Analysis of the responses in the epigenome and transcriptome in the phloem companion cells of Arabidopsis following a change in daylength identifies the flowering-time regulator MRF1.
Abstract
The phloem plays essential roles in the source-to-sink relationship and in long-distance communication, and thereby coordinates growth and development throughout the plant. Here we employed isolation of nuclei tagged in specific cell types coupled with low-input, high-throughput sequencing approaches to analyze the changes of the chromatin modifications H3K4me3 and H3K27me3 and their correlation with gene expression in the phloem companion cells (PCCs) of Arabidopsis(Arabidopsis thaliana) shoots in response to changes in photoperiod. We observed a positive correlation between changes in expression and H3K4me3 levels of genes that are involved in essential PCC functions, including regulation of metabolism, circadian rhythm, development, and epigenetic modifications. By contrast, changes in H3K27me3 signal appeared to contribute little to gene expression changes. These genomic data illustrate the complex gene-regulatory networks that integrate plant developmental and physiological processes in the PCCs. Emphasizing the importance of cell-specific analyses, we identified a previously uncharacterized MORN-motif repeat protein, MORN-MOTIF REPEAT PROTEIN REGULATING FLOWERING1 (MRF1), that was strongly up-regulated in the PCCs in response to inductive photoperiod. The mrf1 mutation delayed flowering, whereas MRF1 overexpression had the opposite effect, indicating that MRF1 acts as a floral promoter.
INTRODUCTION
As a consequence of their sessile life style, plants need to be able to respond to multiple endogenous signals and environmental cues, to ensure optimal growth and reproductive success (Andrés and Coupland, 2012; Cho et al., 2017). This requires efficient coordination between different tissues and organs within the plant. Communication in plants is accomplished by cell-to-cell signaling across the plasma membranes or through plasmodesmata, as well as by long-distance signaling via the vasculature, which consists of xylem and phloem tissues. The mature vessels of the xylem consist of dead cells, whereas the phloem consists of several cell types—phloem sieve elements (SE), phloem companion cells (PCCs), and supportive cells—that are all alive at maturity.
The PCCs in particular have been suggested to play an important role in coordinating physiological processes between the vasculature and other cell types in the leaf. This regulatory function of PCCs can be attributed at least in part to the circadian clock, which enables plants to anticipate dawn and activate the expression of photosynthesis-related genes before the start of the day. The phloem processes circadian rhythms that regulate the temporal and spatial distribution of metabolites and signaling molecules, and thus plays a central role in coordinating growth and developmental processes (Giakountis and Coupland, 2008; Endo et al., 2014; Shimizu et al., 2015). It has also been shown that the circadian clock and carbohydrate signaling feedback onto each other. For example, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and TIMING OF CAB EXPRESSION1 (TOC1), two key components of the central oscillator of the circadian clock, have been found to modulate photosynthetic activity and its metabolic outputs (Dodd et al., 2005; Graf et al., 2010). Sugars, in turn, are able to regulate the expression of the clock genes PSEUDO-RESPONSE REGULATOR7 (PRR7) and CCA1, and mediate long-termresponses of the circadian clock via GIGANTEA (GI; Dalchau et al., 2011), highlighting the tight link between the clock and carbohydrate metabolism in leaves. Importantly, circadian clocks in different cell types and tissues have long been thought to be uncoupled in plants, but recent evidence suggests that the clock in the vasculature also affects circadian clock regulation in other cell types in Arabidopsis (Arabidopsis thaliana) leaves (Endo et al., 2014; Shimizu et al., 2015).
In addition to its function in distributing photoassimilates throughout the plant (De Schepper et al., 2013), the phloem is instrumental in coordinating physiological and developmental processes in distant organs of the plant. The long-distance effects of the phloem can be explained through the transport of signaling molecules, such as RNA and proteins, from the leaves to the organizing centers in the shoot apical meristem (SAM) and root apical meristems. In the meristems, information about the leaf metabolic status is then integrated into the complex genetic networks that regulate growth and developmental processes (Deeken et al., 2008; Giakountis and Coupland, 2008). Probably the most prominent example of such a long-distance signaling process is the induction of flowering in response to changes in daylength (Turck et al., 2008; Song et al., 2015a). In Arabidopsis, inductive long-day (LD) signals are processed in leaves via light signaling and circadian clock networks, resulting in the stable expression of CONSTANS (CO) specifically at the end of LDs (Turck et al., 2008; Fornara et al., 2009). CO promotes flowering by activating the transcription of FLOWERING LOCUS T (FT) and its close homolog TWIN SISTER OF FT (TSF) in the PCCs of the leaf vasculature (Corbesier et al., 2007; Mathieu et al., 2007; Turck et al., 2008). The FT and TSF proteins then function as long-distance signals (florigens) and are transported through SEs to the SAM, where they interact with the basic Leu zipper transcription factor FD to induce flowering (Abe et al., 2005; Wigge et al., 2005; Turck et al., 2008). Export of the FT protein from PCCs to SEs is essential for FT function and is mediated by PCC-specific proteins FT-INTERACTING PROTEIN1 and ALTERED PHLOEM DEVELOPMENT (Liu et al., 2012; Abe et al., 2015). Interestingly, it has been shown that trehalose 6-phosphate (T6P) signaling, which is thought to inform the cell about carbohydrate availability, is strictly required for FT expression in PCCs (Wahl et al., 2013). The integration of information about carbohydrate availability and photoperiod in PCCs emphasizes the importance of this cell type as a signaling hub that integrates a wide range of metabolic and environmental signals to control systemic plant growth and development.
PCCs clearly play a central role in coordinating physiological and developmental processes in response to changes in photoperiod throughout the plant. However, we still do not understand the regulatory processes that control gene expression in this important cell type. One reason for this lack of understanding is that previous analyses have been limited to analyzing the transcriptome (Zhao et al., 2005; Brady et al., 2007; Deeken et al., 2008; Zhang et al., 2008a) and translatome (Mustroph et al., 2009) of PCCs, and the transcriptome and metabolites of the phloem sap (Deeken et al., 2008). Taken together, these studies revealed substantial differences between data sets of the same type generated from shoot PCCs, root PCCs, and phloem sap, but also between transcriptome and translatome data sets from the same tissue, which could in part be explained by mRNA and protein mobility in the phloem tissues (Giakountis and Coupland, 2008; Calderwood et al., 2016). While these PCC-specific analyses offered insights into some of the complex physiological and developmental processes that take place in PCCs, they mostly provided a rather static picture, as they did not investigate the contribution of chromatin modifications to the regulation of gene expression. This is unfortunate, as epigenetic factors such as histone modifications clearly play an important role in regulating transcriptional activities during plant growth and development (Pikaard and Mittelsten Scheid, 2014; Lämke and Bäurle, 2017).
Regulation at the epigenetic level has been shown to be an important factor controlling the expression of circadian clock and flowering-time genes in whole plants and in specific plant organs (Jiang et al., 2011; Yang et al., 2012; Seo and Mas, 2014; Jeong et al., 2015; You et al., 2017). The MYB transcription factors REVEILLE8 (RVE8) and CCA1, for example, control TOC1 expression by balancing H3 hyperacetylation level at the TOC1 promoter (Farinas and Mas, 2011). Similarly, the histone methyltransferase (HMT) SET DOMAIN GROUP2 (SDG2) has been shown to modulate H3K4me3 levels of core circadian clock genes such as CCA1, LATE ELONGATED HYPOCOTYL (LHY), TOC1, PRR7, PRR9, and LUX ARRHYTHMO (LUX; Malapeira et al., 2012; Seo and Mas, 2014). The H3K36 demethylase JUMONJI C DOMAIN-CONTAINING PROTEIN30 (JMJ30), which is repressed by the core circadian oscillators CCA1 and LHY, in turn promotes expression of CCA1 and LHY, presumably through demethylase activity (Jones et al., 2010; Lu et al., 2011). Furthermore, chromatin modifications, for example histone acetylation and histone H3 Lys-4 methylation, on clock genes have been shown to exhibit 24-h rhythmic oscillations that resemble the rhythmic expression of their respective transcripts, indicating a direct association between epigenetic and transcriptional events (Malapeira et al., 2012). The demethylase JMJ30 has also been implicated as a link between the circadian clock and flowering time, as it removes activating H3K36me2 marks from the FT promoter and thereby represses FT expression (Yan et al., 2014). FT expression is also repressed, under noninductive conditions, by deposition of H3K27me3 by Polycomb group (PcG) complex components such as SET DOMAIN GROUP1 (SDG1) on the FT promoter, gene body, and sometimes even the downstream region (Turck et al., 2007; Jiang et al., 2008; Bratzel and Turck, 2015). However, as these analyses have been conducted on complex tissues, it is not known if they accurately reflect the situation in PCCs. Related to this, it should be noted that PCC-specific expression of FT seems to be maintained independently from PcG repression and H3K27me3 (Farrona et al., 2011), but is strongly affected by H3K4me3 at the transcription start site (TSS; Jeong et al., 2009; Yang et al., 2010; Wang et al., 2014).
Here, we employed isolation of nuclei tagged in specific cell types (INTACT; Deal and Henikoff, 2010, 2011; You et al., 2017) to study the temporal dynamics of the transcriptome and of the chromatin modifications H3K4me3 and H3K27me3 in shoot PCCs in response to long-photoperiod signal. We describe the correlation between the two histone modifications and gene expression, depending on the presence of either one or both marks and their relative positions. Changes in H3K4me3 signals were significantly correlated with transcriptional responses, whereas changes in H3K27me3 were not always correlated with changes in gene expression. Emphasizing the importance of cell type-specific analyses, we identified MORN-MOTIF REPEAT PROTEIN REGULATING FLOWERING1 (MRF1), a previously uncharacterized MORN-motif repeat protein that was significantly up-regulated in the PCCs in response to LD and promoted flowering.
RESULTS
Establishment of INTACT for PCCs of Arabidopsis
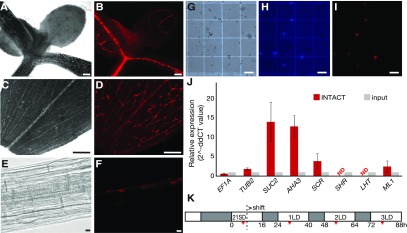
To investigate the epigenomic processes that regulate gene expression in shoot PCCs in response to changes in photoperiod, we prepared an INTACT reporter line using the promoter of the PCC marker gene SUCROSE-PROTON SYMPORTER2 (SUC2; Truernit and Sauer, 1995). The reporter construct is expressed in PCCs (Figures 1A to 1F), enabling affinity-based isolation of nuclei of this cell type from shoot tissues (Figures 1G to 1I). Reverse transcription quantitative PCR (RT-qPCR) demonstrated robust enrichment in the expression of the PCC marker genes SUC2 and ARABIDOPSIS THALIANA ARABIDOPSIS H+-ATPASE (AHA3; DeWitt and Sussman, 1995) in nuclei isolated by INTACT from leaf tissues of 24-d-old LD-grown reporter plants as compared to nuclei before purification (input; Figure 1J). By contrast, genes specifically expressed in xylem (SHORT ROOT, SHR; Cui et al., 2014), mesophyll cells (LYSINE HISTIDINE TRANSPORTER1, LHT1; Hirner et al., 2006), and the epidermis (MERISTEM LAYER1, ML1; Sessions et al., 2002), were not enriched after INTACT (Figure 1J). We did, however, detect weak enrichment of the bundle sheath (BS) cell-specific gene SCARECROW (SCR; Figure 1J; Cui et al., 2014), which may be due to SUC2 having a low level of expression in BS cells, as previously observed by ribosomal profiling (Aubry et al., 2014). Taken together, our results indicate that we have successfully employed INTACT to enrich nuclei from PCCs, possibly with a minor contamination from BS cells.
Figure 1.
Establishment of the INTACT System for the PCCs.
(A) to (B) Microscopy image of the shoot apex of a 7-d-old seedling of the PCC-tagged INTACT line. The shoot apex area was observed (A) in bright field and (B) by mCherry fluorescence. Scale bar = 100 μm.
(C) to (D) Microscopy image of a leaf of the PCC-tagged INTACT line. (C) Leaf area observed in bright field with the main vasculature appearing brighter than mesophyll cells. (D) The RedNTF-tagged PCC nuclei were detected as discontinuous red dots by mCherry fluorescence. Scale bar = 100 μm.
(E) to (F) Microscopy image of root area of a 7-d-old seedling of the PCC-tagged INTACT line. (E) The primary root area was observed in bright field. (F) The nuclear envelope-labeled PCC nuclei were detected by mCherry fluorescence. Scale bar = 10 μm.
(G) to (I) Microscopy images of nuclei purified from the INTACT reporter line. (G) Magnetic beads appear as white spheres in bright field, (H) DNA stained with DAPI is shown in blue, and (I) nuclear envelope domains tagged with RedNTF are detected by mCherry fluorescence. Scale bar = 100 μm.
(J) Relative expression of EF1A, TUB2, SUC2, AHA3, SCR, SHR, LHT1, and ML1 in INTACT-purified PCC-specific nuclei (INTACT) relative to their expression in the input crude nuclei (input) isolated from PCC INTACT reporter lines. Expression of EF1A and TUB2 were used as controls. Error bars = se of the mean values of three biological replications.
(K) Experimental design. Seeds of the INTACT reporter line were geminated on soil and continually grown in SD condition for 21 d and transferred to three consecutive LDs to induce flowering. Light and dark hours are indicated in white and gray, respectively. The time of sample collection (ZT 6 to 7) is indicated with a red bar.
To study the events in PCCs in response to changes in photoperiod, 21-d-old short-day (SD)-grown plants of the PCC INTACT reporter line were shifted to LD conditions. This treatment has previously been shown to result in floral commitment within 2 to 3 d after the shift, as indicated by the expression of early floral homeotic genes such as APETALA1 (AP1; You et al., 2017). Samples were collected 0, 1, 2, and 3 d after the shift to LD at Zeitgeber (ZT) 6 to 7 (Figure 1K). Nuclei were isolated by INTACT as previously in You et al. (2017) and used to study the dynamic changes in histone H3K4me3 and H3K27me3 methylation and gene expression.
RNA sequencing Confirms Enrichment of PCC-Specific Transcripts in INTACT-Purified Nuclei
RNA sequencing (RNA-seq) was performed on the nuclear RNA pool isolated by INTACT from PCCs, before and at 1, 2, and 3 d after the shift to LD (Figure 2). Bioinformatic analyses revealed that the transcriptome data were highly reproducible among the three biological replicates for each time point (Supplemental Figure 1A; Supplemental Data Set 1). The PCC marker genes SUC2 and AHA3 were stably detected in all samples (Figure 2A). By contrast, the marker genes of other tissues, for example JAGGED (JAG), which is expressed in developing leaves (Tsukaya, 2013), the early floral homeotic gene AP1 (Mandel et al., 1992; Schmid et al., 2003), the shoot meristem marker gene SHOOTMERISTEMLESS (STM; Long et al., 1996), the xylem-specific genes SHR and XYLEM CYSTEINE PEPTIDASE1 (XCP1; Funk et al., 2002), and the mesophyll cell-specific gene LHT1, were almost undetectable (Figure 2A). By contrast, ML1 and the BS marker gene SCR were detected at low levels in our RNA-seq data (Figure 2A). Together, these transcriptome-wide results verified the initial RT-qPCR analyses (Figure 1J) and confirmed that the nuclear RNA isolated by INTACT was strongly enriched for mRNAs from PCCs.
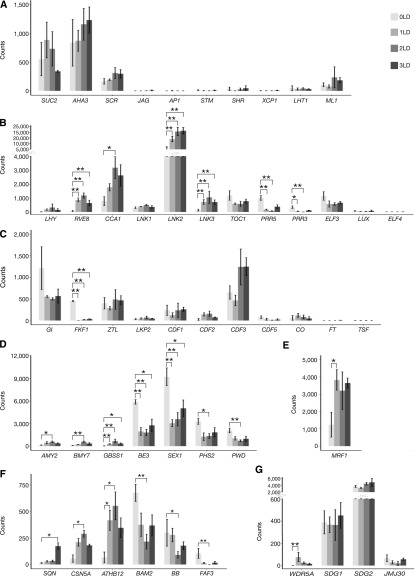
Figure 2.
Differential Gene Expression in the PCCs during LD Induction of Flowering.
Expression is given as count of sequencing reads. Gray gradient colors indicate the time points before (lighter gray) and 1 d (light gray), 2 d (dark gray), and 3 d (darker gray) after the shift to LD, respectively. *P < 0.05, **P < 0.01 (Deseq2 P values corrected for multiple testing using the Benjamini-Hochberg method).
(A) Expression of marker genes for the PCCs (SUC2 and AHA3), bundle sheath (SCR), leaf primordia (JAG), flower primordia (AP1), meristem (STM), xylem (SHR and XCP1), mesophyll cells (LHT1), and epidermis (ML1).
(B) Expression of marker genes for circadian clock, LHY, RVE8, CCA1, LNK1, LNK2, LNK3, TOC1, PRR5, PRR3, ELF3, LUX, and ELF4.
(C) Expression of marker genes for photoperiod flowering pathway, GI, FKF1, ZEITLUPE, LOV KELCH PROTEIN2, CDF1, CDF2, CDF3, CDF5, CO, FT, and TSF.
(D) Expression of important genes involved in carbohydrate metabolism, α-AMYLASE2, β-AMYLASE7, GBSS1, BE3, SEX1, PHS2, and PHOSPHOGLUCAN WATER DIKINASE.
(E) Expression of MRF1.
(F) Expression of selected important genes involved in development, SQUINT, CSN5A, ATHB12, BAM2, BB, and FAF3.
(G) Expression of selected epigenetic modifiers involved in regulation of circadian clock and photoperiodic flowering time, WDR5A, SDG1, SDG2, and JMJ30.
Differential Gene Expression in the PCCs in Response to Photoperiod Changes
To monitor the dynamics of the PCC transcriptome in response to the shift from SD to LD, we performed pairwise comparisons between the four time points. Overall, we detected 356 significantly differentially expressed genes (DEGs), including 200 and 144 genes that were consistently up- or down-regulated, respectively, as well as 12 genes that exhibited more complex expression patterns (Supplemental Data Set 2). Expression of many of the genes that form the central oscillator of the circadian clock in Arabidopsis responded rapidly to the shift from SD to LD (Figure 2B), which most likely reflected a change in phase rather than a change in amplitude (Michael et al., 2008). The transcription factor RVE8, a major clock component that positively regulates the expression of a large fraction of the clock genes(Fogelmark and Troein, 2014), was significantly induced 24 h after the shift to LD (Figure 2B). Similarly, expression of the morning gene CCA1 was significantly increased after shifting to LD, whereas changes in expression of LHY, while following the same general trend as CCA1, were not statistically significant (Figure 2B). In addition, two NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED (LNK) genes, LNK2 and LNK3, which are involved in light regulation of gene expression to control circadian rhythms and photomorphogenic growth responses (Rugnone et al., 2013), were significantly up-regulated (Figure 2B). By contrast, expression of PRR3 and PRR5, which function as repressors of the morning-expressed genes(Fogelmark and Troein, 2014), was significantly down-regulated, and expression of TOC1 followed a similar decreasing trend (Figure 2B). Expression of LUX and EARLY FLOWERING4 (ELF4), which encode components of the evening complex(Fogelmark and Troein, 2014), was almost undetectable in the PCCs, while expression of ELF3 displayed a decreasing trend after shifting to LD, but the expression changes were not statistically significant after correction for multiple testing (Figure 2B).
Next, we examined the expression of the core genes that regulate flowering in response to photoperiod (Figure 2C). Of the genes that make up the photoperiod-dependent flowering pathway, only FLAVIN BINDING KELCH REPEAT F-BOX1 (FKF1) was significantly down-regulated in response to the shift to LD. This finding is in agreement with a previous report (Sawa et al., 2007), which had shown that the expression of FKF1 shifts from the end of SDs (ZT8) to the end of LDs (ZT16) to form a stable complex with GI to promote flowering. The decrease of FKF1 expression in our data set therefore most likely reflects a phase change rather than a change in amplitude. Expression of GI also decreased, but these changes were not statistically significant after correction for multiple testing (Figure 2C). By contrast, the flowering time genes ZEITLUPE, LOV KELCH PROTEIN2, CYCLING DOF FACTOR1 (CDF1), CDF2, and CDF5 did not exhibit any substantial changes in expression after the transfer to LD. Expression of CDF3 showed a similar trend and was increased 2 d and 3 d after the shift to LD but this induction was not statistically significant. Expression of CO was weakly detected without significant change after shifting to LD and expression of the downstream genes FT and TSF was almost undetectable, most likely because the expression of these genes is under diurnal control and was at a minimum at the sampling time (ZT 6 to 7). Significant up-regulation of FT expression usually occurs at the end of the LD (ZT 16; Corbesier et al., 2007). Nevertheless, we were able to detect weak FT expression 3 d after shifting to LD, even at ZT 6 to 7.
Clock components such as CCA1, LHY, PRR5, PRR7, and PRR9, which change in expression in PCCs in response to LD (see above) and diurnal regulation, contribute to metabolic regulation in Arabidopsis (Fukushima et al., 2009; Graf et al., 2010). For example, it has been estimated that ∼30% of primary metabolites and genes encoding enzymes controlling these biosynthetic pathways are under circadian control (Espinoza et al., 2010). In agreement with this, we detected significant expression changes for genes that encode enzymes regulating carbohydrate and starch metabolism, such as α-AMYLASE2, β- AMYLASE7, GRANULE BOUND STARCH SYNTHASE1 (GBSS1), BRANCHING ENZYME3 (BE3), STARCH EXCESS1 (SEX1), α-GLUCAN PHOSPHORYLASE2 (PHS2), and PHOSPHOGLUCAN WATER DIKINASE in PCCs (Figure 2D; Streb and Zeeman, 2012; Feike et al., 2016). In particular, we detect down-regulation of SEX1 and PHS2 mRNA levels at ZT 6 to 7 following the shift to LD, which is in agreement with previous findings (Lu et al., 2005). Furthermore, mutants in SEX1, which encodes an α-glucan water dikinase required for starch degradation, exhibit a late-flowering phenotype (Ortiz-Marchena et al., 2015). In addition, we detected significant changes in expression of genes encoding enzymes involved in energy metabolism, such as the nicotinamide adenine dinucleotide (NAD), uridine diphosphate (UDP), and acetyl-coenzyme A (acetyl-CoA) metabolisms (Table 1; Supplemental Data Set 2).
Table 1. Significantly differentially expressed enzymes catalyzing intermediary energy metabolites after shifting to LD.
| ID | Gene | Expression Change | Annotation |
|---|---|---|---|
| NAD-related | |||
| AT1G20020 | ATLFNR2 | Dec | Ferredoxin-NADP(+)-oxidoreductase 2 |
| AT1G60730 | — | Inc | NAD(P)-linked oxidoreductase superfamily protein |
| AT2G37760 | AKR4C8 | Inc | NAD(P)-linked oxidoreductase superfamily protein |
| UDP-related | |||
| AT1G22370 | AtUGT85A5 | Dec | UDP-glucosyl transferase 85A5 |
| AT1G05680 | UGT74E2 | Inc | UDP-glucosyltransferase, UGT74E2 |
| AT1G32900 | GBSS1 | Inc | UDP-glycosyltransferase superfamily protein |
| AT3G11340 | F11B9.23 | Inc | UDP-glycosyltransferase superfamily protein |
| AT5G05870 | UG766C1 | Inc | UDP-glucosyl transferase 76C1 |
| Acyl-CoA–related | |||
| AT1G03150 | — | Inc | Acyl-CoA n-acyltransferases superfamily protein |
| AT5G65110 | ACX2 | Inc | acyl-CoA oxidase 2 |
Dec, only decreasing during the experiment; Inc, only increasing during the experiment.
Interestingly, we also detected expression changes of several developmentally important genes involved in meristem or floral development in the PCCs (Figure 2F). Significantly decreased expression was detected for BARELY ANY MERISTEM2 (BAM2; Figure 2F), which encodes a CLAVATA (CLV1)-related receptor kinase-like protein, and together with two other BAM genes is required for both shoot and floral meristem function and flowering-time control (DeYoung et al., 2006). Similarly, the E3 ubiquitin ligase BIG BROTHER, which acts as a central negative regulator of Arabidopsis floral organ size and is known to be expressed in proliferating tissues such as shoot and floral meristems and young organs, as well as in the vasculature (Disch et al., 2006), had significantly decreased expression in PCCs (Figure 2F). In addition, expression of FANTASTIC FOUR3 (FAF3), which has previously been shown to be expressed in the vasculature and down-regulated after shifting to LD (Wahl et al., 2010), was reduced in our PCC data set (Figure 2F). FAF3, together with three other FAF proteins, has been implicated in the regulation of shoot meristem size, potentially by repressing WUSCHEL and modulating the WUSCHEL-CLV feedback loop (Wahl et al., 2010). By contrast, expression of SQUINT, which promotes stem cell homeostasis and floral meristem termination through AP2 and CLV signaling (Prunet et al., 2015), was significantly up-regulated in PCCs, as was expression of COP9 SIGNALOSOME5A (CSN5A), which participates in floral organ morphogenesis (Wang et al., 2003; Gusmaroli et al., 2007), and ARABIDOPSIS THALIANA HOMEOBOX12 (ATHB12), which regulates the growth of the inflorescence stem by modulating the expression of GIBBERELLIN 20-OXIDASE1 and GA9 levels (Figure 2F; Son et al., 2010).
We also examined the expression of several chromatin regulators that are known to regulate circadian clock function and/or flowering time (Jiang et al., 2009, 2011; Yang et al., 2010, 2012, 2016; Malapeira et al., 2012; Gu et al., 2013; Wang et al., 2014; Jeong et al., 2015). For example, we detected significant up-regulation of WDR5A, which encodes a core component of the COMplex of Proteins Associated with Set1 (COMPASS)-like histone methylase complex, in PCCs in response to the shift to LD (Figure 2G). WDR5A is preferentially expressed in shoot and root apical regions and in vasculature tissues (Jiang et al., 2009, 2011) where it promotes expression of the potent floral repressor FLOWERING LOCUS C (FLC; Jiang et al., 2009, 2011). However, FLC expression was not changed in our data (Supplemental Data Set 1); thus, increased expression of WDR5A does not appear to affect FLC in PCCs in response to LD. In contrast to WDR5A, the induction of SDG2, which has been suggested to contribute to the regulation of core circadian clock genes by modulating H3K4me3 accumulation and binding of repressors, was not statistically significant after correction for multiple testing (Figure 2G). Similarly, the expression of JMJ30, which represses CCA1, LHY, and FT expression via H3K36me2 demethylation activity (Jones et al., 2010; Yan et al., 2014), and of SDG1, which encodes a core component of the PcG complex that deposits the H3K27me3 repressive mark at the FT locus (Bratzel and Turck, 2015), did not change throughout the experiment (Figure 2G).
In summary, our transcriptomic analyses confirm that INTACT can be employed to successfully capture gene expression changes in PCCs in response to changes in photoperiod. Genes that responded to the shift from SD to LD include important regulators of circadian rhythm, development, metabolism, and chromatin remodeling. In agreement with the individual genes discussed above, gene ontology (GO) enrichment analysis showed that the GO terms in biological process response to external stimulus (GO:0010033), circadian rhythm (GO:0007623), and starch metabolic/catabolic-related terms (GO: 0,005,982 and GO: 0,005,983) were significantly overrepresented among the genes that changed significantly in their expression (Supplemental Data Set 3).
PCC-Specific H3K4me3 and H3K27me3 Profiles
Dynamic changes in gene expression as described above are in part the result of transcriptional regulation at the chromatin level. H3K4me3 and H3K27me3 are canonical epigenetic marks that have been implicated in promoting and repressing gene expression, respectively (Li et al., 2007; Ha et al., 2011). To better understand the regulation of transcription in PCCs in response to changes in photoperiod, we profiled the genome-wide distribution of H3K4me3 and H3K27me3 using the chromatin immunoprecipitation sequencing (ChIP-seq) method. The ChIP-seq data produced using nuclei isolated from PCCs at the four time points in two biological replications showed good replicability (Supplemental Figure 1B).
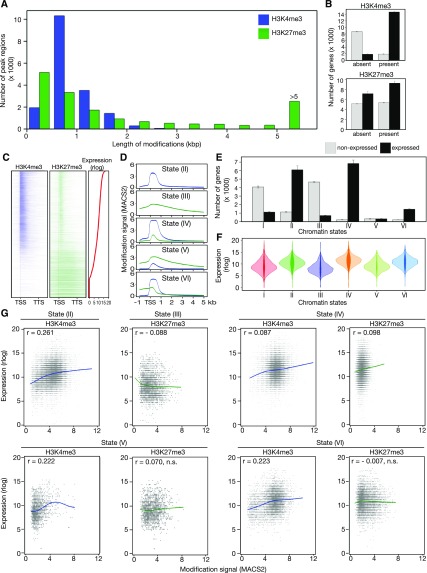
At the four time points analyzed, we detected from 16,712 to 17,462 regions in the genome that carried H3K4me3 marks in the PCCs. Overall, 63.4% (17,248) of all protein coding genes annotated in Arabidopsis (TAIR10 ver. 24) were marked with H3K4me3 at one or more time points. The length of the H3K4me3-marked regions varied between 134 basepair (bp) and 4,950 bp (Figure 3A; Supplemental Figure 2A; Supplemental Data Set 4). In agreement with the role of H3K4me3 as an activating mark of transcription, most genes (on average over time points: 88.7% ± 0.3%; “±” refers to se, n = 4) that carried this mark were expressed in PCCs (fragments per kilobase of transcript per million mapped reads, FPKM > 1). By contrast, only 17.3% ± 0.6% of genes without H3K4me3 on their gene body were found to be expressed in our data set (Figures 3B and 3C). The majority of these regions intersected with genes (97.21% ± 0.04%), and most intersections contained the TSS (75.26% ± 0.34%; Supplemental Figure 2B; Supplemental Data Set 4).
Figure 3.
Correlation of H3K4me3 and H3K27me3 Mark Signals with Gene Expression.
(A) Distribution of length of H3K4me3 and H3K27me3 marks at 0 LD. Blue and green indicate H3K4me3 and H3K27me3, respectively.
(B) Numbers of not-expressed (light gray) and expressed (black) protein coding genes in classes without/with H3K4me3/H3K27me3 marks.
(C) Location of H3K4me3 and H3K27me3 modifications on protein-coding genes in the PCCs at 0 LD. Expressed genes were sorted by expression level (rlog). Genes with no detectable expression were sorted by H3K27me3 signal (H3K27me3/H3 fold change) levels. Blue and green colors indicate H3K4me3 and H3K27me3, respectively.
(D) Mean H3K4me3 and H3K27me3 signals on protein coding genes with division into classes of chromatin states. Blue and green indicate H3K4me3 and H3K27me3, respectively.
(E) Numbers of not-expressed/expressed genes in classes with various chromatin states. Light gray and black indicate not-expressed and expressed protein-coding genes, respectively.
(F) Distributions of gene expression in chromatin state classes.
(G) Correlation between expression signal and modification signal (H3K4me3/H3 and H3K27me3/H3 fold changes) in classes of chromatin states. Significance of r, the rank correlation coefficient, declared at P < 0.001.
Over the four time points analyzed, H3K27me3 was found on 11,507 to 13,919 genes, and the length of the marked regions varied from 172 to 30,724 bp (Figure 3A). Overall, 65.7% (17,879) of the protein-coding genes were marked with H3K27me3 at least at one time point, which is slightly more than for H3K4me3. Most regions intersected with genes (92.77% ± 0.18%), with about half of the intersections containing the TSS (49.29% ± 0.18%; Supplemental Figure 2B; Supplemental Data Set 5). On average over the four time points, the fraction of expressed genes (FPKM > 1) with or without H3K27me3 on their body was 63.3% (±0.9%) and 57.9% (±1.8%), respectively (Figures 3B and 3C). In agreement with our previous study on epigenetic marks at the SAM (You et al., 2017), we observed broad H3K27me3 regions on 51.0% ± 0.6% of transcriptionally silent genes (FPKM < 1), but narrower H3K27me3 peaks on 56.4% ± 2.7% of expressed genes in PCCs (Supplemental Figure 2B).
To characterize the epigenetic landscape of PCCs in more detail, we distinguished six chromatin modification states of protein-coding genes based on the presence and absence of H3K4me3 and H3K27me3 and the overlap of the two marks, as described in You et al. (2017); Supplemental Data Set 6). On average over the four time points, we observed that (I) 19.1% (±0.5%) of protein coding genes had neither H3K4me3 nor H3K27me3 marks (“None” state); (II) 26.6 (±1.8%) had H3K4me3 only; (III) 19.8 (±0.2%) had H3K27me3 only; (IV) 26.0% (±1.6%) exhibited an “embedded” state (E-state) with narrow H3K27me3 marks within broader H3K4me3 domains; (V) 2.4% (±0.2%) exhibited a “harboring” state (H-state) with opposite characteristics to E-state, i.e. broad H3K27me3 regions containing narrow H3K4me3 peaks; and (VI) 6.2% (±0.2%) showed partial overlap of H3K4me3 and H3K27me3 on the gene body (“Other” state). We found that overlapping H3K4me3 and H3K27me3 signals, averaged over categories (IV) to (VI), were most prominent between the TSS (included) and 1,000 bases downstream of the TSS (TSS + 1000 nucleotides; Figure 3D).
Correlations between H3K4me3 and H3K27me3 Modification and Gene Expression in PCCs
We first examined the fractions of expressed and nonexpressed genes within the six chromatin states defined above (Figure 3E). On average over the four time points, the state I genes lacking both H3K4me3 and H3K27me3 were mostly (78.4% ± 0.7%) not expressed (FPKM < 1), and the fraction increased even further (86.7% ± 0.6%) among the state III genes that carried only H3K27me3 on their gene body. By contrast, most (84.2% ± 0.7%) of the state II genes with only H3K4me3 were expressed (FPKM >1). Interestingly, compared with state II genes, the fraction of expressed genes increased to 97% ± 0.1% among the state IV genes, which is defined as E-state, displaying narrow peaks of the supposedly repressive H3K27me3 mark within broader H3K4me3 regions. By contrast, partially overlapping H3K4me3 and H3K27me3 marks, state VI, did not change the fraction of expressed genes (87.3% ≥ 0.6%) in comparison to state II, whereas the presence of broad H3K27me3 regions with narrow H3K4me3 peaks in state V (H-state) strongly decreased the likelihood of expression (50.8% ± 1.2%).
Among the expressed genes (Figure 3F), there were significant differences between different states with respect to mean expression (analysis of variance P value < 0.001), and the E-state (state IV) was associated with the highest expression level, followed by H3K4me3 only (state II), and partial overlapping of H3K4me3 and H3K27me3 (state VI). The H-state (state V) considerably reduced the mean expression level, and the genes covered with broad H3K27me3 regions alone (state III) exhibited the lowest expression. For expressed genes, the H3K4me3 signals were always (in states III to VI) significantly positively correlated with gene expression levels (Figure 3G). Interestingly, H3K27me3 signal was only negatively correlated with expression when H3K27me3 alone was present on the gene body (state III), and we found a significant positive correlation between H3K27me3 signal and expression in the group of genes with E-state (state IV, Figure 3G).
Correlations between Chromatin Modification and Differential Gene Expression Changes
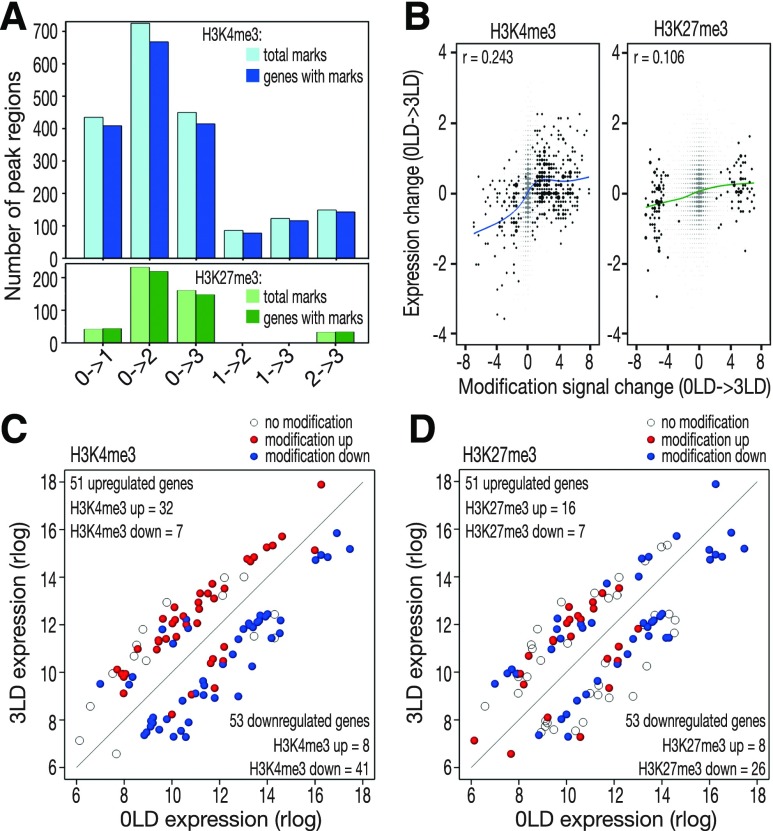
We next investigated the correlation between changes in histone modifications and differential gene expression (Figure 4). Significant changes in H3K4me3 and H3K27me3 modification signals occurred mostly between 0 LD and other time points (Figure 4A; Supplemental Data Set 7 and 8). The signal change for H3K4me3 was positively correlated with expression change in all the six pairwise comparisons (Figure 4B; Supplemental Figure 3A), whereas the change in H3K27me3 was weakly positively correlated with expression change for comparisons of 0 LD with the rest of time points and the comparison of 1 LD to 3 LD (Figure 4B; Supplemental Figure 3B). For genes that were differentially expressed between 0 LD and 3 LD, there was a considerable agreement between the direction of expression change and direction of H3K4me3 signal change (Figure 4C), but not between expression change and H3K27me3 signal change (Figure 4D).
Figure 4.
Correlation between Changes in H3K4me3 and H3K27me3 Marks with Gene Expression.
(A) Total numbers of H3K4me3 (light blue) and H3K27me3 (light green) marks with signal changing significantly in pair-wise comparisons between time points. The darker colors indicate the numbers of genes intersecting with the two histone marks, respectively.
(B) Change of expression signal versus modification signal for genes in comparison between 0 LD and 3 LD. r, the rank correlation coefficients, significant at P < 0.001. Dark points represent genes with significant modification signal change.
(C) Changes of H3K4me3 signal on 104 genes differentially expressed between 0 LD and 3 LD.
(D) Changes of H3K27me3 signal on 104 genes differentially expressed between 0 LD and 3 LD.
For (C) and (D), open circles mark DEGs without modification marks and filled circles indicate DEGs with changes in modification signal between 0 LD and 3 LD. Red, increasing modification signal; blue, decreasing modification signal.
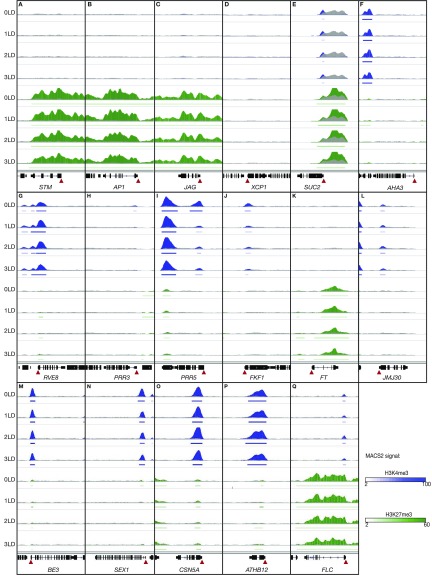
Finally, we examined the H3K4me3 and H3K27me3 modification patterns and their dynamics at selected genomic loci in PCCs (Figure 5). STM, AP1, and JAG displayed strong and even H3K27me3 across their entire promoter and gene body (Figures 5A to 5C), which is in agreement with the finding that these genes were not expressed in PCCs (Figure 2A). The xylem marker gene XCP1 was marked neither with H3K4me3 nor H3K27me3 (state I; Figure 5D), and its expression was almost undetectable in PCCs (Figure 2A). The SUC2 locus in PCCs presented the H-state (Figure 5E), which was associated with even expression throughout the four time points (Figure 2A). By contrast, the second PCC marker gene, AHA3, was marked neither with H3K4me3 nor H3K27me3 (state I) in PCCs (Figure 5F). However, the steady expression of AHA3 in PCCs throughout the four time points (Figure 2A) suggests that expression of this gene could be controlled by other epigenetic marks not included in this study. Of the circadian clock genes, the increased expression of RVE8 after the shift to LD (Figure 2B) was accompanied by acquisition of the E-state at the TSS (Figure 5G). By contrast, the TSSs of both PRR3 and PRR5 were marked with H3K4me3 alone (Figures 5H and 5I). H3K4me3 levels at these two genes significantly decreased after the shift to LD and this decrease was associated with reduced gene expression (Figure 2B). Concerning genes involved in regulating photoperiod-dependent flowering, decreased expression of FKF1 in response to LD was accompanied by decreased H3K4me3 level at the TSS (Figures 2C and 5J). The FT locus was marked broadly with H3K27me3 throughout the experiment (Figure 5K) and expression of FT was almost undetectable at the time of sampling (ZT 6 to 7; Figure 2C), whereas JMJ30 expression was associated with H3K4me3 mark at the TSS, and neither expression nor modification levels changed after shifting to LD (Figures 2G and 5L). Of the selected carbohydrate metabolism genes, BE3, whose expression was significantly down-regulated after shifting to LD (Figure 2D), presented E-state and neither H3K4me3 nor H3K27me3 levels changed throughout the experiment (Figure 5M). By contrast, SEX1 was initially in E-state, but lost the H3K27me3 mark within 24 h after the shift to LD (Figure 5N). Interestingly, this loss of H3K27me3 was accompanied by decreased expression (Figure 2D). Finally, for the selected genes involved in meristem development, changes in expression of CSN5A (Figure 2F) could not be attributed to changes in histone marks, as H3K4me3 levels at the TSS did not change over the course of the experiment (Figure 5O). Similarly, ATHB12, which significantly increased in expression after shifting to LD (Figure 2F), presented unchanged E-state throughout the experiment (Figure 5P).
Figure 5.
Examples of H3K4me3 and H3K27me3 Dynamics in the PCCs during LD Induction of Flowering.
(A) to (T) Integrative Genomics Viewer traces of (A) STM, (B) AP1, (C) JAG, (D) XCP1, (E) SUC2, (F) AHA3, (G) RVE8, (H) PRR3, (I) PRR5, (J) FKF1, (K) FT, (L) JMJ30, (M) BE3, (N) SEX1, (O) CSN5A, (P) ATHB12, and (Q) FLC. H3K4me3, H3K27me3, and H3 signals are depicted in blue, green, and gray, respectively. Horizontal bars indicate regions and the significant enrichment of H3K4me3 and H3K27me3 (according to analysis in MACS2). The TSS is indicated using a red triangle. The Integrative Genomics Viewer vertical data range was set as “0 to 1,000” for all bar chart tracks, except for (E), where the data range was set to “0 to 5,000” due to the large number of ChIP-seq reads mapping to the SUC2 locus.
Taken together, our analyses provide a comprehensive description of the dynamic nature of gene expression and two important histone modifications, H3K4me3 and H3K27me3, in the PCCs in response to change in photoperiod. Our results reveal a good correlation between changes in H3K4me3 modification and gene expression, as well as a weak positive correlation between changes of H3K27me3 modification and gene expression. However, the two chromatin marks are not sufficient to explain epigenetic regulatory mechanisms for all expression changes in the PCCs. This is not surprising, as it is well known that the transcriptome is modulated by a range of histone modifications. However, as the number of nuclei isolated by INTACT is quite small, these analyses focused on two important modifications, H3K4me3 and H3K27me3.
MRF1 Is Expressed in Phloem Tissues and Promotes Flowering
Many of the genes that responded rapidly to the change in daylength in PCCs encode known regulators of the circadian clock, the photoperiod pathway, meristem identity, or starch metabolism (Figures 2B to 2E). However, our data sets also revealed previously uncharacterized photoperiod-responsive genes that might contribute to the regulation of flowering time. One gene that was rapidly and strongly induced in response to LD is AT1G21920, which we now refer to as MRF1 (Figures 2E and 6). MRF1 has been annotated (Araport v11) as a putative histone H3 K4-specific methyltransferase SET7/9 family protein. However, closer inspection revealed that, while the predicted protein contains MORN-repeat motifs similar to the N-terminal regions of the histone-Lys n-methyltransferase SET7/9 in animals, MRF1 appears to lack a proper SET domain (Supplemental Figure 4; Yeates, 2002). It is therefore unlikely that MRF1 functions as a real methyltransferase, and its molecular function should be considered unknown.
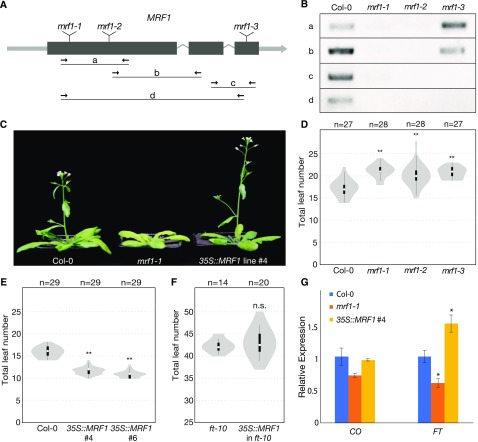
Figure 6.
MRF1 Promotes Flowering.
(A) Gene model of MRF1 showing the positions of mrf1-1, mrf1-2, and mrf1-3 T-DNA insertions and the primer pairs used for RT-PCR analysis.
(B) Detection of MRF1 transcript in wild-type Col-0 plants and homozygous mrf1-1, mrf1-2, and mrf1-3 mutant plants. Plants were grown in 23°C LDs for 8 d.
(C) Flowering-time phenotypes of 4-week-old wild-type Col-0, mrf1-1, and Pro35S:MRF1 plants grown in 23°C LDs. The Pro35S:MRF1 plant is the homozygous T3 generation.
(D) Flowering time of wild-type Col-0 and the three mrf1 mutants grown in 18°C LDs.
(E) Flowering time of wild-type Col-0 and two independent Pro35S:MRF1 homozygous T3 lines grown in 23°C LDs.
(F) Flowering time of ft-10 and Pro35S:MRF1 in ft-10 T1 Plants Grown in 23°C LDs.
For (D) to (F), white circles show the medians, box limits indicate the 25th and 75th percentiles, whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, and polygons represent density estimates of data. Numbers of plants are shown above. Significant differences from the control population, calculated using Student’s t test, are shown above each violin plot. **P < 0.01; n.s., not significant.
(G) Expression of CO and FT in 10-d-old Col-0, mrf1-1, and Pro35S:MRF1 plants grown in 23°C LDs. Samples were harvested at ZT15. Data show the mean of three biological replicates, normalized against TUB2 and relative to Col-0, and error bars indicate the se. Significant differences, calculated using Student’s t test, are shown above. *P < 0.05.
To assess whether MRF1 plays a role in regulating flowering time in Arabidopsis, we obtained three T-DNA mutants: mrf1-1 (GK-271A12) and mrf1-2 (SAIL_609_E11), which carry T-DNA insertions at different positions in the first exon of the gene, and mrf1-3 (SALK_055678), which has an insertion in the last exon (Figure 6A). RT-PCR analysis did not detect substantial expression of MRF1 in mrf1-1 and mrf1-2 plants, indicating that they are strong mutant alleles (Figure 6B). However, partial transcripts were detected in mrf1-3 plants, suggesting that this mutant may retain residual MRF1 activity (Figure 6B). When grown at 23°C under LD conditions, the flowering time phenotypes of the three mrf1 mutants were variable, with mrf1-1 being the only genotype that flowered consistently later than wild-type Col-0 controls (Figure 6C; Supplemental Table 1; Supplemental Data Set 9). We therefore grew the plants at reduced ambient temperatures, which is known to slow down flowering, thereby revealing the phenotype of weak flowering time mutants. At 18°C in LDs, we observed statistically significant late flowering in all three mrf1 mutants (Figure 6D; Supplemental Table 1; Supplemental Data Set 9). Similar results were obtained when the three mrf1 mutants were grown at 16°C in LDs, although mrf1-2 flowered only slightly later than wild-type controls and the difference was not statistically significant (Supplemental Table 1; Supplemental Data Set 9). The consistent late flowering of multiple mrf1 alleles suggests that MRF1 plays a role in promoting flowering, an interpretation that is further supported by the finding that constitutive overexpression of MRF1 resulted in a strong early-flowering phenotype (Figures 6C and 6E; Supplemental Table 1; Supplemental Data Set 9). MRF1 is thus both necessary and sufficient to promote timely flowering in response to inductive photoperiods. The function of MRF1 appears to be FT-dependent, as overexpression of MRF1 in an ft-10 mutant background did not result in early flowering (Figure 6F; Supplemental Table 1; Supplemental Data Set 9). Furthermore, MRF1 seems to exert its function mainly in LDs, as we observed only minor changes in flowering time in the mrf1 mutants in SDs (Supplemental Table 1; Supplemental Data Set 9). However, expression of MRF1 from the PCC-specific SUC2 promoter (Supplemental Figure 5) had no significant effect on flowering time in either LDs or SDs (Supplemental Table 1; Supplemental Data Set 9), indicating that, under the conditions tested, MRF1 expression is also required in other cell types aside from PCCs to promote flowering.
To integrate MRF1 into the existing genetic framework of flowering-time control, we performed RT-qPCR analysis on the known central regulators of the photoperiodic flowering pathway, CO and FT (Figure 6G). We found that FT expression was significantly lower in mrf1-1 mutants and higher in Pro35S:MRF1 plants than in wild-type controls. However, levels of CO expression were not significantly different in these lines, indicating that MRF1 does not act upstream of CO in the photoperiod pathway. Thus, we conclude that MRF1 positively regulates FT to promote flowering under LD conditions.
DISCUSSION
Tissue- and Cell-Type–Specific Chromatin States
In recent years, increasing numbers of studies have revealed that different cell types can exhibit distinct epigenetic features. For example, unique DNA methylation patterns have been reported for different cell types in the root apical meristem (Kawakatsu et al., 2016), and the H3K27me3 and DNA methylation patterns in endosperm nuclei varied in a cell-type–specific and also a parent-of-origin–specific manner (Moreno-Romero et al., 2017). Interestingly, the spatiotemporal expression of Polycomb Repressive Complex 2 genes corresponds with differential H3K27me3 modification in vascular and nonvascular cells in the root (de Lucas et al., 2016). However, the precise mechanisms by which dynamic changes in epigenetic marks contribute to the adjustment of transcriptional programs during plant development, differentiation, and growth in response to environmental changes have only begun to be investigated (de Lucas et al., 2016; Kawakatsu et al., 2016; Moreno-Romero et al., 2017; Palovaara et al., 2017; You et al., 2017; Maher et al., 2018).
We have previously shown that during the transition to flowering at the Arabidopsis SAM, H3K4me3 is a better indicator of change in gene expression than H3K27me3, which is usually considered a repressive epigenetic mark (You et al., 2017). Similar observations have been reported in a recent study that investigated changes of H3K4me3 and H3K27me3 modifications during flower development with temporal and tissue-specific resolution (Engelhorn et al., 2017). However, at the SAM we observed H3K27me3 not only on the gene body, where its presence was indeed indicative of gene silencing, but also in relatively narrow peaks at the TSS of expressed genes (You et al., 2017). In the latter situation, we distinguished between H- and E-states, depending on the relative width of the H3K4me3- and H3K27me3-marked region within ±500 bp around the TSS of expressed genes (You et al., 2017). The E-state, which we defined as narrow H3K427me3 peaks embedded (“E”) in broader H3K4me3-marked regions, was observed for >9,000 genes at the SAM. As this chromatin state was not apparent in an INTACT study on hair and nonhair cells in the root epidermis (Deal and Henikoff, 2011), and was also largely absent from whole seedling data (Sequeira-Mendes et al., 2014; You et al., 2017), the E-state was initially speculated to be specific to the undifferentiated cells at the SAM. However, in this study, we also detected the E-state on >10,000 gene loci in the differentiated PCCs, indicating that this chromatin state is not specific to undifferentiated cells. Furthermore, we found that the relatively great depth of sequencing in our study contributed to the detection of the E-state. This is not surprising as it has been shown that sequencing depth has a significant impact on identification of H3K27me3 marks by ChIP-seq (Jung et al., 2014). Previous studies that defined multiple chromatin states in Arabidopsis were mainly performed by microarray and/or on complex tissues (Deal and Henikoff, 2011; Roudier et al., 2011; Sequeira-Mendes et al., 2014), which might explain why these more specialized chromatin states were not detected, or were at least underestimated, in these earlier studies.
Overlapping epigenetic marks in ChIP-seq data (such as the E-state and H-state) can indicate bivalency where a single histone molecule carries more than one modification. In plants, bivalency has been detected at individual gene loci such as FT and FLC, as well as in whole-genome studies (Jiang et al., 2008; Deal and Henikoff, 2011; Sequeira-Mendes et al., 2014; Engelhorn et al., 2017). It is tempting to speculate that the concomitant detection of H3K4me3 and H3K27me3 on some of the E-state and H-state genes in PCCs could indicate true bivalency. However, it should be noted that the co-occurrence of these two marks in our data might also reflect differences in chromatin modifications between different PCC cells, or between two sister chromosomes in a given cell. It would be very interesting if at least some of the genes in E-state and H-state were bivalently marked, but testing for bivalency would require sequential ChIP, which is not currently feasible given the limited number of nuclei obtained by INTACT.
Interestingly, only ∼1/3 of all genes in E-states in SAM and PCCs are shared between the two tissues. This suggests that, even though the analyses were performed slightly differently between the two tissues, the E-state might be more widespread than previously thought. Similar to the situation described in the SAM (You et al., 2017), the E-state in PCCs was relatively stable. Only 1,787 (22.8%) of the genes that were in E-state in SD (n = 7902) acquired a different chromatin state in response to the shift to LD (Supplemental Data Set 6). Furthermore, the E-state in PCCs did not always correspond to gene expression changes. For example, BE3, CSN5A, and ATHB12, which significantly changed in expression after shifting to LD (Figure 2), had unchanged E-state throughout the course of the experiment (Figures 5M, 5O, and 5P). By contrast, decreased expression of SEX1 was accompanied by the change from initial E-state to state II (H3K4me3 only) within 24 h after the shift to LD (Figure 5N). The most likely explanation for this disconnect between chromatin marks and gene expression is that additional epigenetic modifications, which were not examined in this study due to limited input material, contribute to the regulation of gene expression. Interestingly, the H-state appears to be more stable in PCCs compared with the SAM, where about half of the H-states present in the vegetative meristem resolved, after flowering, into nonoverlapping, partially overlapping, or E-states (You et al., 2017). By contrast, in PCCs only 25% (n = 130) of the genes in H-state in SD (n = 521) changed to other chromatin states in the 3-LD samples (Supplemental Data Set 6), indicating that there are differences in chromatin dynamics between the differentiated PCCs and the largely undifferentiated cells in the SAM. In summary, our findings suggest that the chromatin landscape in Arabidopsis and its dynamic responses to environmental changes might be even more complex than previously thought, and that tissue- and cell-type–specific studies are needed to capture this complexity.
Spatiotemporal Epigenomic Regulation of Gene Expression in PCCs
Transcriptional control is a complex process that is governed in part by epigenetic modifications and chromatin structure, which regulate access of transcription factors to DNA, and a growing number of epigenetic modifiers have been implicated in the regulation of plant growth and development in response to endogenous and environmental signals (Yang et al., 2010, 2016; Gu et al., 2013; Wang et al., 2014; Jeong et al., 2015). Several of these epigenetic modifiers regulate gene expression indirectly by modulating the activity of the circadian clock, and some chromatin dynamics have been shown to follow a circadian rhythm (Jones et al., 2010; Malapeira et al., 2012). This may be the reason why epigenetic modifications at the FT locus did not change significantly throughout the experiment, and why the expression of known epigenetic modifiers of FT was mostly unchanged in PCCs after shifting from SD to LD (Supplemental Figure 6). For example, JMJ30, which has been shown to repress flowering by demethylation of H3K36me2 marks at the FT locus (Yan et al., 2014), is hardly detectable in our samples, but this is in agreement with previous reports demonstrating that expression of JMJ30 is under circadian control and is at its minimum at the time of sampling (ZT 6 to 7; Jones et al., 2010; Lu et al., 2011). JMJ30 expression peaks at the end of LD (ZT 12 to 20) and has the strongest effect on FT expression at ZT16, when FT expression also peaks (Gan et al., 2014; Yan et al., 2014). At all four time points sampled, the FT locus was covered with broad H3K27me3 marks (Supplemental Data Set 5). One possible explanation is that H3K27me3 is under circadian control and is removed and re-established over the course of the day to allow expression of FT only at the end of the day.
Emphasizing the importance of tissue-specific epigenetics, we detected an increase in WDR5A expression (Figure 2G). WDR5A is recruited by FRIGIDA to mediate deposition of H3K4 methylation on the FLC promoter and thus promote FLC expression (Jiang et al., 2009, 2011). In our data set, the increased expression of WDR5A was not associated with any change in expression or chromatin state of FLC in the PCCs (Supplemental Data Set 1; Figure 5Q), suggesting that WDR5A might target other genes than FLC in the PCCs. It is known that the COMPASS-like histone methylase complex, of which WDR5A is a core component, is recruited by basic Leu zipper transcription factors to the promoters of endoplasmic reticulum stress-responsive genes (Song et al., 2015b). Thus, it would be interesting for future research to identify the interacting protein(s) in the PCCs that recruit(s) the increased amount of WDR5A to deposit H3K4me marks for activating specific gene expression in response to photoperiod changes.
A Link between Photoperiodic, Metabolic, and Epigenetic Regulation of Gene Expression
In both the animal and the plant kingdoms, gene expression, metabolism, and chromatin dynamics have been shown to be controlled by the circadian clock, and thus follow a circadian rhythm. Metabolites and epigenetic modulators can in turn directly regulate the nuclear transcription of clock oscillators. Studies in mammalian cells show that many key energy metabolites, such as acetyl-CoA, s-Adenosyl methionine, NAD+, and ATP, are substrates and essential cofactors for epigenetic enzymes such as histone acetyltransferases (HAT) and HMT, and thus modulate gene expression via epigenetic mechanisms (Shahbazian and Grunstein, 2007; Donohoe and Bultman, 2012; Mentch et al., 2015). Emphasizing the importance of tissue-specific studies, analyses of liver tissue revealed a circadian clock-mediated metaboloepigenetic mechanism of transcriptional regulation (Shahbazian and Grunstein, 2007; Donohoe and Bultman, 2012; Mentch et al., 2015). Briefly, the abundance of NAD+ is regulated via circadian rhythms, and the NAD-dependent histone deacetylases sirtuin-1 and -6 regulate the expression of Circadian Locomotor Output Cycles Kaput (CLOCK) and Brain and Muscle Arnt-Like protein-1, the two transcription factors at the core of the circadian transcriptional circuits, by targeting acetylated histone-3 residues at these gene loci (Etchegaray and Mostoslavsky, 2016). Interestingly, the core mammalian circadian oscillator CLOCK is itself also a HAT (Doi et al., 2006).
In plants, the highly complex tissue-specific gene regulation and crosstalk of coordinated photoperiodic, metabolic, and epigenetic networks have not been well-integrated, largely because of the poor accessibility of many plant tissues and cell types, such as the SAM and PCCs. In our PCC-specific data set, we were able to identify several links between these various networks. For example, the expression of WDR5A, a core component of the COMPASS-like histone methylase complex, was significantly up-regulated in response to photoperiod changes (Figure 2G), and the MYB transcription factors RVE8 and CCA1, which are involved in the regulation of H3 acetylation level at the TOC1 locus (Farinas and Mas, 2011), also significantly changed their expression after the shift to LD (Figure 2B). In addition, we detected altered expression of key enzymes involved in energy metabolism after shifting to LD (Figure 2D; Table 1; Supplemental Data Set 2). In Arabidopsis, the redox of NADPH and NADP+, as well as their ratio, has been shown to be driven by circadian rhythm, and a perturbation in the redox status can change expression of both morning and evening clock genes and lead to reinforcement of the circadian clock (Zhou et al., 2015). Recently, acetyl-CoA has been shown to promote histone acetylation, predominantly at H3K27 (Chen et al., 2017). As the 24-h rhythmic oscillation in H3K4me3 and H3 acetylation has been suggested to facilitate transcriptional activation of the central clock oscillators (Malapeira et al., 2012), and because acetyl-CoA and NAD+ are substrates and essential cofactors for HAT and HMT enzymatic activities (Shahbazian and Grunstein, 2007; Donohoe and Bultman, 2012; Mentch et al., 2015), it is conceivable that a circadian clock-mediated metaboloepigenetic mechanism of transcriptional regulation might also exist in planta. However, because plants display more complex metabolic and redox pathways than animals, such a mechanism might be more complex than previously portrayed in proposed models. The PCC-specific INTACT line generated in this study provides the ideal platform to further investigate the dynamic nature of gene expression and epigenetic marks, for example by obtaining a full diurnal series to resolve the high complexity of the coordinated photoperiodic, metabolic, and epigenetic gene regulation pathways with temporal and cell type-specific resolution.
Potential Roles of MRF1 in Regulating Flowering Time
While expression of most epigenetic modifiers was comparable in the SAM and in PCCs(Supplemental Figure 6), one gene, AT1G21920 (MRF1), which has previously been annotated as a putative histone H3 K4-specific methyltransferase SET7/9 family protein, was significantly induced specifically in PCCs within 24 h after the shift to LD (Figure 2E). The locus exhibited stable E-state throughout the experiment (Supplemental Data Set 6). MRF1 mRNA has previously been detected in phloem exudate (Deeken et al., 2008), which is in agreement with our finding that MRF1 is expressed in the phloem tissues.
Analysis of the MRF1 domain structure suggests that it is unlikely to function as a real methyltransferase, as it lacks a proper SET domain, but interestingly MRF1 shares some similarities with phosphatidylinositol-4-phosphate 5-kinases (IPR023610), particularly in two transmembrane helical repeats (Supplemental Figure 4; The UniProt Consortium, 2017). Phosphatidylinositol-4-phosphate 5-kinases are involved in the production of phosphatidylinositol-4,5-bisphosphate, a precursor to inositol-1,4,5-triphosphate and diacylglycerol, two compounds that regulate diverse cellular and developmental processes, including flowering (Ischebeck et al., 2013; Nakamura et al., 2014a). In this context, it is noteworthy that FT has been shown to bind certain phospholipids and that this interaction is important for its flowering-promoting function (Nakamura et al., 2014b). Irrespective of its molecular function, which will need to be determined later, MRF1 clearly participates in the regulation of flowering time and functions upstream of FT.
MRF1 expression has also been shown to be significantly induced in Arabidopsis suspension culture cells in response to Suc starvation (Contento et al., 2004). This study also identified three class II trehalose-6-phosphate synthases (TPS), which have been suggested to regulate TPS1, the main T6P-synthesizing enzyme in Arabidopsis (Paul et al., 2008), that were significantly up-regulated upon Suc starvation. T6P has been suggested to function as a signaling molecule that informs the cell about carbohydrate availability (Lunn et al., 2006; Ponnu et al., 2011). Interestingly, the tps1-2 mutant fails to induce FT in the PCCs even under otherwise inductive LD conditions, indicating that T6P/TPS1 signaling is required for flowering (Wahl et al., 2013). It will be interesting to see if MRF1 is involved in the integration of the T6P/TPS1 signal into the canonical flowering time pathways. T6P signaling has also been implicated in the regulation of diurnal starch synthesis and breakdown (Kolbe et al., 2005; Lunn et al., 2006; Ponnu et al., 2011) and it is interesting to note that genes involved in starch metabolism are among those that respond most quickly to the shift from SD to LD (Figure 2D). The identification of MRF1 as a regulator of flowering in Arabidopsis demonstrates the power of cell-type–specific transcriptomic and epigenomic approaches.
METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accession Col-0 was used as wild type in this study. The homozygous ft-10 (GABI_290e08) plants have been described in Yoo et al. (2005). The mrf1-1 (GK-271A12), mrf1-2 (SAIL_609_E11), and mrf1-3 (SALK_055678) alleles were obtained from the Nottingham Arabidopsis Stock Centre, and homozygous mrf1-1, mrf1-2, and mrf1-3 plants were identified by genotyping using primers designed by the online T-DNA Primer Design Tool (http://signal.salk.edu/tdnaprimers.2.html; Supplemental Table 2).
Seeds were stratified in 0.1% agar at 4°C for 3 d in the dark before sowing, to synchronize germination. All plants were grown on soil or 1/2 MS agar plates containing antibiotics in growth chambers at 23°C with 65% relative humidity. A mixture of Cool White and Gro-Lux Wide Spectrum fluorescent lights, with a fluence rate of 125 to 175 μmol m−2 s−1, was used. LDs were 16 h of light and 8 h of dark, and SDs were 8 h of light and 16 h of dark. In floral induction experiments, plants were grown on soil in SD for 21 d before shifting to LD. Phenotyping of the flowering time of mrf1 mutants was performed under LD conditions at 16°C, 18°C, and 23°C, and under SD conditions at 23°C, respectively.
Molecular Cloning
For preparation of the PCC-tagged INTACT reporter line, the ProSUC2:RedNTF construct was generated by a modified Gateway recombination of the entry plasmid pYY1204 (You et al., 2017) that carries the red nuclear envelope-targeting protein (RedNTF), consisting of the tryptophan-proline-proline domain at the N terminus, followed by red fluorescent protein (mCherry), and the biotin ligase recognition peptide at the C terminus, into a pGREEN-IIS based destination vector (pHW058) that harbors the Arabidopsis SUC2 (AT1G22710) promoter (Mathieu et al., 2007). The Gateway reaction was performed with the Gateway LR clonase II Enzyme mix (Invitrogen). Subsequently, the ProSUC2:RedNTF construct was transformed into homozygous ProUBQ10:BirA lines (You et al., 2017), making use of Agrobacterium tumefaciens strain ASE and the floral dip method (Clough and Bent, 1998). Homozygous PCC-tagged INTACT lines were identified by selective germination on 1/2 MS agar plates containing 50 μg/mL of kanamycin.
The Pro35S:MRF1 line and the ProSUC2:MRF1 transgenic lines were also generated in this study. Briefly, total RNA was extracted from 3-week-old LD-grown seedlings using the RNeasy Mini Kit (Qiagen), and genomic DNA was removed from the RNA samples by DNase I treatment (Thermo Fisher Scientific). First-strand cDNA was synthesized with ThermoScript Reverse Transcriptase (Thermo Fisher Scientific) using oligo(dT) primers, and the full-length MRF1 coding sequence was amplified by PCR using gene-specific primers containing EcoRI and BamHI sites (Supplemental Table 2). The PCR product was cloned into the Gateway entry vector pJLBlue reverse (Mathieu et al., 2007) by a restriction enzyme ligation reaction with T4 DNA ligase (New England Biolabs) to make pYY1601, which was subsequently recombined into two pGREEN-IIS–based destination vectors containing the 35S promoter (pFK210) and SUC2 promotor (pHW058) in front of the attR1-attR2 GateWay recombination cassette, respectively (Mathieu et al., 2007). The Pro35S:MRF1 and ProSUC2:MRF1 constructs were transformed into Col-0 and ft-10 plants using the floral dip method (Clough and Bent, 1998), and the transgenic lines were selected by germination of T1 seeds on soil watered with 0.1% glufosinate (BASTA) and on 1/2 MS agar plates containing 50 μg/mL of kanamycin, respectively. Two independent Pro35S:MRF1 T1 lines (Supplemental Data Set 9) were continued to the T3 generation for further characterization.
Sequences of all primers used in this work are listed in Supplemental Table 2. PCR reactions were performed using Taq polymerase (New England Biolabs), and all constructs were verified by Sanger sequencing after cloning.
Flowering Time Measurement
Flowering time was measured using the total number of leaves at the time of bolting, and in some cases the chronological time to flowering. The rosette leaves were counted when the bolting shoot reached ∼1 cm in length and the cauline leaves were counted when they were all visible on the shoot. The number of days to flowering was counted from the day the seeds were placed in the growth chamber until the opening of the first flower. The significance of differences in flowering time between genotypes was calculated using Student’s t test. Violin plots were generated using the BoxPlotR web tool (Spitzer et al., 2014).
Microscopy
A Zeiss Axioplan 2 imaging system was used for imaging the histological sections and for examining the quality and quantity of purified nuclei after each INTACT experiment using 4′,6-diamidino-2-phenylindole and mCherry filters.
Sample Collection and INTACT Purification of PCC Nuclei
To evaluate the enrichment of PCC-specific nuclei after INTACT, ProSUC2:RedNTF ProUBQ10:BirA seeds were sown directly on soil and germinated in LD. Leaf material from approximately sixty 24-d-old plants per biological replicate was collected, frozen immediately in liquid nitrogen, and stored at −80°C until further use. A quantity of 1.5 g of the collected material was used for INTACT as described in You et al. (2017). Minor modifications compared with the original protocol were the addition of 5 mM dithiothreitol to the nuclear purification buffer and omission of bovine serum albumin during the purification.
The SD to LD shift experiments were performed with three independent biological replicates. For each biological replicate, ∼500 plants were grown in SD, and the 0 LD, 1 LD, 2 LD, and 3 LD samples were collected on the 21st d in SD and 1, 2, and 3 d after the shift to LD. For each sample, shoot material from ∼120 plants was collected and immediately frozen in 50-mL falcon tubes suspended in liquid nitrogen. The samples were collected at ZT 6 to 7 and samples were stored at −80°C before INTACT purification. The INTACT experiments were performed as described in You et al. (2017), with ∼1/3 of the ground tissue used for isolating nonfixed nuclei for transcriptome analysis by the RNA-seq method and the remaining ∼2/3 of the ground tissue used for isolating fixed nuclei and subsequently used to analyze histone modifications by the ChIP-seq method. The purity and yield of intact nuclei were assessed by microscopy and nuclear samples were stored at −80°C before further experiments.
RNA-seq
RNA-seq was performed for three biological replicates from independently grown plants harvested 0 LD, 1 LD, 2 LD, and 3 LD after the shift to LD. For each sample, nuclear RNA was extracted from ∼10,000 nonfixed nuclei using the RNeasy Micro Kit (Qiagen), and further treated with DNase I (0.05 U per μL) for 30 min at 37°C (Thermo Fisher Scientific) to remove any contaminating genomic DNA. Afterwards, the RNA was purified a second time using the RNeasy Micro Kit (Qiagen). Using ∼500 pg RNA, measured using the RNA 6000 Pico Kit (Agilent), double-stranded cDNA was amplified using the SMARTer Ultra Low Input RNA for Illumina Sequencing-HV kit (Clontech). The concentration and yield of the amplified cDNA were determined using the High Sensitivity DNA Kit (Agilent). The RNA sequencing samples were prepared from 1 to 2 ng of amplified double-stranded cDNA using the Low Input Library Prep Kit (Clontech) as described in You et al. (2017). Sequencing libraries were validated using the High Sensitivity DNA Kit (Agilent), and sequencing was performed on an Illumina Hiseq 2000 system. Approximately 30 million 2 × 101-bp paired-end reads that passed the Illumina quality control filter were collected for each sample (Supplemental Data Set 10).
ChIP-seq
The ChIP experiments were performed as described in You et al. (2017). Briefly the chromatin was prepared from ∼10,000 purified fixed nuclei per sample, and sheared to 100–500-bp fragments using a focused-ultrasonicator (Covaris S2 system) as described in You et al. (2017). The sheared chromatin was divided into three aliquots at the ratio of 1:2:3 for immuno-hybridization with anti-H3 antibody (Millipore, Cat.17-10254, Lot. 2,051,404), anti-H3K4me3 antibody (Millipore, Cat. 17-614, Lot.1973237), and anti-H3K27me3 antibody (Active Motif, Cat. 39155, Lot. 25,812,014), respectively. The specificity of the antibodies had been verified previously by immunoblot and dot blot analyses (You et al., 2017). Approximately 50 pg of ChIP DNA per sample was obtained and used to prepare ChIP-seq libraries using the ThruPlex-FD Prep Kit (Rubicon Genomics). The linear amplification and the sequencing samples were monitored by quantitative PCR including SYBR Green I (Invitrogen). Libraries were quantified using the KAPA Library Quantification Kit (KAPABIOSYSTEMS), and further evaluated on a high sensitivity DNA chip (Agilent). Libraries were sequenced on Illumina HiSeq 2000 and Miseq systems for collecting 50-bp single-end or 150-bp paired-end reads. Sequencing depth ranged from 32 to 71 million reads, depending on replicate and antibody used (Sims et al., 2014; Supplemental Data Set 10). Only Read 1 of the 150-bp paired-end reads was used and further trimmed to 50 bps. The ChIP-seq data were collected for each 0 LD, 1 LD, 2 LD, and 3 LD samples from two biological replications (Supplemental Data Set 10).
Transcriptome Data Analysis
Next generation sequencing (NGS) reads were mapped to Arabidopsis reference transcriptome TAIR10 ver. 24, with rRNA regions (2:3471-9557; 3:14197350-14203988) masked, using TopHat 2.0.13 (parameters: no-mixed alignments; up to 20 secondary alignments; no novel junctions; Trapnell et al., 2009). Counts of NGS reads covering transcripts were computed using the function “summarizeOverlaps” (Lawrence et al., 2013) in R. Expressed genes were defined as those having the value of FPKM > 1 at a given time point. Read counts were submitted to differential gene expression analysis in Deseq2 (default parameters; false-discovery rate, FDR < 0.05; Love et al., 2014). Regularized logarithms of read count computed by Deseq2, denoted by rlog, were used to analyze relationships between gene expression level and histone modifications signal.
RT-qPCR Analysis Using INTACT Purified Nuclei
Crude nuclei (input) and INTACT-purified PCC-specific nuclei were prepared from leaf material of 24-d-old LD-grown ProSUC2:RedNTF INTACT reporter line. Total nuclear RNA was isolated from each sample using the RNeasy Micro Kit (Qiagen). DNase I treatment was performed on column during an RNA extraction procedure following the manufacturer’s instructions. RNA was quantified using a 10-fold diluted Qubit RNA BR Assay (Thermo Fisher Scientific) in a Bio-Rad CFX96 machine, and ∼0.1 to 2 ng of RNA was used for cDNA synthesis and amplification using the SMART-seq2 protocol (Picelli et al., 2013). The amplified cDNA was purified using the QIAquick PCR purification Kit (Qiagen) and used as the template for qPCR reactions including SYBR Green I (Roche Life Science) in a Bio-Rad CFX96 machine. The Ct values of each examined genes were normalized to the average CT values of elongation factor 1α (EF1A) and second β-tubulin-encoding (TUB2) house-keeping genes, and the relative gene expression in the INTACT samples over the expression in the input samples was calculated using the ΔΔCT method. Because amplification signals of ML1 and SCR in some INTACT samples did not cross the detection threshold, the CT values were set to 50 to be able to conservatively estimate the differences in expression. qPCR analysis was performed for three biological replicates that were grown at the same time but harvested and processed independently of each other, with three technical repetitions per sample.
RT-PCR Analysis of MRF1 and RT-qPCR Analysis of the Flowering-Time Genes
Whole-seedling samples of wild-type Col-0, mrf1 mutants, ProSUC2:MRF1 and Pro35S:MRF1 lines were collected at ZT15, 8 d after sowing. RNA was extracted using the RNeasy Plant Mini kit (Qiagen), and treated with DNase I (Thermo Fisher Scientific) to remove any contaminating genomic DNA. cDNA was synthesized from 1 μg RNA using the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific) with oligo-dT primers, and the cDNA was used as template in semi-quantitative PCRs (semi-qPCR) with DreamTaq (Thermo Fisher Scientific) or in qPCR reactions using SYBR Green I (Roche Life Science) in a Bio-Rad CFX96 machine. qPCR analysis was performed using three independently grown biological replicates of each genotype with three technical replications per sample. Primer sequences are listed in Supplemental Table 2.
Histone Modification Data Analysis
Fifty-bp single-end NGS reads were mapped to the Arabidopsis reference genome TAIR10 ver. 24 using Bowtie2 2.2.4 (Langmead and Salzberg, 2012; minimum alignment score ‒12.5; uniquely mapped reads only). The sets of aligned H3K27me3 and H3 reads were down-sampled to minimum number of reads, respectively, for peak-calling from the same sequencing depth in all samples (Jung et al., 2014). Significant enrichment regions in H3K4me3 and H3K27me3 samples, relative to H3 control samples, were identified using MACS2 2.1.0 (default settings for H3K4me3, FDR < 0.05; broad-peaks mode for H3K27me3, FDR < 0.1; Zhang et al., 2008b; Kellis et al., 2014). Regions for which the difference between signal levels measured by FPKM(ChIP)/FPKM(H3 control) in the ChIP-seq data collected from the two independently-grown biological replicates of each time point (0 LD, 1 LD, 2 LD, and 3 LD after the shift to LD) constituted “far” outliers (procedure boxplot in Genstat 18; VSN International, https://www.vsni.co.uk/) were filtered out. Consensus regions found as a sum of regions from two biological replicates of the ChIP-seq data collected for each antibody and time point were taken as histone modification marks. Differential modification analysis for H3K4me3 and H3K27me3 marks in pair-wise comparisons of time points was performed using DiffBind (Bioconductor 3.2; minoverlap = 1; FDR < 0.01; Stark and Brown, 2011). The modification marks were intersected with annotated genes to obtain a list of genes with specific coverage by H3K4me3 and H3K27me3. Multiple marks intersecting with a gene were joined into one mark with combined limits and average signal. The mapped reads and modification marks were visualized using the Integrated Genomics Viewer (Robinson et al., 2011).
GO Term Analysis
AmiGO 2 (version 2.5.8) was used for GO analysis (Carbon et al., 2009) to assess the overrepresentation in biological processes of significantly DEGs. Tests were performed using Fisher’s Exact with FDR multiple test correction, and only FDR < 0.05 were considered as significant. To reduce the complexity, redundant child terms based on GO hierarchy were removed from the reports.
Alignment and Domain Analysis
Protein sequences of human SET domain-containing protein7, mouse SET domain-containing protein7, Arabidopsis SDG1, and MRF1 were aligned using the CLC Main Workbench (Version 7.9.1; standard parameters). The sequence and domain information were obtained from The UniProt Consortium (2017).
Accession Numbers
ChIP-seq and RNA-seq data have been deposited in ArrayExpress database at European Bioinformatics Institute as part of the European Molecular Biology Laboratory (EMBL-EBI; www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-5736 and E-MTAB-5737.
Supplemental Data
Supplemental Figure 1. Replicability of RNA-seq data and ChIP-seq data. Related to Figures 2 and 3.
Supplemental Figure 2. Distribution of length of H3K4me3 and H3K27me3 marks, and location of H3K4me3 and H3K27me3 modifications on the protein-coding genes in the PCCs. Related to Figure 3.
Supplemental Figure 3. Correlations between change in expression and changes in histone modifications of protein-coding genes. Related to Figure 4.
Supplemental Figure 4. Domain analysis of MRF1.
Supplemental Figure 5. RT-qPCR verification of the overexpression of MRF1 in Pro35S:MRF1 and ProSUC2:MRF1 transgenic lines.
Supplemental Figure 6. Tissue-specific expression of selected epigenetic histone modifiers regulating circadian clock and flowering.
Supplemental Table 1. Flowering time of mrf1 mutants and transgenic lines.
Supplemental Table 2. Sequences of oligonucleotide primers used in this work.
Supplemental Data Set 1. Gene expression profiles in the PCCs.
Supplemental Data Set 2. DEGs in the PCCs during floral transition.
Supplemental Data Set 3. GO enrichment in biological process of significantly DEGs.
Supplemental Data Set 4. Genes with H3K4me3 modifications in the PCCs at the four time points.
Supplemental Data Set 5. Genes with H3K27me3 modifications in the PCCs at the four time points.
Supplemental Data Set 6. Categories of chromatin states on protein-coding genes in PCCs at the four time points.
Supplemental Data Set 7. Genes with significant H3K4me3 signal changes in pair-wise comparisons.
Supplemental Data Set 8. Genes with significant H3K27me3 signal changes in pair-wise comparisons.
Supplemental Data Set 9. Flowering time of wild-type, mrf1, Pro35S:MRF1, and ProSUC2:MRF1 plants.
Supplemental Data Set 10. Sequencing statistics.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Christa Lanz and Natalie Agarwala for technical assistance, Richard Neher for discussion, and the Poznan Supercomputing and Networking Center for access to their infrastructure for part of the computations.This work was supported by infrastructure grants from VINNOVA (The Swedish Governmental Agency for Innovation Systems, 2016-00504), and The Knut and Alice Wallenberg Foundation (2016.0341) to the Umeå Plant Science Centre; the Max Planck Institute for Developmental Biology, Department of Molecular Biology (postdoctoral fellowship to Y.Y.); the Institutional Strategy Program of the University of Tübingen (Deutsche Forschungsgemeinschaft, ZUK 63); the Polish National Centre for Research and Development ERA-CAPS project “FlowPlast” (ERA-CAPS-1/1/2014 to P.K.); the Deutsche Forschungsgemeinschaft (SCHM 1560/10–1 to M.S.); the Vetenskapsrådet (VR grant 2015-04617 to M.S.); and The Knut and Alice Wallenberg Foundation (2016.0025).
Author Contributions
Y.Y. and M.S. designed the experiments and supervised the work; P.K. supervised the RNA-seq and ChIP-seq analyses; Y.Y. performed most experiments; M.N. assisted in Y.Y.’s experiments; Y.Y. and R.M.B. performed INTACT experiments; Y.Y. and J.E.L. characterized MRF1 functions in flowering time control; Y.Y., J.E.L., and R.M.B. performed the RT-qPCR analysis; Y.Y., A.S., and P.K. analyzed RNA-seq and ChIP-seq data; and Y.Y., J.E.L., R.M.B., P.K., and M.S. wrote the manuscript, with input from other co-authors.
Footnotes
Articles can be viewed without a subscription.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- Abe M., Kaya H., Watanabe-Taneda A., Shibuta M., Yamaguchi A., Sakamoto T., Kurata T., Ausín I., Araki T., Alonso-Blanco C. (2015). FE, a phloem-specific Myb-related protein, promotes flowering through transcriptional activation of FLOWERING LOCUS T and FLOWERING LOCUS T INTERACTING PROTEIN 1. Plant J. 83: 1059–1068. [DOI] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Aubry S., Smith-Unna R.D., Boursnell C.M., Kopriva S., Hibberd J.M. (2014). Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 78: 659–673. [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Bratzel F., Turck F. (2015). Molecular memories in the regulation of seasonal flowering: From competence to cessation. Genome Biol. 16: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood A., Kopriva S., Morris R.J. (2016). Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell 28: 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO HubWeb Presence Working Group (2009). AmiGO: Online access to ontology and annotation data. Bioinformatics 25: 288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., et al. (2017). Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants 3: 814–824. [DOI] [PubMed] [Google Scholar]
- Cho L.H., Yoon J., An G. (2017). The control of flowering time by environmental factors. Plant J. 90: 708–719. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Contento A.L., Kim S.J., Bassham D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135: 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Cui H., Kong D., Liu X., Hao Y. (2014). SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 78: 319–327. [DOI] [PubMed] [Google Scholar]
- Dalchau N., Baek S.J., Briggs H.M., Robertson F.C., Dodd A.N., Gardner M.J., Stancombe M.A., Haydon M.J., Stan G.B., Gonçalves J.M., Webb A.A. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 108: 5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18: 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2011). The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G., Hedrich R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55: 746–759. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Pu L., Turco G., Gaudinier A., Morao A.K., Harashima H., Kim D., Ron M., Sugimoto K., Roudier F., Brady S.M. (2016). Transcriptional regulation of Arabidopsis Polycomb Repressive Complex 2 coordinates cell-type proliferation and differentiation. Plant Cell 28: 2616–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schepper V., De Swaef T., Bauweraerts I., Steppe K. (2013). Phloem transport: a review of mechanisms and controls. J. Exp. Bot. 64: 4839–4850. [DOI] [PubMed] [Google Scholar]
- DeWitt N.D., Sussman M.R. (1995). Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7: 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B.J., Bickle K.L., Schrage K.J., Muskett P., Patel K., Clark S.E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45: 1–16. [DOI] [PubMed] [Google Scholar]
- Disch S., Anastasiou E., Sharma V.K., Laux T., Fletcher J.C., Lenhard M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16: 272–279. [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Doi M., Hirayama J., Sassone-Corsi P. (2006). Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508. [DOI] [PubMed] [Google Scholar]
- Donohoe D.R., Bultman S.J. (2012). Metaboloepigenetics: Interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell. Physiol. 227: 3169–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Shimizu H., Nohales M.A., Araki T., Kay S.A. (2014). Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhorn J., Blanvillain R., Kröner C., Parrinello H., Rohmer M., Posé D., Ott F., Schmid M., Carles C. (2017). Dynamics of H3K4me3 chromatin marks prevails over H3K27me3 for gene regulation during flower morphogenesis in Arabidopsis thaliana. Epigenomes 1: 8. [Google Scholar]
- Espinoza C., Degenkolbe T., Caldana C., Zuther E., Leisse A., Willmitzer L., Hincha D.K., Hannah M.A. (2010). Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One 5: e14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray J.-P., Mostoslavsky R. (2016). Interplay between metabolism and epigenetics: A nuclear adaptation to environmental changes. Mol. Cell 62: 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B., Mas P. (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66: 318–329. [DOI] [PubMed] [Google Scholar]
- Farrona S., Thorpe F.L., Engelhorn J., Adrian J., Dong X., Sarid-Krebs L., Goodrich J., Turck F. (2011). Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feike D., et al. (2016). The starch granule-associated protein EARLY STARVATION1 is required for the control of starch degradation in Arabidopsis thaliana leaves. Plant Cell 28: 1472–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelmark K., Troein C. (2014). Rethinking transcriptional activation in the Arabidopsis circadian clock. PLOS Comput. Biol. 10: e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86. [DOI] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Nakamichi N., Kobayashi M., Hayashi N., Sakakibara H., Mizuno T., Saito K. (2009). Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 106: 7251–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk V., Kositsup B., Zhao C., Beers E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128: 84–94. [PMC free article] [PubMed] [Google Scholar]
- Gan E.S., Xu Y., Wong J.Y., Goh J.G., Sun B., Wee W.Y., Huang J., Ito T. (2014). Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 5: 5098. [DOI] [PubMed] [Google Scholar]
- Giakountis A., Coupland G. (2008). Phloem transport of flowering signals. Curr. Opin. Plant Biol. 11: 687–694. [DOI] [PubMed] [Google Scholar]
- Graf A., Schlereth A., Stitt M., Smith A.M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 107: 9458–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Wang Y., He Y. (2013). Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 11: e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli G., Figueroa P., Serino G., Deng X.W. (2007). Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19: 564–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Ng D.W., Li W.H., Chen Z.J. (2011). Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 21: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirner A., Ladwig F., Stransky H., Okumoto S., Keinath M., Harms A., Frommer W.B., Koch W. (2006). Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18: 1931–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T.,et al. (2013). Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25: 4894–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.J., Yang J., Yi J., An G. (2015). Controlling flowering time by histone methylation and acetylation in Arabidopsis and rice. J. Plant Biol. 58: 203–210. [Google Scholar]
- Jeong J.H., Song H.R., Ko J.H., Jeong Y.M., Kwon Y.E., Seol J.H., Amasino R.M., Noh B., Noh Y.S. (2009). Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One 4: e8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y.Q., Wang Y.H., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb Repressive Complex 2 components. PLoS One 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Gu X., He Y. (2009). Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Kong N.C., Gu X., Li Z., He Y. (2011). Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 7: e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L. (2010). Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 107: 21623–21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.L., Luquette L.J., Ho J.W.K., Ferrari F., Tolstorukov M., Minoda A., Issner R., Epstein C.B., Karpen G.H., Kuroda M.I., Park P.J. (2014). Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res. 42: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Stuart T., Valdes M., Breakfield N., Schmitz R.J., Nery J.R., Urich M.A., Han X., Lister R., Benfey P.N., Ecker J.R. (2016). Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2: 16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M.,et al. (2014). Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA 111: 6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe A., Tiessen A., Schluepmann H., Paul M., Ulrich S., Geigenberger P. (2005). Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. USA 102: 11118–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämke J., Bäurle I. (2017). Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. (2013). Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Liu L., Liu C., Hou X., Xi W., Shen L., Tao Z., Wang Y., Yu H. (2012). FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Knowles S.M., Webb C.J., Celaya R.B., Cha C., Siu J.P., Tobin E.M. (2011). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Gehan J.P., Sharkey T.D. (2005). Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 138: 2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J.E., Feil R., Hendriks J.H., Gibon Y., Morcuende R., Osuna D., Scheible W.R., Carillo P., Hajirezaei M.R., Stitt M. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher K.A.,et al. (2018). Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30: 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapeira J., Khaitova L.C., Mas P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 109: 21540–21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M.A., Gustafson-Brown C., Savidge B., Yanofsky M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277. [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- Mentch S.J., Mehrmohamadi M., Huang L., Liu X., Gupta D., Mattocks D., Gómez Padilla P., Ables G., Bamman M.M., Thalacker-Mercer A.E., Nichenametla S.N., Locasale J.W. (2015). Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J., Santos-González J., Hennig L., Köhler C. (2017). Applying the INTACT method to purify endosperm nuclei and to generate parental-specific epigenome profiles. Nat. Protoc. 12: 238–254. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Andrés F., Kanehara K., Liu Y.C., Dörmann P., Coupland G. (2014a). Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 5: 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Teo N.Z.W., Shui G., Chua C.H.L., Cheong W.-F., Parameswaran S., Koizumi R., Ohta H., Wenk M.R., Ito T. (2014b). Transcriptomic and lipidomic profiles of glycerolipids during Arabidopsis flower development. New Phytol. 203: 310–322. [DOI] [PubMed] [Google Scholar]
- Ortiz-Marchena M.I., Romero J.M., Valverde F. (2015). Photoperiodic control of sugar release during the floral transition: What is the role of sugars in the florigenic signal? Plant Signal. Behav. 10: e1017168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovaara J., Saiga S., Wendrich J.R., van ’t Wout Hofland N., van Schayck J.P., Hater F., Mutte S., Sjollema J., Boekschoten M., Hooiveld G.J., Weijers D. (2017). Transcriptome dynamics revealed by a gene expression atlas of the early Arabidopsis embryo. Nat. Plants 3: 894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.J., Primavesi L.F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59: 417–441. [DOI] [PubMed] [Google Scholar]
- Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10: 1096–1098. [DOI] [PubMed] [Google Scholar]
- Pikaard C.S., Mittelsten Scheid O. (2014). Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6: a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu J., Wahl V., Schmid M. (2011). Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant Sci. 2: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N., Morel P., Champelovier P., Thierry A.M., Negrutiu I., Jack T., Trehin C. (2015). SQUINT promotes stem cell homeostasis and floral meristem termination in Arabidopsis through APETALA2 and CLAVATA signalling. J. Exp. Bot. 66: 6905–6916. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone M.L., Faigón Soverna A., Sanchez S.E., Schlaen R.G., Hernando C.E., Seymour D.K., Mancini E., Chernomoretz A., Weigel D., Más P., Yanovsky M.J. (2013). LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 110: 12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Seo P.J., Mas P. (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J., Aragüez I., Peiró R., Mendez-Giraldez R., Zhang X., Jacobsen S.E., Bastolla U., Gutierrez C. (2014). The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (2002). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100. [DOI] [PubMed] [Google Scholar]
- Shimizu H., Katayama K., Koto T., Torii K., Araki T., Endo M. (2015). Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nat. Plants 1: 15163. [DOI] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. (2014). Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 15: 121–132. [DOI] [PubMed] [Google Scholar]
- Son O., et al. (2010). ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol. 51: 1537–1547. [DOI] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. (2015a). Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z.T., Sun L., Lu S.J., Tian Y., Ding Y., Liu J.X. (2015b). Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl. Acad. Sci. USA 112: 2900–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M., Wildenhain J., Rappsilber J., Tyers M. (2014). BoxPlotR: A web tool for generation of box plots. Nat. Methods 11: 121–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Brown G.D. (2011). DiffBind: Differential binding analysis of ChIP-seq peak data. (Bioconductor).
- Streb S., Zeeman S.C. (2012). Starch metabolism in Arabidopsis. Arabidopsis Book 10: e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45 (D1): D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 3: 564–570. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2013). Leaf development. Arabidopsis Book 11: e0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594. [DOI] [PubMed] [Google Scholar]
- Wahl V., Brand L.H., Guo Y.L., Schmid M. (2010). The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J.E., Stitt M., Schmid M. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Wang X., Feng S., Nakayama N., Crosby W.L., Irish V., Deng X.W., Wei N. (2003). The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15: 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gu X., Yuan W., Schmitz R.J., He Y. (2014). Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell 28: 727–736. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Yan Y., Shen L., Chen Y., Bao S., Thong Z., Yu H. (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 30: 437–448. [DOI] [PubMed] [Google Scholar]
- Yang H., et al. (2012). A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet. 8: e1002664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Howard M., Dean C. (2016). Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. USA 113: 9369–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Jiang D., Jiang J., He Y. (2010). A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 62: 663–673. [DOI] [PubMed] [Google Scholar]
- Yeates T.O. (2002). Structures of SET domain proteins: Protein lysine methyltransferases make their mark. Cell 111: 5–7. [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139: 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Sawikowska A., Neumann M., Posé D., Capovilla G., Langenecker T., Neher R.A., Krajewski P., Schmid M. (2017). Temporal dynamics of gene expression and histone marks at the Arabidopsis shoot meristem during flowering. Nat. Commun. 8: 15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Barthelson R.A., Lambert G.M., Galbraith D.W. (2008a). Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008b). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Craig J.C., Petzold H.E., Dickerman A.W., Beers E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138: 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Wang W., Karapetyan S., Mwimba M., Marqués J., Buchler N.E., Dong X. (2015). Redox rhythm reinforces the circadian clock to gate immune response. Nature 523: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]