Abstract
For exploration of chemical and biological systems, the combined quantum mechanics and molecular mechanics (QM/MM) and machine learning (ML) models have been developed recently to achieve high accuracy and efficiency for molecular dynamics (MD) simulations. Despite its success on reaction free energy calculations, how to identify new configurations on insufficiently sampled regions during MD and how to update the current ML models with the growing database on-the-fly are both very important but still challenging. In this letter, we apply the QM/MM ML method to solvation free energy calculations and address these two challenges. We employ three approaches to detect new data points and introduce the gradient boosting algorithm to reoptimize efficiently the ML model during ML-based MD sampling. The solvation free energy calculations on several typical organic molecules demonstrate that our developed method provides a systematic, robust and efficient way to explore new chemistry using ML-based QM/MM MD simulations.
Graphical Abstract

Introduction
The solvation free energy (SFE) is vital for studying chemical and biological reactions as well as designing drugs.1–4 Many methods of free energy calculations, such as free energy perturbation,5 thermodynamic integration (TI),6 umbrella sampling7 and λ-dynamics,8 have been widely used to calculate SFE from molecular dynamics (MD) simulations.9 On one hand, sufficient MD samplings for free energy calculations require an efficient method to calculate the potential energies of the whole system for at least thousands of configurations. On the other hand, an important ingredient to accurately estimate SFE is the appropriate description of potential energy surface (PES). Although empirical models such as molecular mechanical (MM) force fields can predict the SFE of various small organic molecules successfully,10–14 quantum mechanical (QM) models at the ab initio level are considered to be more general and rigorous. Treating both the solute and solvent molecules using QM method is extremely time-consuming if not impossible. The combined quantum mechanical and molecular mechanical (QM/MM) method, first proposed by Warshel, Karplus and Levitt, considers a tradeoff between accuracy and efficiency for large systems.15,16 In the framework of QM/MM model, the whole system is divided into a small active site (e.g., the solute) treated at the QM level and a relatively large surrounding part (e.g., the solvents) treated at the MM level. Even though the ab initio QM/MM can achieve high accuracy for PES, it is still computationally demanding in long-time MD simulations.17–26 Alternatively, the semiempirical QM/MM (SQM/MM) significantly speeds up electronic structure calculations but sometimes sacrifices the accuracy.27–29
Recently, many kinds of machine learning (ML) techniques such as neural network (NN) representation,30–32 kernel-based Gaussian approximation potential33,34 and support vectormachine35 are being increasingly used for accurate interpolation/prediction of PES.36–39 Inspired by the high-dimensional NN scheme to represent the total potential energy as a sum of atomic energies32 and the Δ-machine learning method to predict the difference between the calculations at two levels,40 Shen and Yang developed a novel method to introduce the machine learning methods to QM/MM models, denoted as QM/MM-NN, to achieve the accuracy of high-level ab initio QM/MM with only the computational cost of low-level SQM/MM.41,42 The highly accurate free energy profiles on aqueous chemical processes such as SN2 and proton transfer reactions can be obtained from direct QM/MM MD simulations combined with QM/MM-NN.
Although there is a growing trend in molecular simulations based on the ML-predicted PES, how to detect new configurations outside of the domain of training data remains challenging. One choice is based on the boundary of input variables of the current ML model, which may miss many new data points encountered during MD.43 Another way is based on the uncertainty analysis on a machine learning ensemble,44,45 which may also lead to failure on MD simulations, according to the results of our SFE calculation. Furthermore, in order to explore new regions of the PES, a robust and efficient method to update the current ML model during MD simulations is also an urgent issue. Several adaptive methods have been reported and can be further classified as “microiteration” and “macroiteration” procedures. In the microiteration approach, the ML model was adjusted immediately to mimic the reference when a new conformation was sampled. Despite the success of the related works such as the adaptive ML framework44,46 and “learn-on-the-fly” technique,47,48 there are still some challenges that should be addressed. Specially, the reconstruction of ML models is usually frequent and expensive, which hampers the efficiency of ML for exploring diverse configurations. In the macroiteration approach, for example, the adaptive QM/MM-NN proposed by Shen and Yang recently,42 the NN construction and MD simulation were performed iteratively. The ML model was unchanged during MD samplings at each iteration cycle and reconstructed using both existing and additional configurations after MD. This approach requires a large number of MD sampling steps before obtaining the final ML model. It may be unavailable for larger systems on which the SQM/MM computational cost becomes considerable. How to explore something new for large systems using ML models, even in the configurational space, is still an open problem without satisfactory solutions.
In this paper, we will address the two aforementioned challenges on the ML-based QM/MM MD for free energy calculations. First, we explore a systematic way for the identification of insufficiently sampled regions during ML-driven MD in order to overcome the limitations on the existing methods based on the boundary of input variables or the uncertainty of machine learning ensembles. Second, we employ the gradient boosting algorithm, which was first reported by Friedman49 and followed by classification and regression applications50 such as the predictions on elimination half-lives51 and RNA-protein interactions,52 to update the QM/MM ML model effectively when a new configuration is detected. We finally perform SFE calculations on several typical organic molecules to validate our new QM/MM ML approaches.
Methods
For the SFE calculations with QM/MM MD,53–56 the total potential energy of the system with QM/MM model we use is57
| (1) |
where λele and λvdw are two switching parameters varying from 0 to 1 for TI, is the Hamiltonian of the QM subsystem, qi is the point charge of MM atom i, VMM(ri) is the electrostatic potential on MM atom i exerted by the QM subsystem, ri is the position of atom i, and is the soft-core potential that can avoid numerical instability problem in MD simulation when λvdw is close to zero (see Simulation Details in SI).58 Applying the same MM force field at two levels, the energy difference ΔE between high-level ab initio QM/MM (labeled as H) and low-level SQM/MM (labeled as L) is represented as41
| (2) |
To properly represent the atomistic environment, two types of descriptors, modified symmetry functions32 and power spectrum,59 were employed as the input variables in this work. The same cutoff function60 is involved in both input variables as
| (3) |
where Rij is the distance between atom i and j, and Rc is the cutoff radius. The interaction is neglected if the distance between two atoms is larger than Rc. Symmetry functions consist of a radial function
| (4) |
where η and Rs are pre-determined hyperparameters, and an angular function
| (5) |
where ζ and λ are hyperparameters, and θijk is the angle centered at atom i. Since the radial and angular symmetry functions are only dependent on QM geometry, the external electrostatic potential generated by surrounding charges is employed as another input feature to describe the influence of MM subsystem, which is written as42
| (6) |
where qj is the point charge on MM atom j. Different from symmetry functions, the input variables of power spectrum are defined as
| (7) |
where n and l are pre-determined integers, m is an integer varying from −l to l, and is constructed as
| (8) |
where α and Rn are hyperparameters, Ylm is the spherical harmonic function to describe the relative positions of i and j using spherical coordinates θ and φ centered at atom i, and ωj is defined as the nuclear charge Zj if atom j belongs to the QM subsystem or defined as λeleqj if atom j belongs to the MM subsystem.61 The original power spectrum59 applied orthonormalized high-order polynomials as radial basis functions, but here we change them to equidistant Gaussian functions together with cutoff functions because they are sensitive and cover wide range of distances, which satisfies the requirements mentioned in the original paper.59 Note that in the framework of modified symmetry functions, two functions, symmetry functions in Eqs 4 and 5 and external electrostatic potentials in Eq 6, are employed to describe the QM and MM subsystem, respectively; while in the framework of power spectrum, QM and MM subsystems are described with the same function as in Eq 7 with different hyperparameters.
Using modified symmetry functions or power spectrum as input variables, the energy difference ΔE in Eq 2 can be predicted with any ML method such as neural network or Gaussian kernel. Here we use the linear regression model, which has successfully described the energy landscape of metal systems with symmetry functions,62,63 as
| (9) |
where xj is the input variables with the total number of p, and aj is the ML parameters that can be obtained using the component-wise gradient boosting algorithm64 combined with the early stopping technique65,66 (see below for more details). Actually, the initial QM/MM ML model can be constructed with any other training method, not restricted to gradient boosting. The forces acting on atom i can be calculated as
| (10) |
where atom i belongs to either QM or MM subsystem, and is the low-level force calculated with SQM/MM. Using Eq 10, MD evolution can be implemented on the ML-predicted PES. The final SFE through TI is calculated as
| (11) |
where the angular bracket denotes an average over the samplings with a fixed λ.
Although many ML predictions are capable of reaching beyond the chemical accuracy,39,46,67,68 both the energy and forces of some configurations in new regions on PES, where the samples in the existing database are insufficient, are unreliable using the current ML model. A systematic and effective approach to detect such new configurations is necessary. Here we applied three different methods based on the boundary of reference energy, the clustering of data points69 and the Gaussian distribution on the density of data space,70 respectively. The first method is new to our best knowledge. The second and third are popular clustering algorithms that have been successfully used to solve classification and regression problems.71–76 For examples, the k-means clustering and Gaussian mixture model have been widely used in molecular simulations, such as enhanced sampling, identifying states, and free energy calculations.77–82 Here we introduce them to the field of ML-driven MD simulations for real-time detection of new configurations, that is, learn-on-the-fly, because of the insufficiency of the database. The additional computational cost on all methods is small.
The first is denoted as the output-check model (Model 1), which corresponds to the boundary of reference energy. This model is based on the minimum and maximum values of the reference energy in the database. If the predicted value for a configuration is smaller than the minimum or larger than the maximum, the configuration will be identified as a new point and added into the database.
The second is denoted as the k-means clustering model (Model 2), in which the kmeans clustering algorithm is employed to classify the existing data points into k numbers of clusters. The initial centroids are selected by the k-means++,83 and then an iterative procedure is performed to determine optimal clusters.69 The database will be extended if the configuration encountered during MD does not belong to any cluster.
The third is denoted as the Gaussian model (Model 3) in which the density of data space is described with multiple Gaussian distributions.70 This model is constructed as follows and more details are illustrated in SI. First, the k-th Gaussian distribution is represented with the mean vector μk and covariance matrix Σk, and the percentage of the k-th Gaussian distribution (denoted as Pk) are initialized using the k-means clustering based on the existing data points. Second, the probability of a configuration xi in the k-th Gaussian distribution, denoted as P(xi ∈ k), is expressed as
| (12) |
where Norm(xi|μk, σk) denotes the k-th Gaussian distribution. The Pk, μk and Σk are then recalculated based on P(xi ∈ k) for all existing configurations in the current database, leading to the updated values of P(xi ∈ k). The iterative procedure is repeated until Pk is unchanged for all Gaussian distributions or the maximum step is reached. Finally, a new configuration x encountered during MD is checked using
| (13) |
and added to the database if P (x) is smaller than the pre-determined threshold.
Once the database is extended during MD simulations on the ML-predicted PES, the ab initio QM/MM potential energy of the additional configuration should be calculated as the reference value, and the current QM/MM ML model should be updated to explore broader regions on PES. Here we applied the component-wise gradient boosting algorithm64 for reoptimization. The feature of gradient boosting is to add some weak models (called as the base learner) to the existing ML model to build a stronger model. No matter how complicated the initial ML model is, only the base learner is necessary to be optimized to correct the errors of the existing model. In other words, the choice on base learner for reoptimization is independent of the previous construction, indicating the high efficiency of reoptimization regardless of the complexity of the initial ML model. Generally, the base learner is very simple, e.g., the linear functions in this work, so the computational cost of on-the-fly reconstructions on ML model during ML-driven MD is low. In contrast, frequent reoptimizations on a complicated ML model with either microiteration44,46–48 or macroiteration42 procedure are usually time-consuming. The detailed procedure of the component-wise gradient boosting algorithm used in this work are illustrated in SI.
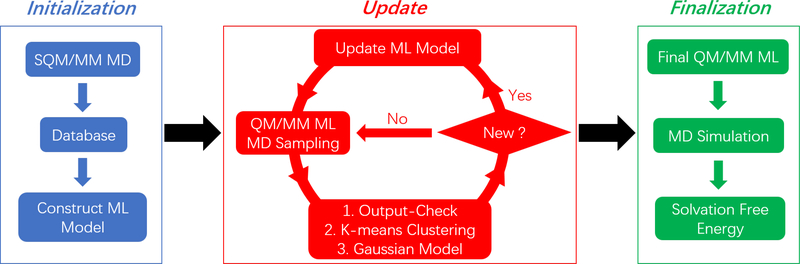
The whole procedure of our method is demonstrated in Figure 1. At the initialization stage, MD simulations with different values of λvdw and λele were performed at the SQM/MM level to construct the initial database. The initial models, including the QM/MM ML model, the output-check model, the k-means clustering model and the Gaussian model, were optimized based on the database. In the update stage, the direct QM/MM MD combined with our ML model was implemented. One of the three approaches, that is, the outputcheck model, the k-means clustering model and the Gaussian model, was employed to check the validity of the predictions. If the configuration was identified as a new data point, the potential energy and forces at this step would be calculated using the low-level SQM/MM model. This data point would be added to the ML database unless another configuration within the previous 100 MD steps had been selected. Then the QM/MM ML model would be updated with the gradient boosting technique. At the final stage, MD simulations were performed with the updated QM/MM ML model, and the final properties of interest such as SFE at the approximate ab initio QM/MM level were calculated using Eq 11.
Figure 1.
The flowchart of solvation free energy calculation with ML-based QM/MM MD simulations. The initialization stage contains SQM/MM MD simulation, construction of initial database, and establishment of initial QM/MM ML models. The update stage contains direct QM/MM MD simulations combined with the ML predictions, determination of new configurations encountered during MD, and reconstruction of ML models. The finalization stage contains MD simulation with the final QM/MM ML model and the solvation free energy calculation.
Results and Discussion
Six small organic molecules, acetic acid, acetamide, acetone, benzene, ethanol and methylamine, were investigated to demonstrate our new approach. The self-consistent charge density functional tight binding with second-order formulation and MIO basis (DFTB2/MIO)84,85 was employed as the low-level SQM model, and the DFT method with the B3LYP hybrid functional86,87 and the 6–31G(d) basis set was employed as the high-level ab initio QM model. First, we randomly selected about one thousand configurations from low-level QM/MM MD trajectories and assigned them as the training and testing sets to construct the initial QM/MM ML model. Second, we screened the optimal hyperparameters for symmetry functions and power spectrum. For symmetry functions, the Rc, Rs and λ were set as 6 Å, 0 Å and 1.0, respectively. The η and ζ are dependent on elements, which are shown in Table S1. For power spectrum, the Rc is 9 Å, and the nmax, lmax, Rn and α are displayed in Table S2. Note that different n ∈ [1,nmax] and l ∈ [0,lmax] can be chosen for QM and MM subsystems. Finally, the linear regression model was optimized using the component-wise gradient boosting algorithm64 combined with the early stopping techniques.65,66 The reliability of the QM/MM ML model is shown in Table 1. The root mean squared errors (RMSEs) for high-level QM/MM potential energies in all cases are lower than 0.7 kcal/mol. The corresponding Q2 values vary from 0.56 to 0.91, most of which are larger than 0.7.
Table 1.
Root Mean Squared Errors (kcal/mol) of Training Set and Testing Set of Symmetry Functions and Power Spectrum with Q2 Values (in Parentheses) for Six Molecules.
| Molecules | Symmetry Functions |
Power Spectrum |
||
|---|---|---|---|---|
| training | testing | training | testing | |
| Acetic acid | 0.60 | 0.57 (0.81) | 0.31 | 0.42 (0.90) |
| Acetamide | 0.57 | 0.54 (0.66) | 0.37 | 0.49 (0.72) |
| Acetone | 0.44 | 0.46 (0.56) | 0.28 | 0.34 (0.75) |
| Benzene | 0.32 | 0.35 (0.82) | 0.20 | 0.26 (0.91) |
| Ethanol | 0.65 | 0.66 (0.70) | 0.36 | 0.53 (0.80) |
| Methylamine | 0.52 | 0.51 (0.83) | 0.55 | 0.68 (0.70) |
The values of solvation free energies were calculated based on MD samplings using three QM/MM models as DFTB2/MIO/MM, B3LYP/6–31G(d)/MM and the initial QM/MM ML model, that is, the DFTB2/MIO/MM model with ML corrections on the basis of the initial training data. All MD samplings using DFTB2/MIO/MM and B3LYP/6–31G(d)/MM models were repeated by 11 times, respectively, to obtain the values of SFEs with standard deviations (see Table S3). More simulation details can be found in SI. As shown in Table 2, the differences of results between DFTB2/MIO/MM and B3LYP/6–31G(d)/MM are significant, varying from 1.6 to 6.1 kcal/mol. The results with QM/MM ML are very close to the reference in the major part of all testing examples, except the simulations on acetamide, acetone and ethanol with symmetry functions and methylamine with power spectrum. Specially, we terminate MD trajectories for acetamide, acetone and ethanol using symmetry functions once a convergence problem on electronic structure calculations takes place, which indicates that some unphysical configurations are encountered because of the insufficient samples on the present ML database.42
Table 2.
Solvation Free Energies (kcal/mol) from MD Simulations with DFTB2/MIO/MM, B3LYP/6–31G(d)/MM and Initial QM/MM ML Models Using Symmetry Functions and Power Spectrum.
| Molecules | DFTB/MM | B3LYP/MM | Initial QM/MM ML |
|
|---|---|---|---|---|
| Symmetry Functions | Power Spectrum | |||
| Acetic acid | −5.0 | −7.5 | −7.0 | −7.8 |
| Acetamide | −9.0 | −12.1 | —a | −11.4 |
| Acetone | −2.3 | −4.3 | —a | −4.3 |
| Benzene | 1.0 | −0.6 | −1.0 | −1.1 |
| Ethanol | −1.0 | −4.8 | —a | −5.0 |
| Methylamine | 0.9 | −5.2 | −5.3 | −43.2 |
MD trajectories are terminated.
To address this issue, we first tried two existing detecting methods, boundary of input variables and uncertainty analysis on a ML ensemble. These methods were first applied to the three cases whose MD trajectories are terminated without updating QM/MM ML models, to validate whether the detecting methods are eligible to select new configurations. With boundary of input variables, MD trajectories of those three cases become normal. However, with uncertainty analysis on a ML ensemble, even though MD trajectories of acetamide and acetone become also normal, the MD trajectory of ethanol is still terminated, since we could find that two ML models have similarly poor predictions on some configurations. Hence, uncertainty analysis on a ML ensemble is not appropriate in this work. Next, the boundary of input variables was applied to all cases and QM/MM ML models were updated on the fly, whose results are displayed in Table S4. All calculated solvation free energies are within the chemical accuracy to the high-level results, except methylamine with power spectrum, which will be explained later. However, even though the percentages of new configurations for symmetry functions are low, the percentages for power spectrum are too high (around 60%), which is unacceptable because such frequent updates, even with gradient boosting, are computationally expensive. Therefore, more effective detecting methods are desirable.
To solve this problem, the QM/MM ML model is updated based on three models, that is, the output-check model, the k-means clustering model and the Gaussian model, respectively. These models for detecting new configurations were also updated every 1, 30 and 50 new data points encountered for output-check, k-means clustering and Gaussian model, respectively. 10 clusters and 10 Gaussian distributions were applied for k-means clustering and Gaussian model, respectively. All MD samplings using QM/MM ML models were repeated by 5 times to obtain average values and standard deviations of SFEs. The results from the updated QM/MM ML models are displayed in Table 3 and Table S3. First, the aforementioned terminated MD trajectories can all be continued using the updated QM/MM ML models. Second, the SFEs calculated with all models are close to the reference values. One exception is the simulations on methylamine with power spectrum. Compared with the results at the ab initio QM/MM level as -5.2 kcal/mol, the final SFE predicted with our updated QM/MM ML models is about -2.5 kcal/mol. However, it is much better than the results of -43.2 kcal/mol using the initial ML database as shown in Table 2. On the other hand, the simulations on methylamine with modified symmetry functions lead to more accurate results, indicating that the errors of power spectrum may result from the higher dimensionality of input variables, which may be more sensitive to small configurational changes and inconsistent with the simple linear regression model. Therefore, for the simplicity ML model, the symmetry functions could be the more appropriate choice as input variables. Finally, in the update stage only the few new configurations are selected by both output-check (≤ 0.4%) and k-means clustering (≤ 2.0%) methods, while the Gaussian model can extract much more new configurations up to 40%. However, only a few cases of the Gaussian model have large percentages and they are much less than those from boundary of input variables. The three models lead to more efficient update, since the number of updating times is the most expensive part for on-the-fly calculations. The accuracy of SFEs using the above three models is similar, so the model extracting lowest percentages of new configurations, that is, output-check model, is suggested among the three models. Since 50 ps of MD simulations are enough in the update stage and the percentages of new configurations are less than 2% for Model 1 and 2 and usually less than 10% for Model 3, the present method appears to be more efficient than the adaptive QM/MM-NN reported by our group recently,42 in which 30–50% configurations were observed as new data points using the initial ML model and 150–250 ps of MD samplings were required for reconstructing the NN model. In addition, the update scheme in this work is more efficient than on-the-fly retraining because of the characteristic of gradient boosting.
Table 3.
Solvation Free Energies (kcal/mol) and Percentages of New Configurations Sampled during MD Simulations (in Parentheses) with QM/MM ML Models (Symmetry Functions and Power Spectrum) Updated Using Output-Check (Model 1), k-Means Clustering (Model 2) or Gaussian Model (Model 3) to Detect New Configurations.
| Molecules | Symmetry Functions |
Power Spectrum |
||||
|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 3 | Model 1 | Model 2 | Model 3 | |
| Acetic acid | −6.8 (0.3%) | −6.7 (1.1%) | −7.0 (20.3%) | −6.9 (0.4%) | −6.8 (1.0%) | −7.3 (6.2%) |
| Acetamide | −11.4 (0.1%) | −11.3 (1.2%) | −11.6 (16.0%) | −10.9 (0.2%) | −10.9 (0.9%) | −11.7 (3.8%) |
| Acetone | −3.6 (0.1%) | −3.9 (1.1%) | −3.8 (5.0%) | −3.9 (0.2%) | −3.8 (1.0%) | −3.9 (5.7%) |
| Benzene | −0.3 (0.2%) | −0.2 (2.0%) | −0.3 (1.8%) | −0.2 (0.3%) | −0.5 (1.6%) | −0.6 (4.4%) |
| Ethanol | −4.6 (0.1%) | −4.6 (1.3%) | −4.3 (40.0%) | −4.0 (0.4%) | −4.0 (1.0%) | −4.6 (9.8%) |
| Methylamine | −3.8 (0.4%) | −4.0 (1.7%) | −4.5 (7.2%) | −2.4 (0.4%) | −2.2 (1.3%) | −2.5 (10.2%) |
Conclusions
In summary, we developed a novel machine learning method combining with additional models to identify new configurations encountered during MD and the gradient boosting algorithm to update the ML models with an increasing database. The solvation free energies were calculated based on the direct QM/MM MD simulations on the ML-predicted PES. Two types of input features, namely, modified symmetry functions and power spectrum, and three approaches to identify new data points, namely, the output-check model, the k-means clustering model and the Gaussian model, were all implemented and employed. We concluded that the ML methods using linear regression and modified symmetry functions should be updated during MD simulations to achieve robust and accurate results. All three approaches to detect new configurations are reliable, and the output-check model is recommended based on efficiency. The ML models with linear regression fitting and power spectrum meet more challenges, so we suggest modified symmetry functions for the simple ML model. However, power spectrum could be employed for more complex ML model such as neural network and it is more systematically generated. The gradient boosting algorithm makes the procedure to update the QM/MM ML models much more efficient. The present machine learning method for direct QM/MM MD simulations not only achieves the high-level ab initio QM/MM accuracy and low-level SQM/MM efficiency simultaneously, but also provides a systematic, robust and effective way to update ML models on-the-fly. Applying this procedure to various complex ML models such as the high-dimensional neural network and Gaussian kernel should be promising for exploring new chemistry using ML-based simulations.
Supplementary Material
Acknowledgements
Financial support from National Institutes of Health (Grant No. R01 GM061870–13) is gratefully acknowledged. The authors are grateful to Dr. Xiangqian Hu for help discussions.
Footnotes
The authors declare no competing financial interest.
Supporting Information
Details of gradient boosting, Gaussian model and MD simulations. Tables S1 and S2 for hyperparameters of symmetry functions and power spectrums, respectively. Table S3 for calculated solvation free energies with standard deviations of MD simulations. Table S4 for calculated solvation free energies and percentages of new configurations with QM/MM ML models updated using boundary of input variables. Figure S1 for comparison of the ML-predicted potential energies with the reference values.
References
- (1).Eisenberg D; McLachlan AD Solvation energy in protein folding and binding. Nature 1986, 319, 199–203. [DOI] [PubMed] [Google Scholar]
- (2).Kollman P Free energy calculations: Applications to chemical and biochemical phenomena. Chem. Rev 1993, 93, 2395–2417. [Google Scholar]
- (3).Shoichet BK; Leach AR; Kuntz ID Ligand solvation in molecular docking. Proteins: Struct., Funct., Bioinf 1999, 34, 4–16. [DOI] [PubMed] [Google Scholar]
- (4).Jorgensen WL The Many Roles of Computation in Drug Discovery. Science 2004, 303, 1813–1818. [DOI] [PubMed] [Google Scholar]
- (5).Zwanzig RW High-Temperature Equation of State by a Perturbation Method. I. Nonpolar Gases. J. Chem. Phys 1954, 22, 1420–1426. [Google Scholar]
- (6).Kirkwood JG Statistical Mechanics of Fluid Mixtures. J. Chem. Phys 1935, 3, 300–313. [Google Scholar]
- (7).Torrie GM; Valleau JP Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys 1977, 23, 187–199. [Google Scholar]
- (8).Kong X; Brooks CL λ-dynamics: A new approach to free energy calculations. J. Chem. Phys 1996, 105, 2414–2423. [Google Scholar]
- (9).Christ CD; Mark AE; van Gunsteren WF Basic ingredients of free energy calculations: A review. J. Comput. Chem 2010, 31, 1569–1582. [DOI] [PubMed] [Google Scholar]
- (10).Morgantini P-Y; Kollman PA Solvation Free Energies of Amides and Amines: Disagreement between Free Energy Calculations and Experiment. J. Am. Chem. Soc 1995, 117, 6057–6063. [Google Scholar]
- (11).Shivakumar D; Deng Y; Roux B Computations of Absolute Solvation Free Energies of Small Molecules Using Explicit and Implicit Solvent Model. J. Chem. Theory Comput 2009, 5, 919–930. [DOI] [PubMed] [Google Scholar]
- (12).Shivakumar D; Williams J; Wu Y; Damm W; Shelley J; Sherman W Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput 2010, 6, 1509–1519. [DOI] [PubMed] [Google Scholar]
- (13).Wang L; Wu Y; Deng Y; Kim B; Pierce L; Krilov G; Lupyan D; Robinson S; Dahlgren MK; Greenwood J; Romero DL; Masse C; Knight JL; Steinbrecher T; Beuming T; Damm W; Harder E; Sherman W; Brewer M; Wester R; Murcko M; Frye L; Farid R; Lin T; Mobley DL; Jorgensen WL; Berne BJ; Friesner RA; Abel R Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J. Am. Chem. Soc 2015, 137, 2695–2703. [DOI] [PubMed] [Google Scholar]
- (14).Boulanger E; Huang L; Rupakheti C; MacKerell AD; Roux B Optimized Lennard-Jones Parameters for Druglike Small Molecules. J. Chem. Theory Comput 2018, 14, 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Warshel A; Karplus M Calculation of ground and excited state potential surfaces of conjugated molecules. I. Formulation and parametrization. J. Am. Chem. Soc 1972, 94, 5612–5625. [Google Scholar]
- (16).Warshel A; Levitt M Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol 1976, 103, 227–249. [DOI] [PubMed] [Google Scholar]
- (17).Friesner RA; Guallar V Ab Initio Quantum Chemical and Mixed Quantum Mechanics/Molecular Mechanics (QM/MM) Methods for Studying Enzymatic Catalysis. Annu. Rev. Phys. Chem 2005, 56, 389–427. [DOI] [PubMed] [Google Scholar]
- (18).Hu H; Yang W Free Energies of Chemical Reactions in Solution and in Enzymes with Ab Initio Quantum Mechanics/Molecular Mechanics Methods. Annu. Rev. Phys. Chem 2008, 59, 573–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hu P; Wang S; Zhang Y How Do SET-Domain Protein Lysine Methyltransferases Achieve the Methylation State Specificity? Revisited by Ab Initio QM/MM Molecular Dynamics Simulations. J. Am. Chem. Soc 2008, 130, 3806–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hu P; Wang S; Zhang Y Highly Dissociative and Concerted Mechanism for the Nicotinamide Cleavage Reaction in Sir2Tm Enzyme Suggested by Ab Initio QM/MM Molecular Dynamics Simulations. J. Am. Chem. Soc 2008, 130, 16721–16728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hu H; Yang W Development and application of ab initio QM/MM methods for mechanistic simulation of reactions in solution and in enzymes. J. Mol. Struct. THEOCHEM 2009, 898, 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Senn HM; Thiel W QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Ed 2009, 48, 1198–1229. [DOI] [PubMed] [Google Scholar]
- (23).Liu M; Wang Y; Chen Y; Field MJ; Gao J QM/MM through the 1990s: The First Twenty Years of Method Development and Applications. Isr. J. Chem 2014, 54, 1250–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Brunk E; Rothlisberger U Mixed Quantum Mechanical/Molecular Mechanical Molecular Dynamics Simulations of Biological Systems in Ground and Electronically Excited States. Chem. Rev 2015, 115, 6217–6263. [DOI] [PubMed] [Google Scholar]
- (25).Chung LW; Sameera WMC; Ramozzi R; Page AJ; Hatanaka M; Petrova GP; Harris TV; Li X; Ke Z; Liu F; Li H-B; Ding L; Morokuma K The ONIOM Method and Its Applications. Chem. Rev 2015, 115, 5678–5796. [DOI] [PubMed] [Google Scholar]
- (26).Lu X; Fang D; Ito S; Okamoto Y; Ovchinnikov V; Cui Q QM/MM free energy simulations: recent progress and challenges. Mol. Simul 2016, 42, 1056–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Åqvist J; Warshel A Simulation of enzyme reactions using valence bond force fields and other hybrid quantum/classical approaches. Chem. Rev 1993, 93, 2523–2544. [Google Scholar]
- (28).Cui Q; Elstner M; Kaxiras E; Frauenheim T; Karplus M A QM/MM Implementation of the Self-Consistent Charge Density Functional Tight Binding (SCC-DFTB) Method. J. Phys. Chem. B 2001, 105, 569–585. [Google Scholar]
- (29).Akimov AV; Prezhdo OV Large-Scale Computations in Chemistry: A Bird’s Eye View of a Vibrant Field. Chem. Rev 2015, 115, 5797–5890. [DOI] [PubMed] [Google Scholar]
- (30).Blank TB; Brown SD; Calhoun AW; Doren DJ Neural network models of potential energy surfaces. J. Chem. Phys 1995, 103, 4129–4137. [Google Scholar]
- (31).Lorenz S; Groß A; Scheffler M Representing high-dimensional potential-energy surfaces for reactions at surfaces by neural networks. Chem. Phys. Lett 2004, 395, 210–215. [Google Scholar]
- (32).Behler J; Parrinello M Generalized Neural-Network Representation of High-Dimensional Potential-Energy Surfaces. Phys. Rev. Lett 2007, 98, 146401. [DOI] [PubMed] [Google Scholar]
- (33).Bartók AP; Payne MC; Kondor R; Csányi G Gaussian Approximation Potentials: The Accuracy of Quantum Mechanics, without the Electrons. Phys. Rev. Lett 2010, 104, 136403. [DOI] [PubMed] [Google Scholar]
- (34).Bartók AP; Csányi G Gaussian approximation potentials: A brief tutorial introduction. Int. J. Quantum Chem 2015, 115, 1051–1057. [Google Scholar]
- (35).Balabin RM; Lomakina EI Support vector machine regression (LS-SVM)—an alternative to artificial neural networks (ANNs) for the analysis of quantum chemistry data? Phys. Chem. Chem. Phys 2011, 13, 11710–11718. [DOI] [PubMed] [Google Scholar]
- (36).Rupp M; Tkatchenko A; Müller K-R; von Lilienfeld OA Fast and Accurate Modeling of Molecular Atomization Energies with Machine Learning. Phys. Rev. Lett 2012, 108, 058301. [DOI] [PubMed] [Google Scholar]
- (37).Hansen K; Montavon G; Biegler F; Fazli S; Rupp M; Scheffler M; von Lilienfeld OA; Tkatchenko A; Müller K-R Assessment and Validation of Machine Learning Methods for Predicting Molecular Atomization Energies. J. Chem. Theory Comput 2013, 9, 3404–3419. [DOI] [PubMed] [Google Scholar]
- (38).Behler J Perspective: Machine learning potentials for atomistic simulations. J. Chem. Phys 2016, 145, 170901. [DOI] [PubMed] [Google Scholar]
- (39).Nguyen TT; Székely E; Imbalzano G; Behler J; Csányi G; Ceriotti M; Götz AW; Paesani F Comparison of permutationally invariant polynomials, neural networks, and Gaussian approximation potentials in representing water interactions through many-body expansions. J. Chem. Phys 2018, 148, 241725. [DOI] [PubMed] [Google Scholar]
- (40).Ramakrishnan R; Dral PO; Rupp M; von Lilienfeld OA Big Data Meets Quantum Chemistry Approximations: The Δ-Machine Learning Approach. J. Chem. Theory Comput 2015, 11, 2087–2096. [DOI] [PubMed] [Google Scholar]
- (41).Shen L; Wu J; Yang W Multiscale Quantum Mechanics/Molecular Mechanics Simulations with Neural Networks. J. Chem. Theory Comput 2016, 12, 4934–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Shen L; Yang W Molecular Dynamics Simulations with Quantum Mechanics/Molecular Mechanics and Adaptive Neural Networks. J. Chem. Theory Comput 2018, 14, 1442–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Behler J First Principles Neural Network Potentials for Reactive Simulations of Large Molecular and Condensed Systems. Angew. Chem. Int. Ed 2017, 56, 12828–12840. [DOI] [PubMed] [Google Scholar]
- (44).Behler J Constructing high-dimensional neural network potentials: A tutorial review. Int. J. Quantum Chem 2015, 115, 1032–1050. [Google Scholar]
- (45).Peterson AA; Christensen R; Khorshidi A Addressing uncertainty in atomistic machine learning. Phys. Chem. Chem. Phys 2017, 19, 10978–10985. [DOI] [PubMed] [Google Scholar]
- (46).Botu V; Ramprasad R Adaptive machine learning framework to accelerate ab initio molecular dynamics. Int. J. Quantum Chem 2015, 115, 1074–1083. [Google Scholar]
- (47).Csányi G; Albaret T; Payne MC; De Vita A “Learn on the Fly”: A Hybrid Classical and Quantum-Mechanical Molecular Dynamics Simulation. Phys. Rev. Lett 2004, 93, 175503. [DOI] [PubMed] [Google Scholar]
- (48).Li Z; Kermode JR; De Vita A Molecular Dynamics with On-the-Fly Machine Learning of Quantum-Mechanical Forces. Phys. Rev. Lett 2015, 114, 096405. [DOI] [PubMed] [Google Scholar]
- (49).Friedman JH Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat 2001, 29, 1189–1232. [Google Scholar]
- (50).Natekin A; Knoll A Gradient boosting machines, a tutorial. Front. Neurorob 2013, 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lu J; Lu D; Zhang X; Bi Y; Cheng K; Zheng M; Luo X Estimation of elimination half-lives of organic chemicals in humans using gradient boosting machine. Biochim. Biophys. Acta, Gen. Subj 2016, 1860, 2664–2671. [DOI] [PubMed] [Google Scholar]
- (52).Jain DS; Gupte SR; Aduri R A Data Driven Model for Predicting RNA-Protein Interactions based on Gradient Boosting Machine. Sci. Rep 2018, 8, 9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Rosta E; Haranczyk M; Chu ZT; Warshel A Accelerating QM/MM Free Energy Calculations: Representing the Surroundings by an Updated Mean Charge Distribution. J. Phys. Chem. B 2008, 112, 5680–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kamerlin SCL; Haranczyk M; Warshel A Progress in Ab Initio QM/MM Free-Energy Simulations of Electrostatic Energies in Proteins: Accelerated QM/MM Studies of pKa, Redox Reactions and Solvation Free Energies. J. Phys. Chem. B 2009, 113, 1253–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Lim H-K; Lee H; Kim H A Seamless Grid-Based Interface for Mean-Field QM/MM Coupled with Efficient Solvation Free Energy Calculations. J. Chem. Theory Comput 2016, 12, 5088–5099. [DOI] [PubMed] [Google Scholar]
- (56).Wang M; Li P; Jia X; Liu W; Shao Y; Hu W; Zheng J; Brooks BR; Mei Y Efficient Strategy for the Calculation of Solvation Free Energies in Water and Chloroform at the Quantum Mechanical/Molecular Mechanical Level. J. Chem. Inf. Model 2017, 57, 2476–2489. [DOI] [PubMed] [Google Scholar]
- (57).Wu P; Hu X; Yang W λ-Metadynamics Approach To Compute Absolute Solvation Free Energy. J. Phys. Chem. Lett 2011, 2, 2099–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Beutler TC; Mark AE; van Schaik RC; Gerber PR; van Gunsteren WF Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chem. Phys. Lett 1994, 222, 529–539. [Google Scholar]
- (59).Bartók AP; Kondor R; Csányi G On representing chemical environments. Phys. Rev. B 2013, 87, 184115. [Google Scholar]
- (60).Behler J Atom-centered symmetry functions for constructing high-dimensional neural network potentials. J. Chem. Phys 2011, 134, 074106. [DOI] [PubMed] [Google Scholar]
- (61).Wang H; Yang W Force Field for Water Based on Neural Network. J. Phys. Chem. Lett 2018, 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Seko A; Takahashi A; Tanaka I Sparse representation for a potential energy surface. Phys. Rev. B 2014, 90, 024101. [Google Scholar]
- (63).Cubuk ED; Malone BD; Onat B; Waterland A; Kaxiras E Representations in neural network based empirical potentials. J. Chem. Phys 2017, 147, 024104. [DOI] [PubMed] [Google Scholar]
- (64).Schmid M; Hothorn T Boosting additive models using component-wise P-Splines. Comput. Stat. Data Anal 2008, 53, 298–311. [Google Scholar]
- (65).Zhang T; Yu B Boosting with early stopping: Convergence and consistency. Ann. Statist 2005, 33, 1538–1579. [Google Scholar]
- (66).Hastie T; Tibshirani R; Friedman JH The elements of statistical learning: data mining, inference, and prediction; New York, NY: Springer, 2009. [Google Scholar]
- (67).Smith JS; Isayev O; Roitberg AE ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci 2017, 8, 3192–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Podryabinkin EV; Shapeev AV Active learning of linearly parametrized interatomic potentials. Comput. Mater. Sci 2017, 140, 171–180. [Google Scholar]
- (69).Hartigan JA; Wong MA Algorithm AS 136: A K-Means Clustering Algorithm. Appl. Stat 1979, 28, 100–108. [Google Scholar]
- (70).Gut A An Intermediate Course in Probability; Springer, New York, NY, 1995; pp 119–148. [Google Scholar]
- (71).Kriegel H-P; Kröger P; Sander J; Zimek A Density-based clustering. WIREs Data Min. Knowl. Discov 2011, 1, 231–240. [Google Scholar]
- (72).Sittel F; Stock G Robust Density-Based Clustering To Identify Metastable Conformational States of Proteins. J. Chem. Theory Comput 2016, 12, 2426–2435. [DOI] [PubMed] [Google Scholar]
- (73).Guan Y; Yang S; Zhang DH Application of Clustering Algorithms to Partitioning Configuration Space in Fitting Reactive Potential Energy Surfaces. J. Phys. Chem. A 2018, 122, 3140–3147. [DOI] [PubMed] [Google Scholar]
- (74).Li X; Curtis FS; Rose T; Schober C; Vazquez-Mayagoitia A; Reuter K; Oberhofer H; Marom N Genarris: Random generation of molecular crystal structures and fast screening with a Harris approximation. J. Chem. Phys 2018, 148, 241701. [DOI] [PubMed] [Google Scholar]
- (75).Jeong W; Lee K; Yoo D; Lee D; Han S Toward Reliable and Transferable Machine Learning Potentials: Uniform Training by Overcoming Sampling Bias. J. Phys. Chem. C 2018, 122, 22790–22795. [Google Scholar]
- (76).Meldgaard SA; Kolsbjerg EL; Hammer B Machine learning enhanced global optimization by clustering local environments to enable bundled atomic energies. J. Chem. Phys 2018, 149, 134104. [DOI] [PubMed] [Google Scholar]
- (77).Maragakis P; van der Vaart A; Karplus M Gaussian-Mixture Umbrella Sampling. J. Phys. Chem. B 2009, 113, 4664–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Hadley KR; McCabe C On the Investigation of Coarse-Grained Models for Water: Balancing Computational Efficiency and the Retention of Structural Properties. J. Phys. Chem. B 2010, 114, 4590–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Hu Y; Hong W; Shi Y; Liu H Temperature-Accelerated Sampling and Amplified Collective Motion with Adiabatic Reweighting to Obtain Canonical Distributions and Ensemble Averages. J. Chem. Theory Comput 2012, 8, 3777–3792. [DOI] [PubMed] [Google Scholar]
- (80).Jain A; Stock G Identifying Metastable States of Folding Proteins. J. Chem. Theory Comput 2012, 8, 3810–3819. [DOI] [PubMed] [Google Scholar]
- (81).Miao Y; Feher VA; McCammon JA Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput 2015, 11, 3584–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Nocito D; Beran GJO Averaged Condensed Phase Model for Simulating Molecules in Complex Environments. J. Chem. Theory Comput. 2017, 13, 1117–1129. [DOI] [PubMed] [Google Scholar]
- (83).Arthur D; Vassilvitskii S K-means++: The Advantages of Careful Seeding. Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms Philadelphia, PA, USA, 2007; pp 1027–1035. [Google Scholar]
- (84).Elstner M; Porezag D; Jungnickel G; Elsner J; Haugk M; Frauenheim T; Suhai S; Seifert G Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 1998, 58, 7260–7268. [Google Scholar]
- (85).Gaus M; Goez A; Elstner M Parametrization and Benchmark of DFTB3 for Organic Molecules. J. Chem. Theory Comput 2013, 9, 338–354. [DOI] [PubMed] [Google Scholar]
- (86).Lee C; Yang W; Parr RG Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [DOI] [PubMed] [Google Scholar]
- (87).Becke AD Density-functional thermochemistry. III. The role of exact exchange. J.Chem. Phys 1993, 98, 5648–5652. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.