A mechanism is established that tunes the hydraulics of leaf tissues to synchronize water supply with day and night cycles, thereby optimizing plant growth.
Abstract
The circadian clock regulates plant tissue hydraulics to synchronize water supply with environmental cycles and thereby optimize growth. The circadian fluctuations in aquaporin transcript abundance suggest that aquaporin water channels play a role in these processes. Here, we show that hydraulic conductivity (Kros) of Arabidopsis (Arabidopsis thaliana) rosettes displays a genuine circadian rhythmicity with a peak around midday. Combined immunological and proteomic approaches revealed that phosphorylation at two C-terminal sites (Ser280, Ser283) of PLASMA MEMBRANE INTRINSIC PROTEIN 2;1 (AtPIP2;1), a major plasma membrane aquaporin in rosettes, shows circadian oscillations and is correlated with Kros. Transgenic expression of phosphodeficient and phosphomimetic forms of this aquaporin indicated that AtPIP2;1 phosphorylation is necessary but not sufficient for Kros regulation. We investigated the supporting role of 14-3-3 proteins, which are known to interact with and regulate phosphorylated proteins. Individual knockout plants for five 14-3-3 protein isoforms expressed in rosettes lacked circadian activation of Kros. Two of these [GRF4 (14-3-3Phi); GRF10 (14-3-3Epsilon)] showed direct interactions with AtPIP2;1 in the plant and upon coexpression in Xenopus laevis oocytes and activated AtPIP2;1, preferentially when the latter was phosphorylated at its two C-terminal sites. We propose that this regulatory mechanism assists in the activation of phosphorylated AtPIP2;1 during circadian regulation of Kros.
INTRODUCTION
Plants have to cope with daily fluctuations in temperature, light, and humidity. They continuously adjust their water status through stomatal regulation and osmotic adjustment and by acting on the water transport capacity (hydraulic conductivity) of their root and shoot tissues. The latter processes are individually controlled by various climatic or endogenous hormonal factors. The circadian clock (Moshelion et al., 2002; Nardini et al., 2005; Takase et al., 2011) also tunes plant tissue hydraulics to synchronize water supply with environmental cycles and thereby optimize growth (Caldeira et al., 2014). Genetic and pharmacological studies have indicated that short-term plant hydraulic regulation is in large part mediated through aquaporin (AQP) water channels (Chaumont and Tyerman, 2014; Maurel et al., 2015). For instance, circadian oscillations of hydraulic conductance of maize (Zea mays) plants, water permeability of rain tree (Samanea saman) leaf motor cells, and water content of Arabidopsis (Arabidopsis thaliana) roots are typically paralleled by fluctuations in the transcript abundance of AQPs (Moshelion et al., 2002; Takase et al., 2011; Caldeira et al., 2014).
The circadian clock, an endogenous, cell-autonomous biological timekeeper that produces outputs with rhythms close to 24 h, functions through a set of transcription-translation feedback loops that drives the rhythmic transcription of core clock genes. Circadian rhythms are also based on numerous temporally regulated posttranslational modifications of the core clock proteins, in particular phosphorylation (Seo and Mas, 2014; Mendoza-Viveros et al., 2017). An uncharacterized nontranscriptional clock mechanism that drives persistent overoxidation rhythms in transcriptionally inactive cells was also reported across taxa (O’Neill and Reddy, 2011; O’Neill et al., 2011). Thus, posttranslational modifications of proteins may constitute a basic and ancient level of timekeeping in modern organisms. Although much emphasis has been placed on global analyses of transcriptome dynamics during circadian oscillations in eukaryotes, two recent genome-wide analyses revealed a broad pattern of circadian-dependent protein phosphorylation that controls the metabolism and physiology of mouse liver (Robles et al., 2017) and >100 proteins involved in photosynthesis, hormone signaling, and membrane transport in Arabidopsis (Choudhary et al., 2015).
When compared with gene regulation, reversible phosphorylation represents a key, highly flexible mode for AQP regulation (Johansson et al., 1998; Törnroth-Horsefield et al., 2006). Yet, its involvement in the diurnal or circadian regulation of tissue hydraulics has remained elusive (Lopez et al., 2003). We previously demonstrated that phosphorylation of the PLASMA MEMBRANE INTRINSIC PROTEIN 2;1 (AtPIP2;1) AQP, at two C-terminal sites (Ser280, Ser283; Johansson et al., 1996) is necessary but not sufficient for the enhancement of Arabidopsis rosette hydraulic conductivity (Kros) under darkness (Prado et al., 2013). In the present work, we show that rather than a light adaptation mechanism, this regulation reflects a genuine circadian rhythmicity and is correlated with oscillations of AtPIP2;1 phosphorylation. We further use this context to investigate the supporting role of 14-3-3 proteins. These phosphosensors are known to interact with phosphorylated client proteins to change their activity, localization, stability, or association with protein complexes (Chevalier et al., 2009). Although 14-3-3 proteins were previously shown to interact with AQPs (Jaspert et al., 2011; Moeller et al., 2016), the present study uncovers a role for these proteins in plant hydraulics via the regulation of AtPIP2;1 activity and water transport in rosettes.
RESULTS
Circadian Oscillation of Kros
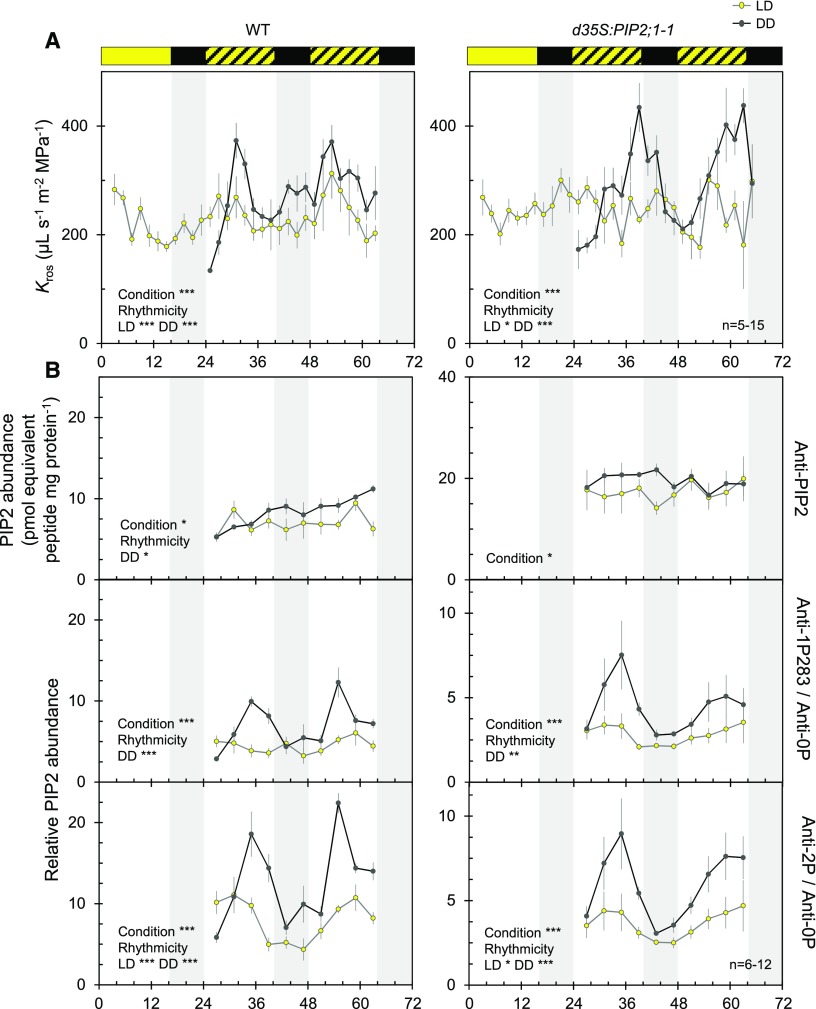
We previously investigated Kros in excised organs by taking pressure chamber measurements (Prado et al., 2013; Prado and Maurel, 2013). When compared with plants grown in the light (L) under a 16-h light/8-h dark (LD) cycle, Arabidopsis wild-type plants exposed to prolonged (13–23 h) darkness showed a stimulation of Kros by ∼50% because of the activity of AtPIP2;1 in leaf veins, for example, bundle sheath and xylem parenchyma cells (Prado et al., 2013). Here, we refined these kinetic measurements and uncovered a daily rhythm of Kros under LD cycles with peaks slightly before midday (Zeitgeber Time [ZT] 31 and ZT53; Figure 1A). To determine the kinetics of a potential dark adaptation mechanism, we also monitored Kros under constant darkness (DD). Surprisingly, spontaneous oscillations of Kros in phase with those of LD plants but with significantly larger amplitude were observed (Figure 1A). Transgenic plants overexpressing AtPIP2;1 under the control of a double 35S CaMV promoter (d35S:PIP2;1-1; Péret et al., 2012) showed fluctuations with no discernible pattern under LD cycles (Figure 1A). By contrast, we observed greater than twofold spontaneous daily oscillations in Kros with a peak before the subjective dusk and thus a slight phase shift compared with wild-type plants under DD conditions (Figure 1A). These measurements reveal pronounced circadian regulation of Kros in Arabidopsis.
Figure 1.
Circadian Rhythms of Kros and AQP Phosphorylation.
(A) Temporal Kros profiles of wild type (left) and d35S:PIP2;1-1 (right) plants under LD (yellow dots) or DD (gray dots) conditions. Yellow and black bars at top indicate light and dark treatments, whereas hatched bars indicate days and subjective days (dark) under LD and DD conditions, respectively. Plants of the DD series were transferred to an independent dark growth chamber at ZT20. This displacement is thought to disturb the plants and induce a slight drop in Kros, as observed at ZT24. Each point represents an average Kros value (±se; 5–15 plants) per 2 h from 3 independent cultures. Asterisks indicate an effect of a condition based on ANOVA with a dynamic linear model or significant rhythmicity, as determine by analysis with CircWave software (*P < 0.05; **P < 0.01; ***P < 0.001).
(B) ELISA analysis of temporal variation of PIP2 protein abundance (top) and AtPIP2;1-AtPIP2;3 C-terminal phosphorylation (four at bottom) in microsomal rosette extracts. Average signals obtained for each genotype and condition using anti-1P283 or anti-2P antibodies were divided by the corresponding signal of the anti-0P antibody. Cumulative data (mean ± se) from 3 biological (plant cultures) and 2 technical repeats. Experimental conditions, conventions and details on statistical tests as in (A).
AQP Phosphorylation Shows Circadian Oscillations in Phase with the Oscillations of Kros
Given that C-terminal di-phosphorylation of AtPIP2;1 is required for dark-induced enhancement of Kros (Prado et al., 2013), we also investigated the kinetics of AQP phosphorylation in rosettes under LD and DD conditions. The first step was to prepare antibodies raised against the nonphosphorylated (anti-0P) or the mono- or di-phosphorylated AtPIP2;1 C terminus (anti-1P280; anti-1P283; anti-2P; Supplemental Figure 1). The AtPIP2;1 C-terminal sequence is shared with the AtPIP2;2 and AtPIP2;3 homologs, which, however, show a much lower abundance in the Arabidopsis rosette (Prado et al., 2013). When used to probe rosette microsomal extracts by protein gel blot analysis, the anti-1P283 and anti-2P antibodies revealed profiles similar to those obtained with a previously prepared generic anti-PIP2 antibody (Santoni et al., 2003). The two antibodies recognized a typical PIP2 band at ∼30 kD in wild-type plant extracts; this signal was reduced and enhanced in pip2;1-2 and d35S:PIP2;1-1, respectively. Multimeric forms at >55 kD were also revealed in the latter extract. The anti-0P and anti-1P280 showed a weaker signal but also detected a PIP2 doublet in d35S:PIP2;1-1 extracts. Thus, the anti-0P, anti-1P280, anti-1P283, and anti-2P antibodies mostly reacted with AtPIP2;1 and possibly its two close homologs in rosette extracts. Next, we performed ELISAs using purified AQP peptides as baits. These assays showed that, although they are not fully phosphorylation site specific, the anti-1P280, anti-1P283, and anti-2P antibodies preferentially recognize the phosphorylated over the nonphosphorylated forms of AtPIP2;1 (Supplemental Figures 1B and 1C). Thus, when the AtPIP2;1 peptides were provided in equal amounts, the nonphosphorylated form accounted for 3–15% of the overall ELISA signal delivered by these antibodies. Following these characterizations, we used the antibodies in ELISAs on microsomal extracts of rosettes sampled over time in wild type and d35S:PIP2;1-1 plants (Figure 1B and Supplemental Figure 2). In both genotypes, the generic anti-PIP2 antibody (Santoni et al., 2003), which probes overall AtPIP2;1-AtPIP2;3 abundance, yielded a signal lacking rhythmicity over two successive days, with similar patterns under LD and DD conditions (Figure 1B). When normalized with respect to the anti-PIP2 signal, the semi-quantitative phosphorylation signals of anti-1P283 and anti-2P showed significant daily rhythms in wild type, with a similar tendency in d35S:PIP2;1-1 (Supplemental Figure 2). Also, a significant difference in phosphorylation (by 1.5- and twofold) between LD and DD conditions was observed for wild type and d35S:PIP2;1-1 plants, respectively. Interestingly, we obtained curves with more pronounced variations by normalizing the anti-1P283 and anti-2P signals with respect to the anti-0P signal (Figure 1B). In this case, significant rhythmicity was detected in both wild type and d35S:PIP2;1-1 plants under LD and DD conditions, with the phosphorylation signals approximately in phase with variations in Kros (Figure 1).
We performed more precise investigation of the relative abundance of non- (0P), mono- (1P), or di-phosphorylated (2P) C-terminal forms of AtPIP2;1 via label-free phosphoproteomics at three representative ZT time points in both genotypes and under LD and DD conditions (Supplemental Figure 3A). The 1P and 2P peptides exhibited a single phosphorylation at Ser280 and a double phosphorylation at Ser280 and Ser283, respectively (Prado et al., 2013; Qing et al., 2016). The measured phosphorylation ratios (Supplemental Figure 3B) were in good agreement with the dramatic peaks in ELISA signals (Figure 1B) observed at ZT35 under DD conditions. Next, we considered the whole set of parallel Kros and AQP phosphorylation kinetic measurements in wild type and d35S:PIP2;1-1 plants. Realizing that phosphorylation of Ser283 is always present together with that of Ser280 (Prak et al., 2008), we observed a significant positive correlation of Kros with AtPIP2;1 di-phosphorylation in wild-type plants, as detected with the anti-2P and anti-1P283 antibodies (Supplemental Figure 4). Similar correlations were observed for the d35S:PIP2;1-1 line provided that phosphorylation signals were reported to the Kros values corresponding to a delayed phase of 2 h (Supplemental Figure 4). These analyses support previous findings obtained using a pip2;1-1 mutant expressing phosphodeficient or phosphomimetic forms of AtPIP2;1, with Ser280 and Ser283 mutated to Ala (double Ser-to-Ala [dSA]) or Asp (double Ser-to-Asp [dSD]), respectively. Analysis of these plants revealed the requirement of C-terminal di-phosphorylation of AtPIP2;1 for dark-induced Kros (Prado et al., 2013). A kinetic re-assessment of these earlier data indicated that, under DD compared with LD conditions, Kros of pip2;1-1 plants expressing the native or dSD forms of AtPIP2;1 was strongly increased at subjective midday and decreased thereafter. By contrast, native pip2;1-1 plants, as well as those expressing the dSA form of AtPIP2;1, showed steady and very similar Kros values between LD and DD conditions at all time points (Supplemental Figure 5). The overall data point to a requirement for AtPIP2;1 phosphorylation for the circadian regulation of leaf hydraulics.
General Regulatory Factors are Necessary for Circadian Regulation of Kros
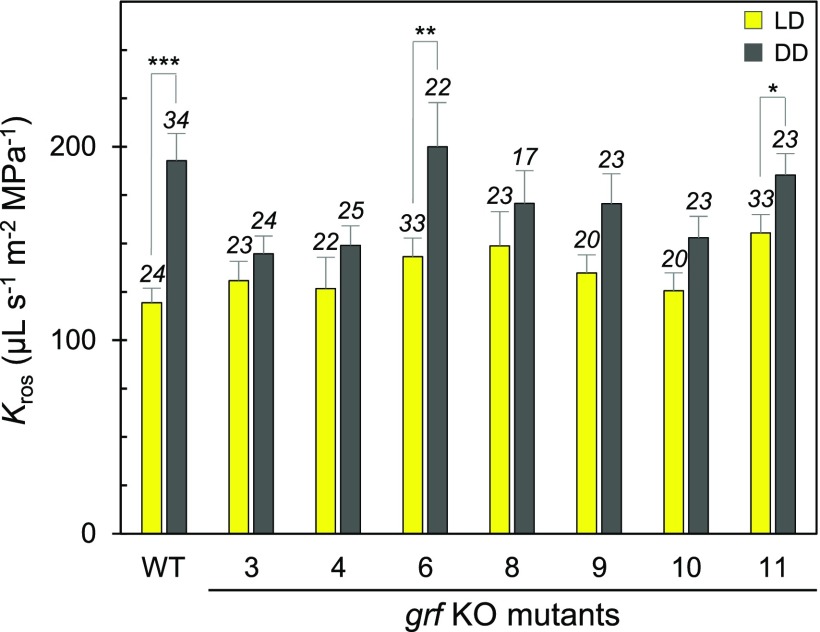
Because PIP phosphorylation is necessary but not sufficient for Kros regulation (Prado et al., 2013), we wondered about the complementary molecular activities that may mediate the circadian activation of phosphorylated AtPIP2;1 in the Arabidopsis rosette. The 14-3-3 proteins, also called General Regulatory Factors (GRFs) in Arabidopsis, are good candidates, because they preferentially bind to and modulate the functions of phosphorylated proteins (Wilson et al., 2016) and they can physically interact with AQPs (Jaspert et al., 2011; Moeller et al., 2016). To address the putative role of plant GRFs in the circadian regulation of leaf AQPs, we identified six isoforms out of the 13 GRFs in Arabidopsis that show significant expression in leaves (Wilson et al., 2016). We then selected the corresponding knockout (KO) mutants (grf3, grf4, grf6, grf8, grf9, and grf10) and a KO mutant for GRF11 (14-3-3 omicron), which is expressed at low levels and was therefore used as a reference, and investigated their Kros responses to prolonged darkness (i.e., an 8-h night prolonged by 4–21 h of darkness; DD; Figure 2; Supplemental Figure 6). In contrast with wild-type plants, grf6 (14-3-3 lambda) and grf11, the five other grf mutants showed attenuated or a lack of enhancement of Kros in DD compared with LD conditions, suggesting that each of the corresponding GRFs somewhat contributes to Kros rhythmicity.
Figure 2.
Kros of Single grf Knockout Mutants.
Wild type and single grf knockout (KO) mutants (numbered) were grown under LD (yellow bars) or DD (gray bars) conditions. Kros measurements were performed over a 17 h-period (between ZT28 and ZT45; see Figure 1) and averaged. Cumulative data (+se) from 6 independent cultures are shown, with the number of plants shown above the bars. Asterisks indicate significant effects of growth conditions, as determined by pairwise comparisons using a Wilcoxon test (*P < 0.05; **P < 0.01; ***P < 0.001).
CoExpression of GRFs and AtPIP2;1 in Xenopus laevis Oocytes Reveals Distinct Activation Capacities
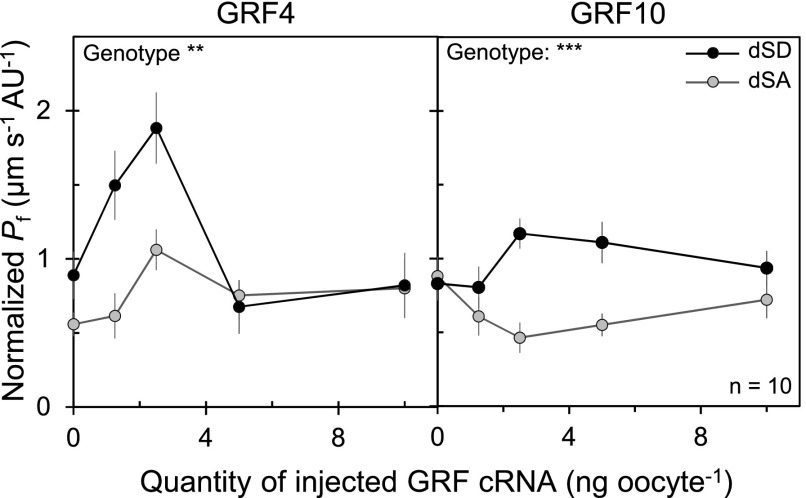
Because the adverse effects of GRF inactivation on Kros oscillation may simply be because of a pleiotropic impact on the circadian clock in leaves, we further investigated the capacity of GRFs to directly target AQP function. Oocytes of the aquatic frog X. laevis were used for functional expression of native, phospho-deficient (dSA) or phospho-mimetic (dSD) forms of AtPIP2;1 fused to green fluorescent protein (GFP; GFP-PIP2;1). We estimated the genuine water transport activity of each form based on the ratio between oocyte osmotic water permeability (Pf) and abundance of the GFP-PIP2;1 form at the oocyte surface, as determined based on peripheral fluorescence intensity (Supplemental Figures 7A and 7B). Resulting normalized Pf was similar for individually expressed native, dSA, or dSD forms of GFP-PIP2;1 (Supplemental Figure 7C). To test the capacity of GRFs to regulate the activity of AtPIP2;1 in a phosphorylation-dependent manner, the seven Arabidopsis GRFs investigated above were individually coexpressed with the dSA or dSD form of GFP-PIP2;1 (Figure 3). dSD GFP-PIP2;1 yielded a significantly higher normalized Pf than dSA GFP-PIP2;1 upon coexpression with GRF3 (14-3-3 Psi), GRF4 (14-3-3 Phi), GRF8 (14-3-3 Kappa), and GRF10 (14-3-3 Epsilon). Notably, the corresponding KO plants also lacked dark-induced activation of Kros (Figure 2). GRF4 and GRF10, which gave the strongest responses in the oocyte assay, were investigated in closer detail. We obtained bell-shaped curves of GFP-PIP2;1 activity as a function of the dose of GRF4 complementary RNA (cRNA) injected in oocytes and, to a lesser extent, of GRF10 cRNA, indicating that a precise stoichiometry of AQP and GRF may be needed for optimal activation, which was in both cases more pronounced with the dSD than the dSA form (Figure 4). The overall data indicate that GRF4 and GRF10 can functionally discriminate and regulate AtPIP2;1 activity according to its phosphorylation status.
Figure 3.
Functional Coexpression of AtPIP2;1 and GRFs in Xenopus Oocytes.
Effects of GRFs on normalized Pf of oocytes expressing dSA (white bars) or dSD (black bars) forms of GFP-PIP2;1. Each oocyte was injected with PIP2;1-GFP cRNA (2.5 ng) and water (H2O) or 2.5 ng cRNA of the indicated GRF. Data are means + se from the indicated number of oocytes from 3 independent batches. Asterisks indicate significant difference, as determined by pairwise comparisons using a Wilcoxon test (*P < 0.05; ***P < 0.001).
Figure 4.
Dose-Dependent Effects of GRF4 and GRF10 on AtPIP2;1 Activity in Xenopus Oocytes.
Dose-dependent effects of GRF4 (left) and GRF10 (right) on normalized Pf of oocytes expressing dSA (gray dots) or dSD (black dots) forms of GFP-PIP2;1. Oocytes were co-injected with the indicated quantity of GRF cRNAs and 2.5 ng of dSA or dSD GFP-PIP2;1 cRNA. The higher normalized Pf seen in dSD compared with dSA oocytes in the absence of GRF4 is probably because of self-activation of dSD or its activation by an endogenous oocyte 14-3-3 protein in some oocyte batches. Representative data from one batch of oocytes are shown, with each point representing mean ± se from 10 cells. The effect of genotype was investigated by ANOVA with a dynamic linear model (**P < 0.01; ***P < 0.001).
The residual activation of dSA GFP-PIP2;1 with GRF4 suggests the presence of another 14-3-3 recognition site in AtPIP2;1 complementary to the two C-terminal phosphorylated Ser. A phosphorylation site located in the second cytosolic loop (Ser121) possibly matches a consensus 14-3-3 binding motif (Wilson et al., 2016). To investigate its role in 14-3-3-dependent activation, we designed dSA forms of GFP-PIP2;1 carrying phosphodeficient (AdSA) or phosphomimetic (DdSA) mutations at Ser121 (Supplemental Figure 8). The lack of activation of the DdSA form of GFP-PIP2;1 (carrying phosphomimetic mutation at Ser121) by GRF4 indicates that this putative interaction site plays a minor functional role, if any.
Activation of AtPIP2;1 by GRF4 and GRF10 Involves Phosphorylation-Dependent Molecular Interactions
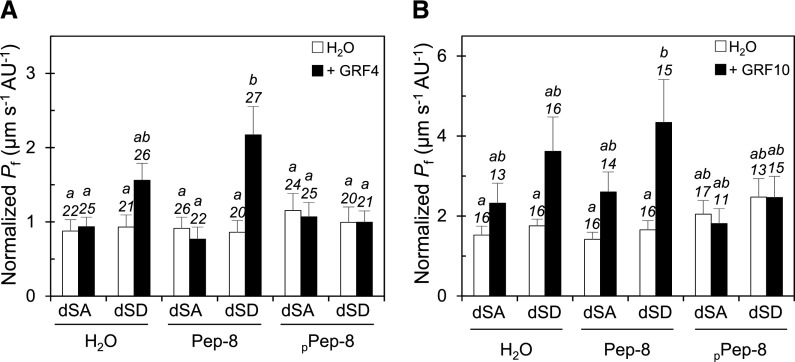
To further define the properties of the activation of AtPIP2;1 by GRF4 and GRF10, we used a synthetic phosphorylated peptide (PPep-8) that mimics the mode I 14-3-3 binding motif (Moorhead et al., 1999), that is, it binds to an amphipathic groove conserved in all 14-3-3 monomers and therefore competes with their phosphorylated interactors. When pre-injected in oocytes for 2 h before water transport measurements, PPep-8 but not its unphosphorylated form (Pep-8) was able to counteract the activation of dSD GFP-PIP2;1 by GRF4 coexpression (Figure 5A). Neither PPep-8 nor Pep-8 had significant effects on the normalized Pf value of oocytes expressing dSD GFP-PIP2;1 alone or dSA GFP-PIP2;1 alone or in combination with GRF4. In these experiments, we also examined the fluorescence signal at the periphery of oocytes expressing dSA or dSD GFP-PIP2;1 (Supplemental Figure 9). Neither coexpression with GRF4 nor co-injection with PPep-8 or Pep-8 had a significant effect on the subcellular localization of the two GFP-PIP2;1 forms. Thus, GRF4 enhances the functionality of GFP-PIP2;1 without altering its targeting or stability at the oocyte plasma membrane. Similar observations were made in oocytes coexpressing the two GFP-PIP2;1 forms and GRF10 and co-injected with PPep-8 or Pep-8 (Figure 5B).
Figure 5.
Effects of Peptide Co-Injection on the Activation of GFP-PIP2;1-induced Water Transport by GRF4 and GRF10.
Oocytes were injected with 2.5 ng cRNA of dSA or dSD GFP-PIP2;1 in the absence (H2O; white bars) or presence of 2.5 ng GRF4 [(A); black bars] or GRF10 [(B); black bars] cRNA. After 2 d, the oocytes were re-injected with the indicated peptide (Pep-8 or pPep-8) or H2O as a negative control and incubated for 2 h before the water transport assays. Data are means + se from the indicated number of oocytes and 2 independent batches. Different letters indicate statistically different values at P < 0.05, as determined by ANOVA with a generalized linear model followed by a Tukey test.
GRF4 and GRF10 Directly Bind to AtPIP2;1
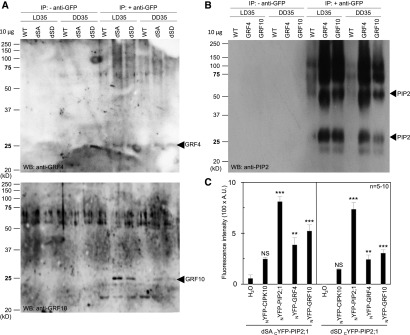
A previous interactomic study (Bellati et al., 2016) indicated that, in the Arabidopsis root, AtPIP2;1 can interact with several GRFs, including GRF10. In the present work, we investigated whether AtPIP2;1 interacts directly with GRF4 and GRF10 in vivo in both Arabidopsis leaves and Xenopus oocytes (Figure 6). First, we performed reverse co-immunoprecipitations using transgenic plants expressing either the dSA or dSD form of GFP-PIP2;1 or GRF4 or GRF10 also tagged with GFP (Figures 6A and 6B). The same anti-GFP antibody was used for immunoprecipitation of protein extracts from wild type or the transgenic lines grown under LD or DD conditions. Subsequent protein gel blot analysis using anti-GRF or anti-PIP2 antibodies revealed specific bands matching the expected size of GRFs (Figure 6A) or PIP2 (Figure 6B) in extracts from GFP-PIP2;1 and GRF-GFP lines, respectively, compared with WT. We further investigated physical interactions between AtPIP2;1 and GRF4 or GRF10 by Bimolecular Fluorescence Complementation (BiFC) in Xenopus oocytes (Figure 6C). Cells were injected with cRNA of dSA or dSD C-terminal portion of Yellow Fluorescent Protein(cYFP)-PIP2;1 in the absence (water) or presence of cRNAs encoding GRF4 or GRF10 fused to N-terminal portion of Yellow Fluorescent Protein (NYFP). Fusions of CBL-INTERACTING PROTEIN KINASE (CIPK10) protein kinase and AtPIP2;1 with NYFP were tentatively used as negative (Waadt et al., 2008) and positive controls, respectively. With respect to oocytes solely expressing dSA or dSD cYFP-PIP2;1 fusions, a significant increase in fluorescence intensity was noticed in the presence of NYFP fusions with GRF4, GRF10 or AtPIP2;1 but not CIPK10. These data point to a physical interaction between dSA or dSD PIP2;1 and GRF4 or GRF10. We also investigated molecular interactions between AtPIP2;1 and GRF4 by performing Fluorescence Lifetime Imaging Microscopy (FLIM) of tobacco (Nicotiana tabacum) leaf epidermal cells coexpressing fluorescent proteins GFP and red fluorescent protein (RFP) fused with AtPIP2;1 (either native or with dSA or dSD mutations) and GRF4, respectively. Förster resonance energy transfer (FRET) efficiency between a GFP-PIP2;1 donor and a RFP-GRF4 acceptor was significantly higher than in control interactions with either an undifferentiated donor (GFP) or acceptor (RFP; Supplemental Table 1). Although the three types of in vivo approaches presented above assessed molecular interactions between AtPIP2;1 and GRF4 or GRF10, no quantitative difference in interaction was detected between the dSA and dSD forms of AtPIP2;1. Because these forms have distinct functionalities when coexpressed with GRF4 or GRF10 (Figures 3 to–5), these results suggest that a molecular interaction with these GRFs is not sufficient per se to alter the activity of AtPIP2;1.
Figure 6.
In Vivo Interaction Between AtPIP2;1 and GRFs.
(A) and (B) Wild type (WT; Col-0) or transgenic plants expressing the dSA or dSD forms of GFP-PIP2;1 (A) or fusions of GFP with GRF4 or GRF10 (B) were cultivated in vitro for 2 weeks before transfer to soil for two additional weeks under LD conditions. Leaves were harvested at ZT35 in plants grown under either condition (LD35, DD35). Immunoprecipitations were performed in the absence (− anti-GFP) or presence (+ anti-GFP) of an anti-GFP antibody. The proteins were further analyzed by protein gel blot (protein gel blot; WB) analysis using anti-GRF4 [(A), top], anti-GRF10 [(A), bottom], or anti-PIP2 (B) antibodies. All lanes were loaded with 10 µg of proteins. Arrowheads indicate the expected molecular weight of monomeric forms of GRF4 or GRF10 (A) or monomeric and dimeric forms of PIP2 (B). The above experiments were repeated twice with similar results.
(C) BiFC analysis of PIP2;1-GRF complexes in oocytes. Oocytes were injected with 2.5 ng cRNA of the dSA or dSD forms of cYFP-PIP2;1 in the absence (H2O) or presence of 2.5 ng cRNA of CIPK10, AtPIP2;1, GRF4, or GRF10 fused to NYFP. Oocytes expressing dSA or dSD and GRF linked by BiFC are shown and compared with oocytes injected with H2O. CIPK10-NYFP and NYFP-PIP2;1 were used as negative and positive controls, respectively, for fluorescence complementation with the forms of cYFP-PIP2;1. Representative data from one batch of oocytes are shown, with each bar representing mean + se from 5 to 10 oocytes. The significance of fluorescence complementation was tested using ANOVA with a generalized linear model followed by a simultaneous test for general linear hypotheses taking the H2O series as a reference (NS, not significant; **P < 0.01; ***P < 0.001). Note that the data sets for dSA cYFP-PIP2;1 and dSA cYFP-PIP2;1 were acquired independently and cannot be compared. A.U., arbitrary units.
Overall, these data show how specific 14-3-3 proteins (GRF4 or GRF10) can activate AtPIP2;1, preferentially in its di-phosphorylated form, via a process involving a conserved site of the 14-3-3 protein required for binding to its client targets. We propose that this regulatory mechanism assists in the activation of phosphorylated AtPIP2;1 during the circadian regulation of Kros.
DISCUSSION
Several studies have described circadian variations in plant tissue hydraulics with parallel fluctuations in AQP transcript abundance (Moshelion et al., 2002; Takase et al., 2011; Caldeira et al., 2014). Yet, reversible phosphorylation is emerging as a major mechanism for the large-scale distribution of timing information and circadian regulation of multiple cellular targets (Choudhary et al., 2015; Robles et al., 2017). By combining immunological and proteomic approaches, we performed rigorous analysis of the circadian oscillations of phosphorylation of a major leaf AQP isoform and established a parallel with clock-regulated leaf hydraulics. Furthermore, we assessed the requirement for phosphorylation of AtPIP2;1 at two C-terminal sites via transgenic expression of phosphodeficient and phosphomimetic forms of this AQP. The functional link between the clock and cycles of tissue hydraulics depending on phosphorylation appears to be particularly relevant for proteins with a slow turnover rate such as AQPs (Luu et al., 2012). The slight phase shift in Kros oscillations observed after AtPIP2;1 overexpression points, however, to additional feedback effects of the transcriptional and/or posttranscriptional components of plant hydraulics. Nevertheless, the common idea that oscillations of AQP transcript abundance as observed in various plant organs (Moshelion et al., 2002; Takase et al., 2011; Caldeira et al., 2014) can directly account for the circadian control of tissue hydraulics will have to be critically reexamined.
Circadian rhythms provide an adaptive advantage to living organisms, allowing them to correctly anticipate daily changes in environmental parameters. In wild-type plants, Kros and AQP phosphorylation peaked around midday, possibly to support water supply and leaf growth under high transpiration demand. Yet, it is somewhat surprising that their oscillation amplitudes were larger under DD than LD conditions. In relation to earlier work in maize (Caldeira et al., 2014) indicating that circadian oscillations in root hydraulics were the highest under low water availability, we propose that the variations in Kros under an LD regime were of low amplitude simply because the plants were grown in hydroponic (i.e., water replete) conditions. We further speculate that, under DD conditions, the lack of a strong transpirational driving force may have required an alternative, osmotically driven transport of water and therefore enhanced transcellular hydraulic conductivities and high Kros. Enhanced tissue hydraulics would thereby support peaks in leaf growth and metabolic activities during subjective days.
AQP phosphorylation, however, is not the only driver of cycling of plant tissue hydraulics. The supporting role of 14-3-3 proteins, which usually interact with and activate phosphorylated proteins, was demonstrated, unraveling a mechanism for plant AQP regulation. Indeed, functional studies in oocytes indicated that at least two plant 14-3-3 protein isoforms (GRF4 and GRF10) are able to regulate AtPIP2;1, preferentially after Ser280 and Ser283 di-phosphorylation. We note, however, that, although GRF10 had no stimulatory effect on dSA AtPIP2;1, a slight GRF4-dependent activation of this form was sometimes observed (Figure 4), although less prominent than for the dSD form. More unexpected was the binding of GRF4 and GRF10 to AtPIP2;1, which occurred irrespective of the phosphorylation status of its C-terminal domain. Because this domain represents an atypical 14-3-3 binding site, other AtPIP2;1 regions may be involved. Overall, our data point to a model whereby the GRF-AtPIP2;1 molecular interaction would either occur at the PIP2;1 C terminus and would be phosphorylation-independent or would involve alternative sites. In either case, the subsequent activation process of AtPIP2;1 would essentially require its C-terminal di-phosphorylation. Moreover, expression monitoring in oocytes suggested that GRF4 and GRF10 activate AtPIP2;1 by acting on pore gating rather than protein trafficking or stability (Moeller et al., 2016). Based on the Kros phenotype of grf4 and grf10 plants, we further propose that 14-3-3 protein-dependent regulation of AtPIP2;1 operates in leaf veins, thereby explaining how Ser280 and Ser283 di-phosphorylation is required but not sufficient for circadian oscillations of Kros (Prado et al., 2013; this work). According to this model, other clock-regulated components, in conjunction with variations in AtPIP2;1 phosphorylation, would help adjust the phase and amplitude of Kros oscillations. In particular, we observed that although four distinct 14-3-3 proteins can individually activate AtPIP2;1 in oocytes, they seem not to be fully redundant in planta. Thus, rhythmic variations in the functionality of these 14-3-3 proteins, possibly through variable phosphorylation (Paul et al., 2012) or stability, may be necessary for both PIP activation and upstream signaling events leading to PIP phosphorylation.
Leaf water status is at the crossroads of the fundamental physiological functions of plants, including growth and adaptation to environmental changes. Clock-generated biological rhythms provide an adaptive advantage (Dodd et al., 2005; Pruneda-Paz and Kay, 2010), presumably by allowing organisms to correctly anticipate daily changes in temperature, light, and humidity (Atamian and Harmer, 2016). The present work provides a refined mechanistic understanding of clock-regulated leaf water status, which may be crucial for genetically manipulating and improving crop performance.
METHODS
Plant Materials and Growth Conditions
All experiments were performed with Arabidopsis (Arabidopsis thaliana) wild type plants or derived transgenic lines with altered AtPIP2;1 or GRF functions. The pip2;1-2 line and plants overexpressing a AtPIP2;1 cDNA under the control of a double enhanced CaMV 35S promoter (d35S:PIP2;11) were previously described (Da Ines et al., 2010; Péret et al., 2012). The d35S:PIP2;1-1 line was derived from the natural WS2 accession and was always compared with proper wild type plants. Transgenic plants expressing GFP-PIP2;1 dSA or dSD or GRF4-GFP or GRF10-GFP (kindly provided by Dr. R. Ferl; Paul et al., 2005, 2012;) under the control of a constitutive 35S promoter were used for co-immunoprecipitations. The SAIL_739_D01 (grf9) and GK-848G03 (grf10) T-DNA insertion lines were obtained from the Nottingham Arabidopsis Stock Center. Homozygous mutations and gene inactivation were verified by genotyping and reverse transcription quantitative PCR (see Supplemental Figure 6) using the primers indicated in Supplemental Table 2. Reverse transcription quantitative PCR was performed according to Czechowski et al., 2005 using At1g13320, At2g28390, At3g01150, and At4g34270 as reference genes. All procedures for plant genotyping were as described (Postaire et al., 2010). The rci1a (grf3), SALK_088321 (grf4), SALK_129554C (grf6), SALK_148929 (grf8), and SALK_123168C (grf11) mutants were described elsewhere (Yang et al., 2013; Catalá et al., 2014; van Kleeff et al., 2014). For physiological or PIP expression analysis, plants were germinated in vitro and grown in hydroponic conditions as described (Postaire et al., 2010) unless otherwise specified in the figure legend. Briefly, sterile seeds were sown on 1/2 Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% Suc (w/v), 0.05% MES (w/v), and 7 g L−1 agar. The seeds were kept at 4°C for 48 h and cultivated in vitro for 10 d under LD conditions (16-h light [250 µmol photons m−2 s−1 with a 1:1 mix of sodium-vapor and hydrargyrum quartz iodide lamps] at 22°C/8-h dark at 21°C). The seedlings were then transferred to a hydroponic culture solution (1.25 mM KNO3, 1.5 mM Ca(NO3)2, 0.75 mM MgSO4, 0.5 mM KH2PO4, 50 µM H3BO3, 12 µM MnSO4, 0.7 µM CuSO4, 1 µM ZnSO4, 0.24 µM Na2MoO4, 50 µM FeEDTA, 100 µM Na2SiO3) and grown under LD for 11 d. The culture solution was replaced weekly. All assays were performed with 21- to 25-d-old plants. For DD conditions, plants were grown under normal LD cycles and maintained in constant dark after a normal night (Prado et al., 2013).
Kros Measurements in Rosettes
Pressure chamber measurements of Kros were performed as described (Postaire et al., 2010; Prado et al., 2013). Briefly, whole rosettes were excised, bathed in a liquid solution (5 mM KNO3, 2 mM MgSO4, 1 mM Ca(NO3)2, 10 mM MES, pH 6.0, adjusted with KOH), and inserted into a pressure chamber with the sectioned hypocotyl adhered tightly to an adaptor pipe using low-viscosity dental paste (Coltene, President Microsystem, Switzerland), the adaptor pipe being pushed through the metal lid of the chamber. Pressure was slowly applied to the chamber using nitrogen gas. Pressurization of the chamber resulted in a flow of liquid entering through the leaf surface and exiting from the hypocotyl section. Prepressurization at 10 min was required to obtain a stabilized sap flow (Jv). The flow rate was proportional to the applied pressure (P). The slope of a Jv(P) plot was normalized to the cumulative surfaces of rosette leaves to calculate the Kros of an individual rosette (in μL s−1 m−2 MPa−1).
Rosette Microsomal Protein Purification
The entire procedure for microsomal protein purification from rosettes was performed at 4°C as described (Gerbeau et al., 2002; Prado et al., 2013). In brief, rosettes of 21-d-old plants (equivalent of 10 g of fresh weight) were ground with a Waring blender (4 × 10 s) in 2 mL g fresh weight (FW)−1 of grinding buffer (500 mM Suc, 50 mm NaF, 20 mm EDTA, 20 mm EGTA, 5 mm β-glycerophosphate, 1 mm phenantroline, 10% glycerol (v/v), 0.6% polyvinylpyrrolidone (w/v), 10 mm ascorbic acid, 1 µm leupeptin, 5 mM DTT, 1 mM stabilized vanadate, 1 mM phenylmethylsulfonyl fluoride, 50 mM Tris, pH 8.0 with 1m MES), filtered through a mesh with 200 µm pores, and centrifuged at 13,000 g for 30 min (JA14 rotor, Beckman Coulter). The supernatant was successively filtered through meshes of 63 and 36 µm pore size, and centrifuged at 30,000 g for 12 min (Ti45 rotor, Beckman Coulter). The resulting pellet was suspended in a minimal volume of conservation buffer (300 mM Suc, 50 mM NaF, 10 mM H3BO3, 9 mM KCl, 5 mM EDTA, 5 mM EGTA, 4.2 µM leupeptine, 1 mM phenylmethylsulfonyl fluoride, 5 mM DTT, 10 mM Tris, pH 8.3) and stored at −60°C. Protein concentration was estimated using a modified Bradford procedure (Stoscheck, 1990).
Purification of Phosphospecific Forms of AtPIP2;1 Peptide Antibodies
With use of an approach comparable with that reported before (Qing et al., 2016), high affinity polyclonal antibodies directed against AtPIP2;1 C-terminal peptides carrying up to two phosphorylated residues were developed by Eurogentec (Seraing). The following peptides harboring the indicated phosphorylated Ser residues (SP) were designed to reduce possible cross-reactivity between antibodies and used to immunize rabbits: 0P, ASGSKSLGSFRSAAN; 1P280, GSKSLGSpFRSAA; 1P283, KSLGSFRSpAANV; 2P, KSLGSpFRSpAANV. Four injections of peptides coupled to Keyhole Limpet hemocyanin were operated at 0, 14, 28, and 56 d from the start using two rabbits per peptide. Blood samples were collected at 0, 38, 66, and 87 d from the start. Each crude polyclonal antiserum was initially purified on an affinity matrix column carrying the corresponding immunizing peptide. For anti-phosphopeptide antibodies, the eluate was subsequently loaded onto a matrix column coupled with a nonmodified peptide sequence, and the flow-through was captured. After each purification, ELISAs were performed to control the titer and the affinity of each of the produced antibodies. The procedure was stopped at this stage for the anti-1P280 antibody. The anti-0P, anti-1P283, and anti-2P antibodies were further purified to reduce cross-reactivity with other peptides by loading the previously collected eluates onto matrix columns coupled with these peptides. Each time, the flow-through was captured and ELISAs were performed to control the titer and the affinity of the purified antibody. Because of sequence similarity between AtPIP2;1, AtPIP2;2, and AtPIP2;3, the four antibodies can indistinctly react with either one of these isoforms.
Immunodetections
Expression of AtPIP2;1 and its homologs in rosettes was probed by ELISA and protein gel blot assays as described previously (Boursiac et al., 2005) with minor modifications. The assays were performed using microsomal extracts and purified antibodies described above diluted at 1:300 for anti-0P and anti-1P280 and 1:3000 for anti-1P283 and anti-2P. We also used an anti-PIP2 antibody raised against a 17-amino-acid C-terminal peptide of AtPIP2;1 and recognizing three out of eight AtPIP2s (AtPIP2;1, AtPIP2;2, AtPIP2;3), as previously described (Santoni et al., 2003). For protein gel blot analysis, proteins from microsomal extracts were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes for 16 h at 35 V and 4°C in transfer buffer (10 mM CAPS, 10% MeOH [v/v], pH 11). Following transfer, the membranes were saturated with Phosphate-Buffered Saline supplemented with Tween and BSA (PBS-TB; 115 mM NaCl, 4 mM Na2HPO4, 16 mM KH2PO4, 3 mM KCl, 1% BSA [w/v], 0.1% Tween 20 [v/v], pH 7.3) for 30 min at room temperature. The membranes were then incubated for 2 h with the anti-PIP antibody, washed twice with PBS-TB for 10 min, and incubated for 2 h with a goat anti-rabbit secondary antibody conjugated to peroxidase (1:20,000, A6154, Sigma-Aldrich). After four additional washes with PBS for 5 min, immunodetection was performed using the chemiluminescent ECL protein gel blotting analysis system (GEHealthcare). Protein gel blotting was also performed to characterize the immunoprecipitation products. The procedure was similar to that described above except that proteins after separation by SDS-PAGE were transferred onto polyvinylidene fluoride membranes for 2 h at 75 V in transfer buffer (25 mM Tris, 192 mM Gly, 1 mM SDS). The membranes were saturated with Tris-Buffered Saline supplemented with Tween (TBST; 0.5 M NaCl, 25 mM Tris, 0.1% Tween 20 [v/v], pH 7) and with 5% (w/v) skimmed milk for 2 h at room temperature, washed three times with TBST, and incubated overnight with antibodies supplemented with 1 mg mL−1 BSA. The membranes were then washed three times with TBST for 20 min and incubated for 1 h with a goat anti-rabbit secondary antibody conjugated to peroxidase (1:5000, ab6721, Abcam). After three additional washes with TBST for 20 min and a final wash with TBS, immunodetection was performed using the same chemiluminescent ECL protein gel blotting analysis system as previously described. Anti-GRF4, anti-GRF10, and anti-PIP2 antibodies were used at dilutions of 1:1000, 1:1000, and 1:5000, respectively. The anti-GRF4 and anti-GRF10 antibodies were kindly provided by Dr. R. Ferl (University of Florida; Alsterfjord et al., 2004).
Immunoprecipitations
Immunoprecipitation (IP) of GFP-tagged proteins was performed at 4°C from 25-d-old plants as previously described (Bellati et al., 2016) with minor modifications. Briefly, leaves were wrapped with Miracloth (Millipore), and leaf proteins were crosslinked twice under a vacuum using 1% formaldehyde (v/v) for 15 min each time. The crosslinking was stopped by performing three successive washes in 125 mm Gly under a vacuum, each for 5 min. The leaves were rinsed twice with water and PBS and ground to a fine powder in liquid nitrogen. The leaf tissue (5 g) was solubilized with 2.1 mL g−1 FW of a buffer containing 300 mm NaCl, 1% Triton X-100 (v/v), 0.5% Na deoxycholate (w/v), 0.1% SDS (w/v), and 100 mm Tris-HCl, pH 8.0, in the presence of cOmplete, EDTA-free Protease Inhibitor Cocktail (Roche) and filtered through Miracloth. After centrifugation at 4500 g for 30 min, the supernatant was again centrifuged at 10,000 g for 15 min at 4°C. IP was performed using the supernatant, an antibody against GFP, and a μMagnetic-Activated Cell Sorting anti-GFP Microbeads kit (Miltenyi Biotec) according to the manufacturer's instructions. Briefly, the sample was incubated at 4°C for at least 1 h with a volume equivalent to 35 μL g−1 FW of an anti-GFP Microbeads solution. The sample was then loaded onto μMagnetic-Activated Cell Sorting columns that were previously conditioned with 200 μL of a lysis buffer (150 mm NaCl, 1% Triton X-100 [v/v], 50 mm Tris-HCl, pH 8). After four washes with 200 μL of a buffer containing 150 mm NaCl and 1% Igepal CA-630 (v/v) and an additional washing step with 100 μL of 20 mm Tris-HCl pH 7.5, the proteins were eluted with 50 μL of elution buffer (50 mm Tris-HCl, pH 6.8, 50 mm DTT, 1% SDS [w/v], 1 mm EDTA, 0.005% bromophenol blue [v/v], 10% glycerol [v/v]). Formaldehyde fixation was reversed by heating the eluted proteins at 100°C for 20 min.
Proteomic Analysis
Proteins and digested peptides were purified as described (Prado et al., 2013) and analyzed with a QTOF mass spectrometer (Maxis; Bruker Daltonics, Bremen, Germany), interfaced with a nano-HPLC Ultimate 3000 (Dionex; Vialaret et al., 2014). The corresponding data were interrogated with Mascot 2.2.07 (Matrix Science, http://www.matrixscience.com) against the Arabidopsis proteome (Tair10, 31,960 entries, http://www.arabidopsis.org/) as previously described (Vialaret et al., 2014). The quantification was based on the manual extraction of the XIC (eXtracted Ion Chromatogram) signal arising from peptides with m/z 554.80, 594.78, and 634.76, and corresponding to unmodified (SLGSFRSAANV), mono-phosphorylated (SLGSpFRSAANV), or di-phosphorylated (SLGSpFRSpAANV) AtPIP2;1 C-terminal peptides, respectively (Prado et al., 2013), and was performed using Data analysis software (Bruker). The relative proportion of each form was calculated per sample. Three independent biological experiments and up to four technical replicates per biological repeat were used.
Plasmid Construction
GFP-PIP2;1 cDNA was amplified from Arabidopsis plants expressing GFP-PIP2;1 fusions (Cutler et al., 2000) and mutated at Ser280 and Ser283 using recombinant PCR with a forward GFP primer (GFP-BglII FW) and mutagenic reverse primers (dSA, dSD) that spanned C-terminal sequences and introduced the required Ser-to-Ala or Ser-to-Asp mutations, respectively (Supplemental Table 2). The forward and reverse primers also incorporated restriction sites for BglII and SpeI, respectively. GFP-PIP2;1 was mutated at residue Ser121 by using a previously mutated dSA GFP-PIP2;1 plasmid as a template and a QuickChange Site-Directed Mutagenesis kit (Stratagene). cDNAs encoding each investigated GRF were amplified by PCR from Col0 plants using specific primers that, in addition, introduced restriction sites at the 5′ and 3′ ends (Supplemental Table 2). All PCR-amplified fragments containing the newly introduced restriction sites were cloned into the pT7Ts oocyte expression vector (Tournaire-Roux et al., 2003). For BiFC assays, we used versions of the oocyte expression vector pGEM-Xho containing halves of YFP called NYFP or CYFP. In brief, NYFP-CIPK10 was amplified from a pSPYNE-CIPK10 construct (Waadt et al., 2008) and subcloned into pGEM-Xho. Similarly, the NYFP-PIP2;1 and CYFP-PIP2;1 constructs were cloned into pGEM-Xho. GRF fragments were amplified by PCR from previously described pT7Ts constructs using forward and reverse primers including restriction sites for ApaI and SpeI, respectively, and substituted for CIPK10 in pGEM-Xho–NYFP-CIPK10 (Supplemental Table 2). Mutated versions of the AtPIP2;1 fragments were amplified by PCR from pT7Ts constructs using forward and reverse primers including restriction sites for SpeI and HindIII, respectively, and substituted for PIP2;1 in pGEM-Xho-CYFP-PIP2;1. The validity of all constructs was checked by restriction and sequence analysis of the full cDNA.
cRNA Production and Expression in Xenopus laevis Oocytes
Capped cRNAs were synthetized from linearized plasmids by in vitro transcription using T7 RNA polymerase. The cRNAs were purified as previously described (Maurel et al., 1993), quantified with a NanoDrop 1000 spectrophotometer (Thermo Fischer Scientific, USA), and checked for integrity by agarose gel electrophoresis. Fully grown (stages V and VI) oocytes were isolated from X. laevis as previously described (Maurel et al., 1993). Briefly, the oocytes were isolated and defolliculated by digestion at 20°C with 2 U mL−1 collagenase A (Roche Diagnostics, Mannheim, Germany) in OR2 solution (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, pH 7.5). After 1 h, the oocytes were washed with OR2 containing 1 mg mL−1 BSA, incubated in K buffer (100 mM K2HPO4, 1 g L−1 BSA, pH 6.5) for 1 h, washed with a Barth’s solution (88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, 0.82 mM MgSO4, 10 mM HEPES-NaOH, pH 7.4) containing 1 mg mL−1 BSA, and resuspended in Barth’s solution. The oocytes were injected with 1.25 to 20 ng of cRNA or diethyl pyrocarbonate–treated water in a volume of 50 nL using a nanopipette (Drummond Scientific Company, Broomall, PA, USA). The oocytes were incubated at 20°C in Barth’s solution supplemented with 50 µg mL−1 gentamycin sulfate for 2 d before analysis.
Osmotic Water Permeability Measurements
Swelling assays were performed as previously described (Maurel et al., 1993) by transferring individual oocytes from an iso-osmotic (200 mosM kg−1) to a hypo-osmotic (40 mosM kg−1) Barth’s solution and monitoring volume changes by video-microscopy (GigE Vision CMOS camera, Phase, Luebeck, Germany). Images were captured at 20-s intervals for 2 min and digitized with ImageJ software (Schneider et al., 2012). The projected surface area of the oocyte was used to calculate its relative volume, based on an ellipsoid shape model. The data were fitted as a linear change in volume over time, in agreement with cell swelling under the initial conditions. The osmotic water permeability coefficient (Pf) of the oocyte plasma membrane was calculated according to the equation:
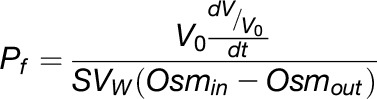 |
where V0 is the initial oocyte volume (9 × 10−4 cm3), V is the volume at time t, Vw is the partial molar volume of water (18 cm3 mol−1), S is the initial oocyte surface area (0.045 cm2), and Osmin and Osmout are the osmolarity within the oocyte and the external medium, respectively.
Confocal Imaging of Oocytes
GFP fluorescence of oocytes expressing GFP constructs was visualized using a Zeiss LSM710 confocal microscope with laser excitation at 488 nm and detection between 510 and 540 nm with a 10× objective lens. The parameters of laser beam intensity, gain, and offset were kept constant throughout a full experiment, allowing subsequent comparisons of fluorescence intensities. The fluorescence intensity at the oocyte surface was quantified as follows using an in-house developed ImageJ macro function. The edge of the oocyte was delineated using a visible light image and was used to define (using a 20 pixel-implement or subtraction) two regions of interest encompassing the overall cell or the inner cell space, respectively. The fluorescence at the plasma membrane and its vicinity was then determined based on the difference between the mean fluorescence intensity of the two regions of interest. For BiFC analysis, oocytes expressing the indicated constructs in fusion with NYFP or CYFP were gently transferred onto a microscope slide in a droplet of Barth’s solution. Oocytes were observed under an inverted Leica SP8 inverted confocal laser scanning microscope with the settings described above except that fluorescence emission was detected at 520–560 nm. Fluorescence intensity at the edge of each cell was averaged using ImageJ along a line of 10 pixels.
Peptide Synthesis
Two purified peptides, H-ARAASAPA-OH (Pep-8) and H-ARAApSAPA-OH (PPep-8), corresponding to the mode I 14-3-3 binding motif (Moorhead et al., 1999) were provided by GeneCust (Luxembourg). The quality of each peptide was assessed by mass spectrometry and HPLC. For the swelling assays, the peptides were injected into oocytes at 1 mg mL−1 in 50 nL (i.e., 50 ng oocyte−1) 2 h before the beginning of Pf measurements.
FLIM Experiments
Transient expression in tobacco (Nicotiana tabacum cv SR1) leaves and FLIM measurements were performed as described (Bellati et al., 2016). Briefly, 4- to 6-week-old plants were infiltrated with Agrobacterium tumefaciens strain GV3101 harboring the desired construct. Tobacco epidermal cells were observed on a portion of ∼25 mm2 of transformed leaf 72 h after infiltration. FRET and FLIM measurements were performed with a multiphoton confocal microscope (ZEISS LSM 780, Göttingen, Germany) using the Time Correlated Single Photon Counting method. GFP was excited at 920 nm with a Ti:Saphir pulsed infra-red laser (Chameleon ULTRA II, COHERENT) for 3 min. The emitted fluorescence was measured using a HPM-100 Hybrid detector (Hamamatsu R10467–40 GaAsP). The laser synchronization and measurement of photon life time were performed using a SPC-830 capture card (BandH). Fluorescence intensity images were used to calculate decay curves per pixel, which were then fitted with different models. A mono-exponential model function was applied to donor samples with only GFP present, whereas a double-exponential model function (without fixing any parameter) was applied to samples containing two fluorophores, GFP/mCherry or GFP/RFP. The FRET efficiency was calculated as FRET = 1 − (GFP lifetime in the presence of mCherry/RFP) / (GFP lifetime expressed alone).
Statistical Analysis
The rhythmicity of time-series data was analyzed with CircWave v1.4 software (Oster et al., 2006; http://www.euclock.org/) by using a linear harmonic regression fit with an assumed period of 24 h. The other statistical tests were performed using a R software (R Core Team, 2018). The effects of environmental conditions in time-series data were investigated by ANOVA with a dynamic linear model, followed by a Tukey test for phosphoproteomics analysis. The effect of genotype was investigated by ANOVA with a generalized linear model followed by a Tukey test or with a dynamic linear model for the dose response analyses. The effect of complementation in BiFC assays was analyzed by ANOVA with a generalized linear model followed by a simultaneous test for general linear hypotheses. Pairwise comparisons and numerical FRET-FLIM analyses were performed using a Wilcoxon test and a Tukey test at P < 0.05, respectively. The statistical analyses are summarized in the Supplemental File.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AtPIP2;1 (AT3G53420), AtPIP2;2 (AT2G37170), AtPIP2;3 (AT2G37180), AtGRF3 (AT5G38480), AtGRF4 (AT1G35160), AtGRF6 (AT5G10450), AtGRF8 (AT5G65430), AtGRF9 (AT2G42590), AtGRF10 (AT1G22300), AtGRF11 (AT1G34760), AtCIPK10 (AT5G58380).
Supplemental Data
Supplemental Figure 1. Immunological characterization of purified phosphospecific antibodies.
Supplemental Figure 2. Circadian rhythms of AQP phosphorylation.
Supplemental Figure 3. Phosphoproteomics analysis of PIP2;1-PIP2;3 C-terminal peptides.
Supplemental Figure 4. Correlation between Kros and AQP phosphorylation.
Supplemental Figure 5. Temporal Kros profiles in plants expressing AtPIP2;1 phosphorylation mutants.
Supplemental Figure 6. Molecular characterization of single grf knockout mutants.
Supplemental Figure 7. Effects of mutations of AtPIP2;1 at Ser280 and Ser283 on normalized Pf of oocytes expressing GFP-PIP2;1.
Supplemental Figure 8. Dose-dependent effects of GRF4 on normalized Pf of oocytes expressing forms of GFP-PIP2;1 with triple mutations at Ser121, Ser280, and Ser283.
Supplemental Figure 9. Effects of GRF4 co-expression and peptide co-injection on fluorescence intensity at the cell periphery in oocytes expressing dSA or dSD forms of GFP-PIP2;1.
Supplemental Table 1.FRET interactions between PIP2;1 and GRF4 measured by FLIM.
Supplemental Table 2. Nucleotide sequences of primers used for BIFC assays, FRET-FLIM, oocyte assays and genotyping of grf knockout mutants.
Supplemental File. Statistical analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the Montpellier Rio Imaging platform (MRI) and the Mass Spectrometry Proteomics Platform for their technical support in the imaging experiments and proteomic measurements, respectively. We thank Dr. Bert de Boer (Vrije Universiteit, Amsterdam, NL) and Dr. Julio Salinas (Centro Investigaciones Biológicas, Madrid, ES) for providing the grf mutant lines. We also thank Dr. J. Kudla (University of Münster, Germany) for the gift of the pSYNE-CIPK10 vector, Dr. R. Ferl (University of Florida, Gainesville, USA) for providing the GRF-GFP transgenic plant lines and anti-GRF antibodies, and Dr. C. Oecking (University of Tübingen, Germany) for the GRF-GFP transgenic plant lines and advice. This work was supported in part by the Agence Nationale de la Recherche (ANR-07-BLAN-0206) and a research contract with Syngenta. K.P. acknowledges a Institut National de la Recherche Agronomique doctoral fellowship (Contrat Jeune Scientifique).
AUTHOR CONTRIBUTIONS
K.P., V.C., and C.M. designed research; K.P. performed most of the experiments; G.L. contributed to proteomics experiments; J.B. performed FRET-FLIM; N.T. contributed new genetic materials; C.T.-R. and A.M. performed the BiFC assays; K.P., V.C., V.S., and C.M. analyzed data; K.P. and C.M. wrote the paper, which was read by all the authors.
Footnotes
Articles can be viewed without a subscription.
References
- Alsterfjord M., Sehnke P.C., Arkell A., Larsson H., Svennelid F., Rosenquist M., Ferl R.J., Sommarin M., Larsson C. (2004). Plasma membrane H(+)-ATPase and 14-3-3 isoforms of Arabidopsis leaves: Evidence for isoform specificity in the 14-3-3/H(+)-ATPase interaction. Plant Cell Physiol. 45: 1202–1210. [DOI] [PubMed] [Google Scholar]
- Atamian H.S., Harmer S.L. (2016). Circadian regulation of hormone signaling and plant physiology. Plant Mol. Biol. 91: 691–702. [DOI] [PubMed] [Google Scholar]
- Bellati J., Champeyroux C., Hem S., Rofidal V., Krouk G., Maurel C., Santoni V. (2016). Novel aquaporin regulatory mechanisms revealed by interactomics. Mol. Cell. Proteomics 15: 3473–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y., Chen S., Luu D.T., Sorieul M., van den Dries N., Maurel C. (2005). Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 139: 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira C.F., Jeanguenin L., Chaumont F., Tardieu F. (2014). Circadian rhythms of hydraulic conductance and growth are enhanced by drought and improve plant performance. Nat. Commun. 5: 5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R., López-Cobollo R., Mar Castellano M., Angosto T., Alonso J.M., Ecker J.R., Salinas J. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26: 3326–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F., Tyerman S.D. (2014). Aquaporins: Highly regulated channels controlling plant water relations. Plant Physiol. 164: 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D., Morris E.R., Walker J.C. (2009). 14-3-3 And FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60: 67–91. [DOI] [PubMed] [Google Scholar]
- Choudhary M.K., Nomura Y., Wang L., Nakagami H., Somers D.E. (2015). Quantitative circadian phosphoproteomic analysis of Arabidopsis reveals extensive clock control of key components in physiological, metabolic, and signaling pathways. Mol. Cell. Proteomics 14: 2243–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O., Graf W., Franck K.I., Albert A., Winkler J.B., Scherb H., Stichler W., Schäffner A.R. (2010). Kinetic analyses of plant water relocation using deuterium as tracer - reduced water flux of Arabidopsis pip2 aquaporin knockout mutants. Plant Biol (Stuttg) 12: 129–139. [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Gerbeau P., Amodeo G., Henzler T., Santoni V., Ripoche P., Maurel C. (2002). The water permeability of Arabidopsis plasma membrane is regulated by divalent cations and pH. Plant J. 30: 71–81. [DOI] [PubMed] [Google Scholar]
- Jaspert N., Throm C., Oecking C. (2011). Arabidopsis 14-3-3 proteins: Fascinating and less fascinating aspects. Front. Plant Sci. 2: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I., Larsson C., Ek B., Kjellbom P. (1996). The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell 8: 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I., Karlsson M., Shukla V.K., Chrispeels M.J., Larsson C., Kjellbom P. (1998). Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell 10: 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez F., Bousser A., Sissoëff I., Gaspar M., Lachaise B., Hoarau J., Mahé A. (2003). Diurnal regulation of water transport and aquaporin gene expression in maize roots: Contribution of PIP2 proteins. Plant Cell Physiol. 44: 1384–1395. [DOI] [PubMed] [Google Scholar]
- Luu D.T., Martinière A., Sorieul M., Runions J., Maurel C. (2012). Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress. Plant J. 69: 894–905. [DOI] [PubMed] [Google Scholar]
- Maurel C., Reizer J., Schroeder J.I., Chrispeels M.J. (1993). The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO J. 12: 2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C., Boursiac Y., Luu D.T., Santoni V., Shahzad Z., Verdoucq L. (2015). Aquaporins in plants. Physiol. Rev. 95: 1321–1358. [DOI] [PubMed] [Google Scholar]
- Mendoza-Viveros L., Bouchard-Cannon P., Hegazi S., Cheng A.H., Pastore S., Cheng H.M. (2017). Molecular modulators of the circadian clock: Lessons from flies and mice. Cell. Mol. Life Sci. 74: 1035–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller H.B., Slengerik-Hansen J., Aroankins T., Assentoft M., MacAulay N., Moestrup S.K., Bhalla V., Fenton R.A. (2016). Regulation of the water channel Aquaporin-2 via 14-3-3θ and -ζ. J. Biol. Chem. 291: 2469–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G., Douglas P., Cotelle V., Harthill J., Morrice N., Meek S., Deiting U., Stitt M., Scarabel M., Aitken A., MacKintosh C. (1999). Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J. 18: 1–12. [DOI] [PubMed] [Google Scholar]
- Moshelion M., Becker D., Biela A., Uehlein N., Hedrich R., Otto B., Levi H., Moran N., Kaldenhoff R. (2002). Plasma membrane aquaporins in the motor cells of Samanea saman: Diurnal and circadian regulation. Plant Cell 14: 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nardini A., Salleo S., Andri S. (2005). Circadian regulation of leaf hydraulic conductance in sunflower (Helianthus annuus L. cv Margot). Plant Cell Environ. 28: 750–759. [Google Scholar]
- O’Neill J.S., Reddy A.B. (2011). Circadian clocks in human red blood cells. Nature 469: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J.S., van Ooijen G., Dixon L.E., Troein C., Corellou F., Bouget F.Y., Reddy A.B., Millar A.J. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H., Damerow S., Hut R.A., Eichele G. (2006). Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J. Biol. Rhythms 21: 350–361. [DOI] [PubMed] [Google Scholar]
- Paul A.L., Sehnke P.C., Ferl R.J. (2005). Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol. Biol. Cell 16: 1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.L., Denison F.C., Schultz E.R., Zupanska A.K., Ferl R.J. (2012). 14-3-3 Phosphoprotein interaction networks - does isoform diversity present functional interaction specification? Front. Plant Sci. 3: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., et al. (2012). Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14: 991–998. [DOI] [PubMed] [Google Scholar]
- Postaire O., Tournaire-Roux C., Grondin A., Boursiac Y., Morillon R., Schäffner A.R., Maurel C. (2010). A PIP1 aquaporin contributes to hydrostatic pressure-induced water transport in both the root and rosette of Arabidopsis. Plant Physiol. 152: 1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado K., Maurel C. (2013). Regulation of leaf hydraulics: from molecular to whole plant levels. Front. Plant Sci. 4: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado K., Boursiac Y., Tournaire-Roux C., Monneuse J.-M., Postaire O., Da Ines O., Schäffner A.R., Hem S., Santoni V., Maurel C. (2013). Regulation of Arabidopsis leaf hydraulics involves light-dependent phosphorylation of aquaporins in veins. Plant Cell 25: 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak S., Hem S., Boudet J., Viennois G., Sommerer N., Rossignol M., Maurel C., Santoni V. (2008). Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: Role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol. Cell. Proteomics 7: 1019–1030. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Kay S.A. (2010). An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15: 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing D., Yang Z., Li M., Wong W.S., Guo G., Liu S., Guo H., Li N. (2016). Quantitative and functional phosphoproteomic analysis reveals that ethylene regulates water transport via the C-terminal phosphorylation of aquaporin PIP2;1 in Arabidopsis. Mol. Plant 9: 158–174. [DOI] [PubMed] [Google Scholar]
- R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/.
- Robles M.S., Humphrey S.J., Mann M. (2017). Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 25: 118–127. [DOI] [PubMed] [Google Scholar]
- Santoni V., Vinh J., Pflieger D., Sommerer N., Maurel C. (2003). A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem. J. 373: 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Mas P. (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck C.M. (1990). Quantitation of protein. Methods Enzymol. 182: 50–68. [DOI] [PubMed] [Google Scholar]
- Takase T., Ishikawa H., Murakami H., Kikuchi J., Sato-Nara K., Suzuki H. (2011). The circadian clock modulates water dynamics and aquaporin expression in Arabidopsis roots. Plant Cell Physiol. 52: 373–383. [DOI] [PubMed] [Google Scholar]
- Törnroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., Neutze R., Kjellbom P. (2006). Structural mechanism of plant aquaporin gating. Nature 439: 688–694. [DOI] [PubMed] [Google Scholar]
- Tournaire-Roux C., Sutka M., Javot H., Gout E., Gerbeau P., Luu D.T., Bligny R., Maurel C. (2003). Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425: 393–397. [DOI] [PubMed] [Google Scholar]
- van Kleeff P.J., Jaspert N., Li K.W., Rauch S., Oecking C., de Boer A.H. (2014). Higher order Arabidopsis 14-3-3 mutants show 14-3-3 involvement in primary root growth both under control and abiotic stress conditions. J. Exp. Bot. 65: 5877–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialaret J., Di Pietro M., Hem S., Maurel C., Rossignol M., Santoni V. (2014). Phosphorylation dynamics of membrane proteins from Arabidopsis roots submitted to salt stress. Proteomics 14: 1058–1070. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wilson R.S., Swatek K.N., Thelen J.J. (2016). Regulation of the regulators: Post-translational modifications, subcellular, and spatiotemporal distribution of plant 14–3-3 proteins. Front. Plant Sci. 7: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.L., Chen W.W., Chen L.Q., Qin C., Jin C.W., Shi Y.Z., Zheng S.J. (2013). The 14-3-3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. New Phytol. 197: 815–824. [DOI] [PubMed] [Google Scholar]