ABSTRACT
A hereditary system that is based on double-helix DNA sequences provides a stable way to store inherited traits and is favored by most life forms on Earth. However, emerging studies on the phenomenon of non-DNA sequence-based inheritance in multiple species, including mammals, urges a rethinking of the nature of hereditary information carriers and the ways in which they encode heritable traits. In this short opinion piece, we discuss how potential hereditary information carriers such as DNA-associated proteins, epigenetic marks, RNAs and organelles could function via distinct and synergistic modes of action in encoding and transmitting ancestral traits, either inherited or acquired from the environment. In particular, we discuss how the 3D structure of chromatin, in addition to its DNA sequence, may represent a higher-order carrier of hereditary information.
KEY WORDS: DNA methylation, Histone mark, RNA, RNA modifications, Epigenetic inheritance, Hereditary information carrier
Summary: Emerging evidence on the phenomenon of non-DNA sequence-based inheritance in multiple species, including mammals, urges a rethinking of the nature of hereditary information carriers and the ways in which they encode heritable traits.
Introduction
The ability to reproduce and pass on heritable information to offspring is a prerequisite of life. An ideal hereditary system is expected to be stable enough to maintain a species while being tolerant of information input and/or modification, thus enabling natural selection and evolution. DNA sequences have long been believed to be the building blocks of hereditary information and are sufficient to allow evolution through the introduction of mutations. However, recent studies have shown non-DNA sequence-based inheritance of a variety of traits across multiple species (e.g. yeast, plants, worms, flies and mammals), including the intergenerational (i.e. in immediate offspring) and transgenerational (i.e. in subsequent generations of offspring) epigenetic inheritance of acquired characteristics from environmental exposure, e.g. diet-induced metabolic disorders, stress-induced behavioral changes (Chen et al., 2016a; Gapp et al., 2014; Gapp et al., 2018; Grandjean et al., 2015; Öst et al., 2014; Rechavi et al., 2014; Sharma et al., 2016; Zhang et al., 2018). These emerging studies have begun to resurrect the once disputed idea of the ‘inheritance of acquired traits’ (favored by Jean-Baptiste Lamarck; see Box 1) and to erode the concept that DNA sequence is the sole hereditary information carrier. These findings also highlight the potential involvement and functional mechanisms of other (epigenetic) hereditary information carriers such as DNA methylation, histone modifications, RNAs, prions and organelles. In this Spotlight article, we do not elaborate on the cases of non-DNA sequence-based inheritance or the methods used to study them, as they have been extensively reviewed elsewhere (Bohacek and Mansuy, 2017; Chen et al., 2016b; Heard and Martienssen, 2014; Miska and Ferguson-Smith, 2016; Perez and Lehner, 2019; Skvortsova et al., 2018). Instead, we would like to initiate a discussion by rethinking what it means to be a hereditary information carrier, how these carriers evolved and the potential modes by which carriers can encode and transmit ancestral traits that are either inherited or acquired from the environment.
Box 1. Lamarckian inheritance: now and then.
In his (1809) book Philosophie Zoologique, Jean-Baptiste Lamarck proposed two key ideas to explain how a species may adapt to its environment and continuously change over evolution: (1) changes in the environment alter an organism's behavior, leading to greater or lesser use of a given structure and/or organ (e.g. a giraffe's neck), which could then result in an increase or decrease in the size of this structure; (2) these adaptive changes are inherited by subsequent generations in a process known as the ‘inheritance of acquired traits’ or ‘Lamarckian inheritance’. From a modern day perspective, Lamarck's original idea regarding the use and disuse of an organ is obviously crude and naïve, but under certain circumstances it may not be entirely wrong. For example, it has been shown that physical exercise can indeed alter the size, metabolism, transcriptome and epigenome of muscle tissue in mammals (Barrès and Zierath, 2016). Yet the real challenge is to understand how altered (hereditary) information in the soma, either induced by environmental changes or not, can be transferred to the germline and passed to future generations without losing fidelity. This is currently difficult to ascertain, especially without knowing the nature of the hereditary information carrier that is involved; this leads to the downfall of ‘Lamarckian inheritance’. Interestingly, Charles Darwin later supported Lamarck's idea of the ‘inheritance of acquired traits’ by proposing his ‘Pangenesis’ theory, which poses that hypothetical particles termed ‘gemmules’ can transfer (hereditary) information between the soma and germline and to subsequent generations (Liu and Chen, 2018). Whether and to what extent the newly appreciated category of hereditary information carriers we discuss here may live up to the vision of Lamarck and Darwin remains to be clarified and is a topic of contemporary debate.
Rethinking inheritance and hereditary information carriers
From RNA to DNA: the ‘RNA world’ hypothesis
The ‘RNA world’ hypothesis (Higgs and Lehman, 2015) posits that, in the early history of life on Earth, RNA first emerged as both a hereditary information carrier and an enzymatic tool (in the form of ribozymes) to orchestrate self-replication, to create perhaps the simplest version of life on Earth (Joyce, 2012). However, sometime during evolution, the RNA world began to embrace an alternative form of hereditary information carrier: DNA. Concurrent with this major shift was the emergence of specialized factors – proteins – as more sophisticated enzymatic tools that could be used, for example, for converting RNA-based hereditary information into a DNA-based version (reverse transcriptase) and for DNA replication (DNA polymerases). This was a major transition point in evolution; scientists today still do not have a clear idea of how this ‘quantum leap’ in hereditary information storage originated. Yet the impact of this transition was huge, as the chemical structure of DNA is more stable than that of RNA and is thus more suitable for the storage and long-term propagation of hereditary information. As such, the use of DNA as the new hereditary information carrier led to the abundance of DNA-based life forms and eventually drove evolution towards the development of more complicated life forms. Today, RNA still serves as the hereditary material for some viruses, which evolve rapidly because RNA is not as faithfully replicated as DNA and bears higher rates of mutation. However, the majority of more complex life forms use DNA as their primary hereditary information carrier.
Building blocks beyond linear DNA sequence
Another major leap in evolution, after the advent of using DNA as a hereditary information carrier, was the transition of life from prokaryotic to eukaryotic cells. This transition was characterized by the development of membrane-bound nuclei; inside these nuclei, the DNA evolved from circular to linear, and began to bind to proteins (e.g. histones, protamines and other anchor proteins). This binding enabled DNA to form regulated secondary structures and to increase its density without affecting its sequence. The formation of such higher-order structures could be also facilitated by modifications on DNA-associated proteins (e.g. histone modifications) and on DNA itself (e.g. DNA methylation), as well as by non-coding RNAs and their associated modifications. Importantly, the resulting secondary structure could represent a new layer of hereditary information.
Outside the membrane-bound nucleus, almost all eukaryotic cells contain mitochondria that contain circular DNA (mtDNA), which possibly originated from the endocytosis of an ancient prokaryote. This DNA serves as another hereditary information carrier. Mitochondrial-encoded information, such as mtDNA, is maternally inherited. Moreover, the state of mitochondria can be affected by the environment (e.g. dysfunction induced by diet) and such states can be inherited by offspring, either dependent or independent of mtDNA (Saben et al., 2016; Wu et al., 2015). Importantly, alterations in the environment, such as nutritional fluctuations, can change various types of metabolites that are closely related to both mitochondrial function and DNA/chromatin modifications (Ma et al., 2018). For example, some metabolites that are generated by mitochondria, such as acetyl-CoA, are needed for histone acetylation, whereas mitochondria-controlled S-adenosyl methionine (SAM) synthesis is essential for both histone and DNA methylation (Matilainen et al., 2017). Thus, the cellular energy state that is mediated by and related to mitochondrial function is inextricably linked to the epigenetic state of the cell.
In addition to DNA, RNA can play a role in assisting the propagation of hereditary information and might contribute to adaptation to certain environments. This idea is supported by findings in worms, in which exogenous small double-strand RNAs (dsRNA) can induce RNA interference (RNAi) and heritable gene silencing that persists for multiple generations (Fire et al., 1998). Moreover, the alteration of endogenous small interfering RNAs (siRNAs) in response to environmental stimuli (i.e. calorie restriction) can persist transgenerationally in the absence of the initial environmental stimuli, which contributes to an altered phenotype (i.e. longevity) in offspring (Rechavi et al., 2014). Under these circumstances, maintenance of the altered gene expression state requires a small RNA amplification system, the RNA-dependent RNA polymerases (RdRPs), as well as chromatin modifying pathways that may interplay with the RNA signal (Rechavi and Lev, 2017). Similar RNA-based hereditary systems exist in yeast and plants, in which an RdRP system is also present; however, other strategies to exert RNA-mediated inheritance may be present in flies and mammals in which such an RdRP system is absent.
Equipped with the above building blocks and hereditary information carriers, eukaryotic cells were likely able to evolve from unicellular into more complex multicellular organisms, whereas prokaryotic cells never achieved this leap in evolution and remained unicellular.
Hereditary information in 3D?
In the majority of multicellular organisms, the germline comprises specialized cells that store and pass on hereditary material to progeny. Recent studies of the 3D structures of eukaryotic genomes in both germline and somatic cells have revealed that chromatin contains genomic regions that are organized into topologically associating domains (TADs), within which DNA preferentially interacts with itself, as well as DNA loops that bridge functional DNA regions (e.g. promoters, enhancers) that are far apart in linear distance (Rowley and Corces, 2018); the establishment of these structures is facilitated by anchor proteins (e.g. CTCF and cohesin), transcription factors, histone marks and non-coding RNAs etc. This suggests that the correct folding of chromatin within the nucleus might be crucial for gene expression and phenotype. Based on this, it would be a reasonable hypothesis to assume that hereditary information in the germline is encoded in 3D, and that the secondary structure of the genome itself, if replicable between generations, could represent a higher-order dimension of hereditary information beyond just its DNA sequence (Fig. 1). Importantly, if hereditary information stored in 3D format also allows for the input of certain modifications from the environment, such a 3D chromatin structure may encode and/or memorize acquired traits from the environment and thus transmit these traits to offspring, thereby mirroring the ancestral environmental exposure.
Fig. 1.
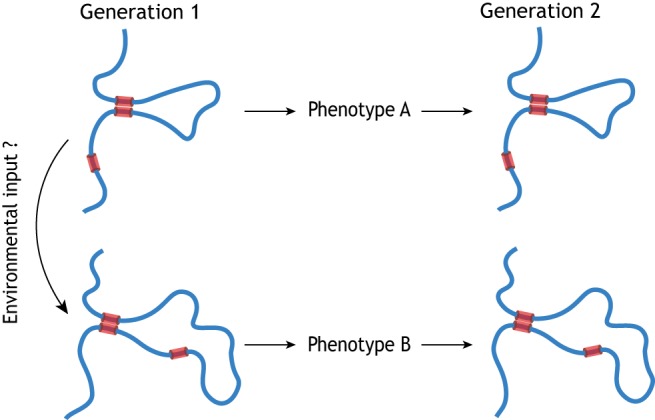
Higher-dimension hereditary information: beyond linear DNA sequence? The schematic depicts a hypothetical self-replication process in which a higher-order chromatin structure (facilitated by anchor proteins, chemical marks on DNA/histones and RNAs etc.) encodes hereditary information and can be modified by environmental input to allow transmission of a modified phenotype across generations. Red boxes represent anchor proteins.
Indeed, recent emerging evidence has shown that altered histone marks and non-coding RNAs, either by genetic alteration of enzymes or environmental input, affect offspring phenotypes across multiple generations without altering DNA sequence (Ciabrelli et al., 2017; Daxinger et al., 2016; Gaydos et al., 2014; Klosin et al., 2017; Öst et al., 2014; Seong et al., 2011; Siklenka et al., 2015; Zhang et al., 2018). For example, it was shown in worms that temperature-induced alterations in H3K9me3 (via the histone methyltransferase SET-25) and in transposon expression can be inherited for multiple generations (Klosin et al., 2017). In flies, a paternal high-sugar diet can induce intergenerational obesity conveyed by H3K9/K27 methyltransferase-dependent chromatin changes (Öst et al., 2014). In addition, transgene-induced phenotypic variation (i.e. eye color) in flies can be determined by Polycomb repressive complex 2 (PRC2)-mediated H3K27me3 levels, which are stably inherited without changing DNA sequence; these molecular events are triggered by 3D contacts between the transgenic and endogenous loci (Ciabrelli et al., 2017). Similar examples exist in mammals. For example, transgenic mice that overexpress the H3K4 demethylase LSD1 (Kdm1a) during spermatogenesis exhibit decreased H3K4me2 in sperm, which results in transgenerationally impaired development and survival in offspring despite the absence of germline LSD1 overexpression (Siklenka et al., 2015). In addition, the deletion of the RNA methyltransferase Dnmt2 (Trdmt1) in mice can diminish RNA-mediated intergenerational transmission of paternally acquired metabolic disorders by disrupting the sperm RNA signature that is induced by a paternal high-fat diet (Zhang et al., 2018). Under these circumstances, altered histone marks and non-coding RNAs may generate as yet unidentified effects on 3D chromatin structure, in addition to their roles in conferring chromatin opening and closing. If such altered 3D chromatin structures represent a type of hereditary information, and could be maintained across generations, they may drive adaptation and Darwinian evolution under the laws of natural selection, similar to natural selection acting on DNA mutation.
Synergistic and distinct coding mechanisms for hereditary information
In many species, especially mammals, extensive reprogramming events erase most epigenetic marks (i.e. DNA methylation and histone modifications) that are carried by germ cells during early embryo development, and this is followed by another round of erasure during fetal germ cell development (Miska and Ferguson-Smith, 2016; Skvortsova et al., 2018). The extent of this germline reprogramming process varies greatly between species, and may be rooted in the relative time over which these species are exposed to a specific environment; species with a short lifespan (i.e. yeast and worm) and those that cannot move freely (i.e. plants) are more likely to be exposed to constant stimuli within their lifespan or between generations, and thus may have developed less extensive germline reprogramming relative to mammals. Given the robust reprogramming of epigenetic marks during development (Eckersley-Maslin et al., 2018), a key issue that underlies the argument of using 3D structures to carry hereditable information in the germline concerns how the disassembly and reassembly of such 3D structures between generations is achieved.
Using genetically engineered transgenes, it has been shown that DNA methylation at the early embryo stage alters chromatin state by up- or downregulating certain histone modifications in adult mice (Hashimshony et al., 2003), which suggests an important role for DNA methylation in regulating chromatin structure. However, whether the germline inherits this type of chromatin alteration, and whether this contributes to the phenotype of offspring, remains unknown. In fact, recent data in mammals appear to suggest against the role of DNA methylation, at least in most cases, as a persistent mark that transfers phenotypes to subsequent generations (Kazachenk, 2018; Radford et al., 2014), which has inspired the search for other modes of action. Interestingly, in recent studies of yeast, worms and mice, transgenerational inheritance of phenotype was shown to involve altered histone marks associated with an altered RNA signal (Rechavi and Lev, 2017; Siklenka et al., 2015; Yu et al., 2018). Although not fully demonstrated, these changes in RNAs may be involved in both establishing the altered chromatin structure and being a consequence of altered chromatin structure. Recent reports have also demonstrated that the zygotic injection of sperm RNAs from male mice that had been exposed to a high-fat diet or mental trauma is sufficient to induce phenotypic changes in the offspring that (partially) mimic the original paternal phenotype following environmental exposure (Chen et al., 2016a; Gapp et al., 2014; Gapp et al., 2018; Grandjean et al., 2015; Zhang et al., 2018). This effect possibly involves the interaction of RNAs (both long and small RNAs) with chromatin structures, in addition to more well-known small RNA-based mechanisms such as post-transcriptional gene silencing (PTGS) at the level of cytoplasmic RNA, and transcriptional gene silencing (TGS) in the nucleus. However, the detailed mechanism that underlies these RNA-mediated epigenetic inheritance phenomena remains largely unknown.
Based on the above findings, we hypothesize that synergistic interactions between different hereditary information carriers – DNA, RNA, chromatin modifications and mitochondria – may be required to achieve a specific 3D chromatin structure in the germline, which in turn is used to encode and transmit complex traits, either inherited or acquired from the environment. A dissection of the modes of action that are exerted by such different hereditary information carriers may lead to a better understanding of how these layers of regulation are possible. Below, we further discuss some principles and concepts that should be considered when evaluating potential hereditary information carriers and their functions in encoding and transmitting heritable traits.
Read-only versus read-written
In general, if a specific trait that has been acquired from the environment is inherited by offspring, the information of such a trait needs to be transferred either directly to the germline or via somatic cells (Chen et al., 2016b), and to be transformed (written) into a form of hereditary information that is carried by the germline and that is readable during offspring development. This process is difficult when considering DNA sequences as the hereditary information carrier because the information stored in genomic DNA sequences is largely ‘read-only’, i.e. it cannot be modified (although multigenerational DNA mutation followed by natural selection may change the DNA sequence and thus bias a trait in a specific environment). However, as certain environmentally induced parental phenotypes (e.g. diet, stress) are indeed known to be recapitulated in immediate offspring (Perez and Lehner, 2019; Skvortsova et al., 2018), this suggests that other hereditary information carriers that are more readily ‘read-and-written’ are involved. For example, DNA methylation and histone marks could be added or erased by specific enzymes, and the loci-specific recruitment of specific enzymes could be mediated by non-coding RNAs (Holoch and Moazed, 2015), such as small RNAs and their associated RNA modifications that are also read-and-written (Zhang et al., 2016). A synergistic interplay between these more labile non-DNA sequence-based epigenetic information carriers enables an attractive mode of action, and provides the potential for hereditary information to be ‘written’ based on environmental input. If such ‘written’ information in the germline can be transmitted to the next generation, it may enable the once-dismissed idea of the ‘inheritance of acquired traits’.
Replication versus reconstruction
The epigenetic reprogramming that is known to occur during early development creates a major barrier in allowing ancestrally ‘written’ information in the germline to be transmitted to offspring. This has also generated a certain degree of skepticism over the idea of the ‘inheritance of acquired traits’ in mammals (Horsthemke, 2018). Indeed, how certain acquired hereditary information escapes the extensive epigenetic reprogramming that occurs during early development to enable intergenerational and transgenerational inheritance remains a central question. In general, preservation and amplification of the original hereditary information that is carried by the germline during embryo development could be achieved either by means of replication or by reconstruction, as discussed previously (Jablonka and Raz, 2009; Miska and Ferguson-Smith, 2016). Essentially, the replication of hereditary information involves directly copying DNA or epigenetic marks; for the latter, this means the original epigenetic marks need to be protected from erasure, as occurs in the case of DNA methylation on certain imprinted genes and repetitive sequences. By contrast, the reconstruction of hereditary information suggests a mode of action wherein epigenetic marks are first erased but are then re-established after extensive epigenetic reprogramming. This reconstruction of epigenetic marks likely involves other molecules, such as those that could act in trans and are not subject to erasure by epigenetic reprogramming. Regulatory RNAs could represent such trans-acting molecules in the germline. Hypothetically, the alteration of DNA methylation and histone marks may trigger specific non-coding RNA expression, and these non-coding RNAs in turn may reconstruct the specific DNA methylation and histone marks after their erasure. Such a regulatory cycle has not yet been demonstrated in mammals but, in our opinion, merits further investigation. The action of non-coding RNAs in mammals may also involve a self-amplification system similar to that involving RdRPs in plants and worms (Rechavi and Lev, 2017), which may be achieved by yet to be discovered protein complexes in mammalian germ cells or the early embryo.
In addition, another special form of RNAs – transcribed transposon elements (e.g. LINE1) – could act as trans-acting factors. These elements can change not only DNA-associated epigenetic information, such as chromatin structure, but also the DNA sequence itself (Rodriguez-Terrones and Torres-Padilla, 2018). Moreover, the expression of transcribed transposon elements can be regulated by germ cell-derived small RNAs such as piRNAs (Siomi et al., 2011) and by tRNA-derived small RNAs (tsRNAs) (Martinez et al., 2017; Schorn et al., 2017; Sharma et al., 2016). Because of these unique features, RNAs, especially small non-coding RNAs, have recently emerged as an important type of hereditary information carriers in mediating non-DNA sequence-based inheritance (Chen et al., 2016b).
Digital versus analog
Another fundamental feature that is observed in non-DNA sequence-based inheritance is that the inherited traits in offspring typically exhibit non-Mendelian patterns with variable penetrance of phenotype (Chen et al., 2016b; Heard and Martienssen, 2014; Miska and Ferguson-Smith, 2016), compared with the relatively stricter Mendelian distribution patterns that are induced by DNA mutation. This could be explained, in part, by the fact that the information stored in a defined segment of DNA sequence in the germline is ‘digital’ (i.e. mutant or wild type), whereas the information that is stored by other epigenetic carriers is usually ‘analog’ (i.e. dose-dependent). Thus, the hereditary information that is carried by DNA sequences (e.g. a single-copy gene that controls eye color) has limited manifestation regarding genotype (+/+, +/−, −/−), which is digital in its nature. The phenotype that is encoded by such a digital signal could be represented as either strong or none, depending on the genotype. On the other hand, hereditary information carried by other forms, such as that encoded by non-coding RNAs, is analog in nature. This is because of the fact that RNA copy numbers can range from zero to tens of thousands, and their function with regard to altering DNA methylation and histone marks is dose-dependent and thus variable. As such, the phenotypic output shows a ‘noisy’ pattern: the magnitude of the phenotype may depend on the strength of signal input, such as the frequency, intensity and timing of an environmental exposure.
In addition to RNA-based analog signals, multi-copy genes such as sperm ribosomal DNA (rDNA) exhibit intrinsic genetic variations, and this also provides a basis for differential levels of DNA methylation at specific loci upon environmental exposure (e.g. dietary exposure). For example, it has been shown that the level of methylation at multi-copy rDNA loci is linearly correlated with the growth phenotype of offspring (Holland et al., 2016). This provides another example of how a hereditary phenotype can be controlled by an analog signal based on multi-copy DNA elements. Interestingly, using a recently improved bioinformatic pipeline (Shi et al., 2018), sperm were found to harbor ribosomal RNA-derived small RNAs (rsRNAs) that are sensitive to dietary exposure (Zhang et al., 2018). The potential link between DNA methylation at rDNA loci and the level of sperm rsRNAs in this context is interesting; their mechanism in regulating offspring phenotype may relate to ribosomal heterogeneity (Genuth and Barna, 2018) during embryo development and warrants further investigation.
Finally, it is noteworthy that, although DNA sequence-coded hereditary information could be thought of as digital information, many DNA-mutation induced phenotypes, according to mouse knockout studies, also show incomplete penetrance of phenotype. The underlying mechanisms may involve the compensation of other functionally redundant and/or modifier genes, but they could also involve epigenetic regulators (DNA methylation, histone modifications, non-coding RNAs) that function as analog signals that interfere with the ‘digital’ (clear) manifestation of a phenotype.
Future perspectives
The evidence for non-DNA sequence-based inheritance is now growing, but the molecular mechanisms that underlie these phenomena remain mostly elusive, especially in mammals. In these cases, the information that is acquired through either heritable epimutations or environmental input that is transmitted to offspring is likely encoded in a manner that goes beyond a linear DNA sequence. In some cases, it is difficult to attribute the underlying mechanism to any of the known hereditary information carriers (Cartier et al., 2018). This suggests that our current understanding of the ‘coding mechanism’ of non-DNA sequence-based inheritance in mammals remains rudimentary at best.
The higher-order level of information that is encoded by 3D chromatin structure, which is established via multiple hereditary information carriers such as DNA methylation, histone modifications and RNAs under their distinctive and synergistic modes of action, begins to form a framework that may lay the foundation for future research. Understanding the cycle of how such higher-order information exerts self-replication while allowing environmentally induced modification holds the key to a deeper understanding of inheritance and life. Spatiotemporal compartmentalization and segregation of signals, such as that mediated by nuclear speckles and cytoplasmic granules (which are known to be facilitated by non-coding RNAs and anchor proteins) (Wan et al., 2018), may provide potential systems that link different hereditary information carriers together. This would allow these carriers to regulate collectively the dynamic disassembly and reassembly of heritable information.
Additional studies, especially in mammals, are much needed in the future to establish the principles of how non-DNA sequence-based information carriers and/or their effects are maintained and relayed throughout development and across generations. It will be especially important to address how different hereditary information carriers are dynamically organized into a higher-order chromatin structure to allow storage and propagation of hereditary information, and to what extent these hereditary information carriers acquire information from parental exposure and contribute to offspring phenotypes that may be adaptive to the ancestral environment. Complete clarification of these issues might be a distant goal but, in our opinion, these topics represent the most exciting challenges in genetic and epigenetic research of our time.
Acknowledgements
We thank Junchao Shi for preparing the figure illustrations, Maya Pahima for language editing and our lab members for insightful discussion on manuscript contents. We apologize for those important works that could not be cited owing to space limitation.
Footnotes
Competing interests
The author declares no competing or financial interests.
Funding
Research related to this paper is supported by the Ministry of Science and Technology of the People's Republic of China (2018YFC1004500 and 2015CB9430000 to Y.Z.), the National Natural Science Foundation of China (31671201 to Y.Z.) and the National Institutes of Health (R01HD092431 to Q.C.). Y.Z. is a fellow of the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2016081). Deposited in PMC for release after 12 months.
References
- Barrès R. and Zierath J. R. (2016). The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat. Rev. Endocrinol. 12, 441-451. 10.1038/nrendo.2016.87 [DOI] [PubMed] [Google Scholar]
- Bohacek J. and Mansuy I. M. (2017). A guide to designing germline-dependent epigenetic inheritance experiments in mammals. Nat. Methods 14, 243-249. 10.1038/nmeth.4181 [DOI] [PubMed] [Google Scholar]
- Cartier J., Smith T., Thomson J. P., Rose C. M., Khulan B., Heger A., Meehan R. R. and Drake A. J. (2018). Investigation into the role of the germline epigenome in the transmission of glucocorticoid-programmed effects across generations. Genome Biol. 19, 50 10.1186/s13059-018-1422-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G. H., Peng H., Zhang X., Zhang Y. et al. (2016a). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397-400. 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- Chen Q., Yan W. and Duan E. (2016b). Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 17, 733-743. 10.1038/nrg.2016.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabrelli F., Comoglio F., Fellous S., Bonev B., Ninova M., Szabo Q., Xuereb A., Klopp C., Aravin A., Paro R. et al. (2017). Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila. Nat. Genet. 49, 876-886. 10.1038/ng.3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L., Oey H., Isbel L., Whitelaw N. C., Youngson N. A., Spurling A., Vonk K. K. and Whitelaw E. (2016). Hypomethylation of ERVs in the sperm of mice haploinsufficient for the histone methyltransferase Setdb1 correlates with a paternal effect on phenotype. Sci. Rep. 6, 25004 10.1038/srep25004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M. A., Alda-Catalinas C. and Reik W. (2018). Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436-450. 10.1038/s41580-018-0008-z [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S. O., Montgomery M. K., Kostas S. A., Driver S. E. and Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E. and Mansuy I. M. (2014). Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 17, 667-669. 10.1038/nn.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapp K., van Steenwyk G., Germain P. L., Matsushima W., Rudolph K. L. M., Manuella F., Roszkowski M., Vernaz G., Ghosh T., Pelczar P. et al. (2018). Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol. Psychiatry, s41380-018-0271-6 10.1038/s41380-018-0271-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Wang W. and Strome S. (2014). Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345, 1515-1518. 10.1126/science.1255023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth N. R. and Barna M. (2018). Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 19, 431-452. 10.1038/s41576-018-0008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V., Fourré S., De Abreu D. A. F., Derieppe M.-A., Remy J.-J. and Rassoulzadegan M. (2015). RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci. Rep. 5, 18193 10.1038/srep18193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T., Zhang J., Keshet I., Bustin M. and Cedar H. (2003). The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 34, 187-192. 10.1038/ng1158 [DOI] [PubMed] [Google Scholar]
- Heard E. and Martienssen R. A. (2014). Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95-109. 10.1016/j.cell.2014.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs P. G. and Lehman N. (2015). The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7-17. 10.1038/nrg3841 [DOI] [PubMed] [Google Scholar]
- Holland M. L., Lowe R., Caton P. W., Gemma C., Carbajosa G., Danson A. F., Carpenter A. A., Loche E., Ozanne S. E. and Rakyan V. K. (2016). Early-life nutrition modulates the epigenetic state of specific rDNA genetic variants in mice. Science 353, 495-498. 10.1126/science.aaf7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D. and Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71-84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B. (2018). A critical view on transgenerational epigenetic inheritance in humans. Nat. Commun. 9, 2973 10.1038/s41467-018-05445-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E. and Raz G. (2009). Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131-176. 10.1086/598822 [DOI] [PubMed] [Google Scholar]
- Joyce G. F. (2012). Bit by bit: the Darwinian basis of life. PLoS Biol. 10, e1001323 10.1371/journal.pbio.1001323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazachenk A. (2018). Identification, characterization, and heritability of murine metastable epialleles: implications for non-genetic inheritance. Cell 175, 1259-1271. 10.1016/j.cell.2018.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T. and Lehner B. (2017). Transgenerational transmission of environmental information in C. elegans. Science 356, 320-323. 10.1126/science.aah6412 [DOI] [PubMed] [Google Scholar]
- Liu Y. and Chen Q. (2018). 150 years of Darwin's theory of intercellular flow of hereditary information. Nat. Rev. Mol. Cell Biol. 19, 749-750. 10.1038/s41580-018-0072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Niu R., Huang T., Shao L. W., Peng Y., Ding W., Wang Y., Jia G., He C., Li C. Y. et al. (2018). N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol., s41556-018-0238-5 10.1038/s41556-018-0238-5 [DOI] [PubMed] [Google Scholar]
- Martinez G., Choudury S. G. and Slotkin R. K. (2017). tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 45, 5142-5152. 10.1093/nar/gkx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen O., Quirós P. M. and Auwerx J. (2017). Mitochondria and epigenetics - crosstalk in homeostasis and stress. Trends Cell Biol. 27, 453-463. 10.1016/j.tcb.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Miska E. A. and Ferguson-Smith A. C. (2016). Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science 354, 59-63. 10.1126/science.aaf4945 [DOI] [PubMed] [Google Scholar]
- Öst A., Lempradl A., Casas E., Weigert M., Tiko T., Deniz M., Pantano L., Boenisch U., Itskov P. M., Stoeckius M. et al. (2014). Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159, 1352-1364. 10.1016/j.cell.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Perez M. F. and Lehner B. (2019). Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143-151. 10.1038/s41556-018-0242-9 [DOI] [PubMed] [Google Scholar]
- Radford E. J., Ito M., Shi H., Corish J. A., Yamazawa K., Isganaitis E., Seisenberger S., Hore T. A., Reik W., Erkek S. et al. (2014). In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 345, 1255903 10.1126/science.1255903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O. and Lev I. (2017). Principles of Transgenerational Small RNA Inheritance in Caenorhabditis elegans. Curr. Biol. CB 27, R720-R730. 10.1016/j.cub.2017.05.043 [DOI] [PubMed] [Google Scholar]
- Rechavi O., Houri-Ze'evi L., Anava S., Goh W. S. S., Kerk S. Y., Hannon G. J. and Hobert O. (2014). Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158, 277-287. 10.1016/j.cell.2014.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D. and Torres-Padilla M. E. (2018). Nimble and ready to mingle: transposon outbursts of early development. Trends Genet. 34, 806-820. 10.1016/j.tig.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Rowley M. J. and Corces V. G. (2018). Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789-800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saben J. L., Boudoures A. L., Asghar Z., Thompson A., Drury A., Zhang W., Chi M., Cusumano A., Scheaffer S. and Moley K. H. (2016). Maternal metabolic syndrome programs mitochondrial dysfunction via germline changes across three generations. Cell Rep. 16, 1-8. 10.1016/j.celrep.2016.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn A. J., Gutbrod M. J., LeBlanc C. and Martienssen R. (2017). LTR-Retrotransposon control by tRNA-derived small RNAs. Cell 170, 61-71.e11. 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K.-H., Li D., Shimizu H., Nakamura R. and Ishii S. (2011). Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145, 1049-1061. 10.1016/j.cell.2011.05.029 [DOI] [PubMed] [Google Scholar]
- Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., Belleannee C., Kucukural A., Serra R. W., Sun F. et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391-396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Ko E.-A., Sanders K. M., Chen Q. and Zhou T. (2018). SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genomics Proteomics Bioinformatics 16, 144-151. 10.1016/j.gpb.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siklenka K., Erkek S., Godmann M., Lambrot R., McGraw S., Lafleur C., Cohen T., Xia J., Suderman M., Hallett M. et al. (2015). Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 350, aab2006 10.1126/science.aab2006 [DOI] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D. and Aravin A. A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246-258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Skvortsova K., Iovino N. and Bogdanovic O. (2018). Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 19, 774-790. 10.1038/s41580-018-0074-2 [DOI] [PubMed] [Google Scholar]
- Wan G., Fields B. D., Spracklin G., Shukla A., Phillips C. M. and Kennedy S. (2018). Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679-683. 10.1038/s41586-018-0132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. L., Russell D. L., Wong S. L., Chen M., Tsai T.-S., St John J. C., Norman R. J., Febbraio M. A., Carroll J. and Robker R. L. (2015). Mitochondrial dysfunction in oocytes of obese mothers: transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development 142, 681-691. 10.1242/dev.114850 [DOI] [PubMed] [Google Scholar]
- Yu R., Wang X. and Moazed D. (2018). Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature 558, 615-619. 10.1038/s41586-018-0239-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Cozen A. E., Liu Y., Chen Q. and Lowe T. M. (2016). Small RNA modifications: integral to function and disease. Trends Mol. Med. 22, 1025-1034. 10.1016/j.molmed.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Shi J., Tuorto F., Li X., Liu Y., Liebers R., Zhang L., Qu Y., Qian J. et al. (2018). Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 20, 535-540. 10.1038/s41556-018-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]


