Abstract
Although mass spectrometry is well-suited to identifying thousands of possible protein post-translational modifications (PTMs), it has historically been biased towards just a few. To measure the entire set of PTMs across diverse proteomes, software must overcome the dual challenges of searching enormous search spaces and distinguishing correct from incorrect spectrum interpretations. Here, we describe TagGraph, a computational tool that overcomes both challenges with an unrestricted string-based search method that is as much as 350-fold faster than existing approaches, and a probabilistic validation model we optimized for PTM assignments. We applied TagGraph to a published human proteomic data set of 25 million mass spectra and tripled confident spectrum identifications compared its original analysis. We identified thousands of modification types on almost one million sites in the proteome. We show new contexts for highly abundant yet understudied PTMs such as proline hydroxylation, and its unexpected association with cancer mutations. By enabling broad PTM characterization TagGraph informs how their functions and regulation intersect.
Editor Summary:
Swift, robust identification of post-translational modifications in MS/MS data sets is enabled by a string-based computational tool.
Introduction
Post translational modifications (PTMs) dynamically modulate the activity, conformation states, localization, interactions, abundance and degradation of almost all proteins encoded by the human genome1,2. However most remain poorly understood since mapping the full breadth of PTM identities and locations across the entire human proteome has remained intractable3. Although mass spectrometry is arguably the best-suited technology for direct, large-scale PTM measurement, most global PTM studies have focused on modifications for which optimized enrichment workflows exist4. Consequently, our current view of PTMs’ collective impact on the human proteome is heavily skewed towards a small fraction of the possible PTM landscape5.
Despite this, PTM-containing peptides are readily detected by routine tandem mass spectrometry (MS/MS) experiments, but are believed to comprise much of the “dark matter” in proteome datasets that consistently evades reliable identification6. Search parameters used by most MS/MS search approaches strike a compromise between the diversity of discoverable modifications, the time needed to interpret MS/MS datasets and the ability to distinguish correct from incorrect assignments. Strategies that constrain the number of proteins being searched, apply protease specificity rules or minimize the allowable types and numbers of PTMs partially address this compromise7. In practice, however, these approaches only marginally decrease search times and still do not distinguish correct from incorrect PTM assignments8.
Here, we describe TagGraph, a computational tool that addresses these limitations in two ways. First, TagGraph leverages accurate de novo mass spectrum interpretations9, efficient string-based indexing10, and a graph-based string reconciliation algorithm to rapidly search MS/MS data sets without restricting the number of proteins, PTMs or protease specificities. Second, by replacing conventional “target-decoy” error estimation11 with a PTM-optimized probabilistic model, we can accurately discover and discriminate high-confidence peptide identifications from large search spaces. Combined, these advances make unconstrained surveys of large MS/MS data sets possible. Our analyses expand the number of known modification sites for some PTM types by as much as 30-fold, particularly those lacking biochemical enrichment techniques. We show that TagGraph enables unbiased and rapid proteome-wide PTM characterization.
Results
Development of TagGraph
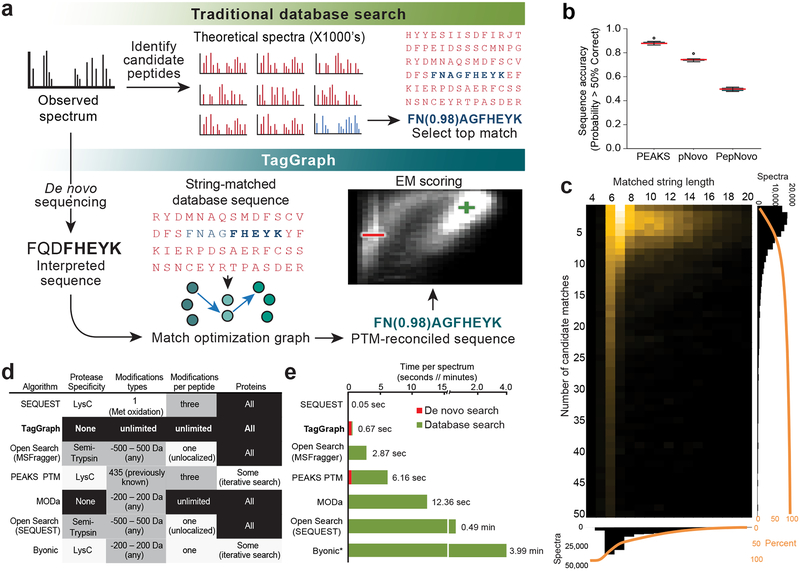
We developed TagGraph (Supplementary Code) to accurately assign peptides bearing multiple unspecified PTMs or amino acid substitutions (Fig. 1a, Supplementary Fig. 1). TagGraph leverages the speed of indexed string matching algorithms by first transforming complex, numeric mass spectra into discrete, unambiguous query strings using de novo peptide sequencing. De novo sequencing algorithms are more sensitive to MS/MS spectra imperfections than traditional database search engines, and therefore have not been widely used for large-scale proteomics analysis. However, we recently showed that they frequently produce long, reasonably accurate sequence predictions from high resolution MS/MS spectra: these predictions were over 50% correct for nearly all interpretable MS/MS spectra (Fig. 1b)9. We therefore suspected that many de novo peptides contain long (e.g., >5 amino acids) substrings that perfectly match the true protein sources of their observed MS/MS spectra. The FM-index data structure10 was developed to facilitate this kind of search. TagGraph uses it to rapidly find a small number of substring-matched candidate peptides from an arbitrarily large, pre-indexed sequence database with no restrictions on protease specificity, post-translational modifications, or sequence variants (Fig. 1c). These candidates are then reconciled against the input de novo sequence using a graph-based alignment algorithm. This strategy lets TagGraph discover and localize multiple PTMs and other sequence alterations that co-occur on a single peptide sequence without anticipating them a priori (Supplementary Note 1). Modification masses localized to specific amino acids are cross-referenced with the Unimod resource3 to suggest the modification’s identity based on mass and amino acid specificity. In this way, TagGraph effectively searches all possible sequence alterations on time scales and search sensitivities commensurate with conventional database search tools (Fig. 1d, 1e, Dataset 1 (https://taggraph.page.link/Datasets)).
Figure 1. TagGraph efficiently manages large proteome search spaces through flexible, long string matches.
a) Traditional versus TagGraph workflow. b) Most de novo-interpreted high-resolution MS/MS spectra are mostly correct. The proportion of analyzed spectra interpreted with over 50% sequencing accuracy by PEAKS56, pNovo57, and PepNovo58 on a data set of 168,391 MS/MS spectra derived from the A375 melanoma cell line (n=12 fractions evaluated in parallel). Central line denotes medians; whiskers indicate the 1.5 interquartile range, and circles indicate data points outside this range. c) TagGraph’s longest matching substring criterion (Supplementary Fig. 1, Supplementary Note 1) yields very few candidate sequences for subsequent refinement and scoring. Heat map depicts the number of candidate peptide matches per spectrum (y-axis) versus the longest-matching substring used for candidate retrieval. Over 50% of all spectra TagGraph considered from the A375 data set were selected from <6 candidates, and over 50% matched with sub-string lengths >6 amino acids. d) TagGraph considered an entirely unrestricted peptide search space (black) with respect to protease specificity, range of modifications, number of modifications per peptide, and number of proteins. The same A375 data set was searched with conventional and expanded database search methods, but with one or more major (white) or modest (grey) space restrictions. Restrictions were selected so as to approach TagGraph’s unrestricted parameters while allowing each search engine to execute without failure (see Online Methods). e) The combined de novo sequencing and TagGraph search times on the A375 data set were 4.3-fold shorter than previously described expanded modification and iterative search strategies, even when the latter were given considerably reduced search spaces (Fig. 1d). Search rates for TagGraph and PEAKS PTM included the preliminary de novo sequencing step using PEAKS (red; 0.51 sec/spectrum). Excluding the de novo step, TagGraph was 17.9-fold faster than the next-ranked search method. *Extrapolated values: Byonic was run on just 1/12 of the A375 data set due to search speed limitations (see Online Methods, Dataset 1 (https://taggraph.page.link/Datasets)).
TagGraph surveys very large sequence spaces that would be impractical to query using traditional database search engines and other flexible search methods. Our strategy contrasts with prior approaches including sequence tag filtering12, iterative search13, precursor mass shift14,15 and spectrum clustering16 (Supplementary Note 2). Compared to an equivalent database search, these approaches reduce the number of peptides considered per mass spectrum (or vice versa for spectrum clustering). However, they are subject to similar speed limitations as they consider larger numbers of amino acid modifications and variants. As a result, most are prone to impractically long search times (Supplementary Note 2). TagGraph addresses this obstacle by shifting the computational burden of generating many numerical pattern comparisons to an efficient string matching and reconciliation procedure. We measured the resulting speed advantage by comparing TagGraph’s execution time to five algorithms designed to consider greatly expanded search spaces12–15,17, and found that none could execute on both the entire data set and the search space TagGraph considered in this comparison. Recognizing that most of these methods were not designed to consider such unrestricted parameters, we provided them with reduced number of spectra, search spaces or both. Nevertheless, TagGraph was 4- to 18-fold faster than any other algorithm, depending on whether we considered its preliminary de novo sequencing step (Fig. 1d, 1e). We note that more restrictive search parameters than the ones tested here could be expected to produce substantially faster search times for some of these prior methods. In contrast, however, TagGraph has just one unrestricted operation mode removing researchers’ need to choose between search speed and search depth.
Hierarchical Bayes Model for decoy-free error estimation
Indexed string searches solve the conflict between search speed and search depth, but exacerbate the challenge of estimating reliable FDRs. The standard target-decoy estimation method we previously developed11 is unsuitable for unconstrained search results since it loses discrimination accuracy as more peptides and PTMs are considered (Supplementary Note 3)8,18,19. Consequently, we developed a probabilistic validation strategy using a hierarchical Bayes Model optimized by expectation maximization (EM) (Supplementary Figs. 2, 3)20. Our model deduces the likelihood that any individual peptide-spectrum match is correctly interpreted based on fourteen quantitative and categorical attributes (Supplementary Note 4.B); of these, half relate specifically to modified peptides. Consequently, our model concurrently discriminates correctly and incorrectly interpreted spectra from both modified and unmodified peptides.
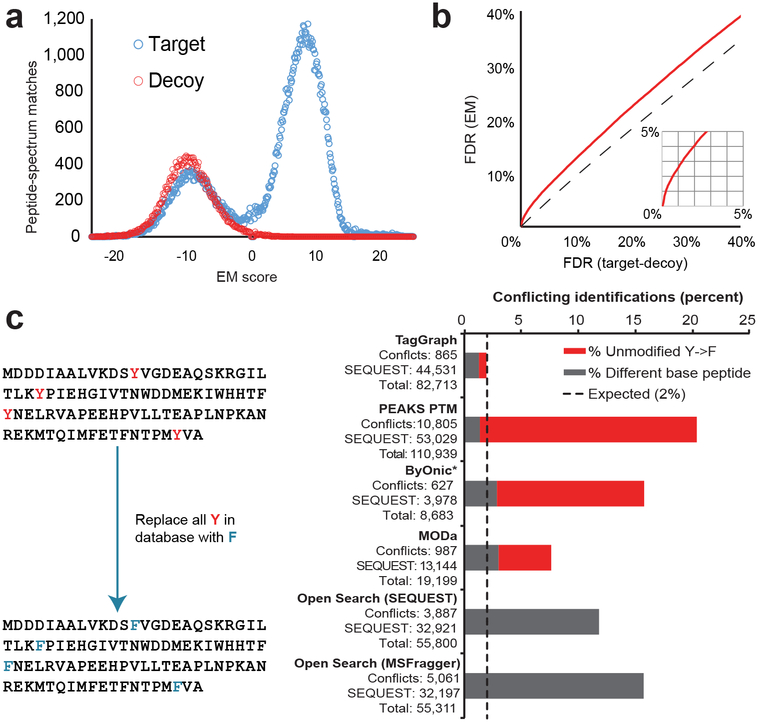
We first evaluated TagGraph’s error model by comparing it to target-(shuffled) decoy database searching (Online Methods) with SEQUEST using the cell line dataset described in Figure 1. We found that the EM-based scoring discriminated decoys from high-confidence identifications (Fig. 2a) while generating FDR estimates which were more conservative than those inferred from target-decoy searches (Fig. 2b, Dataset 2 (https://taggraph.page.link/Datasets)), as expected for a model that discriminates between correct and incorrect PTM assignments. Furthermore, we found that the extent to which SEQUEST and TagGraph disagreed was consistent with the estimated 1% FDR threshold we applied to both (Supplementary Fig. 4a). For the majority of these disagreements, however, TagGraph-generated peptide-spectrum matches were more consistent with correct identifications based on protease specificity, algorithm-assigned scores, and ion assignment (Supplementary Figs. 4b, 5). Twelve other large data sets analyzed in a similar manner yielded comparable observations (Supplementary Analysis 1).
Figure 2. Single model estimates modified and unmodified peptide identification errors without decoys.
Correct and incorrect TagGraph search results were discriminated with a hierarchical Bayes probabilistic model optimized by Expectation Maximization (EM) to score identification confidence and estimate false discovery rates (Supplementary Note 4, Supplementary Figure 2). a) The A375 data set (Fig. 1) was searched against a composite target-decoy sequence database (Online Methods) with TagGraph and evaluated by EM (Dataset 1 (https://taggraph.page.link/Datasets)). Decoys were effectively discriminated with this approach although the EM was blind to the decoy label in this analysis and in 12 other data sets (Supplementary Analysis 1a). b) EM was generally consistent with, but more conservative than target-decoy-based FDR estimates when both were applied to TagGraph results. This is expected considering target-decoy’s inability to distinguish correct and incorrect modifications. These findings were reproduced considering 12 other data sets (Supplementary Analysis 1b). c) The human proteome sequence database was modified, substituting every tyrosine residue with a phenylalanine (mass difference of −15.994915 Da). Unrestricted search error was estimated from the frequencies of “base” peptide sequences (grey) or substituted phenylalanine-containing peptides (red) reported by each algorithm that conflicted with results returned by a standard database search with SEQUEST (Online Methods). Only TagGraph reported results with an empirically calculated error rate close to the expected 1.99% upper limit, assuming each algorithm’s results were filtered to a 1% FDR (Online Methods). Open Search methods using SEQUEST14 and MSFragger15 were not evaluated for incorrect PTM localization because they do not directly localize modifications to specific residue positions. Values are reported for peptide-spectrum matches as provided in Dataset 1 (https://taggraph.page.link/Datasets). *Byonic was run on just 1/12 of the A375 data set due to search speed limitations (see Online Methods).
To further evaluate TagGraph’s error model, we sought to measure how often it misassigns modifications to incorrect amino acid sites. To accomplish this, we replaced all tyrosine residues with phenylalanines (mass difference of one oxygen, 15.9995 Da) in an altered human proteome sequence database (Fig. 2c). We reasoned that an accurate expanded search algorithm should return phenylalanine-containing peptides with an additional oxygen localized to converted phenylalanines. We benchmarked five search methods against TagGraph with this validation tool. Each algorithm’s results were filtered based on target-decoy-based criteria (either the algorithm’s own implementation or a linear discriminant analysis21) or, for TagGraph, the hierarchical Bayes model. The proportion of peptide-spectrum matches containing unmodified phenylalanines at tyrosine positions was used to estimate both sequence- and modification-specific FDRs relative to the 1% predicted FDR. Only TagGraph’s error model reliably discriminated both incorrect “base” peptide sequences and those bearing unmodified altered phenylalanines. TagGraph reported 3.5-fold fewer inconsistencies with the expected search results than the next-best flexible search method (MODa) but with 4.3-fold greater sensitivity (Fig. 2c). These data suggest that both incorrect sequences and modifications may be common when searching multiple types of target-decoy protein databases (Supplementary Fig. 6).
Stringent error tolerance often comes at the expense of overall analysis sensitivity11. However, we found that TagGraph doubled the number of unique peptide identifications relative to SEQUEST, in part, by enabling accurate identification of peptides with any modification state. Once reconciled with Unimod, we found that unanticipated post-isolation modifications accounted for the majority of this increase (50.2% of novel modifications), followed by biologically-regulated post-translational modifications (8.5%) and those with no previous association (2.1%) (Supplementary Fig 4a). This analysis demonstrated TagGraph’s unique ability to characterize modified peptides with the speed, accuracy and sensitivity that is compatible with large-scale proteomic workflows.
To further verify TagGraph’s ability to generate bona fide PTMs, we analyzed 451,655 MS/MS spectra from a published phosphorylation-enriched data set22. We first established a set of high-confidence phosphorylated peptides by searching these spectra with SEQUEST and localizing phosphates to specific amino acid residues with the A-score algorithm23. In parallel, we searched these spectra with the four expanded search approaches demonstrating the greatest throughput: TagGraph, Peaks-PTM, Open Search and MSFragger (Fig. 1e). Comparing TagGraph with PEAKS-PTM, we found similar proportions of phosphorylated peptides (30% vs 31%, respectively), and agreement with high-confidence SEQUEST phosphopeptide identifications (98% vs 99%) (Supplementary Fig. 7, Dataset 3 (https://taggraph.page.link/Datasets)). Furthermore, we found that TagGraph rarely (1–2%) yielded tied peptide candidates for individual spectra (Dataset 4 (https://taggraph.page.link/Datasets), Supplementary Table 1), supporting our scoring model’s ability to discriminate between alternate modification configurations. The Open Search and MSFragger methods produced proportionately fewer phosphopeptide identifications (24 and 22%) which were not inherently localized to specific amino acids due to the general nature of this approach (Supplementary Note 2).
The human proteome’s broad modification landscape
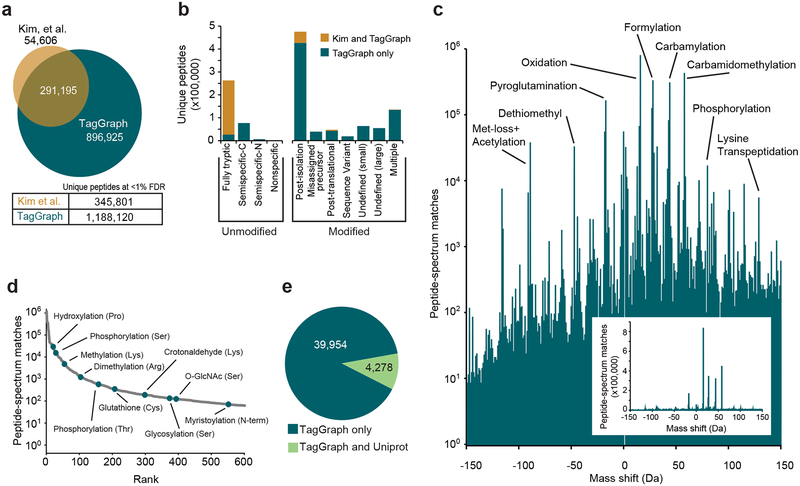
To demonstrate TagGraph’s utility for deep PTM characterization, we re-analyzed a draft human proteome data set24, which is approximately 150 times larger than our initial test data set. Interpreting these 25 million tandem mass spectra – derived from 30 adult and fetal tissues and over 2,000 raw data files – would not be feasible with prior expanded search approaches, but took just six days on a single desktop computer once de novo sequencing was complete (one month of total running time). These data yielded over 1.1 million unique peptides, tripling the number originally reported using traditional database searching (Fig. 3a, Dataset 5 (https://taggraph.page.link/Datasets)). As with the phosphorylation data set described above, the human proteome analysis yielded few tied spectra (Supplementary Table 1, Dataset 6 (https://taggraph.page.link/Datasets)), further supporting our scoring and validation model. Although the analysis presented here focused on identification and FDR estimations for peptides rather than proteins, we nevertheless found evidence for 100 (Adult CD8+ T Cells) to over 600 (Adult Gallbladder) additional proteins per tissue that were not originally reported (Supplementary Fig. 8a, Dataset 7 (https://taggraph.page.link/Datasets)). Several of these were corroborated by histological staining (Supplementary Fig. 8b).
Figure 3. TagGraph extends deep proteome characterization to post-translational modifications.
a) TagGraph confirmed the majority of identifications made by Kim et al.24 (Dataset 5 (https://taggraph.page.link/Datasets)), but also expanded unique peptide identities from the human proteome dataset over three-fold relative to those originally reported. b) Categorical breakdown of unique peptide forms (distinguishing PTMs) identified by TagGraph. As expected, the majority of peptides identified by both TagGraph and Kim et al. correspond to tryptic peptides. Peptides identified by TagGraph but not Kim et al. primarily originated from non-tryptic peptides and peptides with unanticipated modifications. Post-isolation modifications comprised the most prevalent identification category in this dataset. c) Mass shifts (modified amino acid mass – unmodified amino acid mass) corresponding to all modifications identified by TagGraph from the human proteome dataset reveal a complex modification landscape. Numbers of identifications (peptide-spectrum matches) span six orders of magnitude. Despite the presence of several highly abundant post-isolation modifications (e.g., formylation), the depth of the proteomic profiling achieved in this dataset made it possible to characterize lower abundance post-translational modifications. Inset: modification frequencies without log transformation. d) Ranked relative abundances of 2,576 PTM-amino acid combinations, as estimated by the number of spectra bearing each from the human proteome dataset. Ten of these are highlighted; all modifications are represented in Dataset 9 (https://taggraph.page.link/Datasets). e) Over 90% of modification sites (39,954 of 44,232) identified by TagGraph from the human proteome dataset were not among 52,959 previously described in Uniprot. However, the overlap in the sites reported by TagGraph and Uniprot (4,278) is highly significant (p-value = 1e-308, one-tail Fisher’s exact test).
As with our cell line analysis (Supplementary Fig. 4), TagGraph rescued peptides bearing at least one modification that was not considered in the original search (Fig. 3b). A small number of post-isolation modifications (methionine oxidation; N-terminal carbamylation, carbamidomethylation, and formylation) collectively accounted for 38% of modified spectra (Fig. 3c, Table 1), as previously observed12,14,15,25,26. TagGraph rescued other commonly disregarded peptide classes, including semi-specific and non-specific trypsin cleavage, and misassigned monoisotopic precursor masses (Fig. 3b, Supplementary Fig. 9). We note that these general identification classes persisted when we searched low-resolution MS/MS spectra with TagGraph, but the overall identification success rate was reduced two-fold relative to conventional database search (Supplementary Fig. 10, Dataset 8 (https://taggraph.page.link/Datasets)). This coincided with decreased performance of our EM model on this data set (Supplementary Analysis 1). Thus, our ability to probe post-translationally modified proteomes depends on the mass resolution of the underlying MS/MS spectra.
Table 1.
Top 10 Post-isolation, post-translational, and previously uncharacterized amino acid modifications identified from the Kim et al. dataset.
| Categorya | Modificationb | Specificityc | Mass Shiftd | Unique Peptidese | Peptide-Spectrum Matchesf | Sitesg |
|---|---|---|---|---|---|---|
| Post-isolation | Carbamylation | Peptide N-term | +43.01 | 63,302 | 254,382 | 70,207 |
| Formylation | Peptide N-term | +27.99 | 47,329 | 293,863 | 57,371 | |
| Carbamidomethylation | Peptide N-term | +57.02 | 42,310 | 304,898 | 52,899 | |
| Oxidation | Met | +15.99 | 42,193 | 784,001 | 41,115 | |
| Deamidation | Asn | +0.98 | 21,064 | 252,713 | 22,609 | |
| Acetaldehyde | Peptide N-term | +26.02 | 15,016 | 132,086 | 20,402 | |
| Deamidation | Gln | +0.98 | 8,911 | 36,897 | 13,015 | |
| Gln->pyroglutamate | Peptide N-term Gln | −17.03 | 8,372 | 123,277 | 7,966 | |
| Carbamidomethylation | Lys | +57.02 | 7,575 | 40,422 | 12,355 | |
| Carbamylation | Lys | +43.01 | 6,932 | 30,245 | 11,409 | |
| Post-translational | Phosphorylation | Ser | +79.97 | 3,466 | 15,798 | 3,729 |
| Met-loss + Acetyl | Protein N-term Met | −89.03 | 2,391 | 39,799 | 1,705 | |
| Hydroxylation | Pro | +15.99 | 1,901 | 26,348 | 3,079 | |
| Citrullination | Arg | +0.98 | 1,551 | 5,328 | 2,020 | |
| GlyGly | Lys | +114.04 | 871 | 2,403 | 1,613 | |
| Allysine | Lys | −1.03 | 861 | 2,801 | 1,670 | |
| Hydroxylation | Lys | +15.99 | 766 | 3,316 | 1,090 | |
| Cyano | Cys | −32.03 | 601 | 2,591 | 567 | |
| Carboxylation | Glu | +43.98 | 467 | 726 | 913 | |
| Acetylation | Lys | +42.01 | 455 | 1,323 | 1,196 | |
| Previously undefined | Unknown | Peptide N-term | +12.00 | 3,834 | 14,743 | 3,692 |
| Unknown | Peptide N-term | +51.01 | 2,569 | 8,834 | 2,698 | |
| Iron(III)h | Asp, Glu | +52.92 | 2,552 | 9,843 | 1,468 | |
| carbamidomethyl and formyl on same residueh | Peptide N-term | +85.02 | 1,632 | 4,848 | 2,072 | |
| disulfide bondh | Cys with nearby Cys | −116.06 | 1,529 | 9,788 | 1,840 | |
| carbamidomethyl on C-terminus or Argh | Peptide C-term Arg | +57.02 | 1,372 | 3,914 | 3,150 | |
| Unknown | Peptide N-term | +83.04 | 1,280 | 4,632 | 1,948 | |
| carbamidomethyl and pyro-glutamination on same residueh | Peptide N-term Glu | +39.01 | 1,099 | 3,622 | 908 | |
| Unknown | Peptide N-term | +23.98 | 1,059 | 2,919 | 1,381 | |
| reaction of N-terminal carbamidomethyl with internal Meth | Peptide N-term, co-occurs with dethiomethyl modification of internal Met | +105.02 | 957 | 4,907 | 991 |
Top ten modifications of the three indicated categories are shown, ordered by the number of unique peptides identified in the Kim et al. human proteome dataset. Categories assigned based on the likely modification identity, as determined by TagGraph.
Modification identities assigned by TagGraph, based on observed mass shifts, modification specificity, and evidence in the Unimod resource.
Specificity determined from the sites within modified peptdies to which observed mass shifts were assigned.
Mass shift measured from the difference between an observed amino acid residue’s monoisotopic mass and the expected value. Negative values indicate a net mass loss.
Number of unique peptide sequences bearing the annotated modification. Does not include peptide sequences identified with multiple modifications.
Total number of spectra in which indicated modification was confidently identified.
Total number of distinct amino acid residue sites bearing indicated modification.
Hypothetical identity of mass shift
In comparison to the handful of abundant yet biologically irrelevant post-isolation modifications, this extremely deep proteome analysis revealed a much wider array of lesser-abundant PTMs (Fig. 3c,d, Dataset 9 (https://taggraph.page.link/Datasets)). For example, we found N-terminal myristoylation, lysine hydroxylation and arginine dimethylation hundreds to thousands of times in the proteome without requiring the kind of targeted, sample-intensive enrichment procedures that have previously been essential to PTM analysis. This study confirmed 4,278 modifications previously reported in the Uniprot proteomics resource, while extending it by an additional 39,954 (Fig. 3e, Dataset 10 (https://taggraph.page.link/Datasets)). To evaluate the extent to which marginal, incorrect identifications might disproportionately inflate our modification tally, we manually examined 100 diverse modification-bearing peptide assignments from this human proteome dataset, which were assigned scores just at the threshold of acceptance. We found that just 26 were clearly incorrect as deemed by expert scrutiny, coinciding with an overall expected increased FDR for these proportionally rare and marginal results (Supplementary Analysis 1d, Supplementary Analysis 2, Dataset 11 (https://taggraph.page.link/Datasets)). Comparing MS/MS spectra from these datasets to spectra derived from synthetic peptides (Supplementary Analysis 3) further served to validate 75 unexpected, yet high-scoring identifications.
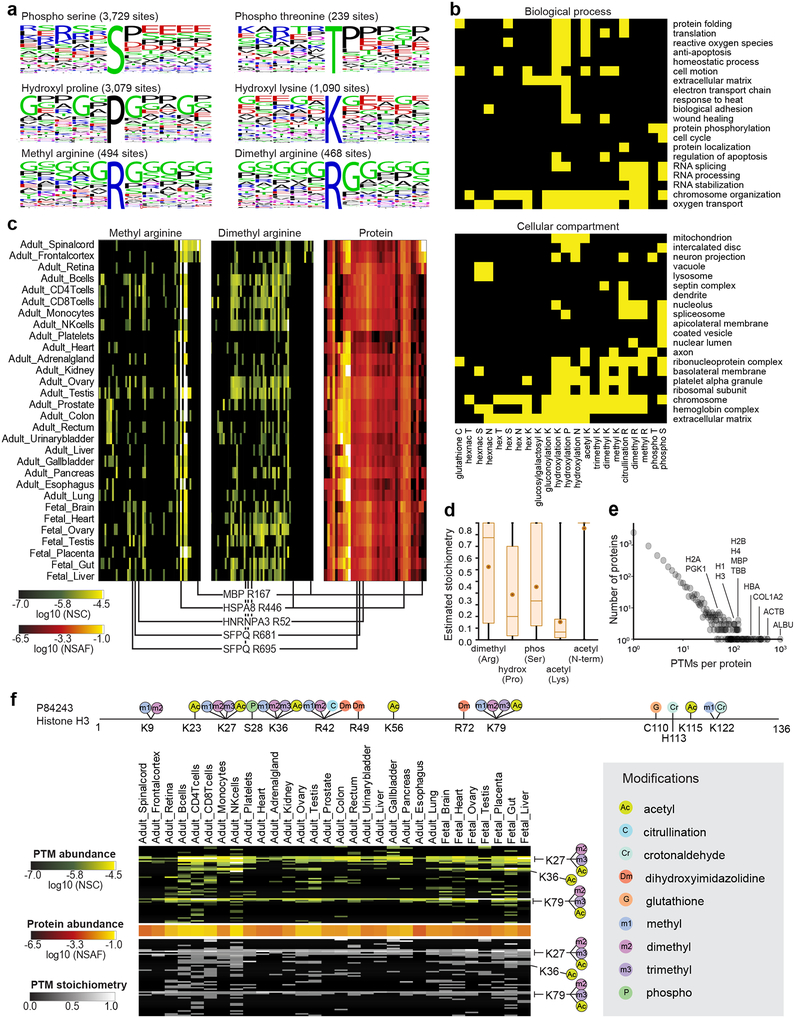
Many PTMs act as reversible switches on protein function. Their enzymatic addition and removal regulates signaling networks, protein binding, and other cellular processes2,27. Although more than 90% of TagGraph-identified PTMs were previously unreported, we found several known PTM-flanking sequence motifs enriched in this dataset (e.g. proline-directed phosphorylation21 and glycine-directed arginine methylation28) (Fig. 4a, Supplementary Fig. 11). We also identified over 200 gene ontologies that were significantly (Benjamini-corrected Fisher’s Exact p-value <0.001) enriched among proteins bearing 22 noteworthy PTMs (Fig. 4b, Supplementary Fig. 12, Dataset 12 (https://taggraph.page.link/Datasets)). This analysis confirmed biological processes known to be regulated by multiple PTMs5 (e.g., acetyl Lys, methyl Lys, phosphorylated Ser regulating chromatin function27). Other processes, such as the cell cycle, were associated with a much more restricted set of PTMs (phosphorylated Ser)29.
Figure 4. TagGraph reveals insights into PTM dynamics, function, and regulation.
a) Sequence logos corresponding with select TagGraph-identified PTMs, as generated by WebLogo59. b) Significantly enriched gene ontologies associated with prevalent post-translational modifications (yellow, 1% FDR (one-tail Fisher’s exact probability, Benjamini-Hochberg corrected)). Ontologies and significances were assigned with the DAVID web tool60. See Dataset 12 (https://taggraph.page.link/Datasets) for the entire list enriched ontologies. Ontologies significantly enriched among post-isolation modifications were excluded to correct for abundance-biased PTM detection (Supplementary Fig. 12). c) Arginine methylation and dimethylation distribution across proteins and tissues. The 64-most abundant monomethylated or dimethylated Arg sites from the entire data set are displayed across the y-axis, along with corresponding protein expression levels (49 proteins). Three modification sites on HNRNPA3 and SFPQ are highlighted for their distinct arginine monomethylation and dimethylation patterns across the tissues, despite demonstrating near uniform protein levels. Methyl modifications on MBP and HSPA8 are highlighted for their tissue specificity and ubiquity (respectively). Proteins were ordered by hierarchical clustering. PTMs were arranged to match their substrate proteins. All methylation sites are reported in Dataset 13 (https://taggraph.page.link/Datasets). d) Stoichiometry distributions vary for different PTMs, giving insight into their regulation and function. Box plots indicate the average (circle) and median (horizontal bar) values, 25th quartile and 75th quartile (box), and minimum and maximum (whiskers) stoichiometry values, measured from n=920 (dimethyl arginine), n=14,888 (hydroxylproline) n=9,287 (phosphoserine), and n=17,816 (n-terminal acetylation) site and tissue combinations. e) Several proteins were found to be heavily modified in this data set. Histogram shows the number of proteins identified with the indicated number of distinct PTMs (site and modification). Of note, 921 distinct PTM sites were identified for human serum albumin. f) TagGraph identified both known and novel Histone H3 PTM sites (Dataset 15 (https://taggraph.page.link/Datasets)); a subset of which are shown. Site positions are numbered excluding the initiating methionine. PTMs circled in black are present in Uniprot. See Dataset 15 (https://taggraph.page.link/Datasets) for more detailed Histone PTM annotations.
We found that 19 of the 22 PTMs represented in Figure 4b were enriched in multiple biological processes or cellular compartments. For example, reversible arginine methylation dynamically regulates proteins involved in RNA splicing and stabilization30, as confirmed by our ontology analysis (Fig. 4b). We observed a relative increase in the mono- and di-methylation site abundances on RNA splicing proteins such as HNRNPA3 and SFPQ in reproductive tissues and lymphocytes (Fig. 4c, Dataset 13 (https://taggraph.page.link/Datasets)), suggesting that these modifications have specific roles in these contexts.
Quantifying PTMs without biochemical enrichment
Since PTMs and their host proteins can be simultaneously quantified by mass spectrometry, we accordingly estimated each PTM’s stoichiometry (Online Methods, Dataset 14 (https://taggraph.page.link/Datasets))22. This method contrasts with previous PTM stoichiometry assays which required metabolic labeling31 or enzymatic removal of a single target PTM class32. Since PTM stoichiometries can have important implications for substrate protein activity and function, deeply sequenced proteome datasets like this are an untapped resource for measuring a wide range of protein regulation. We found that protein N-terminal acetylation demonstrated the most consistently high stoichiometry (95.5%; stdev = 16.7%, Fig. 4d). This is expected, considering the broad and irreversible acetyl group addition, co-translationally catalyzed by N-terminal acetyltransferases33. Conversely, we found that lysine acetylation exhibited low and variable stoichiometry (15.2%; stdev = 22.7%, Fig. 4d), consistent with its heterogeneous representation on histone proteins34, and its possible non-enzymatic origins on abundant cytosolic and mitochondrial proteins35,36. We found that neither PTM abundance nor stoichiometry correlated with substrate protein abundance (Supplementary Fig. 13), supporting the complementary use of both measurements in proteome characterization.
Characterizing multiple PTM types on highly modified proteins
TagGraph identified multiple intersecting PTMs on individual proteins, and on individual residues (Fig 4c, Dataset 10 (https://taggraph.page.link/Datasets)), with hundreds of PTMs identified on proteins such as albumin (921 PTMs) and actin (514 PTMs) (Fig. 4e). Histones are known to undergo extensive and combinatorial modifications to encode epigenetic information27, but deciphering these modifications has previously required individual histone isoform37 or specific modification38 enrichment. Using TagGraph, we identified 273 PTMs across the five major histone proteins, 128 of which were not previously reported (Dataset 15 (https://taggraph.page.link/Datasets)). While we found modifications such as K28 dimethylation and K80 methylation on Histone H3 were both abundant and ubiquitous across the tissues examined here (Fig. 4f), we note several tissue-specific PTMs, such as Histone H4 R56 dimethylation occurring with 25-fold greater abundance in fetal than adult tissues. Twenty-six diverse PTMs showed similarly greater abundance in fetal tissues, suggesting specific roles in development (Dataset 15 (https://taggraph.page.link/Datasets)). Systematically evaluating these modifications in conjunction with the rest of the proteome, and without targeted enrichment techniques, supports TagGraph’s use for this type of secondary analysis.
Enrichment-free discovery and characterization of protein hydroxylation
We found that hydroxylated prolines, tyrosines and lysines were a sizeable (16%) proportion of histone PTMs (Dataset 15 (https://taggraph.page.link/Datasets)), yet only hydroxylated tyrosine was previously described39. Proline hydroxylation is the most abundant modification in the human body40, but just 321 sites have been catalogued41. Unlike more widely studied modifications, no enrichment methods have facilitated its focused analysis. Furthermore, of 11 amino acids capable of becoming hydroxylated3, four (Met, Trp, Phe, His) are often hydroxylated by sample preparation protocols. Thus, true post-translational proline hydroxylation must be distinguished from mislocalized artifacts23. TagGraph’s modification-focused error model allowed us to identify and localize over 10-fold more hydroxyl proline residues than were previously known in humans (Table 1).
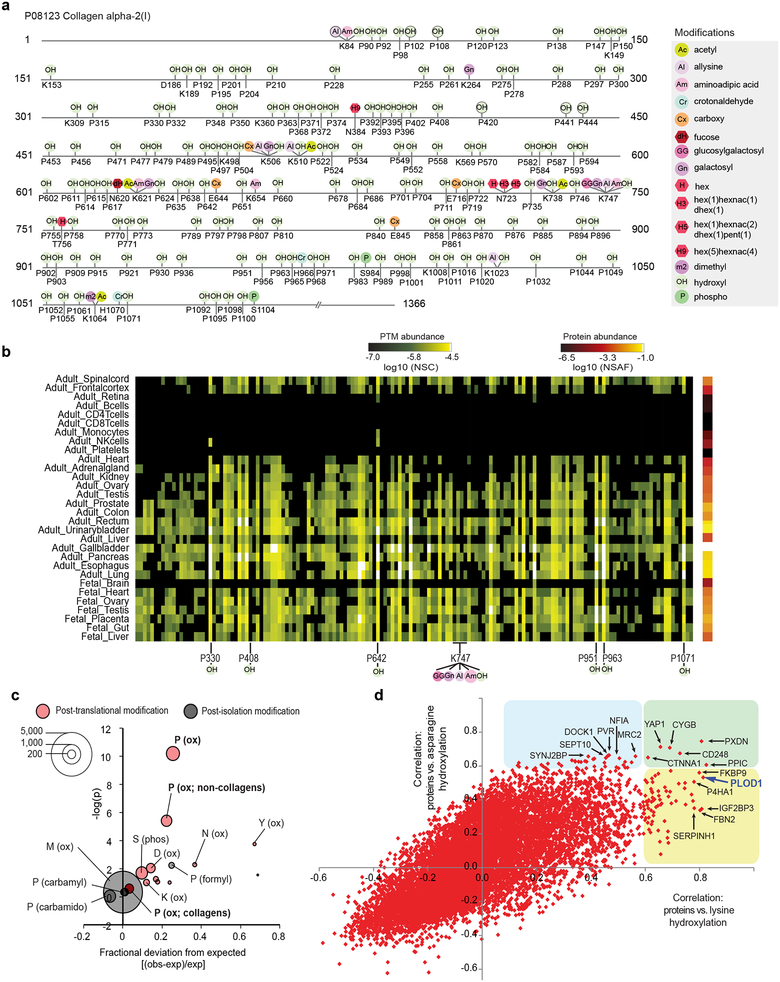
Proline hydroxylation is essential to the role of collagen proteins in maintaining extracellular matrix (ECM) stability. Despite hydroxyl proline comprising over 13% of mammalian collagen by weight42, only 254 sites across all collagens were previously assigned in humans (79% of all charted hydroxyl prolines in the human proteome). TagGraph identified 166 proline hydroxylation sites on COL1A2 alone, just 4 of which were previously described43 (Fig. 5a). While most proline hydroxylation sites were represented across most solid tissues examined here (e.g., P330, P642), several displayed tissue-specific abundance (e.g., P408, restricted to colon, bladder, liver, gallbladder and pancreas) (Fig. 5b). TagGraph identified 14 other PTM types from this single protein, suggesting multiple routes by which PTMs cooperatively regulate collagen structure and function (Dataset 16 (https://taggraph.page.link/Datasets)).
Figure 5. Characterization of hydroxylation, an un-enrichable PTM, enabled by TagGraph.
a) TagGraph extensively expanded known hydroxylation sites across the human proteome; a selection of the most abundant PTMs on COL1A2 are shown as an example; where three proline hydroxylation site were previously reported (P420, P441, P444)43, 166 are shown along with 25 other types of PTMs on this single protein (Dataset 16 (https://taggraph.page.link/Datasets)). b) PTMs identified in (a) were found to vary in abundance across tissues. Many hydroxylations displayed uniform abundance across solid tissues (i.e., P330, P642), whereas others displayed tissue-specific abundance variations (P408). c) Comparison between modification and cancer mutation sites (COSMIC). The size of each bubble indicates the number TagGraph-identified modification sites that were also found to be mutated in sequenced tumors (Dataset 17 (https://taggraph.page.link/Datasets)). The expected value and significance (one-tailed Fisher’s exact test) of this overlap was determined from the background of all peptides confidently identified by TagGraph (Online Methods, Dataset 18). Proline hydroxylation sites significantly overlapped with mutation sites both overall (p =6e-11, Fisher’s exact test; p=1e-9, Bonferroni corrected) and when restricted to non-collagen domain containing proteins (p =4e-6, Fisher’s exact test; p=7e-5, Bonferroni corrected), suggesting that mutating these sites’ PTM capacity plays a role in cancer pathogenesis. d) Correlations between protein abundance and total PTM profiles across tissues (Supplementary Fig. 14) suggest candidate regulatory enzymes and functional associations. Proteins that highly correlated with lysine hydroxylation (x-axis), asparagine hydroxylation (y-axis) or both are highlighted (yellow, blue, or green, respectively). PLOD1 (in purple), the enzyme responsible for lysine hydroxylation in collagen emerged among the proteins most correlated with this modification. Protein expression levels were correlated with PTM stoichiometry across all tissues (Online Methods).
Our analysis extends known proline hydroxylation by nearly 3,000 sites spanning almost 1,000 substrate proteins (Dataset 10 (https://taggraph.page.link/Datasets)). These proteins were significantly enriched for 113 biological processes and 60 cellular compartments (Benjamini-corrected Fisher’s exact p <0.01) (Dataset 12 (https://taggraph.page.link/Datasets)), suggesting that proline hydroxylation could influence many cellular processes beyond matrix homeostasis (Fig. 4b). Noting that tumors exploit some of these same processes during oncogenesis, we hypothesized that proline hydroxylation could play a role in cancer. Associations were previously shown between specific phosphorylation sites and cancer-associated mutations44, and using a similar approach, we examined whether proline hydroxylation significantly intersected with missense somatic cancer mutations catalogued in the COSMIC database45. We found that hydroxylated prolines were 25% more likely to be associated with cancer mutations than expected (p=6e-11, Fisher’s exact test; p<1e-9, Bonferroni corrected), even excluding collagens (22%, p=4e-6 Fisher’s exact test; p<7e-5, Bonferroni corrected) (Fig. 5c, Datasets 17, 18 (https://taggraph.page.link/Datasets)). Methionine oxidation, a common post-isolation modification, was not enriched (p=0.49, Fisher’s exact test), nor were other post-isolation proline modifications (Fig. 5c). Thus, further study of hydroxylation could provide insight into cancer pathogenesis and reveal new therapeutic targets.
As with proline hydroxylation, TagGraph substantially expanded the number of known lysine hydroxylation and asparagine hydroxylation sites by 14- and 4-fold, respectively. To elucidate possible direct protein-PTM interactions, we screened all PTM and protein quantifications for strong correlations across the 30 tissues examined here. We found several that confirmed known functional associations (Fig. 5d, Supplementary Fig. 14). Generally, we found that proteins that were highly correlated with specific modifications did not bear those modifications themselves (Supplementary Fig. 14b). However, they tended to be enriched for the same functional ontologies as the PTMs’ substrates (Supplementary Fig. 14c). Such highly correlated proteins may be candidate PTM-altering enzymes (analogous to kinases) or indirect regulators (analogous to cyclins).
We found over 70 proteins that correlated highly with lysine hydroxylation across all tissues (Fig. 5d). Many of these proteins, such as PXDN and CYGB have known roles in oxygen transport or oxidoreductase activity (Supplementary Fig. 14c), supporting their role in regulating hydroxylation PTMs. Notably, many proteins that correlated with lysine hydroxylation also correlated with asparagine hydroxylation despite no previous evidence linking these PTMs to the same biological context. ECM collagens are the major substrates for lysine hydroxylation. Though asparagine hydroxylation was not previously characterized in the ECM, TagGraph revealed 45 sites on Fibrillin-1 and Fibrillin-2 (Datasets 10 (https://taggraph.page.link/Datasets)), both of which are ECM constituents. These data suggest that proteins correlating with both PTMs may function as general positive regulators of ECM homeostasis through structure-stabilizing hydroxylation40,46.
Discussion
While conventional database searches remain the optimal strategy for restricted protein identification, improvements in de novo sequencing speed47 suggest that the TagGraph strategy can approach real-time data acquisition speeds. We expect this capability will become increasingly important as high-resolution and high-volume mass spectrometers become more widely available. Relatedly, enrichment methods remain the optimal choice for very deep analysis of select modifications, but we expect this requirement will greatly diminish as instrumentation sensitivity improves. Here, we find that given sufficiently deep MS/MS spectra, TagGraph makes otherwise un-enrichable PTMs readily measurable (Fig. 5a,b).
TagGraph incorporates a single-pass probabilistic model that simultaneously evaluates the likelihood that a peptide’s sequence and any modifications it bears are correct (Fig. 2). This component of our approach was essential because current “target-decoy” error estimation methods are inherently blind to amino acid modifications23,48. We demonstrate the accuracy of our error estimations with expert inspection, synthetic peptides, direct comparison to target-decoy error estimations and secondary modification localizations. The re-discovery of known modification sites, motifs, protein-PTM relationships and functional roles for various PTMs further supports the validity of our approach. We expect this model can be further expanded for protein-level error estimation and to incorporate additional mass spectrum and peptide features attributes made possible by new data acquisition methods49.
This kind of high-throughput, unbiased PTM discovery platform is compatible with any proteomics experiment that uses high-resolution tandem-mass spectrometry. We envision applying TagGraph to many types of experiments requiring much larger sequence spaces, including metaproteomics50, alternate start site utilization51 and gene prediction refinement52. By rapidly evaluating many candidate protein and peptide configurations, TagGraph can validate translation products that would confound conventional database search methods. TagGraph’s peptide-focused output maintains compatibility with protein assembly and related post-search computations53, and this flexibility should have direct application to systems that have previously been intractable to large-scale proteome analysis such as the gut microbiome and other complex microbial communities. Finally, by learning essential experimental details directly from input data, TagGraph can help standardize proteomic analyses. This could enable direct comparisons between data sets collected by multiple laboratories, thereby fostering the kind of large-scale collaborations that have transformed the genomics field.
ONLINE METHODS
A375 Dataset
Sample processing
A375 melanoma cells (ATCC)61 were cultured in DMEM supplemented with 10% FCS and antibiotics. Cells were detached by trypsinization, pelleted, washed with PBS and flash frozen in liquid nitrogen. 5×107 flash-frozen A375 cells were thawed on ice and lysed by tip sonication in Urea lysis buffer (8 M Urea, 100 mM NaCl, 50 mM Tris, 1 mM PMSF, 10 μM E-64, 100 nM bestatin, pH 8.2). The cell lysate was reduced (5 mM DTT, 55 °C, 30 min), alkylated (12.5 mM iodoacetamide, room temperature, 1 hr in the dark), and digested overnight with LysC at an enzyme:substrate ratio of 1:100 (37 °C). The resulting peptide mixture was desalted using C-18 Sep-Pak cartridges (Waters), dried using vacuum centrifugation, and resuspended in 10 mM ammonium formate, pH 10 prior to high pH reverse phase (HPRP) separation. HPRP was performed using an Agilent 1100 binary HPLC, delivering a gradient (0%−5% B over 10 min, 5%−35% B over 60 min, 35%−70% B over 15 min, 70% B for 10 min) across an Agilent C-18 Zorbax Extend column. Buffer A was 10 mM ammonium formate, pH 10 and buffer B was 10 mM ammonium formate, 90% acetonitrile, pH 10. Sixty one-minute fractions were collected and concatenated into twelve fractions as described previously62.
Mass Spectrometry
All HPRP fractions were desalted using C-18 Sep-pak cartridges (Waters), vacuum centrifuged, and resuspended in 5% ACN, 5% formic acid at approximately 1 ug/ul. One microgram of each fraction was analyzed by microcapillary liquid chromatography electrospray ionization tandem mass spectrometry (LC-MS/MS) on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an in-house built nanospray source, an Agilent 1200 Series binary HPLC pump, and a MicroAS autosampler (Thermo Fisher Scientific). Peptides were separated on a 125 um ID × 18 cm fused silica microcapillary column with an in-house pulled emitter tip with an internal diameter of approximately 5 um. The column was packed with ProntoSIL C18 AQ reversed phase resin (3 um particles, 200Å pore size; MAC-MOD, Chadds Ford, PA). Each sample was separated by applying a two-step gradient: 7% −25% buffer B over 2h; 25–45% B over 30 min. Buffer A was 0.1% formic acid, 2.5% ACN and buffer B was 0.1% formic acid, 97.5% ACN. The mass spectrometer was operated in a data dependent mode in which a full MS scan was acquired in the Orbitrap (AGC: 5×105; resolution: 6×104; m/z range: 360–1600; maximum ion injection time, 500 ms), followed by up to 10 HCD MS/MS spectra, collected from the most abundant ions from the full MS scan. MS/MS spectra were collected in the Orbitrap (AGC: 2×105; resolution: 7.5×103; minimum m/z: 100; maximum ion injection time, 1000 ms; isolation width: 2 Da; normalized collision energy: 30; default charge state: 2; activation time: 30 ms; dynamic exclusion time: 60 sec; exclude singly-charged ions and ions for which no charge-state could be determined). The mass calibration of the Orbitrap analyzer was maintained to deliver mass accuracies of ±5 ppm without an external calibrant. All raw mass spectrometry data were uploaded to the PRIDE repository54 and assigned the accession ID PXD005912.
Synthetic peptide confirmation data set
In total, 86 synthetic peptides (SpikeTides from JPT Peptide Technologies GmbH) validating various modifications, semi- or non-specific peptide assignments were evaluated. A final pool of all peptides dissolved in 0.1% formic acid was generated and the concentration of each individual peptide was roughly 250 fmol/μL. Two LC-MS/MS runs were performed and the auto-sampler injected 1 μl of the synthetic peptide pool. An ESI-Orbitrap Elite mass spectrometer (Thermo Electron, Bremen, Germany) interfaced with an Eksigent ekspert nanoLC 425 system (Eksigent technologies, Dublin, CA, USA) was used. Peptides were introduced into the mass spectrometer via a fused silica microcapillary column (100 μm inner diameter) ending in an in-house pulled needle tip. The columns were packed in-house to a length of 18 cm with a C18 reversed-phase resin (with Reprosil-Pur C18-AQ resin (3 μm Dr. Maisch, GmbH, Germany). For elution a two-step gradient of 4–25% buffer B (5 % DMSO, 0.2% formic acid and 94.8 % acetonitrile (v/v)) in buffer A (5 % DMSO, 0.2% formic acid in water (v/v)) over 60 min followed by a second phase of 25–45% buffer B over 20 min was used. The LTQ-Orbitrap was operated in data-dependent mode to automatically switch between Orbitrap-MS (from m/z 340 to 1600) and ten MS/MS acquisition. Each FT-MS scan was acquired at 60,000 FWHM nominal resolution settings while the MS/MS spectra were acquired using HCD and at a resolution of 15,000. Precursor ion charge state screening was enabled (charge state 1 rejected) and the normalized collision energy was set to 35%.
The resulting data were analyzed by TagGraph to match mass spectra with their best-matching synthetic peptide sequence. Synthetic-derived and experiment-derived mass spectra (e.g., from the draft human proteome data set) were only compared if spectra were assigned to the same peptide sequence in the same charge state. Of the 86 peptides synthesized, 75 were matched in this manner and used to validate TagGraph peptide-spectrum assignments (Supplementary Analysis 3). The mass tolerance used to match b- and y-ions was 0.1 Daltons.
Phosphorylation–enriched proteome data set
A subset of RAW data from a recent phosphorylation analysis22 were downloaded from the PRIDE data repository using accession ID PXD000612. The re-analysis data presented here were deposited into PRIDE with the accession ID PXD008899.
Draft human proteome data set
All RAW data and database search results from the draft human proteome data set24 were downloaded from the PRIDE data repository using accession PXD000561. Lung-specific, low-resolution RAW data were described in the companion human proteome data set63, and were downloaded from the PeptideAtlas data repository using the accession ID PAe001771. The re-analysis data presented here for the Kim et al data set were deposited in the MassIVE repository and linked with the original source identifier MSV000079514. The Lung-specific low resolution data were deposited into PRIDE with the accession ID PXD008902.
De novo search engine comparisons
We compared the performance of three de novo sequencing algorithms, PEAKS 756, PepNovo+ (ver. 3.1)58, and pNovo (ver 1.1)57 and assayed their ability to generate mostly correct sequence interpretations of high mass accuracy MS/MS spectra9. Each algorithm was used to search the A375 dataset, and the resulting peptide identifications were compared against high confidence peptide spectrum matches obtained from a SEQUEST search of the same dataset as previously described9 (Target-decoy database construction and utilization section, below). Static and differential modifications were set as for the SEQUEST search. Mass tolerance parameters were optimized to achieve maximum sequencing accuracy for each algorithm individually9. PEAKS was run with a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.01 Da. PepNovo+ was run a 0.01 Dalton precursor mass tolerance and 0.05 Da mass tolerance on fragment ions. pNovo was run with a 6 ppm precursor mass tolerance and a 25 ppm fragment ion tolerance. These mass tolerances were previously determined to maximize sequence accuracy corresponding with our instrumentation9.
The accuracy of each de novo algorithm was assessed using the sequence accuracy metric9. For a given de novo peptide-spectrum match and its corresponding high confidence SEQUEST peptide-spectrum match, sequence accuracy represents the fraction of prefix residue masses present in the high confidence SEQUEST match which were also present in the de novo sequence9.
Database search engine comparisons
Algorithm benchmarking with the A375 data set
We assessed several expanded database search algorithms’ abilities to detect undefined modifications, without constraining protease specificity, using the A375 dataset. As a baseline, we searched all 168,391 MS/MS spectra in this dataset with SEQUEST64 (ver. 28 rev 12) using an indexed sequence database comprised of the human proteome (Uniprot, downloaded 12/9/2014)65 plus common contaminants. These sequences were appended by decoy protein sequences constructed as described in TagGraph Parameters section, below. The SEQUEST search was conducted with LysC protease specificity, 50 ppm precursor ion mass tolerance, and 0.5 Da fragment ion mass tolerance. Cysteine carbamidomethylation (+57.021464 Da) was set as a static modification and methionine oxidation (+15.994915 Da) was set as a differential modification. False discovery rates (FDRs) were estimated with a linear discriminant method21.
The A375 dataset was then searched using PEAKS PTM (ver. 7)17, Byonic (ver. 2.5.6)13, ModA (ver. 1.03)12, the Open Search method using SEQUEST14 and MSFragger (build 20170103)15 – four distinct strategies described as being able to either consider relatively large numbers of discrete amino acid modifications, or searching spectra with no a priori constraints on possible modifications. It was not possible to search the entire A375 dataset with any of the above algorithms using completely unconstrained parameters with respect to both modifications and protease specificity: either the algorithms would not execute, or they did not complete within a reasonable amount of time (5 days per RAW data file). Thus, CPU times (Fig. 1e) were calculated using the least restrictive parameters that were compatible with each algorithm. Search times were extrapolated from a limited subset of search results (Byonic) for which searching the entire dataset would be too computationally intensive with similar computational resources used by the other algorithms. As such, reported search times represent a substantial underestimate of the true time needed for each of these algorithms to analyze a sample in a manner equivalent to TagGraph, as described below.
PEAKS PTM:
the A375 dataset was first de novo sequenced with PEAKS using the following settings: 10 ppm precursor ion tolerance and 0.01 Da fragment ion tolerance, cysteine carbamidomethylation as a static modification, and methionine oxidation as a differential modification. The dataset was then analyzed with PEAKS PTM using the same modification and mass tolerances as for the de novo sequencing, LysC enzyme specificity allowing for nonspecific cleavage at both ends of the peptide, and considering all 485 unimod-based modifications curated in PEAKS’s internal PTM database. FDRs were estimated using PEAKS’ internal target-decoy method.
MODa:
the A375 dataset was analyzed with the following settings: 0.05 Da precursor mass tolerance, 0.05 Da fragment ion tolerance, allowing multiple modifications per peptide, no protease specificity, modification size between −200 Da and 200 Da and cysteine carbamidomethylation as a static modification. FDRs were estimated using the “anal_moda.jar” program included in the MODa software package
Byonic:
the A375 dataset was analyzed using the following settings: 10 ppm precursor ion tolerance, 20 ppm fragment ion tolerance, LysC protease specificity, cysteine carbamidomethylation as a static modification, methionine oxidation as a differential modification, wildcard search enabled with a minimum mass of −200 Daltons and a maximum mass of 200 Daltons. FDRs were estimated using Byonic’s internal target-decoy method.
Open Search (SEQUEST):
the A375 dataset was analyzed using the same search parameters as the initial SEQUEST search, except allowing a 500 Da mass window surrounding each precursor and allowing semi-trypsin specificity. FDRs were estimated using the linear discriminant method as described above for SEQUEST.
Open Search (MSFragger):
the A375 dataset was analyzed using the following settings: 500 Da precursor mass tolerance; 10 ppm precursor true tolerance; 20 ppm fragment mass tolerance; no isotope_error correction; semispecific Trypsin digestion; one missed cleavage; clipped N-terminal Methionine; cysteine carbamidomethylation as a static modification; variable oxidized methionine, allow multiple modifications on one residue; allow up to three variable modifications per modification type; allow up to 1000 variable modifications per peptide; require at least 15 fragment ion peaks per MS/MS spectrum. FDRs were estimated using PeptideProphet as originally described15.
For the above algorithms, the target sequence database used was the same as for SEQUEST. PEAKS PTM and Byonic were allowed to use their own internal methods for creating decoy sequences, whereas ModA and both Open Search methods were provided the same concatenated target-decoy sequence database used for TagGraph EM comparisons (see TagGraph Parameters section, below). Open Search with SEQUEST used the same pre-processed sequence index as the conventional SEQUEST search. One CPU thread was used for all searches.
CPU search times (Fig. 1e) were calculated as the sum of the CPU time over all processes spawned by each database search algorithm to analyze the data. Due to computational constraints, it was not possible to run Byonic with semi specific or nonspecific enzyme specificity or on the entire A375 dataset. Thus, we conducted the Byonic search with full LysC specificity (Fig. 1d), and analyzed only the first fraction of the A375 dataset. The estimated CPU time over the entire dataset was extrapolated by multiplying the CPU time recorded from the analysis of a single HPRP fraction by the ratio of the total MS/MS spectra in the dataset (168,391) divided by the number of MS/MS spectra in the single fraction (16,613).
Unrestricted modification discovery from phosphorylation-enriched data set
We compared TagGraph to the highest-performing search methods emerging from Figure 1, PEAKS-PTM, Open Search (SEQUEST) and Open Search (MSFragger) using the Sharma et al phosphorylation-enriched data set22 as well as conventional database search with SEQUEST. We searched 451,655 MS/MS spectra as described for the A375 data set above, noting the following differences:
SEQUEST:
searches were conducted with trypsin protease specificity, and phosphorylation (+79.99633) was set as a differential modification on serine, threonine and tyrosines.
PEAKS PTM:
searches applied trypsin specificity allowing for nonspecific cleavage at both ends of the peptide.
Open Search (SEQUEST):
searches were conducted with trypsin protease specificity.
Open Search (MSFragger):
no changes relative to the A375 data set.
Unrestricted modification discovery from low-resolution MS/MS spectra.
We evaluated TagGraph’s ability to search low-resolution MS/MS spectra, relative to standard database searching with the SEQUEST algorithm. SEQUEST searches of were performed on 336,344 MS/MS spectra corresponding with the lung data set (PeptideAtlas data repository entry PAe001771), as described above except trypsin specificity was required, and fragment ion tolerance was set to null (i.e., 1 Da).
Target-decoy database construction and utilization
For the purpose of comparing EM-based FDR estimations with the standard target-decoy approach (Figure 2a, 2b, Supplementary Analysis 1), we concatenated human proteome sequences with decoy counterparts for target-decoy FDR estimation11. We previously observed that de novo sequencing algorithms occasionally mistake b- and y-ion series, and consequently have a tendency towards returning reversed sequences9. As a result, decoys generated by strict sequence reversal are at risk of systematically overestimating FDRs of TagGraph results: PSMs derived from incorrect, reversed de novo sequence would preferentially match decoy sequences over target sequences.
We addressed this bias by generating decoy sequences in a manner that shuffled amino acid sequences while preserving the distribution of tryptic peptides and frequencies of peptides shared between proteins: (i) protein sequences are first reversed; (ii) the locations of all lysines and arginines in the reversed sequences are fixed, and the intervening sequences are shuffled; (iii) for each unique intervening pre-shuffled peptide sequence, the shuffled variant is cached and all subsequent peptide instances are mapped to the same shuffled variant. This method well-preserved a desired 50–50 target decoy ratio (Fig. 2a, Supplementary Analysis 1a) by maintaining the same number, length, and mass distribution of unique LysC/tryptic peptides in the target and decoy databases, while removing the source of the overestimation bias described above.
FDRs based on the target-decoy strategy (Fig. 2b, and Supplementary Analysis 1b–d) were calculated by summing the number of decoy identifications exceeding a given EM score threshold, dividing this by the total number of identifications exceeding this threshold, and then multiplying this quotient by the “decoy factor” we previously described to account for the slight bias we observed towards decoy identifications11. This factor was estimated as the proportion of low-confidence (EM score <0) target identifications relative to all low-confidence identifications. This factor was on average 1.88, rather than the expected 2.0. Short peptides <8 amino acids in length were excluded from this analysis due to their lower information content.
TagGraph Parameters
TagGraph was used to analyze proteome data sets described above. All available MS/MS were first de novo sequenced using PEAKS as described in the Database search engine comparisons section above and noted below. The resulting peptide sequences and raw mass spectra (mzXML-formatted66) were given as input to TagGraph.
For the A375 dataset, PEAKS was run with a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.01 Da. To account for wider fragment mass variances for several component data sets that comprised the draft Human Proteome, we increased the fragment ion tolerance to 0.05 Da. This parameter was set to 1 Da for the low-resolution MS/MS lung data set. Other parameters were consistent with the A375, phosphorylation and low-resolution lung data set analyses: 10 ppm precursor mass tolerance cysteine carbamidomethylation as a static modification, and methionine oxidation as a differential modification. Unless otherwise specified, de novo sequencing results were searched with TagGraph against the human proteome (Uniprot, downloaded 12/9/2014) plus common contaminants without any additional decoy sequences.
The above database was concatenated with decoy sequences only for searches involving the A375 data set, and for 12 other data sets examined in this report (Target-decoy database construction and utilization section, above): Searching this target-decoy sequence database with TagGraph enabled fair comparison of the CPU times of TagGraph with the other database search algorithms (Fig. 1e) and direct comparisons of FDR estimates (Fig. 2a,b, Supplementary Analysis 1). Otherwise, no decoys were used for TagGraph analyses.
TagGraph empirically optimizes precursor mass tolerances as part of its hierarchical Bayes scoring model. Fragment ion tolerances were initially set to 20 ppm of the precursor mass. We also define the quantity modification mass tolerance, which describes whether a particular modification agrees with one represented in the Unimod resource, or a novel modification specific to the present dataset. This tolerance was set to 0.1 Da. Enzyme specificity was set to LysC for the A375 dataset and Trypsin for the human proteome data set. Although enzyme specificity was considered as a scoring attribute in the hierarchical Bayes model, TagGraph is able to return high-confidence semi specific and nonspecific peptide-spectrum matches regardless of the input enzyme specificity.
For the purposes of calibrating EM- and target-decoy FDR estimations, all returned peptide-spectrum matches were ranked according to their probabilities P(D|+) from highest to lowest, and the inverse (1- P) of these probabilities were summed in order of decreasing rank. FDRs were estimated from EM scores as described in Supplementary Note 4, Equation 2. For all other purposes, we applied a probability threshold of 0.99 (log10 (1-P) ≥ 2), as previously described for parametric error estimation67 – i.e., an EM scoring threshold of 2.0 (Supplementary Analysis 1c,d).
PTM error estimation with amino acid-substituted proteome
Target-decoy based error estimation accuracy declines when applied to peptide modifications and other large search spaces19,68 (Supplementary Note 3). Despite this, all previously described unrestricted search algorithms rely on target-decoy to delineate sets of confidently identified spectra. To assess the degree to which estimated FDRs reflect underlying identification errors, we employed a modified human proteome sequence database in which every tyrosine residue was replaced by a phenylalanine. The mass difference between these residues (15.994915 Da) corresponds with an oxygen atom, and is a frequently observed modification on several residues (e.g., methionine), while distinguishing other unmodified residues (e.g., alanine and serine). Search engines capable of accurate PTM assignment and discrimination should search the tyrosine-substituted database and return phenylalanine-containing peptides modified by an oxygen only on those phenylalanines that were previously tyrosines. They should be able to discriminate these identifications from erroneous ones in which oxidation modifications were assigned to unaltered residues. We analyzed the A375 dataset against this modified sequence database using SEQUEST, PEAKS PTM, Byonic, ModA, and Open Search with SEQUEST and MSFragger. The results from each algorithm were then filtered to a 1% predicted FDR using target-decoy based statistics, or a 0.99 probability threshold for TagGraph (EM score >2.0). Byonic, MODa, and PEAKS PTM were allowed to use their own internal target-decoy based filtering procedures. Search results provided by SEQUEST were filtered using a linear discriminant method21. All results were compared to the initial SEQUEST search results of the unaltered sequences database, which were filtered to a 1% FDR. Identification error was assessed based on the proportion of peptides that conflicted with the high confidence search results with respect to the overall amino acid sequence (base peptide), or to the presence of an unmodified phenylalanine at a former tyrosine position.
The A375 dataset was also analyzed with TagGraph against the phenylalanine-substituted human sequence database. The parameters used were identical to those described in the TagGraph Parameters section above. Results were filtered to a probability value ≥0.99 (EM score >2.0) as described above.
We estimated a 2% maximum expected likelihood of conflicting identifications in Figure 2c as follows: We consider two sets of search results of the same mass spectra such that identifications made in any one set have a 99% likelihood of being correct. Assuming incorrect identifications are randomly distributed within each set of search results, spectra which receive the same identification from both search result sets should have a likelihood of being correct of 0.992 = 0.9801. Accordingly, the likelihood of an incorrect search result returned by either or both searches should be estimated as 1 –, 0.992 = 0.0199.
Abundance calculations
Protein abundance calculation
Protein abundances were calculated using the distributed normalized spectral abundance factor (NSAF) method69. Briefly, the number of spectral counts originating from peptides that uniquely map to single proteins were summed over all proteins identified in an experiment. Spectral counts recorded from peptides that map to multiple proteins were distributed across all such proteins according to the proportion of spectral counts assigned to them from uniquely mapped peptides. Finally, summed spectral counts for each protein were normalized by protein length, and the sum of all protein abundances for each experimental dataset was normalized to one. Protein abundances per tissue were calculated as the average of the individual NSAF for that protein over all experiments performed on that tissue. For the purpose of this analysis, proteins were defined solely by their unique representation in the input sequence file – proteins with equal or overlapping peptide evidence were represented in Dataset 7 (https://taggraph.page.link/Datasets). For protein-level analyses relying on unique protein counts (Figure 4b, 4e; Supplementary Figs. 8, 12, 14) we selected a single highest-abundance entry to represent the indicated gene symbol.
Site abundance and stoichiometry calculations
To compare modification sites between tissues, we quantified the abundance of sites using two methods: normalized spectral counts (NSC) and estimated stoichiometry. For both methods, we first generated a catalog of all confident peptide identifications that span a given modified amino acid position of a protein, regardless of modification state. The total spectral counts corresponding to all peptides containing the amino acid position (ST) and just those corresponding to peptides containing the exact modification on the site of interest (Sm) were calculated for each experimental dataset.
The normalized spectral count of a modification site was calculated as Sm divided by the number of confidently identified peptide-spectrum matches in the experimental dataset. The stoichiometry of the modification was calculated as Sm divided by ST, following a previously described rationale22. Modification site abundances (stoichiometry or NSC) per tissue were calculated as the average of the site abundances over all experiments performed on that tissue. Due to inherent difficulties in accurately reporting very low abundances with spectral counting70, experiments in which no peptides overlapping the site of modification were detected were not included in the average. Thus, the sum of stoichiometries of all modifications at a particular site in a particular tissue may not be normalized. Finally, the abundance, stoichiometry or normalized spectral count of a modification site was set to zero for a particular tissue if the corresponding protein NSAF was zero in that tissue.
Histology images
Eight protein and tissue combinations which were not previously reported24 were arbitrarily selected, and used to query the Human Protein Atlas71 (www.proteinatlas.org). Representative images selected by the web resource’s curators were downloaded and represented in Supplementary Figure 8b. See figure legend for specific image attributions.
Gene ontology analysis
Gene ontology analysis was conducted using the DAVID web portal60. For each post-translational modification of interest, proteins bearing that modification were compiled and input as a gene list. The background list used was the Uniprot human proteome. The resulting gene ontologies were downloaded and a global FDR threshold (Benjamini-Hochberg) of 1% was used as a threshold for determining significantly enriched ontologies.
We observed that many ontologies were broadly enriched across all post-translational modifications and hypothesized that these were simply associated with highly abundant proteins and did not reflect true post-translational modification properties. As a control, we applied the above enrichment analysis and significance criterion to fifteen post-isolation modifications and observed many ontologies that were enriched for all post-isolation modifications considered (Supplementary Fig. 12). These ontologies were excluded from the set of enriched ontologies in the post-translational modification analysis (Fig. 4b, Dataset 12 (https://taggraph.page.link/Datasets)).
COSMIC dataset comparison
The COSMIC database of cancer mutations was downloaded via FTP from the resource’s website45. These mutations were then filtered to keep only missense mutations. To account for slight protein sequence variations between COSMIC and Uniprot, we discarded mutations for which the amino acid residue at the denoted position in the Uniprot protein sequence did not match the non-mutated amino acid identity in the corresponding COSMIC entry.
We tested the significance of any overlap between COSMIC cancer mutations and TagGraph-identified modifications via Fisher’s exact test. Using hydroxylated prolines to illustrate this calculation, we first identified all TagGraph-identified proteins bearing at least one proline hydroxylation and were also represented in the COSMIC database. From these, we tabulated the number of prolines in the protein sequences (31,603); the number of TagGraph-identified hydroxylations among these prolines (3,248); the number of COSMIC-identified mutated prolines (5,005) and the number of mutated sites that were also found to be hydroxylated (645). These values were used to derive the expected number of overlapping mutated and hydroxylated sites by chance (514.4). We observed 25% more overlapping sites than would be predicted by chance: (645–514.4)/514.4 = 25.4%
To guard against biases in background amino acid distributions, overlap statistics were only calculated for proteins on which both cancer mutations and the PTM of interest were detected and only against the background of peptides confidently identified by TagGraph in the human proteome dataset. Using proline hydroxylation as an example, the number of prolines, number of hydroxylation prolines, number of mutated prolines, and number of mutated and hydroxylated prolines were counted only on peptides confidently identified by TagGraph and on proteins containing both cancer mutations and proline hydroxylation. Overlapping residues were then tested for significance via Fisher’s exact test (Dataset 18 (https://taggraph.page.link/Datasets)). This analysis was carried out analogously for other types of hydroxylations (lysine, asparagine, methionine, etc.).
Protein-PTM correlation analysis
Reasoning that many modifications’ abundances and stoichiometries will depend on specific protein-modifying enzymes, we sought to discover functional relationships between post-translational modifications and proteins. We identified highly correlated subsets of modifications and proteins by comparing their abundances across the tissues examined here. Modification site and protein lists were first filtered to include only those identified from at least three tissues. For a particular post-translational modification of interest (e.g., Lysine hydroxylation), the abundance of the modification was averaged across all measured sites from all proteins within each tissue, forming a vector representing the abundance of the modification across all tissues. Similarly, for all identified proteins, the calculated NSAF was used to form an abundance vector of that protein’s expression across all tissues. We next determined the Pearson correlation coefficient between all modification and all protein vectors computed and filtered as described above. The proteins with the largest magnitude correlations (positive or negative) were then considered as candidates having a functional relationship with a modification of interest.
Modification abundance vectors were calculated using both modification stoichiometries and modification-normalized spectral counts. Both types of quantification were used in the correlation analysis, often yielding different results (Supplementary Fig. 14a). However, in both cases, our analysis revealed previously described associations between proteins and post-translational modifications (e.g., arginine methylation and RNA splicing proteins), supporting the validity of this analysis (Supplementary Fig. 14c).
Statistics and reproducibility
Statistical tests were performed using python with packages noted below (Code availability), R version 3.2.2, Microsoft Excel, and web-based tools as noted in main text, Online Methods, Supplementary text, and the Life Sciences Reporting Summary. Details regarding statistical tests are provided in the main text and corresponding figure legends. Numbers of replicates, where applicable, are noted in figure legends. Most statistical calculations, however, describe populations of protein, peptide, and PTM observations made by considering each large-scale proteomic experiment as a single replicate.
Code availability.
The TagGraph algorithm and supporting software can be downloaded via http://sourceforge.net/projects/taggraph. TagGraph was developed under Python ver. 2.7 and makes use of freely available packages including lxml ver 4.2.4; mysqlclient ver 1.3.13; networkX ver 1.1172; NumPy ver 1.10.0; Pympler ver 0.6; pymzml ver 0.7.8; Pyteomics ver 3.5.173; SQLAlchemy ver 1.2.11; and SciPy 1.2.0.
Supplementary Material
Acknowledgements
This work was supported by a Damon Runyon-Rachleff Innovation Award from the Damon Runyon Cancer Research Foundation (DRR-13–11; J.E.E), the W.M. Keck Foundation Medical Research Program (J.E.E.), the Bill and Melinda Gates foundation (OPP1113682; J.E.E.), the National Cancer Institute (U01 CA194389; J.E.E.), the National Institute of Dental and Craniofacial Research (R01DE027750), the Stanford Graduate Fund (A.D.), the Wallenberg Foundation (N.O.), the National Science Foundation Graduate Research Fellowship (C.G.) and Howard Hughes Medical Institute Gilliam Fellowship for Advanced Study (C.G.). We thank P. Khavari for supplying A375 cells and D. Bai (UVA) for use of her fragmentation calculation software. We acknowledge members of the Elias Lab, as well as D. Dill, P. Mallick, T. Meyer, D. Mochley-Rosen, and J. Wysocka for helpful discussions.
Footnotes
Data Availability
Raw and processed data from the A375 cell line (Figs. 1, 2) are available from the PRIDE repository54 (PXD005912). Search results from the phosphorylation-enriched data set (Supplementary Fig. 7) are available from PRIDE (PXD008899). TagGraph search results from the Kim et al Human Proteome data set (Figures 3–5) are available from the MassIVE repository55 with the original accession (MSV000079514). TagGraph search results from the lung-specific low-resolution data set (Supplementary Fig. 10) are available from PRIDE (PXD008902). Dataset files are available from https://taggraph.page.link/Datasets, the above-mentioned data repositories, or directly from the corresponding author.
Competing Interests
The authors declare no competing interests.
REFERENCES
- 1.Eisenhaber B & Eisenhaber F Posttranslational modifications and subcellular localization signals: indicators of sequence regions without inherent 3D structure? Curr. Protein Pept. Sci 8, 197–203 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Nussinov R, Tsai C-J, Xin F & Radivojac P Allosteric post-translational modification codes. Trends Biochem. Sci 37, 447–55 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Creasy DM & Cottrell JS Unimod: Protein modifications for mass spectrometry. Proteomics 4, 1534–6 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y & Jensen ON Modification-specific proteomics: Strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabakaran S, Lippens G, Steen H & Gunawardena J Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med 4, 565–583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skinner OS & Kelleher NL Illuminating the dark matter of shotgun proteomics. Nat. Biotechnol 33, 717–718 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Na S & Paek E Software eyes for protein post-translational modifications. Mass Spectrom. Rev 34, 133–147 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Gupta N, Bandeira N, Keich U & Pevzner PA Target-decoy approach and false discovery rate: when things may go wrong. J. Am. Soc. Mass Spectrom 22, 1111–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devabhaktuni A & Elias JE Application of de novo sequencing to large-scale complex proteomics datasets. J. Proteome Res 15, 732–42 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Ferragina P & Manzini G Opportunistic data structures with applications. in Proceedings 41st Annual Symposium on Foundations of Computer Science 390–398 (IEEE Comput. Soc, 2000). doi: 10.1109/SFCS.2000.892127 [DOI] [Google Scholar]
- 11.Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–14 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Na S, Bandeira N & Paek E Fast multi-blind modification search through tandem mass spectrometry. Mol. Cell. Proteomics 11, M111.010199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bern M, Kil YJ & Becker C Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics Chapter 13, Unit13.20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chick JM et al. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat. Biotechnol 33, 743–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D & Nesvizhskii AI MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 14, 513–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griss J et al. Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nat. Methods 13, 651–656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, He L, Xin L, Shan B & Ma B PeaksPTM: Mass spectrometry-based identification of peptides with unspecified modifications. J. Proteome Res 10, 2930–2936 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Nesvizhskii AI, Vitek O & Aebersold R Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–97 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Fu Y & Qian X Transferred Subgroup False Discovery Rate for Rare Post-translational Modifications Detected by Mass Spectrometry. Mol. Cell. Proteomics 13, 1359–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller A, Nesvizhskii AI, Kolker E & Aebersold R Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem 74, 5383–5392 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Huttlin EL et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma K et al. Ultradeep Human Phosphoproteome Reveals a Distinct Regulatory Nature of Tyr and Ser/Thr-Based Signaling. Cell Rep 8, 1583–1594 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Beausoleil SA, Villén J, Gerber SA, Rush J & Gygi SP A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol 24, 1285–92 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Kim M-S et al. A draft map of the human proteome. Nature 509, 575–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creasy DM & Cottrell JS Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics 2, 1426–34 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Savitski MM, Nielsen ML & Zubarev RA ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics 5, 935–48 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Jenuwein T & Allis CD Translating the histone code. Science (80-.) 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Thandapani P, O’Connor TR, Bailey TL & Richard S Defining the RGG/RG Motif. Molecular Cell 50, 613–623 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Fisher D, Krasinska L, Coudreuse D & Novák B Phosphorylation network dynamics in the control of cell cycle transitions. J. Cell Sci 125, 4703–11 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Guo A et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen JV et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3, ra3 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Wu R et al. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starheim KK, Gevaert K & Arnesen T Protein N-terminal acetyltransferases: When the start matters. Trends in Biochemical Sciences 37, 152–161 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Grunstein M Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Weinert BT et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol 10, 716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner G & Hirschey MD Nonenzymatic Protein Acylation as a Carbon Stress Regulated by Sirtuin Deacylases. Molecular Cell 54, 5–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia B. a, Pesavento JJ, Mizzen C. a & Kelleher NL Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods 4, 487–489 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Xie Z et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics 11, 100–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H, Sabari BR, Garcia BA, David Allis C & Zhao Y SnapShot: Histone modifications. Cell 159, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoulders MD & Raines RT Collagen structure and stability. Annu. Rev. Biochem 78, 929–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res 43, D204–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.NEUMAN RE & LOGAN MA The determination of hydroxyproline. J. Biol. Chem 184, 299–306 (1950). [PubMed] [Google Scholar]
- 43.FIETZEK PP, KUHN K & FURTHMAYR H Comparative Sequence Studies on alpha2-CB2 from Calf, Human, Rabbit and Pig-Skin Collagen. Eur. J. Biochem 47, 257–261 (1974). [DOI] [PubMed] [Google Scholar]
- 44.Reimand J, Wagih O & Bader GD The mutational landscape of phosphorylation signaling in cancer. Sci. Rep 3, 2651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forbes SA et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43, D805–D811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang M et al. Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor. J. Biol. Chem 286, 7648–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma B Novor: Real-Time Peptide de Novo Sequencing Software. J. Am. Soc. Mass Spectrom 26, 1885–1894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fermin D, Walmsley SJ, Gingras A-C, Choi H & Nesvizhskii AI LuciPHOr: algorithm for phosphorylation site localization with false localization rate estimation using modified target-decoy approach. Mol. Cell. Proteomics 12, 3409–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier F et al. Online parallel accumulation – serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics (2018). doi: 10.1101/336743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez CG, Zhang L & Elias JE From mystery to mechanism: can proteomics build systems-level understanding of our gut microbes? Expert Rev. Proteomics 14, (2017). [DOI] [PubMed] [Google Scholar]
- 51.Ingolia NT, Ghaemmaghami S, Newman JRS & Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo S et al. Proteogenomic Database Construction Driven from Large Scale RNA-seq Data. J. Proteome Res 13, 21–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serang O & Noble W A review of statistical methods for protein identification using tandem mass spectrometry. Stat. Interface 5, 3–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vizcaíno JA et al. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9, 4276–83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M et al. Assembling the Community-Scale Discoverable Human Proteome. Cell Syst (2018). doi: 10.1016/j.cels.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma B et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom 17, 2337–42 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Chi H et al. pNovo: de novo peptide sequencing and identification using HCD spectra. J. Proteome Res 9, 2713–24 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Frank A & Pevzner P PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem 77, 964–73 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Crooks GE, Hon G, Chandonia J-M & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–90 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang DW, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Fok JY, Ekmekcioglu S & Mehta K Implications of tissue transglutaminase expression in malignant melanoma. Mol. Cancer Ther 5, 1493–503 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Yang F, Shen Y, Camp DG & Smith RD High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev. Proteomics 9, 129–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilhelm M et al. Mass-spectrometry-based draft of the human proteome. Nature 509, 582–7 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Eng JK, McCormack AL & Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 65.O’Donovan C et al. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief. Bioinform 3, 275–84 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Deutsch E mzML: a single, unifying data format for mass spectrometer output. Proteomics 8, 2776–7 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Kim S, Gupta N & Pevzner PA Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res 7, 3354–3363 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nesvizhskii AI A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Wen Z, Washburn MP & Florens L Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem 82, 2272–81 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Choi H, Fermin D & Nesvizhskii AI Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics 7, 2373–85 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uhlén M et al. Tissue-based map of the human proteome. Science (80-.) (2015). doi: 10.4324/9781315766188 [DOI] [PubMed] [Google Scholar]
- 72.Hagberg AA, Schult DA & Swart PJ Exploring network structure, dynamics, and function using NetworkX. in Proceedings of the 7th Python in Science Conference (SciPy) (2008). doi: 10.1016/j.jelectrocard.2010.09.003 [DOI] [Google Scholar]
- 73.Goloborodko AA, Levitsky LI, Ivanov MV & Gorshkov MV Pyteomics - A python framework for exploratory data analysis and rapid software prototyping in proteomics. J. Am. Soc. Mass Spectrom (2013). doi: 10.1007/s13361-012-0516-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.