Abstract
Point-of-Care (POC) molecular assays improve HIV infant diagnosis and viral load (VL) quantification in resource-limited settings. We evaluated POC performance in Kinshasa (Democratic Republic of Congo), with high diversity of HIV-1 recombinants. In 2016, 160 dried blood samples (DBS) were collected from 85 children (60 HIV−, 18 HIV+, 7 HIV-exposed) and 75 HIV+ adults (65 treated, 10 naive) at Monkole Hospital (Kinshasa). We compared viraemia with Cepheid-POC-Xpert-HIV-1VL and the non-POC-COBAS®AmpliPrep/COBAS®TaqMan®HIV-1-Testv2 in all HIV+, carrying 72.4%/7.2% HIV-1 unique/complex recombinant forms (URF/CRF). HIV-1 infection was confirmed in 14 HIV+ children by Cepheid-POC-Xpert-HIV-1Qual and in 70 HIV+ adults by both Xpert-VL and Roche-VL, identifying 8 false HIV+ diagnosis performed in DRC (4 adults, 4 children). HIV-1 was detected in 95.2% and 97.6% of 84 HIV+ samples by Xpert-VL and Roche-VL, respectively. Most (92.9%) HIV+ children presented detectable viraemia by both VL assays and 74.3% or 72.8% of 70 HIV+ adults by Xpert or Roche, respectively. Both VL assays presented high correlation (R2 = 0.89), but showing clinical relevant ≥0.5 log VL differences in 15.4% of 78 cases with VL within quantification range by both assays. This is the first study confirming the utility of Xpert HIV-1 tests for detection-quantification of complex recombinants currently circulating in Kinshasa.
Introduction
The access to routine molecular tools for early infant HIV-1 diagnosis (EID) and viral load (VL) quantification in children and adults is required for an early antiretroviral treatment failure identification and the prompt linkage to care. It can reduce HIV-associated mortality and morbidity in infected populations1–3. However, most of 37 million HIV-infected individuals live in resource-limited countries with a high number of different circulating HIV-1 variants, high rates of infection and no or limited access to routine HIV monitoring4,5. These settings have insufficient access to laboratory facilities, cold-chain management shortcomings, and difficulties for plasma collection and sample transportation5,6.
Until now, conventional molecular tests for EID and VL needed long procedures conducted in specialized and centralized laboratory settings requiring substantial infrastructure and training, needing turnaround times of several weeks or months7,8. This could increase the risk of loss to clinical follow-up of patients, thus having a negative impact in the HIV treatment cascade9,10.
To improve the linkage to care of HIV-exposed and infected subjects, some new easy to perform molecular assays for EID and VL quantification have been developed: point-of-care or POC assays. They have emerged as potential game-changers for improving EID and antiretroviral therapy (ART) monitoring programs11 since they are simpler, faster (less than 2 hours), automated platforms that do not require as much infrastructure as the conventional lab-based systems8,12. They can be performed directly in health centers and not only in reference laboratories, which favors their use at or near the point-of-care, allowing HIV confirmation, ART initiation, or treatment or adherence interventions quickly after sampling (within about 2 hours). WHO promotes POC use for HIV diagnosis and monitoring in limited-resource settings13, as well as the use of dried blood spots (DBS) instead of plasma as being easier to collect and ship to centralized facilities than plasma14,15. However, most POC HIV assays have not yet been evaluated using well-characterized DBS panels.
HIV genetic variability can affect the success of HIV-1 detection and quantification by molecular assays16–23. However, the performance of most POC and non-POC assays has not been extensively evaluated testing all HIV-1 subtypes and complex recombinants present in countries with high genetic diversity and high rate of HIV infections. This is the case of Kinshasa (Democratic Republic of Congo, DRC), the epicenter of HIV-1 group M epidemic24, were a large number of HIV-1 recombinants are expected24–32. Thus, this study analyzes the efficacy of two POC techniques for EID and VL (Cepheid Xpert HIV-1 Qual and Xpert HIV-1 VL) versus the non-POC Roche CAP/CTM Quantitative VL test v2.0 in the same DBS panel collected from children and adults in Kinshasa, where a large diversity of HIV-1 variants co-circulate.
Material and Methods
From April to November 2016, 160 DBS were collected at Monkole Hospital (Kinshasa, DRC) from 85 children (60 HIV-non infected, 18 HIV-positive, 7 HIV-exposed) and 75 HIV-infected adults (65 treated with clinical suspicion of treatment failure, 10 naive). DBS samples were prepared by spotting 70 µl of venous blood with micropipette, collected by venipuncture in EDTA-anticoagulant tubes into each dot on a Whatman 903 Protein Saver Card (Schleicher & Schuell, Dassel, Germany). Two or three DBS cards were collected per patient. They were dried separately on a drying-rack overnight at room temperature in Monkole Hospital, sealed in a zip-lock plastic bag with desiccant bags and stored at −20 °C until transported in dry ice to the laboratories in Madrid and Pamplona, Spain, where children and adult samples, respectively, were stored at −80 °C until further use.
HIV diagnosis and viraemia quantification
HIV diagnosis was firstly performed in DRC using rapid serological tests: Determine™ HIV-1/2 Ag/Ab (Alere), Double-Check Gold HIV 1&2 (Orgenics) and Uni-Gold HIV (Trinity Biotech) from 18-months old and by Biomerieux 4th generation immunoassay VIDAS® HIV Duo Ultra or exceptionally by molecular Abbott real-time HIV-1 Qualitative in infants under 18-months old. In Madrid, Spain, HIV serological status in the 85 children was confirmed with BioRad GeeniusTM HIV-1/2 confirmatory assay using one DBS dot per patient, as previously reported33. All HIV seropositive and undetermined pediatric DBS by Geenius were then tested by POC Cepheid Xpert Qual (Xpert Qual), which provides a binary “detected”/“not detected” result34. In Navarra, Spain, HIV serostatus was confirmed in all adults by two 4th generation immunoassays: Elecsys® HIV combi PT (Roche) and VIDAS® HIV Duo Quick (bioMerieux).
HIV-1 viremia was quantified using Cepheid Xpert HIV-1 VL (Xpert VL)35 and COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test v2.0 (Roche VL)36 in all HIV+ DBS, both techniques based on real time amplification of HIV genome. All assays were performed using one dot eluted in Xpert Qualitative buffer for Xpert assays or Roche SPEX buffer for Roche-VL as lysis buffer to elute the DBS dots, according to manufacturer’s instructions. GeneXpert® Instrument automates and integrates specimen preparation, HIV-1 total nucleic acids (viral RNA and proviral DNA) extraction and amplification, and detection of the target sequence in specimens using real-time reverse transcriptase PCR (RT-PCR). The systems require the use of single-use disposable GeneXpert® cartridges that hold all the necessary RT-PCR reagents and host the RT-PCR processes.
For statistical analysis of VL data, any viraemia values reported by the system as <40 cp/ml (by Xpert VL) or <20 cp/ml (by Roche VL), lower limit of detection of each assay, were reported as 39 cp/ml or 19 cp/ml, respectively, being considered detected but not quantifiable. We identified treated subjects under therapeutic failure when they present HIV-1 viraemias of 1,000 cp/ml or higher, clinical treatment failure threshold using DBS37. Both HIV-1 VL assays were based on real time PCR, providing an assay-specific cycle threshold (Ct), which inversely correlates with the starting concentration of the viral genome in the infected specimen. Ct values were recorded following DBS VL quantification by both Xpert VL and Roche VL platforms using one DBS dot in each sample.
We provided the number of HIV-1 RNA copies per dot and per plasma milliliter after considering patient’s hematocrit assuming 39% hematocrit for children, 42% for women and 47% for men, according to previous studies38,39. This lead to plasma volumes of 42.7 µl, 40.6 µl and 37.1 µl, respectively, in 70 µl blood collected per dot. The main features of the three molecular HIV assays used in the study are described in Table 1.
Table 1.
Characteristics of molecular assays for HIV-1 diagnosis and VL quantification.
| Qualitative assays (for HIV-1 diagnosis) | Quantitative assays (for HIV-1 viral load quantification) | ||
|---|---|---|---|
| Xpert Qual | Roche VL | Xpert VL | |
| Company | Cepheid | Roche | Cepheid |
| POC molecular assay | Yes | No | Yes |
| Viral targets | 3′end-5′UTR | Gag + LTR | 3′end-5′UTR |
| Sample (according to technical report) | Whole blood (100 µl) DBS (1 dot) | Plasma | Plasma (1 ml) |
| LOD | 203 cp/ml (VQA, whole blood) 278 cp/ml (WHO, whole blood) 531 cp/ml (VQA in DBS) 668 cp/ml (WHO in DBS) | 20 cp/ml (plasma) | 15.3 cp/ml (VCA in plasma) 18.3 cp/ml (WHO in plasma) |
| LOQ | — | 20 cp/ml (plasma) 20 cp/dot (DBS)* | 40 cp/ml (plasma) 40 cp/dot (DBS) |
| Approved for EID using plasma | No | No | No |
| Approved for EID using DBS or whole blood | Yes | No | No |
| Approved for VL using plasma | — | Yes | Yes |
| Approved for VL using DBS | — | No | No |
| Detected HIV-1 groups | M, N and O | M and O | M, N and O |
VL, viral load; EID, early infant HIV-1 diagnosis; POC, point of care; LOD/Q, limit of detection/quantification; DBS, Dried Blood Spots; HIV-1-RNA cp/ml, cp/ml plasma; LTR, long terminal repeats; UTR, untranslated region within viral LTR; Roche VL, COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test v2.0; Xpert Qual, Cepheid Xpert HIV-1 Qual; Xpert VL, Cepheid Xpert HIV-1 VL; VQA: HIV-1 subtype B from viral quality assurance laboratory; WHO: HIV-1 subtype B from WHO 3rd International Standard NIBSC code 10/152 http://www.nibsc.org/documents/ifu/10-152.pdf. Data according to technical reports. LOD Xpert HIV-1 VL and Xpert HIV-1 Qual available61,76. *Data reported by this study.
HIV-1 variant characterization
For HIV-1 variant characterization, RNA was extracted from 2 DBS dots using the NucliSENS easyMAG automated platform (BioMerieux) or manual High-Pure Viral Nucleic Acid (Roche) kit. Viral RNA was amplified in the HIV-1 pol coding region by RT-PCR and nested-PCR using primers designed by WHO40 as previously described41 and/or ANRS42. Viral sequences included the complete HIV-1 protease (PR, codons 1–99), and partial retrotranscriptase (RT, codons 1–335/440) and integrase (IN, codons1–285). PCR amplicons were purified using the Illustra™ ExoProStar 1-Step™ (GE Healthcare Life Sciences, Little Chalfont, UK) and sequenced by Macrogen Inc. (Gasan-dong, Geumchun-gu, Seoul, Korea). HIV-1 variant was characterized by phylogenetic analysis (phy) using MEGA6 with Tamura 3-parameters as the evolutionary model with 1,000 bootstrap resampling. The bootstrap cut-off was set at 70. The tree topology was obtained using Neighbor Joining method. At least two representative HIV-1 sequences of each HIV-1 non-M group (O, P, N), and from each group M variant (9 subtypes, 6 sub-subtypes and 83 CRF available at the moment of the analysis among the 98 described43 were taken as references. Sequences not identified as any known non-M group, group M subtype or CRF by phy were considered HIV-1 group M unique recombinant forms (URF) in pol (URFpol).
Accession numbers
PR, RT and/or IN HIV-1 sequences were submitted to GenBank (www.ncbi.nlm.nih.gov/genbank) with the following accession numbers: MH920378-MH920435.
Statistical analysis
Correlation analysis was performed using the Spearman rank test and linear regression. We calculated the intraclass correlation coefficient (ICC). To determine differences between two viral load assays, the Bland-Altman plot method44 was used. The clinically relevant difference between two VL measurements was considered at 0.5 log10 cp/ml, as described previously45–47. For all analysis, 95% confidence intervals were considered. All statistical analyses were performed using Excel, STATA v11 and GraphPad Prism 6.
Ethical aspects
The project was approved by the Human Subjects Review Committees at Monkole Hospital/University of Kinshasa (Kinshasa, DRC), University Hospital Ramón y Cajal (Madrid, Spain) and University of Navarra (Pamplona, Spain). Informed consent of enrolled adults and of parents or guardians of enrolled children was obtained. All methods were carried out in accordance with relevant guidelines and regulations.
Results
High percentage of false positive diagnosis by rapid serological testing in DRC and delay in infant HIV diagnosis
We evaluated the HIV-1 quantification efficacy of different molecular assays (POC and non-POC) in dried blood carrying different HIV-1 non-B subtypes and complex recombinants, mainly URF. For that purpose, DBS were collected from 85 children (60 HIV-uninfected, 18 HIV-infected, and 7 HIV-exposed) and 75 HIV-infected adults from Kinshasa (DRC) during 2016. The main characteristics of the study subjects are recorded in Table 2. The 160 study subjects were mainly seropositive by rapid serological tests in DRC (58.75%), female (57.5%) and antiretroviral experienced (46.2%). The mean age for HIV diagnosis in DRC was 8.1 (SD 5.38) years old in children and 40.4 (SD 12.13) in adults. The mean age at DBS collection was 9.8 (SD 5.11) years old in children and 46.5 (SD 12.27) in adults. All but one children were born in Kinshasa, 8.2% were orphaned and 6 presented HIV/ Mycobacterium tuberculosis confection. Most adults were female (70.7%)
Table 2.
Characteristics of study population from Kinshasa (DRC) with collected DBS in 2016.
| Children | Adults | Total (%) | |
|---|---|---|---|
| Number | 85 (100%) | 75 (100%) | 160 (100%) |
| Gender (male) | 46 (54.1%) | 22 (29.3%) | 68 (42.5%) |
| Mean age | |||
| At DBS collection (range) | 9.8 (0–18) | 46.5 (24.8–73) | 26.5 (0–73) |
| At HIV diagnosis at DRC (range) | 8.1 (0–16) | 40.4 (0–65.3) | 33.7 (0–65.3) |
| HIV positive status | |||
| By rapid testing in DRC | 18 (21.2%) | 74 (98.7%) | 92 (57.5%) |
| by molecular testing and serology in Spain | 14 (16.5%) | 70 (93.3%) | 84 (52.5%) |
| False positive HIV diagnosis in DRC | 4 (4.7%) | 4 (5.3%) | 8 (5%) |
| ART exposure among 92 HIV+ diagnosed in DRC | |||
| ART naïve | 2 | 10 | 13 |
| ART | 16* | 58* | 74 |
| Unknown | 0 | 7 | 5 |
| ART exposure among 84 with confirmed diagnosis in Spain | |||
| ART naïve | 1 | 9 | 10 |
| ART | 13 | 56 | 69 |
| Unknown | 0 | 5 | 5 |
| HIV-1 viraemia among 69 confirmed HIV+ under ART in DRC | |||
| Not detected only by Roche VL | 0 | 1 | 1 |
| Not detected only by Xpert VL | 0 | 4 | 4 |
| Not detected by both Roche and Xpert VL | 3 | 5 | 8 |
| >1,000 cp/dot by Xpert VL | 3 | 12 | 15 |
| >1,000 cp/ml by Xpert VL* | 12 | 52 | 64 |
| >1,000 cp/dot by Roche VL | 6 | 12 | 18 |
| >1,000 cp/ml by Roche VL* | 12 | 32 | 44 |
| Number of different ART regimens among 69 treated in DRC | |||
| 1 | 10 (76.9%) | 34 (60.7%) | 44 (63.8%) |
| 2 | 2 (15.4%) | 14 (25%) | 16 (23.2%) |
| 3 | 0 | 7 (12.5%) | 7 (10.1%) |
| 4 | 1 (7.7%) | 1 (1.8%) | 2 (2.9%) |
| NRTI experienced | |||
| 3TC | 13 | 58 | 71 |
| AZT | 10 | 46 | 56 |
| TDF | 5 | 27 | 32 |
| DDI | 0 | 3 | 3 |
| ABC | 0 | 4 | 4 |
| NNRTI experience | |||
| NVP | 10 | 43 | 53 |
| EFV | 6 | 29 | 35 |
| PI experience | |||
| LPV/r | 1 | 7 | 8 |
| HIV+ subjects with available pol HIV-1 sequences | 13 (92.8%) | 45 (64.3%) | 58/(69%) |
| HIV-1 variant by phy | |||
| Non-B subtypes | 2/13 (15.4%) | 4/45 (8.9%) | 6 (10.3%) |
| CRF | 2/13 (15.4%) | 8/45 (17.8%) | 10 (17.3%) |
| URF | 9/13 (69.2%) | 33/45 (73.3%) | 42 (72.4%) |
DRC, Democratic Republic of Congo; DBS, dried blood Spot; ART, antiretroviral treatment; NTRI, nucleoside transcriptase reverse inhibitor; NNRTI, non-NRTI; PI, Protease inhibitor; 3TC, Lamivudine; AZT, Zidovudine; TDF, Tenofovir; DDI, Didanosine; ABC, Abacavir; NVP, Nevirapine; EFV, Efavirez; LVP/r, Lopinavir/Ritonavir; Phy, phylogenetic analysis; CRF, circulating recombinant form; URF, unique recombinant form. *Corrected cp/ml plasma considering hematocrit. Among 74 treated subjects in DRC, 3 were false positive children and 2 false positive adults.
Among the 92 children and adults diagnosed as HIV positive in the local laboratory in Congo by rapid tests and 80.4% were antiretroviral (ARV) experienced at sampling. The remaining were ART-naive or with unknown treatment data. NRTI and NNRTI were the most used ARVs in the study cohort, Zidovudine + Lamivudine + Nevirapine being prescribed in 60 treated patients (13 children and 47 adults), mainly as first line therapy (50 cases). Only 8 patients received protease inhibitors (PIs) based treatment with Lopinavir/Ritonavir, while integrase inhibitor use was absent among study subjects. (Table 2).
HIV-1 infection was confirmed in 16.5% of 85 children by Xpert Qual and in 93.3% of 75 HIV+ adults diagnosed in DRC by both Xpert VL and Roche VL. However, we identified false positive HIV diagnosis in DRC after following rapid serology testing algorithm in 4 adults (range 23.8–28.5 years age) and in 4 children (range 5.3–13.1 years age), and 5 of them (2 adults, 3 children) were under unnecessary ART for a mean time of 3.9 years (Table 3). Xpert Qual confirmed the absence of HIV-1 infection in 4 DBS from children erroneously diagnosed as HIV positive in the DRC, as in other 8 HIV negative cases providing undetermined HIV status by Geenius BioRad. The 4 false positive in adults provided negative results by two VL assays (Roche and Xpert), three of them provided negative result by two 4th generation immunoassays (Roche Elecsys® HIV combi PT and bioMerieux VIDAS® HIV Duo Quick), and the remaining case only by VIDAS assay. Thus, the rate of false positive HIV diagnosis among pediatric HIV-exposed population was 22.2% (4/18) and 5.3% (4/75) for HIV-infected adults.
Table 3.
Eight false HIV diagnosis in DRC using rapid serological testing.
| IDSpain | ARV experience at sampling | Time under ART (m) | Democratic Republic of the Congo | Spain | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Determine | Uni-Gold | Double Check | Definitive diagnosis | Age at diagnosis (y) | ElecsysRoche | Vidas Duo Quick | BioRad Geenius | Xpert Qual | Roche VL | Xpert VL | Definitive diagnosis | Age at diagnosis (y) | ||||
| Children | P6 | AZT + 3TC + NVP | 96 | Pos | Pos | Pos | Pos | unknown | — | — | Neg | Neg | — | — | HIV- | 9.11 |
| P12 | AZT + 3TC + NVP | 21 | Pos | Pos | Pos | Pos | 0.55 | — | — | Neg | Neg | — | — | HIV- | 5.36 | |
| P14 | AZT + 3TC + NVP | 22.4 | Pos | Pos | Pos | Pos | 3.57 | — | — | Ind | Neg | — | — | HIV- | 8.64 | |
| N18 | Naive | 0 | Pos | Pos | Pos | Pos | 6.77 | — | — | Ind | Neg | — | — | HIV- | 13.17 | |
| Adults | CUN84 | AZT + 3TC + NVP | 176.4 | Pos | Pos | — | Pos | unknown | Pos | Neg | Neg | — | Neg | Neg | HIV- | 24.7 |
| CUN33 | Naive | 0 | Pos | Pos | — | Pos | 0 (birth) | Neg | Neg | Neg | — | Neg | Neg | HIV- | 28.7 | |
| CUN41 | AZT + 3TC + EFV | 87 | Pos | Pos | — | Pos | 28.5 | Neg | Neg | Neg | — | Neg | Neg | HIV- | unknown | |
| CUN109 | Naive | 0 | Pos | Pos | — | Pos | 51 | Neg | Neg | Neg | — | Neg | Neg | HIV- | 29.07 | |
HIV testing in DRC: Determine, rapid test Determine™ HIV-1/2 Ag/Ab (Alere); Uni-Gold, Uni-Gold HIV (Trinity Biotech) and Double-Check, Double-Check Gold HIV 1&2 (Orgenics). HIV testing in Spain: Elecsys Roche, 4th gen immunoassay Elecsys® HIV combi PT (Roche); VIDAS DUO Quick, 4th gen immunoassay VIDAS® HIV Duo Quick (bioMerieux); BioRad GeeniusTM HIV-1/2; Xpert Qual, Cepheid Xpert Qual; Xpert VL, Cepheid Xpert HIV-1 VL; Roche VL, COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test v2.0. ARV, antiretroviral drugs; AZT, Zidovudine; 3TC, Lamivudine; NVP, Nevirapine; EFV, Efavirenz; m, months; y, years; VL, viral load; dash, not done; Ind, indeterminate; Neg, HIV negative; Pos, HIV positive.
Successful detection of HIV-1 variants in Kinshasa by Xpert VL and Roche VL
Among the 84 HIV+ total samples selected after excluding those 8 DBS samples determined to be false positives, 95.2% could be detected by Xpert VL and 97.6% by Roche VL, the remaining being undetected (Table 4). Among those specimens with detectable HIV RNA, Xpert VL vs. Roche VL could quantify and provide HIV-1 viraemia values (≥40 vs. ≥20 cp/dot or ≥936–1078 vs. ≥468–539 cp/ml plasma depending on hematocrit) for 65 (77.4%) vs. 66 (78.6%) DBS samples, respectively. Most (92.9%) of 14 HIV+ children presented quantifiable viraemia by both Xpert-VL (≥40 cp/dot) and Roche-VL (≥20 cp/dot) assays and 74.3% or 72.8% of 70 HIV+ adults by Xpert or Roche, respectively. Two specimens not detected by Roche were detected by Xpert (<40 cp/ml), while 4 specimens below limit of detection of Xpert could be only detected by Roche, with lower detection limit (<20 cp/ml) (Table 4).
Table 4.
HIV-1 VL quantification in 84 HIV-1+ DBS (14 HIV+ children and 70 HIV+ adults) using two molecular assays.
| Xpert VL (POC assay) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Not detected | Detected not quantified <40 cp/dot | Quantified ≥40 cp/dot | No. DBS | |||||
| 40–1,000 cp/dot | >1,000 cp/dot | Ct mean [range] | ||||||
| Roche VL (Non-POC assay) | Not detected | — | 2 | 0 | 0 | 0 | 2 | |
| Detected not quantified <20 cp/dot | 4 | 9 | 3 | 0 | 34.1 [30.1–36.6] | 16 | ||
| Quantified (≥20 cp/dot) | ≥20–39 cp/dot | 0 | 3 | 7 | 0 | 32.5 [31.8–34.2] | 10 | |
| 40–1,000 cp/dot | 0 | 1 | 25 | 2 | 30.3 [28.2–32.8] | 28 | ||
| >1,000 cp/dot | 0 | 0 | 3 | 25 | 25.2 [20.3–27.4] | 28 | ||
| Ct mean [range] | 0 | 39.6 [33.2–42.6] | 36.3 [33.9–41.8] | 30.2 [23.6–33.6] | — | — | ||
| No. DBS | 4 | 15 | 38 | 27 | — | 84 | ||
| Available pol HIV-1 sequences | nd | 4 (26.7%) | 27 (71%) | 27 (100%) | 58 (69%) | |||
| Carrying complex recombinants (CRF + URF) | nd | 4 | 23 | 25 | — | 52 | ||
| Carrying non-B variants | nd | 0 | 4 | 2 | — | 6 | ||
No., number; VL, HIV-1 viral load POC, point of care; DBS, dried blood sample; cp/dot, HIV-1 RNA copies per DBS dot; nd, not available sequences due to low VL. Ct, VL assay-specific cycle threshold, which inversely correlates with the starting concentration of the viral genome in infected specimen.
High correlation among Xpert and Roche VL assays using DBS
Viral Load results within the quantification range of both assays were available for 78 (92.8%) of 84 HIV+ patients. The POC Cepheid Xpert HIV-1 VL assay showed excellent agreement (ICC = 1) with Roche VL for HIV-RNA quantification. A high and significant correlation was observed among both VL assays (R2 = 0.89, P < 0.001), as shows the estimated regression line (Fig. 1). However, Ct values for HIV-1 quantification were VL assay dependent when quantifying the 78 DBS detected by both VL assays.
Figure 1.
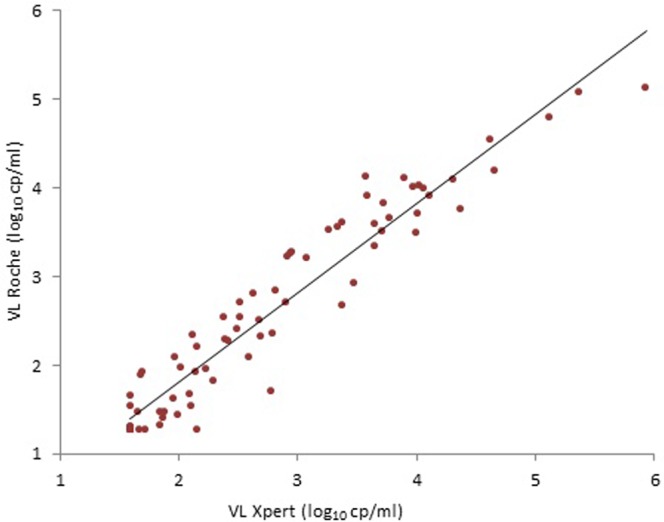
Correlation between Xpert VL and Roche VL assays in 78 HIV+ samples with quantified VL. Scatter plot with a simple linear regression analysis of 78 samples (14 children + 64 adults) which were quantified (VL of ≥40 or ≥20 cp/ml) by both assays. Graphic using log of direct VL from in one DBS dot (HIV-1 RNA copies per dot). VL, Viral load.
Xpert VL provided higher mean Ct (34.75 ± 7.7, range 23.6–42.6) than Roche VL (29.4 ± 3.77, range 20.3–36.6) in the panel, resulting in a mean Xpert VL of 4.22 log10 cp/ml ± SD 1.06 (2.76 log10 cp/dot ± SD 1.16) and mean Roche VL of 4.04 log10 cp/ml ± SD 1.12 (2.57 log10 cp/dot ± SD 1.12).
The similarities between both VL assays were evaluated by the Bland-Altman plot method (Fig. 2). HIV-1 VL overestimation by one of the two assays (Xpert VL or Roche VL) was observed in all but one specimen among the 78 DBS, although the difference was below a clinically relevant threshold of 0.5 log10 cp/ml in most cases (84.6%). The POC Xpert HIV-1 VL assay tended to overestimate HIV-1 VL in 69.2% samples, and the non-POC Roche VL in 29.5% specimens (Fig. 3, Table 4). The overall mean difference in the HIV-1 RNA values obtained by Xpert VL assay and Roche VL was 0.30 log10 cp/dot (95% CI: 0.26 to 0.35 log10 cp/dot) (P < 0.001). However, clinical relevant differences (≥0.5 log VL) ranging from −0.55 to 1.07 were observed in 12 (15.4%) of 78 DBS specimens with VL above detection limit by both assays (Figs 2 and 3), differing across samples and assays. Eleven cases corresponded to Xpert VL use, while only one to Roche VL testing (Table 4).
Figure 2.
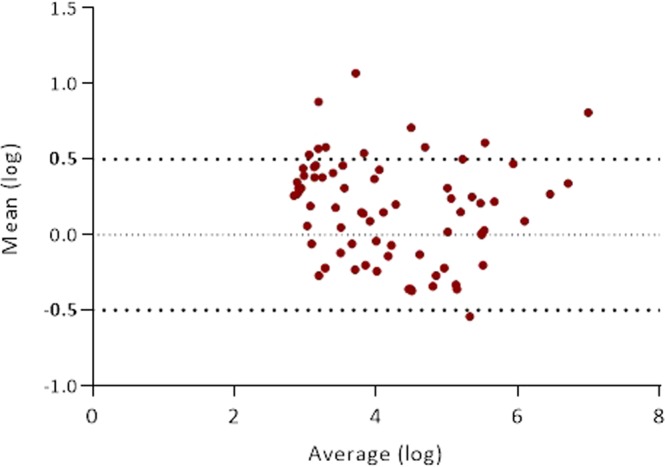
Bland-Altman analysis showing difference vs. average viral load comparing Xpert VL and. Roche VL in 78 HIV+ quantified by both assays. HIV+ samples from 78 patients (14 children, 64 adults) quantified by both assays.
Figure 3.
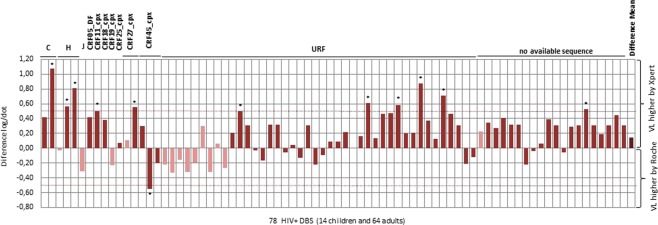
HIV-1 viraemia differences by Xpert VL vs. Roche VL in 78 HIV+ DBS quantified by both assays and HIV-1 variants in 58 samples with available sequence. Light color in pediatric samples. VL, viral load; 58 non-B variants infecting study population: 2C, 3H, 1J, 1CRF05_DF, 1CRF11_cpx, 1 CRF18_cpx, 1CRF19_cpx, 1 CRF25_cpx, 2 CRF27_cpx, 3 CRF45_cpx. The absence of bar in one URF indicates the same VL values using both assays. CRF, circulating recombinant form; URF, unique recombinant form. *Viral load differences >0.5 log.
High percentage of complex recombinants in DRC and impact in VL quantification
HIV-1 pol sequence was recovered from 58 of 84 HIV-1 + individuals (13 children and 45 adults) and studied by phylogeny. Among all 58 viral sequences obtained, we identified 6 (10.4%) non-B subtypes (2C, 3H, 1J), ten (17.2%) CRF (1 CRF05_DF, 1 CRF11_cpx, 1 CRF18_cpx, 1 CRF19_cpx, 1 CRF25_cpx, 2 CRF27_cpx, 3 CRF45_cpx) and 42 (72.4%) URF. Thus, most (89.6%) of 58 obtained HIV-1 sequences were URF or CRF recombinants (Table 2). Neither HIV-2 nor non-M group infections were found in our study population. Both VL tests were able to detect and quantify all variants, including CRF and URF recombinants of the study cohort (Table 4).
The impact of each complex recombinant on VL quantification was unclear (Table 4, Supplementary Table 1, and Fig. 3). Among 42 URF detected, 61.9% provided higher VL by Xpert VL and 35.7% by Roche VL. Among 10 samples ascribed to 7 different CRF, 70% showed higher viraemia by Xpert VL and 30% by Roche VL. Subtypes C and H displayed higher viraemia values by Xpert VL and subtype J by Roche VL.
Discussion
POC test use can improve the clinical management of HIV-infected infants and adults and reduce the delay in diagnosis and in ART failure identification14. Early infant diagnosis is a WHO priority14,48, since it allows early ART to be established and reduces irreversible damage to central nervous and immune systems, viral reservoirs, as well as HIV transmission and morbidity/mortality associated with HIV-1 infection49. A correct early HIV diagnosis is also essential, since false positive HIV tests might result in unnecessary antiretroviral treatment and psychological distress in falsely diagnosed individuals and families50. The use of DBS has been proposed as an alternative sample to plasma/serum, easier to be collected, stored and shipped, very convenient in limited resource countries15,33,37,51. The aim of this study was to evaluate the performance of two POC HIV-1 assays (Xpert-VL and Xpert Qual) for HIV detection and/or quantification using DBS in the DRC, a country with a high HIV-1 diversity including a high prevalence of complex recombinants, mainly URFs.
Although POC molecular testing prevents inappropriate HIV serological diagnosis and is cost-effective52, it is not yet globally adopted in all EID or adult programs48. In the DRC, the National Program of fight against HIV-AIDS (PNLS) recommends performing EID 0–2 days after birth, but POC assays for EID have not been implemented yet within the clinical routine of HIV-exposed infants.
We have evaluated the clinical impact of the lack of routine EID molecular testing in HIV-exposed newborns shortly after birth and of confirmatory serological testing in older children and adults in a cohort in Kinshasa. We have found false HIV diagnoses among 5% of the study participants that lead to unnecessary ART in five HIV uninfected subjects. The high prevalence of false positive diagnosis among HIV-exposed infants less than 18 years old could be explained by the long delay in PCR results from a centralized national laboratory, which were only available at the clinical center in Kinshasa 6 months after the original HIV serological test. Wrong diagnoses may also be a consequence of the absence of a confirmatory molecular test with a second new sample as recommended by WHO14 for infants. In older children and adults, false HIV diagnosis can be due to the local absence of confirmatory serological analysis and the exclusive use of serological rapid testing for HIV diagnosis. Although rapid immunochromatographic test for HIV are recommended in low income countries53, the low HIV prevalence among the general population in the DRC (0.7%)4 may be associated with a lower positive predictive value for these methods. In addition, rapid HIV testing is not appropriate for acute infection diagnosis53. Another aspect that could influence a misdiagnosis is the fact that a subjective reading of rapid HIV tests may speed up the communication of false positive results. The high mean age at HIV diagnosis in DRC in children (8.1 years) from the study cohort would suggest HIV diagnosis delay, since most of them acquired HIV infection by vertical route according to clinical reports.
Results also show that Xpert and Roche molecular tests were superior to fourth generation serological screening assays to identify HIV infection in HIV-exposed children older than 18 months and adults. Moreover, Xpert resolved HIV status and rule out HIV infection in DBS from 10 individuals with undetermined results by Geenius confirmatory assay, in agreement with recent studies54. It could be due to the fact that Xpert HIV-1 Qual can detect HIV-1 infections up to 7–10 days before seroconversion, an average of nine days earlier than a panel of HIV-1 antibody tests and five days earlier than a panel of HIV-1 antigen p24 tests34. Xpert® HIV-1 Qual may play a role in the diagnosis of HIV, either in EID, as a confirmatory test after antibody-based testing, or for the detection of acute HIV infection in antibody negative patients recently infected55.
POC Xpert VL and non-POC VL Roche assays were also suitable in quantifying VL using DBS in the DRC. A high percentage of subjects under ART in the DRC showed VL levels above 1,000 cp/ml by Xpert (92.7%) or Roche (63.8%) (Table 1), the VL threshold defined by WHO to confirm virological failure in low- and middle-income countries and when using DBS in adults and children37. This is the optimal threshold for presenting the lowest percentage of misclassification compared with higher thresholds51. In fact, most patients in the study had a clinical suspicion of virological failure. However, the prevalence of ART failure was overestimated using Roche VL and this threshold, resulting less specific than other VL assays using DBS, as described in previous studies51,56.
Despite the good correlation in VL quantification using Xpert VL with the no-POC Roche VL assay (R2 = 0.89), still 15.4% of 78 samples with VL within the quantification range of both assays presented clinically significant VL differences above 0.5 log10 cp/ml, according to previous studies16,23,57. Thus, we recommend the expanded use of VL in the DRC for an early detection of virological failure as well as the use of the same VL technique for each patient during ART monitoring to reduce potential assay-associated viraemia overestimations, which could be interpreted as virological failure events. This could reduce unnecessary ART regimen switches in these patients, favoring an early clinical response by reinforcing adherence or changing ART regimen if resistant variants are detected before clinical symptoms associated with treatment failure appear. The achievement of the 90-90-90 UNAIDS objectives depend on HIV monitoring, otherwise a future epidemic of HIV resistant strains may occur and delay these objectives in Sub-Saharan Africa58. The finding of assay dependent Ct values for HIV-1 quantification reinforces the risk of establishing a standard Ct cutoff as accurate threshold value to differentiate virological failures in subjects under ART.
The continuous evolution of HIV can hinder diagnosis and complicate clinical practice59. Thus, one of the main challenges for molecular diagnostic and VL assays is to detect and/or quantify different HIV-1 variants correctly. According to the manufacturer’s information, the Xpert® HIV-1 Qual assay has been validated for specimens including groups N, O and M (9 subtypes and recombinants A/E, A/B and AG/GH)60 and the Xpert VL for groups N, O and M (9 subtypes, CRF01_AE, CRF02_AG, and CRF03_AB)61. However, most 98 HIV-1 CRF43 and complex unique recombinant forms (URF) have not been validated yet. Roche VL was evaluated by analysis of HIV-1 group O and group M subtypes A through H from cell culture origin36,62, although it was also able to quantify a number of CRFs16. However, none of them has been evaluated across a large panel of URF variants, as we reported. We demonstrate that POC-Xpert assays and VL and Roche-VL can successfully detect and quantify complex recombinants in pol.
We also provide new data related to the HIV molecular epidemiology in Kinshasa, reporting an extremely high rate of unique inter-subtype recombinants in recently infected populations, although URF prevalence could be underestimated since HIV-1 variants were characterized considering pol gene but not the complete genome. Other authors have also highlighted the extreme diversity of HIV strains circulating in the country with a high presence of URFs and different CRFs, together with a low presence of subtype C28,31.
Some studies have reported clinically-significant differences (>0.5 log) in VL quantification across techniques with the same non-B strains23,63–65. These results would suggest different rates of detection across assays due to genetic variability in the HIV-1 gene region targeted by the assays in incorrectly quantified samples, as previously demonstrated for a CRF02_AG variant63. Previous evaluations supported the efficacy of Xpert technology for the detection and quantification of HIV-1 non-B variants64,66–68, highlighting Xpert utility in areas with a high genetic variability of HIV, such as Sub-Saharan Africa57,69–72.
POC Xpert technology for diagnosis of other diseases such as tuberculosis73, shigellosis74 or Monkeypox virus75 has already been used in the DRC. The National HIV/AIDS Program in the DRC plans to use the existing Xpert machines for HIV EID testing and VL monitoring in order to improve the clinical care of HIV-infected children and adults. Our results demonstrate that Xpert performance is adequate for testing HIV-1 variants currently circulating in the DRC. Thus, due to simplicity, rapid results and good performance, POC Xpert HIV-1 can be useful in the decentralization of EID and VL monitoring from specialized laboratories in the DRC to local hospitals and clinics within their routine clinical care. This will help to reach the ambitious 90-90-90 goals in the country. We also confirmed that DBS could be a suitable sample for Xpert use in the DRC, requiring a minimum volume of blood, favoring molecular testing in infants and low-weight children. In addition, DBS samples offer additional advantages, they are not considered biohazardous once dried and are not as time and temperature sensitive as plasma specimens37.
An important limitation of this study is that we did not compare DBS to plasma due to the lack of paired plasma/DBS specimens collected for each subject in the study population. Moreover, due to the design of the study and sample size, we could not determine the statistical power of possible performance differences across assays in each specific HIV-1 variant. Finally, we could not explore the effect of DBS lysis buffer in viraemia quantification, which could influence DBS VL results according to previous reports76. The main strength of our study is that it shows the first results confirming the utility of POC Xpert HIV-1 tests and Roche VL platform for early HIV-1 diagnosis and for VL quantification of complex recombinants (mainly URF) currently circulating in Kinshasa, the epicenter of HIV-1 group M epidemic and where a large number of complex recombinants cocirculate. To our knowledge, there are no previous studies that have included a large panel of different HIV-1 complex recombinants characterized by phylogenetic analysis during Xpert-POC HIV-1 evaluation for EID and VL. We also report some of the current limitations of HIV diagnosis and monitoring in DRC. Since Xpert assays and DBS use can improve early diagnosis in HIV-exposed infants and early detection of ART failures in countries with complex HIV-1 recombinants and limited infrastructures, as in the DRC, our results could have a direct clinical impact in global HIV diagnosis and monitoring to reach early the 90-90-90 objectives.
Supplementary information
Acknowledgements
We thank patients who participated in the study as well as local professionals responsible for the DBS and data collection of enrolled patients at Monkole hospital in Kinshasa. We also thank to Paul James Erskine Devlin for English editing of manuscript. We thank Patricia Sanz for her technical support during lab processing of DBS samples. This study was funded by fundraising and donations (mainly of Bomberos Ayudan Association), by the Government of Spain (Fondo de Investigación en Salud-FI, grants PI16/01908 and PI18/00904) and the Government of Navarre (grant 045-2015). The study was integrated in the research supported by the Spanish HIV infected Paediatric Cohort (CoRISpe) integrated in the Spanish AIDS Research Network and by Instituto de Salud Carlos III, Spanish Health Ministry (Grant no. RD06/0025-ISCIII-FEDER). This study is also included in the “Subprograma de Inmigración y Salud” from CIBERESP (Spain). Xpert VL cartridges to study DBS samples from adult patients were supplied as a donation from Werfen Company.
Author Contributions
A.H. and G.R. conceived and designed the study, contributed to data analysis and result discussion. A.H. and S.C. collaborated in sample shipping. A.N. selected clinical and epidemiological data from patients in DRC and supervised DBS collection and shipping. M.R. performed the virological analysis of paediatric samples (Xpert VL, Xpert EID, and viral sequencing) and the phylogenetic analysis for variant characterization of the complete study cohort. A.H. supervised phylogenetic analysis. D.B., G.R. and M.F. performed sequencing and HIV-1 viraemia quantification by Roche and Xpert assays from adults and by Roche in children. S.C. collaborated in statistical analysis. A.H. and M.R. designed tables and Figures and wrote the manuscript. A.N., M.F. and S.C. revised the paper and contributed to results discussion. All authors approved the final version.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41963-y.
References
- 1.Newell ML, et al. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet. 2004;364:1236–1243. doi: 10.1016/S0140-6736(04)17140-7. [DOI] [PubMed] [Google Scholar]
- 2.Violari A, et al. Early antiretroviral therapy and mortality among HIV-infected infants. N Engl J Med. 2008;359:2233–2244. doi: 10.1056/NEJMoa0800971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes MD, et al. Monitoring plasma HIV-1 RNA levels in addition to CD4 lymphocyte count improves assessment of antiretroviral therapeutic response. ACTG 241 Protocol Virology Substudy Team. Ann Intern Med. 1997;126:929–938. doi: 10.7326/0003-4819-126-12-199706150-00001. [DOI] [PubMed] [Google Scholar]
- 4.Joint United Nations Programme on HIV/AIDS (UNAIDS). UNAIDS data 2018, http://www.unaids.org/sites/default/files/media_asset/20170720_Data_book_2017_en.pdf (2018). [PubMed]
- 5.Rutstein SE, et al. On the front line of HIV virological monitoring: barriers and facilitators from a provider perspective in resource-limited settings. AIDS Care. 2016;28:1–10. doi: 10.1080/09540121.2015.1058896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bélec L, Bonn JP. Challenges in implementing HIV laboratory monitoring in resource-constrained settings: how to do more with less. Future Microbiol. 2011;6:1251–1260. doi: 10.2217/fmb.11.121. [DOI] [PubMed] [Google Scholar]
- 7.Bourlet T, Memmi M, Saoudin H, Pozzetto B. Molecular HIV screening. Expert Rev Mol Diagn. 2013;13:693–705. doi: 10.1586/14737159.2013.829703. [DOI] [PubMed] [Google Scholar]
- 8.UNITAID, World Health Organization (WHO). HIV/AIDS Diagnostics Technology Landscape, https://unitaid.eu/assets/UNITAID-HIV_Diagnostic_Landscape-4th_edition.pdf (2014).
- 9.Lecher S, et al. Progress with scale-up of HIV Viral Load monitoring - seven Sub-Saharan African countries, January 2015-June 2016. MMWR Morb Mortal Wkly Rep. 2016;64:1287–1290. doi: 10.15585/mmwr.mm6446a3. [DOI] [PubMed] [Google Scholar]
- 10.Roberts T, Cohn J, Bonner K, Hargreaves S. Scale-up of routine viral load testing in resource-poor settings: current and future implementation challenges. Clin Infect Dis. 2016;62:1043–1048. doi: 10.1093/cid/ciw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celletti F, Sherman G, Mazanderani AH. Early infant diagnosis of HIV: review of current and innovative practices. Curr Opin. HIV AIDS. 2017;12:112–116. doi: 10.1097/COH.0000000000000343. [DOI] [PubMed] [Google Scholar]
- 12.Engel N, Pie Pant N. Qualitative research on point-of-care testing strategies and programs for HIV. Expert Rev Mol Diagn. 2015;15:71–75. doi: 10.1586/14737159.2015.960518. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Zaman M. Low-cost tools for diagnosing and monitoring HIV infection in low-resource settings. Bull World Health Organ. 2012;90:914–920. doi: 10.2471/BLT.12.102780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization (WHO). WHO recommendations on the diagnosis of HIV infection in infants and children, http://www.who.int/iris/handle/10665/44275 (2010). [PubMed]
- 15.De Mulder M, Holguín A. Dried blood spots for monitoring HIV infection in Public Health Programs in developing countries. Enferm Infecc Microbiol Clin. 2013;31:100–107. doi: 10.1016/j.eimc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez P, et al. HIV-1 variability and viral load technique could lead to false positive HIV-1 detection and to erroneous viral quantification in infected specimens. J Infect. 2015;71:368–376. doi: 10.1016/j.jinf.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez P, et al. Evaluation of four commercial virological assays for early infant HIV-1 diagnosis using dried blood specimens. Pediatr Res. 2017;81:80–87. doi: 10.1038/pr.2016.183. [DOI] [PubMed] [Google Scholar]
- 18.Maritz J, van Zyl GU, Preiser W. Irreproducible positive results on the Cobas AmpliPrep/Cobas TaqMan HIV-1 Qual test are different qualitatively from confirmed positive results. J Med Virol. 2014;86:82–87. doi: 10.1002/jmv.23811. [DOI] [PubMed] [Google Scholar]
- 19.Sutcliffe CG, Moss WJ, Thuma EP. False positive results in infancy and management of uninfected children receiving antiretroviral therapy. Pediatr Infect Dis. J. 2015;34:607–609. doi: 10.1097/INF.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 20.Feucht UD, Forsyth B, Kruger M. False-positive HIV DNA PCR testing of infants: implications in a changing epidemic. S Afr Med J. 2012;102:149–152. doi: 10.7196/SAMJ.4951. [DOI] [PubMed] [Google Scholar]
- 21.Kageha S, et al. Discrepant test findings in early infant diagnosis of HIV in a national reference laboratory in Kenya: challenges and opportunities for programs. J Trop Pediatr. 2012;58:247–252. doi: 10.1093/tropej/fmr076. [DOI] [PubMed] [Google Scholar]
- 22.Avettand-Fénoël V, et al. Comparative performance of the Biocentric Generic Viral Load, Roche CAP/CTM v1.5, Roche CAP/CTM v2.0 and m2000 Abbott assays for quantifying HIV-1 B and non-B strains: Underestimation of some CRF02 strains. J Clin Virol. 2019;110:36–41. doi: 10.1016/j.jcv.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Holguín A, López M, Molinero M, Soriano V. Performance of three commercial viral load assays, Versant human immunodeficiency virus type 1 (HIV-1) RNA bDNA v3.0, Cobas AmpliPrep/Cobas TaqMan HIV-1, and NucliSens HIV-1 EasyQ v1.2, testing HIV-1 non-B subtypes and recombinant variants. J Clin Microbiol. 2008;46:2918–2923. doi: 10.1128/JCM.02414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worobey M, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potts KE, et al. Genetic diversity of human immunodeficiency virus type 1 strains in Kinshasa, Zaire. AIDS Res Hum Retroviruses. 1993;9:613–618. doi: 10.1089/aid.1993.9.613. [DOI] [PubMed] [Google Scholar]
- 26.Vidal N, et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–10507. doi: 10.1128/JVI.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidal N, Mulanga-Kabeya C, Nzilambi N, Delaporte E, Peeters M. Identification of a complex env subtype E HIV type 1 virus from the Democratic Republic of Congo, recombinant with A, G, H, J, K, and unknown subtypes. AIDS Res Hum Retroviruses. 2000;16:2059–2064. doi: 10.1089/088922200750054800. [DOI] [PubMed] [Google Scholar]
- 28.Yang C, et al. Genetic diversification and recombination of HIV type 1 group M in Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2005;21:661–666. doi: 10.1089/aid.2005.21.661. [DOI] [PubMed] [Google Scholar]
- 29.Vidal N, Mulanga-Kabeya C, Nzilambi N, Delaporte E, Peeters M. Characterization of an old complex circulating recombinant form, CRF27_cpx, originating from the Democratic Republic of Congo (DRC) and circulating in France. AIDS Res Hum Retroviruses. 2008;24:315–321. doi: 10.1089/aid.2007.0241. [DOI] [PubMed] [Google Scholar]
- 30.Vidal N, Bazepeo SE, Mulanga C, Delaporte E, Peeters M. Genetic characterization of eight full-length HIV type 1 genomes from the Democratic Republic of Congo (DRC) reveal a new subsubtype, A5, in the A radiation that predominates in the recombinant structure of CRF26_A5U. AIDS Res Hum Retroviruses. 2009;25:823–832. doi: 10.1089/aid.2008.0283. [DOI] [PubMed] [Google Scholar]
- 31.Djoko CF, et al. High HIV type 1 group M pol diversity and low rate of antiretroviral resistance mutations among the uniformed services in Kinshasa, Democratic Republic of the Congo. AIDS Res Hum Retroviruses. 2011;27:323–329. doi: 10.1089/aid.2010.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward MJ, Lycett SJ, Kalish ML, Rambaut A, Leigh Brown AJ. Estimating the rate of intersubtype recombination in early HIV-1 group M strains. J Virol. 2013;87:1967–1973. doi: 10.1128/JVI.02478-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPhee-Fernández C, et al. HIV-1 infection using dried blood spots can be confirmed by Bio-Rad GeeniusTM HIV 1/2 confirmatory assay. J Clin Virol. 2015;63:66–69. doi: 10.1016/j.jcv.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Cepheid. Xpert® HIV-1 Qual, http://www.cepheid.com/en/cepheid-solutions/clinical-ivd-tests/virology/xpert-hiv-1-qual (2018).
- 35.Cepheid. Xpert® HIV-1 Viral Load, http://www.cepheid.com/en/cepheid-solutions/clinical-ivd-tests/virology/xpert-hiv-1-viral-load (2018).
- 36.Roche. COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test v2.0, http://www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/160530_0147_046_00_public_report_v1_final.pdf (2012).
- 37.World Health Organization (WHO). Technical and operational considerations for implementing HIV viral load testing, http://apps.who.int/iris/bitstream/handle/10665/128121/9789241507578_eng.pdf?sequence=1 (2014).
- 38.Robins EB, Blum S. Hematologic Reference Values for African American Children and Adolescents. Am J Hematol. 2007;82:611–614. doi: 10.1002/ajh.20848. [DOI] [PubMed] [Google Scholar]
- 39.Food and Drug Administration (FDA). Blood Serum Chemistry: Normal Values, https://www.fda.gov/downloads/ICECI/Inspections/IOM/UCM135835.pdf (2015).
- 40.World Health Organization (WHO). WHO manual for HIV drug resistance testing using dried blood spot specimens, http://www.who.int/hiv/topics/drugresistance/dbs_protocol.pdf (2010).
- 41.Holguín A, et al. Transmitted drug-resistance in human immunodeficiency virus-infected adult population in El Salvador, Central America. Clin Microbiol Infect. 2013;19:523–532. doi: 10.1111/1469-0691.12264. [DOI] [PubMed] [Google Scholar]
- 42.France Recherche Nord & Sud SIDA-HIV Hépatites (ANRS) protocol. ANRS AC11 Resistance Study Group PCR and Sequencing Procedures: HIV-1, http://www.hivfrenchresistance.org/anrs-procedures.pdf (2015).
- 43.Los Alamos National Laboratory. Circulating Recombinant Forms (CRFs). HIV sequence database, https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html (2018).
- 44.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. doi: 10.1016/S0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- 45.Saag MS, et al. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 46.Department of Health and Human Services. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents, http://www.aidsinfo.nih.gov/contentfiles/adultandadolescentGL.pdf (2018).
- 47.Sollis KA, et al. Systematic review of the performance of HIV viral load technologies on plasma samples. PloS One. 2014;18(9):85869. doi: 10.1371/journal.pone.0085869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mallampati D, Ford N, Hannaford A, Sugandhi N, Penazzato M. Performance of virological testing for early infant diagnosis: A systematic review. J Acquir Immune Defic Syndr. 2017;1:308–314. doi: 10.1097/QAI.0000000000001387. [DOI] [PubMed] [Google Scholar]
- 49.Cohen MS, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;11:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya R, Barton S, Catalan J. When good news is bad news: psychological impact of false positive diagnosis of HIV. AIDS Care. 2008;20:560–564. doi: 10.1080/09540120701867206. [DOI] [PubMed] [Google Scholar]
- 51.Schmitz ME, et al. Field evaluation of Dried Blood Spots for HIV-1 Viral Load monitoring in adults and children receiving antiretroviral treatment in Kenya: Implications for scale-up in resource-limited settings. J Acquir Immune Defic Syndr. 2017;74:399–406. doi: 10.1097/QAI.0000000000001275. [DOI] [PubMed] [Google Scholar]
- 52.Dunning L, et al. The value of confirmatory testing in early infant HIV diagnosis programmes in South Africa: A costeffectiveness analysis. Plos One. 2017;14(11):1002446. doi: 10.1371/journal.pmed.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan WS, et al. Sensitivity of HIV rapid tests compared with fourth-generation enzyme immunoassays or HIV RNA tests. AIDS. 2018;30:1951–1960. doi: 10.1097/QAD.0000000000001134. [DOI] [PubMed] [Google Scholar]
- 54.Rakovsky A, et al. Diagnosis of HIV-1 infection: Performance of Xpert Qual and Geenius supplemental assays in fourth generation ELISA-reactive samples. J Clin Virol. 2018;101:7–10. doi: 10.1016/j.jcv.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Sanders EJ, et al. Targeted screening of at-risk adults for acute HIV-1 infection in sub-Saharan Africa. AIDS. 2015;29:221–230. doi: 10.1097/QAD.0000000000000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawadogo S, et al. Limited utility of dried-blood and plasma spot-based screening for antiretroviral treatment failure with Cobas Ampliprep/TaqMan HIV-1 version 2.0. J Clin Microbiol. 2014;52:3878–3883. doi: 10.1128/JCM.02063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ceffa S, et al. Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 realtime molecular assays for monitoring HIV-1 viral load and detectingHIV-1 infection. J Virol Methods. 2016;229:35–39. doi: 10.1016/j.jviromet.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Guichet E, et al. High viral load and multidrug resistance due to late switch to second-line regimens could be a major obstacle to reach the 90-90-90 UNAIDS Objectives in Sub-Saharan Africa. AIDS Res Hum Retroviruses. 2016;32:1159–1162. doi: 10.1089/aid.2016.0010. [DOI] [PubMed] [Google Scholar]
- 59.De Oliveira F, et al. Detection of numerous HIV-1/MO recombinants in France. AIDS. 2018;32:1289–1299. doi: 10.1097/QAD.0000000000001814. [DOI] [PubMed] [Google Scholar]
- 60.Cepheid. XPERT® HIV-1 QUAL datasheet, http://www.cepheid.com/administrator/components/com_productcatalog/library-files/5263e4b1cad8733be904f496b070bdd0-82a91c538de2b0d71ac23e5594020052-Xpert-HIV-1-Qual-Datasheet-3041-02.pdf (2018).
- 61.Cepheid. Xpert ® HIV-1 Viral Load Supporting National Programs from High-Throughput Reference Labs to Near-Patient Testing, http://www.cepheid.com/administrator/components/com_productcatalog/library-files/f3d173a26b0ba1763775f9d1c9606297-e9f319c1b7c611fc41fe17cfa76f19ff-Xpert-HIV-1-Viral-Load-Datasheet-3038-02.pdf (2017).
- 62.Roche. COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test Datasheet, https://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/PremarketApprovalsPMAs/ucm092878.pdf (2018).
- 63.Avidor B, et al. Comparison between Roche and Xpert in HIV-1 RNA quantitation: A high concordance between the two techniques except for a CRF02_AG subtype variant with high viral load titters detected by Roche but undetected by Xpert. J Clin Virol. 2017;93:15–19. doi: 10.1016/j.jcv.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 64.Mor O, et al. Evaluation of the RealTime HIV-1, Xpert HIV-1, and Aptima HIV-1 Quant Dx assays in comparison to the NucliSens EasyQ HIV-1 v2.0 assay for quantification of HIV-1 viral load. J Clin Microbiol. 2015;53:3458–3465. doi: 10.1128/JCM.01806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muenchhoff M, et al. Evaluation of the NucliSens EasyQ v2.0 assay in comparison with the Roche Amplicor v1.5 and the Roche CAP/CTM HIV-1 Test v2.0 in quantification of C-clade HIV-1 in plasma. Plos One. 2014;9(8):e103983. doi: 10.1371/journal.pone.0103983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gueudin M, et al. Performance evaluation of the new HIV-1 quantification assay, Xpert HIV-1 Viral Load, on a wide panel of HIV-1 variants. J Acquir Immune Defic Syndr. 2016;15:521–526. doi: 10.1097/QAI.0000000000001003. [DOI] [PubMed] [Google Scholar]
- 67.Bruzzone B, et al. Xpert HIV-1 Viral Load assay and VERSANT HIV-1 RNA 1.5 assay: A performance comparison. J Acquir Immune Defic Syndr. 2017;74:86–88. doi: 10.1097/QAI.0000000000001227. [DOI] [PubMed] [Google Scholar]
- 68.Jordan JA, Plantier JC, Templetonc K, Wu AH. Multi-site clinical evaluation of the Xpert®HIV-1 viral load assay. J Clin Virol. 2016;80:27–32. doi: 10.1016/j.jcv.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 69.Ibrahim M, et al. High sensitivity and specificity of the Cepheid Xpert® HIV-1 Qualitative Point-of-Care test among newborns in Botswana. J Acquir Immune Defic Syndr. 2017;75(5):e128–e131. doi: 10.1097/QAI.0000000000001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gous N, Scott L, Berrie L, Stevens W. Options to expand HIV Viral Load testing in South Africa: Evaluation of the GeneXpert® HIV-1 Viral Load assay. Plos One. 2016;11(12):e0168244. doi: 10.1371/journal.pone.0168244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moyo S, et al. Point-of-Care Cepheid Xpert HIV-1 Viral Load test in rural African communities is feasible and reliable. J Clin Microbiol. 2016;54:3050–3055. doi: 10.1128/JCM.01594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrett NJ, Drain PK, Werner L, Samsunder N, Karim SSA. Diagnostic accuracy of the Point-of-Care Xpert HIV-1 Viral Load assay in a South African HIV clinic. J Acquir Immune Defic Syndr. 2016;72:45–48. doi: 10.1097/QAI.0000000000000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mbonze NB, et al. Xpert® MTB/RIF for smear-negative presumptive TB: impact on case notification in DR Congo. Int J Tuberc Lung Dis. 2016;20:240–246. doi: 10.5588/ijtld.15.0177. [DOI] [PubMed] [Google Scholar]
- 74.Haddar C, Begaud E, Maslin J, Germani Y. Point-of-care tests for the rapid diagnosis of shigellosis. Bull Soc Pathol Exot. 2017;110:1–8. doi: 10.1007/s13149-016-0538-6. [DOI] [PubMed] [Google Scholar]
- 75.Li D, et al. Evaluation of the GeneXpert for human Monkeypox diagnosis. Am J Trop Med Hyg. 2017;96:405–410. doi: 10.4269/ajtmh.16-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Makadzange AT, et al. A simple phosphate-buffered-saline-based extraction method improves specificity of HIV Viral Load monitoring Using Dried Blood Spots. J Clin Microbiol. 2017;55:2172–2179. doi: 10.1128/JCM.00176-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.