Abstract
Cerebrovascular disorders are underlain by perturbations in cerebral blood flow and abnormalities in blood vessel structure. Here, we provide an overview of the current knowledge of select cerebrovascular disorders that are associated with genetic lesions and connect genomic findings with analyses aiming to elucidate the cellular and molecular mechanisms of disease pathogenesis. We argue that a mechanistic understanding of genetic (familial) forms of cerebrovascular disease is a prerequisite for the development of rational therapeutic approaches, and has wider implications for treatment of sporadic (non-familial) forms, which are usually more common.
Keywords: Cerebrovascular disease, Hemorrhagic cerebrovascular disease, Small vessel disease, Genetics, Model organisms
Introduction
Homeostatic signaling between neuronal, glial, and vascular compartments (the “neurovascular unit”) within the adult brain is of fundamental significance for normal physiological functions of the central nervous system (CNS) [1]. The importance of interactions between different cell types is indeed already evident during development, exemplified by the orchestrated interplay between neural and vascular progenitors [2, 3]. Disruptions in neurovascular development lead to diverse disorders underlain by abnormalities in blood vessel structure and perturbations in cerebral blood flow, which is critical for proper neuronal function [4–7].
In this review, we provide a brief history and summarize our current understanding of cerebrovascular diseases that have been clearly associated with underlying genetic lesions (Table 1). Rapidly evolving molecular genetic methodologies led to the identification of causative genes for a wide spectrum of cerebrovascular diseases involving both large- and small-sized vessels, offering the opportunity to investigate the underlying mechanisms using model organisms and in vitro assays [8–11]. We connect current genomic findings with analyses aiming to elucidate the cellular and molecular mechanisms of disease pathogenesis in an attempt to understand how specific genetic lesions cause diverse prototypes of cerebrovascular disorders. We argue that a mechanistic understanding of genetic (familial) cerebrovascular disease is a prerequisite for the development of rational therapeutic approaches, and has wider implications for treatment of sporadic (non-familial) forms, which are usually more common. When applicable, we discuss possible methodological improvements that could advance our understanding of these abnormalities, as well as therapeutic strategies informed by current findings.
Table 1.
Cerebrovascular diseases and associated genes
| Disease | Gene | Chromosome | Inheritance | Function | References | |||
|---|---|---|---|---|---|---|---|---|
| Hemorrhagic disease | ||||||||
| CCM | CCM1/KRIT1 | 7q | AD | Controlling endothelial barrier junctions | Laberge-le Couteulx et al. [44] | |||
| CCM2 | 7p | AD | Controlling endothelial barrier junctions |
Liquori et al. [45] Denier et al. [46] |
||||
| CCM3/PDCD10 | 3q | AD | Induction of apoptosis; broadly expressed |
Bergametti et al. [47] Guclu et al. [48] |
||||
| IA | SOX17 | 8q | – | Maintenance of arterial integrity |
Bilguvar et al. [139] Yasuno et al. [140] |
|||
| CDKN2A-B | 9p | – | Cell growth regulator |
Bilguvar et al. [139] Yasuno et al. [140] Leeper et al. [145] |
||||
| CNNM2 | 10q | – | Unknown | Yasuno et al. [140] | ||||
| KL/STARD13 | 13q | – | Unknown/cell proliferation | Yasuno et al. [140] | ||||
| RBBP8 | 18q | – | Cell cycle progression | Yasuno et al. [140] | ||||
| EDNRA | 4q | – | Vasoconstriction | Yasuno et al. [141] | ||||
| MMD | RNF213 | 17q | AD | Angiogenic response to inflammation |
Kamada et al. [159] Liu et al. [160] |
|||
| GUCY1A3 | 4q | AR | Subunit of the major NO receptor in the vascular wall | Herve et al. [169] | ||||
| BRCC3 | Xq | XR | Proper angiogenesis | Miskinyte et al. [170] | ||||
| Small vessel disease | ||||||||
| CADASIL | NOTCH3 | 19q | AD | Cell fate; stability of vascular smooth muscle cells | Joutel et al. [182] | |||
| CARASIL | HTRA1 | 10q | AR | Serine protease; regulation of TGFβ signaling | Hara et al. [183] | |||
| CAA | APP | 21q | AD | Multiple; primary function unknown | Rovelet-Lecrux et al. [245] | |||
| BRI2 | 13q | AD | Unknown | Vidal et al. [253] | ||||
| CST3 | 20p | AD | Cysteine protease inhibitor | Ghiso et al. [249] | ||||
| TTR | 18q | AD | Transportation of thyroxine and retinol-binding protein | Brett et al. [250] | ||||
| GSN | 9q | AD | Actin filament dynamics | Ghiso et al. [251] | ||||
| CRV | TREX1 | 3p | AD | Maintenance of vascular integrity | Richards et al. [184] | |||
List of diseases, associated genes with chromosomal localization, pattern of inheritance, function of encoded proteins, and main references are given
AD, autosomal dominant; APP, amyloid precursor protein, AR, autosomal recessive; BRCC3, BRCA1/BRCA2-containing complex subunit 3; BRI2, integral membrane protein 2B; CAA, cerebral amyloid angiopathy; CADASIL, cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy; CARASIL, cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy; CCMs, cerebral cavernous malformations; CDKN2A-B, cyclin-dependent kinase inhibitor 2A-B; CNNM2, cyclin M2; CRV, cerebroretinal vasculopathy; CST3, cystatin C; EDNRA, endothelin receptor type A; GSN, gelsolin; GUCY1A3, guanylate cyclase soluble subunit alpha-3; HTRA1, high-temperature requirement A serine peptidase 1; IA, intracranial aneurysms; KL/STARD13, Klotho/StAR related lipid transfer domain containing 13; KRIT1, Krev1 interaction trapped gene 1; MMD, Moyamoya disease; NOTCH3, neurogenic locus notch homolog protein 3; PDCD10, programmed cell death protein 10; RBBP8, retinoblastoma binding protein 8; RNF213, ring finger protein 213; SOX17, SRY-related HMG-box family member 17; TTR, transthyretin; TREX1, three prime repair exonuclease 1; XR, X-linked recessive
Hereditary hemorrhagic cerebrovascular disease
The first known case of a cerebrovascular lesion, a berry-like growth within the frontal lobe, was described in 1854 by von Luschka [12]. More than 70 years later, Cushing [13] and Dandy [14, 15] independently reported extensive series of cerebrovascular malformations, which were subsequently classified into distinct histopathological subtypes (telangiectasias, cerebral cavernous malformations, venous malformations, and arteriovenous malformations) [16, 17]. These occur with a high prevalence (initially estimated to be 4–6% [18–20], but approximately 3% in more recent population-based brain magnetic resonance imaging [MRI] studies [21]) around the globe in all ethnicities, and although they are most often sporadic, familial forms are not infrequent.
Cerebral cavernous malformations (CCMs)
CCMs are common cerebrovascular lesions, also termed cavernomas, in the CNS and, rarely, the retina, with an estimated prevalence of 0.3–0.9% on the basis of large prospective autopsy studies, clinically-based and population-based brain MR images [18, 21–29]. Cavernomas are collections of enlarged, densely packed vascular sinusoids with an abnormal structure lined with a single layer of endothelial cells, lack vessel wall elements, and are embedded in a collagen matrix without intervening brain parenchyma [16, 17]. They can be found in all brain regions and are usually 1–5 cm in size. Due to their primitive vessel architecture and low-pressure blood flow, CCMs tend to leak, causing microhemorrhages and subsequent hemosiderin deposition, leading to a typical MRI signature [30–32] (Fig. 1a). Although mostly clinically silent, depending on location and size, the lesions can cause a broad range of symptoms, including headache, seizures, and focal neurological deficits. A fraction of patients with CCM experience catastrophic, potentially fatal, cerebral hemorrhages [23, 25, 26, 33–35]. Treatment options include observation of asymptomatic lesions, antiepileptic medication, surgical excision of accessible lesions in patients with symptomatic hemorrhage or intractable seizures, and radiosurgery; however, pharmacological therapies that improve outcome are not available.
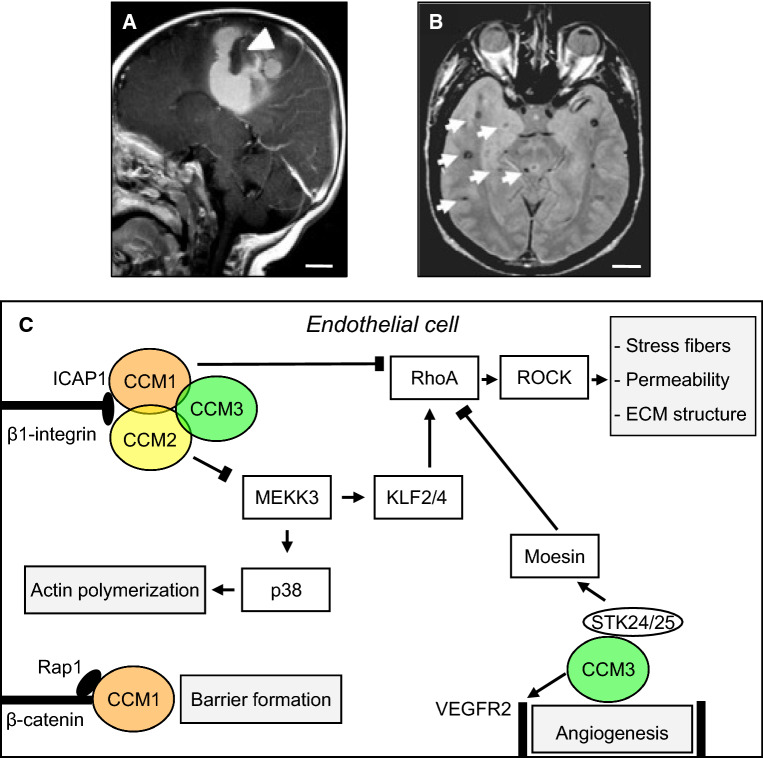
Fig. 1.
Cerebral cavernous malformations (CCMs). a Sagittal magnetic resonance (MR) imaging shows a large cavernoma (arrowhead) with a resultant gross hemorrhage. b Axial MR image of a patient with the genetic form of CCM demonstrates multiple small lesions with a characteristic hemosiderin ring surrounding them (arrows). Scale bars (a, b): 2 cm. Images courtesy of Dr. Murat Gunel (Department of Neurosurgery, Yale School of Medicine and Yale New Haven Hospital). c Summary of select interactors, intracellular pathways, and cellular processes that have been associated with the CCM proteins. The three CCM proteins maintain endothelial function and control angiogenesis. CCM1, CCM2, and CCM3 form a trimeric complex with the scaffolding protein CCM2 acting as a hub. All CCM proteins have an antagonistic effect on the RhoA/ROCK signaling pathway mediated by distinct interactors (arrow-headed lines: activation; bar-headed lines: inhibition). RhoA/ROCK pathway activation in the absence of CCM proteins results in stress fiber formation, increased vascular permeability (possibly explaining why lesions in patients tend to leak causing microhemorrhages) of CCM, and alters the composition of the extracellular matrix (ECM). CCM1 interaction with ICAP1 inhibits β1-Integrin activation and supports Rap1-mediated stabilization of endothelial junctions, thereby contribution to barrier formation. CCM2 interaction with MEKK3 results in decreased p38 activity to modulate actin polymerization. CCM3 stabilizes VEGFR2 signaling to regulate angiogenesis and negatively regulates Rho via its direct interactors STK24/25, which activate moesin. CCM: cerebral cavernous malformation. ECM: extracellular matrix. ICAP1, integrin cytoplasmic domain-associated protein 1; KLF2/4, Krüppel-like factor 2/4; MEKK3, mitogen-activated protein kinase kinase kinase 3; Rap1, Ras-associated protein 1; RhoA, Ras homolog gene family, member A; ROCK, Rho-associated coiled-coil kinase; VEGFR2, vascular endothelial growth factor receptor 2
Both sporadic (more common) and hereditary forms of CCM exist; sporadic lesions are typically solitary [36], whereas familial CCM is characterized by multiple lesions in the setting of a strong family history (Fig. 1b). The hereditary nature of CCM was recognized as early as 1928, when Kufs described a father and daughter presenting with cerebral, retinal, and cutaneous cavernous malformations [37]. Genetic linkage analyses performed in the 1990s mapped three CCM loci, and subsequent efforts identified the causative genes: CCM1/KRIT1 (on chromosome 7q), CCM2 (7p), and CCM3/PDCD10 (3q) [38–49]. Loss of function mutations in these three genes account for 98% of familial CCM disease [8, 17, 50], and although they are generally thought to lead to clinically and radiographically similar disease, familial CCM associated with CCM3 mutations is characterized by exceptionally aggressive clinical phenotypes, greater lesion burden, and more frequent hemorrhages earlier in life [51, 52]. At least some cases of sporadic CCM also involve somatic mutations in these same genes [53]. A single case involving a large genomic rearrangement was recently reported in familial CCM2 [54].
Familial CCM is characterized by autosomal-dominant mode of inheritance and high but incomplete penetrance, estimated to be 53% for CCM1, 19% for CCM2, and 16% for CCM3 [55, 56]. Knudson’s two-hit hypothesis, according to which one mutational event is inherited in the form of a germline mutation, whereas a second is acquired in a subset of somatic cells [57], has been employed to explain CCM inheritance [58–61]. The non-homologous, cytoplasmic CCM proteins (CCM1/KRIT1, CCM2, and CCM3) are expressed in the arterial endothelium in many tissues and organs and are critical for vascular development [62–65]. In addition to endothelial cells, the CCM proteins are expressed in neurons and glia [66–68], an observation potentially also related to the finding that CCM lesions form almost exclusively within the central nervous system [69]. In the setting of treatment, it is necessary to screen for the patient’s family history, as numerous families with CCM have been reported around the globe [43, 70, 71] and in various ethnic populations [71, 72]. The first study describing clinical and radiographic features for 13 Hispanic American families with CCM [72] was indeed published in 1998, accompanied in the same year by the first large series of Caucasian non-Hispanic patients with familial CCM [71]. These and other studies demonstrated that, in contrast to only 10–20% of other ethnic groups, as many as 50% of Hispanic Americans, a population with a strong founder mutation [73, 74], have a first-degree relative with CCM.
The CCM proteins have essential roles in angiogenesis and cardiovascular development and are required for endothelial cell integrity [63–65, 75–80]. In mice, conditional knockout of individual Ccm genes in endothelial cells at early postnatal stages results in acute CCM disease, characterized by vascular malformations in the retina and the cerebellum, two organs undergoing intense postnatal angiogenesis [64, 77, 81–83]. The CCM proteins are also expressed in neurons and astrocytes [66–68], where their role remains largely unknown. CCM3 appears to have critical functions not only in endothelial cells, where it has been implicated in stabilization of VEGFR2 signaling and regulation of angiopoietin 2 exocytosis [65, 84], but also in neural cells. CCM3 is required for neuronal migration and, indirectly, for cerebrovascular development, with its loss from the neural lineage leading to simplified vascularity, diffuse vessel dilation, and cerebrovascular malformations that resemble cavernomas [85, 86]. The emerging role of CCM3 in non-vascular cells implies it functions in multiple components of the neurovascular unit and may help explain why cavernomas, in humans and model organisms alike, only develop within neural context (CNS and retina), despite widespread expression of the CCM proteins in endothelial cells of most organs, where they play critical roles, as demonstrated by the impact of Ccm gene inactivation in model organisms.
The cellular and molecular mechanisms by which CCM protein loss leads to lesion formation remains an active area of investigation. Recent, occasionally conflicting, findings have contributed to our understanding of the pathogenesis of CCM, and accumulating knowledge of CCM protein interactors might help to elucidate CCM disease pathophysiology. The CCM proteins can be found in a dynamic trimeric complex, with CCM2, a scaffolding protein, acting as the hub and interacting directly with CCM1 and CCM3; however, each also interacts with a variety of other signaling, cytoskeletal, and adaptor proteins [60, 87, 88] (Fig. 1c). Several direct interactors for each of the CCM proteins have been identified, including, for CCM1, the small GTPase RAP1, the membrane anchor protein heart of glass 1 (HEG1) [89], and the integrin cytoplasmic domain-associated protein-1 (ICAP1) [90, 91]; for CCM2, MAPK/ERK kinase kinase-3 (MEKK3) (leading to p38 activation) [92, 93]; and for CCM3, the three Germinal Center Kinase III (GCKIII) serine/threonine (STE20) kinases STK24 and STK25 (via which CCM3 interacts with moesin), and MST4, as well as the scaffolding proteins paxillin and striatin (Fig. 1c) [94–100]. Of all these bona fide interactors, MEKK3 [92, 93, 101–104] has emerged as a key player in the formation and progression of CCM lesions. Gain of MEKK3 signaling and upregulation of its targets, the transcription factors KLF2 and KLF4, have been causally linked to CCM pathogenesis and onset and progression of lesions [105–107], preceding endothelial-to-mesenchymal transition underlain by changes in TGF-β/BMP signaling sensitivity [82, 108, 109]. Additional work will be needed to fully elucidate lesion formation at the cellular and molecular level.
What remains well-established, yet mechanistically not fully understood, is that CCM protein disruption leads to a common biochemical defect: activation of the small GTPase RhoA, consistent with early proteomic studies suggesting CCM involvement in cytoskeletal regulation (Fig. 1c) [101]. RhoA activation is a common outcome of CCM protein modulation in cell culture and model organisms [75, 78, 85, 86, 110–115], as well as in surgically resected familial and sporadic CCM lesions [53, 112, 116]. Although the mechanistic relationships leading to RhoA-ROCK activation are still being investigated, they have inspired attempts to explore pharmacological inhibition of RhoA-ROCK signaling as a potential therapeutic strategy for symptomatic CCM patients [111, 117]. A handful of candidate drugs with variable effects have been identified in studies involving cell culture and preclinical models: (a) fasudil, a ROCK inhibitor (approved in Japan and China but not in the US or Europe), which reduces vascular leakiness in Ccm1 and Ccm2 heterozygous mice and lesion burden in a chronic, sensitized model of CCM1, but not CCM2, disease [112, 114, 118]; (b) simvastatin, which stabilizes the endothelium of Ccm2 heterozygous mice [75], yet does not reduce lesion burden in acute (inducible) or chronic models of CCM1 or CCM2 [118, 119]; (c) cholecalciferol (vitamin D3) and tempol (scavenger of superoxide), two repurposed drugs, which decrease lesion burden in an acute model of CCM2 [119]; and (d) the anti-inflammatory drugs sulindac sulfide and sulindac sulfone, which decrease lesion burden in an acute model of CCM3 [120]. More recent findings suggested that sustained inhibition of the mevalonate pathway by inhibition of HMG-CoA reductase and prenylation (respectively, by fluvastatin and the N-bisphosphonate zoledronic acid) is effective in chronic and acute models of CCM3 [83]. It should be noted, however, that these candidate drugs have yet to be tested across all three CCM models, and thus, whether they are broadly effective, even in model organisms, remains an open question.
Intracranial aneurysms (IAs)
IAs are localized vascular bulges that typically form along cerebral arteries due to weakening of the vessel wall, which can become unstable at branch points and eventually dissect (Fig. 2a–c). Rupture of saccular IAs accounts for about 80% of cases of subarachnoid hemorrhage, and is associated with high mortality: many individuals do not survive the first few days after the event, and around 50% die within a month [121, 122]. IA is a multifactorial disorder of poorly understood etiology. Both IA-specific (size, location) and patient-related (smoking, high blood pressure, excessive alcohol consumption, female gender) risk factors are associated with aneurysm rupture [9, 123, 124]. In addition, there is a clear genetic predisposition for IA formation and rupture, first noted in the 1950s with the description of confirmed aneurysms in two members of the same family [125] and receiving increasing attention in the 1960s [126–128]. A number of systemic heritable disorders are frequently associated with IA, namely autosomal-dominant polycystic kidney disease (ADPKD), Ehlers–Danlos syndrome (EDS), Marfan syndrome (MFS), Alagille syndrome, and neurofibromatosis type 1 [129–132]. In particular, the connective-tissue disorder EDS is frequently associated with IA rupture underlain by systemic vessel wall instability. Efforts to elucidate the complex genetics of saccular IA date to 2001 [133, 134]. Although early candidate gene and small case–control studies were not particularly successful, family-based genome-wide linkage analyses identified a large number of significant, very rare variants [135–138], but still not a susceptibility gene, most likely due to complex inheritance patterns of the variants, as well as genetic heterogeneity. On the other hand, population-based, large genome-wide association studies detected numerous single-nucleotide polymorphisms (SNP) as risk factors for IA formation or rupture. These variants occur in a large number within patient cohorts, whereas each polymorphism carries just a small risk for disease development. Nonetheless, the sum of these variants may increase the disease risk dramatically. Intriguingly, the SNPs often map within genomic regions encoding proteins important for connective tissue stability or metabolic vessel function. These genomic loci, including namely SOX17, CDKN2A-CDKN2B, CNNM2, EDNRA, KL/STARD13, and RBBP8 [139–142], have been linked to other vascular disorders (e.g., EDNRA to migraine headaches [143]). In line with these findings, deletion of Sox17 or Cdkn2b in the mouse results in aneurysm-like arterial dilatations in the cerebral vasculature [144, 145].
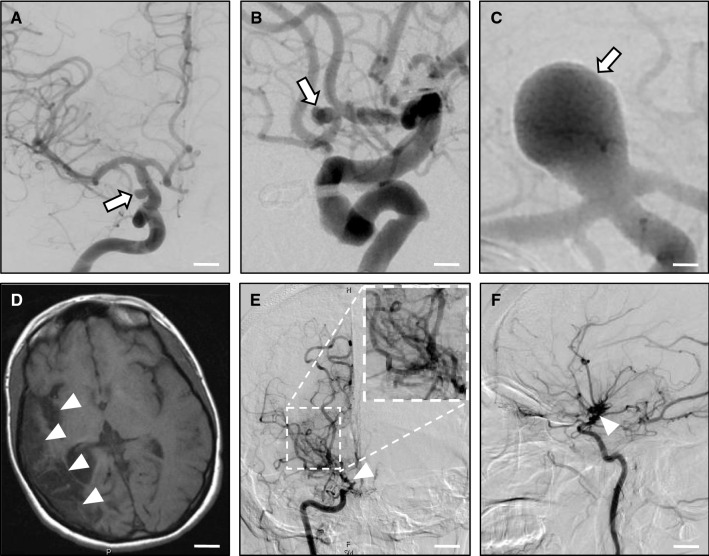
Fig. 2.
Intracranial aneurysms (IA) and Moyamoya disease (MMD). a–c Intracranial aneurysms. Computed tomography (CT) angiography studies demonstrate cerebral aneurysms (arrows) of the supraclinoid portion of the internal carotid artery (a); the anterior communicating artery (b); and the basilar artery (c). Scale bars: a 0.7 cm; b 0.5 cm; c 0.2 cm. d–f Moyamoya disease. MR imaging (d) shows right-sided encephalomalacia (loss of brain tissue after repeated strokes; arrowheads) above the middle cerebral artery, arising from stenosis/occlusion of the right internal carotid artery. Anterior–posterior (e) and lateral (f) CT angiography reveals occlusion of the right internal carotid artery (arrowhead) with a ‘puff-of-smoke’ network of collateral vessels (inlay in E). Scale bars: d 2 cm; e, f 1.3 cm. Images courtesy of Drs. Charles Matouk and Branden Cord (Department of Neurosurgery, Yale School of Medicine and Yale New Haven Hospital)
Although these findings strongly suggest a genetic basis in IA formation and rupture, no diagnostic test for studying specific genetic risk factors neither for IA development nor for IA rupture risk has been established. A larger number of IA cases will have to be studied in multi-center efforts to distinguish between background mutation in the individual [146] and aneurysm-causing aberrations, to improve our understanding of IA formation and rupture.
Moyamoya disease (MMD)
MMD is clinically characterized by repetitive ischemic strokes caused by spontaneous occlusions of the circle of Willis, a circulatory anastomosis localized at the cerebral base that supplies large parts of the brain with blood (Fig. 2d–f). MMD was first described in 1969 [147] in Japan and indicates an isolated cerebral angiopathy; syndromic Moyamoya is associated with additional conditions. Underlying MMD is a progressive stenosis within the distal internal carotid artery in the anterior circle of Willis, leading to development of numerous compensating collaterals, the so-called ‘moyamoya’ vessels (‘moyamoya’ is a Japanese mimetic word describing the ‘puff of smoke’ appearance of collateral vessels on conventional X-ray angiography). Clinical features differ considerably in pediatric and adult patients and include cerebral ischemia in children, and cerebral hemorrhage in adults (although ischemia has been reported in non-Asian adults) [148, 149].
MMD is more common in Far East countries, especially Japan and Korea, with about 16.1 in 100,000 individuals affected in endemic areas [150], where it represents the most common pediatric cerebrovascular disease, with a prevalence of 3 per 100,000 children [151]. Recently, however, the number of diagnosed patients in Europe and North America has been increasing [152]. Environmental factors were once thought to cause MMD [153], yet the personal MMD risk of individuals emigrating from high to low incidence areas [154, 155], considered together with global distribution patterns and the identification of familial forms, strongly suggest a genetic contribution. This notion is supported by findings that MMD is a systemic, rather than brain-limited, disease [156]. Familial forms of MMD appear to have variable, i.e., autosomal-dominant, autosomal-recessive, or X-linked, inheritance.
Autosomal-dominant MMD with incomplete penetrance has been linked to a locus on chromosome 17q25.3 in Japanese families, and subsequently confirmed in additional populations [157, 158]. Linkage analysis and genome-wide association studies demonstrated significant association between MMD and a variant (R4810K) of RNF213, a ring finger protein with ubiquitin ligase and ATPase activities [159–162]; this variant has not been reported in Caucasian populations yet, suggesting variability in the genetic landscape [163]. RNF213 plays a role in angiogenic and inflammatory responses of endothelial cells [160, 164]. Of interest, homozygosity for the R4810K variant leads to MMD with extracranial systemic vasculopathy [165]. Given that around 80% of Japanese MMD patients harbor susceptibility variants of RNF213, 17q25.3 is considered a major disease locus [159, 161], however, the lack of a cerebrovascular phenotype in Rnf213 knockout mice and high prevalence of RNF213 variants in healthy control populations suggest multifactorial etiology [166]. On the other hand, autosomal-recessive MMD with achalasia has been associated with mutations in GUCY1A3 (4q32.1) [167], encoding a subunit of the main receptor for nitric oxide (NO) [168]. In addition, syndromic MMD displaying X-linked recessive inheritance [169] has been associated with loss of BRCC3 (Xq28), a gene essential for angiogenesis [170]. Finally, MMD with no strictly defined pattern of inheritance has been associated with numerous other loci [171–176]. The most recently identified candidate gene, CCER2 on chromosome 19, has been proposed as a potential biomarker of MMD due to brain-specific expression [177]. Additional insight into MMD will be obtained by establishing and investigating patient-derived cell lines and by analysis of coding as well as non-coding RNA, which could also provide potential therapeutic targets [178–180].
Hereditary cerebral small vessel disease (SVD)
Cerebral SVD represent a heterogeneous group of disorders, common in the elderly, that predispose to ischemic or hemorrhagic events, often with white matter, or almost exclusively leptomeningeal, involvement. Clinically, these vascular diseases result in progressive cognitive decline [181]. A minor part of cerebral SVD, including cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) [182]; cerebral autosomal-recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL) [183]; autosomal-dominant retinal vasculopathy with cerebral leukodystrophy [184]; brain SVD with hemorrhage (Collagen 4–related disorder) [185]; familial cerebral amyloid angiopathy (CAA) [186, 187]; and cerebroretinal vasculopathy (CRV) [188], are hereditary and have been associated with specific genetic lesions. Below we review a subset of hereditary cerebral SVD, with emphasis on those most prevalent, extensively studied in patients, and modeled in the laboratory.
CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy)
In 1977, Sourander and Walinder reported a hereditary multi-infarct dementia with autosomal-dominant mode of inheritance in multiple members of a three-generation family [189, 190]. About 20 years earlier, van Bogaert had described a progressive subcortical encephalopathy in two sisters [191], which was verified much later by genetic analysis of old tissue specimens as the first family reported with this condition. Often manifesting in early adulthood, but also increasingly recognized as a disease of late onset and milder course [192–194], CADASIL is characterized by small subcortical infarcts, and underlain by arterial and arteriolar changes leading to vascular cognitive impairment and migraine with aura (Fig. 3a). The disease locus was mapped to chromosome 19q12 [195] and a few years later dominant mutations in NOTCH3 were identified in patients with CADASIL [182]. This discovery linked for the first time the Notch signaling pathway with hereditary disease. CADASIL is now recognized as the most common monogenic form of hereditary ischemic stroke. To date, more than 500 families with the disease have been identified worldwide (the majority in Europe), with more than 230 unique mutations reported (Human Genetic Mutation Database website, www.hgmd.cf.ac.uk), but a clear genotype–phenotype correlation has still to emerge [196, 197]. Mutations mapping to EGF repeats 1–6 have been associated with higher lesion load on MRI, possibly predisposing to a more severe form of the disease [198].
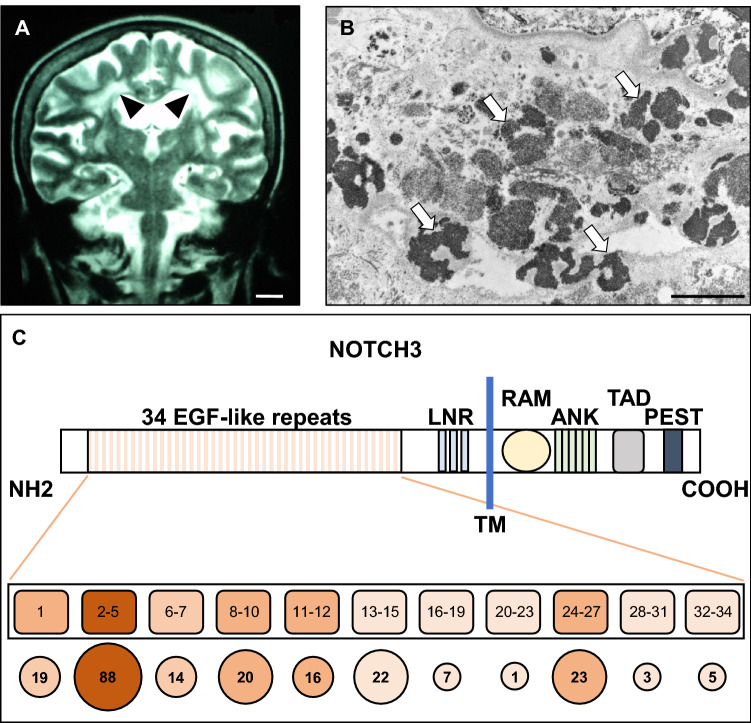
Fig. 3.
CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy). a Coronal MR imaging of a patient with the C455R mutation in NOTCH3 shows characteristic white matter lesions (arrowheads). Image courtesy of Dr. Francisco Lopera (University of Antioquia, Colombia). b Electron micrographs of subcortical white matter of postmortem brains from an individual carrying the C455R mutation shows abundant electron-dense granular osmiophilic (GOM) deposits (arrows). Scale bars: a 1.4 cm; b 2 µm. c Structure of the Notch3 receptor. A schematic of the receptor indicating key structural features. The 34 epidermal growth factor-like (EGF-like) repeats and the 3 Lin12-Notch repeats (LNR) are indicated in the extracellular domain. The transmembrane (TM), and the intracellular RAM, ankyrin repeat region (ANK), TAD, and PEST domains are also shown (top). Distribution of known CADASIL mutations along the EGF-like repeats (bottom). The relative size of circles below the schematic indicates the number of mutations reported for each cluster of EGF repeats
The overall prevalence of CADASIL is unknown (it is estimated to be 1–2/50,000 but likely remains underdiagnosed) [199, 200] and, in addition to the hereditary forms, rare sporadic cases have been reported [201, 202], as have homozygous patients with phenotypes not different from heterozygotes [203–207]. The clinical presentation varies, and includes subcortical ischemic events, cognitive impairment and dementia, migraine with aura (an initial clinical sign in 20–30% of patients), mood disturbances and apathy [192, 208]. Currently, there is no treatment of proven efficacy. The arteriopathy affects mainly the small penetrating cerebral and leptomeningeal arteries and is characterized by thickening of the arterial wall and prominent morphological alterations, and ultimately, loss of vascular smooth muscle cells and pericytes [192, 209–211]. A specific, and indeed pathognomonic, ultrastructural feature of CADASIL is the presence of granular osmiophilic material (a.k.a. GOM) (Fig. 3b) close to the cell surface of smooth muscle cells and pericytes in brain and skin arteries [210, 212–215]; these deposits are mostly extracellular and of variable morphology, size, shape, and osmiophilic density. Their detection by electron microscopy in skin biopsies represents a highly reliable diagnostic method [194, 214]. Extensive accumulation of the Notch3 extracellular domain has been reported in arterial walls, capillaries, and, possibly, GOMs [216–218]. Direct proteomic analysis of blood vessels from postmortem brains of CADASIL patients identified additional components of the GOM, including the proteins clusterin and collagen 18α1/endostatin, as well as various proteins of the extracellular matrix [219]. Further proteomic and immunohistochemical analyses identified two functionally important ECM proteins, tissue inhibitor of metalloproteinases 3 (TIMP3) and vitronectin, which are sequestered in aggregates containing the extracellular domain of Notch3 [218]. The latent TGF-β binding protein 1 (LTBP-1) is also thought to be recruited to these aggregates [220]. Finally, enrichment in brain collagen subtypes has also been reported in the arterial vessel wall of CADASIL patients [142]. The functional, structural, and cell biological consequences of extracellular Notch3 receptor CADASIL-linked mutations remain incompletely understood (for a discussion, see [194, 221]).
An extraordinary feature of CADASIL is the commonality of the disease-causing mutations in the Notch3 receptor (Fig. 3c). They are highly stereotypical, affecting exclusively the extracellular domain and occur in exons 2–24, encoding the EGF-like repeats, with a strong clustering in exons 3 and 4 [222]. More than 95% of the mutations are missense, whereas the remaining are small in-frame deletions or splice site mutations. Importantly, the vast majority of mutations lead to an odd number of cysteine residues within the affected EGF-like repeat; rare cysteine-sparing mutations have been reported in patients with clinical picture considered to be compatible with CADASIL (e.g., [223, 224]). However, the causal relationship between disease and these variants remains a matter of debate [194]. Perhaps surprisingly, given that the mutations were identified more than 20 years ago, the jury is still out, despite considerable effort from many groups using in vitro assays, as well as analyses in model organisms [219, 225–230]. Attaining a molecular understanding of CADASIL pathophysiology is essential if we are to contemplate a rational therapeutic approach to this devastating disease. In particular, determining the nature of the CADASIL-linked mutations is essential as gain or loss of function abnormalities would be treated radically differently. The extraordinary pleiotropy of Notch, including a profound involvement in the biology of adult stem cells, presents also a great challenge, as modulating its activity for therapeutic purposes will be highly dependent on the cellular context and quantitative aspects of signal modulation, carrying thus implicitly the danger of unacceptable toxicities.
CARASIL (cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy)
CARASIL is an ischemic, non-hypertensive, very rare cerebral SVD, with the majority of the approximately 50 known cases reported in Japan [194, 231]. Clinical signs include a stepwise deterioration in brain functions and progressive dementia; the disease is characterized by diffuse white matter changes and multiple lacunar infarctions in the basal ganglia and thalamus. Histological findings include intense atherosclerosis affecting mainly small penetrating arteries; GOM or amyloid deposits are however absent. Biallelic mutations in HTRA1, encoding a serine protease implicated in negative regulation of TGFβ signaling, have been associated with CARASIL [183, 232]; interestingly, heterozygous HTRA1 mutations have also been reported in a family with autosomal-dominant SVD with clinical features distinct from CARASIL and CADASIL [233].
Cerebral amyloid angiopathy (CAA)
CAA, a common clinical entity in elderly patients, is characterized by accumulation of amyloid fibrils, cleavage products of the Amyloid Precursor Protein (APP), within the wall of small- to medium-sized vessels of the brain and leptomeninges [186, 187]. CAA is a major cause of primary intracerebral hemorrhage leading in most cases to cognitive decline, dementia, or death [234–236]. Notably, these symptoms also characterize Alzheimer’s disease (AD), often confounding diagnosis. AD and CAA can be thought as related diseases with overlapping pathogenesis. It is indeed true that most patients with AD have some degree of CAA [237, 238]; however, the clinical features of CAA and AD differ. CAA, being a vascular disorder, is mainly characterized by small intracranial hemorrhages leading to stepwise cognitive decline. AD presents with progressive cognitive decline leading to terminal dementia [239, 240]. Amyloid beta (Aβ) aggregates accumulate in the parenchyma in the form of neuritic plaques in AD pathology, whereas in CAA they form in the walls of small arteries and arterioles.
CAA and its clinical complications are typically prevalent in the elderly: while postmortem series revealed at least moderate CAA in 2.3% of autopsies performed at age 65–74, about 12% of individuals older than 85 years harbored CAA [241], suggesting a significant age-dependent increase in disease prevalence. Overall, most of Aβ-CAA cases are sporadic, although genetic risk factors exist. Of interest, genetic variants implicated in AD, e.g., apolipoprotein E, presenilin 1, α-1 antichymotrypsin, and neprilysin, are also associated with increased risk for CAA [242]. As in AD [243], a high frequency of the ApoE ε4 allele correlates with the severity of CAA [244]. Consequently, CAA and AD do not only share a similar fingerprint regarding their characteristic clinical symptoms, but also similar genetic underpinnings.
In addition to sporadic cases, numerous hereditary forms of CAA have been reported. The pedigrees of the affected families reveal frequently an autosomal-dominant pattern of inheritance, involving APP on chromosome 21 [245]. Mutations mapping close to the cleavage sites of APP result in early-onset AD, whereas those affecting residues 21-23 and 34 of the Aβ peptide are associated with a broad spectrum of CNS diseases, including CAA [246]. Hereditary CAAs are sub-categorized according to their geographical area of prevalence, e.g., the Italian (E693K), Flemish (A692G), and Arctic (E693G) variants [236]. Neuropathological outcomes extend from increased Aβ production to abundant neurofibrillar appearance, with all eventually resulting in Aβ deposits [247, 248]. Although more rarely, CAA can also be characterized (and neuropathologically verified) by depositions of cystatin C (HCHWA-I), diverse transthyretins, or gesolin fragments, which are all associated with mutations in the genes encoding the respective proteins [249–251]. Non-Aβ phenotypes are also most often inherited in an autosomal-dominant pattern. Protein accumulations within the vessel wall represent only the first step in CAA pathogenesis, and are often accompanied by loss of smooth muscle cells, obliterative intimal changes, as well as microaneurysmal dilatations [236]. Animal models for CAA [252, 253] have yet to provide significant insight into the exact mechanisms leading to these structural changes.
Cerebroretinal vasculopathies (CRVs)
CRVs are a collection of severe autosomal-dominant disorders with middle-age onset and 100% penetrance presenting with vasculopathy that predominantly involves the white matter and the retina [188], caused by C-terminal truncating mutations in TREX1, which encodes the 3’ to 5′ exonuclease DNAse III [184]. The truncating mutations do not interfere with enzymatic activity, but disrupt subcellular localization; however, the mechanism(s) by which these mutations lead to microvascular endotheliopathy remain unknown [254].
Conclusion
Many cerebrovascular disorders are associated with genetic lesions; for some, such as CADASIL, a clear monogenic link has been established, while for others, an interplay of genetic susceptibility with environmental and other risk factors contributes to pathogenetic mechanisms leading to disruptions of the cerebral vasculature. Gene discovery in cerebrovascular disease, made possible by advances in genetics and genomics, not only refines our understanding of the basic biological mechanisms governing the development and homeostasis of the neurovascular unit, but has the potential to inform strategies for the development of rational therapeutic approaches. Expanding our understanding of pathophysiology in the context of familial forms, should also help elucidate the mechanisms underlying sporadic forms of disease.
Acknowledgements
We thank Drs. Murat Gunel, Charles Matouk, Branden Cord (Department of Neurosurgery, Yale School of Medicine) and Francisco Lopera (University of Antioquia, Colombia) for sharing MR and CT-angiography images and the two anonymous reviewers for suggestions. P.K. was supported by the German Academic Scholarship Foundation. Work in the Louvi laboratory is supported by the National Institutes of Health (NIH).
References
- 1.Lo EH, Dalkara T, Moskowitz MA. Mechanisms, challenges and opportunities in stroke. Nat Rev Neurosci. 2003;4(5):399–415. doi: 10.1038/nrn1106. [DOI] [PubMed] [Google Scholar]
- 2.Walchli T, Wacker A, Frei K, Regli L, Schwab ME, Hoerstrup SP, Gerhardt H, Engelhardt B. Wiring the Vascular Network with Neural Cues: a CNS Perspective. Neuron. 2015;87(2):271–296. doi: 10.1016/j.neuron.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Eichmann A, Thomas JL. Molecular parallels between neural and vascular development. Cold Spring Harb Perspect Med. 2013;3(1):a006551. doi: 10.1101/cshperspect.a006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attwell D, Buchan AM, Charpak S, Lauritzen M, Macvicar BA, Newman EA. Glial and neuronal control of brain blood flow. Nature. 2010;468(7321):232–243. doi: 10.1038/nature09613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moskowitz MA, Lo EH, Iadecola C. The science of stroke: mechanisms in search of treatments. Neuron. 2010;67(2):181–198. doi: 10.1016/j.neuron.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iadecola C. The pathobiology of vascular dementia. Neuron. 2013;80(4):844–866. doi: 10.1016/j.neuron.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisler K, Nelson AR, Montagne A, Zlokovic BV. Cerebral blood flow regulation and neurovascular dysfunction in Alzheimer disease. Nat Rev Neurosci. 2017;18(7):419–434. doi: 10.1038/nrn.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louvi, A. and Gunel, M., Genetics of Cerebral Cavernous Malformations, in Youmans and Winn Neurological Surgery H.R. Winn, Editor. 2017, Elsevier. p. 3547-3553
- 9.Barak T, Cheng Y, Youngblood MW, Yasuno K, Gunel M. Genetics of intracranial aneurysms. In: Winn HR, editor. Youmans and Winn neurological surgery. Amsterdam: Elsevier; 2018. pp. 3198–3206. [Google Scholar]
- 10.NINDS Stroke Genetics Network (SiGN), International Stroke Genetics Consortium (ISGC) Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 2016;15(2):174–184. doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran D, Karschnia P, Gaillard JR, Karimy JK, Youngblood MW, DiLuna ML, Matouk CC, Aagaard-Kienitz B, Smith ER, Orbach DB, Rodesch G, Berenstein A, Gunel M, Kahle KT. Human genetics and molecular mechanisms of vein of Galen malformation. J Neurosurg Pediatr. 2018;21:1–8. doi: 10.3171/2017.9.PEDS17365. [DOI] [PubMed] [Google Scholar]
- 12.Luschka H. Cavernose Blutgeschwulste des Gehirns. Virch Arch Path Anat. 1854;6(4):457–470. [Google Scholar]
- 13.Cushing H, Bailey P. Tumors arising from the blood vessels of the brain: angiomatous malformations and hemangioblastomas. Springfield: Thomas, C.C.; 1928. [Google Scholar]
- 14.Dandy WE. Arteriovenous aneurysm of the brain. Arch Surg. 1928;17:190–243. doi: 10.1001/archsurg.1928.01140080014002. [DOI] [Google Scholar]
- 15.Dandy WE. Venous abnormalities and angiomas of the brain. Arch Surg. 1928;17:715–793. doi: 10.1001/archsurg.1928.01140110002001. [DOI] [Google Scholar]
- 16.McCormick WF. The pathology of vascular (“arteriovenous”) malformations. J Neurosurg. 1966;24(4):807–816. doi: 10.3171/jns.1966.24.4.0807. [DOI] [PubMed] [Google Scholar]
- 17.Russell DS, Rubenstein LJ. Pathology of tumors of the nervous system. Baltimore: Williams and Wilkins; 1989. [Google Scholar]
- 18.McCormick WF (1984) Pathology of vascular malformations of the brain in Intracranial Vascular Malformations In: Wilson CB, Stein BM (eds), Williams & Wilkins, Baltimore, pp 44–63
- 19.Demick DA. Cerebrovascular malformation causing sudden death. Analysis of three cases and review of the literature. Am J Forensic Med Pathol. 1991;12(1):45–49. doi: 10.1097/00000433-199103000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Berenstein A, Lasjaunias P, ter Brugge KG (2004) Cerebral vascular malformations: incidence, classification, angioarchitecture, and symptomatology of brain arteriovenous and venous malformations, in surgical neuroangiography. Springer: Berlin, Heidelberg, pp 609–694
- 21.Bos D, Poels MM, Adams HH, Akoudad S, Cremers LG, Zonneveld HI, Hoogendam YY, Verhaaren BF, Verlinden VJ, Verbruggen JG, Peymani A, Hofman A, Krestin GP, Vincent AJ, Feelders RA, Koudstaal PJ, van der Lugt A, Ikram MA, Vernooij MW. Prevalence, clinical management, and natural course of incidental findings on brain mr images: the population-based rotterdam scan study. Radiology. 2016;281(2):507–515. doi: 10.1148/radiol.2016160218. [DOI] [PubMed] [Google Scholar]
- 22.Berry RG, Alpers BJ, White JC. The site, structure and frequency of intracranial aneurysms, angiomas and arteriovenous abnormalities. Res Publ Assoc Res Nerv Ment Dis. 1966;41:40–72. [PubMed] [Google Scholar]
- 23.Otten P, Pizzolato GP, Rilliet B, Berney J. 131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies. Neurochirurgie. 1989;35(2):82–83. [PubMed] [Google Scholar]
- 24.Del Jr Curling O, Jr Kelly DL, Elster AD, Craven TE. An analysis of the natural history of cavernous angiomas. J Neurosurg. 1991;75(5):702–708. doi: 10.3171/jns.1991.75.5.0702. [DOI] [PubMed] [Google Scholar]
- 25.Robinson JR, Awad IA, Little JR. Natural history of the cavernous angioma. J Neurosurg. 1991;75(5):709–714. doi: 10.3171/jns.1991.75.5.0709. [DOI] [PubMed] [Google Scholar]
- 26.Sage MR, Brophy BP, Sweeney C, Phipps S, Perrett LV, Sandhu A, Albertyn LE. Cavernous haemangiomas (angiomas) of the brain: clinically significant lesions. Australas Radiol. 1993;37(2):147–155. doi: 10.1111/j.1440-1673.1993.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 27.Vernooij MW, Ikram MA, Tanghe HL, Vincent AJ, Hofman A, Krestin GP, Niessen WJ, Breteler MM, van der Lugt A. Incidental findings on brain MRI in the general population. N Engl J Med. 2007;357(18):1821–1828. doi: 10.1056/NEJMoa070972. [DOI] [PubMed] [Google Scholar]
- 28.Morris Z, Whiteley WN, Longstreth WT, Jr, Weber F, Lee YC, Tsushima Y, Alphs H, Ladd SC, Warlow C, Wardlaw JM, Al-Shahi Salman R. Incidental findings on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ. 2009;339:b3016. doi: 10.1136/bmj.b3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flemming KD, Graff-Radford J, Aakre J, Kantarci K, Lanzino G, Brown RD, Jr, Mielke MM, Roberts RO, Kremers W, Knopman DS, Petersen RC, Jack CR., Jr Population-based prevalence of cerebral cavernous malformations in older adults: mayo clinic study of aging. JAMA Neurol. 2017;74(7):801–805. doi: 10.1001/jamaneurol.2017.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell PG, Jabbour P, Yadla S, Awad IA. Emerging clinical imaging techniques for cerebral cavernous malformations: a systematic review. Neurosurg Focus. 2010;29(3):E6. doi: 10.3171/2010.5.FOCUS10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart BL, Taheri S, Rosenberg GA, Morrison LA. Dynamic contrast-enhanced MRI evaluation of cerebral cavernous malformations. Transl Stroke Res. 2013;4(5):500–506. doi: 10.1007/s12975-013-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan H, Liu T, Wu Y, Thacker J, Shenkar R, Mikati AG, Shi C, Dykstra C, Wang Y, Prasad PV, Edelman RR, Awad IA. Evaluation of iron content in human cerebral cavernous malformation using quantitative susceptibility mapping. Invest Radiol. 2014;49(7):498–504. doi: 10.1097/RLI.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moultrie F, Horne MA, Josephson CB, Hall JM, Counsell CE, Bhattacharya JJ, Papanastassiou V, Sellar RJ, Warlow CP, Murray GD, Al-Shahi Salman R, Scottish Audit of Intracranial Vascular Malformations steering, c., and collaborators Outcome after surgical or conservative management of cerebral cavernous malformations. Neurology. 2014;83(7):582–589. doi: 10.1212/WNL.0000000000000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zabramski JM, Wascher TM, Spetzler RF, Johnson B, Golfinos J, Drayer BP, Brown B, Rigamonti D, Brown G. The natural history of familial cavernous malformations: results of an ongoing study. J Neurosurg. 1994;80(3):422–432. doi: 10.3171/jns.1994.80.3.0422. [DOI] [PubMed] [Google Scholar]
- 35.Zabramski JM, Kalani YS. Natural history of cavernous malformations. In: Winn HR, editor. Youmans and Winn neurological surgery. Amsterdam: Elsevier; 2017. pp. 3537–3546. [Google Scholar]
- 36.Siegel AM, Andermann E, Badhwar A, Rouleau GA, Wolford GL, Andermann F, Hess K. Anticipation in familial cavernous angioma: a study of 52 families from International Familial Cavernous Angioma Study. IFCAS Group. Lancet. 1998;352(9141):1676–1677. doi: 10.1016/S0140-6736(05)61447-X. [DOI] [PubMed] [Google Scholar]
- 37.Kufs H. Uber die heredofamilare Angiomatose des Gehirns und der Retina, ihre Beziehingen zueinander und sur Angiomatose der Haut. Z Neurol Psychiatrie. 1928;113:651–686. doi: 10.1007/BF02884519. [DOI] [Google Scholar]
- 38.Dubovsky J, Zabramski JM, Kurth J, Spetzler RF, Rich SS, Orr HT, Weber JL. A gene responsible for cavernous malformations of the brain maps to chromosome 7q. Hum Mol Genet. 1995;4(3):453–458. doi: 10.1093/hmg/4.3.453. [DOI] [PubMed] [Google Scholar]
- 39.Gunel M, Awad IA, Anson J, Lifton RP. Mapping a gene causing cerebral cavernous malformation to 7q11.2-q21. Proc Natl Acad Sci USA. 1995;92(14):6620–6624. doi: 10.1073/pnas.92.14.6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson EW, Iyer LM, Rich SS, Orr HT, Gil-Nagel A, Kurth JH, Zabramski JM, Marchuk DA, Weissenbach J, Clericuzio CL, Davis LE, Hart BL, Gusella JF, Kosofsky BE, Louis DN, Morrison LA, Green ED, Weber JL. Refined localization of the cerebral cavernous malformation gene (CCM1) to a 4-cM interval of chromosome 7q contained in a well-defined YAC contig. Genome Res. 1995;5(4):368–380. doi: 10.1101/gr.5.4.368. [DOI] [PubMed] [Google Scholar]
- 41.Gil-Nagel A, Dubovsky J, Wilcox KJ, Stewart JM, Anderson VE, Leppik IE, Orr HT, Johnson EW, Weber JL, Rich SS. Familial cerebral cavernous angioma: a gene localized to a 15-cM interval on chromosome 7q. Ann Neurol. 1996;39(6):807–810. doi: 10.1002/ana.410390619. [DOI] [PubMed] [Google Scholar]
- 42.Notelet L, Chapon F, Khoury S, Vahedi K, Chodkiewicz JP, Courtheoux P, Iba-Zizen MT, Cabanis EA, Lechevalier B, Tournier-Lasserve E, Houtteville JP. Familial cavernous malformations in a large French kindred: mapping of the gene to the CCM1 locus on chromosome 7q. J Neurol Neurosurg Psychiatry. 1997;63(1):40–45. doi: 10.1136/jnnp.63.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig HD, Gunel M, Cepeda O, Johnson EW, Ptacek L, Steinberg GK, Ogilvy CS, Berg MJ, Crawford SC, Scott RM, Steichen-Gersdorf E, Sabroe R, Kennedy CT, Mettler G, Beis MJ, Fryer A, Awad IA, Lifton RP. Multilocus linkage identifies two new loci for a mendelian form of stroke, cerebral cavernous malformation, at 7p15-13 and 3q25.2-27. Hum Mol Genet. 1998;7(12):1851–1858. doi: 10.1093/hmg/7.12.1851. [DOI] [PubMed] [Google Scholar]
- 44.Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF, Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23(2):189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 45.Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, Plummer NW, Cannella M, Maglione V, Squitieri F, Johnson EW, Rouleau GA, Ptacek L, Marchuk DA. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73(6):1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, Houtteville JP, Jan M, Lapierre F, Loiseau H, Menei P, Mercier P, Moreau JJ, Nivelon-Chevallier A, Parker F, Redondo AM, Scarabin JM, Tremoulet M, Zerah M, Maciazek J, Tournier-Lasserve E. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet. 2004;74(2):326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M, Tournier-Lasserve E. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76(1):42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guclu B, Ozturk AK, Pricola KL, Bilguvar K, Shin D, O’Roak BJ, Gunel M. Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery. 2005;57(5):1008–1013. doi: 10.1227/01.NEU.0000180811.56157.E1. [DOI] [PubMed] [Google Scholar]
- 49.Liquori CL, Berg MJ, Squitieri F, Ottenbacher M, Sorlie M, Leedom TP, Cannella M, Maglione V, Ptacek L, Johnson EW, Marchuk DA. Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat. 2006;27(1):118. doi: 10.1002/humu.9389. [DOI] [PubMed] [Google Scholar]
- 50.Riant F, Bergametti F, Ayrignac X, Boulday G, Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277(5):1070–1075. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- 51.Riant F, Bergametti F, Fournier HD, Chapon F, Michalak-Provost S, Cecillon M, Lejeune P, Hosseini H, Choe C, Orth M, Bernreuther C, Boulday G, Denier C, Labauge P, Tournier-Lasserve E. CCM3 mutations are associated with early-onset cerebral hemorrhage and multiple meningiomas. Mol Syndromol. 2013;4(4):165–172. doi: 10.1159/000350042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shenkar R, Shi C, Rebeiz T, Stockton RA, McDonald DA, Mikati AG, Zhang L, Austin C, Akers AL, Gallione CJ, Rorrer A, Gunel M, Min W, De Souza JM, Lee C, Marchuk DA, Awad IA. Exceptional aggressiveness of cerebral cavernous malformation disease associated with PDCD10 mutations. Genet Med. 2015;17(3):188–196. doi: 10.1038/gim.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald DA, Shi C, Shenkar R, Gallione CJ, Akers AL, Li S, De Castro N, Berg MJ, Corcoran DL, Awad IA, Marchuk DA. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Hum Mol Genet. 2014;23(16):4357–4370. doi: 10.1093/hmg/ddu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spiegler S, Rath M, Hoffjan S, Dammann P, Sure U, Pagenstecher A, Strom T, Felbor U. First large genomic inversion in familial cerebral cavernous malformation identified by whole genome sequencing. Neurogenetics. 2017;19:55–59. doi: 10.1007/s10048-017-0531-7. [DOI] [PubMed] [Google Scholar]
- 55.Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, Maciazek J, Vicaut E, Brunereau L, Tournier-Lasserve E, Societe Francaise de N Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60(5):550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 56.Choquet H, Pawlikowska L, Lawton MT, Kim H. Genetics of cerebral cavernous malformations: current status and future prospects. J Neurosurg Sci. 2015;59(3):211–220. [PMC free article] [PubMed] [Google Scholar]
- 57.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gault J, Awad IA, Recksiek P, Shenkar R, Breeze R, Handler M, Kleinschmidt-DeMasters BK. Cerebral cavernous malformations: somatic mutations in vascular endothelial cells. Neurosurgery. 2009;65(1):138–144. doi: 10.1227/01.NEU.0000348049.81121.C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gault J, Shenkar R, Recksiek P, Awad IA. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36(4):872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- 60.Draheim KM, Fisher OS, Boggon TJ, Calderwood DA. Cerebral cavernous malformation proteins at a glance. J Cell Sci. 2014;127(Pt 4):701–707. doi: 10.1242/jcs.138388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18(5):919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, Mayo AH, Drakos SG, Marchuk DA, Davis GE, Li DY. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15(2):177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131(6):1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 64.Boulday G, Blecon A, Petit N, Chareyre F, Garcia LA, Niwa-Kawakita M, Giovannini M, Tournier-Lasserve E. Tissue-specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis Model Mech. 2009;2(3–4):168–177. doi: 10.1242/dmm.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Y, Zhang H, Yu L, Gunel M, Boggon TJ, Chen H, Min W. Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci Signal. 2010;3(116):ra26. doi: 10.1126/scisignal.2000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guzeloglu-Kayisli O, Kayisli UA, Amankulor NM, Voorhees JR, Gokce O, DiLuna ML, Laurans MS, Luleci G, Gunel M. Krev1 interaction trapped-1/cerebral cavernous malformation-1 protein expression during early angiogenesis. J Neurosurg. 2004;100(5 Suppl Pediatrics):481–487. doi: 10.3171/ped.2004.100.5.0481. [DOI] [PubMed] [Google Scholar]
- 67.Seker A, Pricola KL, Guclu B, Ozturk AK, Louvi A, Gunel M. CCM2 expression parallels that of CCM1. Stroke. 2006;37(2):518–523. doi: 10.1161/01.STR.0000198835.49387.25. [DOI] [PubMed] [Google Scholar]
- 68.Tanriover G, Boylan AJ, Diluna ML, Pricola KL, Louvi A, Gunel M. PDCD10, the gene mutated in cerebral cavernous malformation 3, is expressed in the neurovascular unit. Neurosurgery. 2008;62(4):930–938. doi: 10.1227/01.neu.0000318179.02912.ca. [DOI] [PubMed] [Google Scholar]
- 69.Plummer NW, Squire TL, Srinivasan S, Huang E, Zawistowski JS, Matsunami H, Hale LP, Marchuk DA. Neuronal expression of the Ccm2 gene in a new mouse model of cerebral cavernous malformations. Mamm Genome. 2006;17(2):119–128. doi: 10.1007/s00335-005-0098-8. [DOI] [PubMed] [Google Scholar]
- 70.Gunel M, Awad IA, Finberg K, Steinberg GK, Craig HD, Cepeda O, Nelson-Williams C, Lifton RP. Genetic heterogeneity of inherited cerebral cavernous malformation. Neurosurgery. 1996;38(6):1265–1271. doi: 10.1097/00006123-199606000-00059. [DOI] [PubMed] [Google Scholar]
- 71.Labauge P, Laberge S, Brunereau L, Levy C, Tournier-Lasserve E. Hereditary cerebral cavernous angiomas: clinical and genetic features in 57 French families. Societe Francaise de Neurochirurgie. Lancet. 1998;352(9144):1892–1897. doi: 10.1016/S0140-6736(98)03011-6. [DOI] [PubMed] [Google Scholar]
- 72.Rigamonti D, Hadley MN, Drayer BP, Johnson PC, Hoenig-Rigamonti K, Knight JT, Spetzler RF. Cerebral cavernous malformations. Incidence and familial occurrence. N Engl J Med. 1988;319(6):343–347. doi: 10.1056/NEJM198808113190605. [DOI] [PubMed] [Google Scholar]
- 73.Hayman LA, Evans RA, Ferrell RE, Fahr LM, Ostrow P, Riccardi VM. Familial cavernous angiomas: natural history and genetic study over a 5-year period. Am J Med Genet. 1982;11(2):147–160. doi: 10.1002/ajmg.1320110205. [DOI] [PubMed] [Google Scholar]
- 74.Gunel M, Awad IA, Finberg K, Anson JA, Steinberg GK, Batjer HH, Kopitnik TA, Morrison L, Giannotta SL, Nelson-Williams C, Lifton RP. A founder mutation as a cause of cerebral cavernous malformation in Hispanic Americans. N Engl J Med. 1996;334(15):946–951. doi: 10.1056/NEJM199604113341503. [DOI] [PubMed] [Google Scholar]
- 75.Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, Mayo AH, Drakos SG, Jones CA, Zhu W, Marchuk DA, Davis GE, Li DY. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15(2):177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, Chen M, Guo L, Lu MM, Zhou D, Kitajewski J, Affolter M, Ginsberg MH, Kahn ML. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15(2):169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan AC, Drakos SG, Ruiz OE, Smith AC, Gibson CC, Ling J, Passi SF, Stratman AN, Sacharidou A, Revelo MP, Grossmann AH, Diakos NA, Davis GE, Metzstein MM, Whitehead KJ, Li DY. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J Clin Invest. 2011;121(5):1871–1881. doi: 10.1172/JCI44393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, Seiler C, Chen M, Cheng L, Xiao J, He J, Pack MA, Sessa WC, Kahn ML. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120(8):2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoruk B, Gillers BS, Chi NC, Scott IC. Ccm3 functions in a manner distinct from Ccm1 and Ccm2 in a zebrafish model of CCM vascular disease. Dev Biol. 2012;362(2):121–131. doi: 10.1016/j.ydbio.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 80.Lisowska J, Rodel CJ, Manet S, Miroshnikova YA, Boyault C, Planus E, De Mets R, Lee HH, Destaing O, Mertani H, Boulday G, Tournier-Lasserve E, Balland M, Abdelilah-Seyfried S, Albiges-Rizo C, Faurobert E. Cerebral Cavernous Malformation 1/2 complex controls ROCK1 and ROCK2 complementary functions for endothelial integrity. J Cell Sci. 2018;1:ics-216093. doi: 10.1242/jcs.216093. [DOI] [PubMed] [Google Scholar]
- 81.Cunningham K, Uchida Y, O’Donnell E, Claudio E, Li W, Soneji K, Wang H, Mukouyama YS, Siebenlist U. Conditional deletion of Ccm2 causes hemorrhage in the adult brain: a mouse model of human cerebral cavernous malformations. Hum Mol Genet. 2011;20(16):3198–3206. doi: 10.1093/hmg/ddr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, Tournier-Lasserve E, Chapon F, Richichi C, Retta SF, Lampugnani MG, Dejana E. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498(7455):492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 83.Nishimura S, Mishra-Gorur K, Park J, Surovtseva YV, Sebti SM, Levchenko A, Louvi A, Gunel M. Combined HMG-COA reductase and prenylation inhibition in treatment of CCM. Proc Natl Acad Sci USA. 2017;114(21):5503–5508. doi: 10.1073/pnas.1702942114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou HJ, Qin L, Zhang H, Tang W, Ji W, He Y, Liang X, Wang Z, Yuan Q, Vortmeyer A, Toomre D, Fuh G, Yan M, Kluger MS, Wu D, Min W. Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nat Med. 2016;22(9):1033–1042. doi: 10.1038/nm.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Louvi A, Chen L, Two AM, Zhang H, Min W, Gunel M. Loss of cerebral cavernous malformation 3 (Ccm3) in neuroglia leads to CCM and vascular pathology. Proc Natl Acad Sci U S A. 2011;108(9):3737–3742. doi: 10.1073/pnas.1012617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Louvi A, Nishimura S, Gunel M. Ccm3, a gene associated with cerebral cavernous malformations, is required for neuronal migration. Development. 2014;141(6):1404–1415. doi: 10.1242/dev.093526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fisher OS, Zhang R, Li X, Murphy JW, Demeler B, Boggon TJ. Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus. FEBS Lett. 2013;587(3):272–277. doi: 10.1016/j.febslet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baranoski JF, Kalani MY, Przybylowski CJ, Zabramski JM. Cerebral cavernous malformations: review of the genetic and protein-protein interactions resulting in disease pathogenesis. Front Surg. 2016;3:60. doi: 10.3389/fsurg.2016.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gingras AR, Puzon-McLaughlin W, Ginsberg MH. The structure of the ternary complex of Krev interaction trapped 1 (KRIT1) bound to both the Rap1 GTPase and the heart of glass (HEG1) cytoplasmic tail. J Biol Chem. 2013;288(33):23639–23649. doi: 10.1074/jbc.M113.462911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum Mol Genet. 2002;11(4):389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 91.Liu W, Draheim KM, Zhang R, Calderwood DA, Boggon TJ. Mechanism for KRIT1 release of ICAP1-mediated suppression of integrin activation. Mol Cell. 2013;49(4):719–729. doi: 10.1016/j.molcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, Horne EA, Dell’Acqua ML, Johnson GL. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5(12):1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 93.Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson GL, Marchuk DA. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14(17):2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 94.Li X, Ji W, Zhang R, Folta-Stogniew E, Min W, Boggon TJ. Molecular recognition of leucine-aspartate repeat (LD) motifs by the focal adhesion targeting homology domain of cerebral cavernous malformation 3 (CCM3) J Biol Chem. 2011;286(29):26138–26147. doi: 10.1074/jbc.M110.211250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ceccarelli DF, Laister RC, Mulligan VK, Kean MJ, Goudreault M, Scott IC, Derry WB, Chakrabartty A, Gingras AC, Sicheri F. CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J Biol Chem. 2011;286(28):25056–25064. doi: 10.1074/jbc.M110.213777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Draheim KM, Li X, Zhang R, Fisher OS, Villari G, Boggon TJ, Calderwood DA. CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J Cell Biol. 2015;208(7):987–1001. doi: 10.1083/jcb.201407129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goudreault M, D’Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, Aebersold R, Raught B, Gingras AC. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteom. 2009;8(1):157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kean MJ, Ceccarelli DF, Goudreault M, Sanches M, Tate S, Larsen B, Gibson LC, Derry WB, Scott IC, Pelletier L, Baillie GS, Sicheri F, Gingras AC. Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. J Biol Chem. 2011;286(28):25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lant B, Yu B, Goudreault M, Holmyard D, Knight JD, Xu P, Zhao L, Chin K, Wallace E, Zhen M, Gingras AC, Derry WB. CCM-3/STRIPAK promotes seamless tube extension through endocytic recycling. Nat Commun. 2015;6:6449. doi: 10.1038/ncomms7449. [DOI] [PubMed] [Google Scholar]
- 100.Xu X, Wang X, Ding J, da Wang C. Crystallization and preliminary crystallographic studies of CCM3 in complex with the C-terminal domain of MST4. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 7):760–763. doi: 10.1107/S1744309112016843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hilder TL, Malone MH, Bencharit S, Colicelli J, Haystead TA, Johnson GL, Wu CC. Proteomic identification of the cerebral cavernous malformation signaling complex. J Proteome Res. 2007;6(11):4343–4355. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- 102.Zhou Z, Rawnsley DR, Goddard LM, Pan W, Cao XJ, Jakus Z, Zheng H, Yang J, Arthur JS, Whitehead KJ, Li D, Zhou B, Garcia BA, Zheng X, Kahn ML. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev Cell. 2015;32(2):168–180. doi: 10.1016/j.devcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang X, Hou Y, Deng K, Zhang Y, Wang DC, Ding J. Structural insights into the molecular recognition between cerebral cavernous malformation 2 and mitogen-activated protein kinase kinase kinase 3. Structure. 2015;23(6):1087–1096. doi: 10.1016/j.str.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 104.Fisher OS, Deng H, Liu D, Zhang Y, Wei R, Deng Y, Zhang F, Louvi A, Turk BE, Boggon TJ, Su B. Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex. Nat Commun. 2015;6:7937. doi: 10.1038/ncomms8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cullere X, Plovie E, Bennett PM, MacRae CA, Mayadas TN. The cerebral cavernous malformation proteins CCM2L and CCM2 prevent the activation of the MAP kinase MEKK3. Proc Natl Acad Sci USA. 2015;112(46):14284–14289. doi: 10.1073/pnas.1510495112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cuttano R, Rudini N, Bravi L, Corada M, Giampietro C, Papa E, Morini MF, Maddaluno L, Baeyens N, Adams RH, Jain MK, Owens GK, Schwartz M, Lampugnani MG, Dejana E. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol Med. 2015;8(1):6–24. doi: 10.15252/emmm.201505433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou Z, Tang AT, Wong WY, Bamezai S, Goddard LM, Shenkar R, Zhou S, Yang J, Wright AC, Foley M, Arthur JS, Whitehead KJ, Awad IA, Li DY, Zheng X, Kahn ML. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532(7597):122–126. doi: 10.1038/nature17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bravi L, Malinverno M, Pisati F, Rudini N, Cuttano R, Pallini R, Martini M, Larocca LM, Locatelli M, Levi V, Bertani GA, Dejana E, Lampugnani MG. Endothelial cells lining sporadic cerebral cavernous malformation cavernomas undergo endothelial-to-mesenchymal transition. Stroke. 2016;47(3):886–890. doi: 10.1161/STROKEAHA.115.011867. [DOI] [PubMed] [Google Scholar]
- 109.Cunha SI, Magnusson PU, Dejana E, Lampugnani MG. Deregulated TGF-beta/BMP signaling in vascular malformations. Circ Res. 2017;121(8):981–999. doi: 10.1161/CIRCRESAHA.117.309930. [DOI] [PubMed] [Google Scholar]
- 110.Crose LE, Hilder TL, Sciaky N, Johnson GL. Cerebral cavernous malformation 2 protein promotes smad ubiquitin regulatory factor 1-mediated RhoA degradation in endothelial cells. J Biol Chem. 2009;284(20):13301–13305. doi: 10.1074/jbc.C900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Borikova AL, Dibble CF, Sciaky N, Welch CM, Abell AN, Bencharit S, Johnson GL. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J Biol Chem. 2010;285(16):11760–11764. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207(4):881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McDonald DA, Shenkar R, Shi C, Stockton RA, Akers AL, Kucherlapati MH, Kucherlapati R, Brainer J, Ginsberg MH, Awad IA, Marchuk DA. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum Mol Genet. 2011;20(2):211–222. doi: 10.1093/hmg/ddq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McDonald DA, Shi C, Shenkar R, Stockton RA, Liu F, Ginsberg MH, Marchuk DA, Awad IA. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke. 2012;43(2):571–574. doi: 10.1161/STROKEAHA.111.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Faurobert E, Rome C, Lisowska J, Manet-Dupe S, Boulday G, Malbouyres M, Balland M, Bouin AP, Keramidas M, Bouvard D, Coll JL, Ruggiero F, Tournier-Lasserve E, Albiges-Rizo C. CCM1-ICAP-1 complex controls beta1 integrin-dependent endothelial contractility and fibronectin remodeling. J Cell Biol. 2013;202(3):545–561. doi: 10.1083/jcb.201303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Richardson BT, Dibble CF, Borikova AL, Johnson GL. Cerebral cavernous malformation is a vascular disease associated with activated RhoA signaling. Biol Chem. 2013;394(1):35–42. doi: 10.1515/hsz-2012-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bond LM, Sellers JR, McKerracher L. Rho kinase as a target for cerebral vascular disorders. Future Med Chem. 2015;7(8):1039–1053. doi: 10.4155/fmc.15.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shenkar R, Shi C, Austin C, Moore T, Lightle R, Cao Y, Zhang L, Wu M, Zeineddine HA, Girard R, McDonald DA, Rorrer A, Gallione C, Pytel P, Liao JK, Marchuk DA, Awad IA. RhoA kinase inhibition with fasudil versus simvastatin in murine models of cerebral cavernous malformations. Stroke. 2017;48(1):187–194. doi: 10.1161/STROKEAHA.116.015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gibson CC, Zhu W, Davis CT, Bowman-Kirigin JA, Chan AC, Ling J, Walker AE, Goitre L, Delle Monache S, Retta SF, Shiu YT, Grossmann AH, Thomas KR, Donato AJ, Lesniewski LA, Whitehead KJ, Li DY. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation. 2015;131(3):289–299. doi: 10.1161/CIRCULATIONAHA.114.010403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bravi L, Rudini N, Cuttano R, Giampietro C, Maddaluno L, Ferrarini L, Adams RH, Corada M, Boulday G, Tournier-Lasserve E, Dejana E, Lampugnani MG. Sulindac metabolites decrease cerebrovascular malformations in CCM3-knockout mice. Proc Natl Acad Sci USA. 2015;112(27):8421–8426. doi: 10.1073/pnas.1501352112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fogelholm R, Hernesniemi J, Vapalahti M. Impact of early surgery on outcome after aneurysmal subarachnoid hemorrhage. A population-based study. Stroke. 1993;24(11):1649–1654. doi: 10.1161/01.STR.24.11.1649. [DOI] [PubMed] [Google Scholar]
- 122.Nieuwkamp DJ, Setz LE, Algra A, Linn FH, de Rooij NK, Rinkel GJ. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex, and region: a meta-analysis. Lancet Neurol. 2009;8(7):635–642. doi: 10.1016/S1474-4422(09)70126-7. [DOI] [PubMed] [Google Scholar]
- 123.Connolly ES, Jr, Choudhri TF, Mack WJ, Mocco J, Spinks TJ, Slosberg J, Lin T, Huang J, Solomon RA. Influence of smoking, hypertension, and sex on the phenotypic expression of familial intracranial aneurysms in siblings. Neurosurgery. 2001;48(1):64–68. doi: 10.1097/00006123-200101000-00011. [DOI] [PubMed] [Google Scholar]
- 124.Ishibashi T, Murayama Y, Urashima M, Saguchi T, Ebara M, Arakawa H, Irie K, Takao H, Abe T. Unruptured intracranial aneurysms: incidence of rupture and risk factors. Stroke. 2009;40(1):313–316. doi: 10.1161/STROKEAHA.108.521674. [DOI] [PubMed] [Google Scholar]
- 125.Chambers WR, Harper BF, Jr, Simpson JR. Familial incidence of congenital aneurysms of cerebral arteries: report of cases of ruptured aneurysms in father and son. J Am Med Assoc. 1954;155(4):358–359. doi: 10.1001/jama.1954.73690220001007. [DOI] [PubMed] [Google Scholar]
- 126.Graf CJ. Familial intracranial aneurysms. J Neurosurg. 1966;25(3):304–308. doi: 10.3171/jns.1966.25.3.0304. [DOI] [PubMed] [Google Scholar]
- 127.Beumont PJ. The familial occurrence of berry aneurysm. J Neurol Neurosurg Psychiatry. 1968;31(4):399–402. doi: 10.1136/jnnp.31.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schievink WI, Schaid DJ, Michels VV, Piepgras DG. Familial aneurysmal subarachnoid hemorrhage: a community-based study. J Neurosurg. 1995;83(3):426–429. doi: 10.3171/jns.1995.83.3.0426. [DOI] [PubMed] [Google Scholar]
- 129.Graf S, Schischma A, Eberhardt KE, Istel R, Stiasny B, Schulze BD. Intracranial aneurysms and dolichoectasia in autosomal dominant polycystic kidney disease. Nephrol Dial Transplant. 2002;17(5):819–823. doi: 10.1093/ndt/17.5.819. [DOI] [PubMed] [Google Scholar]
- 130.Pepin MG, Schwarze U, Rice KM, Liu M, Leistritz D, Byers PH. Survival is affected by mutation type and molecular mechanism in vascular Ehlers-Danlos syndrome (EDS type IV) Genet Med. 2014;16(12):881–888. doi: 10.1038/gim.2014.72. [DOI] [PubMed] [Google Scholar]
- 131.O’Connell D, Kaliaperumal C, Fanning N, Wyse G, Kaar G. Superior cerebellar aneurysm causing subarachnoid haemorrhage in a 17-year-old with alagille syndrome. Br J Neurosurg. 2012;26(2):287–289. doi: 10.3109/02688697.2011.614022. [DOI] [PubMed] [Google Scholar]
- 132.Schievink WI, Riedinger M, Maya MM. Frequency of incidental intracranial aneurysms in neurofibromatosis type 1. Am J Med Genet A. 2005;134A(1):45–48. doi: 10.1002/ajmg.a.30475. [DOI] [PubMed] [Google Scholar]
- 133.Onda H, Kasuya H, Yoneyama T, Takakura K, Hori T, Takeda J, Nakajima T, Inoue I. Genomewide-linkage and haplotype-association studies map intracranial aneurysm to chromosome 7q11. Am J Hum Genet. 2001;69(4):804–819. doi: 10.1086/323614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peters DG, Kassam AB, Feingold E, Heidrich-O’Hare E, Yonas H, Ferrell RE, Brufsky A. Molecular anatomy of an intracranial aneurysm: coordinated expression of genes involved in wound healing and tissue remodeling. Stroke. 2001;32(4):1036–1042. doi: 10.1161/01.STR.32.4.1036. [DOI] [PubMed] [Google Scholar]
- 135.Farnham JM, Camp NJ, Neuhausen SL, Tsuruda J, Parker D, MacDonald J, Cannon-Albright LA. Confirmation of chromosome 7q11 locus for predisposition to intracranial aneurysm. Hum Genet. 2004;114(3):250–255. doi: 10.1007/s00439-003-1044-z. [DOI] [PubMed] [Google Scholar]
- 136.Nahed BV, Seker A, Guclu B, Ozturk AK, Finberg K, Hawkins AA, DiLuna ML, State M, Lifton RP, Gunel M. Mapping a Mendelian form of intracranial aneurysm to 1p34.3-p36.13. Am J Hum Genet. 2005;76(1):172–179. doi: 10.1086/426953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ozturk AK, Nahed BV, Bydon M, Bilguvar K, Goksu E, Bademci G, Guclu B, Johnson MH, Amar A, Lifton RP, Gunel M. Molecular genetic analysis of two large kindreds with intracranial aneurysms demonstrates linkage to 11q24-25 and 14q23-31. Stroke. 2006;37(4):1021–1027. doi: 10.1161/01.STR.0000206153.92675.b9. [DOI] [PubMed] [Google Scholar]
- 138.Ruigrok YM, Wijmenga C, Rinkel GJ, van’t Slot R, Baas F, Wolfs M, Westerveld A, Roos YB. Genomewide linkage in a large Dutch family with intracranial aneurysms: replication of 2 loci for intracranial aneurysms to chromosome 1p36.11-p36.13 and Xp22.2-p22.32. Stroke. 2008;39(4):1096–1102. doi: 10.1161/STROKEAHA.107.495168. [DOI] [PubMed] [Google Scholar]
- 139.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, von Und Zu Fraunberg M, van Duijn CM, van den Berg LH, Mane S, Mason CE, Choi M, Gaal E, Bayri Y, Kolb L, Arlier Z, Ravuri S, Ronkainen A, Tajima A, Laakso A, Hata A, Kasuya H, Koivisto T, Rinne J, Ohman J, Breteler MM, Wijmenga C, State MW, Rinkel GJ, Hernesniemi J, Jaaskelainen JE, Palotie A, Inoue I, Lifton RP, Gunel M. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008;40(12):1472–1477. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, Simon M, Krex D, Arlier Z, Nayak N, Ruigrok YM, Niemela M, Tajima A, von und zu Fraunberg M, Doczi T, Wirjatijasa F, Hata A, Blasco J, Oszvald A, Kasuya H, Zilani G, Schoch B, Singh P, Stuer C, Risselada R, Beck J, Sola T, Ricciardi F, Aromaa A, Illig T, Schreiber S, van Duijn CM, van den Berg LH, Perret C, Proust C, Roder C, Ozturk AK, Gaal E, Berg D, Geisen C, Friedrich CM, Summers P, Frangi AF, State MW, Wichmann HE, Breteler MM, Wijmenga C, Mane S, Peltonen L, Elio V, Sturkenboom MC, Lawford P, Byrne J, Macho J, Sandalcioglu EI, Meyer B, Raabe A, Steinmetz H, Rufenacht D, Jaaskelainen JE, Hernesniemi J, Rinkel GJ, Zembutsu H, Inoue I, Palotie A, Cambien F, Nakamura Y, Lifton RP, Gunel M. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42(5):420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yasuno K, Bakircioglu M, Low SK, Bilguvar K, Gaal E, Ruigrok YM, Niemela M, Hata A, Bijlenga P, Kasuya H, Jaaskelainen JE, Krex D, Auburger G, Simon M, Krischek B, Ozturk AK, Mane S, Rinkel GJ, Steinmetz H, Hernesniemi J, Schaller K, Zembutsu H, Inoue I, Palotie A, Cambien F, Nakamura Y, Lifton RP, Gunel M. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc Natl Acad Sci USA. 2011;108(49):19707–19712. doi: 10.1073/pnas.1117137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Foroud T, Koller DL, Lai D, Sauerbeck L, Anderson C, Ko N, Deka R, Mosley TH, Fornage M, Woo D, Moomaw CJ, Hornung R, Huston J, Meissner I, Bailey-Wilson JE, Langefeld C, Rouleau G, Connolly ES, Worrall BB, Kleindorfer D, Flaherty ML, Martini S, Mackey J, De Los Rios La Rosa F, Brown F, Broderick JP, Investigators FIAS Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke. 2012;43(11):2846–2852. doi: 10.1161/STROKEAHA.112.656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joshi G, Pradhan S, Mittal B. Vascular gene polymorphisms (EDNRA -231 G > A and APOE HhaI) and risk for migraine. DNA Cell Biol. 2011;30(8):577–584. doi: 10.1089/dna.2010.1197. [DOI] [PubMed] [Google Scholar]
- 144.Lee S, Kim IK, Ahn JS, Woo DC, Kim ST, Song S, Koh GY, Kim HS, Jeon BH, Kim I. Deficiency of endothelium-specific transcription factor Sox17 induces intracranial aneurysm. Circulation. 2015;131(11):995–1005. doi: 10.1161/CIRCULATIONAHA.114.012568. [DOI] [PubMed] [Google Scholar]
- 145.Leeper NJ, Raiesdana A, Kojima Y, Kundu RK, Cheng H, Maegdefessel L, Toh R, Ahn GO, Ali ZA, Anderson DR, Miller CL, Roberts SC, Spin JM, de Almeida PE, Wu JC, Xu B, Cheng K, Quertermous M, Kundu S, Kortekaas KE, Berzin E, Downing KP, Dalman RL, Tsao PS, Schadt EE, Owens GK, Quertermous T. Loss of CDKN2B promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. Arterioscler Thromb Vasc Biol. 2013;33(1):e1–e10. doi: 10.1161/ATVBAHA.112.300399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA, Genomes Project, C A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suzuki J, Takaku A. Cerebrovascular “moyamoya” disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol. 1969;20(3):288–299. doi: 10.1001/archneur.1969.00480090076012. [DOI] [PubMed] [Google Scholar]
- 148.Khan N, Takagi Y, Yonekawa Y. Adult Moyamoya disease. In: Winn HR, editor. Youmans and Winn neurological surgery. Amsterdam: Elsevier; 2017. pp. 3166–3174. [Google Scholar]
- 149.Smith ER, Scott RM. Moyamoya disease. In: Winn HR, editor. Youmans and Winn neurological surgery. Amsterdam: Elsevier; 2017. pp. 1766–1772. [Google Scholar]
- 150.Ahn IM, Park DH, Hann HJ, Kim KH, Kim HJ, Ahn HS. Incidence, prevalence, and survival of moyamoya disease in Korea: a nationwide, population-based study. Stroke. 2014;45(4):1090–1095. doi: 10.1161/STROKEAHA.113.004273. [DOI] [PubMed] [Google Scholar]
- 151.Scott RM, Smith ER. Moyamoya disease and moyamoya syndrome. N Engl J Med. 2009;360(12):1226–1237. doi: 10.1056/NEJMra0804622. [DOI] [PubMed] [Google Scholar]
- 152.Kraemer M, Heienbrok W, Berlit P. Moyamoya disease in Europeans. Stroke. 2008;39(12):3193–3200. doi: 10.1161/STROKEAHA.107.513408. [DOI] [PubMed] [Google Scholar]
- 153.Suzuki J, Kodama N. Moyamoya disease—a review. Stroke. 1983;14(1):104–109. doi: 10.1161/01.STR.14.1.104. [DOI] [PubMed] [Google Scholar]
- 154.Graham JF, Matoba A. A survey of moyamoya disease in Hawaii. Clin Neurol Neurosurg. 1997;99(Suppl 2):S31–S35. doi: 10.1016/S0303-8467(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 155.Yamauchi T, Houkin K, Tada M, Abe H. Familial occurrence of moyamoya disease. Clin Neurol Neurosurg. 1997;99(Suppl 2):S162–S167. doi: 10.1016/S0303-8467(97)00054-1. [DOI] [PubMed] [Google Scholar]
- 156.Ikeda E. Systemic vascular changes in spontaneous occlusion of the circle of Willis. Stroke. 1991;22(11):1358–1362. doi: 10.1161/01.STR.22.11.1358. [DOI] [PubMed] [Google Scholar]
- 157.Mineharu Y, Takenaka K, Yamakawa H, Inoue K, Ikeda H, Kikuta KI, Takagi Y, Nozaki K, Hashimoto N, Koizumi A. Inheritance pattern of familial moyamoya disease: autosomal dominant mode and genomic imprinting. J Neurol Neurosurg Psychiatry. 2006;77(9):1025–1029. doi: 10.1136/jnnp.2006.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mineharu Y, Liu W, Inoue K, Matsuura N, Inoue S, Takenaka K, Ikeda H, Houkin K, Takagi Y, Kikuta K, Nozaki K, Hashimoto N, Koizumi A. Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology. 2008;70(24 Pt 2):2357–2363. doi: 10.1212/01.wnl.0000291012.49986.f9. [DOI] [PubMed] [Google Scholar]
- 159.Kamada F, Aoki Y, Narisawa A, Abe Y, Komatsuzaki S, Kikuchi A, Kanno J, Niihori T, Ono M, Ishii N, Owada Y, Fujimura M, Mashimo Y, Suzuki Y, Hata A, Tsuchiya S, Tominaga T, Matsubara Y, Kure S. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet. 2011;56(1):34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- 160.Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, Hashikata H, Matsuura N, Yamazaki S, Toyoda A, Kikuta K, Takagi Y, Harada KH, Fujiyama A, Herzig R, Krischek B, Zou L, Kim JE, Kitakaze M, Miyamoto S, Nagata K, Hashimoto N, Koizumi A. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One. 2011;6(7):e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moteki Y, Onda H, Kasuya H, Yoneyama T, Okada Y, Hirota K, Mukawa M, Nariai T, Mitani S, Akagawa H. Systematic validation of RNF213 coding variants in Japanese patients with moyamoya disease. J Am Heart Assoc. 2015 doi: 10.1161/JAHA.115.001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim EH, Yum MS, Ra YS, Park JB, Ahn JS, Kim GH, Goo HW, Ko TS, Yoo HW. Importance of RNF213 polymorphism on clinical features and long-term outcome in moyamoya disease. J Neurosurg. 2016;124(5):1221–1227. doi: 10.3171/2015.4.JNS142900. [DOI] [PubMed] [Google Scholar]
- 163.Raso A , Biassoni R, Mascelli S, Nozza P, Ugolotti E, DI Marco E, DE Marco P, Merello E, Cama A, Pavanello M, Capra V (2016) Moyamoya vasculopathy shows a genetic mutational gradient decreasing from East to West. J Neurosurg Sci [DOI] [PubMed]
- 164.Ohkubo K, Sakai Y, Inoue H, Akamine S, Ishizaki Y, Matsushita Y, Sanefuji M, Torisu H, Ihara K, Sardiello M, Hara T. Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells. Sci Rep. 2015;5:13191. doi: 10.1038/srep13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fukushima H, Takenouchi T, Kosaki K. Homozygosity for moyamoya disease risk allele leads to moyamoya disease with extracranial systemic and pulmonary vasculopathy. Am J Med Genet A. 2016;170(9):2453–2456. doi: 10.1002/ajmg.a.37829. [DOI] [PubMed] [Google Scholar]
- 166.Sonobe S, Fujimura M, Niizuma K, Nishijima Y, Ito A, Shimizu H, Kikuchi A, Arai-Ichinoi N, Kure S, Tominaga T. Temporal profile of the vascular anatomy evaluated by 9.4-T magnetic resonance angiography and histopathological analysis in mice lacking RNF213: a susceptibility gene for moyamoya disease. Brain Res. 2014;1552:64–71. doi: 10.1016/j.brainres.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 167.Herve D, Philippi A, Belbouab R, Zerah M, Chabrier S, Collardeau-Frachon S, Bergametti F, Essongue A, Berrou E, Krivosic V, Sainte-Rose C, Houdart E, Adam F, Billiemaz K, Lebret M, Roman S, Passemard S, Boulday G, Delaforge A, Guey S, Dray X, Chabriat H, Brouckaert P, Bryckaert M, Tournier-Lasserve E. Loss of alpha1beta1 soluble guanylate cyclase, the major nitric oxide receptor, leads to moyamoya and achalasia. Am J Hum Genet. 2014;94(3):385–394. doi: 10.1016/j.ajhg.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zabel U, Weeger M, La M, Schmidt HH. Human soluble guanylate cyclase: functional expression and revised isoenzyme family. Biochem J. 1998;335(Pt 1):51–57. doi: 10.1042/bj3350051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Herve D, Touraine P, Verloes A, Miskinyte S, Krivosic V, Logeart D, Alili N, Laredo JD, Gaudric A, Houdart E, Metzger JP, Tournier-Lasserve E, Woimant F. A hereditary moyamoya syndrome with multisystemic manifestations. Neurology. 2010;75(3):259–264. doi: 10.1212/WNL.0b013e3181e8ee3f. [DOI] [PubMed] [Google Scholar]
- 170.Miskinyte S, Butler MG, Herve D, Sarret C, Nicolino M, Petralia JD, Bergametti F, Arnould M, Pham VN, Gore AV, Spengos K, Gazal S, Woimant F, Steinberg GK, Weinstein BM, Tournier-Lasserve E. Loss of BRCC3 deubiquitinating enzyme leads to abnormal angiogenesis and is associated with syndromic moyamoya. Am J Hum Genet. 2011;88(6):718–728. doi: 10.1016/j.ajhg.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M. Analysis of class II genes of human leukocyte antigen in patients with moyamoya disease. Clin Neurol Neurosurg. 1997;99(Suppl 2):S234–S237. doi: 10.1016/S0303-8467(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 172.Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M. Linkage analysis of moyamoya disease on chromosome 6. J Child Neurol. 2000;15(3):179–182. doi: 10.1177/088307380001500307. [DOI] [PubMed] [Google Scholar]
- 173.Han H, Pyo CW, Yoo DS, Huh PW, Cho KS, Kim DS. Associations of Moyamoya patients with HLA class I and class II alleles in the Korean population. J Korean Med Sci. 2003;18(6):876–880. doi: 10.3346/jkms.2003.18.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sakurai K, Horiuchi Y, Ikeda H, Ikezaki K, Yoshimoto T, Fukui M, Arinami T. A novel susceptibility locus for moyamoya disease on chromosome 8q23. J Hum Genet. 2004;49(5):278–281. doi: 10.1007/s10038-004-0143-6. [DOI] [PubMed] [Google Scholar]
- 175.Roder C, Peters V, Kasuya H, Nishizawa T, Takehara Y, Berg D, Schulte C, Khan N, Tatagiba M, Krischek B. Polymorphisms in TGFB1 and PDGFRB are associated with Moyamoya disease in European patients. Acta Neurochir (Wien) 2010;152(12):2153–2160. doi: 10.1007/s00701-010-0711-9. [DOI] [PubMed] [Google Scholar]
- 176.Guey S, Grangeon L, Brunelle F, Bergametti F, Amiel J, Lyonnet S, Delaforge A, Arnould M, Desnous B, Bellesme C, Herve D, Schwitalla JC, Kraemer M, Tournier-Lasserve E, Kossorotoff M. De novo mutations in CBL causing early-onset paediatric moyamoya angiopathy. J Med Genet. 2017;54(8):550–557. doi: 10.1136/jmedgenet-2016-104432. [DOI] [PubMed] [Google Scholar]
- 177.Mukawa M, Nariai T, Onda H, Yoneyama T, Aihara Y, Hirota K, Kudo T, Sumita K, Maehara T, Kawamata T, Kasuya H, Akagawa H. Exome sequencing identified CCER2 as a novel candidate gene for Moyamoya disease. J Stroke Cerebrovasc Dis. 2017;26(1):150–161. doi: 10.1016/j.jstrokecerebrovasdis.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 178.Hamauchi S, Shichinohe H, Uchino H, Yamaguchi S, Nakayama N, Kazumata K, Osanai T, Abumiya T, Houkin K, Era T. Cellular functions and gene and protein expression profiles in endothelial cells derived from Moyamoya disease-specific iPS Cells. PLoS One. 2016;11(9):e0163561. doi: 10.1371/journal.pone.0163561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang W, Gao F, Zhao Z, Wang H, Zhang L, Zhang D, Zhang Y, Lan Q, Wang J, Zhao J. Integrated analysis of LncRNA-mRNA co-expression profiles in patients with moyamoya disease. Sci Rep. 2017;7:42421. doi: 10.1038/srep42421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gao F, Yu L, Zhang D, Zhang Y, Wang R, Zhao J. Long noncoding RNAs and their regulatory network: potential therapeutic targets for adult moyamoya disease. World Neurosurg. 2016;93:111–119. doi: 10.1016/j.wneu.2016.05.081. [DOI] [PubMed] [Google Scholar]
- 181.Vinters HV, Zarow C, Borys E, Whitman JD, Tung S, Ellis WG, Zheng L, Chui HC. Review: vascular dementia: clinicopathologic and genetic considerations. Neuropathol Appl Neurobiol. 2018;44(3):247–266. doi: 10.1111/nan.12472. [DOI] [PubMed] [Google Scholar]
- 182.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383(6602):707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 183.Hara K, Shiga A, Fukutake T, Nozaki H, Miyashita A, Yokoseki A, Kawata H, Koyama A, Arima K, Takahashi T, Ikeda M, Shiota H, Tamura M, Shimoe Y, Hirayama M, Arisato T, Yanagawa S, Tanaka A, Nakano I, Ikeda S, Yoshida Y, Yamamoto T, Ikeuchi T, Kuwano R, Nishizawa M, Tsuji S, Onodera O. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N Engl J Med. 2009;360(17):1729–1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- 184.Richards A, van den Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-Labarca ML, Terwindt GM, Kasai Y, McLellan M, Grand MG, Vanmolkot KR, de Vries B, Wan J, Kane MJ, Mamsa H, Schafer R, Stam AH, Haan J, de Jong PT, Storimans CW, van Schooneveld MJ, Oosterhuis JA, Gschwendter A, Dichgans M, Kotschet KE, Hodgkinson S, Hardy TA, Delatycki MB, Hajj-Ali RA, Kothari PH, Nelson SF, Frants RR, Baloh RW, Ferrari MD, Atkinson JP. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39(9):1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 185.Gould DB, Phalan FC, van Mil SE, Sundberg JP, Vahedi K, Massin P, Bousser MG, Heutink P, Miner JH, Tournier-Lasserve E, John SW. Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N Engl J Med. 2006;354(14):1489–1496. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- 186.Gilbert JJ, Vinters HV. Cerebral amyloid angiopathy: incidence and complications in the aging brain. Cerebral hemorrhage. Stroke. 1983;14(6):915–923. doi: 10.1161/01.STR.14.6.915. [DOI] [PubMed] [Google Scholar]
- 187.Vinters HV, Gilbert JJ. Cerebral amyloid angiopathy: incidence and complications in the aging brain. II. The distribution of amyloid vascular changes. Stroke. 1983;14(6):924–928. doi: 10.1161/01.STR.14.6.924. [DOI] [PubMed] [Google Scholar]
- 188.Kolar GR, Kothari PH, Khanlou N, Jen JC, Schmidt RE, Vinters HV. Neuropathology and genetics of cerebroretinal vasculopathies. Brain Pathol. 2014;24(5):510–518. doi: 10.1111/bpa.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sourander P, Walinder J. Hereditary multi-infarct dementia. Morphological and clinical studies of a new disease. Acta Neuropathol. 1977;39(3):247–254. doi: 10.1007/BF00691704. [DOI] [PubMed] [Google Scholar]
- 190.Sourander P, Walinder J. Hereditary multi-infarct dementia. Lancet. 1977;1(8019):1015. doi: 10.1016/S0140-6736(77)92324-8. [DOI] [PubMed] [Google Scholar]
- 191.Van Bogaert L. Encephalopathie sous-corticale progressive (Binswanger): an evolution rapide chez deux soeurs. Med Hellen. 1955;24:961–972. [Google Scholar]
- 192.Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. Cadasil. Lancet Neurol. 2009;8(7):643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 193.Moreton FC, Razvi SS, Davidson R, Muir KW. Changing clinical patterns and increasing prevalence in CADASIL. Acta Neurol Scand. 2014;130(3):197–203. doi: 10.1111/ane.12266. [DOI] [PubMed] [Google Scholar]
- 194.Tikka S, Baumann M, Siitonen M, Pasanen P, Poyhonen M, Myllykangas L, Viitanen M, Fukutake T, Cognat E, Joutel A, Kalimo H. CADASIL and CARASIL. Brain Pathol. 2014;24(5):525–544. doi: 10.1111/bpa.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tournier-Lasserve E, Joutel A, Melki J, Weissenbach J, Lathrop GM, Chabriat H, Mas JL, Cabanis EA, Baudrimont M, Maciazek J, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy maps to chromosome 19q12. Nat Genet. 1993;3(3):256–259. doi: 10.1038/ng0393-256. [DOI] [PubMed] [Google Scholar]
- 196.Adib-Samii P, Brice G, Martin RJ, Markus HS. Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: study in 200 consecutively recruited individuals. Stroke. 2010;41(4):630–634. doi: 10.1161/STROKEAHA.109.568402. [DOI] [PubMed] [Google Scholar]
- 197.Opherk C, Peters N, Herzog J, Luedtke R, Dichgans M. Long-term prognosis and causes of death in CADASIL: a retrospective study in 411 patients. Brain. 2004;127(Pt 11):2533–2539. doi: 10.1093/brain/awh282. [DOI] [PubMed] [Google Scholar]
- 198.Rutten JW, Dauwerse HG, Gravesteijn G, van Belzen MJ, van der Grond J, Polke JM, Bernal-Quiros M, Lesnik Oberstein SA. Archetypal NOTCH3 mutations frequent in public exome: implications for CADASIL. Ann Clin Transl Neurol. 2016;3(11):844–853. doi: 10.1002/acn3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Narayan SK, Gorman G, Kalaria RN, Ford GA, Chinnery PF. The minimum prevalence of CADASIL in northeast England. Neurology. 2012;78(13):1025–1027. doi: 10.1212/WNL.0b013e31824d586c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Razvi SS, Davidson R, Bone I, Muir KW. The prevalence of cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL) in the west of Scotland. J Neurol Neurosurg Psychiatry. 2005;76(5):739–741. doi: 10.1136/jnnp.2004.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Joutel A, Dodick DD, Parisi JE, Cecillon M, Tournier-Lasserve E, Bousser MG. De novo mutation in the Notch3 gene causing CADASIL. Ann Neurol. 2000;47(3):388–391. doi: 10.1002/1531-8249(200003)47:3<388::AID-ANA19>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 202.Coto E, Menendez M, Navarro R, Garcia-Castro M, Alvarez V. A new de novo Notch3 mutation causing CADASIL. Eur J Neurol. 2006;13(6):628–631. doi: 10.1111/j.1468-1331.2006.01337.x. [DOI] [PubMed] [Google Scholar]
- 203.Tuominen S, Juvonen V, Amberla K, Jolma T, Rinne JO, Tuisku S, Kurki T, Marttila R, Poyhonen M, Savontaus ML, Viitanen M, Kalimo H. Phenotype of a homozygous CADASIL patient in comparison to 9 age-matched heterozygous patients with the same R133C Notch3 mutation. Stroke. 2001;32(8):1767–1774. doi: 10.1161/01.STR.32.8.1767. [DOI] [PubMed] [Google Scholar]
- 204.Liem MK, Lesnik Oberstein SA, Vollebregt MJ, Middelkoop HA, van der Grond J, Helderman-van den Enden AT. Homozygosity for a NOTCH3 mutation in a 65-year-old CADASIL patient with mild symptoms: a family report. J Neurol. 2008;255(12):1978–1980. doi: 10.1007/s00415-009-0036-x. [DOI] [PubMed] [Google Scholar]
- 205.Ragno M, Pianese L, Morroni M, Cacchio G, Manca A, Di Marzio F, Silvestri S, Miceli C, Scarcella M, Onofrj M, Trojano L. “CADASIL coma” in an Italian homozygous CADASIL patient: comparison with clinical and MRI findings in age-matched heterozygous patients with the same G528C NOTCH3 mutation. Neurol Sci. 2013;34(11):1947–1953. doi: 10.1007/s10072-013-1418-5. [DOI] [PubMed] [Google Scholar]
- 206.Soong BW, Liao YC, Tu PH, Tsai PC, Lee IH, Chung CP, Lee YC. A homozygous NOTCH3 mutation p.R544C and a heterozygous TREX1 variant p.C99MfsX3 in a family with hereditary small vessel disease of the brain. J Chin Med Assoc. 2013;76(6):319–324. doi: 10.1016/j.jcma.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 207.Vinciguerra C, Rufa A, Bianchi S, Sperduto A, De Santis M, Malandrini A, Dotti MT, Federico A. Homozygosity and severity of phenotypic presentation in a CADASIL family. Neurol Sci. 2014;35(1):91–93. doi: 10.1007/s10072-013-1580-9. [DOI] [PubMed] [Google Scholar]
- 208.Yamamoto Y, Craggs L, Baumann M, Kalimo H, Kalaria RN. Review: molecular genetics and pathology of hereditary small vessel diseases of the brain. Neuropathol Appl Neurobiol. 2011;37(1):94–113. doi: 10.1111/j.1365-2990.2010.01147.x. [DOI] [PubMed] [Google Scholar]
- 209.Fouillade C, Monet-Lepretre M, Baron-Menguy C, Joutel A. Notch signaling in smooth muscle cells during development and disease. Cardiovasc Res. 2012;95:138–146. doi: 10.1093/cvr/cvs019. [DOI] [PubMed] [Google Scholar]
- 210.Dziewulska D, Lewandowska E. Pericytes as a new target for pathological processes in CADASIL. Neuropathology. 2012;32:515–521. doi: 10.1111/j.1440-1789.2011.01290.x. [DOI] [PubMed] [Google Scholar]
- 211.Ghosh M, Balbi M, Hellal F, Dichgans M, Lindauer U, Plesnila N. Pericytes are involved in the pathogenesis of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Ann Neurol. 2015;78(6):887–900. doi: 10.1002/ana.24512. [DOI] [PubMed] [Google Scholar]
- 212.Baudrimont M, Dubas F, Joutel A, Tournier-Lasserve E, Bousser MG. Autosomal dominant leukoencephalopathy and subcortical ischemic stroke. A clinicopathological study. Stroke. 1993;24(1):122–125. doi: 10.1161/01.STR.24.1.122. [DOI] [PubMed] [Google Scholar]
- 213.Ruchoux MM, Guerouaou D, Vandenhaute B, Pruvo JP, Vermersch P, Leys D. Systemic vascular smooth muscle cell impairment in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Acta Neuropathol (Berl) 1995;89(6):500–512. doi: 10.1007/BF00571504. [DOI] [PubMed] [Google Scholar]
- 214.Tikka S, Mykkanen K, Ruchoux MM, Bergholm R, Junna M, Poyhonen M, Yki-Jarvinen H, Joutel A, Viitanen M, Baumann M, Kalimo H. Congruence between NOTCH3 mutations and GOM in 131 CADASIL patients. Brain J Neurol. 2009;132(Pt 4):933–939. doi: 10.1093/brain/awn364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lewandowska E, Dziewulska D, Parys M, Pasennik E. Ultrastructure of granular osmiophilic material deposits (GOM) in arterioles of CADASIL patients. Folia Neuropathol. 2011;49(3):174–180. [PubMed] [Google Scholar]
- 216.Ishiko A, Shimizu A, Nagata E, Takahashi K, Tabira T, Suzuki N. Notch3 ectodomain is a major component of granular osmiophilic material (GOM) in CADASIL. Acta Neuropathol. 2006;112(3):333–339. doi: 10.1007/s00401-006-0116-2. [DOI] [PubMed] [Google Scholar]
- 217.Yamamoto Y, Craggs LJ, Watanabe A, Booth T, Attems J, Low RW, Oakley AE, Kalaria RN. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol. 2013;72(5):416–431. doi: 10.1097/NEN.0b013e31829020b5. [DOI] [PubMed] [Google Scholar]
- 218.Monet-Lepretre M, Haddad I, Baron-Menguy C, Fouillot-Panchal M, Riani M, Domenga-Denier V, Dussaule C, Cognat E, Vinh J, Joutel A. Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain. 2013;136(Pt 6):1830–1845. doi: 10.1093/brain/awt092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Arboleda-Velasquez JF, Manent J, Lee JH, Tikka S, Ospina C, Vanderburg CR, Frosch MP, Rodriguez-Falcon M, Villen J, Gygi S, Lopera F, Kalimo H, Moskowitz MA, Ayata C, Louvi A, Artavanis-Tsakonas S. Hypomorphic notch 3 alleles link notch signaling to ischemic cerebral small-vessel disease. Proc Natl Acad Sci USA. 2011;108(21):E128–E135. doi: 10.1073/pnas.1101964108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kast J, Hanecker P, Beaufort N, Giese A, Joutel A, Dichgans M, Opherk C, Haffner C. Sequestration of latent TGF-beta binding protein 1 into CADASIL-related Notch3-ECD deposits. Acta Neuropathol Commun. 2014;2:96. doi: 10.1186/s40478-014-0096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126(Pt 10):2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, Cruaud C, Maciazek J, Weissenbach J, Bousser MG, Bach JF, Tournier-Lasserve E. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350(9090):1511–1515. doi: 10.1016/S0140-6736(97)08083-5. [DOI] [PubMed] [Google Scholar]
- 223.Mizuno T, Muranishi M, Torugun T, Tango H, Nagakane Y, Kudeken T, Kawase Y, Kawabe K, Oshima F, Yaoi T, Itoh K, Fushiki S, Nakagawa M. Two Japanese CADASIL families exhibiting Notch3 mutation R75P not involving cysteine residue. Intern Med. 2008;47(23):2067–2072. doi: 10.2169/internalmedicine.47.1391. [DOI] [PubMed] [Google Scholar]
- 224.Muino E, Gallego-Fabrega C, Cullell N, Carrera C, Torres N, Krupinski J, Roquer J, Montaner J, Fernandez-Cadenas I. Systematic review of cysteine-sparing NOTCH3 missense mutations in patients with clinical suspicion of CADASIL. Int J Mol Sci. 2017 doi: 10.3390/ijms18091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lundkvist J, Zhu S, Hansson EM, Schweinhardt P, Miao Q, Beatus P, Dannaeus K, Karlstrom H, Johansson CB, Viitanen M, Rozell B, Spenger C, Mohammed A, Kalimo H, Lendahl U. Mice carrying a R142C Notch 3 knock-in mutation do not develop a CADASIL-like phenotype. Genesis. 2005;41(1):13–22. doi: 10.1002/gene.20091. [DOI] [PubMed] [Google Scholar]
- 226.Wallays G, Nuyens D, Silasi-Mansat R, Souffreau J, Callaerts-Vegh Z, Van Nuffelen A, Moons L, D’Hooge R, Lupu F, Carmeliet P, Collen D, Dewerchin M. Notch3 Arg170Cys knock-in mice display pathologic and clinical features of the neurovascular disorder cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Arterioscler Thromb Vasc Biol. 2011;31(12):2881–2888. doi: 10.1161/ATVBAHA.111.237859. [DOI] [PubMed] [Google Scholar]
- 227.Joutel A, Monet-Lepretre M, Gosele C, Baron-Menguy C, Hammes A, Schmidt S, Lemaire-Carrette B, Domenga V, Schedl A, Lacombe P, Hubner N. Cerebrovascular dysfunction and microcirculation rarefaction precede white matter lesions in a mouse genetic model of cerebral ischemic small vessel disease. J Clin Investig. 2010;120(2):433–445. doi: 10.1172/JCI39733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Monet M, Domenga V, Lemaire B, Souilhol C, Langa F, Babinet C, Gridley T, Tournier-Lasserve E, Cohen-Tannoudji M, Joutel A. The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum Mol Genet. 2007;16(8):982–992. doi: 10.1093/hmg/ddm042. [DOI] [PubMed] [Google Scholar]
- 229.Monet-Lepretre M, Bardot B, Lemaire B, Domenga V, Godin O, Dichgans M, Tournier-Lasserve E, Cohen-Tannoudji M, Chabriat H, Joutel A. Distinct phenotypic and functional features of CADASIL mutations in the Notch3 ligand binding domain. Brain J Neurol. 2009;132(Pt 6):1601–1612. doi: 10.1093/brain/awp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Machuca-Parra AI, Bigger-Allen AA, Sanchez AV, Boutabla A, Cardona-Velez J, Amarnani D, Saint-Geniez M, Siebel CW, Kim LA, D’Amore PA, Arboleda-Velasquez JF. Therapeutic antibody targeting of Notch3 signaling prevents mural cell loss in CADASIL. J Exp Med. 2017;214(8):2271–2282. doi: 10.1084/jem.20161715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fukutake T. Cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL): from discovery to gene identification. J Stroke Cerebrovasc Dis. 2011;20(2):85–93. doi: 10.1016/j.jstrokecerebrovasdis.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 232.Nozaki H, Nishizawa M, Onodera O. Features of cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy. Stroke. 2014;45(11):3447–3453. doi: 10.1161/STROKEAHA.114.004236. [DOI] [PubMed] [Google Scholar]
- 233.Verdura E, Herve D, Scharrer E, Amador Mdel M, Guyant-Marechal L, Philippi A, Corlobe A, Bergametti F, Gazal S, Prieto-Morin C, Beaufort N, Le Bail B, Viakhireva I, Dichgans M, Chabriat H, Haffner C, Tournier-Lasserve E. Heterozygous HTRA1 mutations are associated with autosomal dominant cerebral small vessel disease. Brain. 2015;138(Pt 8):2347–2358. doi: 10.1093/brain/awv155. [DOI] [PubMed] [Google Scholar]
- 234.Vinters HV. Cerebral amyloid angiopathy. A critical review. Stroke. 1987;18(2):311–324. doi: 10.1161/01.str.18.2.311. [DOI] [PubMed] [Google Scholar]
- 235.Revesz T, Ghiso J, Lashley T, Plant G, Rostagno A, Frangione B, Holton JL. Cerebral amyloid angiopathies: a pathologic, biochemical, and genetic view. J Neuropathol Exp Neurol. 2003;62(9):885–898. doi: 10.1093/jnen/62.9.885. [DOI] [PubMed] [Google Scholar]
- 236.Biffi A, Greenberg SM. Cerebral amyloid angiopathy: a systematic review. J Clin Neurol. 2011;7(1):1–9. doi: 10.3988/jcn.2011.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Viswanathan A, Greenberg SM. Cerebral amyloid angiopathy in the elderly. Ann Neurol. 2011;70(6):871–880. doi: 10.1002/ana.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Brenowitz WD, Nelson PT, Besser LM, Heller KB, Kukull WA. Cerebral amyloid angiopathy and its co-occurrence with Alzheimer’s disease and other cerebrovascular neuropathologic changes. Neurobiol Aging. 2015;36(10):2702–2708. doi: 10.1016/j.neurobiolaging.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Smith EE. Cerebral amyloid angiopathy as a cause of neurodegeneration. J Neurochem. 2018;144(5):651–658. doi: 10.1111/jnc.14157. [DOI] [PubMed] [Google Scholar]
- 240.Vinters HV. Emerging concepts in Alzheimer’s disease. Annu Rev Pathol. 2015;10:291–319. doi: 10.1146/annurev-pathol-020712-163927. [DOI] [PubMed] [Google Scholar]
- 241.Greenberg SM, Vonsattel JP. Diagnosis of cerebral amyloid angiopathy. Sensitivity and specificity of cortical biopsy. Stroke. 1997;28(7):1418–1422. doi: 10.1161/01.STR.28.7.1418. [DOI] [PubMed] [Google Scholar]
- 242.Nicoll JA, Yamada M, Frackowiak J, Mazur-Kolecka B, Weller RO. Cerebral amyloid angiopathy plays a direct role in the pathogenesis of Alzheimer’s disease. Pro-CAA position statement. Neurobiol Aging. 2004;25(5):589–597. doi: 10.1016/j.neurobiolaging.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 243.Chalmers K, Wilcock GK, Love S. APOE epsilon 4 influences the pathological phenotype of Alzheimer’s disease by favouring cerebrovascular over parenchymal accumulation of A beta protein. Neuropathol Appl Neurobiol. 2003;29(3):231–238. doi: 10.1046/j.1365-2990.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- 244.Premkumar DR, Cohen DL, Hedera P, Friedland RP, Kalaria RN. Apolipoprotein E-epsilon4 alleles in cerebral amyloid angiopathy and cerebrovascular pathology associated with Alzheimer’s disease. Am J Pathol. 1996;148(6):2083–2095. [PMC free article] [PubMed] [Google Scholar]
- 245.Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38(1):24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 246.Revesz T, Holton JL, Lashley T, Plant G, Frangione B, Rostagno A, Ghiso J. Genetics and molecular pathogenesis of sporadic and hereditary cerebral amyloid angiopathies. Acta Neuropathol. 2009;118(1):115–130. doi: 10.1007/s00401-009-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.De Jonghe C, Zehr C, Yager D, Prada CM, Younkin S, Hendriks L, Van Broeckhoven C, Eckman CB. Flemish and Dutch mutations in amyloid beta precursor protein have different effects on amyloid beta secretion. Neurobiol Dis. 1998;5(4):281–286. doi: 10.1006/nbdi.1998.0202. [DOI] [PubMed] [Google Scholar]
- 248.Basun H, Bogdanovic N, Ingelsson M, Almkvist O, Naslund J, Axelman K, Bird TD, Nochlin D, Schellenberg GD, Wahlund LO, Lannfelt L. Clinical and neuropathological features of the arctic APP gene mutation causing early-onset Alzheimer disease. Arch Neurol. 2008;65(4):499–505. doi: 10.1001/archneur.65.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ghiso J, Jensson O, Frangione B. Amyloid fibrils in hereditary cerebral hemorrhage with amyloidosis of Icelandic type is a variant of gamma-trace basic protein (cystatin C) Proc Natl Acad Sci USA. 1986;83(9):2974–2978. doi: 10.1073/pnas.83.9.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Brett M, Persey MR, Reilly MM, Revesz T, Booth DR, Booth SE, Hawkins PN, Pepys MB, Morgan-Hughes JA. Transthyretin Leu12Pro is associated with systemic, neuropathic and leptomeningeal amyloidosis. Brain. 1999;122(Pt 2):183–190. doi: 10.1093/brain/122.2.183. [DOI] [PubMed] [Google Scholar]
- 251.Ghiso J, Haltia M, Prelli F, Novello J, Frangione B. Gelsolin variant (Asn-187) in familial amyloidosis, Finnish type. Biochem J. 1990;272(3):827–830. doi: 10.1042/bj2720827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Herzig MC, Van Nostrand WE, Jucker M. Mechanism of cerebral beta-amyloid angiopathy: murine and cellular models. Brain Pathol. 2006;16(1):40–54. doi: 10.1111/j.1750-3639.2006.tb00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Vidal R, Barbeito AG, Miravalle L, Ghetti B. Cerebral amyloid angiopathy and parenchymal amyloid deposition in transgenic mice expressing the Danish mutant form of human BRI2. Brain Pathol. 2009;19(1):58–68. doi: 10.1111/j.1750-3639.2008.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Stam AH, Kothari PH, Shaikh A, Gschwendter A, Jen JC, Hodgkinson S, Hardy TA, Hayes M, Kempster PA, Kotschet KE, Bajema IM, van Duinen SG, Maat-Schieman MLC, de Jong P, de Smet MD, de Wolff-Rouendaal D, Dijkman G, Pelzer N, Kolar GR, Schmidt RE, Lacey J, Joseph D, Fintak DR, Grand MG, Brunt EM, Liapis H, Hajj-Ali RA, Kruit MC, van Buchem MA, Dichgans M, Frants RR, van den Maagdenberg A, Haan J, Baloh RW, Atkinson JP, Terwindt GM, Ferrari MD. Retinal vasculopathy with cerebral leukoencephalopathy and systemic manifestations. Brain. 2016;139(11):2909–2922. doi: 10.1093/brain/aww217. [DOI] [PMC free article] [PubMed] [Google Scholar]