Abstract
The asthmatic airways are highly susceptible to inflammatory injury by air pollutants such as ozone (O3), characterized by enhanced activation of eosinophilic granulocytes and a failure of immune protective mechanisms. Eosinophil activation during asthma exacerbation contributes to the proinflammatory oxidative stress by high levels of nitric oxide (NO) production and extracellular DNA release. Surfactant protein-D (SP-D), an epithelial cell product of the airways, is a critical immune regulatory molecule with a multimeric structure susceptible to oxidative modifications. Using recombinant proteins and confocal imaging, we demonstrate here that SP-D directly bound to the membrane and inhibited extracellular DNA trap formation by human and murine eosinophils in a concentration and carbohydrate-dependent manner. Combined allergic airway sensitization and O3 exposure heightened eosinophilia and nos2 mRNA (iNOS) activation in the lung tissue and S-nitrosylation related de-oligomerisation of SP-D in the airways. In vitro reproduction of the iNOS action led to similar effects on SP-D. Importantly, S-nitrosylation abolished the ability of SP-D to block extracellular DNA trap formation. Thus, the homeostatic negative regulatory feedback between SP-D and eosinophils is destroyed by the NO-rich oxidative lung tissue environment in asthma exacerbations.
Keywords: eosinophil extracellular traps (EETs), eosinophils, surfactant pulmonary-associated protein D (SP-D)
1 |. INTRODUCTION
Ozone (O3) is a toxic air pollutant formed by chemical reactions between nitric oxides and volatile organic compounds under sunlight. Exposure to O3 activates the innate immune system causing an influx of inflammatory cells into the airways. The asthmatic airways are highly susceptible to inflammatory injury by air pollutants such as O3, likely due to a failure of protective mechanisms. We previously showed that O3 exposure induced exacerbation of allergic airway inflammation in mice,1 the inflammatory changes were mediated by group 2 innate lymphoid cells,2 and the inflammation was predominated by eosinophilic granulocytes (eosinophils) in allergen sensitized but not naïve mice.1
Activation of eosinophils during asthma exacerbation compounds the proinflammatory oxidative stress by high levels of nitric oxide (NO) production3,4 and release of extracellular DNA traps.5 Eosinophils are involved in host defense and inflammation by phagocytosis of pathogens and secretion of toxic mediators, both functions are central to their role as effector cells. Within the last decade, it has become clear that these cells (just like neutrophils) are able to form extracellular DNA traps that significantly contribute to host defense.6,7 However, DNA traps can also contribute to the pathogenesis of chronic infectious, allergic, and autoimmune diseases.8 Extracellular traps consist of DNA fibers and antimicrobial proteins, including eosinophil granule proteins that can bind and kill bacteria,7,9 fungi10 or even viruses11 in the extracellular space. Although extracellular DNA traps are beneficial in host defense against invading microorganism and in response to various biological stimuli, their inappropriate release is highly pro-inflammatory. We have demonstrated the existence of extracellular DNA traps by eosinophils and neutrophils infiltrating the airways of human asthmatics.12 In bronchial biopsies taken from allergic asthmatics, the number of eosinophil extracellular traps (EETs) consisting of DNA and co-localizing with major basic protein, correlated with the number of infiltrating eosinophils. The molecular mechanisms responsible for regulation of extracellular DNA release remain unclear and in dispute.13,14
Surfactant protein-D (SP-D), an epithelial cell product of the airways is a critical immune protective molecule with a multimeric structure susceptible to oxidative modifications. SP-D-deficient mice demonstrate chronic inflammatory alterations indicating a special importance of this molecule in regulating immune homeostasis and the function of the innate immune cells.15 We demonstrated that SPD plays a protective role both in allergen16 or O3-induced17 airway inflammation. Allergen challenge with either Aspergillus fumigatus (Af) or ovalbumin induced increases in SP-D mRNA and protein levels in the mouse lung18 in an IL-4/IL-13/STAT-6 dependent manner while rSP-D significantly inhibited Af-driven Th2 cell activation in vitro.16 Mice lacking SP-D had increased numbers of CD4+ lymphocytes with elevated IL-13 and thymus- and activation-regulated chemokine levels in the lung and showed exaggerated production of IgE and IgG1 following allergic sensitization.16 Further, exposure of mice17 or human volunteers19 to O3 also induced SP-D expression in the lung and in the circulation, respectively. SP-D induction was mediated by IL-6 in type II alveolar epithelial cells.17 We also showed that SP-D can directly bind to immune cells of the lung20 thereby regulating pulmonary immune homeostasis during the inflammatory response.
Several recent studies suggested that the multimeric (native) form of SP-D is susceptible to oxidative post translational modifications and subsequent structural and functional changes.21 Indeed, there is evidence that SP-D may lose its host defense and anti-inflammatory properties in association with oxidation,22 nitration23 or nitrosylation24 of specific N- and C-terminal residues during oxidative stress in the respiratory tract.
In the present study, we aimed to investigate how SP-D is affected in a model of severe combined airway inflammation induced by O3 exposure and allergic airway sensitization in mice. We hypothesized that SP-D regulates eosinophil extracellular DNA trap formation and that this function is modified by oxidative stress.
2 |. MATERIALS AND METHODS
(A detailed description is provided in the online supplement)
2.1 |. Purification of eosinophils
Human eosinophils were isolated with a StemCell Technologies kit to >97% purity from white blood cells separated from Lithium-heparinized blood of healthy donors as previously described.25 Written, informed consent was obtained from all peripheral blood donors. This study was approved by Ethics Committee of the Canton of Bern. Mouse eosinophils were isolated from IL5tg mice from bone marrow using Abs against CD8α, CD19, CD90.2, and Ly-6G (Miltenyi Biotec, Bergisch Gladbach, Germany) and an EasySep Mouse PE Positive Selection Kit (Stemcell Technologies) to >95% purity, assessed by the Hematocolor Set (Merck Millipore).25
Mouse studies were performed in Balb/c, C57BL/6 (wild type, WT) and SP-D−/− mice. O3 and allergen exposure experiments were carried out as described.1,2,16 Mice were sensitized and challenged with Af and unless otherwise indicated, exposed to O3 (3.0 ppm for 2 h) 84 h later, and studied 12 h post O3 exposure. Lung function (Flexivent) measurements (airway responsiveness) were performed in response to inhaled methacholine, as we previously described.2 Bronchoalveolar lavage (BAL) was collected to assess inflammatory cells and for SP-D measurements, and lung tissue was preserved for gene expression studies (Affymetrix Inc). All methods and the Affymetrix database related to the transcriptomics data have been uploaded onto the NCBI Government website. Access the GSE111155 study number at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE111155
BAL differential cell count was performed on stained cytospins (Siemens Diff Quik Stain, Thermo), under light microscopy (×1000, oil) and counting of total live cells (Countess, Thermo Fisher Scientific). Total BAL SP-D was measured by sandwich ELISA using our in-house generated mAbs and polyclonal Abs. SP-D structural changes were assessed by native gel electrophoresis on the BAL cell-free lysate as previously described.26 Optical Density was semi-quantified by ImageJ analysis.
2.2 |. Cell activation and confocal laser scanning microscopy
rSP-D (10 μg/mL) was treated with medium, glucose (10 mM) or goat anti-SP-D Ab (4 μg/mL) (30 min; 37°C) before addition to the cells. Isolated eosinophils, resuspended in X-VIVO™ 15 medium (2.5 × 106/mL) and pre-incubated (30 min) with SP-D, were primed with GM-CSF (mouse) or IL-5 (human) and stimulated with 10−8 M complement factor 5a (C5a). Fixed cells were stained with Hoechst 33342 and mounted in ProLong Gold mounting medium. Extracellular DNA was visualized by cell permeable fluorescent dyes, such as MitoSox Red or Syto 13 prior to fixation. SP-D binding to the cell surface was analyzed by indirect immunofluorescence as previously described.27 Briefly, eosinophils pre-incubated with either SP-D or recombinant S-nitrosothiol (SNO)-SP-D (5 μg/mL, both His-Tagged), were seeded on glass coverslip, fixed with 4% paraformaldehyde and labeled by monoclonal anti-His Ab (1:100; clone HIS.H8, ThermoFisher Scientific) overnight (4°C), and then with Alexa Fluor® 488 conjugated goat anti-mouse secondary Ab (1:400; ThermoFisher Scientific). Images were acquired by LSM 700 (Carl Zeiss Micro Imaging, Jena, Germany; 63×/1.40 Oil) and analyzed by Imaris software (Bitplane AG, Zurich, Switzerland) as previously reported.27
2.3 |. Quantification of released dsDNA in culture supernatants
Released dsDNA was quantified as previously described.27 Briefly, isolated eosinophils (4 × 106/mL) were stimulated and treated with DNase I (2.5 U/mL; Worthington) and proteinase K (0.2 mg/mL; Roche). Reactions were stopped and cells were centrifuged (500 × g, 5 min). Supernatant was transferred to black, glass-bottom 96-well plates (Greiner Bio-One GmbH) and the fluorescent activity of PicoGreen dye bound to dsDNA was excited at 502 nm and the fluorescence emission intensity was measured at 523 nm using a spectrofluorimeter (Spectra-Max M2, Molecular Devices, Biberach an der Riß, Germany), according to instructions (Quant-iT™PicoGreen® Assay Kit).
SNO-SP-D was generated with slight modifications of the technique used by Guo et al.21,24 Recombinant mouse and human SP-D was treated with S-nitrosoglutathione (GSNO; Sigma, 200 nM/10 μg/mL SP-D) followed by Microcon column centrifugation (MW = 30K cutoff, 13,000 × g, 12 min, room temperature), washing (4×, PBS), aliquoting, and freezing until it was used. Western blot as well as native gel electrophoresis were used to assess recovery of recombinant native and GSNO-treated SP-D. S-nitrosylation of recombinant SP-D was also accomplished by NO obtained by iNOS mRNA (nos2) transfection in human embryonic kidney (HEK) 293 cells in vitro.28 After 4 h of mRNA transfection of cells, 3 μg recombinant SP-D was added. After overnight incubation, cell culture supernatants were processed for the biotin switch assay as described.24,29
2.4 |. Biotin switch,24,29 Western blotting, and native gel electrophoresis21
In mouse BAL, culture supernatant or recombinant SP-D or SNO-SP-D samples, nitrosylated cysteines were converted to biotinylated cysteines in a biotin switch assay using biotin-HPDP. Total protein was precipitated, SNO bonds were decomposed by sodium ascorbate, the newly formed thiols were then linked with N-[6-biotinamido)-hexyl]-1-(2-pyridyldithio) propionamide (Pierce). The biotinylated peptides were pulled down with Streptavidin-agarose beads.
2.5 |. Western blotting
Total protein concentrations were determined by bicinchoninic acid (ThermoFisher Scientific). Immunoblotting was performed using goat anti-mouse SP-D Ab (1:2000; R&D Systems) and equal protein (250 ng SP-D and SNO-SP-D and 5 μg of BAL or culture supernatant protein loaded per lane). Results are representative of at least 3 independent experiments. Native protein samples or denatured in reducing Laemmli buffer prior were separated by 10% SDS-PAGE gel electrophoresis. Proteins were blotted on PVDF transfer membrane (Merck Millipore) and subsequently probed. Secondary donkey anti-goat (1:10,000; GE Healthcare Life Sciences) Ab coupled to HRP was used and signals were detected by enhanced chemiluminescence (ECL Western blotting substrate, ThermoFisher Scientific) on photosensitive film (ECL Hyper-film, GE Healthcare Life Sciences).
2.6 |. In-gel digestion and mass spectrometry of recombinant mouse SP-D
Bands were extracted from BAL run on SDS-PAGE gels, digested with trypsin, and analyzed using Nano liquid chromatography–tandem mass spectrometry with Sequest and Scaffold softwares. In additional experiments, recombinant SP-D (3 μg) was treated with air or O3 (3.0 ppm for 30 min), denatured, reduced, and alkylated before in-solution or ingel trypsin digestion with or without PNGase F de-glycosylation and analyzed using TSQ Vantage and LTQ Orbitrap.22,23
2.7 |. Statistical analysis
Mean values (±SEM) are provided. To compare groups, unpaired Student’s t-test was applied and a P-value less than 0.05 was considered to be significant. Two-way ANOVA with Tukey’s multiple comparisons test was used to compare dose responses and time courses. Correlations were performed using regression analysis (Graph Pad Prism 7, Graph Pad Software Inc.).
3 |. RESULTS AND DISCUSSION
3.1 |. SP-D inhibited eosinophil activation and blocked formation of extracellular DNA traps
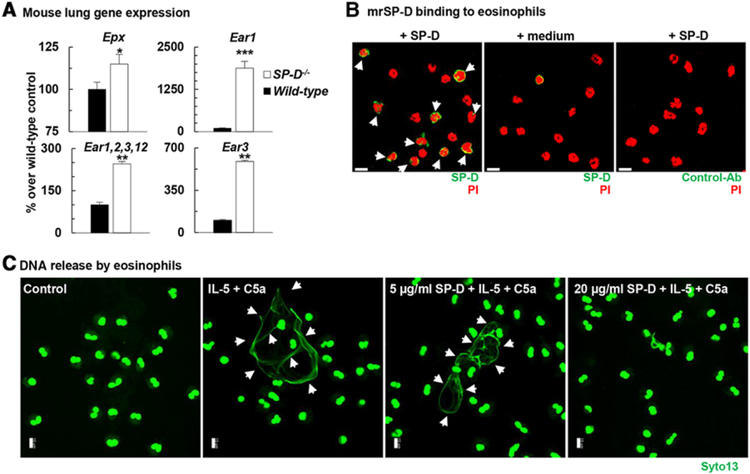
Our laboratory and others previously showed that SP-D deficiency predisposes to inflammation of the airways and lung parenchyma20,30 leading to chronic disease.30–33 Although the role of SP-D in modulating macrophage and dendritic cell function for host defense has been well documented,15,34,35 its regulatory function in allergic airways inflammation and especially on eosinophil activation has been unclear. Under light microscopic examination of the SP-D−/− lung or the BAL, we could not identify eosinophils in non-treated (naïve) mice. To probe deeper the role of SP-D in eosinophil activation in the lung, we studied wild type (C57BL/6) and SP-D−/− mouse lung transcriptomes. The Affymetrix chip (a pre-determined design by Affymetrix) from our mouse lung transcriptome, contained over 45,000 genes including the eosinophil specific epx; ear1; the combined ear1,2,3,12; ear3; ear11; ear2, ear4, and ear5 gene probes. Figure 1A shows that expression for eosinophil peroxidase (epx) and isoforms of eosinophil associated ribonucleases ear1, ear1,2,3,12, and ear3, but not the other above genes (data not shown) were significantly up-regulated in the absence of SP-D in the SP-D−/− mouse lungs. Constitutive gene expression of these granule products reflects activation of eosinophils in the lung tissue or increased eosinophil influx into the lungs36 of SP-D−/− mice.
FIGURE 1. SP-D inhibited eosinophil activation and dsDNA (EET) release.
(A): Total RNA was extracted from naïve wild type (C57BL/6, black bars) and SP-D−/− (white bars) lungs. Eosinophil specific gene expression was studied by Affymetrix microarray. Data were standardized to GAPDH and expressed as arbitrary units (% change from the wild-type control mean value). Mean ± SEM of n = 4–6. *P < 0.05; **P < 0.01; ***P < 0.001 (t-test). (B) Immunofluorescent staining of bound SP-D to the surface of mature mouse eosinophils (white arrows) isolated from bone marrow of IL5tg mice. Scale bars, 10 μm. (C) Human recombinant SP-D blocked the formation of EETs by human eosinophils isolated from peripheral blood of healthy donors. (B and C) Confocal microscopy. Scale bars, 10 μm
To investigate whether SP-D could directly affect activation of these cells, we studied membrane binding of SP-D to the surface of mature mouse eosinophils isolated from bone marrow of IL5tg mice. We have selected the SP-D doses represented in Fig. 1B, and C based on the fact that the physiological SP-D concentration range in the lung is 1–20 μg/mL under non-inflammatory conditions34 and several pilot studies that we conducted on neutrophil extracellular trap formation (data not shown). Figure 1B demonstrates that at 5 μg/mL approximately 60% of eosinophils bound SP-D. SP-D contains a C-type lectin carbohydrate recognition domain on its c-terminus,37 and inhibitory C-type lectin receptors are known to be expressed on several myeloid cell populations, including eosinophils.38 Expression level of these receptors changes by the maturation state of the myeloid cells (unpublished observation). In Figs. 1B and 4G; mouse eosinophils were isolated from bone marrow and could therefore, exhibit differences in their maturation level, leading to differential expression of inhibitory lectin binding receptors, which then could account for the variability of bound SP-D staining.
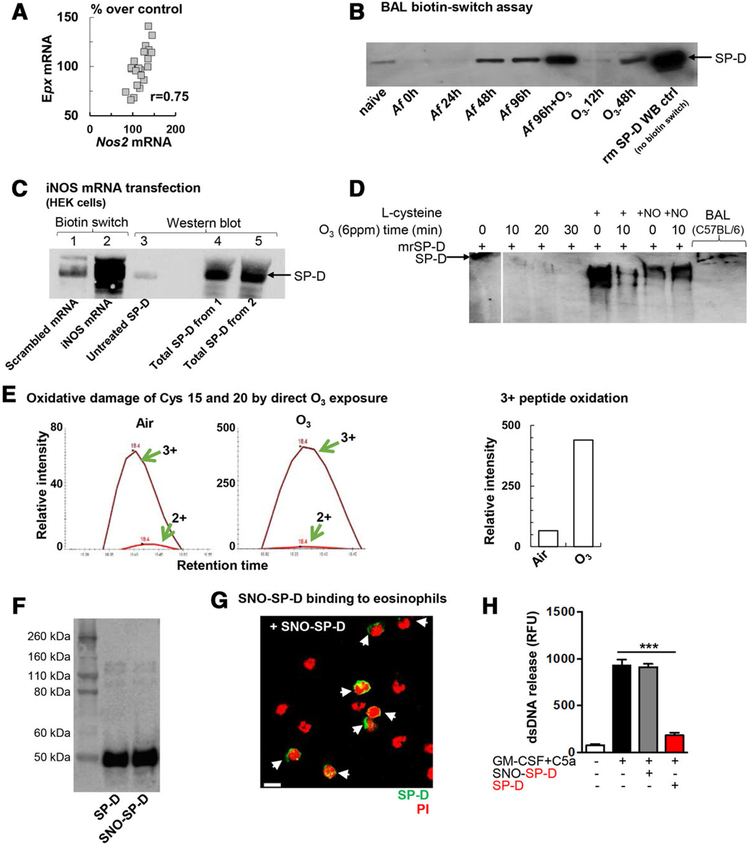
FIGURE 4. S-nitrosylation of the SP-D molecule altered its structure and abolished its inhibitory effects on dsDNA release in a carbohydrate dependent manner.
(A) Correlation between eosinophil peroxidase (Epx) and inducible nitric oxide synthase (Nos2) mRNA in the lung. Data points represent individual Balb/c mice that received the combination of Af sensitization and challenge +O3 exposure (n = 21). (B) Representative blot of n = 3 independent experiments demonstrating S-nitrosylated SP-D in the BAL fluid of allergen and O3 exposed mice by the biotin-switch assay. Mouse recombinant SP-D was added as a Western Blot control without the biotin switch. Naïve: no treatment; O3: ozone exposure (2 ppm for 2h, BAL harvested 12 or 48h later); Af: BAL harvested as indicated; Af+O3: sensitized mice were challenged with Af and received O3 exposure 84 h later. BAL was harvested 96 h after allergen challenge (i.e.,12 h after O3). Rm (recombinant mouse) SP-D: 0.05μg (C): S-nitrosylation of recombinant SP-D with NO obtained by iNOS mRNA translation in HEK cells in vitro. HEK 293 cells were transfected with iNOS mRNA and incubated with rSP-D. After overnight incubation, cell culture supernatants were processed for biotin switch assay. 1, control (scramble transfection)+rSP-D (3 μg)-(biotin switch); 2, iNOS mRNA transfection+ rSP-D (3 μg)-(biotin switch); 3, untreated SP-D (50 ng) –WB ctrl; 4, total SP-D from Sample 1 (WB ctrl, no biotin switch); 5, total SP-D from sample 2 (WB ctrl, no biotin switch). (D) effect of O3 exposure, l-Cys and l-Cys-NO treatment on the structure of recombinant SP-D in vitro. Native gel electrophoresis. (E) Oxidation of peptide SVPNTCTLVMCSPTENGLPGR after in vitro ozone treatment. Recombinant mouse SP-D (3 μg) was de-glycosylated, trypsinized overnight, and then assessed by the TSQ-Vantage mass spectrometer. (F) Immunoblot analysis of denatured recombinant native and oxidized (SNO) SP-D. Results are representative of at least 3 independent experiments. (G) Confocal microscopy. Immunofluorescent staining of bound SNO-SP-D molecules (at 5 μg/mL) on the surface of mature mouse eosinophils isolated from bone marrow of IL5tg mice. Mouse eosinophils were stained with anti-His-Tag Ab. SNO-SP-D binding is indicated by white arrows; scale bar, 10 μm. (H) Quantification of released dsDNA in supernatant of non-activated and activated eosinophils pre-incubated with recombinant SP-D or oxidized SP-D (SNO-SP-D) both at 10 μg/mL. Mean ± SEM of n = 4
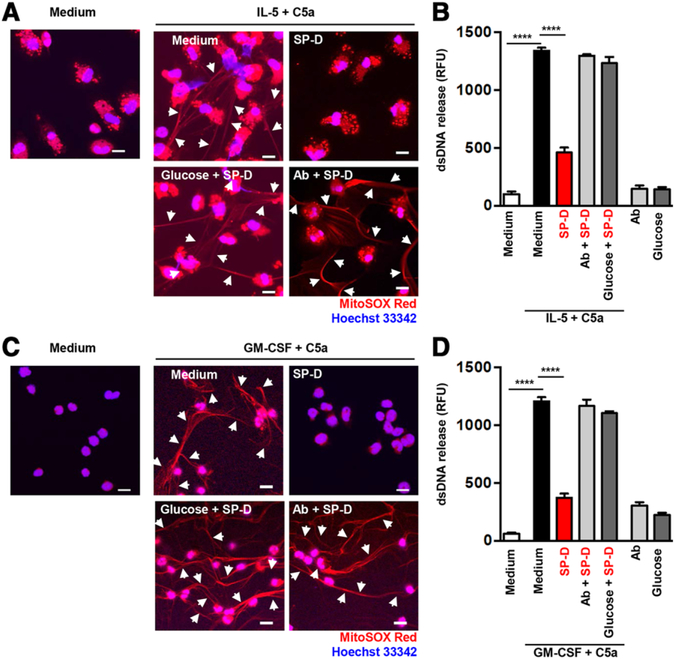
To investigate whether SP-D binding has a functional consequence, we setup in vitro eosinophil activation studies using both mouse and human eosinophils and studied the release of EETs. Following a short-term (45 min) stimulation of eosinophils with IL-5 and C5a, recombinant SP-D inhibited extracellular DNA release in a dose-dependent manner, as assessed by confocal microscopy (Fig. 1C). These results were further corroborated using both purified human peripheral blood eosinophils (Fig. 2A) and mature mouse eosinophils isolated from bone marrow of IL5tg mice (Fig. 2C). Confocal assessment of EET formation was further confirmed by quantification of released dsDNA in the supernatant of activated cells (Fig. 2B and D).7
FIGURE 2. SP-D inhibited extracellular DNA release from human and mouse eosinophils in a carbohydrate dependent manner.
Human blood eosinophils (A and B) and mature mouse eosinophils from bone marrow of IL5tg mice (C and D) were analyzed for EET formation. (A and C) EET formation with or without rSP-D (10 μg/mL) alone or together with a neutralizing anti-SP-D Ab (4 μg/mL) or glucose (10 mM), following short-term stimulation as indicated was assessed by confocal microscopy. White arrows indicate extracellular dsDNA. Bars, 10 μm. (B and D) Quantification of released dsDNA in supernatants of activated eosinophils using PicoGreen fluorescent dye. Data are presented as mean ± SEM of at least 3 independent experiments. ****P < 0.0001
In Fig. 2A human eosinophil granules appear punctate due to the high levels of granule proteins, such as major basic proteins (MBP) and eosinophil peroxidase (EPX), that are known to have autofluorescence (hence the red dots). In contrast, mouse eosinophils comparatively have less eosinophil granule proteins and therefore less auto-fluorescence activity (Fig. 2C). The MitoSOX Red fluorescent dye is a derivative of ethidium bromide (i.e., a DNA binding dye) that is live-cell permeable and is oxidized by superoxide, exhibiting red fluorescence when bound to nucleic acid. MitoSOX is a highly suitable fluorescent dye for the detection of the DNA scaffold of EETs, since it requires DNA in combination with reactive oxygen species to emit a red color. Hoechst 33342, being a weaker staining reagent for released DNA, is here used only to visualize the nuclei of cells after fixation.
We used an anti-SP-D Ab as an experimental control throughout our experiments. Importantly, Ab blockade of SP-D completely reversed its effects on dsDNA release verifying that the inhibition we saw on eosinophil extracellular trap formation was indeed specifically caused by SP-D (Fig. 2A–D). Taken together, these data indicated that lack of SP-D in the lung results in gene activation of eosinophil specific proinflammatory granule products and/or influx of eosinophils into the lungs while presence of SP-D inhibits eosinophil activation by direct cell membrane binding. Our data corroborate previous studies in which SP-D was shown to bind eosinophils in a maltose and Ca++ -dependent manner.39 SP-D also inhibited eotaxin-induced migration, degranulation of eosinophil cationic protein39 and promoted apoptosis upon IL-5 priming in vitro.40–42
SP-D exerts both host-defense and anti-inflammatory functions in the lung. These are not contradictory as they both serve immune protection of the respiratory mucosa through distinct and complementary mechanisms as we and many others have previously shown.15,43,44 Such dual biological role is well illustrated by the relationship of SP-D to extracellular DNA traps, in the absence of pathogens the carbohydrate recognition domain of SP-D is free to ligate inhibitory membrane receptors on the granulocyte surface37 thereby preventing inappropriate release of DNA as shown in Fig. 2 by both human and mouse cells. Figure 2A–D also demonstrate that the SP-D effect on dsDNA release was abolished by disabling the carbohydrate recognition domain with addition of glucose. Studies from other laboratories demonstrated that in the presence of bacteria, SP-D enhanced DNA trapping by simultaneously binding to bacterial carbohydrate ligands and DNA35 and facilitated pathogen and dead cell clearance by macrophages through extracellular DNA traps.35,45 We suggest therefore that occupation of the carbohydrate recognition domain of SP-D by pathogens provides the functional switch that releases the SP-D control of extracellular DNA trap formation and turns the molecule into a host-defense opsonin. DNA trap formation, an important host defense process which is otherwise highly proinflammatory (reviewed by Porto and Stein),8 requires a tight regulation in the lung. Our data supports that SP-D may fulfill such an important regulatory function.
3.2 |. O3 enhanced Af-induced airway eosinophilia and degraded SP-D
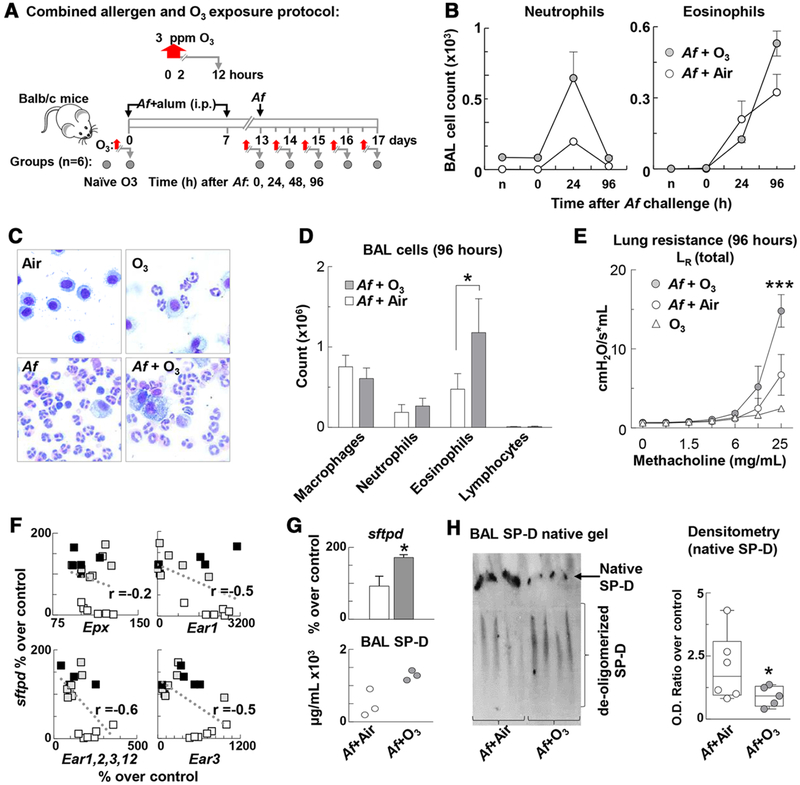
To investigate the in vivo significance of these findings in a model of Th2 high severe asthma where eosinophils are the predominant cell type and responsible for the clinical and pathological changes,46,47 we used a combined model of allergic airway sensitization and O3 exposure.1 Mice were sensitized and challenged with Af, then exposed to O3 at different time points post Af challenge and 12 h post O3 exposure (Fig. 3A). Our time course experiment showed that airway neutrophilia peaked 24 h and airway eosinophilia peaked 96 h after Af challenge. O3 inhalation significantly increased both neutrophil and eosinophil counts (Fig. 3B). A detailed assessment of the differential cell count at this time point (Fig. 3C and D) shows that a doubling of eosinophil numbers in the BAL was associated with a significant increase in lung resistance to methacholine inhalation in the mice that received O3 exposure after Af challenge (Fig. 3E).
FIGURE 3. O3 enhanced Aspergillus fumigatus (Af)-induced airway eosinophilia and degraded SP-D in the BAL.
(A) Balb/c mice (n = 6/group) were sensitized and challenged with Af then exposed to O3. Lung function, airway inflammation, and SP-D expression were determined at the indicated time points (grey circles) post Af challenge and 12 h post O3 exposure. (B) BAL counts of neutrophils and eosinophils were performed on cytospin preparations (Siemens Diff Quik Stain) and counting of total live cells (Countess). (C and D) BAL samples were obtained 96 h after Af challenge of C57BL/6 mice. (C) Photomicrographs of cytospin preparations were taken under immersion oil (1000×) of representative images depicting differential cellular inflammation in response to O3 or Af alone and the combination of these in BAL. (D) Quantitative assessment of macrophages, neutrophils, eosinophils, and lymphocytes using stained cytospins from BAL samples. (E) Airways hyperresponsiveness was assessed to increasing concentrations of nebulized methacholine by Flexivent. Total lung resistance is shown. (D) *P < 0.05 Unpaired t-test; (E): ***P < 0.001 two-way ANOVA with Tukey’s multiple comparisons; (B–E) mean ± SEM of n = 6–8; (F) eosinophil granule product gene expression (studied by an Affymetrix gene chip in the lung) negatively correlated with SP-D mRNA expression. Data points represent group averages (n = 4–5 mice in each groups of C57BL/6 [black squares], Balb/c [grey squares], and SP-D−/− [white squares] mice that received air, ozone, allergen, or a combination of allergen and ozone exposure, one group each, n = 18 groups). (G) Lung tissue SP-D mRNA (sftpd) in Balb/c mice expressed as % over average air exposed control; (top; *P < 0.05; Unpaired t-test; n = 5–6). Total BAL SP-D was measured by ELISA (bottom; n = 3). (H) Native gel electrophoresis of BAL cell-free lysate. Optical Density was assessed by ImageJ analysis. Data are presented as median and interquartile ranges n = 5 *P < 0.05, Unpaired t-test
To investigate how eosinophil granule product gene expression (a marker of eosinophil number and/or activation in the lung) is related to SP-D mRNA expression during inflammatory changes, we analyzed an Affymetrix gene chip of the lung of C57BL/6, Balb/c, and SP-D−/− mice that received air, O3, Af, or a combination exposure of both O3 and Af as described. These results suggested a negative correlation between SP-D and eosinophil granule product mRNA expression (Fig. 3F) that was driven by the inclusion of SP-D−/− mice showing the highest eosinophil granule product gene expression of the three strains. Based on these results and the findings depicted in Figs. 1 and 2, we hypothesized that presence of more SP-D would convey more protection against inflammatory changes in the lung. To study this, we investigated Balb/c mice that showed a prominent increase in eosinophil influx into the airways upon the combined exposure to allergen and O3 (Fig. 3D). Contrary to our hypothesis, heightened airway inflammation in the allergen and O3 exposed mice was in fact associated with significantly elevated SP-D both at the mRNA (Fig. 3G top panel) and the protein level (3G bottom panel). Thus, increased level of SP-D expression did not simply equal better anti-inflammatory protection. To resolve this conundrum, we went ahead and looked into the structural changes of SP-D upon allergen and O3 treatment of the mice by native gel electrophoresis (Fig. 3H). The multimeric (native) structure of SP-D is known to be susceptible to chemical modifications by free radicals during airway inflammation22–24 and such change can result in a loss of its immune protective properties.21,24,26,48,49 Indeed, while O3 enhanced mRNA as well as total SP-D protein expression in the asthmatic mice, the proportion of highly oligomerized SP-D was in fact significantly decreased in these animals (Fig. 3H right panel). Such structural modification occurred in allergen challenged asthma patients26 and in the highly inflamed lung of mice24 and it was associated with a nitric oxide rich milieu and inducible nitric oxide synthase iNOS (nos2) activation.48
3.3 |. S-nitrosylation of the SP-D molecule altered its structure and abolished its inhibitory effects on dsDNA release
Eosinophils have been identified in asthmatic airway inflammation as a major source of iNOS3,4,50 and exhaled NO has become the hallmark of severe high eosinophilic asthma.51,52 In our model, there was a positive correlation between the genes encoding the eosinophil granule product eosinophil peroxidase (epx) and inducible nitric oxide synthase (nos2) in the lung of mice that received the combination of Af and O3 exposure (Fig. 4A) suggesting that greater number of eosinophils or extent of activation is associated with increased presence of the enzyme responsible for NO production. NO was suggested to chemically modify SP-D in a process called S-nitrosylation.21,24 The extent of this modification in SP-D recovered from the BAL of mice is illustrated in Fig. 4B by the biotin switch assay29 (with the exception of the last band which is a Western blot control of untreated mouse recombinant SP-D, run without biotin switch). The role of iNOS was confirmed by an in vitro experiment where we transfected HEK cells with nos2 (or an irrelevant) mRNA and added mouse recombinant SP-D that was then assessed by biotin switch (Fig. 4C). The role of cysteine residues in maintaining the integrity of the multimeric SPD structure is essential. Cys15 and 20 were demonstrated to be particularly important34,53 and susceptible to S-nitrosylation.24 The role of cysteine residues in the direct effects of oxidative modifications by O3 were investigated in vitro (Fig. 4D). As shown before,21,24 addition of L-nitrocysteine resulted in de-oligomerisation of the native recombinant SP-D molecule. However, we also saw such effect upon direct in vitro exposure of SP-D to O3. Mass spectroscopic assessment of O3-treated SP-D revealed that Cys15 and 20 were oxidized (Fig. 4E and Supplementary Fig. 1) suggesting that in addition to S-nitrosylation, direct oxidation by O3 can also affect cysteine residues in SP-D. To study the functional significance of such chemical alterations, we treated SP-D with GSNO. After treatment, we compared SNO-SPD to untreated SP-D in a reduced gel (Fig. 4F) to ensure recovery of similar amounts. We also tested binding of SNO-SP-D molecules on the surface of mature mouse eosinophils (Fig. 4G). We found similar binding intensity to untreated SP-D (Fig. 1B) suggesting that S-nitrosylation did not affect eosinophil membrane binding. However, inhibition of dsDNA release into the supernatant of activated eosinophils was completely lost when cells were treated with SNO-SP-D (Fig. 4H).
In summary, our study showed that SP-D directly bound to and inhibited extracellular DNA-trap release by human and murine eosinophils in a dose- and carbohydrate-dependent manner. In a mouse model, combined allergic airway sensitization and O3 exposure induced heightened airway eosinophilia, nos2 mRNA activation in the lung tissue, and S-nitrosylation related de-oligomerisation of SP-D in the BAL. In vitro reproduction of the iNOS action in a cell culture expression system, led to similar effects on SP-D. Direct S-nitrosylation abolished the ability of SP-D to block extracellular DNA trap formation. We propose that the homeostatic negative regulatory feedback between SP-D and eosinophils is unraveled by the NO-rich oxidative lung tissue environment in asthma exacerbations.
Supplementary Material
ACKNOWLEDGMENTS
Swiss National Science Foundation to S.Y. (#31003A_173215) and H.U.S. (#310030_166473) and R21AI116121 to A.H.; C.H.F. is funded by T32ES007059; N.G. is PhD student of the Graduate School of Cellular and Biomedical Sciences of the University of Bern; Images were acquired on equipment supported by the Microscopy Imaging Centre of the University of Bern. Dr. Paul Axelsen (University of Pennsylvania) is gratefully acknowledged for his generous mentoring, guidance in the experimental design, and troubleshooting of the SP-D mass spectrometry studies. Dr. Carol Anne Ogden and Janssen are gratefully acknowledged for providing the Affymetrix gene expression analysis of the mouse lung tissue.
Abbreviations:
- Af
Aspergillus fumigatus
- BAL
bronchoalveolar lavage
- C5a
complement factor 5a
- EET
eosinophil extracellular trap
- Epx
eosinophil peroxidase gene
- GSNO
S-Nitrosoglutathione
- HEK
Human embryonic kidney
- O3
ozone
- PNGase F
peptide:N-glycosidase F
- SNO
S-Nitrosothiol
- SP-D
surfactant protein-D
- WT
wild type
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional information may be found online in the supporting information tab for this article.
REFERENCES
- 1.Kierstein S, Krytska K, Sharma S, et al. Ozone inhalation induces exacerbation of eosinophilic airway inflammation and hyperresponsiveness in allergen-sensitized mice. Allergy. 2008;63:438–446. [DOI] [PubMed] [Google Scholar]
- 2.Yang Q, Ge MQ, Kokalari B, et al. Group 2 innate lymphoid cells mediate ozone-induced airway inflammation and hyperresponsiveness in mice. J Allergy Clin Immunol. 2016;137:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yates DH. Role of exhaled nitric oxide in asthma. Immunol Cell Biol. 2001;79:178–190. [DOI] [PubMed] [Google Scholar]
- 4.Xu W, Ghosh S, Comhair SA, et al. Increased mitochondrial argi-nine metabolism supports bioenergetics in asthma. J Clin Invest. 2016;126:2465–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueki S, Tokunaga T, Fujieda S, et al. Eosinophil ETosis and DNA traps: a new look at eosinophilic inflammation. Curr Allergy Asthma Rep. 2016;16:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevaert E, Zhang N, Krysko O, et al. Extracellular eosinophilic traps in association with Staphylococcus aureus at the site of epithelial barrier defects in patients with severe airway inflammation. J Allergy Clin Immunol. 2017;139:1849–1860 e6. [DOI] [PubMed] [Google Scholar]
- 7.Yousefi S, Gold JA, Andina N, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. [DOI] [PubMed] [Google Scholar]
- 8.Porto BN, Stein RT. Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front Immunol. 2016;7:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. [DOI] [PubMed] [Google Scholar]
- 10.Urban CF, Reichard U, Brinkmann V, Zychlinsky A. Neutrophil extra-cellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 2006;8:668–676. [DOI] [PubMed] [Google Scholar]
- 11.Jenne CN, Wong CH, Zemp FJ, et al. Neutrophils recruited to sites of infection protect from virus challenge by releasing neutrophil extracellular traps. Cell Host Microbe. 2013;13:169–180. [DOI] [PubMed] [Google Scholar]
- 12.Dworski R, Simon HU, Hoskins A, Yousefi S. Eosinophil and neutrophil extracellular DNA traps in human allergic asthmatic airways. J Allergy Clin Immunol. 2011;127:1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon D, Simon HU, Yousefi S. Extracellular DNA traps in allergic, infectious, and autoimmune diseases. Allergy. 2013;68:409–416. [DOI] [PubMed] [Google Scholar]
- 14.Yousefi S, Simon HU. NETosis—does it really represent nature’s “suicide bomber”?. Front Immunol. 2016;7:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haczku A Protective role of the lung collectins surfactant protein A and surfactant protein D in airway inflammation. J Allergy Clin Immunol. 2008;122:861–879. quiz 880–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haczku A, Cao Y, Vass G, et al. IL-4 and IL-13 form a negative feedback circuit with surfactant protein-D in the allergic airway response. J Immunol. 2006;176:3557–3565. [DOI] [PubMed] [Google Scholar]
- 17.Kierstein S, Poulain FR, Cao Y, et al. Susceptibility to ozone-induced airway inflammation is associated with decreased levels of surfactant protein D. Respir Res. 2006;7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haczku A, Atochina EN, Tomer Y, et al. Aspergillus fumigatus-induced allergic airway inflammation alters surfactant homeostasis and lung function in BALB/c mice. Am J Respir Cell Mol Biol. 2001;25:45–50. [DOI] [PubMed] [Google Scholar]
- 19.Alexis NE, Lay JC, Haczku A, et al. Fluticasone propionate protects against ozone-induced airway inflammation and modified immune cell activation markers in healthy volunteers. Environ Health Perspect. 2008;116:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge MQ, Kokalari B, Flayer CH, et al. Cutting edge: role of NK cells and surfactant protein D in dendritic cell lymph node homing: effects of ozone exposure. J Immunol. 2016;196:553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atochina-Vasserman EN. S-nitrosylation of surfactant protein D as a modulator of pulmonary inflammation. Biochim Biophys Acta. 2012;1820:763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crouch EC, Hirche TO, Shao B, et al. Myeloperoxidase-dependent inactivation of surfactant protein D in vitro and in vivo. J Biol Chem. 2010;285:16757–16770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matalon S, Shrestha K, Kirk M, et al. Modification of surfactant protein D by reactive oxygen-nitrogen intermediates is accompanied by loss of aggregating activity, in vitro and in vivo. FASEB J. 2009;23:1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo CJ, Atochina-Vasserman EN, Abramova E, et al. S-nitrosylation of surfactant protein-D controls inflammatory function. PLoS Biol. 2008;6:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Germic N, Stojkov D, Oberson K, Yousefi S, Simon HU. Neither eosinophils nor neutrophils require ATG5-dependent autophagy for extracellular DNA trap formation. Immunology. 2017;152:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atochina-Vasserman EN, Winkler C, Abramova H, et al. Segmental allergen challenge alters multimeric structure and function of surfactant protein D in humans. Am J Respir Crit Care Med. 2011;183:856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amini P, Stojkov D, Wang X, et al. NET formation can occur independently of RIPK3 and MLKL signaling. Eur J Immunol. 2016;46:178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kariko K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:PL1. [DOI] [PubMed] [Google Scholar]
- 30.Sims MW, Tal-Singer RM, Kierstein S, et al. Chronic obstructive pulmonary disease and inhaled steroids alter surfactant protein D (SP-D) levels: a cross-sectional study. Respir Res. 2008;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay RM, Grainge CL, Lau LC, Barber C, Clark HW, Howarth PH. Airway surfactant protein D deficiency in adults with severe asthma. Chest. 2016;149:1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa H, Ledford JG, Mukherjee S, et al. Surfactant protein D attenuates sub-epithelial fibrosis in allergic airways disease through TGF-beta. Respir Res. 2014;15:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng OZ, Palaniyar N. NET balancing: a problem in inflammatory lung diseases. Front Immunol. 2013;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason RJ, Greene K, Voelker DR. Surfactant protein A and surfactant protein D in health and disease. Am J Physiol. 1998;275: L1–L13. [DOI] [PubMed] [Google Scholar]
- 35.Douda DN, Jackson R, Grasemann H, Palaniyar N. Innate immune collectin surfactant protein D simultaneously binds both neutrophil extracellular traps and carbohydrate ligands and promotes bacterial trapping. J Immunol. 2011;187:1856–1865. [DOI] [PubMed] [Google Scholar]
- 36.Sharma SK, Almeida FA, Kierstein S, et al. Systemic FasL neutralization increases eosinophilic inflammation in a mouse model of asthma. Allergy. 2012;67:328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardai SJ, Xiao YQ, Dickinson M, et al. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. [DOI] [PubMed] [Google Scholar]
- 38.Redelinghuys P, Brown GD. Inhibitory C-type lectin receptors in myeloid cells. Immunol Lett. 2011;136:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Bredow C Hartl D, Schmid K, Schabaz F, Brack E, Reinhardt D, Griese M. Surfactant protein D regulates chemotaxis and degranulation of human eosinophils. Clin Exp Allergy. 2006;36:1566–1574. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan L, Madan T, Kamal N, et al. Recombinant surfactant protein-D selectively increases apoptosis in eosinophils of allergic asthmatics and enhances uptake of apoptotic eosinophils by macrophages. Int Immunol. 2008;20:993–1007. [DOI] [PubMed] [Google Scholar]
- 41.Mahajan L, Pandit H, Madan T, et al. Human surfactant protein D alters oxidative stress and HMGA1 expression to induce p53 apoptotic pathway in eosinophil leukemic cell line. PLoS One. 2013;8:e85046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahajan L, Gautam P, Dodagatta-Marri E, Madan T, Kishore U. Surfactant protein SP-D modulates activity of immune cells: proteomic profiling of its interaction with eosinophilic cells. Expert Rev Proteomics. 2014;11:355–369. [DOI] [PubMed] [Google Scholar]
- 43.Forbes LR, Haczku A. SP-D and regulation of the pulmonary innate immune system in allergic airway changes. Clin Exp Allergy. 2010;40:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SPD, orchestrate innate immunity in the lung. J Clin Invest. 2002;109:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palaniyar N, Nadesalingam J, Reid KB. Innate immune collectins bind nucleic acids and enhance DNA clearance in vitro. Ann N Y Acad Sci. 2003;1010:467–470. [DOI] [PubMed] [Google Scholar]
- 46.Peters SP, Busse WW. New and anticipated therapies for severe asthma. J Allergy Clin Immunol Pract. 2017;5:S15–S24. [DOI] [PubMed] [Google Scholar]
- 47.Stokes JR, Casale TB. Characterization of asthma endotypes: implications for therapy. Ann Allergy Asthma Immunol. 2016;117:121–125. [DOI] [PubMed] [Google Scholar]
- 48.Guo C, Atochina-Vasserman E, Abramova H, et al. Role of NOS2 in pulmonary injury and repair in response to bleomycin. Free Radic Biol Med. 2016;91:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atochina-Vasserman EN, Beers MF, Gow AJ. Review: chemical and structural modifications of pulmonary collectins and their functional consequences. Innate Immun. 2010;16:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes PJ, Liew FY. Nitric oxide and asthmatic inflammation. Immunol Today. 1995;16:128–130. [DOI] [PubMed] [Google Scholar]
- 51.Katial RK, Bensch GW, Busse WW, et al. Changing paradigms in the treatment of severe asthma: the role of biologic therapies. J Allergy Clin Immunol Pract. 2017;5:S1–S14. [DOI] [PubMed] [Google Scholar]
- 52.Agrawal S, Townley RG. Role of periostin, FENO, IL-13, lebrikzumab, other IL-13 antagonist and dual IL-4/IL-13 antagonist in asthma. Expert Opin Biol Ther. 2014;14:165–181. [DOI] [PubMed] [Google Scholar]
- 53.Brown-Augsburger P, Hartshorn K, Chang D, et al. Site-directed muta-genesis of Cys-15 and Cys-20 of pulmonary surfactant protein D. Expression of a trimeric protein with altered anti-viral properties. J Biol Chem. 1996;271:13724–13730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.