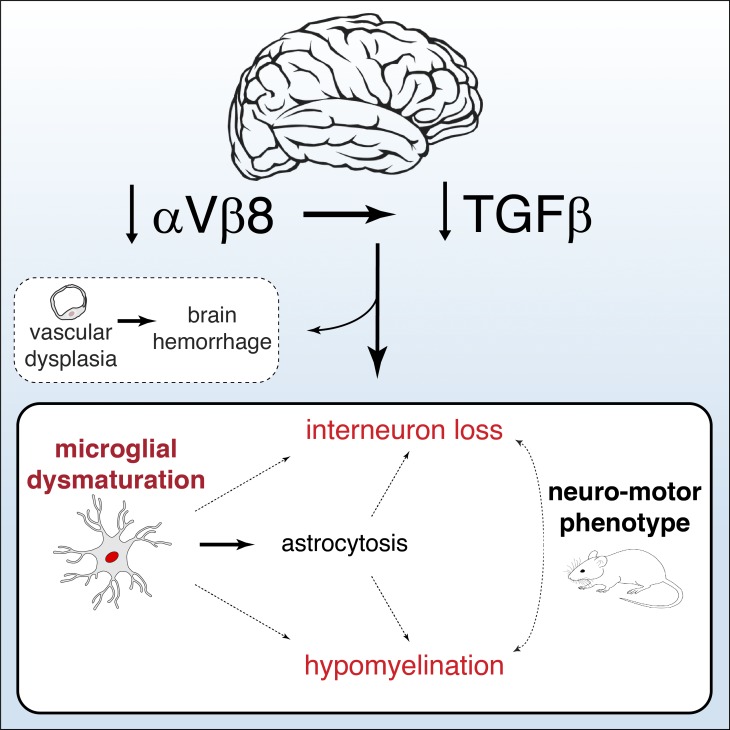
Arnold et al. demonstrate critical roles for both integrin αVβ8 and TGFβ in the maturation of microglia and characterize the effects that microglia deficient in TGFβ signaling have on brain development.
Abstract
Microglia play a pivotal role in the coordination of brain development and have emerged as a critical determinant in the progression of neurodegenerative diseases; however, the role of microglia in the onset and progression of neurodevelopmental disorders is less clear. Here we show that conditional deletion of αVβ8 from the central nervous system (Itgb8ΔCNS mice) blocks microglia in their normal stepwise development from immature precursors to mature microglia. These “dysmature” microglia appear to result from reduced TGFβ signaling during a critical perinatal window, are distinct from microglia with induced reduction in TGFβ signaling during adulthood, and directly cause a unique neurodevelopmental syndrome characterized by oligodendrocyte maturational arrest, interneuron loss, and spastic neuromotor dysfunction. Consistent with this, early (but not late) microglia depletion completely reverses this phenotype. Together, these data identify novel roles for αVβ8 and TGFβ signaling in coordinating microgliogenesis with brain development and implicate abnormally programmed microglia or their products in human neurodevelopmental disorders that share this neuropathology.
Graphical Abstract
Introduction
Microglia derive from common yolk sac myeloid precursors, then expand and differentiate in the central nervous system (CNS) during the early postnatal period in mice (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015). Early microglial differentiation occurs simultaneously with neuronal and macroglial (astrocyte, oligodendrocyte) differentiation (Matcovitch-Natan et al., 2016), and microglia-deficient mice have disrupted neural and glial development (Cunningham et al., 2013; Shigemoto-Mogami et al., 2014; Squarzoni et al., 2014; Hagemeyer et al., 2017; Wlodarczyk et al., 2017), suggesting tight developmental coordination.
TGFβ is a multifunctional cytokine important for development and functioning of many cell types in different organs and with broad activities including modulation of cell survival, differentiation, apoptosis, and cellular activation. Due to its otherwise promiscuous nature, TGFβ signaling needs to be directed with temporal and spatial precision. This is accomplished in large part by integrin-mediated activation of TGFβ, which is normally sequestered in the extracellular matrix in a latent form. For example, αVβ6 and αVβ8 on skin keratinocytes activate TGFβ, which signals to Langerhans cells to maintain their epithelial residence (Mohammed et al., 2016), and αVβ8 on dendritic cells activates TGFβ, which induces Th17 T cell differentiation (Travis et al., 2007). In the CNS, αVβ8 on neuroepithelial cells activates TGFβ, which signals to vascular endothelium and is required for embryonic cerebrovascular morphogenesis (Arnold et al., 2014). Despite the known roles for TGFβ in neural (Brionne et al., 2003; Yi et al., 2010; He et al., 2014) and glial development (Palazuelos et al., 2014; Stipursky et al., 2014) and reports identifying potential roles for TGFβ in microglial differentiation and/or homeostasis (Brionne et al., 2003; Butovsky et al., 2014; Bohlen et al., 2017), the mechanisms controlling TGFβ activation and signaling to microglia are unknown.
Here, we present evidence that integrin αVβ8 (expressed on neuroepithelial lineage cells) regulates TGFβ signaling to microglia. In the absence of this signaling, microglia are developmentally arrested and persistently activated. The presence of these “dysmature” microglia (and not just the absence of mature microglia) during a critical postnatal window is necessary and sufficient to disrupt oligodendrocyte development, cause interneuron loss, and lead to severe neuromotor dysfunction. These data therefore identify an important mechanism by which the CNS microenvironment coordinates microglial differentiation with the development of neurons and other glial cells and detail the downstream neurodevelopmental consequences that occur when microglia are developmentally arrested and activated due to reduced αVβ8 signaling in the brain or reduced TGFβ signaling in microglia themselves.
Results
αVβ8 and TGFβ signaling to microglia
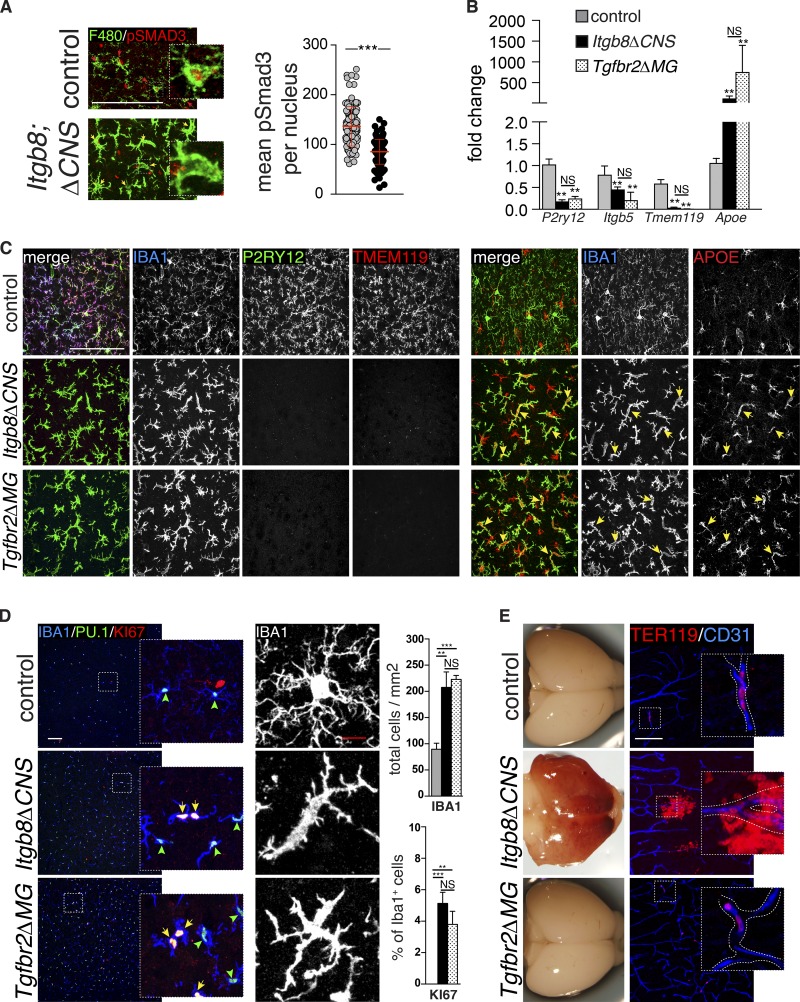
We previously documented a reduction in active TGFβ in the brains of Itgb8ΔCNS mice, a finding consistent with the known role of αVβ8 in activation of latent TGFβ1 (Arnold et al., 2014). In theory, any or all CNS cell types, including neural and macroglial lineages, vascular cells, and microglia, could be affected by reduced levels of activated TGFβ in these mice. To directly assess whether TGFβ signaling is affected in microglia from Itgb8ΔCNS mice, we immunostained brain sections for phospho-SMAD3 (pSMAD3; the major downstream transcription factor activated by TGFβ signaling) and cell type–specific markers for myeloid cells (F4/80), neurons (NEUN), astrocytes (SOX9), or oligodendrocytes (OLIG2; Fig. 1 A and Fig. S1). Fluorescent intensity mapping (Arnold et al., 2014) revealed high levels of pSMAD3 in F4/80+ microglia from control mice and reduced pSMAD3 staining intensity in microglia from adult Itgb8ΔCNS mice (Fig. 1 A). pSMAD3 staining intensity was also relatively high in neurons, astrocytes, and oligodendrocytes, but not significantly reduced in these cells in Itgb8ΔCNS mice (Fig. S1), suggesting alternative or compensating mechanisms to maintain canonical TGFβ signaling in these cells. TGFβ1 signaling influences the expression of several genes in microglia, including P2ry12, Tmem119, and ApoE (Butovsky et al., 2014). Differential gene expression in adult microglia sorted from Itgb8ΔCNS mice was nearly identical to that reported in Tgfb1 knockout mice (Butovsky et al., 2014; Fig. 1 B), consistent with reduced TGFβ signaling. This was confirmed at the protein level by immunostaining (Fig. 1 C): APOE (normally expressed on astrocytes) was highly up-regulated in microglia of Itgb8ΔCNS mice compared with controls, while P2RY12 and TMEM119 were both significantly down-regulated, and apparently absent, in adult Itgb8ΔCNS mutant mice. To further characterize microglia in Itgb8ΔCNS mice, we immunostained brain sections and performed flow cytometry using markers of myeloid cell proliferation and activation. This revealed a profound increase in the density and proliferative state of IBA1+ PU.1+ cells (Fig. 1 D) and strong up-regulation of activation markers, MHCII, lysosomal-associated protein, CD68, and F4/80 in Itgb8ΔCNS mutants compared with age-matched controls (Fig. S2). Consistent with an activated state, microglia in Itgb8ΔCNS mice took on a fusiform morphology with retracted dendrites (Fig. 1 D). Flow cytometry showed that most brain myeloid cells expressed the resident microglial marker, CD39, with minimal contribution from LY6C+/CCR2+ infiltrating monocytes (Fig. S2), indicating that loss of αVβ8 led to these cellular changes in endogenous microglia. Consistent with this, bone marrow transplant (BMT) experiments revealed negligible infiltration of bone marrow–derived GFP-labeled myeloid cells from the circulation into the CNS of mutant mice (Fig. S3).
Figure 1.
αVβ8 and TGFβ signaling to microglia. (A) Immunostaining for pSMAD3 (red) in microglia (F4/80, green) reveals reduced pSMAD3 staining intensity (yellow arrows) in most microglia from Itgb8ΔCNS mice compared with intense staining (red arrows) in controls; quantified on right: pSMAD3 staining intensity within individual microglia (arbitrary units) documents reduced microglia-specific pSMAD3. See Fig. S1 for pSMAD staining intensity in other CNS cell types. (B) Transcriptional profiling (quantitative PCR) of sorted microglia documents alterations in several TGFβ-dependent genes, confirmed by immunostaining (C). See Fig. S2 for sorting strategy. Note expression of APOE in IBA1-negative astrocytes. (D) Increased number and proliferation (KI67+) and activated morphology in IBA1+PU.1+ microglia from Itgb8ΔCNS and Tgfbr2ΔMG mice; quantified on right (we did not observe KI67+IBA1+ cells in the cortex of control animals at this time point). (E) Developmental brain hemorrhage in Itgb8ΔCNS but not Tgfbr2ΔMG mice, as evidenced by gross examination and immunostaining for red blood cells (TER119, red) outside of blood vessels (CD31, blue). Bars, 50 µm (A); 100 µm (C–E). Error bars indicate SE. **P < 0.005; ***P < 0.0005. Student’s t test (A) or ANOVA with Tukey’s post hoc test (B and D). n = 4 animals (P60) for all groups.
The above results suggest that reduced TGFβ-SMAD3 signaling in microglia from Itgb8ΔCNS mutant mice causes the observed microglial phenotype: activation, reduced expression of canonical microglia markers, and increased expression of APOE. However, because Itgb8ΔCNS mice develop severe perinatal brain hemorrhage, a consequence of reduced TGFβ signaling to endothelial cells (Fig. 1 E; Arnold et al., 2014), we thought it possible that the microglial phenotype could be secondary to hemorrhage and downstream inflammation. To directly evaluate the autonomous effects of reduced TGFβ signaling in microglia, we generated mice with myeloid-specific conditional deletion of the primary TGFβ receptor, Tgfbr2, using Cx3cr1 (Cx3c chemokine receptor 1) Cre mice, hereafter termed Tgfbr2ΔMG mice (see Materials and methods). We found that microglia from Tgfbr2ΔMG are nearly identical in morphology and gene expression compared with microglia in Tgfb1 (Butovsky et al., 2014) and Itgb8ΔCNS mutant mice (Fig. 1, B–D; and Fig. S2). Recombination analysis demonstrated efficient and specific recombination of microglia in Tgfbr2ΔMG mutants and Cre-positive controls (not shown). Importantly, these microglial changes occurred in the absence of the developmental vasculopathy and brain hemorrhage observed in Itgb8ΔCNS and Tgfb1 mutants (Fig. 1 E; Arnold et al., 2014). Together, these data suggest that integrin αVβ8 directly or indirectly influences TGFβ-SMAD3 signaling to microglia, which is, in turn, required by these cells to suppress activation and promote or maintain the expression of canonical microglial markers. The lack of brain hemorrhage in Tgfbr2ΔMG mutants compared with Itgb8ΔCNS and Tgfb1 mutant mice further suggests that the microglial phenotype observed in these mice is due to reduced TGFβ signaling in microglia and is not secondary to hemorrhage.
Brain abnormalities underlie spastic motor deficits in mice with deficient αVβ8 or TGFβ signaling to microglia
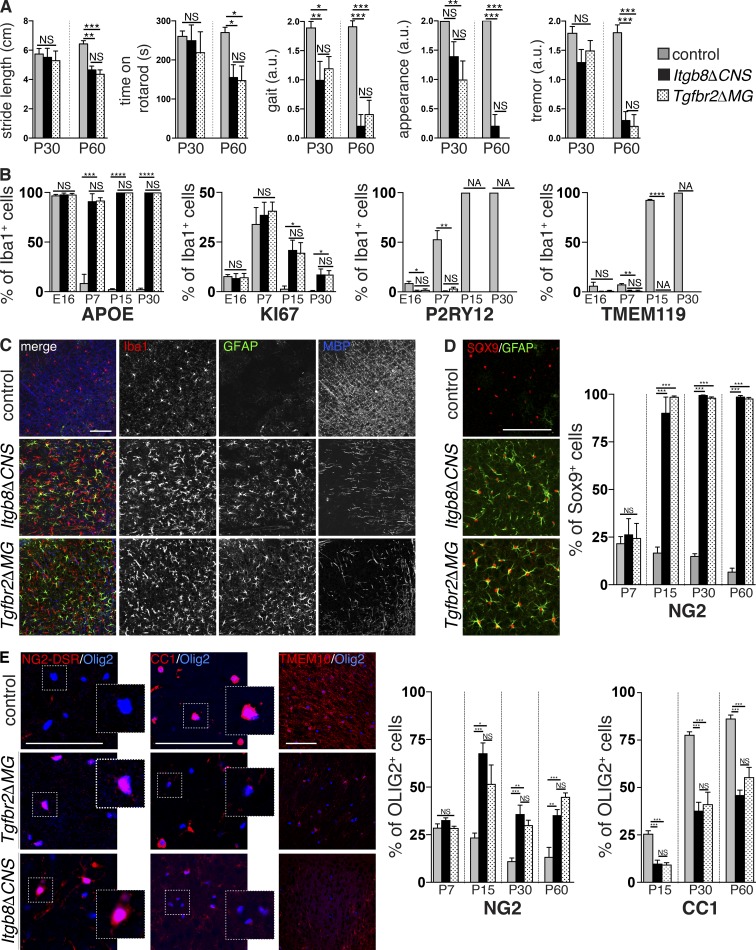
We and others previously reported that Itgb8ΔCNS mice develop a severe neuromotor syndrome characterized by spasticity, motor deficits, gait disturbance, and seizure-like activity, ultimately resulting in premature death (Fig. 2 A and Videos 1, 2, and 3; Proctor et al., 2005; Mobley et al., 2009). Because these mice develop severe perinatal brain hemorrhage (Fig. 1 E; Arnold et al., 2014), we thought these neurological symptoms could either be secondary to hemorrhage and associated tissue damage and inflammation or directly due to microglial abnormalities. To address this, we compared the neurobehavioral phenotypes in Itgb8ΔCNS mice (with brain hemorrhage) with Tgfbr2ΔMG mice (no brain hemorrhage). Both mutant mice developed spasticity and gate disturbance at postnatal day (P) 30 and pronounced upper motor neuron abnormalities by P60 (Fig. 2 A). Therefore, Tgfbr2ΔMG mutant mice virtually phenocopied Itgb8ΔCNS mice, despite having no evidence of hemorrhage, suggesting that spastic motor deficits in these mice can entirely be explained by reduced TGFβ signaling in microglia.
Figure 2.
Motor deficits and glial abnormalities in mice with deficient αVβ8 or TGFβ signaling to microglia. (A) Identical neuromotor symptoms in adult (P30–60) Itgb8ΔCNS and Tgfbr2ΔMG mice including waddling unstable gait with shortened stride length, reduced time on rotarod, unkempt appearance, and tremors. See also Videos 1 and 2. (B) Persistent expression of APOE and KI67 and reduced expression of P2RY12 and TMEM119 over time in microglia from Itgb8ΔCNS and Tgfbr2ΔMG mice. See accompanying Fig. S4. (C) Overlapping astrogliosis, microgliosis, and reduced MBP staining in P30 Itgb8ΔCNS and Tgfbr2ΔMG mice compared with controls. (D) Increased percentage of GFAP+SOX9+ astrocytes in Itgb8ΔCNS and Tgfbr2ΔMG mice over time; quantified on right. See accompanying Fig. S4. (E) Increased percentage of OLIG2+NG2-DSR+ OPC, and reduced mature OLIG2+CC1+ oligodendrocytes over time (quantified on right) and reduced staining for mature myelin marker TMEM10 in Itgb8ΔCNS and Tgfbr2ΔMG mice compared with controls. See accompanying Fig. S4. Bars, 100 µm. Error bars indicate SE. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001. Student’s t test. n = 4 animals for all groups. Behavioral analysis: ANOVA with Tukey’s post hoc test; n = 6 animals for all groups. a.u., arbitrary units.
We next evaluated a time course of pathological changes in the brain associated with these neurological impairments in both Itgb8ΔCNS and Tgfbr2ΔMG mice. We first assessed the expression of microglia markers that were dysregulated in adult Tgfbr2ΔMG mice, assuming that major changes would be evident in the cells from which we deleted Tgfbr2. Indeed, microglia in both Itgb8ΔCNS and Tgfbr2ΔMG mice were significantly affected early in postnatal development, with down-regulation of P2RY12 and TMEM119 by P7 and up-regulation of APOE and KI67 by P15 (Figs. 2 B and Fig. S4). Subsequently, we observed signs of astrocyte activation (increased expression of glial fibrillary acidic protein [GFAP] in SOX9 [SRY-box 9]+ astrocytes) and reduced myelin basic protein (MBP) staining (a marker of myelinating oligodendrocytes), overlapping areas with abnormal (fusiform) microglia (Fig. 2, C and D; and Fig. S4). These data suggest that reduced αVβ8 or TGFβ signaling to microglia leads to abnormalities in these cells, followed by downstream activation of astrocytes and impairments in MBP expression.
In humans and animal models, developmental brain injury/inflammation is accompanied by astrocyte and microglial activation, and hypomyelination that is a consequence of oligodendrocyte maturational arrest—an increase in the number of oligodendrocyte progenitors (OPCs) that fail to terminally differentiate into mature myelinating oligodendrocytes (Fancy et al., 2011; Favrais et al., 2011; Buser et al., 2012; Scafidi et al., 2014). Itgb8ΔCNS and Tgfbr2ΔMG mutants displayed more than a twofold increase in the percentage of OLIG2 (oligodendrocyte lineage transcription factor 2)+ NG2 (neuron-glial antigen 2)+ OPCs in subcortical white matter (Fig. 2 E) and strong reductions in markers of mature oligodendrocytes (OLIG2+CC1+cells) and myelinating fibers (MBP, TMEM10 [transmembrane protein 10]), which appeared disorganized (Fig. 2 E). This delay in oligodendrocyte development was apparent by P15 and persisted into adulthood (P60; Fig. 2 E and Fig. S4), coincident with astrocyte activation (Fig. 2 D and Fig. S4) and preceded by microglial changes (Fig. 2 B and Fig. S4).
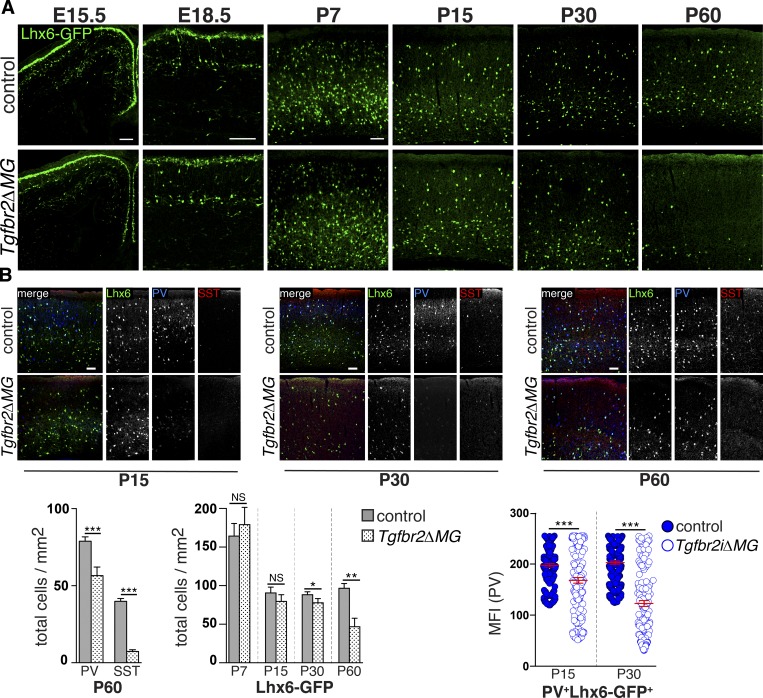
Gray matter abnormalities, including loss of cortical GABAergic interneurons, are observed in patients with white matter injury, hypomyelination, and seizures (Robinson et al., 2006), and a recent study in an animal model of white matter injury (neonatal hypoxia) suggested that OPC maturational arrest may be mediated in part by reduced GABAergic signaling secondary to loss of interneurons (Zonouzi et al., 2015). Based on these reports, we examined whether myeloid-specific deletion of Tgfbr2 might affect cortical interneuron development. We found that the densities of cortical SST (somatostatin)+ and PV (parvalbumin)+ interneurons were significantly reduced in adult (P60) Tgfbr2ΔMG mutants (Fig. 3). PV and SST expression is activity and maturation dependent (Komitova et al., 2013). Therefore, the observed reduction in PV and SST cells could indicate loss of interneurons or a down-regulation of marker expression. To distinguish between these possibilities, we used an Lhx6 (LIM homeobox protein 6)-GFP reporter line that constitutively marks most median ganglionic eminence–derived GABAergic interneurons (Cobos et al., 2006). Comparing Tgfbr2ΔMG mutants with controls, we found no appreciable difference in the numbers or distribution of Lhx6-GFP+ interneurons in the cortex at embryonic day (E) 15.5, E18.5, P7, P15, or P30 (Fig. 3 A), suggesting there are no major defects in the production, tangential migration, or late gestational laminar positioning of these cells due to conditional deletion of Tgfbr2 in microglia. However, cortical Lhx6-GFP+ interneurons in Tgfbr2ΔMG mutants had reduced PV staining intensity at P15 and P30, and there was a subsequent reduction in the number Lhx6-GFP+ cells by P60 (Fig. 3), possibly indicating a block in the functional maturation of GABAergic interneurons before the loss of these cells. Taken together, our results suggest that microglial abnormalities due to loss of TGFβ signaling lead to early postnatal astrocyte activation, oligodendrocyte maturational arrest and delayed differentiation with subsequent loss of cortical GABAergic interneurons. A similar cellular phenotype was observed in Itgb8ΔCNS mice (Fig. S4). Notably, all observed cellular changes occurred before or during the early progression of the behavioral and motor symptoms, suggesting a causal relationship.
Figure 3.
Interneuron abnormalities in mice with deficient αVβ8 or TGFβ signaling to microglia. (A) Delayed loss of Lhx6-GFP+ cortical interneurons in Tgfbr2ΔMG; quantification below. (B) Reduced PV expression in Lhx6-GFP+ interneurons at P15 and P30, before the reduction in the numbers of these cells; quantification below. See also Fig. S4. Bars, 100 µm. Error bars indicate SE. *P < 0.05; **P < 0.005; ***P < 0.0005. Student’s t test. n = 4 animals for all groups. MFI, mean fluorescence intensity.
Presence of TGFβ signaling–deficient microglia (and not absence of mature microglia) drives neuromotor phenotype
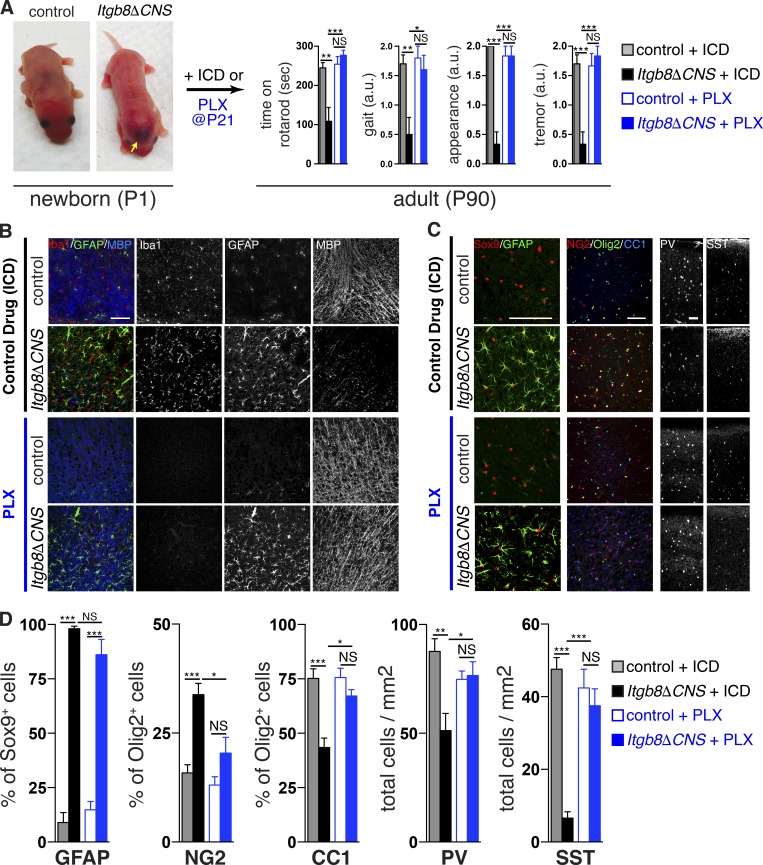
The developmental neurobehavioral phenotype in Itgb8ΔCNS and Tgfbr2ΔMG mice is associated with both the absence of mature homeostatic microglia and with the presence of abnormal microglia, leading us to question which of these might cause the observed neuropathological changes. We posited that postnatal depletion of microglia in Itgb8ΔCNS mice could differentiate these two possibilities: if lack of homeostatic microglia during a critical window leads to the phenotype (loss of function), then depletion would have no effect on the phenotype, whereas if the phenotype is due to the presence of abnormal microglia (gain of function), then depletion of these cells would be protective. To test this, whole litters of mice containing at least one Itgb8ΔCNS mouse with brain hemorrhage (visualized through the skin of newborn pups) were randomized to receive either PLX5622 chow (a potent and selective CSF1R inhibitor known to rapidly deplete microglia) or inactive control drug (ICD) chow, upon weaning (P21; Fig. 4 A). As previously demonstrated (Acharya et al., 2016), PLX5622 administration reduced microglia numbers, resulting in nearly complete depletion by P45 (not shown) and beyond P60 (Fig. 4 B). Remarkably, microglia depletion protected Itgb8ΔCNS mice from developing the behavioral abnormalities seen in mice treated with control chow (Fig. 4 A). Mice treated with PLX5622 also had apparently normal oligodendrocyte development (MBP staining; OPC and mature oligodendrocyte numbers) and interneuron numbers (PV and SST staining) compared with untreated control and Itgb8ΔCNS mice (Fig. 4, B–D). Control (Itgb8flox/+;NestinCre) mice treated with PLX5622 had comparatively normal oligodendrocyte development and interneuron numbers and performed similarly in behavioral tests to mice given control diet, suggesting that PLX5622 has no independent effects on these glial or neuronal populations, as has been previously shown (Elmore et al., 2014) and consistent with the absence of major neuromotor disturbance in Csf1r−/− (Erblich et al., 2011; Nandi et al., 2012) and Pu.1−/− (Beers et al., 2006) mice, which lack all CNS myeloid cells. Interestingly, astrocytosis (indicated by increased percentage of GFAP+ Sox9+ astrocytes) in Itgb8ΔCNS mice was persistent despite microglial depletion (Fig. 4, B–D). Late treatment of Itgb8ΔCNS mice with PLX5622 after P60 had no observable effect on the neuromotor symptoms (not shown). Together, these data indicate that (1) the presence of abnormal microglial (and not the absence of mature microglia) in Itgb8ΔCNS mice is necessary and sufficient to drive oligodendrocyte abnormalities, interneuron loss, and associated neuromotor abnormalities; (2) brain hemorrhage in these mice has no obvious independent effects on the development of these abnormalities; and (3) there is a critical time window during which depletion of abnormal microglia can protect from or reverse the neuromotor phenotype.
Figure 4.
Presence of abnormal microglia (and not absence of mature microglia) drives neuromotor phenotype in Itgb8ΔCNS mice. (A–D) Itgb8ΔCNS pups and controls were identified at birth, randomized to receive either ICD or PLX5622, then evaluated for neuromotor symptoms (A) and associated neuropathology (B–D) 90 d later (P90). Compared with Itgb8ΔCNS mice treated with control drug and littermate control mice treated with either ICD or PLX5622, Itgb8ΔCNS mice treated with PLX5622 had normalization of oligodendrocyte abnormalities (B, MBP staining; C, Olig2/NG2/CC2 staining) and interneuron deficiencies (C, PV/SST staining), but persistence of reactive astrocytes (C, Sox9/GFAP staining). Note overlapping staining of GFAP and MBP in B, which shows normalization of MBP staining despite persistent GFAP staining in Itgb8ΔCNS mice treated with PLX5622. (D) Quantification of cellular phenotypes in Itgb8ΔCNS and control mice treated with either ICD or PLX5622. Bars, 100 µm. Error bars indicate SE. *P < 0.05; **P < 0.005; ***P < 0.0005. Behavioral analysis and cell counting: ANOVA with Tukey’s post hoc test; n = 4 animals. a.u., arbitrary units; PLX, PLX5622.
Microglial dysmaturation in the absence of TGFβ signaling
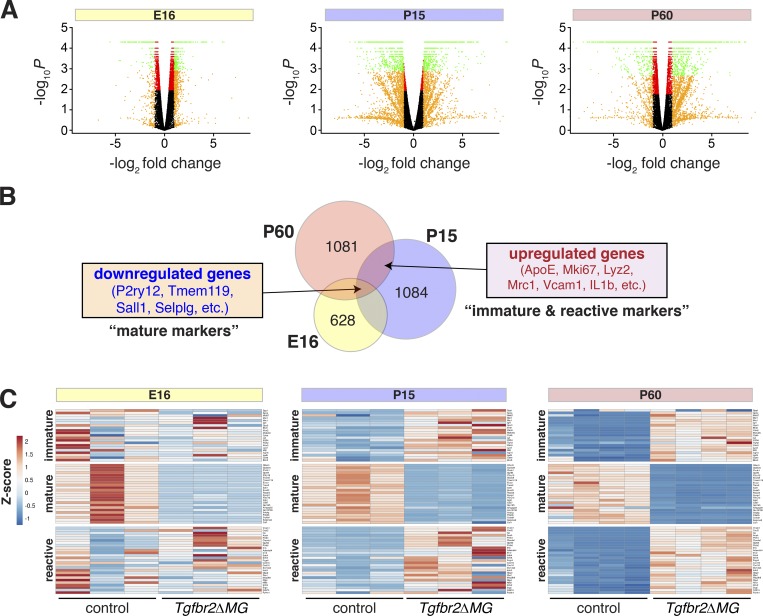
Microglia follow a stepwise developmental program demarcated by groups of genes differentially expressed at sequential ages (Bennett et al., 2016; Matcovitch-Natan et al., 2016). For instance, microglial markers Tmem119, Itgb5, and P2ry12 are most highly expressed in adolescent and adult mice, while Ki67 and ApoE are markers of immature microglia. In Itgb8ΔCNS and Tgfbr2ΔMG mice, these markers are apparently reversed (Fig. 1 and Fig. S4) and activation markers are increased in these cells (Fig. S2). To better understand microglia alterations over time in Tgfbr2ΔMG mice, we performed expression profiling by RNA-sequencing microglia purified from the brains of Tgfbr2ΔMG mice versus controls at three time points: E16, P15, and P60 (Fig. 5 and Table S2). Consistent with other reports (Matcovitch-Natan et al., 2016), we observed large numbers of genes differentially expressed across embryonic and postnatal time points, delineating distinct developmental phases (Fig. 5). Comparing Tgfb2ΔMG mice with littermate controls, we observed a trend toward progressively more differentially expressed genes over time (Fig. 5 A). Interestingly, the most highly differentially expressed genes (>1.5-fold difference) common to all three time points were generally down-regulated, while most up-regulated genes were common to P15 and P60 (and generally not differentially expressed at E16; Fig. 5 B). Most down-regulated genes are markers of mature microglia, while up-regulated genes are normally expressed in either immature or reactive microglia. Based on this, we annotated our gene profiles according to three gene lists generated from previously published transcriptomic data including developmental profiles of microglia (mature and immature genes; Bennett et al., 2016; Matcovitch-Natan et al., 2016) and microglia from adult and neonatal mice exposed to LPS (reactive genes; Bennett et al., 2016; Hirbec et al., 2018; Table S1). Organized this way, our analysis confirms an apparent failure of microglia in Tgfbr2ΔMG mutants to express markers of maturation, as was found in Tgfb1−/− mice (Butovsky et al., 2014). Unique to our dataset, we also discovered many genes normally expressed only in immature or reactive microglia that were highly up-regulated in microglia from Tgfbr2ΔMG mice at P15 and P60. Taken together, these data indicate that TGFβ signaling is required for the stepwise development of microglia and that microglia deficient in TGFβ signaling are highly activated and overexpress markers of immature microglia. We have termed these microglia “dysmature.”
Figure 5.
Microglial dysmaturation in the absence of TGFβ signaling. (A) Volcano plots of differentially expressed genes (RNA sequencing) in microglia from Tgfbr2ΔMG versus litter mate controls at indicated time points (n = 3–4 per genotype). Green, P < 0.05 and log2 fold change >1; red, P < 0.05; orange, log2 fold change >1; black, P > 0.05. (B) Venn diagrams of overlaps of differentially expressed genes from E16 (yellow), P15 (blue), and P60 (pink) time points. (C) Heat map representation of differential expression Z statistics for indicated datasets. Genes are divided into immature, mature, and reactive groups based on similarity to published data (see Table S1).
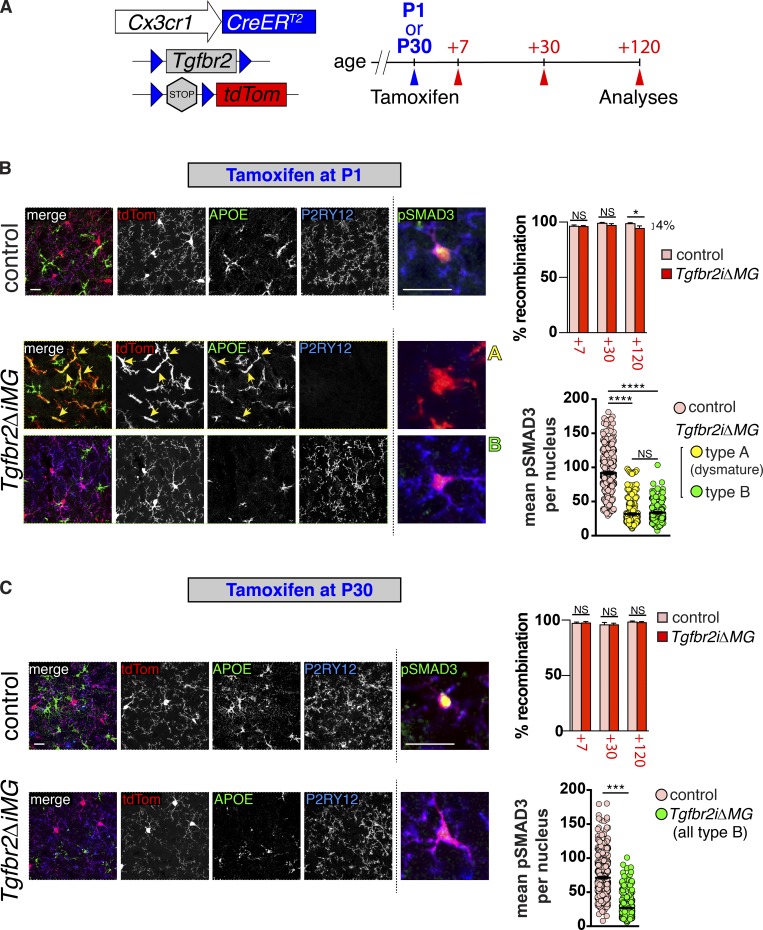
To directly evaluate microglial dependence on TGFβ signaling during development, we generated mice capable of tamoxifen-inducible Tgfbr2 gene deletion directed under the endogenous Cx3cr1 promotor (Yona et al., 2013) and with a fluorescent tdTomato Cre recombinase reporter (Tgfbr2ΔiMG mice) to follow recombination events (Fig. 6 A). We administered tamoxifen to target gene deletion during early microglial maturation (P1) or after microglia are fully mature (P30), then examined Cre reporter (tdTomato) expression and the microglial phenotype 7, 30, and 120 d later (Fig. 6, B and C). Flow cytometry analysis at these time points revealed nearly complete recombination of CD45+ F4/80+ microglia in both mutants (Tgfbr2fl/fl;Cx3cr1CreERT2/+;R26LSLtdTom) and controls (Tgfbr2fl/+;Cx3cr1CreERT2/+;R26LSLtdTom) 7, 30, and 120 d after tamoxifen administration. We did observe a small percentage (∼4%) of nonrecombined microglia in Tgfbr2ΔiMG mutants induced on P1, 120 d after tamoxifen administration, whereas ∼100% of microglia were recombined in littermate controls using an identical induction strategy, and ∼100% of microglia were recombined at all time points assessed in controls and mutants induced on P30. Cx3cr1CreERT2 recombines resident myeloid cells, as well as circulating progenitors; however, recombined circulating monocytes are only present for ∼7 d, after which they die and are replaced by nonrecombined bone marrow–derived cells (Yona et al., 2013). This suggests that circulating monocytes are a potential source of these nonrecombined microglia in Tgfbr2ΔiMG mutants induced on P1. The low percentage of these cells observed in Tgfbr2ΔiMG mutants is consistent with the relatively low number of circulation-derived Ly6C+/CCR2+ cells observed in Itgb8ΔCNS and Tgfbr2ΔMG (constitutive Cx3cr1Cre) mice (Fig. S2) and the low CNS chimerism observed when Tgfbr2ΔMG mice are transplanted with GFP-labeled WT bone marrow (Fig. S3).
Figure 6.
Microglial dysmaturation due to loss of TGFβ signaling is critically time dependent. (A) Diagram of Tgfbr2 gene inactivation and mutant analysis. Tamoxifen was injected at P1 for early induction in B or at P30 for adult induction in C. Mice were sacrificed and analyzed at indicated time points after tamoxifen injection. (B and C) Staining of cortical brain sections from Tgfbr2iΔMG and Cre+ control littermates 120 d after tamoxifen administration following early (B) or adult (C) induction. Left panels: tdTomato recombination reporter (tdTom, red) marks all microglia and immature (APOE, green) and mature (P2RY12, blue) microglia markers to identify type A (dysmature) or type B microglia, respectively. Right panels (insets from Fig. S5): pSMAD3 (green) coimmunostaining reveals reduction in pSMAD3 staining intensity in type A (dysmature) microglia compared with type B microglia and controls. Percent (%) recombination (upper right graphs in B and C) based on % F4/80+, CD45+, CD11b+ cells that are also tdTomato positive (cells isolated and analyzed by flow cytometry as in Fig. S3). Mean pSMAD3 per nucleus (lower right graphs in B and C) based on fluorescent intensity of individual recombined (tdTom+) microglia coexpressing P2RY12 (blue, control and type B cells), or lacking P2RY12 expression (type A cells). Bars, 100 µm or 25 µm (pSMAD3, right panels). Error bars indicate SE. *P < 0.05; ***P < 0.005; ****P < 0.0005. ANOVA with Tukey’s post hoc test (B); Student’s t test (C); n = 4 animals for each group.
We next immunostained brain sections from mice 120 d after tamoxifen administration (given on P1 or P30) to examine the phenotype in microglia with and without TGFBR2-mediated signaling during or after the perinatal developmental window (Fig. 6, B and C). This analysis revealed two general microglial phenotypes in Tgfbr2ΔiMG mutants (Fig. 6 B): “type A” microglia that were tdTomato+ P2RY12neg APOE+ and had fusiform/activated microglial morphology identical to dysmature microglia from Tgfbr2ΔMG mice with constitutive Cx3cr1Cre activity and “type B” microglia that were tdTomato+ P2RY12+ APOEneg and had a quiescent morphology. Brains from Tgfbr2ΔiMG mutants induced on P1 and analyzed on P120 contained both microglial phenotypes (A and B) with distinct patches of each type distributed randomly throughout the brain (Fig. 6 B and Fig. S5). (It is interesting to note that there was little intermixing of microglia subtypes. See Discussion for further explanation.) In contrast, deletion of Tgfbr2 from microglia in adult mice (induced on P30, analyzed on P150) resulted in no obvious changes to microglia compared with controls; all observed microglia in mutants were type B (Fig. 2 C and Fig. S5). We reasoned that type B microglia could represent cells that were recombined at the R26LSLtdT locus, but not at the Tgfbr2 locus, and/or cells that incurred deletion of Tgfbr2 after they had achieved maturation. To distinguish these possibilities, we assessed pSMAD3 activity on a single-cell basis within each microglia type (Fig. 6 B and Fig. S5). This analysis revealed a significant reduction in pSMAD3 in most recombined cells, compared with control cells, suggesting that most type B microglia incurred Tgfbr2 gene deletion and had down-regulation of TGFβ-mediated signaling after they had already matured. Consistent with this, Tgfbr2 (exon 2) mRNA was significantly reduced in both induction strategies based on quantitative PCR. Of note, neither model in which Tgfbr2 was deleted from microglia postnatally showed signs of neurological deficits (gate abnormalities, spasticity, or seizure-like activity), suggesting that either insufficient “dysmature” microglia were present to cause these phenotypes or that a distinct phenotype results in mice with reduced TGFβ signaling in microglia during embryonic time points. Future experiments should differentiate these possibilities. Taken together, these data demonstrate that TGFβ signaling is required for microglial maturation but that an induced reduction in TGFβ signaling in microglia after they have matured has little effect on their homeostatic phenotype or on neuromotor functioning. It was recently suggested that TGFβ is constitutively required for microglial survival (Bohlen et al., 2017). The persistence of recombined microglia with an immature phenotype and the low level of replacement by nonrecombined circulating myeloid cells suggest that this is not a major feature of TGFβ signaling to microglia in vivo.
Discussion
In this paper, we demonstrate critical roles for integrin αVβ8 and TGFβ signaling to direct the maturation of microglia. In both Itgb8ΔCNS and Tgfbr2ΔMG mice, microglia are developmentally arrested and hyper-activated. We show that the presence of these dysmature microglia (and not just the absence of mature microglia) leads to the loss of GABAergic interneurons and abnormalities in oligodendrocyte development, pathology underlying disordered movement, and spasticity. This phenotype is apparently due to loss of TGFβ signaling in microglia because microglia-specific deletion of Tgfbr2 (required for TGFβ signaling) completely recapitulates the Itgb8ΔCNS phenotype. This is a distinctly developmental disorder because deletion of Tgfbr2 from immature microglia causes the phenotype, whereas deletion of Tgfbr2 after they have matured does not, and because early depletion of microglia in Itgb8ΔCNS mice is protective, whereas late depletion is not.
Together with our previous work, the experiments reported here reveal two essential roles for integrin αvβ8 and TGFβ signaling in CNS development: regulated brain vascular sprouting and stabilization (Arnold et al., 2014) and the coordination of microglial maturation with early brain development (this report). Because these processes are spatially and temporally overlapping, the major effects of Itgb8 or Tgfb1 deletion are tightly intertwined: both Itgb8ΔCNS and Tgfb1−/− mice have developmental brain hemorrhage and postnatal microglial maturational arrest and activation. Using mice with endothelial cell–specific (Arnold et al., 2014) or microglial lineage–specific conditional deletion of Tgfbr2, we have disentangled these phenotypes and demonstrate that the CNS microenvironment appears to use a common molecular mechanism (αVβ8–TGFβ signaling) to drive the specification and maturation of both blood vessels and microglia, but that the functional consequences of loss of either outcome are distinct from one another. The close similarities between Itgb8ΔCNS, Tgfb1−/−, and Tgfbr2ΔMG mice, evidence of reduced pSMAD3 signaling in microglia in Itgb8ΔCNS mice, and the observation that microglia depletion abrogates the observed phenotype in these mice support a model by which αVβ8 activates TGFβ and signals directly to microglia. This model is consistent with αVβ8’s principal biological function—latent TGFβ binding and activation. That said, we did not directly prove this model here (e.g., rescue microglia and neuromotor abnormalities in Itgb8ΔCNS by re-expression of activated TGFβ) and cannot fully rule out the possibility that these phenotypes are merely coincidental.
TGFβ is a pleotropic cytokine important for development and functioning of many cell types in different organs. In the brain, TGFβ has been shown to have direct effects on neuronal and glial populations (Brionne et al., 2003; Yi et al., 2010; He et al., 2014; Palazuelos et al., 2014). We therefore originally assumed that the CNS abnormalities and behavioral phenotype observed in Itgb8 and Tgfb1 mutant mice (Brionne et al., 2003; Butovsky et al., 2014) were a consequence of either reduced TGFβ signaling to individual neuro/glial populations or secondary to in utero brain hemorrhage. In our studies, we found that TGFβ signaling (pSMAD3) was active in both glia and neurons in control animals but, surprisingly, was reduced only in microglia in Itgb8ΔCNS mice. Consistent with this finding, the entire neuromotor phenotype observed in Itgb8ΔCNS mice, absent vascular dysplasia and hemorrhage, is recapitulated in mice with conditional deletion of Tgfbr2 only in microglia, whereas conditional deletion of Tgfbr2 in neuro-glia lineage cells (Tgfbr2fl/fl;NestinCre mice) causes no apparent neuromotor abnormalities (Nguyen et al., 2011). These data suggest that presentation of activated TGFβ by αVβ8 is specific to microglia, as opposed to more regional presentation to a “field” of nearby cells. The downstream temporal accumulation of cellular abnormalities in these mutants suggests oligodendrocyte and interneuron abnormalities are caused by dysmature microglia and/or reactive astrocytes. Microglial depletion in Itgb8ΔCNS reversed or prevented these cellular abnormalities despite continued increased expression of the astrocyte activation marker (GFAP), suggesting that reactive astrocytes might not directly contribute to the phenotype. However, GFAP is only one marker of reactive astrocytes and its expression is not necessarily correlated to astrocyte function (Liddelow et al., 2017). Consistent with our work, others have shown that microglial activation can directly or indirectly influence early GABAergic interneuron lamination, and abnormal myelination has been attributed to reactive microglia (Miron et al., 2013; Beckmann et al., 2018), reactive astrocytes (Hammond et al., 2015), and reduced GABAergic input from interneurons (Zonouzi et al., 2015). It was also recently shown that reactive astrocytes (induced by activated microglia) can directly kill neurons and mature oligodendrocytes (Liddelow et al., 2017). Taken together, our data illustrate the concept that early brain development requires the coordinated maturation of microglia, neurons and macroglia and reveal that αVβ8 and TGFβ signaling to microglia is central to this process.
Butovsky et al. (2014) reported that mice lacking expression of Tgfb1 in the CNS also have marked changes in CNS myeloid cells. In contrast to our findings, their mice had a more heterogeneous CNS myeloid compartment with reduced numbers of IBA1+, F4/80+, and CD39+ cells and increased numbers of CD39−LY6C+ cells. They proposed that loss of Tgfb1 expression in the brain impaired survival of microglia and that these cells were progressively replaced by infiltrating monocytes. This model was supported by Bohlen et al. (2017), who showed that microglia in culture depend on TGFβ for survival. Using BMT and lineage tracing methods, we confirmed the presence of a minor population of circulation-derived microglia in our models. However, as these are a minor population of cells, our data suggest that TGFβ is not a critical in vivo survival cue for microglia in adult mice. Rather, we observed that cell-autonomous reductions in TGFβ signaling in microglia early in development produce populations of dysmaturemicroglia, which persist well into adulthood. Interestingly, early postnatal deletion of Tgfbr2 resulted in mosaics of dysmature (type A) and mature (type B) microglia (type A), with little spatial intermixing of subtypes. What drives the dimensions (shape, size, and location) of these populations is not immediately obvious (their boundaries do not conform to neuroanatomical regions or other physical constraints) but could be due to clonal expansion (individual clones generated at the time of Tgfbr2 gene deletion give rise to “patches” of either type A or type B microglia) or regional programing (local environmental cues established within the CNS parenchyma program microglia to conform to a particular subtype). We are currently exploring these alternative hypotheses.
Lund et al. (2018) recently described a demyelinating disorder in adult mice in which the spinal cord microglial compartment was replaced with peripherally derived monocyte-lineage cells with conditional deletion of Tgfbr2. Their report raises the possibility that the neurological phenotypes observed in Itgb8ΔCNS and Tgfbr2ΔMG mice are due to direct effects from CNS-infiltrating myeloid cells or indirect effects (e.g., secreted cytokines) from myeloid cells in the peripheral immune system, as opposed to developmental arrest of resident microglia. In support of our model, we found limited evidence for significant extraneural myeloid/monocyte CNS infiltration in these mice. The transcriptional profile of mutant myeloid cells in the paper by Lund et al. (2018) is distinct from that of our cells, and the associated neurological symptoms are quite different (they report stereotyped lower motor neuron symptoms, such as flaccid paralysis, whereas our models have upper motor neuron dysfunction [spasticity] and seizure-like activity). We also note that Itgb8ΔCNS mice delete Itgb8 only in the CNS compartment and therefore should not directly affect the peripheral immune system (including both circulating monocytes and CNS boundary macrophages). Consistent with this, deletion of Tgfbr2 from the peripheral myeloid compartment using LysMCre does not cause similar microglia changes or obvious downstream neurobehavioral abnormalities (Lund et al., 2018). Taken together, these findings, in our view, best support a model in which resident microglia lacking TGFβ signaling early in development lead to the observed microglia phenotype and downstream neurological consequences. Our results therefore highlight essential developmental aspects of TGFβ signaling in brain microglia.
TGFβ drives microglial ontogeny through the selection of brain-specific Smad3-Pu.1 enhancer elements (Gosselin et al., 2014), and PU.1 binds gene regions that regulate the expression of several αVβ8-TGFβ–dependent microglia-specific genes including P2ry12, Slc2a5 (maturation markers), Igf1, Cybb (early development markers), CD68, and Vcam1 (reactive markers; Satoh et al., 2014). Consistent with this, we found reduced phosphorylation of SMAD3 in microglia from Itgb8ΔCNS and Tgfbr2ΔiMG mice and associated dysregulation of these genes. The transcriptomes of microglia from mice with myeloid-specific conditional deletion of Mafb (Mafbfl/fl;Csf1rCre; Matcovitch-Natan et al., 2016) or Sall1 (Sall1fl;Sal1CreERT2; Buttgereit et al., 2016; two important transcription factor genes down-regulated in microglia from Itgb8 and Tgfbr2 mutant mice) were remarkably similar to that from Tgfbr2ΔMG mice, suggesting common gene regulator networks. However, neither of these mouse mutants were reported to have neurological sequelae similar to those observed in Itgb8, Tgfb1, or Tgfbr2 mutant mice. It will be important to determine if the lack of neurological symptoms in these mice is due to late/adult deletion (as in Sall1 mutants) or due to compensation from other transcription factors (e.g., cMAF is known to compensate for MAFB; SALL3 can compensate for SALL1).
Clinical implications of αVβ8 and TGFβ signaling and microglial maturation
As in mice, αVβ8 is widely expressed in the developing and mature CNS in humans (Zhang et al., 2016). The striking similarities between the phenotype and neuropathology we describe in Itgb8ΔCNS and Tgfbr2ΔMG mice and those seen in humans with developmental brain injury resulting in disordered movement, developmental disabilities, and seizures (disorders colloquially characterized as cerebral palsy) suggest that dysmature and persistently activated myeloid cells might drive the progression of motor symptoms after birth in these disorders. Therefore, it is possible that neuropathology and motor abnormalities in some developmental brain disorders might not simply be inevitable consequences of a pre- or perinatal insult, as has been widely assumed, but could be treatable by therapies that reverse or ameliorate the consequences of microglial activation or dysmaturation in the CNS. The protective effects of postnatal pharmacologic microglial depletion strongly support this concept. In future studies, it will be critical to determine what products or activities of dysmature microglia are responsible for the phenotypes we observed. Interestingly, many of the genes up-regulated in dysmature microglia from Tgfbr2ΔMG mice (e.g., Spp1, Igf1, GPR65, Gpx3, Msr1, and CD44) are differentially expressed in “myelinogenic” microglia during brain development and in demyelinating disease (experimental autoimmune encephalomyelitis; Krasemann et al., 2017; Wlodarczyk et al., 2017). The hyaluronan receptor CD44 seems a particularly relevant target: CD44 is up-regulated in gliotic lesions from humans with white matter injury and hypomyelination (Buser et al., 2012), and transgenic overexpression of CD44 leads to oligodendrocyte maturational arrest in mice (Back et al., 2005).
During the preparation of this manuscript, Kotlarz et al. (2018) described the phenotypes resulting from three individuals with biallelic loss-of-function mutations in the TGFB1 gene leading to reduced TGFB1 bioavailability and reduced downstream SMAD2/3 signaling in immune system cells. The neurological phenotype in these patients is highly similar to that observed in Itgb8 and Tgfbr2 mutant mice reported here: early-onset global developmental delay or regression with spasticity, hyper-reflexia, abnormal tone, epilepsy, and magnetic resonance imaging showing cortical atrophy, periventricular leukomalacia, and delayed or reduced myelination. Interestingly, increased levels of IL-1b (a reactive microglia marker highly up-regulated in dysmature microglia from TgfbrΔMG mice) were present in the cerebrospinal fluid from patient 1 in their report. It is tempting to speculate that the white matter disease and development of neurological symptoms observed in these patients is due to the presence of TGFβ signaling–deficient dysmature microglia.
Microglial abnormalities similar to those found in Tgfbr2ΔMG mice have been reported in a number of other neurodevelopmental disorders (Derecki et al., 2012; Rademakers et al., 2012; Gupta et al., 2014) and recently in neurodegenerative conditions including Alzheimer’s disease (Griciuc et al., 2013; Hong et al., 2016; Wang et al., 2016), amyotrophic lateral sclerosis (Butovsky et al., 2012; Krasemann et al., 2017), frontotemporal dementia (Lui et al., 2016), and neurodegenerative Langerhans cell histiocytosis (Mass et al., 2017). Our work should therefore stimulate investigation of possible roles for perturbed TGFβ signaling within microglia in a variety of currently untreatable or poorly treatable diseases of the CNS.
Materials and methods
Experimental model and subject details
Mice
All mice were maintained in mixed (C57/Bl6; FVB) background. There were no observable differences found between male or female mice in any of our outcomes studied. All mouse strains have been previously reported other than Cx3cr1Cre. Animal husbandry and procedures were performed according to University of California, San Francisco (UCSF), guidelines under Institutional Animal Care and Use Committee (IACUC)–approved protocols.
Itgb8ΔCNS (Itgb8flox/flox;nestinCre)
These mice have conditional deletion of Itgb8 (targeting exon 4; Proctor et al., 2005) from all neuroepithelial lineage cells using nestinCre (Tronche et al., 1999). Littermate Itgb8flox/+;nestinCre mice were used for controls. These mice have been previously described (Proctor et al., 2005). Note that Itgb8 is highly expressed in neurons and macroglia (astrocytes and oligodendrocytes) but not microglia or endothelial cells in the CNS (Zhang et al., 2016). Importantly, the nestinCre line we used in this and previous studies recombines nearly 100% of neural and macroglia (astroglial and oligodendroglial) progenitors embryonically/perinatally and does not recombine vascular cells (endothelium, vascular smooth muscle cells, and pericytes) or microglia.
Tgfbr2ΔMG (Tgfbr2flox/flox;Cx3cr1Cre)
These mice have conditional deletion of Tgfbr2 (targeting exon 2; Chytil et al., 2002) from microglial cells using Cx3cr1Cre (036395-UCD; Mutant Mouse Resource and Research Centers). Cx3cr1Cre mice were created in the GENSAT project in collaboration with the Intramural Program of the National Institute of Mental Health. The transgenic mice express Cre recombinase under the RP24-285B17 mouse genomic bacterial artificial chromosome (BAC) at the ATG transcription initiation codon of Cx3cr1 gene so that expression is driven by the regulatory sequences of the mouse gene. Littermate Tgfbr2flox/+;Cx3cr1Cre mice were used for controls.
NG2-DSR
The NG2-DSR reporter mouse was purchased from The Jackson Laboratory (Tg(Cspf4-DsRed.T1)1Akik/J). These transgenic mice express an optimized RFP variant (DsRed.T1) under the control of the mouse NG2 (Cspg4) promoter/enhancer, labeling NG2 OPCs in the CNS.
Lhx6-BAC-GFP (GENSAT)
The Lhx6-GFP reporter mouse was a generous gift from the laboratory of John Rubenstein (UCSF, San Francisco, CA), created in the GENSAT project. The transgenic mice express enhanced GFP (EGFP) under the RP23-2D16 mouse genomic BAC at the ATG transcription initiation codon of the Lhx6 gene so that expression of the reporter gene is driven by the regulatory sequences of the mouse gene.
Ai14(RCL-tdT)-D
The Ai14 Cre reporter mouse was purchased from The Jackson Laboratory (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J). These mice harbor a targeted mutation of the Gt(ROSA)26Sor locus with a loxP-flanked STOP cassette preventing transcription of a CAG promoter-driven RFP variant (tdTomato). TdTomato is expressed following Cre-mediated recombination (Madisen et al., 2010).
CCR2-RFP
CCR2-RFP reporter mice were purchased from The Jackson Laboratory (B6.129(Cg)-Ccr2tm2.1Ifc/J). These mice have a monomeric RFP sequence replacing the coding sequence of the chemokine (C-C motif) receptor 2 (Ccr2) gene and is useful to track CCR2+ monocyte recruitment to sites of inflammation.
UBC-GFP
UBC-GFP reporter mice were purchased from The Jackson Laboratory (C57BL/6-Tg(UBC-GFP)30Scha/J). These transgenic mice express EGFP under the direction of the ubiquitin C promoter in all tissues/cells and are useful to track, in irradiated hosts, cells derived from transplanted bone marrow.
Tgfbr2iΔMG (Tgfbr2flox;Cx3cr1CreERT2/+;R26LSLtdTom)
These mice have tamoxifen-inducible conditional deletion of Tgfbr2 (targeting exon 2; Chytil et al., 2002) from microglial cells using Cx3cr1CreERT2, which was purchased from The Jackson Laboratory (B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J). These mice express a Cre-ER fusion protein from endogenous Cx3cr1 promoter/enhancer elements. The Ai14 Cre reporter mouse line (R26LSLtdTom allele) was crossed into these mice to monitor gene recombination and facilitate lineage tracing.
Inducible genetic experiments
For myeloid-specific loss-of-function and lineage tracing experiments, Tgfbr2fl/fl;Cx3cr1CreERT2/+;R26LSLtdTom and littermate Tgfbr2fl/+;Cx3cr1CreERT2/+;R26LSLtdTom controls were used. For P1, Tgfbr2 gene inactivation pups were injected transcutaneously into the gastrum (visualized by “milk bubble”) with 50 µl of tamoxifen solution (Sigma; T5648–1G, 1 mg/ml, generated by diluting a 10-mg/ml tamoxifen stock solution in 1:4 ethanol:corn oil with corn oil) once daily on P1, P2, and P3 using a hypodermic 27-G needle. For P30 induction, 50 µl of 20 mg/ml tamoxifen solution was administered by gavage for three doses every other day. Inducible genetic experiments were performed under UCSF IACUC-approved guidelines.
PLX5622 treatment
PLX5622 and ICDs were provided by Plexxikon and administered via food ad libitum (1,200 mg/kg).
BMT
Eight 12-wk-old UBC-GFP mice were used as bone marrow donors. 4-wk-old Itgb8DCNS mice (Itgb8flox/flox;NestinCre and Itgb8flox/+;NestinCre littermate controls) and Tgfbr2ΔMG mice (Tgfbr2flox/flox;Cx3cr1Cre and Tgfbr2flox/+;Cx3cr1Cre littermate controls) were used as recipients for cell transplantation. Recipient mice were irradiated with 900 rads, split dose, 3 h apart using a cesium source. Purified donor cells (4 × 106) from bone marrow were injected intravenously with 200,000 spleen helper cells, and hematopoietic reconstitution was monitored in the peripheral blood based on GFP expression. Recipients with ≥1% donor chimerism were considered reconstituted. Transplanted mice were kept on antibiotic-containing food for 2 wk. All mice were maintained at UCSF in accordance with IACUC-approved protocols.
Immunofluorescence and confocal microscopy
Postnatal mice were perfusion fixed in 4% paraformaldehyde (PFA) in PBS, then post-fixed in 4% PFA at 4°C overnight and stored at 4°C in PBS. Embryos at indicated time points were rinsed in PBS, fixed in 4% PFA at 4°C overnight, and stored at 4°C in PBS. For thin sectioning, fixed brains/embryos were transferred to 30% sucrose in PBS overnight, embedded in optimal cutting temperature compound (Sakura Finetek; 4583), and then cryosectioned at 25 µm onto slides or 60 µm for free-floating sections. Cryosections were permeabilized and blocked with 0.3% Triton X-100, 1% BSA, and 5% donkey serum in PBS. Sections were incubated with primary antibodies overnight at 4°C, then with fluorophore-conjugated secondary antibodies (Abcam) and mounted with Prolong Gold (Invitrogen). Microscopic images were captured on a Zeiss LSM5 Pascal microscope and compiled using ImageJ. We used the primary and secondary antibodies listed in Table S3. Unless noted in the text, images and quantification are from layers 1–3 in the motor or somatosensory cortex.
Cell counting
Four to five animals per genotype were used to examine the cellular marker expression for each time point. Sectionsused for all cell-counting experiments were coronal and 25-µm thick, taken between approximately the bregma area +0.5 to −0.1. 10× images (for interneuron counting) or 20× images (for all other cells) were taken from the area between and including the somatosensory cortex (S1HL) and the cingulate cortex (Cg1; subcortical white matter), including both motor areas M1 and M2. Four to five nonadjacent sections were counted per animal. Quantification of pSMAD3 immunofluorescent staining was determined using stained cryosections from four mutants and four controls at P60. Four randomly chosen confocal images from each sample were taken using the same confocal settings. ImageJ software was used to quantify the number of microglia cell nuclei (DAPI-positive, F4/80-positive), and the intensity of pSMAD3 staining in each microglia cell nuclei, per image.
Microglia/myeloid cell isolation and analysis
Mice were transcardially perfused with ice-cold PBS and brains were dissected. Single-cell suspensions were prepared and centrifuged over a 30%/70% discontinuous Percoll gradient (GE Healthcare), mononuclear cells were isolated from the interface, and total cell count determined. Isolated cells were labeled with fluorophore-conjugated monoclonal antibodies and sorted in a BD FACS Aria III. Flow cytometric analyses were performed on a FACS Verse or FACS Aria III using the FACSDiva 8.0 software (BD Biosciences) and data analyzed using the FlowJo v10.0.7 (Tree Star). Appropriate antibody IgG isotype controls (BD Biosciences) were used for all staining. Macrophages from the spleen were sorted after nonenzymatic disaggregation and were identified as LY6C+ F4/80hi cells for comparison.
Real-time RT-PCR expression analysis
For in vivo transcriptional characterization of isolated microglia, CD11b+F4/80+CD45+ (tdTomato+ in Cx3cr1Cre mice with Ai14 reporter) DAPI-excluded cells were sorted into MCDB-131 complete medium and RNA was immediately extracted using RNeasy Plus Micro Kit (Qiagen; 74034). RNA (50–300 ng) was reverse transcribed using Superscript III Reverse transcription (Life Technologies; 18080044) according to the manufacturer’s instructions and cDNA was quantified with Fast SYBR Green Master Mix (Life Technologies; 4385612) in a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Fluorescence was interpreted relative to Gapdh housekeeping gene expression and quantified using the ΔCt method to obtain relative expression or the ΔΔCt method for fold-change values, as indicated. A full list of oligonucleotide sequences is listed in Table S3.
RNA isolation and sequencing
Microglia were isolated from Tgfbr2ΔMG mice (Tgfbr2flox/flox;Cx3cr1Cre and Tgfbr2flox/+;Cx3cr1Cre littermate controls) at E16, P15, and P60. Total RNA was isolated and purified from sorted cells using the RNeasy Plus Micro Kit (Qiagen; 74034) following the manufacturer's instructions. RNA integrity was determined using a Bioanalyzer. Samples were poly-A primed and amplified with the Clontech SMART-Seq ultra low input kit. Single-end × SR50 sequencing was performed using the Illumina HiSeq4000 system. RNA sequencing analysis was done using R-Studio, and Clustvis was using to perform with a false discovery rate of 0.05. Raw sequencing data are deposited in the Gene Expression Omnibus (GEO) under accession no. GSE124868.
Measurements of locomotor activity
Hind limb stride length was measured by the method adapted by Zhang et al. (2007). Hind paws of control, Itgb8ΔCNS, and Tgfbr2ΔMG mice were wetted with ink. Animals were then placed on a strip of 3MM filter paper (4.5 cm wide, 40 cm long). Stride lengths were measured as the distance between two hind paw prints. Mice were placed on a 6-cm diameter rod A Rotarod device (Ugo Basile) accelerated from 0 to 40 rpm over 5 min. Mice received three 5-min trials with a 2-h intertrial interval. The amount of time spent on the rod before falling off was measured.
Observational outcome measures
The following scoring system was adapted from Wang et al. (2015).
“Gait”: A score for walking gait was given using a three-point observational scoring system. “2” indicated normal gait; “1” indicated broad-based hind limbs and waddling while walking; “0” indicated severe abnormalities with inability to ambulate, dragging limbs, severe tremor.
“Appearance”: A score for appearance of general condition was given using a three-point observational scoring system to assess coat condition, appearance of eyes, and body stance. “2” indicated clean shiny coat, clear eyes, and normal stance; “1” indicated dull coat, ungroomed appearance, dull eyes, and hunched stance; “0” indicated piloerection, crusted eyes, and kyphosis.
“Tremor”: A score was given for tremor or spastic movements using a three-point observational scoring system. “0” indicated no tremor; “1” indicated mild intermittent tremor, made worse when feet were lifted; “2” indicated constant tremor and uncontrollable spastic movements.
Quantification and statistical analysis
For statistical analyses, data distribution was assumed to be normal. Data are presented as mean ± SE (SEM). P values were defined using Student’s t test for paired comparisons and ANOVA for group-wise comparisons, with Tukey’s post hoc test analysis to compare individual groups. Statistics were generated using GraphPad Prism 6 software. Four or more animals/samples were used for all experiments (n ≥ 4). Controls for experiments with control, Itgb8ΔCNS, and Tgfbr2ΔMG mice were not significantly different in any parameter measured and were therefore grouped together and used as a collective “control.” Data collection and analysis were performed blind to the conditions of the experiments. Also, data for each experiment were collected and processed randomly, and animals were assigned to various experimental groups randomly as well. All n and P values and statistical tests are indicated in figure legends.
Online supplemental material
Fig. S1 shows pSMAD3 expression in the brains of control and Itgb8ΔCNS mice. Fig. S2 shows expression of activation and circulatory monocyte markers in Itgb8ΔCNS and Tgfbr2ΔMG microglia. Fig. S3 assesses origin of microglia in Itgb8ΔCNS and Tgfbr2ΔMG mice. Fig. S4 shows expression time course of glial and interneuron markers in Itgb8ΔCNS and Tgfbr2ΔMG mice. Fig. S5 assesses expression of pSMAD3 and microglia markers in Tgfbr2iΔMG mice. Table S1 is a curated list of mature, immature, and reactive microglia genes. Table S2 lists differentially expressed genes from Tgfbr2ΔMG microglia. Table S3 lists the reagents used in our study. Videos S1, S2, and S3 document neurological deficits in Tgfbr2ΔMG mice.
Acknowledgments
We thank Ben Barres, Patrick McQuillen, Zena Vexler, Donna Ferriero, Daniel Vogt, Gabriel Mckinsey, and John Rubenstein for critical discussions and David Julius (P2RY12; UCSF, San Francisco, CA) and Paul Worley (TMEM10; Johns Hopkins University, Baltimore, MD) for generous gifts of antibodies.
This work was supported by the National Institutes of Health National Institute of Child Health and Human Development grant K12HD047349 (T.D. Arnold) and National Institutes of Health National Heart, Lung, and Blood Institute grant R37 HL53949 (D. Sheppard).
The authors declare no competing financial interests.
Author contributions: T.D. Arnold, L.R. Reichardt, and D. Sheppard conceived and planned experiments. T.D. Arnold, C.O. Lizama, K.M. Cautivo, L. Lin , H. Qui, C. Liu, and N. Santander performed experiments. All authors helped analyze and evaluate data. T.D. Arnold and D. Sheppard wrote the manuscript with critiques from other authors.
References
- Acharya M.M., Green K.N., Allen B.D., Najafi A.R., Syage A., Minasyan H., Le M.T., Kawashita T., Giedzinski E., Parihar V.K., et al. 2016. Elimination of microglia improves cognitive function following cranial irradiation. Sci. Rep. 6:31545 10.1038/srep31545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold T.D., Niaudet C., Pang M.F., Siegenthaler J., Gaengel K., Jung B., Ferrero G.M., Mukouyama Y.S., Fuxe J., Akhurst R., et al. 2014. Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking αVβ8-TGFβ signaling in the brain. Development. 141:4489–4499. 10.1242/dev.107193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S.A., Tuohy T.M., Chen H., Wallingford N., Craig A., Struve J., Luo N.L., Banine F., Liu Y., Chang A., et al. 2005. Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nat. Med. 11:966–972. 10.1038/nm1279 [DOI] [PubMed] [Google Scholar]
- Beckmann N., Giorgetti E., Neuhaus A., Zurbruegg S., Accart N., Smith P., Perdoux J., Perrot L., Nash M., Desrayaud S., et al. 2018. Brain region-specific enhancement of remyelination and prevention of demyelination by the CSF1R kinase inhibitor BLZ945. Acta Neuropathol. Commun. 6:9 10.1186/s40478-018-0510-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers D.R., Henkel J.S., Xiao Q., Zhao W., Wang J., Yen A.A., Siklos L., McKercher S.R., and Appel S.H.. 2006. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 103:16021–16026. 10.1073/pnas.0607423103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. 2016. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 113:E1738–E1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J., Bennett F.C., Tucker A.F., Collins H.Y., Mulinyawe S.B., and Barres B.A.. 2017. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron. 94:759–773.e8. 10.1016/j.neuron.2017.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brionne T.C., Tesseur I., Masliah E., and Wyss-Coray T.. 2003. Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron. 40:1133–1145. 10.1016/S0896-6273(03)00766-9 [DOI] [PubMed] [Google Scholar]
- Buser J.R., Maire J., Riddle A., Gong X., Nguyen T., Nelson K., Luo N.L., Ren J., Struve J., Sherman L.S., et al. 2012. Arrested preoligodendrocyte maturation contributes to myelination failure in premature infants. Ann. Neurol. 71:93–109. 10.1002/ana.22627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Siddiqui S., Gabriely G., Lanser A.J., Dake B., Murugaiyan G., Doykan C.E., Wu P.M., Gali R.R., Iyer L.K., et al. 2012. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 122:3063–3087. 10.1172/JCI62636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17:131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A., Lelios I., Yu X., Vrohlings M., Krakoski N.R., Gautier E.L., Nishinakamura R., Becher B., and Greter M.. 2016. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17:1397–1406. 10.1038/ni.3585 [DOI] [PubMed] [Google Scholar]
- Chytil A., Magnuson M.A., Wright C.V.E., and Moses H.L.. 2002. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 32:73–75. 10.1002/gene.10046 [DOI] [PubMed] [Google Scholar]
- Cobos I., Long J.E., Thwin M.T., and Rubenstein J.L.. 2006. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb. Cortex. 16(Suppl 1):i82–i88. 10.1093/cercor/bhk003 [DOI] [PubMed] [Google Scholar]
- Cunningham C.L., Martínez-Cerdeño V., and Noctor S.C.. 2013. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 33:4216–4233. 10.1523/JNEUROSCI.3441-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki N.C., Cronk J.C., Lu Z., Xu E., Abbott S.B., Guyenet P.G., and Kipnis J.. 2012. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 484:105–109. 10.1038/nature10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore M.R., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., et al. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erblich B., Zhu L., Etgen A.M., Dobrenis K., and Pollard J.W.. 2011. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 6:e26317 10.1371/journal.pone.0026317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy S.P., Harrington E.P., Yuen T.J., Silbereis J.C., Zhao C., Baranzini S.E., Bruce C.C., Otero J.J., Huang E.J., Nusse R., et al. 2011. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat. Neurosci. 14:1009–1016. 10.1038/nn.2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favrais G., van de Looij Y., Fleiss B., Ramanantsoa N., Bonnin P., Stoltenburg-Didinger G., Lacaud A., Saliba E., Dammann O., Gallego J., et al. 2011. Systemic inflammation disrupts the developmental program of white matter. Ann. Neurol. 70:550–565. 10.1002/ana.22489 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., et al. 2015. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 518:547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F., et al. 2014. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 159:1327–1340. 10.1016/j.cell.2014.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A., Serrano-Pozo A., Parrado A.R., Lesinski A.N., Asselin C.N., Mullin K., Hooli B., Choi S.H., Hyman B.T., and Tanzi R.E.. 2013. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 78:631–643. 10.1016/j.neuron.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Ellis S.E., Ashar F.N., Moes A., Bader J.S., Zhan J., West A.B., and Arking D.E.. 2014. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun. 5:5748 10.1038/ncomms6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer N., Hanft K.M., Akriditou M.A., Unger N., Park E.S., Stanley E.R., Staszewski O., Dimou L., and Prinz M.. 2017. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134:441–458. 10.1007/s00401-017-1747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R., McEllin B., Morton P.D., Raymond M., Dupree J., and Gallo V.. 2015. Endothelin-B receptor activation in astrocytes regulates the rate of oligodendrocyte regeneration during remyelination. Cell Reports. 13:2090–2097. 10.1016/j.celrep.2015.11.002 [DOI] [PubMed] [Google Scholar]
- He Y., Zhang H., Yung A., Villeda S.A., Jaeger P.A., Olayiwola O., Fainberg N., and Wyss-Coray T.. 2014. ALK5-dependent TGF-β signaling is a major determinant of late-stage adult neurogenesis. Nat. Neurosci. 17:943–952. 10.1038/nn.3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H., Marmai C., Duroux-Richard I., Roubert C., Esclangon A., Croze S., Lachuer J., Peyroutou R., and Rassendren F.. 2018. The microglial reaction signature revealed by RNAseq from individual mice. Glia. 66:971–986. 10.1002/glia.23295 [DOI] [PubMed] [Google Scholar]
- Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A., et al. 2016. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 352:712–716. 10.1126/science.aad8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komitova M., Xenos D., Salmaso N., Tran K.M., Brand T., Schwartz M.L., Ment L., and Vaccarino F.M.. 2013. Hypoxia-induced developmental delays of inhibitory interneurons are reversed by environmental enrichment in the postnatal mouse forebrain. J. Neurosci. 33:13375–13387. 10.1523/JNEUROSCI.5286-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz D., Marquardt B., Barøy T., Lee W.S., Konnikova L., Hollizeck S., Magg T., Lehle A.S., Walz C., Borggraefe I., et al. 2018. Human TGF-β1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat. Genet. 50:344–348. 10.1038/s41588-018-0063-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.S., Peterson T.C., et al. 2017. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 541:481–487. 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.Y., Shang Y., Oldham M.C., Martens L.H., Gao F., et al. 2016. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 165:921–935. 10.1016/j.cell.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund H., Pieber M., Parsa R., Grommisch D., Ewing E., Kular L., Han J., Zhu K., Nijssen J., Hedlund E., et al. 2018. Fatal demyelinating disease is induced by monocyte-derived macrophages in the absence of TGF-β signaling. Nat. Immunol. 19:1–7. 10.1038/s41590-018-0091-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E., Jacome-Galarza C.E., Blank T., Lazarov T., Durham B.H., Ozkaya N., Pastore A., Schwabenland M., Chung Y.R., Rosenblum M.K., et al. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 549:389–393. 10.1038/nature23672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P., et al. 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 353:aad8670 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- Miron V.E., Boyd A., Zhao J.W., Yuen T.J., Ruckh J.M., Shadrach J.L., van Wijngaarden P., Wagers A.J., Williams A., Franklin R.J.M., et al. 2013. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci. 16:1211–1218. 10.1038/nn.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley A.K., Tchaicha J.H., Shin J., Hossain M.G., and McCarty J.H.. 2009. Beta8 integrin regulates neurogenesis and neurovascular homeostasis in the adult brain. J. Cell Sci. 122:1842–1851. 10.1242/jcs.043257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J., Beura L.K., Bobr A., Astry B., Chicoine B., Kashem S.W., Welty N.E., Igyártó B.Z., Wijeyesinghe S., Thompson E.A., et al. 2016. Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat. Immunol. 17:414–421. 10.1038/ni.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S., Gokhan S., Dai X.M., Wei S., Enikolopov G., Lin H., Mehler M.F., and Stanley E.R.. 2012. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev. Biol. 367:100–113. 10.1016/j.ydbio.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.L., Lee Y.J., Shin J., Lee E., Park S.O., McCarty J.H., and Oh S.P.. 2011. TGF-β signaling in endothelial cells, but not neuroepithelial cells, is essential for cerebral vascular development. Lab. Invest. 91:1554–1563. 10.1038/labinvest.2011.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazuelos J., Klingener M., and Aguirre A.. 2014. TGFβ signaling regulates the timing of CNS myelination by modulating oligodendrocyte progenitor cell cycle exit through SMAD3/4/FoxO1/Sp1. J. Neurosci. 34:7917–7930. 10.1523/JNEUROSCI.0363-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor J.M., Zang K., Wang D., Wang R., and Reichardt L.F.. 2005. Vascular development of the brain requires beta8 integrin expression in the neuroepithelium. J. Neurosci. 25:9940–9948. 10.1523/JNEUROSCI.3467-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R., Baker M., Nicholson A.M., Rutherford N.J., Finch N., Soto-Ortolaza A., Lash J., Wider C., Wojtas A., DeJesus-Hernandez M., et al. 2012. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 44:200–205. 10.1038/ng.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S., Li Q., Dechant A., and Cohen M.L.. 2006. Neonatal loss of gamma-aminobutyric acid pathway expression after human perinatal brain injury. J. Neurosurg. 104(6, Suppl):396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J., Asahina N., Kitano S., and Kino Y.. 2014. A comprehensive profile of ChIP-Seq-Based PU. 1/Spi1 target genes in microglia. Gene Regul. Syst. Bio. 8:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafidi J., Hammond T.R., Scafidi S., Ritter J., Jablonska B., Roncal M., Szigeti-Buck K., Coman D., Huang Y., McCarter R.J. Jr., et al. 2014. Intranasal epidermal growth factor treatment rescues neonatal brain injury. Nature. 506:230–234. 10.1038/nature12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y., Hoshikawa K., Goldman J.E., Sekino Y., and Sato K.. 2014. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34:2231–2243. 10.1523/JNEUROSCI.1619-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P., Oller G., Hoeffel G., Pont-Lezica L., Rostaing P., Low D., Bessis A., Ginhoux F., and Garel S.. 2014. Microglia modulate wiring of the embryonic forebrain. Cell Reports. 8:1271–1279. 10.1016/j.celrep.2014.07.042 [DOI] [PubMed] [Google Scholar]
- Stipursky J., Francis D., Dezonne R.S., Bérgamo de Araújo A.P., Souza L., Moraes C.A., and Alcantara Gomes F.C.. 2014. TGF-β1 promotes cerebral cortex radial glia-astrocyte differentiation in vivo. Front. Cell. Neurosci. 8:393 10.3389/fncel.2014.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis M.A., Reizis B., Melton A.C., Masteller E., Tang Q., Proctor J.M., Wang Y., Bernstein X., Huang X., Reichardt L.F., et al. 2007. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 449:361–365. 10.1038/nature06110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., and Schütz G.. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23:99–103. 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Wang J., Wegener J.E., Huang T.-W., Sripathy S., De Jesus-Cortes H., Xu P., Tran S., Knobbe W., Leko V., Britt J., et al. 2015. Wild-type microglia do not reverse pathology in mouse models of Rett syndrome. Nature. 521:E1–E4. 10.1038/nature14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarczyk A., Holtman I.R., Krueger M., Yogev N., Bruttger J., Khorooshi R., Benmamar-Badel A., de Boer-Bergsma J.J., Martin N.A., Karram K., et al. 2017. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36:3292–3308. 10.15252/embj.201696056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.J., Barnes A.P., Hand R., Polleux F., and Ehlers M.D.. 2010. TGF-beta signaling specifies axons during brain development. Cell. 142:144–157. 10.1016/j.cell.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pho V., Bonasera S.J., Holtzman J., Tang A.T., Hellmuth J., Tang S., Janak P.H., Tecott L.H., and Huang E.J.. 2007. Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat. Neurosci. 10:77–86. 10.1038/nn1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., et al. 2016. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 89:37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonouzi M., Scafidi J., Li P., McEllin B., Edwards J., Dupree J.L., Harvey L., Sun D., Hübner C.A., Cull-Candy S.G., et al. 2015. GABAergic regulation of cerebellar NG2 cell development is altered in perinatal white matter injury. Nat. Neurosci. 18:674–682. 10.1038/nn.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]