Although the unfolded protein response is known as a stress response in plants, it also functions in normal plant development.
Abstract
The unfolded protein response (UPR) is activated in plants in response to endoplasmic reticulum stress and plays an important role in mitigating stress damage. Multiple factors act in the UPR, including the membrane-associated transcription factor, BASIC LEUCINE ZIPPER 17 (bZIP17), and the membrane-associated RNA splicing factor, INOSITOL REQUIRING ENZYME1 (IRE1). We have analyzed an Arabidopsis (Arabidopsis thaliana) ire1a ire1b bzip17 triple mutant, with defects in stress signaling, and found that the mutant is also impaired in vegetative plant growth under conditions without externally applied stress. This raised the possibility that the UPR functions in plant development in the same manner as it does in responding to stress. bZIP17 is mobilized to the nucleus in response to stress, and through the analysis of a mobilization-defective bZIP17 mutant, we found that to support normal plant development bZIP17 must be capable of mobilization. Likewise, through the analysis of ire1 mutants defective in either protein kinase or RNase activities, we found that both must be operative to promote normal development. These findings demonstrate that the UPR, which is associated with stress responses in plants, also functions under unstressed conditions to support normal development.
The unfolded protein response (UPR) is elicited by the accumulation of misfolded proteins in the endoplasmic reticulum (ER), a condition defined as ER stress (Urano et al., 2000). In general, the UPR in plants can be induced by adverse environmental conditions or by treatment with ER stress agents, such as tunicamycin or dithiothreitol (DTT). However, ER stress can also be induced in the absence of external stressors, such as under certain physiological or developmental conditions in which the demand for protein folding exceeds the capacity of the folding machinery. For example, ER stress is induced in animals when β-lymphocytes differentiate into plasma cells and produce high levels of IgGs (Reimold et al., 2001). In plants, the UPR is provoked by the heavy demand in the anther tapetal cells to synthesize and secrete materials comprising the pollen coat (Deng et al., 2016).
In response to ER stress, the conditions in the ER are communicated to the nucleus through the UPR signaling pathway (Walter and Ron, 2011). This results in an up-regulation of genes involved in protein import, folding, export, and quality control. Signaling is mediated by signal transducers that constitute two arms of the UPR signaling pathway in plants (Howell, 2013; Bao and Howell, 2017). One arm involves membrane-associated transcription factors, such as BASIC LEUCINE ZIPPER 17 (bZIP17) and bZIP28, and the other arm involves an RNA splicing factor, INOSITOL REQUIRING ENZYME1 (IRE1). In response to ER stress, bZIP17 and/or bZIP28 are mobilized and transported to the Golgi, where they are processed by Golgi-resident proteases, which release their transcription factor domains [bZIP17(p) and/or bZIP28(p)] into the cytoplasm for further import into the nucleus. The other arm of the UPR signaling pathway involves IRE1, for which there are two isoforms in Arabidopsis (Arabidopsis thaliana), IRE1a and IRE1b (Koizumi et al., 2001). In response to ER stress, IRE1 is activated and splices bZIP60 mRNA, creating a frameshift such that the mRNA encodes a form of bZIP60(s) that is transportable to the nucleus (Deng et al., 2011). bZIP17(p), bZIP28(p), and bZIP60(s) can homodimerize or heterodimerize to activate stress response genes in the nucleus (Liu and Howell, 2010). IRE1 has another activity independent of its bZIP60 RNA splicing function, and that is to degrade other mRNAs in response to stress through a process called regulated IRE1-dependent RNA degradation (RIDD; Hollien and Weissman, 2006; Hollien et al., 2009). In Arabidopsis, RIDD primarily attacks mRNAs encoding secreted proteins that are being translated by ribosomes on the ER (Mishiba et al., 2013).
This report largely focuses on the membrane-bound transcription factor bZIP17, which functions in one branch of the UPR. Loss-of-function bzip17 mutants have no observable growth phenotype under normal conditions and have only a modest salt-sensitive root growth phenotype when grown on 150 mM NaCl (Liu et al., 2007b). The salt sensitivity of bzip17 was complemented by introduction of 35S:bZIP17 into the bzip17 mutant background. Overexpression of a constitutively active, truncated form of bZIP17 (35S:bZIP17ΔC) in a wild-type background produced seedlings that were growth inhibited, while overexpression of full-length bZIP17 (35S:bZIP17) had no effect (Liu et al., 2008). Thus, overexpression of an activated form of bZIP17 in a wild-type background results in a marked phenotype, while the loss-of-function mutation in bZIP17 has no effect under unstressed conditions and results in only mild sensitivity to the presence of salt.
Kim et al. (2018) generated multiple mutants involving bZIP17 and observed considerable growth inhibition in the double bzip17 bzip28 mutant, from which they concluded that bZIP17 plays a pivotal role in vegetative development, with functional redundancy to bZIP28. In this report, we have extended those observations by knocking out both arms of the UPR signaling pathway and demonstrating that bZIP17 has profound effects on vegetative development when it occurs in conjunction with debilitating mutations in IRE1a and IRE1b, particularly in IRE1b. These findings reveal that the UPR is active during normal plant development and that mutations that affect the UPR in response to stress also impact its role in normal development. Thus, the UPR signaling pathway must be competent to respond to stress in order to support plant development under unstressed conditions.
RESULTS
Mutant Analysis
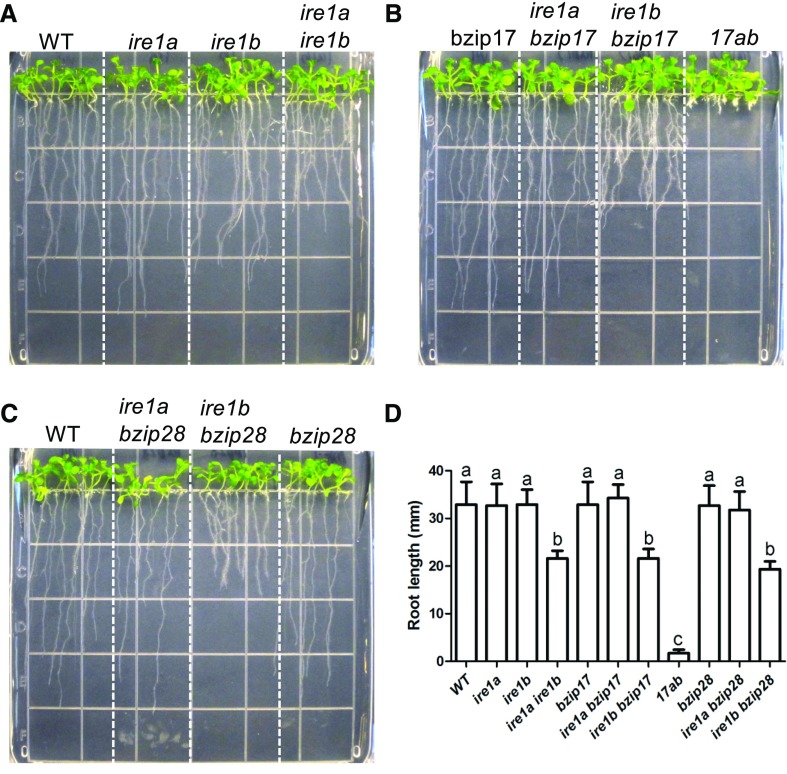
Single mutants in the RNA splicing arm of the UPR signaling pathway, such as ire1a or ire1b, or in the membrane-associated transcription factor arm involving bzip28 or bzip17, have little effect on the root growth of seedlings under unstressed conditions (Fig. 1). On the other hand, various UPR multiple mutants such as the ire1a ire1b double mutant have pronounced defects in root growth (Fig. 1, A and D; Deng et al., 2013; Chen et al., 2014). Among the double mutants, ire1b in conjunction with membrane-associated transcription factor mutants, such as bzip17 or bzip28, slowed root growth (Fig. 1, B–D), as was also shown for the double ire1b bzip28 mutant by Deng et al. (2013). Although ire1a in conjunction with bzip17 or bzip28 seemed to have little effect on root growth, bzip17 in combination with ire1b in the double bzip17 ire1b mutant clearly retarded root growth. The root growth defects in the triple ire1a ire1b bzip17 mutant, which we call 17ab, were far more severe, resulting in seedlings with short, stubby roots (Fig. 1, B and D). Even though the ire1b bzip17 and ire1b bzip28 double mutants are inhibited in primary root growth, the mutations appear to stimulate lateral root formation. Although the effects in double mutants were not as severe as in the triple mutant, the mutations are not leaky. We verified this by genotyping and showing that the genes are interrupted by T-DNA insertions (Supplemental Fig. S1), and we found in previous studies that the accumulation of full-length transcripts was undetectable in all three single mutant lines (Liu et al., 2007a, 2007b; Bao et al., 2018).
Figure 1.
Root elongation assay for UPR mutants. A to C, Various single and higher order mutants of UPR signaling pathway components (ire1a, ire1b, bzip17, and bzip28) were grown vertically on one-half-strength Murashige and Skoog (MS) medium with a 16-h-light/8-h-dark cycle for 7 d, and representative images are shown. D, Root lengths at 7 d of the different genotypes shown in A to C. The bars are means ± sd of three independent experiments. One-way ANOVA with Tukey’s method was used to examine significant differences at P < 0.01 in pairwise comparison and classified by a, b, or c. WT, Wild type.
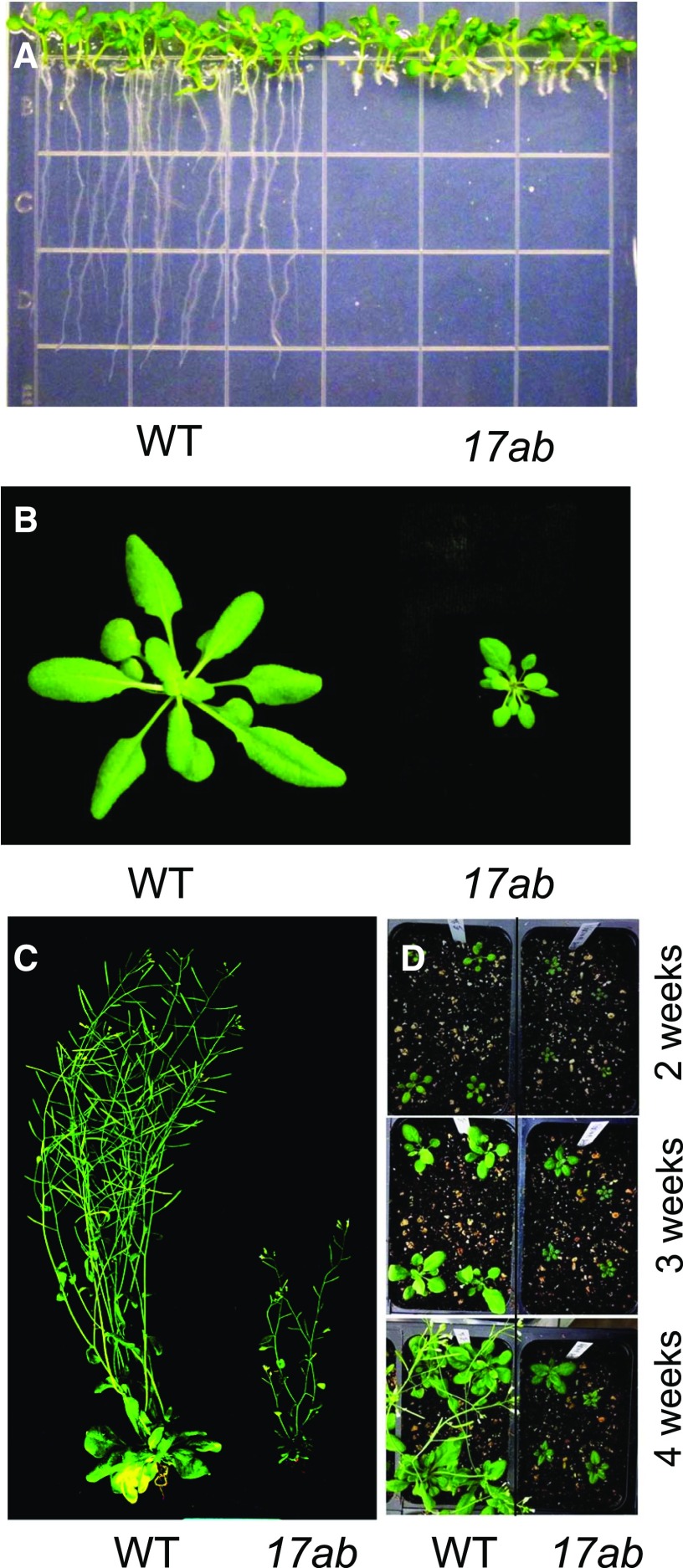
To better understand the role of bZIP17 in plant development, we focused on the triple mutant, 17ab. We were not able to analyze the triple ire1a ire1b bzip28 mutant because we have been unable to recover it, and therefore deemed it not viable. The 17ab triple mutant exhibits profound effects on shoot growth as well as root growth in unstressed seedlings and plants (Fig. 2, A, B, and D). The mutant plants are dwarf, with smaller but otherwise normal rosette leaves. The 17ab plants produce inflorescences that appear morphologically normal, but they are also stunted (Fig. 2C) and were late to flower in comparison with the wild type (Fig. 2D). Note the increased number of rosette leaves when the 17ab plants start bolting, an indicator of delayed flowering (Supplemental Fig. S2).
Figure 2.
Phenotypes of the 17ab mutant at different growth stages. A, 17ab and wild-type (WT) seedlings were grown vertically on one-half-strength MS medium with a 16-h-light/8-h-dark cycle for 7 d. B and C, 17ab and wild-type plants were grown in soil under short-day (8 h of light and 16 h of dark; B) and long-day (16 h of light and 8 h of dark; C) conditions for 45 d. Rosette images in B were digitally abstracted for comparison. D, 17ab and wild-type plants were grown in soil under long-day conditions for 2, 3, and 4 weeks, and representative images are shown.
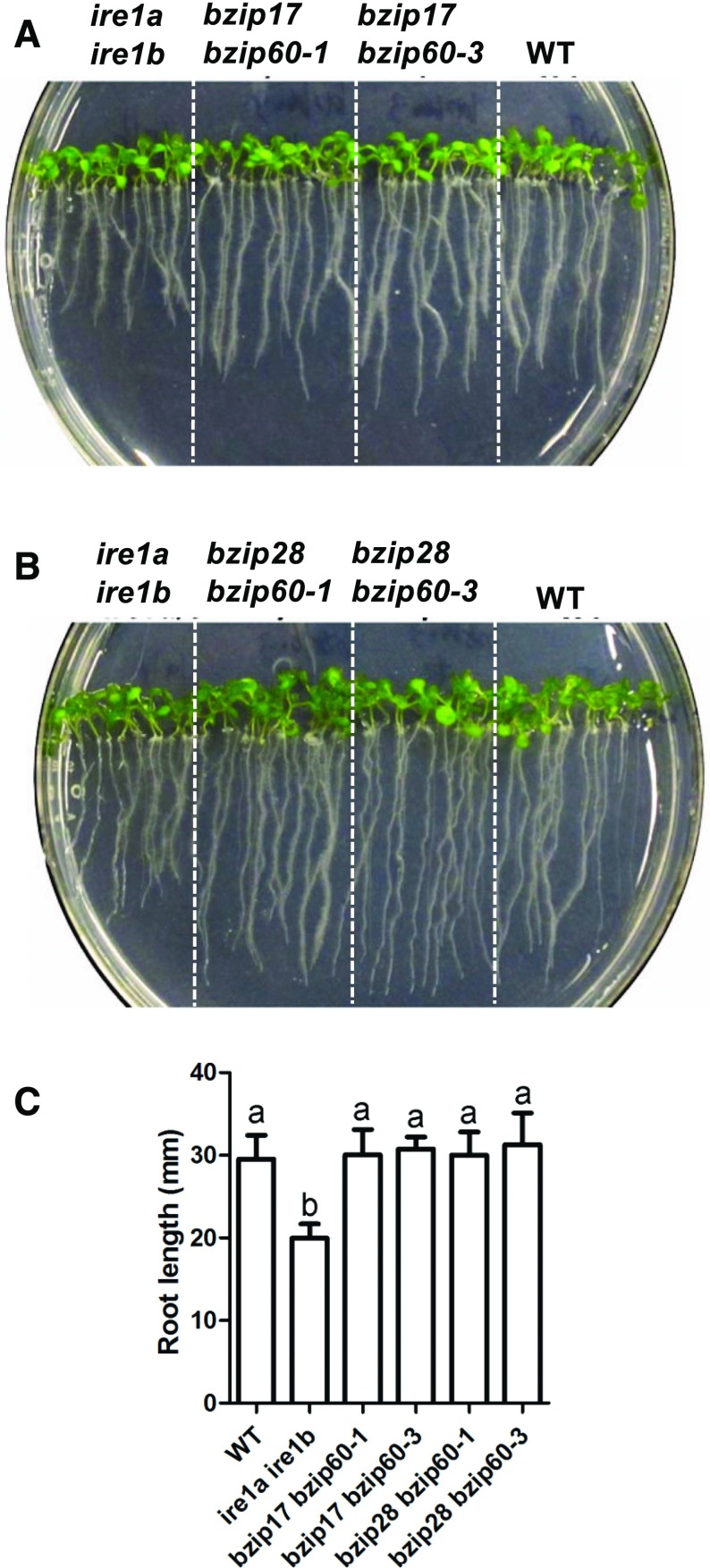
In genetic terms, bZIP60 is immediately downstream from IRE1, and its RNA transcript is the principal target of IRE1’s RNA splicing activity. Therefore, we determined whether the double bzip17 bzip60 and bzip28 bzip60 mutants had root growth inhibition phenotypes comparable to 17ab (Fig. 3, A and B). Two alleles of bZIP60 were tested, because bzip60-1 is considered to be hypomorph while bzip60-3 is a null mutant (Bao et al., 2018). Neither allele in conjunction with bzip17 or bzip28 in double mutants had any significant effect on root elongation in seedlings (Fig. 3, A and B) or on shoot phenotype (data not shown). These findings argue that IRE1a and IRE1b function in the support of root growth in ways other than the splicing of bZIP60 mRNA.
Figure 3.
Root elongation assay for bZIP60 mutants. A and B, Wild-type (WT), ire1a ire1b, bzip17 bzip60, and bzip28 bzip60 seedlings were grown vertically on one-half-strength MS medium with a 16-h-light/8-h-dark cycle for 7 d. C, Root lengths at 7 d for different genotypes shown in A and B. The data are means ± sd of three independent experiments. One-way ANOVA with Tukey’s method was used to examine significant differences at P < 0.01 in pairwise comparison and classified by a or b.
Complementation Analysis
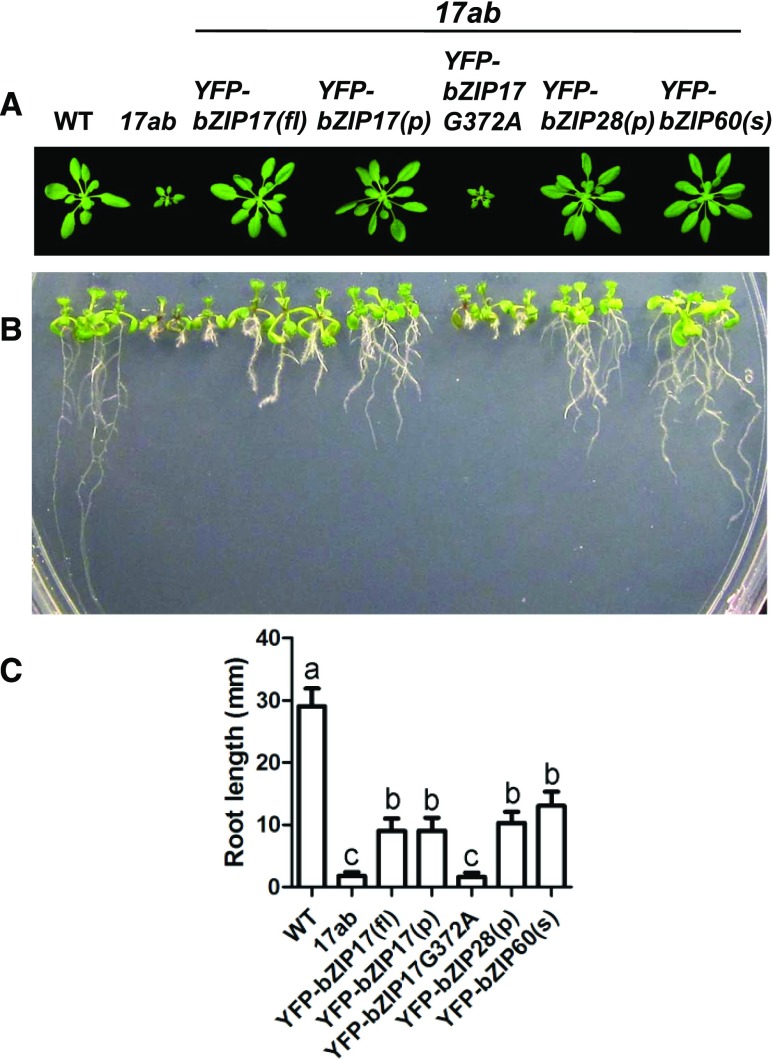
We used vegetative shoot and root growth to further assess the effects of UPR mutants on plant development (Fig. 4). To do so, we conducted complementation experiments by introducing yellow fluorescent protein (YFP)-tagged UPR transcription factors driven by the octopine synthase-containing super promoter (Li et al., 2001; Lee et al., 2007) into 17ab to determine whether they could rescue the mutant phenotype. We showed that the activated, tagged forms of these bZIP transcription factors, YFP-bZIP17(p), YFP-bZIP28(p), and YFP-bZIP60(s), are successfully targeted to the nucleus (Supplemental Fig. S3). We found that the processed form of YFP-bZIP17(p) and its unprocessed, full-length form, YFP-bZIP17(fl), rescued 17ab’s dwarf-shoot and short-root phenotypes, although the rescue only partially restored normal root growth (Fig. 4). To our surprise, we observed that the processed form of YFP-bZIP28(p) and the spliced form of YFP-bZIP60(s) also rescued shoot growth and partially restored root growth in the triple mutant. This finding indicates that, when overexpressed, YFP-bZIP17(p), YFP-bZIP28(p), and YFP-bZIP60(s) can perform similar functions in promoting shoot and root growth.
Figure 4.
Complementation of 17ab with various UPR constructs. The YFP-tagged constructs include full-length bZIP17(fl); the processed form, bZIP17(p); the bZIP17 point mutant bZIP17G372A, which prevents processing; and the processed and activated forms, bZIP28(p) and bZIP60(s), respectively. A, Wild-type (WT), 17ab, and different transgenic plants were grown under short-day conditions (8 h of light and 16 h of dark). Rosette images were digitally abstracted for comparison. B, Seedlings representing different genotypes were grown vertically on one-half-strength MS medium with a 16-h-light/8-h-dark cycle for 7 d, and a representative image is shown. C, Root lengths for the seedlings shown in A and B. The data are means ± sd of three independent experiments. One-way ANOVA with Tukey’s method was used to examine significant differences at P < 0.01 in pairwise comparison and classified by a, b, or c.
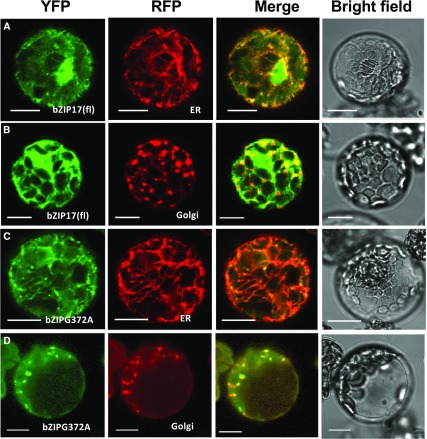
An important issue to be addressed is whether bZIP17 has to be mobilized to the nucleus to rescue 17ab’s mutant phenotype under unstressed conditions. Given the perception that bZIP17 and bZIP28 are inactive in unstressed cells, it is not clear where these factors act during normal development. To test whether bZIP17 must be mobilized and transported to the nucleus to support normal development, we attempted to rescue 17ab with a mutant form of bZIP17 that interferes with its mobilization. bZIP17 is processed in the Golgi by regulated intramembrane proteolysis involving SITE-2-PROTEASE (S2P; Che et al., 2010). S2P cleaves transmembrane domains, which are recognized as substrates for S2P proteolysis by the presence of a helix-breaking Gly residue in the middle of the transmembrane domain (Fluhrer et al., 2012). In related studies, the helix-breaking Gly in bZIP28 was replaced by an Ala to produce bZIP28G329A (Srivastava et al., 2012). The mutation allowed bZIP28 to exit the ER in response to stress but disrupted its processing, leading to its retention in the Golgi. We made a comparable mutation in bZIP17 called bZIP17G372A and tested its ability to complement 17ab. We found that YFP-bZIP17G372A was expressed at levels comparable to YFP-bZIP17(fl; Supplemental Fig. S4). However, YFP-bZIP17G372A was incapable of rescuing 17ab (Fig. 4), which we interpret to mean that bZIP17 must be mobilized to the nucleus for it to support normal shoot development.
To determine whether YFP-bZIP17G372A is transportable, we assessed its subcellular localization in protoplasts in response to stress. As controls, we demonstrated that YFP-bZIP17(fl) relocated to the nucleus in response to stress conferred by treatment with 2 mM DTT (Fig. 5, A and B). However, YFP-bZIP17G372A did not relocalize to the nucleus but appeared to accumulate in punctate structures, many of which colocalized with a Golgi marker (Fig. 5, C and D). Thus, we interpret this to mean that YFP-bZIP17G372A can be mobilized out of the ER in response to stress but that its pathway to the nucleus is blocked in the Golgi. Under unstressed conditions, we do not see evidence for the relocation of YFP-bZIP17(fl) or YFP-bZIP17G372A to the nucleus, although the resolution of the technique would not have detected the movement of a small amount of these factors (Supplemental Fig. S5).
Figure 5.
Subcellular localization of mobilization-defective bZIP17. Arabidopsis leaf protoplasts were cotransformed with YFP-bZIP17(fl) or YFP-bZIPG372A and with either RFP-tagged ER markers (A and C) or Golgi markers (B and D; Nelson et al., 2007). Protoplasts were treated overnight with 2 mM DTT to induce ER stress, and protoplasts were observed by confocal microscopy. Bars = 10 μm.
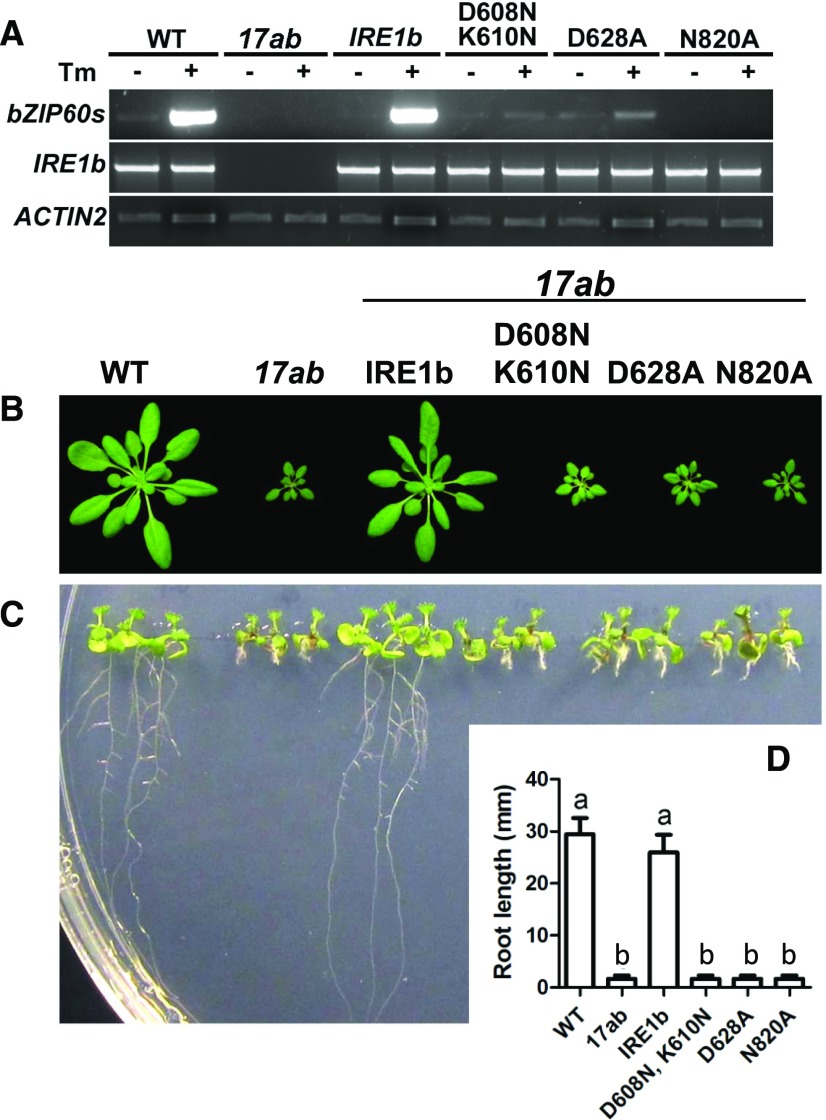
IRE1 is a dual protein kinase/RNase, so we attempted to determine which of its activities are required for normal development under nonstressed conditions by assessing the capabilities of various IRE1 constructs to rescue shoot and root growth in 17ab. To perform this analysis, we tried to complement 17ab with site-specific IRE1b mutants that inactivate various IRE1 functions (Deng et al., 2013). Both D608N K610N and D628A interfere with the protein kinase activity of IRE1, and N820A knocks out its RNase activity. All three mutations prevent the splicing of bZIP60 mRNA in response to stress (Fig. 6A). We observed that none of the mutants rescued the dwarf phenotypes of 17ab (Fig. 6, B–D), meaning that the protein kinase and RNase functions of IRE1 are needed to restore normal growth. The protein kinase and RNase activities contribute to the RNA splicing capabilities of IRE1, and yet we showed above that bZIP60, whose RNA is the splicing target for IRE1, is not required for normal development under unstressed conditions. We interpret this to mean that the RIDD activity of IRE1, and not its bZIP60 splicing activity, is a major contributor to normal development.
Figure 6.
Complementation of 17ab with various mutant forms of IRE1b. IRE1b, IRE1bD608N, K610N, IRE1bD628A (the latter two are protein kinase dead), and IRE1bN820A (RNase dead) were introduced into the 17ab mutant background by agrobacteria-mediated transformation. A, Reverse transcription (RT)-PCR analysis of bZIP60(s) (spliced form of bZIP60 mRNA) and IRE1b in the wild type (WT), 17ab, and different transgenic plants. ACTIN2 was used as a loading control. B, The wild type, 17ab, and different transgenic plants were grown under short-day conditions (8 h of light and 16 h of dark) for 45 d. Rosette images were digitally abstracted for comparison. C, Seedlings of different genotypes listed above were grown vertically on one-half-strength MS medium with a 16-h-light/8-h-dark cycle for 7 d. D, Root lengths of the different genotypes shown in B and C. Data represent means ± sd of three independent experiments. One-way ANOVA with Tukey’s method was used to examine significant differences at P < 0.01 in pairwise comparison and classified as a or b.
RNA Sequencing Analysis
RNA sequencing (RNAseq) analysis was used to diagnose the plant growth syndrome exhibited by the 17ab mutant. Genes up-regulated in the mutant compared with the wild type may represent genes with RNA transcripts that are normally degraded by the RIDD activity of IRE1 in the wild type under unstressed conditions (Supplemental Table S1; Supplemental Fig. S6). On the other hand, the genes that are down-regulated can be considered to represent the genes normally activated by bZIP17, IRE1a or IRE1b, individually or collectively, under unstressed conditions (Supplemental Table S2; Supplemental Fig. S6).
Of the 163 genes that are up-regulated by more than 2-fold in the comparison between 17ab and the wild type, 74, or 45.4%, encode proteins predicted to have signal peptides according to the SignalP server (http://www.cbs.dtu.dk/services/SignalP/). That is a considerable enrichment over the 17.6% of genes, genome wide, that encode proteins with putative signal peptides in Arabidopsis (Arabidopsis Genome Initiative, 2000). These data strongly implicate IRE1’s RIDD activity, which preferentially attacks the mRNAs of secreted proteins (Mishiba et al., 2013). In terms of Gene Ontology categories (according to the Gene Ontology term enrichment tool in TAIR; Supplemental Fig. S7), the set of up-regulated genes is enriched 2.83-fold for those that encode proteins predicted to be extracellular (P = 1.89E−11), with the most prominent molecular functions being β-glucosidase (P = 6.95E−7) and peroxidase activities (P = 1.94E−6).
Only 27 genes were down-regulated (discounting IRE1 and bZIP17) more than 2-fold, but of these, only two, or 7.7%, were predicted to have signal peptides, which is less than half of the frequency of genes genome wide in Arabidopsis that encode proteins with putative signal peptides. As for biological process, there was a 27-fold enrichment in photosynthesis genes compared with genome wide (P = 7.81E−8), and none of the 27 genes represent canonical UPR genes.
DISCUSSION
It seems logical to assume that that the UPR would not have a role in normal development when plants are not being subject to applied stress, because ER stress activates the UPR and, therefore, the UPR would be expected to be inactive under unstressed conditions. In fact, the plants in this study are being coddled, growing in the lab at room temperature under well-watered conditions, quite without applied stress. These conditions would not be expected to launch the UPR, to activate IRE1, and to mobilize the membrane-associated transcription factors bZIP17 and bZIP28. However, is it possible that even under these conditions, the UPR functions at a low level, in a minor tissue or during a brief period of development in unstressed seedlings, below the level of their usual means of detection.
In fact, there is evidence of low-level activity of the UPR under unstressed conditions in Arabidopsis. Che et al. (2010) reported that bZIP17 was mobilized, transported through the Golgi apparatus, and trafficked to the nucleus in unstressed seedlings. In an immunoblot analysis of extracts from unstressed Arabidopsis seedlings, they demonstrated the presence of three forms of bZIP17: (1) a full-length, endo H-sensitive form located in the ER; (2) an endo H-insensitive form located in the Golgi; and (3) a truncated form located in the nucleus. The endo H-sensitive form located in the ER was the most prominent form in unstressed seedlings, but in response to stress the pattern changed and the truncated form in the nucleus became more prominent. However, small amounts of the truncated and the endo H-insensitive forms were found in unstressed cells, indicating low-level transport of bZIP17 to the Golgi and on to the nucleus in unstressed seedlings. This could potentially be sufficient to activate the genes we identified by RNAseq as possible bZIP17 targets under nonstressed conditions.
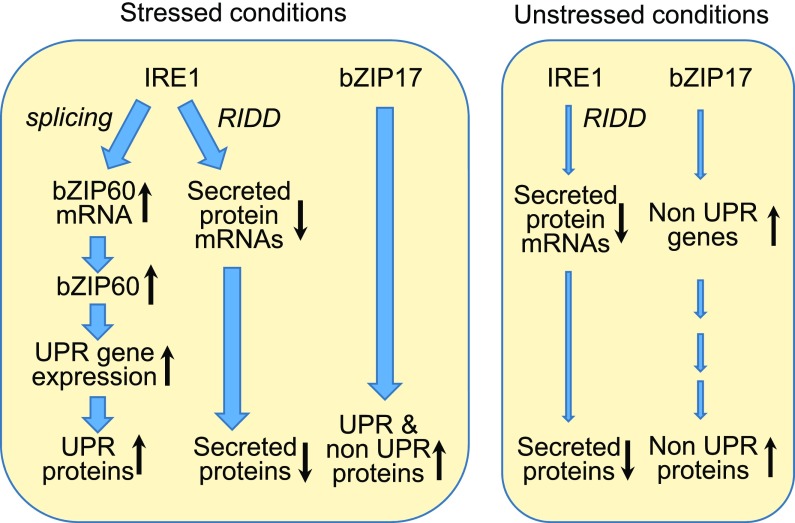
Our studies are consistent with the hypothesis that the UPR contributes to normal development in Arabidopsis by operating at low levels or in limited tissues in much the same manner as it does in responding to ER stress (Fig. 7). That conclusion is reached because components of the UPR have to be competent to function under stress conditions in order to support normal development under unstressed conditions. We show that normal development of 17ab can be restored by transgenic expression of IRE1b, but it cannot be rescued by IRE1b mutants with defects in the RNase or protein kinase domains of the protein. Likewise, the triple mutant can be rescued by transgenic expression of bZIP17. However, it cannot be rescued by a mutant of bZIP17 with a mutation in its transmembrane domain that interferes with bZIP17 processing (i.e. proteolytic cleavage by S2P). In other words, if bZIP17 cannot be processed as it is in response to stress, then it cannot rescue the growth defect in 17ab under unstressed conditions. Thus, the components of the UPR signaling pathway have to be competent to respond to ER stress (i.e. able to be activated) in order to support normal development in the absence of applied stress. A major difference between the role of the UPR in normal development and its response to stress is the apparent lack of involvement of bZIP60 in normal development. Single bZIP60 mutants or double bZIP60 mutants in combination with either bZIP17 or bZIP28 appear to be normal under our unstressed conditions and as reported by Kim et al. (2018).
Figure 7.
Proposed model for the function of IRE1 and bZIP17 under stressed and unstressed conditions. Under stressed conditions, IRE1 up-regulates the expression of the canonical UPR genes in a bZIP60-dependent manner. Under these conditions, the RIDD activity of IRE1 is also activated, leading to the degradation of the RNAs of many secreted protein genes. In addition, bZIP17 is mobilized and functions along with bZIP60 to up-regulate canonical and noncanonical UPR genes. Under unstressed conditions, the RIDD activity of IRE1 presumably leads to the degradation of unidentified mRNAs, but likely those encoding secreted protein. bZIP17 is also modestly active, leading to the production of some noncanonical UPR proteins.
An interesting sidelight is that 17ab can be rescued by transgenic expression of a processed form of bZIP28(p) and a spliced form of bZIP60(s). bZIP17 and bZIP28 are thought to have overlapping functions; however, a triple mutant involving bZIP28 is not viable, while the one involving bZIP17 is. The difference in viability could be matter of degree. According to the eFP browser (http://bar.utoronto.ca), bZIP28 (AT3G10800) is more highly expressed than bZIP17 (AT2G40950). A knockout in bZIP28, therefore, might be more severe than a knockout of bZIP17, because the levels of bZIP17 may be inadequate to support growth in a bZIP28 mutant. Alternatively, there might be some vital gene that only bZIP28 can regulate and bZIP17 cannot. Unfortunately, the reciprocal experiment of rescuing a bZIP28-containing triple mutant with bZIP17 cannot easily be performed, since the bZIP28 triple mutant is not viable. The rescue of 17ab by the spliced form of bZIP60(s) is curious, since the double bzip60 bzip17 mutant does not affect normal development. However, it should be pointed out that in a yeast two-hybrid system, bZIP17, bZIP28, and bZIP60 both homodimerize and heterodimerize with each other (Liu and Howell, 2010) and have overlapping target genes. However, the difference between the knockout mutants and the complementation experiments might be due to the difference in expression of the genes or the differences in the degree of activation of these factors in unstressed plants.
To better understand the 17ab mutant syndrome, we conducted a transcriptome analysis of the mutant under unstressed conditions. In comparison with the wild type, the up-regulated genes in the mutant are highly enriched for secreted proteins. This is consistent with the possible role of IRE1’s RIDD activity in promoting normal development under unstressed conditions. As stated above, we have shown that that IRE1’s bZIP60 mRNA splicing is not required for normal development, because the bzip17 bzip60 double mutant is normal. Although the splicing of bZIP60 mRNA is not required for normal development under unstressed conditions, the RNase activity of IRE1 is required. That was demonstrated by the inability to rescue 17ab with an RNase-inactive IRE1b. The fact that it does not rescue argues that IRE1’s RIDD function, but not its mRNA splicing activity, is required for normal development. Thus, when the IRE1 genes are knocked out in 17ab, RIDD-targeted mRNAs accumulate. RIDD targets are largely mRNAs that encode secreted proteins, many of which traffic to membranes, membranous organelles, and the cell walls. Therefore, during normal development under unstressed conditions, these mRNAs are likely held in check by the RIDD activity of IRE1, and the degradation or turnover of these RNAs is required for normal development.
This, however, does not explain the role of bZIP17 in normal development. bZIP17 is a positive-acting transcription factor that up-regulates the expression of both canonical and noncanonical UPR genes in response to stress (Liu et al., 2007b; Che et al., 2010; Kim et al., 2018). Therefore, one would expect to see the down-regulation of bZIP17 transcriptional targets in the 17ab mutant. There are not many genes substantially down-regulated in 17ab compared with the wild type under unstressed conditions. However, those that are down-regulated are not canonical UPR genes. This is similar to the finding made by Kim et al. (2018), who showed in the bzip17 bzip28 double mutants that the down-regulated differentially expressed genes (DEGs) under unstressed conditions are not the canonical UPR targets. Many of the DEGs in their study were associated with auxin regulation or cell growth, while in our study many were associated with chloroplast functions.
In other organisms, such as Caenorhabditis elegans, a functioning UPR is required for normal development (Shen et al., 2001). C. elegans has a single IRE1 gene, and a knockdown or a knockout mutant of IRE1 seems to have no effect on normal development. However, C. elegans has a homolog of the ER protein kinase called PEK. Although the single pek-1 mutant, like ire1, has no noticeable effect on development, the ire1 pek-1 double mutant arrests development at the L2 larval stage. The arrested larvae show vacuolization of intestinal cells, prompting Shen et al. (2001) to suggest that the excessive demand for protein synthesis in intestinal cells may induce endogenous ER stress. More recent studies in C. elegans indicate that IRE1 on its own, independent of X-BOX BINDING PROTEIN1 (XBP1), is required under nonstressed conditions for neural development, in particular polymodal sensory neuron arborization (Salzberg et al., 2017).
The UPR is also required for normal development in Drosophila melanogaster. One demonstration of this requirement is the fact that compromises in the XBP arm of the UPR signaling pathway contribute to retinal degeneration in D. melanogaster (Ryoo et al., 2007). In an effort to explain the role of the UPR in normal D. melanogaster development, Sone et al. (2013) developed a high-gain reporter to visualize the splicing of XBP1 mRNA under normal physiological conditions. The gain of the reporter was heightened by engineering it with a hydrophobic region to better facilitate its interaction with the XBP1 mRNA splicing machinery. With the high-gain reporter, they were able to detect XBP1 RNA splicing in the brain, gut, Malpighian tubules, larval trachea, and the male reproductive organ of D. melanogaster.
As described in the introduction, the UPR transcription factor Xbp1 in mice is required during normal development to generate plasma cells from B-lymphocytes (Reimold et al., 2001). Also, mouse lymphoid chimeras generated by injecting XBP-1−/− embryonic stem cells into mouse blastocysts developed normal spleens and lymph nodes but had low levels of all immunoglobin subtypes. In culture, Reimold et al. (2001) found that the levels of immunoglobulins in the supernatants from XBP-1-deficient B cells were much lower than in controls, even though the cells could be activated to proliferate and to undergo cell surface activation marker expression and class switch recombination. In general, ablation of IRE1α or XBP1 in animals causes a wide variety of dysfunctions, such as severe abnormalities in exocrine pancreatic acinar cells, plasma B cells, zymogenic Paneth cells, and Chief cells in the gastrointestinal tract (Hur et al., 2012).
In the other arm of the UPR signaling pathway, Activating Transcription Factor6 (ATF6) in animals, a homolog of bZIP17 and bZIP28 in plants, is a membrane-associated transcription factor. ATF6 is mobilized from the ER in response to ER stress, translocated to the Golgi, and processed to release its transcription factor component, which is subsequently imported into the nucleus (Ye et al., 2000; Shen et al., 2002; Nadanaka et al., 2006). ATF6 has also been shown to function in normal animal development and has been implicated in brain, muscle, cartilage, bone, uterine, and lens development as well as in adipogenesis (Hillary and FitzGerald, 2018).
Thus, the UPR plays important developmental roles in various organisms under conditions without the application of external stress. In some cases, the stress appears to be endogenous, such as in cells that are heavy secretors. But in other cases, there is no apparent cause for the induction of stress. The requirement for the UPR signaling factors to be competent to respond to stress in order to support normal development suggests that the UPR signaling factors are activated during normal development in a manner similar to their response to stress, except at a low level. What low-level expression actually accomplishes has yet to be determined but should be the subject of further study.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All plants used in this study are Arabidopsis (Arabidopsis thaliana) Columbia-0. Plants were grown under long days (16 h of light and 8 h of dark) or short days (8 h of light and 16 h of dark) in Percival growth chambers at 22°C (day) or 18°C (night). Mutant accessions are bzip17 (SALK_104326), bzip28-2 (SALK_132285), bzip60-1 (SALK_050203), bzip60-3 (GK-326A12; Bao et al., 2018), ire1a (SALK_018112), and ire1b (SAIL_238_F07). T-DNA insertions were confirmed by genomic PCR using the T-DNA border primer and gene-specific primers (Supplemental Table S3). ACTIN2 was used as an internal control.
Vector Construction and Plant Transformation
To generate the constructs for 17ab mutant complementation with the bZIP transcriptional factors, open reading frames with different lengths or variants were inserted into a modified pCAMBIA1300-Super-YFP vector via BglII and SalI restriction sites, using specific primers listed in Supplemental Table S1. Constructs for 17ab mutant complementation with different IRE1b variants were described before (Deng et al., 2013). All constructs were introduced into Agrobacterium tumefaciens strain GV3101 and then into Arabidopsis plants by floral dip (Clough and Bent, 1998). Transgenic plants were screened on one-half-strength MS medium (Murashige and Skoog, 1962) medium supplemented with 50 mg L−1 hygromycin or 50 mg L−1 kanamycin, and resistant transformants were selected.
Confocal Microscopy
For colocalization analysis, combinations of equal amounts (10 µg) of plasmids (1 µg µL−1) encoding YFP-bZIP17(fl) and YFP-bZIP17G372A with red fluorescent protein (RFP) tag and ER or Golgi markers (Nelson et al., 2007) were used for cotransformation. Transiently transformed Arabidopsis protoplasts were analyzed by confocal microscopy (Zeiss LSM 510 META) at excitation and emission wavelengths of 520 and 550 nm for YFP and 584 and 607 nm for mRFP.
RNA Extraction and RT-PCR
To analyze gene transcripts in the complementation lines, RT-PCR was performed with RNA samples isolated from 10-d-old seedlings grown on one-half-strength MS medium treated with or without 2 mM DTT. Primers used in this study are listed in Supplemental Table S3.
RNAseq and Data Analysis
Wild-type and 17ab seeds were germinated, and seedlings were grown vertically on 100-mm × 100-mm square plates (Fisher Scientific) on one-half-strength MS medium for 10 d. Triplicate samples were immediately ground, and total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen; catalog no. 74904). RNAseq libraries were prepared and subjected to paired end sequencing with read length of 250 bp. Sequences were aligned to the TAIR10 genome, and DEGs were identified using NOISeq. NOISeq is an R package divided into three blocks: (1) count data quality control; (2) filtering of low-count features, normalization, and batch effect correction; and (3) DEG analysis (Tarazona et al., 2015). The method differs from other RNAseq analysis methods in that it is data adaptive and nonparametric. The method does not have a strong dependency on sequencing depth for differential expression calls, and because of that the method reduces the incidents of false positives as the number of reads grows (Tarazona et al., 2011). We applied the method to our data using the criterion of fold change ≥ 2 and a divergence probability ≥ 0.8, in which a divergence probability for identical distributions is 0.
Statistical Analysis
One-way ANOVA with Tukey’s method was used to verify significant differences between genotypes in root growth assays.
Data Availability
Raw RNAseq data and the relevant processed data for RNAseq analysis were deposited in the National Center for Biotechnology Information Gene Expression Omnibus with accession number GSE120672. Accession numbers of each gene are listed in Supplemental Table S1, and the relevant gene sequences and detailed information for each gene can be found at TAIR (www.arabidopsis.org).
Accession Numbers
Sequence data for the genes described in this article can be found in the TAIR database (https://www.arabidopsis.org/index.jsp) under the following accession numbers: bZIP17 (AT2G40950), bZIP28 (AT3G10800), bZIP60 (AT1G42990), IRE1a (AT2G17520), and IRE1b (AT5G24360).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotype analysis of the 17ab triple mutant.
Supplemental Figure S2. Rosette leaf number at the time of bolting.
Supplemental Figure S3. Activated forms of the bZIP transcription factors target the nucleus.
Supplemental Figure S4. Expression of the introduced bZIP17 transgenes.
Supplemental Figure S5. Subcellular localization of bZIP17 and bZIP17G372A without stress treatment.
Supplemental Figure S6. RT-PCR analysis on selected genes to validate the RNAseq results.
Supplemental Figure S7. Gene Ontology in the cell components category for genes up-regulated in the comparison between 17ab and the wild type.
Supplemental Table S1. Genes up-regulated greater than 4-fold in the 17ab mutant.
Supplemental Table S2. Genes down-regulated greater than 2-fold in the 17ab mutant.
Supplemental Table S3. Primers used in this study.
Footnotes
This work was supported by the National Science Foundation (IOS 1353867 to D.C.B. and S.H.H.).
Articles can be viewed without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bao Y, Howell SH (2017) The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front Plant Sci 8: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Pu Y, Yu X, Gregory BD, Srivastava R, Howell SH, Bassham DC (2018) IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy 14: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Bussell JD, Zhou W, Estavillo GM, Pogson BJ, Smith SM (2010) Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci Signal 3: ra69. [DOI] [PubMed] [Google Scholar]
- Chen Y, Aung K, Rolčík J, Walicki K, Friml J, Brandizzi F (2014) Inter-regulation of the unfolded protein response and auxin signaling. Plant J 77: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA 110: 19633–19638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Quilichini TD, Dong H, Bao Y, Horner HT, Howell SH (2016) IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J 88: 193–204 [DOI] [PubMed] [Google Scholar]
- Fluhrer R, Martin L, Klier B, Haug-Kröper M, Grammer G, Nuscher B, Haass C (2012) The α-helical content of the transmembrane domain of the British dementia protein-2 (Bri2) determines its processing by signal peptide peptidase-like 2b (SPPL2b). J Biol Chem 287: 5156–5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary RF, FitzGerald U (2018) A lifetime of stress: ATF6 in development and homeostasis. J Biomed Sci 25: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107 [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477–499 [DOI] [PubMed] [Google Scholar]
- Hur KY, So JS, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Iwawaki T, Glimcher LH, Lee AH (2012) IRE1α activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med 209: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Yamaguchi-Shinozaki K, Shinozaki K (2018) ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol 176: 2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127: 949–962 [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Kononov ME, Bassuner B, Frame BR, Wang K, Gelvin SB (2007) Novel plant transformation vectors containing the superpromoter. Plant Physiol 145: 1294–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gong ZZ, Koiwa H, Niu XM, Espartero J, Zhu XP, Veronese P, Ruggiero B, Bressan RA, Weller SC, et al. (2001) Bar-expressing peppermint (Mentha × piperita L. var. Black Mitcham) plants are highly resistant to the glufosinate herbicide Liberty. Mol Breed 8: 109–118 [Google Scholar]
- Liu JX, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007a) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007b) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Howell SH (2008) Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ 31: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Mishiba K, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y, Koizumi N (2013) Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA 110: 5713–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nadanaka S, Yoshida H, Sato R, Mori K (2006) Analysis of ATF6 activation in Site-2 protease-deficient Chinese hamster ovary cells. Cell Struct Funct 31: 109–116 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature 412: 300–307 [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Domingos PM, Kang MJ, Steller H (2007) Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J 26: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg Y, Coleman AJ, Celestrin K, Cohen-Berkman M, Biederer T, Henis-Korenblit S, Bülow HE (2017) Reduced insulin/insulin-like growth factor receptor signaling mitigates defective dendrite morphogenesis in mutants of the ER stress sensor IRE-1. PLoS Genet 13: e1006579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3: 99–111 [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107: 893–903 [DOI] [PubMed] [Google Scholar]
- Sone M, Zeng X, Larese J, Ryoo HD (2013) A modified UPR stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell Stress Chaperones 18: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Chen Y, Deng Y, Brandizzi F, Howell SH (2012) Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J 70: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: A matter of depth. Genome Res 21: 2213–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Furió-Tarí P, Turrà D, Pietro AD, Nueda MJ, Ferrer A, Conesa A (2015) Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res 43: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D (2011) The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6: 1355–1364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw RNAseq data and the relevant processed data for RNAseq analysis were deposited in the National Center for Biotechnology Information Gene Expression Omnibus with accession number GSE120672. Accession numbers of each gene are listed in Supplemental Table S1, and the relevant gene sequences and detailed information for each gene can be found at TAIR (www.arabidopsis.org).