Abstract
Aims/hypothesis
Epigenetic mechanisms may play an important role in the aetiology of type 2 diabetes. Recent epigenome-wide association studies (EWASs) identified several DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels. Here we present a systematic review of these studies and attempt to replicate the CpG sites (CpGs) with the most significant associations from these EWASs in a case–control sample of the Lifelines study.
Methods
We performed a systematic literature search in PubMed and EMBASE for EWASs to test the association between DNA methylation and type 2 diabetes and/or glycaemic traits and reviewed the search results. For replication purposes we selected 100 unique CpGs identified in peripheral blood, pancreas, adipose tissue and liver from 15 EWASs, using study-specific Bonferroni-corrected significance thresholds. Methylation data (Illumina 450K array) in whole blood from 100 type 2 diabetic individuals and 100 control individuals from the Lifelines study were available. Multivariate linear models were used to examine the associations of the specific CpGs with type 2 diabetes and glycaemic traits.
Results
From the 52 CpGs identified in blood and selected for replication, 15 CpGs showed nominally significant associations with type 2 diabetes in the Lifelines sample (p < 0.05). The results for five CpGs (in ABCG1, LOXL2, TXNIP, SLC1A5 and SREBF1) remained significant after a stringent multiple-testing correction (changes in methylation from −3% up to 3.6%, p < 0.0009). All associations were directionally consistent with the original EWAS results. None of the selected CpGs from the tissue-specific EWASs were replicated in our methylation data from whole blood. We were also unable to replicate any of the CpGs associated with HbA1c levels in the healthy control individuals of our sample, while two CpGs (in ABCG1 and CCDC57) for fasting glucose were replicated at a nominal significance level (p < 0.05).
Conclusions/interpretation
A number of differentially methylated CpGs reported to be associated with type 2 diabetes in the EWAS literature were replicated in blood and show promise for clinical use as disease biomarkers. However, more prospective studies are needed to support the robustness of these findings.
Electronic supplementary material
The online version of this article (10.1007/s00125-017-4497-7) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: DNA methylation, Epigenome-wide association studies, Glucose, Glycated haemoglobin, Systematic review, Type 2 diabetes
Introduction
Type 2 diabetes mellitus is a complex metabolic disease, of which the prevalence worldwide is growing rapidly. According to recent data, globally 415 million people are estimated to have type 2 diabetes [1]. Hallmarks of type 2 diabetes include chronically elevated blood glucose levels due to decreased insulin secretion from pancreatic beta cells and insulin resistance in different tissues [2].
In addition to well-known risk factors for type 2 diabetes such as being overweight, unhealthy lifestyle, metabolic alterations, previous diagnosis of gestational diabetes, or a family history of cardiovascular disease (CVD) or type 2 diabetes [3], genetic susceptibility to the disease is also important, with heritability estimates ranging from 20% to 80% [4, 5]. To date, genome-wide association studies (GWASs) have identified at least 75 loci associated with type 2 diabetes [6]. However, these genetic variants explain only 10–15% of disease heritability, suggesting a major role for environmental and lifestyle factors [6, 7].
To identify the missing component of type 2 diabetes pathogenesis, researchers have started to examine the role of epigenetics in the disease aetiology. Epigenetics refers to DNA alterations that lead to differences in gene expression without changing the DNA sequence. These epigenetic changes can be influenced by the environment and may cause differences in disease susceptibility between individuals [8].
Initially, epigenetic studies used a candidate gene approach to identify DNA methylation changes in known type 2 diabetes susceptibility genes. With the advances in measurement technology, approaches have shifted towards epigenome-wide association studies (EWASs), allowing novel biomarkers for complex diseases to be found. Development of type 2 diabetes requires perturbation of multiple biological mechanisms in different organs, including pancreas, liver, skeletal muscle and adipose tissue [9]. EWASs using those tissues would provide a comprehensive insight into the disease aetiology; however, access to such samples is not possible on a large scale. Therefore, most EWASs have been conducted using whole blood [10].
Here, we present an overview of recent human EWASs investigating DNA methylation changes associated with type 2 diabetes and/or glycaemic traits represented by fasting glucose and HbA1c levels. Moreover, we discuss the EWASs findings and the strengths and limitations of different approaches. To validate methylation loci identified in the reviewed EWASs, we also performed a replication study in blood samples of 100 diabetic and 100 control individuals selected from a Dutch population-based Lifelines study [11]. Next, we investigated whether differential DNA methylation patterns as previously identified in pancreas, liver and adipose tissue were also reflected in blood.
Methods
Literature search
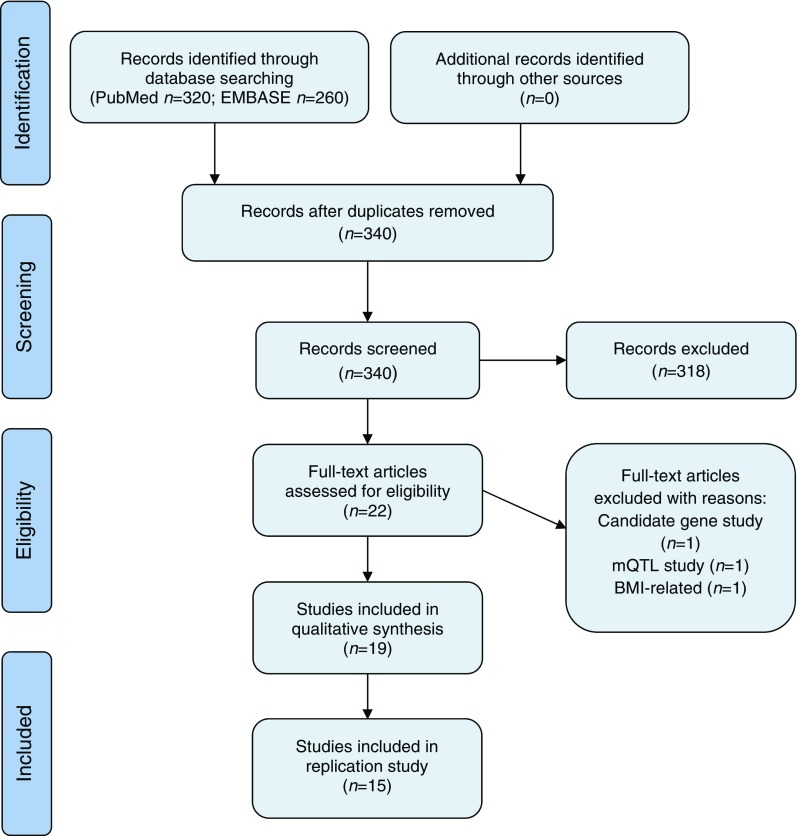
The systematic review was conducted according to the PRISMA and MOOSE guidelines. We searched PubMed and EMBASE for relevant studies investigating DNA methylation associated with type 2 diabetes or fasting glucose and HbA1c levels, up to 26 April 2017. The search strategy, inclusion and exclusion criteria are provided in the electronic supplementary material (ESM Methods). Ultimately, 22 publications were selected for whole-text evaluation. Three studies were excluded (Fig. 1), resulting in a total of 19 studies included in the review.
Fig. 1.
PRISMA 2009 flow chart of the literature search performed up to 26 April 2017
Replication analyses: selection of CpG sites
For the replication analyses, four additional studies were excluded: one that only indirectly investigated association with type 2 diabetes [12] and three that used a different platform from the Illumina array [13–15]. Thus, 15 studies were included for replication analysis (Fig. 1). For further CpG sites (CpGs) selection, we applied a study-specific Bonferroni correction for multiple testing for EWASs results (p value < 0.05/(the number of CpGs analysed)), even if a different multiple-testing correction was used by the authors of the original manuscript. This was done to avoid false positive results from the studies that used lenient significance thresholds.
Lifelines case–control sample
Lifelines is a prospective population-based cohort to study health and health-related behaviours of 167,729 individuals living in the North of the Netherlands [16]. Details on clinical examination and biochemical measurements have been described elsewhere [16]. In short, a standardised protocol was used to obtain blood pressure and anthropometric measurements such as height, weight and waist circumference. Blood was collected in the fasting state, between 08:00 and 10:00 h. On the same day, fasting blood glucose and HbA1c were measured.
For this study we used a case–control sample selected from the baseline of the Lifelines study (all unrelated and European ancestry samples, n = 13,436) [11]. Four groups were selected based on the following criteria (n = 50 for each group):
type 2 diabetes patients without CVD complications;
type 2 diabetes patients with CVD complications;
non-diabetic control participants, with no history of CVD risk factors, and age- and sex-matched to groups 1 and 2;
healthy, normal-weight control participants (BMI < 25), additionally obtained from available methylation dataset to increase the power of the study.
In total, we included 100 type 2 diabetic individuals and 100 control individuals. Diagnosis was based on self-reported disease and/or use of blood-glucose lowering medication, or an elevated fasting blood glucose ≥ 7.0 mmol/l at examination. Individuals with CVD complications had a CVD history defined as self-reported myocardial infarction, stroke, angina pectoris or vascular intervention.
DNA methylation methodology
DNA was isolated from fasting whole blood samples. Next, 500 ng of genomic DNA was bisulphite modified using the EZ DNA Methylation kit (Zymo Research, Irvine, CA, USA) and hybridised to Illumina 450K arrays (San Diego, CA, USA) according to the manufacturer’s protocols. Data were generated by the Genome Analysis Facility of UMCG, the Netherlands (www.rug.nl/research/genetics/genomeanalysisfacility/). Quality control (QC) and normalisation steps are described in detail elsewhere [17] and in ESM Methods. In short, the QC pipeline developed by Touleimat and Tost was used with background correction and probe type normalisation [18]. Then, normalised β values were logit-transformed into M values for downstream analysis, since they have been shown to perform better in studies with smaller sample sizes [19].
Statistical analysis
All analyses were performed using R-studio software (version 3.3.0; https://www.rstudio.com; https://www.r-project.org) and the limma package. Linear regression model 1 included age, sex, measured blood cell composition (percentage of basophilic granulocytes, eosinophilic granulocytes, neutrophilic granulocytes, lymphocytes and monocytes), plate number and position on the plate as covariates. Additionally, we adjusted for other covariates in models 2–6: (2) model 1 + BMI; (3) model 1 + medication use and newly diagnosed diabetes; (4) model 1 + smoking status and education level; (5) model 1 + presence of cardiovascular complications; (6) model 2 + education level. In addition to the adjustment for measured cell type composition, we estimated cell types based on the Houseman method [20] and compared results. We also performed sensitivity analyses using the model 1 in smaller groups: (1) 50 type 2 diabetes individuals without complications compared only with 50 age- and sex-matched control individuals; and (2) 100 type 2 diabetes individuals with and without complications compared only with 50 age- and sex-matched control individuals. To determine whether the methylation levels at replicated top hits were correlated with type 2 diabetes risk factors, we calculated Pearson correlation coefficients based on methylation β values. We used a strict analysis-specific Bonferroni correction for multiple testing (p value < 0.05/(the number of CpGs selected for replication)).
Results
Recent discoveries
Our search strategy retrieved 19 EWASs investigating DNA methylation associated with type 2 diabetes or glycaemic traits (Fig. 1), including 16 studies focusing on type 2 diabetes as outcome (Table 1) and four studies focusing on glycaemic traits (Table 2), with one study listed twice [25]. We assessed the quality of included studies using the Newcastle–Ottawa scale for observational studies (details in ESM Methods) [36]. Seventeen out of 20 studies (one listed twice) were assessed to have a low or medium risk of bias and only three studies were evaluated to have high risk of bias (data not shown). In the majority of the reviewed studies, an array-based methodology was employed in the discovery phase: two using the 27K and 13 using the 450K Illumina array. Only one study used whole-genome bisulphite sequencing, which is considered a gold standard in methylation studies [14]. Most of the blood-based studies (ten out of 19) were performed in larger sample sizes (n = 6 – Z2000) than studies in pancreas, liver, skeletal muscle and adipose tissue (n = 12–100). The EWASs were conducted in different ethnic groups: Europeans, Indian Asians, Mexican Americans, and Ashkenazi Jews [21, 24, 25, 28]. Despite the differences in ethnicity and study design, some CpGs such as those in the ABCG1, TXNIP and SREBF1 genes were reported in multiple blood-based studies [21, 23–25, 33, 34]. There was no clear overlap in significant CpGs across tissues, but some studies reported a significant correlation between the level of methylation at specific CpGs in blood and liver [21] or in blood and pancreas [12].
Table 1.
Characteristics of EWASs associated with type 2 diabetes
| Referencea | Population for DNA methylation analysis | Female/male | Design | Tissue | Method | Covariates included in analysis | Multiple-testing correctionb | Top findings | No. of CpGs included in replication studyc |
|---|---|---|---|---|---|---|---|---|---|
| Chambers et al, 2015 [21]a | 1074 incident type 2 diabetes patients, 1590 controls (Indian Asians); 1141 Europeans for replication |
P 352/722 C 426/1083 |
Longitudinal (nested case–control) | Blood | 450k for discovery; pyrosequencing and 450k for replication | Age, sex, intensity values from 450k control probes, batch, measured and imputed (Houseman method) cell count, 5 PC | Discovery p < 5 × 10−7
Meta-analysis Bonferroni |
5 DMS replicated in independent cohorts: TXNIP gene (cg19693031); SREBF1 gene (cg11024682); PHOSPHO1 gene (cg02650017); SOCS3 gene (cg18181703); ABCG1 gene (cg06500161) | 5 |
| Al Muftah et al, 2016 [22]a | 30 T2D patients, 93 controls from 15 families of Qatari descent; 810 female twins from TwinsUK for replication |
72/51 | Cross-sectional (case–control) | Blood | 450k | Age, sex, smoking status, cell count (Houseman method); BMI as confounder | Bonferroni | 1 DMS: DQX1 gene (cg06721411) replicated in TwinsUK | 1 |
| Soriano-Tarraga et al, 2016 [23]a | 151 T2D patients and 204 controls from IS cohort; BISMAR_2 (59 T2D patients and 108 controls); REGICOR (63 T2D patients and 582 controls) for replication |
P 61/90 C 88/116 |
Cross-sectional (case–control) | Blood | 450k | Age, sex, smoking, hyperlipidaemia, BMI, cell count (Houseman method) | Bonferroni | 1 DMS: TXNIP gene (cg19693031) replicated in 2 independent cohorts | 1 |
| Florath et al, 2016 [24]a | 153 T2D patients and 835 controls; 87 T2D patients and 527 controls for replication |
P 56 & 59% male (controlled or poorly controlled T2D) C 49% male |
Cross-sectional (case–control) | Blood | 450k | Age, sex, BMI, smoking, batch, cell count (Houseman method) | FDR < 5% Bonferroni for replication |
39 DMS associated with T2D in discovery cohort, after replication in independent cohort 1 DMS remain significant: TXNIP (cg19693031) | 1 |
| Kulkarni et al, 2015 [25]a | 850 pedigreed Mexican Americans (174 T2D patients) | 536/314 | Cross-sectional (case–control) | Blood | 450k | Age, sex, BMI, cell count (Jaffe method) | Bonferroni | Overall 51 DMS associated with T2D status; 19 with fasting glucose level and 24 with HOMA-IR | 51 |
| Yuan et al, 2014 [26]a | 27 monozygotic twin pairs from TwinsUK (17 pairs T2D-discordant, 3 pairs T2D concordant and 7 healthy pairs); 42 unrelated T2D cases and 221 controls for replication | 23 pairs/4 pairs | Cross-sectional (twins study) | Blood (white blood cells) | MeDIP-seq 450k for replication |
Age, sex, BMI, beadchip, bisulphite conversion efficiency, (family as a random effect) | FDR < 5% | 1 DMR: MALT1 gene (ch18:56336501-56337000), replicated using Illumina 450k array (cg24182998), replicated reached Bonferroni threshold (0.05/20 = 0.0025) | 1 |
| Matsha et al, 2016 [13] | 3 T2D patients, 3 pre-diabetes, 3 controls (age, sex, BMI and duration of residence matched) | All female | Cross-sectional (case–control) | Blood | MeDIP sequencing | – | q = 10−2 | 450,142 DMRs were statistically significant in all samples, among others associated with cell surface receptor signalling, inflammatory pathways, glucose transport, muscle and pancreas development genes, insulin signalling | 0, not an Illumina array |
| Toperoff et al, 2012 [15] | Ashkenazi Jews: 710 T2D patients and 459 controls were assembled in 4 age-matched pools | Males fraction around 50% in all 4 pools | Cross-sectional with multistep design (case–control) | Blood | Microarray-based assay for methylation levels; sequencing of bisulfite converted DNA pools | Sex and lymphocyte percentage | FDR | 6 DMRs were found in LD blocks. After replication and multiple hypothesis testing 13 out of 93 CpGs located in 6 DMRs showed significant case–control difference. Among them the nearest genes were: THADA, JAZF1, SLC30A8, TCF7L2, KCNQ1 and FTO. CpG site near FTO showed small (3.35%) but significant hypomethylation of cases vs controls | 0, not an Illumina array |
| Bacos et al, 2016 [12] | 87 non-diabetic donors for discovery phase and 112 individuals from Danish Family Study | 34/53 | Cross-sectional association with age | Pancreatic islets and whole blood | 450k genome-wide and 4 sites replicated using pyrosequencing in blood | Sex, BMI, HbA1c, bisulphite treatment, days in culture and islet purity | FDR < 5% | Ageing was significantly associated with altered DNA methylation at 241 sites in pancreas; almost 60% of sites were found also in other studies in blood; 4 most significant sites (FHL2, ZNF518B, GNPNAT1 and HLTF) were selected for follow-up analysis and they showed functional effects on beta cells or association with T2D risk. Higher methylation of those sites was associated with lower risk of T2D development during progression into T2D (mean 10.8 years; Botnia Prospective Study) | 0, indirect association with diabetes |
| Volkov et al, 2017 [14] | 6 T2D donors and 8 control donors | P 3/3 C 4/4 |
Cross-sectional (case–control) | Pancreatic islets | WGBS | – | – | Average methylation level was 75.9%, 25,820 DMRs were identified in T2D pancreatic islets, while 13,696 were hypermethylated and 12,124 were hypomethylated. 692 DMRs had a methylation difference > 10%, the highest in regions annotated to ARX and TFAM genes | 0, not an Illumina array |
| Dayeh et al, 2014 [27]a | 15 T2D donors and 34 control donors | P 5/10 C 12/22 |
Cross-sectional (case–control) | Pancreatic islets | 450k | Age, sex, BMI, batch, islet purity, day of culture | FDR < 5% | 1649 DMS (853 genes and 561 intragenic) with at least 5% difference in methylation between diabetic and non-diabetic donors | 15 |
| Volkmar et al, 2012 [28]a | 5 T2D donors and 11 non-diabetic donors matched by age, BMI and ethnicity; 12 T2D patients and 12 age- and BMI-matched controls for replication |
– | Cross-sectional (case–control) | Pancreatic islets | 27k | – | p < 0.01 and 15% group-wise difference on methylation level | 276 DMS (254 genes) were found, 96% were hypomethylated | 0 |
| Nilsson et al, 2014 [29]a | 14 monozygotic twins discordant for T2D; Cohort 2: 28 T2D patients/28 controls (unrelated) | P 5/9 C 5/9 |
Cross-sectional (twins study) | Adipose tissue | 450k | BMI, glucose | FDR < 15% | In twins 23,470 DMS were found, none passed FDR correction In Cohort 2 15,627 DMS (7046 genes) were found after a FDR correction DNA methylation of 266 sites, corresponding to 103 genes, was significantly associated with expression in the discordant twins at q < 0.15 |
0 |
| Ribel-Madsen et al, 2012 [30]a | 12 Danish monozygotic twins discordant for T2D | P 6/6 C 6/6 |
Cross-sectional (twins study) | Skeletal muscle (11 pairs) Adipose tissue (5 pairs) |
27k | – | Padj (Westfall–Young resampling method p < 0.001) | 1 DMS in skeletal muscle: IL8 gene and 7 DMS in adipose tissue: ZNF668; HSPA2; C8orf31; CD320; TWIST1; MYO5A | 0 |
| Kirchner et al, 2016 [31]a | 8 obese T2D men, 7 obese non-diabetic controls and 7 non-obese metabolically healthy control individuals | All male | Cross-sectional (case–control) | Liver | 450k | – | FDR < 25% | 2255 DMS (1388 genes) were found (T2D obese compared with non-obese control individuals) | 0 |
| Nilsson et al, 2015 [32]a | 35 T2D patients and 60 control individuals | P 18/17 C 43/17 |
Cross-sectional (case–control) | Liver | 450k | Age, sex, BMI, NASH diagnosis, degree of steatosis | FDR < 5% | 251 DMS (167 genes) were found mostly hypomethylated (94%) in those with T2D. A decrease in folate levels in T2D patients was observed, which could explain decreased methylation in the human liver in diabetes | 3 |
aStudies included into replication study to provide CpGs, which passed strict Bonferroni correction threshold
bMultiple-testing threshold calculated originally by the authors of particular study
c0 in last column means zero CpG sites passed study-specific Bonferroni correction threshold
C, controls; DMS, differentially methylated sites; DMR, differentially methylated region; FDR, false discovery rate; HOMA-IR, homeostatic model assessment; IS, ischemic stroke; LD, linkage disequilibrium; NASH, non-alcoholic steatohepatitis; P, patients; PC, principal component; T2D, type 2 diabetes; 27k, Infinium HumanMethylation27 BeadChip; 450k, Infinium HumanMethylation450 BeadChip
Table 2.
Characteristics of EWASs associated with glycaemic traits
| Referencea | Population for DNA methylation analysis | Female/male | Design | Tissue | Method | Covariates included into analysis | Multiple-testing correctionb | Top findings | No. of CpGs included in replication studyc |
|---|---|---|---|---|---|---|---|---|---|
| Kriebel et al, 2016 [33] a | 1448 non-diabetic individuals (fasting glucose, 2-h glucose, HbA1c); 1440 non-diabetic individuals (fasting insulin, HOMA-IR); 617 non-diabetic individuals (2-h insulin) |
47.1% male | Cross-sectional | Blood | 450k | FDR | Age, sex, cell count (Houseman method), smoking, BMI | Total of 31 CpGs were found to be associated with phenotypic traits: 5 DMS associated with fasting glucose (including ABCG1, CPT1A), 1 with 2-h glucose, 8 with fasting insulin, 26 with HOMA-IR and none with HbA1c. Using a different model, adjustment for BMI resulted in ~30% reduction in effect size, suggesting BMI had a confounding effect | 3 for fasting glucose |
| Kulkarni et al, 2015 [25] a | 850 pedigreed Mexican Americans (174 T2D patients) | 536/314 | Cross-sectional (case–control) |
Blood | 450k | Bonferroni | Age, sex, BMI, cell count (Jaffe method) | 51 CpGs were significantly associated with T2D, 19 with fasting glucose and 24 with HOMA-IR | 19 for fasting glucose |
| Hidalgo et al, 2014 [34] | 544 healthy individuals in discovery stage, 293 in replication stage | 286/258 | Cross-sectional | Blood | 450k | Bonferroni | Age, sex, study site, 4 PC, insulin, glucose | 1 DMS associated with fasting insulin and HOMA-IR: ABCG1 (cg06500161); marginally significant site also in ABCG1 (cg1881899, p = 3.36 × 10−6), associated with HOMA-IR only. No DMS associated with fasting glucose | 0 (no information about insulin levels in Lifelines) |
| Ronn et al, 2015 [35] a | 96 healthy male participants for discovery stage, 94 healthy female participants for validation stage, 2 separate EWASs performed | 96 male 94 female |
Cross-sectional association with age, BMI and HbA1c | Adipose tissue | 450k | FDR < 5%, q < 0.05 | Sex, family number; pedigree, age, BMI | 711 DMS associated with HbA1c were found in the male cohort with most significant negative correlation at ANKRD11 gene; 7 DMS associated with HbA1c were found in the female cohort, none of which were significantly associated with HbA1c level in the male cohort | 11 from male cohort |
aStudies included into replication study to provide CpGs, which passed strict Bonferroni correction threshold
bMultiple-testing threshold calculated originally by the authors of particular study
c0 in last column means zero CpG sites passed study-specific Bonferroni correction threshold
C, controls; DMS, differentially methylated sites; DMR, differentially methylated region; FDR, false discovery rate; HOMA-IR, homeostatic model assessment; NASH, non-alcoholic steatohepatitis; P, patients; PC, principal component; T2D, type 2 diabetes; 27k, Infinium HumanMethylation27 BeadChip; 450k, Infinium HumanMethylation450 BeadChip
Study design
The majority of the reviewed EWASs (18 out of 19) used a cross-sectional design, in which phenotype and DNA methylation profile were measured at the same time point either in unrelated individuals (type 2 diabetic and healthy control participants, 15 studies) or in twin pairs, discordant for type 2 diabetes (three studies) (Tables 1, 2). Strengths of this approach typically include a large study population selected from ongoing cohorts and the possibility to adjust for existing confounders like BMI or smoking. However, a cross-sectional approach cannot establish whether the difference in methylation preceded the onset of type 2 diabetes.
Tissue
Blood: The interpretation of blood-based EWASs results can be difficult, because many top hits from EWASs are known genes from immune response and inflammatory pathways, which can be mediated by the blood cell composition and, thus, do not reflect true associations with type 2 diabetes. Six out of ten blood-based studies used the reference-based estimation methods by Houseman [20] or Jaffe [37] to adjust for confounding effects of cell composition. Results from the majority of those studies indicate that differentially methylated sites in the TXNIP, ABCG1, CPT1A and SREBF1 genes are associated with type 2 diabetes and glycaemic traits [21, 23–25, 33, 34].
Pancreas: The pancreas plays a key role in maintaining normoglycaemia through insulin secretion in response to blood glucose elevation [9]. In addition to the ten EWASs performed in blood, four of the included studies examined the association between DNA methylation in pancreas and type 2 diabetes. These studies were conducted in a limited number of individuals (n = 16 to 87) [27, 28] and no overlap in identified CpGs was found between the studies when considering specific multiple-testing corrections applied by the authors (FDR < 5% [12, 27]; p < 0.01 and 15% group-wise difference on methylation [28]). Interestingly, one study used whole-genome bisulphite sequencing (WGBS) and identified over 25,000 differentially methylated regions across the whole genome, suggesting large changes in methylome associated with type 2 diabetes [14].
Liver: Another important organ in glucose metabolism is the liver where, in diabetic individuals, suppression of hepatic glucose output by insulin is reduced, contributing to hyperglycaemia [38]. The exact pathophysiology causing liver insulin resistance is still unknown, suggesting a role for epigenetic mechanisms. We found two EWASs performed in liver tissue (Table 1) using rather small sample sizes (n = 15 [32] and 95 [31]). The majority of CpGs showing a significant methylation difference from these two studies were hypomethylated in individuals with type 2 diabetes compared with control individuals (92% and 94%, FDR < 25% and FDR < 5%, respectively). No overlap was found between liver and blood-based results of EWASs, suggesting that significant CpGs from liver EWASs might be tissue specific.
Adipose tissue: Pathogenesis of glucose intolerance is also associated with adipocyte metabolism and altered fat topography [39]. Among the reviewed studies, three EWASs were performed in adipose tissue: two investigating an association with type 2 diabetes (one study with five twin pairs and another with unrelated individuals, n = 95) and one investigating an association with HbA1c level (96 healthy male, 94 healthy female participants) [29, 30, 35]. We observed no overlap (manually checked) in the top 100 CpGs from the two studies focusing on type 2 diabetes [29, 30].
Ethnicity
In 2013, the highest diabetes prevalence was observed in the North American and Caribbean region (around 11%), while the lowest was in the African region (around 5.7%) [40], suggesting differences in prevalence between ethnic groups. In the recent EWAS, the total risk of developing type 2 diabetes was three times higher in Indian Asians than in Europeans, regardless of differences in adiposity, physical activity and glycaemic values [21]. The authors estimated that 32% of the unexplained risk for future type 2 diabetes among Indian Asians compared with controls was associated with a higher methylation score based on the top five markers at TXNIP, ABCG1, SREBF1, SOCS3 and PHOSPHO1 [21]. A family-based study of 859 Mexican Americans showed that the degree of methylation at top regions including TXNIP, ABCG1 and SAMD12 genes and two intragenic regions accounted for 7.8% of the heritability of type 2 diabetes in Mexican Americans [25]. An EWAS performed in an Arab population showed that around 10% of methylation sites with FDR < 1% had median heritability of 0.7, supporting previous findings [22, 41]. These differences in DNA methylation between ethnic groups can be partly explained by their genetic ancestry, but also environmental and lifestyle factors may contribute to the variation, while some methylation loci (TXNIP or ABCG1) were found in populations with divergent ethnic backgrounds [21, 23–25].
Replication study
Selected CpGs
From the 19 studies included in the review, we selected 15 studies (Fig. 1). A list of CpGs robustly associated with type 2 diabetes or glycaemic traits was compiled based on the application of a stringent study-specific multiple-testing correction threshold to avoid false positive results (see Methods). After the removal of duplicates, we obtained a list of 100 unique CpGs (ESM Table 1) identified in peripheral blood (52 for type 2 diabetes and 21 for fasting glucose), pancreas (15 for type 2 diabetes), adipose tissue (ten for HbA1c blood level) and liver (two for type 2 diabetes).
Study population
We investigated which of the above-mentioned EWASs findings, both in blood and in other tissues, could be replicated in blood samples from the Lifelines case–control sample (for clinical characteristics see Table 3 and ESM Table 2). Individuals with type 2 diabetes were older, had a significantly higher BMI, waist–hip ratio and blood pressure, as well as higher levels of HbA1c, fasting glucose and triacylglycerols compared with control individuals. We observed no differences in socioeconomic status represented by level of education between type 2 diabetic and control participants (Table 3).
Table 3.
Baseline characteristics of the study sample of type 2 diabetic individuals and healthy individuals from the Lifelines cohort (n = 198)
| Type 2 diabetic individuals (n = 100) | Control individuals (n = 98)a | p value | |
|---|---|---|---|
| Sex (M) (n, %) | 52 (52) | 44 (44.9) | 0.44 |
| Age (years) | 62 (53–69) | 50 (46–63) | 3 × 10−8 |
| BMI (kg/m2) | 30.8 ± 4.7 | 25.3 ± 3.6 | < 2.2 × 10−16 |
| Waist (cm) | 105.3 ± 12.4 | 89.2 ± 11.0 | < 2.2 × 10−16 |
| Waist–hip ratio | 0.98 ± 0.08 | 0.9 ± 0.08 | 1.1 × 10−10 |
| Fasting statusb | 98 (98) | 97 (99) | 0.57 |
| Biochemical measurements | |||
| HbA1c (%) | 6.6 (6.4–8.5) | 5.6 (5.3–5.7) | < 2.2 × 10−16 |
| HbA1c (mmol/l) | 49 (45.8–55.3) | 37.5 (35.3–39) | < 2.2 × 10−16 |
| Fasting glucose (mmol/l)c | 7.4 (6.4–8.5) | 4.9 (4.6–5.3) | < 2.2 × 10−16 |
| Triacylglycerol (mmol/l) | 1.4 (1.1–1.9) | 1.0 (0.7–1.2) | 2.2 × 10−8 |
| HDL-cholesterol (mmol/l) | 1.2 ± 0.32 | 1.54 ± 0.4 | 1.6 × 10−8 |
| LDL-cholesterol (mmol/l) | 2.8 ± 0.9 | 3.5 ± 0.9 | 3 × 10−7 |
| Total cholesterol (mmol/l) | 4.5 ± 1.0 | 5.3 ± 1.0 | 1.1 × 10−7 |
| Systolic BP (mmHg) | 135 ± 18 | 122 ± 11 | 4.2 × 10−9 |
| Diastolic BP (mmHg) | 76 ± 9 | 73 ± 7 | 6.7 × 10−3 |
| Education level (n, %)c | |||
| Low Intermediate High |
55 (59) 22 (24) 16 (17) |
34 (37) 28 (30) 30 (33) |
0.2 |
| Insulin use (n, %) | 10 (10) | 0 (0) | – |
| Oral blood glucose lowering drugs (n, %) | 51 (51) | 0 (0) | – |
| Lipid lowering drugs (n, %) | 60 (60) | 1 (1) | – |
Normal distribution assessment based on histograms and probability–probability plots
Data are shown as mean ± SD for normally distributed variables, as median and 25th and 75th quintile for not normally distributed variables and as number of individuals (%) for categorical variables
p values are obtained from Student’s t test for normally distributed variables or from Mann–Whitney U test for not normally distributed variables and χ 2 square for categorical variables. Significant p values < 0.05
aTwo controls were excluded because of a sex mismatch (between actual data and methylation data)
bFasting status data apply to all biochemical blood measurements presented in the table
cFasting glucose value missing for one individual; education level missing for seven individuals
Association with type 2 diabetes: blood-specific CpGs
First, we analysed the 52 CpGs associated with type 2 diabetes in blood (ESM Table 1). In our Lifelines sample, five out of 52 included CpGs showed significant associations with type 2 diabetes (the Bonferroni-adjusted p < 0.0009 (0.05/52 CpGs)), including the loci in the ABCG1, LOXL2, TXNIP, SLC1A5 and SREBF1 genes (see a short description in ESM Box 1). This number increased to 15 CpGs when using the nominal significance level (p < 0.05) (Table 4). In agreement with previous studies, we observed hypermethylation in the loci at the ABCG1 and SREBF1 genes and hypomethylation in TXNIP, LOXL2 and SLC1A5 in type 2 diabetic compared with control individuals. Also, all nominally significant associations showed the same direction of effect as in the original EWASs. After adjustment for BMI, only the CpG site in ABCG1 remained significantly associated with type 2 diabetes, while for all other CpGs effect sizes became smaller and were no longer significant (ESM Fig. 1). Based on β values from regression analysis, we concluded that associations between significant CpGs and type 2 diabetes are partly explained by BMI (BMI accounted for 5–70% of variance, data not shown). Additional adjustment for other factors (see Methods) demonstrated that these covariates had only a relatively small impact on effect sizes and p values (ESM Table 3). Furthermore, we performed a sensitivity analysis on subsamples (see Methods), in which only the CpGs in TXNIP (50 vs 49) and ABCG1 (100 vs 49) reached the significance threshold (p < 0.0009), suggesting lack of power compared with the total group comprising 198 samples (data not shown). We also examined, for the 15 nominally significant CpGs, whether the differences in methylation were influenced by the occurrence of complications in diabetic individuals. We found no significant difference between individuals with and without complications (ESM Table 4). Finally, to check the effect of inflammation, we also adjusted the analysis for C-reactive protein (CRP) level and found no difference in the outcome (data not shown).
Table 4.
Significant differentially methylated CpGs for type 2 diabetes as originally identified in blood and replicated in the Lifelines type 2 diabetes EWAS sample in blood (n = 198)
| Illumina ID | CHR | MAPINFO | Gene name | Location in gene | Location in CpG island | Mean methylation (%) | Model 1 | Model 1 + BMI | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta methylation (%) | p value | Delta methylation (%) | p value | |||||||
| cg06500161a | 21 | 43656587 | ABCG1 | Body | Shore | 60.9 | 3 | 2.9 × 10−7 | 2.39 | 3 × 10−4 |
| cg24531955a | 8 | 23154691 | LOXL2 | 3′UTR | Open sea | 25.4 | −1.99 | 1.6 × 10−4 | −1.63 | 6 × 10−3 |
| cg19693031 | 1 | 145441552 | TXNIP | 3′UTR | Open sea | 69.5 | −3.6 | 2.5 × 10−4 | −2.68 | 1.5 × 10−2 |
| cg02711608a | 19 | 47287964 | SLC1A5 | 1stExon | Shelf | 20.1 | −1.81 | 3.2 × 10−4 | −1.26 | 2.7 × 10−2 |
| cg11024682a | 17 | 17730094 | SREBF1 | Body | Shelf | 44.6 | 1.88 | 5.5 × 10−4 | 1.04 | 8 × 10−2 |
| cg07960624 | 8 | 119208486 | SAMD12 | 3′UTR | Open sea | 39.7 | −2.3 | 4.8 × 10−3 | −1.59 | 9 × 10−2 |
| cg03497652 | 16 | 4751569 | ANKS3 | Body | Open sea | 55.5 | 1.86 | 9.7 × 10−3 | 1.79 | 3 × 10−2 |
| cg19266329 | 1 | 145456128 | POLR3GL b | – | Open sea | 60.9 | −1.77 | 1 × 10−2 | −0.98 | 0.20 |
| cg22909677 | 6 | 109172312 | ARMC2 | 5′UTR | Shelf | 80.4 | 1.11 | 1.2 × 10−2 | 1.04 | 0.07 |
| cg08309687a | 21 | 35320596 | ATP5O b | – | Open sea | 56.7 | −2.61 | 1.5 × 10−2 | −0.92 | 0.36 |
| cg26804423a | 7 | 8201134 | ICA1 | Body | Open sea | 63.8 | 1.39 | 1.5 × 10−2 | 0.78 | 0.23 |
| cg13199639 | 6 | 33360495 | KIFC1 | Body | Shore | 11.7 | −0.91 | 1.9 × 10−2 | −0.49 | 0.34 |
| cg15962267 | 5 | 138612986 | SNHG4 | Body | Shelf | 69.9 | −1.29 | 2.9 × 10−2 | −0.79 | 0.22 |
| cg03725309a | 1 | 109757585 | SARS | Body | Shore | 17.9 | −1.06 | 3.4 × 10−2 | −0.72 | 0.19 |
| cg10919522a | 14 | 74227441 | C14orf43 | 5′UTR | Shore | 31.4 | −1.42 | 4 × 10−2 | −0.42 | 0.56 |
No CpGs originally identified in other tissues were replicated in the present study looking at methylation in blood
Delta methylation is based on β values; p values are from analyses based on M values
Abbreviations: CHR, chromosome; MAPINFO, position on the chromosome; Shore, 0–2 kb from CpG island; Shelf, 2–4 kb from CpG island; Open sea, more than 4 kb from CpG island
Significant p values below 9.6 × 10−4 based on Bonferroni calculation
aCpGs also found to be associated with BMI in recently published EWAS [42]
bClosest genes were POLR3GL (108 bp downstream) and ATP5O (32,438 bp upstream)
Next, we investigated whether the five replicated type 2 diabetes-associated CpGs are also correlated with glycaemic and lipid phenotypes of healthy individuals (n = 98, Table 5). The methylation level at the ABGC1 site was significantly and positively correlated with age, fasting glucose and triacylglycerols, while the methylation levels of the TXNIP and SLC1A5 CpGs was negatively correlated with age. The methylation level at SREBF1 was positively correlated with both fasting glucose and lipid levels. No significant correlation with BMI was found in healthy individuals.
Table 5.
Correlations between DNA methylation (β values) of five replicated CpGs with type 2 diabetes risk factors in healthy individuals in Lifelines sample (n = 98)
| ABCG1 | LOXL2 | TXNIP | SLC1A5 | SREBF1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | p value | r | p value | r | p value | r | p value | r | p value | |
| Age | 0.31 | 1.7 × 10−3 | −0.17 | 8 × 10−2 | −0.11 | 3.4 × 10−2 | −0.27 | 6.6 × 10−3 | 0.45 | 4.4 × 10−6 |
| Fasting glucose | 0.31 | 1.9 × 10−3 | −0.09 | 0.33 | −0.15 | 0.14 | −0.01 | 0.86 | 0.21 | 3.5 × 10−2 |
| Triacylglycerol | 0.25 | 1.3 × 10−2 | −0.11 | 0.26 | −0.17 | 9 × 10−2 | −0.13 | 0.18 | 0.23 | 2.2 × 10−2 |
| Total cholesterol | 0.15 | 0.14 | −0.10 | 0.32 | −0.11 | 0.24 | 0.03 | 0.75 | 0.44 | 6.7 × 10−6 |
| LDL-cholesterol | 0.15 | 0.13 | 0.11 | 0.25 | −0.14 | 0.16 | −0.03 | 0.71 | 0.41 | 2.8 × 10−5 |
| HDL-cholesterol | −0.07 | 0.46 | 0.04 | 0.65 | 0.09 | 0.37 | 0.27 | 6.4 × 10−3 | 0.07 | 0.44 |
| BMI | 0.19 | 0.065 | −0.1 | 0.35 | −0.12 | 0.26 | −0.16 | 0.1 | 0.15 | 0.12 |
r = Pearson’s correlation coefficient
Significant p values < 0.05
Associations with type 2 diabetes: other tissue-specific CpGs
In addition to the 52 CpGs associated with type 2 diabetes in blood, we also analysed 17 CpGs that were associated with type 2 diabetes in pancreas and liver to test whether DNA methylation in metabolically active tissues may be reflected in DNA methylation in blood. No significant associations were found for any of these CpGs in blood samples (all p > 0.1).
Associations with glycaemic traits
Finally, we tested the CpGs previously shown to be associated with fasting glucose and HbA1c levels. In blood samples from the 98 healthy individuals, we replicated the association between CpGs in the CCDC57 and ABCG1 genes and fasting glucose level at nominal significance (p < 0.05, Table 6). Interestingly, after adjustment for BMI, two more CpGs, located in MDN1 and FLAD1 genes reached nominal significance (Table 6). We found no significant association between the level of HbA1c and DNA methylation at any of the ten CpGs identified in adipose tissue.
Table 6.
Significant differentially methylated CpGs for fasting glucose replicated in healthy control individuals from the Lifelines type 2 diabetes EWAS subsample (n = 98)
| Illumina ID | CHR | MAPINFO | Gene name | Location in gene | Location in CpG island | Mean methylation (%) | Model 1 | Model 1 + BMI | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta methylation (%) | p value | Delta methylation (%) | p value | |||||||
| cg06500161 | 21 | 43656587 | ABCG1 | Body | Shore | 59.1 | 1.82 | 6.8 × 10−3 | 1.68 | 1.6 × 10−3 |
| cg06715330 | 17 | 80158206 | CCDC57 | Body | Open sea | 81.3 | −1.82 | 0.01 | −2.05 | 6.6 × 10−3 |
| cg16809457 | 6 | 90399677 | MDN1 | Body | Open sea | 56.6 | 1.71 | 0.08 | 2.08 | 0.04 |
| cg16097041 | 1 | 154965544 | FLAD1 | 3′UTR | Open sea | 59.4 | 1.27 | 0.09 | 1.61 | 0.04 |
Delta methylation is based on β values; p values are based on M values
Significant p values < 0.05
CHR, chromosome; MAPINFO, position on the chromosome; Shore, 0–2 kb from CpG island; Shelf, 2–4 kb from CpG island; Open sea, more than 4 kb from CpG island
The EWASs for other metabolically relevant traits
Since high BMI and dyslipidaemia are well-known risk factors for type 2 diabetes and are commonly observed in diabetic individuals [43], we compared the results from our replication study with the results from recent EWASs studying DNA methylation related to adiposity and blood lipids [42, 44–46]. We found a large overlap between CpGs that are significantly associated with BMI and triacylglycerol levels, and those that are associated with type 2 diabetes and fasting glucose (ESM Table 5).
Discussion
In this study, we first comprehensively reviewed recently published EWASs investigations of DNA methylation patterns associated with type 2 diabetes and glycaemic traits. The potential use of DNA methylation as biomarker for type 2 diabetes is frequently reported in the literature, mostly using cross-sectional approaches. A more ideal setting for testing biomarkers would be to capture changes in the methylation profile prior to disease onset. A longitudinal study design would allow for this, since it provides measurements of methylation at multiple time points in the same individual, thereby capturing the epigenetic dynamics during life. However, due to higher costs and study duration, such EWASs are scarce, especially for complex diseases. To date, only one longitudinal EWAS study focusing on type 2 diabetes has been published, identifying five CpGs associated with disease onset in Indian Asians during the follow-up period [21], two of which (the CpGs in ABCG1 and PHOSPHO1) were replicated in a prospective study [47]. In our analysis we replicated three CpGs from the longitudinal study (i.e. ABCG1, TXNIP and SREBF1) indicating that those differences in methylation can also be captured in a cross-sectional study, for example, due to the stability of methylation level after disease onset. These CpGs represent potential predictive biomarkers for type 2 diabetes susceptibility.
Another issue concerns the inconsistency in EWASs methylation levels across tissues and whether blood can serve as a proxy tissue to capture these patterns. Changes in DNA methylation have been reported for different tissues like pancreas, liver, skeletal muscle or adipose tissue relevant in type 2 diabetes (ESM Table 1) [27, 31, 32, 48, 49]. The overlap in those results is limited, suggesting that the majority of the identified DNA methylation loci are tissue specific. However, some studies reported an overlap in disease-specific and age-related differentially methylated CpGs between blood and other relevant tissues. In recent EWASs, around 60% of the methylation changes associated with age in pancreatic islets also occur in blood, including FHL2, KLF14, FAM123C and GNPNAT1, all genes known to be associated with type 2 diabetes or insulin secretion [12]. Chambers et al reported that two out of five tested CpGs (in TXNIP and SOCS3) were differentially methylated in liver and reflected in blood [21]. Interestingly, another recent study showed hypermethylation at a CpG located in the SREBF1 gene in pancreatic cells and blood from type 2 diabetic individuals, and hypomethylation at the TXNIP locus in pancreatic islets, skeletal muscle and blood, which is directionally consistent with our findings in blood [47]. Taken together, these data indicate that some methylation changes found in the other tissues can be mirrored in blood. However, in our study we did not replicated the CpGs from the liver, pancreas and adipose tissue EWASs. This may be due to the small discovery sample sizes, the relatively small sample size of our replication study and/or reflect tissue-specific methylation patterns.
Epigenetic changes can be either a cause or a consequence of disease or an indirect contributing factor through environmental exposures that can affect both epigenome and type 2 diabetes risk [50]. Multiple factors can affect DNA methylation, such as environmental exposures [51], psychosocial [52] and genetic factors [53], together explaining the variance in DNA methylation levels between individuals. Also, accumulating data indicate that interactions between genetics and epigenetics influence gene expression levels in relevant metabolic traits, leading to the development of complex diseases [54, 55]. Recently, genetic ancestry and ethnicity is also shown to influence the methylation level [41]. Between the EWASs reviewed, we observed an overlap for a number of CpGs (TXNIP, ABCG1, SOCS3, SREBF1 and CPT1A) from EWASs performed in blood samples from Europeans, Indian Asians, Mexican Americans and Arabs, suggesting an association of DNA methylation with type 2 diabetes at these sites, irrespective of ethnic, social and environmental differences. Moreover, this finding highlights the usefulness of data sharing to create opportunities to perform meta-analyses, as is common practice for genome-wide association studies (GWASs).
In this study, we replicated five CpGs in blood, from which four reside in the genes previously shown to be associated with type 2 diabetes (ABCG1, LOXL2, SLC1A5, SREBF1) (ESM Box 1). Another replicated CpG site is TXNIP (cg19693031), which is shown to be hypomethylated in type 2 diabetes [21, 23–25]. Expression of TXNIP has been linked to glucose levels (ESM Box 1). Despite its important function in type 2 diabetes pathogenesis, TXNIP was not identified as one of the susceptibility genes in recent GWAS studies for type 2 diabetes [6]. These data suggest that DNA methylation is the major mechanism of controlling TXNIP expression, thereby affecting glucose homeostasis.
Blood cell composition can influence EWAS analyses and outcomes. There are several ways to avoid potential confounding effects of the cell composition, such as adjustment for direct measured cell count or reference-based cell count (e.g. the Houseman method [20]). In our analysis we observed no difference in effect sizes for the CpGs showing a significant association when using either the Houseman method or the measured cell count approach for adjustment, suggesting that these two methods may be used interchangeably (data not shown). Especially in studies in which information on blood cell composition is not available, methods such as the Houseman approach are essential.
It has been recently shown that methylation changes of the CpGs located in SREBF1, ABCG1 and CPTA1 were not only associated with type 2 diabetes but also with BMI [42, 44, 46]. Therefore, we compared our results with those from recent EWASs for adiposity and other relevant metabolic phenotypes [42, 44, 46]. We observed a substantial overlap between BMI and triacylglycerol-related CpGs, and CpGs associated with type 2 diabetes and glycaemic traits. Approximately 60% to 70% of diabetic individuals show some lipid abnormalities, which are associated with insulin resistance. The observed overlap in EWASs results could be explained by the fact that hypertriacylglycerolaemia leads to elevated non-esterified fatty acid levels, which in turn could induce insulin resistance and beta cell dysfunction [56]. Next, recent findings from the EWASs for adiposity indicate that adiposity determines methylation level at the majority of the identified loci [42] and that the methylation changes in blood might in part be a consequence of the alterations in lipid and glucose metabolism associated with BMI. In this EWAS, 62 of the 187 BMI methylation loci were associated with incidence of type 2 diabetes, and the BMI methylation risk score, calculated based on those CpGs, predicted future development of type 2 diabetes [42]. Together, this supports the hypothesis that BMI accounts partly for the association between DNA methylation and type 2 diabetes.
Overall, we conclude that a number of differentially methylated CpGs associated with type 2 diabetes in the published EWASs can be replicated in blood and show promise as disease biomarkers. Our data indicate that BMI partly explains the associations between DNA methylation and type 2 diabetes (i.e. only five out of 15 CpGs remained significant after adjustment for BMI). Whether these markers can be used as biomarkers for type 2 diabetes in a clinical practice requires further investigation. We recommend that more longitudinal studies are performed to confirm the robustness of these markers and to identify additional potential markers.
Electronic supplementary material
(PDF 840 kb)
Acknowledgements
The authors wish to acknowledge all participants of the Lifelines Cohort Study and everyone involved in the set-up and implementation of the study. We thank the Genome Analysis Facility of the University of Groningen, University Medical Center Groningen, for performing the Illumina 450K methylation array experiments.
Abbreviations
- CpG
Cytosine-phosphate-guanine
- CVD
Cardiovascular disease
- EWAS
Epigenome-wide association study
- GWAS
Genome-wide association study
- WGBS
Whole-genome bisulphite sequencing
Funding
This work was supported by the National Consortium for Healthy Ageing (NCHA) (NCHA NGI Grant 050-060-810), a grant from the National Institute for Public Health and the Environment (RIVM) and the Ministry of Health, Welfare and Sports of the Netherlands (S/132005), the European Union’s Seventh Framework programme (FP7/2007-2013) through the Biobank Standardisation and Harmonisation for Research Excellence in the European Union (BioSHaRE-EU) project, grant agreement 261433, and by the Dutch Diabetes Foundation (Diabetes Funds Junior Fellowship grant 2013.81.1673 to JVvVO).
Data availability
Data are available upon request from the authors.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
JVvVO and EW designed and implemented the study and drafted the manuscript. HS, ML and AMWS contributed significantly to study design. EW and MJB analysed the data. JVvVO, HLL and BHRW were involved in data acquisition. EW, JVvVO, ML, AMWS, HS and MJB contributed to the interpretation of the data. ML, AMWS and HS critically reviewed the manuscript for important intellectual content. All authors reviewed and approved the final manuscript. EW and JVvVO are the guarantors of the study.
References
- 1.The International Diabetes Federation (IDF) IDF Diabetes Atlas, 7th edition. Available from http://www.diabetesatlas.org/. Accessed 4 April 2016
- 2.Fonseca VA. Defining and characterizing the progression of type 2 diabetes. Diabetes Care. 2009;32(Suppl 2):S151–S156. doi: 10.2337/dc09-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Expert Committee on the Diagnosis and Classification of Diabetes Mellitus Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 2003;26(Suppl 1):S5–S20. doi: 10.2337/diacare.26.2007.s5. [DOI] [PubMed] [Google Scholar]
- 4.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham offspring study. Diabetes. 2000;49:2201–2207. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 5.Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia. 1999;42:139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- 6.Kwak SH, Park KS. Recent progress in genetic and epigenetic research on type 2 diabetes. Exp Mol Med. 2016;48:e220. doi: 10.1038/emm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 9.Kahn SE, Cooper ME, Del Prato S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet. 2014;383:1068–1083. doi: 10.1016/S0140-6736(13)62154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA methylation in whole blood: uses and challenges. Curr Environ Health Rep. 2015;2:145–154. doi: 10.1007/s40572-015-0050-3. [DOI] [PubMed] [Google Scholar]
- 11.Scholtens S, Smidt N, Swertz MA, et al. Cohort profile: LifeLines, a three-generation cohort study and biobank. Int J Epidemiol. 2015;44:1172–1180. doi: 10.1093/ije/dyu229. [DOI] [PubMed] [Google Scholar]
- 12.Bacos K, Gillberg L, Volkov P, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsha TE, Pheiffer C, Humphries SE, Gamieldien J, Erasmus RT, Kengne AP. Genome-wide DNA methylation in mixed ancestry individuals with diabetes and prediabetes from South Africa. Int J Endocrinol. 2016;2016(2016):3172093. doi: 10.1155/2016/3172093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkov P, Bacos K, Ofori JK, et al. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66:1074–1085. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 15.Toperoff G, Aran D, Kark JD, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klijs B, Scholtens S, Mandemakers JJ, Snieder H, Stolk RP, Smidt N. Representativeness of the LifeLines cohort study. PLoS One. 2015;10:e0137203. doi: 10.1371/journal.pone.0137203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonder MJ, Kasela S, Kals M, et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics. 2014;15:860. doi: 10.1186/1471-2164-15-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Touleimat N, Tost J. Complete pipeline for Infinium® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang J, Widschwendter M, Teschendorff AE. A comparison of feature selection and classification methods in DNA methylation studies using the Illumina Infinium platform. BMC Bioinf. 2012;13:59. doi: 10.1186/1471-2105-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers JC, Loh M, Lehne B, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–534. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Muftah WA, Al-Shafai M, Zaghlool SB, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenetics. 2016;8:13. doi: 10.1186/s13148-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soriano-Tarraga C, Jimenez-Conde J, Giralt-Steinhauer E, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet. 2016;25:609–619. doi: 10.1093/hmg/ddv493. [DOI] [PubMed] [Google Scholar]
- 24.Florath I, Butterbach K, Heiss J, et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia. 2016;59:130–138. doi: 10.1007/s00125-015-3773-7. [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni H, Kos MZ, Neary J, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24:5330–5344. doi: 10.1093/hmg/ddv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan W, Xia Y, Bell CG, et al. An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat Commun. 2014;5:5719. doi: 10.1038/ncomms6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dayeh T, Volkov P, Salo S, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkmar M, Dedeurwaerder S, Cunha DA, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson E, Jansson PA, Perfilyev A, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 30.Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchner H, Sinha I, Gao H, et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab. 2016;5:171–183. doi: 10.1016/j.molmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson E, Matte A, Perfilyev A, et al. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J Clin Endocrinol Metab. 2015;100:E1491–EE501. doi: 10.1210/jc.2015-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kriebel J, Herder C, Rathmann W, et al. Association between DNA methylation in whole blood and measures of glucose metabolism: KORA F4 study. PLoS One. 2016;11:e0152314. doi: 10.1371/journal.pone.0152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hidalgo B, Irvin MR, Sha J, et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes. 2014;63:801–807. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24:3792–3813. doi: 10.1093/hmg/ddv124. [DOI] [PubMed] [Google Scholar]
- 36.Wells, GA. Shea, B. O'Connell, et al. (2000) The Newcastle–Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available from www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 16 July 2017
- 37.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Defronzo RA. Banting Lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58:773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bays H, Mandarino L, DeFronzo RA. Role of the adipocyte, free fatty acids, and ectopic fat in pathogenesis of type 2 diabetes mellitus: peroxisomal proliferator-activated receptor agonists provide a rational therapeutic approach. J Clin Endocrinol Metab. 2004;89:463–478. doi: 10.1210/jc.2003-030723. [DOI] [PubMed] [Google Scholar]
- 40.Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014;103:137–149. doi: 10.1016/j.diabres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Galanter JM, Gignoux CR, Oh SS, et al. Differential methylation between ethnic sub-groups reflects the effects of genetic ancestry and environment esposures. eLife. 2017;6:e20532. doi: 10.7554/eLife.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bays HE, Chapman RH, Grandy S. The relationship of body mass index to diabetes mellitus, hypertension and dyslipidaemia: comparison of data from two national surveys. Int J Clin Pract. 2007;61:737–747. doi: 10.1111/j.1742-1241.2007.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dekkers KF, van Iterson M, Slieker RC, et al. Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17:138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demerath EW, Guan W, Grove ML, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeiffer L, Wahl S, Pilling LC, et al. DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet. 2015;8:334–342. doi: 10.1161/CIRCGENETICS.114.000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dayeh T, Tuomi T, Almgren P, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics. 2016;11:482–488. doi: 10.1080/15592294.2016.1178418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nitert MD, Dayeh T, Volkov P, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronn T, Volkov P, Davegardh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 51.Ho SM, Johnson A, Tarapore P, Janakiram V, Zhang X, Leung YK. Environmental epigenetics and its implication on disease risk and health outcomes. ILAR J. 2012;53:289–305. doi: 10.1093/ilar.53.3-4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borghol N, Suderman M, McArdle W, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10-2011-12-1-r10. doi: 10.1186/gb-2011-12-6-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsson AH, Volkov P, Bacos K, et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014;10:e1004735. doi: 10.1371/journal.pgen.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkov P, Olsson AH, Gillberg L, et al. A genome-wide mQTL analysis in human adipose tissue identifies genetic variants associated with DNA methylation, gene expression and metabolic traits. PLoS One. 2016;11:e0157776. doi: 10.1371/journal.pone.0157776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parhofer KG. Interaction between glucose and lipid metabolism: more than diabetic dyslipidemia. Diabetes Metab J. 2015;39:353–362. doi: 10.4093/dmj.2015.39.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 840 kb)
Data Availability Statement
Data are available upon request from the authors.