Abstract
Introduction:
Understanding pathways and mechanisms of drug binding to receptors is important for rational drug design. Remarkable advances in supercomputing and methodological developments have opened a new era for application of computer simulations in predicting drug-receptor interactions at an atomistic level.
Areas covered:
Gaussian accelerated molecular dynamics (GaMD) is a computational enhanced sampling technique that works by adding a harmonic boost potential to reduce energy barriers. GaMD enables free energy calculations without the requirement of predefined collective variables. GaMD has proven useful in biomolecular simulations, in particular the prediction of drug-receptor interactions. Here, we review recent GaMD simulation studies that elucidated pathways of drug binding to proteins, including the G-protein-coupled receptors and HIV protease.
Expert Opinion:
GaMD is advantageous for enhanced simulations of, amongst many biological processes, drug binding to target receptors. Compared with conventional molecular dynamics, GaMD speeds up biomolecular simulations by orders of magnitude. GaMD enables routine drug binding simulations using personal computers with GPUs or common computing clusters. GaMD and, more broadly, enhanced sampling simulations are expected to dramatically increase our capabilities to determine the mechanisms of drug binding to a wide range of receptors in the near future. This will greatly facilitate computer-aided drug design.
Keywords: Gaussian accelerated molecular dynamics, enhanced sampling, drug pathways, GPCRs, HIV protease, computer-aided drug design
1. Introduction
Successful design of a drug nowadays often takes more than a decade and has become increasingly expensive1. To reduce the drug design cost, it is important to understand the pathways and mechanisms of drug binding to target receptors. The pharmaceutical industry would benefit immensely from studies of the protein’s druggable binding sites and drug pathways. While experimental techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) and site-directed mutagenesis can be used to probe drug binding sites in target receptors2–5, the experiments are expensive and do not provide a detailed picture of drug pathways. This is often limited by the temporal and spatial resolution of the experimental techniques. Computer simulations, on the other hand, can be applied to overcome such limitations and identify drug pathways at an atomistic level6–9. This has been enabled by remarkable advances in both supercomputing and methodological developments.
Molecular dynamics (MD) is a computer simulation technique that models the time evolution of biomolecules based on atomic force fields10. MD simulations determine the trajectory of biomolecules by solving the Newton’s equations of motion for all atoms in the system. MD simulations can be used to calculate thermodynamic free energies that characterize the behavior of biomolecules at or near equilibrium8, as well as kinetic pathways and reaction rates of biomolecules11. MD simulations are able to explore biomolecular structural transition pathways, such as protein folding and drug binding, and identify stable configurations of biomolecules.
Using a specialized supercomputer Anton, the DE Shaw Research Group has successfully captured drug/ligand binding to the Src protein kinase9, the β1- and β2-adrenergic receptors (β1AR and β1AR)7 and the M2 and M3 muscarinic receptors12 through microsecond-timescale conventional MD (cMD) simulations. The De Fabritiis group has been able to capture binding of benzamidine to the trypsin enzyme by running hundreds-of-nanosecond cMD simulations combined with Markov State Models13. However, due to limited simulation time scales, cMD may suffer from insufficient sampling and is not able to simulate complete binding pathways of slow ligand binders12. In this regard, enhanced MD methods are useful to help address the challenge.
During the last several decades, many enhanced MD methods have been developed as reviewed in previous articles14–17. Enhanced MD simulations have been applied to investigate the drug binding pathways, especially for G-protein-coupled receptors (GPCRs). Random acceleration MD (RAMD) has been applied to explore ligand unbinding pathways from rhodopsin18 and the β2AR19. Steered MD simulations have been performed to characterize ligand dissociation from the β1AR and β2AR20. Metadynamics has been successfully applied to calculate free energy profiles of drug-receptor interactions21–24 and characterize drug binding kinetics such as the binding and unbinding rates and drug residence time25, 26. Accelerated MD (aMD) simulations have been able to capture various ligand binding to the M3 muscarinic receptor6. A potential scaled-MD-based method has been implemented to predict the residence times of activator molecules in glucokinase. These approaches can be important for drug design and treating type two diabetes mellitus27. These enhanced MD techniques have greatly extended the capabilities of MD for studying both drug binding and unbinding processes. However, enhanced sampling methods including steered MD and metadynamics require predefined collective variables16, 28, which could lead to constrained sampling of the drug pathways. On the other hand, unconstrained enhanced sampling methods including RAMD and aMD do not need collective variables, but they often suffer from large energetic noise that precludes proper energetic reweighting and free energy calculations17, 28. Therefore, further method developments are needed to characterize drug binding pathways more accurately.
2. Gaussian Accelerated Molecular Dynamics
Gaussian accelerated molecular dynamics (GaMD) is built upon the development of aMD, which was introduced by Voter to accelerate infrequent transitions in solids by boosting the potential energy in regions other than the barriers29 and later innovated by Hamelberg et al. for simulating biomolecular dynamics30. The boost potential in aMD is able to smooth potential energy surfaces and thus accelerate transitions between low-energy states. Because aMD does not require predefined collective variables, it can sample molecular conformations without a priori knowledge or constraints. aMD has been successfully demonstrated on solid materials31, lipids32 and both globular and membrane proteins33–35. However, aMD is known to suffer from large energetic noise during reweighting36. While the aMD boost potential can be sufficiently low for proper reweighting of simulations on small systems such as alanine dipeptide and fast-folding proteins37, it is typically tens-to-hundreds of kilocalories per mole in simulations of normal proteins, which does not allow accurate reweighting for free energy calculations.
GaMD has been developed to achieve both unconstrained enhanced sampling and proper energetic reweighting for free energy calculations of large biomolecules such as proteins38. GaMD accelerates protein conformational transitions and ligand binding by orders of magnitude39, 40. Furthermore, because the boost potential follows a Gaussian distribution, we can properly recover the original free energy profiles of biomolecules through “Gaussian approximation”. GaMD allows us to more accurately characterize conformational changes of the fast-folding proteins38, 41, GPCRs42–45, CRISPR-Cas946, 47, T cell receptor signaling protein48, virus enzymes49, 50, bacterial biosynthesis enzymes51, antifungal proteins52, plant ion channel53, human dystonia related protein54 and other biomolecules39. Without the need to set predefined collective variables as in aMD, GaMD is particularly advantageous for studying complex protein-protein interactions45, 48, 54 and ligand binding processes38, 41, 42, 50, 53, 55. Here, we review the applications of GaMD for elucidation of drug pathways. GaMD simulations have so far been applied to determine pathways of drug binding to the GPCRs and HIV protease. GaMD has proven to be a promising tool for prediction of drug–receptor interactions. Future applications of GaMD to a wider range of target receptors will help us to understand functional mechanisms of various drug molecules. GaMD will thus facilitate computer-aided drug design.
In GaMD, a harmonic boost potential is applied to smooth the potential energy surface and reduce system energy barriers38 (Figure 1). Since details of the method has been described extensively in previous studies38, 39, 41, a brief summary is provided here. Consider a system with N atoms at positions . When the system potential is lower than a reference energy E, the modified potential of the system is calculated as:
| (1) |
where k is a harmonic force constant. The two parameters E and k can be determined based on three enhanced sampling principles as described earlier38. E needs to be set in a range:
| (2) |
where Vmax and Vmin are the system minimum and maximum potential energies. To ensure that Eqn. (2) is valid, let us define , then . The standard deviation of needs to be small enough to ensure accurate energetic reweighting37: , where and are the average and standard deviation of the system potential energies, is the standard deviation of with as a user-specified upper limit for accurate reweighting. When E is set to the lower bound E=Vmax, according to Eqn. (2), can be calculated as:
| (3) |
Alternatively, when the threshold energy E is set to its upper bound , is set to:
| (4) |
if is found to be between 0 and 1. Otherwise, is calculated using Eqn. (3).
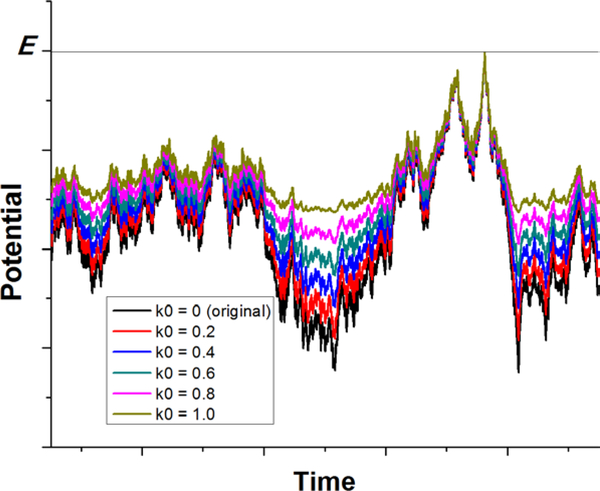
Figure 1:
Schematic illustration of Gaussian accelerated molecular dynamics (GaMD): biomolecular potential energy surface is smoothed by adding a harmonic boost potential that follows a Gaussian distribution. The original potential energy surface is shown in black. The modified potential energy surfaces obtained after adding the boost potential with different effective harmonic constants k0 are shown in red (0.2), blue (0.4), cyan (0.6), purple (0.8), and yellow (1.0). Adapted with permission from Y. Miao, V.A. Feher, J.A. McCammon, Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11(8), 3584–3595.
GaMD has been implemented in the AMBER and NAMD software packages38, 41. GaMD currently provides three options to apply the boost potential: (1) only the total potential boost , (2) only dihedral potential boost , and (3) the dual potential boost (both and ). The dual-boost simulation generally provides higher acceleration than the other two types of simulations for enhanced sampling40. The GaMD acceleration parameters comprise of the threshold energy values and the effective harmonic force constants, and for the total and dihedral potential boost, respectively.
In practice, initial energy minimization and equilibration on systems of interest follow standard MD simulation39. GaMD simulation then proceeds in three stages: (1) short cMD, (2) GaMD equilibration, and (3) GaMD production. During the first stage of short cMD, system potential statistics (including the minimum, maximum, average and standard deviation) are collected to calculate the GaMD acceleration parameters. In the second stage of GaMD equilibration, the system potential statistics are updated to recalculate the GaMD acceleration parameters on the fly, which are expected to level off (e.g., the effective harmonic force constants, and approach 1) near the end of GaMD equilibration. Note that the potential statistics are not collected or updated during a small set of steps called preparation steps at the onset of cMD and equilibration stage, where system are allowed to adapt to the simulation environment39. In the third stage of GaMD production, boost potential is applied to the system with GaMD acceleration parameters fixed. Simulation frames and the corresponding boost potential values are saved for analysis.
Since GaMD does not require system-dependent collective variables, it is easy to set simulation parameters that are generally transferrable to different systems. Statistics of system potential energies are collected from short cMD and updated with GaMD equilibration. Then it is straightforward to calculate the boost potential and apply it to the system in the GaMD production simulation. The GaMD input parameters mainly involve the number of simulation steps needed for cMD, GaMD equilibration and GaMD production, while the other parameters can be set to the default values as demonstrated through previous successful simulations39. Notably, the reference energy E can be set to the lower bound, i.e., E=Vmax. The upper limits of the standard deviation of the total and dihedral boost potentials ( and ) are set to 6.0 kcal/mol, which are used to automatically calculate force constants of the boost potentials. Table 1 summarizes parameters of GaMD simulations performed on drug binding to protein targets including the HIV protease, the M2 and M3 muscarinic GPCRs and adenosine A1 receptor (A1AR).
Table 1:
Parameters of GaMD simulations performed on drug binding to target receptors.
| Systema | Program | Boostb | cMD (ns) | GaMD Equilibration (ns) | GaMD Production (ns) |
|---|---|---|---|---|---|
| HIV Protease | AMBER | Dual | 10 | 50 | 2500, 2500, 2436, 2434, 1445, 500, 500, 500, 500, 500 |
| M2-QNB | AMBER | Dual | 10 | 50 | 400, 400, 400 |
| M2-ARC | AMBER | Dual | 10 | 50 | 2030, 1520, 1190, 650, 650, 650, 620, 600, 600, 540 |
| M2-IXO | AMBER | Dual | 10 | 50 | 1050, 1000, 940, 500, 500, 500, 500, 480, 450, 410 |
| M3 | NAMD | Dual | 10 | 50 | 400, 300, 300 |
| A1AR | AMBER | Dual | 2 | 50 | 500, 500, 500, 500, 500 |
| NAMD | Dual | 2 | 50 | 300, 300, 300, 300, 300 | |
| NAMD | Dihedral | 2 | 50 | 200, 200, 200, 200, 200 | |
QNB is the 3-quinuclidinyl-benzilate inverse agonist, ARC is the arecoline partial agonist and IXO is the iperoxo full agonist of the M2 muscarinic receptor.
The “Dual” and “Dihedral” stand for dual-boost and dihedral-boost GaMD simulations, respectively.
With GaMD simulations, Python toolkit “PyReweighting” is available for energetic reweighting to analyze boost potential distribution and calculate free energy profiles37. Importantly, because the boost potential follows a Gaussian distribution, the original free energy profiles of biomolecules can be recovered through “Gaussian approximation” or cumulant expansion to the second order38. GaMD solves the energetic noise problem encountered in the previous aMD method36. GaMD has been compared with the previous aMD on the performance of smoothing the potential energy surface and energetic reweighting38. Taking the T4-lysozyme as a protein model, higher average boost potential was applied in the GaMD simulation (36.36 kcal/mol) than in the aMD simulation (29.85 kcal/mol). However, the GaMD boost potential exhibited narrower distribution with smaller standard deviation in the GaMD simulation (4.72 kcal/mol) than in the aMD simulation (6.78 kcal/mol). Anharmonicity of the boost potential distribution was significantly reduced from 6.21 × 10−3 in the aMD simulation to 1.39 × 10−3 in the GaMD simulation. Moreover, standard deviations of the modified potential energies were significantly smaller in the GaMD simulation than in aMD simulation. The protein potential energy surface was thus smoother in GaMD than in aMD. Provided the smoother potential energy surface as well as the narrower distribution and lower anharmonicity of the boost potential, GaMD allows for more efficient enhanced sampling and improved free energy calculations for proteins and other large biomolecules.
3. Ligand Binding to the HIV Protease
The HIV protease is an important target for drug design to treat HIV infection50, 56. It is a retroviral aspartyl protease with two glycine rich flaps. The two flaps play a key role in gating access of ligands to the protein active site. In X-ray crystal structures, the two flaps adopt a semi-open conformation in the ligand-free (apo) form of the protein, but a closed conformation in the ligand-bound (holo) protein57, 58 (Figure 2A). In addition, the protein flaps are able to sample an open conformational state59. Recently, Huang et al. investigated ligand binding to the HIV protease through long timescale MD simulations, which revealed important insights into the ligand binding mechanism56. However, in a 14 μs Anton cMD simulation, the sampling ligand XK263 (a cyclic urea inhibitor) could not reach the protein active site with a minimum root-mean-square deviation (RMSD) of 5.73 Å compared with the X-ray conformation.
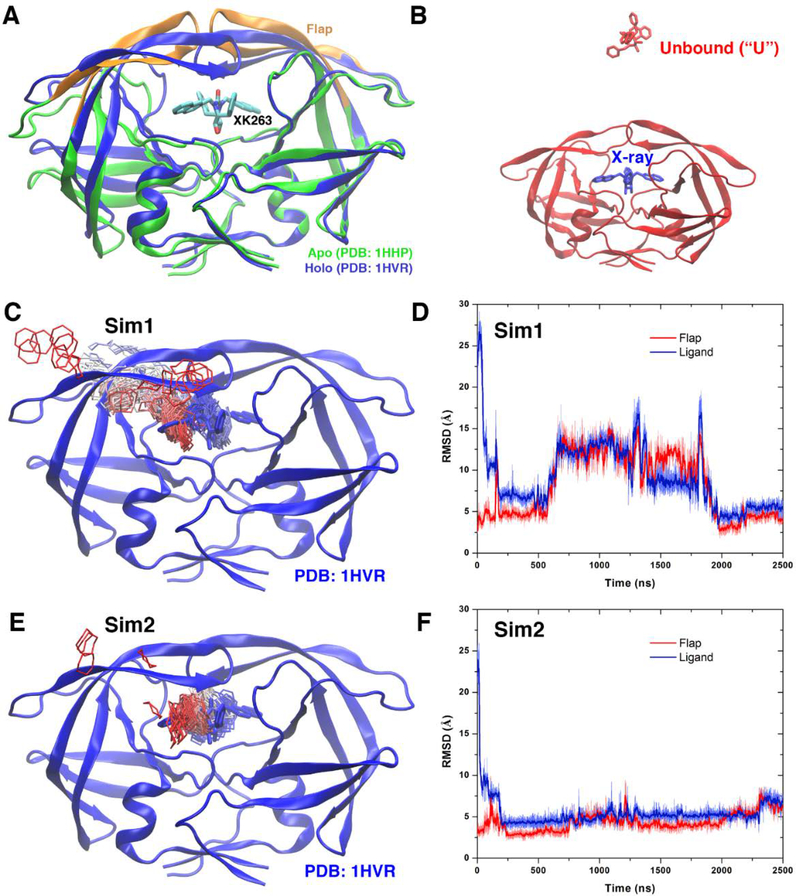
Figure 2:
(A) X-ray structures of the HIV protease in the apo form (PDB: 1HHP, green) and holo form that is bound by the XK263 ligand (PDB: 1HVR, blue). The ligand is shown as sticks and the protein as ribbons. Protein residues Lys43-Tyr59 in the flaps of two protein monomers are colored orange. (B) Simulation starting structure of the HIV protease in which the ligand molecule (red sticks) is placed ∼20 Å from the protein surface in the unbound (“U”) state. A virtual ligand molecule of the X-ray conformation is colored blue for reference. (C) During the “Sim1” GaMD trajectory, the XK263 ligand molecule binds to the active site of the HIV protease within 2500 ns, for which the center ring of XK263 is represented by lines and colored by simulation time in a red-white-blue (RWB) color scale. The 1HVR X-ray conformation is colored blue for reference. (D) Root-mean-square deviations (RMSDs) of the ligand molecule and protein flaps relative to the 1HVR X-ray conformation are plotted for “Sim1”. Thick lines depict the running average over 5 ns time windows. (E and F) Ligand binding pathway and ligand and flap RMSDs obtained from the “Sim2” GaMD trajectory, respectively. Adapted with permission from Miao Y, Huang Y-mM, Walker RC, McCammon JA, Chang C-eA. Ligand binding pathways and conformational transitions of the HIV protease. Biochemistry. 2018;57(9):1533–41.
To overcome sampling limitation of the cMD simulation, GaMD enhanced simulations were applied to capture complete ligand binding to the HIV protease50. Specifically, ten independent GaMD simulations lasting 500−2500 ns were performed starting from the apo HIV protease in the semi-open conformation, with XK263 placed ~20 Å away from the protein (Figure 2B). During two of the GaMD simulations lasting 2500 ns, XK263 was observed to successfully bind the protein active site with a minimum RMSD of only 2.26 Å relative to the X-ray structure. In the first successful simulation Sim 1 (Figures 2C and 2D), XK263 initially attached to one of the protein flaps and then entered the active site, which induced protein flaps to open. The flaps significantly rearranged their conformation to accommodate the ligand. Finally, the protein flaps switched back to closed conformation and the ligand was locked in the protein active site. In the second successful GaMD simulation Sim 2 (Figures 2E and 2F), the ligand reached a minimum RMSD of 2.74 Å. The two protein flaps opened for relatively shorter time and smaller magnitude before changing back to closed ligand-bound conformation. Both of these simulations captured complete ligand binding to the HIV proteases within 2500 ns simulation time. The GaMD simulations thus provided significantly improved sampling compared with the previous 14 μs Anton cMD simulation.
Furthermore, correlations were found between ligand binding and conformational changes of the HIV protease. Two intermediate conformational states were identified from free energy profiles obtained from the GaMD simulations. In the intermediate-1 state, the two protein flaps adopted an open conformation. XK263 moved close to one of the protein flaps, forming nonpolar interactions with residues Val32’, Ile47’, Ile50’, Pro81’ and Val82’. Oxygen atoms of the ligand formed further hydrogen bonds with protein residues Asp25’ and Ile50’. In the intermediate-2 state, while one protein flap remained open, the other closed towards the active site55. Therefore, the protein underwent significant conformational changes via distinct intermediate states during ligand binding. The GaMD simulations revealed the mechanism of highly dynamic ligand binding in the HIV protease50.
4. Drug pathways of G-protein-coupled receptors (GPCRs)
GPCRs represent the largest family of human membrane proteins. Due to the critical roles in various physiological functions, they are the primary targets of about one third of currently marketed drugs8. GPCRs transduce cellular signals often by binding of extracellular ligands, such as hormones, neurotransmitters and chemokines8. In this regard, we have performed GaMD simulations to study mechanisms of drug binding in several prototypical GPCRs.
4.1. M2 Muscarinic GPCR
The M2 muscarinic acetylcholine receptor is one of the five subtypes of muscarinic GPCRs. It is widely distributed in mammalian tissues and involved in crucial cardiac and neural functions. Activation of the M2 receptor results in a decrease in the human heart rate and reduction in the heart contractile forces. The M2 receptor has thus been targeted for treating abnormal heart rate, heart failure and neurological diseases. X-ray crystal structures have been determined for the M2 receptor bound by the 3-quinuclidinyl-benzilate (QNB) inverse agonist60 and in complex with the iperoxo (IXO) full agonist and a G-protein mimetic nanobody Nb9–861. The first X-ray structure of allosteric ligand-bound GPCR has also been obtained for the M2 receptor with the LY211926 compound bound in the receptor extracellular vestibule61.
In a recent study, GaMD was applied to simulate the M2 receptor that was bound by different ligands, including QNB, IXO and a partial agonist arecoline (ARC)42. In the absence of the G protein-mimetic nanobody Nb9–8, the QNB inverse agonist with ~0.06 nM high binding affinity was tightly bound to the receptor orthosteric site. In contrast, the full and partial agonists with lower affinities (~5 μM for ARC and ~0.01 μM for IXO) exhibited significantly higher fluctuations. IXO could move out of the orthosteric pocket and to the receptor extracellular vestibule42. During a 2030 ns GaMD simulation, not only did ARC escape out of the orthosteric pocket, but it also dissociated completely from the receptor and rebound to the receptor repeatedly (Figure 3A). The partial agonist exited and entered the receptor orthosteric pocket through opening between the extracellular loops 2 and 3 (ECL2 and ECL3) as depicted by Figures 3B–3H. The drug pathway will be defined more quantitatively through structural clustering of the diffusing ligand and free energy calculations in the following. To our knowledge, it was for the first time that both drug dissociation and binding was captured in one single all-atom GPCR simulation, which demonstrates the exceptional enhanced sampling power of GaMD.
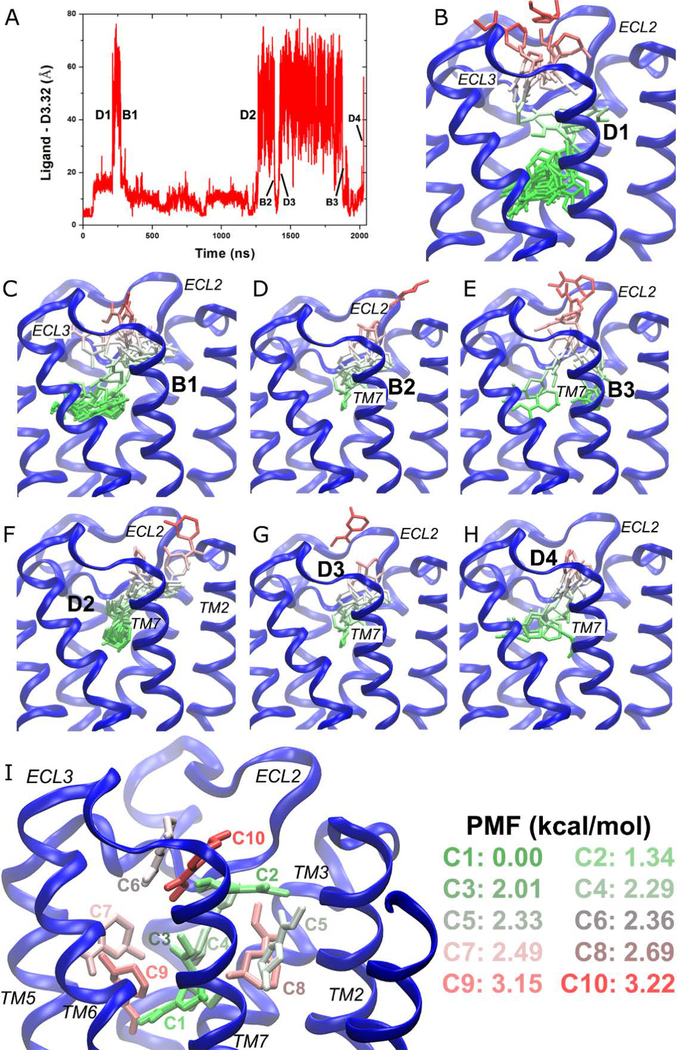
Figure 3:
GaMD simulations revealed pathways of dissociation and binding of the arecoline (ARC) partial agonist in the M2 muscarinic GPCR: (A) timecourse of the distance between ARC and residue Asp1033.32 of the M2 receptor during 2030 ns GaMD simulation. Four dissociation and three binding events are labeled. (B–H) Schematic representations of the ligand pathways during (B) “D1,” (C) “B1,” (D) “B2,” (E) “B3,” (F) “D2,” (G) “D3,” and (H) “D4.” The receptor is represented by blue ribbons and the ligand by sticks colored by the position along the membrane normal. (I) Ten lowest energy structural clusters of ARC that are labeled and colored in a GWR scale according to the PMF values. Adapted with permission from Y. Miao, J.A. McCammon, Graded activation and Free Energy Landscapes of a Muscarinic G-Protein-Coupled Receptor. Proc. Natl. Acad. Sci. U. S. A. 2016, 113(43), 12162–12167.
With the GaMD simulation, snapshots of the diffusing ligand were clustered and the PyReweighting toolkit37 was applied to compute free energies of the ligand clusters. Ten clusters with the lowest free energies were identified for the partial agonist ARC in the M2 receptor (Figure 3I). Cluster “C1” with the zero energy minimum was located at the orthosteric site, whereas cluster “C2” with the second lowest energy was at the center of the extracellular vestibule between ECL2 and transmembrane (TM) helix 7. Moreover, “C1” at the orthosteric site and “C2” in the extracellular vestibule were connected by two clusters of higher energies in the receptor, “C3” and “C4”. Another two clusters “C5” and “C8” were found in the cavity formed by extracellular domains of TM3/TM2/TM7. Two clusters “C7” and “C9” filled the cavity formed by the TM4/TM5/TM6 extracellular domains. Importantly, the extracellular opening between ECL2/ECL3 had two energetically favored clusters, i.e., “C6” and “C10”. Therefore, clusters “C1” ↔ “C3” ↔ “C4” ↔ “C2” ↔ “C10” ↔ “C6” represent an energetically preferred pathway for dissociation and binding of ARC. IXO followed the same pathway for dissociation42. This route was found to be a favored pathway for drug binding of β2AR as well7. Moreover, GaMD identified the orthosteric pocket and extracellular vestibule as the two low-energy binding sites of ARC, which was consistent with previous binding assay experiments. These experiments suggested that partial agonists have two or more binding sites at the M2 receptor62. In summary, GaMD unprecedentedly captured both drug dissociation and binding in a single all-atom GPCR simulation and elucidated the drug pathways. Such information will be extremely valuable for drug design of GPCRs.
4.2. M3 Muscarinic GPCR
The M3 muscarinic GPCR is highly expressed in human epithelial and endothelial cells, especially in the exocrine glands and smooth muscle tissues63. It is implicated in human diabetes and cancers in the brain, breast, colon, ovary, lung and skin. The X-ray crystal structure of the M3 receptor has been determined in an inactive state bound to the tiotropium (TTP) antagonist12. While the X-ray structure provided important insights into the receptor-antagonist interactions, the mechanism and pathways of agonist binding remain unknown.
To shed light onto the agonist binding pathways, GaMD was applied to simulate the M3 receptor, with the endogenous agonist acetylcholine initially placed far away from the receptor (Figure 4A)41. Three independent GaMD simulations lasting 300 – 400 ns were performed. While the agonist visited only the receptor extracellular vestibule in two 300 ns GaMD simulations, it entered the receptor and bound to the receptor orthosteric site in a 400 ns GaMD simulation (Figures 4B and 4C). The agonist reached a minimum RMSD of 2.0 Å at the orthosteric site compared to a reference binding pose obtained from Glide docking.
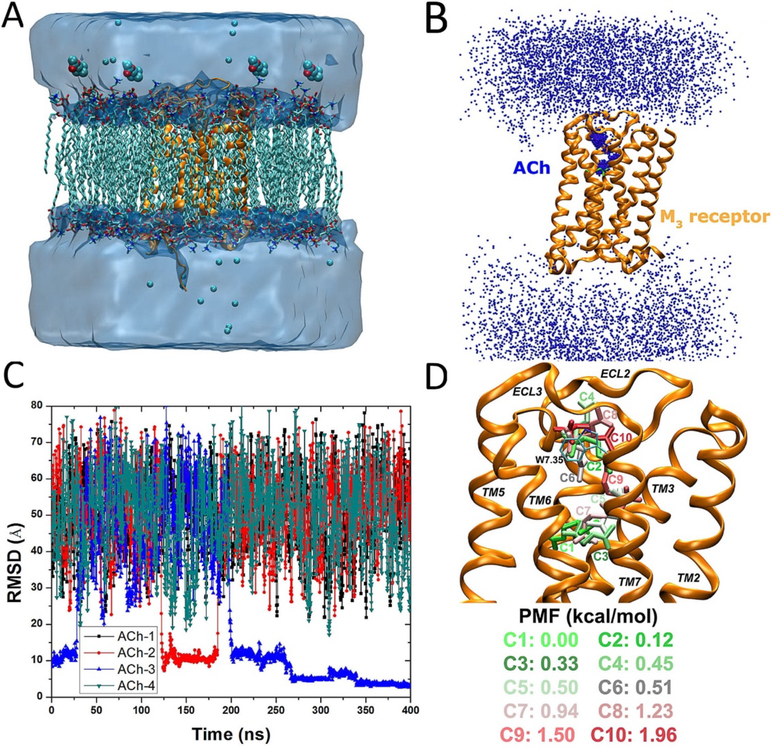
Figure 4:
GaMD simulations captured binding of the acetylcholine (ACh) endogenous agonist to the M3 muscarinic GPCR: (A) schematic representation of the computational model, in which the receptor is shown in ribbons (orange), lipid in sticks, ions in small spheres, and four ligand molecules in large spheres, (B) trace of the ACh (the N atom in blue dots) diffusing in the bulk solvent and bound to the M3 receptor (orange ribbons), in which the Glide docking pose of ACh is shown in green sticks, (C) the RMSD of the diffusing ACh relative to the Glide docking pose calculated from the 400 ns GaMD simulation, and (D) ten lowest energy structural clusters of ACh that are labeled and colored in a green–white–red (GWR) scale according to the PMF values obtained from reweighting of the GaMD simulation. Reproduced with permission from Y.T. Pang, Y. Miao, Y. Wang, J.A. McCammon, Gaussian Accelerated Molecular Dynamics in NAMD. J. Chem. Theory Comput. 2017, 13(1), 9–19.
Furthermore, the DBSCAN algorithm64 was applied to cluster the diffusing ligand, followed by energetic reweighting37, 38 to identify low-energy poses and binding pathway of the agonist in the M3 receptor. Ten different clusters with the lowest free energies were shown in Figure 4D. The lowest energy cluster “C1” was at the orthosteric site. The second lowest energy cluster “C2” was located in extracellular vestibule between ECL2/ECL3, where the positively charged N atom of the agonist formed cation–π interactions with residue Trp5257.35. Similarly, cluster “C3” was identified in the orthosteric pocket but with relatively higher energy. Clusters “C4”, “C6”, “C8” and “C10” were located in the extracellular vestibule. Similar to the M2 receptor, the M3 receptor adopted the same energetically preferred pathway for agonist binding from the ECL2/ECL3 opening to the orthosteric site.
4.3. Adenosine A1 Receptor
Four subtypes of class A GPCRs (the A1, A2A, A2B and A3) mediate the effects of adenosine and play key roles in physiological functions65. In particular, the adenosine A1 receptor (A1AR) has served as an important drug target for treating cardiovascular diseases, ischemia-reperfusion injury and neuropathic pain65. Because adenosine receptors are widely distributed throughout the human body, off-target side-effects have greatly hindered the development of agonists as effective drugs66. Therefore, it is appealing to design selective allosteric modulators that regulate responsiveness of the A1AR to endogenous adenosine in local regions of its production67, 68. However, structural information has been very scarce for guiding design of allosteric modulators of the A1AR. The mechanisms of allosteric modulator binding to the receptor remain unknown, which undermines the success of structure-activity relationship studies and greatly increases the cost of drug design68.
GaMD implemented in the NAMD and AMBER software packages was applied to simulate spontaneous binding of two prototypical positive allosteric molecules (PAMs), PD8172369 and VCP1770, to the A1AR55. The first X-ray crystal structure of A1AR71 was used. The antagonist was replaced by the 5’-N-ethylcarboxamidoadenosine (NECA) agonist. During the GaMD simulations, both PD81723 and VCP17 PAMs were observed to bind an allosteric site formed by ECL2 of the A1AR, which was in excellent agreement with the mutagenesis experimental data5, 72. Receptor residues that were identified within 5 Å of the bound PAMs in the GaMD simulations were shown to be important for PAM binding in the mutagenesis experiments. Mutations of these residues into alanine significantly changed the PAM binding affinity, binding cooperativity between the PAM and NECA agonist, the receptor efficacy and/or the functional cooperativity between the PAM and NECA agonist5, 72. Therefore, the GaMD simulations and mutation experiments combined provide a structural basis for binding of PAMs at the ECL2 allosteric site in the A1AR.
Additionally, the GaMD simulations showed that the PAMs enhanced agonist binding at the receptor orthosteric site. The NECA agonist moved slightly with PAMs bound at the ECL2 allosteric site. In the absence of PAM binding at the ECL2, the agonist explored a significantly larger conformational space and could even dissociate from the A1AR. The NECA agonist exited through the receptor opening between ECL2 and ECL3. This energetically preferred pathway connecting the orthosteric site and ECL2/ECL3 opening is consistent with findings on the M242 and M3 muscarinic receptors41 as described above. An antagonist ZM241385 dissociated from the A2AAR through the same pathway as observed in the Anton simulations using temperature accelerated MD73. Mutations of residues near the ECL2/ECL3 opening changed dissociation rates of an antagonist ligand from the A2AAR73 and PAM binding to the A1AR (putatively at the ECL2) changed kinetic rates of antagonist and agonist ligands at the A1AR74, 75. Therefore, the GaMD simulations are consistent with previous experimental73–75 and computational6, 7, 12 studies, suggesting that orthosteric ligands of class A GPCRs bind to target receptors from the ECL2/ECL3 opening at the extracellular opening to the orthosteric site as an energetically preferred pathway.
4.4. Mu Opioid Receptor
The mu opioid receptor (MOR) is a key GPCR that is expressed in neural structure and spinal cords. The MOR has served as an important drug target, especially for treating pain and neurological diseases. X-ray structures of the MOR have been determined in both active (bound by agonist BU72) and inactive (bound by antagonist β-funaltrexamine). But extensive research efforts are still needed for understanding the receptor functional mechanisms and designing effective drug molecules.
Wang and Chan76 applied GaMD to study the binding mechanisms of agonist BU72 and antagonist β-funaltrexamine in the MOR. PMF profiles calculated from five individual 1000 ns GaMD simulations identified one major and one minor energy barrier for both the agonist and antagonist ligands. According to root-mean–squared fluctuations (RMSFs) calculated from the GaMD simulations, Lys209–Phe221 and Ile301–Cys321 of the MOR were identified as 34 key residues for binding of the two ligands in receptor active site. When the ligand was bound, the MOR adopted an open conformation with greater binding pocket surface area (BPSA). In contrast, the MOR changed to a closed conformation with decreased BPSA in the absence of ligand binding76. In addition, Liao and Wang44 applied GaMD simulations to investigate the inactivation and activation mechanisms of the MOR. The simulations revealed four conformational states (including the active, inactive and two intermediate states) of the MOR.
5. Conclusions
Pharmacologically important proteins such as the HIV protease and GPCRs are implicated in viral infection and numerous human diseases including the diabetes, heart failure, asthma, arthritis, renal diseases and neurological disorders. Despite remarkable advances, significant research efforts are still needed for us to understand the structural dynamics and functional mechanisms of these proteins, which will ultimately facilitate rational drug design of the protein targets. In particular, understanding the mechanisms and pathways of drug binding to a receptor for effective drug design and biomedical research is crucial. Due to limited temporal and spatial resolution, it has been difficult to determine drug pathways using experimental techniques. In contrast, computer simulations are able to overcome these limitations and reveal drug pathways at an atomistic level.
Based on unconstrained enhanced sampling, GaMD has proven useful in capturing spontaneous drug binding to different target receptors, in addition to studies of large-scale conformational changes in proteins and other important biomolecules39. In particular, GaMD has been successfully applied to elucidate the drug pathways in the HIV protease and a number of different GPCRs as summarized in this study. With all-atom description for systems of interest and classical force fields, GaMD is generally applicable to any drug-receptor binding systems. More applications of GaMD in elucidation of drug pathways are thus expected in the future.
6. Expert Opinion
GaMD is a robust computational method for simultaneous unconstrained enhanced sampling and free energy calculations of biomolecular dynamics, in particular drug binding to target receptors. GaMD is able to speed up biomolecular simulations by orders of magnitude and greatly reduce our computational cost. GaMD does not require predefined collective variables, which is especially advantageous for simulating “free” drug binding processes. Furthermore, by adding a boost potential that follows a Gaussian distribution, cumulant expansion to the second order (Gaussian approximation) can be applied to reweight GaMD simulations for free energy calculations. The free energy profiles allow us to identify low-energy states and binding pathways of the drug molecules.
Since GPCRs are primary targets of about one third of currently marketed drugs, they serve as excellent model systems for computational studies of drug binding. Computer simulations using cMD successfully captured drug binding to GPCRs, but they needed to be performed on the microsecond timescales and required an expensive and specialized supercomputer Anton7. In comparison, GaMD enhanced simulations were able to capture drug binding to GPCRs within hundreds of nanoseconds41, 55, which can be routinely done using personal computers with GPUs or common computing clusters. Microsecond GaMD simulations were able to capture both drug dissociation and binding in a muscarinic GPCR42. Thus, GaMD greatly reduces our computational cost for studying drug binding to target receptors.
In addition, GaMD is particularly advantageous for elucidation of drug pathways based on simultaneous unconstrained enhanced sampling and free energy calculations. Notably, a number or different enhanced MD methods have been applied to study ligand binding and dissociation processes of GPCRs. Ligand dissociation from β2AR was successfully simulated in a previous RAMD study, but it was difficult to capture rebinding of the ligand as limited by the method. Similar probability was found for the ligand to exit through the ECL2/ECL3 and ECL2/TM2/TM7 openings19. In another study, steered MD obtained free energy profiles of ligand dissociation from the β1AR and β2AR. The simulations suggested that the two routes “may serve indistinguishably for ligand entry and exit”. This was contradictory to more recent finding obtained from long-timescale Anton cMD simulations7. Moreover, the ligand was constrained to predetermined channels and could not follow the “real” pathways7. In comparison, GaMD is a powerful unconstrained enhanced sampling technique that allows us to characterize the drug pathways more accurately through simulation-derived free energy profiles. GaMD has shown that the pathway connecting the orthosteric site and the ECL2/ECL3 opening is energetically favorable for ligand binding to the M2 and M3 muscarinic receptors39, 41, 42 and A1AR39, 41, 42, 55. This route appears to be a common pathway for drug binding to class A GPCRs. Among the unconstrained enhanced sampling methods that do not require predefined collective variables, replica exchange simulations have also been successfully applied to predict protein-ligand binding structures77–79. Our recent study showed improved enhanced conformational sampling of proteins was achieved by combining GaMD with replica exchange (i.e., rex-GaMD)80. It is subject to future studies whether rex-GaMD can further help with sampling the drug pathways. In summary, GaMD serves as a promising tool for the prediction of drug–receptor interactions and will facilitate computer-aided drug design and discovery.
Article Highlights.
Compared with conventional molecular dynamics, GaMD is a robust enhanced sampling technique that accelerates computer simulations by orders of magnitude.
GaMD is advantageous for elucidation of drug pathways based on simultaneous unconstrained enhanced sampling and free energy calculations.
GaMD simulations have successfully captured ligand binding to the HIV protease with a minimum RMSD of 2.26 Å compared with the X-ray structure.
GaMD simulations have revealed pathways of both dissociation and binding of a partial agonist in the M2 muscarinic GPCR and agonist binding to the M3 muscarinic receptor.
GaMD simulations have elucidated binding modes of prototypical allosteric modulators and provided important insights into allostery in the adenosine A1 receptor.
Acknowledgements
This work was supported in part by the American Heart Association (Award 17SDG33370094) and the College of Liberal Arts and Sciences at the University of Kansas.
References
- 1.Stromgaard K, Krogsgaard-Larsen P, Madsen U. Textbook of drug design and discovery: CRC Press, 2016. [Google Scholar]
- 2.Kallen J, Spitzfaden C, Zurini MG, Wider G, Widmer H, Wüthrich K, et al. Structure of human cyclophilin and its binding site for cyclosporin A determined by X-ray crystallography and NMR spectroscopy. Nature 1991;353(6341):276. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson KA, Costanzi S. New insights for drug design from the X-ray crystallographic structures of G-protein-coupled receptors. Molecular pharmacology 2012;82(3):361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan L, Appelman C, Smith AR, Yu J, Larsen S, Marquez RT, et al. Natural product (−)‐gossypol inhibits colon cancer cell growth by targeting RNA‐binding protein Musashi‐1. Mol Oncol 2015;9(7):1406–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen AT, Vecchio EA, Thomas T, Nguyen TD, Aurelio L, Scammells PJ, et al. Role of the Second Extracellular Loop of the Adenosine A1 Receptor on Allosteric Modulator Binding, Signaling, and Cooperativity. Molecular pharmacology 2016. December;90(6):715–25. [DOI] [PubMed] [Google Scholar]
- 6.Kappel K, Miao Y, McCammon JA. Accelerated molecular dynamics simulations of ligand binding to a muscarinic G-protein-coupled receptor. Quarterly reviews of biophysics 2015;48(4):479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, et al. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proceedings of the National Academy of Sciences 2011;108(32):13118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao Y, McCammon JA. G-protein coupled receptors: advances in simulation and drug discovery. Current opinion in structural biology 2016;41:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan Y, Kim ET, Eastwood MP, Dror RO, Seeliger MA, Shaw DE. How does a drug molecule find its target binding site? Journal of the American Chemical Society 2011;133(24):9181–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nature Structural and Molecular Biology 2002;9(9):646. [DOI] [PubMed] [Google Scholar]
- 11.Wong CF. Molecular simulation of drug-binding kinetics. Molecular Simulation 2014;40(10–11):889–903. [Google Scholar]
- 12.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 2012;482(7386):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buch I, Giorgino T, De Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc Natl Acad Sci 2011;108(25):10184–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christen M, Van Gunsteren WF. On searching in, sampling of, and dynamically moving through conformational space of biomolecular systems: A review. Journal of computational chemistry 2008;29(2):157–66. [DOI] [PubMed] [Google Scholar]
- 15.Spiwok V, Sucur Z, Hosek P. Enhanced sampling techniques in biomolecular simulations. Biotechnology advances 2015;33(6):1130–40. [DOI] [PubMed] [Google Scholar]
- 16.Abrams C, Bussi G. Enhanced Sampling in Molecular Dynamics Using Metadynamics, Replica-Exchange, and Temperature-Acceleration. Entropy-Switz 2014. January;16(1):163–99. [Google Scholar]
- 17.Miao Y, McCammon JA. Unconstrained Enhanced Sampling for Free Energy Calculations of Biomolecules: A Review. Mol Simulat 2016;42(13):1046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Duan Y. Chromophore channeling in the G-protein coupled receptor rhodopsin. J Am Chem Soc 2007;129(22):6970–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T, Duan Y. Ligand entry and exit pathways in the β2-adrenergic receptor. J Mol Biol 2009;392(4):1102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González A, Perez-Acle T, Pardo L, Deupi X. Molecular basis of ligand dissociation in β-adrenergic receptors. Plos One 2011;6(9):e23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provasi D, Bortolato A, Filizola M. Exploring molecular mechanisms of ligand recognition by opioid receptors with metadynamics. Biochemistry-Us 2009;48(42):10020–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun HY, Chen PC, Li D, Li YY, Hou TJ. Directly Binding Rather than Induced-Fit Dominated Binding Affinity Difference in (S)- and (R)-Crizotinib Bound MTH1. J Chem Theory Comput 2016. February;12(2):851–60. [DOI] [PubMed] [Google Scholar]
- 23.Sun HY, Li YY, Tian S, Wang JM, Hou TJ. P-loop Conformation Governed Crizotinib Resistance in G2032R-Mutated ROS1 Tyrosine Kinase: Clues from Free Energy Landscape. Plos Comput Biol 2014. July;10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleh N, Ibrahim P, Saladino G, Gervasio FL, Clark T. An Efficient Metadynamics-Based Protocol To Model the Binding Affinity and the Transition State Ensemble of G-Protein-Coupled Receptor Ligands. J Chem Inf Model 2017. May 22;57(5):1210–17. [DOI] [PubMed] [Google Scholar]
- 25.Sun HY, Li YY, Shen MY, Li D, Kang Y, Hou TJ. Characterizing Drug Target Residence Time with Metadynamics: How To Achieve Dissociation Rate Efficiently without Losing Accuracy against Time-Consuming Approaches. J Chem Inf Model 2017. August;57(8):1895–906. [DOI] [PubMed] [Google Scholar]
- 26.Casasnovas R, Limongelli V, Tiwary P, Carloni P, Parrinello M. Unbinding Kinetics of a p38 MAP Kinase Type II Inhibitor from Metadynamics Simulations. J Am Chem Soc 2017. April 5;139(13):4780–88. [DOI] [PubMed] [Google Scholar]
- 27.Mollica L, Theret I, Antoine M, Perron-Sierra Fo, Charton Y, Fourquez J-M, et al. Molecular dynamics simulations and kinetic measurements to estimate and predict protein–ligand residence times. J Med Chem 2016;59(15):7167–76. [DOI] [PubMed] [Google Scholar]
- 28.Zuckerman DM. Equilibrium sampling in biomolecular simulations. Annu Rev Biophys 2011;40:41–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voter AF. Hyperdynamics: Accelerated molecular dynamics of infrequent events. Physical Review Letters 1997;78(20):3908. [Google Scholar]
- 30.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. The Journal of chemical physics 2004;120(24):11919–29. [DOI] [PubMed] [Google Scholar]
- 31.Perez D, Uberuaga BP, Shim Y, Amar JG, Voter AF. Accelerated molecular dynamics methods: introduction and recent developments. Annu Rep Comp Chem 2009;5:79–98. [Google Scholar]
- 32.Wang Y, Markwick PR, de Oliveira CAF, McCammon JA. Enhanced lipid diffusion and mixing in accelerated molecular dynamics. J Chem Theory Comput 2011;7(10):3199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce LC, Salomon-Ferrer R, Augusto F. de Oliveira C, McCammon JA, Walker RC. Routine access to millisecond time scale events with accelerated molecular dynamics. J Chem Theory Comput 2012;8(9):2997–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao Y, Feixas F, Eun C, McCammon JA. Accelerated molecular dynamics simulations of protein folding. Journal of computational chemistry 2015;36(20):1536–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao Y, Nichols SE, Gasper PM, Metzger VT, McCammon JA. Activation and dynamic network of the M2 muscarinic receptor. Proceedings of the National Academy of Sciences 2013;110(27):10982–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen T, Hamelberg D. A statistical analysis of the precision of reweighting-based simulations. The Journal of chemical physics 2008;129(3):034103. [DOI] [PubMed] [Google Scholar]
- 37.Miao Y, Sinko W, Pierce L, Bucher D, McCammon JA. Improved reweighting of accelerated molecular dynamics simulations for free energy calculation. J Chem Theory Comput 2014;10(7):2677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao Y, Feher VA, McCammon JA. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation. Journal of chemical theory and computation 2015;11(8):3584–95.** Introduction of the Gaussian accelerated molecular dynamcis (GaMD) method
- 39.Miao Y, McCammon JA. Gaussian Accelerated Molecular Dynamics: Theory, Implementation and Applications. Annu Rep Comp Chem 2017;13:231–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao Y Acceleration of Biomolecular Kinetics in Gaussian Accelerated Molecular Dynamics. J Chem Phys 2018;149(7):072308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang YT, Miao Y, Wang Y, McCammon JA. Gaussian Accelerated Molecular Dynamics in NAMD. J Chem Theory Comput 2017. January 10;13(1):9–19.** Implementation of GaMD in the NAMD package and GaMD simulations successfully captured spontaneous agonist binding to a muscarinic GPCR.
- 42.Miao Y, McCammon JA. Graded activation and free energy landscapes of a muscarinic G-protein–coupled receptor. Proc Natl Acad Sci 2016 October 10, 2016;113(43):12162–67.** Both drug dissociation and binding was, for the first time, captured in a single all-atom GPCR simulation using GaMD.
- 43.Wang YT, Chan YH. Understanding the molecular basis of agonist/antagonist mechanism of human mu opioid receptor through gaussian accelerated molecular dynamics method. Sci Rep 2017. August 10;7(1):7828.* GaMD simulations revealed mechanisms of agonist and antagonsit binding in the mu opioid receptor.
- 44.Liao JM, Wang YT. In silico studies of conformational dynamics of Mu opioid receptor performed using gaussian accelerated molecular dynamics. J Biomol Struct Dyn 2018. January 7:1–12. [DOI] [PubMed] [Google Scholar]
- 45.Miao Y, McCammon JA. Mechanism of the G-Protein Mimetic Nanobody Binding to a Muscarinic G-Protein-Coupled Receptor. Proc Natl Acad Sci 2018;115(12):3036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc Natl Acad Sci 2017 July 11, 2017;114(28):7260–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ricci CG, Chen JS, Miao Y, Jinek M, Doudna JA, McCammon JA, et al. Molecular mechanism of off-target effects in CRISPR-Cas9. bioRxiv 2018:421537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, et al. Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding. Cell 2018. July 26;174(3):672–87 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuang CH, Chiou SJ, Cheng TL, Wang YT. A molecular dynamics simulation study decodes the Zika virus NS5 methyltransferase bound to SAH and RNA analogue. Sci Rep 2018. April 20;8(1):6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao Y, Huang YM, Walker RC, McCammon JA, Chang CA. Ligand Binding Pathways and Conformational Transitions of the HIV Protease. Biochemistry-Us 2018. March 6;57(9):1533–41.* Microsecond-timescale GaMD simulations successfully captured ligand binding to the HIV protease, accompanied by large-scale conformational changes in the protein.
- 51.Venkatramani A, Gravina Ricci C, Oldfield E, McCammon JA. Remarkable similarity in Plasmodium falciparum and Plasmodium vivax geranylgeranyl diphosphate synthase dynamics and its implication for antimalarial drug design. Chem Biol Drug Des 2018. June;91(6):1068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Utesch T, de Miguel Catalina A, Schattenberg C, Paege N, Schmieder P, Krause E, et al. A Computational Modeling Approach Predicts Interaction of the Antifungal Protein AFP from Aspergillus giganteus with Fungal Membranes via Its gamma-Core Motif. mSphere 2018. October 3;3(5):e00377–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Wang N, Miao Y, Hauser F, Rappel W-J, McCammon JA, et al. Identification of SLAC1 anion channel residues required for CO2/bicarbonate sensing and regulation of stomatal movements. Proc Natl Acad Sci 2018;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salawu EO. The Impairment of TorsinA’s Binding to and Interactions With Its Activator: An Atomistic Molecular Dynamics Study of Primary Dystonia. Front Mol Biosci 2018;5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miao Y, Bhattarai A, Nguyen AT, Christopoulos A, May LT. Structural Basis for Binding of Allosteric Drug Leads in the Adenosine A1 Receptor. Sci Rep-Uk 2018;In review.* GaMD simulations captured spontaneous binding of allosteric drug leads to the adenosine A1 receptor and the simulation predicted binding poses were highly consistent with mutation experimental data.
- 56.Huang Y-mM, Raymundo MAV, Chen W, Chang C-eA. Mechanism of the association pathways for a pair of fast and slow binding ligands of HIV-1 protease. Biochemistry-Us 2017;56(9):1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spinelli S, Liu Q, Alzari P, Hirel P, Poljak R. The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochimie 1991;73(11):1391–96. [DOI] [PubMed] [Google Scholar]
- 58.Lam P, Jadhav PK, Eyermann CJ, Hodge CN, Ru Y, Bacheler LT, et al. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science 1994;263(5145):380–84. [DOI] [PubMed] [Google Scholar]
- 59.Hornak V, Okur A, Rizzo RC, Simmerling C. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proc Natl Acad Sci 2006;103(4):915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 2012;482(7386):547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 2013. November 20;504(7478):101–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Redka DyS, Heerklotz H, Wells JW. Efficacy as an intrinsic property of the M2 muscarinic receptor in its tetrameric state. Biochemistry-Us 2013;52(42):7405–27. [DOI] [PubMed] [Google Scholar]
- 63.Matsui M, Motomura D, Karasawa H, Fujikawa T, Jiang J, Komiya Y, et al. Multiple functional defects in peripheral autonomic organs in mice lacking muscarinic acetylcholine receptor gene for the M3 subtype. Proc Natl Acad Sci 2000;97(17):9579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ester M, Kriegel H-P, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd; 1996; 1996. p. 226–31. [Google Scholar]
- 65.Jacobson KA, Gao Z-G. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov 2006;5(3):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiesman WF, Elzein E, Zablocki J. A1 adenosine receptor antagonists, agonists, and allosteric enhancers Adenosine Receptors in Health and Disease: Springer; 2009:25–58. [DOI] [PubMed] [Google Scholar]
- 67.Romagnoli R, G Baraldi P, A Tabrizi M, Gessi S, A Borea P, Merighi S. Allosteric enhancers of A1 adenosine receptors: state of the art and new horizons for drug development. Curr Med Chem 2010;17(30):3488–502. [DOI] [PubMed] [Google Scholar]
- 68.Goblyos A, Ijzerman AP. Allosteric modulation of adenosine receptors. Bba-Biomembranes 2011. May;1808(5):1309–18. [DOI] [PubMed] [Google Scholar]
- 69.Bruns RF, Fergus JH. Allosteric enhancement of adenosine A1 receptor binding and function by 2-amino-3-benzoylthiophenes. Mol Pharmacol 1990;38(6):939–49. [PubMed] [Google Scholar]
- 70.Imlach WL, Bhola RF, May LT, Christopoulos A, Christie MJ. A positive allosteric modulator of the adenosine A1 receptor selectively inhibits primary afferent synaptic transmission in a neuropathic pain model. Molecular pharmacology 2015:mol. 115.099499. [DOI] [PubMed] [Google Scholar]
- 71.Glukhova A, Thal DM, Nguyen AT, Vecchio EA, Jörg M, Scammells PJ, et al. Structure of the adenosine A1 receptor reveals the basis for subtype selectivity. Cell 2017;168(5):867–77. e13. [DOI] [PubMed] [Google Scholar]
- 72.Peeters MC, Wisse LE, Dinaj A, Vroling B, Vriend G, IJzerman AP. The role of the second and third extracellular loops of the adenosine A1 receptor in activation and allosteric modulation. Biochem Pharmacol 2012. July 1;84(1):76–87. [DOI] [PubMed] [Google Scholar]
- 73.Guo D, Pan AC, Dror RO, Mocking T, Liu R, Heitman LH, et al. Molecular Basis of Ligand Dissociation from the Adenosine A2A Receptor. Molecular pharmacology 2016. May;89(5):485–91. [DOI] [PubMed] [Google Scholar]
- 74.Guo D, Venhorst SN, Massink A, van Veldhoven JP, Vauquelin G, AP IJ, et al. Molecular mechanism of allosteric modulation at GPCRs: insight from a binding kinetics study at the human A1 adenosine receptor. British journal of pharmacology 2014. December;171(23):5295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo D, Heitman LH, Ijzerman AP. Kinetic Aspects of the Interaction between Ligand and G Protein-Coupled Receptor: The Case of the Adenosine Receptors. Chemical Reviews 2016 2016/April/18. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y-T, Chan Y-H. Understanding the molecular basis of agonist/antagonist mechanism of human mu opioid receptor through gaussian accelerated molecular dynamics method. Sci Rep-Uk 2017;7(1):7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kokubo H, Tanaka T, Okamoto Y. Ab Initio Prediction of Protein-Ligand Binding Structures by Replica-Exchange Umbrella Sampling Simulations. J Comput Chem 2011. October;32(13):2810–21. [DOI] [PubMed] [Google Scholar]
- 78.Ostermeir K, Zacharias M. Accelerated flexible protein-ligand docking using Hamiltonian replica exchange with a repulsive biasing potential. Plos One 2017. February 16;12(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang K, Chodera JD, Yang YZ, Shirts MR. Identifying ligand binding sites and poses using GPU-accelerated Hamiltonian replica exchange molecular dynamics. J Comput Aided Mol Des 2013. December;27(12):989–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang YM, McCammon JA, Miao Y. Replica Exchange Gaussian Accelerated Molecular Dynamics: Improved Enhanced Sampling and Free Energy Calculation. J Chem Theory Comput 2018. April 10;14(4):1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]