Abstract
The microbiome modulates host immune function across the gastrointestinal tract, peripheral lymphoid organs and central nervous system. In this review, we highlight emerging evidence that microbial effects on select immune phenotypes arise developmentally, where the maternal and neonatal microbiome influence immune cell ontogeny in the offspring during gestation and early postnatal life. We further discuss roles for the perinatal microbiome and early life immunity in regulating normal neurodevelopmental processes. In addition, we examine evidence that abnormalities in microbiota-neuroimmune interactions during early life are associated with altered risk for neurological disorders in humans. Finally, we conclude by evaluating the potential implications of microbiota-immune interventions for neurological conditions. Continued progress toward dissecting mechanistic interactions between the perinatal microbiota, immune system and nervous system may uncover fundamental insights into how developmental interactions across physiological systems inform later-life health and disease.
eTOC Blurb
Mounting evidence suggests that communication between indigenous microbes, leukocytes and neurons begins during early life. Pronovost and Hsiao review evidence suggesting that interactions between the early life microbiome and the immune system are important for neurodevelopment and that alterations in microbiome-neuroimmune communication may predispose individuals to neurological diseases.
Introduction
The microbiome, immune system and nervous system are each comprised of specialized cells that actively monitor the environment, sense and interpret physical and chemical stimuli, and coordinate with other cells to respond and adapt to alterations in host physiology. Research from the past decade highlights intricate interactions between the three systems; indigenous microbes, leukocytes and neurons have the capacity to communicate with each other via shared molecular signaling factors, including neurotransmitters, secondary metabolites and cytokines, and their receptors. Mounting evidence suggests that communication across the functional systems begins during early life. Indeed, the maternal and neonatal microbiome regulate hundreds of soluble biochemical signals that impact host physiology, including immune function and neurodevelopment. The nature of these early life influences can have lasting impact on later life health and predisposition to neurological disease. Herein we discuss these lines of evidence highlighting microbiome-immune-neural interactions during early development. We focus particularly on the gut microbiome, using the term “maternal” to refer to indigenous microbes colonizing the mother during pregnancy, “neonatal” to refer to the offspring microbiome at birth through the first weeks of postnatal life, “perinatal” to refer to both maternal or neonatal microbiome, and “early life” to refer to offspring gestational development through weaning.
The maternal and neonatal microbiome
Pregnancy is accompanied by a myriad of changes in maternal health and lifestyle, which together shape the composition and activity of the maternal microbiome. In a study of the gut microbiota from 91 pregnant women of varied health statuses, first trimester mothers exhibited profiles that were clearly distinguishable from those of matched non-pregnant controls (Koren et al., 2012). However, in a case-control study of 40 women sampled weekly during and after pregnancy, these reported shifts in the composition of the gut microbiota were not observed; rather, profiles of microbiota from the maternal distal gut appeared relatively stable throughout gestation, with only a trending decrease in alpha diversity over time and no difference in taxonomic profiles (DiGiulio et al., 2015). Another study of gut microbiota from 7 mothers over the perinatal period highlighted the possibility that the metabolic activity of the gut microbiota may be altered during pregnancy, despite no changes in microbiota composition (Jost et al., 2014). These inconsistencies highlight the need to finely resolve the question of whether there are inherent alterations in the maternal microbiota based on biological consequences of pregnancy itself, or whether individual state- and context-dependent factors shape the maternal microbiota. Toward the former, additional well-controlled investigations of large cohorts of individuals over the course of pregnancy are warranted. There is a wealth of evidence supporting the latter, where maternal body weight (Collado et al., 2008), probiotic treatment (Lindsay et al., 2013; Vitali et al., 2012), antibiotic exposure (Stokholm et al., 2014), and diet (Chu et al., 2016) among numerous other factors, are associated with alterations in the human maternal microbiome.
Despite the complexity and variability of the human maternal microbiome, a preponderance of studies in both humans and animal models indicate that the maternal microbiome is important for conditioning the metabolic environment of the developing embryo, seeding the offspring’s microbiome during the birthing process and further influencing offspring development. The birth mode and first contacts with microbial communities from maternal body sites, including the skin and reproductive tract, as well as the surrounding environment are considered primary determinants of the neonatal microbiome. Maturation of the gut microbiome occurs over approximately the first three years of life, in which phylogenetic diversity gradually increases through infancy. In a case study that collected sixty fecal samples over an infant’s first 2.5 years of life, the gut microbiome was dominated by Firmicutes bacteria at birth, and progressively acquired Proteobacteria followed by Bacteroidetes. These major ecological shifts in the infant microbiome were associated with developmental milestones and nutritional transitions (Koenig et al., 2011). Differences in the skin, nasal, and oral microbiota were observed between newborns born by vaginal versus cesarean delivery (Chu et al., 2017). Early treatment with antibiotics delayed the maturation of the microbiota in infancy (Bokulich et al., 2016). In a population study of more than 776,000 Danish births, antibiotic use before and during pregnancy correlated with increased risk of offspring susceptibility to infection in childhood (Miller et al., 2018). During this period, the diversification of the microbiome coincides with the maturation of the host, supporting active interest in host-microbial interactions that impact normal development, particularly of the immune system. Altogether, these studies suggest that the maternal microbiome shapes the composition of the neonatal microbiome and early life immune function in the offspring.
Effect of the maternal and neonatal microbiome on immune development
The microbiome exhibits intimate interactions with immune cells across all mucosal sites. The intestinal microbiome is comprised of trillions of bacteria juxtaposed against layers of mucous and a villous epithelium that covers specialized lymphoid structures important for immune surveillance. Select bacterial species of the gut microbiome communicate with innate and adaptive immune cells across this interface, impacting immune homeostasis in gut-associated lymphoid tissues (GALT), as well as in peripheral lymphoid organs and resident immune cells in the nervous system. The extent of microbial influences on adult immune functions in the intestine, periphery and brain have been the subject of thorough recent reviews (Belkaid and Harrison, 2017; Fung et al., 2017; Macpherson et al., 2017; Rooks and Garrett, 2016; Thaiss et al., 2016).
Microbiome modulation of host immune status raises the important question of whether microbial effects on immunity arise developmentally or whether they are controlled by persistent, active signaling between the microbiota and immune cells during later postnatal life. Recent studies illuminate critical time windows during which the maternal and neonatal microbiome impact immune development and function (Figure 1, Table 1).
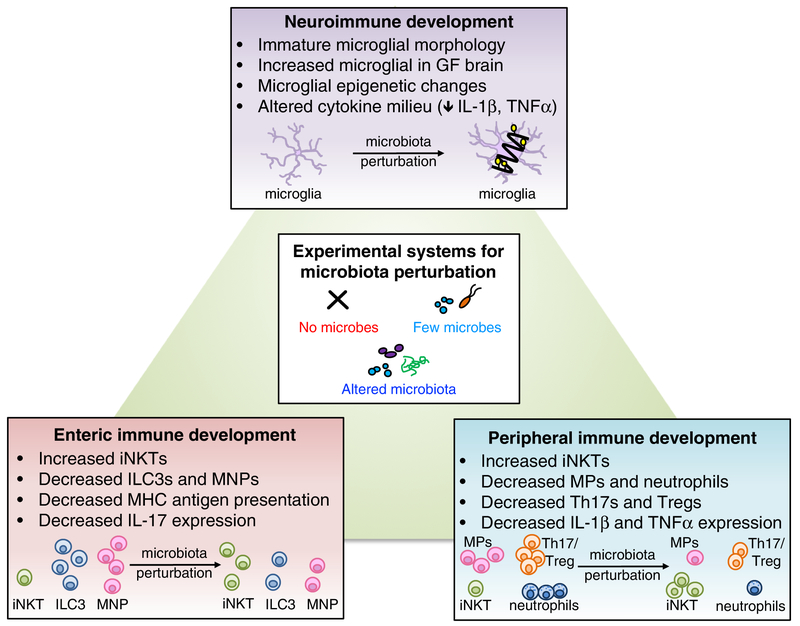
Figure 1. Roles for the microbiome in neuro-, peripheral and enteric immune development.
Microbiota perturbations, including elimination, reduction, or alteration of endogenous microbes, lead to altered immune development in multiple tissue sites. In the brain, absence of the microbiome alters morphological and transcriptional features of brain-resident microglia. Additionally, the cytokine milieu of the brain is altered in the absence of microbes. In the periphery, particular bacteria such as Lactobacillus modulate memory T cells, and microbial metabolites such as the short-chain fatty acid, butryate, regulate Treg cell populations. Depletion of the microbiome results in peripheral immune dysfunction, reducing macrophages (MP) and neutrophils and increasing iNKT cells. Altered diversity of the microbiota also impacts immune development. When microbes are substantially reduced, both Th17 and Treg cell numbers decrease. Conversely, alteration of microbiota composition, as with increased prevalence of Bacteroides fragilis, increases Th17 and Treg cells. In the intestine, transient maternal colonization with non-replicating E. coli, or with altered Schaedler flora, increases ILC3s, while ablation of the microbiota reduces gut ILC3s, mononuclear phagocytes, Paneth cell activity, MHC antigen presentation, and mast cell gene expression.
Table 1.
: The microbiome shapes enteric-, peripheral- and neuro-immune development.
| Species (Strain) | Microbiome Perturbation | Description | Reference |
|---|---|---|---|
| Enteric Immunity | |||
| Mice (C57BL/6) | Abx treatment E15-continuous (ampicillin, gentamycin, metronidazole, vancomycin, neomycin; ampicillin, gentamycin, vancomycin); GF mice | Decreased: small intestine IL-17A transcripts; small intestine IL-17+ ILC3s | Deshmukh et al.,2014 |
| Mice (NOD, H2g7) | Abx treatment E0-continuous (streptomycin, colistin, ampicillin; vancomycin only) | Decreased: colonic and ileal CD45+CD4+TCRαβ+IL-17+ cells; colonic, Peyer’s patches and MLN IL-17A transcript | Candon et al., 2015 |
| Mice (NOD/Caj) | Abx treatment E0-birth (neomycin, polymyxin B, streptomycin) | Decreased: MLN CD4+IFNγ+ and CD8+IFNγ+ cells; T cell IL-17 and IFNγ production; APC tolerance Increased: MLN CD4+CD25+Foxp3+cells |
Hu et al., 2015 |
| Mice (NOD) | Abx treatment E0-E4 + E14-E18 (metronidazole, neomycin, polymyxin) | Decreased: Peyer’s patches CD3+CD4+CD25+, CD4+CD69+CD25+ and CD4+CD62L+ cells Increased: MLN CD3+ and CD3+CD8+ cells; Peyer’s patches CD4+CD62L+ cells |
Tormo-Badia et al., 2014 |
| Mice (C57BL/6J) | Abx treatment E0-continuous (vancomycin; streptomycin) | Decreased: colonic CD4+CD25+Foxp3+ cells (vancomycin) | Russell et al., 2012 |
| Mice (Swiss Webster; C57BL/6) | GF mice; microbe exposure at P0 or P35 | Increased: colonic iNKT cells; colitis-associated colonic IL-4, IL-13, IL-1β; greater colonic Cxcl16* *corrected by microbe exposure at P0 |
Olszak et al., 2012 |
| Mice (C57BL/6J) | GF mice; colonization with non-replicating E. coli HA107 every 4 days E4-E15 | Increased: small intestine lamina propria NKp46+ ILC3s, CD11c+F4/80+ mononuclear cells (following gestation colonization) Altered small intestine qene expression |
Gomez de Aguero et al., 2016 |
| Pigs (Landrace X Large white X Duroc) | Abx treatment P0-continuous (ampicillin, gentamycin, metronidazole) | Decreased: small intestine IL-1β and IL-8; Increased: colonic goblet cells Altered small intestine gene expression |
Jensen et al., 2014 |
| Rats (Sprague-Dawley) | Abx treatment P7-continuous (Clamoxyl solution) | Altered small intestine and colonic gene expression | Schumann et al.,2005 |
| Mice (C57BL/6J) | GF mice; conventionalized (adult) or conventionally raised | Temporal-specific effects of colonization on colonic gene expression | El Aidy et al., 2013 |
| Mice (C57BL/6; Balb/c; Swiss Webster) | GF mice | IgG2b and IgG3 antibodies generated against microbiota and transmitted to offspring | Koch et al., 2016 |
| Peripheral Immunity | |||
| Mice (NOD/Caj) | Abx treatment E0-P0 (neomycin, polymyxin B, streptomycin) | Decreased: splenic CD11c+ IFNγ+ cells, CD11b+IL-12+ and CD11b+IL-17+ cells; splenic APC response to LPS Increased: splenic CD11b+IL-10+ cells |
Hu et al., 2015 |
| Mice (Swiss Webster; C57BL/6) | GF mice; microbe exposure at P0 or P35 | More lung iNKT cells*; more BALF eosinophils and IgE; greater serum CXCL16 and lung Cxcl16* *corrected by microbe exposure during pregnancy |
Olszak et al., 2012 |
| Mice (C57BL/6) | Abx treatment E18-P14 (ampicillin, vancomycin, metronidazole, neomycin) | Decreased: skin IL-1β andTNF-α | Nakamura et al.,2012 |
| Mice (C57/BL6) | Abx treatment E15-continuous (ampicillin, gentamycin, metronidazole, vancomycin, neomycin; ampicillin, gentamycin, vancomycin); GF mice | Decreased: circulating and bone marrow (LY6G+) neutrophils; plasma G-CSF (P3-P14) | Deshmukh et al.,2014 |
| Mice (C57BL/6J) | Abx treatment final 3–5 days of gestation (ampicillin and clindamycin) | Decreased: splenic CD8+IFNγ+ cells; Bcl-2 post-infection; splenic CD11chiMHCIIhi and CD11b−CD103+ cells; reduced splenic NK1.1+CD49b+ cells Increased: PD-1 post-infection |
Gonzalez-Perez et al., 2016 |
| Mice (C57BL/6J) | Abx treatment final 3–5 days of gestation-continuous (ampicillin, streptomycin, clindamycin) | Decreased: frequency of splenic OT-I IFNγ+/TNF-α+ and CD8+Erk2/pErk1/2 cells; CD8+ T cell IFNγ production | Gonzalez-Perez and Lamouse-Smith 2017 |
| Mice (NOD/Caj) | Abx treatment E0-birth (neomycin; vancomycin) | Decreased: splenic CD11b+CD80+, CD11b+CD86+, CD11c+CD80+ and CD11c+CD86+ cells; IFNγ production Differential effects of abx treatment in peripheral APCs, CD4+, CD8+cells | Hu et al., 2016 |
| Rhesus Macaque | Breast-fed versus nursery-fed microbiota | Decreased: circulating Th17 and CD4+CCR4+cells Altered circulating CD4+ and CD8+ T cell gene expression |
Ardeshir et al., 2014 |
| Human | Lactobacillus in vaginal microbiota | Decreased: CD4+CD45RO+ T cells; CBMC culture IL-12 (lacking Lactobacillus in vaginal microbiota) | Stencel-Gabriel et al., 2009 |
| Human | Differential neonatal microbiota compositions | Decreased: TLR4 mRNA in PBMCs (Bacteroides fragilis-containing samples) Increased: salivary IgA (Bifidobacterium-containing samples) |
Sjogren et al., 2009 |
| Human | Caesarean section-delivered neonatal microbiota | Decreased: blood CXCL10 and CXCL11 (Caesarean section birth and reduced Bacteroidetes) | Jakobsson et al.,2014 |
| Mice (C57BL/6) | Abx treatment adult (ampicillin, vancomycin, neomycin, metronidazole) | Decreased: peripheral blood B cells, CD4+ and CD8+ T cells; thymic and splenic weight; bone marrow and common lymphoid progenitors; HSCs; granulocytes and B cells Increased: T cells |
Josefsdottir et al.,2017 |
| Mice (C57BL/6) | GF mice | Decreased: splenic F4/80hiCD11b+ and F4/80loCD11b+ cells; splenic myelopoiesis bone marrow neutrophils | Khosravi et al., 2014 |
| Rats (Wistar, male) | SCFA treatment adult cultured neutrophils (acetate, propionate, butyrate) | Decreased: neutrophil TNF-α, CINC-2αβ and nitrite | Vinolo et al., 2011 |
| Mice (C57BL/6) | Abx treatment adult (metronidazole, vancomycin, ampicillin, kanamycin); SCFA treatment adult (butyrate, acetate, propionate) | Decreased: Foxp3+ Tregs (GF and Abx) Increased: Foxp3+ Tregs and acetylation of Foxp3 locus with SCFA treatment |
Arpaia et al., 2013 |
| Mice (C57BL/6) | SCFA treatment adult cultured bone marrow cells (acetate, propionate, butyrate, nicotinate) | Decreased: bone marrow DCs with SCFA treatment; PU.1 and RelB expression | Singh et al., 2010 |
| Mice (C57BL/6) | Abx treatment adult (metronidazole and vancomycin); n-butyrate treatment adult | Decreased: bone marrow-derived macrophage Nos2, Il6, Il12a and Il12b (butyrate) Increased: bone marrow-derived macrophage H3 acetylation (butyrate) |
Chang et al., 2014 |
| Mice (C57BL/6) | SCFA treatment adult (acetate, propionate) | Decreased: allergic airway inflammation and CD4+CD44+ T cells (propionate); CD11bhi CD40, PD-L2, IL-4, IL-5 and IL-10 expression (propionate) Increased: bone marrow Lin−c-KitloFlt3+ precursors and Lin−c-KithiFlt3+ precursors |
Trompette et al.,2014 |
| Mice (IQI; C57BL/6) | GF mice; SCFA treatment adult naive CD4+ T cells (acetate, propionate, butyrate) | Decreased: colonic neuropilin-1−Foxp3+, neuropilin-1+Foxp3+, CD103−Foxp3+ and CD103+Foxp3+ Tregs Increased: colonic Foxp3+Tregs (propionate, butyrate) Altered Foxp3 locus (butyrate) |
Furusawa et al.,2013 |
| Mice (Balb/c; Swiss Webster) | GF mice; SCFA treatment (acetate, propionate, butyrate) | Decreased: colonic Foxp3+ cells (GF) Increased: colonic Foxp3+ cells and Foxp3 and IL-10 expression (propionate); IL-10+Foxp3+ Tregs (propionate, acetate, butyrate) |
Smith et al., 2013b |
| Mice (C57BL/6J) | GF mice; Abx treatment adult (vancomycin, ampicillin, kanamycin, metronidazole) | Altered intestinal ILC gene transcription and epigenetic landscape | Gury-BenAri et al., 2016 |
| Neuroimmunity | |||
| Mice (C57BL/6) | GF mice | Altered newborn and adult microglial gene expression | Matcovitch-Natan et al., 2016 |
| Mice (C57BL/6) | GF mice adult; SCFA treatment (acetate, propionate, butyrate) | Decreased: response to LPS and LCMV (rescue with SCFAs) Increased: Iba1+ microglia (GF brain) Altered microglial morphology and gene expression profiles |
Erny et al., 2015 |
| Mice (C57BL/6J) | GF mice | Microglial transcriptomic profiles are altered by the microbiome in a temporal- and sex-specific manner; Increased: mid-gestation GF brain Iba1+ microglia; E18.5 GF male brain Iba1+ microglia; P20 GF female brain Iba1+ microglia Altered GF microglia epigenetic landscape |
Thion et al., 2018 |
| Mice (Swiss Webster) | GF mice | Decreased: neonatal brain IL-1β andTNF-α; cell death in neonatal ARC Increased: neonatal PVN and CA1 oriens cell death; neonatal hypothalamus, PVN, CA1 oriens and ARC Iba1+ microglia Altered microglial morphology |
Castillo-Ruiz et al., 2018 |
Abbreviations: Abx, antibiotic; APC, antigen presenting cell; ARC, arcuate nucleus; BALF, bronchioalveolar lavage fluid; Bcl-2, B cell lymphoma 2; CBMC, cord blood mononuclear cells; CINC-2αβ, cytokine-induced neutrophil chemoattractant-2αβ; DC, dendritic cell; Flt3, FMS-related tyrosine kinase 3; G-CSF, granulocyte-colony stimulating factor; GF, germ-free; HSC, hematopoietic stem cells; Iba-1, ionized calcium binding adapter molecule-1; IL, interleukin; ILC3, innate lymphoid cell 3; IFNγ, interferon-gamma; iNKT, invariant natural killer cell; LCMV, lymphocytic choriomeningitis virus; LPS, lipopolysaccharide; MHCII, major histocompatibility complex II; MLN, mesenteric lymph node; PBMC, peripheral blood mononuclear cells; PD-1, programmed cell death protein 1; PVN, paraventricular nucleus; SCFA, short-chain fatty acid; TCR, T cell receptor; TNF-α, tumor necrosis factor-alpha
Enteric immune system
Studies employing depletion of the maternal gut microbiome reveal an important role for perinatal gut microbes in regulating distributions of enteric immune cells. Pregnant mice that were treated with ampicillin, gentamicin, vancomycin, metronidazole and neomycin starting in late gestation [embryonic day (E)15] and through the post-partum period yielded offspring with deficiencies in small intestinal interleukin(IL)-17 transcript and lamina proprial IL-17-producing cells (Deshmukh et al., 2014). These alterations were seen as early as postnatal day (P)3 and continued through P14, and correlated with substantial reductions in richness and taxonomic diversity of the gut microbiota. Similar reductions in IL-17 were observed in neonatal mice raised germ-free (GF), suggesting that the immune impairments were due to alterations in the perinatal microbiome rather than confounding off-target effects of antibiotics. Three additional studies conducted in the non-obese diabetes (NOD) mouse model reported that maternal microbiome depletion by antibiotics reduces neonatal IL-17-producing cells (Candon et al., 2015; Hu et al., 2015; Tormo-Badia et al., 2014). Pregnant NOD mice treated with neomycin, streptomycin and polymyxin from conception through delivery similarly yielded offspring with reduced diversity of the gut microbiota and reduced IL-17- and interferon(IFN)γ-producing cluster of differentiation (CD)4+ and CD8+ cells in the mesenteric lymph nodes and Peyer’s patches (Hu et al., 2015). These were also associated with increases in CD4+CD25+forkhead box P3 (Foxp3)+ regulatory T (Treg) cells in the mesenteric lymph node and spleen. Analogous changes were seen in response to maternal exposure to a different regimen of antibiotics (Candon et al., 2015; Tormo-Badia et al., 2014), suggesting that the effects could be broadly generalizable to reductions in maternal bacterial load and diversity. These effects of maternal microbiome depletion on promoting neonatal immunosuppression correlated with altered resistance to endophenotypes of diabetes that appear to vary depending on the specific neonatal microbiota changes observed. Consistent with this, another study involving maternal and neonatal vancomycin treatment of wildtype mice revealed reductions in cluster of differentiation (CD)4+CD25+ forkhead box P3 (Foxp3)+ Tregs in offspring colon (Russell et al., 2012) which contrasted the increases seen after treatment of pregnant NOD mice with a cocktail of antibiotics (Hu et al., 2015). Overall, these studies reveal that the perinatal gut microbiome is capable of altering early life enteric immune homeostasis.
Additional studies employing temporally restricted conventionalization reveal distinct roles for the neonatal versus maternal microbiome in regulating early life immune status. Neonatal mice raised GF exhibited increased levels of invariant natural killer T cells (iNKTs) in the colonic lamina propria, a phenomenon dependent on increased expression of chemokine (C-X-C motif) ligand 16 (CXCL16) (Olszak et al., 2012). These elevations in colonic iNKTs and CXCL16 were abrogated by conventionalization just before birth, but not by conventionalization of adult GF mice, revealing an early critical period during which the gut microbiota impacts intestinal iNKT levels. These effects of the neonatal microbiota on colonic iNKT numbers were important for later life disease susceptibility. Adult offspring subjected to neonatal microbiota depletion exhibited increased morbidity in response to models of oxazolone-induced colitis and ovalbumin-induced allergic asthma, both in a CD1d- and iNKT-dependent manner. Consistent with a role for the neonatal microbiome regulating enteric immune function, early postnatal antibiotic treatment consistently altered intestinal expression of immune-related genes across host species (Gomez de Aguero et al., 2016; Jensen et al., 2014; Schumann et al., 2005). For example, Sprague-Dawley rats treated orally with amoxicillin analogue from P7 to 17 exhibited reduced expression of genes relevant to Paneth cell activity and major histocompatibility complex (MHC)-based antigen presentation, and upregulation of genes involved in mast cell function. Moreover, pigs treated orally with gentamycin, ampicillin and metronidazole from birth through P5 exhibited altered small intestinal expression of genes related to pathways for the complement cascade, cytokine signaling and innate immunity (Jensen et al., 2014). Consistent with this, GF mice exhibited numerous immune-related changes in intestinal gene expression that were restored only by colonization at birth, but not by microbiota conventionalization during adulthood, pointing to the importance of the early life microbiota in guiding immune-related transcriptional programs (El Aidy et al., 2013). These studies examining early postnatal antibiotic-treatment and microbiota conventionalization support the notion that the neonatal microbiota alters immune development.
Approaches to selectively manipulate the maternal microbiota are challenging because maternal antibiotic treatment and GF rearing result in confounding neonatal microbiome alterations due to vertical transmission of the disrupted maternal microbiota to the offspring. However, one study achieved gestation-restricted bacterial colonization by transiently colonizing pregnant GF dams with a non-replicating strain (HA107) of E. coli (Gomez de Aguero et al., 2016). Compared to GF controls, GF mice born to E. coli-colonized dams displayed increased frequencies and absolute numbers of small intestinal NKp46+RORγt+ type 3 innate lymphoid cells (ILC3s), with corresponding increases in intestinal F4/80+CD11c+ mononuclear cells. In addition to maternal microbiota-dependent increases of particular innate immune cells, offspring of maternally colonized dams exhibited altered intestinal transcriptional profiles compared to GF controls, characterized by elevated expression of genes encoding antimicrobial peptides and proteins important for metabolism of microbial metabolites. Interestingly, these immune alterations were observed by P14 and continued after weaning, revealing long-term effects of transient maternal exposure to gut bacteria on offspring immune profiles. Similar changes in postnatal innate immunity were also seen in offspring of mothers colonized with a limited consortium of gut bacteria called altered Schaedler’s flora, suggesting that the effects were not unique to maternal gestational colonization particularly with E. coli species. Interestingly, there were no overt effects of maternal gestational colonization on distributions of T and B cells across lymphoid organs. The findings of the study provide a fundamental proof-of-concept that the maternal microbiome impacts postnatal immune homeostasis in offspring. However, some limitations include the use of a single bacterial species, the transient nature of maternal colonization, and differences in adaptive immune development between rodents and humans or non-human primates. Interestingly, the study went on to demonstrate that the effects of maternal gestational colonization on offspring NKp46+ ILC3s, but not CD11c+F4/80+ induced mononuclear cells, were mediated by serum transfer of maternal antibodies and microbial metabolites such as indole-derivative ligands of the aryl hydrocarbon receptor (AhR). Consistent with this, an independent study revealed that maternal immunoglobulin (Ig)G and IgA are generated against the microbiota through mechanisms that are dependent on toll-like receptor (TLR) signaling and independent of T cell function (Koch et al., 2016). These maternal antibodies were transmitted to developing offspring and important for dampening T follicular helper responses and germinal center B cell responses in mesenteric lymph nodes by P25. Altogether, these findings raise the interesting possibility that offspring immunity may be differentially modulated by the maternal microbiome, and further highlight the need for additional studies that evaluate critical periods for maternal and neonatal microbiome contributions to immunomodulation.
Peripheral immune system
The influences of the perinatal microbiome on offspring immune development extend beyond the gastrointestinal tract, as many phenotypes observed in the GALT are also seen in peripheral tissues. Like profiles seen in the gut, lungs from GF mice also exhibited elevated iNKT cell levels, which were restored by neonatal, but not adult, microbiota conventionalization (Olszak et al., 2012). In a mouse model of autoimmunity based on the NLRP3 inflammasome mutation R258W, the maternal microbiota was required for neonatal IL-1β and tumor necrosis factor-α (TNFα) responses in the skin (Nakamura et al., 2012). Interestingly, neonates born to antibiotic-treated mothers did not exhibit symptoms associated with NLRP3 R258W-mediated skin inflammation. In another study, late gestational and early postnatal antibiotic treatment yielded murine offspring with decreased abundance of circulating neutrophils (Deshmukh et al., 2014). Interestingly, reconstitution of the neonatal gut microbiota of antibiotic-treated mice with age-matched conventional microbiota from P3, 5, 7, or 14 completely reversed this neutrophil deficiency. However, only a partial restoration in host survival against E. coli-induced sepsis was observed, suggesting a contribution of the maternal microbiota or immediate postnatal microbiota (before P3) to host resistance to infection. Similarly, dams treated with antibiotics during gestation and lactation yielded pups with deficient production of IFNγ by CD8+ T cells and altered distributions of dendritic cell (DC) and NK cell subsets in response to in vitro or in vivo viral challenge (Gonzalez-Perez et al., 2016). This impaired IFNγ response was partially restored by treating P15 mice with lipopolysaccharide (LPS), raising the possibility that endogenous LPS from the neonatal microbiome may be required for proper CD8+ T cell function (Gonzalez-Perez and Lamouse-Smith, 2017). Overall, these studies highlight the potential for the perinatal microbiome to induce changes in peripheral immune responses across several organ systems.
The diversity in microbiota-associated immune phenotypes during early life highlight the need to advance from broad manipulations of the microbiome, such as antibiotic depletion and GF rearing, toward identifying mechanisms by which specific microbes and microbial products impact offspring immune development. In some cases, the lack of consistency seen across microbiota depletion studies may be due in part to the particular regimen of antibiotic treatment employed, host species- and strain-specific variations in autochthonous microbiome, and differences in diet, among various other factors that impact microbial and immune homeostasis. For example, treatment of pregnant NOD mice with neomycin, which targets Gram-negative bacteria, yielded offspring with decreased relative frequencies of CD11b+ and CD11c+ antigen presenting cells expressing costimulatory molecules CD86 and CD80 in both spleen and mesenteric lymph node. In contrast, treating pregnant NOD mice with vancomycin, which targets Gram-positive bacteria, yielded no detectable changes in offspring CD11b+CD80+ and CD11b+CD86+ antigen presenting cells (Hu et al., 2016). Indeed, specific bacteria indigenous to the maternal microbiota may mediate changes in immune function seen in offspring. Breast-fed rhesus macaque offspring showed distinct differences in microbiota composition at six-months of age compared to nursery-fed controls, which correlated with elevated Th17 cell levels in the blood (Ardeshir et al., 2014). Roles for the maternal and early neonatal microbiota on early life immune regulation have also been implicated in humans, as the presence of Lactobacillus in the maternal vaginal microbiota was strongly correlated with increased CD4+ CD45RO+ T cells and IL-12 levels in neonatal cord blood (Stencel-Gabriel et al., 2009). Higher prevalence of Bifidobacterium at one month of age was associated with increased concentrations of salivary secretory IgA at 6 months (Sjogren et al., 2009). The same study reported an inverse correlation between early colonization of Bacteroides fragilis and LPS-induced CCL4 and IL-6 production by peripheral blood mononuclear cells. Caesarean section-born infants had reduced Bacteroidetes, which correlated with reduced Th1-associated chemokines, CXCL10 and CXCL11 (Jakobsson et al., 2014). Further research is needed to determine causal effects of select members of the gut microbiota on early life immunity.
The ability of the perinatal microbiome to modulate widespread immune distributions and responses may be due in part to microbial effects on immune cell ontogeny. Proliferating Ki67+ bone marrow hematopoietic progenitors were markedly reduced in antibiotic-treated animals compared with those from SPF controls, through signal transducer and activator of transcription 1 (Stat1)-dependent mechanisms (Josefsdottir et al., 2017). Reductions in circulating neutrophils in response to late gestational and early postnatal antibiotic treatment were attributed to deficits in Ly6G+ bone marrow progenitors and impaired granulopoiesis (Deshmukh et al., 2014). Similarly, GF mice exhibited impaired peripheral myelopoiesis, characterized by reduced F4/80+CD11b+ macrophages in the spleen and bone marrow (Khosravi et al., 2014). Exactly how the perinatal microbiome induces persistent changes in hematopoiesis, immune differentiation and function remains poorly understood. However, there is increasing evidence that select microbes and microbial products can regulate immune programming by altering epigenomic landscapes. In particular, the short chain fatty acid (SCFA) and histone deacetylase (HDAC) inhibitor butyrate derives largely from bacterial fermentation and promotes anti-inflammatory responses in neutrophils (Vinolo et al., 2011), DCs (Arpaia et al., 2013; Singh et al., 2010) and macrophages (Chang et al., 2014; Trompette et al., 2014). Direct effects of butyrate on acetylation of the Foxp3 locus are reported to contribute to microbial modulation of Treg cell differentiation (Furusawa et al., 2013; Smith et al., 2013). In addition, depletion of the gut microbiota and GF rearing resulted in global changes in histone modifications and corresponding transcriptional alterations across distinct small intestinal ILC subsets (Gury-BenAri et al., 2016). While many of these microbial influences appear to occur through active signaling during adulthood, whether the perinatal microbiome contributes to epigenetic programming of immune function and the extent to which microbial influences on immune epigenetics can be passed transgenerationally requires future investigation.
Neuroimmune system
Consistent with the finding that the gut microbiota regulates peripheral myelopoeisis, several recent studies reveal that manipulating the gut microbiota alters development of brain-resident microglia of the neuroimmune system. Microglia arise from erythromyeloid cells in the yolk-sac and are distinct from peripheral macrophages based on their unique gene expression profiles, particularly colony stimulating factor 1 receptor (CSF1R), DNAX activation protein of 12 kDa (DAP12), interferon regulatory factor 8 (IRF8), and transcription factor PU.1 (Prinz and Priller, 2014). Additional features distinguish microglia from macrophages, in that they are long-lived in the brain, able to self-renew and less motile despite highly dynamic processes. Murine microglia have recently been reported to undergo defined transcriptional transitions during development (Matcovitch-Natan et al., 2016), characterized by three distinct stages of maturation: early- (midgestation; E10.5 to E14), pre- (late gestation to early postnatal; E14 to P9), and adult- (four weeks of age and on) microglia. Importantly, human fetal microglia exhibited developmental transcriptomic changes similar to those observed in murine fetal microglia, suggesting that microglial developmental patterns are conserved across different species. To determine whether the microbiome impacts microglial development, transcriptomes were profiled in newborn and adult microglia from GF mice compared to conventionally-colonized controls. Across two independent studies, adult microglia from GF mice exhibited decreased expression of genes relevant to inflammation and immune defense, such as Cst7, P2ry13, Mcm5 and Tpi1 (Erny et al., 2015; Matcovitch-Natan et al., 2016). In addition, gene expression signatures of adult stage microglia, such as Neurl3, Bre and Relb, were downregulated in microglia from GF mice, suggesting that GF rearing impairs microglial maturation. Consistent with this, microglia from adult GF mice exhibited reduced stellate morphology in vivo, denoting an immature histological phenotype with corresponding impairments in morphological and transcriptional responses to viral and bacterial immune challenge (Erny et al., 2015). Similar abnormalities in cell morphology and gene expression were observed in microglia from adult mice treated orally with antibiotics, suggesting that alterations in microglia may be induced postnatally. Moreover, abnormalities in microglia could be restored by adult conventionalization of GF mice with a complex microbiota or by treating adult GF mice chronically with SCFAs. Overall these findings reveal that microbiota depletion and GF rearing impairs microglial development, in a postnatally inducible and reversible manner.
Although the conventionalization and metabolite supplementation experiments reveal that microglial status can be modulated in microbiota-depleted mice during adulthood, three independent studies highlight a role for the maternal microbiota in guiding embryonic microglial development. Gene expression changes observed in microglia from GF mice compared to conventional controls were more pronounced in adult microglia relative to newborn microglia (Matcovitch-Natan et al., 2016). These findings suggest that the microbiome is important for microglial development from the pre- to adult phenotype. Nonetheless, 240 genes were differentially expressed even in newborn microglia from GF mice, suggesting that the maternal microbiome may guide microglial maturation during prenatal development. Consistent with this, 19 differentially expressed genes were detected in microglia harvested from GF vs conventional mice at E14.5, suggesting a modest effect of the maternal microbiome on microglial progenitors during mid-gestation (Thion et al., 2018). However, numerous genes were differentially expressed in microglia isolated from E18.5 GF versus conventionally-colonized mice, particularly in males (1,216 differentially-expressed genes) compared to females (20 differentially expressed genes), revealing a sexually dimorphic effect of the maternal microbiome on microglial development. This sexual dimorphism in microbiota-dependent microglial maturation aligns with several reported sex differences in innate and adaptive immune responses (Klein and Flanagan, 2016).
In addition to exhibiting microglia with altered gene expression, offspring from GF dams displayed increased numbers of brain microglia beginning in embryonic ages (Castillo-Ruiz et al., 2018; Thion et al., 2018). Increases in microglial density and ramifications were observed across the somatosensory cortex, striatum and pre-optic area of E14.5, E16.5 and E18.5 embryos of GF mothers, compared to those from conventionally-colonized control mothers. Notably, a sex bias toward males was only detected for the somatosensory cortex at E18.5. Despite their elevated regional density, microglia from GF mice exhibited altered morphology during gestation (Castillo-Ruiz et al., 2018) and early postnatal development (Thion et al., 2018), which was consistent with an immature phenotype analogous to the reported morphology of adult GF microglia (Erny et al., 2015). This immature phenotype was coincident with reduced expression of the cytokines IL-1β and TNFα in the GF neonatal mouse hind- and midbrain (Castillo-Ruiz et al., 2018). Overall, these studies reproducibly demonstrate that the microbiota regulates microglial gene expression, morphology and function, and these effects arise early during embryonic development.
Precisely how the microbiome regulates the antenatal development of microglia remains unclear, but the findings from adult intervention studies suggest that the effects may be specific to particular microbial communities and microbial products (Erny et al., 2015). Epigenomic profiling of microglia from embryos of GF compared to conventionally colonized dams revealed global changes in chromatin accessibility that correlated with transcriptional alterations (Matcovitch-Natan et al., 2016). These changes in microglial epigenetic profiles may align with the reported effects of SCFAs on histone acetylation across various immune cell subtypes and the ability of chronic SCFA supplementation to correct microglial impairments seen in adult GF mice (Erny et al., 2015). Continued studies are needed to elucidate the signaling pathways from the maternal microbiome to microglial progenitors that underlie their postnatal morphology, densities, transcriptional profiles and cellular responses in the brain. In addition, influences of the microbiota on cellular functions of microglia, including early life phagocytosis and synaptic pruning, are warranted.
Early life immune function and normal neurodevelopment
Interactions between the immune system and nervous system during gestation and the neonatal period are important for guiding normal neurodevelopment. Neurodevelopment involves a precisely orchestrated series of events that transpire from gestation through the postnatal period. Following differentiation of neural stem cells in early gestation, neural progenitor cells (NPCs) from the ventricular zone proliferate and migrate to pattern the major structures of the brain (Kriegstein and Alvarez-Buylla, 2009; Stiles and Jernigan, 2010). Several signaling factors traditionally acknowledged for their roles in the immune system are increasingly recognized for their fundamental roles in the developing brain, including TLR ligands, cytokines, complement proteins and MHC (Figure 2). In addition, maintaining maternal and early life immune homeostasis is integral to ensuring proper neurodevelopment (Table 2). Growing evidence that the maternal and neonatal microbiome regulate immune development raise the intriguing question of whether microbial modulation of early life immunity impacts brain development. Such interactions between the microbiome, immune system and nervous system particularly during early life could provide insight into potential mechanisms underlying the striking effects of microbiome depletion, GF rearing and microbial dysbiosis, on various host behaviors and neurophysiologies [reviewed in (Vuong et al., 2017)].
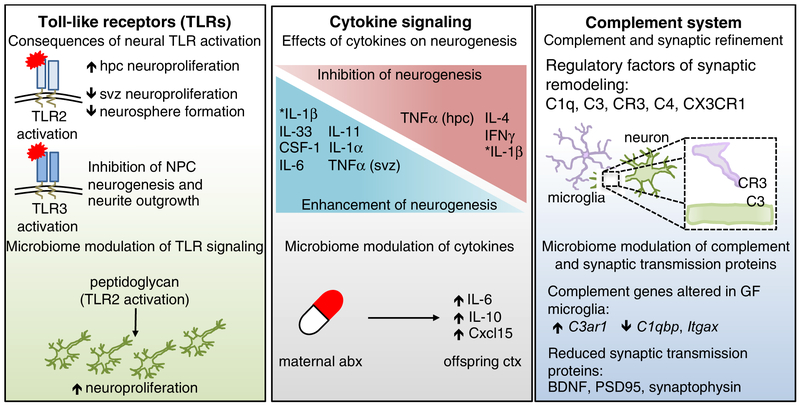
Figure 2. Canonical immune molecules play fundamental roles in normal neurodevelopment.
TLRs are expressed in every major cell type of the brain, and exhibit temporally distinct expression patterns that are critical for normal neurodevelopment. Activation of TLRs can have opposing effects on neuroproliferation and neurogenesis. For example, TLR2 activation increased hippocampal (HPC) neuroproliferation, but reduced neuroproliferation in the subventricular zone (SVZ). Activation of TLR3 resulted in reduced neural progenitor cell neurogenesis and neurite outgrowth, highlighting the differential effects of TLR stimulation in the developing brain. Fetal TLR2 activation by the bacterial cell wall component, peptidoglycan, increased hippocampal neuroproliferation. The developing brain is also highly dependent on a balanced cytokine milieu, as different cytokines can have opposing effects on neurogenesis. Maternal treatment with antibiotics resulted in sustained increases in IL-6, IL-10 and Cxcl15 in the cortex of offspring. Further, cytokines induced by maternal immune activation (MIA), were dependent on the maternal microbiota, such as increased prevalence of segmented filamentous bacteria (SFB), which led to exacerbated behavioral deficits in offspring. The complement system is critical for synaptic editing and refinement of neural circuits. Recognition of neuronal C3 by microglial-expressed C3R is important for this process, as are C1q and C4. Mice lacking the fractalkine receptor, CX3CR1, also exhibit disrupted synaptic editing. GF microglia exhibited altered gene expression of complement-associated genes, such as increased C3ar1 and decreased C1qbp and Itgax. Proteins essential for synaptic transmission, such as BDNF, synaptophysin, and PSD95, are also reduced in microbiota-depleted mice. Many other molecules, such as the aryl hydrocarbon receptor (AhR) and MHCI impact neurodevelopment as well (Bryceson et al., 2005; Glynn et al., 2011; Kimura et al., 2016; Latchney et al., 2013; Zohar et al., 2008). Deletion of AhR reduced cell proliferation and neuronal differentiation in the dentate gyrus (DG). Reduction in surface-expressed MHCI resulted in increased synapse density. Several MHCI receptors are expressed in the brain, including PirB, CD3z, Ly49 and KIR.
Table 2.
: The microbiome modulates canonical immune factors and neurodevelopment.
| Species (Strain) | Microbiome Perturbation | Effect on Host Brain | Reference |
|---|---|---|---|
| Toll-like receptor (TLR)/Neurogenesis | |||
| Mice (C57BL/6) | GF mice | Decreased: NF-κB signaling in HEK293T cells with GF serum | Clarke et al, 2010 |
| Mice (C57BL/6) | Peptidogylcan-teichoic acid injected at E10 or E15 | Decreased: Spatial- and novel object-recognition (E10) Increased: cortical neuroproliferation; NPC neuroproliferation |
Humann et al, 2016 |
| Mice (Swiss Webster) | GF mice; conventionalization at P21 | Increased: hippocampal neurogenesis (GF and GF-conventionalized) | Ogbonnaya et al, 2015 |
| Mice (C57BL/6) | Abx treatment adult (ampicillin + sulbactum, vancomycin, imipenem + cilastatin, metronidazole) | Decreased: hippocampal neurogenesis; novel object recognition; brain CCR2+Ly6Chi monocytes | Mohle et al, 2016 |
| Cytokines | |||
| Mice (Balb/c) | Abx treatment E0.5-P21 | Increased: brain transcripts arginine vasopressin receptor 1b), IL-6, Cxcl15 and IL-10 (females only) Sex-specific alterations of behavior | Leclercq et al, 2017 |
| Mice (C57BL/6J) | GF mice colonized with pre-term human microbiota at E15 | Decreased: brain NeuN expression (GF, low-weight pre-term colonization), brain Olg2 expression (GF), brain MBP (low-weight colonization), brain IGF-1 (GF, low-weight pre-term colonization, 4 weeks) Increased: brain transcripts IL1β and TNFα(GF), brain IGF-1 (GF, 2 weeks) Altered brain gene expression |
Lu et al, 2018 |
| Mice (C57BL/6) | GF mice; MIA induced E12.5 (poly(I:C)) | Decreased: MIA-associated behavioral deficits; Th17 cell-dependent behavioral phenotypes Increased: SFB-dependent plasma and ileal IL-17a |
Kim et al, 2017 |
| Mice (C57BL/6) | GF mice; MIA induced E12.5 (poly(I:C)) |
Cortical patches associated with MIA | Shin Yim et al, 2017 |
| Mice (C57BL/6) | MIA induced E11.5 and E12.5 (poly(I:C)); co-housing | Taconic-associated microbiota associated with MIA-induced behavioral deficits and serum IL-17a | Lammert et al, 2018 |
| Mice (C57BL/6J) | E12.5 injection of IL-6or IFNγ | IL-6 induced behavioral abnormalities and gene expression changes | Smith et al, 2007 |
| Mice (C57BL/6N) | MIA induced at E12.5 (poly(I:C)) | Increased: MIA-induced fetal brain II6, Stat3 and Myc expression; MIA-induced fetal brain IL-4, IL-6 and IP-10 | Wu et al, 2017 |
| Mice | GF mice; colonization with human microbiota | Differential expression of cecal cytokines in GF mice colonized with human first-trimester or third-trimester fecal samples | Koren et al, 2012 |
| Complement System | |||
| Mice (C57BL/6) | GF mice (neonatal and adult) | Altered microglial gene expression, including complement factors C3ar1, C1qbp and Itgax *supplemental data | Matcovitch-Natan et al., 2016 |
| Mice (NMRI) | GF mice adult | Decreased: anxiety-like behavior; brain BDNF, synaptophysin and PSD-95 Increased: spontaneous motor activity Altered brain gene expression |
Diaz Heijtz et al., 2011 |
| Human | Ulcerative colitis microbiota | Increased: IL-12 and complement genes with ulcerative colitis microbiota | Morgan et al., 2015 |
| Mice (C57BL/6J) | Abx treatment P28-P56 (ampicillin, vancomycin, neomycin, metronidazole; mesenteric ischemia/reperfusion | Decreased: mesenteric ischemia/reperfusion associated intestinal C3 | Yoshiya et al., 2011 |
| Mice (C57BL/6) | GF mice adult | Decreased: skin gene expression C3, C1qa, C1qb and C3ar1 | Meisel et al., 2018 |
| Mice (C57BL/6 and DBA/2J) | Subcutaneous LPS adult | Increased: retinal gene expression of TLR-4, Myd88, c-JUN, TRIF, IRF3 and C1qb following LPS | Astafurov et al., 2014 |
Abbreviations: Abx, antibiotic; BDNF, brain-derived neurotrophic factor; GF, germ-free; IFNγ, interferon-gamma; IGF-1, insulin-like growth factor-1; IL, interleukin; IRF3, interferon transcription regulatory factor 3; LPS, lipopolysaccharide; MBP, myelin basic protein; MIA, maternal immune activation; NPC, neural progenitor cell; Olg2, oligodendrocyte transcription factor 2; PSD-95, post-synaptic density protein-95; SFB, segmented filamentous bacteria; TLR-4, toll-like receptor 4; TNF-α, tumor necrosis factor-alpha; TRIF, TIR-domain-containing adapter-inducing interferon-β
Toll-like receptor signaling on neuronal proliferation
TLRs are evolutionarily conserved proteins that enable innate immune recognition of microbial components. TLRs detect a wide array of microbial constituents, including dsRNA (TLR3), LPS (TLR4), flagellin (TLR5), CpG-DNA (TLR9) and triacylated or diacylated lipopeptides (TLR2/1 or TLR2/6) (Kawai and Akira, 2011; O’Neill et al., 2013). Within the last decade, roles for TLRs have been found to extend beyond innate immune recognition of microorganisms.
TLRs are expressed in all subtypes of brain-resident cells. The full repertoire of TLRs is expressed by brain microglia, while a more limited subset of TLRs have been detected in neurons (TLR2, 3, 4, 7, 8, 9), astrocytes (TLR2, 3, 9) and oligodendrocytes (TLR2, 3) (Hanke and Kielian, 2011). Particular TLRs exhibit temporally controlled brain expression patterns, pointing to their functional importance during critical periods of neurodevelopment (Barak et al., 2014). For example, TLR3 is the most highly expressed during embryonic development (Kaul et al., 2012), during which TLR3 signaling exerts a negative regulatory effect on neurogenesis (Lathia et al., 2008). Stimulation of E12.5 NPCs with the TLR3 ligand polyinosinic:polycytidylic acid [poly(I:C)] and adaptor molecule IFNβ reduced their proliferative capacity (Lathia et al., 2008). Another study reported that TLR3 is expressed particularly in neuronal growth cones, where activation of TLR3 by poly(I:C) inhibits neurite outgrowth (Cameron et al., 2007). Consistent with this, mice lacking TLR3 exhibited elevated levels of proliferative cell nuclear antigen+ (PCNA) cells in the dorsal telencephalon at E13.5 but not E16.5, highlighting the effects of TLR3 signaling on neurogenesis that correspond with its temporal patterns of gene expression.
These temporal and spatial effects of TLR3 activation on neurodevelopment are similarly seen with TLRs that detect bacterial products. Like TLR3, TLR2 is expressed in the mouse cortex during embryonic and early postnatal stages of development. Stimulating cortical NPCs with endogenous or synthetic TLR2 ligands inhibited neurosphere formation (Okun et al., 2010), wherein neural proliferation is assessed based on the ability of stem cells from the adult brain to form proliferative clusters of cells in vitro (Reynolds et al., 1992; Reynolds and Weiss, 1996). Moreover, in utero intracerebral administration of TLR2 ligands sufficiently reduced the number of proliferative BrdU+ cells in the subventricular zone and enlarged ventricular size. Another study points to the contrasting effects of TLR2 and TLR4 signaling on adult NPC proliferation and differentiation (Rolls et al., 2007). TLR2-deficiency reduced NPC differentiation to β-III tubulin+ neurons, whereas TLR4-deficiency increased proliferation of NPCs (Rolls et al., 2007). Overall, these studies raise the question of whether early life exposures to microbe-associated molecular patterns (MAMPs) can alter neuronal proliferation and specification via temporally-restricted and spatially-localized TLR signaling.
One key question is whether these effects of TLR activation during early development primarily reflect detrimental consequences of microbial infection on neurogenesis or whether endogenous TLR ligands from the indigenous microbiome can also elicit responses. Supporting the former, offspring born to dams challenged with LPS in mid-gestation exhibited reduced doublecortin+ (Dcx) neurons and T-box brain protein 2+ intermediate NPCs in the dentate gyrus. While LPS reportedly does not cross the placental barrier, co-injection of an IL-6 neutralizing antibody abrogated the detrimental influences on neurogenesis (Mouihate, 2016), suggesting indirect effects of inflammatory factors on neuronal function. Many groups have also reported direct roles for TLR signaling on adult neurogenesis in conditions that do not involve MAMP-mediated immune activation. For example, in a mouse model of cerebral ischemic stroke, neuronal differentiation was reduced in the ipsilesional cortex of TLR4−/− mice (Moraga et al., 2014). In addition, TLR4 expression and BrdU-incorporation increased in NPCs during the acute phase following traumatic brain injury (Ye et al., 2014).
One study to date has examined effects of microbiota-related TLR activation on early neurogenesis. Peptidoglycan, a cell wall component of Gram-positive bacteria, is a known ligand and activator of TLR2 (Aliprantis et al., 1999; Yoshimura et al., 1999). Modest levels of peptidoglycan can be detected in the circulation under homeostatic conditions, and peptidoglycan produced by the endogenous microbiota is sufficient to prime systemic innate immune cells (Clarke et al., 2010). In a model where pregnant mice were injected intravenously with fluorescently labeled peptidoglycan-teichoic acid complex to mimic low endogenous levels, the bacterial cell wall components traversed the placental barrier and entered the fetal brain itself (Humann et al., 2016). Fetal brains exposed in utero to peptidoglycan-teichoic acid complex exhibited enhanced hippocampal neuroproliferation in the absence of any evidence of cell death. This phenotype was not observed in fetal brains from TLR2-deficient mice, suggesting direct signaling of bacterial cell wall components through neuronal TLR2. Consistent with this, NPCs stimulated in vitro with peptidoglycan-teichoic acid complex exhibited elevated neuroproliferation, as measured by BrdU incorporation, FACS-dependent cell quantitation and neurite profiling. Interestingly, transient exposure to bacterial cell wall components in utero led to alterations in adult spatial learning and working memory that were dependent on the timing of embryonic exposure (Humann et al., 2016).
Additional studies suggest that the microbiome also regulates adult hippocampal neurogenesis through pathways that depend on critical developmental periods. Mice reared GF exhibited increased adult hippocampal neurogenesis, as measured by increased relative abundance of proliferating BrdU+ cells in the dorsal hippocampus (Ogbonnaya et al., 2015). Interestingly, colonizing GF mice with a conventional mouse microbiome post-weaning had no impact on the phenotype, suggesting that the microbiome during early life suppresses later life hippocampal neurogenesis. Another report suggests that microbial modulation of adult neurogenesis is mediated by peripheral immunoregulation. Adult mice treated chronically with antibiotics exhibited reduced proliferating neuronal progenitors, labeled by BrdU+Dcx+NeuN+ in the subgranular zone of the dentate gyrus, which correlated with deficits in brain, blood and bone marrow Ly6C(hi) monocytes (Mohle et al., 2016). CCR2-deficient mice or mice treated with anti-CCR2 antibody to deplete monocytes sufficiently decreased hippocampal neurogenesis, whereas adoptive transfer of Ly6C(hi) monocytes in vivo or co-culture of NPCs with Ly6C(hi) monocytes promoted neurogenesis. These results suggest that microbial modulation of peripheral monocyte levels mediate microbial effects on adult hippocampal neurogenesis. The specific microbial products involved and the prospective role for TLR signaling in this model remain unclear. Considered together, these studies emphasize the need to investigate how MAMPs contributed by the microbiome influence neurodevelopment, both pre- and postnatally.
Cytokine signaling on neurogenesis
Cytokines canonically known to instruct systemic- and tissue-specific immune responses are expressed by neurons, microglia and astrocytes during normal neurodevelopment. Fundamental roles for cytokines in the brain have been the subject of extensive prior review (Deverman and Patterson, 2009). Here, we highlight influences of cytokines during various stages of early neurogenesis from neuronal proliferation and differentiation to axonal pathfinding. We also evaluate emerging evidence that modulation of cytokines by the perinatal microbiome may alter brain development in ways that impact later life behavior.
As in the immune system, cytokines in the brain exert diverse influences on neurogenesis. For example, IL-4 and IFNγ each inhibited proliferation of mouse embryonic NPCs (Ahn et al., 2015), whereas IL-34 and colony-stimulating factor-1 (CSF-1) each enhanced neuronal proliferation, consistent with their high expression in the early postnatal brain (Nandi et al., 2012). IL-6 promoted neurogenesis in human fetal striatal NPCs (Johansson et al., 2008), which was consistent with the ability of IL-6 to increase neurogenesis of mouse fetal subventricular zone NPCs in vivo (Islam et al., 2009). For other cytokines, such as IL-1β and TNFα, results have been variable and sometimes conflicting (Gutierrez et al., 2008; O’Keeffe et al., 2008). In human fetal hippocampal NPC cultures, IL-1β decreased numbers of Dcx+ neuroblasts (Zunszain et al., 2012). In contrast, single stereotaxic injections of IL-1β into the hippocampus had no effect on levels of Dcx+ progenitors, but increased neuronal proliferation, as measured by numbers of BrdU+ neurons. This suggests that IL-1β promotes neuronal expansion or differentiation, rather than progenitor self-renewal (Seguin et al., 2009). For TNFα, low doses enhanced proliferation of mouse neonatal subventricular zone NPCs, whereas high doses induced apoptosis (Bernardino et al., 2008). TNFα reduced neurite outgrowth in co-cultures of mouse embryonic NPCs with astrocytes (Neumann et al., 2002) and similarly reduced neuronal proliferation of human fetal hippocampal NPCs when co-cultured with astrocytes (Chen et al., 2013). These inhibitory effects of TNFα on neurite outgrowth were restricted to critical periods of late embryonic and early postnatal development (Nolan et al., 2014). Overall, these studies reveal that cytokines elicit varied effects on neuronal proliferation that depend on several factors, such as the gestational age, brain region, cell type, internal state and external context.
A few reports have also highlighted roles for cytokines in guiding early neuronal differentiation. IL-1β decreased differentiation of rat neonatal dentate gyrus NPCs into serotonergic neurons (Zhang et al., 2013), whereas IL-11, IL-1α and IL-1β each promoted differentiation of rat fetal mesencephalic NPCs into dopaminergic neurons (Ling et al., 1998). The latter finding is further corroborated by evidence from an in vivo study wherein intrastriatal injection of IL-1β promoted outgrowth of dopaminergic neurons in the ventral tegmental area (Wang et al., 1994). Together, this evidence suggests that particular cytokines can contribute to early neuronal fate specification.
Emerging evidence implicates the maternal and neonatal microbiome in cytokine regulation and neurodevelopment. Treating pregnant mice with low-dose penicillin during late pregnancy and through early postnatal life yielded offspring with reduced diversity of the gut microbiota and increased expression of IL-6, IL-10 and Cxcl15 in the frontal cortex, compared to vehicle-treated controls (Leclercq et al., 2017). These alterations corresponded with abnormalities in adult stress-induced behaviors and sociability, revealing that early life depletion of the gut microbiota leads to later life alterations in brain cytokine levels and behavior. Another report utilized a human microbiota transplantation approach to examine effects of the preterm infant microbiome on neurodevelopment in mice. Pregnant GF mice colonized with human preterm infant microbiota on E15 yielded offspring with reduced cortical expression of NeuN, a neuronal marker, by P14 that persisted through P28 (Lu et al., 2018). These effects were correlated with dysregulation of cortical expression of genes important for neurotransmission and elevated expression of IL-1β, TNF and Nos1, suggesting that dysbiosis of the human infant microbiome can impair neurodevelopment via modulation of brain neurochemical and neuroinflammatory cytokine signaling.
Further studies have revealed causal roles for maternal microbiota-dependent cytokine regulation in directing fetal neurodevelopment and later life behaviors. Pregnant mice injected systemically with poly(I:C) or LPS experienced transient elevations of pro-inflammatory cytokines, including IL-6, IL-1β and IL-17, across the maternal blood, placenta and fetal brain (Careaga et al., 2017). The composition of the maternal microbiota influenced the severity of this inflammatory response, where the presence of Segmented Filamentous Bacterium in the maternal microbiota promoted the inflammatory response and mediated downstream disruptions to brain development and behavior (Kim et al., 2017). In particular, early life induction of IL-17 is reported to be both necessary and sufficient for yielding adult offspring with impaired neuronal differentiation, characterized by patches of deficient parvalbumin expression by cortical interneurons (Shin Yim et al., 2017). IL-6 and IL-17, as regulated by the maternal microbiota, have each been reported as necessary and sufficient for causing later life behavioral abnormalities in offspring of immune activated mothers (Kim et al., 2017; Lammert et al., 2018; Smith et al., 2007). Interestingly, maternal and placental cytokine signaling in response to immune activation was required for downstream induction of fetal brain cytokine responses (Wu et al., 2017). Supporting an important role for the maternal microbiome in regulating maternal immune status, transplantation of fecal microbiota from pregnant women into GF mice sufficiently altered intestinal cytokine profiles and maternal physiology (Koren et al., 2012). Taken together, these findings support a cascade of cellular and molecular interactions that link the maternal microbiome to maternal immune responses, maternal and fetal brain cytokine profiles, neurodevelopment and behavior. Uncovering how brain cytokines are regulated, and the extent to which changes in the maternal and neonatal microbiome are involved, is integral to understanding the molecular bases of neurodevelopment.
Complement proteins in synaptic refinement
The complement system is an ancient and evolutionarily conserved arm of the innate immune system that facilitates humoral and cell-mediated clearance of pathogenic microbes. This system is divided into three arms: the classical, alternative and lectin pathways (Ricklin et al., 2010; Ricklin et al., 2016; Sarma and Ward, 2011). In the classical pathway, the C1q protein on innate immune cells recognizes foreign or apoptotic cells, and triggers a signal cascade that includes complement proteins C3 and C4 and ultimately leads to the assembly of a pore-like protein complex, called the membrane attack complex, on target cells. While originally discovered for its ability to lyse target cells, several recent discoveries reveal that the classical complement cascade is also fundamental for normal neurodevelopment, mediating neural circuit refinement during early postnatal life [reviewed in (Stephan et al., 2012)]. During neurogenesis, the fetus generates an excess of synaptic connections between neurons, which are removed in an activity-dependent process called synaptic pruning to create precise networks of neural circuits. The retinogeniculate system is particularly useful for evaluating synaptic pruning, as removal of unnecessary synapses is required to generate discrete, nonoverlapping axonal projections from each eye to its proper relay neurons. Mice deficient in complement proteins C1q, C3 or C4, exhibited reduced synaptic pruning of the retinogeniculate system (Sekar et al., 2016; Stevens et al., 2007). During early development, C3 protein was localized to axons of retinal geniculate cells, which was dependent on upstream complement proteins C1q and C4 (Bialas and Stevens, 2013; Sekar et al., 2016) mirroring the sequence of the complement cascade in the innate immune system. Interestingly, microglia were the only resident brain cells to express CR3, the receptor for C3 protein, suggesting that microglia actively prune synapses tagged by C3. Indeed, microglia-mediated phagocytosis of synapses was dependent on signaling between CR3 and C3 (Schafer et al., 2012), where mice lacking CR3, C3 or the fractalkine receptor (CX3CR1) exhibited reduced microglia and increased structural synapses in the visual system as well as the hippocampus during early postnatal life (Paolicelli et al., 2011). In addition to CR3, microglia express various other complement protein receptors, suggesting that there are additional complement-related interactions that regulate microglial function (Stephan et al., 2012). Notably, neurons and astrocytes also express various complement proteins and receptors, indicating that there are other functions for the complement system within the brain aside from guiding microglia-dependent synapse elimination. Indeed, complement has been implicated in several neurological functions during the embryonic and early postnatal period, including neurogenesis and neuronal migration.
Recent studies suggest that the gut microbiome modulates expression of complement proteins by brain-resident microglia and the peripheral complement system. Transcriptomic comparisons of newborn microglia from GF versus conventionally-colonized mice revealed differential expression of many complement-related genes. Newborn microglia from GF mice exhibited increased expression of C3a receptor-1 (C3ar1) and decreased expression of C1q binding protein (C1qbp) and integrin subunit alpha X (Itgax) relative to conventional controls (Matcovitch-Natan et al., 2016). Whether these transcriptional signatures result in microbiota-dependent alterations in complement-dependent functions in microglia, such as synaptic pruning, remains unclear. Consistent with this possibility, proteins important for synaptic transmission, such as brain-derived neurotrophic factor (BDNF), synaptophysin and postsynaptic density protein 95 (PSD95), were decreased in brains of GF mice compared to conventional controls (Diaz Heijtz et al., 2011), suggesting microbiota-dependent alterations in synaptic structure.
Microbiome alterations have also been associated with dysregulation of complement proteins in peripheral sites. Intestinal biopsies from ulcerative colitis patients compared to controls displayed elevated expression of genes related to the complement cascade and IL-12 pathway, which correlated with altered relative abundance of select bacterial taxa of the gut microbiota (Morgan et al., 2015). Consistent with this, depletion of the gut microbiota altered complement expression in the gut, as antibiotic treatment reduced C3 in the intestinal epithelium following mesenteric ischemia/reperfusion injury in mice (Yoshiya et al., 2011). In the skin, associations between the microbiome and complement are also evident; expression of genes involved in the complement cascade, including C3, C1qa, C1qb, and C3ar1 were reduced in cutaneous tissue from GF mice compared to conventional controls (Meisel et al., 2018). Moreover, subcutaneous administration of LPS increased C1qb transcript levels and altered microglial phenotype in a mouse model of optic nerve axon degeneration (Astafurov et al., 2014). Taken together, these studies demonstrate the capacity for the microbiome and microbial products to modulate complement-associated gene expression across different cell types and tissue compartments. Continued research is warranted to examine functional consequences of microbiome-dependent modulation of key pathways for neuroimmune communication, such as complement, cytokines and TLR signaling, and how such signaling between the early life microbiome, immune system and nervous system impacts neurodevelopment.
Early life microbiome-immune interactions and risk for neurological disease
Interactions between the microbiome, immune system and nervous system during gestation and early postnatal life have important implications for many neurological diseases (Figure 3). Neurodevelopmental disorders, such as autism spectrum disorder and schizophrenia, are co-morbid with immune dysregulation across the gut, periphery and brain, and are associated with many immune-related genetic and environmental risk factors (Vuong and Hsiao, 2017). Both early life immune activation and microbial dysbiosis are regarded as susceptibility factors for the etiopathogenesis of neurobehavioral issues that underlie the diagnostic features of the disorders (Vuong et al., 2017). In particular, large epidemiological studies, combined with numerous clinical and case studies, reveal that maternal viral or bacterial infection during pregnancy and elevated levels of maternal pro-inflammatory cytokines are associated with increased risk for autism and schizophrenia in the offspring (Brown, 2012). In animal models of this risk factor, maternal injection of pregnant mice, rats or monkeys with such antigens as poly(I:C) or LPS sufficiently yielded offspring with neuropathological and neurobehavioral endophenotypes of autism and schizophrenia (Careaga et al., 2017). Inhibiting maternal IL-1β, IL-6 or IL-17 signaling using neutralizing antibodies or mice deficient for their respective cytokine or cytokine receptors prevented the development of brain and behavioral abnormalities in the offspring, whereas injecting recombinant pro-inflammatory cytokine instead of microbial antigen was sufficient to induce the brain and behavioral abnormalities (Kim et al., 2017; Lammert et al., 2018; Smith et al., 2007). Consistent with the ability of the gut microbiota to regulate immune homeostasis, differences in the composition of the gut microbiome were important for determining the severity of inflammatory responses to immune activation and the downstream consequences on fetal brain development and adult behavior. Notably, manipulation of the offspring microbiome at weaning was sufficient to correct only a subset of behavioral abnormalities in the model (Hsiao et al., 2013), and adult treatment exhibited limited efficacy. This points to the importance of gestational and early postnatal critical periods during which microbiome interactions with host immunity and physiology can modulate behavioral symptoms of neurodevelopmental disease.
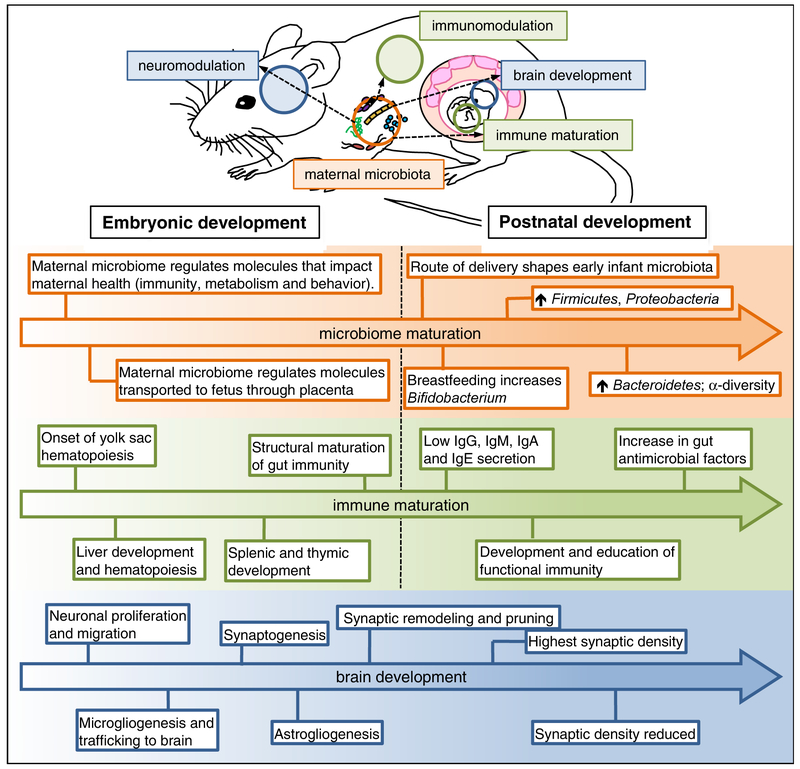
Figure 3. The maternal microbiome influences fetal immune and brain development.
The maternal microbiome produces microbe-associated molecular patterns and secondary metabolites that can impact the developing fetus. Several factors influence the initial acquisition of microbes, including the route of delivery and breast-feeding. The initial infant gut microbiota is dominated by Firmicutes and Proteobacteria, and experiences diet-related shifts, such as the increase in Bifidobacterium following breast-feeding, and Bacteroidetes following a transition to solid foods. Bacterial alpha-diversity increases, eventually reaching complexity similar to the adult. Antimicrobial factors in the gut increase concurrent to the changing infant microbiome. Maturation of the immune system also begins in the fetal period and persists into postnatal development. Hematopoiesis first occurs in the yolk sac and continues in the fetal liver. Lymphoid organs, such as the spleen and thymus, follow, and immune circulating immune cells are also found in the developing fetus. Structural maturation of gut immunity is evident just before birth, although immune function is reduced during this phase. The fetus is capable of producing low levels of immunoglobulins, such as IgG, but relies heavily on maternal immunoglobulins. Following birth, acquisition of immunoglobulins is facilitated through breast-milk. Brain development occurs concurrently over defined stages, starting with the proliferation and migration of neuronal progenitor cells. Shortly after, immature macrophages from the yolk sac migrate to the central nervous system where they mature to resident microglial cells. Shortly after, wiring of the brain circuitry begins with the onset of synaptogenesis. During the late gestational period, astrocytes develop to facilitate the increasing metabolic demand for brain growth. During the late fetal period, and into early postnatal development, synaptic remodeling and pruning occurs, with studies demonstrating a clear role for microglial mediated synaptic editing as well as astrocyte-mediated pruning of the synapse.
Early life microbiome-immune interactions with the developing nervous system may also be relevant to select neurodegenerative and neuropsychiatric diseases, such as Alzheimer’s disease and major depressive disorder, which are associated with microbiome abnormalities, immune dysregulation and early risk factors that predispose to later life disease (Borenstein et al., 2006; Milgrom et al., 2008; Modgil et al., 2014; Tremlett et al., 2017). In the APPSWE/PS1DeltaE9 mouse model for Alzheimer’s disease (Minter et al., 2017), early life exposure to antibiotics for one week led to persistent changes in microbiota composition, with increased Lachnospiraceae and decreased S24–7 later in life and decreases in IL-2, IL-3, and stem cell factor (SCF) in the cerebrospinal fluid. Interestingly, early life antibiotic exposure and alterations in the gut microbiome reduced adult amyloid-β plaque burden in the cortex and hippocampus which coincided with fewer plaque-associated, ramified microglia and astroglia, and a greater abundance of Tregs. In addition, several animal models of prenatal and early postnatal stress have consistently revealed stress-induced alterations in the microbiome, with reductions in Lactobacillus spp in particular, that correlate with stress-induced immunological and behavioral abnormalities relevant to anxiety and depression (Mackos et al., 2017). Transplantation of vaginal microbiomes from maternally stressed animals into C-sectioned offspring from control mothers sufficiently induced alterations in hypothalamic gene expression (Jasarevic et al., 2018). In another study, supplementing stressed animals with Lactobacillus reuteri improved behavioral impairments in the forced swim task, used to measure features of depression-related “despair” (Marin et al., 2017). These studies suggest that the microbiome can tune neuroimmune interactions and neurodevelopment to alter predisposition to later-life symptoms of neurological disease.
Future Directions
Research over the past decade has led to the understanding that intimate interactions between the nervous system, immune system and microbiome are important for guiding brain development and function. This realization offers the opportunity to investigate basic mechanisms underlying microbiome-neuroimmune interactions to reveal fundamental insights into how disparate organ systems communicate with one another to orchestrate complex biological functions. Foundational studies have employed gnotobiotic colonization and antibiotic depletion approaches to examine contributions of the early life microbiome to later-life health and disease. In many cases, however, microbiome interventions applied maternally yield alterations in both the maternal and offspring microbiome, rendering it difficult to clearly distinguish gestational versus neonatal influences on offspring outcomes. In addition, the extent to which select microbial taxa contribute to functional outcomes remains poorly understood.
Further efforts are needed to clearly define the temporal contributions of specific members of the maternal versus neonatal microbiome to host physiology. Existing approaches for conventionalization, depletion, and transplantation may be applied with improved temporal resolution. Gnotobiotic approaches, including c-section, cross-fostering and selective colonization at birth, may aid in evaluating influences of select microbial taxa on the host and further inform the identification of keystone species that are functionally important for development. In addition, advances in targeted ecological engineering of microbiomes and synthetic modification of symbiotic microorganisms may enable microbiome modulation with finer temporal and functional control during critical developmental time periods. Such approaches could also improve spatial control of gut microbes from different tissue compartments. This is an important need as current tools for manipulating the microbiome, such as antibiotic treatment and gnotobiotic colonization, perturb microbiomes across various host sites, and methods for selectively manipulating the gut, skin, vaginal or lung microbiomes are lacking. Advancements in these areas promise to illuminate how homeostatic host-microbiome interactions align with the developmental processes that span gestation through the postnatal period.
Many pioneering studies have demonstrated effects of the maternal and neonatal microbiome on immune and neuronal outcomes, offering the opportunity to identify molecular and cellular mechanisms that mediate early microbiome-neuroimmune communication and long-term consequences of microbiome-neuroimmune interactions on host physiology. Advances in functional genomic, metabolomic, proteomic and lipidomic technologies have revealed numerous molecules in the host that are derived from or modulated by the microbiome. However, functional roles for many such microbial molecules in the host remain unknown. In addition, how active signaling between the microbiome and host cells during early life confers long-lasting effects on host biology remains unclear. Additional efforts are needed to define the identities of microbial and microbially modulated molecules, and to further determine their effects on host physiology. Particular attention to developmental programming of cellular function is warranted, including investigation into microbial influences on host epigenetic modifications and the timescales of their impacts. Longitudinal investigation of host-microbial signaling will also be useful, including the application of benchmark tools for metabolic tracing, cellular lineage tracking and intravital imaging. Continued research into the molecular and cellular bases for how the maternal and neonatal microbiomes interact with the immune and nervous systems will facilitate the understanding of fundamental principles that govern the organization of integrative biological systems.
The maternal and neonatal microbiome is increasingly recognized as an important regulator of health and predisposition to later-life disease. However, much of the evidence supporting causal effects of the early life microbiome on symptoms of disease derive from animal models that utilize reductionist approaches, such as axenic animals and high dose-antibiotic treatment, to demonstrate proof of concept. While such studies are critical for informing basic mechanisms of microbiome-host interactions, the ability to directly translate these findings to clinical applications is limited. Continued efforts to better model clinically relevant microbiome perturbations and risk factors for disease will be important toward informing growing interest in microbiome-based clinical interventions. Of particular interest are effects of environmental factors, such as micronutrients, xenobiotics and stress, on the maternal and neonatal microbiomes. For example, consideration should be given to apply pharmacologically relevant dosages of antibiotics to animal models and to evaluate potential off-target effects of the antibiotics used for microbiome perturbation. In addition, further studies are needed to elucidate how such interactions influence key developmental processes, such as placentation, blood brain barrier formation, biochemical availability and transport across organ systems. Overall, foundational knowledge regarding microbiome-neuroimmune interactions could shed light on new targets for neuromodulation and inform novel strategies for treating symptoms of neurological disorders. A clear mechanistic and functional understanding of how specific microbes, immune cells and neurons signal and respond to each other is paramount to evaluating the safety, efficacy and promise of such endeavors.
Acknowledgements
Preparation of this review was supported by the Ruth L. Kirschstein National Research Service Award AI007323 (G.N.P.) and fellowships from the Alfred P. Sloan Foundation and Packard Foundation (E.Y.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Ahn J, Lee J, and Kim S (2015). Interferon-gamma inhibits the neuronal differentiation of neural progenitor cells by inhibiting the expression of Neurogenin2 via the JAK/STAT1 pathway. Biochem Biophys Res Commun 466, 52–59. [DOI] [PubMed] [Google Scholar]
- Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, and Zychlinsky A (1999). Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285, 736–739. [DOI] [PubMed] [Google Scholar]
- Ardeshir A, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, Van Rompay KK, Lynch SV, and Hartigan-O’Connor DJ (2014). Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med 6, 252ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, and Rudensky AY (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astafurov K, Elhawy E, Ren L, Dong CQ, Igboin C, Hyman L, Griffen A, Mittag T, and Danias J (2014). Oral microbiome link to neurodegeneration in glaucoma. PLoS One 9, e104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak B, Feldman N, and Okun E (2014). Toll-like receptors as developmental tools that regulate neurogenesis during development: an update. Front Neurosci 8, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Harrison OJ (2017). Homeostatic Immunity and the Microbiota. Immunity 46, 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardino L, Agasse F, Silva B, Ferreira R, Grade S, and Malva JO (2008). Tumor necrosis factor-alpha modulates survival, proliferation, and neuronal differentiation in neonatal subventricular zone cell cultures. Stem Cells 26, 2361–2371. [DOI] [PubMed] [Google Scholar]
- Bialas AR, and Stevens B (2013). TGF-beta signaling regulates neuronal C1q expression and developmental synaptic refinement. Nature neuroscience 16, 1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, A DL, Wu F, Perez-Perez GI, Chen Y, et al. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8, 343ra382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein AR, Copenhaver CI, and Mortimer JA (2006). Early-life risk factors for Alzheimer disease. Alzheimer Dis Assoc Disord 20, 63–72. [DOI] [PubMed] [Google Scholar]
- Brown AS (2012). Epidemiologic studies of exposure to prenatal infection and risk of schizophrenia and autism. Dev Neurobiol 72, 1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson YT, Foster JA, Kuppusamy SP, Herkenham M, and Long EO (2005). Expression of a killer cell receptor-like gene in plastic regions of the central nervous system. J Neuroimmunol 161, 177–182. [DOI] [PubMed] [Google Scholar]
- Cameron JS, Alexopoulou L, Sloane JA, DiBernardo AB, Ma Y, Kosaras B, Flavell R, Strittmatter SM, Volpe J, Sidman R, and Vartanian T (2007). Toll-like receptor 3 is a potent negative regulator of axonal growth in mammals. J Neurosci 27, 13033–13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, Milani C, Ventura M, Bach JF, and Chatenoud L (2015). Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PloS one 10, e0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga M, Murai T, and Bauman MD (2017). Maternal Immune Activation and Autism Spectrum Disorder: From Rodents to Nonhuman and Human Primates. Biol Psychiatry 81, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ruiz A, Mosley M, George AJ, Mussaji LF, Fullerton EF, Ruszkowski EM, Jacobs AJ, Gewirtz AT, Chassaing B, and Forger NG (2018). The microbiota influences cell death and microglial colonization in the perinatal mouse brain. Brain Behav Immun 67, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, and Medzhitov R (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America 111, 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Xu D, Lan X, Jia B, Sun L, Zheng JC, and Peng H (2013). A novel role of the STAT3 pathway in brain inflammation-induced human neural progenitor cell differentiation. Curr Mol Med 13, 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, and Aagaard KM (2017). Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 23, 314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Meyer KM, Prince AL, and Aagaard KM (2016). Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function. Gut Microbes 7, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, and Weiser JN (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 16, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Isolauri E, Laitinen K, and Salminen S (2008). Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr 88, 894–899. [DOI] [PubMed] [Google Scholar]
- Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, Kolls JK, Weiser JN, and Worthen GS (2014). The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 20, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman BE, and Patterson PH (2009). Cytokines and CNS development. Neuron 64, 61–78. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, and Pettersson S (2011). Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Goltsman DS, Wong RJ, Shaw G, et al. (2015). Temporal and spatial variation of the human microbiota during pregnancy. Proceedings of the National Academy of Sciences of the United States of America 112, 11060–11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S, Hooiveld G, Tremaroli V, Backhed F, and Kleerebezem M (2013). The gut microbiota and mucosal homeostasis: colonized at birth or at adulthood, does it matter? Gut microbes 4, 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 18, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Olson CA, and Hsiao EY (2017). Interactions between the microbiota, immune and nervous systems in health and disease. Nature neuroscience 20, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. [DOI] [PubMed] [Google Scholar]
- Glynn MW, Elmer BM, Garay PA, Liu XB, Needleman LA, El-Sabeawy F, and McAllister AK (2011). MHCI negatively regulates synapse density during the establishment of cortical connections. Nat Neurosci 14, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, and Lamouse-Smith ES (2016). Maternal Antibiotic Treatment Impacts Development of the Neonatal Intestinal Microbiome and Antiviral Immunity. J Immunol 196, 3768–3779. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez G, and Lamouse-Smith ES (2017). Gastrointestinal Microbiome Dysbiosis in Infant Mice Alters Peripheral CD8(+) T Cell Receptor Signaling. Front Immunol 8, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. (2016). The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell 166, 1231–1246 e1213. [DOI] [PubMed] [Google Scholar]
- Gutierrez H, O’Keeffe GW, Gavalda N, Gallagher D, and Davies AM (2008). Nuclear factor kappa B signaling either stimulates or inhibits neurite growth depending on the phosphorylation status of p65/RelA. J Neurosci 28, 8246–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke ML, and Kielian T (2011). Toll-like receptors in health and disease in the brain: mechanisms and therapeutic potential. Clin Sci (Lond) 121, 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jin P, Peng J, Zhang X, Wong FS, and Wen L (2016). Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun 72, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong FS, and Wen L (2015). Maternal Antibiotic Treatment Protects Offspring from Diabetes Development in Nonobese Diabetic Mice by Generation of Tolerogenic APCs. J Immunol 195, 4176–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humann J, Mann B, Gao G, Moresco P, Ramahi J, Loh LN, Farr A, Hu Y, Durick-Eder K, Fillon SA, et al. (2016). Bacterial Peptidoglycan Traverses the Placenta to Induce Fetal Neuroproliferation and Aberrant Postnatal Behavior. Cell host & microbe 19, 901. [DOI] [PubMed] [Google Scholar]
- Islam O, Gong X, Rose-John S, and Heese K (2009). Interleukin-6 and neural stem cells: more than gliogenesis. Mol Biol Cell 20, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, and Andersson AF (2014). Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63, 559–566. [DOI] [PubMed] [Google Scholar]
- Jasarevic E, Howard CD, Morrison K, Misic A, Weinkopff T, Scott P, Hunter C, Beiting D, and Bale TL (2018). The maternal vaginal microbiome partially mediates the effects of prenatal stress on offspring gut and hypothalamus. Nat Neurosci. [DOI] [PubMed] [Google Scholar]
- Jensen ML, Thymann T, Cilieborg MS, Lykke M, Molbak L, Jensen BB, Schmidt M, Kelly D, Mulder I, Burrin DG, and Sangild PT (2014). Antibiotics modulate intestinal immunity and prevent necrotizing enterocolitis in preterm neonatal piglets. Am J Physiol Gastrointest Liver Physiol 306, G59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson S, Price J, and Modo M (2008). Effect of inflammatory cytokines on major histocompatibility complex expression and differentiation of human neural stem/progenitor cells. Stem Cells 26, 2444–2454. [DOI] [PubMed] [Google Scholar]
- Josefsdottir KS, Baldridge MT, Kadmon CS, and King KY (2017). Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 129, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T, Lacroix C, Braegger C, and Chassard C (2014). Stability of the maternal gut microbiota during late pregnancy and early lactation. Curr Microbiol 68, 419–427. [DOI] [PubMed] [Google Scholar]
- Kaul D, Habbel P, Derkow K, Kruger C, Franzoni E, Wulczyn FG, Bereswill S, Nitsch R, Schott E, Veh R, et al. (2012). Expression of Toll-like receptors in the developing brain. PLoS One 7, e37767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, and Akira S (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650. [DOI] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, and Mazmanian SK (2014). Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15, 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, and Huh JR (2017). Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura E, Ding Y, and Tohyama C (2016). AhR signaling activation disrupts migration and dendritic growth of olfactory interneurons in the developing mouse. Sci Rep 6, 26386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, and Flanagan KL (2016). Sex differences in immune responses. Nat Rev Immunol 16, 626–638. [DOI] [PubMed] [Google Scholar]
- Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, and Barton GM (2016). Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell 165, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, and Ley RE (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108 Suppl 1, 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, and Alvarez-Buylla A (2009). The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32, 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert CR, Frost EL, Bolte AC, Paysour MJ, Shaw ME, Bellinger CE, Weigel TK, Zunder ER, and Lukens JR (2018). Cutting Edge: Critical Roles for Microbiota-Mediated Regulation of the Immune System in a Prenatal Immune Activation Model of Autism. J Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchney SE, Hein AM, O’Banion MK, DiCicco-Bloom E, and Opanashuk LA (2013). Deletion or activation of the aryl hydrocarbon receptor alters adult hippocampal neurogenesis and contextual fear memory. J Neurochem 125, 430–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Okun E, Tang SC, Griffioen K, Cheng A, Mughal MR, Laryea G, Selvaraj PK, ffrench-Constant C, Magnus T, et al. (2008). Toll-like receptor 3 is a negative regulator of embryonic neural progenitor cell proliferation. J Neurosci 28, 13978–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Mian FM, Stanisz AM, Bindels LB, Cambier E, Ben-Amram H, Koren O, Forsythe P, and Bienenstock J (2017). Low-dose penicillin in early life induces long-term changes in murine gut microbiota, brain cytokines and behavior. Nature communications 8, 15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay KL, Walsh CA, Brennan L, and McAuliffe FM (2013). Probiotics in pregnancy and maternal outcomes: a systematic review. J Matern Fetal Neonatal Med 26, 772–778. [DOI] [PubMed] [Google Scholar]
- Ling ZD, Potter ED, Lipton JW, and Carvey PM (1998). Differentiation of mesencephalic progenitor cells into dopaminergic neurons by cytokines. Exp Neurol 149, 411–423. [DOI] [PubMed] [Google Scholar]
- Lu J, Lu L, Yu Y, Cluette-Brown J, Martin CR, and Claud EC (2018). Effects of Intestinal Microbiota on Brain Development in Humanized Gnotobiotic Mice. Scientific reports 8, 5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackos AR, Maltz R, and Bailey MT (2017). The role of the commensal microbiota in adaptive and maladaptive stressor-induced immunomodulation. Hormones and behavior 88, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, de Aguero MG, and Ganal-Vonarburg SC (2017). How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol 17, 508–517. [DOI] [PubMed] [Google Scholar]
- Marin IA, Goertz JE, Ren T, Rich SS, Onengut-Gumuscu S, Farber E, Wu M, Overall CC, Kipnis J, and Gaultier A (2017). Microbiota alteration is associated with the development of stress-induced despair behavior. Scientific reports 7, 43859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. [DOI] [PubMed] [Google Scholar]
- Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, Kim B, Brestoff JR, Tyldsley AS, Zheng Q, et al. (2018). Commensal microbiota modulate gene expression in the skin. Microbiome 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgrom J, Gemmill AW, Bilszta JL, Hayes B, Barnett B, Brooks J, Ericksen J, Ellwood D, and Buist A (2008). Antenatal risk factors for postnatal depression: a large prospective study. J Affect Disord 108, 147–157. [DOI] [PubMed] [Google Scholar]
- Miller JE, Wu C, Pedersen LH, de Klerk N, Olsen J, and Burgner DP (2018). Maternal antibiotic exposure during pregnancy and hospitalization with infection in offspring: a population-based cohort study. Int J Epidemiol. [DOI] [PubMed] [Google Scholar]
- Minter MR, Hinterleitner R, Meisel M, Zhang C, Leone V, Zhang X, Oyler-Castrillo P, Zhang X, Musch MW, Shen X, et al. (2017). Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1DeltaE9 murine model of Alzheimer’s disease. Scientific reports 7, 10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modgil S, Lahiri DK, Sharma VL, and Anand A (2014). Role of early life exposure and environment on neurodegeneration: implications on brain disorders. Transl Neurodegener 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohle L, Mattei D, Heimesaat MM, Bereswill S, Fischer A, Alutis M, French T, Hambardzumyan D, Matzinger P, Dunay IR, and Wolf SA (2016). Ly6C(hi) Monocytes Provide a Link between Antibiotic-Induced Changes in Gut Microbiota and Adult Hippocampal Neurogenesis. Cell reports 15, 1945–1956. [DOI] [PubMed] [Google Scholar]
- Moraga A, Pradillo JM, Cuartero MI, Hernandez-Jimenez M, Oses M, Moro MA, and Lizasoain I (2014). Toll-like receptor 4 modulates cell migration and cortical neurogenesis after focal cerebral ischemia. FASEB J 28, 4710–4718. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Kabakchiev B, Waldron L, Tyler AD, Tickle TL, Milgrom R, Stempak JM, Gevers D, Xavier RJ, Silverberg MS, and Huttenhower C (2015). Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol 16, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouihate A (2016). Prenatal Activation of Toll-Like Receptor-4 Dampens Adult Hippocampal Neurogenesis in An IL-6 Dependent Manner. Front Cell Neurosci 10, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Franchi L, Kambe N, Meng G, Strober W, and Nunez G (2012). Critical role for mast cells in interleukin-1beta-driven skin inflammation associated with an activating mutation in the nlrp3 protein. Immunity 37, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Gokhan S, Dai XM, Wei S, Enikolopov G, Lin H, Mehler MF, and Stanley ER (2012). The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev Biol 367, 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Schweigreiter R, Yamashita T, Rosenkranz K, Wekerle H, and Barde YA (2002). Tumor necrosis factor inhibits neurite outgrowth and branching of hippocampal neurons by a rho-dependent mechanism. J Neurosci 22, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan AM, Collins LM, Wyatt SL, Gutierrez H, and O’Keeffe GW (2014). The neurite growth inhibitory effects of soluble TNFalpha on developing sympathetic neurons are dependent on developmental age. Differentiation 88, 124–130. [DOI] [PubMed] [Google Scholar]
- O’Keeffe GW, Gutierrez H, Pandolfi PP, Riccardi C, and Davies AM (2008). NGF-promoted axon growth and target innervation requires GITRL-GITR signaling. Nat Neurosci 11, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Golenbock D, and Bowie AG (2013). The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol 13, 453–460. [DOI] [PubMed] [Google Scholar]
- Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, and O’Leary OF (2015). Adult Hippocampal Neurogenesis Is Regulated by the Microbiome. Biological psychiatry 78, e7–9. [DOI] [PubMed] [Google Scholar]
- Okun E, Griffioen KJ, Son TG, Lee JH, Roberts NJ, Mughal MR, Hutchison E, Cheng A, Arumugam TV, Lathia JD, et al. (2010). TLR2 activation inhibits embryonic neural progenitor cell proliferation. J Neurochem 114, 462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, and Blumberg RS (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- Prinz M, and Priller J (2014). Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci 15, 300–312. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Tetzlaff W, and Weiss S (1992). A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci 12, 4565–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, and Weiss S (1996). Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol 175, 1–13. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, and Lambris JD (2010). Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Reis ES, and Lambris JD (2016). Complement in disease: a defence system turning offensive. Nat Rev Nephrol 12, 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls A, Shechter R, London A, Ziv Y, Ronen A, Levy R, and Schwartz M (2007). Toll-like receptors modulate adult hippocampal neurogenesis. Nat Cell Biol 9, 1081–1088. [DOI] [PubMed] [Google Scholar]
- Rooks MG, and Garrett WS (2016). Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, and Finlay BB (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 13, 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma JV, and Ward PA (2011). The complement system. Cell Tissue Res 343, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann A, Nutten S, Donnicola D, Comelli EM, Mansourian R, Cherbut C, Corthesy-Theulaz I, and Garcia-Rodenas C (2005). Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics 23, 235–245. [DOI] [PubMed] [Google Scholar]
- Seguin JA, Brennan J, Mangano E, and Hayley S (2009). Proinflammatory cytokines differentially influence adult hippocampal cell proliferation depending upon the route and chronicity of administration. Neuropsychiatr Dis Treat 5, 5–14. [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. (2016). Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Yim Y, Park A, Berrios J, Lafourcade M, Pascual LM, Soares N, Yeon Kim J, Kim S, Kim H, Waisman A, et al. (2017). Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 549, 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, Offermanns S, and Ganapathy V (2010). Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem 285, 27601–27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren YM, Tomicic S, Lundberg A, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E, and Jenmalm MC (2009). Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy 39, 1842–1851. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, and Garrett WS (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Li J, Garbett K, Mirnics K, and Patterson PH (2007). Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci 27, 10695–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Gabriel K, Gabriel I, Wiczkowski A, Paul M, and Olejek A (2009). Prenatal priming of cord blood T lymphocytes by microbiota in the maternal vagina. Am J Reprod Immunol 61, 246–252. [DOI] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, and Stevens B (2012). The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci 35, 369–389. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Stiles J, and Jernigan TL (2010). The basics of brain development. Neuropsychol Rev 20, 327–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm J, Schjorring S, Eskildsen CE, Pedersen L, Bischoff AL, Folsgaard N, Carson CG, Chawes BL, Bonnelykke K, Molgaard A, et al. (2014). Antibiotic use during pregnancy alters the commensal vaginal microbiota. Clin Microbiol Infect 20, 629–635. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Zmora N, Levy M, and Elinav E (2016). The microbiome and innate immunity. Nature 535, 65–74. [DOI] [PubMed] [Google Scholar]
- Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G, et al. (2018). Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell 172, 500–516 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo-Badia N, Hakansson A, Vasudevan K, Molin G, Ahrne S, and Cilio CM (2014). Antibiotic treatment of pregnant non-obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand J Immunol 80, 250–260. [DOI] [PubMed] [Google Scholar]
- Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, and Waubant E (2017). The gut microbiome in human neurological disease: A review. Ann Neurol 81, 369–382. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, and Marsland BJ (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20, 159–166. [DOI] [PubMed] [Google Scholar]
- Vinolo MA, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, and Curi R (2011). Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem 22, 849–855. [DOI] [PubMed] [Google Scholar]
- Vitali B, Cruciani F, Baldassarre ME, Capursi T, Spisni E, Valerii MC, Candela M, Turroni S, and Brigidi P (2012). Dietary supplementation with probiotics during late pregnancy: outcome on vaginal microbiota and cytokine secretion. BMC microbiology 12, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong HE, and Hsiao EY (2017). Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol Psychiatry 81, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong HE, Yano JM, Fung TC, and Hsiao EY (2017). The Microbiome and Host Behavior. Annu Rev Neurosci 40, 21–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bankiewicz KS, Plunkett RJ, and Oldfield EH (1994). Intrastriatal implantation of interleukin-1. Reduction of parkinsonism in rats by enhancing neuronal sprouting from residual dopaminergic neurons in the ventral tegmental area of the midbrain. J Neurosurg 80, 484–490. [DOI] [PubMed] [Google Scholar]
- Wu WL, Hsiao EY, Yan Z, Mazmanian SK, and Patterson PH (2017). The placental interleukin-6 signaling controls fetal brain development and behavior. Brain, behavior, and immunity 62, 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Xu H, Zhang X, Li Z, Jia Y, He X, and Huang JH (2014). Association between toll-like receptor 4 expression and neural stem cell proliferation in the hippocampus following traumatic brain injury in mice. Int J Mol Sci 15, 12651–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, and Golenbock D (1999). Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol 163, 1–5. [PubMed] [Google Scholar]
- Yoshiya K, Lapchak PH, Thai TH, Kannan L, Rani P, Dalle Lucca JJ, and Tsokos GC (2011). Depletion of gut commensal bacteria attenuates intestinal ischemia/reperfusion injury. Am J Physiol Gastrointest Liver Physiol 301, G1020–1030. [DOI] [PubMed] [Google Scholar]
- Zhang K, Xu H, Cao L, Li K, and Huang Q (2013). Interleukin-1beta inhibits the differentiation of hippocampal neural precursor cells into serotonergic neurons. Brain Res 1490, 193–201. [DOI] [PubMed] [Google Scholar]
- Zohar O, Reiter Y, Bennink JR, Lev A, Cavallaro S, Paratore S, Pick CG, Brooker G, and Yewdell JW (2008). Cutting edge: MHC class I-Ly49 interaction regulates neuronal function. J Immunol 180, 6447–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunszain PA, Anacker C, Cattaneo A, Choudhury S, Musaelyan K, Myint AM, Thuret S, Price J, and Pariante CM (2012). Interleukin-1beta: a new regulator of the kynurenine pathway affecting human hippocampal neurogenesis. Neuropsychopharmacology 37, 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]