Abstract
The genetic basis of autism spectrum disorder (ASD) is known to consist of contributions from de novo mutations in variant-intolerant genes. We hypothesize that rare inherited structural variants in cis-regulatory elements (CRE-SVs) of these genes also contribute to ASD. We investigated this by assessing evidence for natural selection and transmission distortion of CRE-SVs in whole genomes of 9,274 subjects from 2,600 families affected by ASD. In a discovery cohort of 829 families, structural variants were depleted within promoters and UTRs, and paternally-inherited CRE-SVs were preferentially transmitted to affected offspring and not to their unaffected siblings. The association of paternal CRE-SVs was replicated in an independent sample of 1,771 families. Our results suggest that rare inherited non-coding variants predispose children to ASD, with differing contributions from each parent.
Microarray and exome sequencing studies over the past decade have demonstrated that de novo protein-altering variants contribute to approximately 25% of cases of autism spectrum disorder (ASD) (1, 2). Much of the allelic spectrum of ASD genetics has been unexplored, particularly variants that lie outside of protein coding sequences of genes. Recent studies have made great progress in identifying regulatory elements throughout the genome (3, 4). The next challenge is to identify ASD risk variants affecting genetic regulatory elements. However, deleterious cis-regulatory variants are not easily distinguishable from the vast background of neutral variation in the genome. Therefore initial applications of whole genome sequencing (WGS) in ASD have so far been underpowered to detect the association of rare cis-regulatory single nucleotide variants (SNVs) with ASD (5-7).
Structural variants (SVs), such as deletions, duplications, insertions and inversions (8), are more likely than SNVs to impact gene regulation because of their potential to disrupt or rearrange functional elements in the genome. Recent WGS efforts led by the 1000 Genomes consortium and our group have revealed thousands of rare SVs in each genome that were previously undetectable with microarray or exome sequencing technologies (8, 9).
Here we investigate the contribution of cis-regulatory SVs (CRE-SVs) to autism in three stages: (1) selection of target functional categories based on evidence of SV-intolerance; (2) association tests of cis-regulatory elements in a primary WGS dataset; and (3) pre-registered replication in an independent cohort.
Our discovery dataset consisted of whole genome sequencing (mean coverage = 42.6) of 829 families, comprising 880 affected, 630 unaffected individuals, and their parents (table S1). A majority of the subjects in the discovery sample were selected on the basis that they had previously screened negative for de novo loss of function mutations or large copy number variants from exome sequencing (2) and microarray (10) studies. The ascertainment of this sample was therefore designed to eliminate the well-established categories of genetic risk and thereby to enrich for novel inherited and non-coding risk variants.
We developed a pipeline for genome wide analysis of SV that consisted of complementary methods for SV discovery (fig. S1). A key innovation was the development of SV2, a support-vector machine (SVM) based software for accurately estimating genotype likelihoods from short read WGS data, which enabled accurate genotyping of SVs in families with a detection limit of ≥100 bp (11). An average of 3,746 SVs were detected per individual, including biallelic deletion, tandem duplications, inversions, four classes of complex SV, and four families of mobile element insertion (summarized in figures S2, S3 and table S2). The overall false discovery rate (FDR) was estimated from Illumina 2.5M SNP array data to be 4.2% for deletions, 9.4% for duplications (fig. S4, table S3). SVs were also validated through Nanopore whole genome sequencing of three individuals at a mean coverage of 7-9X (table S3). Private deletions and duplications >100 bp in length displayed low Mendelian-error rates and 50% transmission to offspring (fig. S4).
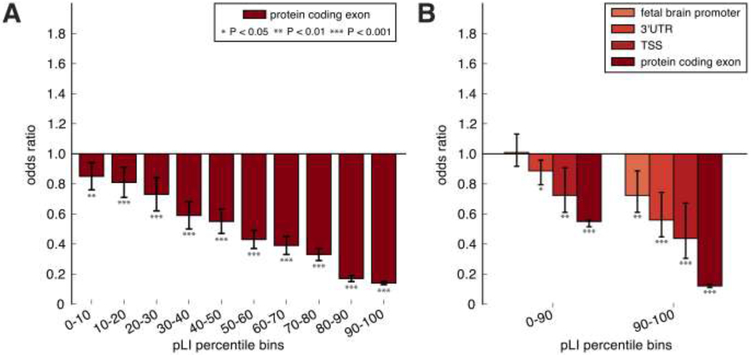
Measures of functional constraint that are based on population data are useful metrics for predicting the pathogenicity of rare variants. For example, genes that display strong negative selection against loss-of-function variants in the general population, as assessed by the exome aggregation consortium (ExAC) (12), are highly enriched in de novo mutations in children with ASD (13), and the vast majority of known autism genes display loss of function intolerance scores (pLI) above the 90th percentile for all genes (OR = 17.6; Fisher’s Exact P = 7.3×10−30; table S4 and fig. S5). Furthermore, we show here that the intolerance of genes to exonic deletions is correlated with the SNP-based pLI measure of functional constraint (fig. 1A-B).
Figure 1. Selection of target functional categories based on deletion intolerance.
Bar charts illustrating functional elements that show depletions in deletions relative to random permutations, stratified by deciles of gene variant-intolerance (pLI) as estimated by the ExAC consortium. A) Protein-coding deletions. B) Cis regulatory elements deletions. Odds ratios calculated based on observed counts versus expected based on permutation. Stars indicate the level of significance in the permutation analysis; whiskers represent 95% confidence intervals. TSS = transcription start site.
We reasoned therefore, that SV intolerance would be a valid criterion for defining categories of functional elements to be tested for disease association in this study. As our measure of SV intolerance, we tested the observed depletion of SVs within functional elements relative to random distributions of SVs generated by two types of permutation (14), one in which SVs were shuffled throughout the genome randomly and a second based on a model in which the correlation of SVs to genome features (GC content, coverage, low-complexity repetitive elements, and segmental duplications) was accounted for (15). SV depletion was assessed in functional elements grouped by categories such as exons, UTRs, promoters, cis-regulatory RNAs, enhancers, evolutionarily conserved and human accelerated regions (28 categories in total, described in table S5). SVs were each assigned to a single category according to the order listed above; for example a SV that disrupts an exon, a UTR, and an enhancer simultaneously would be classified as “exonic”. Genes were also defined in advance as “intolerant”, based on an EXAC pLI score > 90th percentile (fig S5). SV depletion was tested for the 28 categories, and analysis was stratified by SV type (deletion or duplication) and by loss-of-function intolerance (pLI) above or below the 90th percentile, a total of 104 tests.
Functional elements that showed significant evidence of SV depletion among intolerant genes (pLI ≥90th percentile; Benjamini Hochberg FDR Q<0.01; OR<1) were selected as our target categories (fig 1B, table S5). Nearly identical results were obtained with both random models in the discovery sample and in an independent cohort from the 1000 Genomes project (table S5; fig. S6). Categories that showed depletion of SVs relative to simulations included exons (OR= 0.18; P < 0.0001), TSSs (OR= 0.45; P < 0.0001) and 3’UTRs (OR= 0.57; P < 0.0001) and promoter annotations derived from fetal brain tissue (fetal brain promoters) from the epigenome roadmap (OR = 0.73; P = 0.0011), and the depletion of CREs was restricted to intolerant genes (fig 1B, table S5). Functional elements were further collapsed into “cis regulatory” and “coding and non-coding” categories respectively, and we included one non-depleted category “intron” as a control, resulting in a total of 10 target categories.
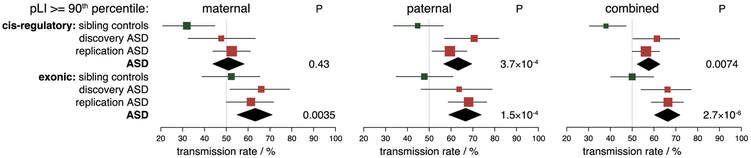
Focusing on the target functional categories above, family based association was tested using a group-wise transmission/disequilibrium test (TDT), applying it to private variants (autosomal parent allele frequency = 0.0003) assuming a dominant model of transmission. We confirmed a 50% parental transmission rate for deletions and duplications overall across a range of sizes (table S6). In variant-intolerant genes (pLI≥90th percentile), protein coding deletions were over-transmitted to cases (54/83; transmission rate = 65.1%; P = 0.002), but not to controls (26/57, transmission rate = 45.6%; P = 0.54; figure 2, table S6). Paternally inherited CRE-SVs (fetal-brain promoters, TSSs or 3’UTRs) of intolerant genes were over-transmitted to cases (39/55; transmission rate = 70.9 %; P = 0.0013), whereas maternal CRE-SVs were not significantly associated with ASD (21/44; transmission rate = 47.7%). The above associations were significant after correction for 20 tests (10 categories of SVs tested for each parent separately, table S6). Validation of cis-regulatory and exonic SVs was performed where possible using Nanopore sequencing, PCR or an in-silico SNV based approach (see methods). 96% (150/156) of SVs were validated with 100% genotype concordance SV2 (table S7).
Figure 2. Parental transmission of private cis-regulatory and exonic SVs to cases and sibling controls.
Rate of transmission from parents to offspring was tested for SVs that disrupt cis-regulatory elements or exons of variant-intolerant genes (pLI > 90th percentile). Whiskers represent the 95% confidence intervals. Effect sizes for CRE-SVs in all four cohorts individually is provided in figure S7).
The primary hypothesis to be tested in the replication sample (association of paternally inherited CRE-SVs) was pre-registered in the form of a preprint describing the analytic details and results of our primary analysis (16). We then replicated the association by applying our pipeline to an independent sample of 6,105 genomes from 1,771 families (17). The association of rare (allele frequency ≤ 0.0003) paternally-transmitted CRE-SVs was significant in the replication sample (65/109; transmission rate = 59.6%; P = 0.027). Also consistent with our primary results, maternally-transmitted CRE-SVs were not associated with ASD and inherited coding variants from both parents were associated with ASD (fig. 2, table S6).
In the combined dataset of 2,600 families, the association of paternal CRE-SVs was significant (P = 3.7×10−4) after correction for 20 tests. Consistent with a paternal-origin effect, CRE-SVs in cases were inherited more frequently from fathers (104 paternal, 74 maternal; Binomial P = 015). All private cis-regulatory and exonic variants in intolerant genes are given in table S7. The median lengths of cis-regulatory and exonic SVs were 2,920 bp (interquartile range IQR = 396-8,282bp) and 17,261bp (IQR = 4,390-112,251bp) respectively.
The smaller effect size observed in the replication sample (over-transmission of 59.6%, compared to 70.6% in the discovery sample) could be explained by a combination of factors including chance or true differences in the genetic architecture between samples. Cohorts did not differ dramatically in the numbers of trios and concordant sib pair (multiplex) families (table S1), thus, family structure is unlikely to have an influence. As mentioned above, selection of families for a subset of the discovery sample (SSC1) was designed to enrich for novel inherited and non-coding risk variants. Thus, ascertainment could in part explain why the SSC1 had the largest effect size of all individual cohorts (fig. S7).
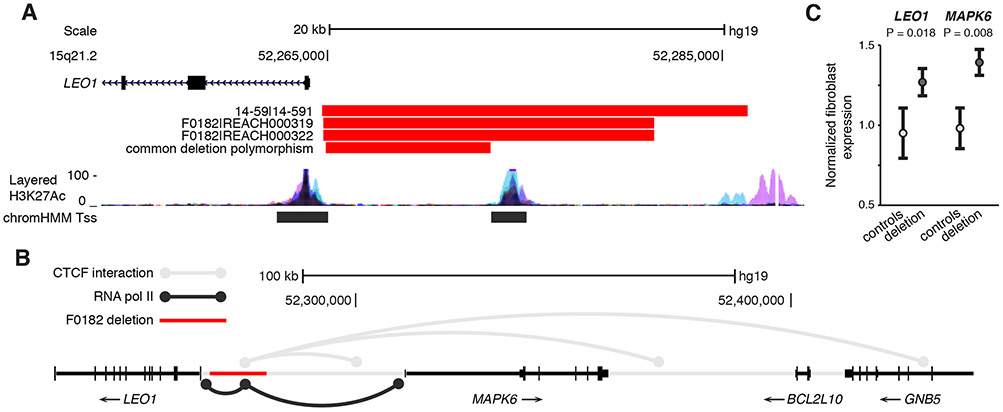
Recurrent CRE-SVs disrupting intolerant genes were observed in cases, including CNTN4, LEO1, RAF1, and MEST (table S7; permutation P = 0.0036). Two de novo LoF variants disrupting LEO1 (18, 19) have been observed in a combined exome dataset of ASD and developmental delay from 20 studies, a higher rate of LoF variants than would be expected by chance (expected n = 0.1; P = 0.0025) (14). Both LEO1 deletions eliminate an upstream regulatory element that has a chromatin signature associated with an active transcription start site (fig. 3A) (20). A smaller 8.7kb deletion polymorphism (parent allele frequency = 0.011) was detected within this region, but this variant does not disrupt any annotated functional elements. The deletions were fine-mapped by Nanopore single-molecule sequencing of long PCR products (fig. S8). Published chromatin interactions associated with transcription factors CTCF and RNA polymerase II mapped by ChIA-PET (21, 22) revealed this upstream cis-regulatory element to be a focal point for long range chromatin interactions associated with transcription (fig. 3B). Expression of LEO1 and the neighboring MAPK6 was higher in fibroblast cell lines from two deletion carriers compared to lines from three non-carrier controls (LEO1 T test P = 0.018; MAPK6 P = 0.008; fig. 3C; table S8).
Figure 3. Recurrent promoter deletions of LEO1 derepress expression.
A) Paternally inherited deletions of the LEO1 promoter were detected in three affected individuals, one trio (14-59) and one concordant sib pair (F0182). A common deletion polymorphism (parent allele frequency = 0.011) is also present in this locus. B) Chromatin interactions associated with transcription factors RNA Polymerase II and CTCF based on ChIA-PET data suggests that the cis-regulatory element upstream of LEO1 disrupted by both rare deletions (F0182 deletion shown here) serves as a focal point for the spatially organized transcription of LEO1 and MAPK6. C) mRNA expression of LEO1 and MAPK6 in fibroblast lines derived from two deletion carriers (REACH00319 and REACH000322) compared to three control lines. Whiskers represent 95% Confidence Intervals. Layered H3K27Ac = Histone 3 lysine 27 acetylation (an active promoter associated mark) in seven cell types from ENCODE. ChromHMM Tss = predicted transcription start site based on chromatin signatures in multiple cell types from the Epigenomics Roadmap Project (20).
As follow up to our previous studies of de novo SVs (9), we detected de novo mutations in the discovery sample, including 104 deletions, 19 duplications, 2 inversions, 8 complex SVs and 32 mobile element insertions (MEIs) (fig. S9; table S9). The majority (68%) of phased de novo SVs originated from the father (binomial test P = 0.038; table S9), comparable to the bias observed for SNVs and indels (23). We also confirm that de novo SNVs and indels cluster in proximity to de novo SV breakpoints (permutation P = 0.0029; table S10; fig. S10) (9). ASD cases did not display higher SV mutation rates than sibling controls (fig. S11) (9). Considering only the subset of the discovery sample that had not been characterized previously (REACH), gene disrupting de novo variants were significantly enriched in cases (7.2% in ASD versus 2.1% in controls; permutation P = 9.2×10−5; an excess of 5.1% in cases.
Based on this study, we estimate that rare inherited cis-regulatory and coding SVs contribute in 0.77% (95% CI - 0.39-1.13) and 1.21% (95% CI - 0.76-1.62) of cases respectively, and inherited known pathogenic SVs not accounted for above (table S11) contribute in another 1.9% of cases. As expected, the contribution of de novo coding SVs is substantial (5.1%); however no de novo CRE-SVs were detected in cases in the discovery sample (table S9).
Here we demonstrate that rare SVs that disrupt CREs confer risk for ASD, and this association is concentrated among genes that are highly dosage sensitive. The contribution of CRE-SVs that we observe consists exclusively of inherited variants that are carried by a parent. This result is consistent with non-coding variants having moderate effects on gene function and disease risk. We find no evidence for a contribution of de novo CRE-SVs, in contrast to anecdotal findings from previous studies (5, 7). We cannot exclude the possibility that de novo CRE-SVs contribute to ASD; however, we can conclude that they are extremely rare.
CRE-SVs exhibited a significant paternal-origin effect. This result was unexpected and contrasts with a simpler genetic model (24) in which inherited genetic risk is transmitted predominantly from mothers due the reduced vulnerability of females to ASD. Previous studies have shown a maternal bias for inherited truncating variants in genes that were previously implicated from studies of de novo mutation (25-27). In our study, the contribution of exonic variants to risk was similar for paternal and maternal SVs, suggesting that a maternal origin bias might be restricted to genes that have the most extreme dosage sensitivity. Taken together, our findings indicate that parent-of-origin effects on genetic risk for ASD are more complex than we previously thought, and the allelic spectrum of variants differs between the maternal and paternal genomes.
We propose three possible mechanisms to explain the observed paternal-origin effect of CRE-SVs, the first is a “bilineal two-hit model”, in which inherited risk is attributable to a combination of two risk variants: a maternally-inherited coding variant of large effect and a paternally-inherited CRE variant of moderate effect. This bilineal model predicts that a paternal bias might also be evident for other variants of moderate effect including hypomorphic missense alleles or LoF variants in genes with a moderate degree of intolerance. While this paper was under review, a genetic study of common variation in ASD families reported suggestive evidence of a paternal bias for variants of modest effect (28), a result that lends support to a bilineal model.
An alternative explanation for a paternal-origin effect is an epigenetic mechanism. For example, deletion of CREs can lead to de-repression of imprinted genes (29). However, an epigenetic mechanism could only explain our results if non-canonical imprinting of regulatory elements is widespread. Such a phenomenon has not been described, but we cannot rule out this possibility. A third potential mechanism to explain parent-of-origin effects could be a type of “meiotic drive”, in which allele-specific selection occurs differently in paternal and maternal germ cells. However, this mechanism is also unlikely given that there are few known examples of gene drive in humans and their effects appear to be quite weak at the population level (30).
Due to the greater potential of SVs to impact gene function and regulation relative to SNVs and indels, this class of genetic variation has historically proven effective for illuminating new components of the genetic architecture of disease. Our findings provide a further demonstration of the utility of SV analysis for characterizing the genetic regulatory elements that influence risk for ASD.
Supplementary Material
Acknowledgments:
We would like to thank the families who volunteered for the study. We would also like to thank W. Pfeiffer, M. Tatineni, A. Majumdar, S. Strande, R. Hawkins, the San Diego Supercomputer Center, and Amazon Web Services for hosting the computing infrastructure necessary for completing this project. This study was supported by grants to JS from the NIH (MH076431, MH113715) and Simons Foundation Autism Research Initiative (#275724) and by a gift to JS from the Beyster Family Foundation. Support to J.S. and K.K.V. was also provided from the ASD enlight foundation. Funding for K.P. is from the NIMH (R01MH110558). Funding for E.C. is from the NIMH (R01MH110558,I-P50-MH081755) and a Simons Foundation Grant. L.M.I. was supported by NIH (R21 MH104766, R01 MH105524, and R01 MH109885) and in part by the Simons Foundation grant #345469. S.W.S. Holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at the University of Toronto and the Hospital for Sick Children. Funding for B.C. is from MINECO (SAF2015-68341-R), AGAUR (2014-SGR-0932), La Marató de TV3 (092020) and The European Commission H2020 Programme MiND (643051). A.H. and M.J.A. received grant support from the Institute Carlos III (FIS PI11/00620) and Mutua Terrassa (FMT grant BE062). Funding for C.T. was provided by La Marato′ de TV3’ (092010), Postdoctoral Fellowships were provided to WB from the Autism Science Foundation and to MLK from the Canadian Institutes of Health Research. A T32 training grant to D.A. from the NIH (GM008666). Funding for collection of fibroblast cell lines was provided by a grant (IT1-06611) to J.G.G from CIRM. A.R.M. is supported by a NARSAD and NIH grants R01MH108528 and R01MH109885. We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman). We would like the thank Genome Canada and the Centre for Applied Genomics (TCAG) for contributing the replication dataset from the Autism Speaks MSSNG cohort (www.mss.ng).
Footnotes
Competing Interests: J.S. declares that a patent has been issued to the Cold Spring Harbor Laboratory by the US Patent and Trademark Office on genetic methods for the diagnosis of autism (patent number 8554488). W.M.B., B.K., A.T., J.C.V are employed by Human Longevity Inc. Y.Y., E.H., S. J., and D.J.T. work for Oxford Nanopore Technologies Inc.
Data and Materials Availability: The data reported in this paper are archived at the National Database for Autism Research (DOI:10.15154/1340302), including the structural variant callset, and raw sequence (FASTQ), alignment (BAM) and variant call (VCF) files from the REACH cohort. We appreciate obtaining access to Simons Simplex Collection genomic and phenotypic data on SFARI Base. Approved researchers can obtain the SSC population dataset described in this study (https://sfari.org/resources/autism-cohorts/simons-simplex-collection) by applying at https://base.sfari.org.
References and Notes:
- 1.Sebat J et al. , Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iossifov I et al. , The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rands CM, Meader S, Ponting CP, Lunter G, 8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet 10, e1004525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellis M et al. , Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A 111, 6131–6138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doan RN et al. , Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner TN et al. , Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am J Hum Genet 98, 58–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner TN et al. , Genomic Patterns of De Novo Mutation in Simplex Autism. Cell 171, 710–722 e712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudmant PH et al. , An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandler WM et al. , Frequency and Complexity of De Novo Structural Mutation in Autism. Am J Hum Genet 98, 667–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders SJ et al. , Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antaki D, Brandler WM, Sebat J, SV2: Accurate Structural Variation Genotyping and De Novo Mutation Detection from Whole Genomes. Bioinformatics, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M et al. , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson EB et al. , Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet 48, 552–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebat, Materials and methods are available as supplementary materials.>
- 15.Ruderfer DM et al. , Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat Genet 48, 1107–1111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandler WM et al. , Paternally inherited noncoding structural variants contribute to autism. bioRxiv, (2017). [Google Scholar]
- 17.RK CY et al. , Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 20, 602–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Rubeis S et al. , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deciphering Developmental Disorders S, Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roadmap Epigenomics C et al. , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G et al. , Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Z et al. , CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611–1627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelson JJ et al. , Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X et al. , A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A 104, 12831–12836 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iossifov I et al. , Low load for disruptive mutations in autism genes and their biased transmission. Proc Natl Acad Sci U S A 112, E5600–5607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krumm N et al. , Excess of rare, inherited truncating mutations in autism. Nat Genet 47, 582–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B et al. , CNV analysis in Chinese children of mental retardation highlights a sex differentiation in parental contribution to de novo and inherited mutational burdens. Sci Rep 6, 25954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye K et al. , Measuring shared variants in cohorts of discordant siblings with applications to autism. Proc Natl Acad Sci U S A 114, 7073–7076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorvaldsen JL, Duran KL, Bartolomei MS, Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12, 3693–3702 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffreys AJ, Neumann R, Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet 31, 267–271 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Loken E, Gelman A, The Statistical Crisis in Science. American Scientist 102, 460 (2014). [Google Scholar]
- 32.Bejerano G et al. , Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Khurana E et al. , Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 342, 1235587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R et al. , A haplotype-based framework for group-wise transmission/disequilibrium tests for rare variant association analysis. Bioinformatics 31, 1452–1459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.