Abstract
Fatty acid translocase cluster of differentiation (CD36) is a multifunctional membrane protein that facilitates the uptake of long-chain fatty acids. Lipophagy is autophagic degradation of lipid droplets. Accumulating evidence suggests that CD36 is involved in the regulation of intracellular signal transduction that modulates fatty acid storage or usage. However, little is known about the relationship between CD36 and lipophagy. In this study, we found that increased CD36 expression was coupled with decreased autophagy in the livers of mice treated with a high-fat diet. Overexpressing CD36 in HepG2 and Huh7 cells inhibited autophagy, while knocking down CD36 expression induced autophagy due to the increased autophagosome formation in autophagic flux. Meanwhile, knockout of CD36 in mice increased autophagy, while the reconstruction of CD36 expression in CD36-knockout mice reduced autophagy. CD36 knockdown in HepG2 cells increased lipophagy and β-oxidation, which contributed to improving lipid accumulation. In addition, CD36 expression regulated autophagy through the AMPK pathway, with phosphorylation of ULK1/Beclin1 also involved in the process. These findings suggest that CD36 is a negative regulator of autophagy, and the induction of lipophagy by ameliorating CD36 expression can be a potential therapeutic strategy for the treatment of fatty liver diseases through attenuating lipid overaccumulation.
Keywords: cluster of differentiation 36, adenosine monophosphate-activated protein kinase, lipid droplets, autophagy, hepatic steatosis, β-oxidation
Nonalcoholic fatty liver disease (NAFLD) is the most common cause of liver disease worldwide, with prevalence estimates ranging from 25% to 45% (1). NAFLD is considered to be the liver manifestation of metabolic syndrome, which encompasses a wide spectrum beginning with steatosis (2, 3). The cluster of differentiation 36 (CD36) belongs to the scavenger receptor family, also known as fatty acid translocase, that facilitates the uptake of long-chain fatty acids (LCFAs) and is widely expressed on the surface of many cell types in vertebrates (4, 5). CD36 recognizes different ligands and may promote different intracellular signaling pathways. This multifunctional membrane glycoprotein has been studied extensively in relation to its role in the uptake of LCFAs, which are involved in NAFLD. Hepatic CD36 expression is significantly elevated in animal models and patients with NAFLD and is regarded as positively correlated with liver fat content and insulin resistance (6).
Accumulating evidence indicates that CD36 is not only a fatty acid transporter but also an essential regulator of intracellular fatty acid homeostasis. Recent studies have found that CD36 is involved in fatty acid oxidation by the activation of adenosine monophosphate-activated protein kinase (AMPK) (7), suggesting that it modulates fatty acid storage or usage. It has also been reported to function as a pattern recognition receptor that conducts intracellular signals and activates inflammatory pathways such as Toll-like receptor, NF-κB, and c-Jun N-terminal kinase signals to control the chronic metabolic inflammatory response (8–11), confirming its significance in the pathogenesis of nonalcoholic steatohepatitis.
Cell autophagy maintains organelle quality control by engulfing and degrading damaged intracellular components and cytoplasmic contents such as proteins (12), glycogen (13), and lipid droplets (14, 15). Impaired autophagy has been linked to the initial development of hepatic steatosis and the progression of steatosis to liver injury (14, 16, 17). The inhibition of autophagy by genetic knockdown of the autophagy gene Atg5 or pharmacological inhibition with an autophagy inhibitor significantly increased cellular triglyceride (TG) content (14, 17); thus, autophagy plays a central role in the breakdown of hepatic lipid droplet-stored TG and cholesterol by lipophagy (17). This is an alternative pathway of lipid metabolism acting via the lysosomal degradative pathway of autophagy, degrading lipid droplet TG and cholesterol by lysosomal acidic hydrolases. The free fatty acids generated by lipophagy from the breakdown of TGs then fuel cellular rates of mitochondrial β-oxidation. Decreased liver lipophagy aggravates hepatic lipid overaccumulation and an increased incidence of NAFLD (14). Although CD36 has been confirmed to significantly contribute to the CD5L-mediated macrophage autophagy (18), the relationship between CD36 and autophagy/lipophagy is largely unknown. The potential role of CD36 in regulating lipophagy has never been addressed in NAFLD.
AMPK, a serine-threonine kinase, functions as an energy and metabolic sensor in maintaining metabolic homeostasis (19). The activation of AMPK occurs primarily through phosphorylation of its catalytic α subunit at the Thr172 residue by liver kinase B1 (LKB1) or by Ca2+/calmodulin-dependent protein kinase kinase β (20, 21). In addition to the role of AMPK as a regulator of energy homeostasis, growing evidence has implicated AMPK in the regulation of various physiologic and pathologic pathways such as lipid metabolism, mitochondrial biogenesis, gene expression, and protein synthesis (22, 23). In the autophagy process, AMPK directly phosphorylates unc-51-like autophagy activating kinase 1 (ULK1) at sites Ser317, Ser555, andSer777 and phosphorylates Beclin1 at site S91/S94 to activate the proautophagy Vps34 complex, which is critical for its function in autophagy (19, 24–26). Moreover, AMPK indirectly activates autophagy by suppressing the activity of the mammalian target of rapamycin (mTOR) complex 1, whose high activity prevents the activation of ULK1 by phosphorylating ULK1 at Ser757 and disrupts the interaction between ULK1 and AMPK (25). Interestingly, the role of CD36 in enhancing fatty acid oxidation appears to be linked to CD36 AMPK interregulation (27). CD36 was shown to be important for coordinating the dynamic protein interactions within a molecular complex consisting of the CD36 partner tyrosine kinase Fyn, the AMPK kinase LKB1, and AMPK. CD36 expression maintains AMPK quiescence by allowing Fyn to access and phosphorylate LKB1, promoting its nuclear sequestration away from AMPK. LCFA binding to CD36 activates AMPK within minutes via its ability to dissociate Fyn from the complex as CD36 is internalized into LKB1-rich vesicles. An earlier work from our group found that CD36 translocation to the plasma membrane of hepatocytes was associated with low AMPK activity and accompanied by low hepatic fatty acid oxidation, which may also result from the increased LKB1 phosphorylation (11). These studies lead to the reasonable speculation that CD36 may be associated with autophagy through the AMPK pathway.
In this study, we aimed to investigate the regulatory action of CD36 in autophagy/lipophagy and its underlying molecular mechanism in vitro and in vivo. Our results demonstrate that the hepatocyte CD36 has a negative role in the regulation of lipophagy through an AMPK-dependent pathway. This suggests that correction of the autophagy deficiency by ameliorating CD36 expression in hepatocytes may be a novel strategy for the treatment of NAFLD.
MATERIALS AND METHODS
Chemicals and reagents
Palmitic acid (PA) (Sigma-Aldrich, St. Louis, MO) was dissolved in 10% BSA with a stock concentration of 5 mM, aliquoted, and stored at −20°C. Chloroquine (CQ) (Sigma-Aldrich) inhibits lysosomal acidification and therefore prevents autophagy by blocking autophagosome fusion and degradation, which was dissolved in double-distilled water with a stock concentration of 50 mM and stored at 4°C. Wortmannin (Sigma-Aldrich), a phosphoinositide 3-kinase (PI3K) inhibitor, was dissolved in DMSO with a stock concentration of 400 µM at −80°C. Compound C (CC) (Selleck Chemicals, Houston, TX) was dissolved in DMSO and stored at −80°C. BODIPY 492/502 (Invitrogen, Carlsbad, CA) was dissolved in DMSO and stored at −20°C. LysoTracker was purchased from Life Technology (Grand Island, NY).
Cell culture
Hepatocytes (HepG2 and Huh7 cells) were maintained in DMEM (HyClone, Logan, UT) supplemented with 10% FBS (Natocor, Corcovado, Argentina). Cells were cultured at 37°C in a humidified atmosphere with 5% CO2 (v/v). HepG2 and Huh7 stable cell lines were constructed with the lentivirus constructs, including GV341 empty vector and GV341 vector containing CD36, respectively. Stable cell lines were also cultured according to the abovementioned methods. Western blotting, quantitative PCR (qPCR), immunofluorescence, lipid droplet staining, and autophagic flux analysis in cells were performed following treatment with 0.5 mM PA for 24 h.
Experimental animals
CD36−/− mice created on a C57BL/6J background were provided by Maria Febbraio (Lerner Research Institute, Cleveland, OH). Eight-week-old male C57BL/6J or CD36−/− mice were fed a normal chow diet (NCD; n = 6) or high-fat diet (HFD; n = 6) for 14 weeks. In the experiment of reconstructing CD36 expression in CD36−/− mice, the 8-week-old male CD36−/− mice were injected with lentivirus empty vector (n = 6) or lentivirus vector containing CD36 (n = 6) in the tail vein and kept on the HFD for 8 weeks. All mice were housed in a temperature-controlled environment and on a 12 h light/dark cycle with free access to diet and water. Before being euthanized, mice with free access to water were deprived of food overnight. Animal treatment conformed to the guidelines of the Institutional Animal Care and Use Committee of Chongqing Medical University. All animals received care according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Gene silencing
Knockdown of CD36 in HepG2 cells was achieved by using a reverse siRNA transfection procedure performed in six-well plates. Therefore, for each well to be transfected, 5 μl Lipofectamine RNAiMAX (Thermo Fisher Scientific, Eugene, OR) were mixed with 500 μl Opti-MEM (Thermo Fisher Scientific) and combined with 25 pmol siRNA (GenePharma, Shanghai, China). The transfection mixture was incubated at room temperature for 20 min. HepG2 cells were harvested in complete growth medium without antibiotics and diluted so that 2 ml contained the appropriate number of cells to give 30% to 50% confluence 24 h after plating. Cell suspensions were mixed with the transfection mixture and incubated. CD36-knockdown Huh7 cells were achieved by using a forward transfection procedure according to the product specification.
Western blot analysis
Cells and mouse liver tissue were lysed with RIPA lysis buffer. Whole-cell extracts containing 30 μg protein per lane were dissolved on an 8% or 12% acrylamide gel and blotted wetly onto a PVDF membrane (Immobilon-P; 0.2 or 0.45 μm; Merck Millipore, Darmstadt, Germany). Membranes were blocked with 3% BSA for 1 h at room temperature and probed with specific antibodies overnight at 4°C [anti-LC3B: 1:2000 (Sigma-Aldrich); anti-p62: 1:5000 (Abcam, Cambridge, UK); anti-CD36: 1:2000 (Novus Biologicals, Centennial, CO); anti-CPT1a: 1:1000 (Proteintech, Rosemont, IL); anti-AMPK: 1:1000 (CST, Danvers, MA); anti-Phospho-AMPKα (Thr172): 1:1000 (CST); anti-LKB1: 1:3000 (Abcam); anti-mTOR: 1:1000 (CST); anti-Phospho-mTOR (Ser2448): 1:1000 (CST); anti-FOXO1: 1:500 (Merck Millipore); anti-Phospho-FOXO1 (Ser256): 1:1000 (CST); anti-ATG7: 1:1000 (CST); anti-ULK1: 1:1000 (CST); anti-Phospho-ULK1 (Ser555): 1:1000 (CST); anti-Beclin1: 1:1000 (CST); anti-Phospho-Beclin1 (Ser93): 1:1000 (CST); and anti-β actin: 1:3000 (Proteintech)]. Detection was achieved by using appropriate HRP-conjugated secondary antibodies (1:8000; Proteintech) in conjunction with the ECL reagent (Clarity Western ECL; Bio-Rad Laboratories, Hercules, CA). Afterwards, membranes were exposed, and signals were quantified using an Image Analyzer.
Immunofluorescence assay
Cells were seeded on glass coverslips prior to treatment. Following treatment, cells were washed in PBS, fixed for 25 min in 4% formaldehyde, and washed three times. Cells were then permeabilized in 100% methanol at −20°C for 10 min, washed, and blocked in normal goat serum for 1 h. Cells were then incubated with primary antibody overnight at 4°C, washed with PBS, and then incubated for 2 h at room temperature with fluorescence-conjugated secondary antibodies. Cells were washed three times and then treated with DAPI for 5 min. This was followed by three washes with PBS. For experiments involving lipid droplet staining, cells were stained with a fluorescent lipid dye, BODIPY 492/502, at a 1:1000 dilution for 30 min at room temperature before DAPI staining. Coverslips were visualized using a Leica confocal microscope.
Autophagic flux analysis
HepG2 or Huh7 cells were treated with siCD36 or siNeg and grown for 72 h. During the last 24 h of incubation prior to the experiment, the cells were either untreated or treated with 10 µM CQ. Autophagy was assessed by combinatory detection of the autophagosome formation marker LC3II, along with p62. In another experiment to detect autophagic flux, HepG2 cells were first transduced with siCD36 or siNeg in a confocal dish. Twenty-four hours after the first transduction, the cells were then transduced with monomeric red fluorescent protein (mRFP)-GFP-LC3 adenoviral vectors (HanBio Technology, Shanghai, China). The principle of the assay is based on the different pH stability of red and green fluorescent proteins. The enhanced GFP signal could be quenched under the acidic condition (pH <5) inside the lysosome, whereas the mRFP signal did not change significantly in acidic conditions. In red- and green-merged images, autophagosomes are shown as yellow puncta, while autolysosomes are shown as red puncta. An enhancement of both yellow and red puncta in cells indicate that autophagic flux is increased, while autophagic flux is blocked when only yellow puncta are increased without alteration of red puncta, or when both yellow and red puncta are decreased in cells (28, 29). HepG2 cells were incubated in 1 ml growth medium with the adenoviruses for 2 h at 37°C, and the growth medium was replaced with fresh medium. Experiments were performed 48 h after the second transduction. LC3 puncta were examined with a Leica confocal microscope.
Electron microscopy
Fresh tissue was placed in 4% glutaraldehyde overnight at 4°C. Ultrathin sections were cut and then stained. Images were acquired on a transmission electron microscope. For the quantification of autophagy, autophagic vacuoles (defined as autophagosomes, double-membraned structures surrounding cytoplasmic material, and autolysosomes, lysosomes containing cytoplasmic material) were counted.
Measurements of oxygen consumption rate
Oxygen consumption was measured on a Seahorse XF24 analyzer at 37°C. The Seahorse XF Cell Mito Stress Test Kit, XFe24 FluxPak mini, and XF Base Medium were purchased from Agilent Technologies (Santa Clara, CA). Forty-eight hours before assays, 40,000 cells of siCD36- or siNeg-treated HepG2 cells were seeded on a Seahorse XF24 analyzer plate. To study the effects of fat treatment on cells, media were replaced with 0.2% BSA and 0.5 mM PA 24 h before analysis. On the day of the assay, media were replaced with the Seahorse assay media. The oxygen consumption rate was measured in the basal state and after oligomycin treatment, FCCP treatment, and rotenone/antimycin A treatment. Basal respiration and maximal respiration were calculated respectively. ATP production was calculated by subtracting the minimum rate measurement after oligomycin injection from the last rate measurement in the basal state. Following the assay, cells were lysed, and protein content was taken for normalization.
TG content assays
The assay was performed with a TG assay kit according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Immunocytochemistry
Prior to treatment, cells were seeded on glass coverslips. Following treatment, cells were washed in PBS, fixed for 15 min in precooled acetone, and washed three times. Cells were then permeabilized in 0.5% Triton X-100 for 10 min and washed. Endogenous peroxidases were inactivated using 3% H2O2, followed by blocking with goat serum. Cells were incubated overnight at 4°C with transcription factor EB (TFEB) antibody (1:100; Proteintech). Cells were then washed and incubated for 45 min with secondary antibody. Cytochemical reactions were performed using a diaminobenzidine kit, and cells were counterstained with hematoxylin. Images were captured using a Zeiss microscope (Jena, Germany).
qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Then, 1 μg total RNA was reverse-transcribed to obtain cDNA using PrimeScript RT Reagent Kit (TaKaRa, Kusatsu, Japan) according to the manufacturer’s protocol. qPCR was performed using SYBR Green PCR Mix kit (TaKaRa). Specific primer sequences used for qPCR are listed in Table 1. The housekeeping gene of β-actin was used for normalization in cultured cells, whereas 18 s ribosomal RNA expression was used for normalization in mouse liver tissue. Fold change was calculated using 2−ΔΔCt.
TABLE 1.
qPCR primer sequences
| Genes | Sequences |
| Human CD36 | Forward: 5′- CTTTGGCTTAATGAGACTGGGAC -3′ |
| Reverse: 5′- GCAACAAACATCACCACACCA -3′ | |
| Human ULK1 | Forward: 5′- AGCACGATTTGGAGGTCGC-3′ |
| Reverse: 5′- GCCACGATGTTTTCATGTTTCA-3′ | |
| Human Beclin1 | Forward: 5′- CTCCTGGGTCTCTCCTGGTT-3′ |
| Reverse: 5′- TGGACACGAGTTTCAAGATCC-3′ | |
| Human Atg5 | Forward: 5′- ACTGTCCATCTGCAGCCAC -3′ |
| Reverse: 5′- TGCAGAAGAAAATGGATTTCG -3′ | |
| Human Atg7 | Forward: 5′- ATTGCTGCATCAAGAAACCC -3′ |
| Reverse: 5′- GAGAAGTCAGCCCCACAGC -3′ | |
| Human Atg12 | Forward: 5′- CCATCACTGCCAAAACACTC -3′ |
| Reverse: 5′- TTGTGGCCTCAGAACAGTTG -3′ | |
| Human Atg16l1 | Forward: 5′- AACGCTGTGCAGTTCAGTCC -3′ |
| Reverse: 5′- AGCTGCTAAGAGGTAAGATCCA -3′ | |
| Human Atg16l2 | Forward: 5′- TGGACAAGTTCTCAAAGAAGCTG -3′ |
| Reverse: 5′- CCTCAGTGCGACCAGTGAT -3′ | |
| Human FOXO1 | Forward: 5′-TCGTCATAATCTGTCCCTACACA -3′ |
| Reverse: 5′-CGGCTTCGGCTCTTAGCAAA -3′ | |
| Human β-actin | Forward: 5′-GTTGTCGACGACGAGCG -3′ |
| Reverse: 5′-GCACAGAGCCTCGCCTT -3′ | |
| Mouse ULK1 | Forward: 5′- AAGTTCGAGTTCTCTCGCAAG-3′ |
| Reverse: 5′- ACCTCCAGGTCGTGCTTCT-3′ | |
| Mouse Beclin1 | Forward: 5′-GGCGAGTTTCAATAAATGGC-3′ |
| Reverse: 5′-CCAGGAACTCACAGCTCCAT-3′ | |
| Mouse CPT1a | Forward: 5′-TGGCATCATCACTGGTGTGTT-3′ |
| Reverse: 5′-GTCTAGGGTCCGATTGATCTTTG-3′ | |
| Mouse 18sRNA | Forward: 5′- TCGAGGCCCTGTAATTGGAA-3′ |
| Reverse: 5′- CCCTCCAATGGATCCTCGTT-3′ |
Statistical analysis
All cell culture experiment data represent at least three independent experiments and are expressed as means ± SDs. The difference between two groups was statistically analyzed using Student’s t-test in GraphPad Prism version 5. Statistical tests of significance are given in the figure legends.
RESULTS
CD36 expression in the liver of mice treated with the HFD was increased and LC3II, an autophagosome marker, was decreased
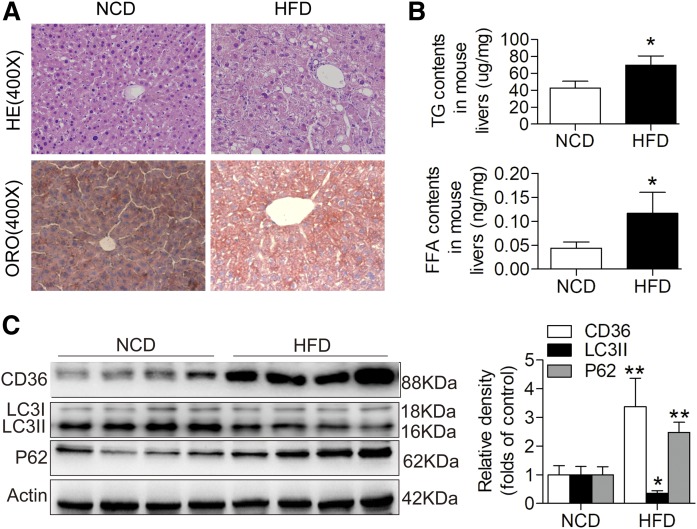
Eight-week-old C57BL/6 mice were fed the HFD for 14 weeks to induce the NAFLD model. Hematoxylin and eosin staining and Oil Red O staining showed evident steatosis in the livers of mice fed the HFD compared with the NCD (Fig. 1A). TG and free fatty acid contents were significantly higher in the livers of mice fed the HFD compared with those fed the NCD (Fig. 1B). To determine CD36 expression and the status of autophagy in NAFLD, we performed Western blotting to detect CD36 and LC3II in mice with liver steatosis and corresponding controls and found that the expression of CD36 was increased and LC3II level was significantly reduced in the mice with liver steatosis. Furthermore, we observed an increase in the protein levels of SQSTM1/p62, one of the selective substrates of autophagy, which was commonly used as a marker of autophagic flux (Fig. 1C). These data suggested that there may be a negative correlation between the expression of CD36 and autophagy in vivo.
Fig. 1.
Hepatic CD36 expression is increased and autophagy is decreased in response to HFD treatment in mice. A: HE staining (original magnification: ×400) and ORO staining (original magnification: ×400). B: TG and FFA levels in mouse liver tissues. C: Representative Western blotting analysis of CD36, LC3II, and p62 expression in mouse livers. Data are presented as means ± SDs, n = 6. *P < 0.05 and **P < 0.01 versus NCD groups. FFA, free fatty acid; HE, hematoxylin and eosin staining; ORO, Oil Red O.
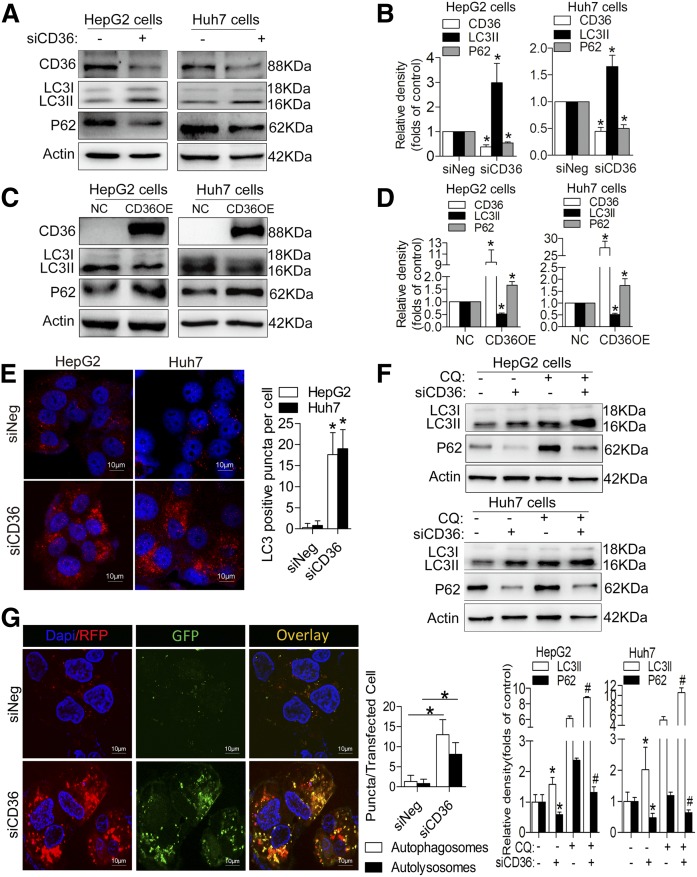
Downregulation of CD36 increased autophagy and upregulation of CD36 inhibited autophagy in HepG2 and Huh7 cells
Next, we transfected HepG2 and Huh7 cells with siCD36s to knock down CD36 expression and observed an increase in LC3II protein and a decrease in p62 level under the PA treatment conditions (Fig. 2A, B). We also observed an increase in LC3II protein and a decrease in p62 level after downregulating the CD36 expression in the HepG2 cells without PA treatment (supplemental Fig. S1A). Meanwhile, we constructed CD36 overexpressing (OE) stable cell lines in both HepG2 and Huh7 cells, finding that CD36 OE in both cells with PA treatment reduced LC3II expression and increased p62 accumulation (Fig. 2C, D). To further confirm autophagy changes, we detected the LC3 puncta following immunofluorescence staining. An increase in LC3 puncta was observed in siCD36 HepG2 and Huh7 cells with PA treatment (Fig. 2E). Because the increase in LC3II levels by immunoblot analysis could be due to either the induced formation or the decreased clearance of autophagosomes, we treated siCD36 and siNeg HepG2 and Huh7 cells with the lysosomal inhibitor CQ in the presence of PA. We observed increased LC3II and decreased accumulation of p62 in both CD36 knockdown cells, and we observed a relative increase in LC3II level and decreased p62 after using CQ when these same cells were compared with siNeg cells (Fig. 2F). These data suggested that an increase in autophagosome formation had occurred in CD36 knockdown cells, thereby inducing autophagic flux. To further support this finding, we transfected siCD36 and siNeg HepG2 cells with mRFP-GFP-LC3 adenoviral vectors that fluoresce both red and green (yellow) in autophagosomes and only red in autolysosomes and then treated them with 0.5 mM PA for 24 h. We observed an increase in the numbers of both yellow and red puncta in siCD36 HepG2 cells compared with control cells, suggesting an increase in autophagosome formation (Fig. 2G). Taken together, these data supported the notion that autophagosome induction was due to increased formation rather than decreased clearance in the CD36 knockdown cells.
Fig. 2.
Downregulation of CD36 increases autophagy and upregulation of CD36 inhibits autophagy in HepG2 and Huh7 cells with PA treatment. A, B: LC3II and p62 protein levels after CD36 knockdown. C, D: LC3II and p62 levels after CD36 overexpression. E: Immunofluorescence staining with LC3 antibody. Fifty cells were used for statistical analysis of the number of LC3-positive puncta per cell. F: LC3II and p62 protein levels after treatment with CQ in siNeg and siCD36 HepG2 or Huh7 cells. G: Analysis of double-fluorescent mRFP-enhanced GFP-LC3 fusion protein expression to detect autophagic flux (yellow puncta represent autophagosomes and red puncta represent autolysosomes) in siNeg and siCD36 HepG2 cells. Fifty cells were used for the statistical analysis of the number of autophagosomes and autolysosomes per transfected cell. Data are presented as means ± SDs, n = 3; for A, B, E, and G, *P < 0.05 versus siNeg groups; for C and D, *P < 0.05 versus NCD groups; and for F, *P < 0.05 versus siNeg cells without CQ. #P < 0.05 versus siNeg cells with CQ.
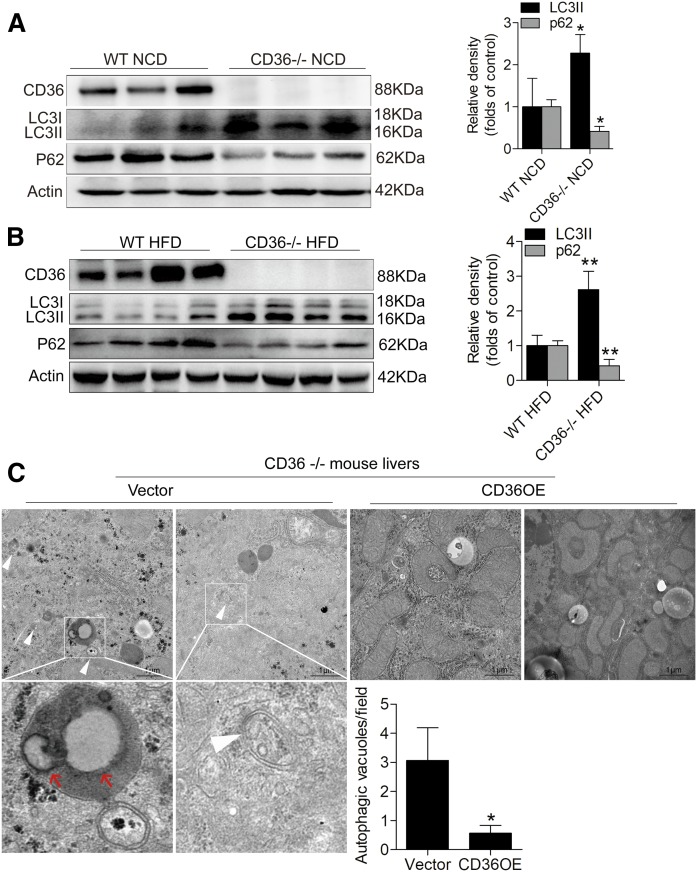
CD36 knockout in mice increased liver autophagy, and its reconstruction in CD36 knockout mice reduced autophagy
To further confirm the relationship between CD36 and autophagy in vivo, we detected CD36, LC3II, and p62 protein levels in the livers of CD36 knockout mice (CD36−/− mice) and WT mice. In both the NCD and HFD conditions, we observed a very significant increase in LC3II expression and a pronounced reduction in p62 level in the livers of CD36−/− mice compared with WT mice (Fig. 3A, B). This suggested that the deficiency of CD36 vigorously promoted autophagy. After reconstructing CD36 expression with lentivirus vectors (CD36 OE) in CD36−/− mice, ultrastructural analysis by electron microscopy showed a decrease in numbers in autophagic vacuoles (defined to include autophagosomes and autolysosomes) in the livers, and there were more autophagosomes and autolysosomes in the livers of CD36−/− mice injected with lentivirus empty vector (Fig. 3C). Interestingly, in the livers of CD36−/− mice injected with lentivirus empty vector, we found that autolysosomes engulfed lipid droplets. Thus, we determined that lipophagy occurred in CD36−/− mouse livers.
Fig. 3.
CD36 knockout in vivo increases autophagy of mouse livers and its reconstruction in CD36 knockout mice decreases autophagy. A: Western blotting analysis of LC3II and p62 levels in CD36−/− and WT mice fed the NCD. B: Western blotting analysis of LC3II and p62 levels in CD36−/− and WT mice fed the HFD. C: Ultrastructural analysis of the livers of the CD36−/− mice treated with lentivirus empty vector or lentivirus vector containing CD36. The white arrowhead represents autophagic vacuoles (defined to include autophagosomes and autolysosomes), and the red arrowhead represents intralysosomal lipids. Twenty fields were used for the statistical analysis of the number of autophagic vacuoles per field. Data are presented as means ± SDs, n = 6. *P < 0.05 and **P < 0.01 versus respective control groups.
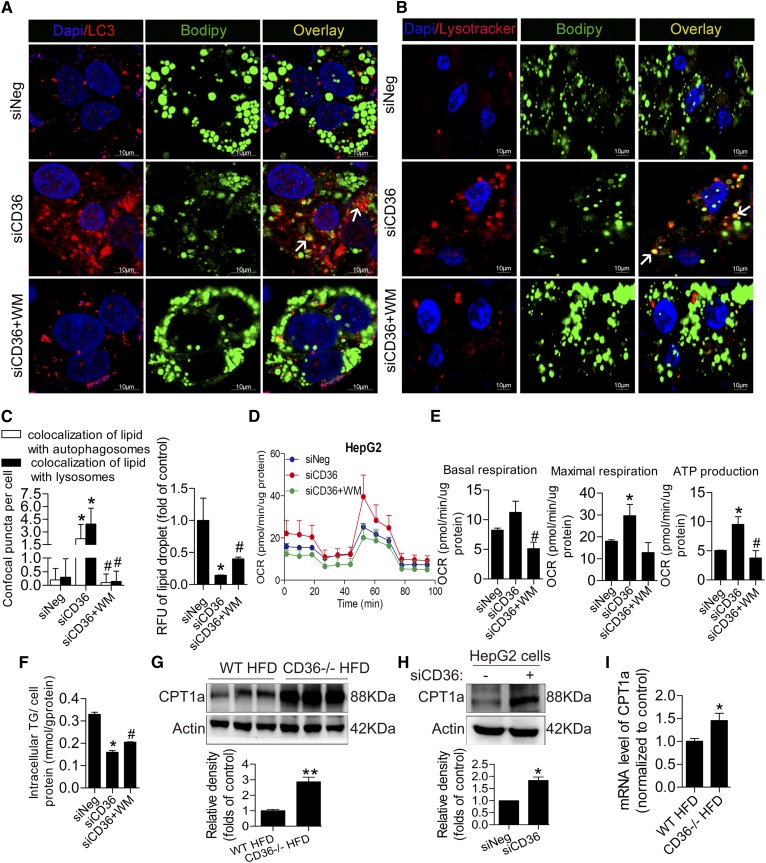
Lipophagy was induced by CD36 knockdown in HepG2 cells, followed by enhanced β-oxidation, while inhibition of autophagy with wortmannin reduced lipophagy and β-oxidation
CD36 knockdown in HepG2 cells exhibited a significant increase in autophagy coupled with a decrease in PA-treated lipid droplets. Although the reduction of intracellular lipid content may be partially due to decreased fatty acid uptake, increased β-oxidation of fatty acid is also important for reducing abnormal lipid accumulation. Lipophagy engulfing lipid droplets has been linked to reduced lipid content and increased β-oxidation. In this connection, we noted an increase in autophagosomal (LC3/BODIPY) and lysosomal (LysoTracker/BODIPY) lipids in siCD36 HepG2 cells after loading 0.5 mM PA for 24 h but a disappearance in siCD36 HepG2 cells treated with wortmannin (Fig. 4A–C). Furthermore, we measured the oxygen consumption rate and found that CD36 knockdown in HepG2 cells also increased basal respiration, maximal respiration, and ATP production compared with control HepG2 cells when pretreated with lipids, and the inhibition of autophagy reversed the situation (Fig. 4D, E). These data suggested that CD36 knockdown increased lipophagy and β-oxidation, subsequently contributing to reducing lipid accumulation, and the inhibition of autophagy decreased lipophagy and β-oxidation, thereby aggravating lipid accumulation. To further confirm the effect of CD36-mediated lipophagy on lipid content, we detected the intracellular TG level in HepG2 cells of the three groups after PA treatment. The results showed that CD36 knockdown decreased TG content, and the inhibition of autophagy in siCD36 HepG2 cells increased TG content (Fig. 4F). Moreover, the protein level of CPT1a, a rate-limiting enzyme for fatty acid oxidation, was increased in CD36 −/− mice fed both the HFD and NCD (Fig. 4G, supplemental Fig. S1B). With or without PA treatment, CD36 knockdown increased CPT1a protein expression (Fig. 4H, supplemental Fig. S1C). The mRNA level of CPT1a was increased in the livers of CD36−/− mice fed the HFD compared with WT mice (Fig. 4I). These data suggested that the deletion of CD36 increased fatty acid oxidation.
Fig. 4.
Knockdown of CD36 increases lipophagy and β-oxidation and inhibition of autophagy reduces lipophagy and β-oxidation. A–C: Autophagosomal (LC3/BODIPY) and lysosomal (LysoTracker/BODIPY) colocalization with lipids under PA treatment. Thirty cells were used for the statistical analysis of the number of confocal puncta per cell. D, E: Oxygen consumption rate in HepG2 cells. F: Intracellular TG content in HepG2 cells with PA treatment. G: Western blotting analysis of CPT1a in WT and CD36−/− mice fed the HFD. H: CPT1a protein level in HepG2 cells with PA treatment. I: CPT1a mRNA level in WT and CD36−/− mice fed the HFD. Data are presented as means ± SDs; n = 3 for A–F and H, *P < 0.05 versus siNeg groups and #P < 0.05 versus siCD36 groups; n = 6 for G and I, *P < 0.05 and **P < 0.01 versus WT HFD groups.
The AMPK pathway was activated in CD36 knockdown hepatocytes and CD36 knockout mouse livers
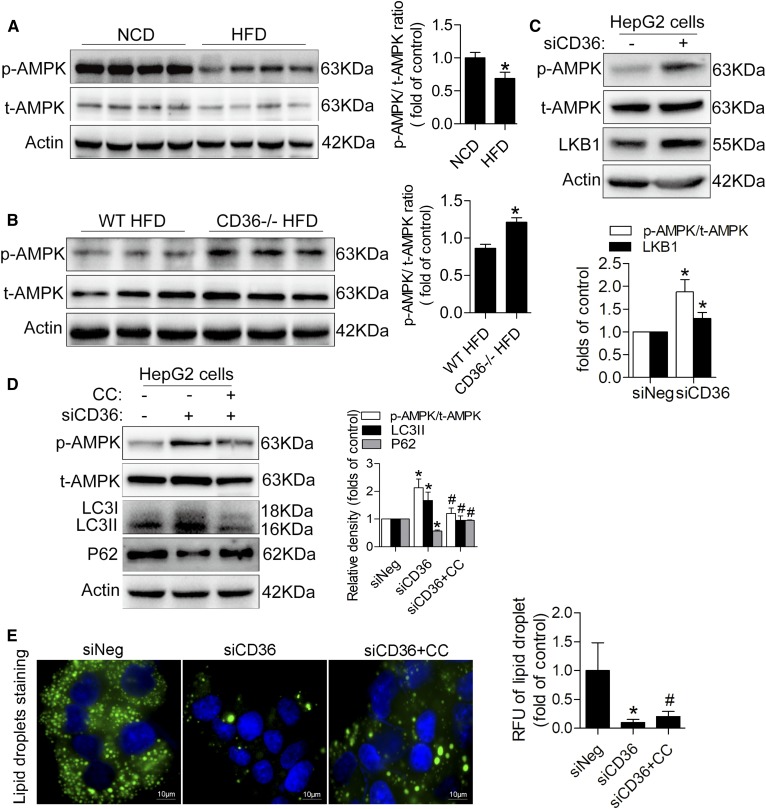
We next investigated classical signaling pathways known to regulate autophagy. Phosphorylation of AMPK at site Thr172 in the livers of mice fed the HFD was lower than that of mice fed the NCD (Fig. 5A). The livers of CD36−/−mice fed the HFD showed a significant increase in the phosphorylation of AMPK, suggesting that the knockout of CD36 activated the AMPK pathway (Fig. 5B). To further confirm the AMPK pathway, we detected p-AMPK (Thr172) and total AMPK protein levels in siCD36 HepG2 and control cells. Compared with the control group, there was a significant increase of p-AMPK (Thr172) in siCD36 HepG2 cells whether in the presence of PA or in the absence of PA (Fig. 5C, supplemental Fig. S1D), confirming that the AMPK pathway was activated in vitro. The serine/threonine kinase LKB1 is one of the major upstream kinases that activate AMPK by phosphorylating the Thr172 residue in AMPKα. We found that CD36 knockdown cells showed a higher LKB1 protein level, which was consistent with AMPK activity (Fig. 5C). The autophagy markers LC3II and p62 were reversed when AMPK was inhibited by the AMPK inhibitor CC in siCD36 HepG2 cells under the condition of PA treatment (Fig. 5D), which was accompanied by a rebound of lipid accumulation (Fig. 5E).
Fig. 5.
AMPK signaling is mechanistically involved in CD36-regulated autophagy. A: Representative Western blotting analysis of p-AMPK and total AMPK levels in NCD and HFD mouse livers. B: p-AMPK and total AMPK levels in CD36−/− and WT mice fed the HFD. C: p-AMPK, total AMPK, and LKB1 levels in HepG2 cells with PA treatment. D: p-AMPK, total AMPK, LC3II, and p62 levels in siCD36 HepG2 cells with or without CC under PA treatment. E: Lipid droplets in siCD36 HepG2 cells with or without CC under PA treatment. Data are presented as means ± SDs; n = 6 for A,*P < 0.05 versus the NCD group; n = 6 for B, *P < 0.05 versus the WT HFD group; n = 3 for C–E, *P < 0.05 versus siNeg groups; and #P < 0.05 versus siCD36 groups.
Reducing CD36 expression increased autophagy through the phosphorylation of ULK1/Beclin1 in vitro
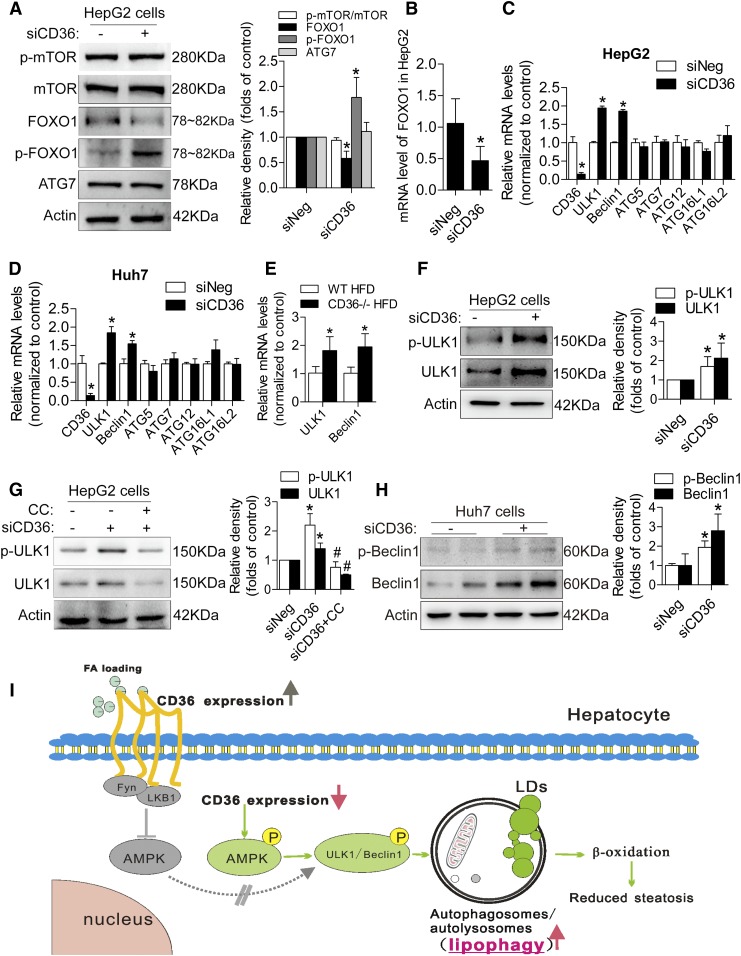
AMPK promotes autophagy partly by suppressing mTOR or increasing the transcriptional activity of FOXO1. However, we found that siCD36 HepG2 cells showed no noticeable difference in p-mTOR and mTOR levels compared with siNeg cells (Fig. 6A). In addition, the protein and mRNA expressions of the transcription factor FOXO1 were not increased in siCD36 HepG2 cells, and the phosphorylation of FOXO1 was increased in siCD36 HepG2 cells, but the interacting protein ATG7 was not increased (Fig. 6A, B). Moreover, the mRNA screening results of several ATGs showed that ULK1 and Beclin1 were induced in siCD36 HepG2 and Huh7 cells (Fig. 6C, D). The results from CD36−/− mouse liver tissue also confirmed this finding (Fig. 6E). We tested the protein expression and phosphorylation level of ULK1 and Beclin1 in siCD36 HepG2 and Huh7 cells. CD36 knockdown in HepG2 cells significantly increased ULK1 expression and phosphorylation (Ser555) (Fig. 6F), and the inhibition of AMPK with CC reversed the expression and phosphorylation level of ULK1 (Fig. 6G). Meanwhile, knockdown of CD36 in Huh7 cells induced Beclin1 expression and phosphorylation (Ser93) (Fig. 6H), suggesting that CD36 knockdown induced autophagy through the AMPK-ULK1/Beclin1 pathway. Furthermore, we also observed an induced nuclear TFEB after downregulating CD36 expression, suggesting that an induction in nuclear TFEB may also contribute to the increased autophagy (supplemental Fig. S1E).
Fig. 6.
Reducing CD36 expression increased autophagy through the phosphorylation of ULK1/Beclin1 in the presence of PA. A: Representative Western blotting analysis of p-mTOR, mTOR, FOXO1, p-FOXO1, and ATG7 levels in siCD36 HepG2 cells. B: mRNA level of FOXO1 in siCD36 HepG2 cells. C, D: mRNA levels of several important ATGs, including ULK1, Beclin1, ATG5, ATG7, ATG12, ATG16L1, and ATG16L2, in siCD36 HepG2, and Huh7 cells. E: mRNA levels of ULK1 and Beclin1 in CD36−/− and WT mice fed the HFD. F: p-ULK1 and total ULK1 protein levels in siCD36 HepG2 cells. G: p-ULK1 and total ULK1 protein levels in siCD36 HepG2 cells with or without CC. H: p-Beclin1 and total Beclin1 protein levels in siCD36 Huh7 cells. Data are presented as means ± SDs; n = 3 for A–D and F–H, *P < 0.05 versus siNeg groups; n = 6 for E, *P < 0.05 versus WT HFD group; #P < 0.05 versus siCD36 group; I: The proposed model of CD36-mediated lipophagy.
DISCUSSION
Lipid overaccumulation is the hallmark of NAFLD. CD36 is an important membrane glycoprotein facilitating fatty acid uptake (4, 30). In addition to the function of fatty acid translocase, CD36 plays an important role in mediating signal transduction, acting as a regulator of the inflammatory response and modulating fatty acid β-oxidation, which contributes to the maintenance of cellular fatty acid homeostasis (7). Impaired autophagy in the liver has been connected with disorders of lipid overaccumulation (14). In fatty livers, a variety of factors can regulate autophagy, such as insulin resistance, oxidative stress, excess TGs, and free fatty acids (16, 31, 32). However, the relationship between CD36 and liver autophagy is still unknown. Our study is the first to demonstrate that CD36 negatively regulates autophagy in mouse livers and hepatocytes.
Autophagy is characterized as a selective, bulk degradative system engulfing cytosolic materials, and selective autophagy regulates hepatic metabolic pathways (33). Lipophagy is a form of selective autophagy that pinches off part of a lipid droplet and fuses it with a lysosome (14). Lipophagy promotes lipolysis, which is subsequently catabolized by β-oxidation to produce energy and ketone bodies so could function as a hepatocyte defense mechanism against NAFLD. Enhancement of autophagy using rapamycin, carbamazepine, and other pharmaceutical agents alleviates liver steatosis (31, 34–36). Our study demonstrates that CD36 knockdown in hepatocytes upregulates autophagy, which reduces lipid accumulation by facilitating β-oxidation of fatty acids. We have provided evidence for a novel autophagy-dependent mechanism of CD36 knockdown that attenuates PA-induced lipid accumulation. Moreover, we have shown that the inhibition of autophagy in hepatocytes is involved in lipid accumulation by perturbing β-oxidation of fatty acid.
The mechanism of increased autophagy in CD36 knockdown hepatocytes and CD36 knockout mice is probably multifactorial. Our results reveal that CD36 knockdown/knockout is associated with AMPK activation in hepatocytes and mouse livers. These observations are consistent with the report that the reduction of CD36 expression induces AMPK activation (7). Stimulation of AMPK pathways is a proautophagic classical mechanism that occurred both in vitro and in vivo. Our data reveal that AMPK activation plays a significant role in CD36 knockdown-induced autophagy and subsequent attenuated lipid accumulation.
AMPK activation is one of the major mechanisms accounting for autophagy induction. AMPK promotes autophagy partly by suppressing the mTOR pathway, a negative regulator of autophagy that phosphorylates ULK1 at Ser757. Our data show no obvious change of p-mTOR and mTOR protein levels, indicating that the mTOR pathway may not be involved in CD36 knockdown-induced autophagy. Mammalian FOXO transcription factors, including FOXO1, FOXO3a, FOXO4, and FOXO6, function critically in the regulation of metabolism, cell cycle arrest, and oxidative stress resistance (37). FOXO1 is also largely known as an important transcription factor to induce autophagy (38). The activation of AMPK can regulate the activation of FOXO1 (39, 40). However, our results reveal that CD36 knockdown in HepG2 cells reduces the mRNA and protein expression of FOXO1 but increases the level of p-FOXO1. Several studies have reported that cytoplasmic p-FOXO1 is inactivated (37, 39). However, Wang et al. (41) noted that p-FOXO1 is located in the cytoplasm of iNKs and interacts with ATG7, leading to the induction of autophagy, which was independent of the transcriptional activity of FOXO1. ATG7 is an E1-like enzyme activating the ubiquitin-like molecule ATG12 and driving the conjugation of ubiquitin-like molecule LC3 to the lipid phosphatidylethanolamine (42). Therefore, we detected ATG7 expression. However, our data show no obvious ATG7 change, suggesting that FOXO1 transcriptional function and the interaction between p-FOXO1 and ATG7 may not be involved in CD36 knockdown-induced autophagy. We detected several important autophagy-related genes in HepG2 and Huh7 cells and mouse livers. The results showed that ULK1 and Beclin1 mRNA expressions were increased, implying that a transcriptional mechanism may be involved in the upregulation of autophagy. The ATG1/ULK1 complex plays a crucial role in autophagy induction, integrating signals from upstream sensors such as AMPK and transducing them to the downstream autophagy pathway. The protein kinase activity of ULK1 is required for its autophagic function (43). Research shows that in most cases, AMPK activation promotes autophagy by directly binding to and phosphorylating ULK1 at multiple residues such as Ser317, Ser555, and Ser777, and AMPK can also indirectly promote autophagy by reducing the inhibition of mTOR on ULK1 activity through phosphorylating ULK1 at Ser757 (24, 25, 44). Our data showed that phosphorylation of ULK1 at Ser555 and the protein expression of ULK1 in HepG2 were significantly increased after CD36 knockdown. CC administration attenuated the CD36 knockdown-induced increase in ULK1 expression and phosphorylation. In Huh7 cells, protein level and phosphorylation of Beclin1 were increased, which is critical for Vps34 complex formation to induce autophagy. These results suggest that CD36 knockdown activates autophagy through the AMPK-ULK1/Beclin1 signaling pathway in hepatocytes. TFEB is a key positive regulator of autophagy and lysosome biogenesis, and nuclear TFEB induction may contribute to the increased autophagy. Some reports suggest that the nuclear localization of TFEB is directly regulated by mTOR to activate autophagic gene expression (45). Another study reported that ezetimibe induced nuclear TFEB translocation in tsc2−/− mouse embryonic fibroblasts and ezetimibe could still induce autophagy through AMPK activation and TFEB nuclear translocation, related to the MAPK/ERK, and independent of the mTOR pathway. (46). In this study, nuclear TFEB was induced after downregulating CD36 expression. However, the change of mTOR was not observed. Whether the induced nuclear TFEB is involved in the increased autophagy and more details about the pathways of TFEB participating in or AMPK-TFEB axis in CD36-mediated autophagy need further study.
Given that CD36 plays an important role in modulating fatty acid β-oxidation, which acts as a contributor to maintain cellular fatty acid homeostasis, our results provide evidence for a novel lipophagy-dependent mechanism by which the downregulation of CD36 attenuates cellular lipid overaccumulation by promoting β-oxidation of fatty acids. CD36 is a fatty acid translocase, which facilitates the uptake of LCFA. The cell uptake of saturated and unsaturated fatty acids also affects autophagy (47). Exposing cells to saturated fatty acids induced the canonical, BECN1/PI3K-dependent autophagy pathway, and the unsaturated fatty acid oleate triggered autophagic responses that were independent of the BECN1/PI3K complex. Accordingly, we speculated that the uptake of fatty acids in siCD36 cells and CD36−/− mice might also be involved in autophagy. We have also performed experiments to detect autophagy after downregulating CD36 expression in hepatocytes without PA treatment and in CD36−/− mice fed the NCD, and similar results were obtained compared with under PA treatment and in CD36−/− mice fed the HFD. Our main aim was to investigate the regulatory action of CD36 on autophagy/lipophagy in cell and mouse models of NAFLD. We observed that lipophagy occurred in the CD36 knockdown cells with PA treatment and in the livers of CD36−/− mice fed the HFD. The putative effects of blocking lipid uptake on autophagy in siCD36 cells and CD36−/− mice need further experimental exploration.
In conclusion, we have demonstrated that knockdown of CD36 in hepatocytes induces autophagy due to the increased autophagosome formation in autophagic flux, while CD36 overexpression inhibits autophagy. Lipophagy is increased through an AMPK-ULK1/Beclin1 pathway in CD36 knockdown hepatocytes, which contributes to the increased β-oxidation and steatosis alleviation (Fig. 6I). In addition to the fatty acid uptake function of CD36, CD36 is a novel negative regulator of autophagy, and the reduced autophagy is a new mechanism for lipid accumulation in hepatocytes and livers of mice. These findings suggest that attenuating CD36 expression, which enhances the clearance of fatty acids by the induction of lipophagy in addition to the reduction of fatty acid uptake, could be a potential therapeutic strategy for fatty liver diseases, as well as other metabolic disorders.
Supplementary Material
Acknowledgments
We thank Maria Febbraio for providing the CD36−/− mice.
Footnotes
Abbreviations:
- AMPK
- adenosine monophosphate-activated protein kinase
- ATG
- autophagy-related gene
- CC
- compound C
- CD36
- cluster of differentiation 36
- CPTa1
- carnitine palmitoyltransferase 1A
- CQ
- chloroquine
- FOXO
- forkhead box class O
- HFD
- high-fat diet
- LCFA
- long-chain fatty acid
- LKB1
- liver kinase B1
- mRFP
- monomeric red fluorescent protein
- mTOR
- mammalian target of rapamycin
- NAFLD
- nonalcoholic fatty liver disease
- NCD
- normal chow diet
- OE
- overexpressing
- PA
- palmitic acid
- PI3K
- phosphoinositide 3-kinase
- qPCR
- quantitative PCR
- siCD36
- siRNA targeting CD36
- TFEB
- transcription factor EB
- TG
- triglyceride
- UNK1
- unc-51-like autophagy activating kinase
This work was supported by National Natural Science Foundation of China Grants 81873569, 81570517, and 31571210; National Key R&D Program of China Grant 2018YFC1312700; Science and Technology Research Program of Chongqing Municipal Education Commission Grant KJZD-K201800401; and Program for Innovation Team of Higher Education in Chongqing Grant CXTDX201601015. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Rinella M. E. 2015. Nonalcoholic fatty liver disease: a systematic review. JAMA. 313: 2263–2273. [DOI] [PubMed] [Google Scholar]
- 2.Shen J., Chan H. L., Wong G. L., Choi P. C., Chan A. W., Chan H. Y., Chim A. M., Yeung D. K., Chan F. K., Woo J., et al. 2012. Non-invasive diagnosis of non-alcoholic steatohepatitis by combined serum biomarkers. J. Hepatol. 56: 1363–1370. [DOI] [PubMed] [Google Scholar]
- 3.Pais R., Charlotte F., Fedchuk L., Bedossa P., Lebray P., Poynard T., Ratziu V., and Group L. S.. 2013. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J. Hepatol. 59: 550–556. [DOI] [PubMed] [Google Scholar]
- 4.Hoosdally S. J., Andress E. J., Wooding C., Martin C. A., and Linton K. J.. 2009. The human scavenger receptor CD36: glycosylation status and its role in trafficking and function. J. Biol. Chem. 284: 16277–16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong S., Zhao L., Wang Y., Zhang C., Liu J., Wang P., Zhou W., Yang P., Varghese Z., Moorhead J. F., et al. 2017. Cluster of differentiation 36 deficiency aggravates macrophage infiltration and hepatic inflammation by upregulating monocyte chemotactic protein-1 expression of hepatocytes through histone deacetylase 2-dependent pathway. Antioxid. Redox Signal. 27: 201–214. [DOI] [PubMed] [Google Scholar]
- 6.Miquilena-Colina M. E., Lima-Cabello E., Sanchez-Campos S., Garcia-Mediavilla M. V., Fernandez-Bermejo M., Lozano-Rodriguez T., Vargas-Castrillon J., Buque X., Ochoa B., Aspichueta P., et al. 2011. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 60: 1394–1402. [DOI] [PubMed] [Google Scholar]
- 7.Samovski D., Sun J., Pietka T., Gross R. W., Eckel R. H., Su X., Stahl P. D., and Abumrad N. A.. 2015. Regulation of AMPK activation by CD36 links fatty acid uptake to β-oxidation. Diabetes. 64: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silverstein R. L., and Febbraio M.. 2009. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2: re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy D. J., Kuchibhotla S., Westfall K. M., Silverstein R. L., Morton R. E., and Febbraio M.. 2011. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc. Res. 89: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart C. R., Stuart L. M., Wilkinson K., van Gils J. M., Deng J., Halle A., Rayner K. J., Boyer L., Zhong R., Frazier W. A., et al. 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L., Zhang C., Luo X., Wang P., Zhou W., Zhong S., Xie Y., Jiang Y., Yang P., and Tang R.. 2018. CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J. Hepatol. 69: 705–717. [DOI] [PubMed] [Google Scholar]
- 12.Ezaki J., Matsumoto N., Takedaezaki M., Komatsu M., Takahashi K., Hiraoka Y., Taka H., Fujimura T., Takehana K., and Yoshida M.. 2011. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 7: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotoulas O. B., Kalamidas S. A., and Kondomerkos D. J.. 2004. Glycogen autophagy. Microsc. Res. Tech. 64: 10–20. [DOI] [PubMed] [Google Scholar]
- 14.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., and Czaja M. J.. 2009. Autophagy regulates lipid metabolism. Nature. 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z., and Klionsky D. J.. 2010. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuo Y., Yamashina S., Sonoue H., Arakawa A., Nakadera E., Aoyama T., Uchiyama A., Kon K., Ikejima K., and Watanabe S.. 2014. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol. Res. 44: 1026–1036. [DOI] [PubMed] [Google Scholar]
- 17.Amir M., and Czaja M. J.. 2011. Autophagy in nonalcoholic steatohepatitis. Expert Rev. Gastroenterol. Hepatol. 5: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjurjo L., Amezaga N., Aran G., Naranjo-Gomez M., Arias L., Armengol C., Borras F. E., and Sarrias M. R.. 2015. The human CD5L/AIM-CD36 axis: a novel autophagy inducer in macrophages that modulates inflammatory responses. Autophagy. 11: 487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihaylova M. M., and Shaw R. J.. 2011. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woods A., Johnstone S. R., Dickerson K., Leiper F. C., Fryer L. G. D., Neumann D., Schlattner U., Wallimann T., Carlson M., and Carling D.. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13: 2004–2008. [DOI] [PubMed] [Google Scholar]
- 21.Woods A., Dickerson K., Heath R., Hong S. P., Momcilovic M., Johnstone S. R., Carlson M., and Carling D.. 2005. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2: 21–33. [DOI] [PubMed] [Google Scholar]
- 22.Guigas B., and Viollet B.. 2016. Targeting AMPK: from ancient drugs to new small-molecule activators. EXS. 107: 327–350. [DOI] [PubMed] [Google Scholar]
- 23.Hardie D. G., Schaffer B. E., and Brunet A.. 2016. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 26: 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laker R. C., Drake J. C., Wilson R. J., Lira V. A., Lewellen B. M., Ryall K. A., Fisher C. C., Zhang M., Saucerman J. J., Goodyear L. J., et al. 2017. Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat. Commun. 8: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J., Kundu M., Viollet B., and Guan K. L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J., Kim Y. C., Fang C., Russell R. C., Kim J. H., Fan W., Liu R., Zhong Q., and Guan K. L.. 2013. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 152: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abumrad N. A., and Goldberg I. J.. 2016. CD36 actions in the heart: lipids, calcium, inflammation, repair and more? Biochim. Biophys. Acta. 1861: 1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N., Chen Y., Chen H., Hong J., Zou W., and Fang J. Y.. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 170: 548–563.e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou C., Zhong W., Zhou J., Sheng F., Fang Z., Wei Y., Chen Y., Deng X., Xia B., and Lin J.. 2012. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy. 8: 1215–1226. [DOI] [PubMed] [Google Scholar]
- 30.Knøsgaard L., Kazankov K., Birkebæk N. H., Holland-Fischer P., Lange A., Solvig J., Hørlyck A., Kristensen K., Rittig S., Vilstrup H., et al. 2016. Reduced sCD36 following weight loss corresponds to improved insulin sensitivity, dyslipidemia and liver fat in obese children. Eur. J. Clin. Nutr. 70: 1073–1077. [DOI] [PubMed] [Google Scholar]
- 31.Park H. W., Park H., Semple I. A., Jang I., Ro S. H., Kim M., Cazares V. A., Stuenkel E. L., Kim J. J., and Kim J. S.. 2014. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat. Commun. 5: 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Li P., Fu S., Calay E. S., and Hotamisligil G. S.. 2010. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueno T., and Komatsu M.. 2017. Autophagy in the liver: functions in health and disease. Nat. Rev. Gastroenterol. Hepatol. 14: 170–184. [DOI] [PubMed] [Google Scholar]
- 34.Lin C-W., Zhang H., Li M., Xiong X., Chen X., Chen X., Dong X. C., and Yin X-M.. 2013. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J. Hepatol. 58: 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha R. A., Farah B. L., Singh B. K., Siddique M. M., Li Y., Wu Y., Ilkayeva O. R., Gooding J., Ching J., and Zhou J.. 2014. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology. 59: 1366–1380. [DOI] [PubMed] [Google Scholar]
- 36.Sun L., Zhang S., Yu C., Pan Z., Liu Y., Zhao J., Wang X., Yun F., Zhao H., and Yan S.. 2015. Hydrogen sulfide reduces serum triglyceride by activating liver autophagy via the AMPK-mTOR pathway. Am. J. Physiol. Endocrinol. Metab. 309: E925–E935. [DOI] [PubMed] [Google Scholar]
- 37.Hedrick S. M., Hess Michelini R., Doedens A. L., Goldrath A. W., and Stone E. L.. 2012. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12: 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta A., Molkentin J. D., and Yutzey K. E.. 2009. FoxO transcription factors promote autophagy in cardiomyocytes. J. Biol. Chem. 284: 28319–28331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou J., Hong L., Luo C., Li Z., Zhu Y., Huang T., Zhang Y., Yuan H., Hu Y., Wen T., et al. 2016. Metformin inhibits estrogen-dependent endometrial cancer cell growth by activating the AMPK-FOXO1 signal pathway. Cancer Sci. 107: 1806–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Zhao H., Jin Q., You W., Cheng H., Liu Y., Song E., Liu G., Tan X., Zhang X., et al. 2018. Resveratrol induces apoptosis and inhibits adipogenesis by stimulating the SIRT1-AMPKalpha-FOXO1 signalling pathway in bovine intramuscular adipocytes. Mol. Cell. Biochem. 439: 213–223. [DOI] [PubMed] [Google Scholar]
- 41.Wang S., Xia P., Huang G., Zhu P., Liu J., Ye B., Du Y., and Fan Z.. 2016. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nat. Commun. 7: 11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murrow L., Malhotra R., and Debnath J.. 2015. ATG12–ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17: 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong P. M., Puente C., Ganley I. G., and Jiang X.. 2013. The ULK1 complex. Autophagy. 9: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong J., Gu H., Zhao L., Wang L., Liu P., Wang F., Xu H., and Zhao T.. 2018. Phosphorylation of ULK1 by AMPK is essential for mouse embryonic stem cell self-renewal and pluripotency. Cell Death Dis. 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martina J. A., Chen Y., Gucek M., and Puertollano R.. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S. H., Kim G., Han D. H., Lee M., Kim I., Kim B., Kim K. H., Song Y. M., Yoo J. E., Wang H. J., et al. 2017. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 13: 1767–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niso-Santano M., Malik S. A., Pietrocola F., Bravo-San Pedro J. M., Marino G., Cianfanelli V., Ben-Younes A., Troncoso R., Markaki M., Sica V., et al. 2015. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 34: 1025–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.