Abstract
Oxazolidinones are synthetic antibiotics used for treatment of infections caused by Gram-positive bacteria. They target the bacterial protein synthesis machinery by binding to the peptidyl transferase centre (PTC) of the ribosome and interfering with the peptidyl transferase reaction. Cadazolid is the first member of quinoxolidinone antibiotics, which are characterized by combining the pharmacophores of oxazolidinones and fluoroquinolones, and it is evaluated for treatment of Clostridium difficile gastrointestinal infections that frequently occur in hospitalized patients. In vitro protein synthesis inhibition by cadazolid was shown in Escherichia coli and Staphylococcus aureus, including an isolate resistant against linezolid, the prototypical oxazolidinone antibiotic. To better understand the mechanism of inhibition, we determined a 3.0 Å cryo-electron microscopy structure of cadazolid bound to the E. coli ribosome in complex with mRNA and initiator tRNA. Here we show that cadazolid binds with its oxazolidinone moiety in a binding pocket in close vicinity of the PTC as observed previously for linezolid, and that it extends its unique fluoroquinolone moiety towards the A-site of the PTC. In this position, the drug inhibits protein synthesis by interfering with the binding of tRNA to the A-site, suggesting that its chemical features also can enable the inhibition of linezolid-resistant strains.
Introduction
In all living cells, ribosomes are responsible for translating the genetic information from mRNAs to proteins. Due to this central role in protein synthesis ribosomes are frequently targeted by antibiotics that specifically inhibit bacterial translation by binding to the prokaryotic 70S but not eukaryotic 80S ribosomes1,2. Many of the clinically used antibiotic classes (chloramphenicols, macrolides, lincosamides, streptogramins and oxazolidinones) inhibit protein synthesis by binding to the peptidyl transferase centre (PTC), the catalytic centre of the ribosome, located in the large ribosomal subunit (50S subunit in prokaryotes), and to the nascent polypeptide exit tunnel of the 50S subunit3. The mechanism of how these antibiotics inhibit protein synthesis by interfering with the peptidyl transferase reaction or preventing the diffusion of the nascent polypeptide chain through the ribosomal tunnel are quite well understood based on biochemical, genetic and structural studies3–6.
Among the antibiotics that interfere with the peptidyl transferase reaction, oxazolidinones have been in the focus of antibacterial discovery research for many years, and two drugs have been approved for the treatment of infections due to susceptible Gram-positive pathogens7,8. Linezolid (LZD) was the first clinically used antibiotic of this class, but a wide variety of oxazolidinone analogues were designed in recent years to improve efficiency against resistant strains9,10.
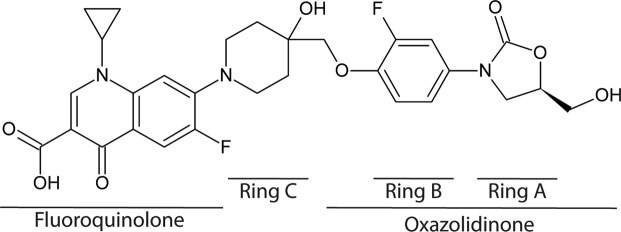
Cadazolid (CDZ, Fig. 1) belongs to a new class of antibiotics termed quinoxolidinones and is in development for the treatment of Clostridium difficile infections (CDI). CDI is a major health care issue due to its morbidity and mortality, mainly for elderly hospitalized patients11, and due to the occurrence of hypervirulent drug-resistant strains11,12. The molecule combines elements of an oxazolidinone with a fluoroquinolone moiety, resulting in unique biological and physicochemical properties. CDZ showed potent in vitro activity against C. difficile13 and has an antibacterial spectrum largely limited to Gram-positive bacteria, while activity against Gram-negative bacteria is weak or not detectable14, consistent with the observed limited impact on bacteria of the normal gut microflora in an in vitro human gut model15. In phase 2 clinical trials in CDI, CDZ demonstrated clinical cure rates similar to vancomycin with lower recurrence rates16. Two phase 3 clinical trials have been completed recently (NCT01983683, NCT01987895)17. The propensity of spontaneous resistance development in vitro is low, and CDZ retained activity against quinolone-resistant as well as linezolid-resistant strains and did not select for strains with significantly increased minimum inhibitory concentrations (MIC) for fluoroquinolones or LZD, indicating an absence of cross resistance14,18. CDZ is therefore a particularly interesting new drug in the fight against multidrug-resistant pathogens.
Figure 1.
Chemical structure of cadazolid (CDZ) with the names of the different moieties indicated below.
In this study, we determined the cryo-electron microscopy (cryo-EM) structure of an E. coli 70S ribosome in complex with mRNA, a P-site tRNA and CDZ, resolved at 3.0 Å. We observe that CDZ binds to the same binding pocket of the 50S subunit as LZD in the vicinity of the PTC4,19. The structure reveals CDZ-specific differences in binding and explains the potency of CDZ against linezolid-resistant strains.
Results
Inhibition of protein synthesis
In previous studies, we established that CDZ leads to a reduced de novo protein synthesis and that translation can be inhibited in vitro in C. difficile13. To expand the understanding about the protein synthesis inhibition, cell-free coupled transcription/translation assays were performed with extracts prepared from E. coli and two S. aureus strains.
CDZ potently inhibited in vitro translation in cell-free extracts from E. coli and from S. aureus linezolid-susceptible and -resistant strains (IC50 0.24–0.33 µM; Table 1, Supplementary Fig. S1). In contrast, LZD was significantly less potent (IC50 2.6 µM) and its activity was further reduced when using an extract from the S. aureus linezolid-resistant strain (IC50 7.03 µM). In S. aureus, IC50 values were mirrored by the minimum inhibitory concentrations (MICs), which suggests that inhibition of protein synthesis is the primary mode of action in these strains. Furthermore, in contrast to LZD, the activity of CDZ is hardly affected by the 23S rRNA mutation G2576U or the methylation by Cfr (Supplementary Table S1 and Fig. S2). Not surprisingly, in E. coli the LZD and CDZ potency against a wt-strain is very low or is not measurable, however inhibitory activity can be detected in a tolC mutant that lacks the outer membrane drug efflux protein TolC (Table 1). Therefore, to characterize the binding of CDZ to the bacterial ribosome, compare its binding mode to the one of LZD and to determine the role of the quinolone moiety, we elucidated the structure of the E. coli 70S ribosome in complex with CDZ.
Table 1.
In vitro transcription/translation assays for cadazolid and linezolid against E. coli wt ATCC 25922; S. aureus wt ATCC 29213; S. aureus S1 (LZDR). The minimal inhibitory concentrations are also shown.
| Cadazolid | Linezolid | ||
|---|---|---|---|
| E. coli ATCC 25922 | Average (range)a | 0.24 (0.13–0.34) | 4.03 (1.92–5.39) |
| MICb | 8/< = 0.063c | >32/8d | |
| S. aureus ATCC 29213 | Average (range) | 0.32 (0.31–0.35) | 2.61 (1.12–4.15) |
| MIC | 0.5 | 2 | |
| S. aureus S1 LZDR d | Average (range) | 0.33 (0.25–0.44) | 7.03 (6.16–9.16) |
| MICc | 1 | 32 | |
aIC50 in µM (average value and range of three independent experiments).
bMIC in µg/ml (median value of at least 3 determinations).
cMeasured against E. coli MG1655 tolC::kan.
dHomozygous G2576U in 23S rRNA.
Cryo-EM structure of CDZ in complex with the bacterial ribosome
E. coli 70S ribosomes incubated with CDZ (Fig. 1), mRNA and Phe-tRNA(Phe) and tRNA(fMet)i were analysed by single particle cryo-EM. During the analysis, two dominant subpopulations were found in the sample corresponding to the 70S in complex with either two (P- and E-site) or three (A-, P-, and E-site) tRNAs (Supplementary Fig. S3). Both particle subsets were refined independently. The subset containing two tRNAs in P and E-sites of the ribosome revealed the presence of the quinoxolidinone antibiotic and was refined to higher resolution by focusing refinement on the 50S subunit, resulting in reconstructions reaching overall resolutions of 3.0 Å (Fig. 2, Supplementary Table S2 and Figs S3 and S4). The maps were interpreted by docking and refining an E. coli 50S subunit extracted from a crystal structure of the 70S ribosome (PDB: 4YBB20).
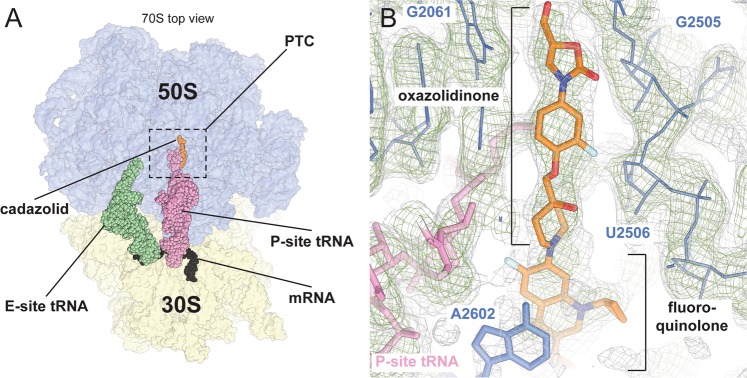
Figure 2.
Overview of the E. coli 70S with bound tRNAs, mRNA and CDZ. (a) For orientation, the docked models of the 30S and 50S subunits are shown in yellow and blue, respectively. The mRNA and the P- and E-site tRNAs are shown as spheres in black, magenta and green. (b) Details of cadazolid and its surrounding area in the experimental 3.0 Å cryo-EM map. Two contour levels are shown in green and grey mesh.
In the focused reconstruction we could unambiguously identify the density of the CCA-acyl end of a charged P-site tRNA and CDZ bound in the vicinity of the PTC (Fig. 2A,B and Supplementary Fig S4A). To verify the identity of the P-site tRNA in the decoding centre of the 30S subunit, we additionally performed a refinement focused on the small subunit, which resulted in a map resolved to 3.2 Å. Inspection of the anticodon stem loop and the codon-anticodon bases confirmed the identity of the P-site tRNA as a tRNA(fMet)i at an AUG start codon (Supplementary Fig. S5). We observed density for the amino acid attached to the 3′ end of the tRNA bound to the P-site. Since it is likely that only a fraction of tRNA(fMet)i purified from E. coli (tRNA Probes, LLC) was aminoacylated, the presence of the amino acid attached to the tRNA suggests that charged tRNAs preferentially bound to the 70S ribosomes (Supplementary Fig. S4A).
Binding of cadazolid to the PTC
CDZ is composed of a system with five rings of which the A- and B-rings are found in all oxazolidinones, while the C-ring links this pharmacophore to the fluoroquinolone moiety (Fig. 1). CDZ is bound deeply in a pocket within the PTC. The quality of the density allowed unambiguous modelling of this part of the compound as well as the PTC area in the vicinity. Less-well defined density for the fluoroquinolone moiety of CDZ was also observed at lower contour levels, indicating intrinsic flexibility. The fluoroquinolone moiety is oriented towards the empty A-site tRNA binding pocket of the PTC, where it could be reasonably modelled in two alternative conformations: one with the cyclopropyl group pointing towards the P-site tRNA and one pointing towards residue A2507 of the rRNA (Fig. 2B and Supplementary Fig S4B).
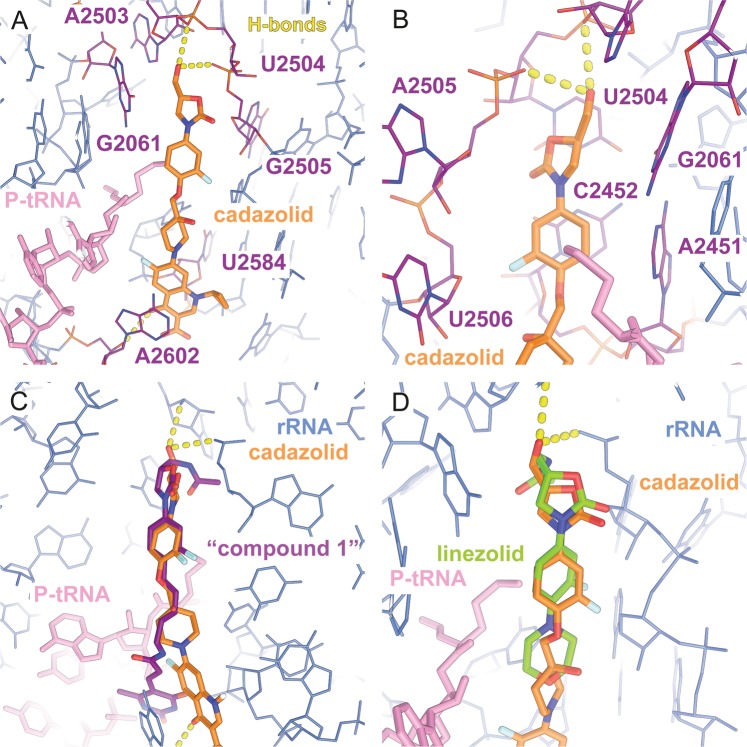
Overall, the oxazolidinone moiety of CDZ binds to the same pocket and in a similar orientation as other oxazolidinone antibiotics, with the A- and B-rings forming the main interface next to the polypeptide exit tunnel at the PTC (Fig. 3A–D)4,19,21,22. The fluorophenyl B-ring is stacking onto residue C2452 while being sandwiched between residues U2506 and A2451 (Fig. 3B). The position of the B-ring is similar to the one observed in a structure of a Haloarcula marismortui (Hm) 50S-LZD complex19 and the position predicted based on crosslinking studies and molecular modelling simulations using LZD with an E. coli model23. The 2-oxazolidone A-ring is bound to the PTC pocket in a similar fashion as oxazolidinones, in the close vicinity of G2061, stacking against U2504 with its hydroxymethyl tail oriented towards A2503 (Fig. 3A). However, this hydroxymethyl tail, which is significantly shorter than the acetamide tail found in LZD (Fig. 3D), is positioned within hydrogen-bonding distance to the backbone of either A2503 (3.5 Å) or G2505 (2.9 Å) (Fig. 3A).
Figure 3.
CDZ bound to the PTC pocket and superposition with structures of two other antibiotics of the oxazolidinone family. (a) Detailed view of CDZ and its interactions with the ribosome. CDZ is shown in orange, the P-site tRNA in pink and the rRNA in blue. The residues forming the pocket where CDZ binds are highlighted in purple. CDZ is within hydrogen-bonding distance of the backbones of A2503 and U2504 (yellow dashes). (b) Close-up view of the A- and B-rings of CDZ. The B-ring is stacking onto residue C2452 while being sandwiched between residues U2506 and A2451. (c) Overlay of CDZ with “compound 1”10,21 or (d) LZD19. The B rings of both oxazolidinones coincide with ring B of CDZ, but the compounds differ in the ring A and C substitutions. “Compound 1”, the tail of which also interacts with A2602, is a hybrid of an oxazolidinone antibiotic and sparsomycin.
Mechanism of translation inhibition by cadazolid
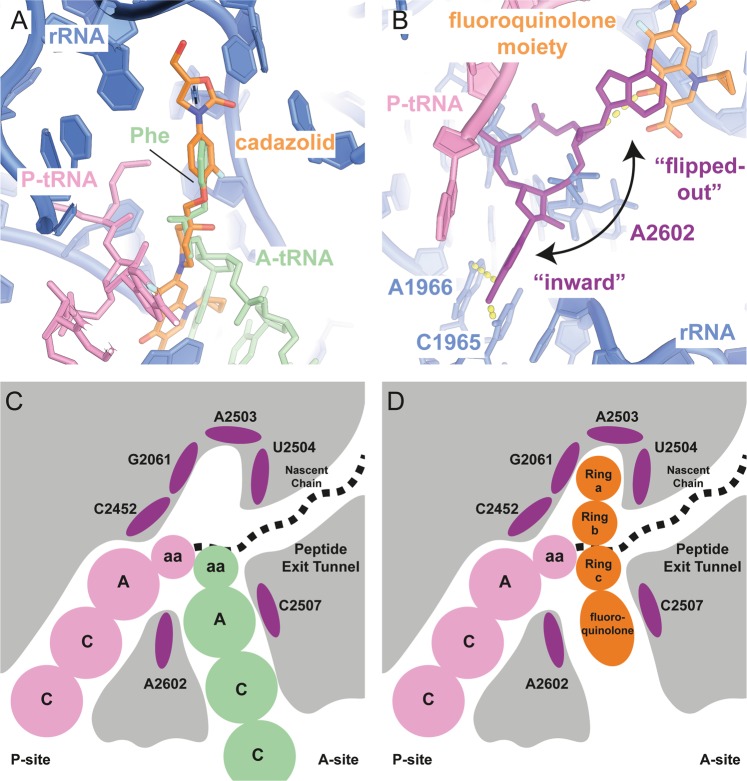
The specific mechanism of inhibition of translation by CDZ can be explained by the positions of the B-ring and the orientation of the fluoroquinolone moiety, which, together, would clash with a 3′ terminal nucleotide of an aminoacylated A-site tRNA (Fig. 4A) and hence prevent the elongation of the polypeptide chain by steric hindrance (Fig. 4C,D).
Figure 4.
Observed mechanism of translation inhibition by cadazolid. (a) Overlay of an A-site tRNA (PDB 1VY451) (light green) and CDZ (orange). Ring B and the fluoroquinolone moiety clash with an aminoacylated A-site tRNA, thereby preventing binding. (b) The two observed conformations of A2602. The fluoroquinolone moiety stacks with the “flipped-out” conformation to stabilize the PTC in a post-translocation conformation. (c,d) Schematic representation of the PTC with an A-site tRNA (c) or CDZ (d).
The fluoroquinolone moiety interacts with the backbone of U2584 and with one of the two alternative conformations observed for A2602 (Fig. 4B and Supplementary Fig. S4C). In the “inward” conformation, A2602 forms a non-standard base pair with C1965 and is positioned too far away from the antibiotic to become engaged in an interaction. However, in the other conformation, A2602 is “flipped-out”, thereby stacking on top of the fluoroquinolone moiety. A related stacking interaction had also been found in a complex with sparsomycin bound to the PTC24 as well as in a complex between the Hm 50S and “compound 1”, where the stacking partners are oriented at a different angle21 (Fig. 3C). Moreover, this highly conserved but flexible adenosine residue has been shown in a wide range of conformations, depending on the state of the ribosomes25–27, and was shown to be crucial for the peptidyl transferase reaction24,28,29. Interestingly, it was proposed that this base was linked to translocation of the tRNA30. By stacking with the base, the fluoroquinolone moiety likely stabilizes the P-site tRNA in a post-translocation state conformation resulting in a rigid and stably inhibited complex.
Compared to LZD, the acetamidomethyl tail on the A-ring is replaced by a hydroxymethyl group in CDZ, which is able to reach the bottom of the pocket to form a hydrogen bond with either A2503 or G2505. Together with the additional contacts between A2602 and the fluoroquinolone moiety, this could contribute to the overall higher potency of CDZ compared to LZD13,18.
Previous docking experiments revealed that changing the acetamide substituent at the C-5 position of the oxazolidinone ring in linezolid to a smaller hydroxymethyl substituent in tedizolid, a related oxazolidinone antibiotic, allowed the compound to bind ribosomes that either contained single nucleotide changes in the domain V of the 23S rRNA or modifications of A2503 through methylation by Cfr31–34. Interestingly, the smaller hydroxymethyl substituent on the oxazolidinone ring of CDZ would still allow binding of CDZ to ribosomes when A2503 is methylated, in agreement with the mode of binding observed in our cryo-EM structure (Supplementary Fig. S2).
Additionally, mutations in ribosomal protein uL3, which is located close to the CDZ binding pocket, were reported to be responsible for LZD resistance in S. aureus32,35 and were structurally linked to differences in the G2505 conformation36, resulting in a tighter binding pocket that would lead to a clash with LZD. A related mutation was found in C. difficile linezolid-resistant but CDZ-sensitive strains18. This mutation likely also causes reduction of the oxazolidinone binding pocket, showing that CDZ can still bind to a constricted pocket, most likely due to its shorter hydroxymethyl tail.
In the same study, a lysine-to-asparagine mutation in ribosomal protein uL4 was found in C. difficile with increased MIC and IC50 for CDZ. Despite being located 15 Å away from the bound CDZ, this residue is only separated from the antibiotic by one nucleotide, G2061, whose base lines the side of the pocket opposite to G2505. Interestingly, these mutants also have an increased MIC for LZD, indicating that this face of the pocket interacts with both LZD and CDZ in a similar fashion.
Discussion
Cadazolid belongs to the new quinoxolidinone class of antibiotics that has been shown to inhibit both DNA and protein synthesis in vitro and in cellular assays. In C. difficile, CDZ predominantly acts by inhibition of protein synthesis, via its oxazolidinone pharmacophore which is also present in LZD, rather than by inhibition of DNA gyrase and topoisomerase IV14. We expanded our analyses and showed that CDZ is also potently inhibiting protein translation in vitro in extracts from E. coli and S. aureus. Mechanistic and structural studies suggested that oxazolidinones interfere with binding of the substrate tRNAs to the A-site and that tRNA bound to the P-site increases the affinity of binding of LZD to the ribosome19,34. These observations were further supported by the identification of mutations in the rRNA that confer resistance to LZD31–34. Indeed, extracts from S. aureus S1 that contain the G2576U mutation in the 23S rRNA gene were less susceptible to the inhibition by LZD. Still, we observed a large MIC shift suggesting that other mutations contributed to the resistance phenotype. (summarized in32).
In contrast, the activity of CDZ was not affected by any of those mutations that led to resistance to LZD. The activity against a panel of clinical S. aureus isolates either containing a mutation in the ribosomal RNA gene harbouring a methylase that is able to modify the 23S ribosomal RNA (cfr) was not reduced compared to wild-type isolates. Previously, only a single mutation in ribosomal protein uL4, which is located in proximity to the CDZ binding pocket, occurred in C. difficile during in vitro selection with CDZ18.
Our reconstruction of the E. coli 70S in complex with mRNA, E- and P-site tRNAs and CDZ reveals important mechanistic differences in the binding mode of CDZ, although the antibiotic is related to other compounds of the oxazolidinone family (Fig. 3C)4,19. The high potency of CDZ can now be explained structurally by the tight fitting of the rather rigid oxazolidinone moiety into the bottom of the pocket close to the PTC together with the role of CDZ in stabilization of the PTC in a P-site tRNA occupied/post-translocation state. Additionally, the binding mode of CDZ to the ribosomes observed in our structure rationalizes the activity against LZD-resistant bacterial isolates.
Materials and Methods
Bacterial strains and antimicrobial agents
Strains were obtained from ATCC (25922, 29213), Public Health England (formerly Health Protection Agency, Centre for Infections), London, GB (S. aureus S1), Merck (formerly Novagen) (E. coli BL21(DE3)) or are from the Actelion strain collection. Cadazolid (CDZ, ACT-179811) was synthesized at Actelion Pharmaceuticals Ltd. Linezolid (LZD) was obtained from commercial sources (AK Scientific, LZD 70412).
Determination of the MIC
The MICs were determined using the micro dilution method recommended by the Clinical and Laboratory Standards Institute (CLSI).
In vitro transcription/translation (IVTT) assays
The cell-free protein synthesis reactions were carried out based on a procedure published for E. coli37 and adapted for S. aureus as described previously for C. difficile13. To improve expression from extracts derived from S. aureus, the firefly luciferase reporter gene was resynthesized with codons optimized for S. aureus at GeneArt AG (Regensburg, Germany) and cloned into plasmid pSP-luc_NF Fusion (Promega), replacing the existing luciferase gene. In addition, a 180-bp-long BglII-HindIII promoter fragment of gene abrB310 derived from Clostridium acetobutylicum was synthesized and cloned upstream of the codon-optimized luciferase gene for efficient translation, leading to plasmid pSPAbrlac-luc_Saur. S. aureus bacterial extracts were prepared according to a simplified E. coli procedure using a FastPrep®-24 instrument from Lucerna Chem AG (Cat No. 6004-500) and lysing matrix B (Lucerna Chem, Cat No. 116911100; silica spheres < 100 µm).
Test compounds diluted in 8% DMSO were added to reaction buffer containing bacterial extract and SP6 RNA polymerase, yielding final concentrations of 100, 20, 4, 0.8, 0.16, 0.04, 0.008, 0.0016, and 0 µM. Samples were mixed and pre-incubated for 10 min at RT before 100 ng plasmid pBestluc (E. coli) or 500 ng plasmid pSPAbrlac-luc_Saur (S. aureus) was added. After mixing, the plate was incubated at RT for 90 min. The luminescence was immediately measured after addition of the luciferase substrate Bright Glo (Promega, E2610) using a Tecan SpectralFluor Plus Reader (GeniosPro) instrument (parameters: integration time 1000 ms; intensity normal). Fifty percent inhibitory concentrations (IC50s) were calculated from the data.
Ribosome purification
The ribosomes were purified from E. coli BL21(DE3) strain as described previously38 using associative conditions for the final gradient (20 mM HEPES-NaOH pH 7.4, 6.5 mM MgCl2, 0.5 mM EDTA, 60 mM NH4Cl, 5 mM DTT, 10–40% (w/v) sucrose).
Complex formation
Due to low aqueous solubility of CDZ, the drug was dissolved in 100% DMSO resulting in a 7.85 mM solution, which was then diluted with the final buffer (20 mM HEPES-KOH pH 7.4, 10 mM MgCl2, 0.5 mM EDTA, 60 mM NH4Cl) to achieve a CDZ concentration of 1 mM. To assure proper binding, the 70S ribosomes (1.8 µM final concentration) were incubated overnight at 4 °C with an excess of CDZ (final concentration 10 µM) to allow the fairly large drug to have time to access the PTC and reach its binding equilibrium.
A 70S ribosome in complex with a P-site tRNA was then generated as described previously39. The mRNA was chemically synthetized (Dharmacon) with the sequence 5′-GCAAGGAGGUAAAAAUGUUUAAA-3′39. Incubated 70S ribosomes were first diluted to 180 nM while keeping the CDZ concentration constant. The mRNA (0.5 µM final concentration) was added to the 70S ribosome, and the mixture was incubated for 5 min at 37 °C. Next, the initiator tRNA(Met)i (2 µM final concentration) (tRNA Probes, LLC) was added and the mixture was incubated for 30 min at 37 °C, followed by the addition of Phe-tRNA(Phe) (2 µM final concentration) (tRNA Probes, LLC) and another 30 min incubation at 37 °C.
EM sample preparation and data collection
The grids were prepared using a FEI Vitrobot Mk4 (FEI Company), whose chamber was set to 100% humidity and 4 °C. 5 µL of the mixture prepared above was added to Quantifoil R2/2 holey carbon grids, which were previously coated with an additional thin layer of carbon and glow-discharged for 40 seconds with a current of 25 mA (negative). After 30 seconds incubation in the chamber, the excess liquid was blotted away for 11–24 seconds.
EM data processing and reconstruction
One grid was used for data collection with a FEI Titan Krios cryo-transmission electron microscope (FEI company) operating at 300 KeV and equipped with a Falcon II direct electron detector (FEI company). For every hole of the holey carbon, four movies were collected at a magnification of 130’000x resulting in a pixel size of 1.079 Å/pixel, and the total dose was set to 40 electrons/Å2, resulting in 8328 movies.
The resulting movie frames were aligned using a patch-based algorithm (25 patches) and dose weighted using MotionCor240. The contrast transfer function (CTF) parameters of the aligned micrographs were estimated using GCTF41, and the good micrographs were selected according to their CTF and micrograph quality. The particles of the resulting micrographs were picked using BATCHBOXER from the EMAN 1.9 package42. The ~773500 particles were then extracted and processed using Relion 2.043. After initial 2D classification to remove the bad particles and the dissociated 50S ribosomal subunits, the 70S particles were refined according to the “gold standard”44 (Supplementary Fig. S3). These ~609000 aligned particles were then used for initial 3D classification to remove free 50S ribosomal subunits, immediately followed by a 3D classification with a mask around the intersubunit space to categorize for tRNA occupancy.
In this last classification, two main sets were visible: a first ribosomal complex containing tRNAs in its P- and E-sites, and a second complex containing tRNAs in its A-, P-, and E-sites. An additional classification was performed on the dataset containing two tRNAs to separate the different ratchet states of the ribosome, and the most populated one was selected. The two sets of particles were then refined independently reaching an overall resolution of 3.1 Å for the subpopulation with the two tRNAs and 2.9 Å for the subpopulation containing three tRNAs. The set containing two tRNAs was then subjected to a per-particle CTF estimation (GCTF41) followed by a refinement with a mask around the 50S subunit, post-processed and sharpened resulting in an overall resolution of 3.0 Å (Supplementary Fig. S6). Lastly, the same subset was refined with a mask around the 30S, reaching an overall resolution of 3.2 Å.
Modelling, refinement and validation of the 50S large ribosomal subunit
First, an atomic model of E. coli 50S ribosomal subunit from the crystal structure of the 70S ribosome (PDB: 4YBB20) was docked into the EM density of the large subunit using UCSF Chimera45. The model was refined in reciprocal space using phenix.refine46 into the map as described previously47. After refinement, the differences between the EM map and the model were corrected by inspection of the Fobs-Fcalc difference Fourier maps using the programs O48 and COOT49. After rebuilding of the 50S subunit, inspection of the remaining difference density revealed the presence of an isolated CDZ molecule and a P-site tRNA in the PTC (Supplementary Fig. S4).
To model the CDZ molecule, coordinates were obtained in SDF format from the PubChem server (http://pubchem.ncbi.nlm.nih.gov) and, after conversion to PDB, were fed into PRODRG50 to obtain a topology file suitable for real and reciprocal space refinement. The CDZ model was fitted into the density using the real space fitting algorithm in COOT. The CCA-3′ termini of the tRNAs were built by docking and adjusting the corresponding model from a Thermus thermophilus 70S crystal structure (PDBs: 4QCM and 4QCN51).
The completed model was then refined to convergence by real space refinement using phenix.real_space_refine52. Validation of the atomic coordinates resulted in an optimal weighting of the model geometry versus the structure factors at a weight value of 1.4 (Supplementary Table S1). The estimated resolution of the model vs. map Fourier shell correlation (FSC) according to the FSC = 0.5 criterion coincided well with the resolution of the map established from the Fourier half set FSC correlation at the gold-standard criterion of 0.143 (Supplementary Fig. S6).
Figure generation
Figures showing cryo-EM maps and the final model were generated using UCSF Chimera45 and PyMOL (The PyMOL Molecular Graphics System Version 1.8.4.0 Schrodinger).
Supplementary information
Acknowledgements
We would like to thank the ETH Zürich scientific center for optical and electron microscopy (ScopeM) for access to electron microscopy equipment and are indebted to Peter Tittmann, Philippe Pfaff and Susanne Schröder for technical support. Also, we thank Nenad Ban for his support in designing the experiments and critical comments and Roland Lange for intellectual input and critical discussions.
Author Contributions
A.S. carried out ribosome purification and complex reconstitutions. A.S. and D.Bo. performed sample preparation and cryo-EM data collection analysis. A.S., D.Bo. and M.L. carried out data processing and reconstruction. M.L. and D.Bu. performed modelling. P.C. carried out in vitro transcription-translation assays. H.H.L. was responsible for microbiological data. H.H.L, G.R. and D.R. contributed to the design of the experiments. G.R. and D.R. provided intellectual input. D.R. was responsible for the overall oversight of the project.
Data Availability
The final model of the 50S ribosomal subunit containing the CDZ molecule as well as the CCA-fMet of the P-site tRNA is available from the Protein Data Base (PDB) under accession number 6QUL. The cryo-EM map of whole 70S ribosome, as well as the reconstruction from the focused refinement around the 50S and 30S ribosomal subunits are deposited in the Electron Microscopy Data Base (EMDB) under the accession number 4639, 4638 and 4641, respectively.
Competing Interests
P.C., D.Bu., H.H.L., G.R. and D.R. were employees of Actelion Pharmaceuticals Ltd until June 2017, P.C., G.R. and D.R. are current employees and P.C., H.H.L., G.R. and D.R. own equity of Idorsia Pharmaceuticals Ltd. A.S., M.L. and D.B. were supported by a research grant from Actelion Pharmaceuticals Ltd.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42155-4.
References
- 1.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 2.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 4.Wilson DN, et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu Rev Biochem. 2018;87:451–478. doi: 10.1146/annurev-biochem-062917-011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 7.Crotty MP, Krekel T, Burnham CA, Ritchie DJ. New Gram-Positive Agents: the Next Generation of Oxazolidinones and Lipoglycopeptides. J Clin Microbiol. 2016;54:2225–2232. doi: 10.1128/JCM.03395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moellering RC., Jr. New approaches to developing antimicrobials for resistant bacteria. J Infect Chemother. 2003;9:8–11. doi: 10.1007/s10156-002-0213-7. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Cheng X, Zhou Y, Xi Z. Sparsomycin-linezolid conjugates can promote ribosomal translocation. Chembiochem. 2011;12:2801–2806. doi: 10.1002/cbic.201100508. [DOI] [PubMed] [Google Scholar]
- 10.Zhou JC, et al. Design at the atomic level: Generation of novel hybrid biaryloxazolidinones as promising new antibiotics. Bioorg Med Chem Lett. 2008;18:6179–6183. doi: 10.1016/j.bmcl.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 12.Dubberke E. Clostridium difficile infection: the scope of the problem. J Hosp Med. 2012;7(Suppl 3):S1–4. doi: 10.1002/jhm.1916. [DOI] [PubMed] [Google Scholar]
- 13.Locher HH, et al. Investigations of the mode of action and resistance development of cadazolid, a new antibiotic for treatment of Clostridium difficile infections. Antimicrob Agents Chemother. 2014;58:901–908. doi: 10.1128/AAC.01831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locher HH, et al. In vitro and in vivo antibacterial evaluation of cadazolid, a new antibiotic for treatment of Clostridium difficile infections. Antimicrob Agents Chemother. 2014;58:892–900. doi: 10.1128/AAC.01830-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chilton CH, et al. In vitro activity of cadazolid against clinically relevant Clostridium difficile isolates and in an in vitro gut model of C. difficile infection. J Antimicrob Chemother. 2014;69:697–705. doi: 10.1093/jac/dkt411. [DOI] [PubMed] [Google Scholar]
- 16.Louie T, et al. Multicenter, Double-Blind, Randomized, Phase 2 Study Evaluating the Novel Antibiotic Cadazolid in Patients with Clostridium difficile Infection. Antimicrob Agents Chemother. 2015;59:6266–6273. doi: 10.1128/AAC.00504-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerding, D. N. et al. Cadazolid for the treatment of Clostridium difficile infection: results of two double-blind, placebo-controlled, non-inferiority, randomised phase 3 trials. Lancet Infect Dis, 10.1016/S1473-3099(18)30614-5 (2019). [DOI] [PubMed]
- 18.Caspers, P. et al. Different Resistance Mechanisms for Cadazolid and Linezolid in Clostridium difficile Found by Whole-Genome Sequencing Analysis. Antimicrob Agents Chemother61, 10.1128/AAC.00384-17 (2017). [DOI] [PMC free article] [PubMed]
- 19.Ippolito JA, et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 20.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22:336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou JC, et al. Design at the atomic level: Design of biaryloxazolidinones as potent orally active antibiotics. Bioorg Med Chem Lett. 2008;18:6175–6178. doi: 10.1016/j.bmcl.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Yang KL, et al. Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis. Nucleic Acids Res. 2017;45:10884–10894. doi: 10.1093/nar/gkx785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach KL, et al. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell. 2007;26:393–402. doi: 10.1016/j.molcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/S0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 25.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 26.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 27.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/S1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 28.Amort M, et al. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007;35:5130–5140. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2 ‘ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Molecular Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 31.Locke JB, et al. Structure-activity relationships of diverse oxazolidinones for linezolid-resistant Staphylococcus aureus strains possessing the cfr methyltransferase gene or ribosomal mutations. Antimicrob Agents Chemother. 2010;54:5337–5343. doi: 10.1128/AAC.00663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long KS, Vester B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother. 2012;56:603–612. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw KJ, Barbachyn MR. The oxazolidinones: past, present, and future. Ann N Y Acad Sci. 2011;1241:48–70. doi: 10.1111/j.1749-6632.2011.06330.x. [DOI] [PubMed] [Google Scholar]
- 34.Wilson DN. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci. 2011;1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 35.Locke JB, Hilgers M, Shaw KJ. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother. 2009;53:5275–5278. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belousoff, M. J. et al. Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome. MBio8, 10.1128/mBio.00395-17 (2017). [DOI] [PMC free article] [PubMed]
- 37.Kim TW, et al. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol. 2006;126:554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 38.Bingel-Erlenmeyer R, et al. A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature. 2008;452:108–111. doi: 10.1038/nature06683. [DOI] [PubMed] [Google Scholar]
- 39.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 40.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K, Gctf, Real-time CTF. determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 43.Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife5, 10.7554/eLife.18722 (2016). [DOI] [PMC free article] [PubMed]
- 44.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greber BJ, et al. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 48.Jones TA. Interactive electron-density map interpretation: from INTER to O. Acta Crystallogr D Biol Crystallogr. 2004;60:2115–2125. doi: 10.1107/S0907444904023509. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 51.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final model of the 50S ribosomal subunit containing the CDZ molecule as well as the CCA-fMet of the P-site tRNA is available from the Protein Data Base (PDB) under accession number 6QUL. The cryo-EM map of whole 70S ribosome, as well as the reconstruction from the focused refinement around the 50S and 30S ribosomal subunits are deposited in the Electron Microscopy Data Base (EMDB) under the accession number 4639, 4638 and 4641, respectively.