Abstract
Background
We have recently described a peculiar feature of the promoters in two Drosophila Tc1-like elements, Bari1 and Bari3. The AT-richness and the presence of weak core-promoter motifs make these promoters, that we have defined “blurry”, able to activate transcription of a reporter gene in cellular systems as diverse as fly, human, yeast and bacteria. In order to clarify whether the blurry promoter is a specific feature of the Bari transposon family, we have extended this study to promoters isolated from three additional DNA transposon and from two additional LTR retrotransposons.
Results
Here we show that the blurry promoter is also a feature of two vertebrate transposable elements, Sleeping Beauty and Hsmar1, belonging to the Tc1/mariner superfamily. In contrast, this feature is not shared by the promoter of the hobo transposon, which belongs to the hAT superfamily, nor by LTR retrotransposon-derived promoters, which, in general, do not activate transcription when introduced into non-related genomes.
Conclusions
Our results suggest that the blurry promoter could be a shared feature of the members of the Tc1/mariner superfamily with possible evolutionary and biotechnological implications.
Electronic supplementary material
The online version of this article (10.1186/s13100-019-0155-6) contains supplementary material, which is available to authorized users.
Keywords: Horizontal gene transfer; Promoter; Tc1/mariner transposons; Luciferase assay, transcriptional regulation, transposition, D; Melanogaster, S; Cerevisiae, H; Sapiens, E; Coli
Background
Transposable elements (TEs) are widespread genetic elements that have played a fundamental role in genome evolution [1], contributing to generating diversity, both at small and large scale [2], and to evolving new functions through molecular domestication [3] or exaptation [4]. Their ubiquitous presence in the genomes of extant species suggests an ancient history dating back to early living organisms, as well as an extraordinary ability to overcome the canonical genetic barriers between species, the latter being an intrinsic feature of the horizontal gene transfer process (HGT).
TEs, like other genes, are usually transmitted from parents to offspring, and propagate in the population. In parallel to one or several bursts of genomic expansion TEs are subjected to mutational load, denoting their a neutral mode of evolution. The overall absence of selection acting on TEs establishes a kind of “genomic homeostasis”, i.e. a balance in terms of numbers of functional and non-autonomous TE copies. When mutant copies overcome functional ones, the TE is destined to extinction in the genome in which it resides, an effect that could be also translated at the species level [5, 6]. Horizontal Transposon Transfer (HTT) is an important mechanism that mobile genetic elements undertake to escape extinction.
Tc1 and mariner are two related DNA transposon families, part of the Tc1/mariner/pogo/IS630 (typically referred to as Tc1/mariner in short) superfamily. They were first identified in nematode [7] and insect [8] genomes respectively, and are both characterized by an open reading frame encoding a transposase flanked by two terminal inverted repeats (TIRs) and TA dinucleotides representing duplicated target sites [9]. Previous studies have identified Tc1-like elements in a variety of animals and fungi [10], as well as in the parasitic amoebozoa Entamoeba invadens [11] and in plants [12]. The ubiquitous presence of Tc1/mariner-like elements in the genomes of virtually all extant eukaryotic species and the phylogenetic inconsistencies found in many cases studied [13, 14] support the hypotheses that they are ancient components of the eukaryotic genomes and could have also spread by means of HTT [15].
The establishment of a TE in a new genome after a HTT event depends on the expression of the TE-encoded genes required for element mobilization and propagation. Considering that expression of such genes relies to a large extent on the activity of their own promoters, it can be predicted that the greater the evolutionary distance between the donor and recipient species involved in the HTT event, the lesser the chance that the promoter could be recognized by transcription factors of the new host. Translated in terms of transposition efficiency, insufficient expression of transposition-related proteins would mean that the TE will be “dead on arrival” in the new host unless other surviving strategies are adopted.
We have recently reported that the promoter elements isolated from two related Drosophila Tc1-like transposons, Bari1 and Bari3, are functional in evolutionarily distant genomic backgrounds [16]. These transposons carry an AT-rich “blurry promoter” with divergent or no sharply predictable core-promoter motifs, which is able to drive transcription when transplanted into an unrelated genetic and genomic context. On the basis of our previously reported results, we hypothesized that this feature could be important for the success of HTT events of such genetic elements. The TF/TFBS (Transcription Factor/Transcription Factor Binding Site) recognition is indeed the result of complex co-evolution processes [17] that does not allow promoters to work properly in divergent genetic backgrounds. Our previous results [16], based on the comparison of the promoters of the Bari1 and Bari3 transposons with that of an LTR (Long Terminal Repeat) retrotransposon (the copia element), suggested that the blurry promoter is a peculiarity of the Tc1 family.
Here, in order to further investigate the existence of the blurry promoter in Tc1-like elements from other organisms and to assess whether mariner-like elements also have similar feature, we extended our study to the promoters of two additional members of the Tc1/mariner superfamily [Sleeping Beauty (SB) from fish and Hsmar1 from human] and two additional LTR retrotransposons (Tirant and ZAM) from D. melanogaster. Furthermore, we included in this study the promoter of hobo, a member of the hAT superfamily (named after the hobo, Activator and Tam3 founding elements [18]). The hAT superfamily represents an ideal outgroup to assess whether Class II elements, not related to the Tc1/mariner superfamily and able to undertake HTT [19–22], have blurry promoters that allow them to survive once transferred into distant genomes.
Our results suggest that the blurry promoter is a feature shared by the elements of the Tc1/mariner superfamily tested in this study, while the promoters of hobo, Tirant and Zam are usually functional within a limited range of genomic environments strictly related to the species of origin.
Results
Description of the sequences analyzed and outlined experimental strategy
SB and Hsmar1 were used as representative elements to gain insights into promoter features of the Tc1/mariner superfamily. The promoter of the hobo element was chosen as an outgroup to Class II of transposons, whereas Tirant and ZAM were used as representative elements of the LTR retrotransposons belonging to the Ty3-gypsy superfamily. For the sake of comparison, the results obtained from the present work have been integrated with the results obtained in our recent study [16] concerning the promoters of Bari1, Bari3 (two Tc1-like elements) and copia (a LTR retrotransposon of the Ty1-copia superfamily).
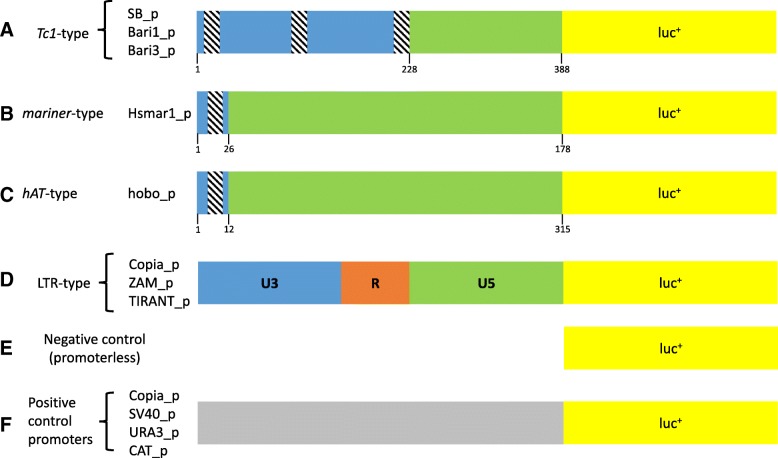
The promoters of LTR retrotransposons usually maps within the 5′ LTR of the elements [23], whereas the promoters of Tc1/mariner elements are commonly located in the 5′ terminus of the element and, as previously suggested, within the intervening sequences between the TIR and the ATG codon of the transposase open reading frame (ORF) [16] [24]. We cloned fragments of the test TEs predicted to contain the promoters (Table 1) into reporter constructs. The 5′ terminal sequences of the SB transposon assayed in this study (hereafter SB_p) contain three binding sites, two essential and one dispensable, for the respective transposase [25] and also contains an intervening sequence, in which the transposon’s endogenous promoter has been previously mapped [24] (Fig. 1a). The Hsmar1 5′ terminal sequence tested (hereafter referred as Hsmar1_p), contains a single, 19-bp-long, transposase binding sequence mapping to the nucleotides 7–25 relative to the sequence tested [26] and a 152-bp-long flanking sequence (Fig. 1b). The hobo sequence tested spans the first 315 bp of the canonical element and consists of the 5′ TIR of 12 bp and an intervening sequence of 303 bp (Fig. 1c).
Table 1.
Key features of the promoters analyzed in this study. The length of the tested fragment, the accession number of the reference element and its source organism, and the AT/GC content are listed for each element. References to previous works reporting the promoter characterization are also provided
| Element | Cloned fragment (bp) | Reference element | Source Organism | AT% | GC% | Promoter characterization |
|---|---|---|---|---|---|---|
| SB | 388 | L48685.1 [27] | T. albonubes | 63,66 | 36.34 | [24], This study |
| Bari1 | 377 | X67681.1 [28] | D. melanogaster | 66,05 | 33.95 | [16] [29] |
| Bari3 | 356 | CH933806 [30] | D. mojavensis | 65,23 | 34.77 | [16] [31] |
| Hsmar1 | 178 | U52077.1 [32] | H. sapiens | 70,79 | 29,21 | [33], This study |
| hobo | 315 | M69216.1 [18] | D. melanogaster | 51,43 | 48,57 | This study |
| Tirant | 416 | X93507.1 [34] | D. melanogaster | 54,81 | 45,19 | This study |
| ZAM | 472 | AJ000387.1 [35] | D. melanogaster | 55,08 | 44,92 | This study |
| copia | 276 | X02599.1 [36] | D. melanogaster | 72,46 | 27,54 | [16, 37] |
Fig. 1.
Overall scheme of the expression cassettes tested in this study. The tested sequences of Tc1-like, mariner-like and hobo elements (a, b and c) are divided into a blue region containing the transposase binding sites (depicted as dashed boxes) and sequences situated between the upstream TIRs and the transposase ORFs in these transposons (green region). These fragments are directly fused to a luciferase reporter gene (yellow). The LTR-retrotransposon sequences (d) are depicted with their canonical U3-R-U5 structure (more details are given in the main text). The organization of the negative (e) and positive (f) construct are also reported. Drawings are not in scale
The expression plasmids, carrying the luciferase reporter gene under the transcriptional control of the tested promoters, were either transiently (i.e. transfections into human HeLa cells and Drosophila S2R+ cells) or stably expressed (i.e. transformation in S. cerevisiae and E. coli) at the episomal level (see Material and Methods section). We used a promoter-less luciferase cassette as a negative control for background expression correction in each experiment (Fig. 1e). In addition, we used plasmids constitutively expressing luciferase under the control of strong species-specific promoters as positive controls, which also served as references to quantify the transposon-derived promoters’ activity (Fig. 1f).
Activities of TE promoters in human cells
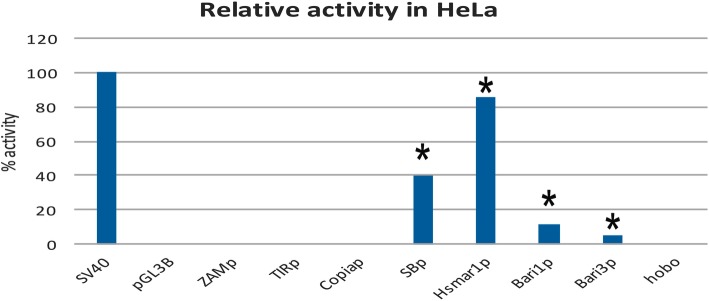
We first tested the promoter activity of SB and Hsmar1 in human HeLa cells (Fig. 2). As expected, and consistent with previously reported data [24, 33], SB_p and Hsmar1_p were able to drive reporter expression in cultured vertebrate cells (Fig. 2). In our hands, the SB_p has approximately 40% activity compared to the SV40 promoter. Similarly, Hsmar1_p is a strong promoter in HeLa cells (~ 80% activity when compared to SV40_p). As reported earlier [16], Bari promoters drive transcription of the reporter gene in HeLa cells. Conversely, the hobo promoter does not show any detectable activity in human cells. Likewise, the promoters of the retrotransposons copia ZAM and Tirant showed no detectable reporter activation over the background defined by the negative control (Fig. 2). These results suggest that the promoters isolated from the Drosophila hobo, Tirant and Zam elements do not support trans-Phylum transcription, i.e. in vertebrate cells.
Fig. 2.
Promoter analysis in HeLa cells. Relative promoter activity, expressed as corrected mean RLU values, relative to the positive control (SV40 promoter, set to 100) and to the promoter-less construct (set to 0). Promoters with activity significantly different from the promoter-less reporter cassette are marked with an asterisk. Actual values of each experiment set are shown in Additional file 5: Figure S1
Transcriptional activity of transposon promoters in insect cells
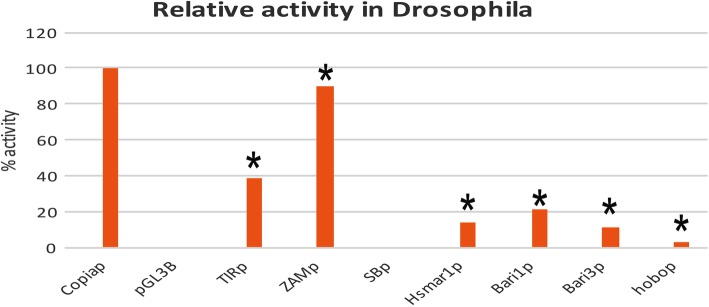
We next tested the ability of Hsmar1_p and SB_p to drive trans-Phylum transcription in insect cells by performing the luciferase assay in S2R+ Drosophila cells, using as a reference and positive control the pGL3B/copia expression vector. Whereas SB_p did not show an activity significantly different from the negative control, Hsmar1_p activity was roughly 14% compared to the copia promoter, suggesting that Hsmar1_p is an active promoter in Drosophila cells (Fig. 3). This result mirrors the ability of Bari promoters to drive trans-Phylum transcription human HeLa cells (Fig. 2).
Fig. 3.
Promoter analysis in S2R+ cells. Relative promoter activity, expressed as corrected mean RLU values, relative to the positive control (copia promoter, set to 100) and to the promoter-less construct (set to 0). Promoters with activity significantly different from the promoter-less reporter cassette are marked with an asterisk. Actual values of each experiment set are shown in Additional file 6: Figure S2. The statistical significance of promoter activity against the promoter-less cassette is shown. **P < 0.005; ***P < 0.001
The hobo promoter displayed significant, although weak, promoter activity in Drosophila, consistent with being an endogenous element of the fly genome [38].
Moreover, and consistently with our previous report, both Bari promoters were active in driving reporter gene expression in Drosophila. Finally, due to their retroviral nature, the promoters from ZAM and Tirant displayed an expected intense transcriptional activation of the reporter, as compared to the copia promoter (Fig. 3).
Promoter analysis in yeast
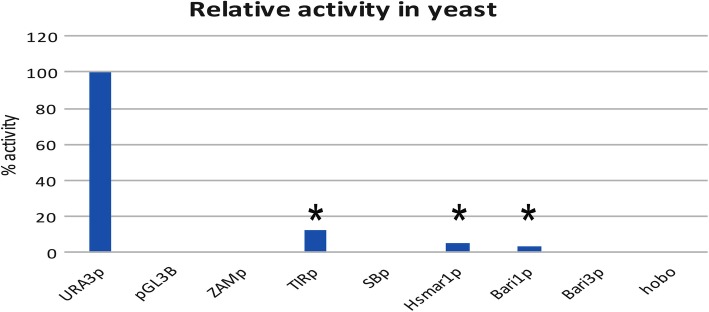
In S. cerevisiae the activity of TE promoters was compared to the URA3 promoter and to the promoter-less construct. To this end, the same sequences tested in animal cells (i.e. human and insect cells) were sub-cloned into pFL39, a low-copy centromeric yeast vector that allowed an easy clonal analysis in S. cerevisiae. Interestingly, and similarly to what was previously found for Bari1_p, the activity of Hsmar1_p was about 4% of the strong URA3 promoter, suggesting that it is a weak, but active promoter in yeast (Fig. 4). We also observed a promoter activity associated with the TIR_p sequence, at the level of 14% of the URA3 promoter, a feature not shared by the hobo promoter and by the other LTR-derived promoters tested in this study (Fig. 4).
Fig. 4.
Promoter analysis in yeast cells. Relative promoter activity, expressed as corrected mean RLU values, relative to the positive control (URA3 promoter, set to 100) and to the promoter-less construct (set to 0). Promoters with activity significantly different from the promoter-less reporter cassette are marked with an asterisk. Actual values of each experiment set are shown in Additional file 7: Figure S3. The statistical significance of promoter activity against the promoter-less cassette is shown. *P < 0.05; **P < 0.005
Promoter analysis in bacteria
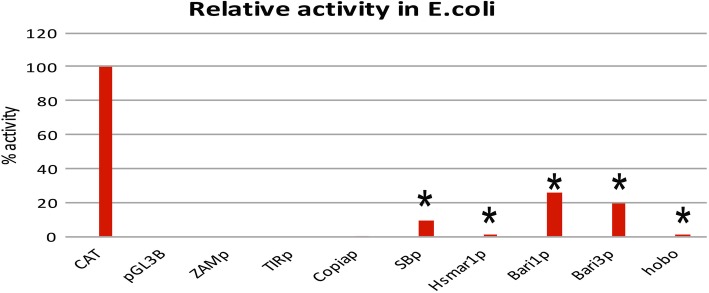
Luciferase assays conducted in E. coli showed that the activity of Hsmar1_p and hobo_p are very low when compared to the CAT promoter (0.15% and 0,42% respectively), but still significantly different from the promoter-less vector. Contrarily, SB_p displayed a strong activity, representing roughly 17% of the CAT promoter activity (Fig. 5). These results suggest that SB_p is more similar to the Bari promoters in E. coli (Fig. 5). No significant promoter activity has been observed to the tested sequences isolated from LTR-retrotransposons, which suggest the poor ability of these sequences to drive transcription in cells of a different domain of life.
Fig. 5.
Promoter analysis in E. coli cells. Relative promoter activity, expressed as corrected mean RLU values, relative to the positive control (CAT promoter, set to 100) and to the promoter-less construct (set to 0). Promoters with activity significantly different from the promoter-less reporter cassette are marked with an asterisk. Actual values of each experiment set are shown in Additional file 8: Figure S4. The statistical significance of promoter activity against the promoter-less cassette is shown. ***P < 0.001
Eukaryotic core promoter motifs in test TEs
With the aim of identifying the core promoter sequences that could explain the observed behavior of the sequences tested in this study, we combined a bioinformatics approach with expressed sequence tags (ESTs) mapping and scientific literature data mining. We scanned the sequences of SB_p, Hsmar1_p, hobo_p, ZAM_p, TIR_p and Copia_p using matrices describing the TATA-box, the initiator element (InR) and the downstream promoter element (DPE), three DNA motifs commonly found in eukaryotic promoters. The results obtained using relaxed parameters (Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 3: Table S3) show that it is not possible to identify all of the searched motifs in the Tc1/mariner elements. Indeed, SB_p lacks predictable Inr motifs, whereas two of the predicted TATA boxes and two of the predicted DPEs fall into the sequence previously described to contain the promoter [24].
The transcriptional start site (TSS) of Hsmar1 can be inferred from a group of ESTs that arise from Hsmar1 relics in the human genome (Additional File 4) that allow mapping of the TSS at position 130 in Hsmar1_p. The matrix-scan analysis performed on Hsmar1_p revealed two TATA boxes falling downstream the transposase binding site region, and mapping 56 and 28 bp upstream the inferred TSS. No Inr and DPE motifs can be detected using the matrix-scan approach.
The matrix-scan analysis of the hobo promoter reveals a high scoring TATA-box motif at position 105–116, which is in accordance with the TATA-box annotated in the GenBank record (M69216.1), and low-scoring Inr and DPE motifs at position 156–162 and 305–309 respectively. A CAAT box (position 49–52) is also annotated in the GenBank file. No transcripts arising from within the analyzed hobo sequence could be found in the database, in accordance with the reported low transcriptional activity of hobo in D. melanogaster [39].
The promoters of LTR-retrotransposons are usually located within specific LTR sub-regions. The LTRs of Ty3 and Ty1 elements usually contain three functional elements, namely U3 (Unique 3′ region), R (Repeated sequence) and U5 (Unique 5′ region) [40]. The R region starts invariably at the transcription start site and ends at the polyadenylation site. Therefore, the R sequence must contain the Inr motif and possibly the DPE motif, whereas the TATA box should be expected to lay in the U3 region. Previous studies have unveiled the U3-R-U5 structure of the ZAM element [35, 41, 42], which identified a 22-bp R region (nucleotides 326 to 347). A predicted TATA-box (Additional file 1: Table S1) maps at nucleotides 313–317, thus falling into the U3 region. The Inr of ZAM is defined by the TSS (also supported by a group of ESTs identified using BLASTn, see Additional File 4) across or near the R region start, although no sharp Inr can be predicted using our approach with the applied cutoff parameters (Additional file 2: Table S2). A potential DPE motif maps 24 bp downstream the Inr. The relative orientation and spacing of the three core promoter motifs are compatible, although sub-optimal, with the expected configuration of an ideal eukaryotic promoter [43].
To our knowledge, no data is available from the scientific literature concerning Tirant TSS. As a preliminary attempt to map the minimal promoter region of Tirant, we conducted a BLASTn search against the D. melanogaster ESTs database. As shown in Additional File 4, two different subsets of ESTs support two TSSs in the head and in embryos, thus predicting the possible U3-R boundary. One of the predicted TATA boxes and Inr motifs have compatible spacing (TATA-box at -28 bp, DPE at + 48 with respect to the Inr) with this predicted U3-R configuration.
It has been assessed by previous studies that the copia promoter lies within bases 70–276 [37]. Despite its wide use in expression vectors, little and contrasting information is available about the organization of its core-promoter motifs [37, 44, 45]. Our core-promoter motif prediction fits, in part, with the TATA box predicted at position 83–94, and the Inr (position 118–124) lying 6 bases upstream the Inr [45], and a DPE (position 179–183) outside the expected sequence spacing respect to the Inr motif.
Taken together, these results suggest that LTR-derived promoters and hobo_p might have a more structured organization compared to the promoters derived from Tc1/mariner elements, which more often have divergent, or sometimes lack, basal core-promoter elements, but still are able to drive transcription of a downstream gene.
Discussion
TEs are special genomic components with an intrinsic ability to move within the same genome and across different genomes. This latter feature is more prominent for Class II transposons (i.e. DNA transposons), and particularly for Tc1/mariner elements [22]. However, the chance for survival of a TE upon HTT relies on several bottlenecks and on the possibility to express the transposase gene(s) [13].
The experimental procedure of transfection/transformation, routinely applied in many laboratories, mimics the naturally occurring HGT (and HTT) process, where the donor and receiving cells might be from different species. Thus, studying the activity of transcriptional regulatory sequences, using evolutionarily distant recipient cells in vitro, could allow us to infer how the tested sequences would determine the activation of gene expression after HGT. We have recently shown that the promoters isolated from Bari1 and Bari3, two Drosophila Tc1-like transposons [46], are able to drive transcription of a downstream reporter gene in unrelated cellular environments, a feature not shared by the promoter of the copia retrotransposon. Based on this evidence, we have proposed that Tc1-like elements contain blurry promoters, i.e. AT-rich sequences containing weak core-promoter motifs [16].
In the present study, with the aim of validating the blurry promoter hypothesis and assess if this could be a feature shared with other TE families, we have analyzed the promoter region of three DNA transposons (SB, Hsmar1 and hobo) and two LTR retrotransposons (ZAM and Tirant). Our analyses suggest that the promoter region of SB (a Tc1-like element) and Hsmar1 (a mariner-like element) share structural and functional similarity with the promoter of Bari elements. First, a base usage analysis of the promoters from Tc1/mariner transposons studied reveals that they are all AT-rich in sequence. Conversely, the promoter of hobo, which belongs to the hAT superfamily, and the promoters of LTR-retrotransposons, with the exception of copia (72% AT), have an unbiased base content (Table 1). Second, the core-promoter search in the tested sequences suggests that the Tc1/mariner elements are less structured, might lack several core-promoter motifs and/or they might be unevenly spaced (Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 3: Table S3). Third, the promoter of Tc1/mariner elements is the ability to activate gene transcription in distant hosts, a distinctive feature of the transposon’s “blurry” promoters. Blurry promoters could predispose certain TEs to be more successful in HTT than other coding and noncoding DNA sequences [21], thereby contributing to the success of TEs in establishing a foothold in a new genome upon a HTT event. Here, we show that this feature is not distinctive of the two Bari transposon promoters, nor limited to transposons of the Drosophila genus, but it can be extended at least to other members of the Tc1/mariner superfamily.
SB and Hsmar1 are two inactive transposable elements found in vertebrates, whose transposase genes have been reconstructed through in vitro site directed mutagenesis [33, 47]. Thousands of SB and Hsmar1 relics can be found in extant salmonid fish and primate genomes respectively. For both transposons, the terminal sequences were found to be the most conserved in sequence, i.e. less divergent due to the accumulation of disrupting mutations. Indeed, the SB_p sequence used in this study was derived from a Tc1-like element isolated from the White Cloud Mountain fish, Tanichthys albonubes [47], whereas the Hsmar1_p sequence is part of a consensus assembled from the human genome [33]. The results obtained in this study clearly show that similarly to what was observed for the promoter of Bari elements, SB_p and Hsmar1_p are able to drive transcription in distant genomes.
Unlike SB_p and Hsmar1_p, neither the LTR-derived promoters nor hobo_p displayed significant activity in HeLa cells. While it would be expected that vertebrate promoters could be active in a vertebrate non-resident cellular environment, it should be noted that primates and fishes (Chondrichthyes) shared their last common ancestor more than 460 Mya (source http://www.timetree.org/). Despite their ancient relationship, the transcriptional machinery in fishes and humans are apparently able to cross-recognize cis-acting sequences, suggesting that some TFs and TFBSs are interchangeable in these two lineages. This observation is also supported by several studies in which fish promoters were tested in human cells [48, 49].
The transfection of SB_p and Hsmar1_p into Drosophila cells (Fig. 3) mimics an inter-phyla HTT event, from vertebrates to insects. Our results show that, unlike SB_p, Hsmar1_p is an active promoter in Drosophila and its activity is comparable to that of Bari3_p. Although we could detect a promoter activity significantly different from the promoter-less control the hobo_p promoter is a weak promoter in Drosophila (Fig. 2). Previous works have highlighted that hobo transcriptional activity can only be detected in the germline, with very poor (or absent) somatic expression in Drosophila [39], suggesting strong transcriptional regulation of transposase expression. The S2R+ cell line used in this study is indeed of somatic origin [50] thus explaining the observed low promoter activity in our assay.
Even more strikingly, we have observed promoter activity of SB_p in bacteria and of Hsmar1_p in yeast (Fig. 4). Transformation into yeast and bacteria of vertebrate sequences could be seen as laboratory recapitulations of inter-Kingdom HTT events (i.e. HTT events involving animals and fungi) and inter-Domain HTT events, (i.e. HTT event involving a eukaryote and a prokaryote). While promoters from Tc1/mariner like transposons are generally active upon inter-Kingdom or inter-Domain transfer, the LTR-derived promoters appear to be less prone to support transcription in distantly related hosts (Figs. 4 and 5). However, some of the non-Tc1/mariner elements tested in this study are also able to drive gene expression outside their native host genetic environment (Figs. 4 and 5). This observation implies that many other types of TEs could be able to activate transcription of their transposition-related enzymes when they move from the species of origin to an unrelated one, but in a more narrow range of species if compared to Tc1/mariner elements. In conclusion, the results presented in this study involving the promoters from TEs of different types (i.e. 4 Tc1/mariner, 1 hAT and 3 LTR-retrotransposons elements), suggest that Tc1/mariner elements are more promiscuous in activating transcription in distantly related hosts, a feature that could be linked to their enhanced success in the HTT process [21, 22].
Possible impact of HGT of active promoters on genes, genomes and adaptation
Despite being inactive as transposons, several thousands of SB and Hsmar1 relics, most of which contain the promoter sequences, can be found in the respective host genomes, suggesting possible impact on gene regulation in their native hosts. For example, transposon insertions can affect host gene expression by juxtaposition of their transcriptional regulatory sequences to endogenous transcription units, thereby overriding their physiological transcriptional program. D. melanogaster provides significant examples of transcriptional perturbation of host genes [51–55] due to potent TE-related cis-acting regulatory sequences, such as enhancers, silencers or insulators, mainly associated with LTR-retrotransposons [56–61]. Our results suggest that promoters of the analyzed TEs act as weak transcriptional activators, especially when they move to distant hosts. For instance, the promoters of Hsmar1 and hobo have a very low, although statistically significant, activity in E. coli. This could still have a biological significance, especially in the absence of additional repressive mechanisms that acted on the TE in the species of origin, leading to a successful invasion of the new genome.
In natural ecosystems, the relationships between different lineages have been established during evolution. Environmental perturbations could be seen as genetic stressors by the community members, which would react promptly especially with the acquisition of novel traits allowing them to persist in the same environment or to colonize new environments. The role of HGT/HTT in the determination of gene flow in ecological niches is well recognized [62]. In this context, genome colonization by TEs could be a critical contributor to eliciting complex genetic changes occurring in ecological niches of special interest, such as intestine and soil. Investigations of TE-associated promoters will help understanding the potential invasiveness of the TE in new genomes and the potential ability to rewire pre-existing transcriptional circuitries, leading to adaptation in a given eco-niche.
Potential biotechnological application of TE- related promoters
To date, few promoters of viral origin are known to drive transcription in multiple hosts. As an example the 35S promoter of the CaMV virus, a strong constitutive plant promoter [63], displays its activity in virtually all organisms, from higher vertebrates to prokaryotes [64] [65]. Similarly, although not so strikingly, the promoter of the crustacean IHHNV virus, is able to drive the transcription of reporter genes in insects and fish cells [66]. Given the non-viral origin of TEs, these sequences, especially Class II transposons, have been recently highly regarded in the development of biotechnological tools, especially in the field of gene therapy [67]. Moreover, some TE-related regulatory sequences (e.g. the copia promoter) have been used to develop commercially available in vitro gene expression tools.
The observation that the promoters of some transposon families display transcriptional activation of downstream genes in multiple cellular hosts enables the development of expression vectors based on blurry promoters. Enabling ectopic expression in many cellular model systems using a single recombinant construct could be useful in basic research to accelerate industrial production processes. The weakness of the promoters described here could limit their use in such applications. However, this could be a desirable feature to express genes closer to their physiological levels. To better assess the strength of TE-derived promoters a comparison with weaker promoters will be needed. Further investigations in the field of TEs and mutagenesis of preliminary characterized TE- related promoters could hopefully lead to a wider application of TE-derived regulatory sequences for the development of new biotechnological tools with a broader range of transcriptional activation in multiple hosts.
Conclusion
This study demonstrates that, similar to the Bari elements isolated from Drosophila, the SB (from fish) and Hsmar1 (human) transposons contain transcriptionally promiscuous “blurry” promoters. The blurry promoter could be a feature widely shared among the members of the Tc1/mariner superfamily.
The results obtained in this work confirm our previous hypothesis that the promoters of TEs from the Tc1/mariner superfamily could contribute to the success and spread of these mobile elements and might represent one of the keys towards the full understanding of the complex phenomenon of HTT.
Given the high number of annotated and characterized TEs, we believe that other elements could also carry promiscuous promoters. Continuing effort put into the study of TE’s regulatory sequences, would enable the discovery of additional peculiar features that could be used in biotechnological applications and would allow a significant advancement in the field of TE biology.
Materials and methods
S2R+ cells were passaged in Schneider’s Insect Medium supplemented with 10% FBS, 1% penicillin/streptomycin, and maintained at 25 °C. HeLa cells were maintained at 37 °C with 5% CO2 in Dulbecco’s Minimum Essential Medium supplemented with 10% FBS, 200 mM glutamine, 1% penicillin/streptomycin. Saccharomyces cerevisiae, strain BMA64-1A (MATa leu2–3112 his3–11,15 trp1∆ ade2–1 ura3–1), was cultured on Synthetic Complete medium [68] supplemented with 2% glucose as carbon source. Escherichia coli cultures, strain DH5alpha, were grown on selective LB medium supplemented with ampicillin.
The transposable elements’ fragments analyzed in this study have been amplified using the primers listed in Table 2 and cloned into the pGL3B vector. The promoter of hobo, ZAM and Tirant were amplified from the D. melanogaster reference strain genomic DNA (y1; cn1, bw1, sp1). The entire expression cassettes (i.e. promoter-luc + SV40 terminator) were cut out from the pGL3B vector and sub-cloned into pFL39 vector [69] using either KpnI and BamHI or the SacI and SalI restriction sites. The pFL39 vector is a centromeric yeast plasmid that allows selection of stably transformed clones on selective media lacking tryptophan. The constructs containing Bari1 and Bari3 promoters, as well as the control promoters used in this study (namely Copia_p, URA3_p, and CAT_p), have been described in [16]. Sequence verification of all the cloned fragments was performed at the BMR-Genomics (Padova, Italy) using the Luc278_rev sequencing primer.
Table 2.
List of primers used in this study
| SB_f | CTCGAGCAGTTGAAGTCGGAAGTTTACATACACTTAGGTTGGAG |
|---|---|
| SB_r | CCATGGATGTTTTTGGCGTCTTCCATGATGTCAAGCAAAGAGGCACTG |
| Hsmar1_f | CTCGAGTTAGGTTGGTGCAAAAGTAATTGC |
| Hsmar1_r | CCATGGAGTCTAAAATAAACATAAAATAAACA |
| Tirant_LTR_f | CTCGAGGGAGTTACCACCCCACCCCCTA |
| Tirant_LTR_r | AGATCTCAGTTAAGTCCGTGATCGAGGGT |
| ZAM_LTR_f | CTCGAGTACCGACCCATCGGTACCATAC |
| ZAM_LTR_r | CCATGGGCGCAGTTACCTCCGGGGAGTCT |
| hobo_prom_UP | GATCCTCGAGCAGAGAACTGCAAGGGTGGCACT |
| hobo_prom_LOW | GATCCCATGGTTGACTCGACTACCTACGAGA |
| Luc278_rev | GCCCAACACCGGCATAAAGAATT |
Transfections were performed in 6-well plates using TransIt LT1 (Mirus Bio, Madison, WI), and 1 μg of the appropriate plasmid or co-transfected with the Renilla luciferase construct (pRL-SV40; Promega, Madison, WI, USA), according to the manufacturer instructions. Yeast transformation was performed using the TRAFO methods described in [70]. Bacteria transformation was performed using standard transformation protocol and chemically competent cells [71]. Luciferase activities measurements were recorded on GLOMAX 20/20 Luminometer (Promega, Madison, WI, USA) 24 h post-transfection. S. cerevisiae and E. coli transformants were assayed in the log-phase. We recorded three independent luciferase activity measurements per sample and the average value taken as sample measure. Experiments were made at least in triplicates.
RLUs obtained for the tested promoters, were corrected for the RLU value of the respective negative control, and normalized to the total protein content measured using the Bradford Assay or to the Renilla luciferase measure.
The normalized luciferase activity of positive controls, were arbitrarily set to 100, whereas the negative controls were set to zero in each experiment. T-student test was carried out to assess the statistical significance of the differences observed between the transposons’ promoter tested and the respective promoter-less construct (H0: normalized RLU measurement is not different between test promoter and negative control promoter).
Positional weight matrices used to map the TATA-box, Inr (Initiator Element) and DPE (Downstream Promoter Element) core-promoter motifs were retrieved at YAPP Eukaryotic Core Promoter Predictor (www.bioinformatics.org/yapp/cgi-bin/yapp.cgi; last accessed June 2018). Matrix scan analysis was performed using Regulatory Sequence Analysis Tools (RSAT) (http://pedagogix-tagc.univ-mrs.fr/rsat/RSAT_portal.html; last accessed July 2018) [72]).
BLAST searches were performed at the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against appropriate databases.
Additional files
Table S1. TATA box motif prediction. Pval cutoff 10exp-2; origin = START; only direct strand results are shown (DOCX 180 kb)
Table S2. Inr motif prediction. Pval cutoff 10exp-2; origin = START; only direct strand results are shown (DOCX 115 kb)
Table S3. DPE motif prediction. Pval cutoff 10exp-2; origin = START; only direct strand results are shown (DOCX 151 kb)
Accessions of cDNA/ESTs supporting TSS predicted in the sequences tested (TXT 643 bytes)
Figure S1. Luciferase-promoter assay in HeLa cells. Individual promoter-luciferase assay results in HeLa cells. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. *P < 0.05; **P < 0.005; ***P < 0.001 (PDF 168 kb)
Figure S2. Luciferase-promoter assay in S2R+ cells. Individual promoter-luciferase assay results in S2R+ cells of D. melanogaster. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. **P < 0.005; ***P < 0.001 (PDF 174 kb)
Figure S3. Luciferase-promoter assay in S. cerevisiae cells (BMA64-1A). Individual promoter-luciferase assay results in yeast. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. *P < 0.05; **P < 0.005 (PDF 216 kb)
Figure S4. Luciferase-promoter assay in bacteria (DH5alpha cells). Individual promoter-luciferase assay results in DH5alpha cells. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. ***P < 0.001 (PDF 154 kb)
Acknowledgements
We thank Prof. Ruggiero Caizzi for helpful critical comments on this work. We are also grateful to the Reviewers for their thoughtful comments and efforts towards improving our manuscript.
Funding
No specific funds were received for this project.
Availability of data and materials
All the materials mentioned and used in this work will be made available upon request.
Abbreviation
- EST
Expressed Sequence Tags
- HGT
Horizontal Gene Transfer
- HTT
Horizontal Transposon Transfer
- LTR
Long Terminal Repeat
- ORF
Open Reading Frame
- R
Repeated sequence
- TEs
Transposable Elements
- TF
Transcription Factor
- TFBS
Transcription Factor Binding Site
- TIRs
Terminal Inverted Repeats
- TSS
Transcriptional Start Site
- U3
Unique 3′ region
- U5
Unique 5′ region
Authors’ contributions
AP, PL, AG, CMTM, RMM performed cell cultures and promoter-luciferase assays. AP, PL, CM, OW, RMM generated plasmids. RMM analyzed data. ZI, RMM wrote the manuscript. RMM conceived and designed the analyses. All Authors read and approved the final version the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The Authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Antonio Palazzo, Email: antonio.palazzo@uniba.it.
Patrizio Lorusso, Email: p.lorusso25@studenti.uniba.it.
Csaba Miskey, Email: Csaba.Miskey@pei.de.
Oliver Walisko, Email: Oliver.Walisko@pei.de.
Andrea Gerbino, Email: andrea.gerbino@uniba.it.
Carlo Marya Thomas Marobbio, Email: carlomarya.marobbio@uniba.it.
Zoltán Ivics, Email: Zoltan.Ivics@pei.de.
René Massimiliano Marsano, Phone: +390805442241, Email: renemassimiliano.marsano@uniba.it.
References
- 1.Gray YHM. It takes two transposons to tango:transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16(10):461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 2.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jangam D, Feschotte C, Betran E. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 2017;33(11):817–831. doi: 10.1016/j.tig.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joly-Lopez Z, Bureau TE. Exaptation of transposable element coding sequences. Curr Opin Genet Dev. 2018;49:34–42. doi: 10.1016/j.gde.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Hartl DL, Lozovskaya ER, Nurminsky DI, Lohe AR. What restricts the activity of mariner-like transposable elements. Trends Genet. 1997;13(5):197–201. doi: 10.1016/s0168-9525(97)01087-1. [DOI] [PubMed] [Google Scholar]
- 6.Miskey C, Izsvak Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell Mol Life Sci. 2005;62(6):629–641. doi: 10.1007/s00018-004-4232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babity JM, Starr TV, Rose AM. Tc1 transposition and mutator activity in a Bristol strain of Caenorhabditis elegans. Mol Gen Genet. 1990;222(1):65–70. doi: 10.1007/BF00283024. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson JW, Medhora MM, Hartl DL. Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci U S A. 1986;83(22):8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plasterk RH, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15(8):326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 10.Robertson HM. Mobile DNA II: American Society of Microbiology. 2002. Evolution of DNA transposons in eukaryotes. [Google Scholar]
- 11.Pritham EJ, Feschotte C, Wessler SR. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol Biol Evol. 2005;22(9):1751–1763. doi: 10.1093/molbev/msi169. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Yang G. Tc1-like transposable elements in plant genomes. Mob DNA. 2014;5:17. doi: 10.1186/1759-8753-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6(1):57–71. [PubMed] [Google Scholar]
- 14.Loreto EL, Carareto CM, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100(6):545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- 15.Capy P, Langin T, Bigot Y, Brunet F, Daboussi MJ, Periquet G, et al. Horizontal transmission versus ancient origin: mariner in the witness box. Genetica. 1994;93(1–3):161–170. doi: 10.1007/BF01435248. [DOI] [PubMed] [Google Scholar]
- 16.Palazzo A, Caizzi R, Viggiano L, Marsano RM. Does the promoter constitute a barrier in the horizontal transposon transfer process? Insight from Bari transposons. Genome Biol Evol. 2017;9(6):1637–1645. doi: 10.1093/gbe/evx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tugrul M, Paixao T, Barton NH, Tkacik G. Dynamics of transcription factor binding site evolution. PLoS Genet. 2015;11(11):e1005639. doi: 10.1371/journal.pgen.1005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvi BR, Hong TJ, Findley SD, Gelbart WM. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, activator, and Tam3. Cell. 1991;66(3):465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 19.Bernardo LP, Loreto EL. Hobo-brothers elements and their time and place for horizontal transfer. Genetica. 2013;141(10–12):471–478. doi: 10.1007/s10709-013-9746-1. [DOI] [PubMed] [Google Scholar]
- 20.Simmons GM. Horizontal transfer of hobo transposable elements within the Drosophila melanogaster species complex: evidence from DNA sequencing. Mol Biol Evol. 1992;9(6):1050–1060. doi: 10.1093/oxfordjournals.molbev.a040774. [DOI] [PubMed] [Google Scholar]
- 21.Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25(9):537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert C, Feschotte C. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev. 2018;49:15–24. doi: 10.1016/j.gde.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havecker ER, Gao X, Voytas DF. The diversity of LTR retrotransposons. Genome Biol. 2004;5(6):225. doi: 10.1186/gb-2004-5-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walisko O, Schorn A, Rolfs F, Devaraj A, Miskey C, Izsvak Z, et al. Transcriptional activities of the sleeping beauty transposon and shielding its genetic cargo with insulators. Mol Ther. 2008;16(2):359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- 25.Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in sleeping beauty transposition. J Biol Chem. 2002;277(37):34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 26.Roman Y, Oshige M, Lee YJ, Goodwin K, Georgiadis MM, Hromas RA, et al. Biochemical characterization of a SET and transposase fusion protein, Metnase: its DNA binding and DNA cleavage activity. Biochemistry. 2007;46(40):11369–11376. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivics Z, Izsvak Z, Minter A, Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci U S A. 1996;93(10):5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caizzi R, Caggese C, Pimpinelli S. Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics. 1993;133(2):335–345. doi: 10.1093/genetics/133.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palazzo A, Marconi S, Specchia V, Bozzetti MP, Ivics Z, Caizzi R, et al. Functional characterization of the Bari1 transposition system. PLoS One. 2013;8(11):e79385. doi: 10.1371/journal.pone.0079385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moschetti R, Chlamydas S, Marsano RM, Caizzi R. Conserved motifs and dynamic aspects of the terminal inverted repeat organization within Bari-like transposons. Mol Gen Genomics. 2008;279(5):451–461. doi: 10.1007/s00438-008-0324-7. [DOI] [PubMed] [Google Scholar]
- 31.Palazzo A, Moschetti R, Caizzi R, Marsano RM. The Drosophila mojavensis Bari3 transposon: distribution and functional characterization. Mob DNA. 2014;5:21. doi: 10.1186/1759-8753-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson HM, Zumpano KL. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205(1–2):203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- 33.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, et al. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol Cell Biol. 2007;27(12):4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viggiano L, Caggese C, Barsanti P, Caizzi R. Cloning and characterization of a copy of Tirant transposable element in Drosophila melanogaster. Gene. 1997;197(1–2):29–35. doi: 10.1016/s0378-1119(97)00217-5. [DOI] [PubMed] [Google Scholar]
- 35.Leblanc P, Desset S, Dastugue B, Vaury C. Invertebrate retroviruses: ZAM a new candidate in D.Melanogaster. EMBO J. 1997;16(24):7521–7531. doi: 10.1093/emboj/16.24.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emori Y, Shiba T, Kanaya S, Inouye S, Yuki S, Saigo K. The nucleotide sequences of copia and copia-related RNA in Drosophila virus-like particles. Nature. 1985;315(6022):773–776. doi: 10.1038/315773a0. [DOI] [PubMed] [Google Scholar]
- 37.Sinclair JH, Burke JF, Ish-Horowicz D, Sang JH. Functional analysis of the transcriptional control regions of the copia transposable element. EMBO J. 1986;5(9):2349–2354. doi: 10.1002/j.1460-2075.1986.tb04503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3(12):RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blackman RK, Grimaila R, Koehler MM, Gelbart WM. Mobilization of hobo elements residing within the decapentaplegic gene complex: suggestion of a new hybrid dysgenesis system in Drosophila melanogaster. Cell. 1987;49(4):497–505. doi: 10.1016/0092-8674(87)90452-1. [DOI] [PubMed] [Google Scholar]
- 40.Arkhipova IR, Mazo AM, Cherkasova VA, Gorelova TV, Schuppe NG, Llyin YV. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986;44(4):555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- 41.Meignin C, Dastugue B, Vaury C. Intercellular communication between germ line and somatic line is utilized to control the transcription of ZAM, an endogenous retrovirus from Drosophila melanogaster. Nucleic Acids Res. 2004;32(13):3799–3806. doi: 10.1093/nar/gkh708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS One. 2008;3(2):e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339(2):225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sneddon A, Flavell AJ. The transcriptional control regions of the copia retrotransposon. Nucleic Acids Res. 1989;17(11):4025–4035. doi: 10.1093/nar/17.11.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkhipova IR, Ilyin YV. Control of transcription of Drosophila retrotransposons. Bioessays. 1992;14(3):161–168. doi: 10.1002/bies.950140305. [DOI] [PubMed] [Google Scholar]
- 46.Palazzo A, Lovero D, D’Addabbo P, Caizzi R, Marsano RM. Identification of Bari Transposons in 23 Sequenced Drosophila Genomes Reveals Novel Structural Variants, MITEs and Horizontal Transfer. PLoS ONE. 2016;11(5):e0156014. [DOI] [PMC free article] [PubMed]
- 47.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91(4):501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 48.Bernardi E, Deflorian G, Pezzimenti F, Diaz VM, Mione M, Blasi F. Characterization of the regulatory region of the zebrafish Prep1.1 gene: analogies to the promoter of the human PREP1. PLoS One. 2010;5(12):e15047. doi: 10.1371/journal.pone.0015047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenner S, Venkatesh B, Yap WH, Chou CF, Tay A, Ponniah S, et al. Conserved regulation of the lymphocyte-specific expression of lck in the Fugu and mammals. Proc Natl Acad Sci U S A. 2002;99(5):2936–2941. doi: 10.1073/pnas.032680599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273(48):32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- 51.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci U S A. 1996;93(18):9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desset S, Conte C, Dimitri P, Calco V, Dastugue B, Vaury C. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol Biol Evol. 1999;16(1):54–66. doi: 10.1093/oxfordjournals.molbev.a026038. [DOI] [PubMed] [Google Scholar]
- 53.Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc Natl Acad Sci U S A. 2003;100(23):13436–13441. doi: 10.1073/pnas.2333111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7(4):597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Chung H, Bogwitz MR, McCart C, Andrianopoulos A, Ffrench-Constant RH, Batterham P, et al. Cis-regulatory elements in the accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175(3):1071–1077. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376(6540):533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 57.Cai HN, Levine M. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 1997;16(7):1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson S, Matyunina LV, McDonald JF. An enhancer region within the copia untranslated leader contains binding sites for Drosophila regulatory proteins. Gene. 1998;209(1–2):239–246. doi: 10.1016/s0378-1119(98)00048-1. [DOI] [PubMed] [Google Scholar]
- 59.Conte C, Dastugue B, Vaury C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol Cell Biol. 2002;22(6):1767–1777. doi: 10.1128/MCB.22.6.1767-1777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minervini CF, Ruggieri S, Traversa M, D'Aiuto L, Marsano RM, Leronni D, et al. Evidences for insulator activity of the 5'UTR of the Drosophila melanogaster LTR-retrotransposon ZAM. Mol Gen Genomics. 2010;283(5):503–509. doi: 10.1007/s00438-010-0529-4. [DOI] [PubMed] [Google Scholar]
- 61.Bire S, Casteret S, Piegu B, Beauclair L, Moire N, Arensbuger P, et al. Mariner transposons contain a silencer: possible role of the Polycomb repressive complex 2. PLoS Genet. 2016;12(3):e1005902. doi: 10.1371/journal.pgen.1005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andam CP, Carver SM, Berthrong ST. Horizontal gene flow in managed ecosystems. Annu Rev Ecol Evol Syst. 2015;46(1):121–143. [Google Scholar]
- 63.Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313(6005):810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- 64.Fromm M, Taylor LP, Walbot V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc Natl Acad Sci U S A. 1985;82(17):5824–5828. doi: 10.1073/pnas.82.17.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seternes T, Tonheim TC, Myhr AI, Dalmo RA. A plant 35S CaMV promoter induces long-term expression of luciferase in Atlantic salmon. Sci Rep. 2016;6:25096. doi: 10.1038/srep25096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhar AK, Lakshman DK, Natarajan S, Allnutt FC, van Beek NA. Functional characterization of putative promoter elements from infectious hypodermal and hematopoietic necrosis virus (IHHNV) in shrimp and in insect and fish cell lines. Virus Res. 2007;127(1):1–8. doi: 10.1016/j.virusres.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Hudecek M, Izsvák Z, Johnen S, Renner M, Thumann G, Ivics Z. Going non-viral: the sleeping beauty transposon system breaks on through to the clinical side. Crit Rev Biochem Mol Biol. 2017;52(4):355–380. doi: 10.1080/10409238.2017.1304354. [DOI] [PubMed] [Google Scholar]
- 68.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 69.Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. Cerevisiae/E. Coli shuttle vectors. Yeast. 1991;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 70.Gietz RD. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2014;1205:1–12. doi: 10.1007/978-1-4939-1363-3_1. [DOI] [PubMed] [Google Scholar]
- 71.Green R, Rogers EJ. Chemical transformation of E. Coli. Methods Enzymol. 2013;529:329–336. doi: 10.1016/B978-0-12-418687-3.00028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Medina-Rivera A, Defrance M, Sand O, Herrmann C, Castro-Mondragon JA, Delerce J, et al. RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015;43(W1):W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. TATA box motif prediction. Pval cutoff 10exp-2; origin = START; only direct strand results are shown (DOCX 180 kb)
Table S2. Inr motif prediction. Pval cutoff 10exp-2; origin = START; only direct strand results are shown (DOCX 115 kb)
Table S3. DPE motif prediction. Pval cutoff 10exp-2; origin = START; only direct strand results are shown (DOCX 151 kb)
Accessions of cDNA/ESTs supporting TSS predicted in the sequences tested (TXT 643 bytes)
Figure S1. Luciferase-promoter assay in HeLa cells. Individual promoter-luciferase assay results in HeLa cells. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. *P < 0.05; **P < 0.005; ***P < 0.001 (PDF 168 kb)
Figure S2. Luciferase-promoter assay in S2R+ cells. Individual promoter-luciferase assay results in S2R+ cells of D. melanogaster. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. **P < 0.005; ***P < 0.001 (PDF 174 kb)
Figure S3. Luciferase-promoter assay in S. cerevisiae cells (BMA64-1A). Individual promoter-luciferase assay results in yeast. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. *P < 0.05; **P < 0.005 (PDF 216 kb)
Figure S4. Luciferase-promoter assay in bacteria (DH5alpha cells). Individual promoter-luciferase assay results in DH5alpha cells. Top and bottom whiskers: maximum and minimum values of each samples respectively. Top and bottom of boxes: 75th and 25th percentile of the samples respectively. Line through the boxes: median of each sample. X markers: mean of each samples. The statistical significance of promoter activity against the promoter-less cassette is shown. ***P < 0.001 (PDF 154 kb)
Data Availability Statement
All the materials mentioned and used in this work will be made available upon request.