Abstract
Epstein‐Barr virus (EBV) is a human tumor virus and is etiologically linked to various malignancies. Certain EBV‐associated diseases, such as Burkitt lymphomas and nasopharyngeal carcinomas, are endemic and exhibit biased geographic distribution worldwide. Recent advances in deep sequencing technology enabled high‐throughput sequencing of the EBV genome from clinical samples. Rapid cloning and sequencing of cancer‐derived EBV genomes, followed by reconstitution of infectious virus, have also become possible. These developments have revealed that various EBV strains are differentially distributed throughout the world, and that the behavior of cancer‐derived EBV strains is different from that of the prototype EBV strain of non‐cancerous origin. In this review, we summarize recent progress and future perspectives regarding the association between EBV strain variation and cancer.
Keywords: Epstein‐Barr virus, geographic distribution, nasopharyngeal carcinoma, sequencing, strain variation
Abbreviations
- FR
family of repeats
- HPV
human papillomavirus
- IR1
internal repeat 1
- LCL
lymphoblastoid cell line
- ORF
open reading frame
- SNP
single nucleotide polymorphism
- ORF
open reading frame
- T/NK lymphoma
T‐/natural killer cell lymphoma
- BAC
bacterial artificial chromosome
- EBER
EBV‐encoded RNA
- EBNA
EBV nuclear antigen
- EBVaGC
EBV‐associated gastric cancer
- EBV
Epstein‐Barr virus
- IR
internal repeat
- LMP
latent membrane protein
- NPC
nasopharyngeal carcinomas
1. INTRODUCTION
Epstein‐Barr virus asymptomatically infects more than 95% of healthy adults worldwide. However, EBV is also an oncogenic herpesvirus associated with various neoplastic diseases, such as lymphoproliferative diseases in immunocompromised patients, various lymphomas and epithelial cancers.1, 2 EBV‐associated malignancies show a highly unusual geographic distribution in the world.3 The biased distribution of EBV‐associated diseases is, at least partially, explained by differences in the host (human) genetic background, environmental factors (climate, prevalence of malaria) and dietary habits.
The development of high‐throughput sequencing technologies enabled sequencing of EBV genomes derived from a wide variety of clinical samples, such as tumor biopsy samples and saliva.4 The sequencing results have raised the hypothesis that certain EBV‐associated diseases are caused by specific EBV strains. Several review articles have focused on EBV strain variation in relation to diseases.5, 6, 7, 8, 9 Here, we summarize the current knowledge of EBV strain variation and its relationship with cancer.
2. EBV STRAIN VARIATION: GEOGRAPHIC OR DISEASE‐SPECIFIC?
African Burkitt lymphoma, from which EBV was originally isolated,10 is most common in equatorial Africa and designated as “endemic Burkitt lymphoma” (Figure 1). Endemic Burkitt lymphomas are nearly 100% EBV‐positive,11 whereas many Burkitt lymphomas in non‐endemic areas are EBV‐negative.3 Epidemiological and serological studies indicate that EBV infection has a causal relationship with endemic Burkitt lymphoma. Endemic Burkitt lymphomas are invariably associated with an immunosuppressed state caused by chronic malaria infection.11 Thus, endemic Burkitt lymphomas are most likely caused by environmental factors.
Figure 1.
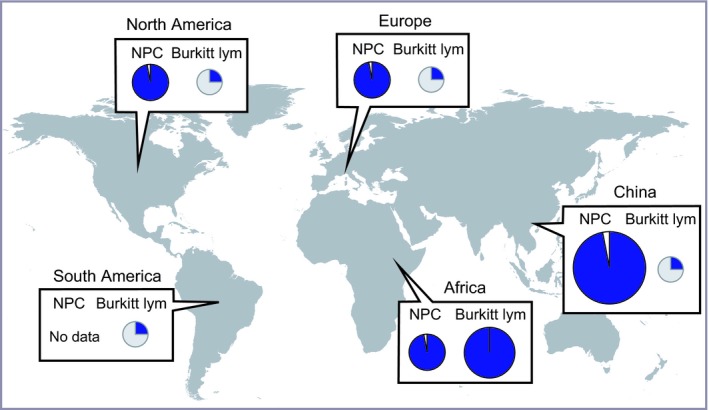
Epstein‐Barr virus (EBV) association frequencies of nasopharyngeal carcinomas (NPC) and Burkitt lymphomas are presented for different geographic areas. The sizes of the circles roughly represent the disease incidence relative to that in other geographic regions. Adapted from Chang et al3 and Cohen et al.13 Blue means EBV‐positive. Burkitt lym, Burkitt lymphoma
Nasopharyngeal carcinoma is an EBV‐associated epithelial carcinoma, and is common in South China and Southeast Asia (Figure 1).12 More than 97% of NPC are EBV‐positive,13 and the tight link between NPC and EBV is observed worldwide.3 Several predisposing factors have been postulated, including host immune‐related genes such as human leukocyte antigens,14 and environmental factors such as dietary habits (i.e. eating salted fish).15 In addition, genetic and epigenetic aberrations of host cells drive NPC development and progression.16 On the other hand, it has long been known that distinct EBV strains are predominant in areas with a high incidence of NPC.17 This suggests that EBV strain variation contributes to the endemic nature of NPC.
Another EBV‐associated disease with a distinct geographic distribution pattern is EBV‐associated T/NK lymphoma. Although T/NK lymphoma is a rare disease, it occurs with relatively high frequency in East Asia including Japan and Korea,18, 19 where the incidence of NPC is relatively low. Thus, NPC endemic areas do not coincide with areas of relatively high incidence of T/NK lymphomas, although it is possible that they substantially overlap. The reason why EBV is associated with different malignancies in different areas within Asian countries is not clear. One possible explanation is differences in host genetic backgrounds; however, another explanation is differences in EBV strains that colonize different areas in Asia.
Another malignancy causally associated with EBV infection is EBVaGC.20 EBVaGC account for approximately 9% of all gastric cancers,13 and show no biased geographic distribution. The EBV infection causes CpG island methylation of host chromosomes and EBV genome DNA methylation as well.21 Chromosomal CpG methylation may trigger epithelial carcinogenesis through silencing of tumor suppressor genes.21, 22 There is a possibility that EBVaGC are caused by oncogenic variants of EBV, which are functionally distinct from those derived from lymphoid cells.
3. EBV GENOME STRUCTURE AND SEQUENCING
3.1. EBV genome
Epstein‐Barr virus has a linear dsDNA genome of approximately 175 kb (Figure 2), which is circularized in latently infected cells. The prototypical strain B95‐8 was the first complete EBV genome sequenced using a conventional strategy (i.e. subcloning followed by Sanger sequencing) (GenBank accession no. X01555.2).23 Subsequently, the sequence of a putative “wild‐type” EBV strain was artificially created by repairing the 12‐kb defect of the B95‐8 strain (Figure 2)24 with the corresponding sequence of the Raji strain EBV25 (EBV‐wt,26 GenBank accession no. NC_007605.1). The EBV genome encodes more than 80 ORF, several non‐coding RNA (EBER1 and EBER2),1 and 44 mature miRNA.27
Figure 2.
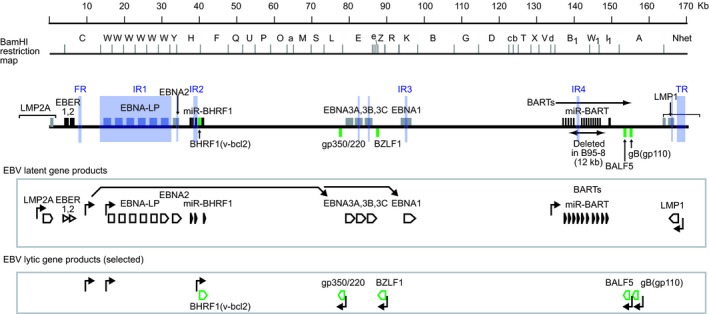
Schematic illustration of the Epstein‐Barr virus (EBV) genome and viral gene products. A linear EBV genome, viral genes, non‐coding RNA (EBV‐encoded RNA [EBER] and BART), and viral microRNA genes (miR‐BHRF1 and miR‐BART) are illustrated. The BamHI restriction map is based on the sequence of the SNU‐719 strain of EBV.46 The scale of DNA sizes is at the top. EBV latent gene products and lytic gene products (selected, green) are illustrated below. Repetitive sequences are shaded in purple. Viral gene promoters are indicated with arrows. Details of mRNA splicing are not faithfully represented. EBNA, EBV nuclear antigen; FR, family of repeats; IR, internal repeats; LMP, latent membrane protein; TR, terminal repeats
Viral genes that are specifically expressed in latently infected cells are called “EBV latent genes”.28 Among latent gene products, four EBNA (EBNA1, EBNA2, EBNA3A and EBNA3C) and LMP1 are essential for B‐cell transformation.1 Viral genes that are specifically expressed during the productive replication cycle are called “EBV lytic genes” (Figure 2). Lytic genes encode viral transcription factors (e.g. BZLF1), a viral DNA polymerase (BALF5) and associated factors, and viral glycoproteins (e.g. gp350/220 and gp110) and structural proteins (capsid and tegument proteins) (Figure 2).
The EBV genome has various repetitive sequences scattered throughout the viral genome, such as within the coding regions of viral latent proteins or near the viral replication origins (Figure 2).1 Short‐read deep sequencing cannot determine the sequences of these repetitive regions (see below).
3.2. A new era of sequencing: The hybrid capture approach
A new experimental strategy that was recently developed to enable high‐throughput EBV sequencing29, 30 uses a hybrid capture protocol analogous to human exome sequencing to enrich viral DNA sequences (Figure 3). In this method, overlapping 120‐mer biotinylated RNA baits, designed by tiling across the EBV reference genome, are used for viral DNA enrichment,29 followed by short‐read sequencing (such as Illumina HiSeq) and de novo sequence assembly.
Figure 3.
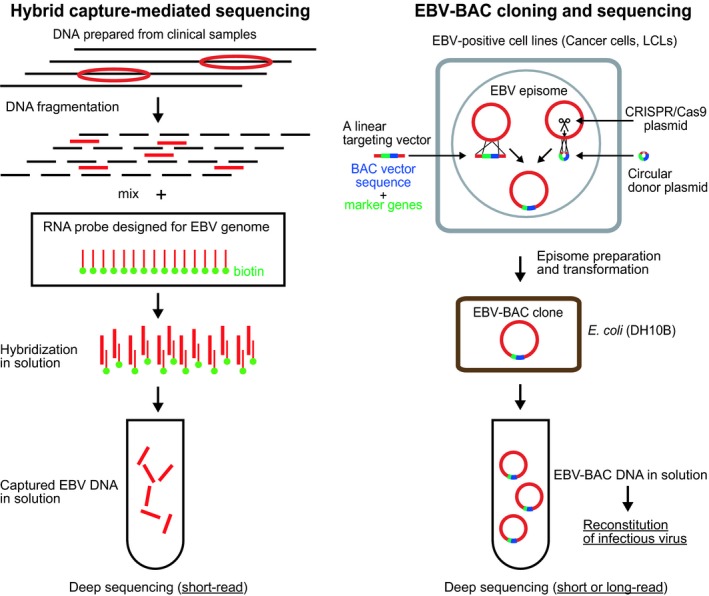
Experimental strategies to sequence whole Epstein‐Barr virus (EBV) genomes. (Left) For the hybrid capture‐mediated sequencing, the prepared DNA, consisting of host cell DNA (black) and viral DNA (red), were fragmented and hybridized with biotin (green)‐labeled RNA probes spanning the EBV genome. Captured EBV DNA were then subjected to deep sequencing using a short‐read sequencer. (Right) For EBV‐bacterial artificial chromosome (BAC) cloning, a linear targeting vector was transfected into EBV latently infected cells to obtain homologous recombinants. Alternatively, a circular donor plasmid and a CRISPR/Cas9 plasmid were co‐transfected into the cells to increase the homologous recombination efficiency.46, 47 Episomal DNA were prepared and used to transform bacterial cells and obtain EBV‐BAC clones
This new technology marks a new era of EBV genome sequencing. Until 2013, EBV whole genome sequences available from GenBank were limited to less than 10 strains (B95‐8, EBV‐wt, GD1,31 AG876,32 GD2,33 HKNPC1,34 and Akata and Mutu35) (Figure 4). The number dramatically increased after 2014, when the hybrid capture method was developed. The method was applied to readily determine eight EBV strains derived from NPC biopsy specimens.30 The same experimental strategy was used by the group of Farrell and colleagues to sequence 71 EBV strains derived from various areas of the world.36 The sequenced materials include various EBV disease‐derived cell lines, spontaneously arising LCL, NPC tumor and biopsy samples, and saliva. Many of the Asian strains in the dataset were from Southern China, an NPC endemic area, and a few were from Japan and South Korea. This analysis clearly separated Asian EBV strains from those derived from the rest of the world.
Figure 4.
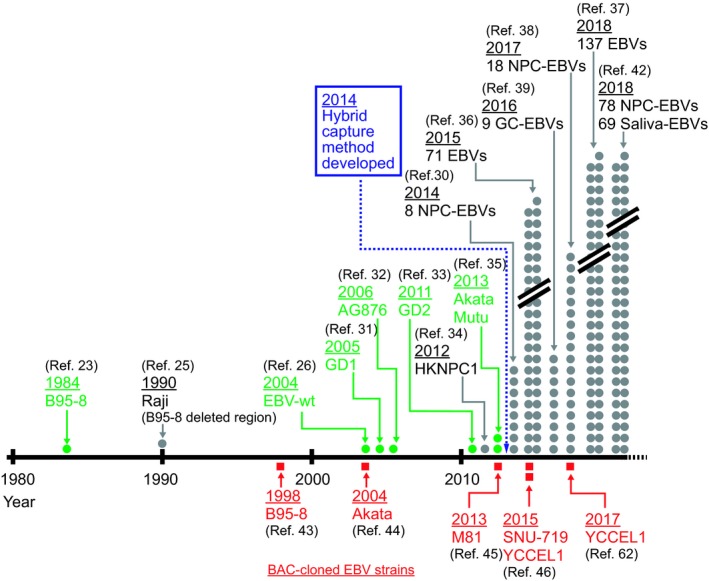
Epstein‐Barr virus (EBV) sequencing milestones and the recent explosive increase in EBV whole genome sequences. Green dots indicate EBV strains whose sequences were completely determined, whereas gray circles indicate those in which genome sequences were determined except for the repetitive regions. Red squares indicate EBV strains that were cloned as bacterial artificial chromosome (BAC) clones and completely sequenced. GC, gastric cancer. References (Ref.) are indicated. NPC, nasopharyngeal carcinomas
The number of available EBV sequences is increasing exponentially (Figure 4), and approximately 200 EBV genomes have been sequenced by the group of Farrell and colleagues.36, 37 Eighteen additional EBV strains derived from NPC biopsy specimens38 and nine EBV strains derived from biopsy specimens of Chinese EBVaGC patients39 were sequenced by other groups.
One drawback of the hybrid capture approach is that repetitive sequences (Figure 2) cannot be determined because the method uses short‐read sequencers.36 Most of the repetitive regions therefore remain to be sequenced. Another drawback is that sequence assembly becomes difficult when multiple EBV strains are present in the material, which is often the case in the saliva of healthy individuals40, 41 and that of infectious mononucleosis patients.42
3.3. Cloning of the EBV genome and sequencing
To circumvent the problems associated with the hybrid capture method, the DNA of each EBV genome can be cloned prior to sequencing. Cloning of EBV genomes can be achieved by inserting a BAC vector sequence into EBV episomes in EBV latently infected cells through homologous recombination (Figure 3). The experimental strategy was pioneered by the group of Hammerschmidt and colleagues43 using the prototype B95‐8 strain as the starting material, followed by BAC cloning of the Japanese Burkitt lymphoma‐derived Akata strain by our group.44 The experimental strategy was recently applied to the Chinese NPC‐derived EBV M81 strain,45 which had not been sequenced previously. The obtained M81 EBV‐BAC was sequenced using a short‐read sequencer and Sanger sequencing‐mediated gap filling. Phylogenetic analysis indicated that M81 is closely related to other NPC strains (GD2 and HKNPC1), whereas it is distant from other strains (B95‐8 and Mutu).45
We recently improved the EBV‐BAC cloning efficiency using CRISPR/Cas9‐mediated genome editing technology to introduce a single cut at the transgene insertion site and stimulate homologous recombination (Figure 3).46, 47 A long‐read sequencer (PacBio Menlo Park, CA, USA) was then used to obtain “complete” EBV genome sequences, including the repetitive regions, of two gastric cancer‐derived EBV strains (SNU‐719 and YCCEL1). Sequence comparison of SNU‐719 and YCCEL1 indicates that, although they are not identical, they belong to the same group of Asian type EBV.
Currently, EBV‐BAC cloning can only be applied to established EBV‐positive cell lines. In addition, converting the strategy into a high‐throughput method is difficult. However, one advantage of EBV‐BAC cloning is that it can be used to clone multiple strains that coexist in a single specimen. In addition, purified EBV‐BAC clone DNA can be used to reconstitute infectious viruses and examine their phenotypes (Figure 3, see below).
4. EBV GENOME HETEROGENEITY
4.1. Overview of viral genome heterogeneity
A methodology formerly used to detect EBV strain variation is detection of restriction fragment length polymorphisms by means of Southern blot.17, 40 Sanger sequencing of specific viral genes (e.g. LMP1, EBNA1, BZLF1) was later used to detect sequence diversity. Genome‐wide analysis is becoming possible based on the results of high‐throughput sequencing.
The first major EBV variants identified were type 1 (type A) and type 2 (type B).1 Type 1 EBV (e.g. B95‐8, GD1 and Akata) is the main EBV strain prevalent worldwide; type 2 EBV (e.g. AG876 and P3HR‐1) is as abundant as type 1 EBV in sub‐Saharan Africa. Type 1 and type 2 EBV encode different EBNA2 genes, with only 54% amino acid sequence identity.48 A recent whole genome sequencing study confirmed that EBNA2 and EBNA3 are the only genes that can distinguish type 1 and type 2 EBV strains.36
Various SNP are identified in viral latent and lytic genes. EBV latent genes are more heterogeneous than viral lytic genes.36, 49 Although relatively little attention has been paid to the heterogeneity of viral lytic genes, it is becoming clear that they play significant roles in EBV‐mediated tumorigenesis.
Relatively large deletions within the EBV genome have also been detected in B95‐8 strain24 and in several Burkitt lymphoma‐derived strains.50
In the following sections, we summarize recent findings of viral genome heterogeneity related to cancer.
4.2. Viral latent gene heterogeneity and deletions in cancer
The essential roles of viral latent proteins in EBV‐mediated transformation have prompted an extensive analysis of latent gene heterogeneity, especially that of LMP1.5, 6, 7, 8, 9 LMP1 genes are classified into six patterns, namely China 1, China 2, Alaskan, Mediterranean, NC (North Carolina) and B95‐8.51 The functional differences between the LMP1 of epithelial cancer‐derived EBV (CAO‐LMP1, belonging to China 1 type) and lymphoid‐derived EBV (B95‐8 type) are well‐characterized.52 NPC‐derived CAO‐LMP1 is less cytostatic than B95‐8 LMP1 (Figure 5).53 Such functional difference is attributed to amino acid differences in the transmembrane domain of LMP1, and is not related to the 30‐bp deletion in the carboxy‐terminal cytoplasmic domain.53 Functional differences in LMP1 most likely contribute to differences in viral phenotypes, although this remains to be experimentally verified.
Figure 5.
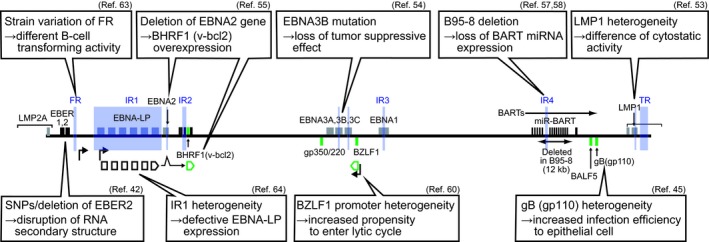
Summary of Epstein‐Barr virus (EBV) gene heterogeneity possibly affecting EBV‐mediated carcinogenesis. References (Ref.) are indicated. EBER, EBV‐encoded RNA; EBNA, EBV nuclear antigen; FR, family of repeats; IR, internal repeats; LMP1, latent membrane protein 1; SNP, single nucleotide polymorphism; TR, terminal repeats
It was previously demonstrated that EBNA3B acts as a tumor suppressor (Figure 5).54 Recombinant EBV lacking the EBNA3B gene shows higher B‐cell‐transforming ability than the wild type, and EBV‐transformed B cells lacking the EBNA3B gene expand more rapidly in mice than those having the EBNA3B gene. EBNA3B mutations were detected in various EBV‐positive human lymphoma samples,54 suggesting that EBV strains with EBNA3B gene mutations are selected in these tumors.
Deletion of the EBNA2 gene is commonly found in Burkitt lymphoma‐derived cell lines, such as P3HR‐1 and Daudi (Figure 5).50 The EBV genome lacking the EBNA2 gene shows a specific latent gene expression pattern55 characterized by the expression of BHRF1 (a viral counterpart of cellular blc‐2), which confers anti‐apoptotic activity to Burkitt lymphoma cells (Figure 5). This suggests that the EBV genome with deletion of the EBNA2 gene is selected to confer growth advantages during Burkitt lymphomagenesis.
Another well‐characterized EBV alteration is the region deleted in the B95‐8 strain (Figure 5). The B95‐8 deletion removes most of the BART miRNA sequences and one lytic origin of replication (Figure 2). Although this deletion is apparently dispensable for EBV‐mediated B‐cell transformation, it may affect EBV‐mediated epithelial carcinogenesis, as BART miRNA are highly expressed in EBV‐positive NPC and EBVaGC.56 A pair of recombinant viruses with and without BART miRNA regions was used to identify NDRG1 (N‐myc downstream‐regulated gene 1), an epithelial cell‐specific metastasis suppressor, as a possible target of BART miRNA.57 Another study demonstrated that BART miRNA of M81 strain EBV play multiple roles following infection of primary B lymphocytes, such as suppression of lytic replication and suppression of tumor growth in mice.58 However, because of the lack of an appropriate infection system for epithelial cells, the behavior of the M81 virus during epithelial cell infection remains to be clarified.
A recent case‐control study compared EBV sequences isolated from saliva (of 142 population carriers) with those isolated from primary NPC biopsy samples (62 patients).42 Cluster analysis identified five subgroups (type 1 A‐C and type 2 A‐B) in population carriers, and type 1A and 1B were predominant in NPC patients. A genome‐wide association study identified a panel of NPC‐associated SNP and insertion/deletions. The most significant polymorphism is a 4‐base deletion polymorphism downstream of the EBER2 gene (Figure 5). The result may be related to previous findings that EBER2, but not EBER1, contributes to the efficiency of EBV‐mediated transformation.59
4.3. Viral lytic gene heterogeneity in cancer
Research interest has focused on a viral BZLF1 gene, a master regulator of viral lytic replication.1 BZLF1 is highly polymorphic both for its coding sequence and promoter region.7, 60
When the NPC‐derived M81 strain was reconstituted and used to immortalize B lymphocytes, M81 EBV spontaneously replicated at unusually high levels in established LCL.45 This is consistent with the observation that NPC patients have unusually high antibody titers against EBV lytic proteins.2 The propensity to enter lytic infection is believed to enhance the malignant potential in animal models of lymphomagenesis.61 A recent study further demonstrated that sequence variation of the BZLF1 promoter region of M81 EBV contributes to the property to enter the lytic cycle.60
The M81 virus and other epithelial cancer‐derived EBV strains show high propensities to infect epithelial cells.45, 62 Epitheliotropism is apparently advantageous for the virus to increase the chance to infect the nasopharyngeal epithelium. The viral gB protein (gp110) of the M81 strain is at least partly responsible for the enhanced tropism toward epithelial cells (Figure 5).45
4.4. Other heterogeneities possibly affecting carcinogenesis
Subtle variations of repetitive sequences, such as those of FR63 and IR1,64 can significantly affect viral phenotypes. The IR1 heterogeneity generates a modestly defective EBNA‐LP and has a detrimental impact on B‐cell transforming ability (Figure 5).64
Sequence variations of EBV genes also result in amino acid epitope exchanges, which should have a significant impact on EBV‐specific T‐cell immunity.65 EBV latency‐associated gene products, especially EBNA3, are targets of immune recognition during persistent infection. A higher frequency of SNP and a higher ratio of non‐synonymous amino acid changes in latency‐associated genes suggest that these genes are evolving under positive selection to escape host immunity (Figure 5).36 Sequence polymorphisms of LMP2A, an EBV‐encoded B‐cell receptor mimic, have been detected in clinical isolates from various diseases, but their pathogenetic roles remain unclear.6
5. CONCLUDING REMARKS
Numerous efforts have been made to determine whether cancer‐specific EBV variants exist. However, no definitive answers have been obtained to date. In the case of HPV, HPV 16 and 18 are well‐characterized as high‐risk HPV.66 The EBV genome (175 kb) is more than 20‐fold larger than that of HPV (~8000 bp) and encodes approximately 10‐fold more ORF. The identification of high‐risk EBV is difficult because multiple viral gene heterogeneities likely cooperate in the acquisition of oncogenic potential. In addition, there is no experimental system to investigate phenotypic changes caused by heterogeneities.
Nasopharyngeal carcinomas researchers started conducting case‐control studies to identify high‐risk EBV that are tightly associated with NPC. Future studies should focus on verifying the biological significance of EBV sequence variations. It is now possible to establish new EBV‐positive NPC cell lines directly from patient NPC tissues.67 CRISPR/Cas9‐mediated rapid cloning method46, 47 can then be applied to the cell lines. EBV strain variation in EBVaGC is also being investigated,39 and EBV‐associated T/NK lymphomas should be a future target.
Knowledge about EBV strain variation is important for clinical applications. The identification of high‐risk groups of people who are predisposed to NPC should have significant impact on the early diagnosis of NPC. In addition, this knowledge is important for developing EBV vaccines and anti‐EBV T‐cell therapy to prevent the attenuation of antiviral T‐cell immunity caused by viral strain variation.
CONFLICTS OF INTEREST
The authors have no conflict of interest.
NOTE
After this review was submitted, a paper was published online, highlighting a pathogenic link between intragenic EBV deletions and EBV‐associated neoplastic disorders, including T/NK lymphomas. (Okuno Y, Murata T, Sato Y, et al. Defective Epstein‐Barr virus in chronic active infection and haematological malignancy. Nat Microbiol. 2019. https://doi.org/10.1038/s41564‐018‐0334‐0).
ACKNOWLEDGMENTS
This work was funded by Japan Society for the Promotion of Science (JSPS) KAKENHI (15K14391, T. Kanda) and by the Japan Agency for Medical Research and Development (Japanese Initiative for Progress of Research on Infectious Disease for Global Epidemic, 17fm0208016, T. Kanda).
Kanda T, Yajima M, Ikuta K. Epstein‐Barr virus strain variation and cancer. Cancer Sci. 2019;110:1132–1139. 10.1111/cas.13954
REFERENCES
- 1. Kieff E, Rickinson AB. Epstein‐Barr virus and its replication In: Knipe DM, Howley PM, eds. Fields Virology, 5th edn Philadelphia: Lippincott Williams & Wilkins; 2007:2603‐2654. [Google Scholar]
- 2. Rickinson AB, Kieff E. Epstein‐Barr virus In: Knipe DM, Howley PM, eds. Fields Virology, 5th edn Philadelphia: Lippincott Williams & Wilkins; 2007:2655‐2700. [Google Scholar]
- 3. Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. The extent of genetic diversity of Epstein‐Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res. 2009;143:209‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwok H, Chiang AK. From conventional to next generation sequencing of Epstein‐Barr virus genomes. Viruses. 2016;8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi SJ, Jung SW, Huh S, Cho H, Kang H. Phylogenetic comparison of Epstein‐Barr virus genomes. J Microbiol. 2018;56:525‐533. [DOI] [PubMed] [Google Scholar]
- 6. Farrell PJ. Epstein‐Barr virus strain variation. Curr Top Microbiol Immunol. 2015;390:45‐69. [DOI] [PubMed] [Google Scholar]
- 7. Feederle R, Klinke O, Kutikhin A, Poirey R, Tsai MH, Delecluse HJ. Epstein‐Barr virus: from the detection of sequence polymorphisms to the recognition of viral types. Curr Top Microbiol Immunol. 2015;390:119‐148. [DOI] [PubMed] [Google Scholar]
- 8. Neves M, Marinho‐Dias J, Ribeiro J, Sousa H. Epstein‐Barr virus strains and variations: geographic or disease‐specific variants? J Med Virol. 2017;89:373‐387. [DOI] [PubMed] [Google Scholar]
- 9. Tzellos S, Farrell PJ. Epstein‐Barr virus sequence variation‐biology and disease. Pathogens. 2012;1:156‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. 1964;1:702‐703. [DOI] [PubMed] [Google Scholar]
- 11. Molyneux EM, Rochford R, Griffin B, et al. Burkitt's lymphoma. Lancet. 2012;379:1234‐1244. [DOI] [PubMed] [Google Scholar]
- 12. Mahdavifar N, Ghoncheh M, Mohammadian‐Hafshejani A, Khosravi B, Salehiniya H. Epidemiology and inequality in the incidence and mortality of nasopharynx cancer in Asia. Osong Public Health Res Perspect. 2016;7:360‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen JI, Fauci AS, Varmus H, Nabel GJ. Epstein‐Barr virus: an important vaccine target for cancer prevention. Sci Transl Med. 2011;3:107 fs107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hildesheim A, Wang CP. Genetic predisposition factors and nasopharyngeal carcinoma risk: a review of epidemiological association studies, 2000‐2011: Rosetta Stone for NPC: genetics, viral infection, and other environmental factors. Semin Cancer Biol. 2012;22:107‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liebowitz D. Nasopharyngeal carcinoma: the Epstein‐Barr virus association. Semin Oncol. 1994;21:376‐381. [PubMed] [Google Scholar]
- 16. Dai W, Zheng H, Cheung AK, Lung ML. Genetic and epigenetic landscape of nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5:16. [DOI] [PubMed] [Google Scholar]
- 17. Abdel‐Hamid M, Chen JJ, Constantine N, Massoud M, Raab‐Traub N. EBV strain variation: geographical distribution and relation to disease state. Virology. 1992;190:168‐175. [DOI] [PubMed] [Google Scholar]
- 18. Kimura H. EBV in T‐/NK‐cell tumorigenesis In: Kimura H, ed. Kawaguchi YM, Y. Human Herpesviruses: Springer; 2018:459‐475. [Google Scholar]
- 19. Kimura H, Ito Y, Kawabe S, et al. EBV‐associated T/NK‐cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood. 2012;119:673‐686. [DOI] [PubMed] [Google Scholar]
- 20. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaneda A, Matsusaka K, Aburatani H, Fukayama M. Epstein‐Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445‐3450. [DOI] [PubMed] [Google Scholar]
- 22. Fukayama M, Kunita A, Kaneda A. Gastritis‐infection‐cancer sequence of Epstein‐Barr virus‐associated gastric cancer. Adv Exp Med Biol. 2018;1045:437‐457. [DOI] [PubMed] [Google Scholar]
- 23. Baer R, Bankier AT, Biggin MD, et al. DNA sequence and expression of the B95‐8 Epstein‐Barr virus genome. Nature. 1984;310:207‐211. [DOI] [PubMed] [Google Scholar]
- 24. Raab‐Traub N, Dambaugh T, Kieff E. DNA of Epstein‐Barr virus VIII: B95‐8, the previous prototype, is an unusual deletion derivative. Cell. 1980;22:257‐267. [DOI] [PubMed] [Google Scholar]
- 25. Parker BD, Bankier A, Satchwell S, Barrell B, Farrell PJ. Sequence and transcription of Raji Epstein‐Barr virus DNA spanning the B95‐8 deletion region. Virology. 1990;179:339‐346. [DOI] [PubMed] [Google Scholar]
- 26. de Jesus O, Smith PR, Spender LC, et al. Updated Epstein‐Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J Gen Virol. 2003;84:1443‐1450. [DOI] [PubMed] [Google Scholar]
- 27. Klinke O, Feederle R, Delecluse HJ. Genetics of Epstein‐Barr virus microRNAs. Semin Cancer Biol. 2014;26C:52‐59. [DOI] [PubMed] [Google Scholar]
- 28. Kanda T. EBV‐encoded latent genes. Adv Exp Med Biol. 2018;1045:377‐394. [DOI] [PubMed] [Google Scholar]
- 29. Depledge DP, Palser AL, Watson SJ, et al. Specific capture and whole‐genome sequencing of viruses from clinical samples. PLoS ONE. 2011;6:e27805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwok H, Wu CW, Palser AL, et al. Genomic diversity of Epstein‐Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol. 2014;88:10662‐10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeng MS, Li DJ, Liu QL, et al. Genomic sequence analysis of Epstein‐Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J Virol. 2005;79:15323‐15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dolan A, Addison C, Gatherer D, Davison AJ, McGeoch DJ. The genome of Epstein‐Barr virus type 2 strain AG876. Virology. 2006;350:164‐170. [DOI] [PubMed] [Google Scholar]
- 33. Liu P, Fang X, Feng Z, et al. Direct sequencing and characterization of a clinical isolate of Epstein‐Barr virus from nasopharyngeal carcinoma tissue by using next‐generation sequencing technology. J Virol. 2011;85:11291‐11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwok H, Tong AH, Lin CH, et al. Genomic sequencing and comparative analysis of Epstein‐Barr virus genome isolated from primary nasopharyngeal carcinoma biopsy. PLoS ONE. 2012;7:e36939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin Z, Wang X, Strong MJ, et al. Whole‐genome sequencing of the Akata and Mutu Epstein‐Barr virus strains. J Virol. 2013;87:1172‐1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palser AL, Grayson NE, White RE, et al. Genome diversity of Epstein‐Barr virus from multiple tumor types and normal infection. J Virol. 2015;89:5222‐5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Correia S, Bridges R, Wegner F, et al. Sequence variation of Epstein‐Barr virus: viral types, geography, codon usage, and diseases. J Virol. 2018;92:pii: e01132‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tu C, Zeng Z, Qi P, et al. Genome‐wide analysis of 18 Epstein‐Barr viruses isolated from primary nasopharyngeal carcinoma biopsy specimens. J Virol. 2017;91:e00301‐e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y, Yang W, Pan Y, Ji J, Lu Z, Ke Y. Genome‐wide analysis of Epstein‐Barr virus (EBV) isolated from EBV‐associated gastric carcinoma (EBVaGC). Oncotarget. 2016;7:4903‐4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sitki‐Green D, Covington M, Raab‐Traub N. Compartmentalization and transmission of multiple epstein‐barr virus strains in asymptomatic carriers. J Virol. 2003;77:1840‐1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walling DM, Brown AL, Etienne W, Keitel WA, Ling PD. Multiple Epstein‐Barr virus infections in healthy individuals. J Virol. 2003;77:6546‐6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hui KF, Chan TF, Yang W, et al. High risk Epstein‐Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int J Cancer. 2018. 10.1002/ijc.32049 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 43. Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein‐Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A. 1998;95:8245‐8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kanda T, Yajima M, Ahsan N, Tanaka M, Takada K. Production of high‐titer Epstein‐Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J Virol. 2004;78:7004‐7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsai MH, Raykova A, Klinke O, et al. Spontaneous lytic replication and epitheliotropism define an Epstein‐Barr virus strain found in carcinomas. Cell Rep. 2013;5:458‐470. [DOI] [PubMed] [Google Scholar]
- 46. Kanda T, Furuse Y, Oshitani H, Kiyono T. Highly efficient CRISPR/Cas9‐mediated cloning and functional characterization of gastric cancer‐derived Epstein‐Barr virus strains. J Virol. 2016;90:4383‐4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yajima M, Ikuta K, Kanda T. Rapid CRISPR/Cas9‐mediated cloning of full‐length Epstein‐Barr virus genomes from latently infected cells. Viruses. 2018;10:E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein‐Barr virus DNA may encode Epstein‐Barr nuclear antigen 2. Proc Natl Acad Sci U S A. 1984;81:7632‐7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGeoch DJ, Gatherer D. Lineage structures in the genome sequences of three Epstein‐Barr virus strains. Virology. 2007;359:1132‐5. [DOI] [PubMed] [Google Scholar]
- 50. Kelly G, Bell A, Rickinson A. Epstein‐Barr virus‐associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat Med. 2002;8:1098‐1104. [DOI] [PubMed] [Google Scholar]
- 51. Edwards RH, Seillier‐Moiseiwitsch F, Raab‐Traub N. Signature amino acid changes in latent membrane protein 1 distinguish Epstein‐Barr virus strains. Virology. 1999;261:79‐95. [DOI] [PubMed] [Google Scholar]
- 52. Dawson CW, Eliopoulos AG, Blake SM, Barker R, Young LS. Identification of functional differences between prototype Epstein‐Barr virus‐encoded LMP1 and a nasopharyngeal carcinoma‐derived LMP1 in human epithelial cells. Virology. 2000;272:204‐217. [DOI] [PubMed] [Google Scholar]
- 53. Johnson RJ, Stack M, Hazlewood SA, et al. The 30‐base‐pair deletion in Chinese variants of the Epstein‐Barr virus LMP1 gene is not the major effector of functional differences between variant LMP1 genes in human lymphocytes. J Virol. 1998;72:4038‐4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. White RE, Ramer PC, Naresh KN, et al. EBNA3B‐deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J Clin Invest. 2012;122:1487‐1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kelly GL, Milner AE, Baldwin GS, Bell AI, Rickinson AB. Three restricted forms of Epstein‐Barr virus latency counteracting apoptosis in c‐myc‐expressing Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 2006;103:14935‐14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lo AK, Dawson CW, Jin DY, Lo KW. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J Pathol. 2012;227:392‐403. [DOI] [PubMed] [Google Scholar]
- 57. Kanda T, Miyata M, Kano M, Kondo S, Yoshizaki T, Iizasa H. Clustered microRNAs of the Epstein‐Barr virus cooperatively downregulate an epithelial cell‐specific metastasis suppressor. J Virol. 2015;89:2684‐2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin X, Tsai MH, Shumilov A, et al. The Epstein‐Barr virus BART miRNA cluster of the M81 strain modulates multiple functions in primary B cells. PLoS Pathog. 2015;11:e1005344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu Y, Maruo S, Yajima M, Kanda T, Takada K. Epstein‐Barr virus (EBV)‐encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV‐induced B‐cell growth transformation. J Virol. 2007;81:11236‐11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bristol JA, Djavadian R, Albright ER, et al. A cancer‐associated Epstein‐Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog. 2018;14:e1007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ma SD, Hegde S, Young KH, et al. A new model of Epstein‐Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J Virol. 2011;85:165‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsai MH, Lin X, Shumilov A, et al. The biological properties of different Epstein‐Barr virus strains explain their association with various types of cancers. Oncotarget. 2017;8:10238‐10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ali AK, Saito S, Shibata S, Takada K, Kanda T. Distinctive effects of the Epstein‐Barr virus family of repeats on viral latent gene promoter activity and B‐lymphocyte transformation. J Virol. 2009;83:9163‐9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ba Abdullah MM, Palermo RD, Palser AL, et al. Heterogeneity of the Epstein‐Barr virus (EBV) major internal repeat reveals evolutionary mechanisms of EBV and a functional defect in the prototype EBV strain B95‐8. J Virol. 2017;91:e00920–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cirac A, Stutzle S, Dieckmeyer M, et al. Epstein‐Barr virus strain heterogeneity impairs human T‐cell immunity. Cancer Immunol Immunother. 2018;67:663‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Howley PM, Lowy DR. Papillomaviruses In: Knipe DM, Howley PM, eds. Fields Virology, 5th edn Philadelphia: Lippincott Williams & Wilkins; 2007:2299‐2354. [Google Scholar]
- 67. Lin W, Yip YL, Jia L, et al. Establishment and characterization of new tumor xenografts and cancer cell lines from EBV‐positive nasopharyngeal carcinoma. Nat Commun. 2018;9:4663. [DOI] [PMC free article] [PubMed] [Google Scholar]


