDS protein sample buffer, with or without dithiothreitol (DTT) as the reducing agent, were denatured at 95°C for 8 min and then separated by 4–15% SDS–PAGE and transferred onto PVDF membranes. Blots were then incubated with indicated antibodies. ECL reagent (Thermo Scientific) was applied for immunoblotting.
Immunofluorescence assay
PMA‐differentiated THP‐1 cells were seeded on coverslips in 24‐well plates. Cells were treated with FITC‐2′3′‐cGAMP (FITC‐cGAMP) for indicated times. Stimulated cells were then fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature (RT). Cells were subsequently blocked and permeabilized in blocking buffer (2% BSA, 0.2% Triton X‐100 in PBS) for 30 min, followed by staining with the indicated antibody for 1 h at RT followed by staining with corresponding Alexa Fluor 555‐ or Alexa Fluor 647‐labeled anti‐rabbit, anti‐mouse, or anti‐sheep antibodies (Life Technologies) for 30 min at RT. Images were acquired using a Leica TCS SP8 confocal laser microscopy system (Leica Microsystems).
Cell stimulation
Murine BMDMs (2 × 106/ml), THP‐1 cells (1.5 × 106/ml), and RAW264.7 cells (2 × 106/ml) were seeded in 6‐well plates and transfected with poly(dA:dT), poly (I:C), or ISD using Lipofectamine 3000 (Invitrogen) according to manufacturer's instructions. CDNs were exogenously added to the culture medium or delivered to the cytosol by the addition of digitonin (10 μg/ml). HEK293T cells were transfected with indicated plasmids for 24 h for further analysis of type I IFN responses or for signaling. The inhibitors employed were 3‐MA (5 mM), BafA1 (1 μM), and ciliobrevin D (50 μM).
Real‐time quantitative reverse transcription PCR
RNA was isolated with TRIzol reagent as described by the manufacturer (Invitrogen). RNA (1 μg) was used to generate cDNA via the iScript cDNA Synthesis Kit (Bio‐Rad), and real‐time quantitative PCR was performed using Power SYBR green (Applied Biosystems) in a Roche LC480 thermocycler. The average threshold cycle of quadruplicate reactions was employed for all subsequent calculations using the ∆∆Ct method. Gene expression was normalized to glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH). Real‐time quantitative reverse transcription PCR (qRT–PCR) data were average from at least three independent experiments, with two technical replicates per experiment. Primer sequences were from PrimerBank and listed as follows: h‐GAPDH forward, 5′‐GGAGCGAGATCCCTCCAAAAT‐3′, h‐GAPDH reverse, 5′‐GGCTGTTGTCATACTTCTCATGG‐3′; h‐IFNB1 forward, 5′‐ATGACCAACAAGTGTCTCCTCC‐3′, h‐IFNB1 reverse, 5′‐GGAATCCAAGCAAGTTGTAGCTC‐3′; h‐IL6 forward, 5′‐ACTCACCTCTTCAGAACGAATTG‐3′, h‐IL6 reverse,5′‐CCATCTTTGGAAGGTTCAGGTTG‐3′; m‐Gapdh forward, 5′‐AGGTCGGTGTGAACGGATTTG‐3′, m‐Gapdh reverse, 5′‐TGTAGACCATGTAGTTGAGGTCA‐3′; m‐Ifnb1 forward, 5′‐CAGCTCCAAGAAAGGACGAAC‐3′, m‐Ifnb1 reverse, 5′‐GGCAGTGTAACTCTTCTGCAT‐3′; m‐Il6 forward, 5′‐TAGTCCTTCCTACCCCAATTTCC‐3′; m‐Il6 reverse, 5′‐TTGGTCCTTAGCCACTCCTTC‐3′.
IFNβ measurement
Cell culture supernatants were removed from cells stimulated with indicated ligands and centrifuged for detection of IFNβ (PBL Interferon Source) by ELISA according to manufacturer's instructions.
cGAMP uptake assay
HEK293T cells and PMA‐differentiated THP‐1 cells were pretreated with DMSO or different inhibitors (Dynasore, 10 μM; CPZ, 10 μM; DMA, 100 μM; Poly I, 50 μg/ml; Mannans, 1 mg/ml) for 30 min and then kept on ice for 10 min followed by stimulation with FITC‐ecGAMP or FITC‐icGAMP in prewarmed Opti‐MEM medium (Thermo Fisher Scientific) for indicated times in the presence of DMSO or indicated inhibitors. Cells were washed and analyzed using a BD LSR II flow cytometer. Frequencies of FITC+ cells were calculated.
Binding kinetics of cGAS to 2′3′‐cGAMP
Label‐free binding kinetics were measured with a microarray‐compatible optical biosensor oblique‐incidence reflectivity difference (OI‐RD) scanning microscope 26. Each microarray experiment consisted of 2′3′‐cGAMP and purified cGAS protein with each printed in triplicate on an epoxy‐functionalized glass slide (CapitalBio Corporation, China) at concentrations of 10 mM and 1.33 μM. Six identical microarrays were fabricated on each glass slide. The printed glass slide then was assembled into a fluidic cartridge, with each microarray housed in a separate chamber. Before the binding reaction proceeded, the slide was washed in situ with a flow of 1 × PBS to remove excess unbound samples. Subsequently, blocking was performed with 7,600 nM of BSA (Sigma‐Aldrich) in 1 × PBS for 30 min. PBS (1×) was passed through a reaction chamber at a flow rate of 0.01 ml/min for 9 min to acquire a baseline reading. The PBS then was quickly replaced with GST‐labeled cGAS solution at a flow rate of 2 ml/min. The flow rate then was reduced to 0.01 ml/min, and the microarray was incubated in the cGAS solution for 31 min. This constituted the association phase of the reaction. The cGAS solution was then quickly replaced with 1 × PBS at a flow rate of 2 ml/min, and the flow rate then was reduced to 0.01 ml/min to allow dissociation of cGAS for 40 min. This was the dissociation phase of the reaction. By repeating the cGAS binding reactions at concentrations of 248, 124, and 62 nM on separate fresh microarrays, binding curves of purified cGAS with 2′3′‐cGAMP were determined at three concentrations. Reaction kinetic rate constants were estimated by fitting the binding curves globally using a 1‐to‐1 Langmuir reaction model 26.
SAXS data collection and analysis
SAXS data from cGAS samples were collected at the P12 beamline EMBL‐Hamburg, Petra‐III ring, DESY, Germany, using a Pilatus 2 M detector (Dectris) covering the momentum transfer range 0.002 < s < 4.989 nm−1, where s = 4π sin(θ)/λ (where 2θ is the scattering angle and λ = 1.24 Å is the X‐ray wavelength). A sample‐detector distance of 3.1 m and an exposure time of 1 s were employed. For each SAXS measurement, 90 μl of affinity‐purified protein sample was loaded onto a Superdex 200 10/300 GL SEC column (GE Healthcare). Samples eluting from the SEC were directed to the SAXS flow cell for scattering measurements. For each individual sample, the scattering profiles over the elution peak were averaged and used for further analysis. Buffer scattering profiles were obtained from the SAXS frames collected prior to the sample elution peaks to allow for background subtraction.
Model‐free parameters
The extrapolated forward scattering (I(0)) and radius of gyration (Rg) were determined using PRIMUS from the ATSAS suite 56. The indirect Fourier transformation approach of the program GNOM was used to determine the pair distance distribution function and the maximum particle dimensions D max 57 (Appendix Table S1).
Structural modeling against SAXS data
Ab initio models were reconstructed from the scattering data using the simulated annealing‐based bead modeling program DAMMIF 58. Ten independent reconstructions were averaged to generate a representative model with the program DAMAVER 59 In addition, the average DAMMIF ab initio model was used to calculate an excluded volume of the particle, VDAM, from which an independent MW estimate can be derived (empirically, MMDAM ~ VDAM/2). Additionally, the MW estimates were derived from the scattering data based on the hydrated volume Vp computed using Porod analysis 60. Resolutions of the ab initio model ensembles were computed using a Fourier shell correlation (FSC)‐based approach (Appendix Table S1) 61.
The program EOM was employed for SAXS‐based structural modeling to test whether the experimental data could be fitted by an ensemble of structures 62. In the case of the cGAS apo sample, a pool of structures with a flexible N‐terminus (amino acid residues 1–160) modeled with coarse‐grained residues was generated using the atomic X‐ray crystallographic structure of human apo‐cGAS as a starting point (amino acid residues 161–522, PDB: 4O68). The modeling of cGAS dimer with bound 2′3′‐cGAMP was based on the human cGAS dimer X‐ray crystallographic structure with the cGAMP (PDB: 4O67). Similarly, the N‐terminus (amino acid residues 1–160) was allowed to be flexible. Using the genetic algorithm GAJOE, we tested fitting of the experimental scattering data with selected ensembles from individual pools of monomers for apo‐cGAS and dimers for cGAS with cGAMP. 10,000 were generated for each pool and the maximum ensemble size was restricted to 50 entities.
Statistical analysis
Statistical analysis was performed by two‐tailed Student's t‐test or one‐way ANOVA followed by Dunnett's post hoc test or two‐way ANOVA followed by Tukey's post hoc test or Mann–Whitney U‐test using GraphPad Prism 7 (GraphPad Software). All data are expressed as mean + SD of the averages of technical replicates from indicated number of independent experiments. Differences with values of P < 0.05 were considered statistically significant.
Author contributions
HL, AD, and SHEK conceived and designed the study and wrote the manuscript. HL, AD, MKo, and SHEK designed the experiments and performed data analysis. HL performed most of the experiments with help from XW, FW, SL and MM. PM‐A generated KD cells, with help from MKl and UG‐B, and performed quantitative RT–PCR. GP generated cGAS complemented THP‐1 cells and performed quantitative RT–PCR. YF and CZ performed the label‐free biomolecular interaction assay to detect the binding kinetics of cGAS to 2′3′‐cGAMP. H‐JM, RH, A‐BK, DO‐M and KH provided technical help. BG provided helpful discussions. AT and MK performed the structural modeling of cGAS. All authors commented on the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
The authors thank Nayoung Kwak (Max Planck Institute for Infection Biology, Berlin, Germany) for the purification of cGAS and performing size exclusion chromatography assay; B. Opitz (Charité, Berlin, Germany), V. Hornung (Ludwig‐Maximilians‐Universität München, Germany), Skip Virgin (Washington University School of Medicine in St. Louis, MO, USA), L. Jin (Albany Medical College, Albany, USA), and D.M. Monack (Stanford University, Stanford, USA) for providing reagents and experimental tools; and L. Lozza (Max Planck Institute for Infection Biology, Berlin, Germany) and V. Brinkmann (Max Planck Institute for Infection Biology, Berlin, Germany) for assistance with flow cytometry and confocal microscopy, respectively. The authors greatly thank H. Su (Shanghai Pulmonary Hospital, Tongji University, Shanghai, China) for assistance with the design of the photograph for synopsis. The authors gratefully acknowledge M.L. Grossman and S. Sibaei for excellent editorial assistance They are supported by intramural funding of the Max Planck Society to S.H.E.K. and received grants from National Natural Science Foundation of China (81200003, 8170006 and 81370108) to H.L. H.L. is also sponsored by the Shanghai Pujiang Program (16PJ1408600) and the Shanghai Medical and Health Services Outstanding Youth Talent Program (2017YQ078). M. Kolbe acknowledges grant support from the European Union's Seventh Framework Programmes (EU‐FP7/2007‐2013), ERC Grant‐No. 311374. Materials may be requested upon signing a material transfer agreement.
EMBO Reports (2019) 20: e46293
Contributor Information
Anca Dorhoi, Email: anca.dorhoi@fli.de.
Stefan HE Kaufmann, Email: kaufmann@mpiib-berlin.mpg.de.
Data availability
The SAXS data of apo‐cGAS (code SASDEP9; https://www.sasbdb.org/data/SASDEP9/) and the cGAS/cGAMP complex (code SASDEQ9; https://www.sasbdb.org/data/SASDEQ9/) were deposited in the small‐angle scattering biological databank (SASDB).
References
- 1. Iwasaki A, Medzhitov R (2010) Regulation of adaptive immunity by the innate immune system. Science 327: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ et al (2009) A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic‐di‐GMP. J Exp Med 206: 1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burdette DL, Monroe KM, Sotelo‐Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE (2011) STING is a direct innate immune sensor of cyclic di‐GMP. Nature 478: 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL (2011) MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic‐di‐AMP and cyclic‐di‐GMP. J Immunol 187: 2595–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee JH, Bishai WR (2015) A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat Med 21: 401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woodward JJ, Iavarone AT, Portnoy DA (2010) c‐di‐AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328: 1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W et al (2012) The helicase DDX41 recognizes the bacterial secondary messengers cyclic di‐GMP and cyclic di‐AMP to activate a type I interferon immune response. Nat Immunol 13: 1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xia P, Wang S, Xiong Z, Zhu X, Ye B, Du Y, Meng S, Qu Y, Liu J, Gao G et al (2018) The ER membrane adaptor ERAdP senses the bacterial second messenger c‐di‐AMP and initiates anti‐bacterial immunity. Nat Immunol 19: 141–150 [DOI] [PubMed] [Google Scholar]
- 10. McFarland AP, Luo S, Ahmed‐Qadri F, Zuck M, Thayer EF, Goo YA, Hybiske K, Tong L, Woodward JJ (2017) Sensing of bacterial cyclic dinucleotides by the oxidoreductase RECON promotes NF‐kappaB activation and shapes a proinflammatory antibacterial state. Immunity 46: 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai X, Chiu YH, Chen ZJ (2014) The cGAS‐cGAMP‐STING pathway of cytosolic DNA sensing and signaling. Mol Cell 54: 289–296 [DOI] [PubMed] [Google Scholar]
- 12. Danilchanka O, Mekalanos JJ (2013) Cyclic dinucleotides and the innate immune response. Cell 154: 962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ (2013) Pivotal roles of cGAS‐cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341: 1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen W, Kuolee R, Yan H (2010) The potential of 3′,5′‐cyclic diguanylic acid (c‐di‐GMP) as an effective vaccine adjuvant. Vaccine 28: 3080–3085 [DOI] [PubMed] [Google Scholar]
- 15. Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y et al (2007) Bacterial c‐di‐GMP is an immunostimulatory molecule. J Immunol 178: 2171–2181 [DOI] [PubMed] [Google Scholar]
- 16. Blaauboer SM, Mansouri S, Tucker HR, Wang HL, Gabrielle VD, Jin L (2015) The mucosal adjuvant cyclic di‐GMP enhances antigen uptake and selectively activates pinocytosis‐efficient cells in vivo . Elife 4: e06670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V (2013) cGAS produces a 2′‐5′‐linked cyclic dinucleotide second messenger that activates STING. Nature 498: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, Rehwinkel J (2015) Viruses transfer the antiviral second messenger cGAMP between cells. Science 349: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marina‐Garcia N, Franchi L, Kim YG, Hu Y, Smith DE, Boons GJ, Nunez G (2009) Clathrin‐ and dynamin‐dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol 182: 4321–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mansour MK, Schlesinger LS, Levitz SM (2002) Optimal T cell responses to Cryptococcus neoformans mannoprotein are dependent on recognition of conjugated carbohydrates by mannose receptors. J Immunol 168: 2872–2879 [DOI] [PubMed] [Google Scholar]
- 21. Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y (1991) Bafilomycin A1, a specific inhibitor of vacuolar‐type H(+)‐ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 266: 17707–17712 [PubMed] [Google Scholar]
- 22. Mauvezin C, Neufeld TP (2015) Bafilomycin A1 disrupts autophagic flux by inhibiting both V‐ATPase‐dependent acidification and Ca‐P60A/SERCA‐dependent autophagosome‐lysosome fusion. Autophagy 11: 1437–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seglen PO, Gordon PB (1982) 3‐Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 79: 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saiga H, Nieuwenhuizen N, Gengenbacher M, Koehler AB, Schuerer S, Moura‐Alves P, Wagner I, Mollenkopf HJ, Dorhoi A, Kaufmann SH (2015) The recombinant BCG DeltaureC:hly vaccine targets the AIM2 inflammasome to induce autophagy and inflammation. J Infect Dis 211: 1831–1841 [DOI] [PubMed] [Google Scholar]
- 25. Lau L, Gray EE, Brunette RL, Stetson DB (2015) DNA tumor virus oncogenes antagonize the cGAS‐STING DNA‐sensing pathway. Science 350: 568–571 [DOI] [PubMed] [Google Scholar]
- 26. Landry JP, Fei Y, Zhu X (2012) Simultaneous measurement of 10,000 protein‐ligand affinity constants using microarray‐based kinetic constant assays. Assay Drug Dev Technol 10: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam CA, Zhang X, Chen ZJ (2014) The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch‐like conformational changes in the activation loop. Cell Rep 6: 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kranzusch PJ, Lee ASY, Wilson SC, Solovykh MS, Vance RE, Berger JM, Doudna JA (2014) Structure‐guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158: 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, Gray NS, Mekalanos JJ, Kranzusch PJ (2018) Structure of the human cGAS‐DNA complex reveals enhanced control of immune surveillance. Cell 174: 300–311 e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barber GN (2014) STING‐dependent cytosolic DNA sensing pathways. Trends Immunol 35: 88–93 [DOI] [PubMed] [Google Scholar]
- 31. Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N (2015) STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV et al (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347: aaa2630 [DOI] [PubMed] [Google Scholar]
- 33. Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524–530 [DOI] [PubMed] [Google Scholar]
- 34. Roossien DH, Miller KE, Gallo G (2015) Ciliobrevins as tools for studying dynein motor function. Front Cell Neurosci 9: 252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.(2018) Nuclear cGAS blocks DNA repair to drive tumorigenesis. Cancer Discov 8: 1506 [Google Scholar]
- 36. Otto G (2018) Inhibition by nuclear cGAS. Nat Rev Mol Cell Biol 19: 752–753 [DOI] [PubMed] [Google Scholar]
- 37. Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, Jiang Y, Fei Y, Zhu C, Tan R et al (2018) Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563: 131–136 [DOI] [PubMed] [Google Scholar]
- 38. Monroe KM, McWhirter SM, Vance RE (2010) Induction of type I interferons by bacteria. Cell Microbiol 12: 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE (2013) The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 3: 1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, Liu W et al (2015) STING agonist formulated cancer vaccines can cure established tumors resistant to PD‐1 blockade. Sci Transl Med 7: 283ra52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ et al (2015) Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep 11: 1018–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abdul‐Sater AA, Tattoli I, Jin L, Grajkowski A, Levi A, Koller BH, Allen IC, Beaucage SL, Fitzgerald KA, Ting JP et al (2013) Cyclic‐di‐GMP and cyclic‐di‐AMP activate the NLRP3 inflammasome. EMBO Rep 14: 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jakobsen MR (2018) ERAdP standing in the shadow of STING innate immune signaling. Nat Immunol 19: 105–107 [DOI] [PubMed] [Google Scholar]
- 44. Canesso MCC, Lemos L, Neves TC, Marim FM, Castro TBR, Veloso ES, Queiroz CP, Ahn J, Santiago HC, Martins FS et al (2017) The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal Immunol 11: 820–834 [DOI] [PubMed] [Google Scholar]
- 45. Ahn J, Son S, Oliveira SC, Barber GN (2017) STING‐dependent signaling underlies IL‐10 controlled inflammatory colitis. Cell Rep 21: 3873–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, de Maziere A, Klumperman J, Schlatter M, Delamarre L, Mellman I (2014) Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509: 240–244 [DOI] [PubMed] [Google Scholar]
- 47. Sigal N, Kaplan Zeevi M, Weinstein S, Peer D, Herskovits AA (2015) The human P‐glycoprotein transporter enhances the type I interferon response to Listeria monocytogenes infection. Infect Immun 83: 2358–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tao J, Zhang XW, Jin J, Du XX, Lian T, Yang J, Zhou X, Jiang Z, Su XD (2017) Nonspecific DNA binding of cGAS N terminus promotes cGAS activation. J Immunol 198: 3627–3636 [DOI] [PubMed] [Google Scholar]
- 49. Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, Severac D, Dubois E, Schwartz O, Bennasser Y, Benkirane M (2017) HEXIM1 and NEAT1 long non‐coding RNA form a multi‐subunit complex that regulates DNA‐mediated innate immune response. Mol Cell 67: 387–399 e5 [DOI] [PubMed] [Google Scholar]
- 50. Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T et al (2009) Atg9a controls dsDNA‐driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA 106: 20842–20846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ablasser A, Schmid‐Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V (2013) Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503: 530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G et al (2015) Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science 349: 1232–1236 [DOI] [PubMed] [Google Scholar]
- 53. Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid‐Burgk JL, Schmidt T, Hornung V, Cole ST et al (2015) Mycobacterium tuberculosis differentially activates cGAS‐ and inflammasome‐dependent intracellular immune responses through ESX‐1. Cell Host Microbe 17: 799–810 [DOI] [PubMed] [Google Scholar]
- 54. Storek KM, Gertsvolf NA, Ohlson MB, Monack DM (2015) cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J Immunol 194: 3236–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou Y, He C, Yan D, Liu F, Liu H, Chen J, Cao T, Zuo M, Wang P, Ge Y et al (2016) The kinase CK1varepsilon controls the antiviral immune response by phosphorylating the signaling adaptor TRAF3. Nat Immunol 17: 397–405 [DOI] [PubMed] [Google Scholar]
- 56. Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM et al (2017) ATSAS 2.8: a comprehensive data analysis suite for small‐angle scattering from macromolecular solutions. J Appl Crystallogr 50: 1212–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Svergun DI (1992) Determination of the regularization parameter in indirect‐transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- 58. Franke D, Svergun DI (2009) DAMMIF, a program for rapid ab‐initio shape determination in small‐angle scattering. J Appl Crystallogr 42: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small‐angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Porod G (1982) Small‐angle X‐ray scattering. London: Academic Press; [Google Scholar]
- 61. Tuukkanen AT, Kleywegt GJ, Svergun DI (2016) Resolution of ab initio shapes determined from small‐angle scattering. IUCrJ 3: 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tria G, Mertens HD, Kachala M, Svergun DI (2015) Advanced ensemble modelling of flexible macromolecules using X‐ray solution scattering. IUCrJ 2: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Data Availability Statement
The SAXS data of apo‐cGAS (code SASDEP9; https://www.sasbdb.org/data/SASDEP9/) and the cGAS/cGAMP complex (code SASDEQ9; https://www.sasbdb.org/data/SASDEQ9/) were deposited in the small‐angle scattering biological databank (SASDB).


