Abstract
The Major Histocompatibility complex (MHC) class I-related molecule, MR1, presents vitamin B metabolites from bacteria and yeast to Mucosal-associated invariant T (MAIT) cells. Despite the evolutionary conservation of MR1, we do not know whether different allele variants of MR1 exist within the nonhuman primate (NHP) populations that are commonly used for biomedical research. In this study, we identified 21 distinct MR1 nucleotide sequences representing 32 different alleles across five different NHP populations. The majority of the alleles conferring amino acid changes (allele variants) were found in or near the alpha-1 domain of the mature MR1 protein. We expressed four of the most commonly observed MR1 allele variants in 293T cells, and we found that each variant could present bacterial metabolites on the cell surface. We successfully induced cytokine production in macaque MAIT cells stimulated with 293T cells expressing the four most common MR1 allele variants, demonstrating the usefulness of these cell lines to study MAIT cell activity. Our data suggests that MR1 is not monomorphic, but that there are multiple MR1 alleles in NHPs. The materials we describe here will be valuable for characterizing differences in MR1 antigen presentation and MAIT cell function in NHPs.
Keywords: MHC class I-related molecule, nonhuman primate, population genetics, sequencing analysis
Introduction:
Mucosal Associated Invariant T (MAIT) cells play a key role in innate anti-microbial immunity. MAIT cells recognize vitamin B metabolites produced by certain bacteria and yeast. These metabolites are presented by MR1 molecules on the surface of cells that have internalized or been infected by these pathogens (Kjer-Nielsen et al. 2012; López-Sagaseta et al. 2013a; López-Sagaseta et al. 2013b). MR1 shares structural similarity to classical MHC class I molecules and non-classical CD1 molecules (Hashimoto et al. 1995; Kjer-Nielsen et al. 2012; López-Sagaseta et al. 2013a). A semi-invariant T cell receptor (TCR) present on MAIT cells detects the metabolites presented by MR1 molecules (Gold et al. 2010; Huang et al. 2005; López-Sagaseta et al. 2013a; Reantragoon et al. 2012). Upon recognition of antigen, MAIT cells secrete cytokines and cytolytic enzymes to destroy bacterially infected cells in an MR1-dependent manner (Dias et al. 2016; Gold et al. 2010; Kjer-Nielsen et al. 2012; Kurioka et al. 2015; Le Bourhis et al. 2010; Le Bourhis et al. 2013; López-Sagaseta et al. 2013a; López-Sagaseta et al. 2013b). MR1 knockout mice are more susceptible to certain bacterial infections, further implicating the importance of MAIT cells to an effective antibacterial immune response (Georgel et al. 2011; Meierovics et al. 2013).
While MR1 and classical MHC class I molecules have similar structure, MR1 has been described as monomorphic, whereas the MHC is incredibly polymorphic (Awad et al. 2018; Godfrey et al. 2015; Shiina et al. 2017; Treiner et al. 2003). MR1 has been reported to be one of the most highly conserved genes between humans and other mammalian species (Huang et al. 2009; Parra-Cuadrado et al. 2001; Riegert et al. 1998; Tsukamoto et al. 2013; Yamaguchi et al. 1997). However, given that the genomic organization of the MHC in NHPs is more complex than the human MHC (Daza-Vamenta et al. 2004; Fukami-Kobayashi et al. 2005; Watanabe et al. 2007), it is possible that the MR1 locus may also be more complex.
While some inter-species diversity exists in the MR1 gene (Greene et al. 2017; Parra-Cuadrado et al. 2001), very few studies to date have focused on the identification of intra-species allele variants of MR1. Two studies have found MR1 polymorphisms in humans (Parra-Cuadrado et al. 2000; Seshadri et al. 2017), including the association of an intronic polymorphism with increased susceptibility to Tuberculosis disease (Seshadri et al. 2017). Furthermore, at least one polymorphism has been identified in some nonhuman primates leading to a truncated MR1 molecule (Parra-Cuadrado et al. 2001). Thus, it is reasonable to consider that other polymorphisms in MR1 may exist in NHPs and influence the outcomes of important and expensive biomedical studies.
NHPs are physiologically relevant for studying many human viral and bacterial infections, as well as testing vaccine efficacy (Kaushal et al. 2012). Here, we show that among five NHP populations, at least 21 distinct MR1 nucleotide sequences exist, representing 32 alleles. We also assess surface expression of the four most common MR1 allele variants when loaded with the vitamin B9 (folic acid) derivative 6-Acetyl Formylpterin, as well as metabolites derived from paraformaldehyde-fixed E.coli. We find that 293T cells expressing any of the four most common MR1 allele variants can be loaded with fixed E.coli and induce cytokine production from macaque MAIT cells in vitro. Overall, our findings expand our knowledge of MR1 genetic diversity in NHPs, and provide useful tools for future studies of MR1 activation of MAIT cells in NHPs during pathogenic infections.
Compliance with Ethical Standards:
The marmosets used in this study were housed at the Wisconsin National Primate Research Center (WNPRC). They were cared for by staff at the Wisconsin National Primate Research Center using protocols approved by the University of Wisconsin Graduate School Animal Care and Use Committee.
Materials and Methods:
Animals and cells.
For all marmoset samples, fresh blood was obtained using approved protocols as mentioned above, and peripheral blood mononuclear cells (PBMC) were isolated by application to a 4mL sodium citrate CPT tube (BD biosciences, San Jose, CA). The buffy coat was removed, cells were washed twice in RPMI+10%FBS, and pelleted for RNA isolation. The PBMC were frozen and then used for RNA isolation as described below.
Cryopreserved samples from 15 Mauritian cynomolgus macaques (Macaca fascicularis; MCM), 10 Asian cynomolgus macaques (Macaca fascicularis; ACM), 10 Pigtailed macaques (Macaca nemestrina; PTM), and 10 Indian Rhesus Macaque (Macaca mulatta; RM) were collected as part of previous studies (Ellis-Connell et al., 2018; Gooneratne et. al., 2014; Harris et. al., 2013; Sutton et. al. 2016; Sutton et. al. 2018). Their cryopreserved PBMC were used in this study.
Cell lines and reagents.
293T cells (ATCC) were grown in 293T media containing DMEM/High Glucose (Thermo Fisher Scientific; Waltham, MA); 10% Fetal Bovine Serum (FBS; Thermo Fisher Scientific), Antibiotic/Antimycotic (Thermo Fisher Scientific), 2mM L-glutamine (Thermo Fisher Scientific), and 10mM HEPES buffer (Thermo Fisher Scientific). 293T cells stably expressing MR1 variants were grown in 293T media supplemented with 300 μg/mL Zeocin (InvivoGen, San Diego, CA). Acetyl-6-formylpterin (Ac-6-FP; Schircks Laboratories; Switzerland) was solubilized in 0.01N NaOH according to company protocols. MAIT cells isolated from macaque PBMC were cultured in RPMI (Thermo Fisher Scientific) supplemented with 10% FBS (R10), 4mM L-glutamine, antibiotic/antimycotic, 25ng/mL human IL-7 (Biolegend, San Diego, CA), and 100 Units/mL of human IL-2 (Peprotech, Rocky Hill, NJ).
RNA Isolation.
RNA was isolated from fresh and frozen PBMC pellets using a Maxwell® 16 LEV simplyRNA Cells Kit (Promega; Fitchburg, WI) according to the manufacturer’s protocols. Isolation was performed using a Maxwell16 instrument according to the manufacturer’s protocols.
cDNA Synthesis and MR1 amplification.
To synthesize cDNA from RNA samples, an AccuScript High Fidelity First-Strand cDNA Synthesis Kit (Agilent Technologies; Santa Clara, CA) was used according to the manufacturer’s protocol using oligo (dT) primers. After cDNA synthesis, the MR1 cDNA from individual macaque and marmoset samples were amplified using Phusion DNA polymerase (New England Biolabs [NEB]; Ipswich MA) and the following primers that were designed based on the cynomolgus macaque MR1 mRNA sequence (accession number NM_001284968) and marmoset MR1 mRNA sequence (accession number XM_008984893.2): Macaque MR1 FWD; 5’-ACA GGT ACC ACC ATA GGG GAA CTG ATG GTG-3’; Macaque MR1 REV; 5’-ACA GAA TTC TCA TCG ATC TGG TGT CGG-3’. Marmoset MR1 FWD; 5’-ACA GGT ACC ACC ATA GGT CAA CTG ATG GTG-3’; Marmoset MR1 REV; 5’-ACA GAA TTC TCA TCG ATC TGG TGT TGG-3’. PCR amplicons were separated on a 1% agarose gel, and then gel purified using a Qiagen minelute gel extraction kit (Qiagen; Germantown, MD).
MR1 plasmid construction.
The PCR amplified MR1 gene products for each individual were ligated into the pCRII-Blunt TOPO® vector using a Zero Blunt® TOPO® PCR Cloning Kit (Life Technologies; Carlsbad CA) according to the manufacturer’s protocol. Ligations were transformed into TOP10® OneShot Bacteria. DNA from six to eight individual colonies per sample were purified using the QIAprep Spin miniprep kit (Qiagen).
The MR1 allele variants that were identified were subcloned into the pTRACER-EF/V5-His A vector (Thermo Fisher; Waltham, MA) using EcoRI (NEB) and KpnI (NEB) cloning sites. Human MR1 (accession no. U22963.1) was synthesized (Genscript; Piscataway, NJ) and ligated into pTRACER-EF/V5-HisA vector using the EcoRI and KpnI cloning sites.
Transfection of MR1 plasmid vectors into 293T cells.
The pTRACER-EF/V5-HisA constructs expressing the different MR1 allele variants were stably expressed in 293T cells. Briefly, each plasmid was transfected into 293T cells using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. Stably transfected cells were selected with 300 μg/mL Zeocin. The pTRACER-EF/V5-HisA plasmid also encodes GFP, so stable expression could be identified by flow cytometry.
Surface expression of MR1 allele variants in 293T cells.
Stimulation of 293T cells stably transfected with human and NHP MR1 plasmid expression vectors was performed as previously described (Dias et al. 2016). Briefly, cells stably expressing MR1 were plated in 24-well plates at approximately 2e5 cells/well. Twenty-four hours later, cells were loaded with either 50 μM of Acetyl-6-Formylpterin (Ac-6-FP; Schircks Laboratories) or media control for 16 hours at 37°C. Alternatively, cells were incubated with 200 colony forming units (CFU) per one 293T cell of E.coli that was fixed for exactly three minutes with 2% paraformaldehyde for 24 hours. After incubation, cells were stained with LIVE/DEAD Near IR (Thermo Fisher Scientific) and an anti-MR1-PE antibody (clone 26.5 BD Biosciences; San Jose, CA) and fixed with 2% paraformaldehyde (PFA). Flow cytometry was performed on a BD LSR II (Becton Dickinson; Franklin Lakes, NJ), and the data were analyzed using FlowJo software for Macintosh (version 9.9.3).
MAIT cell activation assay.
A MAIT cell activation assay was performed using an adaptation of protocols previously described (Dias et al. 2016). Briefly, frozen peripheral blood mononuclear cells (PBMC) were thawed from previously collected blood samples from Mauritian cynomolgus macaques. The cells were incubated in RPMI media containing IL-7 and IL-2, as described above, for either 3 (i.e. unlicensed) or 24 hours to ‘license’ them, as previously described (Dias et al. 2016). MAIT cells were isolated from PBMC by staining cells with TCRVα7.2-PE (Table 1, BD biosciences) for 30 minutes, followed by purification using an anti-PE microbead kit according to the manufacturer’s protocol (Miltenyi Biotec; Auburn, CA). Isolated MAIT cells were then incubated with 293T target cells.
Table 1.
Staining panel for MAIT cells in MAIT activation assay.
| Antibody | Clone | Fluorochrome | Cat.No. | Company |
|---|---|---|---|---|
| CD3 | SP34–2 | AlexaFluor700 | 557917 | BD Biosciences |
| CD4 | SK3 | BV711 | 563020 | BD Biosciences |
| CD8 | RPA-T8 | Pacific Blue | 558207 | BD Biosciences |
| CD107a | H4A3 | BV605 | 328633 | Biolegend |
| IFNγ | B27 | BV510 | 563287 | BD Biosciences |
| TNFα | Mab11 | PerCP Cy5.5 | 502926 | BD Biosciences |
| TCRVα7.2 | 3C10 | PE | 351706 | Biolegend |
| LIVE/DEAD | ----- | Near InfraRed | L10119 | Thermo Fisher Scientific |
| Mamu MR1–5OPRU tetramer | ----- | APC | ---- | * |
The Mamu-MR1–5OPRU monomer was provided by the NIH tetramer Core Facility, Emory University, Atlanta, GA.
293T cells stably transfected with a plasmid expressing the MR1*01:01, MR1*01:02, MR1*01:03, and MR1*01:04 allele variants were split into 96-well plates at approximately 10,000 cells/well. Twenty-four hours later, cells were fed 0, 10, 30, 50, or 100 colony-forming units (CFU) of E.coli per 293T cell. The E.coli was fixed with 2% paraformaldehyde for three minutes and washed extensively with PBS to remove excess paraformaldehyde. After incubation for 24 hours with fixed E.coli, the licensed and unlicensed macaque MAIT cells were delivered to each well at a 1:1 effector:target ratio. Cells were incubated for 16 hours at 37°C in the presence of 2uM monensin (Biolegend; San Diego, CA), 5μg/mL Brefeldin A (Biolegend), and CD107a-BV605 (Table 1). Cells were then stained with the Mamu MR1–5OPRU tetramer (developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie, produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne (Corbett et al. 2014)) in R10+500nM Dasatinib (Thermo Fisher Scientific) for 1 hour. Cells were washed once with 1X Phosphate Buffered Solution (PBS; Thermo Fisher Scientific) and stained for live cells using the LIVE/DEAD NearIR staining kit (Thermo Fisher Scientific) at a 1:4000 dilution in 1XPBS. Cells were washed once with PBS containing 10%Fetal Bovine Serum and then stained with the CD3, CD4, and CD8 antibodies (Table 1). Cells were fixed in 2% paraformaldehyde for 20 minutes, then stained with IFNγ and TNFα (Table 1). Flow cytometry was performed on a BD LSR II (Becton Dickinson; Franklin Lakes, NJ), and the data were analyzed using FlowJo software for Macintosh (version 9.9.3).
MR1 sequencing analysis.
DNA was prepared for sequencing analysis using a minor adaptation of the Illumina Nextera® XT Library preparation kit (Illumina; San Diego, CA). Briefly, the kit was followed according to company protocols but 20% of the DNA input and reagent volumes were used. Library preparation products were purified using Agencourt AMPure XP beads (Beckman Coulter) at a 1:2 PCR product:bead ratio. Samples were then pooled, denatured, and sequenced on an Illumina Miseq® according to the manufacturer’s protocol.
Fastq reads were extracted from the data set, then initially mapped to either the Cynomolgus macaque MR1 mRNA reference sequence (accession number NM_001284968) or the Indian rhesus macaque MR1 mRNA reference sequence (accession number XM_015122468) in Geneious (Biomatters, Ltd.). From this analysis, a full-length contig was generated representing a macaque consensus MR1 nucleotide sequence. Reads spanning the macaque MR1 open reading frame sequence were extracted, translated, and unique amino acid sequences were predicted for each individual sequence compared to the translated macaque MR1 consensus sequence. For identification of MR1 alleles, 4–8 plasmids were sequenced per animal. A minimum of two plasmids for any given individual had to contain the same nucleotide sequence to be confirmed as an allele.
Results:
Identification of 21 distinct MR1 nucleotide sequences in nonhuman primates.
To analyze the sequence diversity of the MR1 gene amongst NHPs, we cloned and sequenced MR1 from five different NHP populations: Mauritian cynomolgus macaques (Macaca fascicularis; MCM), Asian cynomolgus macaques (Macaca fascicularis; ACM), Indian rhesus macaques (Macaca mulatta; RM), pigtailed macaques (Macaca nemestrina; PTM), and common marmosets (Callithrix jacchus). These species were chosen for the analysis of MR1 as they have been used frequently as model systems for human diseases, such as Human Immunodeficiency Virus (HIV) and Mycobacterium tuberculosis (Mtb) infection (Gardner and Luciw 2008; Kaushal et al. 2012; Letvin et al. 1983; London et al. 1983; Misra et al. 2013; Peña and Ho 2015; Vinton et al. 2016). For each sample, mRNA was reverse transcribed and then the MR1 cDNA was amplified with gene-specific primers. Amplified MR1 cDNA was cloned into the pCRII-Blunt TOPO® vector, and individual DNA clones were isolated and sequenced.
We initially mapped Fastq reads generated for each plasmid to the published cynomolgus macaque (accession number NM_001284968) and rhesus macaque MR1 (accession number XM_015122468) mRNA sequences to construct full-length contigs for MR1. From this analysis, we generated a macaque MR1 nucleotide consensus sequence. Each NHP MR1 sequence was then aligned to this nucleotide ‘Consensus,’ and MR1 alleles were identified and named relative to this sequence. The alleles were named in a format as follows, with three numbers appearing after MR1: MR1*0X:0X:0X. The first number after the asterisk (MR1*0X:0X:0X) indicated alleles in which there were nonsynonymous changes at sites that were previously shown to play a key role in the function of MR1. Because none of the alleles we identified have changes in these critical residues(Corbett et al. 2014; Eckle et al. 2014; Kjer-Nielsen et al. 2012; López-Sagaseta et al. 2013a; López-Sagaseta et al. 2013b; Reantragoon et al. 2012), all of the alleles were named MR1*01:0X:0X. The second number after the first colon (MR1*01:0X:0X), indicated nonsynonymous changes leading to alterations in amino acids in the MR1 protein outside known important residues. Finally, the third number after the second colon (MR1*01:0X:0X), indicated synonymous nucleotide changes relative to the parent allele. From this analysis, we identified 21 distinct MR1 nucleotide sequences across all five NHP populations (Table 2).
Table 2.
Nonhuman primate distinct MR1 nucleotide variants.
| Allele Name | Nonsynonymous nucleotide changes (Rel. to MR1*01:01:01 ‘Consensus’) |
Synonymous nucleotide changes (Rel. to MR1*01:01:01 ‘Consensus’) |
|---|---|---|
| MR1*01:01:01 | ---- | ---- |
| MR1*01:01:02 | G-->A (69) | |
| MR1*01:01:03 | T-->C (216); C-->T (888) | |
| MR1*01:01:04 | T-->C (216) | |
| MR1*01:01:05 | T-->C (363); C-->T (573) | |
| MR1*01:02:01 | C-->G (177) | ---- |
| MR1*01:02:02 | G-->A (774) | |
| MR1*01:02:03 | A-->G (144) | |
| MR1*01:02:04 | A-->G (144); C-->T (330); C-->T (888) | |
| MR1*01:02:05 | A-->G (144); C-->T (330) | |
| MR1*01:02:06 | C-->T (888) | |
| MR1*01:03:01 | CA-->TG (87–88) | ---- |
| MR1*01:03:02 | C-->T (888) | |
| MR1*01:04:01 | C-->T (221) | ---- |
| MR1*01:04:02 | T-->C (363) | |
| MR1*01:04:03 | T-->C (363); C-->T (573) | |
| MR1*01:05:01 | G-->A (25) | T-->C (363); C-->T (543) |
| MR1*01:06:01 | C-->T (17) | ---- |
| MR1*01:07:01 | A-->G (46); C-->G (177) | A-->G (69); C-->T (330) |
| MR1*01:08:01 | C-->A (676) | ---- |
| MR1*01:09:01 | G-->A (928) | T-->C (216); T-->C (363); T-->C (888); A-->C (964) |
Detection of distinct MR1 alleles within each NHP population.
We next characterized each of the MR1 nucleotide sequences within each NHP population. When a nucleotide sequence was detected in a given population, it was given a unique prefix based on the NHP population in which it was observed: Mauritian and Asian cynomolgus macaque (MCM and ACM) MR1 alleles were given the prefix Mafa- (Table 3), rhesus macaques (RM) were given the prefix Mamu- (Table 4), pigtailed macaques (PTM) were given the prefix Mane- (Table 5), and marmosets were given the prefix Caja- (Table 6). For all of the macaque populations, we observed 100% nucleotide sequence identity for alleles with the same name, but a different prefix (Tables 3–5). All marmoset MR1 alleles contained one synonymous nucleotide difference at position 999, with an adenine present, rather than a guanine when compared with the macaque MR1 nucleotide consensus sequence (Table 6). Overall, we identified a total of 32 species-specific MR1 alleles.
Table 3.
Cynomolgus macaque (Macaca fascicularis) MR1 alleles.
| Allele name | Non-synonymous nucleotide changes (rel. to consensus) | Synonymous nucleotide changes (rel. to consensus) | Accession number | No. of animals in which allele was detected | |
|---|---|---|---|---|---|
| MCM* | ACM** | ||||
| Mafa-MR1*01:01:01 | ---- | ---- | MH678588 | 8/15 | 4/10 |
| Mafa-MR1*01:01:02 | G-->A (69) | MH697408 | 1/15 | 2/10 | |
| Mafa-MR1*01:01:03 | T-->C (216); C-->T (888) | MH697409 | 0/15 | 2/10 | |
| Mafa-MR1*01:01:04 | T-->C (216) | MH697411 | 0/15 | 1/10 | |
| Mafa-MR1*01:02:01 | C-->G (177) | ---- | MH697414 | 5/15 | 2/10 |
| Mafa-MR1*01:02:02 | G-->A (774) | MH697416 | 8/15 | 1/10 | |
| Mafa-MR1*01:02:03 | A-->G (144) | MH697417 | 0/15 | 1/10^ | |
| Mafa-MR1*01:02:05 | A-->G (144); C-->T (330) | MH697419 | 0/15 | 1/10^ | |
| Mafa-MR1*01:02:06 | C-->T (888) | MH697421 | 0/15 | 1/10^ | |
| Mafa-MR1*01:03:01 | CA-->TG (87–88) | ---- | MH703787 | 0/15 | 1/10 |
| Mafa-MR1*01:04:03 | C-->T (221) | T-->C (363); C-->T (573) | MH703793 | 1/15 | 1/10 |
| Mafa-MR1*01:07:01 | A-->G (46); C-->G (177) | C-->T (330) | MH703797 | 0/15 | 1/10 |
| Mafa-MR1*01:09:01 | G-->A (928) | T-->C (216); T-->C (363); T-->C (888); A-- >C (964) | MH703799 | 0/15 | 1/10^ |
MCM; Mauritian cynomolgus macaque;
ACM; Asian cynomolgus macaque;
indicated allele was only observed in 2 plasmids
Table 4.
Rhesus macaque (Macaca mulatta) MR1 alleles.
| Allele name | Non-synonymous nucleotide changes (rel. to consensus) | Synonymous nucleotide changes (rel. to consensus) | Accession number | Number of RM* in which allele was detected |
|---|---|---|---|---|
| Mamu-MR1*01:01:01 | ---- | ---- | MH697406 | 1/10 |
| Mamu-MR1*01:01:03 | T-->C (216); C-->T (888) | MH697410 | 4/10 | |
| Mamu-MR1*01:02:01 | C-->G (177) | ---- | MH697415 | 2/10 |
| Mamu-MR1*01:03:01 | CA-->TG (87–88) | ---- | MH703788 | 6/10 |
| Mamu-MR1*01:06:01 | C-->T (17) | ---- | MH703796 | 1/10^ |
RM; Rhesus macaque;
indicated allele was only observed in 2 plasmids
Table 5.
Pigtailed macaque (Macaca nemestrina) MR1 alleles.
| Allele name | Non-synonymous nucleotide changes (rel. to consensus) | Synonymous nucleotide changes (rel. to consensus) | Accession number | Number of PTM* in which allele was detected |
|---|---|---|---|---|
| Mane-MR1*01:01:01 | ---- | ---- | MH697407 | 7/10 |
| Mane-MR1*01:01:05 | T-->C (363); C-->T (573) |
MH697412 | 1/10^ | |
| Mane-MR1*01:03:01 | CA-->TG (87–88) | ---- | MH703789 | 3/10 |
| Mane-MR1*01:04:03 | C-->T (221) | T-->C (363); C-->T (573) |
MH703794 | 2/10 |
| Mane-MR1*01:05:01 | G-->A (25) | T-->C (363); C-->T (543) |
MH703795 | 2/10 |
| Mane-MR1*01:08:01 | C-->A (676) | ---- | MH703798 | 1/10^ |
PTM; Pigtailed macaque;
the indicated allele was observed in only 2 plasmids
Table 6.
Common marmoset (Callithrix jacchus) MR1 alleles.
| Allele name | Non-synonymous nucleotide changes (rel. to consensus) | Synonymous nucleotide changes (rel. to consensus) | Accession number | Number of Marmosets in which allele was detected |
|---|---|---|---|---|
| Caja-MR1*01:01:01 | ---- | G-->A (999) | MH678587 | 1/8 |
| Caja-MR1*01:02:01 | C-->G (177) | G-->A (999) | MH697413 | 2/8 |
| Caja-MR1*01:02:04 | A-->G (144); C-->T (330); C-->T (888); G-->A (999) | MH697418 | 1/8^ | |
| Caja-MR1*01:03:01 | CA-->TG (87–88) | G-->A (999) | MH703786 | 1/8 |
| Caja-MR1*01:03:02 | C-->T (888); G-->A (999) | MH697420 | 1/8 | |
| Caja-MR1*01:04:01 | C-->T (221) | G-->A (999) | MH703790 | 1/8^ |
| Caja-MR1*01:04:02 | T-->C (363); G-->A (999) | MH703791 | 1/8 | |
| Caja-MR1*01:04:03 | T-->C (363); C-->T (573); G-->A (999) | MH703792 | 4/8 |
indicated allele was only observed in 2 plasmids
Each group of NHPs exhibited different frequencies of the identified MR1 alleles (Tables 3–6) among the animals we examined. To state that an MR1 allele was present in a given animal, it had to be identified in at least two of the plasmids that were sequenced for that animal. Anywhere from 5 alleles (RM; Table 4) to 13 alleles (ACM; Table 3) were found among each individual population of animals. We did not determine whether an animal was homozygous or heterozygous for different MR1 alleles, but rather, we focused simply on the existence of polymorphisms in the MR1 sequences. We found that across all of the macaque and marmoset populations examined, the MR1*01:01:01 allele that was identical to the MR1 nucleotide ‘Consensus’ was the most frequently observed. At least one copy of the MR1*01:01:01 nucleotide sequence was observed in 21 out of 53 total animals that were used in this analysis. Among each individual NHP population, the most frequently observed allele varied. For example, the MR1*01:01:01 sequence was the most frequently observed allele in the pigtailed macaque (7/10; Table 5), Mauritian (8/15; Table 3), and Asian cynomolgus macaque (4/10; Table 3) populations. The MR1*01:03:01 and MR1*01:04:03 sequences were the most frequently observed for rhesus macaque (6/10; Table 4) and marmoset (4/8; Table 6) populations, respectively. It is possible that these frequencies would be different if we had examined hundreds of animals across each species.
Overall, the striking similarities between marmoset and macaque MR1 alleles agree with the findings of others that the MR1 gene is highly conserved across species (Greene et al, 2017; Huang et al. 2009; Riegert et al. 1998; Tsukamoto et al. 2013; Yamaguchi et al. 1997). However, the fact that up to 13 unique MR1 alleles could be found within a small population of animals is indicative that MR1 may have greater genetic diversity than previously thought.
Nine MR1 alleles confer amino acid changes in NHP populations.
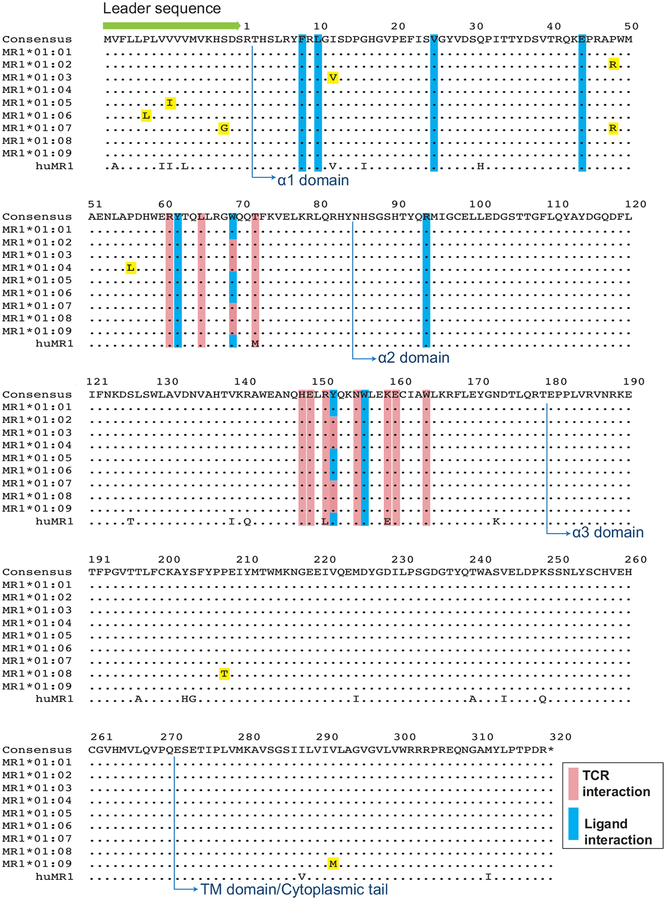
From the MR1 nucleotide ‘Consensus’ sequence, we determined the predicted amino acid translation. The most frequent amino acid residues that appeared globally at each position across all MR1 sequences are shown in Figure 1 and listed as the ‘Consensus’ (top line). We identified nine unique sequences with nonsynonymous differences (termed MR1*01:01 through MR1*01:09) that were observed in at least one of the five NHP species. MR1*01:01 was identical to the ‘Consensus’ macaque MR1 sequence at the amino acid level (Fig. 1). Three of the variants had changes in amino acid residues in the α1 domain of MR1 (I12V, P48R, and P56L). Three of the variants had amino acid changes in the leader sequence of the immature MR1 protein (S-3G/P48R, V-10I, and P-13L). One variant had an amino acid change in the α3 domain (P208T), and one variant had an amino acid change in the transmembrane domain/cytoplasmic tail of MR1 (V292M; Fig. 1). None of the identified variants had changes in residues that are known to affect the interaction of MR1 with the MAIT T cell receptor (Fig. 1, pink bars), or the ability of MR1 to present vitamin B or riboflavin derivatives (Fig. 1, blue bars) (Corbett et al. 2014; Eckle et al. 2014; Kjer-Nielsen et al. 2012; López-Sagaseta et al. 2013a; López-Sagaseta et al. 2013b; Reantragoon et al. 2012). We furthermore compared the newly identified macaque MR1 amino acid variant sequences to the human MR1 amino acid sequence (Fig. 1, bottom row; Accession number AAC50174.1). Two macaque MR1 allele variants (MR1*01:03 and MR1*01:05) contained amino acids identical to human MR1 at positions 12 (Valine; MR1*01:03) and −10 (Isoleucine; MR1*01:05). Amino acid differences in the T-cell receptor (TCR) interaction domain (pink bars) between human and macaque MR1 have been previously described in (Greene et al., 2017)
Fig. 1. Macaque MR1 consensus sequence and identified MR1 allele variants and comparison with human MR1.
The macaque MR1 consensus sequence (top row, letters) represents the most common amino acid found at each position for the MR1 variants detected in the study. Amino acid differences between each MR1 variant (termed MR1*01:01 through MR1*01:09) and the consensus are shown as letters highlighted in yellow. The bottom row indicates the human MR1 amino acid sequence (accession number AAC50174.1) and letters represent amino acid differences between the macaque MR1 consensus sequence and human MR1. Also shown are the leader sequence (green arrow), the alpha-1, 2, and 3 domains (blue arrows with text), and the transmembrane (TM) domain and cytoplasmic tail (blue arrows with text). Residues have been highlighted that have been shown previously to interact with MAIT T-cell receptor (TCR) residues (pink bars) or metabolite ligands (blue bars) (Corbett et al. 2014; Eckle et al. 2014; López-Sagaseta et al. 2013a; Reantragoon et al. 2012).
Expression and functional analysis of macaque MR1 allele variants in 293T cells.
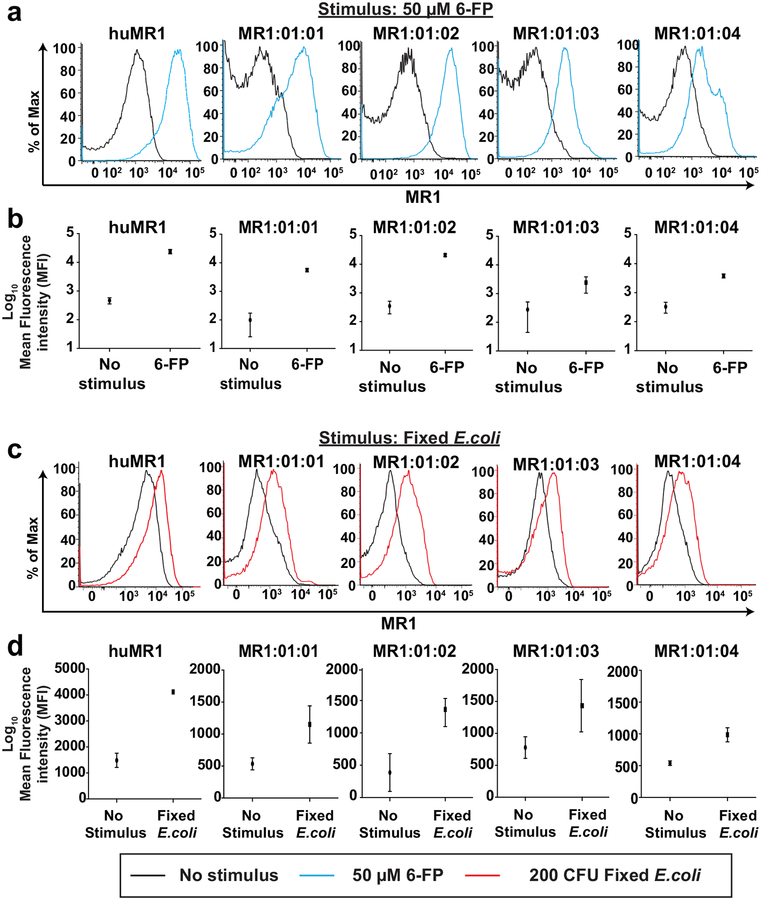
We wanted to determine if each MR1 variant could be presented on the cell surface. To do this, we subcloned the most commonly observed MR1 allele variants into the pTRACER-EF/V5-HisA vector, and then stably expressed them in 293T cells. We treated each 293T-MR1 cell line with 50 μM Acetyl-6-Formyl pterin (Ac-6-FP), which binds to MR1 and stabilizes it on the cell surface (Eckle et al. 2014). 293T cells expressing human MR1 were used as a positive control. MR1 surface expression, as measured by the mean fluorescence intensity (MFI) of the anti-MR1-PE antibody, increased by 1–3 logs relative to unstimulated controls for each macaque MR1 variant (Fig. 2a–b). The MR1*01:01 and MR1*01:02 variants exhibited the highest fold increase in surface expression (Fig. 2b). We additionally expressed the other five MR1 allele variants (MR1*01:05 through MR1*01:09) in 293T cells as well, and found that they all were detected on the cell surface at similar levels in this assay (data not shown).
Fig. 2. Macaque MR1 variants are able to present bacterial metabolites on the cell surface.
a, 293T cells stably expressing human MR1 (huMR1) and the four most common macaque MR1 variants were incubated with either media control (no stimulus, black lines in histograms) or 50μM 6-Acetylformylpterin (Ac-6-FP; blue lines in histograms) for 16 hours to measure upregulation in MR1 cell surface expression. Samples were stained for MR1 surface expression as indicated in methods and flow cytometric analysis was performed. Shown is a representative image of three independent experiments. b, The average mean fluorescence intensity (MFI) in MR1 surface expression across three experiments was determined for the experiment described in (a) for cell lines stimulated with media control or Ac-6-FP. c, The cell lines described in (a) were incubated for 24 hours with either media control (no stimulus, black lines) or 200 Colony forming units (CFU) per 293T cell of E.coli that was fixed for exactly three minutes with 2% paraformaldehyde (200 CFU E.coli, red lines). Flow cytometry was performed as described in (a). Shown is a representative image of three independent experiments. d, The average mean fluorescence intensity (MFI) in MR1 surface expression across three experiments was determined for the experiment described in (c) for cell lines stimulated with media control or 200 CFU E.coli
We next assessed the ability of each allele variant to process bacterial metabolites and present them on the cell surface. We fed paraformaldehyde-fixed E.coli (200 Colony Forming Units per 293T cell) to each of the 293T cells stably expressing macaque MR1 variants. Surface expression of MR1 was measured after 24 hours of stimulation. We detected increased MR1 surface expression in all cell lines fed E.coli compared to unstimulated controls (Fig. 2c), but these increases were markedly lower than when cells were treated with Ac-6-FP (Fig. 2b). Similar observations were made for 293T cells expressing the other MR1 allele variants (data not shown). These differences were most likely attributed to recycling of the MR1 molecules away from the cell surface during E.coli stimulation, which cannot occur during Ac-6-FP stimulation (Eckle et al. 2014; Harriff et al. 2016).
The four most common macaque MR1 allele variants can all stimulate cytokine production from macaque MAIT cells.
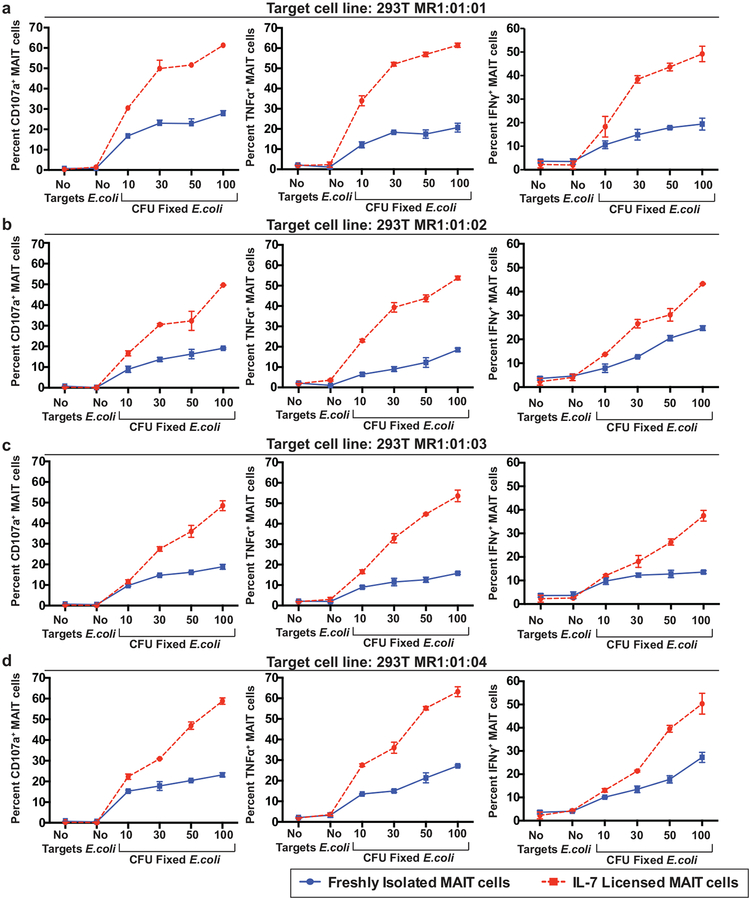
We wanted to determine if the 293T cells expressing the four most common macaque MR1 allele variants could be used to characterize the function of NHP MAIT cells. We fed the 293T cells expressing MR1*01:01 (Fig 3a), MR1*01:02 (Fig 3b), MR1*01:03 (Fig 3c), and MR1*01:04 (Fig 3d) increasing microbial doses of fixed E.coli for 24 hours, similar to assays previously designed to activate human MAIT cells in vitro (Dias et al. 2016). Effector MAIT cells were isolated from Mauritian cynomolgus Macaques using an anti-TCRVα7.2 antibody. We found approximately 97% of the MAIT cells isolated by this method were successfully stained with the MR1-tetramer provided by the NIH Tetramer Core Facility (data not shown). We either licensed the MAIT cells using IL-7 as previously described (Dias et al. 2016; Leeansyah et al. 2015) or used freshly isolated MAIT cells for this assay. The licensed (Fig. 3, red dashed lines) or unlicensed (Fig. 3, blue lines) MAIT cells were incubated with the target cell lines at a 1:1 effector to target ratio (Fig. 3a–d). We then measured expression of cytokines (IFN-γ and TNF-α) or degranulation (CD107a surface expression) in the MAIT cells. We found that all target cell lines alone with no E.coli induced minimal CD107a or cytokine production from macaque MAIT cells (Fig. 3a–d). In contrast, co-incubation of MAIT cells with target cells that were loaded with increasing doses of E.coli metabolites led to dose-dependent increases in production of CD107a, TNF-α, and IFN-γ in licensed, as well as unlicensed MAIT cells to a lesser degree (Fig. 3a–d). We did not detect large differences in cytokine or CD107a production from MAIT cells incubated with the different MR1 allele variants. Overall, we conclude that the four most common macaque MR1 allele variants can fairly equally present antigen and stimulate MAIT cell cytokine production in a microbial dose-dependent manner.
Fig. 3. 293T cell lines expressing the four most common macaque MR1 allele variants loaded with E.coli metabolites can activate macaque MAIT cells.
in a microbial dose-dependent manner a-d, 293T cells expressing MR1*01:01 (a); MR1:01:02 (b); MR1:01:03 (c); or MR1:01:04 (d) were incubated overnight with the indicated doses of fixed E.coli. The next day, the cells were mixed at a 1:1 ratio with macaque MAIT cells that were isolated as indicated in the methods and had been incubated with media containing 50ng/mL IL-7 overnight (licensed MAIT cells, red dashed lines) or freshly isolated (unlicensed MAIT cells, blue lines). After 5 hours, MAIT cells were stained as described in Table 1, and the expression of CD107a (left panels), TNFα (middle panels), and IFNγ (right panels) was determined by flow cytometric analysis. Shown is a representative image of three independent experiments.
DISCUSSION
Even though the MR1 molecule is structurally related to MHC class I molecules, it is much less genetically polymorphic. It is widely accepted that MR1 is an evolutionarily conserved molecule that presents vitamin B metabolites to MAIT cells (Huang et al. 2009; Riegert et al. 1998; Tsukamoto et al. 2013; Yamaguchi et al. 1997). Even so, there are differences between human and NHP MR1 consensus sequences at important residues that are known to contact the TCR on MAIT cells (Greene et al. 2017). These species-specific differences greatly affect the reagents that can be used to study MAIT cells in NHPs, but they also call into question whether additional MR1 polymorphisms within each species may impact antigen presentation, MAIT cell function and abundance, and the host immune response to bacterial pathogens. One example of this includes a recent description of an intronic MR1 polymorphism in humans that was associated with susceptibility to TB disease (Seshadri et al. 2017). In addition, multiple isoforms of human and mouse MR1 have been observed (Riegert et al. 1998). Therefore, it is not unreasonable to suppose that additional polymorphisms outside of solely the TCR contact sites or ligand binding sites can influence the host immune response.
In this study, we found 21 unique MR1 nucleotide sequences, which represented 32 MR1 alleles across five NHP populations. Predicted translation of these 21 unique nucleotide sequences was represented by nine different amino acid variants of MR1 (Fig. 1). None of the amino acid differences were located in known antigen binding or TCR contact sites (Corbett et al. 2014; López-Sagaseta et al. 2013a; López-Sagaseta et al. 2013a; Reantragoon et al. 2012). Some of the new alleles were present only at very low frequencies (Tables 3–6, indicated by the ^ symbol); further work should be performed in the future to validate these rarely observed alleles.
Even though the nucleotide differences that we observed in the MR1 alleles were not located in sites that are known to be important for MAIT cell function, it is possible these variants are linked to changes in introns that may affect the expression of MR1 molecules in an individual. For example, in a study of MR1 gene association with TB disease in human subjects, a correlation was found between a polymorphism in an intron in the coding region of MR1 and susceptibility to tuberculosis disease (Seshadri et al. 2017). This polymorphism was not known to directly affect antigen presentation or TCR interaction, but was associated with the ability of the MR1 molecule to be expressed within cells (Seshadri et al. 2017). Differential MR1 expression could impact the development of MAIT cells within each animal population, or affect the ability of MAIT cells from certain NHP populations to develop optimal responses to bacterial disease. Indeed, it has been observed that innate immune cells, such as iNKT cells, exhibit highly variable frequencies across NHP populations in the peripheral blood (Rout et al. 2012). Similarly, we have also observed greatly varying MAIT cell frequencies across the NHP populations we used in this study (data not shown). Studying how genetic MR1 variation impacts the development of MAIT cells could be of future interest.
The materials we generated in this study can also be a useful resource to study the ex-vivo activity of NHP MAIT cells. Given the differences between human and NHP MR1 molecules (Greene et al. 2017), it is unlikely that 293T cells expressing human MR1 will be sufficient for measuring the function of MAIT cells isolated from NHPs. To test the function of NHP MR1, we expressed four of the most commonly observed MR1 variants in 293T cells and we successfully loaded them with Ac-6-FP or E.coli metabolites. Each of the MR1 variants was detected on the cell surface compared to controls not loaded with ligand (Fig. 2). We then used these 293T-MR1 expressing cells lines loaded with paraformaldehyde-fixed E.coli to successfully stimulate MAIT cells isolated from macaques (Fig. 3), similar to an assay previously described for human MAIT cells (Dias et al. 2016). Overall, these macaque MR1-expressing cell lines will be valuable in future studies investigating changes in the function of NHP MAIT cells during different disease conditions, such as during SIV and/or M.tuberculosis infection.
There are relatively few ligands for which the crystal structure of the MR1 binding pocket has been determined and the ligand interaction residues have been resolved (Corbett et al. 2014; Eckle et al. 2014; López-Sagaseta et al. 2013b; Reantragoon et al. 2012). It is plausible that other currently unknown ligands may interact with a different composition of residues within or near the MR1 binding pocket, when compared to the ligand 5-OP-RU. Thus, the variants we detected could contribute to antigen presentation in ways that we do not yet understand. We identified a variant (MR1*01:02) that replaced a structurally rigid amino acid, proline (P), at position 48 with a basic amino acid, arginine (R). Basic residues (R9, K43, R94) comprise part of the binding pocket for MR1 for riboflavin-based ligands (Corbett et al. 2014; Eckle et al. 2014; Kjer-Nielsen et al. 2012; López-Sagaseta et al. 2013a). Furthermore, the lysine at position 43 has proven to be critical for riboflavin-based ligand interaction (Corbett et al. 2014; Eckle et al. 2014; Kjer-Nielsen et al. 2012; López-Sagaseta et al. 2013a). One caveat of our study as shown here is that we overexpressed all of these MR1 allele variants; this could abrogate subtle differences in antigen presentation that might affect their ability to present bacterial metabolites if expressed at endogenous levels. While we did not detect significant differences in antigen presentation of 293T cells expressing MR1*01:02 compared to other macaque MR1 variants using Ac-6-FP or fixed E.coli (Fig. 2), nor in the ability of the 293T MR1*01:02-expressing cell line to activate macaque MAIT cells using E.coli metabolites (Fig. 3); the close proximity of the P48R change to the ligand binding pocket may be relevant for interaction of K43 with yet-to-be identified MR1 ligands. It is also possible that while we did not observe any differences in the ability to present E.coli metabolites or Ac-6-FP on the cell surface in our 293T-overexpression systems (data not shown), the amino acid differences in the signal peptide of MR1 (S-3G, MR1*01:07; P-13L, MR1*01:06; V-10I, MR1*01:05) may alter intracellular trafficking, as the MR1 molecule exhibits a complex trafficking pathway through the endoplasmic reticulum and endosomal compartments (Harriff et al. 2016; Huang et al. 2008; McWilliam et al. 2016; McWilliam and Villadangos 2017). Together, the location of the observed amino acid variants within these newly identified MR1 variants relative to known MR1 sequences may warrant further investigation.
Overall, the MR1 allele variants that we identified in this study expand our knowledge of the intra-species diversity of the MR1 gene for NHPs used in biomedical research. One potential drawback from our study was that a subset of the NHP samples were obtained from animals that were a part of breeding colonies. While we took care to select animals that did not share a common parent, this did not eliminate the possibility that the animals were distant relatives. This consideration makes it even more remarkable that we detected so many unique alleles. It is entirely possible that wild macaque and marmoset populations may have even greater MR1 sequence diversity. Furthermore, it should thus be considered that MR1 may have more genetic diversity across the human population than previously appreciated.
Acknowledgements:
The research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459–01 and RR020141–01. We also thank members of the Wisconsin National Primate Research Center; a facility supported by grants P51RR000167 and P51OD011106. We thank Dane Gellerup and Katie Zarbock for technical support with sequencing experiments. We thank Julie Karl for help with the proper genetic nomenclature for the identified the MR1 alleles. We thank Sidonia Eckle and Erin Adams for their helpful comments and suggestions with regards to analyzing MR1 allele structure and function. We thank Dr. Stephen Kent (University of Melbourne; Melbourne, Australia) and Dr. JoAnne Flynn (University of Pittsburgh; Pittsburgh, Pennsylvania, USA) for providing frozen pigtailed macaque and Asian cynomolgus macaque PBMC samples, respectively. The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. This study was funded in part by the National Institutes of Health NIH R21AI127127–01, and also in part by an NIH NIAID contract HHSN272201600007C.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
Ethical Approval:
All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
References
- Awad W, Le Nours J, Kjer-Nielsen L, McCluskey J, and Rossjohn J (2018) Mucosal-associated invariant T cell receptor recognition of small molecules presented by MR1. Immunol Cell Biol [DOI] [PubMed] [Google Scholar]
- Corbett AJ et al. (2014) T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509: 361–365. [DOI] [PubMed] [Google Scholar]
- Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, and Geraghty DE (2004) Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14: 1501–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J, Sobkowiak MJ, Sandberg JK, and Leeansyah E (2016) Human MAIT-cell responses to Escherichia coli: activation, cytokine production, proliferation, and cytotoxicity. J Leukoc Biol 100: 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle SB et al. (2014) A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med 211: 1585–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis-Connell AL et al. (2018) ALT-803 Transiently Reduces Simian Immunodeficiency Virus Replication in the Absence of Antiretroviral Treatment. J Virol 92(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Shiina T, Anzai T, Sano K, Yamazaki M, Inoko H, and Tateno Y (2005) Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci U S A 102: 9230–9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MB, and Luciw PA (2008) Macaque models of human infectious disease. ILAR J 49: 220–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P, Radosavljevic M, Macquin C, and Bahram S (2011) The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol 48: 769–775. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, and Moody DB (2015) The burgeoning family of unconventional T cells. Nat Immunol 16: 1114–1123. [DOI] [PubMed] [Google Scholar]
- Gold MC et al. (2010) Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol 8: e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooneratne SL et al. (2014) Linking pig-tailed macaque major histocompatibility complex class I haplotypes and cytotoxic T lymphocyte escape mutations in simian immunodeficiency virus infection. J Virol 88: 14310–14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JM et al. (2017) MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol 10: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriff MJ, Karamooz E, Burr A, Grant WF, Canfield ET, Sorensen ML, Moita LF, and Lewinsohn DM (2016) Endosomal MR1 Trafficking Plays a Key Role in Presentation of Mycobacterium tuberculosis Ligands to MAIT Cells. PLoS Pathog 12: e1005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M et al. (2013). Acute-phase CD8 T cell responses that select for escape variants are needed to control live attenuated simian immunodeficiency virus. J Virol 87: 9353–9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Hirai M, and Kurosawa Y (1995) A gene outside the human MHC related to classical HLA class I genes. Science 269: 693–695. [DOI] [PubMed] [Google Scholar]
- Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, Fremont DH, and Hansen TH (2005) Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem 280: 21183–21193. [DOI] [PubMed] [Google Scholar]
- Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant AJ, Fremont DH, Lantz O, and Hansen TH (2008) MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med 205: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, Lantz O, and Hansen TH (2009) MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci U S A 106: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal D, Mehra S, Didier PJ, and Lackner AA (2012) The non-human primate model of tuberculosis. J Med Primatol 41: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L et al. (2012) MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491: 717–723. [DOI] [PubMed] [Google Scholar]
- Kurioka A et al. (2015) MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol 8: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L et al. (2013) MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog 9: e1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L et al. (2010) Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol 11: 701–708. [DOI] [PubMed] [Google Scholar]
- Leeansyah E, Svärd J, Dias J, Buggert M, Nyström J, Quigley MF, Moll M, Sönnerborg A, Nowak P, and Sandberg JK (2015) Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection. PLoS Pathog 11: e1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Aldrich WR, King NW, Blake BJ, Daniel MD, and Hunt RD (1983) Experimental transmission of macaque AIDS by means of inoculation of macaque lymphoma tissue. Lancet 2: 599–602. [DOI] [PubMed] [Google Scholar]
- London WT, Sever JL, Madden DL, Henrickson RV, Gravell M, Maul DH, Dalakas MC, Osborn KG, Houff SA, and Gardner MB (1983) Experimental transmission of simian acquired immunodeficiency syndrome (SAIDS) and Kaposi-like skin lesions. Lancet 2: 869–873. [DOI] [PubMed] [Google Scholar]
- López-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, Van Rhijn I, Saghatelian A, and Adams EJ (2013a) The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci U S A 110: E1771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sagaseta J, Dulberger CL, McFedries A, Cushman M, Saghatelian A, and Adams EJ (2013b) MAIT recognition of a stimulatory bacterial antigen bound to MR1. J Immunol 191: 5268–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam HE et al. (2016) The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat Immunol 17: 531–537. [DOI] [PubMed] [Google Scholar]
- McWilliam HEG, and Villadangos JA (2017) How MR1 Presents a Pathogen Metabolic Signature to Mucosal-Associated Invariant T (MAIT) Cells. Trends Immunol 38: 679–689. [DOI] [PubMed] [Google Scholar]
- Meierovics A, Yankelevich WJ, and Cowley SC (2013) MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci U S A 110: E3119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra A, Thippeshappa R, and Kimata JT (2013) Macaques as model hosts for studies of HIV-1 infection. Front Microbiol 4: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Cuadrado JF, Gómez del Moral M, García-Pavía P, Setién F, and Martínez-Naves E (2001) Characterization of the MHC class I-related MR1 locus in nonhuman primates. Immunogenetics 53: 643–648. [DOI] [PubMed] [Google Scholar]
- Parra-Cuadrado JF, Navarro P, Mirones I, Setién F, Oteo M, and Martínez-Naves E (2000) A study on the polymorphism of human MHC class I-related MR1 gene and identification of an MR1-like pseudogene. Tissue Antigens 56: 170–172. [DOI] [PubMed] [Google Scholar]
- Peña JC, and Ho WZ (2015) Monkey models of tuberculosis: lessons learned. Infect Immun 83: 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R et al. (2012) Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med 209: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert P, Wanner V, and Bahram S (1998) Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol 161: 4066–4077. [PubMed] [Google Scholar]
- Rout N, Greene J, Yue S, O’Connor D, Johnson RP, Else JG, Exley MA, and Kaur A (2012) Loss of effector and anti-inflammatory natural killer T lymphocyte function in pathogenic simian immunodeficiency virus infection. PLoS Pathog 8: e1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri C, Thuong NT, Mai NT, Bang ND, Chau TT, Lewinsohn DM, Thwaites GE, Dunstan SJ, and Hawn TR (2017) A polymorphism in human MR1 is associated with mRNA expression and susceptibility to tuberculosis. Genes Immun 18: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Blancher A, Inoko H, and Kulski JK (2017) Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 150: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MS et al. (2016) Vaccination with Live Attenuated Simian Immunodeficiency Virus (SIV) Protects from Mucosal, but Not Necessarily Intravenous, Challenge with a Minimally Heterologous SIV. J Virol 90: 5541–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MS et al. (2018) Acute-phase CD4+ T cell responses targeting invariant viral regions are associated with control of live-attenuated simian immunodeficiency virus. J Virol: In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, and Lantz O (2003) Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422: 164–169. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Deakin JE, Graves JA, and Hashimoto K (2013) Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics 65: 115–124. [DOI] [PubMed] [Google Scholar]
- Vinton C, Wu F, Rossjohn J, Matsuda K, McCluskey J, Hirsch V, Price DA, and Brenchley JM (2016) Mucosa-Associated Invariant T Cells Are Systemically Depleted in Simian Immunodeficiency Virus-Infected Rhesus Macaques. J Virol 90: 4520–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A et al. (2007) A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics 89: 402–412. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Hirai M, Kurosawa Y, and Hashimoto K (1997) A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem Biophys Res Commun 238: 697–702. [DOI] [PubMed] [Google Scholar]