Remote-controlled manipulation of gene editing was achieved in vivo.
Abstract
As an RNA-guided nuclease, CRISPR-Cas9 offers facile and promising solutions to mediate genome modification with respect to versatility and high precision. However, spatiotemporal manipulation of CRISPR-Cas9 delivery remains a daunting challenge for robust effectuation of gene editing both in vitro and in vivo. Here, we designed a near-infrared (NIR) light–responsive nanocarrier of CRISPR-Cas9 for cancer therapeutics based on upconversion nanoparticles (UCNPs). The UCNPs served as “nanotransducers” that can convert NIR light (980 nm) into local ultraviolet light for the cleavage of photosensitive molecules, thereby resulting in on-demand release of CRISPR-Cas9. In addition, by preparing a single guide RNA targeting a tumor gene (polo-like kinase-1), our strategies have successfully inhibited the proliferation of tumor cell via NIR light–activated gene editing both in vitro and in vivo. Overall, this exogenously controlled method presents enormous potential for targeted gene editing in deep tissues and treatment of a myriad of diseases.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR)–associated protein 9 (Cas9) technology, which depends on an engineered single guide RNA (sgRNA) for site recognition to correct gene mutations via nonhomologous end joining or homology-directed repair, provides a flexible platform to revolutionize the treatment of diseases (1–3). Nevertheless, the CRISPR-Cas9 system suffers from a formidable problem owing to the simultaneous delivery of two macromolecules: Cas9 (ca. 160 kDa) and sgRNA (more than 100 base pairs) (4–6). Although it has been successfully used as a vehicle to deliver the CRISPR-Cas9 system, viral delivery, including adeno-associated virus, lentivirus, and adenovirus (7–10), still falters, since viral vectors may lead to carcinogenesis, insertional mutagenesis, and immunogenicity (11). To address this issue, researchers recently have shown tremendous enthusiasm for the march toward the exploration of nonviral vectors (4). To date, various vehicles, including gold nanoparticles (12), black phosphorus (13), metal-organic frameworks (14), graphene oxide (15), polymeric nanoparticles (16, 17), and other nanomaterials (18, 19), have been achieved availably to deliver CRISPR-Cas9 in vitro or in vivo. However, on-demand release of CRISPR-Cas9 for the precise remote temporal and spatial control over genome editing remains elusive.
Over decades, on-demand gene/protein/drug delivery based on stimuli-responsive nanomaterial design has been extensively exploited in nanomedicine (20, 21). Among the multifarious control strategies, photo regulation has proved to be an ideal noninvasive option by which release of bioactive molecules can be readily monitored with high spatiotemporal precision, without introducing chemical contaminants (22). Until now, several photoresponsive molecules have been developed, such as azobenzene derivatives (23), spiropyran derivatives (24), and a group of photosensitive molecules containing o-nitrobenzyl moieties (25). These molecules can readily undergo photoisomerization or the ester bond cleavage when exposed to ultraviolet (UV) light. Howbeit, UV light demonstrates limited penetration depth in vivo because of substantial absorption by skin and underlying soft tissues. Furthermore, it would damage tissues and may even lead to oncogenic scars under prolonged exposure (26). Research on optical window in biological applications shows that tissues seem to be most transparent to near-infrared (NIR) range (700 to 1000 nm), which can deeply penetrate the body (even up to 3.2 cm) (27–29). Thereby, the excavation of NIR light–triggered systems will be a potential and desirable strategy in biological applications.
As emerging nanotransducers, lanthanide-doped upconversion nanoparticles (UCNPs) have been developed for the conversion of low-energy NIR radiation (800 or 980 nm) to high-energy UV or visible light, which offers an appealing avenue to remotely manipulate biomolecules and control biological processes (30, 31). Notable applications include biosensor in cells or tissues (32, 33), dynamic cell regulation (34, 35), and flexible therapeutic platforms (36, 37). Inspired by these achievements, we designed a UCNP-based CRISPR-Cas9 delivery system for cancer therapy in this work. As illustrated in Fig. 1, CRISPR-Cas9 was covalently anchored on UCNPs by photocleavable 4-(hydroxymethyl)-3-nitrobenzoic acid (ONA) molecules (denoted as UCNPs-Cas9) and then coated with polyethylenimine (PEI) to assist endosomal escape (18) (denoted as UCNPs-Cas9@PEI). By preparing an sgRNA that targeted a cancer therapeutic gene, polo-like kinase 1 (PLK-1), we experimentally realized the inhibition of tumor progression both in vitro and in vivo through the NIR-controllable release of CRISPR-Cas9.
Fig. 1. Design of the UCNP-based CRISPR-Cas9 delivery system for NIR light–controlled gene editing.
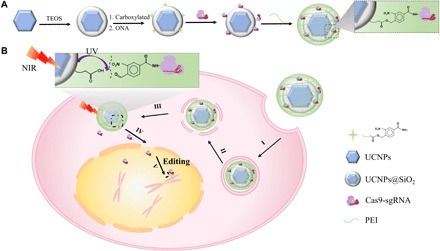
(A) Preparation of UCNPs-Cas9@PEI. (B) NIR-triggered delivery of Cas9-sgRNA to the nucleus of the cell for gene editing: (I) attachment to the cell membrane, (II) endocytosis, (III) endosome escape, (IV) release from particles and step into the nucleus, and (V) search for the target DNA locus and initiate the DNA double-strand break for genome editing.
RESULTS
Design of NIR-controlled release vehicle for delivery of Cas9-sgRNA
As shown in the transmission electron microscopy (TEM) image (Fig. 2A), the oleate-capped UCNPs, which were synthesized according to the previous protocols (32, 37), appeared considerably monodispersed with hexagon-shaped plates. The d-spacing value of the UCNPs coincided with a single hexagonal-phase crystal of 0.52 nm (fig. S2). To improve water solubility and biocompatibility, we coated 6-nm-thick silica shells on the surface of UCNPs, as shown in Fig. 2B (UCNPs@SiO2; fig. S3, A and B). Compared with that of UCNPs, the fluorescence intensity of UCNPs@SiO2 merely decreased (fig. S3C). Then, upon treatment with a carboxylation reagent, the carboxylic-functionalized nanoparticles (UCNPs@SiO2-COOH) were obtained. These surface modification processes can be confirmed by ζ potential measurements (Fig. 2E) and Fourier transform infrared (FTIR) spectrum (fig. S3D). When excited by a 980-nm laser beam, the UCNPs@SiO2-COOH colloidal solutions displayed purple-blue photoluminescence (fig. S3C, inset). The emission fluorescence peaks, which were assigned to the 1I6 → 3F4 and 1D2 → 3H6 transition from Tm3+, overlapped with the UV absorption band of the ONA linker (Fig. 2D), thus enabling Förster resonance energy transfer. A very recent report suggests that the blue light of UCNPs with a power density of ~0.063 mW/mm2 can be detected upon a 980-nm CW laser of 2.0 W, even located at ~4.2 mm below a mouse skull (37), which laid a solid foundation for the implementation of delivery in deep tissues.
Fig. 2. Characterization of the nonviral nanovehicles.
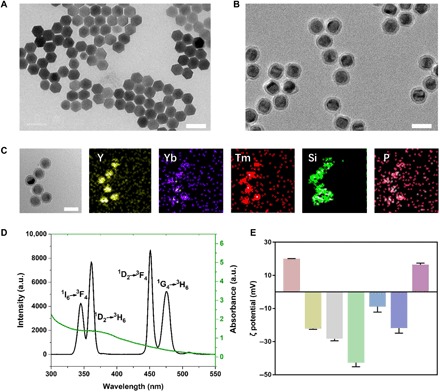
TEM images of (A) oleate-stable UCNPs, (B) UCNPs@SiO2, and (C) UCNPs-Cas9@PEI and its EDX elemental mapping. Scale bars, 50 nm. (D) Fluorescence spectrum of the UCNPs activated by a 980-nm laser (black line) and UV-vis absorption spectrum of 4-(hydroxymethyl)-3-nitrobenzoic acid (ONA) (green line). a.u., arbitrary units. (E) The ζ potentials of Cas9 protein, Cas9/sgRNA, UCNPs@SiO2, UCNPs@SiO2-COOH, UCNPs@SiO2-ONA, UCNPs-Cas9, and UCNPs-Cas9@PEI (from left to right).
To achieve remote NIR-controlled gene editing, we used UV-photocleavable ONA molecules to assemble the UCNP–CRISPR-Cas9 complexes. The ONA molecules were synthesized as reported previously (38) and modified on UCNPs@SiO2-COOH via ester bond (UCNPs@SiO2-ONA). The 1H NMR (nuclear magnetic resonance) (400 MHz) spectra showed that ONA was successfully obtained (fig. S3E), and the absorbance in the UV region after reaction with ONA (fig. S3F) indicated the successful attachment of ONA on the surface of UCNPs. Afterward, we covalently conjugated Cas9 proteins on UCNPs@SiO2-ONA via carbodiimide cross-linker chemistry and incubated them with sgRNA to form the complexes (UCNPs-Cas9). To enhance endosomal escape, we applied PEI to coat the UCNPs-Cas9. To confirm the fabrication of UCNPs-Cas9@PEI, we adopted TEM mapping, and the results showed that UCNPs-Cas9@PEI contains lanthanide ion dopants (Tm, Y, and Yb), Si, and P (Fig. 2C), which originated in UCNPs, SiO2, and the phosphate backbone in the CRISPR-Cas9 system, respectively. In addition, the ζ potentials at each step have also been measured for further determination, which revealed that positively charged Cas9 (+20.02 ± 0.18 mV) would become negatively charged after the addition of sgRNA (−22.22 ± 0.34 mV) and that the ζ potential from a negative value (UCNPs-Cas9, −21.80 ± 3.08 mV) to a positive value upon the addition of PEI (UCNPs-Cas9@PEI, +14.96 ± 1.35 mV).
NIR light–controlled release of CRISPR-Cas9
To examine the NIR response of our system, we exposed two groups of UCNPs@SiO2-ONA solution to NIR and UV light, and then the absorption spectra of the supernatant after centrifugation were distinctly measured. As shown in fig. S4B, the UV absorption bands in 300 to 450 nm increased along with the UV irradiation time, attributed to the amount of ONA released from the nanoparticles (34). As expected, the spectral changes after NIR irradiation revealed a similar trend, indicating that the UCNPs have availably generated photons in the UV regions upon irradiation with NIR laser and, therefore, triggered the photocleavage reaction (fig. S4C). While the similarly sized SiO2-ONA was exposed to NIR laser, no observable absorption change in 300 to 450 nm was detected (fig. S4, D and E), thereby confirming the importance of UCNPs for the cleavage. To demonstrate that CRISPR-Cas9 could be efficiently released from UCNPs irradiated by NIR light, we also treated UCNPs-Cas9 with UV and NIR, as did previous experiments. Similarly, we acquired the data closely paralleling previous results (Fig. 3A and fig. S4F). To further confirm that stimulating UCNPs could induce the release of cas9/sgRNA from the surface, we also labeled Cas9 in UCNPs-Cas9 with Cy3 for light-controlled releasing test (fig. S4, G and H). Red fluorescence was obtained after irradiation of NIR, which was consistent with our anticipation as well.
Fig. 3. NIR light–controlled release of Cas9 for gene editing.
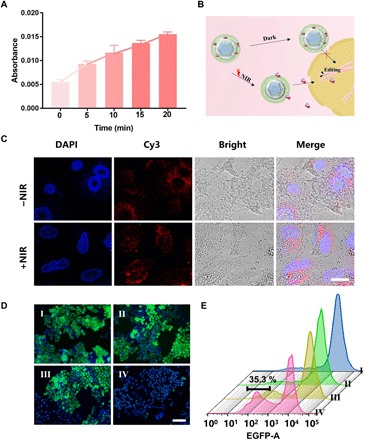
(A) UV-vis absorption (280 nm) of the supernatant of UCNPs-Cas9 with different irradiation times by a 980-nm laser. (B) Schematic illustration of internalization of Cas9 that enters the nuclei and triggered by NIR light for gene editing. (C) CLSM images of cells incubated with UCNPs-Cas9@PEI for 6 hours with or without 980-nm irradiation (red, Cas9 labeled with Cy3; blue, nuclei stained with DAPI). Scale bar, 10 μm. (D) Fluorescence microscopy images of KB cells treated with different formulations (I, NIR only; II, UCNPs@PEI + NIR; III, UCNPs-Cas9@PEI without NIR; IV, UCNPs-Cas9@PEI + NIR). Green, EGFP; blue, nuclei stained with DAPI. Scale bar, 100 μm. (E) Flow cytometry (FCM) analysis of EGFP disruption in cells treated with (I) NIR only, (II) UCNPs@PEI + NIR, (III) UCNPs-Cas9@PEI without NIR, and (IV) UCNPs-Cas9@PEI + NIR. Bars represent mean ± SD (n = 5). The EGFP silencing efficiencies of UCNPs-Cas9@PEI and UCNPs-Cas9@PEI + NIR were 35.3 and 6.9%, respectively.
We further investigated the ability of photoresponse for controlling the release of target molecules in A549 cells (a human lung adenocarcinoma cell line). Cas9 protein was also labeled with Cy3 to facilitate the observation (Fig. 3B). UCNPs-Cas9@PEI bound to the cell surface only within 2 hours of incubation, and then particles entered the cytosol and motivated the red fluorescence signal from Cas9 protein in the cell while the incubation time increased to 4 hours (fig. S5). Upon NIR irradiation, the colocalization of red fluorescence with 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclei manifested that the particles were eventually localized to the nuclei at 6 hours (Fig. 3C and fig. S5). In contrast, without the irradiation of NIR light, the red fluorescence signal can only be observed in the cytoplasm owing to the endocytic pathways of UCNPs-Cas9@PEI and scarcely appeared in nuclei even after 12 hours (Fig. 3C and fig. S5). Thereby, our nanocarriers have potential for NIR light–activated genome editing both in cells and in deep tissues.
NIR light–activated genome editing in vitro
Before investigating its capability for genome editing, the biocompatibility of UCNPs-Cas9@PEI as well as the biosafety of NIR laser (980 nm), especially the upconverted UV light emitted from UCNPs, should be carefully evaluated. A549 cells were cultured with different doses of UCNPs-Cas9@PEI, and their viability was assessed with the Cell Counting Kit-8 (CCK-8) assay after 48 hours. As shown in fig. S6, cell viability was not susceptible to NIR, and scarce reduction in cell viability was observed when the cells were treated with UCNPs-Cas9@PEI (<50 μg/ml). Subsequently, we exposed cells incubated with UCNPs-Cas9@PEI (50 μg/ml) at different NIR (2.0 W/cm2) exposure times to detect the toxicity of the UV light inspired from UCNPs. The results suggested no substantial attenuation in the cell viability of the treated A549 cells under the said conditions (fig. S6), which might be attributed to the converted UV light being partially absorbed by o-nitrobenzyl groups and the fact that the UV light produced by the UCNPs was localized radiation, which appeared weaker than the conventional whole-cell irradiation by direct UV light (34, 39). In addition, the absorption of the 980-nm laser by water would increase the temperature; hence, the short interval irradiation (30-s break after 30-s irradiation) was used in our experiment to solve the problem. To obtain a safe concentration of UCNPs for more cell lines, we determined the viability of KB (human oral epidermoid carcinoma) cells under the same conditions. Compared with A549 cells, few differences were observed in KB cells.
We then examined the ability of UCNPs-Cas9@PEI for remote NIR-controlled genome editing. In this work, the sgRNA that targeted the coding region of the enhanced green fluorescent protein (EGFP) was chosen because the mutation frequency can match the EGFP expression in cells and shift to a readable green fluorescence signal. First, EGFP-transfected KB cells were cultured in pure medium (with NIR light irradiation), medium containing UCNPs-Cas9@PEI (with or without NIR light irradiation), and free UCNPs (with NIR light irradiation), respectively. On day 3, the fluorescence intensities of the cell samples were determined by confocal laser scanning microscopy (CLSM). As preconceived, the CRISPR-Cas9 system would be effectively delivered into cultured cells, bypassed karyotheca, and accessed the cell nucleus for activation of genome editing if NIR light was applied, which was confirmed by the remarkable fluorescent quenching in cells treated with UCNPs-Cas9@PEI upon NIR radiation (Fig. 3D and fig. S7). For benchmarking, a slight decrease (or no decrease) in green fluorescence signal was detected in cells treated with other groups (Fig. 3D and fig. S7). Flow cytometry (FCM) analysis of KB cells treated with different formulations (as previously carried out) brought about similar experimental results (Fig. 3E and fig. S7), which further identified the NIR-controlled genome editing in vitro. In addition, the control group (CRISPR-Cas9@PEI) was set here to figure out that it was the combination of UCNPs and PEI that led to the effective delivery.
In vitro study of NIR light–activated gene therapy
On the basis of the evidence of successful knock-outs of the EGFP gene, we next checked the inhibition of tumor proliferation through NIR-responsive release of CRISPR-Cas9 in cells (Fig. 4A). As previously reported (12, 40), the PLK-1 gene has been proven to be relevant to many different types of cancers, and depletion of the protein encoded by this gene can markedly inhibit cell proliferation and induce apoptosis. Hence, the PLK-1 gene in A549 cells was chosen as the targeted gene in this experiment. A549 cells were treated with UCNPs-Cas9@PEI and irradiated by 980-nm NIR light for 20 min (30-s break after 30-s irradiation) before further culturing. On day 3, the viability of cells was evaluated by the CCK-8 assay. As depicted in Fig. 4D, UCNPs-Cas9@PEI with NIR irradiation was much associated with cytotoxicity to A549 cells compared with other groups. CLSM images of the cells also revealed that cells after treatment with UCNPs-Cas9@PEI upon NIR radiation (2.0 W/cm2) for 20 min showed the weakest green intensity of calcein-AM (stained live cells) and the highest red intensity of propidium iodide (PI)–stained cells (stained dead cells) among all experimental groups (Fig. 4, E to I, and fig. S8), coinciding with the results of the CCK-8 assay. In addition, FCM also revealed similar results. To determine whether the apoptosis largely resulted from the suppressed expression of PLK-1 protein, we lysed cells to extract proteins for Western blot. As shown in Fig. 4B, PLK-1 protein of the cells (UCNPs-Cas9 + NIR) was lower than that of the control group. We then quantified the frequency of mutations using the SURVEYOR assay. The assay revealed that the target gene in the cells (UCNPs-Cas9 + NIR) was successfully knocked out with a high mutation frequency (Fig. 4C). By contrast, A549 cells incubated with UCNPs exposed to NIR light and A549 cells incubated with UCNPs-Cas9@PEI without NIR light irradiation displayed mere mutation frequencies (Fig. 4C), further suggesting the importance of NIR as a switch for this controllable gene therapy.
Fig. 4. NIR-mediated gene editing induced apoptosis of A549 cells.
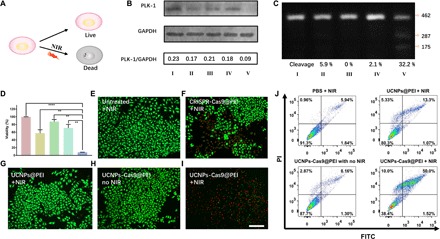
(A) Schematic illustration of the cancer cell killing triggered by NIR. (B) Western blot assay. (C) T7EI assay. (I) Pure media + NIR, (II) CRISPR-Cas9@PEI + NIR, (III) UCNPs@PEI + NIR, (IV) UCNPs-Cas9@PEI without NIR, and (V) UCNPs-Cas9@PEI + NIR. (D) CCK-8 analysis of A549 cells treated with pure media only, CRISPR-Cas9@PEI + NIR, UCNPs@PEI + NIR, UCNPs-Cas9@PEI without NIR, and UCNPs-Cas9@PEI + NIR (from left to right). CLMS images of cells treated by (E) pure media + NIR, (F) CRISPR-Cas9@PEI + NIR, (G) UCNPs@PEI + NIR, (H) UCNPs-Cas9@PEI without NIR, and (I) UCNPs-Cas9@PEI + NIR. Green, calcein-AM; red, PI. Scale bar, 200 μm. (J) Flow cytometry assay of cell apoptosis. The mean value was analyzed using t test (n = 5). **P < 0.01 represents significant difference and ****P < 0.0001 represents highly significant difference.
Therapeutic effect of NIR light-triggered Cas9 release in vivo
To test the efficacy of UCNPs-Cas9@PEI in vivo, we first experimentally studied its biodistribution in xenograft nude mice model bearing A549 cells. As shown in fig. S9A, a high level of Y3+ was detected in the tumor tissues but a low level was found in other tissues after in situ injection. The biodistribution of UCNPs-Cas9@PEI by intravenous injection was further evaluated, and the massive accumulation in liver and spleen indicated that UCNPs-Cas9@PEI presumably triggered the clearance of the reticuloendothelial system (41, 42). We also studied the pharmacokinetics of our UCNPs via intravenous administration. The elimination half-life (t½ = 1.0177 hours) was similar to that in the previous report (t½ = 0.9489 hours) (42).
Next, the tumors in mice were treated with different formulations via intratumoral injection after growing to ca. 80 mm3 (Fig. 5A). Derived from a previous in vitro study, mice were irradiated by NIR light (2.0 W/cm2) for 20 min every other day. After 20 days, the tumor sizes in both the control group [phosphate-buffered saline (PBS), with NIR light irradiation] and the UCNPs-Cas9@PEI group without NIR light irradiation had sharply grown, indicating that these formulations held limited inhibition efficacy, whereas in the UCNPs-Cas9@PEI group with NIR irradiation, the progression of tumors was gradually delayed over this period, with 74% of the tumor volume of the PBS control group on day 20 (Fig. 5B). We also collected and weighed the tumors after euthanasia of mice (Fig. 5, C and E), which further confirmed their controlled therapeutic effect in this work. Subsequently, histologic analysis of the tumor sections was performed. After staining with hematoxylin and eosin (H&E), the tumor tissues demonstrated a decreased cell density after administration of UCNPs-Cas9@PEI + NIR (Fig. 5F). The in situ TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) assays have indicated an apparent enhancement of massive cell apoptosis after treatment with UCNPs-Cas9@PEI upon NIR radiation (Fig. 5F). Conversely, a negligible tumor growth–suppressing effect in other groups was obtained. Moreover, caspase-3 labeling for apoptosis and KI67 antigen labeling for cell proliferation displayed significant differences after NIR radiation as well (Fig. 5F), which verified the NIR light–controlled therapeutic effect.
Fig. 5. The drug administration in a xenograft model of A549 cells.
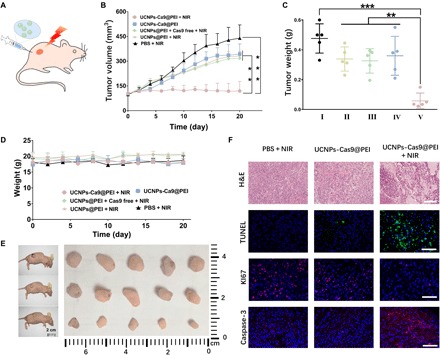
(A) Schematic diagram of the working mechanism of remote-controlled oncotherapy in vivo. (B) Tumor sizes after different treatments, as indicated. (C) Weights of tumors treated by (I) PBS + NIR, (II) UCNPs@PEI + NIR, (III) UCNPs@PEI + Cas9 free + NIR, (IV) UCNPs-Cas9@PEI, and (V) UCNPs-Cas9@PEI + NIR (from left to right). (D) Changes in mouse body weight after different treatments, as indicated. (E) Images of tumor with PBS + NIR (top row), UCNPs-Cas9@PEI without light (middle row), and UCNPs-Cas9@PEI + NIR (bottom row). (F) H&E staining (scale bar, 200 μm), TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) staining (scale bar, 100 μm), KI67 antigen staining (scale bar, 100 μm), and caspase-3 staining (scale bar, 100 μm) of tumor tissues. The mean value was analyzed using t test (n = 5). **P < 0.01 represents significant difference and ***P < 0.001 represents highly significant difference.
To detect the potential toxic side effects of UCNPs-Cas9@PEI–based NIR light–mediated gene treatment, we performed histology analysis of major organs from different groups of mice. Moreover, the results indicated no noticeable abnormality or appreciable organ damage (fig. S9). In addition, no lethality or significant drop in body weight for UCNPs-Cas9@PEI was observed (Fig. 5C). By analyzing urinary and intestinal excretion, we found that the excreted concentrations of lanthanide ion (Y3+) steadily decreased over time via both intravenous and intratumoral administration, which suggested that the UCNPs would be cleared possibly through both fecal and renal excretions (fig. S9). Overall, our approach can apparently control CRISPR-Cas9 release from the UCNPs-Cas9@PEI by NIR light for gene therapy and also show safety as well as high potency.
DISCUSSION
Despite immense efforts exerted on the CRISPR-Cas9, controllable delivery remains a challenge for effective achievement of gene editing both in vitro and in vivo. Our UCNPs-Cas9@PEI platform can be effectively internalized by cells via endocytosis pathways, which is followed by endosomal escape and cytosolic releases of the loaded UCNPs-Cas9 in the cytoplasm. When exposed to NIR light, UCNPs can emit local UV light and trigger the cleavage of the photocaged linkers. Hence, Cas9 could be released from the surface of UCNPs and thereby proceed to the nucleus for gene editing. Besides, by targeting a marker of cancer gene (PLK-1 gene), our platform has successfully inhibited cancer cell proliferation and tumor growth. Our strategies underline the remote-operated nature of the noninvasive NIR light governing CRISPR-Cas9 release, as well as the growing demand to develop more sophisticated nonviral delivery for gene editing. Collectively, anchored Cas9 protein via photo-dependent small molecules on UCNPs endows itself with targeted editing, which would intensively enhance the appeal of CRISPR-Cas9 technology and encourage the developed smart delivery in nanomedicine.
Nevertheless, owing to the lack of property of targeting tumors, a relatively small number of the nanovehicles were observed in the tumor tissues via intravenous administration (fig. S9A); thus, not enough Cas9 accumulated in tumor to trigger gene editing by NIR. Hence, the proof of concept is done in such a way that UCNPS-cas9@PEI allows gene editing, but only after local administration to accessible tumors for which surgery may, however, be preferred. In addition, further research is needed to develop similar UCNPs that can target tumoral or metastatic nodules after systemic administration.
MATERIALS AND METHODS
Materials
LnCl3⋅6H2O (Ln = Tm, Yb, and Y) was purchased from Energy Chemical Reagent Co. Ltd. (Shanghai, China). N-(trimethoxysilylpropyl)ethylenediaminetriacetate trisodium salt (TEDATS) was acquired from Gelest Inc. (America). Oletic acid, tetraethyl orthosilicate (TEOS), and (3-aminopropyl)trimethoxysilane were obtained from Sigma-Aldrich. PEI, dicyclohexylcarbodiimide (DCC), and 4-bromomethyl-3-nitrobenzoic acid (BNA) were purchased from Aladdin (Shanghai, China). PI and calcein-AM dye were acquired from Yeasen Biology (Shanghai, China). DAPI was purchased from Sangon Biotech (Shanghai) Co. Ltd. All aqueous solutions were prepared using ultrapure water (18.2 MU, Milli-Q; Millipore). The commercial antibodies used in this study at the indicated concentrations were purchased from Abcam. Cas9 proteins and the sgRNA transcription kit were obtained from Eastinno Biotechnology Co. Ltd. (Ningbo, China) and Inovogen Tech. Co. Ltd. (Beijing, China), respectively.
Measurements and characterizations
The UV-visible spectra were acquired by a microplate photometer (Multiskan FC; Thermo Fisher Scientific Inc.). The morphology of nanoparticles was detected by TEM (JEM-200CX; JEOL Ltd.). ζ potentials of all samples were recorded by electrophoretic light scattering (Brookhaven Instruments Corporation, Holtsville, NY, USA). FTIR was performed using an FTIR spectrometer (NEXUS870; Nicolet). 1H NMR (400 MHz) spectra were carried out on a spectrometer (DRX500; Bruker) using dimethyl sulfoxide as the solvent. Upconversion fluorescence emission spectra were acquired on a fluorescence spectrometer (F-4600; Hitachi) equipped with an external adjustable 980-nm NIR laser (optical parametric oscillator, Continuum Surelite, USA) as the excitation source.
Synthesis of photosensitive molecule (ONA)
The ONA was synthesized according to a previous report (34). Briefly, 1.01 g (3.88 mmol) of BNA and 2.68 g (13.6 mmol) of Na2CO3 were dissolved in a mixture of H2O and acetone (1:1, 30 ml). After 5 hours of reflux, acetone was evaporated and diethyl ether was applied to extract the resulting produce twice. Afterward, the pH value of the obtained solution should be regulated below 1 by the addition of concentrated HCl for yellow precipitate, which then redissolved in ethyl acetate (20 ml). The aqueous phases were separated from the organic one and then extracted with ethyl acetate (2 × 20 ml). Subsequently, the organic phase was combined and washed with H2O (1 × 20 ml). Last, MgSO4 was used to remove water, and the resulting solution was evaporated to dryness under reduced pressure.
Preparation of UCNPs@SiO2
UCNPs were prepared as reported previously with little modification (32, 37). Typically, 2 mmol of LnCl3 (Ln = Y, Yb, and Tm with a molar ratio of 78:20:2) was added to a 250-ml three-necked flask containing 15 ml of oleic acid and 35 ml of ODE (1-octadecene). After heating to 150°C for 45 min, the solution appeared light yellow transparent. The solution was slowly cooled down to 50°C while stirring; afterward, 25 ml of methanol containing NH4F (8 mmol) and NaOH (5 mmol) was pipetted into the solution. To remove waste gas, the mixture was heated to 100°C for 30 min. Then, the reaction temperature was increased to 290°C under N2 atmosphere. After 1.5 hours, the heating mantle was removed, and the reaction mixture started to cool down while stirring. When it reached room temperature, the mixture was washed by pouring 20 ml of ethanol solution three times (6000 rpm for 5 min) and collected in 20 ml of cyclohexane. Ethanol (4 ml) was added into 4 ml of the as-synthesized UCNPs; then, the solution was centrifuged at 14,000 rpm for 5 min. After this, the nanoparticles were redispersed into a mixture of 4 ml of HCl aqueous solution (2 M) and 4 ml of ethanol by sonication. To remove excess acid, we washed the acid-treated UCNPs three times with ethanol. The ligand-free UCNPs were thereafter dispersed into 4 ml of deionized water for further use as follows: To coat silica, we added 4 ml of the ligand-free UCNPs slowly into 18 ml of polyvinylpyrrolidone aqueous solution (50 mg/ml). Then, the solution was sonicated for 20 min and further stirred for 1 hour. After this, the solution was mixed with 80 ml of ethanol, followed by 20 min of ultrasonic treatment and 2 hours of stirring. Before additional 20 min of sonication, 3.2 ml of ammonium was added into the solution to adjust pH. Subsequently, 80 μl of TEOS was added to initiate the growth of silica on the surface of UCNPs. The reaction solution was stirred continuously for 12 hours at room temperature.
Design of UCNPs-Cas9@PEI
The obtained UCNPs@SiO2 (100 mg) was treated with 20 μl of TEDATS for 4.5 hours to acquire the carboxylic group first (UCNPs@SiO2-COOH). Then, the nanoparticles were transferred into 10 ml of dry tetrahydrofuran, including 100 μg of ONA for esterification catalyzed by DCC and 4-dimethylaminopyridine. After 12 hours, the nanoparticles were washed three times with tetrahydrofuran and then resolved in tetrahydrofuran with 200 μg of DCC to further incubate for activation of the carboxyl group. After 8 hours, the particles were collected and transferred into PBS. At the same time, Cas9 protein and sgRNA (1:2) were mixed in PBS for 5 min to form the CRISPR-Cas9 system. Afterward, the CRISPR-Cas9 system was mixed with the particles overnight at 4°C. After centrifugation, PEI was coated onto the UCNPs-Cas9 at a PEI:sgRNA weight ratio of 5:1 and needed further equilibration at room temperature for 5 min. The nanovehicles were further diluted to the concentration of Cas9 (0.36 μg/ml) (before centrifugation) in the in vitro experiment. To visualize the internalization process of UCNPs-Cas9@PEI by CLSM (LSM 710, Zeiss), the Cas9 was conjugated with Cy3-NHS.
Construction of sgRNA
The sgRNA used in this study was prepared with the help of an sgRNA in vitro transcription kit purchased from Inovogen Tech. Co. Ltd. The necessary positive primers containing targeted genes were obtained from Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China) and are as follows: sgRNA (EGFP), 5′-TAATACGACTCACTATAGGGGGGCGAGGAGCTGTTCACCGGTTTC AGAGCTATGCTGGA-3′; sgRNA (PLK-1), 5′-TAATACGACTCACTATAGGGTACCTACGGCAAATTGTGCTGTTTCA GAGCTATGCTGGA-3′. The underlined parts represent targeted genes.
Development of the nanovehicle for NIR-responsive CRISPR-Cas9 delivery
ONA was covalently attached to UCNPs@SiO2-COOH via esterification to obtain UCNPs@SiO2-ONA. After treatment with DCC, UCNPs@SiO2-ONA and the CRISPR-Cas9 system were bound together. To assist endosomal escape, PEI coated the above nanoparticles via electrostatic absorption (see the Supplementary Materials for details).
The CRISPR-Cas9 release stimulated by NIR light
The NIR-controlled release of the obtained UCNPs@SiO2-ONA and UCNPs-Cas9 was investigated upon radiation by the 980-nm NIR laser (Changchun Laser Optoelectronics Tech Co. Ltd). The UV absorption spectra of those solutions were recorded by a microplate photometer (Multiskan FC; Thermo Fisher Scientific Inc.). For control, UV radiation was manipulated to study the controlled release of molecules.
Cell culture
Human oral epidermoid carcinoma KB cells and the human lung cancer cell line A549 were obtained from the Institute of Chinese Academy of Medical Sciences (Shanghai, China). They were all cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin in an incubator where the temperature was set at 37°C with a 5% CO2 humidified atmosphere.
Evaluation of cell viability
Here, cell viability was quantitatively evaluated by (i) CCK-8, (ii) LIVE/DEAD Viability/Cytotoxicity Kit, and (iii) YF 488-annexin V and PI Apoptosis Kit (U.S. Everbright Inc., Suzhou, China). Briefly, cells were planted in a 96-well plate (5 × 103 per well) and then cultured at 37°C and 5% CO2 in an incubator. After 24 hours, different treatments (including the incubation of UCNPs, UCNPs-Cas9@PEI at different concentrations and laser irradiation times, Cas9@PEI, etc.) were conducted and further cultured for an additional 72 hours. Then the cell viability can be obtained by three methods as follows: (i) CCK-8 solution (10 μl/100 μl medium) was added to each well. After incubation for another 1.5 hours, OD450 value (absorbance at 450 nm) with background subtraction at 650 nm was detected by a microplate photometer (Multiskan FC; Thermo Fisher Scientific Inc.). Cell viability (%) was obtained by calculating the ratio of OD450–650 of the cells under targeted conditions to those of untreated control cells. (ii) LIVE/DEAD Viability/Cytotoxicity Kit was used as a protocol, followed by analysis with a confocal laser microscope (CLSM 710; Carl Zeiss, Germany). (iii) Cells were planted in a 24-well plate treated as before. The YF 488–annexin V and PI Apoptosis Kit was applied as a protocol. The results were obtained by a flow cytometer (BD FACSVerse; BD Biosciences).
Imaging of cellular uptake of UCNPs-Cas9@PEI
A549 cells were planted into 96-well plates (5 × 103 cells per well) in DMEM (containing 10% FBS and 1% penicillin-streptomycin). After 24 hours, medium was replaced with that including target particles, in which the protein concentration was fixed at 0.5 μg per well. Nuclei were stained with 1.0 mg/l of DAPI. In addition, all the CRISPR-Cas9 proteins applied in this experiment were labeled with Cy3. After 4 hours of incubation, a 980-nm pulsed laser with a power density of 2.0 W/cm2 was triggered for 20 min (30-s break after 30-s irradiation). The process of cellular uptake at different times was observed with a confocal laser microscope (CLSM 710; Carl Zeiss, Germany).
EGFP gene disruption assay
The EGFP gene disruption by virtue of NIR-released Cas9 was assayed using FCM. The KB cells with the EGFP gene integrated into the genome were implanted into six-well plates (1 × 105 cells per well) and cultured for another 24 hours before they were used. After incubation with different formulations for 6 hours, fresh media were adopted to replace the medium before and the cells were continued to culture for 72 hours. The cells were digested, collected by incubation, and immediately detected with FCM (LSR Fortessa; BD Biosciences).
Western blot analysis
After 3 days of treatment under different conditions (pure media and media including UCNP@PEI + NIR, CRISPR-Cas9@PEI + NIR, UCNPs-Cas9@PEI, and UCNPs-Cas9@PEI + NIR), cells in six-well plates (5 × 105 cells per well, before treatment) were washed with PBS twice and lysed on ice for 15 min with gentle stirring. After centrifugation at 12,000 rpm, a BCA protein assay kit (Sangon Biotech Co. Ltd., Shanghai, China) was applied to measure the total protein concentration of the supernatant cell lysates. Then, equal amounts of total protein (50 μg) were analyzed on 8% SDS–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. The samples were blocked with the QuickBlock Blocking Buffer (Beyotime, China) for 1 hour at room temperature. After that, the membranes were incubated with 1000× dilutions of anti–glyceraldehyde-3-phosphate dehydrogenase and anti–PLK-1 rabbit polyclonal antibody (Sangon Biotech Co. Ltd., Shanghai, China), respectively, overnight, and immersed with the conjugated secondary antibody (Sangon Biotech Co. Ltd., Shanghai, China) at room temperature for 1 hour. Last, the membranes were washed five times in Tris-Buffered Saline with 0.02% Tween 20 buffer (5 min each) and targeted proteins were visualized by W-TMB (Sangon Biotech Co. Ltd., Shanghai, China).
SURVEYOR assay to detect genomic modifications
After being treated with different formulations, the genomic DNA of A549 cells was harvested on day 3 using Dzup Genomic DNA Isolation Reagent (Sangon Biotech Co. Ltd., Shanghai, China) according to the manufacturer’s instructions. The sgRNA-targeted genomic locus was amplified with the High Fidelity PCR Master Mix (Sangon Biotech Co. Ltd., Shanghai, China) using the following primers: GGTGCTGCGAATGGTTGTGG and CAGCCTCCTCCAAATTCCAGC. To reduce nonspecific amplifications, the touchdown polymerase chain reaction (PCR) program [(92°C for 15 s and 74°C for 60 s) for 5 cycles, (95°C for 15 s and 72°C for 60 s) for 5 cycles, (95°C for 15 s and 70°C for 60 s) for 5 cycles, (95°C for 15 s and 68°C for 60 s) for 25 cycles, and 68°C for 5 min] was used. After purifying by gel extraction, indel formation efficiencies were detected according to the T7 Endonuclease I Kit (Viewsolid Biotech, Beijing, China). The digested DNA was analyzed using 2% agarose gel electrophoresis. Indel formation efficiencies were calculated by Image J.
Tumor xenografts
Animal procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee of the Nanjing Drum Tower Hospital, the Affiliated Hospital of Nanjing University Medical School. Cultured cells were digested by incubation with trypsin-EDTA solution (Sangon Biotech Co. Ltd., Shanghai, China) and then harvested by centrifugation (1000 rpm for 5 min). After being resuspended in sterile PBS, cells (108 cells per mouse) were subcutaneously implanted into about 18 g of female Balb/c nude mice for further use.
Detection of tumor suppression in vivo
Tumor-bearing mice (tumor size of ca. 80 mm3) were randomly divided into five groups of which each was treated with PBS + NIR, UCNPs-Cas9@PEI, UCNPs-Cas9@PEI + NIR, UCNPs-Cas9@PEI (without NIR), and UCNPs-Cas9@PEI + NIR (100 μl, 3.5 mg/ml, n = 5). The formulations above were adopted through intratumor injection every 3 days. The 980-nm pulsed laser (power density, 2.0 W/cm2) was applied as a vertical beam into the tumor sites aligned underneath for 25 min every time (30-s break after 30-s irradiation). The administration was monitored for up to 20 days, and during that time, the body weights and tumor volumes were recorded for further analysis, and then animals were euthanized on day 20. The tumor volumes were calculated as follows: volume = (length × width2)/2.
H&E and TUNEL staining
For histology, major organs and tumors that stem from mice after different treatments were harvested. After fixing in 10% paraformaldehyde, the collected tumors and organs were embedded in paraffin, sectioned into about 4 μm, and lastly stained with H&E. Moreover, TUNEL Apoptosis Assay Kit (Beyotime Biotechnology) was applied to determine tumor cell apoptosis on tumor slices. The histology was performed at the Nanjing Drum Tower Hospital. The samples were observed using the Invitrogen EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific).
Statistical analysis
Data were presented as means ± SD. For the representativeness of the qualitative images shown, at least two experiments were carried out in duplicate. For differences between two groups, **P < 0.01 was considered significant while ***P < 0.001 was considered highly significant.
Supplementary Material
Acknowledgments
Funding: This work was financially supported by the National Natural Science Foundation of China (21874066, 81601632, 61804076, and 21771150), the Natural Science Foundation of Jiangsu Province (BK20160616 and BK20180700), the Fundamental Research Funds for Central Universities, the Shuangchuang Program of Jiangsu Province, and the Thousand Talents Program for Young Researchers. Author contributions: Y.S. supervised the project and conceived the ideas. Y.P. and J.Y. conducted the main experiments and analyzed the results. Y.P. and Y.S. wrote the manuscript. X. Luan, X. Liu, and L.S. assisted in the animal experiments. X. Li conducted the SURVEYOR assay. J.Y. and T.H. assisted in the cell lysis experiments. T.H., Y.W., and Y.L. assisted in discussing mechanisms. Y.W., Y.L., and Y.S. assisted in revising the manuscript. All authors read and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaav7199/DC1
Fig. S1. Schematic for the synthesis of UCNPs-Cas9.
Fig. S2. The structure of UCNPs confirmed by HRTEM and SAED pattern.
Fig. S3. The surface modification process of UCNPs.
Fig. S4. Photocleavage reaction triggered by NIR light on the surface of UCNPs.
Fig. S5. Cellular internalization of UCNPs-Cas9@PEI monitored by confocal microscopy.
Fig. S6. The viability of cells treated under different conditions.
Fig. S7. The assay of EGFP expression in cells.
Fig. S8. Cell viabilities observed by CLSM images.
Fig. S9. The biocompatibility analysis of UCNPs-Cas9@PEI.
REFERENCES AND NOTES
- 1.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E., A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M., RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H.-X., Li M., Lee C. M., Chakraborty S., Kim H.-W., Bao G., Leong K. W., CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 117, 9874–9906 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Han X., Liu Z., Zhao L., Wang F., Yu Y., Yang J., Chen R., Qin L., Microfluidic cell deformability assay for rapid and efficient kinase screening with the CRISPR-Cas9 system. Angew. Chem. Int. Ed. 55, 8561–8565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han X., Liu Z., Jo M. C., Zhang K., Li Y., Zeng Z., Li N., Zu Y., Qin L., CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci. Adv. 1, e1500454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding Q., Strong A., Patel K. M., Ng S.-L., Gosis B. S., Regan S. N., Cowan C. A., Rader D. J., Musunuru K., Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 115, 488–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., Zetsche B., Shalem O., Wu X., Makarova K. S., In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T., Wei J. J., Sabatini D. M., Lander E. S., Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty S., Ji H., Kabadi A. M., Gersbach C. A., Christoforou N., Leong K. W., A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep. 3, 940–947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nault J.-C., Datta S., Imbeaud S., Franconi A., Mallet M., Couchy G., Letouzé E., Pilati C., Verret B., Blanc J.-F., Balabaud C., Calderaro J., Laurent A., Letexier M., Bioulac-Sage P., Calvo F., Zucman-Rossi J., Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 47, 1187–1193 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Wang P., Zhang L., Zheng W., Cong L., Guo Z., Xie Y., Wang L., Tang R., Feng Q., Hamada Y., Gonda K., Hu Z., Wu X., Jiang X., Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed. 57, 1491–1496 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Zhou W., Cui H., Ying L., Yu X.-F., Enhanced cytosolic delivery and release of CRISPR/Cas9 by black phosphorus nanosheets for genome editing. Angew. Chem. Int. Ed. 57, 10268–10272 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Alsaiari S. K., Patil S., Alyami M., Alamoudi K. O., Aleisa F. A., Merzaban J. S., Li M., Khashab N. M., Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J. Am. Chem. Soc. 140, 143–146 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Yue H., Zhou X., Cheng M., Xing D., Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale 10, 1063–1071 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Luo Y.-L., Xu C.-F., Li H.-J., Cao Z.-T., Liu J., Wang J.-L., Du X.-J., Yang X.-Z., Gu Z., Wang J., Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano 12, 994–1005 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Li L., Song L., Liu X., Yang X., Li X., He T., Wang N., Yang S., Yu C., Yin T., Wen Y., He Z., Wei X., Su W., Wu Q., Yao S., Gong C., Wei Y., Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 11, 95–111 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Sun W., Ji W., Hall J. M., Hu Q., Wang C., Beisel C. L., Gu Z., Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. 54, 12029–12033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao L., Jiahua W., Weihuai G., Yulei H., Zhongchun T., Lijia H., Jiali T., Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 5, 1700611 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng W., Chen W., Clement S., Guller A., Zhao Z., Engel A., Goldys E. M., Controlled gene and drug release from a liposomal delivery platform triggered by X-ray radiation. Nat. Commun. 9, 2713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mura S., Nicolas J., Couvreur P., Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 12, 991–1003 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Bechet D., Couleaud P., Frochot C., Viriot M.-L., Guillemin F., Barberi-Heyob M., Nanoparticles as vehicles for delivery of photodynamic therapy agents. Trends Biotechnol. 26, 612–621 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Bandara H. M., Burdette S. C., Photoisomerization in different classes of azobenzene. Chem. Soc. Rev. 41, 1809–1825 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Klajn R., Spiropyran-based dynamic materials. Chem. Soc. Rev. 43, 148–184 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Shao Q., Xing B., Photoactive molecules for applications in molecular imaging and cell biology. Chem. Soc. Rev. 39, 2835–2846 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Wang C., Tao H., Cheng L., Liu Z., Near-infrared light induced in vivo photodynamic therapy of cancer based on upconversion nanoparticles. Biomaterials 32, 6145–6154 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Frangioni J. V., In vivo near-infrared fluorescence imaging. Curr. Opin. Chem. Biol. 7, 626–634 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Boulnois J.-L., Photophysical processes in recent medical laser developments: A review. Lasers Med. Sci. 1, 47–66 (1986). [Google Scholar]
- 29.Chen G., Shen J., Ohulchanskyy T. Y., Patel N. J., Kutikov A., Li Z., Song J., Pandey R. K., Ågren H., Prasad P. N., (α-NaYbF4:Tm3+)/CaF2core/shell nanoparticles with efficient near-infrared to near-infrared upconversion for high-contrast deep tissue bioimaging. ACS Nano 6, 8280–8287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong H., Du S.-R., Zheng X.-Y., Lyu G.-M., Sun L.-D., Li L.-D., Zhang P.-Z., Zhang C., Yan C.-H., Lanthanide nanoparticles: From design toward bioimaging and therapy. Chem. Rev. 115, 10725–10815 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Chen G., Qiu H., Prasad P. N., Chen X., Upconversion nanoparticles: Design, nanochemistry, and applications in theranostics. Chem. Rev. 114, 5161–5214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng R., Xie X., Vendrell M., Chang Y.-T., Liu X., Intracellular glutathione detection using MnO2-nanosheet-modified upconversion nanoparticles. J. Am. Chem. Soc. 133, 20168–20171 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Chen Z., Liu Z., Li Z., Ju E., Gao N., Zhou L., Ren J., Qu X., Upconversion nanoprobes for efficiently in vitro imaging reactive oxygen species and in vivo diagnosing rheumatoid arthritis. Biomaterials 39, 15–22 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Li W., Wang J., Ren J., Qu X., Near-infrared upconversion controls photocaged cell adhesion. J. Am. Chem. Soc. 136, 2248–2251 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Yan Z., Qin H., Ren J., Qu X., Photocontrolled multidirectional differentiation of mesenchymal stem cells on an upconversion substrate. Angew. Chem. Int. Ed. 130, 11352–11357 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Lucky S. S., Muhammad Idris N., Li Z., Huang K., Soo K. C., Zhang Y., Titania coated upconversion nanoparticles for near-infrared light triggered photodynamic therapy. ACS Nano 9, 191–205 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Chen S., Weitemier A. Z., Zeng X., He L., Wang X., Tao Y., Huang A. J. Y., Hashimotodani Y., Kano M., Iwasaki H., Parajuli L. K., Okabe S., Teh D. B. L., All A. H., Tsutsui-Kimura I., Tanaka K. F., Liu X., McHugh T. J., Near-infrared deep brain stimulation via upconversion nanoparticle–mediated optogenetics. Science 359, 679–684 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Eisenführ A., Arora P. S., Sengle G., Takaoka L. R., Nowick J. S., Famulok M., A ribozyme with michaelase activity: Synthesis of the substrate precursors. Bioorg. Med. Chem. 11, 235–249 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Jayakumar M. K. G., Idris N. M., Zhang Y., Remote activation of biomolecules in deep tissues using near-infrared-to-UV upconversion nanotransducers. Proc. Natl. Acad. Sci. U.S.A. 109, 8483–8488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Cárcer G., Wachowicz P., Martínez-Martínez S., Oller J., Méndez-Barbero N., Escobar B., González-Loyola A., Takaki T., El Bakkali A., Cámara J. A., Jiménez-Borreguero L. J., Bustelo X. R., Cañamero M., Mulero F., de los Ángeles Sevilla M., Montero M. J., Redondo J. M., Malumbres M., Plk1 regulates contraction of postmitotic smooth muscle cells and is required for vascular homeostasis. Nat. Med. 23, 964–974 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Wang Z., Zhang Y., Ju E., Liu Z., Cao F., Chen Z., Ren J., Qu X., Biomimetic nanoflowers by self-assembly of nanozymes to induce intracellular oxidative damage against hypoxic tumors. Nat. Commun. 9, 3334–3347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z., Li Z., Liu J., Gu S., Yuan Q., Ren J., Qu X., Long-circulating Er3+-doped Yb2O3 up-conversion nanoparticle as an in vivo X-ray CT imaging contrast agent. Biomaterials 33, 6748–6757 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaav7199/DC1
Fig. S1. Schematic for the synthesis of UCNPs-Cas9.
Fig. S2. The structure of UCNPs confirmed by HRTEM and SAED pattern.
Fig. S3. The surface modification process of UCNPs.
Fig. S4. Photocleavage reaction triggered by NIR light on the surface of UCNPs.
Fig. S5. Cellular internalization of UCNPs-Cas9@PEI monitored by confocal microscopy.
Fig. S6. The viability of cells treated under different conditions.
Fig. S7. The assay of EGFP expression in cells.
Fig. S8. Cell viabilities observed by CLSM images.
Fig. S9. The biocompatibility analysis of UCNPs-Cas9@PEI.


