Abstract
Background
Osimertinib (AZD9291), a third‐generation EGFR‐tyrosine kinase inhibitor, can effectively prolong survival in non‐small cell lung cancer (NSCLC) patients with EGFR mutations, particularly T790M mutations; however, acquired resistance to AZD9291 is inevitable, thus exploration of the targets of resistance is urgent.
Methods
Considering the important role of circular RNAs (circRNAs) in cancers, we established AZD9291‐resistant NSCLC cell lines (H1975/AZDR and HCC827/AZDR) and used microarray analysis to determine the circRNA expression profiles of the cells. The H1975/AZDR and HCC827/AZDR cell lines were induced by gradually increasing the drug concentration. CircRNA microarray expression profiles were obtained from H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells and validated by quantitative reverse transcription PCR. Expression data were analyzed bioinformatically.
Results
The H1975/AZDR and HCC827/AZDR cell lines were successfully established. The half‐maximal inhibitory concentration and the invasion ability of H1975/AZDR and HCC827/AZDR cells were significantly enhanced. The proliferation rates of H1975/AZDR and HCC827/AZDR were much lower than H1975 and HCC827. Microarray analysis identified 15 504 circRNAs differentially expressed in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells. Among them, 7966 were upregulated and 7538 were downregulated more than two‐fold. We predicted the possible miRNAs of the top dysregulated circRNAs. Furthermore, Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the most modulated circRNAs regulate several cancers and cancer‐related pathways.
Conclusion
Our results reveal that circRNAs may play a role in NSCLC AZD9291 resistance and might be a promising molecular target candidate for gene therapy.
Keywords: AZD9291, circular RNA, microarray, NSCLC, resistance
Introduction
Lung cancer is the leading cause of cancer‐related death worldwide. Non‐small cell lung cancer (NSCLC) is one of the most common types of lung cancer, accounting for approximately 80% of all cases. At present, the main treatments for NSCLC are surgical excision, chemotherapy, radiotherapy, targeted molecular therapy, and biological immunotherapy. Among these, EGFR‐targeted therapy for NSCLC is gaining in prominence and has become a research hotspot worldwide.1, 2 Osimertinib (AZD9291) is a third‐generation EGFR‐tyrosine kinase inhibitor (TKI) for the treatment of advanced NSCLC and has an obvious effect on EGFR mutation (exon 18, 19, 21) and EGFR T790M mutation, which is often present in NSCLC patients with acquired EGFR‐TKI resistance.3, 4 AZD9291 has effectively prolonged the survival of advanced NSCLC patients, but acquired resistance to AZD9291 is inevitable. Recently, the EGFR C797S mutation has been detected in AZD9291‐resistant NSCLC patients, at an incidence of 17–20%.5, 6 Although research of fourth‐generation EGFR‐TKIs for the treatment of EGFR C797S mutation has been conducted, the complex mechanisms of AZD9291 resistance have not been determined, thus the problem of third‐generation EGFR‐TKI (AZD9291) resistance remains. Therefore it is crucial to identify more molecular targets for gene therapy of patients with AZD9291‐resistance.
Circular RNAs (CircRNAs) are a novel class of endogenous noncoding RNAs (ncRNAs) that are mainly formed by RNA splicing of the 5′ end of upstream exon and the 3′ end of downstream exon.7 Originally, circRNAs were regarded as byproducts of splicing mistakes or gene rearrangements, but accumulating studies have proved that circRNAs may be crucial players in a variety of human tumors.8, 9, 10 circRNAs are currently recognized as one of the most promising new stars in the RNA family. Experts have confirmed that circRNAs can interact with microRNAs (miRNAs) and function as sponges to stop miRNAs from regulating gene expression through a circRNA‐miRNA‐messenger RNA (mRNA) pathway.11, 12 CircRNAs are more suitable as tumor biomarkers and provide new ideas for targeted molecular therapy because they are stable, tissue specific, and are competing endogenous RNAs (ceRNAs). CircRNAs such as circ_ITCH,13 circ_0013958,14 and circ_10087615 have been discovered and proven to be closely related to occurrence, development, tumor node metastasis (TNM) staging, pathological grade, and lymph node metastasis in lung cancer. Recent studies of circRNAs in the mechanisms of cancer drug resistance have been limited; related reports were only discovered in gemcitabine‐resistant pancreatic ductal adenocarcinoma and chemotherapeutic drug‐resistant lung, breast, and colon cancer cells.16, 17, 18, 19 To date, no studies have investigated the association between circRNA profiles in NSCLC with acquired resistance to third‐generation EGFR‐TKIs.
Considering the important role of circRNAs in cancers, we hypothesized that changes in circRNA levels might be associated with AZD9291 resistance in NSCLC. We aimed to analyze the drug resistance mechanism in NSCLC, screen the circRNAs related to drug resistance, and provide an experimental model for overcoming cancer drug resistance in order to find new molecular targets for the therapy of AZD9291‐resistant NSCLC patients and provide a solid foundation and effective tool for targeted drug therapy of lung cancer.
Methods
Cell culture and main reagents
H1975 and HCC827 lung adenocarcinoma cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). These cells were cultured in RPMI‐1640 medium supplemented with 10% fetal bovine serum (FBS), 100 μg/mL penicillin, and 100 μg/mL streptomycin. The cultures were maintained in an incubator with 5% CO2 at 37°C under standard culture conditions.
Establishment of AZD9291‐resistant H1975 and HCC827 cell lines
We first established AZD9291‐resistant H1975 and HCC827 cell lines (H1975/AZDR and HCC827/AZDR) by gradually increasing concentrations of AZD9291 for 72 hours with a recovery period between treatments. AZD9291 was dissolved in dimethyl sulfoxide before being added into the complete cell culture medium. A total of 1 × 106 cells/ml of H1975 or HCC827 cells were seeded in a six‐well plate and incubated at 37°C in a 5% CO2 incubator in RPMI‐1640 medium containing AZD9291. The initial concentrations of AZD9291 were started with a concentration equal to the half‐maximal inhibitory concentration (IC50) of H1975 and HCC827 cells: 93 nM for H1975 and 21 nM for HCC827. After a cycle of AZD9291 treatment, only a small percentage of cells remained. Once cells had resumed normal growth and returned to 80% confluence under the light microscope, the next cycle began. The drug concentration was gradually increased for the next cycle until the H1975 and HCC827 cells could survive and proliferate in a cell culture system containing 10 μM AZD9291. After six months, the H1975/AZDR and HCC827/AZDR cell lines were finally successfully established. The newly established H1975/AZDR and HCC827/AZDR cell lines were maintained in a medium containing 10μM AZD9291, while the parental H1975 and HCC827 cell lines were cultured in drug‐free medium.
Calculation of curve‐fitting half‐maximal inhibitory concentration
H1975, HCC827, H1975/AZDR, and HCC827/AZDR cell lines were seeded at 1 × 104 cells/well in 96‐well plates, and different concentrations of AZD9291 were added. The concentration gradients of AZD9291 were 0, 0.001, 0.01, 0.1, 1, and 10 uM, respectively; five replicate wells were used for each concentration. Cells were cultured for 72 hours in a humidified incubator containing 5% CO2 at 37°C. The absorbance of each well was measured at 490 nm by methyl thiazolyl tetrazolium (MTT) assay and the survival rate (%) was calculated according to the following formula:
Survival rate (%) = mean optical density (OD) of the experimental group/mean OD of the control group × 100.
The drug concentration‐survival curve was drawn with concentration of AZD9291 as the abscissa and survival rate as the vertical axis. IC50 was calculated using SPSS version 17.0, and the drug resistance index (RI) was calculated according to the following formula:
Methyl thiazolyl tetrazolium cell proliferation assay
H1975, HCC827, H1975/AZDR, and HCC827/AZDR cell lines were seeded at 1 × 104 cells/well in 96‐well plates. Five replicate wells were used for each analysis. After the cells were incubated for 1, 2, 3, 4, and 5 days, respectively, the viability of the cells was measured by MTT assay following the kit protocol (CT01; Millipore, Billerica, MA, USA). The growth curve was drawn with time as the abscissa and absorbance value as the vertical axis.
Cell invasion assay
For the invasion assays, we used an 8 μm pore polycarbonate membrane Boyden chamber insert in a Transwell apparatus (Millipore) to measure cell motility. The H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells were treated with trypsin/ethylene‐diamine‐tetraacetic acid solution separately and washed once with serum‐containing RPMI‐1640 medium. A total of 1 × 105 cells in 0.2 ml serum‐free RPMI‐1640 medium were seeded on a Transwell apparatus. Each insert was precoated with 45 μg Matrigel (BD Biosciences, San Jose, CA, USA). The chambers were then incubated for 24 hours at 37°C in a 5% CO2 incubator in culture medium with 10% FBS in the bottom chambers. The cells on the upper surface were scraped and washed away, whereas the invaded cells on the lower surface were fixed in 90% precooling methanol for 10 minutes, stained with 0.1% crystal violet for 10 minutes, and then rinsed in phosphate buffered saline and subjected to microscopic inspection. Finally, the invasion values were obtained by counting three fields per membrane. Experiments were independently repeated in triplicate.
RNA Extraction
Total RNA was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA quality and concentration were determined from OD260/280 readings using the NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Montchanin, DE, USA) and assessed via 1% gel electrophoresis, respectively.
Microarray data analysis
The total RNA sample was digested with Rnase R (Epicentre Inc., Madison, WI, USA) to eliminate linear RNAs and enrich circular RNAs. Subsequently, each purified RNA sample was amplified and transcribed into complementary RNA (cRNA) by applying a random priming method (Arraystar Super RNA Labeling Kit; Arraystar Inc., Rockville, MD, USA). The labeled cRNAs were then hybridized onto the circRNAs and incubated for 17 hours at 65°C in an Agilent Hybridization Oven (Agilent Technologies, Santa Clara, CA, USA). Finally, the arrays were washed, fixed, and scanned to images using the Agilent Scanner G2505C. Data analysis was performed using Agilent Feature Extraction software version 11.0. Expression data were quantile normalized and processed using the R software limma package. The significant differentially expressed circRNAs between H1975, HCC827, H1975/AZDR, and HCC827/AZDR cell lines were screened through fold change, P value, and raw intensity. The required microarray information has been imputed into the Gene Expression Omnibus (serial number: GSE48885).
Annotation and function prediction of circRNA and bioinformatics analysis
Interactions between circRNAs/miRNAs were predicted using Arraystar's miRNA target prediction software based on TargetScan and miRanda. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted to evaluate the circRNA attributes in pathways.
Quantitative real‐time PCR (qRT‐PCR)
Quantitative real‐time (qRT) PCR was used to validate the microarray data. The total extracted RNAs of the HCC827/AZDR cells were reverse transcribed using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer's instructions. The complementary DNA template was amplified by qRT‐PCR using SYBR Premix Ex Taq (TaKaRa) according to the manufacturer's instructions. Divergent primers of 5 circRNAs (3 upregulated, 2 downregulated), were screened to design primers. Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as an internal control. The melting curve was drawn to ensure the specificity of primers. QRT‐PCR reactions and data collection were conducted using the StepOne Plus Real‐Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The data was normalized to the GAPDH expression level and presented as the average from three experiments. The relative expression was calculated using the 2−ΔΔCt method, where Ct represents the threshold cycle.
Statistical analysis
SPSS version 17.0 was used for data analysis and GraphPad Prism 5.0 for plotting. Wilcoxon matched pairs tests were used to compare the circRNA expression levels in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cell lines. P < 0.05 was considered statistically significant.
Results
Acquisition of AZD9291‐resistant H1975 and HCC827 cell lines
The AZD9291‐resistant cell line (H1975/AZDR) was established from the parental H1975 cell line by gradually increasing concentrations of AZD9291 from 93 nM to 10 uM for six months. The HCC827/AZDR cell line was established from the parental HCC827 cell line by gradually increasing the concentrations of AZD9291 from 21 nM to 10uM for six months. The IC50 values of AZD9291 for H1975 and H1975/AZDR were 93 nM and 3.416 uM, respectively (Fig 1a). The IC50 values of AZD9291 for HCC827 and HCC827/AZDR were 21 nM and 1.567 uM, respectively (Fig 1b). The resistance indices of H1975/AZDR and HCC827/AZDR cells were 36.73 and 74.62, respectively. The ability of cell proliferation from days 1 to 5 was determined by MTT assay. The proliferation rates of H1975 and H1975/AZDR cells increased sharply until day 4, while the proliferation rate of H1975/AZDR was much lower than H1975 cells (Fig 1c). The growth rates of HCC827 and HCC827/AZDR cells were accelerated from days 3 to 5, and the growth rate of HCC827/AZDR was much lower than HCC827 cells (Fig 1d).
Figure 1.
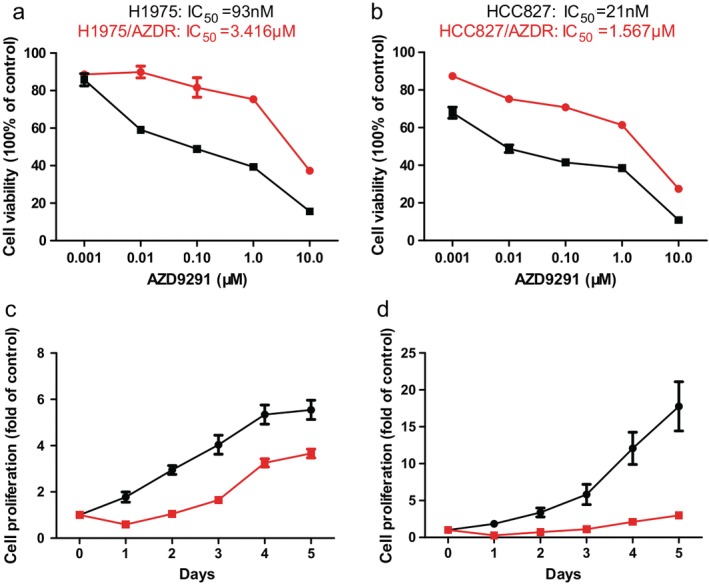
Acquisition of H1975 and HCC827 cells resistant to AZD9291. The drug sensitivity of (a) H1975 ( ) and H1975/AZDR (
) and H1975/AZDR ( ) and (b) HCC827 (
) and (b) HCC827 ( ) and HCC827/AZDR (
) and HCC827/AZDR ( ) cells was detected by methyl thiazolyl tetrazolium (MTT) assay. The viability and proliferation of (c) H1975 (
) cells was detected by methyl thiazolyl tetrazolium (MTT) assay. The viability and proliferation of (c) H1975 ( ) and H1975/AZDR (
) and H1975/AZDR ( ) and (d) HCC827 (
) and (d) HCC827 ( ) and HCC827/AZDR (
) and HCC827/AZDR ( ) cells was detected by MTT assay. IC50, half maximal inhibitory concentration.
) cells was detected by MTT assay. IC50, half maximal inhibitory concentration.
Morphologically, we found that H1975/AZDR and HCC827/AZDR exhibited a mesenchymal‐like phenotype, including spindle‐shaped and elongated appearance, loss of cell–cell junctions, and polarity, which was remarkably different from the parental H1975 and HCC827 cells (Fig 2). These morphological features of H1975/AZDR and HCC827/AZDR indicated promoted cell migration and invasion. We further analyzed the invasion ability of H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells by Transwell cell invasion assay. A significantly higher number of H1975/AZDR and HCC827/AZDR cells traversed the cell‐permeable membrane than H1975 and HCC827 cells (Fig 3). The invasion ability of H1975/AZDR and HCC827/AZDR cells was significantly enhanced. These morphological and functional changes may suggest that drug‐resistant cell lines were successfully established.
Figure 2.
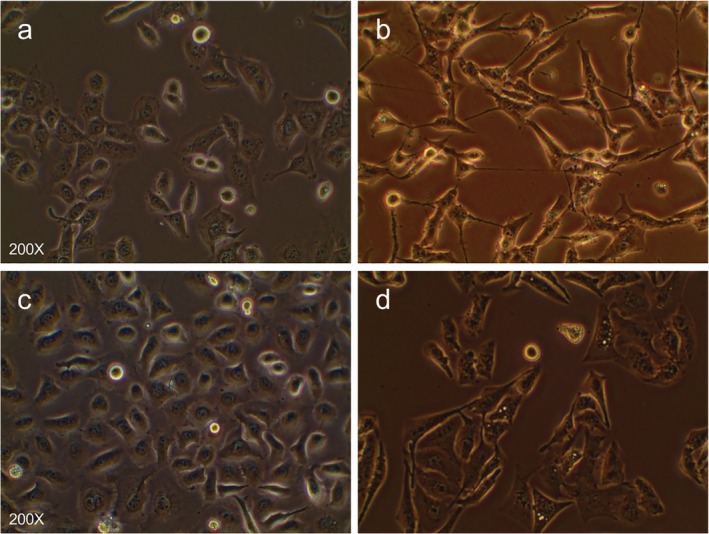
The morphological differences between AZD9291‐resistant non‐small cell lung cancer (NSCLC) and parental NSCLC cell lines were observed under a light microscope. (a) H1975, (b) H1975/AZDR, (c) HCC827, and (d) HCC827/AZDR cells.
Figure 3.
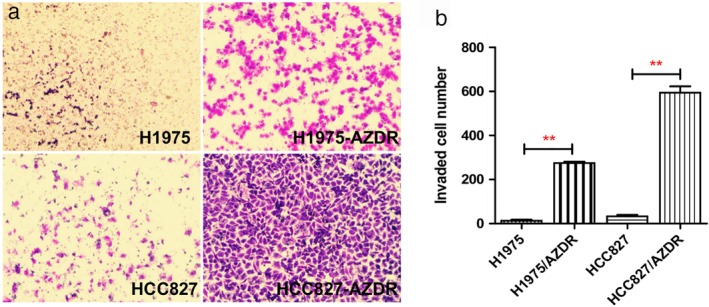
Transwell invasion assay was used to examine the invasion ability of non‐small cell lung cancer (NSCLC) cells. (a) A higher number of H1975/AZDR and HCC827/AZDR cells invaded than H1975 and HCC827 cells. (b) The invasion ability of H1975/AZDR and HCC827/AZDR cells was significantly enhanced. (*P < 0.05 and **P < 0.01 compared to H1975 and HCC827 cells).
Overview of circRNA expression profiles in non‐small cell lung cancer sensitive and resistant cells
In total, 15 504 circRNAs were differentially expressed between the H1975, HCC827, H1975/AZDR, and HCC827/AZDR cell lines (fold change [FC] ≥ 2.0; P < 0.05), including 7966 upregulated and 7538 downregulated circRNAs. A box plot shows the distribution of circRNA intensities in the six samples (Fig 4a). As shown in Figure 4a, the distribution of normalized intensities was almost the same in all of the tested samples. The differentially expressed circRNAs between groups were displayed in scatter plots (Fig 4b). A Volcano plot exhibited statistically significant differences in circRNA expression (Fig. 4c). A clustered heat map showed upregulation or downregulation of circRNAs (Fig 4d). The top 10 upregulated and downregulated circRNAs sorted by FC values are summarized in Table 1. It should be noted that the top five circRNAs were upregulated more than 400‐fold, and three circRNAs were downregulated more than 100‐fold. These circRNAs were located in a variety of genomic locations, including sex chromosomes. Subgroup analysis of the genomic distribution showed that chr7 is the primary region for host genes of differentially expressed circRNAs, representing 8.7% of host genes, followed by 8.26% for chr2, and 7.77% for chr1 (Fig 5).
Figure 4.
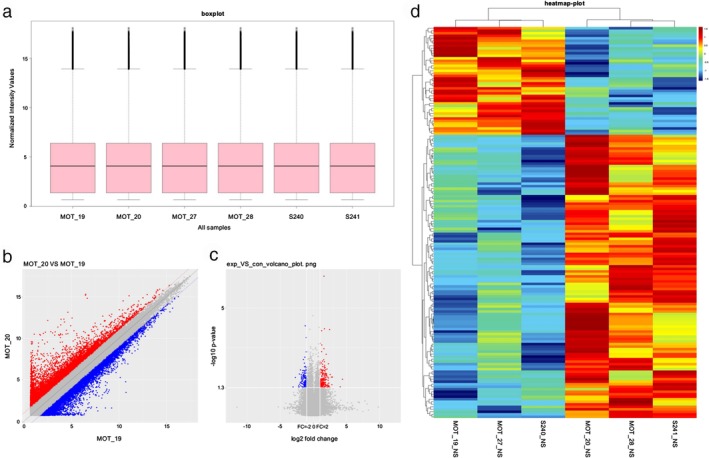
Different expression profiles of circular RNAs (circRNAs) in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells. (a) A box plot was used to assess the distribution of circRNA intensity in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells. (b) A scatter plot revealed microarray data distribution of circRNAs between H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells. The red points show that the circRNAs were upregulated more than two‐fold. The blue points show that the circRNAs were downregulated more than two‐fold. (c) A Volcano plot exhibits significantly dysregulated circRNAs in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells. The horizontal lines represent two‐fold up and down‐expressed circRNAs, while the vertical lines indicate P = 0.05. Red and blue squares mark differentially expressed circRNAs in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells (P < 0.05). (d) Unsupervised hierarchical clustering analysis of the significant differentially expressed circRNAs between H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells. Each column represents the expression profile of a sample; each row corresponds to a circRNA. The color scale varies from red to blue: red refers upregulated circRNAs, and blue to downregulated circRNAs.
Table 1.
The top 10 upregulated and downregulated circRNAs in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cells screened by fold change and P value
| circRNA | P | Regulation | FC (abs) | Chrom | Strand | GeneSymbol |
|---|---|---|---|---|---|---|
| hsa_circ_0043632 | 0.000119 | up | 798.058 | chr17 | − | KRT17 |
| hsa_circ_0048856 | 0.043567 | up | 682.925 | chr19 | − | C3 |
| hsa_circ_0043634 | 0.016589 | up | 449.643 | chr17 | − | KRT17 |
| hsa_circ_0002130 | 0.002189 | up | 446.606 | chr19 | − | C3 |
| hsa_circ_0019088 | 0.009256 | up | 444.628 | chr10 | − | ANKRD1 |
| hsa_circ_0050581 | 0.039654 | down | 200.009 | chr19 | + | FXYD3 |
| hsa_circ_0023302 | 0.031115 | down | 154.768 | chr11 | + | MYEOV |
| hsa_circ_0069997 | 0.032701 | down | 127.056 | chr4 | + | AREG |
| hsa_circ_0050580 | 0.024477 | down | 92.882 | chr19 | + | FXYD3 |
| hsa_circ_0069996 | 0.047489 | down | 64.549 | chr4 | + | AREG |
circRNA, circular RNA; FC, fold change.
Figure 5.
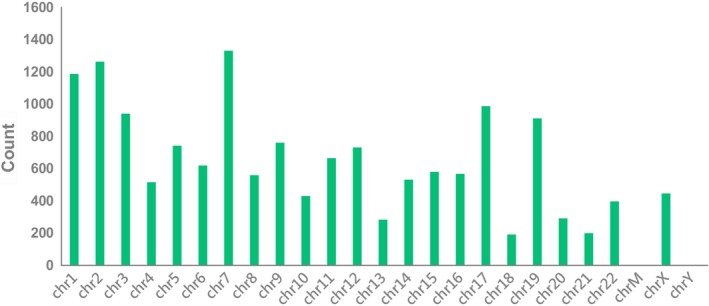
Genomic distribution of differently expressed circular RNAs (circRNAs) in H1975, HCC827, H1975/AZDR, and HCC827/AZDR cell lines.
Validation of the differentially expressed circRNAs by qRT‐PCR
The expression variations of circRNAs was compared between qRT‐PCR and microarray analysis results. Three upregulated and two downregulated differentially expressed circRNAs were validated by qRT‐PCR in HCC827 and HCC827/AZDR cells. The qRT‐PCR results of the five tested circRNAs were consistent with the microarray data (Fig 6). This result suggests that the circRNAs identified by microarray were reliable and worthy of further investigation.
Figure 6.
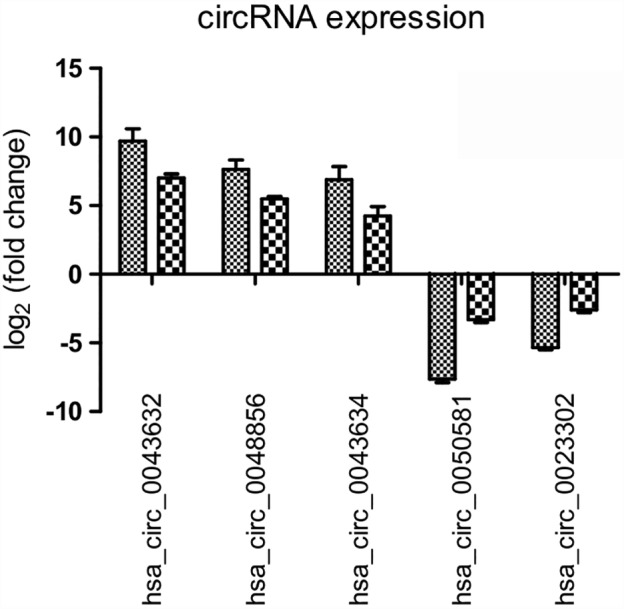
Validation of microarray data by quantitative reverse transcriptase (RT)‐PCR. Five differentially expressed circular RNAs (circRNAs) were validated by qRT‐PCR. The heights of the columns in the chart represent the mean expression value of log2‐fold changes.  , Array;
, Array;  , PCR.
, PCR.
Prediction of circRNA‐miRNA‐mRNA associations
Because the functions of circRNAs remain poorly understood and circRNAs can function as miRNA sponges or inhibitors, we predicted the circRNA‐miRNA‐mRNA axis in cancer‐related pathways. We analyzed the microRNA response elements associated with the top dysregulated circRNAs to identify any circRNA‐miRNA connections predicted from TargetScan and miRanda. The targeted miRNAs were ranked according to their mirSVR scores, and the five miRNAs with the highest mirSVR score for each circRNA were identified for further analysis. The targeted miRNAs of three upregulated and two downregulated differentially expressed circRNAs are listed in Table 2. From Table 2 we can see that hsa_circ_0043632 and hsa_circ_0043634 can bind to the same miR‐6861‐3p, suggesting that different circRNAs may synergistically regulate the activity of specific miRNAs and exert biological roles by indirectly regulating miRNA target genes.
Table 2.
miRNA binding sites predict upregulated circRNAs
| circRNA | miRNA binding sites | ||||
|---|---|---|---|---|---|
| MRE1 | MRE2 | MRE3 | MRE4 | MRE5 | |
| hsa_circ_0043632 | hsa‐miR‐6861‐3p†2 | hsa‐miR‐492†2 | hsa‐miR‐4743‐5p†2 | hsa‐miR‐6829‐3p†2 | hsa‐miR‐6778‐3p†2 |
| hsa_circ_0048856 | hsa‐miR‐4279†2 | hsa‐miR‐221‐5p†2 | hsa‐miR‐8073†2 | hsa‐miR‐1256†2 | hsa‐miR‐4691‐5p†2 |
| hsa_circ_0043634 | hsa‐miR‐4483†3 | hsa‐miR‐1293†3 | hsa‐miR‐6861‐3p†2 | hsa‐miR‐4530†2 | hsa‐miR‐492†2 |
| hsa_circ_0050581 | hsa‐miR‐6722‐5p†1 | hsa‐miR‐4641†1 | hsa‐miR‐4707‐3p†1 | hsa‐miR‐4258†1 | hsa‐miR‐652‐3p†1 |
| hsa_circ_0023302 | hsa‐miR‐765†7 | hsa‐miR‐6804‐3p†6 | hsa‐miR‐8085†5 | hsa‐miR‐6731‐5p†5 | hsa‐miR‐6878‐5p†5 |
Represents the number of microRNA (miRNA) binding sites. circRNA, circular RNA.
Gene ontology and pathway analyses
GO and KEGG pathway analyses were performed for circRNA host genes to evaluate the roles of the differentially expressed circRNAs. Predicted GO terms and pathways with P values < 0.05 were chosen and ranked according to enrichment scores. For the host genes of circRNAs (Fig 7), the most significantly enriched biological process terms included voluntary musculoskeletal movement, transfer RNA catabolic process, and gene silencing by miRNA. The most significantly enriched cellular component terms were related to DNA replication, RNA induced silencing complex loading, micro‐ribonucleoprotein complex, and RNA modification. The most significantly enriched molecular function terms were linked to DNA replication origin binding, 3′‐5’ DNA helicase activity, and DNA topoisomerase activity. The top 30 predicted pathways are shown in Figure 8. Among these pathways, some are directly linked to cancer pathogenesis, such as the non‐small cell lung cancer, glioma, prostate cancer, and renal cell carcinoma signaling pathways. The p53 signaling, mTOR signaling, ErbB signaling and DNA replication pathways; focal adhesion; cell cycle; and adherens junction play an important role in NSCLC drug resistance.
Figure 7.
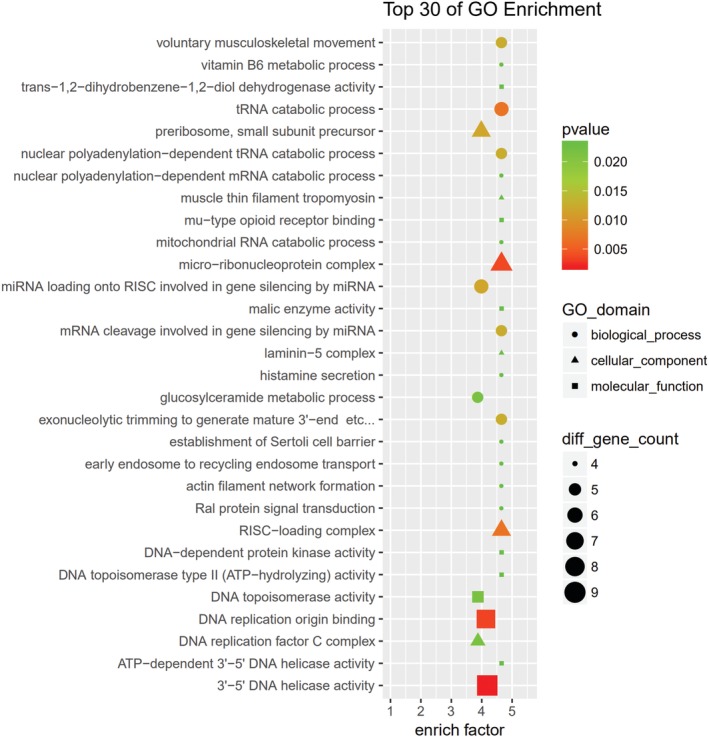
The top 30 Gene Ontology (GO) terms from biological process, cellular component, and molecular function. The size of each circle indicates the number of circRNAs in biological process terms. The size of each triangle indicates the number of circRNAs in cellular component terms. The size of each square indicates the number of circRNAs in molecular function terms. The color of the circle, triangle, and square indicates the P value.
Figure 8.
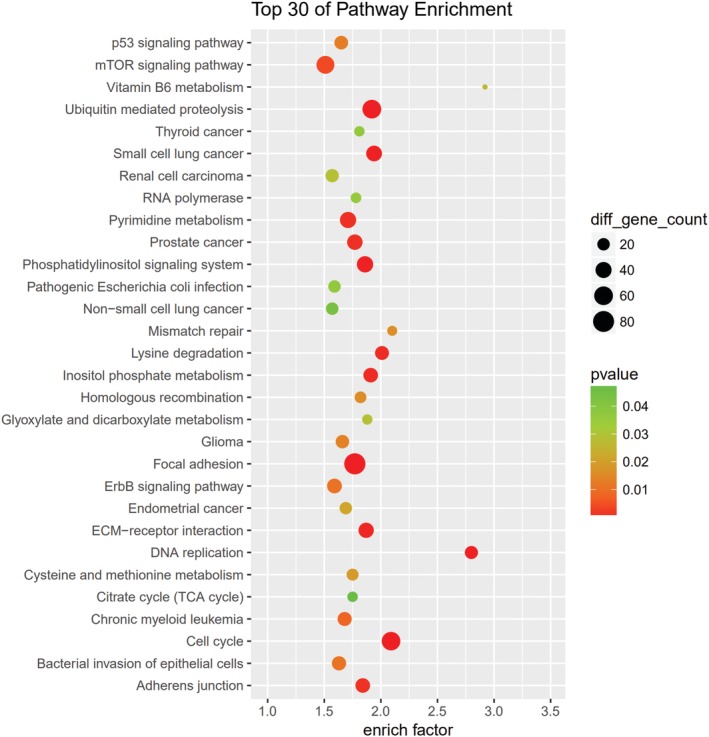
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted to determine the involvement of target genes in different biological pathways. The size of each circle indicates the number of circular RNAs (circRNAs). The color of the circle indicates the P value. The larger the circle and the lower the P value, the more enriched and meaningful the pathway.
Discussion
Tumor drug resistance has always been a difficult problem in the clinical treatment of tumors. A large number of studies have attempted to search for drug resistance‐related genes through the expression profiles of drug‐resistant cell lines. One of the most common methods for establishing drug‐resistant cell line models is to gradually increase the drug concentration16, 18, 19, 20, 21 or prolong the acting time of a constant concentration of drug,22 so that drug‐resistant cell lines with reduced drug sensitivity (e.g. IC50) are obtained. Mechanisms of drug resistance and drug resistance‐related genes can be studied by comparing genes that are differentially expressed between resistant and parental cell lines. However, there is currently no uniform standard to determine whether the establishment of a drug‐resistant cell line model is successful.
In a previous study, the paclitaxel‐resistant A549 cell line (A549/Taxol) was generated by treating A549 cells with gradually increasing concentrations of paclitaxel for six months.18 A549/Taxol cells appeared to be spindle‐shaped and elongated, and the IC50 value of A549/Taxol was 72.87 times higher than that of the A549.18 The AZD9291‐resistant cell line PC9/AZDR was established by culturing in gradually increasing concentrations of AZD9291 (from 10 nM to 1 μM) for approximately six months, and the PC9/AZDR cells were also resistant to other EGFR‐TKIs, such as CO1686, afatinib, gefitinib, and erlotinib.20 The AZD9291‐resistant cell line HCC827/AZDR was generated by selection under increasing gradients of AZD9291 (from 0.02 to 0.16 μM) for six months.21 The IC50 value of HCC827/AZDR cells was 315.8 times higher than that of HCC827 cells. Morphologically, HCC827/AZDR exhibited a mesenchymal‐like phenotype including spindle‐like appearance, loss of cell–cell junctions, and polarity.21
In the present study, the AZD9291‐resistant H1975 and HCC827 cell lines (H1975/AZDR and HCC827/AZDR) were induced by gradually increasing the concentrations of AZD9291 from 93 nM to 10 uM for six months. Our H1975/AZDR and HCC827/AZDR cells also revealed a mesenchymal‐like phenotype, such as spindle shape and loss of cell–cell adhesion. The IC50 and the invasion ability of H1975/AZDR and HCC827/AZDR cells were significantly enhanced. The proliferation rates of H1975/AZDR and HCC827/AZDR were much lower than H1975 and HCC827, which may be a result of differential expression of some cell cycle regulatory molecules between parental and resistant lines. Further research will be performed to investigate the relevant cell cycle regulatory mechanisms.
CircRNAs were recently identified as novel functional ncRNAs involved in several cancers and can function as potential tumor markers.23, 24 CircRNAs, such as circ_ITCH,13 circ_0013958,14 circ_100876,15 and circ_CCDC6625 have been proven to be closely related to occurrence, development, TNM staging, pathological grade, lymph node metastasis, and drug resistance in lung cancer. Circ_ITCH was significantly decreased in lung cancer tissues and inhibited the proliferation of lung cancer cells by downregulating oncogenic miR‐7 and miR‐214 and enhancing its parental gene, ITCH.13 Upregulation of hsa_circ_0013958 could promote proliferation and invasion and inhibit apoptosis of lung adenocarcinoma through the hsa_circ_0013958/miR‐134/cyclin D1 pathway, and therefore can function as a potential noninvasive biomarker for the early detection and screening of lung adenocarcinoma.14 CircRNA_100876 is significantly elevated in NSCLC and has a close relationship with lymph node metastasis, tumor staging, and overall survival, and may serve as a potential prognostic biomarker and therapeutic target for NSCLC.15 CircRNA CCDC66 is highly expressed in EGFR‐resistant H1975 cells and is regulated by HGF/c‐Met to increase epithelial–mesenchymal transition (EMT) and the drug resistance of lung adenocarcinoma cells.25 Recent studies of circRNA profiles in the mechanism of cancer drug resistance have been limited; related reports were only discovered in gemcitabine‐resistant pancreatic ductal adenocarcinoma and chemotherapeutic drug‐resistant lung, breast, and colon cancer cells.16, 17, 18, 19 To date, no studies have investigated circRNA profiles in NSCLC with acquired resistance to third‐generation EGFR‐TKIs.
In the present study, a circRNA microarray was first used to acquire circRNA expression profiles in AZD9291‐resistant NSCLC cells. In total, 15 504 circRNAs were significantly dysregulated circRNAs (FC ≥ 2.0; P < 0.05), including 7966 upregulated and 7538 downregulated circRNAs. The upregulated circRNAs, hsa_circ_0043632 and hsa_circ_0043634, are spliced from KRT17, which is upregulated in highly metastatic cancer,26 and has been used as pathological diagnostic marker of breast cancer27 and skin squamous carcinoma.28 ANKRD1 overexpression, which splices hsa_circ_0019088, is associated with EMT features, anti‐apoptosis, and resistance to second and third‐generation EGFR‐TKIs in NSCLC.29 FXYD3, which splices hsa_circ_0050581 and hsa_circ_0050580, is overexpressed in several common cancers and may be a marker of resistance to cancer treatments.30 Silencing of FXYD3 in breast cancer cells amplifies the effects of doxorubicin and γ‐radiation on cell survival.30 AREG splices hsa_circ_0069997 and hsa_circ_0069996 and mediates the invasion, migration, stemness, and drug resistance of malignant tumors.31, 32 Hsa_circ_0023302 is spliced from MYEOV, which promotes the proliferation, invasion, and migration of colorectal cancer.33 However, controversy remains when applying the research results of drug‐resistant cell models to guide clinical application.34, 35 The differential expression in cancer cells may primarily reflect the response to drug treatment, and is not necessarily related to anticancer drug resistance.34 Therefore, the clinical relevance of drug‐resistant gene markers screened from drug‐resistant cell models needs to be further verified in clinical tissue samples.
Regarding the mechanism of circRNAs, circRNAs can interact with miRNAs and function as sponges to arrest miRNA in regulating gene expression through a circRNA‐miRNA‐mRNA pathway. Our results showed that the top upregulated hsa_circ_0043632 may act as sponge of miR‐6861‐3p, miR‐492, miR‐4743‐5p, miR‐6829‐3p, and miR‐6778‐3p. Among them, miR‐492 is reported to be involved in the cell proliferation, cell cycle, migration, overall survival, and chemotherapy resistance of several tumors, such as cervical squamous cell and hepatocellular carcinomas, and breast and colon cancers.36, 37, 38, 39 TIMP2, as a direct miR‐492 target,36 is critical for modulating cancer cell migration and invasion.40, 41 Further research needs to be carried out to verify whether hsa_circ_0043632 regulates the proliferation, migration, invasion, and AZD9291‐resistance of NSCLC through the hsa_circ_0043632/miR‐492/TIMP2 axis.
In the present study, we investigated the functions of dysregulated circRNAs through KEGG analyses. Among the pathways associated with the dysregulated circRNAs, the P53, mTOR, and focal adhesion signaling pathways have previously been demonstrated as pivotal pathways in cancer cells’ response to the AZD9291‐resistance of NSCLC,42, 43, 44, 45 indicating that dysregulated circRNAs may be of great significance in cellular responses to drug resistance. Huang et al. revealed that the loss of p53 is consistently associated with acquired resistance to EGFR inhibitors, and restoration of p53 was sufficient to reverse EGFR inhibitor resistance.42 The mTOR pathway is widely involved in the regulation of cell proliferation, growth, apoptosis and other cell functions, and mTOR activation is often associated with tumor resistance, occurrence, and development in a variety of tumors.46 Research has shown that inhibition of the mTOR pathway could regulate tumor drug resistance through autophagy induction.47 Whether dysregulated circRNAs can regulate the proliferation, apoptosis, migration, invasion, autophagy, and AZD9291 resistance of NSCLC needs to be further investigated.
To the best of our knowledge, this is the first study to report the aberrant expression of circRNAs in drug resistance of NSCLC to AZD9291. Our results provide promising potential molecular targets for the therapy of NSCLC patients and provide a solid foundation and effective tool for molecular targeted drug therapy of NSCLC. Further research is required to determine the effects of dysregulated circRNAs on AZD9291‐resistant NSCLC.
Disclosure
No authors report any conflict of interest.
Acknowledgments
This article was supported by the National Natural Science Foundation of China (81601995, 81301996), the Shanghai Science and Technology Committee (18411966100), the Zhejiang Provincial Natural Science Foundation of China (LY16H160022), Nurture Projects for Basic Research of Shanghai Chest Hospital (2018YNJCM04), the Wu Jieping Medical Foundation (320.6750.17525), the Open Fund of Zhejiang Provincial Top Key Discipline of Pharmacology (YKFJ2‐001), and the Department of Health of Zhejiang Provincial Government (2013KYA070).
References
- 1. Wang B, Zuo Z, Li F, Yang K, Du M, Gao Y. Gefitinib versus docetaxel in treated non‐small‐cell lung cancer: A meta‐analysis. Open Med 2017; 12: 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bollinger MK, Agnew AS, Mascara GP. Osimertinib: A third‐generation tyrosine kinase inhibitor for treatment of epidermal growth factor receptor‐mutated non‐small cell lung cancer with the acquired Thr790Met mutation. Oncol Pharm Pract 2018; 24 (5): 379–88. [DOI] [PubMed] [Google Scholar]
- 3. Yu HA, Arcila ME, Rekhtman N et al. Analysis of tumor specimens at the time of acquired resistance to EGFR‐TKI therapy in 155 patients with EGFR‐mutant lung cancers. Clin Cancer Res 2013; 19 (8): 2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Janne PA, Yang JC, Kim DW et al. AZD9291 in EGFR inhibitor‐resistant non‐small‐cell lung cancer. N Engl J Med 2015; 372 (18): 1689–99. [DOI] [PubMed] [Google Scholar]
- 5. Rood JJM, van Bussel MTJ, Schellens JHM, Beijnen JH, Sparidans RW. Liquid chromatography‐tandem mass spectrometric assay for the T790M mutant EGFR inhibitor osimertinib (AZD9291) in human plasma. J Chromatogr B, Analyt Technol Biomed Life Sci 2016; 1031: 80–5. [DOI] [PubMed] [Google Scholar]
- 6. Cross DA, Ashton SE, Ghiorghiu S et al. AZD9291, an irreversible EGFR TKI, overcomes T790M‐mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014; 4 (9): 1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashwal‐Fluss R, Meyer M, Pamudurti NR et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 2014; 56 (1): 55–66. [DOI] [PubMed] [Google Scholar]
- 8. Li P, Chen H, Chen S et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer 2017; 116 (5): 626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo JN, Li J, Zhu CL et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther 2016; 9: 7451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xia W, Qiu M, Chen R et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep 2016; 6: 35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hansen TB, Jensen TI, Clausen BH et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495 (7441): 384–8. [DOI] [PubMed] [Google Scholar]
- 12. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015; 12 (4): 381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA‐ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta‐catenin pathway. Biomed Res Int 2016; 2016: 1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu X, Wang X, Wei S et al. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J 2017; 284 (14): 2170–82. [DOI] [PubMed] [Google Scholar]
- 15. Yao JT, Zhao SH, Liu QP et al. Over‐expression of CircRNA_100876 in non‐small cell lung cancer and its prognostic value. Pathol Res Pract 2017; 213 (5): 453–6. [DOI] [PubMed] [Google Scholar]
- 16. Gao D, Zhang X, Liu B et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 2017; 9 (9): 1175–88. [DOI] [PubMed] [Google Scholar]
- 17. Xiong W, Ai YQ, Li YF et al. Microarray analysis of circular RNA expression profile associated with 5‐fluorouracil‐based chemoradiation resistance in colorectal cancer cells. Biomed Res Int 2017; 2017: 8421614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu N, Chen S, Liu Y et al. bioinformatics analysis of differentially expressed circRNAs in taxol‐resistant non‐small cell lung cancer cells. Cell Physiol Biochem 2018; 48 (5): 2046–60. [DOI] [PubMed] [Google Scholar]
- 19. Shao F, Huang M, Meng F, Huang Q. Circular RNA Signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front Pharmacol 2018; 9: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ku BM, Choi MK, Sun JM et al. Acquired resistance to AZD9291 as an upfront treatment is dependent on ERK signaling in a preclinical model. PLoS One 2018; 13 (4): e0194730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu J, Zhao X, He D et al. Loss of EGFR confers acquired resistance to AZD9291 in an EGFR‐mutant non‐small cell lung cancer cell line with an epithelial‐mesenchymal transition phenotype. J Cancer Res Clin Oncol 2018; 144 (8): 1413–22. [DOI] [PubMed] [Google Scholar]
- 22. Bauer JA, Ye F, Marshall CB et al. RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells. Breast Cancer Res 2010; 12 (3): R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meng S, Zhou H, Feng Z et al. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017; 16 (1): 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018; 25 (1): 1–7. [DOI] [PubMed] [Google Scholar]
- 25. Joseph NA, Chiou SH, Lung Z et al. The role of HGF‐MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial‐to‐mesenchymal transition of lung adenocarcinoma cells. J Hematol Oncol 2018; 11 (1): 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ide M, Kato T, Ogata K, Mochiki E, Kuwano H, Oyama T. Keratin 17 expression correlates with tumor progression and poor prognosis in gastric adenocarcinoma. Ann Surg Oncol 2012; 19 (11): 3506–14. [DOI] [PubMed] [Google Scholar]
- 27. Sorlie T, Perou CM, Tibshirani R et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98 (19): 10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moll R, Divo M, Langbein L. The human keratins: Biology and pathology. Histochem Cell Biol 2008; 129 (6): 705–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takahashi A, Seike M, Chiba M et al. Ankyrin repeat domain 1 overexpression is associated with common resistance to afatinib and osimertinib in EGFR‐mutant lung cancer. Sci Rep 2018; 8 (1): 14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu CC, Teh R, Mozar CA, Baxter RC, Rasmussen HH. Silencing overexpression of FXYD3 protein in breast cancer cells amplifies effects of doxorubicin and gamma‐radiation on Na(+)/K(+)‐ATPase and cell survival. Breast Cancer Res Treat 2016; 155 (2): 203–13. [DOI] [PubMed] [Google Scholar]
- 31. Shimada H, Abe S, Kohno T et al. Loss of tricellular tight junction protein LSR promotes cell invasion and migration via upregulation of TEAD1/AREG in human endometrial cancer. Sci Rep 2017; 7: 37049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tung SL, Huang WC, Hsu FC et al. miRNA‐34c‐5p inhibits amphiregulin‐induced ovarian cancer stemness and drug resistance via downregulation of the AREG‐EGFR‐ERK pathway. Oncogene 2017; 6 (5): e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lawlor G, Doran PP, MacMathuna P, Murray DW. MYEOV (myeloma overexpressed gene) drives colon cancer cell migration and is regulated by PGE2. J Exp Clin Cancer Res 2010; 29: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gillet JP, Calcagno AM, Varma S et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti‐cancer drug resistance. Proc Natl Acad Sci U S A 2011; 108 (46): 18708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma SV, Haber DA, Settleman J. Cell line‐based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer 2010; 10 (4): 241–53. [DOI] [PubMed] [Google Scholar]
- 36. Liu M, An J, Huang M et al. MicroRNA‐492 overexpression involves in cell proliferation, migration, and radiotherapy response of cervical squamous cell carcinomas. Mol Carcinog 2018; 57 (1): 32–43. [DOI] [PubMed] [Google Scholar]
- 37. Yu G, Xiao Q, Ma XP et al. miR‐492G>C polymorphism (rs2289030) is associated with overall survival of hepatocellular carcinoma patients. Tumour Biol 2016; 37 (7): 8961–72. [DOI] [PubMed] [Google Scholar]
- 38. Shen F, Cai WS, Feng Z et al. MiR‐492 contributes to cell proliferation and cell cycle of human breast cancer cells by suppressing SOX7 expression. Tumour Biol 2015; 36 (3): 1913–21. [DOI] [PubMed] [Google Scholar]
- 39. Peng L, Zhu H, Wang J et al. MiR‐492 is functionally involved in oxaliplatin resistance in colon cancer cells LS174T via its regulating the expression of CD147. Mol Cell Biochem 2015; 405 (1–2): 73–9. [DOI] [PubMed] [Google Scholar]
- 40. Guan H, Li W, Li Y et al. MicroRNA‐93 promotes proliferation and metastasis of gastric cancer via targeting TIMP2. PLoS One 2017; 12 (12): e0189490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yi X, Guo J, Guo J et al. EZH2‐mediated epigenetic silencing of TIMP2 promotes ovarian cancer migration and invasion. Sci Rep 2017; 7 (1): 3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang S, Benavente S, Armstrong EA, Li C, Wheeler DL, Harari PM. p53 modulates acquired resistance to EGFR inhibitors and radiation. Cancer Res 2011; 71 (22): 7071–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zheng R, Jiang H, Li J, Liu X, Xu H. Polyphyllin II restores sensitization of the resistance of PC‐9/ZD cells to gefitinib by a negative regulation of the PI3K/Akt/mTOR signaling pathway. Curr Cancer Drug Targets 2017; 17 (4): 376–85. [DOI] [PubMed] [Google Scholar]
- 44. Sun Z, Li Q, Zhang S et al. NVP‐BEZ235 overcomes gefitinib‐acquired resistance by down‐regulating PI3K/AKT/mTOR phosphorylation. OncoTargets Ther 2015; 8: 269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murakami Y, Sonoda K, Abe H et al. The activation of SRC family kinases and focal adhesion kinase with the loss of the amplified, mutated EGFR gene contributes to the resistance to afatinib, erlotinib and osimertinib in human lung cancer cells. Oncotarget 2017; 8 (41): 70736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: From basic research to clinical application. Am J Cancer Res 2015; 5 (5): 1602–9. [PMC free article] [PubMed] [Google Scholar]
- 47. Chen S, Rehman SK, Zhang W, Wen A, Yao L, Zhang J. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta 2010; 1806 (2): 220–9. [DOI] [PubMed] [Google Scholar]


