Abstract
In specialized cell types, lysosome-related organelles support regulated secretory pathways, whereas in nonspecialized cells, lysosomes can undergo fusion with the plasma membrane in response to a transient rise in cytosolic calcium. Recent evidence also indicates that lysosome secretion can be controlled transcriptionally and promote clearance in lysosome storage diseases. In addition, evidence is also accumulating that low concentrations of cyclodextrins reduce the cholesterol-storage phenotype in cells and animals with the cholesterol storage disease Niemann-Pick type C, via an unknown mechanism. Here, we report that cyclodextrin triggers the secretion of the endo/lysosomal content in nonspecialized cells and that this mechanism is responsible for the decreased cholesterol overload in Niemann-Pick type C cells. We also find that the secretion of the endo/lysosome content occurs via a mechanism dependent on the endosomal calcium channel mucolipin-1, as well as FYCO1, the AP1 adaptor, and its partner Gadkin. We conclude that endo-lysosomes in nonspecialized cells can acquire secretory functions elicited by cyclodextrin and that this pathway is responsible for the decrease in cholesterol storage in Niemann-Pick C cells.
Keywords: cellular cholesterol, cholesterol/trafficking, endocytosis, Niemann-Pick disease, phospholipids/trafficking, lysobisphosphatidic acid, bis(monoacylglycero)phosphate, endosomes, secretory lysosomes
Niemann-Pick type C (NPC) is an autosomal recessive lysosomal storage disorder (LSD) characterized at the cellular level by the accumulation of cholesterol and other lipids in late endocytic compartments (1). In NPC, the traffic of LDL-derived cholesterol from lysosomes to the ER is defective, which impairs not only cholesterol esterification, but also the feedback transcriptional regulation of cholesterol metabolism via the SREBP pathway (2–4). The disease is caused by loss-of-function mutations in either one of the two late endosome/lysosome proteins NPC1 (5), a multispanning protein of the limiting membrane, and NPC2 (6), a globular protein present in the lumen. Solid evidence shows that both proteins bind cholesterol (7, 8), and structural and mutagenesis data suggest that cholesterol is transferred from NPC2 to NPC1, thereby facilitating export from the organelle (9–12). However, the precise mechanism of export remains unclear. Similarly, it is not clear how cholesterol is transferred from late endosome/lysosomes to reach the regulatory pool in the ER. Several direct routes have been proposed (13), but recent findings indicate that the plasma membrane may be the primary destination of LDL-derived cholesterol before reaching the ER (14). This view is consistent with the earlier notion that cholesterol rapidly equilibrates between the plasma membrane and the regulatory ER pool (15).
There is no approved treatment against NPC, with the exception of miglustat, which delays but does not arrest the progression of the disease (16). Over the last decade, cyclodextrins are emerging as a possible therapeutic strategy. These cyclic oligosaccharides clear cholesterol storage and reestablish the feedback regulation in cultured cells and NPC mice (17–21). They also improve neurological symptoms and survival in NPC animal models (22, 23) and were shown to decrease the neurological progression of the disease in phase 1–2 trials in NPC patients (24). However, the mechanism of cyclodextrin action remains poorly understood.
In recent years, it has become apparent that secretory endosomes or lysosomes (25) may play an important role in LSDs. Indeed, activated or overexpressed transcription factors of the transcription factor EB (TFEB) family can revert storage in Pompe disease and other LSDs by stimulating the secretion of the endo/lysosome content (26–28). This pathway is distinct from the acute and transient calcium-induced process involved in plasma-membrane repair (29–31). Similarly, storage in cells from NPC and other LSDs can be reverted by δ-tocopherol treatment (32) or activation of BK channels (33), which also promote endo/lysosome-mediated secretion. Interestingly, the secretion of endo/lysosome storage materials seems to depend on the activation of the lysosomal cation channel mucolipin-1 (MCOLN1), which is itself responsible for the LSD mucolipidosis type 4 when mutated (34). Endosome- or lysosomes-mediated secretion is impaired in MCOLN1 mutant cells (35), while activating mutations in the channel increase the rate of this exocytic pathway (36).
Here, we provide direct evidence that hydroxypropyl-cyclodextrin (HPCD) promotes the secretion of the endo/lysosome content via a mechanism that requires MCOLN1. Our data indicate that the process also depends on FYCO1, a protein involved in endo-lysosome motility toward the periphery (37, 38), as well as the AP1 adaptor and its partner Gadkin, which have both been implicated in the secretion of endo-lysosomes (30). Finally, we show that this pathway mediates the cyclodextrin-induced decrease in cholesterol storage within late endocytic compartments of NPC cells.
METHODS
Cell cultures, HPCD treatment, and siRNA transfections
HeLa-MZ and BHK-21 cells were maintained as described (39). The near-haploid human cell line HAP1 as well as MCOLN1 KO HAP1 cells were obtained from Horizon Discovery (Cambridge, UK). HeLa and BHK cells are not on the list of commonly misidentified cell lines maintained by the International Cell Line Authentication Committee. Our HeLa-MZ cells were authenticated by Microsynth (Balgach, Switzerland), which revealed 100% identity to the DNA profile of the cell line HeLa (ATCC: CCL-2) and 100% identity over all 15 autosomal short tandem repeats (STRs) to Microsynth’s reference DNA profile of HeLa. BHK cells could not be authenticated because no scientific consortium has agreed yet on the set of hamster markers, and, therefore, no databank is available to match cell lines. Cells are mycoplasma-negative as tested by GATC Biotech (Konstanz, Germany). For siRNA transfection, HeLa cells were plated in 6 cm dishes 6 h before transfection in antibiotic-free medium in order to reach 70% confluency. Cells were then transfected with siRNA (140 pmol in 700 µl Optimem) and siRNA max (7 µl in 700 µl Optimem) following the manufacturer’s instructions. Twenty hours after transfection, cells were split at a 1:5 dilution. Cells were treated with HPCD [Sigma, catalog no. H107, lot no. 090M0140V; average degree of substitution: 5 (mol hydroxypropyl/mol β-cyclodextrin), purity 100%] diluted in medium from a 90 mM stock solution in water. The experiments always ended 72 h after transfection. The following siRNAs were used: AP1µ1 (Qiagen, catalog no. Hs_AP1M1_1), ARL8b (Dharmacon, catalog no. L-020294-01), FYCO1 (Dharmacon, catalog no. L-014350-01), Gadkin (Qiagen, catalog no. Hs_C4orf16_5), Mcoln1 (Dharmacon, catalog no. M-006281-00), NPC1 (Qiagen, catalog no. Hs_NPC1_4), RAB3a (Dharmacon, catalog no. 009668-00), RAB8 (Dharmacon, catalog no. M-003905-00), RAB11a (Dharmacon, catalog no. L-004726-00), RAB27a (Dharmacon, catalog no. L-010388-00), and SYT7 (Dharmacon, catalog no. L-004306-02).
Fluorescent automated microscopy
Transfections with siRNAs (WT HeLa-MZ cells; NPC1 KO clone A3, CRISPR/Cas9; and NPC2 KO clone D11, CRISPR/Cas9) were performed with Lipofectamine RNAiMax (Life Technologies AG, Basel, Switzerland) using the manufacturer’s instructions in a 96-well plate from IBIDI (reference no. 250210) (6,000 cells/well). After 36 h, cells were treated or not with HPDC (0.1% wt/vol) for 36 h and then fixed in 3% paraformaldehyde for 20 min. Cells were stained with anti-LAMP1 Ab (D2D11-XP rabbit ®mAb catalog no. 9091, Cell Signaling) (1:300) and treated with RNase (200 μg/ml) followed by propidium iodide (PI) (5 μg/ml), filipin (50 μg/ml), and Alexa Fluor 647 anti-rabbit secondary Abs (1:400). Cells were imaged using the ImageXpress Micro XL Confocal automated microscope (Molecular Devices LLC, Sunnyvale, CA). Images were quantified using Meta Xpress image-analysis software (Molecular Devices, San Jose, CA). The Custom Module editor software (Molecular Devices) was used to first segment the image and generate relevant masks (PI for the nucleus, PI at a lower threshold for the whole cell, and LAMP1 for late endosomes/lysosomes), which were then applied on the fluorescent images to extract relevant measurements. The filipin signal was quantified in the LAMP1 mask and then expressed as total integrated intensity per cell.
LAMP1 uptake and analysis
HeLa cells were plated in 96-well plates (10,000 cells/well). After 24 h, cells were incubated with anti-LAMP1 Abs (D2D11) (1:300) in full medium with or without HPCD (0.1%) for 1 h, washed 3 times with PBS and fixed in 3% paraformaldehyde for 20 min. Cells were stained with PI (5 μg/ml) and Cy2-labeled anti-rabbit secondary Abs (1:400). Images were acquired with the IXM™ microscope (Molecular Devices LLC) and quantified using Meta Xpress Software. Cells were segmented, and the total integrated intensity of the LAMP1 signal per cell was measured.
Cholesterol esterification assay
Cholesterol esterification was measured by incorporation of [3H] oleate into newly synthesized cholesteryl oleate as previously described (40). Briefly, HeLa cells in 6 cm dishes were starved in 10% LPDS-containing medium for 24 h and then labeled for 12 h with [9,10-3H]oleic acid (OA) (45 Ci/mmol, 7.5 μCi/point) complexed with FA-free BSA, for 14 h in 10% FCS-containing medium. After lipid extraction, lipids were separated by TLC, and [3H]-labeled lipids were visualized and quantified using a cyclone Phosphorimager with a tritium Sensitive Screen (PerkinElmer).
Phospholipid analysis by 2D-TLC
HeLa cells were plated in 10 cm dishes, metabolically labeled with [32P]Pi (80 µCi/point) for 24 h, and chased for 14 h before HPCD addition. At the end of the treatment, post-nuclear supernatants were prepared, and lipids were extracted with the Folch method (41). To analyze the release of phospholipids in the medium, lipids were also extracted from the medium. Lipids were separated by 2D-TLC (42). [32P]-labeled phospholipids were visualized and quantified using a cyclone Phosphorimager. Lysobisphosphatidic acid (LBPA) was identified by comigration with 18:1/18:1 LBPA standard (Echelon Inc, Salt Lake City).
Cholesterol quantification
The cholesterol content of cells and subcellular fractions was quantified enzymatically using the Amplex Red kit (Molecular Probes) (43). Late endosome fractions were prepared from BHK cells by flotation in sucrose gradients (44). Cells or subcellular fractions were lysed with lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% Triton X-100), and 5 µg of proteins/point (total cells) or 1 µg of protein/point were used to quantify cholesterol in a 96-well plate format as described in the kit protocol.
RT-PCR
Total RNA was extracted using the RNeasy Mini Kit from Qiagen (reference no. 74104) from monolayers of HeLa, according to the manufacturer’s recommendation. cDNA synthesis was carried out using a SuperScript™ VILO™ cDNA Synthesis Kit (Life Technologies AG) from 250 ng of total RNA. mRNA expression was evaluated using SsoAdvanced SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) with 10 ng of cDNA with specific primers of interest on a CFX Connect real-time PCR Detection System (Bio-Rad). Relative amounts of mRNA were calculated by comparative CT analysis with 18S rRNA used as internal control. All primers are QuantiTect primer from Qiagen (Hilden, Germany). Primers were as follows: AP1AR (QT00054439), AP1M1 (QT00012215), ARL10C (QT00044758), FYCO1 (QT00037009), KIF5B (QT00046585), MCOLN1 (QT00094234), PLEKHM2 (QT02307109), RAB11A (QT00218491), RAB27A (QT00040054), RAB3A (QT00007490), RAB8A (QT00002485), SYT7 (QT00086975), SYTL4 (QT00088662), RRN18S (QT00199367), LDLR_1 (QT00045864), and HMGCR (QT00004081).
Lipid mass spectrometry
Lipids were extracted as previously described (45) with minor modifications. Briefly, cells were grown on 10 cm plastic plates before washing with, and then scraped into, cold PBS, followed by centrifugation for 5 min at 300 g at 4°C. Cell pellets were resuspended in 100 μl of cold water before addition of 360 μl of methanol and lipid internal standards. Next, 1.2 ml of 2-methoxy-2-methylpropane was added, and the samples followed by 1 h shaking at room temperature. A total of 200 μl of water was added to induce phase separation, and the upper phase was collected. Total phosphates were quantified with an ammonium molybdate colorimetric assay (46). Dried lipid samples were redissolved in chloroform-methanol (1:1, vol/vol). Separation was performed using a HILIC Kinetex column (2.6 μm, 2.1 × 50 mm2) on a Shimadzu Prominence UFPLC xr system (Tokyo, Japan): mobile phase A was acetonitrile:methanol 10:1 (vol/vol) containing 10 mM ammonium formate and 0.5% formic acid, and mobile phase B was deionized water containing 10 mM ammonium formate and 0.5% formic acid. The elution of the gradient began with 5% B at a 200 µl/min flow. The gradient increased linearly to 50% B over 7 min, then the elution continued at 50% B for 1.5 min, and finally the column was reequilibrated for 2.5 min. The sample was injected in 2 µl of chloroform:methanol (1:2, vol/vol). Data were acquired in full-scan mode at high resolution on a hybrid Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany). The system was operated at 240,000 resolution (m/z 400) with an automatic gain control set at 1.0E6 and one microscan set at 10 ms maximum injection time. The heated electrospray source HESI II was operated in positive mode at a temperature of 90°C and a source voltage at 4.0 kV. Sheath gas and auxiliary gas were set at 20 and 5 arbitrary units, respectively, while the transfer capillary temperature was set to 275°C. The mass spectrometry data were acquired with LTQ Tuneplus2.7SP2 and treated with Xcalibur 4.0QF2 (Thermo Fisher Scientific). Lipid identification was carried out with Lipid Data Analyzer II (LDA version 2.5.2, IGB-TUG Graz University) (47). Peaks were identified by their respective retention time, m/z, and intensity. Instruments were calibrated to ensure a mass accuracy lower than 3 ppm. Data visualization was improved with the LCMSexplorer web tool hosted at Ecole Polytechnique Fédérale de Lausanne (https://gecftools.epfl.ch/lcmsexplorer).
Analysis by focused ion beam-scanning electron microscopy
HeLa cells treated or not with HPCD were incubated with 15 nM BSA-gold for 4 h at 37°C, rinsed once with PBS, and fixed for 3 h on ice using 2.5% glutaraldehyde/2% paraformaldehyde in buffer A (0.15 M cacodylate and 2 mM CaCl2). Then, cells were extensively washed on ice in buffer A, then incubated 1 h on ice in 2% osmium tetroxide and 1.5% potassium Ferro cyanide in buffer A, and, finally, rinsed 5 times for 3 min in distilled water at room temperature. Cells were then incubated 20 min at room temperature in 0.1 M thiocarbohydrazide, which had been passed through a 0.22 µm filter, and extensively washed with water. Samples were incubated overnight at 4°C protected from light in 1% uranyl-acetate, washed in water, further incubated in 20 mM Pb aspartame for 30 min at 60°C, and, finally, washed in water after this last contrasting step. Samples were dehydrated in a graded series ethanol, embedded in hard Epon, and incubated for 60 h at 45°C and then for 60 h at 60°C. A small block was cut using an electric saw, and the block was incubated approximatively 30 min in 100% xylene in order to remove the plastic left. Finally, the block was mounted on a pin, coated with gold, and inserted into the chamber the HELIOS 660 Nanolab DualBeam SEM/FIB microscope (FEI Co., Eindhoven, The Netherlands). Regions of interest (ROIs) were prepared using focused ion beam (FIB) and ROI set to be approximatively 20 µm wide. During the acquisition process, the thickness of the FIB slice between each image acquisition was 10 nm. For endosomal density quantification, gold-containing endosomes were identified, image intensity was inverted, and the mean intensity of endosome was measured and divided by the intensity of the cytoplasm for each slice. Results were analyzed using GraphPad Prism. The Gaussian distribution of the data were tested using the Kolmogorov-Smirnov test (with Dallal-Wilkinson-Lillie for P value). As it was a non-Gaussian distribution, a two-tailed nonparametric Mann-Whitney U test was used in order to compare conditions.
β-Hexosaminidase, LDH, and LIPA enzymatic assays
β-Hexosaminidase and lactate dehydrogenase (LDH) enzymatic activities were measured from culture supernatant (100 µl) total cell lysate (5 µg) or light membrane fractions (2.5 µg) as described (30). Acidic lipase (LIPA) activity was measured from light membrane fractions (5 µg) using 4-methylumbelliferyl oleate (4-MUO) as substrate (48). Sample diluted in 25 µl of lysis buffer was mixed with 25 µl substrate solution (4-MUO 2 mg/ml in Triton X-100 4%) and 100 µl assay buffer (NaOAc 200 mM, pH 5.5, and Tween 80 0.02%). Reaction was incubated for 30–60 min at 37°C and stopped by adding 100 ml of Tris 1 M, pH 8.8. Fluorescence of the reaction product 4-methylumbelliferone (excitation 365 nm, emission 460 nm) was measured in a spectrofluorimeter and compared with a standard curve.
CRISPR/Cas9 NPC1 and NPC2 KO cell lines
Guide sequences to produce NPC1 and NPC2 KO cells were obtained using the CRISPR design tool (49): NPC1: forward (fwd): CACCGCCAAGGGCCAGGCCGCGAG; reverse (rev): AAACCTCGCGGCCTGGCCCTTGGCG; and NPC2: fwd: TAATACGACTCACTATAGGTCCTTGAACTGCACC; rev: TTCTAGCTCTAAAACAACCGGTGCAGTTCAAGGA. The sequences were used to insert the target sequence into the pX330 vector using Golden Gate Assembly (New England Biolabs) and transfected into cells. KO clones were isolated by serial dilution and confirmed by RT-PCR, Western blotting, and filipin staining.
Western blotting
Protein determination (50) and gel electrophoresis (51) were described. In Western blot analysis, the following primary Abs were used: Cathepsin D (BD Biosciences, catalog no. 610801), EEA-1 (BD Biosciences, catalog no. 610457), Lamp-1 (Cell Signaling, catalog no. 9091), MCOLN1 (Abcam, catalog no. 28508), NPC1 (Novus Biologicals, catalog no. NB400-148), and tubulin (Sigma, catalog no. T9026). Abs were incubated overnight at 4°C and secondary HRP-linked Abs for 50 min at room temperature. Antigens were visualized using the Western bright quantum reagent (Advansta, Menlo Park, CA) with the FUSION solo Image Station; band intensities were measured on nonsaturated raw images with FUSION solo image-analysis software.
RESULTS
Cyclodextrin reverts the NPC phenotype
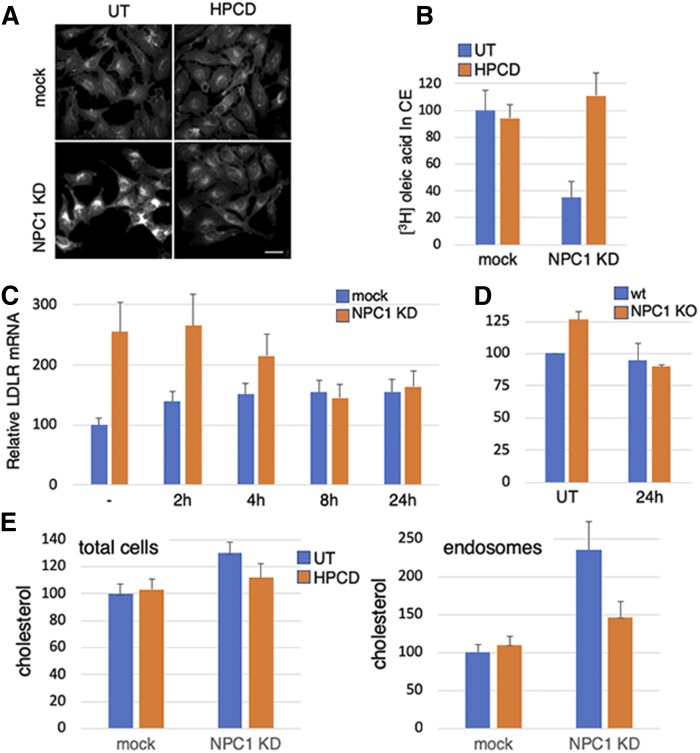
NPC1 was depleted with siRNAs in HeLa cells to provide an experimental system readily amenable to biochemical and cellular manipulations and with a reduced danger of adaptive drift that may occur in patient cells. NPC1 knockdown (KD) cells exhibited the characteristic hallmarks of NPC. Indeed, cholesterol accumulated in late endosomes, as revealed by filipin staining in fluorescence microscopy (Fig. 1A) and by enzymatic quantification in total cells and subcellular fractions (Fig. 1E). As expected, KD cells also exhibited a reduced capacity to esterify LDL-derived cholesterol (Fig. 1B) and to regulate the transcription of cholesterol-dependent genes like the LDL receptor (LDLR) (Fig. 1C). Treatment with HPCD was able to correct the effects of NPC1 KD on cholesterol subcellular distribution (Fig. 1A, E), esterification (Fig. 1B), and transcriptional regulation (Fig. 1C), while having essentially no effect in mock-transfected cells. Treatment with HPCD had no effect on NPC1 KD efficiency (supplemental Fig. S1A). Moreover, we generated an NPC1 KO cell line using CRISPR/Cas9 and confirmed that HPCD was also able to correct the transcriptional regulation defect in these KO cells (Fig. 1D). Much like others (19), we used HPCD at a low dose (0.1% or 0.7 mM), when compared with the dose (≈10 mM) used to extract cholesterol from membranes. A dose-response curve shows that 0.7 mM HPCD is optimal to decrease cholesterol accumulation in endosomes and to restore 3H-OA incorporation in cholesteryl esters in HeLa cells (supplemental Fig. S1B, C). This concentration is also similar to the concentration measured in the cerebrospinal fluid of NPC patients after intracerebroventricular injection of HPCD (52)—a treatment that reduced cholesterol accumulation in NPC patients and decreased the progression of neurological symptoms in clinical trials (53). Although HPCD reduced endosomal cholesterol in NPC1 KD cells (Fig. 1A, E), the drug only marginally affected total cellular cholesterol, presumably because low doses of cyclodextrins do not cause net cholesterol extraction, but instead facilitate redistribution within the cell. Together, these observations strengthen the notion that HPCD is able to revert the NPC phenotype in both KD and KO cells, presumably by redistributing cholesterol from endosomes to other membranes (18, 19).
Fig. 1.
A–C: HeLa cells were mock-transfected or transfected with anti-NPC1 siRNA for 72 h. When indicated in A, cells were treated for the last 24 h with 0.7 mM HPCD and fixed and stained with filipin (bar: 20 μm). In B, 36 h after transfection, cells were starved for 24 h in 10% LPDS and then labeled with [3H]OA for 12 h in 10% FCS. When indicated, 0.7 mM HPCD was added 12 h after starvation and maintained during OA incorporation. In C, cells were treated before the end of the 72 h with 0.7 mM HPCD, as indicated (all samples were treated equally with siRNAs, and the time of HPCD treatment was adjusted accordingly). Total mRNA was extracted, and LDLR mRNA was quantified by RT-PCR. D: HeLa NPC1 KO cells were treated for 24 h with 0.7 mM HPCD, and LDLR mRNA was quantified as in C. E: BHK cells were transfected as in A and treated or not for the last 24 h with 0.7 mM HPCD. A late endosomal fraction was prepared, and unesterified cholesterol was quantified (total cells and fractions) enzymatically. B–D: Data are expressed as a percentage of the mock control. UT, untreated.
HPCD stimulates endo-lysosome secretion
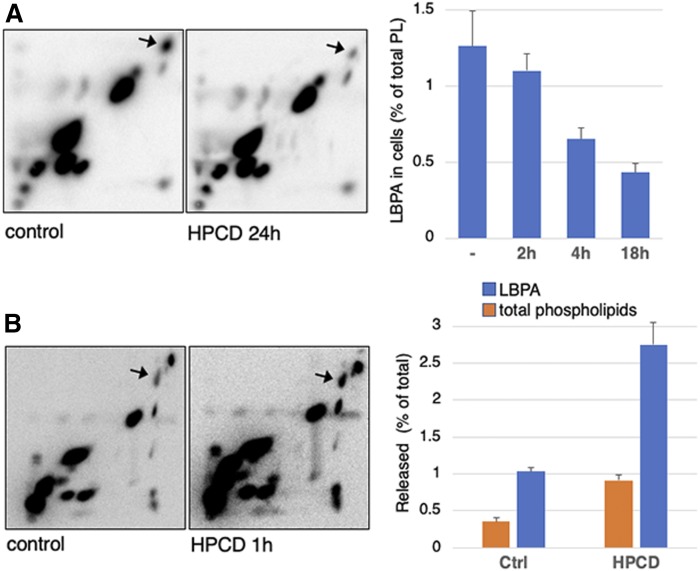
We previously found that the intralumenal vesicles of multivesicular late endosomes contain LBPA/bis(monoacylglycero)phosphate, an atypical phospholipid that is not detected in other organelles (54, 55). LBPA is functionally linked to cholesterol, because interfering with LBPA phenocopies NPC (56, 57). Strikingly, intracellular levels of LBPA were strongly decreased in HPCD-treated cells (40% of control after 18 h), whereas other phospholipids remained essentially unaffected (Fig. 2A). Cellular phospholipids released in the medium could be unambiguously quantified after metabolic labeling with 32P. Concomitant with decreased cellular levels, 32P-LBPA was released into the culture medium, reaching 2.5–3% of the total cellular content within 1 h, when compared with <1% for other 32P-phospholipids (Fig. 2B). The analysis of LBPA released into the medium was not feasible after longer incubations, likely because of its high instability in the extracellular environment (58).
Fig. 2.
A: HeLa cells were labeled with [32P]Pi for 24 h, chased for 14 h, and then treated or not with 0.7 mM HPCD (as in Fig. 1). Lipids were extracted, separated by 2D-TLC, and visualized by autoradiography. The position of LBPA (arrow) was identified by comigration with pure synthetic LBPA as standard. Right: LBPA quantified in 2D-TLCs at different time after HPCD treatment and expressed as a percentage of total 32P-phospholipids (PL). B: Cells labeled as in A were treated or not with 0.7 mM HPCD for 1 h. The lipids were extracted from medium and separated by 2D-TLC. Right shows the quantification of LBPA and total phospholipids in medium. Data are expressed as a percentage of total cellular LBPA or phospholipids, extracted, and quantified in the same experiments.
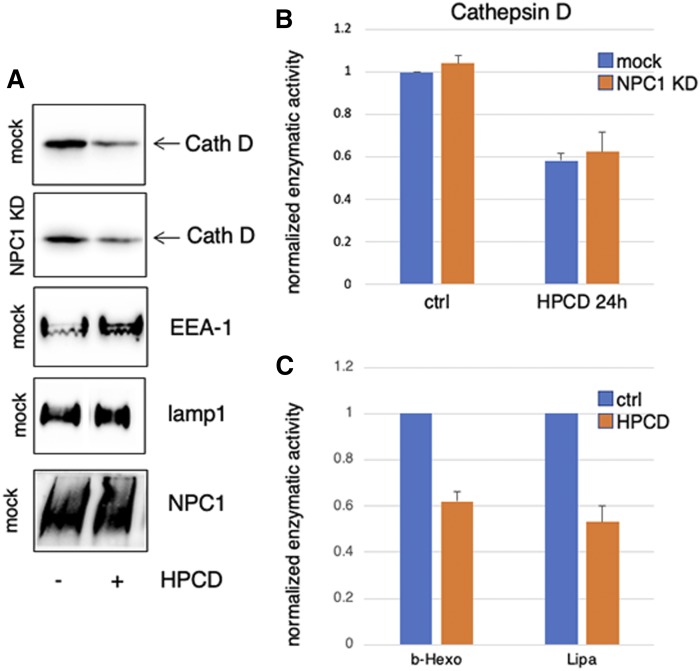
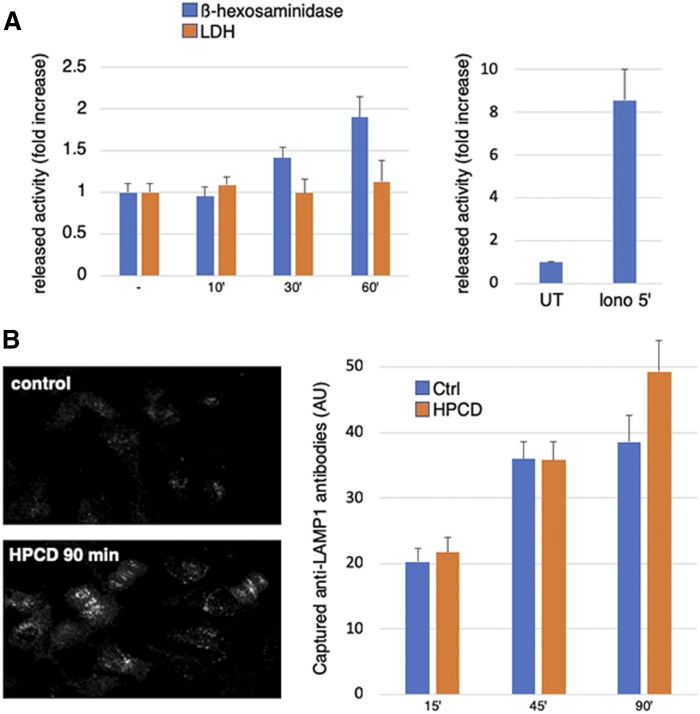
Much like LBPA, treatment with HPCD for 18 h strongly reduced the total cellular amounts of the aspartyl endopeptidase cathepsin D (Fig. 3A), to ≈60% of controls (Fig. 3B). Interestingly, cathepsin D was reduced by HPCD to a similar extent in control (mock-treated) cells and in NPC1 KD cells (Fig. 3A, B), indicating that the mechanism controlling cathepsin D decrease is not affected by NPC1 depletion. In control cells, HPCD also reduced the levels of two other lysosomal hydrolases, β-hexosaminidase and LIPA, to the same extent (Fig. 3C) as cathepsin D (Fig. 3B). By contrast, the drug had no effect on the total cellular amounts of two endo-lysosomal membrane proteins, LAMP1 and NPC1, itself or on the early endosome marker EEA1 (Fig. 3A). Concomitant with decreased cellular amounts, β-hexosaminidase appeared in the medium of HPCD-treated cells in a time-dependent fashion (Fig. 4A). This was not due to some deleterious effects of the treatment, because HPCD did not increase the release of the cytosolic enzyme LDH. Lysosomal enzyme release occurred with relatively slow kinetics and low efficiency, when compared with acute secretion triggered by a transient raise in cytosolic calcium (Fig. 4A). However, in marked contrast to calcium-induced secretion (30), the process continued over hours, eventually depleting half of the total cellular amounts after 24 h (Fig. 3C).
Fig. 3.
A, B: HeLa cells mock-transfected or transfected with anti-NPC1 siRNA for 72 h and treated with 0.7 mM HPCD, and the indicated proteins were analyzed by Western blotting in A. Cathepsin D (Cath D) in A was quantified (B) by scanning the blots in three independent experiments. C: The cellular amounts of LIPA and β-hexosaminidase (b-Hexo) were quantified enzymatically. Data in B and C are normalized to the amounts present in the untreated controls (ctrl).
Fig. 4.
A: HeLa cells washed with PBS and then incubated in serum-free medium were stimulated or not for the indicated time with 0.7 mM HPCD (right) or with ionomycin 5 μM (Iono 5′) for 5 min (left). Then hexosaminidase or LDH activity was measured in the medium and in total cell lysates. The enzyme activity in the medium is normalized to the total activity in lysates. The released enzyme activity is expressed as fold increase compared with untreated (UT) cells. B: HeLa cells in 96-well plates were incubated for the indicated times with lamp1 Ab at 37°C in the presence or not of 0.7 mM HPCD. Cells were than fixed, labeled with Cy3 secondary Ab and analyzed by automated microscopy. The average intensity of the fluorescence signal per cell (arbitrary units; AU) is shown in right.
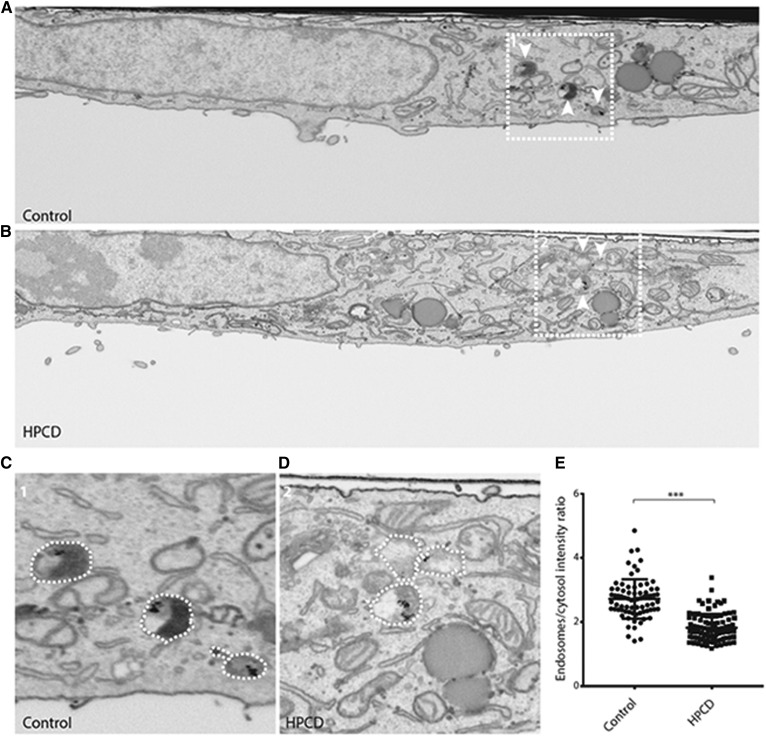
A 3D analysis by FIB scanning electron microscopy (FIB-SEM) showed that treatment with low concentrations of HPCD for 18 h did not affect the morphology of the Golgi complex or mitochondria (supplemental Fig. S2), consistent with the notion that HPCD has no detrimental effects on the overall cell organization. However, late endosomes labeled with BSA-gold endocytosed for 4 h (at the end of the HPCD incubation period) appeared markedly less electron-dense (Fig. 5B, D, low and high magnification), when compared with controls (Fig. 5A, C, low and high magnification). Densitometric quantification confirmed that the lumen of HPCD-treated endosomes was significantly lighter than controls (Fig. 5E). Presumably, the endosomal content had been released into the medium upon HPCD treatment, fully consistent with our analysis of HPCD-mediated LBPA (Fig. 2B) and lysosomal enzyme release (Fig. 4A).
Fig. 5.
HeLa cells treated (B, D) or not (A, C) with HPCD were incubated with 15 nm BSA-gold for 4 h at 37°C and processed for FIB-SEM. Shown are whole-cell slice micrographs (A, B) and high-magnification views of the boxed areas (C, D). White arrowheads indicate BSA-gold positive endosomes. The density of endosomes in was quantified by densitometry, and data are expressed relative to the cytoplasm intensity; controls: two cells, 74 endosomes; HPCD-treated cells: two cells, 95 endosomes. *** P < 0.0001.
Together, these observations suggest that HPCD-induced lysosome enzyme release occurs via endo-lysosome secretion. If so, one may expect the major endo-lysosome membrane protein LAMP1 to appear transiently on the plasma membrane upon endo-lysosome fusion (25, 30). After HPCD treatment, LAMP1 levels hardly changed at the plasma membrane, presumably because LAMP1 was efficiently reendocytosed (59), the secretory process occurring with slow kinetics (Fig. 4A). However, in cells incubated with anti-LAMP1 Abs, HPCD increased the intracellular accumulation of Abs captured at the plasma membrane after binding to their antigen (Fig. 4B). Together, these data demonstrate that HPCD triggers the secretion of the endo-lysosome content.
Mechanism of HPCD-dependent endo-lysosome secretion
In order to better characterize the mechanisms involved in HPCD-mediated endo-lysosome secretion, we used RNA interference (RNAi) to deplete proteins involved in related pathways in various cellular systems. These include: i) RAB27A, secretory pathway of lysosome-related organelles (LROs), including pigmentation and immunity (25, 60); ii) RAB11A, secretion of exosomes (together with RAB27A) derived from intralumenal vesicles of multivesicular endosomes (61–63); iii) RAB8A, selective delivery of endosomal cholesterol to the plasma membranes (64); iv) RAB3A, lysosome positioning and plasma membrane repair (65); v) synaptotagmin 7 (Syt7) (29), the µ1 subunit of AP1 adaptor and its partner Gadkin (30), fusion of endo-lysosomes with the plasma membrane; vi) the motor protein kinesin-1 (KIF5B) and ARL8B, microtubule-dependent movement of endosomes to the cell periphery (66, 67); vii) FYCO1, ER-endosome contact sites, and kinesin-mediated motility of lysosomes to the cell periphery (37, 38); and viii) MCOLN1, an endo-lysosomal calcium channel, exocytosis, and clearance of storage materials in LSDs (26, 28).
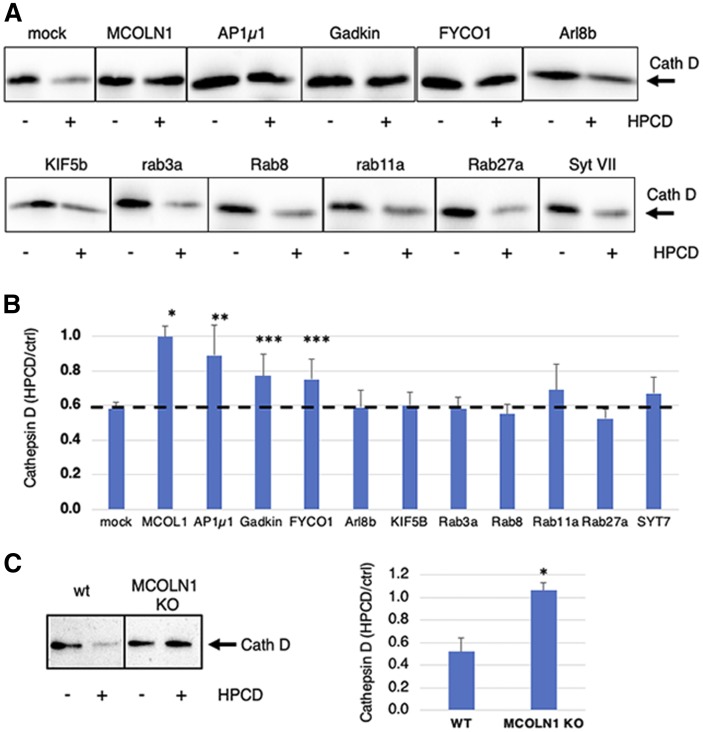
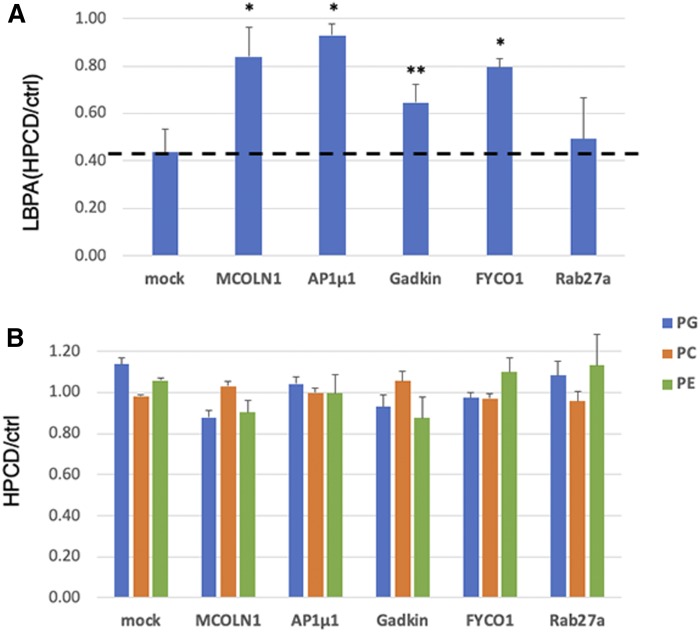
After depletion by RNAi (supplemental Fig. S3), cells were treated with HPCD. Cell-associated cathepsin D was analyzed by Western blotting (Fig. 6A and supplemental Fig. S4A), and amounts were quantified (Fig. 6B). In cells treated with nontargeting siRNAs, cathepsin D was decreased by HPCD to 50%, as expected (Fig. 3). In marked contrast, the amount of cell-associated enzyme was essentially unaffected by HPCD after MCOLN1 depletion with siRNAs (Fig. 6A and supplemental Fig. S3B) or after MCOLN1 KO in the near-haploid human cell line HAP1 (Fig. 6C and supplemental Fig. S4B), suggesting that lysosome enzyme release was essentially abrogated without this channel. Treatment with HPCD had no effect on the expression level of MCOLN1 mRNA and protein (supplemental Fig. S3B, C). The depletion of the µ1 chain of AP1, Gadkin, and FYCO1 also reduced cathepsin D secretion, but effects were less pronounced, whereas the depletion of other regulators was without effect. Much like with cathepsin D, HPCD failed to reduce cell-associated LBPA after the depletion of MCOLN1, AP1µ1, or FYCO1, and to a lesser extent after Gadkin depletion (Fig. 7A). By contrast, HPCD did not affect any other phospholipid, including phosphatidylglycerol (PG), phosphatidylcholine (PC), or phosphatidylethanolamine (PE), whether in control cells or after depletion of any of these factors (Fig. 7B). Together, these data indicate that HPCD-stimulated endo-lysosome secretion is controlled by the calcium channel MCOLN1, together with adaptor protein µ1 and its partner Gadkin, and by the ER-endosome contact site protein FYCO1 involved in endosome translocation.
Fig. 6.
A: HeLa cells were transfected with the indicated siRNA for 72 h and treated or not with HPCD (as in Fig. 1). Cells were lysed, and cathepsin D (Cath D) was analyzed by Western blotting. B: Quantification of cathepsin D normalized to the corresponding β-tubulin signal (shown in supplemental Fig. S5). Data are expressed as a ratio of the HPCD-treated samples over the corresponding control (ctrl) for each KD condition. C: HAP1 cells, WT and MCOLN1 KO, were treated or not with HPCD (as in Fig. 1) for 24 h, and cathepsin D was analyzed by Western blotting. In right, data quantified as in Fig. 3 are expressed as a percentage of the MCOLN1 KO signal. (n : 3 independent experiments). * P < 0.0001; ** P < 0.01; *** P < 0.02.
Fig. 7.
HeLa cells were transfected with the indicated siRNA for 72 h and treated or not with HPCD (as in Fig. 1). The lipids were extracted and quantified by LC/MS. A: All isoforms of LBPA with different acyl chain composition were quantified and normalized to the total phospholipid content. B: All isoforms of PG, PC, and PE were quantified and normalized as in A. Data are expressed as a ratio of HPCD-treated samples over the corresponding control (ctrl) for each KD condition. (n = 3 independent experiments). * P < 0.02; ** P < 0.05.
MCOLN1-dependent endo-lysosome secretion is necessary for cholesterol clearance in NPC cells
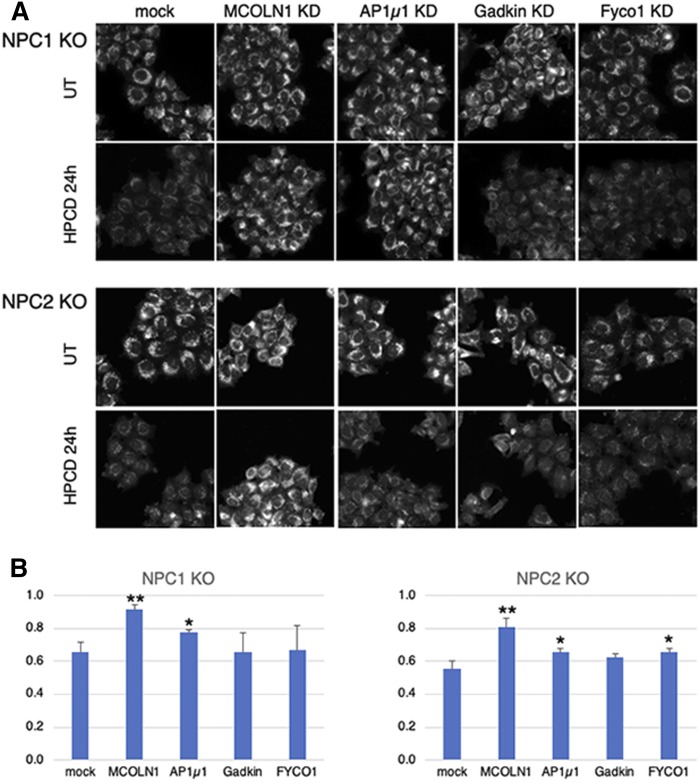
Next, we investigated whether the same mechanism also mediates the clearance of endosomal cholesterol in NPC KD cells. To this end, we used NPC1 and NPC2 KO cell lines generated using CRISPR/Cas9, which both showed strong cholesterol accumulation in late endocytic compartments (Fig. 8A). Treatment of NPC1 and NPC2 KO cells with HPCD massively reduced cholesterol storage (Fig. 8A). Quantification by automated microscopy confirmed that, although HPCD did not affect the major endo-lysosomal protein LAMP1 in NPC1 or NPC2 KO cells and in WT cells, the drug reduced the cholesterol content of LAMP1-containing compartments by half in NPC1 and NPC2 KO cells (Fig. 8B and supplemental Fig. S5), much like in NPC1 KD cells (Fig. 1A).
Fig. 8.
NPC1 and NPC2 KO HeLa cells were plated in 96-well plates, transfected with the indicated siRNA for 72 h, and treated or not with 0.7 mM HPCD for the last 36 h. Cells were fixed, stained with filipin, as well as anti-LAMPl Abs and PI (used for segmentation; not shown), and analyzed by automated microscopy. A: Representative images are shown for filipin. B: The filipin intensity in A was quantified in a lampl mask, and the average filipin intensity per cell was calculated. Data are expressed as the ratio between HPCD-treated and untreated (UT) cells for each condition. Means represent three independent experiments. * P < 0.05; ** P < 0.005.
The depletion of MCOLN1 by RNAi in NPC1 and NPC2 KO cells had no significant effects on the amounts of LAMP1 or on the cholesterol content of LAMP1-contanining endo-lysosomes (Fig. 8A, B and supplemental Fig. S5). Remarkably, however, MCOLN1 KD in NPC1 and NPC2 KO cells abrogated the effects of HPCD on cholesterol accumulation (Fig. 8A, B and supplemental Fig. S5), consistent with the effects of MCOLN1 KD on endo-lysosome secretion in untreated, healthy cells (Fig. 6A). Depletion of AP1µ1 had a smaller, but significant, effect on HPCD-mediated reduction of cholesterol storage in both NPC1 and NPC2 KO cells (Fig. 8A, quantification in B). The difference may be due to differential effects of HPCD on cholesterol-laden endosomes in NPC1 or NPC2 cells versus endosomes in control cells. In conclusion, our data show that the calcium channel MCOLN1 is directly involved in the regulation of cyclodextrin-mediated endo-lysosome secretion, perhaps together with AP1 and its partner Gadkin, which play a role in calcium-mediated endo-lysosome secretion (30) and the ER-endosome contact site protein FYCO1 (37).
DISCUSSION
In the NPC lysosomal storage disease, mutation of NPC1 or NPC2 causes the accumulation of LDL-derived cholesterol in late endocytic compartments, but the functions of the NPC1 and NPC2 proteins remain incompletely understood (68). The accumulation of storage materials in NPC endosomes eventually leads to a traffic jam and a collapse of endosomal membrane dynamics (69, 70), accompanied by defects both in cholesterol movement to the ER and transcriptional regulation of cholesterol metabolism (2–4). Compelling evidence now shows that prolonged incubations with low concentrations of cyclodextrins reduce the cholesterol-storage phenotype and restore cholesterol-dependent transcriptional regulation (68, 71), while cyclodextrin also decreases the progression of neurological symptoms in clinical trials with NPC1 patients (24). The mechanism of action of these cyclic oligosaccharides, however, remains poorly understood. Methyl-β-cyclodextrin was reported to restore the autophagy flux in NPC1 cells through activation of AMP-activated protein kinase (72). Cyclodextrins are also thought to act intracellularly after endocytosis as a fluid phase tracer (18, 72), perhaps bypassing the requirement for the NPC2 protein in the endosome lumen (18). These studies and our work (this paper) collectively establish that, in the presence of serum, low doses of cyclodextrins do not cause net cholesterol extraction, but instead facilitate redistribution within the cell; after exocytosis of NPC endosomes, cholesterol is presumably incorporated into the plasma membrane or released and recaptured by cells and eventually redistributed intracellularly. This is best illustrated by the restoration of cholesterol esterification and SREBP-mediated feedback regulation in the ER (19) (and Fig. 1). It has been reported that very short (60 min vs. 18–24 h in our study) incubations with high doses of cyclodextrin (2% vs. 0.1% in our study) cause calcium-dependent lysosome exocytosis (73). These high concentrations, however, will cause massive cholesterol depletion from plasma membranes, which may trigger a response to plasma-membrane damage (74).
We find that prolonged incubations of control cells with low doses of cyclodextrin cause the secretion of the endo-lysosomal content. Compelling evidence for this secretory pathway comes first from the observed decrease in cellular LBPA content and increase in the medium, because LBPA is only detected in late endosomes (54–56). We also find that prolonged incubation with low doses of cyclodextrin causes significant secretion of lysosomal enzymes leading to partial depletion from cells after 18–24 h. Specialized cell types contain LROs, which have the capacity to undergo fusion with the plasma membrane and to secrete their content in a regulated fashion (25, 75). Nonspecialized cells also possess the capacity to trigger the acute fusion of endocytic compartments with the plasma membrane in response to a transient rise in cytosolic calcium, presumably as a membrane-repair pathway (29–31). Given its slow release rate and prolonged duration, our data argue that the cyclodextrin-mediated secretory pathway is distinct from the calcium-mediated pathway and reminiscent of TFEB-induced clearance of lysosomal content (26–28). This view is also reinforced by our findings that cyclodextrin-mediated secretion is insensitive to the depletion of Syt7 and RAB27a, involved in calcium-mediated secretion.
Similarly to TFEB-induced secretion, we find that the secretory pathway elicited by cyclodextrin depends on the endo/lysosomal calcium channel MCOLN1, which is responsible for the LSD mucolipidosis type 4 when mutated (34). Previous studies showed that MCOLN1 is involved in the secretion of endo-lysosomal content, because secretion is impaired in MCOLN1 mutant cells (35) and stimulated by activating MCOLN1 mutations (36). It should be noted that is not known whether MCOLN1 function is related to the role of cytosolic calcium in membrane repair. We find that the secretion of endo/lysosomes also depends on the RAB7 effector FYCO1 (37), involved in endosome translocation to the cell periphery (38) and in clearance of α-synuclein aggregates (76). Finally, we find that cyclodextrin-induced secretion of endo/lysosomes also requires the adaptor complex AP1 and its partner protein Gadkin, which are both involved in endo/lysosomes secretion (30). Future work will be necessary to unravel the role of MCOLN1 and the mechanism presumably linking this channel to FYCO1 and AP1/Gadkin.
Our data argue that the same mechanism operates in the clearance of storage cholesterol in NPC cells because depletion of MCOLN1 strongly inhibits the HPCD-induced clearance in NPC cells. Depletion of AP1µ1 and FYCO1 had a relatively modest, but still significant, effect in NPC cells. One may speculate that the NPC machinery itself plays a role in this secretory pathway, perhaps via ER-endosome contact sites (38, 77). Alternatively, residual lysosomal secretion without these factors suffices to promote cholesterol clearance, perhaps because of some other compensatory mechanisms operating in the NPC1/NPC2 KO background.
In conclusion, our data demonstrate that cyclodextrins trigger the secretion of endo/lysosomes via a pathway dependent on the calcium channel MCOLN1, as well as FYCO1 and the AP1 adaptor and its partner Gadkin. Our data also demonstrate that cyclodextrin clears the endo/lysosomal content of NPC endosomes by stimulating the same MCOLN1-dependent pathway. We conclude that endo-lysosomes in nonspecialized cells can acquire secretory functions and that this pathway, elicited by cyclodextrin, is responsible for redistribution of cholesterol and decrease in storage in NPC cells. Our findings also indicate the potential benefit of agents that promote lysosome secretion as possible future strategy to treat NPC and possibly other LSDs, given the limitations of currently available therapies.
Supplementary Material
Footnotes
Abbreviations:
- FIB
- focused ion beam
- HPCD
- hydroxypropyl-cyclodextrin
- KD
- knockdown
- LBPA
- lysobisphosphatidic acid
- LDH
- lactate dehydrogenase
- LDLR
- LDL receptor
- LIPA
- acidic lipase
- LSD
- lysosomal storage disorder
- MCOLN1
- mucolipin-1
- NPC
- Niemann-Pick type C
- OA
- oleic acid
- PI
- propidium iodide
- RNAi
- RNA interference
- SEM
- scanning electron microscopy
- Syt7
- synaptotagmin 7
- TFEB
- transcription factor EB
Financial support from the Swiss National Science Foundation, the NCCR in Chemical Biology, and LipidX from the Swiss SystemsX.ch Initiative, evaluated by the Swiss National Science Foundation (J G.) is gratefully acknowledged. F.V. was supported by a fellowship from the National Niemann-Pick Disease Foundation. The authors declare no competing interests.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Vanier M. T. 2010. Niemann-Pick disease type C. Orphanet J. Rare Dis. 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pentchev P. G., Comly M. E., Kruth H. S., Vanier M. T., Wenger D. A., Patel S., and Brady R. O.. 1985. A defect in cholesterol esterification in Niemann-Pick disease (type C) patients. Proc. Natl. Acad. Sci. USA. 82: 8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liscum L., and Faust J. R.. 1987. Low density lipoprotein (LDL)-mediated suppression of cholesterol synthesis and LDL uptake is defective in Niemann-Pick type C fibroblasts. J. Biol. Chem. 262: 17002–17008. [PubMed] [Google Scholar]
- 4.Kristiana I., Yang H., and Brown A. J.. 2008. Different kinetics of cholesterol delivery to components of the cholesterol homeostatic machinery: implications for cholesterol trafficking to the endoplasmic reticulum. Biochim. Biophys. Acta. 1781: 724–730. [DOI] [PubMed] [Google Scholar]
- 5.Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., Gu J., Rosenfeld M. A., Pavan W. J., Krizman D. B., et al. . 1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 277: 228–231. [DOI] [PubMed] [Google Scholar]
- 6.Naureckiene S., Sleat D. E., Lackland H., Fensom A., Vanier M. T., Wattiaux R., Jadot M., and Lobel P.. 2000. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 290: 2298–2301. [DOI] [PubMed] [Google Scholar]
- 7.Infante R. E., Radhakrishnan A., Abi-Mosleh L., Kinch L. N., Wang M. L., Grishin N. V., Goldstein J. L., and Brown M. S.. 2008. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 283: 1064–1075. [DOI] [PubMed] [Google Scholar]
- 8.Xu S., Benoff B., Liou H. L., Lobel P., and Stock A. M.. 2007. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J. Biol. Chem. 282: 23525–23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., and Infante R. E.. 2009. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 137: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Saha P., Li J., Blobel G., and Pfeffer S. R.. 2016. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl. Acad. Sci. USA. 113: 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M. L., Motamed M., Infante R. E., Abi-Mosleh L., Kwon H. J., Brown M. S., and Goldstein J. L.. 2010. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 12: 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong X., Qian H., Zhou X., Wu J., Wan T., Cao P., Huang W., Zhao X., Wang X., Wang P., et al. . 2016. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell. 165: 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfisterer S. G., Peranen J., and Ikonen E.. 2016. LDL-cholesterol transport to the endoplasmic reticulum: current concepts. Curr. Opin. Lipidol. 27: 282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Infante R. E., and Radhakrishnan A.. 2017. Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. eLife. 6: e25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange Y., Ye J., and Steck T. L.. 2004. How cholesterol homeostasis is regulated by plasma membrane cholesterol in excess of phospholipids. Proc. Natl. Acad. Sci. USA. 101: 11664–11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson M. C., Mengel E., Vanier M. T., Schwierin B., Muller A., Cornelisse P., Pineda M., and NPC Registry Investigators . 2015. Stable or improved neurological manifestations during miglustat therapy in patients from the international disease registry for Niemann-Pick disease type C: an observational cohort study. Orphanet J. Rare Dis. 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B., Ramirez C. M., Miller A. M., Repa J. J., Turley S. D., and Dietschy J. M.. 2010. Cyclodextrin overcomes the transport defect in nearly every organ of NPC1 mice leading to excretion of sequestered cholesterol as bile acid. J. Lipid Res. 51: 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenbaum A. I., Zhang G., Warren J. D., and Maxfield F. R.. 2010. Endocytosis of beta-cyclodextrins is responsible for cholesterol reduction in Niemann-Pick type C mutant cells. Proc. Natl. Acad. Sci. USA. 107: 5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abi-Mosleh L., Infante R. E., Radhakrishnan A., Goldstein J. L., and Brown M. S.. 2009. Cyclodextrin overcomes deficient lysosome-to-endoplasmic reticulum transport of cholesterol in Niemann-Pick type C cells. Proc. Natl. Acad. Sci. USA. 106: 19316–19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson C. D., Ali N. F., Micsenyi M. C., Stephney G., Renault S., Dobrenis K., Ory D. S., Vanier M. T., and Walkley S. U.. 2009. Chronic cyclodextrin treatment of murine Niemann-Pick C disease ameliorates neuronal cholesterol and glycosphingolipid storage and disease progression. PLoS One. 4: e6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camargo F., Erickson R. P., Garver W. S., Hossain G. S., Carbone P. N., Heidenreich R. A., and Blanchard J.. 2001. Cyclodextrins in the treatment of a mouse model of Niemann-Pick C disease. Life Sci. 70: 131–142. [DOI] [PubMed] [Google Scholar]
- 22.Vite C. H., Bagel J. H., Swain G. P., Prociuk M., Sikora T. U., Stein V. M., O’Donnell P., Ruane T., Ward S., Crooks A., et al. . 2015. Intracisternal cyclodextrin prevents cerebellar dysfunction and Purkinje cell death in feline Niemann-Pick type C1 disease. Sci. Transl. Med. 7: 276ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B., Turley S. D., Burns D. K., Miller A. M., Repa J. J., and Dietschy J. M.. 2009. Reversal of defective lysosomal transport in NPC disease ameliorates liver dysfunction and neurodegeneration in the npc1−/− mouse. Proc. Natl. Acad. Sci. USA. 106: 2377–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ory D. S., Ottinger E. A., Farhat N. Y., King K. A., Jiang X., Weissfeld L., Berry-Kravis E., Davidson C. D., Bianconi S., Keener L. A., et al. . 2017. Intrathecal 2-hydroxypropyl-beta-cyclodextrin decreases neurological disease progression in Niemann-Pick disease, type C1: a non-randomised, open-label, phase 1–2 trial. Lancet. 390: 1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks M. S., Heijnen H. F., and Raposo G.. 2013. Lysosome-related organelles: unusual compartments become mainstream. Curr. Opin. Cell Biol. 25: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina D. L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J. A., Sardiello M., et al. . 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 21: 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samie M. A., and Xu H.. 2014. Lysosomal exocytosis and lipid storage disorders. J. Lipid Res. 55: 995–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martina J. A., Diab H. I., Lishu L., Jeong A. L., Patange S., Raben N., and Puertollano R.. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7: ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy A., Caler E. V., and Andrews N. W.. 2001. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 106: 157–169. [DOI] [PubMed] [Google Scholar]
- 30.Laulagnier K., Schieber N. L., Maritzen T., Haucke V., Parton R. G., and Gruenberg J.. 2011. Role of AP1 and Gadkin in the traffic of secretory endo-lysosomes. Mol. Biol. Cell. 22: 2068–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews N. W., Almeida P. E., and Corrotte M.. 2014. Damage control: cellular mechanisms of plasma membrane repair. Trends Cell Biol. 24: 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu M., Liu K., Swaroop M., Porter F. D., Sidhu R., Firnkes S., Ory D. S., Marugan J. J., Xiao J., Southall N., et al. . 2012. delta-Tocopherol reduces lipid accumulation in Niemann-Pick type C1 and Wolman cholesterol storage disorders. J. Biol. Chem. 287: 39349–39360. [Erratum. 2013. J. Biol. Chem. 288: 296.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong X. Z., Sun X., Cao Q., Dong G., Schiffmann R., and Dong X. P.. 2016. BK channel agonist represents a potential therapeutic approach for lysosomal storage diseases. Sci. Rep. 6: 33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boudewyn L. C., and Walkley S. U.. Current concepts in the neuropathogenesis of mucolipidosis type IV. J. Neurochem. Epub ahead of print May 16, 2018; doi: 10.1111/jnc.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaPlante J. M., Sun M., Falardeau J., Dai D., Brown E. M., Slaugenhaupt S. A., and Vassilev P. M.. 2006. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 89: 339–348. [DOI] [PubMed] [Google Scholar]
- 36.Dong X. P., Wang X., Shen D., Chen S., Liu M., Wang Y., Mills E., Cheng X., Delling M., and Xu H.. 2009. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J. Biol. Chem. 284: 32040–32052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pankiv S., Alemu E. A., Brech A., Bruun J. A., Lamark T., Overvatn A., Bjorkoy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188: 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raiborg C., Wenzel E. M., Pedersen N. M., Olsvik H., Schink K. O., Schultz S. W., Vietri M., Nisi V., Bucci C., Brech A., et al. . 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520: 234–238. [DOI] [PubMed] [Google Scholar]
- 39.Morel E., and Gruenberg J.. 2007. The p11/S100A10 light chain of annexin A2 is dispensable for annexin A2 association to endosomes and functions in endosomal transport. PLoS One. 2: e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chamoun Z., Vacca F., Parton R. G., and Gruenberg J.. 2013. PNPLA3/adiponutrin functions in lipid droplet formation. Biol. Cell. 105: 219–233. [DOI] [PubMed] [Google Scholar]
- 41.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 42.Sobo K., Chevallier J., Parton R. G., Gruenberg J., and van der Goot F. G.. 2007. Diversity of raft-like domains in late endosomes. PLoS One. 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amundson D. M., and Zhou M.. 1999. Fluorometric method for the enzymatic determination of cholesterol. J. Biochem. Biophys. Methods. 38: 43–52. [DOI] [PubMed] [Google Scholar]
- 44.Aniento F., Emans N., Griffiths G., and Gruenberg J.. 1993. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J. Cell Biol. 123: 1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott C. C., Vossio S., Vacca F., Snijder B., Larios J., Schaad O., Guex N., Kuznetsov D., Martin O., Chambon M., et al. . 2015. Wnt directs the endosomal flux of LDL-derived cholesterol and lipid droplet homeostasis. EMBO Rep. 16: 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loizides-Mangold U., David F. P., Nesatyy V. J., Kinoshita T., and Riezman H.. 2012. Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels. J. Lipid Res. 53: 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartler J., Trotzmuller M., Chitraju C., Spener F., Kofeler H. C., and Thallinger G. G.. 2011. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 27: 572–577. [DOI] [PubMed] [Google Scholar]
- 48.Sheriff S., Du H., and Grabowski G. A.. 1995. Characterization of lysosomal acid lipase by site-directed mutagenesis and heterologous expression. J. Biol. Chem. 270: 27766–27772. [DOI] [PubMed] [Google Scholar]
- 49.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 52.Jiang H., Sidhu R., Fujiwara H., De Meulder M., de Vries R., Gong Y., Kao M., Porter F. D., Yanjanin N. M., Carillo-Carasco N., et al. . 2014. Development and validation of sensitive LC-MS/MS assays for quantification of HP-beta-CD in human plasma and CSF. J. Lipid Res. 55: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tortelli B., Fujiwara H., Bagel J. H., Zhang J., Sidhu R., Jiang X., Yanjanin N. M., Shankar R. K., Carillo-Carasco N., Heiss J., et al. . 2014. Cholesterol homeostatic responses provide biomarkers for monitoring treatment for the neurodegenerative disease Niemann-Pick C1 (NPC1). Hum. Mol. Genet. 23: 6022–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi T., Stang E., Fang K. S., de Moerloose P., Parton R. G., and Gruenberg J.. 1998. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 392: 193–197. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi T., Beuchat M. H., Chevallier J., Makino A., Mayran N., Escola J. M., Lebrand C., Cosson P., and Gruenberg J.. 2002. Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 277: 32157–32164. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi T., Beuchat M. H., Lindsay M., Frias S., Palmiter R. D., Sakuraba H., Parton R. G., and Gruenberg J.. 1999. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1: 113–118. [DOI] [PubMed] [Google Scholar]
- 57.Chevallier J., Chamoun Z., Jiang G., Prestwich G. D., Sakai N., Matile S., Parton R. G., and Gruenberg J.. 2008. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 283: 27871–27880. [DOI] [PubMed] [Google Scholar]
- 58.Chevallier J., Sakai N., Robert F., Kobayashi T., Gruenberg J., and Matile S.. 2000. Rapid access to synthetic lysobisphosphatidic acids using P(III) chemistry. Org. Lett. 2: 1859–1861. [DOI] [PubMed] [Google Scholar]
- 59.Kornfeld S., and Mellman I.. 1989. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5: 483–525. [DOI] [PubMed] [Google Scholar]
- 60.Fukuda M. 2013. Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic. 14: 949–963. [DOI] [PubMed] [Google Scholar]
- 61.Savina A., Vidal M., and Colombo M. I.. 2002. The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 115: 2505–2515. [DOI] [PubMed] [Google Scholar]
- 62.Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., et al. . 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12: 19–30; sup 11–13. [DOI] [PubMed] [Google Scholar]
- 63.Abrami L., Brandi L., Moayeri M., Brown M. J., Krantz B. A., Leppla S. H., and van der Goot F. G.. 2013. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Reports. 5: 986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanerva K., Uronen R. L., Blom T., Li S., Bittman R., Lappalainen P., Peranen J., Raposo G., and Ikonen E.. 2013. LDL cholesterol recycles to the plasma membrane via a Rab8a-Myosin5b-actin-dependent membrane transport route. Dev. Cell. 27: 249–262. [DOI] [PubMed] [Google Scholar]
- 65.Encarnação M., Espada L., Escrevente C., Mateus D., Ramalho J., Michelet X., Santarino I., Hsu V. W., Brenner M. B., Barral D. C., et al. . 2016. A Rab3a-dependent complex essential for lysosome positioning and plasma membrane repair. J. Cell Biol. 213: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21: 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J. S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33: 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosenbaum A. I., and Maxfield F. R.. 2011. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. J. Neurochem. 116: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simons K., and Gruenberg J.. 2000. Jamming the endosomal system: lipid rafts and lysosomal storage diseases. Trends Cell Biol. 10: 459–462. [DOI] [PubMed] [Google Scholar]
- 70.Liscum L. 2000. Niemann-Pick type C mutations cause lipid traffic jam. Traffic. 1: 218–225. [DOI] [PubMed] [Google Scholar]
- 71.Vance J. E., and Karten B.. 2014. Niemann-Pick C disease and mobilization of lysosomal cholesterol by cyclodextrin. J. Lipid Res. 55: 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dai S., Dulcey A. E., Hu X., Wassif C. A., Porter F. D., Austin C. P., Ory D. S., Marugan J., and Zheng W.. 2017. Methyl-beta-cyclodextrin restores impaired autophagy flux in Niemann-Pick C1-deficient cells through activation of AMPK. Autophagy. 13: 1435–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen F. W., Li C., and Ioannou Y. A.. 2010. Cyclodextrin induces calcium-dependent lysosomal exocytosis. PLoS One. 5: e15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hissa B., Pontes B., Roma P. M., Alves A. P., Rocha C. D., Valverde T. M., Aguiar P. H., Almeida F. P., Guimaraes A. J., Guatimosim C., et al. . 2013. Membrane cholesterol removal changes mechanical properties of cells and induces secretion of a specific pool of lysosomes. PLoS One. 8: e82988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luzio J. P., Hackmann Y., Dieckmann N. M., and Griffiths G. M.. 2014. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 6: a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saridaki T., Nippold M., Dinter E., Roos A., Diederichs L., Fensky L., Schulz J. B., and Falkenburger B. H.. 2018. FYCO1 mediates clearance of alpha-synuclein aggregates through a Rab7-dependent mechanism. J. Neurochem. 146: 474–492. [DOI] [PubMed] [Google Scholar]
- 77.Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185: 1209–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.