Histone posttranslational modifications play fundamental roles in the regulation of double-stranded DNA break (DSB) repair. RNF20/RNF40-mediated monoubiquitination of histone H2B on lysine 120 (H2Bub) has been suggested as a potential mediator of DSB repair, although the nature and function of this posttranslational modification remain enigmatic.
KEYWORDS: B cell, double-stranded DNA repair, H2BK120, RNF20, RNF40, class switch recombination, histone ubiquitination, homologous recombination, nonhomologous recombination
ABSTRACT
Histone posttranslational modifications play fundamental roles in the regulation of double-stranded DNA break (DSB) repair. RNF20/RNF40-mediated monoubiquitination of histone H2B on lysine 120 (H2Bub) has been suggested as a potential mediator of DSB repair, although the nature and function of this posttranslational modification remain enigmatic. In this report, we demonstrate that RNF20 and RNF40 are required for DSB repair leading to homologous recombination (HR) and class switch recombination, a process driven by nonhomologous end joining (NHEJ), in mouse B cells. These findings suggest a role for RNF20 and RNF40 in DSB repair proximal to NHEJ/HR pathway choice and likely in the signaling of DSBs. We found that DSBs led to a global increase in H2Bub but not the transcription-associated posttranslational modifications H3K4me3 and H3K79me2. We also found that H2AX phosphorylation was dispensable for H2Bub and that ATM and ATR jointly regulate ionizing radiation (IR)-induced H2Bub. Together, our results suggest that RNF20, RNF40, and H2Bub may represent a novel pathway for DSB sensing and repair.
INTRODUCTION
Maintaining genome integrity is paramount to cell survival and growth, as unchecked DNA damage or improper repair can lead to cell death or genetic aberrations, such as chromosomal translocations, telomere fusions, and aneuploidy (1). Threats to genome integrity can be exogenous, such as UV light exposure or cigarette smoke (2, 3), or endogenous, such as collapse of replication forks (4). Threats such as DNA mutations and double-stranded breaks (DSBs) can even be programmed, such as during V(D)J recombination (5, 6) and class switch recombination (CSR) (7) in B cells or during meiosis in gametes (8).
Histone posttranslational modifications (PTMs) play a pivotal role in the regulation of DSB repair. For example, phosphorylation of histone variant H2AX at serine 139, known as γH2AX, is a classical marker of DSB formation detected within minutes of a DSB forming (9). γH2AX accumulates at DSBs to amplify the DNA damage response (DDR) through recruitment of the Mre11-Rad-Nbs (MRN) complex, ATM, and MDC1 (10–13). This leads to the recruitment of E3 ubiquitin ligases RNF8 (14–17) and RNF168 (18, 19), which polyubiquitinate histone H2A on lysine 15 (H2AK15ub). BRCA1/RAP80 and 53BP1 compete for binding to H2AK15ub in order to commit DSB repair to homologous recombination (HR) or nonhomologous end joining (NHEJ), respectively (17, 20). Therefore, histone PTMs help orchestrate an effective and tailored response to DSBs.
H2B lysine 20 monoubiquitination (H2BK120ub, or H2Bub) and deubiquitination have been put forth as histone PTMs that may be involved in DSB repair, although their function in transcription has been more thoroughly characterized. H2B was first shown to be ubiquitinated in Saccharomyces cerevisiae by the E3 ubiquitin-conjugating enzyme Rad6 (21, 22) and deubiquitinated by the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex (23, 24). In higher eukaryotes, H2B is ubiquitinated by the mammalian paralogues of Bre1, known as RNF20 and RNF40 (25, 26), and deubiquitinated by the mammalian SAGA complex deubiquitination module, composed of Eny2, Atxn7, Atxn7L3, and the ubiquitin-specific protease Usp22 (27, 28). H2B is ubiquitinated and deubiquitinated during transcription in yeast and human, illustrating the conserved nature of H2Bub regulation (24, 29). In resting cells, H2Bub localizes to gene bodies (29–31), as a result of the recruitment of the histone chaperone FACT and subsequent RNF20/40-mediated H2B ubiquitination, leading to the displacement of H2A/H2B dimers to allow for RNA polymerase II elongation past nucleosomes (32).
The role of RNF20, RNF40, and H2B ubiquitination in the DNA damage response is markedly less clear. We and others have observed that DSB-inducing agents, such as ionizing radiation (IR) or radiomimetic drugs, induce a global increase in H2Bub levels, suggesting a role for H2Bub in DSB sensing and/or repair (33–35). Like γH2AX, ubiquitinated H2B broadly accumulates proximal to DSBs (33, 35, 36). Curiously, H2BK120 appears to be acetylated rather than ubiquitinated within a 1-kb window of the DSB and ubiquitinated outside that window (36). Unlike γH2AX, little is understood about the downstream effectors of H2Bub.
Furthermore, the mechanism(s) by which RNF20 and RNF40 activity and/or DSB-induced H2Bub is regulated and the consequences thereof remain elusive. A previous study suggested that phosphorylation of RNF20 and RNF40 by ATM is required for their ubiquitin ligase activity (34), although that study mostly visualized H2B ubiquitination at single time points rather than overall kinetics after DNA damage. Another study presented evidence that RNF20 and RNF40 are required for CtIP- and NBS1-dependent end resection and HR by assessing RAD51 and RPA foci (35). This finding supports a role for RNF20 and RNF40 in HR but is seemingly at odds with a separate observation that RNF20 and RNF40 are required for repair of I-SceI-induced DSBs by NHEJ, which requires repression of resection (34, 35). To date, the functions of RNF20 and RNF40 remain much better characterized in HR than in NHEJ. Others have suggested that H2B ubiquitination may help decondense chromatin to allow repair proteins to access the DNA (34, 37); however, it is unclear whether this increased accessibility could be simply due to a physical interaction between repair factors and H2Bub. Altogether, the many hypotheses regarding the precise role of RNF20, RNF40, and H2Bub in DSB repair call for further examination.
We previously demonstrated that Eny2 and Usp22, components of the SAGA deubiquitinase module that remove ubiquitin from H2B, are required for CSR and repair of activation-induced cytidine deamidase (AID)- and IR-mediated DSBs (33, 38). Since this phenotype could be merely due to abnormally high levels of H2Bub affecting other DSB repair pathways, we investigated whether the addition of ubiquitin by RNF20 and RNF40 might be required for CSR and DSB repair. In this report, we show that RNF20 and RNF40 are required for distal end joining of DSBs leading to CSR, establishing a role for these factors in NHEJ. We also found that RNF20 and RNF40 are required for DSB repair by HR, suggesting a function for RNF20 and RNF40 in the DNA damage response proximal to NHEJ/HR pathway choice. We demonstrate that RNF20/RNF40-mediated H2B ubiquitination does not require H2AX and, by extension, γH2AX. Changes in H2B ubiquitination levels, whether increased by IR or decreased by RNF20 or RNF40 knockdown, did not demonstrably affect H2B acetylation levels. Interestingly, we found that ATM and ATR jointly regulate IR-induced H2B ubiquitination, suggesting a redundant function in a putative H2Bub pathway between the two kinases. Our study suggests that RNF20, RNF40, and H2Bub may constitute a novel axis of DSB repair.
RESULTS
RNF20 and RNF40 are required for class switch recombination.
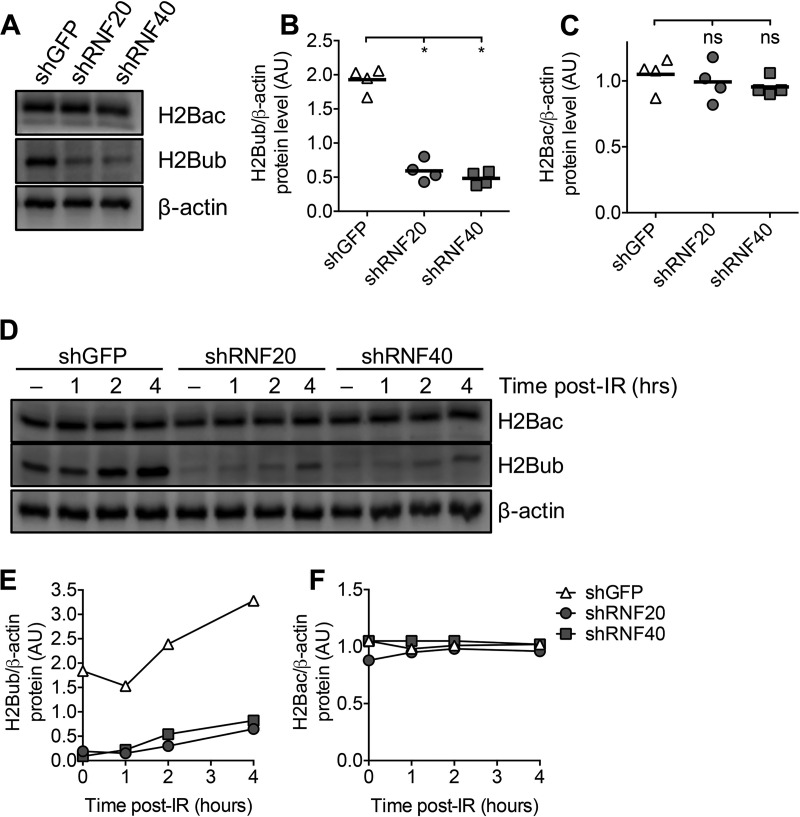
To assess a potential role for RNF20 and RNF40 in DSB repair, we used short hairpin RNAs (shRNAs) to knock down Rnf20 and Rnf40 mRNAs in the CH12F3-2 mouse B lymphoma (CH12) cell line. We had previously attempted to knock out RNF20 and RNF40 in CH12 cells using CRISPR/Cas9 gene editing but were unable to do so (data not shown), suggesting that RNF20 and RNF40 are essential for cell survival. Other studies were also unable to knock out RNF20 or RNF40 in mouse models or human cell lines (39, 40). We found that shRNA knocked down Rnf20 and Rnf40 mRNAs to ∼30% of mRNA levels compared to those in short hairpin green florescent protein (shGFP) control cells (Fig. 1A). shRNA knockdown of RNF20 and RNF40 also reduced H2Bub levels in resting cells (Fig. 1B). Therefore, shRNA led to reduced RNF20 and RNF40 expression and function in CH12 cells.
FIG 1.
RNF20 and RNF40 are required for class switch recombination (CSR). (A) Quantification of Rnf20 and Rnf40 mRNAs in shRNF20 and shRNF40 CH12 cells, respectively, by qPCR. Values were calculated using the Pfaffl method, normalizing expression to that in shGFP CH12 cells. (B) Detection of H2Bub expression in shRNF20 and shRNF40 CH12 cells by Western blotting. (C) Quantification of Rnf20 and Rnf40 mRNAs in shRNF20 and shRNF40 WEHI-279 cells, respectively, by qPCR. Values were calculated using the Pfaffl method, normalizing expression to that in shGFP WEHI-279 cells. (D) Quantification of CSR to IgA in control, shRNF20, and shRNF40 CH12 cells as measured by flow cytometry 2 days after cytokine stimulation with αCD40, IL-4, and TGF-β (CIT). (E) Quantification of Aicda mRNA in shRNF20 and shRNF40 CH12 cells at steady state or 1 day post-CIT. (F) AID protein in shRNF20 and shRNF40 CH12 cells at steady state or 1 day post-CIT. The white line indicates stitching of different lanes from the same blot. (G) Densitometry analysis of AID protein levels relative to H2B in control, shRNF20, and shRNF40 CH12 cells at steady state or 1 day post-CIT. AU, arbitrary units. (H and I) Quantification of germ line transcription of switch regions μ (Sμ) (H) and α (Sα) (I) at steady state or 1 day post-CIT by qPCR. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CH12 cells undergo CSR in vitro from IgM to IgA upon cytokine stimulation with αCD40, interleukin-4 [IL-4], and transforming growth factor β (TGF-β) (CIT) (41). Because CSR requires the formation of distal DSBs mediated by AID at the immunoglobulin heavy chain (Igh) locus, which are then predominantly rejoined by NHEJ (42), CSR is a useful proxy for DSB repair by NHEJ. To determine whether RNF20 and RNF40 are required for CSR, we measured IgA switching in control, shRNF20, and shRNF40 CH12 cells after CIT stimulation by flow cytometry. We found that knockdown of RNF20 or RNF40 in CH12 cells led to a defect in CSR (Fig. 1D), suggesting that these factors are required for efficient NHEJ.
Several intermediate steps are required to facilitate CSR in vivo and in CH12 cells. CD40 engagement results in transcription and expression of AID (43), which mediates DSB formation in Igh. To determine whether the defect in CSR in shRNF20 and shRNF40 CH12 cells was due to reduced AID expression, we measured Aicda mRNA and AID protein by quantitative PCR (qPCR) and Western blotting, respectively. We found that RNF20 and RNF40 knockdown led to a decrease in CIT-induced Aicda transcription (Fig. 1E) and AID protein (Fig. 1F and G). CSR also requires cytokine-induced transcription of the noncoding switch regions, known as germ line transcription, in order to expose single-stranded DNA for AID to deaminate and mediate DSBs (44). We found that germ line transcription levels of switch regions μ (Sμ) and α (Sα) were reduced in shRNF20 and shRNF40 CH12 cells (Fig. 1H and I).
These data suggest that the defect in CSR in shRNF20 and shRNF40 CH12 cells could be a consequence of reduced overall transcription, since ubiquitination of H2B by RNF20 and RNF40 is important for transcription. Therefore, we assessed whether transcription of DSB repair genes was globally reduced in shRNF20 and shRNF40 CH12 cells. We did not observe significant changes in the transcription of various housekeeping and DNA damage repair genes in shRNF20 and shRNF40 CH12 cells whether at steady state or in response to ionizing radiation (IR)-induced DNA damage (Fig. 2). This finding suggests that RNF20 and RNF40 knockdown cells do not exhibit a global reduction in transcription and that Aicda and germ line transcription is particularly sensitive to RNF20/RNF40 levels.
FIG 2.
RNF20 and RNF40 knockdown does not grossly affect transcription of DSB repair genes at steady state or after IR. mRNA levels of RNF20 (Rnf20) (A), RNF40 (Rnf40) (B), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Gapdh) (C), β-actin (Bact) (D), p53 (tp53) (E), p21 (tp21) (F), MDM2 (Mdm2) (G), ATM (Atm) (H), DNA-PKcs (Prkdc) (I), 53BP1 (Trp53bp1) (J), and BRCA1 (Brca1) (K) in shGFP, shRNF20, and shRNF40 CH12 cells at steady state or 4 h after 8 Gy IR as measured by qPCR.
Hence, the precise function of RNF20 and RNF40 in CSR could be to regulate the expression of CSR-associated transcripts and/or to repair DSBs mediated by AID, which we next explored.
RNF20 and RNF40 are required for DSB repair leading to CSR in multiple mouse B cell lines.
To pinpoint whether RNF20 and RNF40 are specifically required for DSB repair during CSR, we assessed whether rejoining of distal DSBs leading to CSR was impaired in RNF20 and RNF40 knockdown cells. To do so, we induced CSR from IgM to IgA in CH12 cells independent of AID expression or activity using CRISPR/Cas9 (33, 45). Briefly, we generated DSBs outside the repetitive switch regions using CRISPR/Cas9 and relevant single guide RNAs (sgRNAs) (Fig. 3A). Recombination between Cas9-induced DSBs outside switch regions μ (S′μ) and α (S′α) leads to class switching from IgM to IgA, which can be quantified by flow cytometry. Therefore, Cas9 can induce CSR without transcription of Aicda or the switch regions, allowing us to determine whether RNF20 and RNF40 are specifically required for DSB repair leading to CSR. Because CH12 cells are capable of undergoing CSR in vitro from IgM to IgA upon cytokine stimulation (41), we eliminated any DSBs mediated by AID or background IgA expression by inducing Cas9-mediated switching to IgA in Aicda−/− CH12 cells.
FIG 3.
RNF20 and RNF40 are required for Cas9-mediated switching and homologous recombination. (A) Schematic of the mouse immunoglobulin heavy chain (Igh) locus and Cas9/sgRNA targeting for CSR. Constant regions Cμ, Cγ, and Cα are preceded by repetitive switch regions Sμ, Sγ, and Sα, respectively. sgRNAs (depicted as black bars) were designed to target outside these switch regions. Recombination between DSBs generated at S′μ and S′γ results in class switching to IgG1 (left), while recombination between DSBs generated at S′μ and S′α results in class switching to IgA (right). The schematic is not to scale. (B) Quantification of Cas9-mediated switching to IgA in Aicda−/− CH12 cells and representative flow plots. (C to E) Cas9-mediated switching to IgG1 in CH12 cells and representative flow plots (C), Cas9-mediated switching to IgA in WEHI-279 cells and representative flow plots (D), and I-SceI-mediated homologous recombination in DR-GFP CH12 cells and representative flow plots (E) following control, RNF20, or RNF40 knockdown as measured by flow cytometry at 3 days posttransfection. FSC, forward scatter. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We found that shRNF20 and shRNF40 CH12 cells exhibited a defect in Cas9-mediated switching to IgA (Fig. 3B), suggesting that RNF20 and RNF40 are required for long-range DSB joining. To determine whether this defect was isotype specific, we also induced Cas9-mediated switching to IgG1 by using sgRNAs targeting outside switch regions μ and γ1 (S′γ1) in shRNF20 and shRNF40 CH12 cells (Fig. 3A). Knockdown of RNF20 and RNF40 also led to a defect in Cas9-mediated switching to IgG1 (Fig. 3C). We further verified that this defect in end joining was not cell line specific by knocking down RNF20 and RNF40 in WEHI-279 cells (Fig. 1C), another mouse B lymphoma IgM-expressing cell line. Like we observed in CH12 cells, WEHI-279 cells expressing shRNF20 and shRNF40 exhibit a defect in Cas9-mediated switching to IgA (Fig. 3D). Altogether, we conclude that RNF20 and RNF40 are required for the DSB repair phase of CSR to two isotypes in two B cell lines.
RNF20 and RNF40 are required for homologous recombination.
CSR is largely driven by NHEJ, and alternative end joining (A-EJ) to a lesser extent, but not homologous recombination (HR), because HR-mediated repair of AID-dependent DSBs in the switch regions would in theory restore the wild-type Igh locus and prevent CSR. Therefore, CSR cannot suggest a function for RNF20 and RNF40 in HR.
To determine whether RNF20 and RNF40 are required for HR, we induced DSBs using the yeast megaendonuclease I-SceI in shRNF20 and shRNF40 CH12 cells harboring the classical DR-GFP repair substrate (46). HR-mediated repair of an I-SceI-induced DSB in the DR-GFP substrate results in restoration of a functional GFP coding sequence, leading to GFP expression quantified by flow cytometry (46). We found that repair of I-SceI-induced DSBs by HR was reduced in shRNF20 and shRNF40 CH12 cells compared to shLacZ control cells (Fig. 3E), in agreement with data from previous studies in human cells (35). This result suggests that RNF20 and RNF40 are required for HR.
Altogether, our data suggest that RNF20 and RNF40 are required for distal end joining leading to CSR as well as HR, hinting toward a role for these factors in DSB repair in the early stages of DSB repair, prior to pathway choice between NHEJ and HR.
Knockdown of RNF20 or RNF40 does not affect cell proliferation dynamics.
Because our data suggest that RNF20 and RNF40 are required for DSB repair, we postulated that RNF20 and RNF40 knockdown might affect cell growth after DNA damage. We assessed cell proliferation by counting control, shRNF20, and shRNF40 CH12 cells in the steady state or with CIT stimulation, which triggers CSR in vitro via AID-mediated DSBs. Surprisingly, we found that knockdown of RNF20 or RNF40 did not affect cell proliferation in the steady state (Fig. 4A) or with CIT stimulation (Fig. 4B). This result suggests that RNF20 and RNF40 are dispensable for cell proliferation under normal conditions or DNA damage. This result also suggests that the defect in CSR observed in shRNF20 and shRNF40 CH12 cells is not due to a reduction in cell growth.
FIG 4.
RNF20 and RNF40 knockdown does not significantly perturb cellular proliferation dynamics. (A and B) Proliferation of shGFP, shRNF20, and shRNF40 CH12 cells in the steady state (A) and after stimulation with CIT (B). (C) Representative flow cytometry plots depicting cell cycle phases of shGFP, shRNF20, and shRNF40 CH12 cells in the steady state and after CIT stimulation. EdUlo PIlo cells (lower left gate) are in G0/G1 phase, EdUhi cells (upper gate) are in S phase, and EdUlo PIhi cells (lower right gate) are in G2/M phase. (D and E) Quantification of shGFP, shRNF20, and shRNF40 CH12 cells in G0/G1, S, and G2/M phases in the steady state (D) and after CIT stimulation (E). Comparisons are not statistically significant unless specifically denoted.
We hypothesized that RNF20 and RNF40 might be required to repair DSBs in a specific phase of the cell cycle. For instance, AID activity and the resulting DSBs in the Igh locus are largely confined to the G1 phase of the cell cycle (47). To assess whether knockdown of RNF20 or RNF40 affects specific phases of the cell cycle in the absence or presence of DSBs, we performed 5-ethynyl-2′-deoxyuridine (EdU)–propidium iodide (PI) cell cycle analysis in shRNF20 and shRNF40 CH12 cells in the steady state or during AID-mediated CSR. We found that the cell cycle profiles of control, shRNF20, and shRNF40 CH12 cells were largely similar (Fig. 4C and D). While CIT stimulation enriched the fraction of cells in G1 phase, as expected, the cell cycle profiles of control, shRNF20, and shRNF40 CH12 cells undergoing CSR were also largely similar (Fig. 4C and E). Together, our data suggest that RNF20 and RNF40 do not affect cell proliferation dynamics, even in the presence of DNA damage.
Ionizing radiation induces a global increase in H2Bub but not other transcription-associated histone PTMs.
We and others have demonstrated that DSBs generated by IR and other genotoxic agents lead to increased global H2Bub (33–35), suggesting that ubiquitination of H2B by RNF20 and RNF40 may function in DSB repair. Since H2Bub marks transcribed genes, we wondered if IR merely increased transcription in CH12 cells, leading to more H2Bub. Therefore, we measured the expression of H3K4me3 and H3K79me2, two other histone PTMs that are found in gene bodies (48), after IR. Of note, RNF20/RNF40-mediated H2B ubiquitination is thought to be required for H3K4 and H3K79 methylation by cross talk (49–51). We found that IR robustly induced a dramatic increase in H2Bub levels but not H3K4me3 and H3K79me2 levels (Fig. 5A and F). To determine whether this observation was specific to IR, we tested whether other DSB-inducing agents would increase global H2Bub, H3K4me3, and H3K79me2 levels. We found that inducing DNA damage with camptothecin, a chemical inhibitor of topoisomerase I, led to a global increase in H2Bub (Fig. 6A and B) but not H3K4me3 or H3K79me2 (Fig. 6A, C, and D) levels. Likewise, chemical inhibition of topoisomerase II by etoposide led to increased H2Bub (Fig. 6E and F) but had no effect on H3K4me3 or H3K79me2 (Fig. 6E, G, and H). Taken together, our findings suggest that DSBs specifically induce H2B ubiquitination rather than a general increase in transcription-associated PTMs. Moreover, DSB-induced H2Bub does not seem to lead to increased H3K4me3 or H3K79me2 cross talk.
FIG 5.
Ionizing radiation (IR) induces a dramatic increase in H2Bub but not other transcription-associated histone PTMs. (A) H2Bub, H3K4me3, and H3K79me2 expression in the nuclear fraction of wild-type CH12 cells at various time points after 8 Gy IR detected by Western blotting. (B) H2Bub, H3K4me3, and H3K79me2 expression in the nuclear fraction of shGFP, shRNF20, and shRNF40 CH12 cells at steady state. (C to E) Densitometry analysis of H2Bub (C), H3K4me3 (D), and H3K79me2 (E) in shGFP, shRNF20, and shRNF40 CH12 cells at steady state. (F to J) Same as panels A to E but instead normalized to histone H3 levels. ns, not significant; *, P < 0.05.
FIG 6.
Camptothecin and etoposide induce a dramatic increase in H2Bub but not H3K4me3 or H3K79me2. (A) H2Bub, H3K4me3, and H3K79me2 expression in wild-type CH12 cells pulsed with 5 μM camptothecin (CPT) for 1 h. (B to D) Densitometry analysis of H2Bub (B), H3K4me3 (C), and H3K79me2 (D) at the indicated time points after camptothecin treatment relative to β-actin. (E) H2Bub, H3K4me3, and H3K79me2 expression in wild-type CH12 cells pulsed with 25 μM etoposide (ETP) for 1 h. (F to H) Densitometry analysis of H2Bub (F), H3K4me3 (G), and H3K79me2 (H) at the indicated time points after etoposide treatment relative to β-actin.
Previous studies found that knockdown of RNF20 or RNF40 in human cells and Bre1/Rad6 in yeast led to decreased H3K4me3 and H3K79me2 (25, 26, 35). To test whether this was the case in CH12 cells, we assessed H3K4me3 or H3K79me2 levels in shRNF20 and shRNF40 CH12 cells at steady state. As expected, shRNF20 and shRNF40 CH12 cells exhibit a significant reduction in global H2Bub (Fig. 5B, C, G, and H). However, knockdown of RNF20 or RNF40 did not affect H3K4me3 or H3K79me2 expression in CH12 cells (Fig. 5D, E, I, and J). Taken together, our findings suggest that neither IR nor RNF20/RNF40 knockdown induces wholesale changes in transcription-associated histone PTMs, allowing us to investigate a role for RNF20, RNF40, and H2Bub specifically in DSB repair.
IR-induced γH2AX formation is independent of RNF20, RNF40, and H2Bub.
Histone variant H2A.X is phosphorylated within minutes of a DSB, and this is one of the earliest events in DSB signaling and repair. The γH2AX signal decreases as DSBs are repaired. Thus, the accumulation and resolution of γH2AX are classical marks of DSB formation and repair. Similarly, RNF20 and RNF40 monoubiquitinate histone H2B in response to DSBs (33–35), although H2Bub appears several hours, rather than minutes, after DSB formation (Fig. 5A). While H2B and H2AX are both histones that undergo DSB-induced PTMs, the relationship between these two PTMs remains unclear (34, 35). We previously found that deficiency in the SAGA complex factor Eny2 or Usp22 led to a reduction in γH2AX ionizing radiation-induced foci (IRIF) in CH12 cells and mice (33, 38), suggesting that H2Bub levels can impact γH2AX after DNA damage. However, this result might be indirect due to increased global H2Bub in these mutant cells, which could in turn interfere with phosphorylation of H2AX (see Discussion).
To determine whether IR-induced H2B ubiquitination is required for γH2AX formation, we irradiated shRNF20 and shRNF40 CH12 cells and measured γH2AX and H2Bub kinetics by Western blotting (Fig. 7A). Consistent with previous findings, IR induced a dramatic increase in global H2Bub in control CH12 cells, which was reduced in shRNF20 and shRNF40 CH12 cells (Fig. 7B). However, IR-induced γH2AX kinetics were unchanged in control, shRNF20, or shRNF40 CH12 cells (Fig. 7C). This result suggests that RNF20, RNF40, and H2Bub are not required for IR-induced γH2AX accumulation and resolution.
FIG 7.
IR-induced γH2AX and H2Bub can arise independently of each other. (A) γH2AX and H2Bub expression in shGFP, shRNF20, and shRNF40 CH12 cells at various time points after 8 Gy IR detected by Western blotting. “−,” steady state. (B and C) Densitometry analysis of H2Bub (B) and γH2AX (C) protein levels in shGFP, shRNF20, and shRNF40 CH12 cells at the indicated time points relative to β-actin. (D) Schematic illustrating the sgRNA targeting strategy for CRISPR/Cas9-mediated H2afx gene knockout in CH12 cells. (E) RT-PCR of H2afx and Gapdh mRNAs from H2afx+/+ and H2afx−/− CH12 cells. (F) AID-mediated CSR to IgA in two H2afx+/+ and four H2afx−/− CH12 clones. Cells were harvested and analyzed by flow cytometry 2 days after cytokine stimulation with αCD40, IL-4, and TGF-β. (G) γH2AX and H2Bub expression in representative H2afx+/+ and H2afx−/− CH12 clones at various time points after 8 Gy IR detected by Western blotting. “−,” steady state. (H and I) Densitometry analysis of γH2AX (H) and H2Bub (I) protein levels from two H2afx+/+ and four H2afx−/− CH12 clones at the indicated time points relative to β-actin. Statistical comparisons between control and experimental groups were conducted using 2-way analysis of variance (ANOVA). Comparisons are not statistically significant unless specifically denoted. **, P < 0.01; ****, P < 0.0001.
IR-induced H2Bub occurs independently of H2AX and γH2AX.
We next considered whether the inverse was true, that is, whether IR-induced γH2AX was required for IR-induced H2B ubiquitination. To do so, we used CRISPR/Cas9 gene editing to largely delete the only exon of H2afx in CH12 cells (Fig. 7D). We generated four H2afx−/− CH12 clones with the expected deletion, completely inhibiting H2afx mRNA expression (Fig. 7E). Consistent with the pivotal role for γH2AX in signaling after DSB formation (52), H2afx−/− CH12 cells exhibit a defect in CSR (Fig. 7F).
To determine whether H2AX/γH2AX is required for IR-induced H2Bub, we irradiated H2afx+/+ and H2afx−/− CH12 cells and visualized γH2AX and H2Bub kinetics by Western blotting (Fig. 7G). As expected, H2afx−/− cells do not exhibit IR-induced γH2AX (Fig. 7H). However, H2AX deficiency did not affect IR-induced H2Bub formation (Fig. 7I). Thus, our data suggest that IR-induced γH2AX is not required for IR-induced H2Bub. Taken together, these results suggest that IR-induced γH2AX and IR-induced H2Bub occur independently of each other.
IR induces a global increase in H2Bub but not H2Bac.
H2BK120 can be acetylated (H2Bac) rather than ubiquitinated. The purpose of H2B acetylation is poorly understood, particularly in relation to DSB repair. The identity of the H2B histone acetyltransferase is also unclear. A recent study examined the relationship between H2B acetylation and ubiquitination near sites of programmed DSBs (36). That study found that H2B was acetylated within a 1-kb window but ubiquitinated within 1 to 5 kb of a programmed DSB, suggesting that there may be a temporal switch from ubiquitination to acetylation in response to DSBs. This is supported by another previous study illustrating elevated H2Bac levels at promoters shortly after transcription initiation, which is later replaced by elevated H2Bub levels (53).
We first assessed the proposed ubiquitination-to-acetylation switch at H2BK120 by testing whether H2Bub is a prerequisite posttranslational modification for H2Bac. To do so, we measured H2Bac levels in shRNF20 and shRNF40 CH12 cells in the steady state and after IR (Fig. 8A). While RNF20 and RNF40 knockdown led to a significant reduction in global H2Bub levels (Fig. 8B), as expected, we did not notice any accompanying changes in global H2Bac levels (Fig. 8C). Thus, we did not find evidence that H2B ubiquitination is required for H2B acetylation.
FIG 8.
IR induces RNF20/RNF40-mediated H2B ubiquitination but not acetylation. (A) H2Bub and H2Bac protein levels in shGFP, shRNF20, and shRNF40 CH12 cells at steady state detected by Western blotting. (B and C) Densitometry analysis of H2Bub (B) and H2Bac (C) protein levels in shGFP, shRNF20, and shRNF40 CH12 cells at steady state relative to β-actin. (D) H2Bub and H2Bac protein levels in shGFP, shRNF20, and shRNF40 CH12 cells at the indicated time points after IR. “−,” steady state. (E and F) Densitometry analysis of H2Bub (E) and H2Bac (F) protein levels in shGFP, shRNF20, and shRNF40 CH12 cells at the indicated time points after IR relative to β-actin. ns, not significant; *, P < 0.05.
Several studies have described the dramatic increase in H2B ubiquitination after DNA damage (34, 35). Clouaire et al. also recently demonstrated that H2Bac is enriched within 1 kb of DSBs (36). To determine whether H2B acetylation levels also dramatically increase after DNA damage, we assessed global H2B acetylation levels after IR by Western blotting (Fig. 8D). We found that IR induced a global increase in H2Bub but not H2Bac (Fig. 8E and F). Static H2Bac levels were also observed after IR during RNF20 and RNF40 knockdown (Fig. 8E and F). This finding suggests that DSBs trigger RNF20 and RNF40 ubiquitin ligase activity to a more profound extent than H2B K120 acetyltransferase activity.
Together, our results suggest that reducing H2Bub levels via RNF20 or RNF40 knockdown does not reduce H2Bac levels, and increasing H2Bub levels with IR does not increase H2Bac levels. We therefore postulate that the H2B ubiquitination response to DNA damage is much more dramatic than H2B acetylation and that H2Bub is not required for H2Bac. However, our experimental methods do not address potential cross talk between H2Bub and H2Bac at the mononucleosomal level or as a function of proximity to DSBs, which should be assessed with more-precise methods, such as chromatin immunoprecipitation (ChIP) or coimmunoprecipitation.
IR-induced H2B ubiquitination is independent of H2AX kinases ATM and DNA-PKcs.
We postulated that while IR-induced γH2AX and H2Bub kinetics may be independent of each other, they may be regulated by the same DSB-sensing kinases. In response to a DSB, H2AX is phosphorylated by ATM and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (54, 55). Indeed, these kinases function early in DSB repair to amplify the DNA damage response through phosphorylation of H2AX as well as hundreds of other targets (56). Of note, a previous study presented evidence that phosphorylation of RNF20 and RNF40 by ATM is required for their ubiquitin ligase activity (34). Thus, we reasoned that these major players in γH2AX-dependent DSB repair might also be required for IR-induced H2B ubiquitination.
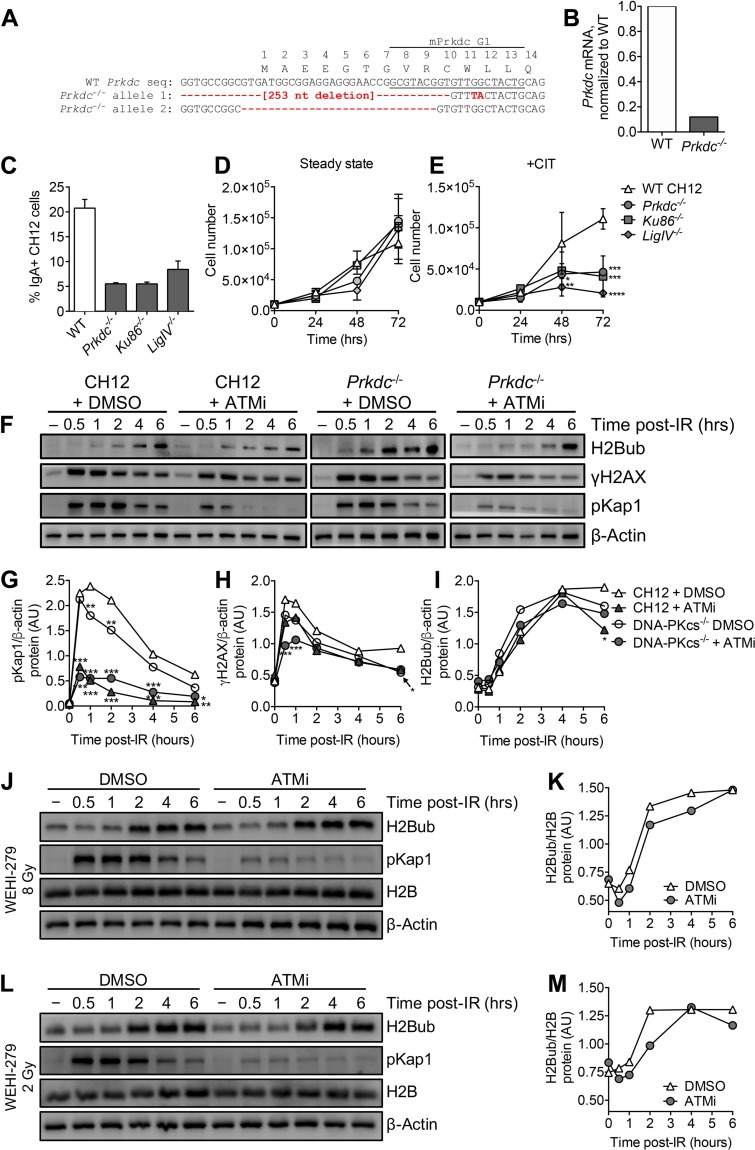
To investigate a potential role for ATM and DNA-PKcs (Prkdc) in IR-induced H2B ubiquitination, we inhibited ATM activity using a small-molecule inhibitor (KU55933 [here denoted ATMi]) (57) and generated Prkdc−/− CH12 cells using CRISPR/Cas9 gene editing. We identified a Prkdc−/− CH12 clone with deletions spanning the start codon in both Prkdc alleles (Fig. 9A). Prkdc mRNA was reduced by over 90% in Prkdc−/− CH12 cells (Fig. 9B). As expected, Prkdc−/− CH12 cells exhibit a defect in AID-mediated CSR (Fig. 9C) and reduced cell proliferation in the presence of DSBs (Fig. 9D and E), like CH12 cells deficient in other core NHEJ factors, Ku86 and DNA ligase IV (58–62).
FIG 9.
ATM and DNA-PKcs are dispensable for IR-induced H2Bub kinetics. (A) Sequenced Prkdc alleles showing Cas9-mediated deletions spanning the start codon. Amino acid residues and position numbers are indicated above the Prkdc nucleotide (nt) sequence. The Prkdc sgRNA target sequence (data not shown) is also indicated. (B) Quantification of Prkdc mRNA in wild-type (WT) and Prkdc−/− CH12 cells by qPCR. (C) AID-mediated CSR to IgA in wild-type, Prkdc−/−, Ku86−/−, and LigIV−/− CH12 cells. Cells were harvested and analyzed by flow cytometry 2 days after cytokine stimulation with αCD40, IL-4, and TGF-β (CIT). (D and E) Proliferation of wild-type, Prkdc−/−, Ku86−/−, and LigIV−/− CH12 cells in the steady state (D) and after stimulation with CIT (E). (F) H2Bub, γH2AX, and pKap1 expression at various time points after 8 Gy IR in four treatment groups of CH12 cells (DMSO treated, ATM inhibited, DNA-PKcs deficient, and ATM inhibited plus DNA-PKcs deficient) detected by Western blotting. “−,” steady state. (G to I) Densitometry analysis of pKap1 (G), γH2AX (H), and H2Bub (I) protein levels at the indicated time points relative to β-actin. (J) H2Bub and pKap1 expression at various time points after 8 Gy IR in DMSO- and ATMi-treated WEHI-279 cells. “−,” steady state. (K) Densitometry analysis of H2Bub protein levels at the indicated time points after 8 Gy IR relative to H2B. (L) H2Bub and pKap1 expression at various time points after 2 Gy IR in DMSO- and ATMi-treated WEHI-279 cells. (M) Densitometry analysis of H2Bub protein levels at the indicated time points after 2 Gy IR relative to H2B. Statistical comparisons between control and experimental groups were conducted using 2-way ANOVA. Comparisons are not statistically significant unless specifically denoted. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To determine whether ATM and/or DNA-PKcs was required for IR-induced H2Bub, we treated wild-type or Prkdc−/− CH12 cells with dimethyl sulfoxide (DMSO) or ATMi to create four treatment groups, DMSO treated, ATM inhibited, DNA-PKcs deficient, and ATM inhibited plus DNA-PKcs deficient, prior to irradiation (Fig. 9F). To validate the efficacy of ATMi, we assessed DSB-induced phosphorylation of Kap1 (pKap1), another substrate for ATM (63). As expected, IR-induced pKap1 levels were reduced in ATMi-treated CH12 cells (Fig. 9G). Interestingly, ATM inhibition and DNA-PKcs deficiency, together or alone, did not ablate IR-induced γH2AX kinetics in CH12 cells (Fig. 9H). Residual DSB-induced γH2AX in the presence of combined ATM and DNA-PK inhibitors was previously observed in human U2OS cells (64), perhaps owing to phosphorylation by other kinases, such as ATR (65). Importantly, we found that neither chemical inhibition of ATM, DNA-PKcs deficiency, nor combined treatment affected IR-induced H2Bub (Fig. 9I). We also found that ATM inhibition had no effect on IR-induced H2Bub in WEHI-279 cells treated with a high dose (Fig. 9J and K) or a low dose (Fig. 9L and M) of irradiation. Quadrupling the dosage of ATMi from 5 to 20 μM in CH12 cells did not reduce IR-induced H2Bub levels either (data not shown). These findings suggest that ATM and DNA-PKcs are dispensable for IR-induced H2B ubiquitination.
IR-induced H2B ubiquitination jointly requires ATM and ATR.
A previous report demonstrated that inhibition of ATM and ATR with caffeine suppressed IR-induced H2Bub (35). Although we did not find evidence that ATM inhibition alone was sufficient to reduce IR-induced H2Bub, we wondered whether ATM and ATR were jointly required for IR-induced H2Bub. Thus, we tested whether inhibition of ATM and ATR together could suppress IR-induced H2Bub using caffeine treatment (Fig. 10A). We validated the efficiency of ATM and ATR inhibition by assessing IR-induced phosphorylation of Kap1 and Chk1, respectively (Fig. 10B and C) (66). Consistent with data from the above-mentioned report, we found that caffeine treatment of CH12 cells almost completely inhibited IR-induced H2Bub (Fig. 10D).
FIG 10.
Simultaneous inhibition of ATM and ATR inhibits IR-induced H2Bub. (A) H2Bub, pChk1, and pKap1 protein levels in H2O- and caffeine-treated CH12 cells at the indicated time points after IR. (B to D) Densitometry analysis of pChk1 (B), pKap1 (C), and H2Bub (D) protein levels at various time points after IR relative to H2B. (E) H2Bub, pChk1, and pKap1 protein levels in DMSO-, ATRi-, and ATRi-plus-ATMi-treated CH12 cells at the indicated time points after IR. (F to H) Densitometry analysis of pChk1 (F), pKap1 (G), and H2Bub (H) protein levels at various time points after IR relative to H2B. Statistical comparisons between control and experimental groups were conducted using 2-way ANOVA. Comparisons are not statistically significant unless specifically denoted. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To test whether caffeine-mediated inhibition of H2Bub was specifically due to combined inhibition of ATM and ATR, we also treated CH12 cells with the ATR inhibitor VE-821 (ATRi) alone or with ATRi plus ATMi and measured IR-induced H2Bub by Western blotting (Fig. 10E). Consistent with our observations from caffeine treatment, only ATRi-plus-ATMi-treated CH12 cells exhibited a decrease in IR-induced H2Bub (Fig. 10F to H). Together, our data suggest that ATM and ATR jointly regulate IR-induced H2Bub.
DISCUSSION
In this study, we found that RNF20 and RNF40 are required for DSB repair in CH12 mouse B cells. Knockdown of RNF20 and RNF40 leads to a defect in CSR (Fig. 1D). We found that RNF20 and RNF40 knockdown impaired distal end joining leading to CSR as well as defective repair of a single I-SceI-induced DSB leading to HR (Fig. 3), suggesting that RNF20 and RNF40 probably function early in the DDR, before pathway choice between NHEJ and HR. If so, this would be consistent with roles played by other histone PTMs, such as γH2AX or H2AK15ub, in signaling repair after DSBs.
Interestingly, we found that IR-induced H2B ubiquitination jointly requires ATM and ATR (Fig. 10) but not each kinase individually (Fig. 9 and 10). This result is consistent with data from a previous report illustrating that inhibition of ATM and ATR with caffeine ablated IR-induced H2Bub in HeLa cells (35). However, we did not find evidence that inhibition of ATM alone is sufficient to inhibit IR-induced H2Bub, in contrast to data from another previous study (34). One advantage of our study was in visualizing the kinetics of H2Bub rather than a single time point, allowing us to examine the relationship between ATM and H2Bub with increased clarity. For instance, that previous study visualized H2Bub within 1 h of inducing DSBs, whereas we typically did not observe a striking increase in global H2Bub levels until 2 to 6 h after IR in CH12 cells (Fig. 9F) or primary mouse B cells (38).
Previous studies have suggested that the IR-induced DNA damage response is primarily regulated by ATM, with a smaller, or perhaps delayed, contribution by ATR (67, 68). Future studies could address whether ATM and ATR regulate IR-induced H2Bub in specific phases of the cell cycle. ATM has been shown to form foci throughout the cell cycle (68), while ATR foci seem to arise mostly in G1 and S phases (68, 69). In addition, since ATR and ATM are both phosphatidylinositol 3-kinases (PI 3-kinases) that phosphorylate targets at SQ/TQ motifs during the DNA damage response, it is likely that these factors regulate IR-induced H2Bub through their kinase activity. Indeed, a screen investigating redundant phosphorylation targets of ATR and ATM identified RNF20, but not RNF40, as a potential substrate (70). A potential follow-up study could mutate SQ/TQ motifs in endogenous RNF20 and assess the effect of these mutations on IR-induced H2Bub.
In this study, we found that IR-induced H2B ubiquitination is not required for normal IR-induced γH2AX kinetics (Fig. 7A to C). Previous studies found that RNF20 knockdown or H2BK120R substitution led to a delayed disappearance of γH2AX and 53BP1 IRIF (34, 35), suggesting that RNF20, RNF40, and H2Bub are required for optimal γH2AX kinetics. We did not find any evidence for this in our study (Fig. 7A to C). One explanation for the discrepancy between our respective findings could be technical differences in measuring γH2AX by Western blotting versus immunofluorescence. Future studies could clarify this discrepancy by measuring γH2AX kinetics at a single DSB by ChIP. Furthermore, we observed distinct kinetic profiles of IR-induced γH2AX versus IR-induced H2Bub. While γH2AX accumulation can be detected by Western blotting within minutes after irradiation, a global increase in H2Bub can be visualized only within hours (Fig. 5A). Indeed, in normal CH12 cells, expression of IR-induced γH2AX and pKap1 has already peaked by the time when IR-induced H2Bub becomes evident (Fig. 5F and Fig. 7A and G). However, we demonstrate that γH2AX is not required for normal IR-induced H2Bub kinetics (Fig. 7G to I). This suggests that despite the earlier appearance of γH2AX than of H2Bub, γH2AX is not a prerequisite PTM for H2Bub after DNA damage. Taken together, these data lead us to hypothesize that RNF20- and RNF40-mediated H2B ubiquitination in response to DSBs may represent an independent yet uncharacterized pathway for DSB signaling.
Still, γH2AX- and H2Bub-dependent pathways could influence one another. For example, we previously found that elevated H2Bub levels in both Usp22−/− CH12 cells and Usp22−/− primary mouse B cells were associated with reduced γH2AX IRIF (33, 38). Perhaps ubiquitination of H2B, which is elevated in Usp22−/− B cells, precludes γH2AX formation within the same nucleosome, possibly due to steric hindrance (see below). This is consistent with the observation that the H2Bub level begins to increase after γH2AX expression has already peaked. Another possibility is that elevated H2Bub levels in Usp22−/− B cells could block recruitment or accessibility of ATM or DNA-PKcs to phosphorylate H2AX. Future studies are required to assess how the dynamic regulation of H2Bub after DSBs might affect γH2AX kinetics.
The exact purpose of H2B ubiquitination by RNF20 and RNF40 in response to DSBs remains poorly understood. Several studies have attempted to explain the functional consequence of H2B ubiquitination after DNA damage. One potential function could be to recruit a specific subset of DSB repair factors to the lesion. Colocalization of ATM, MDC1, or 53BP1 with γH2AX was found to be unaffected by RNF20 depletion in HeLa cells (35). Perhaps H2Bub binds DSB repair factors that act separately from γH2AX-mediated repair. Another potential role for H2Bub in DSB repair is to promote or preclude other histone PTMs within mononucleosomes containing H2Bub. Ubiquitin is a large, bulky PTM almost one-half the molecular weight of H2B itself and is therefore likely to disrupt nucleosome structure. A recent report screened for histone PTMs that allowed or disallowed H2Bub within the same nucleosome (48). That study found that ubiquitination of H2B was hindered in nucleosomes containing histone variant H2A.Z or acetylated H2A tails. It would be interesting to determine whether nucleosomes containing γH2AX at DSB sites also contain H2Bub, given that expression levels of these two PTMs peak at different times after DSB formation (Fig. 7A). Finally, H2Bub itself could act by relaxing chromatin, as previously suggested (37), in order to promote repair.
Several studies have proposed that RNF20/RNF40-dependent H2Bub is a prerequisite for H3K4 and K79 methylation in order to promote transcription (49–51). We found that DSBs induced an increase in H2Bub but not H3K4me3 or H3K79me2 (Fig. 5A and F), in agreement with data from a previous study (34). This suggests that H2B ubiquitination in response to DSBs might play a role during DSB repair that is either modulated from its function during transcription or entirely distinct. The localization of DSB-induced H2Bub within the chromatin could provide valuable insight into the potential dual roles of this PTM during transcription and DSB repair. One previous study found that H2Bub localized to an I-SceI-induced DSB in the exogenous DR-GFP substrate in HeLa cells, which is actively transcribed (35). Curiously, H3K4me3 was also enriched at the break site. We also previously found increased H2Bub levels at switch regions during CSR, which are also actively transcribed and are the substrates for AID-mediated DNA damage (33). In another study, inhibition of transcription in HeLa cells led to a rapid loss of global H2Bub, but H3K4me3 levels remained unchanged (29). The relationship between the roles of H2Bub in transcription and DSB repair and their effects on PTM cross talk would be a rich area for future study.
In conclusion, we find that RNF20 and RNF40 play critical roles in the repair of DSBs leading to both CSR and HR, suggesting a role for RNF20, RNF40, and H2Bub in the signaling and subsequent repair of DSBs. Furthermore, the independence of IR-induced H2Bub from canonical DSB repair pathways governed by ATM and γH2AX suggests that H2B ubiquitination may represent a novel pathway in the orchestration of DSB repair.
MATERIALS AND METHODS
Cell culture.
CH12 and WEHI-279 cells were cultured in RPMI 1640 medium with l-glutamine supplemented with 10% fetal bovine serum, 5% NCTC 109, 50 μM β-mercaptoethanol, and penicillin-streptomycin. HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin. All cells were cultured at 37°C with 5% CO2.
Production of lentivirus and transduction with short hairpin RNAs.
HEK293T cells were cultured in tissue culture plates to 70% confluence 1 day prior to polyethyleneimine transfection with lentiviral packaging plasmids and the pLKO.1 or pLKO_TRC018 plasmid encoding shRNA expression cassettes (data not shown) (RNAi Consortium). Lentiviral culture supernatants were harvested at 2 to 3 days posttransfection and overlaid on target cells in complete medium containing 8 μg/ml Polybrene. Transduced cells were selected at 24 h posttransduction in complete medium containing 1 μg/ml puromycin or 2 mg/ml hygromycin for 3 to 6 days.
Quantitative PCR and reverse transcription-PCR of mRNA transcripts.
Total RNA was isolated from cells using TRIzol (Life Technologies)-chloroform extraction according to the manufacturer’s instructions and stored in diethyl pyrocarbonate (DEPC)-treated water. cDNA was reverse transcribed using Maxima H Minus reverse transcriptase (ThermoFisher) according to the manufacturer’s instructions. For quantitative PCR (qPCR), gene-specific mRNA transcripts were amplified from cDNA using SYBR Fast qPCR master mix (Kapa Biosystems) and relevant primers in a CFX96 Touch real-time PCR detection system (data not shown). Amplification curves, melting curves, and threshold cycle (CT) values were visualized using CFX Manager 3.0 (Bio-Rad). Relative quantification of mRNA transcripts was performed using the Pfaffl method (71). For reverse transcription-PCR (RT-PCR), transcripts were amplified from cDNA with Taq polymerase using primers mGAPDH F and mGAPDH R or primers mH2afx F and mH2afx R (data not shown) and visualized by gel electrophoresis.
AID-mediated CSR.
CH12 cells were resuspended to a concentration of 200,000 cells/ml in medium containing 2 μg/ml αCD40, 10 ng/ml interleukin-4 (IL-4), and 1 ng/ml transforming growth factor β (TGF-β). Cells were harvested at 2 days poststimulation and stained with anti-IgA–phycoerythrin (PE) prior to analysis of IgA expression by flow cytometry.
Cas9-mediated switching.
CH12 cells were resuspended in electroporation buffer containing 4 μg each of pX330 (Addgene plasmid 42230) encoding Cas9 and relevant sgRNAs (data not shown) and electroporated in a Bio-Rad gene pulser apparatus with the following parameters: 325 V, 975 μF, ∞ Ω, and a 4-mm gap. Cells were harvested at 3 days postelectroporation and stained with anti-IgA-PE or anti-IgG1–fluorescein isothiocyanate (FITC) fluorescently labeled antibodies. Cells were analyzed for IgA or IgG1 expression using a BD FACSCalibur or BD LSR II instrument at the Faculty of Medicine Flow Cytometry Facility (University of Toronto, Toronto, Ontario, Canada).
I-SceI-mediated homologous recombination assay.
Five micrograms of pDR-GFP (Addgene plasmid 26475) was previously stably transfected into CH12 cells (33). DR-GFP CH12 cells were transduced with shRNAs, selected in hygromycin, and electroporated with 5 μg of pCBASceI (Addgene plasmid 26477) or no plasmid as a control (“mock”), using the same parameters as the ones stated above. Following electroporation, cells were immediately placed on ice for 5 min and then plated in complete medium. Cells were harvested at 3 days postelectroporation and analyzed for GFP expression by flow cytometry.
Measuring cell growth.
Cells were seeded at an initial density of 100,000 cells/ml in 96-well tissue culture plates. Cells were counted at 24, 48, and 72 h postseeding using a hemocytometer and trypan blue exclusion.
EdU-PI assay for cell cycle analysis.
In the steady state or 24 h after CIT stimulation, 500,000 cells were pulsed in 10 μM 5-ethynyl-2′-deoxyuridine (EdU) (Click-iT EdU Alexa Fluor 647 imaging kit, catalogue number C10340; ThermoFisher) for 1.5 h at 37°C. Cells were then fixed in 2% formaldehyde and permeabilized in a saponin-based buffer. Cells were incubated in a click chemistry reaction cocktail containing copper sulfate, ascorbic acid, and Alexa Fluor 647-azide for 30 min. Cells were washed and counted prior to incubation with 1.5 mg propidium iodide per 100,000 cells. Cell cycle fractions were analyzed by flow cytometry.
Cell irradiation for induction of genome-wide DNA damage.
CH12 and WEHI-279 cells were resuspended to a concentration of 500,000 cells/ml in complete medium and irradiated using a Nordion Gammacell 1000 irradiator at the Division of Comparative Medicine (University of Toronto). Cells were immediately incubated at 37°C until they were harvested at the indicated time points.
Camptothecin- and etoposide-induced genome-wide DNA damage.
CH12 cells were pulsed in 5 μM camptothecin (catalogue number 7689-03-4; Sigma) or 25 μM etoposide (catalogue number 2200S; New England BioLabs) for 1 h at 37°C in complete medium and then exchanged into normal complete medium. Cells were incubated with an equivalent volume of DMSO as a control treatment.
Western blotting.
Cells were harvested and resuspended in TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8]) with protein loading dye and then incubated at 95°C for 10 min. Cell lysates were electrophoresed on 14% SDS-PAGE gels at 150 V using a Power Pac 1000 instrument (Bio-Rad). Electrophoresed proteins were transferred onto nitrocellulose membranes with a 0.2-μm pore size. Following transfer, membranes were blocked in 5% bovine serum albumin in Tris-buffered saline–Tween (TBST) and then incubated with primary and secondary antibodies prior to colorimetric detection using a MicroChemi 4.2 instrument (DNR Bio-Imaging Systems). Densitometry analysis of Western blot bands was performed using ImageJ (NIH).
Nuclear fractionation.
Cells were lysed in a hypotonic buffer containing 0.05% NP-40 to release the cytoplasmic fraction and isolate nuclei. Nuclei were then lysed in a low-salt buffer containing 1% Triton X-100, resuspended in HCl to release acid-soluble factors, and neutralized with an equivalent volume of Tris base. Protein loading dye was added to the resulting nuclear fraction, followed by incubation at 95°C for 10 min.
CRISPR/Cas9-mediated genome editing.
Cells were incubated in electroporation buffer containing 4 μg of the pX330 plasmid encoding Cas9 and sgRNA targeting Prkdc or H2afx (data not shown) and electroporated using the same parameters as the ones stated above. Single clones were obtained by limiting dilution in 96-well tissue culture plates. Clones were screened for indels using a mismatch cleavage assay as done previously (33). Reduction in mRNA expression was verified by qPCR or RT-PCR. Alleles were sequenced at The Centre for Applied Genomics (TCAG) (Toronto, Ontario, Canada) (data not shown).
Chemical inhibition of ATM and/or ATR.
For ATM inhibition, CH12 and WEHI-279 cells were incubated with 5 and 10 μM KU55933 (catalogue number 3544; Tocris), respectively, for 1 h prior to IR. Cells were incubated with an equivalent volume of DMSO as a control treatment. For ATR inhibition, CH12 cells were incubated with 20 μM VE-821 (catalogue number S8007; Selleck Chemicals) or an equivalent volume of water as a control treatment for 1 h prior to IR. For combination ATM and ATR inhibition, CH12 cells were incubated with 20 μM KU55933 and 20 μM VE-821 or 25 mM caffeine for 1 h prior to IR.
Antibodies.
The following antibodies were used for flow cytometry or Western blotting: β-actin (catalogue number A2066; Sigma), H2B (catalogue number ab1790; Abcam), H2BK120ub (catalogue number 05-1312; EMD Millipore), γH2AX (catalogue number 05-636; EMD Millipore), H2BK120ac (catalogue number 39120; Active Motif), pKap1 (catalogue number 4127; Cell Signaling), H3 (catalogue number ab1791; Abcam), H3K4me3 (catalogue number ab8580; Abcam), H3K79me2 (catalogue number ab3594; Abcam), goat anti-mouse IgA–PE (catalogue number 1040-09; SouthernBiotech), and goat anti-mouse IgG1–FITC (catalogue number 406606; BioLegend).
Statistics.
Statistical comparisons were made using the Mann-Whitney test unless otherwise noted in the figure legends.
ACKNOWLEDGMENTS
We are grateful to the Martin laboratory, Daniel Durocher, and Grant Brown for helpful discussions about this work, to Kefei Yu for the Aicda−/−, Ku86−/−, and LigIV−/− CH12 cells, and to Maria Jasin for the DR-GFP substrate.
C.C.S. is supported by an Ontario graduate scholarship. This research is supported by a grant from the Canadian Institutes of Health Research (grant number PJT-153307) to A.M.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Chapman JR, Taylor MRG, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 2.Asami S, Manabe H, Miyake J, Tsurudome Y, Hirano T, Yamaguchi R, Itoh H, Kasai H. 1997. Cigarette smoking induces an increase in oxidative DNA damage, 8-hydroxydeoxyguanosine, in a central site of the human lung. Carcinogenesis 18:1763–1766. doi: 10.1093/carcin/18.9.1763. [DOI] [PubMed] [Google Scholar]
- 3.Kripke ML, Cox PA, Alast LG, Yarosho DB. 1992. Pyrimidine dimers in DNA initiate systemic immunosuppression in UV-irradiated mice. Proc Natl Acad Sci USA 89:7516–7520. doi: 10.1073/pnas.89.16.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanada K, Budzowska M, Davies SL, Van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 5.Schatz DG, Oettinger MA, Baltimore D. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 6.Oettinger MA, Schatz DG, Gorka C, Baltimore D. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 8.Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375–384. doi: 10.1016/S0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 9.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 10.Lavin MF. 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 11.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, Van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Elledge SJ. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A 104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 19.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AMR, Durocher D. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 20.Fradet-Turcotte A, Canny MD, Escribano-Díaz C, Orthwein A, Leung CCY, Huang H, Landry M-CC, Kitevski-Leblanc J, Noordermeer SM, Sicheri F, Durocher D. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robzyk K, Recht J, Osley MA. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 22.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11:267–274. doi: 10.1016/S1097-2765(02)00802-X. [DOI] [PubMed] [Google Scholar]
- 23.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, Grant PA. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 24.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NCT, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Hake SB, Roeder RG. 2005. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Zhu B, Zheng Y, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. 2005. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schüle R, Takeyama KI, Kato S, Tora L, Devys D. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnet J, Wang C-Y, Baptista T, Vincent SD, Hsiao W-C, Stierle M, Kao C-F, Tora L, Devys D. 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davie JR, Murphy LC. 1990. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 29:4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- 31.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 32.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran S, Haddad D, Li C, Le MX, Ling AK, So CC, Nepal RM, Gommerman JL, Yu K, Ketela T, Moffat J, Martin A. 2016. The SAGA deubiquitination module promotes DNA repair and class switch recombination through ATM and DNAPK-mediated γH2AX formation. Cell Rep 15:1554–1565. doi: 10.1016/j.celrep.2016.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang S-Y, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC-T, Chen DJ, Oren M, Shiloh Y. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DVNP, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, Iacovoni JS, Rowicka M, Ginalski K, Côté J, Legube G. 2018. Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol Cell 72:250–262. doi: 10.1016/j.molcel.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. 2011. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Irrazabal T, So CC, Berru M, Du L, Lam E, Ling AK, Gommerman JL, Pan-Hammarström Q, Martin A. 2018. The H2B deubiquitinase Usp22 promotes antibody class switch recombination by facilitating non-homologous end joining. Nat Commun 9:1006. doi: 10.1038/s41467-018-03455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Tarcic O, Pateras IS, Cooks T, Shema E, Kanterman J, Ashkenazi H, Boocholez H, Hubert A, Rotkopf R, Baniyash M, Pikarsky E, Gorgoulis VG, Oren M. 2016. RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep 14:1462–1476. doi: 10.1016/j.celrep.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura M, Kondo S, Sugai M, Nazarea M, Imamura S, Honjo T. 1996. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol 8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 42.Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, Nussenzweig MC, Chen J. 2004. 53BP1 is required for class switch recombination. J Cell Biol 165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. 2004. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFκB. Int Immunol 16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 44.Stavnezer-Nordgren J, Sirlin S. 1986. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J 5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling AK, So CC, Le MX, Chen AY, Hung L, Martin A. 2018. Double-stranded DNA break polarity skews repair pathway choice during intrachromosomal and interchromosomal recombination. Proc Natl Acad Sci U S A 115:2800–2805. doi: 10.1073/pnas.1720962115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrader CE, Guikema JEJ, Linehan EK, Selsing E, Stavnezer J. 2007. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol 179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 48.Wojcik F, Dann GP, Beh LY, Debelouchina GT, Hofmann R, Muir TW. 2018. Functional crosstalk between histone H2B ubiquitylation and H2A modifications and variants. Nat Commun 9:1394. doi: 10.1038/s41467-018-03895-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 50.Ng HH, Xu RM, Zhang Y, Struhl K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 51.Sun ZW, Allis CD. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 52.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gatta R, Dolfini D, Zambelli F, Imbriano C, Pavesi G, Mantovani R. 2011. An acetylation-mono-ubiquitination switch on lysine 120 of H2B. Epigenetics 6:630–637. doi: 10.4161/epi.6.5.15623. [DOI] [PubMed] [Google Scholar]
- 54.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Löbrich M, Jeggo PA. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 64:2390–2396. doi: 10.1158/0008-5472.CAN-03-3207. [DOI] [PubMed] [Google Scholar]
- 55.Falck J, Coates J, Jackson SP. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 56.Shiloh Y, Ziv Y. 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 57.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NMB, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GCM. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 58.Han L, Yu K. 2008. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J Exp Med 205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nussenzweig A, Chen C, Soares VDC, Sanchez M, Sokol K, Nussenzweig MC, Li GC, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 60.Nussenzweig A, Sokol K, Burgman P, Li L, Li GC. 1997. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci U S A 94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, Jackson SP. 1995. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 62.Manis JP, Dudley D, Kaylor L, Alt FW. 2002. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity 16:607–617. doi: 10.1016/S1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 63.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM-and KAP-1 dependent pathway. Nat Cell Biol 8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 64.Caron P, Choudjaye J, Clouaire T, Bugler B, Daburon V, Aguirrebengoa M, Mangeat T, Iacovoni JS, Álvarez-Quilón A, Cortés-Ledesma F, Legube G. 2015. Non-redundant functions of ATM and DNA-PKcs in response to DNA double-strand breaks. Cell Rep 13:1598–1609. doi: 10.1016/j.celrep.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward IM, Chen J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 66.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev 14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 67.Myers JS, Cortez D. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams KE, Medhurst AL, Dart DA, Lakin ND. 2006. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamper AM, Rofougaran R, Watkins SC, Greenberger JS, Beumer JH, Bakkenist CJ. 2013. ATR kinase activation in G1 phase facilitates the repair of ionizing radiation-induced DNA damage. Nucleic Acids Res 41:10334–10344. doi: 10.1093/nar/gkt833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Z, Elledge SJ, Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 71.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]