Abstract
Host–microbiome interactions constitute key determinants of host physiology, while their dysregulation is implicated in a wide range of human diseases. The microbiome undergoes diurnal variation in composition and function, and this in turn drives oscillations in host gene expression and functions. In this review, we discuss the newest developments in understanding circadian host–microbiome interplays, and how they may be relevant in health and disease contexts. We summarize the molecular mechanisms by which the microbiome influences host function in a diurnal manner, and inversely describe how the host orchestrates circadian rhythmicity of the microbiome. Furthermore, we highlight the future perspectives and challenges in studying this new and exciting facet of host–microbiome interactions. Finally, we illustrate how the elucidation of the microbiome chronobiology may pave the way for novel therapeutic approaches.
Keywords: circadian, diurnal, microbiome, rhythm
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Molecular Biology of Disease
Glossary
- BMAL1
Brain and muscle ARNT‐like protein
- CLOCK
circadian locomotor output cycles kaput
- E. aerogenes
Enterobacter aerogenes
- GF
germ‐free
- HFD
high‐fat diet
- ILC
innate lymphoid cell
- linc‐RNA
large intergenic non‐coding RNAs
- NAD
nicotinamide adenine dinucleotide
- NLR
nod‐like receptor
- PER
period
- SCN
suprachiasmatic nucleus
- SFB
segmented filamentous bacterium
- SPF
specific pathogen‐free mice
- TLR
Toll‐like receptor
Introduction
The circadian clock
Diurnal changes of the environment, termed circadian to highlight their circa diem or approximate 24‐h cycling period, have shaped life's evolution on Earth 1, 2, 3, 4, 5. The ability to anticipate circadian environmental changes conferred an evolutionary advantage 3, 4, 5. Under this evolutionary pressure, living organisms on Earth have developed molecular mechanisms that allowed them to synchronize biological processes with the changing time of day, and to time them with associated light and darkness conditions. These molecular mechanisms termed circadian clocks have unique properties that separate them from other oscillatory processes. Circadian clocks are defined as self‐sustained, temperature‐compensated, and entrainable oscillators. Structurally, circadian clocks consist of an input pathway that collects environmental cues and subsequently transmits them to the central oscillator that generates a 24‐h rhythm based on the signals from the input pathway. This is linked to output pathways that synchronize circadian time between the central clocks and the periphery, controlling various metabolic, physiological, and behavioral processes 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18. In mammals, circadian clocks feature a two‐tier hierarchical structure: a (i) master or “central” clocks that are located in the suprachiasmatic nucleus (SCN) of the hypothalamus that in turn orchestrates and synchronizes (ii) “peripheral” clocks within each cell of the body (Fig 1). At its core lies a negative feedback loop with a period of ∼24 h controlled by CLOCK (circadian locomotor output cycles kaput) and ARNTL (aryl hydrocarbon receptor nuclear translocator‐like protein 1), which regulate the rhythmic expression of up to 15% of the entire transcriptome of each mammalian cell 19. Prokaryotic circadian clocks on the other hand are much less understood and have primarily been studied in light‐responsive cyanobacteria. However, recent studies have identified circadian clocks also in light‐independent prokaryotes 20. In a remarkably simple and elegant system in cyanobacteria, a circadian clock of just three proteins (KaiA, KaiB, and KaiC) can be sustained even in the absence of transcription, although transcriptional feedback is required for its stability 21, 22, 23, 24. Circadian clocks of light‐nonresponsive bacteria residing in diurnally changing environments are much less studied. In a pioneering study, Paulose et al 20 have described a circadian oscillator of Enterobacter aerogenes, a member of the human gut microbial community, also termed the human gut microbiome. The circadian oscillator of E. aerogenes is entrained by the human pineal and gastrointestinal hormone melatonin, having the same period as the human central circadian clocks of approximately 24 h. Furthermore, it is temperature compensated within the range of human body temperatures and it also regulates swarming activity of E. aerogenes. Bioinformatic analyses have revealed that circadian clocks of E. aerogenes are similar to those of cyanobacteria 20. This suggests that other members of the human gut microbiome may also possess inherent circadian clocks. Indeed, the human gastrointestinal tract constitutes a diurnally changing environment, in which a multitude of circadian factors, including temperature, pH, nutrient availability, and gut motility, would make an adaptive circadian clock beneficial for some gut bacteria.
Figure 1. The two‐tier hierarchical structure of mammalian circadian clocks.
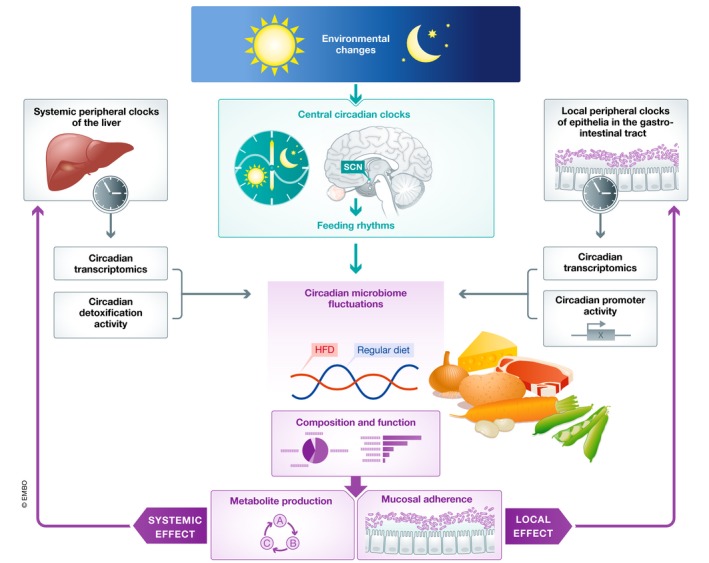
The gut microbiome undergoes diurnal changes in composition and function that are entrained by feeding rhythms. In turn, microbiome circadian fluctuations of the microbiome modulate circadian activity of peripheral mammalian clocks, both locally (right), through mucosal attachment, and systemically (left), through the production of metabolites.
Diurnally shifting host–microbiome interactions may also bear disease‐relevant importance. Disruption of the circadian clock became a hallmark of modern lifestyle brought about by the invention of electric light that made shift‐work widespread and the increase in global traveling activity across time zones resulting in jet‐lag (Fig 2). Chronic disruption of the circadian clock has been associated with a multitude of diseases, including obesity, diabetes, cancer, cardiovascular, psychiatric, and neurodegenerative diseases, and susceptibility to infection 6, 25, 26, 27. Notably, these very conditions have been also associated with an aberrant composition of the intestinal microbiome community, termed microbiome dysbiosis, which is characterized by a sharp drop in diversity and reduced colonization resistance 28, 29, 30, 31 (Fig 2). The association between circadian clock disruption and dysbiosis described in human studies can be recreated in animal models. Indeed, circadian clock disruption, either through genetic impairment of core clock components or their environmental disarrangement in rodent models, leads to gut microbiome dysbiosis 32, 33. Dysbiosis contributes to the adverse metabolic consequences of clock disruption, as antibiotic treatment ameliorates obesity and glucose intolerance in mice with environmental circadian clock disruption, while microbiome transfer from jet‐lagged mice and humans into germ‐free mice fully recapitulates metabolic disease manifestations in a new host 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45. These observations demonstrate that diseases induced by circadian clock disruption can be mediated by aberrations in microbiome composition and function. They also suggest that the microbiome might constitute a previously unrecognized link between the rise of microbiome‐modulated diseases and misalignment between body clock and geophysical time.
Figure 2. Disruption of the circadian clock as a hallmark of modern lifestyle.
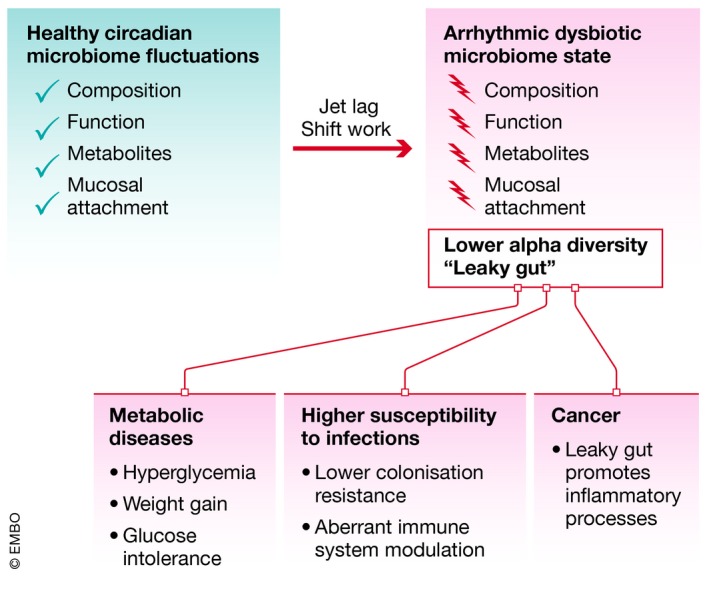
Disruption of circadian microbiome fluctuations results in the establishment of an arrhythmic dysbiotic microbiome state, promoting the development or progression of several diseases, including metabolic syndrome, infections, and cancer.
The gut microbiome
The human body is colonized by a diverse community of microorganisms, termed the human microbiome. The complexity and composition of microbiomes colonizing various barrier tissues of the human body varies, even more so between individuals 46, 47. Recent years have seen a remarkable progress in our understanding of the role the human microbiome plays in health and disease (for comprehensive reviews see 48, 49, 50, 51, 52, 53). Briefly, the “forgotten organ” of gut microbiome was shown to be involved in almost all facets of human health: in the maturation and continued education of the host immune response 54, as an effector of host metabolism, including energy harvest from food 55 and changing host propensity toward weight gain 56, as a contributor to host metabolic homeostasis 57, but also in providing protection against pathogen overgrowth and influencing host‐cell proliferation 58 and gut vascularization 59, regulating neurologic signaling 60 and bone density 61, providing a source of energy biogenesis 62, biosynthesizing vitamins 63 and neurotransmitters 60, metabolizing bile salts, thereby reacting to or modifying drugs, and eliminating exogenous toxins 64, 65. The importance of the gut microbiome for human health is exemplified by observations that a disrupted microbiome exacerbates immune disorders such as auto‐inflammatory colitis 66, while a healthy microbiome can be used therapeutically to cure life‐risking infections 67. While the gut microbiome strongly influences gastrointestinal homeostasis, in addition to local effects, the microbiome may exert its influence systemically. One such example is featured in a mouse model of autism, in which transfer of a healthy microbiome ameliorated some neurological features of this disease 68. All of the above‐mentioned microbiome‐mediated effects on human health are subject to microbiome composition. One of the processes that shapes microbiome configuration is aging 69. The initial colonization of an infant by the microbiome occurs at birth, upon passing through the birth canal. Subsequently, the composition of the neonatal microbiome is primarily shaped by microbial transfer from maternal microbiome communities, including the ones colonizing skin and oral cavity. The neonatal gut microbiome is also strongly influenced by lactic compounds, which promote the growth of advantageous bacteria. As a result of the microbial transfer from the mother, the microbiome of the infant increases in heterogeneity and diversity 70, which in turn 70 leads to enhanced immune maturity and specificity 71. Upon reaching adulthood, microbiome composition stabilizes, with a retention rate of approximately 60% over 5 years 72. However, the composition of the microbiome maintains a rate of change well into old age, with distinct microbiomes noted in elderly subjects 73. The factors that affect long‐term microbiome stability include perturbations as exposure to major dietary changes 74, 75, drugs, antibiotics 76, and food supplements 76, 77. For example, a drastic change in the amount of dietary fiber consumption results in significant microbiome composition rearrangements within 24 h that last for several weeks 74. In addition to diet content, a major change in overall nutritional caloric value also results in a systemic and persisting change of the microbiotic Firmicutes‐to‐Bacteroidetes ratio 78. The effects of the microbiome mentioned above are governed by its composition and function, which is dynamic and potentially amenable to change in contrast to host genetics, making it a promising hub for therapeutic targeting.
Mechanisms of circadian host–microbiome crosstalk
The human–microbiome meta‐organism constitutes a multi‐domain ecosystem, in which the interaction of eukaryotic and prokaryotic symbionts must not only be precisely synchronized with one another, but also with the environmental changes over the course of a day.
Modulation of the gut microbiome by circadian clocks of the host
In recent years, a new facet of host–microbiome interactions has been identified in the gut, consisting of circadian fluctuations in the composition and function of the intestinal microbiome 33. The relative abundance of 20% of all microbiome species in both mice and humans undergoes diurnal fluctuations. As a result, time of day‐specific configurations of the intestinal microbiota can be identified (Fig 1). For example, relative abundance of the commensal genus Lactobacillus declines during the active phase and increases during the resting phase in both mice and humans 33, 35. Interestingly, this phenomenon does not appear to be dependent on a particular microbiota configuration, as diurnal microbiome oscillations can be observed in mice and humans that feature inter‐individual variability in the composition of their baseline commensal gut bacteria. In addition to compositional fluctuations, microbiome communities appear to be performing specific tasks predominantly during one or the other time‐period: detoxification and chemotaxis during the resting phase, and energy harvest, DNA repair, and cell growth during the active phase of the host (Fig 1). Furthermore, the gut microbiome exhibits rhythmic biogeographic localization, adherence to the intestinal epithelium, and rhythmic metabolite secretion (Fig 1). These diurnal fluctuations have both local and systemic consequences for the host: Through rhythmic adherence to the intestinal epithelium, the microbiome is able to modulate chromatin and transcriptional oscillations in intestinal cells, while far‐reaching metabolites allow the microbiome to shape hepatic circadian gene expression and detoxification reactions 33, 79, 80 (Fig 1). These microbiome oscillations depend on the cues from the host's circadian clocks, as mice lacking two major components of the mammalian molecular clock, PER1 and PER2, do not feature daily rhythmicity in bacterial composition and function 33. This indicates that the activity of the meta‐organism is synchronized through entrainment of microbial oscillations by circadian clocks of the host. The exact mechanism behind microbiome compositional and functional oscillations remains unknown. This synchronization may confer a range of evolutionary advantages for the host–microbiome meta‐organism, including colonization resistance toward pathogenic species and the detoxification of noxious xenobiotics. The probability of an encounter of the mentioned noxious xenobiotics and pathogenic species is not constant through the day, suggesting that anticipatory circadian mechanisms may play an important role in maintaining homeostasis. Among the most important diurnal environmental changes is nutrient availability. Indeed, circadian clock machinery and metabolic pathways are closely intertwined, with the NAD pathway being directly coupled to core components of mammalian clocks 81, 82, 83, 84. The interconnectivity of metabolic pathways and the circadian clock is necessary to synchronize the anabolic and catabolic pathways of energy turnover with diurnal variations in nutrient intake. Indeed, time of nutrient consumption is a central driver of peripheral body clocks: Feeding in restricted periods of the light–dark cycle uncouples peripheral clocks from the central master clock in the SCN; mice with a genetically impaired circadian clock not only lose most transcriptome oscillations, but also their strict nocturnal feeding pattern. However, restricting access to food to the dark part of the cycle rescues, at least in part, loss of transcriptional and behavioral rhythms in these mice, suggesting that oscillatory processes exist which are entrained by feeding rhythms, and can persist in the absence of a functional circadian clock 85, 86. This paradigm of food‐entrainable clocks is crucial when understanding the host's circadian interaction with the intestinal microbiome. Restricting food access in mice to the dark or light phase induced a phase shift in microbiome oscillations, while the overall number of oscillatory commensals remained constant. Furthermore, the loss of bacterial oscillations in arrhythmic Per1/2‐deficient mice was restored by subjecting them to scheduled feeding 33. Therefore, feeding times are a dominant driver of the temporal orchestration of microbiome activity (Fig 1). Supporting the notion that microbiome circadian changes are controlled by central circadian clocks, Wu et al 87 recently described the effects of exposure to light on circadian microbiome fluctuations.
In addition to the time of consumption, the type of food also has an effect on the clock machinery. Mice fed a high‐fat diet exhibit attenuated clock gene oscillations, alterations in locomotor activity rhythms, and massive reprogramming of the circadian transcriptome 88, 89. In addition, high‐fat diet alters the temporal organization of the microbiome: Cyclic bacteria species are not identical between mice on HFD and controls, and the overall number of cyclic bacteria species in mice on HFD is reduced 35. The type of food therefore seems to determine the extent and amplitude of microbiome oscillations (Fig 1). Interestingly, time‐restricted feeding rescues the adverse metabolic effects of high‐fat feeding on the host 34, suggesting that not the content of the diet per se, but rather the mistiming of feeding is driving the onset of metabolic disease. Therefore, apart from the entrainment of the host circadian clocks, feeding rhythmicity also shapes the symbiotic community of microorganisms colonizing the mammalian intestine.
The impact of circadian microbiome fluctuations on the host
The complex network of host–microbiome interactions is in many cases bilateral. As such, the host impacts the diurnal activity of the gut microbiome through its clock machinery and feeding behavior (see above), while the diurnally shifting microbiome conversely impacts the host circadian activity. Indeed, different members of the microbiota impact the host in varying ways, depending on their location, metabolic activity, abundance, and other factors. The microbiome was shown to be essential for driving host transcriptomic oscillations in the intestine and in the liver, as complete ablation using antibiotics or germ‐free mice leads to a change of expression patterns of circadian transcripts in the host 79 (Fig 1). Interestingly, in the absence of the microbiome, de novo transcriptomic oscillations are generated. Mechanistically, both contact‐dependent and contact‐independent interactions between the host and the microbiome regulate these phenomena. Contact‐dependent interactions involve adherence of commensals of the mucosal microbiome such as segmented filamentous bacteria (SFB) to intestinal epithelial cells 79. Interestingly, mucosal‐associated bacteria undergo robust diurnal oscillations in both composition and function 79, including their capacity to degrade mucus, which allows other members of the microbiota to dwell in this region of the intestine 90. Indeed, rhythmic attachment of SFB per se induces robust transcriptomic oscillations in colonic epithelial cells 79, while the rhythmically varying number of non‐adhering rat SFB induced significantly weaker transcriptomic oscillations 79. How bacterial proximity induces these oscillations remains to be elucidated, but intestinal epithelial cells and myeloid cells, such as dendritic cells, express a large array of pattern recognition receptors like Toll‐like receptors (TLRs) or NOD‐like receptors (NLRs), which bind conserved microbial molecules and thus may directly detect microbes in their vicinity. Apart from direct attachment and PRR ligands, microbial metabolites may exert local and systemic effects on the host by modulating the function of epithelial, immune, and other cells. Indeed, circadian fluctuations of many metabolites are dependent on the microbiome, leading to transcriptomic and consequently functional oscillations in the intestine and peripheral sites 79. Rhythmic oscillations of the intestinal microbiome are associated with similar oscillations in serum metabolite levels 79. These in turn, drive circadian gene expression patterns in the liver, impacting many important functions, including oxidative phosphorylation and other catabolic pathways 79. Furthermore, administration of bacterial metabolites at specific time points directly entrained peripheral host clocks 91, suggesting that bacteria are an important factor in maintaining host circadian rhythmicity in general (Fig 1).
Equally interesting is the mechanism by which the gut microbiome globally influences the transcriptome. It is becoming increasingly clear that the microbiota has a major role in regulating the epigenetic landscape in host cells, which in turn leads to changes in gene expression patterns. The gut microbiota produces key micronutrients for DNA modifications including B vitamins, which are needed for DNA methylation 92, while changes in the gut microbiome in mice deficient for specific PRRs such as TLR2 lead to differences in DNA methylation patterns 93. Furthermore, the gut microbiota was shown to provide several cofactors needed for enzymes involved in epigenetic modifications such as acetyl‐CoA and NAD+ 94. For histone‐deacetylases (HDAC) in particular, gut microbiome‐derived short‐chain fatty acids such as butyrate or propionate were shown to be key regulators of activity 95, which in turn can have many downstream effects in mediating human disease, such as type 2 diabetes or obesity 96. One key mechanism by which the microbiome regulates host circadian transcriptomic oscillations is through the modulation of the chromatin landscapes. Indeed, the epigenetic landscape of genes with a circadian expression pattern in intestinal epithelial cells is profoundly affected by antibiotic treatment, including gain and loss of histone modifications such as H3K4me3, H3K27 acetylation at active promoters, and H3K27 acetylation at enhancers 79. Overall rhythmicity of histone marks is not affected by depletion of the microbiome, but the loci undergoing oscillations in histone marks are significantly changed, consistent with the changes in transcriptomic oscillations observed under similar conditions 79. Apart from directly interacting with the host, commensals may also indirectly interact with it by suppressing the colonization of pathogens through direct secretion of anti‐bacterial molecules such as bacteriocins or indirectly by competing for the same nutrients. In addition, following their capacity to influence the inflammatory response described above, some commensals may modulate inflammation to prevent favorable conditions for pathogen colonization 97. Despite these advances, many of the molecular mechanisms of host–microbiome interactions remain unknown and more research is necessary to unravel more about how they are regulated in health and disease.
Impact of host–microbiome circadian crosstalk on health and disease
The rhythmic adaptation to environmental fluctuations over the course of a day extends to the entire meta‐organism of both host and symbiotic communities in health and disease. In this section, we will highlight examples of how diurnal host–microbiome interactions impact host physiology and risk of disease.
Infection
Circadian rhythmicity in general has long been recognized as a major factor controlling the host response to infection 98, and disruption of the circadian cycle was found to lead to increased susceptibility to viral 99, as well as bacterial infections 100, including foodborne pathogens 101. However, while significant evidence is emerging that circadian rhythmicity of the microbiota regulates host metabolism, understanding how commensal microbial‐intrinsic clocks influence host resistance to infection remains unclear. The microbiota contributes to the host's resistance to infection by a number of distinct mechanisms. This includes colonization resistance, where commensals occupy distinct biological niches and thereby prevent pathogens from establishing themselves in the microbial ecosystem 97 (Fig 2). In addition, in some instances members of microbiota, e.g., of the Bacteroides family, will directly inhibit pathogen growth by secretion of metabolites such as the short‐chain fatty acid propionate 102. Indeed, microbiota‐derived metabolites were shown to modulate susceptibility to bacterial infections, e.g., through disruption of intracellular pH homeostasis of the pathogen 102, and of viral infection, e.g., through boosting of type I interferon signaling 103, both in the intestinal tract and in peripheral organs. Another major mechanism by which the microbiota contributes to host resistance to infection is the crosstalk with the immune system (Fig 2) 104. Microbial components induce production of anti‐microbial peptides by intestinal epithelial cells 105, promote activation of innate immune cells such as ILCs 106, and induce adaptive T‐ 107 and B‐cell responses 108. Furthermore, the microbiome is essential for a proper development in particular of myeloid cells, which in turn promotes resistance to bacterial infection 109.
While it has not been shown to date that circadian rhythmicity of the microbiome affects susceptibility to infection, there is circumstantial evidence that this may be the case. First, mice featuring a disrupted circadian cycle have a reduced microbiome diversity 33, which in turn was shown to decrease resistance to infection 110. Furthermore, given the well‐known effect of the microbiome on the immune system and increased susceptibility to infection of jet‐lagged mice 111, it is very likely that the absence of circadian rhythmicity affects infection susceptibility in this case (Fig 2). Finally, levels of specific bacterial metabolites, which are known to mediate infection resistance 112, are dysregulated in mice with a disrupted circadian cycle 91 (Fig 2). It is therefore possible, but remains to be investigated, that the well‐established circadian variations of microbiota‐derived metabolite levels will influence infection resistance in these cases. In addition, fluctuations in commensal composition will potentially affect colonization resistance to bacterial pathogens due to the changing availability of a suitable niche (Fig 2). However, no studies to date have addressed the impact of circadian rhythmicity of the microbiota on susceptibility to infection in the intestinal tract or peripheral sites, so it remains to be seen to what extent this phenomenon regulates infection resistance (Fig 2).
Metabolic disease
Disruption of circadian clocks has long been associated with a range of metabolic disorders including obesity 113, 114, 115, 116, 117, 118, 119, 120, metabolic syndrome 114, 121, 122, 123, 124, and type II diabetes 29, 114, 115, 117, 118, 120, 122, 124, 125, 126, 127, 128. Recently, microbiome dysbiosis has emerged as the mechanistic link connecting metabolic diseases and circadian clock disruption (Fig 2). Indeed, mice with either genetic or environmental impairment of circadian clocks develop a number of metabolic derangements similar to human metabolic syndrome: higher weight, hyperglycemia, insulin resistance, and higher body fat composition, compared to circadian clock‐sufficient controls 33, 35, 129, 130. Importantly, antibiotic treatment protected mice from developing circadian clock disruption‐mediated metabolic syndromes. Mechanistically, circadian clock disruption triggers a compositional and functional change in the gut microbiome community, leading to the dysbiotic microbiome state. Microbial transfer of this dysbiotic community from either mice or humans into germ‐free mice fully recapitulates metabolic derangements of the donor 33, 129. These observations demonstrate that metabolic diseases induced by circadian clock disruption can be contributed by aberrations in microbiome composition and function. They also suggest that the microbiome might constitute a previously unrecognized link between the rise of metabolic diseases and misalignment between the circadian clock and the geophysical time.
Liver function
The liver is a key organ orchestrating metabolic homeostasis with multiple functions that oscillate in response to food intake 131, 132, 133, 134, 135, 136, 137. Through the portal vein that connects the liver to the gastrointestinal tract, the liver is exposed to the influence of the gut microbiome. Perturbations of the gut microbiome, coupled with disturbances in gut barrier function, have been associated with common liver disorders, such as non‐alcoholic fatty liver disease 138, 139, 140, non‐alcoholic steatohepatitis 139, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, alcoholic liver disease 139, 140, 142, 145, 147, 148, 150, 151, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, and liver cirrhosis 165, 166, 167, 168, 169, 170. Thus, the microbiome serves as a modulator of liver rhythmic functions. Microbiome transfer into germ‐free (GF) mice resulted in PPARgamma‐mediated activation of new oscillatory transcriptional programs in the liver. Inversely, treatment of specific pathogen‐free mice (SPF) with broad‐spectrum antibiotics suppressed PPARgamma‐driven transcription in the liver, underscoring the essential role of gut microbes in clock reprogramming and hepatic circadian homeostasis 137, 171. One of the ways by which the microbiome orchestrates hepatic transcriptome oscillations is through modulation of the generation and secretion of microbiome‐derived metabolites. Microbial metabolites, specifically short‐chain fatty acids, directly modulate diurnal expression of circadian clock genes in hepatocytes 129, while germ‐free and antibiotic‐treated mice display altered daily oscillation of clock gene expression in the liver 79, 172. In addition to aberrant clock gene expression in the absence of microbiome, global rhythmicity of the liver transcriptome undergoes a profound rearrangement with up to 70% de novo oscillating transcripts 79, 173. Interestingly, while SPF male and female mice feature sex‐specific rhythmic liver transcriptome programs, these changes are largely attenuated in GF mice 173. These results highlight the microbiome as being critical for maintaining the homeostatic rhythmic functions of the liver. Indeed, the capacity of the liver for the detoxification of acetaminophen (acetyl‐para‐aminophenol APAP) varied with time of day and depended on the presence of the microbiome. Remarkably, administration of APAP to SPF mice during the active period resulted in much greater damage to the liver compared to treatments during the resting period, while there was no time‐specific difference in antibiotic‐treated or germ‐free groups 79. Collectively, this suggests that homeostatic microbiota rhythms and microbiota‐mediated maintenance of the circadian transcriptome are necessary to maintain normal diurnal activity in hepatic drug metabolism.
Cancer
There is evidence that both the microbiota and disruptions to the host circadian rhythm play a role in cancer pathogenesis. For both colorectal 174 and lung cancers 175, jet‐lag was found to promote carcinogenesis in mice and humans and it is likely that disruption of microbial rhythmicity in addition to the host clock will influence disease development (Fig 2). Furthermore, there is evidence that apart from modulating malignancy directly or indirectly through modulation of chronic inflammation, the microbiota may modulate the response to chemotherapy 176 and irradiation therapy 177. Whether treatment with chemotherapeutic agents, immune‐modulating drugs, or irradiation is dependent on or influenced by microbial diurnal rhythmicity remains to be investigated in the future. However, due to the well‐established roles of circadian rhythms and the microbiome in controlling inflammation it is likely that fluctuating microbiota‐derived metabolite levels may impact anti‐cancer immunity, and thus the response to “checkpoint blockade”. Indeed, the phenomenon of the “leaky gut”, which is associated with microbiota‐mediated modulation of anti‐tumor therapies due to their cell proliferation suppressive effects, is also induced by environmental circadian disturbances 176, 178, 179, 180, 181, 182, 183, 184. More research is necessary to unravel the relationship between microbiome rhythmicity, composition and function, and the molecular mechanisms promoting cancer development.
Conclusions and future perspectives
Many questions regarding the understanding of how circadian microbial clocks regulate host physiology in the steady state and in inflammation remain unanswered (Box 1), and represent a fascinating area of research in years to come (Fig 3).
Box 1:In need of answers.
Does the host directly regulate microbial clocks other than by food intake? And if yes, what are the underlying molecular mechanisms?
What is the relationship between aging, shifts in the circadian rhythm and the microbiome?
How are diurnal post‐translational modifications regulated by the microbiome?
How are non‐coding elements of the genome such as microRNAs regulated by the diurnal rhythmicity of the microbiota?
How is diurnal rhythmicity of the microbiome regulated at mucosal surfaces other than the intestine?
How does microbial circadian rhythmicity affect the response to microbiota‐based therapies such as probiotics?
Figure 3. Time constitutes an underappreciated but important dimension of host–microbiome interactions.
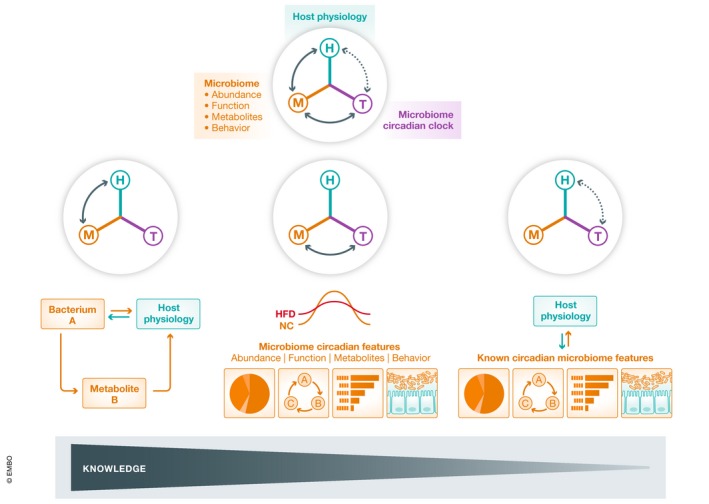
A plethora of factors involving bacterial species, bacterial metabolites, and specific bacterial behaviors have been shown to affect host physiology, and at the same time, many of them have been demonstrated to exhibit circadian oscillations. Thus, the intersection of classical microbiome studies and temporal biology holds great potential for future investigation.
Expanding the role of circadian diurnal rhythmicity to physiological and disease conditions
The diurnally changing microbiome and its impact on the host may influence other microbiome‐associated physiological states and diseases. For example, aging is associated with distinct changes in the gut microbiome 69, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204. Moreover, the microbiome has been implicated to modulate various aspects of aging including longevity 205, and very old individuals were found to feature a microbiota composition enriched for “health‐promoting” commensals 206. While the microbiota is relatively stable in adulthood 72, the aging process is associated with a change in an individual's microbiome composition, which is thought to be mainly driven by environmental effects such as the use of antibiotics, changes in nutrition, and the development of chronic illnesses, including metabolic syndrome and inflammatory diseases 73, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204. The microbial diversity decreases significantly with age 207, while the inter‐individual variability in microbiota increases significantly in elderly compared to younger adults 208. Some of these effects are in part due to a shift in the circadian rhythm as people age. Indeed, it is well established that sleep patterns change with age, with older individuals generally exhibiting shorter and lighter periods of sleep 200. As sleep has been directly linked to regulating the microbiota, it is very likely that the shift in circadian rhythmicity with age has a significant impact on the microbiome. For the human commensal E. aerogenes, it was shown that the sleep hormone melatonin directly regulates swarming motility in the intestinal lumen 20, providing a first insight into how host rhythmicity may in turn control the functions of specific members of the microbiota. Microbiome‐derived metabolites fluctuating in a circadian manner, such as short‐chain fatty acids 91, in turn control the host clock in the intestinal epithelial cells, liver, and possibly other peripheral tissues (Fig 3). As described above, food intake is a major driver of circadian rhythmicity in microbiome composition and function, and it is likely that shifting meal habits commonly observed with increasing age will contribute to differences in metabolism between young adults and elderly. Due to the combined effects of metabolic changes and changes in the circadian rhythm in older individuals, these fluctuations in metabolite levels will likely have a significant impact on the aging process (Fig 3).
An example of a set of diseases potentially modifiable by the shifting gut microbiome–host interface is neurodegenerative diseases. While there is increasing evidence for an involvement of the microbiome in the development of neurological diseases including multiple sclerosis 209, 210 and Parkinson's disease 211, it remains largely unclear to what extent circadian variations in microbiome composition or function contributes to this phenomenon (Fig 3). There is some indication that time‐restricted feeding can modulate disease worsening in Huntingdon's disease 212, and as food uptake is known to orchestrate gut microbiome fluctuations 33, it is very likely that some of the effects seen in this model can be attributed to fluctuating microbial products (Fig 3). More work is necessary to determine whether human commensals such as Bacteroides fragilis, which have been shown to modulate neurodevelopmental disorders 68, fluctuate in a circadian manner. It has been suggested that microbial metabolites play an important role in regulating host physiology at peripheral sites to the intestine, including the brain. Indeed, the “gut–brain axis” is now recognized as an important interaction network where the microbiome directly influences the peripheral and central nervous system 213.
Exploring further mechanisms of circadian microbiome control of host physiology
It is clear that more mechanistic evidence is required for global understanding of the diurnally shifting host–microbiome interface. Open questions include the cell types at mucosal surfaces or peripheral sites that are directly regulated by microbial factors, and how this modulation relates to their circadian variation in gene expression patterns and function over the course of a day. A second interesting yet not fully addressed question is the regulation of the molecular mechanisms driving diurnally shifting microbiome‐mediated impacts on the host. While it has been shown that multiple host transcriptional fluctuations are driven by the microbiome, more needs to be done to understand the mechanisms driving these changes in gene expression, including epigenetic regulation. A third interesting unmet question involves the potential regulation of host post‐translational modifications by the shifting microbiome, including hourly changing protein phosphorylation, ubiquitination, and sumoylation. Such mechanisms may contribute to a mechanistic understanding of observations linking the diurnally oscillating microbiome to host changes in cell cycle, secretion of exosomes, and more. A fourth unanswered key question, and an exciting area of future research, is the elucidation of mechanisms by which the circadian microbiota interacts with the immune system. As briefly described above, the bidirectional relationship of the microbiome and immunity is a very important component of host–microbiome interactions, and understanding how circadian fluctuations in microbiome composition and function control inflammation and key immune capacities, will be of great interest in future years. Examples of these topics include elucidation of possible diurnal microbial regulation of the activation of innate immunity, induction of T‐cell responses, and establishment of memory (Fig 3). It will be necessary to disentangle the role of host circadian control of the immune responses and microbiome diurnal rhythmicity utilizing immune cell specific core circadian clock‐knockout mice. Equally important is to unravel to which extent the diurnally rearranging microbiome contributes to the regulation or promotion of inflammation, possibly influencing resistance to infection and driving immune activation versus tolerance. In addition, further studies are necessary to investigate a role of the immune system in synchronizing bacterial intrinsic clocks with host circadian rhythms, and its relationship with food uptake.
Dissecting the role of specific members of the circadian microbiota
More research is also needed to uncover relationships between specific members of the microbiota, their rhythmicity in abundance or function, and particular host phenotypes. In particular, it will be important to understand which are the essential components in the circadian microbiota driving oscillations in gene expression and function in the host. Furthermore, the analysis of the microbiome oscillations in composition and function needs to be extended to microbiomes at other sites, in particular other barrier tissues such as the skin. The elucidation of the site‐specific circadian regulation of host–microbiome interactions will be crucial to understand their role in microbiome‐mediated diseases in peripheral tissues, including local fluctuations in microbial metabolites, and time‐dependent localization of specific members of the microbiota (i.e., mucosal versus luminal). In this respect, one of the key challenges includes the development of methods to assess fluctuations in microbial composition and function at sites featuring a low biomass. In addition, another major question to be answered in this context is to what extent the regulation of bacterial intrinsic clocks is dependent on the environment (i.e., tissue). Furthermore, a crucial question to be answered is whether the host directly regulates the bacterial clock in certain disease contexts apart from the fluctuations induced by nutrient availability during food uptake.
Studying circadian microbiome rhythmicity in humans—translational aspects
Finally, many important questions remain unanswered regarding the applicability of the findings largely obtained in animal models to humans, potentially enabling future therapeutic exploitation. While it is challenging to directly show causal relationships between diurnal changes in the microbiome and effects on the human host, recent studies have provided some evidence into this direction 27, 33, 214, 215. The microbiome in humans was also suggested to vary in composition and function across the course of a day, although variations in transit and excretion times make such interpretations more complex in the human as compared to the rodent setting 33. In addition, circadian disruption, such as the jet‐lag induced by repeated re‐adaptation to different time zones, leads to metabolic derangements once the human microbiota is transferred to germ‐free mice 33. Although it provides only a glimpse of how the circadian human microbiota controls host physiology, it may nonetheless be a starting point to mechanistically decode the microbiome‐mediated effects of circadian disturbances in shift‐work and jet‐lag on human physiology. Indeed, shift‐work has long been associated with increased risk to develop multiple diseases, including obesity, diabetes, and cancer 27, 214, 215. Therefore, establishing the links between this phenomenon and the impact on the human microbiota will be an essential and exciting step toward identifying new therapeutic targets and preventing or ameliorating the adverse health effects of chronic circadian microbiome‐mediated alterations. Furthermore, from a translational viewpoint, the key question to be addressed will be whether the timing of application of microbiota‐based therapeutics, such as pre‐, pro‐, and post‐biotics, will affect their efficacy. Based on the emerging evidence of microbial circadian rhythmicity, it is possible that there will be diurnal variation of the response to these therapeutic approaches. Chronopharmacological studies of microbiota‐based therapeutics would pave the way toward the possibility of manipulation of host peripheral clocks. This in turn can provide potential means to correct dysregulation of the host circadian rhythm associated with aging or chronic illnesses, such as metabolic syndrome, through the modification of the diurnally shifting microbiome.
Conflict of interest
EE is a paid consultant at DayTwo and BiomX. None of the work reviewed here is related to these or any other commercial entity.
Acknowledgements
We thank the Elinav Lab for fruitful discussions and apologize to those authors whose works could not be cited due to space limitations. S.P.N. was supported by an Early Postdoctoral Fellowship of the Swiss National Science Foundation and an EMBO Long‐term Fellowship. E.E. is supported by Y. and R. Ungar; the Abisch Frenkel Foundation for the Promotion of Life Sciences; the Gurwin Family Fund for Scientific Research; the Leona M. and Harry B. Helmsley Charitable Trust; the Crown Endowment Fund for Immunological Research; the Else Kroener Fresenius Foundation; the estate of J. Gitlitz; the estate of L. Hershkovich; the Benoziyo Endowment Fund for the Advancement of Science; the Adelis Foundation; J. L. and V. Schwartz; A. and G. Markovitz; A. and C. Adelson; the French National Center for Scientific Research (CNRS); D. L. Schwarz; The V.R. Schwartz Research Fellow Chair; L. Steinberg; J.N. Halpern; A. Edelheit, and by grants funded by the European Research Council; a Marie Curie Integration grant; the German–Israeli Foundation for Scientific Research and Development; the Israel Science Foundation; the Minerva Foundation; the Rising Tide Foundation; the Helmholtz Foundation; and the European Foundation for the Study of Diabetes. E.E. is a senior fellow, Canadian Institute of Advanced Research (CIFAR) and an international scholar, The Bill & Melinda Gates Foundation, and Howard Hughes Medical Institute (HHMI).
EMBO Reports (2019) 20: e47129
See the Glossary for abbreviations used in this article.
References
- 1. Farre EM, Liu T (2013) The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol 16: 621–629 [DOI] [PubMed] [Google Scholar]
- 2. Merrow M, Maas MF (2009) Circadian clocks: evolution in the shadows. Curr Biol 19: R1042–R1045 [DOI] [PubMed] [Google Scholar]
- 3. Millar AJ (2016) The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu Rev Plant Biol 67: 595–618 [DOI] [PubMed] [Google Scholar]
- 4. Roenneberg T, Merrow M (2002) “What watch?…such much!” Complexity and evolution of circadian clocks. Cell Tissue Res 309: 3–9 [DOI] [PubMed] [Google Scholar]
- 5. Shindey R, Varma V, Nikhil KL, Sharma VK (2016) Evolution of robust circadian clocks in Drosophila melanogaster populations reared in constant dark for over 330 generations. Naturwissenschaften 103: 74 [DOI] [PubMed] [Google Scholar]
- 6. Brown SA, Kowalska E, Dallmann R (2012) (Re)inventing the circadian feedback loop. Dev Cell 22: 477–487 [DOI] [PubMed] [Google Scholar]
- 7. Buhr ED, Takahashi JS (2013) Molecular components of the mammalian circadian clock. Handb Exp Pharmacol 217: 3–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fitzgerald GA, Yang G, Paschos GK, Liang X, Skarke C (2015) Molecular clocks and the human condition: approaching their characterization in human physiology and disease. Diabetes Obes Metab 17(Suppl 1): 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brandstaetter R (2004) Circadian lessons from peripheral clocks: is the time of the mammalian pacemaker up? Proc Natl Acad Sci USA 101: 5699–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown SA, Zumbrunn G, Fleury‐Olela F, Preitner N, Schibler U (2002) Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12: 1574–1583 [DOI] [PubMed] [Google Scholar]
- 11. Hastings M, Maywood ES (2000) Circadian clocks in the mammalian brain. BioEssays 22: 23–31 [DOI] [PubMed] [Google Scholar]
- 12. Husse J, Eichele G, Oster H (2015) Synchronization of the mammalian circadian timing system: light can control peripheral clocks independently of the SCN clock: alternate routes of entrainment optimize the alignment of the body's circadian clock network with external time. BioEssays 37: 1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ikonomov OC, Stoynev AG, Shisheva AC (1998) Integrative coordination of circadian mammalian diversity: neuronal networks and peripheral clocks. Prog Neurobiol 54: 87–97 [DOI] [PubMed] [Google Scholar]
- 14. Saini C, Suter DM, Liani A, Gos P, Schibler U (2011) The mammalian circadian timing system: synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol 76: 39–47 [DOI] [PubMed] [Google Scholar]
- 15. Susaki EA, Stelling J, Ueda HR (2010) Challenges in synthetically designing mammalian circadian clocks. Curr Opin Biotechnol 21: 556–565 [DOI] [PubMed] [Google Scholar]
- 16. Ueda HR (2007) Systems biology of mammalian circadian clocks. Cold Spring Harb Symp Quant Biol 72: 365–380 [DOI] [PubMed] [Google Scholar]
- 17. Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S (2005) System‐level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37: 187–192 [DOI] [PubMed] [Google Scholar]
- 18. Ukai‐Tadenuma M, Kasukawa T, Ueda HR (2008) Proof‐by‐synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol 10: 1154–1163 [DOI] [PubMed] [Google Scholar]
- 19. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320 [DOI] [PubMed] [Google Scholar]
- 20. Paulose JK, Wright JM, Patel AG, Cassone VM (2016) Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS One 11: e0146643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teng SW, Mukherji S, Moffitt JR, de Buyl S, O'Shea EK (2013) Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science 340: 737–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson CH, Stewart PL, Egli M (2011) The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys 40: 143–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson CH, Zhao C, Xu Y, Mori T (2017) Timing the day: what makes bacterial clocks tick? Nat Rev Microbiol 15: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Golombek DA, Casiraghi LP, Agostino PV, Paladino N, Duhart JM, Plano SA, Chiesa JJ (2013) The times they're a‐changing: effects of circadian desynchronization on physiology and disease. J Physiol Paris 107: 310–322 [DOI] [PubMed] [Google Scholar]
- 26. Bechtold DA, Gibbs JE, Loudon AS (2010) Circadian dysfunction in disease. Trends Pharmacol Sci 31: 191–198 [DOI] [PubMed] [Google Scholar]
- 27. Archer SN, Laing EE, Moller‐Levet CS, van der Veen DR, Bucca G, Lazar AS, Santhi N, Slak A, Kabiljo R, von Schantz M et al (2014) Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci USA 111: E682–E691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sears CL, Garrett WS (2014) Microbes, microbiota, and colon cancer. Cell Host Microbe 15: 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D et al (2012) A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60 [DOI] [PubMed] [Google Scholar]
- 30. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F (2013) Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103 [DOI] [PubMed] [Google Scholar]
- 31. Cox LM, Blaser MJ (2013) Pathways in microbe‐induced obesity. Cell Metab 17: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A (2014) Circadian disorganization alters intestinal microbiota. PLoS One 9: e97500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thaiss CA, Zeevi D, Levy M, Zilberman‐Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N et al (2014) Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159: 514–529 [DOI] [PubMed] [Google Scholar]
- 34. Chaix A, Zarrinpar A, Miu P, Panda S (2014) Time‐restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 20: 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zarrinpar A, Chaix A, Yooseph S, Panda S (2014) Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 20: 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X, Devaraj S (2018) Gut microbiome in obesity, metabolic syndrome, and diabetes. Curr Diab Rep 18: 129 [DOI] [PubMed] [Google Scholar]
- 37. Fava F, Gitau R, Griffin BA, Gibson GR, Tuohy KM, Lovegrove JA (2013) The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short‐chain fatty acid excretion in a metabolic syndrome ‘at‐risk’ population. Int J Obes (Lond) 37: 216–223 [DOI] [PubMed] [Google Scholar]
- 38. Feng X, Uchida Y, Koch L, Britton S, Hu J, Lutrin D, Maze M (2017) Exercise prevents enhanced postoperative neuroinflammation and cognitive decline and rectifies the gut microbiome in a rat model of metabolic syndrome. Front Immunol 8: 1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferrarese R, Ceresola ER, Preti A, Canducci F (2018) Probiotics, prebiotics and synbiotics for weight loss and metabolic syndrome in the microbiome era. Eur Rev Med Pharmacol Sci 22: 7588–7605 [DOI] [PubMed] [Google Scholar]
- 40. Kovatcheva‐Datchary P, Arora T (2013) Nutrition, the gut microbiome and the metabolic syndrome. Best Pract Res Clin Gastroenterol 27: 59–72 [DOI] [PubMed] [Google Scholar]
- 41. Mazidi M, Rezaie P, Kengne AP, Mobarhan MG, Ferns GA (2016) Gut microbiome and metabolic syndrome. Diabetes Metab Syndr 10: S150–S157 [DOI] [PubMed] [Google Scholar]
- 42. Parekh PJ, Balart LA, Johnson DA (2015) The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol 6: e91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shapiro H, Suez J, Elinav E (2017) Personalized microbiome‐based approaches to metabolic syndrome management and prevention. J Diabetes 9: 226–236 [DOI] [PubMed] [Google Scholar]
- 44. Tutkova M, Ruda‐Kucerova J (2018) Microbiome in connection with metabolic syndrome and the therapeutic potential of its influencing. Ceska Slov Farm 67: 71–80 [PubMed] [Google Scholar]
- 45. Ussar S, Fujisaka S, Kahn CR (2016) Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol Metab 5: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Human Microbiome Project C (2012) Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lloyd‐Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG et al (2017) Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buford TW (2017) (Dis)Trust your gut: the gut microbiome in age‐related inflammation, health, and disease. Microbiome 5: 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dinan TG, Cryan JF (2017) The microbiome‐gut‐brain axis in health and disease. Gastroenterol Clin North Am 46: 77–89 [DOI] [PubMed] [Google Scholar]
- 50. Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK (2011) Understanding the role of gut microbiome‐host metabolic signal disruption in health and disease. Trends Microbiol 19: 349–359 [DOI] [PubMed] [Google Scholar]
- 51. Ihekweazu FD, Versalovic J (2018) Development of the pediatric gut microbiome: impact on health and disease. Am J Med Sci 356: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liang D, Leung RK, Guan W, Au WW (2018) Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog 10: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shreiner AB, Kao JY, Young VB (2015) The gut microbiome in health and in disease. Curr Opin Gastroenterol 31: 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fulde M, Hornef MW (2014) Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev 260: 21–34 [DOI] [PubMed] [Google Scholar]
- 55. Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G (2010) Transfer of carbohydrate‐active enzymes from marine bacteria to Japanese gut microbiota. Nature 464: 908–912 [DOI] [PubMed] [Google Scholar]
- 56. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR et al (2013) Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 [DOI] [PubMed] [Google Scholar]
- 58. Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SW, Muller M, Kleerebezem M, van der Meer R (2015) Gut microbiota facilitates dietary heme‐induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc Natl Acad Sci USA 112: 10038–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Ostergren‐Lunden G, Petersen LC, Ruf W, Backhed F (2012) Tissue factor and PAR1 promote microbiota‐induced intestinal vascular remodelling. Nature 483: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY (2015) Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161: 264–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I et al (2012) Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Canfora EE, Jocken JW, Blaak EE (2015) Short‐chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 11: 577–591 [DOI] [PubMed] [Google Scholar]
- 63. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez‐Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP et al (2012) Human gut microbiome viewed across age and geography. Nature 486: 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga‐Thie GM, Ackermans MT, Serlie MJ, Oozeer R et al (2012) Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143: 913–916 e917 [DOI] [PubMed] [Google Scholar]
- 65. Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK (2009) Pharmacometabonomic identification of a significant host‐microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA 106: 14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Elinav E, Strowig T, Kau AL, Henao‐Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI et al (2011) NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rohlke F, Stollman N (2012) Fecal microbiota transplantation in relapsing Clostridium difficile infection. Therap Adv Gastroenterol 5: 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF et al (2013) Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155: 1451–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO (2007) Development of the human infant intestinal microbiota. PLoS Biol 5: e177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, Dahl DB, Herman D, Wang M, Donovan SM, Chapkin RS (2012) A metagenomic study of diet‐dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol 13: r32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J (2012) The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol 3: 425–447 [DOI] [PubMed] [Google Scholar]
- 72. Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL et al (2013) The long‐term stability of the human gut microbiota. Science 341: 1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, Corthier G, Furet JP (2009) The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9: 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R et al (2011) Linking long‐term dietary patterns with gut microbial enterotypes. Science 334: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Suez J, Zmora N, Zilberman‐Schapira G, Mor U, Dori‐Bachash M, Bashiardes S, Zur M, Regev‐Lehavi D, Ben‐Zeev Brik R, Federici S et al (2018) Post‐antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174: 1406–1423 e1416 [DOI] [PubMed] [Google Scholar]
- 77. Zmora N, Zilberman‐Schapira G, Suez J, Mor U, Dori‐Bachash M, Bashiardes S, Kotler E, Zur M, Regev‐Lehavi D, Brik RB et al (2018) Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174: 1388–1405 e1321 [DOI] [PubMed] [Google Scholar]
- 78. Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- 79. Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, David E, Winter DR, Gury‐BenAri M, Tatirovsky E et al (2016) Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167: 1495–1510 e1412 [DOI] [PubMed] [Google Scholar]
- 80. Thaiss CA, Zeevi D, Levy M, Segal E, Elinav E (2015) A day in the life of the meta‐organism: diurnal rhythms of the intestinal microbiome and its host. Gut Microbes 6: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328 [DOI] [PubMed] [Google Scholar]
- 82. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone‐Corsi P (2008) The NAD+‐dependent deacetylase SIRT1 modulates CLOCK‐mediated chromatin remodeling and circadian control. Cell 134: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone‐Corsi P (2009) Circadian control of the NAD+ salvage pathway by CLOCK‐SIRT1. Science 324: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C et al (2009) Circadian clock feedback cycle through NAMPT‐mediated NAD+ biosynthesis. Science 324: 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S (2009) Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA 106: 21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wu G, Tang W, He Y, Hu J, Gong S, He Z, Wei G, Lv L, Jiang Y, Zhou H et al (2018) Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem Biophys Res Commun 501: 16–23 [DOI] [PubMed] [Google Scholar]
- 88. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J (2007) High‐fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6: 414–421 [DOI] [PubMed] [Google Scholar]
- 89. Eckel‐Mahan KL, Patel VR, de Mateo S, Orozco‐Solis R, Ceglia NJ, Sahar S, Dilag‐Penilla SA, Dyar KA, Baldi P, Sassone‐Corsi P (2013) Reprogramming of the circadian clock by nutritional challenge. Cell 155: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ramanan D, Cadwell K (2016) Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe 19: 434–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, Haraguchi A, Ikeda Y, Fukuda S, Shibata S (2018) Gut microbiota‐derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep 8: 1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yamada K, Gherasim C, Banerjee R, Koutmos M (2015) Structure of human B12 trafficking protein CblD reveals molecular mimicry and identifies a new subfamily of nitro‐FMN reductases. J Biol Chem 290: 29155–29166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kellermayer R, Dowd SE, Harris RA, Balasa A, Schaible TD, Wolcott RD, Tatevian N, Szigeti R, Li Z, Versalovic J et al (2011) Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll‐like receptor 2‐knockout mice. FASEB J 25: 1449–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Paul B, Barnes S, Demark‐Wahnefried W, Morrow C, Salvador C, Skibola C, Tollefsbol TO (2015) Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenetics 7: 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ciarlo E, Heinonen T, Herderschee J, Fenwick C, Mombelli M, Le Roy D, Roger T (2016) Impact of the microbial derived short chain fatty acid propionate on host susceptibility to bacterial and fungal infections in vivo . Sci Rep 6: 37944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Remely M, Aumueller E, Merold C, Dworzak S, Hippe B, Zanner J, Pointner A, Brath H, Haslberger AG (2014) Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 537: 85–92 [DOI] [PubMed] [Google Scholar]
- 97. Mullineaux‐Sanders C, Suez J, Elinav E, Frankel G (2018) Sieving through gut models of colonization resistance. Nat Microbiol 3: 132–140 [DOI] [PubMed] [Google Scholar]
- 98. Curtis AM, Bellet MM, Sassone‐Corsi P, O'Neill LA (2014) Circadian clock proteins and immunity. Immunity 40: 178–186 [DOI] [PubMed] [Google Scholar]
- 99. Ehlers A, Xie W, Agapov E, Brown S, Steinberg D, Tidwell R, Sajol G, Schutz R, Weaver R, Yu H et al (2018) BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol 11: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, Saer B, Begley N, Poolman T, Pariollaud M et al (2014) An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med 20: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A (2013) Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 341: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K et al (2018) A gut commensal‐produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 24: 296–307.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ et al (2017) The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Thaiss CA, Zmora N, Levy M, Elinav E (2016) The microbiome and innate immunity. Nature 535: 65–74 [DOI] [PubMed] [Google Scholar]
- 105. Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman‐Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y et al (2015) Microbiota‐modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163: 1428–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M (2014) Microbiota‐dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343: 1249288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM et al (2013) Innate lymphoid cells regulate CD4+ T‐cell responses to intestinal commensal bacteria. Nature 498: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F et al (2014) TLR5‐mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41: 478–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK (2014) Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15: 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A et al (2015) Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517: 205–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tognini P, Murakami M, Sassone‐Corsi P (2017) Interplay between microbes and the circadian clock. Cold Spring Harb Perspect Biol 10: a028365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Trompette A, Gollwitzer ES, Pattaroni C, Lopez‐Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, Marsland BJ (2018) Dietary fiber confers protection against flu by shaping Ly6c(−) patrolling monocyte hematopoiesis and CD8(+) T cell metabolism. Immunity 48: 992–1005 e1008 [DOI] [PubMed] [Google Scholar]
- 113. Ando H, Kumazaki M, Motosugi Y, Ushijima K, Maekawa T, Ishikawa E, Fujimura A (2011) Impairment of peripheral circadian clocks precedes metabolic abnormalities in ob/ob mice. Endocrinology 152: 1347–1354 [DOI] [PubMed] [Google Scholar]
- 114. Barandas R, Landgraf D, McCarthy MJ, Welsh DK (2015) Circadian clocks as modulators of metabolic comorbidity in psychiatric disorders. Curr Psychiatry Rep 17: 98 [DOI] [PubMed] [Google Scholar]
- 115. Challet E (2013) Circadian clocks, food intake, and metabolism. Prog Mol Biol Transl Sci 119: 105–135 [DOI] [PubMed] [Google Scholar]
- 116. Coomans CP, Lucassen EA, Kooijman S, Fifel K, Deboer T, Rensen PC, Michel S, Meijer JH (2015) Plasticity of circadian clocks and consequences for metabolism. Diabetes Obes Metab 17(Suppl 1): 65–75 [DOI] [PubMed] [Google Scholar]
- 117. Delezie J, Challet E (2011) Interactions between metabolism and circadian clocks: reciprocal disturbances. Ann N Y Acad Sci 1243: 30–46 [DOI] [PubMed] [Google Scholar]
- 118. Gimble JM, Sutton GM, Bunnell BA, Ptitsyn AA, Floyd ZE (2011) Prospective influences of circadian clocks in adipose tissue and metabolism. Nat Rev Endocrinol 7: 98–107 [DOI] [PubMed] [Google Scholar]
- 119. Paschos GK (2015) Circadian clocks, feeding time, and metabolic homeostasis. Front Pharmacol 6: 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Scott EM (2015) Circadian clocks, obesity and cardiometabolic function. Diabetes Obes Metab 17(Suppl 1): 84–89 [DOI] [PubMed] [Google Scholar]
- 121. He B, Chen Z (2016) Molecular targets for small‐molecule modulators of circadian clocks. Curr Drug Metab 17: 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mukherji A, Kobiita A, Damara M, Misra N, Meziane H, Champy MF, Chambon P (2015) Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc Natl Acad Sci USA 112: E6691–E6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sancar G, Brunner M (2014) Circadian clocks and energy metabolism. Cell Mol Life Sci 71: 2667–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM (2006) Characterization of peripheral circadian clocks in adipose tissues. Diabetes 55: 962–970 [DOI] [PubMed] [Google Scholar]
- 125. Ando H (2013) [Circadian clocks and lifestyle‐related diseases]. Rinsho Byori 61: 1044–1050 [PubMed] [Google Scholar]
- 126. Berenbaum F, Meng QJ (2016) The brain‐joint axis in osteoarthritis: nerves, circadian clocks and beyond. Nat Rev Rheumatol 12: 508–516 [DOI] [PubMed] [Google Scholar]
- 127. Challet E (2014) Circadian clocks and energy metabolism in rodents. Biol Aujourdhui 208: 269–274 [DOI] [PubMed] [Google Scholar]
- 128. Heden TD, Kanaley JA (2018) Syncing exercise with meals and circadian clocks. Exerc Sport Sci Rev 47: 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N et al (2015) Effects of diurnal variation of gut microbes and high‐fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17: 681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jiang P, Turek FW (2018) The endogenous circadian clock programs animals to eat at certain times of the 24‐hour day: what if we ignore the clock? Physiol Behav 193: 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. de Goede P, Sen S, Su Y, Foppen E, Poirel VJ, Challet E, Kalsbeek A (2018) An ultradian feeding schedule in rats affects metabolic gene expression in liver, brown adipose tissue and skeletal muscle with only mild effects on circadian clocks. Int J Mol Sci 19: E3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Gatfield D, Schibler U (2008) Circadian glucose homeostasis requires compensatory interference between brain and liver clocks. Proc Natl Acad Sci USA 105: 14753–14754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Radziuk JM (2013) The suprachiasmatic nucleus, circadian clocks, and the liver. Diabetes 62: 1017–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Saini C, Liani A, Curie T, Gos P, Kreppel F, Emmenegger Y, Bonacina L, Wolf JP, Poget YA, Franken P et al (2013) Real‐time recording of circadian liver gene expression in freely moving mice reveals the phase‐setting behavior of hepatocyte clocks. Genes Dev 27: 1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Schibler U (2003) Circadian rhythms. Liver regeneration clocks on. Science 302: 234–235 [DOI] [PubMed] [Google Scholar]
- 136. Sosniyenko S, Parkanova D, Illnerova H, Sladek M, Sumova A (2010) Different mechanisms of adjustment to a change of the photoperiod in the suprachiasmatic and liver circadian clocks. Am J Physiol Regul Integr Comp Physiol 298: R959–R971 [DOI] [PubMed] [Google Scholar]
- 137. Tognini P, Murakami M, Liu Y, Eckel‐Mahan KL, Newman JC, Verdin E, Baldi P, Sassone‐Corsi P (2017) Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab 26: 523–538 e525 [DOI] [PubMed] [Google Scholar]
- 138. Lin EA, Barlow GM, Mathur R (2014) The microbiome in non‐alcoholic fatty liver disease: associations and implications. Ann Gastroenterol 27: 181–183 [PMC free article] [PubMed] [Google Scholar]
- 139. Nobili V, Putignani L, Mosca A, Chierico FD, Vernocchi P, Alisi A, Stronati L, Cucchiara S, Toscano M, Drago L (2018) Bifidobacteria and lactobacilli in the gut microbiome of children with non‐alcoholic fatty liver disease: which strains act as health players? Arch Med Sci 14: 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Saltzman ET, Palacios T, Thomsen M, Vitetta L (2018) Intestinal microbiome shifts, dysbiosis, inflammation, and non‐alcoholic fatty liver disease. Front Microbiol 9: 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Aqel B, DiBaise JK (2015) Role of the gut microbiome in nonalcoholic fatty liver disease. Nutr Clin Pract 30: 780–786 [DOI] [PubMed] [Google Scholar]
- 142. Bischoff SC (2017) The intestinal microbiome and metabolic diseases: from obesity to diabetes and nonalcoholic steatohepatitis. Internist (Berl) 58: 441–448 [DOI] [PubMed] [Google Scholar]
- 143. Boursier J, Diehl AM (2016) Nonalcoholic fatty liver disease and the gut microbiome. Clin Liver Dis 20: 263–275 [DOI] [PubMed] [Google Scholar]
- 144. Hara E (2015) Relationship between obesity, gut microbiome and hepatocellular carcinoma development. Dig Dis 33: 346–350 [DOI] [PubMed] [Google Scholar]
- 145. Leung DH, Yimlamai D (2017) The intestinal microbiome and paediatric liver disease. Lancet Gastroenterol Hepatol 2: 446–455 [DOI] [PubMed] [Google Scholar]
- 146. Neuman MG, French SW, Zakhari S, Malnick S, Seitz HK, Cohen LB, Salaspuro M, Voinea‐Griffin A, Barasch A, Kirpich IA et al (2017) Alcohol, microbiome, life style influence alcohol and non‐alcoholic organ damage. Exp Mol Pathol 102: 162–180 [DOI] [PubMed] [Google Scholar]
- 147. Panasevich MR, Peppler WT, Oerther DB, Wright DC, Rector RS (2017) Microbiome and NAFLD: potential influence of aerobic fitness and lifestyle modification. Physiol Genomics 49: 385–399 [DOI] [PubMed] [Google Scholar]
- 148. Puri P, Sanyal AJ (2018) The intestinal microbiome in nonalcoholic fatty liver disease. Clin Liver Dis 22: 121–132 [DOI] [PubMed] [Google Scholar]
- 149. Schnabl B, Brenner DA (2014) Interactions between the intestinal microbiome and liver diseases. Gastroenterology 146: 1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wankhade UD, Zhong Y, Kang P, Alfaro M, Chintapalli SV, Thakali KM, Shankar K (2017) Enhanced offspring predisposition to steatohepatitis with maternal high‐fat diet is associated with epigenetic and microbiome alterations. PLoS One 12: e0175675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Woodhouse CA, Patel VC, Singanayagam A, Shawcross DL (2018) Review article: the gut microbiome as a therapeutic target in the pathogenesis and treatment of chronic liver disease. Aliment Pharmacol Ther 47: 192–202 [DOI] [PubMed] [Google Scholar]
- 152. Zhu L, Baker RD, Baker SS (2015) Gut microbiome and nonalcoholic fatty liver diseases. Pediatr Res 77: 245–251 [DOI] [PubMed] [Google Scholar]
- 153. Adolph TE, Effenberger M, Tilg H (2018) How our understanding of the microbiome has changed the way we look at liver diseases. Biomark Med 12: 1203–1206 [DOI] [PubMed] [Google Scholar]
- 154. Adolph TE, Grander C, Moschen AR, Tilg H (2018) Liver‐microbiome axis in health and disease. Trends Immunol 39: 712–723 [DOI] [PubMed] [Google Scholar]
- 155. Aitbaev KA, Murkamilov IT, Fomin VV (2017) Liver diseases: the pathogenetic role of the gut microbiome and the potential of treatment for its modulation. Ter Arkh 89: 120–128 [DOI] [PubMed] [Google Scholar]
- 156. Capurso G, Lahner E (2017) The interaction between smoking, alcohol and the gut microbiome. Best Pract Res Clin Gastroenterol 31: 579–588 [DOI] [PubMed] [Google Scholar]
- 157. Castano‐Rodriguez N, Mitchell HM, Kaakoush NO (2017) NAFLD, Helicobacter species and the intestinal microbiome. Best Pract Res Clin Gastroenterol 31: 657–668 [DOI] [PubMed] [Google Scholar]
- 158. Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK et al (2017) Gut microbiome‐based metagenomic signature for non‐invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab 25: 1054–1062 e1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Maruvada P, Leone V, Kaplan LM, Chang EB (2017) The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22: 589–599 [DOI] [PubMed] [Google Scholar]
- 160. Omer E, Atassi H (2017) The microbiome that shapes us: can it cause obesity? Curr Gastroenterol Rep 19: 59 [DOI] [PubMed] [Google Scholar]
- 161. Puri P, Liangpunsakul S, Christensen JE, Shah VH, Kamath PS, Gores GJ, Walker S, Comerford M, Katz B, Borst A et al (2018) The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 67: 1284–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Szabo G (2018) Gut‐liver axis beyond the microbiome: how the fungal mycobiome contributes to alcoholic liver disease. Hepatology 68: 2426–2428 [DOI] [PubMed] [Google Scholar]
- 163. Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R (2018) Publisher correction: the gut‐liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 15: 785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R (2018) The gut‐liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 15: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Fukui H (2017) Gut microbiome‐based therapeutics in liver cirrhosis: basic consideration for the next step. J Clin Transl Hepatol 5: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Jacobs JP, Dong TS, Agopian V, Lagishetty V, Sundaram V, Noureddin M, Ayoub WS, Durazo F, Benhammou J, Enayati P et al (2018) Microbiome and bile acid profiles in duodenal aspirates from patients with liver cirrhosis: the microbiome, microbial markers and liver disease study. Hepatol Res 48: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Lu H, Qian G, Ren Z, Zhang C, Zhang H, Xu W, Ye P, Yang Y, Li L (2015) Alterations of Bacteroides sp., Neisseria sp., Actinomyces sp., and Streptococcus sp. populations in the oropharyngeal microbiome are associated with liver cirrhosis and pneumonia. BMC Infect Dis 15: 239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L et al (2014) Alterations of the human gut microbiome in liver cirrhosis. Nature 513: 59–64 [DOI] [PubMed] [Google Scholar]
- 169. Roderburg C, Luedde T (2014) The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes 5: 441–445 [DOI] [PubMed] [Google Scholar]
- 170. Schulz C, Schutte K, Kropf S, Schmitt FC, Vasapolli R, Kliegis LM, Riegger A, Malfertheiner P (2016) RiMINI – the influence of rifaximin on minimal hepatic encephalopathy (MHE) and on the intestinal microbiome in patients with liver cirrhosis: study protocol for a randomized controlled trial. Trials 17: 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Murakami M, Tognini P, Liu Y, Eckel‐Mahan KL, Baldi P, Sassone‐Corsi P (2016) Gut microbiota directs PPARgamma‐driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep 17: 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Montagner A, Korecka A, Polizzi A, Lippi Y, Blum Y, Canlet C, Tremblay‐Franco M, Gautier‐Stein A, Burcelin R, Yen YC et al (2016) Hepatic circadian clock oscillators and nuclear receptors integrate microbiome‐derived signals. Sci Rep 6: 20127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Weger BD, Gobet C, Yeung J, Martin E, Jimenez S, Betrisey B, Foata F, Berger B, Balvay A, Foussier A et al (2018) The mouse microbiome is required for sex‐specific diurnal rhythms of gene expression and metabolism. Cell Metab 29: 362–382.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wang X, Ji A, Zhu Y, Liang Z, Wu J, Li SQ, Meng S, Zheng XY, Xie LP (2015) A meta‐analysis including dose‐response relationship between night shift work and the risk of colorectal cancer. Oncotarget 6: 25046–25060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden MG, Jacks T (2016) Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab 24: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP et al (2018) Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science 359: 91–97 [DOI] [PubMed] [Google Scholar]
- 177. Cui M, Xiao H, Luo D, Zhang X, Zhao S, Zheng Q, Li Y, Zhao Y, Dong J, Li H et al (2016) Circadian rhythm shapes the gut microbiota affecting host radiosensitivity. Int J Mol Sci 17: E1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Summa KC, Voigt RM, Forsyth CB, Shaikh M, Cavanaugh K, Tang Y, Vitaterna MH, Song S, Turek FW, Keshavarzian A (2013) Disruption of the circadian clock in mice increases intestinal permeability and promotes alcohol‐induced hepatic pathology and inflammation. PLoS One 8: e67102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Bashiardes S, Tuganbaev T, Federici S, Elinav E (2017) The microbiome in anti‐cancer therapy. Semin Immunol 32: 74–81 [DOI] [PubMed] [Google Scholar]
- 180. Biancheri P, Divekar D, Watson AJM (2018) Could fecal transplantation become part of PD‐1‐based immunotherapy, due to effects of the intestinal microbiome? Gastroenterology 154: 1845–1847 [DOI] [PubMed] [Google Scholar]
- 181. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC et al (2018) Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science 359: 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF (2018) The commensal microbiome is associated with anti‐PD‐1 efficacy in metastatic melanoma patients. Science 359: 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Santoni M, Piva F, Conti A, Santoni A, Cimadamore A, Scarpelli M, Battelli N, Montironi R (2018) Re: Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Eur Urol 74: 521–522 [DOI] [PubMed] [Google Scholar]
- 184. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP et al (2015) Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science 350: 1079–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Candela M, Biagi E, Brigidi P, O'Toole PW, De Vos WM (2014) Maintenance of a healthy trajectory of the intestinal microbiome during aging: a dietary approach. Mech Ageing Dev 136–137: 70–75 [DOI] [PubMed] [Google Scholar]
- 186. Chotirmall SH, Burke CM (2015) Aging and the microbiome: implications for asthma in the elderly? Expert Rev Respir Med 9: 125–128 [DOI] [PubMed] [Google Scholar]
- 187. Conley MN, Wong CP, Duyck KM, Hord N, Ho E, Sharpton TJ (2016) Aging and serum MCP‐1 are associated with gut microbiome composition in a murine model. PeerJ 4: e1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Desai SN, Landay AL (2018) HIV and aging: role of the microbiome. Curr Opin HIV AIDS 13: 22–27 [DOI] [PubMed] [Google Scholar]
- 189. Ebersole JL, Kirakodu SS, Novak MJ, Orraca L, Martinez JG, Cunningham LL, Thomas MV, Stromberg A, Pandruvada SN, Gonzalez OA (2016) Transcriptome analysis of B cell immune functions in periodontitis: mucosal tissue responses to the oral microbiome in aging. Front Immunol 7: 272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Fischer N, Relman DA (2018) Clostridium difficile, aging, and the gut: can microbiome rejuvenation keep us young and healthy? J Infect Dis 217: 174–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Heintz C, Mair W (2014) You are what you host: microbiome modulation of the aging process. Cell 156: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Hughes GM, Leech J, Puechmaille SJ, Lopez JV, Teeling EC (2018) Is there a link between aging and microbiome diversity in exceptional mammalian longevity? PeerJ 6: e4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Kiecolt‐Glaser JK, Wilson SJ, Madison A (2018) Marriage and gut (microbiome) feelings: tracing novel dyadic pathways to accelerated aging. Psychosom Med 10.1097/PSY.0000000000000647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Kong F, Deng F, Li Y, Zhao J (2018) Identification of gut microbiome signatures associated with longevity provides a promising modulation target for healthy aging. Gut Microbes 10.1080/19490976.2018.1494102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lamming DW (2014) Hot topics at the intersection of aging and energetics: diabetes/insulin resistance, sirtuins, and the microbiome. F1000Res 3: 257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Lewy T, Hong BY, Weiser B, Burger H, Tremain A, Weinstock G, Anastos K, George MD (2018) Oral microbiome in HIV‐infected women: shifts in the abundance of pathogenic and beneficial bacteria are associated with aging, HIV load, CD4 count, and antiretroviral therapy. AIDS Res Hum Retroviruses 10.1089/AID.2017.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Maldonado‐Contreras A, Gerstein RM (2017) Editorial: no Trp, no B: surprising connectivity of diet, microbiome, aging, and adaptive immunity. J Leukoc Biol 101: 807–809 [DOI] [PubMed] [Google Scholar]
- 198. Nagpal R, Mainali R, Ahmadi S, Wang S, Singh R, Kavanagh K, Kitzman DW, Kushugulova A, Marotta F, Yadav H (2018) Gut microbiome and aging: physiological and mechanistic insights. Nutr Healthy Aging 4: 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Ni Lochlainn M, Bowyer RCE, Steves CJ (2018) Dietary protein and muscle in aging people: the potential role of the gut microbiome. Nutrients 10: E929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Paschos GK, FitzGerald GA (2017) Circadian clocks and metabolism: implications for microbiome and aging. Trends Genet 33: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Shoemark DK, Allen SJ (2015) The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer's disease. J Alzheimers Dis 43: 725–738 [DOI] [PubMed] [Google Scholar]
- 202. Steves CJ, Bird S, Williams FM, Spector TD (2016) The microbiome and musculoskeletal conditions of aging: a review of evidence for impact and potential therapeutics. J Bone Miner Res 31: 261–269 [DOI] [PubMed] [Google Scholar]
- 203. Xiao Y, Dong J, Yin Z, Wu Q, Zhou Y, Zhou X (2018) Procyanidin B2 protects against d‐galactose‐induced mimetic aging in mice: metabolites and microbiome analysis. Food Chem Toxicol 119: 141–149 [DOI] [PubMed] [Google Scholar]
- 204. Zapata HJ, Quagliarello VJ (2015) The microbiota and microbiome in aging: potential implications in health and age‐related diseases. J Am Geriatr Soc 63: 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Matsumoto M, Kurihara S, Kibe R, Ashida H, Benno Y (2011) Longevity in mice is promoted by probiotic‐induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS One 6: e23652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, Consolandi C, Quercia S, Scurti M, Monti D et al (2016) Gut microbiota and extreme longevity. Curr Biol 26: 1480–1485 [DOI] [PubMed] [Google Scholar]
- 207. O'Toole PW, Jeffery IB (2015) Gut microbiota and aging. Science 350: 1214–1215 [DOI] [PubMed] [Google Scholar]
- 208. Claesson MJ, Cusack S, O'Sullivan O, Greene‐Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G et al (2011) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 108(Suppl 1): 4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE et al (2017) Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci USA 114: 10719–10724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL et al (2017) Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA 114: 10713–10718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V et al (2016) Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 167: 1469–1480 e1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Wang HB, Loh DH, Whittaker DS, Cutler T, Howland D, Colwell CS (2018) Time‐restricted feeding improves circadian dysfunction as well as motor symptoms in the Q175 mouse model of Huntington's disease. eNeuro 5: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Carabotti M, Scirocco A, Maselli MA, Severi C (2015) The gut‐brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol 28: 203–209 [PMC free article] [PubMed] [Google Scholar]
- 214. Buxton OM, Cain SW, O'Connor SP, Porter JH, Duffy JF, Wang W, Czeisler CA, Shea SA (2012) Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med 4: 129ra143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Suwazono Y, Dochi M, Sakata K, Okubo Y, Oishi M, Tanaka K, Kobayashi E, Kido T, Nogawa K (2008) A longitudinal study on the effect of shift work on weight gain in male Japanese workers. Obesity (Silver Spring) 16: 1887–1893 [DOI] [PubMed] [Google Scholar]


