Abstract
Background:
The current system to predict the outcome of smokers with Bladder cancer (BLCA) is insufficient due to complex genomic and transcriptomic heterogeneities. This study aims to identify serum metabolite associated genes related to survival in this population.
Methods:
We performed liquid chromatography-mass spectrometry (LC-MS) based targeted metabolomic analysis for >300 metabolites in serum obtained from two independent cohorts of BLCA never smokers, smokers, healthy smokers, and healthy never smokers. A subset of differential metabolites was validated using Biocrates absoluteIDQ p180 kit. Genes associated with differential metabolites were integrated with a publicly available cohort of TCGA to obtain an intersecting signature specific for BLCA smokers.
Results:
40 metabolites (FDR <0.25) were identified to be differential between BLCA never smokers and smokers. Increased abundance of amino acids (tyrosine, phenylalanine, proline, serine, valine, isoleucine, glycine, asparagine) and taurine were observed in BLCA smokers. Integration of differential metabolomic gene signature and transcriptomics data from TCGA cohort revealed an intersection of 17 genes that showed significant correlation with patient survival in BLCA smokers. Importantly, Catechol-O-Methyltransferase (COMT), Iodotyrosine Deiodinase (IYD), and Tubulin Tyrosine Ligase (TTL) showed a significant association with patient survival in publicly available BLCA smokers datasets and did not have any clinical association in never smokers.
Conclusions:
Serum metabolic profiling of BLCA smokers revealed dysregulated amino acid metabolism. It provides a distinct gene signature that shows a prognostic value in predicting BLCA smoker survival.
Impact:
Serum metabolic signature derived genes act as a predictive tool for studying the BLCA progression in smokers.
Introduction
Urinary bladder cancer (BLCA) is the 9th most common malignant disease and the 13th most common cause of cancer death worldwide (1). Nearly 1.3 million people worldwide and 600,000 in the United States (US) alone are diagnosed with BLCA each year contributing to a significant healthcare burden. BLCA has a high recurrence rate and requires lifelong surveillance incurring major medical expenses (2–4). Occupational exposure to carcinogens has been long associated with increased BLCA risk. Tobacco smoke contains more than 60 carcinogens causing at least 18 different types of cancer including BLCA. Tobacco smoking is the best-established risk factor for BLCA, with a population attributable risk for smoking of 50% for BLCA (5). More than 30% of people in the US and Europe have a history of smoking (6), based on which BLCA patients can be categorized into former and current smokers. Current smoking also increases the risk of recurrence, drug resistance, and significantly increases the risk of death in BLCA compared to former and never smokers (7, 8). Previous studies have suggested that smoking status and the quantity of smoking is associated with higher tumor grade and stage in BLCA patients (9).
Xenobiotic metabolizing enzymes expressed in the bladder detoxify the genotoxic compounds and excrete them through urine. It is known that alterations of xenobiotic enzymes are associated with BLCA development (10, 11). Environmental toxins, including the carcinogens from cigarettes, are cleared by xenobiotic pathways and pass through the body’s excretion system. Over time, these products accumulate on the walls of the bladder which in turn changes the genetic composition of the bladder wall. Our group has previously shown that tobacco-specific carcinogens alter metabolic pathways and the expression levels of xenobiotic enzymes in BLCA (12). Diagnostically, patients with BLCA are evaluated using cystoscopy, an endoscopic procedure performed by urologist (13). Also, cells in the urine (cytology) are inspected by trained cytopathologist to determine the presence of cancer lesions in the bladder (14). However, the sensitivity for bladder tumor detection with urine cytology surveillance is low. Non-invasive procedures are needed for facilitating diagnostic, therapeutic, and prognostic strategies. Unfortunately, to our knowledge, there are no diagnostic or surveillance serum or plasma associated metabolic biomarkers which have been identified in BLCA smokers.
The advent of metabolic profiling with liquid chromatography-mass spectrometry (LC-MS) has opened the possibilities to identify dysregulated metabolic pathways and contributing markers. These, in turn, can serve as prognostic and diagnostic biomarkers in the identification of disease and its pathogenesis (15). While prior studies have successfully utilized metabolomics for biomarker identification in BLCA sera (10, 16, 17), serum metabolic profiling for BLCA smokers is lacking. In this study, we applied targeted LC-MS approach to delineate serum-based biomarkers that differentiate BLCA patients through smoking history (current smokers, former smokers) from never smoking BLCA patients. This study provides initial evidence of serum-based metabolite prognostic markers in BLCA smokers.
Materials and Methods
Study population and serum profiling by LC-MS
In this study, serum samples from BLCA (n=67) were used from two independent cohorts: (1) National Cancer Institute (NCI, n=59) (2) Augusta university (n=8), Georgia Cancer Center and healthy case controls (n=20) from Baylor College of Medicine procured by a prior written informed consent under Institute review board (IRB) approved protocols. NCI patient cohorts are enrolled in a clinical study with monotherapy cabozantinib.
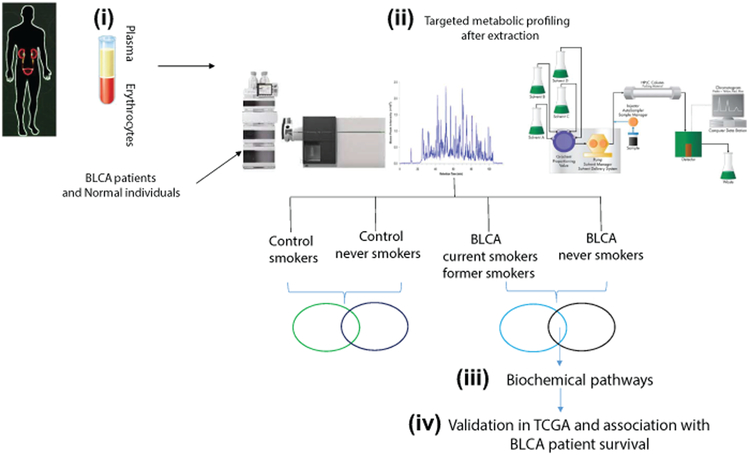
A flow chart depicting the study overview, sample processing, and analysis are illustrated in Figure 1.
Figure 1. Overview of global serum metabolic profiling in BLCA patients and normal individuals.
The methodology involves four important steps: (i) Serum extraction from blood samples of BLCA patients and normal individuals, (ii) Filtration, metabolic extraction, and targeted metabolic profiling by LC-MS, (iii) Identification of differential metabolites and deciphering the biochemical pathways involving the upregulated metabolites, and (iv) Validation of genes involved in regulating the biochemical pathways and their clinical association in TCGA cohorts.
Targeted metabolomics using mass spectrometry Reagents and internal standards
High-performance liquid chromatography (HPLC)-grade ammonium acetate from Sigma, acetonitrile, methanol, chloroform, and water were procured from Burdick & Jackson (Morristown, NJ). Mass spectrometry-grade formic acid was purchased from Sigma-Aldrich (St Louis, MO).) Metabolite standards and internal standards, including N-acetyl Aspartic acid-d3, tryptophan-15N2, sarcosine-d3, glutamic acid-d5, thymine-d4, gibberellic acid, trans-zeatin, jasmonic acid, 15N anthranilic acid, and Testosterone-d3, were purchased from Sigma-Aldrich (St. Louis, MO).
Sample preparation for mass spectrometry and metabolomics analysis
Serum samples were stored at −80°C until the processing. Metabolites were extracted from 40 µl of serum, control serum pool was used as quality controls, and the extraction procedure is as follows: Briefly, each plasma sample was resuspended in 750 µl of ice-cold methanol: water (4:1) containing 20 µl spiked internal standard mix. Ice-cold chloroform and water were added in a 3:1 ratio for a final proportion of 4:3:2 methanol:chloroform:water. Both the organic and aqueous layers were transferred into a new tube, dried, and resuspended with 50:50 methanol:water. The resuspended sample was then deproteinized using a 3kDa molecular filter; (Amicon ultracel-3K Membrane; Millipore Corporation, Billerica, MA) and the filtrate was dried under vacuum (Genevac EZ-2plus, NY). Prior to MS, the dried extracts were re-suspended in 50 µl of 1:1 methanol and water and were subjected to LC-MS.
Separation of Metabolites
Two different analytical methods were used for the separation of targeted metabolomics to measure >300 metabolites.
Method 1). ESI positive mode the HPLC column (Waters X-bridge Amide 3.5 µm, 4.6 × 100 mm (PN: 186004868, Waters Milford, MA) was used. Mobile phase A and B were 0.1% formic acid in water and acetonitrile respectively. Gradient: 0–3 min-85 % B; 3–12 min- 30 % B, 12–15 min-2 % B, 16 min- 95% -B, followed by re-equilibration at the end of the gradient 23 min to the initial starting condition 85% B. Flow rate: 0.3 ml/min. Method 2) In ESI negative mode the HPLC column (Waters X-bridge Amide 3.5 µm, 4.6 × 100 mm (PN: 186004868, Waters, Milford, MA). Mobile phase A and B were 20 mM ammonium acetate in 95% acetonitrile and 5% water (pH 9.0) and 100% acetonitrile respectively. Gradient:0–3 min-85% B, 3–12 min-30% B, 12–15 min- 2% -B, 15–16 min- 85% B followed by re-equilibration at end of the gradient 23 min to the initial starting condition of 85% B. Flow rate: 0.3 ml/min.
Data acquisition through LC/MS Analysis
LC-MS analysis was performed using 6490 triple quadrupole mass spectrometer coupled to an Agilent 1290 series HPLC system (Agilent Technologies, Santa Clara, CA) with single reaction monitoring (SRM). This LC system is equipped with a degasser, binary pump, thermostatted auto sampler, and column oven. This SRM-based measurement of relative metabolite levels used normal phase chromatographic separation. 10 µl of resuspended samples were injected and analyzed using source parameters as follows: Gas temperature- 250 °C; Gas flow- 14 l/min; Nebulizer - 20psi; Sheath gas temperature - 350 °C; Sheath gas flow- 12 l/min; Capillary - 3000 V positive and 3000 V negative; Nozzle voltage- 1500 V positive and 1500 V negative. Approximately 8–11 data points were acquired per each detected metabolite.
Quantification of metabolites using the Biocrates AbsoluteIDQ kit p180
Metabolite concentrations were obtained using the AbsoluteIDQ kit p180 according to manufacturer’s instructions on a QTRAP 6500 LC/MS/MS System. 10 μl of the internal standard (ISTD) solution was added to each well of the 96-well plate, 10 μl of the serum samples, quality control (QC) samples, blank, and calibration standard were added to the appropriate wells. The plate was then dried, the samples were derivatized with phenyl isothiocyanate for the amino acids and biogenic amines. The samples were dried using GeneVac Vaccum system at 37°C and eluted with 5 mM ammonium acetate in methanol. They were then were diluted with 50: 50 methanol: water for LC-MS analysis. The LC column was Agilent Zorbax Eclipse XDB C18,3.0 ×100mm, 3.5 µm. Mobile phase A and B were 0.2% formic acid in water and acetonitrile respectively. The injection volume was 10 µl, and the flow rate was 0.5 ml/min. The samples were integrated and analyzed in AB Sciex Analyst Software to their specific labeled internal standards provided by the kit plate and exported the results file(.rdb). The plate was validated, and concentrations were calculated, exported by using this results file in MetIQ software. The final concentrations exported were in µM units.
Statistical Analysis
Agilent MassHunter Workstation Software - Quantitative analysis was used for manual review of chromatograms. Peak area integration was accessed based on the retention time. The normalization of each metabolite peak area was done by the peak area of the spiked ISTD, and then the data were log2 transformed, per method basis. For differential analysis, two-way sample t-tests were conducted, and ANOVA was performed for three-way analysis. Differential metabolites were identified by adjusting the p-values for multiple testing at a False Discovery Rate (FDR) threshold of <0.25.
Integration of metabolomics and transcriptomics and survival analysis
The panel of altered metabolites between smokers and never smokers was mapped to their corresponding genes using the Kyoto Encyclopedia of Genes and Human Metabolic Data Base (KEGG/HMDB), leading to the identification of 374 associated genes. We used transcriptomic profiles of BLCA were obtained from the Riester (18) and The Cancer Genome Atlas (TCGA) (19) with associated clinical information on BLCA smoking patients. TCGA RNA expression data is based on mRNA-Seq whereas Riester RNA expression data is microarray-based. Next, we generated gene signatures of BLCA smokers versus never smokers using TCGA data. Following this methodology, we identified the common genes from metabolomics and transcriptomics and identified 17 intersecting genes. To avoid dataset-specific bias, we compared the 17 smoking signature genes by smoker status in the Riester dataset and used this fold change signature for patient survival in both TCGA patient cohort (n= 365 patients having smoking status) and Riester cohorts (n= 93 patients having smoking status) by using Kaplan Meier curves. We also analyzed the complete 17 gene signature for survival status in the same cohort.
Validation of identified genes in patient tissues and mouse xenografts
6–8 week old male NOD-SCID beige mice were injected subcutaneously with 1.5 × 106 UMUC3 BLCA cells. The control group (n=3) mice with xenografts are maintained in normal cages. The experimental group (n=3) was exposed to cigarette smoke from two cigarettes (Marlboro) every day for a period of 4 weeks. The tumor volume was measured every four days using vernier calipers and were dissected after reaching a volume of 1500mm3. The dissected tumor were flash frozen and store at −80°C until analysis.
10 mg of control and smoke exposed mouse BLCA tissues were digested with a homogenizer in 500µL Trizol (Ambion). Equal volumes of chloroform were added to the digested tissue and centrifuged at 15000rpm for 15 mins. Next, the aqueous phase was taken in a new tube, and an equal volume of isopropanol was added and kept at −20°C for 30 min. Later, the lysate was loaded on QIAGEN column (QIAGEN RNeasy kit) for RNA purification. RNA was quantified using a Biotek plate reader. cDNA was transcribed from 1µg of RNA by using qScript cDNA Supermix (Quantabio). Primers for the genes were obtained from Invitrogen and their sequences are available upon request. Real-time PCR was performed using SYBR Green Master mix (Lifetech).
Results
Study population characterstics
The clinical-pathologic characteristics are summarized in Table 1. Nearly, 95% of patients have T4 stage BLCA. More than 80% of the patients enrolled are LVI positive and are older than 50 years of age. We characterized the metabolome of BLCA never smokers (39%) and smokers (former 46%; current 15%) and age-matched case-control serum (Table 1). In these two cohorts, the current smokers consume a range between <20 cigarette pack per year (n=5) and >20 pack per year (n=5). The cessation period for former smokers is around 2–20 years. Serum samples from healthy individuals (n=20) were procured from Baylor College of Medicine (BCM) out of which 55% (n=11) are smokers, and 45% (n=9) are never smokers. The characteristics of the healthy individuals are provided in Table 1.
Table 1.
Baseline characteristics of BLCA patients and healthy individuals involved in this study.
| BLCA Serum samples | ||
|---|---|---|
| BLCA Patients Characteristics | ||
| Ethnicity | African American (n=17; 25%) | |
| European American (n=49; 74%) | ||
| Unknown (n=1; 1%) | ||
| Smoking status | n=26 (39%; Never smoker) | |
| n=31 (46%; Former-smoker) | ||
| n=10 (15%; Current smoker) | ||
| Gender | n= 46(69%; Male) | |
| n=21(31%; Female) | ||
| Cohorts | NCI (n= 59; 88%) | |
| Georgia (n =8; 12%) | ||
| T-stage | Ta=1(1%) | |
| T1=1(1%) | ||
| T2=2(2%) | ||
| T3=1(1%) | ||
| T4=62(95%) | ||
| Age | ≤50 years= 8(12%) | |
| >50 years= 59 (88%) | ||
| LVI* | LVI-Positive=59(89%) | |
| LVI-Negative=8(11%) | ||
| Number of packets per year | Current Smokers | |
| <20Pack/year=5(50%) | ≥20Pack/year=5(50%) | |
| Duration of cessation | Former Smokers | |
| Quit <20years ago=9(29%) | Quit ≥20years ago=22(71%) | |
| Case Control serum samples | ||
| Characteristics | ||
| Ethnicity | African American (n=14; 70%) | |
| European American (n=3; 15%) | ||
| Hispanic (n=3; 15%) | ||
| Smoking status | n=9 (45%; Never smoker) | |
| n=7 (35%; Former-smoker) | ||
| n=4 (20%; Current smoker) | ||
| Gender | n=10 (50%; Male) | |
| n=10 (50%; Female) | ||
Lymphovascular invasion
Identification of altered metabolites and associated pathways
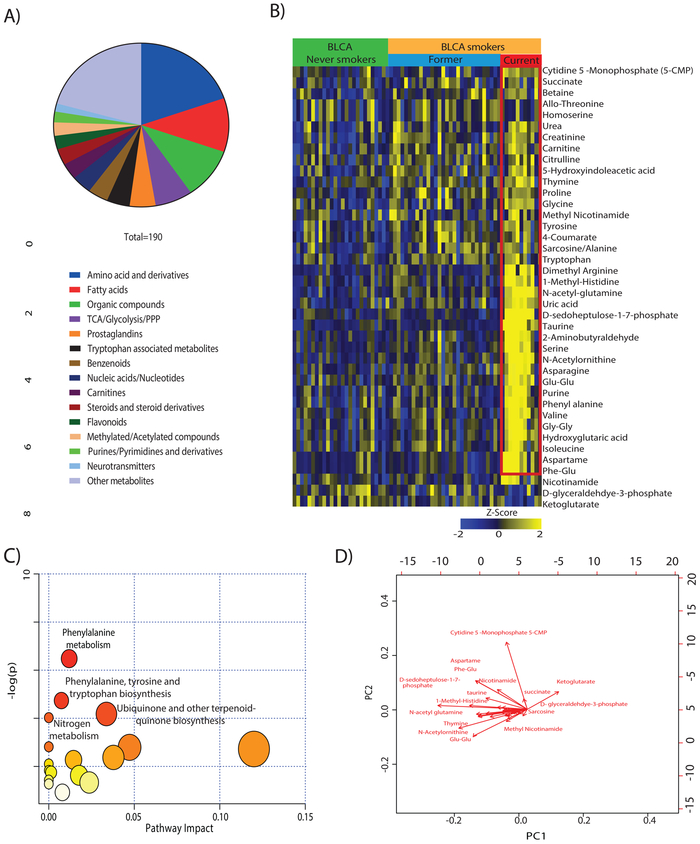
We targeted >300 metabolites (Supplementary Table 1) and detected 190 metabolites of different classes (aminoacid and derivatives, free fatty acids, TCA, Glycolysis/PPP pathway metabolites, nucleic acids/nucleotides) in serum samples (Figure 2A). A repetitive analysis of pooled serum samples served as controls to ascertain the reproducibility and robustness of the profiling platform for both positive and negative ionization (Supplementary Figure 1A and 1B) and show <20 Coefficient of Variations for each metabolite. To identify the smoke specific serum metabolite differences among control, and BLCA smokers, two independent analyses were performed in which the groups were compared (i.e.healthy control never-smokers vs. healthy control smokers, and BLCA never smokers vs. BLCA smokers). The comparison between control smokers (n= 11) and never smokers (n=9) did not yield any differential metabolites (Supplementary Figure 2). Interestingly, we have identified 40 metabolites (out of 190) that were significantly differential in BLCA smokers compared to BLCA never smokers. In the 40 differential metabolites, 37 were upregulated, and three were downregulated (Figure 2B). Further, most of the differential metabolites show higher levels in BLCA current smokers in comparison to BLCA former smokers (refer to the red box in Figure 2B). We performed additional analysis to identify the metabolic signature specific to current smokers. A total of 62 differential metabolites were identified in this analysis (Supplementary Figure 3A). The metabolic signature was compared to altered metabolic pool in Figure 2A, and we identified 34 metabolites were specific to BLCA current smokers (Supplementary Figure 3A, Indicated in red font). Pathway analysis was performed using Metaboanalyst 4.0 (20) for the differential metabolites between BLCA never smokers and BLCA smokers. We identified pathways that are involved in phenylalanine metabolism, tyrosine and tryptophan biosynthesis, and nitrogen metabolism (Figure 2C). To graphically represent the differential metabolites and their coefficient of variance between BLCA smokers and BLCA never smokers, we used metaboanalyst 4.0 for the depiction of variability in a biplot. The length of arrow in the bi-plot represent the degree of variation of each metabolite. Significant changes of the differential metabolites are illustrated in the biplot (Figure 2D). Observations indicate that these metabolic pathways could be dysregulated in BLCA smokers compared to never smokers.
Figure 2. Metabolic profiling of BLCA Serum from smokers and non-smokers and identification of key altered pathways.
(A) Pie chart illustrating the different classes of metabolites obtained in global targeted metabolic profiling of serum samples from BLCA patients. A distinct color denotes each group of metabolites, and the arc length of each slice represents the percentage of that group in the obtained profile. (B) Heat map visualization of 40 significantly altered metabolites in BLCA smokers compared to never smokers (FDR=<0.25). Shades of yellow and blue represent upregulation and downregulation of metabolites respectively, relative to median metabolic levels. (C) Topology analysis of dysregulated metabolic pathways in association with BLCA smokers. The size of bubble area denotes the impact of each pathway, with a color representing the significance from highest in red to lowest in white. (D) Bi-plot showing the upregulated differential metabolites in BLCA smokers and never smokers.
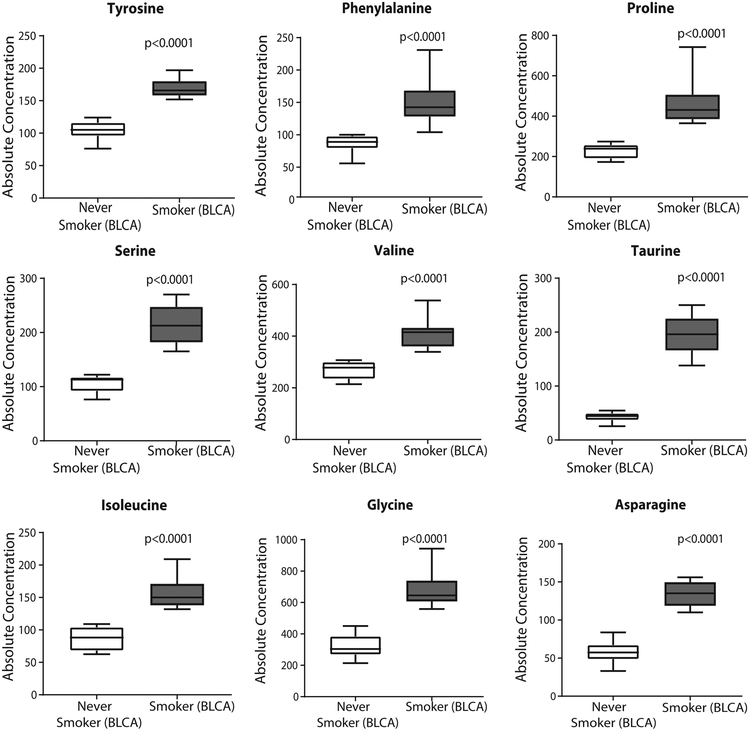
A subset of differential metabolites was validated using Biocrates absoluteIDQ p180 kit. However, the kit provides only a limited number of overlapping metabolites, therefore we could quantify only 9 out of the 40 differential metabolites. Interestingly, the glucogenic amino acids (proline, serine, valine, glycine, asparagine) were found to be high in BLCA smokers compared to never smokers. Also consistent with our earlier findings, phenylalanine, isoleucine, and tyrosine was also upregulated in BLCA smokers. The concentration of taurine, which is involved in the cysteine sulfinic acid pathway is higher in BLCA smokers with respect to never smokers (Figure 3).
Figure 3. Validation of a subset of metabolites using Biocrates absoluteIDQ p180kit.
Box plots showing significant upregulation of amino acids and taurine in BLCA smokers.
Identification of a smoker-specific gene signature by integration of metabolomics and transcriptomics
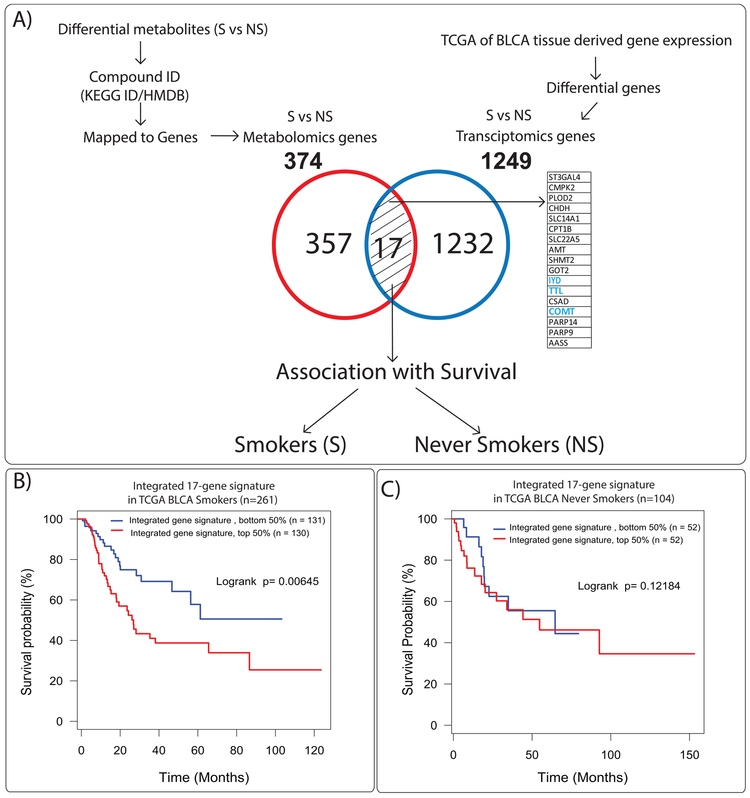
We next integrated our metabolomics data with publicly available transcriptomics data using the TCGA (19) and Riester (18) cohorts in which a majority of the patients are high-grade BLCA (greater than 90%). Interestingly, our serum samples used for metabolite analysis are also high-grade BLCA (greater than 95%, Table 1). The panel of altered metabolites between BLCA smokers and never smokers were mapped to their corresponding genes using KEGG/HMDBs that led to the identification of 374 associated genes. Using TCGA (19), which contains patient smoking information, we found 1249 genes were significantly altered between BLCA smokers and never smokers (Figure 4A). The union of BLCA smoker gene signatures from the metabolomics mapped gene set and TCGA-based gene set revealed 17 common genes (Figure 4A). We checked the expression of these 17 genes for clinical association in BLCA smokers and never smokers. In this analysis, the time is the standard event time. Time is zero when observation began and measured to the last observed time point, and the analysis is the overall survival of the patient (21, 22).
Figure 4. Serum based metabolomics and transcriptomics integration strategy.
(A) (i) Corresponding KEGG/HMDB ids of the 40 differential metabolites were mapped to genes. A total of 374 genes were obtained. (ii) These were further intersected with 1249 genes that were significantly changed between BLCA never smokers and smokers in the TCGA cohort resulting in a focused 17 gene signature. Kaplan-Meier survival plots of the 17 gene signature in BLCA cohorts (B) TCGA BLCA smokers (C) TCGA BLCA never smokers. BLCA smoking patients with a higher signature score (n = 130, Log-rank p = 0.00645) in TCGA.
For generation of this analysis, first, we analyzed the 17 genes in Riester cohort between BLCA smokers and never smokers. Second, we have applied the Riester gene fold change in TCGA (n=261 smokers and 104 never smokers) and Riester (n=75 smokers and 18 never smokers) cohorts. We found that high expression of this 17 gene signature is associated with poor survival in only BLCA smokers in TCGA (Figure 4B) cohort. A similar analysis was applied on Riester cohort by classifying the data to bottom/top 5% to 95%, and we have observed that 17 gene signature was significantly associated with poor survival for bottom 35% and top 65% analysis (Supplementary Figure 3B). Survival analysis in BLCA never smokers did not show any prognostic significance in either TCGA (Figure 4C) or Riester datasets (Supplementary Figure 3C).
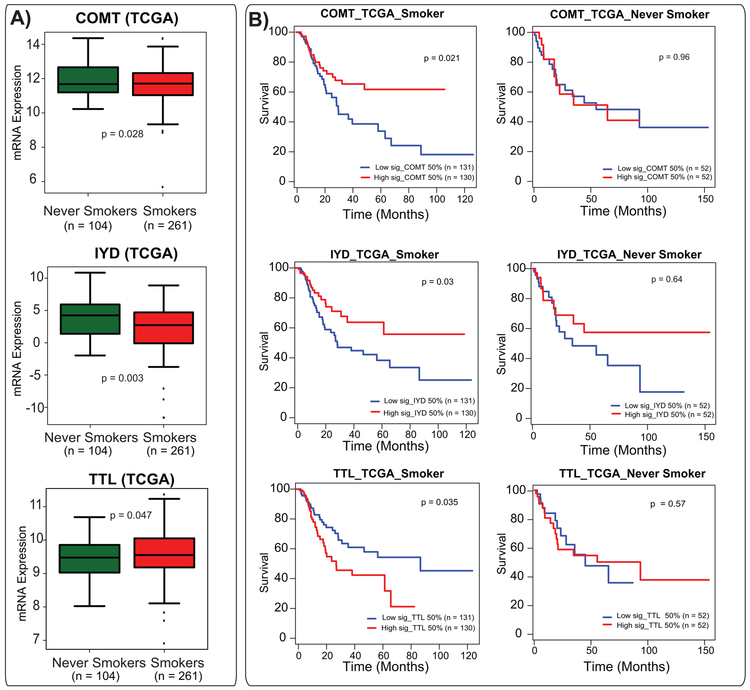
To obtain additional insights, we further investigated the expression of these 17 (Figure 4A) individual genes in BLCA smokers and never-smokers in the TCGA cohort. Here, 11 out of 17 genes (Supplementary Table 2) show a significant difference between BLCA smokers and never smokers in TCGA. Cytidine Uridine Monophosphate Kinase 2 (CMPK2), Poly(ADP-Ribose) Polymerase Family Member 9 (PARP9) and 14 (PARP14), Tubulin Tyrosine Ligase (TTL), Procollagen-Lysine, and 2-Oxoglutarate 5-Dioxygenase 2 (PLOD2) show higher expression levels in TCGA smokers, whereas Catechol-O-Methyltransferase (COMT), Iodotyrosine Deiodinase (IYD), Amino Methyltransferase (AMT), Aspartate Aminotransferase (GOT2), Solute Carrier Family 14 Member 1 (SLC14A1), CMP-N-Acetylneuraminate-beta-Galactosamide-Alpha-2,3-Sialyltransferase (ST3GAL4), Cysteine Sulfinic Acid Decarboxylase (CSAD) were downregulated (Figure 5, Supplementary Figure 4, Supplementary Table 2). To obtain additional intuitions into these 11 genes, we next performed individual gene survival analysis in the TCGA cohort. We found that two down-regulated genes (COMT and IYD) have a significant clinical association (Figure 5A and B). We next looked into the 4 upregulated genes (out of 11) in the same cohort and found that TTL had clinical associations with poor survival. Notably, COMT, IYD, and TTL showed a significant poor survival in TCGA. Kaplan-Meier survival analysis of COMT and IYD showed that low expression had significantly poor survival in TCGA BLCA smokers and did not show any significant difference in never smokers. BLCA smokers with high expression of TTL showed the worst prognosis compared to BLCA never smokers (Figure 5A and B).
Figure 5. mRNA alterations and clinical association of COMT, IYD, and TTL in publicly available cohort.
(A) Expression levels of COMT, IYD, and TTL genes in TCGA cohort. (B) Survival analysis of these three genes in TCGA BLCA smokers cohort and their corresponding survival analysis in TCGA BLCA never smokers cohort.
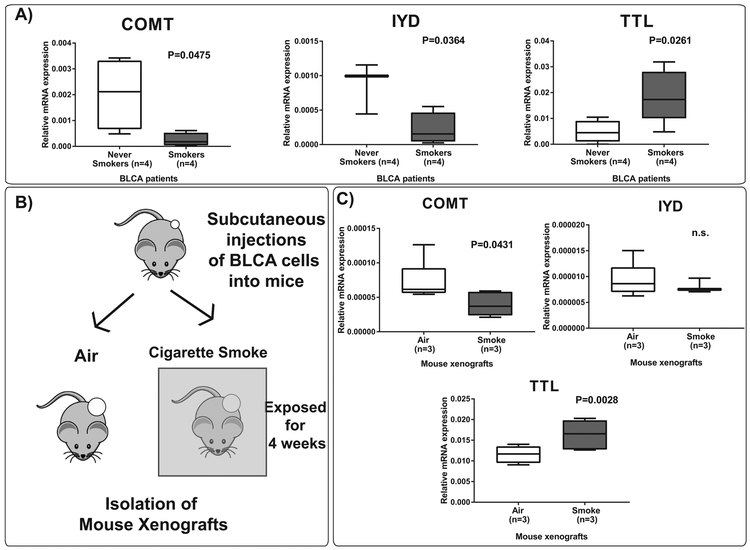
We further validated the mRNA expression levels of these three genes in RNA isolated from BLCA smokers and BLCA never smokers patient tissues by real-time PCR. We observed increased levels of TTL and downregulation of COMT and IYD in BLCA smokers compared to never smokers (Figure 6A). We developed a mouse xenograft model in which NOD-SCID beige mice were injected with UMUC3 BLCA cell line subcutaneously. Next, the mice were exposed to two cigarettes smoke a day for 4 weeks (Figure 6B). Tumors were isolated from these mice, and we analyzed mRNA for the above genes in these tumors. Consistent with the patient data, we observed upregulation of TTL and downregulation of COMT and IYD expression levels in the cigarette smoke exposed xenografts (Figure 6C).
Figure 6. mRNA expression levels of COMT, IYD, and TTL in BLCA patients and mouse xenografts.
(A) Expression levels of COMT, IYD, and TTL genes in BLCA never smokers and BLCA smokers. (B) Schematic diagram representing the mouse xenograft model. UMUC3 cells were subcutaneously injected into NOD-SCID mice. The mice4 were then exposed to cigarette smoke for a period of 4 weeks. (C) Expression levels of COMT, IYD, and TTL in mouse xenografts exposed to cigarette smoke compared to air.
Discussion
Tobacco smoke is a major risk factor for the development of BLCA (23). Further, BLCA patients who smoke while undergoing treatment have decreased recurrence-free survival outcomes compared to their nonsmoking counterparts (8). Previously, we and others have reported metabolic profiling of BLCA using different mass spectrometry approaches (12, 24–27) and identified dysregulated metabolic pathways and gene signatures associated with them. However, to the best of our knowledge, no serum based metabolic profiling was performed in BLCA smokers although it has been studied in other types of cancers and diseases (28–31). The current study focused on the identification of metabolic fingerprint that is unique to BLCA smokers when compared to never smokers. Our targeted mass spectrometry analysis resulted in detection of 190 metabolites of different classes: amino acids and its derivatives, fatty acids, organic compounds, metabolites involved in TCA/Glycolysis/PPP pathway and nucleic acid synthesis.
Forty differential metabolites between BLCA smokers and never smokers were identified. Most of these were amino acids suggesting the intricate dynamics of protein metabolism in the pathogenesis of BLCA smokers. One interesting observation is the upregulation of glycine, valine, isoleucine, and proline in the serum metabolic profile of BLCA smokers. These amino acids with leucine form the major constituents of elastin - a protein required for blood vessel formation (32, 33). Glycine amino acid makes nearly one-quarter of the elastin peptide sequence. This suggests that in BLCA smokers might have increased angiogenesis indicative of aggressive tumor formation.
Our data from serum metabolite profiling from BLCA smokers demonstrated increased serum asparagine levels. It has been shown that blocking the production of asparagine with a drug L-asparaginase and exposing the mice on a low aspargine diet greatly reduced the tumor metastasis (34). Thus increased asparagine levels may potentially be associated with tumor aggression in BLCA smokers. Another observation is an increase in the levels of taurine in BLCA smokers. Taurine has been shown to act as a possible urine biomarker for detection of non-muscle invasive BLCA (35), but it was not established in BLCA serum smokers (36).
The integration of metabolic signature to the TCGA cohort led to the enrichment of 17 gene signature that is specific to BLCA smokers. We found that high expression of this 17 gene signature is associated with poor survival in only BLCA smokers in both TCGA and Riester datasets. The gene signatures of the upregulated (TTL) or downregulated genes (COMT, IYD) showed a significant association with poor survival. COMT is a catalytic enzyme is involved in the methylation of various endobiotic and xenobiotic substances (37, 38). Cigarette smoking can lead to exposure of the body organs to a high number of xenobiotic substances, and this may lead to disruption of the methylation process of endogenous substrates due to a lack of labile methyl groups. This phenomenon could be one of the reasons why the levels of COMT decrease in BLCA smokers.
Another enzyme that is significantly downregulated in BLCA smokers is IYD. The major role of this enzyme is to remove iodide from iodinated tyrosine residues in the thyroid gland (39). The removed iodide is recycled for the synthesis of thyroid hormones (40), which are required for the maintenance of homeostasis by regulating metabolic rate, protein expression, and body temperature (41). One of the constituents of cigarette smoke is cyanide, which is converted to the anti-thyroid agent, thiocyanate. This agent decreases the absorption of iodine into the thyroid and lowers the production of thyroid hormones involved in maintaining body homeostasis (42). It could be inferred that smoking leads to a reduction of absorption of iodine perhaps due to reduced levels of IYD. A case study on one elderly patient suffering from primary BLCA reported thyroid metastases (43). The cytosolic enzyme TTL is involved in post-translational modification of alpha-tubulin. Alpha-tubulin in assembled microtubules is detyrosinated at C-terminus. During microtubule disassembly, TTL plays an important role in restoring the tyrosine residues, thus participating in a cycle of tubulin detyrosination and tyrosination (44). It has been previously shown that cigarette smoke increases alpha-tubulin disassembly (45), which could result in increasing levels of tyrosine residues and hence increase in TTL expression levels. Consistent with this observation, tyrosine is higher in BLCA smokers compare to BLCA never smokers. We have examined the expression of the genes TTL, IYD, and COMT independently in BLCA tissue samples and found that the expression levels concurs with observations from TCGA cohort. A validation on a large number of BLCA tissues and also stage-specific expression analysis of these genes would render significant analytical power for assessing the potential prognostic value of these genes.
In conclusion, we identified critical alterations of serum metabolites that could be a consequence of BLCA disease progression in smokers using advanced LC-MS-based metabolomics in combination with bioinformatics. The integrated study of these platforms of BLCA smokers over never smokers provided numerous novel insights into disease biology and delineated multiple potential opportunities for therapeutic intervention. Importantly, a set of these gene signatures especially COMT, IYD, and TTL in BLCA smokers may play an important role in BLCA tumor progression, and this could provide potential prognostic value and future targets for therapeutic intervention.
Supplementary Material
Acknowledgments
This research was fully supported by NIH/NCI R01CA220297 (N.P), and NIH/NCI R01CA216426 (N.P.) and American Cancer Society (ACS) Award 127430-RSG-15–105-01-CNE (N.P.), partially supported by the following grants: NIH/NCI U01 CA167234 (A.S.K), UO1 CA179674 (A.S.K), as well as funds from Alkek Center for Molecular Discovery (A.S.K.). This project was also supported by the Agilent Technologies Center of Excellence (COE) in Mass Spectrometry at Baylor College of Medicine, Metabolomics Core, Prostate Cancer Foundation, Dianna Helis Adrianne Melvin Helis Foundation, Brockman Foundation, Population Sciences Biorepository core at Baylor College of Medicine with funding from the NIH (P30 CA125123), CPRIT Proteomics and Metabolomics Core Facility (N.P., A.S.K,), (RP170005), and Dan L. Duncan Cancer Center. We want to thank Maharajni Perla, Akhil Mandalapu for their input on the analysis.
Footnotes
Conflict of interest statement: Authors do not have any conflict of interest
References:
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2.Latini DM, Lerner SP, Wade SW, Lee DW, Quale DZ. Bladder cancer detection, treatment and outcomes: opportunities and challenges. Urology. 2010;75:334–9. [DOI] [PubMed] [Google Scholar]
- 3.Brown ML, Riley GF, Schussler N, Etzioni R. Estimating health care costs related to cancer treatment from SEER-Medicare data. Med Care. 2002;40:104–17. [DOI] [PubMed] [Google Scholar]
- 4.Avritscher EB, Cooksley CD, Grossman HB, Sabichi AL, Hamblin L, Dinney CP, et al. Clinical model of lifetime cost of treating bladder cancer and associated complications. Urology. 2006;68:549–53. [DOI] [PubMed] [Google Scholar]
- 5.Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. JAMA. 2011;306:737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease C, Prevention. Vital signs: current cigarette smoking among adults aged >/=18 years--United States, 2005–2010. MMWR Morb Mortal Wkly Rep. 2011;60:1207–12. [PubMed] [Google Scholar]
- 7.Ehdaie B, Furberg H, Zabor EC, Ostroff JS, Shariat SF, Bochner BH, et al. Impact of smoking status at diagnosis on disease recurrence and death in upper tract urothelial carcinoma. BJU Int. 2013;111:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen RJ, Ho YS, Guo HR, Wang YJ. Long-term nicotine exposure-induced chemoresistance is mediated by activation of Stat3 and downregulation of ERK1/2 via nAChR and beta-adrenoceptors in human bladder cancer cells. Toxicol Sci. 2010;115:118–30. [DOI] [PubMed] [Google Scholar]
- 9.Sturgeon SR, Hartge P, Silverman DT, Kantor AF, Linehan WM, Lynch C, et al. Associations between bladder cancer risk factors and tumor stage and grade at diagnosis. Epidemiology. 1994;5:218–25. [DOI] [PubMed] [Google Scholar]
- 10.Lukas C, Selinski S, Prager HM, Blaszkewicz M, Hengstler JG, Golka K. Occupational bladder cancer: Polymorphisms of xenobiotic metabolizing enzymes, exposures, and prognosis. J Toxicol Environ Health A. 2017;80:439–52. [DOI] [PubMed] [Google Scholar]
- 11.Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res. 2011;71:7376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piyarathna DWB, Rajendiran TM, Putluri V, Vantaku V, Soni T, von Rundstedt FC, et al. Distinct Lipidomic Landscapes Associated with Clinical Stages of Urothelial Cancer of the Bladder. Eur Urol Focus. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aaronson DS, Walsh TJ, Smith JF, Davies BJ, Hsieh MH, Konety BR. Meta-analysis: does lidocaine gel before flexible cystoscopy provide pain relief? BJU Int. 2009;104:506–9. [DOI] [PubMed] [Google Scholar]
- 14.van der Aa MN, Steyerberg EW, Bangma C, van Rhijn BW, Zwarthoff EC, van der Kwast TH. Cystoscopy revisited as the gold standard for detecting bladder cancer recurrence: diagnostic review bias in the randomized, prospective CEFUB trial. J Urol. 2010;183:76–80. [DOI] [PubMed] [Google Scholar]
- 15.Bujak R, Struck-Lewicka W, Markuszewski MJ, Kaliszan R. Metabolomics for laboratory diagnostics. J Pharm Biomed Anal. 2015;113:108–20. [DOI] [PubMed] [Google Scholar]
- 16.Bansal N, Gupta A, Mitash N, Shakya PS, Mandhani A, Mahdi AA, et al. Low- and high-grade bladder cancer determination via human serum-based metabolomics approach. J Proteome Res. 2013;12:5839–50. [DOI] [PubMed] [Google Scholar]
- 17.Cao M, Zhao L, Chen H, Xue W, Lin D. NMR-based metabolomic analysis of human bladder cancer. Anal Sci. 2012;28:451–6. [DOI] [PubMed] [Google Scholar]
- 18.Riester M, Taylor JM, Feifer A, Koppie T, Rosenberg JE, Downey RJ, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care. 2004;8:389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeegers MP, Tan FE, Dorant E, van Den Brandt PA. The impact of characteristics of cigarette smoking on urinary tract cancer risk: a meta-analysis of epidemiologic studies. Cancer. 2000;89:630–9. [DOI] [PubMed] [Google Scholar]
- 24.Sahu D, Lotan Y, Wittmann B, Neri B, Hansel DE. Metabolomics analysis reveals distinct profiles of nonmuscle-invasive and muscle-invasive bladder cancer. Cancer Med. 2017;6:2106–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues D, Jeronimo C, Henrique R, Belo L, de Lourdes Bastos M, de Pinho PG, et al. Biomarkers in bladder cancer: A metabolomic approach using in vitro and ex vivo model systems. Int J Cancer. 2016;139:256–68. [DOI] [PubMed] [Google Scholar]
- 26.Shao CH, Chen CL, Lin JY, Chen CJ, Fu SH, Chen YT, et al. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget. 2017;8:38802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Rundstedt FC, Rajapakshe K, Ma J, Arnold JM, Gohlke J, Putluri V, et al. Integrative Pathway Analysis of Metabolic Signature in Bladder Cancer: A Linkage to The Cancer Genome Atlas Project and Prediction of Survival. J Urol. 2016;195:1911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song H, Peng JS, Dong-Sheng Y, Yang ZL, Liu HL, Zeng YK, et al. Serum metabolic profiling of human gastric cancer based on gas chromatography/mass spectrometry. Braz J Med Biol Res. 2012;45:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan B, Huang J, Zhang C, Hu X, Gao M, Shi A, et al. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod Rheumatol. 2016;26:914–22. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Li W, Zou Q, Yin L, Du Y, Gu J, et al. Serum metabolomic profiling of human gastric cancer and its relationship with the prognosis. Oncotarget. 2017;8:110000–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walejko JM, Kim S, Goel R, Handberg EM, Richards EM, Pepine CJ, et al. Gut microbiota and serum metabolite differences in African Americans and White Americans with high blood pressure. Int J Cardiol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrhovski B, Weiss AS. Biochemistry of tropoelastin. Eur J Biochem. 1998;258:1–18. [DOI] [PubMed] [Google Scholar]
- 33.Kielty CM, Sherratt MJ, Shuttleworth CA. Elastic fibres. J Cell Sci. 2002;115:2817–28. [DOI] [PubMed] [Google Scholar]
- 34.Knott SRV, Wagenblast E, Khan S, Kim SY, Soto M, Wagner M, et al. Erratum: Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;556:135. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava S, Roy R, Singh S, Kumar P, Dalela D, Sankhwar SN, et al. Taurine - a possible fingerprint biomarker in non-muscle invasive bladder cancer: A pilot study by 1H NMR spectroscopy. Cancer Biomark. 2010;6:11–20. [DOI] [PubMed] [Google Scholar]
- 36.Grotenhuis AJ, Ebben CW, Aben KK, Witjes JA, Vrieling A, Vermeulen SH, et al. The effect of smoking and timing of smoking cessation on clinical outcome in non-muscle-invasive bladder cancer. Urol Oncol. 2015;33:65 e9–17. [DOI] [PubMed] [Google Scholar]
- 37.Zhou SS, Zhou YM, Li D, Lun YZ. Dietary methyl-consuming compounds and metabolic syndrome. Hypertens Res. 2011;34:1239–45. [DOI] [PubMed] [Google Scholar]
- 38.Fontana L, Delort L, Joumard L, Rabiau N, Bosviel R, Satih S, et al. Genetic polymorphisms in CYP1A1, CYP1B1, COMT, GSTP1 and NAT2 genes and association with bladder cancer risk in a French cohort. Anticancer Res. 2009;29:1631–5. [PubMed] [Google Scholar]
- 39.Moreno JC, Visser TJ. Genetics and phenomics of hypothyroidism and goiter due to iodotyrosine deiodinase (DEHAL1) gene mutations. Mol Cell Endocrinol. 2010;322:91–8. [DOI] [PubMed] [Google Scholar]
- 40.Querido A, Stanbury JB, Kassenaar AA, Meijer JW. The metabolism of iodotyrosines. III. Di-iodotyrosine deshalogenating activity of human thyroid tissue. J Clin Endocrinol Metab. 1956;16:1096–101. [DOI] [PubMed] [Google Scholar]
- 41.Murphy DB, Wallis KT, Machlin PS, Ratrie H 3rd, Cleveland DW. The sequence and expression of the divergent beta-tubulin in chicken erythrocytes. J Biol Chem. 1987;262:14305–12. [PubMed] [Google Scholar]
- 42.Braverman LE, He X, Pino S, Cross M, Magnani B, Lamm SH, et al. The effect of perchlorate, thiocyanate, and nitrate on thyroid function in workers exposed to perchlorate long-term. J Clin Endocrinol Metab. 2005;90:700–6. [DOI] [PubMed] [Google Scholar]
- 43.Mirjalili SM, Hashemipour S, Salehi S, Kazemifar AM, Madani PS. Thyroid metastasis of bladder transitional cell carcinoma. Malays J Pathol. 2016;38:65–70. [PubMed] [Google Scholar]
- 44.Erck C, MacLeod RA, Wehland J. Cloning and genomic organization of the TTL gene on mouse chromosome 2 and human chromosome 2q13. Cytogenet Genome Res. 2003;101:47–53. [DOI] [PubMed] [Google Scholar]
- 45.Borgas D, Chambers E, Newton J, Ko J, Rivera S, Rounds S, et al. Cigarette Smoke Disrupted Lung Endothelial Barrier Integrity and Increased Susceptibility to Acute Lung Injury via Histone Deacetylase 6. Am J Respir Cell Mol Biol. 2016;54:683–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.