Abstract
The Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census (CGC) is an expert-curated description of the genes driving human cancer, used as a standard in cancer genetics across basic research, medical reporting and pharmaceutical development. After a major expansion and complete re-evaluation, the 2018 CGC describes in detail the effect of 719 cancer-driving genes. Recent expansion includes functional and mechanistic descriptions of how each gene contributes to disease generation, described in terms of the key cancer hallmarks and the impact of mutations on gene and protein function. These functional characteristics depict the extraordinary complexity of cancer biology, and suggest multiple cancer-related functions for many genes, which are often highly tissue- or tumour stage-dependent. The 2018 CGC encompasses a second-tier, describing an expanding list of genes (currently 145) from more recent cancer studies which show supportive but less detailed indications of a role in cancer.
Introduction
The Cancer Gene Census1 (CGC) is a key resource within the Catalogue of Somatic Mutations in Cancer (COSMIC), comprising a long-term ongoing effort to catalogue and describe all genes with causal impact in human cancer. To encompass every driver gene across all human cancers, the CGC combines somatically mutated genes (showing detailed mutation patterns in COSMIC), with mutant genes primarily inherited across families causing cancer predisposition syndromes. Since it was begun in 2004, a continuous curation approach to the scientific literature has grown this resource into a comprehensive description of over 700 genes, detailing how each gene contributes to disease causation. As it has grown, the CGC has become a standard in cancer research, and is used across the world in medical reporting (for instance in Genomics England clinical reports2), pharmaceutical development3,4 and a wide range of algorithm and tool developments and benchmarking (e.g. Oncotator5; SomInaClust6; OncodriveROLE7; OncodriveCLUST8).
Available for download and for on-line exploration at: https://cancer.sanger.ac.uk/census, the CGC comprises evidence-based, manually-curated summaries of 719 cancer-driving genes (version v86, August 2018) and brings together the expertise of cancer scientists, a dedicated curation team, and the comprehensive resources of the COSMIC database [https://cancer.sanger.ac.uk/cosmic]9,10. To ensure the accuracy and confidence of these data, a conservative approach is adopted to adding new genes to the CGC; genes are only included when evidence for a gene’s involvement is clear and unequivocal (Box 1).
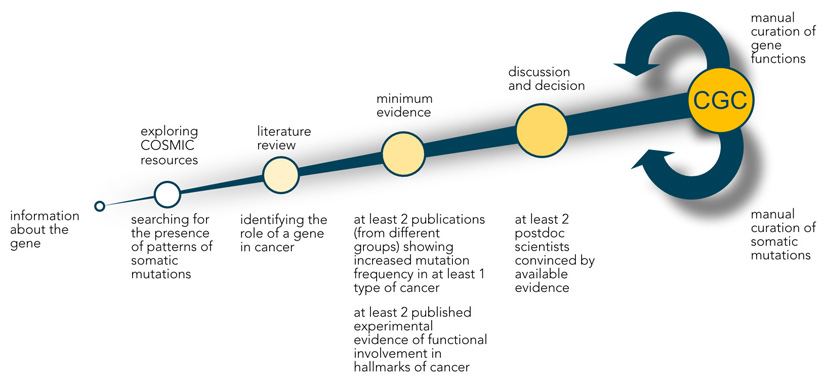
Box1. Curation process used for the Cancer Gene Census.
-
Choosing candidate genes
Candidate genes are selected from the literature including the conference abstracts, large systematic screens, personal communications, and analyses of COSMIC data.
-
Exploration of COSMIC resources
COSMIC data is analysed to determine the presence of patterns of somatic mutations and increased frequency of somatic mutations in cancer.
-
Literature review
All available literature is reviewed to identify the role of a gene in cancer.
-
Minimum evidence
-
• at least 2 publications (from different groups) showing increased mutation frequency in at least 1 type of cancer
• at least 2 publications (from different groups) showing experimental evidence of functional involvement in hallmarks of cancer that allow for functional classification of the gene as an oncogene, tumour suppressor gene or fusion partner
-
-
Independent assessment, discussion and decision
• at least 2 postdoctoral scientists convinced by available evidence
• gene classified as fulfilling the criteria for Tier 1 or Tier 2 Cancer Gene Census (CGC) gene
Inclusion in CGC
Continuous manual curation of somatic mutations from targeted screens and gene functions for Tier 1 CGC genes, collecting more evidence for Tier 2 CGC genes
Figure.
In this review, we will specify which attributes determine a gene’s inclusion into the CGC and how genes could be classified with regard to these attributes in order to better characterise their involvement in oncogenesis. We will also describe the new structure of the CGC which now encompasses two Tiers, as well as a new expansion of the CGC, which describes functional characteristics of Tier 1 cancer genes. Finally, we will explore how this broad characterisation of cancer gene function demonstrates the complexity of cancer genetics, which factors can influence the effect of gene dysfunction in different cancer types, and how this knowledge allows investigation of new ways to target known cancer genes by precision oncology.
Defining cancer genes
During oncogenic transformation, the intracellular regulatory network is disturbed, leading eventually to cell reprogramming that promotes unregulated proliferation and adaptation to the tissue environment. Many cellular processes can be disrupted to promote oncogenesis, and somatic or germline genetic mutations are a major and primary causative factor11. These genetic events qualitatively or quantitatively alter the function of genes and proteins, and in consequence alter the cellular processes in which these proteins participate.
Knowledge of which mutations affect a gene’s function and what consequence they induce allows the generation of a mechanistic and functional causal chain of events leading to neoplasm. This knowledge is being successfully applied to design therapies specifically targeting proteins altered by somatic and germline mutations to drive cancer, with well-known examples including Vemurafenib, an inhibitor of V600E mutant BRAF12 and Olaparib, which induces synthetic lethality [G] in cancer cells with mutations in BRCA1 or BRCA213.
While a wide range of mutations and their consequences have been described across human cancer, driver mutations will typically result in either dysfunction, usually via protein structure change, or dysregulation, altering regulatory signals that control gene expression, or complete abrogation, when the whole TSG is deleted. Therefore, to understand the impact of mutation on disease progression it has to be determined which genes have functions that can be altered to potentially drive or inhibit tumour development.
The CGC has been built to address these challenges and support the world’s wide variety of cancer research. It describes genes functionally associated with hallmarks of cancer14, 15 and characterised (in most cases) by somatic or germline mutations in their coding regions, which change the resulting proteins sequences and affect their function.
The entire CGC is under constant scrutiny to provide an up-to-date resource of the impact of protein dysfunction or dysregulation caused by gene mutation. Each gene has been classified across three categories: oncogene, tumour suppressor gene (TSG) and/or fusion gene, depending on its somatic mutation profile and functional role in oncogenesis.
To classify a gene as an oncogene, evidence is required that the activity of the gene product can drive cancer and that alterations resulting in gain-of-function[G] occur in cancer samples. In contrast, to identify a TSG, loss-of-function [G] alterations are sought in tumours, with experimental evidence reporting a cancer suppressive function of the wild-type gene product.
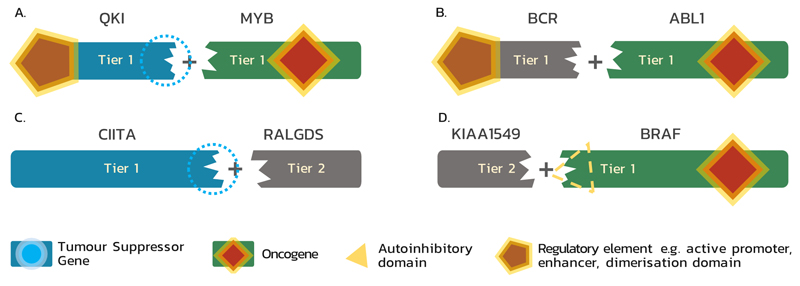
Gene fusions form a specific class of genetic alteration associated with specific functional consequences. They usually arise as a consequence of genomic structural rearrangements and involve two distinct genes, forming a new chimeric gene with novel or dysregulated function16; many oncogenic fusions can be described as an independent neomorphic oncogene. The CGC describes how fusion events impact the function of each fusion partner and the contribution of each partner towards the transforming capability of the fusion (Fig. 1). The consequence of gene fusions sometimes allows each partner to be classified as having TSG or oncogene function, and these genes are identified as such in the CGC (Fig. 1A). Alternatively, one partner may constitute only a minor genetic element, contributing only a regulatory feature or structural domain, but not functioning as an oncogene or TSG on its own (Fig1B-D). Therefore, to incorporate these genes that are crucial for the gain of transforming capacity by the fusion proteins, but which cannot be classified as TSGs nor oncogenes on their own, this third functional category of the CGC was created.
Figure 1. Functional classes of genes involved in fusions and their classification within the Cancer Gene Census.
(A) As a result of gene fusion, a tumour suppressor gene (TSG) may lose its suppressive abilities and a proto-oncogene may be transformed into an oncogene; e.g. QKI-MYB: in angiocentric glioma MYB is activated by truncation and the influence of the QKI enhancer; QKI loses its TSG function105. (B) Genes that, after fusion, upregulate an oncogene through donation of regulatory element (e.g. active promoter, enhancer, or activating domain) are included in Tier 1; e.g. BCR-ABL1: in this fusion (also known as the Philadelphia chromosome), BCR, which is neither TSG nor oncogene, simply provides an oligomerisation domain, which enables constitutive activation of ABL119. (C) A fusion partner which only deactivates a TSG by disrupting its sequence is classified as a Tier 2 Cancer Gene Census (CGC) gene if it is recurrently involved in a fusion; e.g. CIITA-RALGDS: in Hodgkin lymphoma this fusion results in the N-terminal part of CIITA, which loses its TSG function, fused in an out-of-frame fashion to RALGDS, which is neither TSG nor oncogene in this case 106. (D) A fusion may also result in hyperactivation of an oncogene due to loss of an autoinhibitory domain, which is replaced by a fusion partner; e.g. KIAA1549-BRAF: in pilocytic astrocytoma the N-terminal BRAF autoregulatory domain25 is lost in the fusion protein, resulting in constitutively active BRAF expressed under the control of the KIAA1549 promoter107. A fusion gene, such as KIAA1549, acting solely and recurrently through replacing a functional fragment of the other partner is included into Tier 2.
Building a broader perspective
The CGC comprises two Tiers, depending on the strength of evidence supporting each gene’s involvement in oncogenesis.
Tier 1 of the Cancer Gene Census
To classify into Tier 1, a gene must possess a documented and reproducible activity relevant to cancer, along with evidence of mutations in cancer that change the activity of the gene product in a way that promotes oncogenic transformation. Although only somatic mutations from cancer samples and cell lines are catalogued in COSMIC, the CGC additionally uses information about germline mutations and their consequences. We also acknowledge somatic mutation patterns across cancer samples gathered in COSMIC and verify where they are concordant with the gene role determined during CGC curation. TSGs often show a broad range of inactivating mutations, whilst dominant oncogenes usually demonstrate well defined hotspots of missense or inframe indel mutations11. Presence of such patterns alongside functional evidence is required to include a gene into Tier 1 of the CGC (Fig. 2, Box 1), and as such these genes have the most extensive evidence of a role in cancer. Currently Tier 1 includes 574 genes (Fig. 3A).
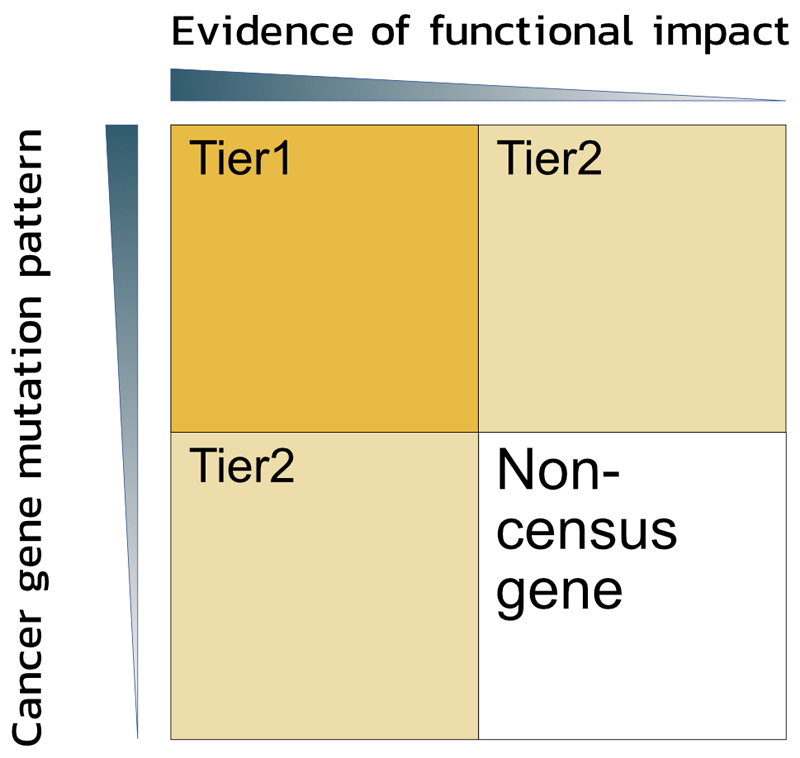
Figure 2. Tiers of the Cancer Gene Census.
Genes are classified into either Tier 1 or Tier 2 of the Cancer Gene Census (CGC) based on two criteria: a) the evidence of functional involvement in oncogenesis via impact on hallmarks of cancer, and b) the presence of patterns of somatic mutations in cancer samples that are concordant with the gene function determined by the literature curation (i.e. gene fusions; highly recurrent missense mutation in oncogenes; or a high proportion of inactivating mutations in tumour suppressor genes (TSGs)). Only strong evidence from both functional and mutational analyses qualify a gene to Tier 1. Genes with mutational patterns typical for cancer drivers but not functionally characterised, as well as genes with published mechanistic description of their involvement in cancer but without a proof of being somatically mutated in cancer comprise Tier 2 of the CGC.
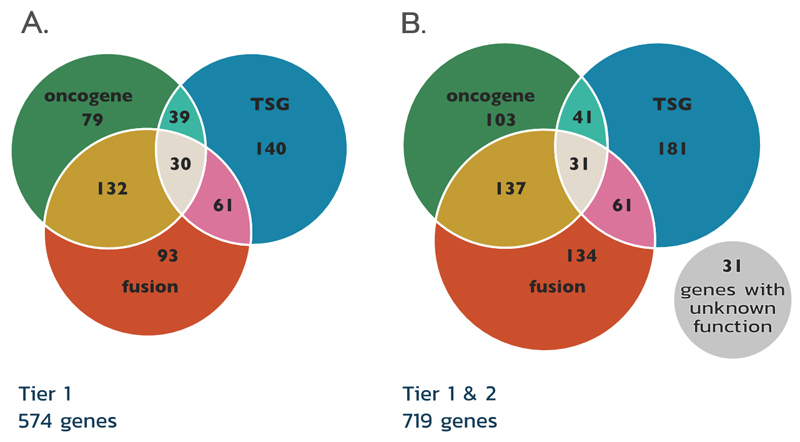
Figure 3. Quantification of the 3 classes of cancer genes in the Cancer Gene Census tiers.
The figure shows Tier 1 (A), and both Tier 1 and Tier 2 combined (B). The role of Cancer Gene Census (CGC) genes in cancer differs, depending on disease type as demonstrated by the overlap between annotation sets. In Tier 1 69 genes can act as tumour suppressor genes (TSGs) or oncogenes. The majority of genes involved in gene fusions promote cancer by gaining oncogenic or losing tumour suppressing activity. However, about one third of the genes that act as fusion partners acts exclusively through modifying the function of their fusion partner. Numbers correspond to the number of genes in each of the categories.
A fusion partner gene is included into Tier 1 when its altered function is proven to drive oncogenesis, including when it provides a partner with regulatory elements, such as an active promoter or enhancer (for example, TMPRSS217, IGH18) or dimerisation domain (for example, BCR19, ERC120).
Tier 2 of the Cancer Gene Census
Tier 2 is a new section of the CGC, describing genes with more recently identified roles in oncology, and consists of genes with strong indications of a role in cancer but with less strong mechanistic or functional evidence. Included in Tier 2 are genes with mutation patterns typical for oncogenes or TSGs but which have less well-established functional evidence in the scientific literature. Similarly, genes with strong published evidence for a function in cancer but unclear mutation patterns or known to be dysregulated for instance solely by epigenetic means (for example by changes to promoter methylation) are also included in Tier 2. For instance, KAT7, which encodes a histone acetyltransferase involved in maintaining pluripotency and self-renewal of embryonic stem cells21, and for which there is evidence of a tumour suppressing role in acute myeloid leukaemia (AML)22,has only evidence of one recurrent missense mutation (D109G) in renal carcinoma provided by one study23 and only few frameshift or nonsense mutations that could support tumour suppressing activity24. As such, this was placed into Tier 2 of the CGC.
Tier 2 additionally incorporates fusion gene partners of unclear consequence, but which disrupt the sequence of a known oncogenic partner, giving it transforming ability (Fig. 1D). An example of such behaviour is KIAA1549, a fusion partner of BRAF in glial and glioneuronal neoplasms. The KIAA1549–BRAF fusion protein is present in 66% of pilocytic astrocytomas and has the active C-terminal kinase domain of BRAF without the N-terminal autoregulatory domain thus resulting in constitutively active BRAF expressed under the control of the KIAA1549 promoter25 (Fig. 1D). While the oncogenic activity of BRAF in this fusion is well recognised, and the frequent recurrence of this rearrangement demonstrates that it is an important event in these malignancies, the mode of action of the KIAA1549 coding gene is unclear in this case.
The current CGC (August 2018, v86) describes 719 genes across two Tiers (Figure 3B). 554 are clearly classified as TSG and/or oncogene, however there is substantial overlap with genes playing different roles between tissues, disease states and different environmental stresses. Below we discuss some of the striking observations that are only possible via such a broad deeply descriptive curation effort as the CGC.
Describing functions of cancer genes
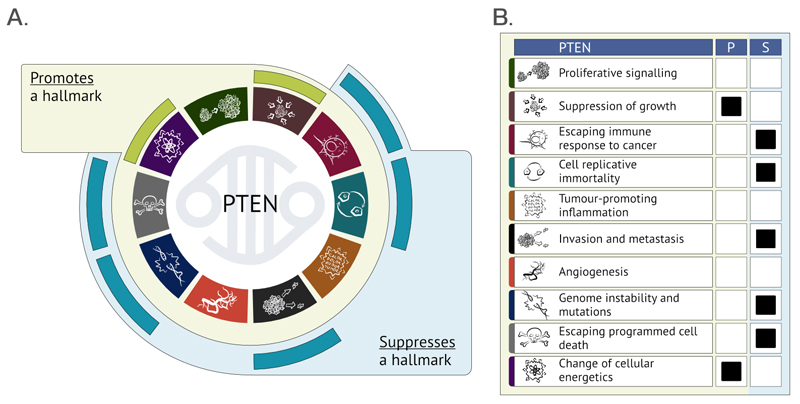
To determine the functional role of each CGC gene in oncogenesis, extensive literature review is performed (and is continually underway) to include new discoveries in cancer genetics. Experimental data from published literature is evaluated and curated in order to describe the oncogenic impact of each gene, and the 10 hallmarks of cancer15 are used to standardise functional categorisation for each gene. Widely accepted as the 10 major attributes of oncogenesis, the cancer hallmarks allow for precise but still concise cancer-focused functional classifications of gene activity. Substantially expanding the descriptive content of the CGC, the new hallmark pages (Fig. 4), developed in collaboration with the Open Targets platform4, combine both mechanistic and functional descriptions of cancer gene activity with manually curated evidence from the literature. Functional annotations define whether each gene in its wild-type form has a stimulating or suppressive effect on individual cancer hallmarks, and provide immediate access to the relevant literature source via PubMed. These annotations are curated from a continually expanding range of review and research articles, currently encompassing over 1600 articles and growing rapidly. New graphics have been created to ensure this information is easily accessible online and simple to interpret (Fig. 4).
Figure 4. Graphical summary of hallmarks of cancer-related functions of PTEN presented on the CGC website.
(A) New gene summaries concisely integrate manually curated information on the impact of proteins (in this case PTEN)108 coded by cancer genes on the hallmarks of cancer in simple graphical form. If the wild-type (WT) protein function promotes a process related to cancer, it is marked within the green outer ring. The protein suppressing a hallmark of cancer in its WT form is marked within the blue outer ring. [modified from Hanahan & Weinberg, Cell, 2011]15
(B) The alternative presentation of the summary of PTEN functions shows how an alteration resulting in gain- or loss-of-function of a gene may impact each of the hallmarks. As WT PTEN promotes (P) growth suppression, an inactivating PTEN mutation will lead to increased proliferation of mutant cells. (S = suppresses)
In addition to the 10 hallmarks of cancer, further descriptions take into account roles in additional biological processes that are relevant for cancer but not covered by the hallmarks, such as cell division, differentiation, global regulation of gene expression, senescence and impact of mutation on gene or protein function. The functional description of how each mutated gene and protein causes cancer is a continual and ongoing process, with approximately half the CGC Tier 1 genes already described in both functional and mechanistic terms at the time of this review. To avoid misrepresenting a gene’s roles, functional summaries are added only when manual curation compiles enough information to illustrate the full known spectrum of each gene’s functions.
Functional complexity of cancer genes
Recent expansion of the CGC to include functional annotations allows for a deeper and broader insight into genetic and physiological causes of cancer. In the new hallmark pages, all possible ways a gene could be involved in oncogenesis are comprehensively overviewed; no indications are made to rank the importance of each function as this can change rapidly according to cancer type and stage, and can be different for different mutations in the gene. Curation aims to be comprehensive, combining all literature evidence for a gene’s involvement in relevant cellular processes; even when infrequently affected, these processes may still contribute toward cancer. This creates a robust and broad perspective on the function of each gene in cancer and can highlight unexpected and targetable aspects of their activity.
Most of the genes comprising Tier 1 of the CGC have a long track record of published evidence on their function. Although in many cases the majority of research is focused on just one of a gene’s many functions, this exhaustive curation approach describes a broader range of their functionality. Analysis of this information shows that the traditional distinction of TSG and oncogene is often not sufficient to represent the complexity of cancer genetics, and that objective curation of all genetic functionality provides unique insights into how each gene can contribute differently to multiple forms of human cancer.
Different roles via distinct hallmarks
One of the most striking features observed by examining the hallmark pages is that very few genes are limited to impacting just a single hallmark of cancer. Indeed, many CGC genes have the ability to both promote and suppress cancer through their impact on multiple different hallmarks of cancer. This functional complexity originates from the well-known fact that many genes have multiple functions and are involved in numerous biological pathways with variable relevance for the development of diverse types of cancer. In consequence, different genetic alterations affecting the same gene may result in differing cellular dysfunction. Many well-known cancer genes belong to this category, contradicting the simplistic assumption implied by the traditional oncogene or TSG gene classification that only inactivating mutations are pathogenic if a gene is described as a TSG or only gain-of-function mutations are worth investigation in known oncogenes. The functional annotation of CGC genes enables identification of the genes that do not follow this simple pattern.
A good example of such gene is ATR, usually assumed to be a TSG through its core function of cell cycle checkpoint kinase and regulator of DNA damage repair, but it may also exhibit antiapoptotic activity at mitochondria in response to UV irradiation26. In the cell, ATR occurs in two isoforms: the trans-isomer is responsible for the cell cycle checkpoint pathway and DNA repair coordination in the nucleus26, while the cis-isomer localises to the cytoplasm and suppresses cytochrome C release in the mitochondria, negatively regulating apoptosis. During isomerisation (performed by PIN1), pro-survival cis-ATR is converted to the trans-isoform and relocated to the nucleus. Exposure to UV light inactivates PIN1 and thus shifts the balance between the two isoforms towards the cis-isomer, delaying apoptosis27. This apoptotic response delay, which allows the nucleotide excision repair (NER) [G] system to revert the UV-induced DNA damage, can potentially be hijacked by the cancer cell, which may be the reason for frequent overexpression and amplification of ATR in multiple cancers28.
As another example, RB129, an archetypal TSG, may in certain circumstances also support tumour development. Extensively researched and described as a negative regulator of the cell cycle, RB1 is usually inactivated by truncating mutations that lead to uncontrolled cell divisions in cancers. However, in its hyperphosphorylated form, the RB1 protein sequesters and deactivates a pro-apoptotic nuclear phosphoprotein ANP32A, inhibiting apoptosis30,31. This antiapoptotic function may explain the observation in muscle-invasive bladder cancer patients that RB1 expression is associated with higher resistance to radiotherapy32.
In contrast to ATR and RB1, which are most often described in the context of their tumour suppressing activity, the gene RAC1 is best known as a positive regulator of angiogenesis and metastasis33 as well as proliferation34, 35 in cancers. Again, in opposition to this cancer promoting role, RAC1 has also been shown to protect from UV-light-induced skin carcinogenesis as a protein necessary for the induction of effective DNA damage response36.
These few examples begin to suggest that cancer arises through complex multidirectional modulation of cellular processes associated with hallmarks of cancer rather than through simple promotion of all hallmarks. They also demonstrate that certain cellular processes important for oncogenic transformation are connected not only through global coordination of relevant pathways, but on a very basic level of one gene having multiple functions associated with these processes. It is also clear that dysfunction of the same gene can be a driving event in one type of cancer, whilst opposite or meaningless in the other. While it has been understood for some time that a gene may play context-dependent roles across cancers, it is only through large-scale deep curation, which underpins the CGC, that sheer extent of this phenomenon can be appreciated.
Mutation-specific effects
Clearly the role of any particular gene in a specific cancer type is determined by the mutations affecting that gene, as well as selective pressures on the cell carrying that mutation.
Neomorphic oncogenes are the most notable examples of how a specific genetic change may define the role of a gene in oncogenesis. Rather than suppressing or potentiating existing functions of the gene, neomorphic mutations result in a gain of a completely new function that can alter signalling and metabolic pathways within the cell and change the structure of cellular networks37.
For example, the protein encoded by PIK3CA, the PI3K catalytic subunit p110α, normally binds to p85, which stabilises it and controls its enzymatic activity. This control of PIK3CA activation is abrogated by the E545K mutation in the helical domain of the protein. This mutation enables the abnormal interaction between p110α and insulin receptor substrate 1 (IRS1), which also stabilises p110α, but in a manner independent of signals controlling p85, resulting in constitutive activation of the PI3K pathway and increased cell proliferation, survival, and motility38. Another highly recurrent PIK3CA mutation – H1047R – increases the protein activity through alteration of its catalytic site, without generating new protein functions or interactions, but also resulting in upregulation of PI3K signalling. However, these two mutants have slightly different phenotypic impact, with the helical domain mutant (E545K) giving the cells additional metastatic capacity39.
As another example, the IDH1 gene encodes a Krebs cycle enzyme, isocitrate dehydrogenase, that catalyses NADPH-dependent reversible decarboxylation of isocitrate to α-ketoglutarate. A single substitution in this gene, R132H, alters this activity. Due to the decreased affinity of this mutated enzyme to bind isocitrate and increased affinity to NADPH caused by the substitution, α-ketoglutarate is reduced to 2-hydroxyglutarate (2HG) during the catalytic cycle40. This 2HG acts as a competitive inhibitor of α-ketoglutarate-regulated enzymes, including histone demethylases and the TET family of 5-methlycytosine (5mC) hydroxylases and thus reshapes the epigenetic landscape of the cell41, and this has been shown to promote gliomas and AML42, 43. A similar effect was observed in cancer cells carrying mutations in IDH2, which occur more frequently in AML than do IDH1 mutations42.
As it is illustrated by these cases, different alterations of the same gene may have distinct impacts on oncogenesis and they may result in new physiological processes that drive cancer. This demonstrates the need for new resources that would describe the impact to oncology not just of each gene, but also each mutation. Due to the large number of data involved (currently over 5 million coding mutations are described in COSMIC), several bioinformatic approaches are ongoing. However, expert manual curation is an essential support for such broad in silico techniques as vast qualitative information is locked in the scientific literature. Future combinations of the CGC, COSMIC and in silico methods may prove the best ways to navigate genetic impact across cancer, as well as guide future therapeutic research via disease-specific functional descriptions as detailed here.
Tissue-dependent roles
Whilst the type of genetic change is an important factor determining protein dysfunction, in numerous cases it is the tissue or cell type in which the transformation occurs that also defines how a gene will drive tumour development44. Tissue specificity arises as a result of distinct gene expression, chromatin organisation, or regulation by endocrine and paracrine signalling and determines which functions provide a given cell type with a growth advantage. As this effect could be achieved through activation or inactivation of a given gene, the cell type may also determine the type of genetic alterations (i.e. gain- or loss-of-function) that are observed in that gene in cancer samples from that tissue.
A surprising number of genes described in the CGC (currently numbering 72), have been shown, when mutated, to possess either tumour promoting or suppressing activity in different tissues. This diverse impact may reflect a variety of tissue-specific microenvironmental dependencies.
Solid and haematological tumours may also involve the same gene in different ways, as evidenced by the occurrence of different mutations affecting different gene functions in different cancer types. For example, DNM2 is a TSG in T-cell-acute lymphoblastic leukaemia (T-ALL), where loss-of-function mutations in its GTPase domain inhibit endocytosis of the interleukin 7 (IL-7) receptor. The increased density of the surface IL-7 receptor restrains the differentiation of leukaemic cells, driving the disease45. Conversely, this same gene is frequently overexpressed in advanced stages of prostate cancer, where it is associated with poor prognosis46. In vitro experiments using pancreatic cancer cells have shown that DNM2 promotes invasion by stabilising VAV1, a RAC1 guanine nucleotide exchange factor that promotes RAC1 activation, leading to enhanced cancer cell migration and invasive capacity47. The importance of these two distinct functions for transformation might be determined by the differences between the two cell types: IL-7 signalling is most likely not crucial in pancreatic or prostate cell transformation, while invasion by leukaemic cells must also be controlled by mechanisms distinct to solid tumours, taking into account their innate ability to anchorage-independent survival and inherent presence within circulatory system.
Having a function that is highly dependent on the biological context particularly applies to genes acting on a broader, cellular level, for instance genes that encode proteins modifying multiple targets, leading to a global change of chromatin structure or gene expression patterns. DROSHA, which encodes a protein responsible for maturation of miRNAs48 is often inactivated in Wilms’ tumour [G] by the E1147K missense somatic mutation49. This alteration causes lower tumour-suppressing miRNA production through a dominant-negative mechanism50. As these miRNA products modulate the levels of multiple mRNAs (which vary substantially between tissues), it is very likely that in other cancers the role of DROSHA will be different. Indeed, in non-small cell lung cancer (NSCLC), DROSHA is frequently amplified, correlating with a reduced survival rate51 indicating a likely oncogenic function for this gene and its miRNA products. It should be noted that even though the dysfunction of DROSHA drives cancer, the mechanism of this process is not related to any of the “classic” hallmarks of cancer15. For that reason, on the hallmark pages we also describe a gene’s participation in cancer-driving genome-wide regulation of gene expression through changes of global epigenetic patterns and ncRNA biosynthesis52.
Similarly, ubiquitination alters the levels and activity of multiple proteins53 and its impact is also tissue-dependent. For example, mutations that inactivate the BIRC3 ubiquitin ligase induce transformation of NSCLC cells54, while in glioblastoma the upregulation of BIRC3 ubiquitin ligase activity enables escape from apoptosis and is associated with worse outcome55.
In the same way, due to the wide spectrum of changes to the cellular metabolism introduced by genes encoding regulators of hormone responses (ESR156,57, TBL1XR158,59), or epigenetic modifiers (CHD460,61, KDM6A62,63, TET164,65), the role of these genes in cancer is highly tissue-specific. An excellent example of a gene that plays roles in both hormonal and epigenetic regulation, as well as in both tumour promotion and suppression is NCOR2. In response to hormones including oestrogens, androgens, thyroid hormones, and signalling molecules such as retinoic acid66–68, NCOR2 recruits histone deacetylase 3 (HDAC3) to chromatin and promotes histone deacetylation, which changes the gene expression patterns in the cell69. Disruption of NCOR2-driven deacetylation results in global histone H4 lysine 5 (H4K5) hyperacetylation and DNA damage70, and loss of this protein has been detected in non-Hodgkin lymphoma patient samples and shown to promote transformation of immortalised fibroblasts71 and prostate cancer cells66. In contrast, in oestrogen-dependent breast cancer cells, NCOR2 activity is essential for estradiol-induced progression through the G1/S transition and its loss results in apoptosis68. NCOR2 additionally represses expression of proapoptotic genes and delays DNA damage-induced caspase activation68,72, which can positively drive oestrogen-dependent breast cancer development.
Cellular responses can be further modified by external factors, such as pathogens, leading to alteration or even inversion of cancer gene function. The NER proteins ERCC4 and ERCC5 protect the genome from damage and their germline mutations are associated with Xeroderma pigmentosum (group F and G respectively) syndromes involving an increased susceptibility to skin cancers73,74. However, in gastric epithelium infected by Helicobacter pylori the activity of these genes is directly responsible for infection-associated DNA double-strand breaks. Contact of the epithelial cells with H. pylori results in activation of the NF-κB transcription factor by bacterial cag (cytotoxin-associated gene) proteins in epithelial β1 integrin-dependent manner. Activated NF-κB binds to its specific promoters and recruits ERCC4 and ERCC5 NER endonucleases75. These two endonucleases have been implicated in transcription-associated generation of transient DNA breaks that help achieving optimal chromatin looping for the efficient transcription of RARB gene in HeLa cells76. The interaction of NF-κB constantly activated by bacterial proteins, and ERCC4 and ERCC5 stimulates the expression of antiapoptotic genes75. Additionally, multiple nicks are generated in the local DNA by ERCC4 and ERCC5, turning into DNA double-strand breaks when closely spaced. These are subsequently repaired through error-prone non-homologous end-joining75. This demonstrates the need for careful interpretation of mutated ERCC4 and ERCC5 variants in gastric cancer, since these proteins can either remove or, in the presence of H. pylori pathogens, generate somatic mutations driving gastric cancer.
Tumour stage matters
The development of many malignant solid tumours includes an initial in situ stage, before the tumour invades surrounding tissues and becomes metastatic. These phases are characterised by slightly different requirements for cellular processes: for example, increased cell motility gives limited advantage to tumour cells during initial transformation, while it is crucial for invasion. It is also becoming clear that the activity of genes protecting normal cells from oncogenic transformation may actually be beneficial for an invasive tumour, whilst genes promoting initial stages of transformation may limit metastasis. The transcription factor FOXA1 illustrates this well. Involved in estrogen and androgen signalling77, it promotes cell proliferation in multiple breast cancer78 and prostate cancer cell lines79,80 but inhibits epithelial-to-mesenchymal transition (EMT) [G] in prostate80, pancreatic81 and breast82 cancer cells. While FOXA1 hyperactivity drives the tumour growth in situ, the inhibition of EMT reduces the invasive capability of the tumour and it is likely that this must be overcome to enable metastasis.
Some TSGs may also promote invasion and metastasis when active in later stages of tumourigenesis. For instance, APC, in addition to repressing the WNT signalling pathway (limiting cell proliferation and self-renewal capacity83), can also promote cell migration in the intestinal epithelium of transgenic mice84. A direct impact of this activity on cancer metastasis hasn’t been shown yet, but relatively frequent overexpression of APC in certain cancer types85 suggests that this possibility should be further investigated. FAS, widely known as an apoptosis inducer86, has also been shown to promote metastasis via increased cell motility in gastric cancer cell lines and mouse models87 as well as cell proliferation in mouse models of ovarian cancer and liver cancer88 and cancer cell replicative immortality in breast, ovary, colon, liver, and brain cancer cell lines89.
BRCA1, a DNA damage repair and cell cycle control protein90 serves in the nucleus as a TSG. Somatic and germline mutations inactivating BRCA1 are amongst the best characterised alterations in breast and ovarian cancers90. Inactivation of BRCA1 promotes the early stages of oncogenic transformation by enabling accumulation of DNA damage and thus increasing the probability of oncogenes and TSGs acquiring somatic mutations. Mutations that disrupt or delete the C-terminal domain of BRCA1, including one of the most frequent germline mutations associated with familial breast cancer, a single nucleotide insertion resulting in a frameshift in the protein, c.5382insC (p.Q1756fs), cause sequestration of the protein in the cytoplasm, with the consequence that BRCA1 cannot perform its canonical role of DNA repair protein in the nucleus91. In addition, increased cytoplasmic levels of mutant BRCA1 can enhance the invasive and metastatic capabilities of breast cancer cells in vitro, and are associated with increased metastasis in patients with breast cancer aged over 4092. Further, the expression of a splice variant of BRCA1 lacking the C-terminal domain (BRCA1-IRIS) has been associated with more aggressive types of breast cancer93. This is an example of an alteration in the same gene, resulting in inactivation of one of its functions but upregulation of the other, with both dysfunctions driving different stages of tumour development. A similar combination of TSG and pro-metastatic activity has been described in spindle-assembly checkpoint gene BUB1B94,95. In addition to its well-described tumour-suppressing properties96, BUB1B can suppress anoikis [G]94. This enables anchorage-independent survival and growth, and thus promotes metastasis in lung adenocarcinomas94. Additionally, DDX3X97,98,99 and SPOP100,101 also act as TSGs during the initial stages of transformation but as drivers of metastasis in later stages.
Context is the key
Clearly the impact of certain hallmarks of cancer may be dramatically different depending on multiple factors across genetic alteration, tissue of origin, environmental and micro-environmental factors and even tumour stage. For instance, apoptosis is widely recognised as one of the main activities typically associated with TSGs and protecting from malignant transformation102, but apoptosis can also accelerate tumour evolution and clonal expansion by creating niches within the tumour microenvironment that can be repopulated by more aggressive sub-clones103. Cessation of initiated apoptosis may also stimulate oncogenesis. The caspase protein family are known for their role in apoptotic signalling, but if cell death is initiated and then inhibited by other factors, caspases can drive cellular transformation through stimulating cell proliferation and cleaving the DNA104. Similarly, inactivation of DNA repair is key in cancer promotion, but a certain level of functional DNA repair is necessary to enable tumour survival, a phenomenon that underlies the efficacy of PARP inhibitors in cancers with a BRCA-related DNA repair defect (synthetic lethality)13.
Easy access to broad information about the whole spectrum of gene functions via the CGC, while intended to help understand the mechanisms of oncogenesis, may also help characterise potential targets for therapy as research reveals the complexity of gene and tumour interactions. This increasing clarity, in combination with information about genetic alterations provided by COSMIC, should help identify new potential therapeutic targets, specific for cancer type, stage, and its genetic profile, and predict possible consequences of these new therapeutic approaches.
Perspectives and challenges
Understanding the biological foundations of cancer and molecular processes involved in oncogenesis is crucial for development of new diagnostic and therapeutic procedures that would allow for better prevention and treatment of the disease. Data integrated in the CGC including the examples shown in this review confirm that cancer cannot be explained exclusively by germline and somatic mutations acting on a single cell level, as extracellular factors including pathogens or the microenvironment of the tissue may modify the development of cancer. However, knowledge of which genes are active, how mutations cause dysfunction of these genes, and how this can drive the functional hallmarks of cancer, is essential for understanding the cellular changes during oncogenesis. The CGC is intended as guidance through this complex web of information, built as a systematic and exhaustive integration of all available data on the genes that have been found to have functional genetic impact across all cancer types. Strongly evidence-based using referenced literature, it defines the role of each gene in disease progression, and describes which cellular processes are affected.
Functional annotations are exclusively focused on cancer driving processes and provide three layers of information granularity. On the simplest level, annotations indicate how each gene is involved in a hallmark of cancer. This information could be used for a quick overview of gene functions, or for determining how the hallmarks are affected in a patient sample or cohort. When combined with curation of disease-specific mutation profiles (from the COSMIC database), the CGC allows for fully controlled construction of functionally related gene sets for focused functional investigations and variant interpretations. On a deeper level, the gene functions that affect each of the hallmarks as well as the impact of mutations on protein functions are concisely described to support deeper research into specific gene functions. Finally, all information is transparently referenced, and all curated details can be independently scrutinised for additional details.
Curating the functional descriptions of CGC genes is a long-term ongoing exercise and the descriptions are updated when manual curation compiles enough information to illustrate the full known spectrum of cancer-related gene functions. At the time of this review 258 of 574 Tier 1 CGC genes have been functionally characterised and this resource will be expanded and updated every 3 months.
Future development of the CGC will focus on expanding Tier 1 and Tier 2, as well as on enhancing the coverage of mutation-related dysfunction across human cancers. The great challenge for the future is to further integrate this data with information about how specific forms of mutation alter relationships across genetic interaction networks (i.e. pathways) in every tissue and how these may be affected by extracellular factors to drive disease.
Glossary.
Anoikis – a form of programmed cell death triggered in anchorage-dependent cells by detachment of the cell from the extracellular matrix.
Epithelial-to-mesenchymal transition (EMT) – a process in which epithelial cells lose cell polarity and cell-cell adhesion, accompanied by increased migratory and invasive capacities; it occurs during embryogenesis, fibrosis and wound healing, but may also be an early event in cancer metastasis.
Gain-of-function mutation – mutation resulting in an altered gene product with intensified activity or with a new biological function (neomorphic mutation).
Loss-of-function mutation – mutation resulting in an altered gene product with lower or no biological function.
Nucleotide excision repair (NER) – a DNA repair mechanism removing DNA damage induced by UV light – mostly thymine dimers – and using the complementary undamaged strand as a template to repair the damage.
Synthetic lethality – a combination of genetic and induced effects (eg, by a therapeutic) working together to induce cell death, where any single one of these effects is non-lethal.
Wilms’ tumour – another name for nephroblastoma, a malignant embryonal neoplasm of the kidney.
Table of Contents Summary.
This Review discusses the 2018 Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census (CGC), an expert-curated description of human cancer genes, which has recently been expanded to include functional descriptions of how each gene contributes to cancer.
Acknowledgments
The authors would like to thank Dr John Tate, who created the web pages presenting the functional descriptions of cancer genes. We also thank Dr Claire Rye, Nidhi Bindal, and Chris Ramshaw as well as the whole COSMIC and Open Targets teams for testing and improving these pages.
This work was supported by the Wellcome Trust grant [206194] and by the Open Targets grant [OTAR007].
Footnotes
Author contributions
Z. S. researched data for the article, substantially contributed to the discussion of content, and wrote, reviewed and edited the article. S. B., C. G. C. and S. A. W. researched data, reviewed and edited the article. I. D. substantially contributed to the discussion of content and reviewed and edited the article. S. A. F. substantially contributed to the discussion of content, and wrote, reviewed and edited the article.
Competing interests
The authors declare no competing interests.
Reviewer information
Nature Reviews Cancer thanks F. Ciccarelli, J. Korbel and the other anonymous reviewer for their contribution to the peer review of this work.
References
- 1.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [This publication describes the first version of the CGC, presenting 291 genes causally implicated in cancer and characterising their alterations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GenomicsEngland. Whole Genome Analysis – 100,000 Genomes Project Cancer Programme. 2017 https://www.genomicsengland.co.uk/information-for-gmc-staff/cancer-programme/genome-analysis/
- 3.Patel MN, Halling-Brown MD, Tym JE, Workman P, Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat Rev Drug Discov. 2013;12:35–50. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 4.Koscielny G, et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 2017;45:D985–D994. doi: 10.1093/nar/gkw1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos AH, et al. Oncotator: Cancer Variant Annotation Tool. Hum Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Eynden J, Fierro AC, Verbeke LP, Marchal K. SomInaClust: detection of cancer genes based on somatic mutation patterns of inactivation and clustering. BMC Bioinformatics. 2015;16:125. doi: 10.1186/s12859-015-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeder MP, Rubio-Perez C, Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveROLE classifies cancer driver genes in loss of function and activating mode of action. Bioinformatics. 2014;30:i549–i555. doi: 10.1093/bioinformatics/btu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 9.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes SA, et al. COSMIC: High-Resolution Cancer Genetics Using the Catalogue of Somatic Mutations in Cancer. Curr Prot Hum Genet. 2016;10.11 doi: 10.1002/cphg.21. [This publication describes COSMIC and provides protocols for access and data analysis.] [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, et al. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [This review describes alterations to genes, and signalling and metabolic pathways driving cancer, identified through whole-genome sequencing of cancer samples.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaherty KT, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-Ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin. 2011;61:31–49. doi: 10.3322/caac.20095. [This study describes the principles of PARP inhibitor-dependent synthetic lethality in BRCA-depleted cancers and its implications for cancer therapy.] [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [This review describes an improved model of the hallmarks that define cancers and malignant transformation.] [DOI] [PubMed] [Google Scholar]
- 16.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 17.Cerveira N, et al. TMPRSS2-ERG Gene Fusion Causing ERG Overexpression Precedes Chromosome Copy Number Changes in Prostate Carcinomas and Paired HGPIN Lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian E, et al. In Multiple Myeloma, 14q32 Translocations Are Non-Random Chromosomal Fusions Driving High Expression Levels of the Respective Partner Genes. Genes Chromosomes Cancer. 2014;53:549–557. doi: 10.1002/gcc.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat Struct Biol. 2002;9:117–120. doi: 10.1038/nsb747. [DOI] [PubMed] [Google Scholar]
- 20.Nakata T, Yokota T, Emi M, Minami S. Differential expression of multiple isoforms of the ELKS mRNAs involved in a papillary thyroid carcinoma. Genes Chromosomes Cancer. 2002;35:30–37. doi: 10.1002/gcc.10095. [DOI] [PubMed] [Google Scholar]
- 21.Seong KM, et al. The histone acetyltransferase Myst2 regulates Nanog expression, and is involved in maintaining pluripotency and self-renewal of embryonic stem cells. FEBS Lett. 2015;589:941–950. doi: 10.1016/j.febslet.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 22.Sauer T, et al. MYST2 acetyltransferase expression and Histone H4 Lysine acetylation are suppressed in AML. Exp Hematol. 2015;43:794–802.e794. doi: 10.1016/j.exphem.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.COSMIC. Gene Analysis: KAT7. 2018 https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=KAT7.
- 25.Jones DTW, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Awasthi P, Foiani M, Kumar A. ATM and ATR signaling at a glance. J Cell Sci. 2015;128:4255–4262. doi: 10.1242/jcs.169730. [DOI] [PubMed] [Google Scholar]
- 27.Hilton BA, et al. ATR Plays a Direct Antiapoptotic Role at Mitochondria Which Is Regulated by Prolyl Isomerase Pin1. Mol Cell. 2015;60:35–46. doi: 10.1016/j.molcel.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.COSMIC. Gene Analysis: ATR. 2018 https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=ATR#tissue.
- 29.Knudson AG. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adegbola O, Pasternack GR. Phosphorylated Retinoblastoma Protein Complexes with pp32 and Inhibits pp32-mediated Apoptosis. J Biol Chem. 2005;280:15497–15502. doi: 10.1074/jbc.M411382200. [DOI] [PubMed] [Google Scholar]
- 31.Indovina P, Pentimalli F, Casini N, Vocca I, Giordano A. RB1 dual role in proliferation and apoptosis: Cell fate control and implications for cancer therapy. Oncotarget. 2015;6:17873–17890. doi: 10.18632/oncotarget.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agerbaek M, Alsner J, Marcussen N, Lundbeck F, von der Maase H. Retinoblastoma protein expression is an independent predictor of both radiation response and survival in muscle-invasive bladder cancer. Br J Cancer. 2003;89:298–304. doi: 10.1038/sj.bjc.6601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: An Emerging Therapeutic Option for Targeting Cancer Angiogenesis and Metastasis. Mol Cancer Ther. 2013;12:1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh A, et al. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene. 2004;23:9369–9380. doi: 10.1038/sj.onc.1208182. [DOI] [PubMed] [Google Scholar]
- 35.Hofbauer SW, et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood. 2014;123:2181–2188. doi: 10.1182/blood-2013-08-523563. [DOI] [PubMed] [Google Scholar]
- 36.Deshmukh J, Pofahl R, Haase I. Epidermal Rac1 regulates the DNA damage response and protects from UV-light-induced keratinocyte apoptosis and skin carcinogenesis. Cell Death Dis. 2017;8:e2664. doi: 10.1038/cddis.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takiar V, Ip CKM, Gao M, Mills GB, Cheung LWT. Neomorphic mutations create therapeutic challenges in cancer. Oncogene. 2016;36:1607. doi: 10.1038/onc.2016.312. [This review describes the neomorphic mutations in cancer, including the best-known examples, and indicates potential therapeutic challenges associated with this class of mutations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Y, et al. Gain of interaction with IRS1 by p110α helical domain mutants is crucial for their oncogenic functions. Cancer Cell. 2013;23:583–593. doi: 10.1016/j.ccr.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang H, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA PI3K. Cancer Res. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M Gagné L, Boulay K, Topisirovic I, Huot M-É, Mallette FA. Oncogenic Activities of IDH1/2 Mutations: From Epigenetics to Cellular Signaling. Trends Cell Biol. 2017;27:738–752. doi: 10.1016/j.tcb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzymatic activity that converts α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider G, Schmidt-Supprian M, Rad R, Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17:239–253. doi: 10.1038/nrc.2017.5. [This article presents a perspective on the molecular, cellular, systemic and environmental determinants of organ-specific tumorigenesis and the mechanisms of context-specific oncogenic signalling outputs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay CS, et al. Loss-of-function mutations of Dynamin 2 promote T-ALL by enhancing IL-7 signalling. Leukemia. 2016;30:1993–2001. doi: 10.1038/leu.2016.100. [DOI] [PubMed] [Google Scholar]
- 46.Xu B, et al. The significance of dynamin 2 expression for prostate cancer progression, prognostication, and therapeutic targeting. Cancer Med. 2014;3:14–24. doi: 10.1002/cam4.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Razidlo GL, et al. Dynamin 2 Potentiates Invasive Migration of Pancreatic Tumor Cells through Stabilization of the Rac1 GEF Vav1. Dev Cell. 2013;24:573–585. doi: 10.1016/j.devcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 49.Torrezan GT, et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun. 2014;5 doi: 10.1038/ncomms5039. 4039. [This work describes somatic mutations in the miRNA processing protein DROSHA, which drives cancer through genome-wide alteration of gene expression patterns.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakheja D, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumors. Nat Commun. 2014;2:4802–4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Czubak K, et al. High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget. 2015;6:23399–23416. doi: 10.18632/oncotarget.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.COSMIC. Cancer Gene Census. Hallmarks of Cancer: DROSHA. 2018 https://cancer.sanger.ac.uk/cosmic/census-page/DROSHA.
- 53.Dwane L, Gallagher WM, Ní Chonghaile T, O'Connor DP. The Emerging Role of Non-traditional Ubiquitination in Oncogenic Pathways. J Biol Chem. 2017;292:3543–3551. doi: 10.1074/jbc.R116.755694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamato A, et al. Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor-κB-activating potential. Cancer Sci. 2015;106:1137–1142. doi: 10.1111/cas.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D, et al. BIRC3 is a novel driver of therapeutic resistance in Glioblastoma. Sci Rep. 2016;6 doi: 10.1038/srep21710. 21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations as a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu WH, et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683–693. doi: 10.1053/j.gastro.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 58.Wu X, et al. Nuclear TBLR1 as an ER corepressor promotes cell proliferation, migration and invasion in breast and ovarian cancer. Am J Cancer Res. 2016;6:2351–2360. [PMC free article] [PubMed] [Google Scholar]
- 59.Daniels G, et al. TBLR1 as an AR coactivator selectively activates AR target genes to inhibit prostate cancer growth. Endocr Relat Cancer. 2014;21:127–142. doi: 10.1530/ERC-13-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Y, et al. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene. 2014;33:2157–2168. doi: 10.1038/onc.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans. 2013;41:777–782. doi: 10.1042/BST20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benyoucef A, et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–668. doi: 10.4161/epi.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang H, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110:11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neri F, et al. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene. 2014;34 doi: 10.1038/onc.2014.356. 4168. [DOI] [PubMed] [Google Scholar]
- 66.Godoy AS, et al. Altered corepressor SMRT expression and recruitment to target genes as a mechanism that change the response to androgens in prostate cancer progression. Biochem Biophys Res Commun. 2012;423:564–570. doi: 10.1016/j.bbrc.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Privalsky ML. The Role of Corepressors in Transcriptional Regulation by Nuclear Hormone Receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 68.Blackmore JK, et al. The SMRT Coregulator Enhances Growth of Estrogen Receptor-α-Positive Breast Cancer Cells by Promotion of Cell Cycle Progression and Inhibition of Apoptosis. Endocrinology. 2014;155:3251–3261. doi: 10.1210/en.2014-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 2003;22:3403–3410. doi: 10.1093/emboj/cdg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhaskara S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song L, et al. Alteration of SMRT Tumor Suppressor Function in Transformed Non-Hodgkin Lymphomas. Cancer Res. 2005;65:4554–4561. doi: 10.1158/0008-5472.CAN-04-4108. [DOI] [PubMed] [Google Scholar]
- 72.Scafoglio C, Smolka M, Zhou H, Perissi V, Rosenfeld MG. The Co-Repressor SMRT Delays DNA Damage-Induced Caspase Activation by Repressing Pro-Apoptotic Genes and Modulating the Dynamics of Checkpoint Kinase 2 Activation. PLoS One. 2013;8:e59986. doi: 10.1371/journal.pone.0059986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene. 2015;569:153–161. doi: 10.1016/j.gene.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emmert S, Schneider TD, Khan SG, Kraemer KH. The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:1443–1452. doi: 10.1093/nar/29.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartung Mara L, et al. H. pylori-Induced DNA Strand Breaks Are Introduced by Nucleotide Excision Repair Endonucleases and Promote NF-κB Target Gene Expression. Cell Rep. 2015;13:70–79. doi: 10.1016/j.celrep.2015.08.074. [This article describes how the infection of gastric epithelium cells by Helicobacter pylori may lead to generation of potentially oncogenic DNA double-strand brakes by nucleases normally involved in DNA repair.] [DOI] [PubMed] [Google Scholar]
- 76.Le May N, Fradin D, Iltis I, Bougnères P, Egly J-M. XPG and XPF Endonucleases Trigger Chromatin Looping and DNA Demethylation for Accurate Expression of Activated Genes. Mol Cell. 2012;47:622–632. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 77.Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaguchi N, et al. FoxA1 as a lineage-specific oncogene in luminal type breast cancer. Biochem Biophys Res Commun. 2008;365:711–717. doi: 10.1016/j.bbrc.2007.11.064. [DOI] [PubMed] [Google Scholar]
- 79.Grasso CS, et al. The Mutational Landscape of Lethal Castrate Resistant Prostate Cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin H-J, Zhao JC, Ogden I, Bergan R, Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73:3725–3736. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is Essential for the Epithelial-to-Mesenchymal Transition in Pancreatic Cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bernardo GM, et al. FOXA1 Represses the Molecular Phenotype of Basal Breast Cancer Cells. Oncogene. 2013;32:554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanson CA, Miller JR. Non-traditional roles for the Adenomatous Polyposis Coli (APC) tumor suppressor protein. Gene. 2005;361:1–12. doi: 10.1016/j.gene.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 85.COSMIC. Gene Analysis: APC. 2018 https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=APC#tissue.
- 86.Nagata S. Fas ligand-induced apoptosis. Annu Rev Genet. 1999;33:29–55. doi: 10.1146/annurev.genet.33.1.29. [DOI] [PubMed] [Google Scholar]
- 87.Yang Y, et al. Fas Signaling Promotes Gastric Cancer Metastasis through STAT3-Dependent Upregulation of Fascin. PLoS One. 2015;10:e0125132. doi: 10.1371/journal.pone.0125132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L, et al. CD95/Fas promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ceppi P, et al. CD95 and CD95L promote and protect cancer stem cells. Nat Commun. 2014;5:5238–5238. doi: 10.1038/ncomms6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 91.Rodriguez JA, Au WWY, Henderson BR. Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res. 2004;293:14–21. doi: 10.1016/j.yexcr.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 92.Santivasi WL, et al. Association between cytosolic expression of BRCA1 and metastatic risk in breast cancer. Br J Cancer. 2015;113:453–459. doi: 10.1038/bjc.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shimizu Y, et al. BRCA1-IRIS Overexpression Promotes Formation of Aggressive Breast Cancers. PLOS One. 2012;7:e34102. doi: 10.1371/journal.pone.0034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen H, et al. Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma. Genes Cancer. 2015;6:106–118. doi: 10.18632/genesandcancer.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsuura S, et al. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am J Med Genet A. 2006;140A:358–367. doi: 10.1002/ajmg.a.31069. [DOI] [PubMed] [Google Scholar]
- 96.Kapanidou M, Lee S, Bolanos-Garcia VM. BubR1 kinase: protection against aneuploidy and premature aging. Trends Mol Med. 2015;21:364–372. doi: 10.1016/j.molmed.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 97.Chao C-H, et al. DDX3, a DEAD Box RNA Helicase with Tumor Growth–Suppressive Property and Transcriptional Regulation Activity of the p21waf1/cip1 Promoter, Is a Candidate Tumor Suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 98.Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2014;34 doi: 10.1038/onc.2014.190. 2790. [DOI] [PubMed] [Google Scholar]
- 99.Botlagunta M, et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27:3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gan W, et al. SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell. 2015;59:917–930. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao W, Zhou J, Deng Z, Gao Y, Cheng Y. SPOP promotes tumor progression via activation of beta-catenin/TCF4 complex in clear cell renal cell carcinoma. Int J Oncol. 2016;49:1001–1008. doi: 10.3892/ijo.2016.3609. [DOI] [PubMed] [Google Scholar]
- 102.Delbridge ARD, Valente LJ, Strasser A. The Role of the Apoptotic Machinery in Tumor Suppression. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Labi V, Erlacher M. How cell death shapes cancer. Cell Death Dis. 2015;6:e1675. doi: 10.1038/cddis.2015.20. [This review describes the role of apoptosis as an anti-cancer defence mechanism, but also as a process fuelling evolution of cancer and promoting the expansion of more aggressive sub-clones.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pérez-Garijo A. When dying is not the end: Apoptotic caspases as drivers of proliferation. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 105.Bandopadhayay P, et al. MYB-QKI rearrangements in Angiocentric Glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48:273–282. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Steidl C, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin A, et al. BRAF Alterations in Primary Glial and Glioneuronal Neoplasms of the Central Nervous System With Identification of 2 Novel KIAA1549:BRAF Fusion Variants. J Neuropathol Exp Neurol. 2012;71:66–72. doi: 10.1097/NEN.0b013e31823f2cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.COSMIC. Cancer Gene Census. Hallmarks of Cancer: PTEN. 2018 https://cancer.sanger.ac.uk/cosmic/census-page/PTEN.