Significance
In the course of certain chronic infections, Pseudomonas aeruginosa can rewire its quorum-sensing (QS) circuits such that the master transcription regulator LasR is no longer required for the subservient transcription regulator RhlR to function. Rather than LasR-dependent growth on casein, growth of such a clinical isolate on casein depends on RhlR. The social dynamics during growth of this isolate differ from those of LasR-positive P. aeruginosa. Our findings advance the fundamental understanding of QS circuits in P. aeruginosa and indicate that RhlR, rather than LasR, might be an appropriate target for therapeutic intervention of chronic P. aeruginosa infections.
Keywords: acyl-homoserine lactone, chronic infection, cystic fibrosis, Pseudomonas quinolone signal, social evolution
Abstract
The opportunistic bacterial pathogen Pseudomonas aeruginosa has a layered acyl-homoserine lactone (AHL) quorum-sensing (QS) system, which controls production of a variety of extracellular metabolites and enzymes. The LasRI system activates genes including those coding for the extracellular protease elastase and for the second AHL QS system, RhlRI. Growth of P. aeruginosa on casein requires elastase production and LasR-mutant social cheats emerge in populations growing on casein. P. aeruginosa colonizes the lungs of individuals with the genetic disease cystic fibrosis (CF), and LasR mutants can be isolated from the colonized lungs; however, unlike laboratory-generated LasR mutants, many of these CF isolates have functioning RhlR-RhlI systems. We show that one such mutant can use the RhlR-RhlI system to activate expression of elastase and grow on casein. We carried out social-evolution experiments by growing this isolate on caseinate and, as with wild-type P. aeruginosa, elastase-negative mutants emerge as cheats, but these are not RhlR mutants; rather, they are mutants that do not produce the non-AHL Pseudomonas quinolone signal (PQS). Furthermore, we generated a RhlRI mutant and showed it had a fitness defect when growing together with the parent. Apparently, RhlR QS and PQS collude to support growth on caseinate in the absence of a functional LasR. Our findings provide a plausible explanation as to why P. aeruginosa LasR mutants, but not RhlR mutants, are common in CF lungs.
The opportunistic human pathogen Pseudomonas aeruginosa is a model for studies of quorum sensing (QS) (1). There are two complete acyl-homoserine lactone (AHL) QS signal synthase and receptor circuits that together activate expression of hundreds of P. aeruginosa genes (2). The lasI and lasR genes code for a N-3-oxo-dodecanoyl-l-homoserine lactone (3OC12-HSL) synthase and receptor, respectively. In the well-studied strains P. aeruginosa PAO1 and PA14, the LasI-LasR circuit activates a number of genes, including those encoding extracellular proteases, and the second QS circuit, RhlI-RhlR (2). The rhlI and rhlR genes code for a N-butanoyl-l-homoserine lactone (C4-HSL) synthase and receptor, respectively. The RhlR regulon consists of a number of genes and overlaps with the LasR regulon; for example, the promoter of lasB, which codes for the extracellular protease elastase, has binding sites for both LasR and RhlR, with LasR showing the strongest lasB activation (3, 4). There is also a non-AHL signaling system called the Pseudomonas quinolone signal (PQS) system. The PQS is 2-heptyl-3-hydroxy-4-quinolone (5), and the PQS synthesis operon is responsible for production of several additional 2-alkylquinolones related to PQS (6). The transcriptional activator of the pqs operon is coded by pqsR (also termed mvfR) (7). The genes affected by PQS and the two AHL signals are overlapping (8, 9).
Growth of P. aeruginosa on proteins like caseinate requires AHL QS for activation of the elastase gene, lasB, and genes for other extracellular proteases (10, 11). These proteases are public goods, and individual cells in a population can benefit from them whether or not they produce them. In groups of cells growing on casein, LasR mutants have a fitness advantage over the wild type. The mutants are cheats (12, 13) and can reach 30–40% of the population in laboratory experiments where the wild type is transferred daily with casein as the sole source of carbon and energy (12, 14). Among genes that the RhlR-RhlI system activates is the hcn operon, which codes for cyanide production. RhlR-RhlI also increases cyanide resistance. Wild-type P. aeruginosa can police cheats during growth on caseinate via cyanide production, which selectively impairs growth of LasR-mutant cheats (15).
Experiments with animal infection models have established that P. aeruginosa LasR QS mutants are attenuated for virulence (16, 17). Thus, it is a curiosity that in individuals with chronic cystic fibrosis (CF) lung infections, P. aeruginosa with LasR-mutant variants can be readily isolated (18). Many LasR-mutant isolates from CF children colonized with P. aeruginosa remain AHL-responsive through the RhlI-RhlR system (19, 20). Furthermore, RhlR-mutant isolates from infected CF lungs are rare and are found specifically in hypermutable strains very late in the course of chronic CF infections (20). Here, we describe experiments with a CF clinical isolate that has a lasR mutation and an active RhlRI QS circuit. This isolate shows RhlRI–dependent growth on caseinate, and protease-deficient mutants arise during extended growth on caseinate; however, these mutants retain a functional RhlRI circuit. We show that this clinical isolate has a fitness advantage during coculture with a RhlR mutant.
Results
The CF Isolate P. aeruginosa E80 Is a LasR Mutant That Can Grow on Caseinate.
We chose to study P. aeruginosa E80 because, unlike many other P. aeruginosa clinical isolates, it proved to be amenable to genetic manipulation and because we found that it had a 4-bp deletion in lasR, which created a frame shift starting at amino acid residue 107 of the 239-aa LasR polypeptide (revealed by analysis of the complete genome sequence and confirmed by sequencing a PCR-amplified lasR). A previous publication showed that the RhlRI system in many LasR-mutant clinical isolates had escaped dependence on LasR and produced relatively high levels of C4-HSL, pyocyanin, and elastase (19). Thus, we tested strain E80 and showed, like other clinical isolates, it also produced C4-HSL, extracellular protease, and high levels of pyocyanin (Fig. 1). We then asked whether E80 was capable of growth with sodium caseinate as the sole source of carbon and energy and found that it had this capability (SI Appendix, Fig. S1). From an inoculum of bacteria grown in 3-(N-morpholino)propanesulfonic acid (Mops)-buffered LB (LB-Mops broth), there was a lag with growth on casein broth; evident growth occurred at 2–3 d after inoculation. Growth was evident after 1 d in subsequent transfers.
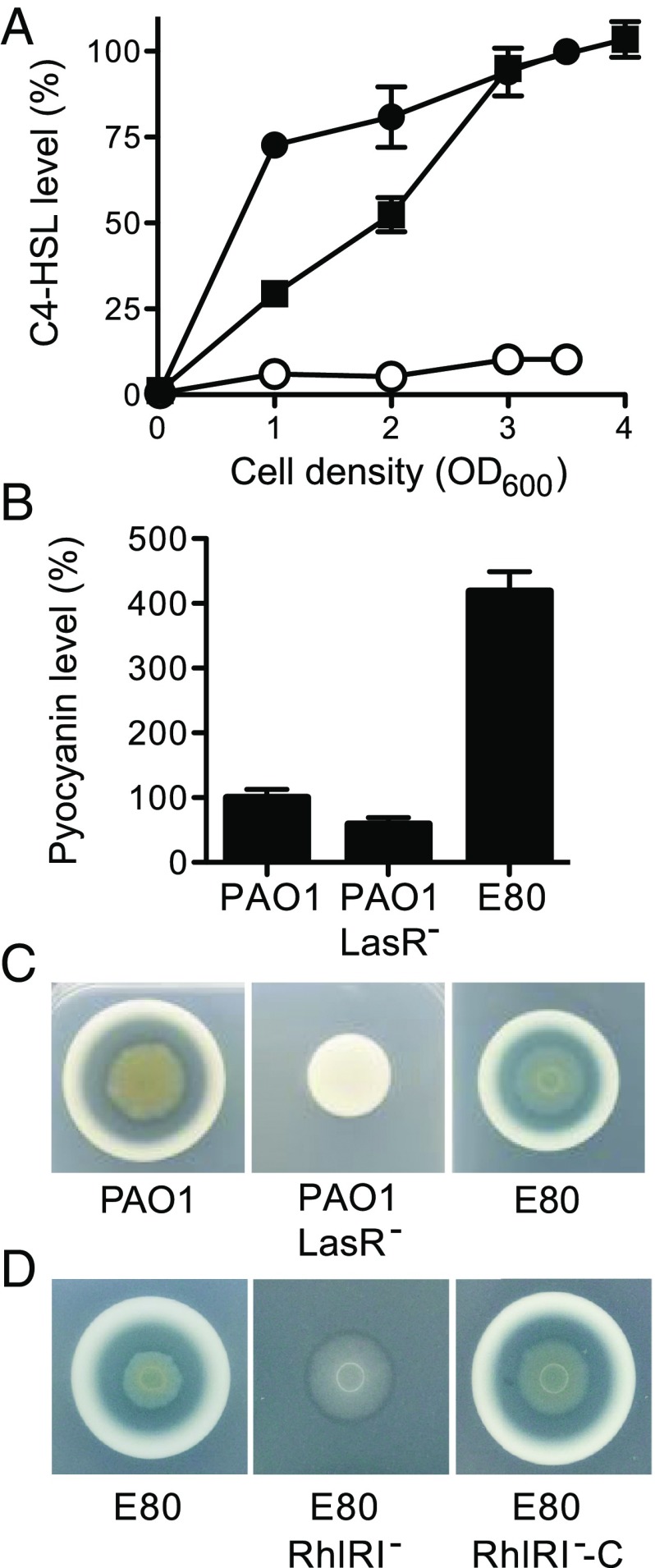
Fig. 1.
Production of C4-HSL, pyocyanin, and protease by the LasR− mutant clinical isolate E80 and E80 RhlRI− mutant. (A) Relative levels of C4-HSL in P. aeruginosa E80 (black squares), PAO1 (black circles), and a PAO1 LasR− mutant (white circles) at different times during cell growth. Levels are normalized to the maximum amount of C4-HSL in PAO1 cultures. The data are means ± SD (bars) of four independent experiments. (B) Relative pyocyanin levels of the indicated strain. The pyocyanin levels produced by strain PAO1 are defined as 100%. Pyocyanin was measured after 24 h of growth in LB-Mops broth. Data represent means and SDs of six replicates. (C and D) Protease production on casein agar. High levels of extracellular proteases result in a zone of clearing around a colony, with a white zone of partial casein degradation at the periphery of the zone of clearing. The PAO1 LasR mutant and the E80 RhlRI mutant produce relatively little protease, as indicated by the lack of a zone of clearing. There is only a small zone of partial casein digestion. The images are of colonies after about 30 h of incubation.
Growth of P. aeruginosa E80 on Caseinate Is RhlR Dependent.
Because there is a RhlR-binding site in the promoter region of lasB (4), and the RhlRI system was active in E80 (Fig. 1A), we hypothesized that growth of this clinical isolate on casein required RhlRI. As a test of the hypothesis, we constructed a RhlRI mutant (Materials and Methods and SI Appendix, Fig. S2) and found that it did not grow in caseinate broth, it made little or no protease, and grew poorly on caseinate agar plates (Fig. 1D). The RhlRI-mutant defects were complemented by introduction of rhlRI at a neutral site on the chromosome (Fig. 1D).
Protease-Deficient Mutants Emerge Rapidly During Growth of P. aeruginosa E80 on Caseinate.
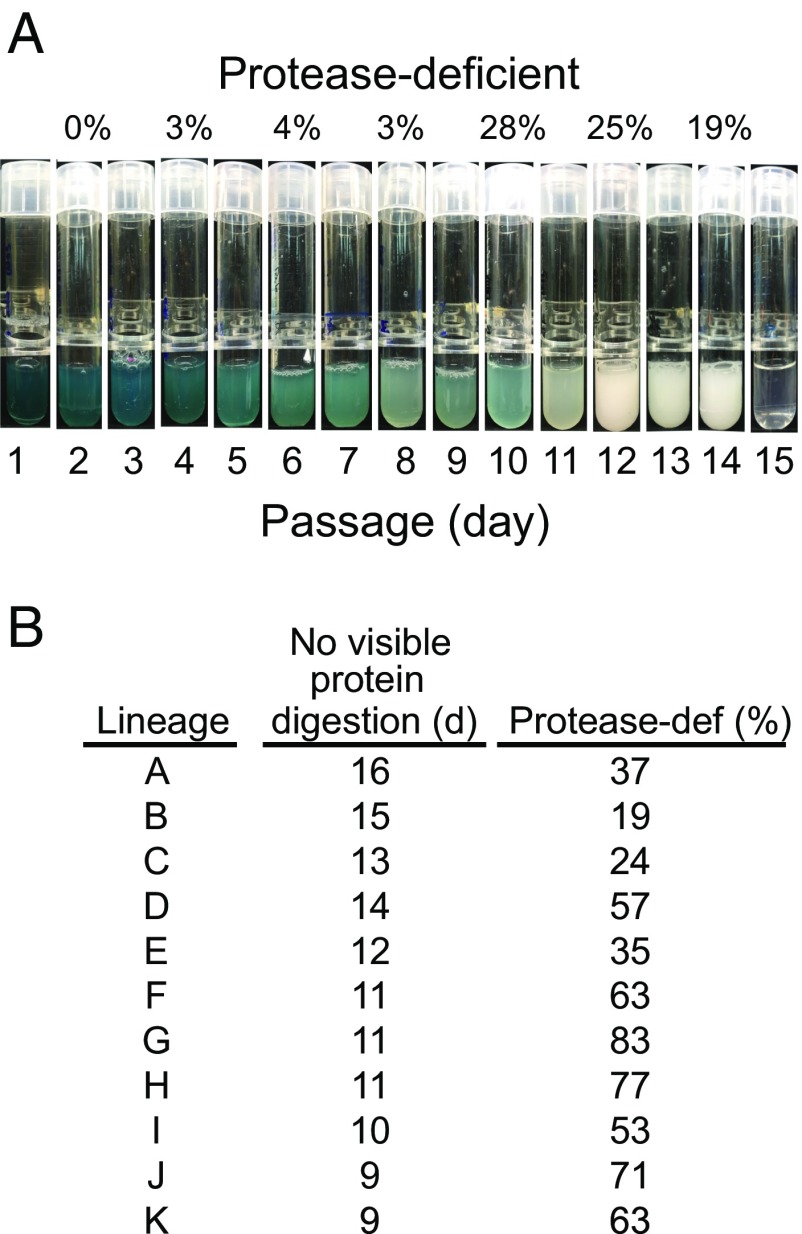
The ability of E80 to grow on caseinate made it possible to ask whether social cheats emerge during RhlRI-dependent growth on this protein source in a fashion similar to the emergence of LasR-mutant cheats of the commonly studied P. aeruginosa PAO1 (12). Control experiments with LasR wild-type PAO1 showed emergence of protease-deficient mutants only after about 7 wk of daily transfer. Protease-deficient mutants emerged in E80 cultures as early as 4–6 d of daily transfer. These protease-deficient mutants did not grow individually in casein broth; therefore, we refer to them as freeloaders because they require the presence of protease-positive cooperators to persist in serial passage experiments on caseinate. In 11 separate experiments, protease-deficient freeloader abundance increased rapidly, and growth ceased to occur on transfer after 9–16 d depending on the culture (Fig. 2). As an example, one of the social-evolution experiments is shown in Fig. 2A. During the first several days, there was heavy growth and copious production of pyocyanin. By day 10, there was relatively little pyocyanin produced, and the caseinate was only partially degraded such that the culture looked milky white. On the last transfer, there was no apparent growth.
Fig. 2.
Emergence of protease-deficient freeloaders during daily passage of strain E80 in casein broth. (A) An example of one serial transfer experiment (lineage B). Each photographic image was captured after 1 d of growth. The relative abundance of protease-deficient freeloaders (as a percentage of the total population) in culture samples just before transfer (50 μL) is shown above the indicated tubes, and the daily passage number is shown below the tubes. Cultures that only partially degrade casein have a milky white appearance due to protein precipitation. (B) Summary of results from 11 independent daily transfer experiments (A through K), including the transfer day (d) that no longer showed protein digestion and the percentage of freeloaders (protease-def) detected at the 2-d interval before the final day.
Characterization of Protease-Deficient Social Freeloaders Emerging During Growth in Caseinate Broth.
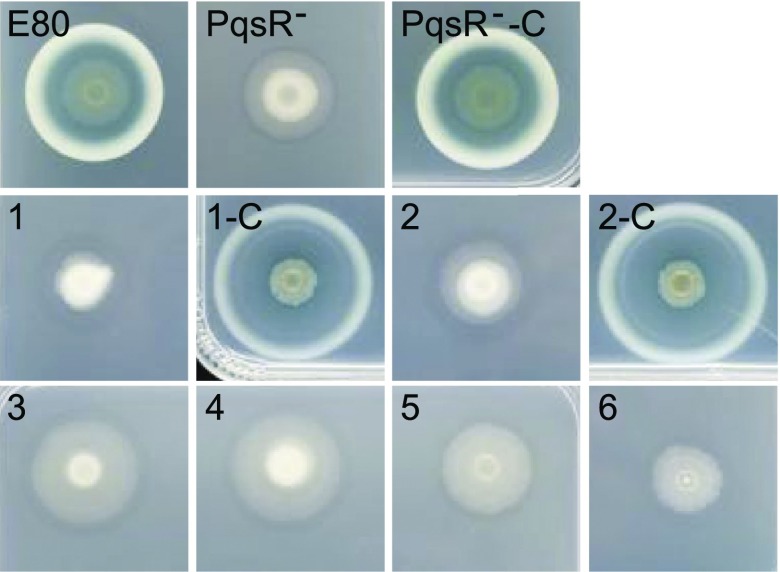
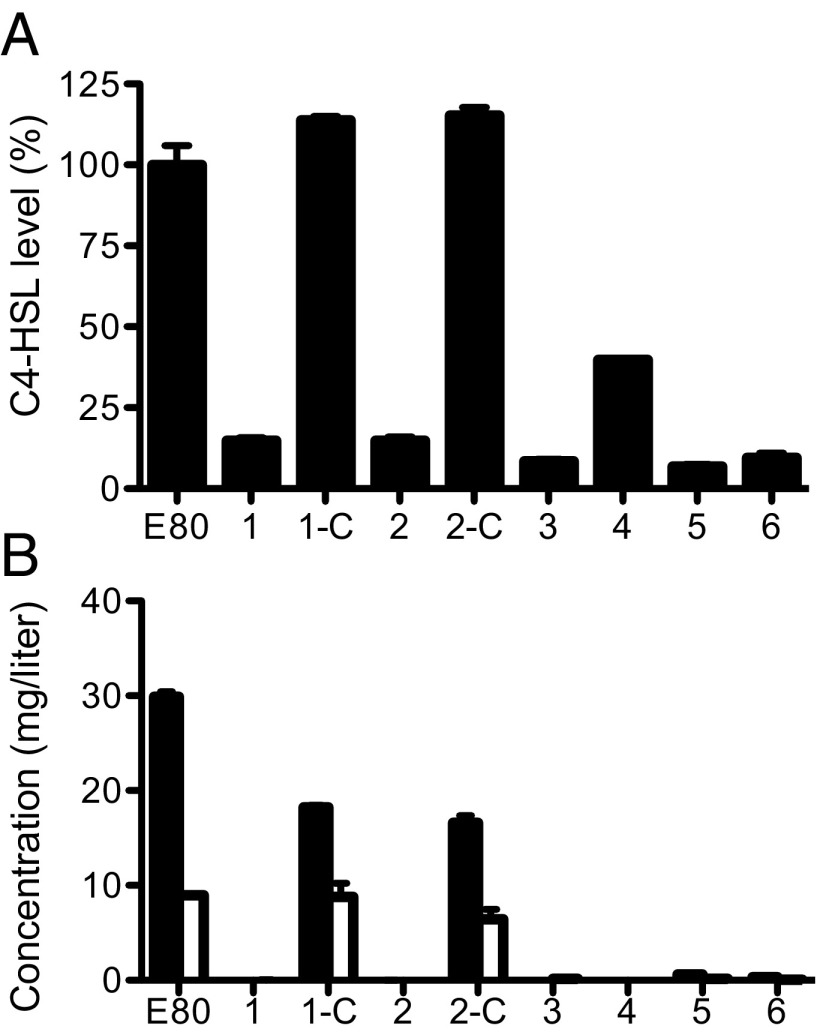
We further characterized six protease-negative isolates from four independent social-evolution experiments. When grown on caseinate agar plates, the agar under colonies of all six isolates was cloudy. By comparison, the E80 parent created a large zone of clearing around a colony, which was surrounded by a milky ring where caseinate degradation was incomplete (Fig. 3). Apparently, there is a small amount of extracellular protease produced by all six isolates, but it is greatly reduced compared with the parent strain E80. The freeloader isolates also did not produce pyocyanin. By analogy to LasR P. aeruginosa PAO1 mutants, which arise during extended growth on casein, we hypothesized that the freeloaders would be RhlR mutants. However, we showed by sequencing that the rhlI-rhlR genomic region in all six freeloaders was identical to that in E80. Although the rhlI-rhlR region was unaltered in the protease-deficient freeloaders, all made substantially less C4-HSL than the parent strain E80 (Fig. 4A).
Fig. 3.
The relationship between PQS and protease production in strain E80. The top row shows an E80 colony, an E80 PqsR mutant, and the PqsR mutant complemented with a functional pqsR (PqsR−-C) in the att site. The middle and bottom rows show colonies of E80 freeloaders (1–6) and colonies of freeloaders 1 and 2 complemented with pqsR in the chromosomal att site (1-C and 2-C).
Fig. 4.
The E80 protease-deficient freeloaders show reduced levels of C4-HSL and 2-alkylquinolone production. (A) Relative C4-HSL levels in cultures of strain E80, E80 freeloaders (1–6), and E80 freeloaders 1 and 2 complemented with a chromosomal copy of pqsR (1-C and 2-C). Values are normalized to E80 levels. (B) Concentrations of PQS (black) and HHQ (white) produced in the indicated strains. For both A and B, data are the means of four replicates ± SD.
We next performed a whole-genome sequencing analysis of the wild-type E80 and the six protease-deficient mutants discussed above. Four of the six had a mutation in pqsR (Table 1). The pqsR gene codes for the transcriptional activator of pqs genes involved in the synthesis of PQS and other 2-alkylquinolones. This finding of pqsR mutations in four of the six cheats led us to hypothesize that PQS or other alkylquinolones might be required in addition to RhlRI for growth of E80 on casein. Thus, we measured PQS and another pqs synthesis gene product 2-heptyl-4-hydroxyquinolone (HHQ) production by E80 and all six cheats. Wild-type E80 made substantial amounts of PQS and HHQ, whereas these 2-alkylquinolones were not produced in detectable amounts by any of the freeloaders (Fig. 4B). We also generated a pqsR-deletion mutant and showed, like the freeloaders, it produced very little protease. We complemented the pqsR mutation in two of the freeloaders (1, 2) and the pqsR-deletion mutant, and all complemented strains produced high levels of protease (Fig. 3). These experiments indicate PQS and/or other 2-alkylquinolones are required for RhlR–dependent growth of E80 on caseinate and that the protease-deficient mutants arising during growth on caseinate are PQS-deficient. The deficiency was often, but not always, the result of a pqsR mutation.
Table 1.
Summary of pqsR variants present in E80 protease-deficient isolates that emerged during growth in caseinate
| Strain | pqsR genotype | Variation |
| 1 | Δ371–374 | Frame-shift deletion |
| 2 | Δ109–121 | Frame-shift deletion |
| 3 | T811G | Tyr-271 Asp |
| 4 | C101T | Ala-34 Val |
| 5 | Wild type | No variation |
| 6 | Wild type | No variation |
Relative Fitness of Wild-Type E80, a PqsR Mutant, and a RhlR Mutant.
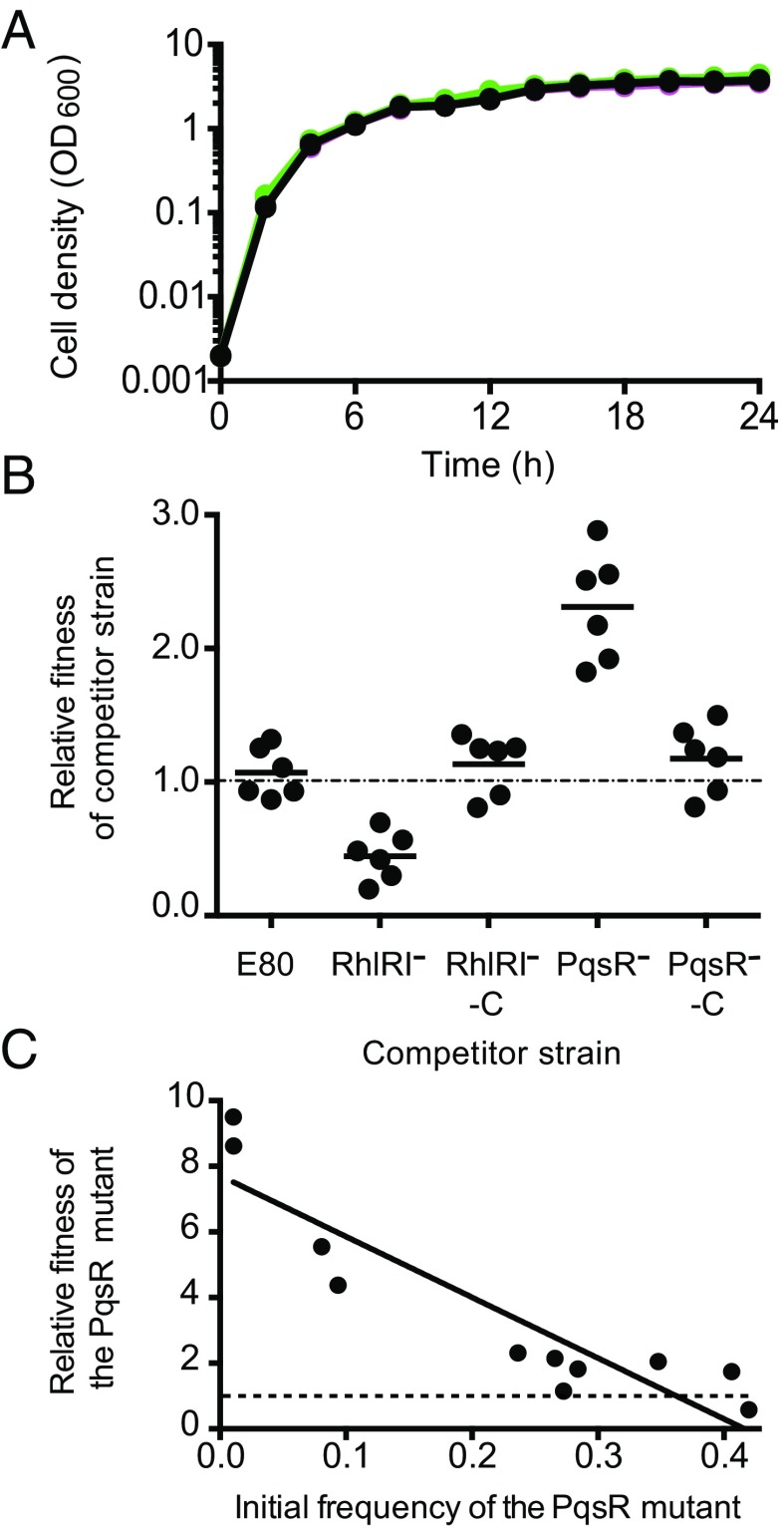
Neither the RhlR nor the PqsR mutants were able to grow in caseinate broth; however, the PqsR mutants showed a small amount of caseinate degradation on agar plates (Fig. 3), whereas the RhlRI mutant did not (Fig. 1D). Thus, it would seem logical that RhlR mutants should be more efficient invaders in E80 social-evolution experiments than PqsR mutants, but surprisingly it is PQS mutants that emerge during growth of strain E80 in caseinate broth. We hypothesized that RhlRI mutants must have a fitness defect in competition with wild type or a PQS synthesis mutant. As an initial assessment of fitness, we monitored growth of E80, an E80 PqsR mutant, and the E80 RhlRI mutant individually in LB-Mops broth. The RhlRI mutant, PQS mutant, and the wild-type growth curves were indistinguishable (Fig. 5A). To assess fitness more directly, we performed competition experiments in LB-Mops broth (Fig. 5B) and casein broth (SI Appendix, Fig. S3). In competition with the E80 wild type, the RhlRI mutant was disadvantaged, whereas the PQS mutant had a strong advantage. These data are consistent with our finding that PQS mutants, and not RhlR mutants, emerge during continuous growth on casein.
Fig. 5.
Growth and competition of the PqsR mutant, the RhlRI mutant, and the parent strain E80. (A) Growth of E80 (black), the RhlRI− mutant (magenta), and the PqsR− mutant (green) in LB-Mops broth. Cell density was measured as OD600, and data are the means of three biological replicates ± SD. (B) Competition experiments showing the competitive indexes of competitor strains against the parent strain (E80 or E80-Gm) after 24 h in LB-Mops broth. Each symbol represents the outcome of an individual experiment; the solid lines represent means for each group. The starting percentage of the competitors was 10%. The RhlRI mutant was less fit than the parent, and the PqsR− mutant was more fit than the parent. (C) The relative fitness of a PqsR mutant exhibits a negative frequency dependence. Data are from 24-h casein broth cultures. The outcomes above the dashed line indicate the competitor had a fitness advantage and below indicate the parent E80 had a fitness advantage.
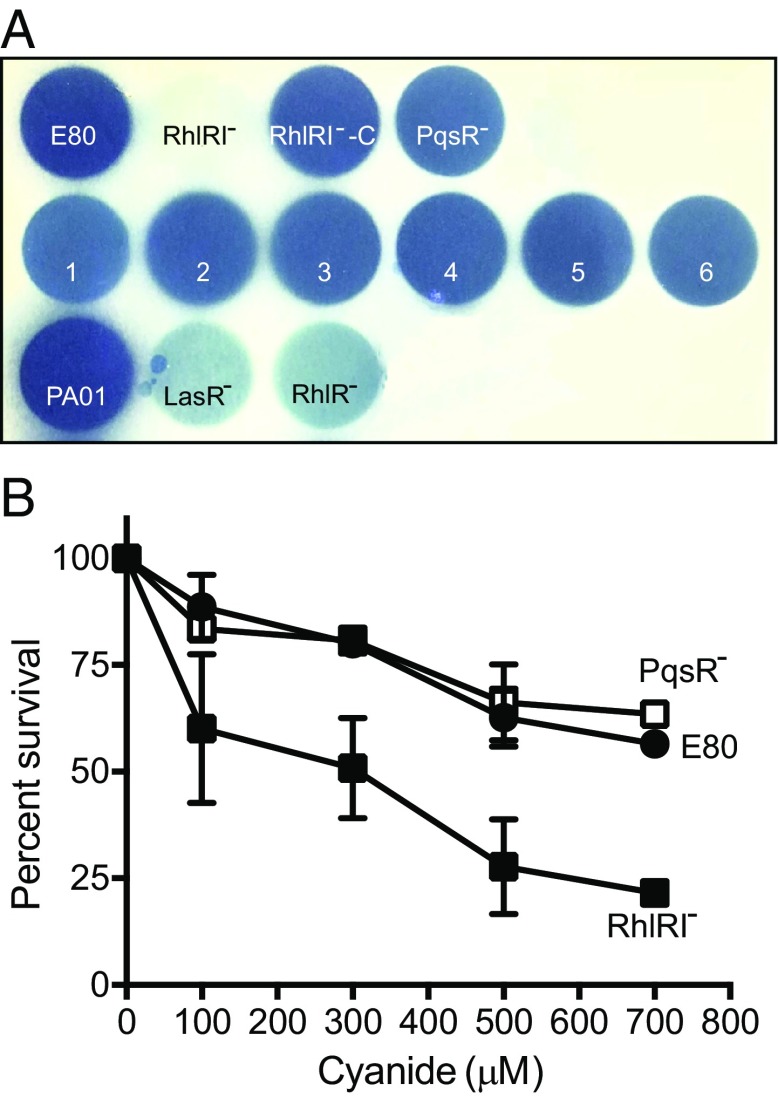
We suspected that RhlRI-dependent cyanide production and cyanide insensitivity might explain the fitness defect of the RhlRI mutant by analogy to the cyanide policing of P. aeruginosa PAO1 QS mutants (15). Thus, we monitored cyanide production by E80 and E80 RhlRI and PQS mutants (Fig. 6A). E80 showed strong cyanide production, the RhlRI mutant did not produce detectable levels of cyanide, but both E80 and the E80 PqsR mutants (including freeloaders 1–4) produced cyanide (Fig. 6A). We showed the RhlRI mutant was more sensitive to cyanide than the E80 parent and PqsR-mutant strains (Fig. 6B). These data suggest the fitness defect of the RhlRI mutant is mediated, at least in part, through cyanide policing.
Fig. 6.
Cyanide production by and sensitivity of a variety of P. aeruginosa strains and mutants. (A) Cyanide was monitored as described in Materials and Methods. The blue coloration of the filter paper reflects cyanide production. The top row is the parent strain E80 and various E80 mutants. The middle row is protease-deficient mutants 1–6, and the bottom row is strain PAO1, a PAO1 LasR mutant, and a PAO1 RhlR mutant shown for comparison. (B) Cyanide-killing curves for E80, the PqsR mutant, and the RhlRI mutant. Results are normalized to the number of cfu without cyanide. Data are the means of three replicates ± SD.
The PqsR Mutant Can Cheat During Growth with E80 on Casein.
P. aeruginosa PAO1 LasR mutants have a frequency-dependent fitness advantage over their parent, with relative fitness decreasing as the mutants become more common. This behavior of the mutants fits the definition of a social cheat (13). To test whether the PqsR mutant is a social cheat when grown together with its parent on casein, we measured its fitness over a range of different frequencies and observed a negative frequency dependence (Fig. 5C), as expected of a cheat. We assume that in the social-evolution experiments (Fig. 2) the protease-deficient mutants are also cheats.
Discussion
P. aeruginosa is an opportunistic pathogen that causes acute or chronic infections (21, 22). Early work showing AHL QS mutants were attenuated for virulence in a variety of animal models (16, 17) led to efforts to target the master QS transcription factor LasR for therapeutic development, particularly for chronic infections (23, 24). Subsequently, it became clear that LasR mutants are common in a variety of chronic infections (18, 25). CF lungs have been the most intensely studied chronic infection with respect to LasR (19). The reason for LasR-mutant emergence in these infections remains a topic of some controversy. There is evidence that such mutants gain an advantage over the wild type for a nutrient available in the infected lungs; they could be social cheats, or perhaps QS is needed only to initiate an infection (19, 26). Regardless, recent work has shown that many LasR mutants isolated from the lungs of children with CF have an active RhlRI system (19), and RhlR mutants from CF patients are not common and occur at very late stages of infection (20). LasR mutants in CF can evolve a RhlR-dependent QS system, independent of the LasR system. Some genes controlled by RhlR must be important in the context of CF infections, and for some reason, RhlR-mutant cheats do not have a fitness advantage in CF lung infections. Obviously, LasR is not an ideal therapeutic target but RhlR might be. In fact, small-molecule inhibitors of RhlR have already been discovered (27, 28). Therefore, we initiated our studies on a LasR-mutant CF isolate to better understand the biology involved in its LasR-independent function.
The CF clinical isolate E80 was capable of growth on casein, and the growth was dependent on RhlRI. The promoter of lasB, which codes for elastase, possesses both a LasR- and a RhlR-binding site. LasR is the dominant activator of lasB in the well-studied P. aeruginosa PAO1. We examined the sequence of the lasB promoter region in CF clinical isolate E80, and there were no obvious differences from the PAO1 lasB promoter that might explain RhlR-dependent growth of strain E80. Another possibility is that there are sufficiently high levels of active RhlR in strain E80 to drive lasB expression independent of LasR.
It is of obvious interest to understand what might be the course of P. aeruginosa QS evolution in chronic CF lung infections. Because E80 showed growth in broth containing casein as the sole carbon and energy source, we were able to perform social-evolution experiments similar to those with strain PAO1 reported previously (12, 14, 15). Daily transfer of PAO1 in casein leads to the emergence of LasR-mutant cheats, which produce very little extracellular protease activity. We also observed emergence of strain E80 mutants that produced very little extracellular protease activity. These protease-deficient mutants rapidly increased in abundance, overtaking the cooperating protease producers, and a population crash ensued. In experiments with strain PAO1, cheats emerge, but they are sufficiently restrained so that crashes are not observed (12, 14, 15). We believe that the instability of E80 growth in our social-evolution experiments may result from a relatively inefficient growth on casein compared with wild-type PAO1 such that only low abundances of cheats can be tolerated, a hypothesis supported by the negative frequency dependence of a PqsR mutant during growth with its parent E80 on casein (Fig. 5C). Furthermore, work with PAO1 revealed that the wild type policed cheats. The PAO1 wild type produced cyanide and was less sensitive to this toxin than the LasR-mutant cheats (15), and we showed different levels of cyanide production and sensitivity between the E80 wild type and the RhlRI mutant (Fig. 6).
To our surprise, the protease-deficient mutants from our social-evolution experiments were not RhlR QS mutants. Rather, the six mutants we studied were deficient in production of the non-AHL signal PQS and the related 2-alkylquinolone HHQ (Fig. 4). Four of the six cheats had mutations in the transcription activator gene pqsR, which is required for PQS production. We were unable to identify the mutations in the other two mutants that resulted in a PQS-negative phenotype. In this regard, a recent publication on the pqsA–E operon and its relationship to RhlR attracted our interest. Whereas pqsA, -B, -C, and -D are required for PQS synthesis, pqsE is not (6, 29, 30). Apparently, pqsE codes for an enzyme that catalyzes synthesis of an alternate ligand for RhlR (31). Among the RhlR-activated genes in P. aeruginosa, some are activated by C4-HSL–bound RhlR, some by RhlR bound to the alternate and so-far unidentified ligand, and some are activated by RhlR bound to either C4-HSL or the alternate ligand. It may be that the PQS-negative phenotype is a surrogate marker for the lack of the alternate ligand in the protease-deficient mutants.
To address the question of why we did not identify RhlR-mutant cheats of our clinical isolate and to try to begin to understand why RhlR mutants are rarely observed among P. aeruginosa isolates from infected lungs of CF patients, we constructed an E80 RhlRI mutant. This mutant was outcompeted by the wild-type E80 in coculture, while the E80 parent was outcompeted by the PqsR mutant (Fig. 5B and SI Appendix, Fig. S3). This can explain why RhlR mutants do not emerge in our social-evolution experiments, and it provides a plausible explanation for why they are not often isolated from infected lungs of CF patients. We show that the wild type and the PQS mutant produce cyanide and the RhlRI mutant is particularly sensitive to this poison (Fig. 6). Of course, we have so far studied just one single CF isolate, and there are likely other courses of evolution among other isolates. Interestingly, there is precedence for LasR−PQS− isolates from CF patients (19, 32); however, we do not yet know what AHL QS-dependent functions might be involved in maintaining chronic P. aeruginosa infections.
In trying to incorporate the current state of knowledge about QS evolution during the course of a chronic CF lung infection, we propose the following hypothesis: initial colonization of the CF lung is by P. aeruginosa with a functional LasI-LasR circuit. Over time, LasR mutants emerge for reasons that are not fully understood. They are in mixed populations with LasR–wild-type bacteria, and thus there is ample local C4-HSL. As the mutants increase in abundance, there is an increasing impact on colonization, and variants like E80, which override the RhlRI dependence on LasR, emerge. These variants produce an as yet unknown factor or factors involved in colonization and dependent on a functional RhlRI system. That the variants have a fitness advantage when growing with RhlRI mutants provides an explanation for why RhlR mutants are rarely isolated from the lungs of CF patients. There remain many unanswered questions. Obviously, many aspects of this hypothetical course of P. aeruginosa evolution rely on our work with a single isolate. We would like to know what are the RhlR-dependent factors we hypothesize to be of importance in chronic infections. The hypothesis predicts that unlike P. aeruginosa PAO1 LasR mutants, LasR-null CF isolates with functional RhlRI QS should be virulent, whereas RhlR mutants of these isolates will be attenuated for virulence. If this hypothesis stands up to further testing, then perhaps RhlR might be a target for development of antipseudomonal therapeutics to treat chronic infections.
During the final stages of preparation of this article, we (33) and another group (34) describe the emergence of RhlRI-dependent growth of P. aeruginosa PAO1 mutants on casein. In the report by Oshri et al. (34), the mutants with a phenotype similar to ours grew out of inoculated casein broth after over a month of incubation. Our group found the mutants emerged after only a few days when strain PAO1 was used to inoculate casein broth with added C4-HSL or an added C4-HSL–producing variant (33). We imagine the situation in CF lung infections might be like the latter case where wild-type P. aeruginosa could be feeding LasR-mutant cells C4-HSL. Both of these reports tracked the variant phenotype to null mutations in mexT, which codes for a transcription repressor of the MexEF-OprN efflux pump genes and other genes.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions.
All strains and mutants used in this study are described in SI Appendix, Table S1. Generally, bacteria were grown in either LB-Mops broth (35), low-salt LB broth, or minimal broth plus 1% sodium caseinate (casein broth) (14). For plating, we used LB agar, casein agar (casein broth with 1.5% agar), Vogel Bonner minimal agar, or skim milk agar (14). For cyanide experiments, we used 2% peptone agar plates (36). Gentamicin was used for mutant selection at 20 μg/mL. Bacteria were grown in 3-mL volumes with shaking in 14-mm Falcon tubes at 37 °C, unless otherwise specified.
Strain Construction.
To complement mutations, we integrated wild-type copies of genes carried on mini-Tn7 vectors into the att site as described elsewhere (37). We deleted genes by using the suicide vector pEXG2 as described previously (38). For all mutants, with the exception of the RhlRI mutant, we deleted only the ORF. The RhlRI mutant had a rhlR ORF deleted and the deletion extending into the rhlI promoter (SI Appendix, Fig. S2). Plasmids and primers used are described in SI Appendix, Table S2. We introduced plasmids to P. aeruginosa E80 by conjugation as described for P. aeruginosa PAO1 (37, 38) with the following modifications: strain E80 cells were grown in low-salt LB broth. Cells were harvested by centrifugation when the cultures reached an OD at 600 nm (OD600) of about 0.8. Cells were washed twice with fresh low-salt LB broth. Conjugal mating mixtures were incubated at 30 °C for at least for 20 h.
QS Signal Measurements.
To measure C4-HSL, bacteria were grown in LB-Mops broth to an OD (OD600) of 3.5, after which, the cultures were extracted with acidified ethyl acetate. The ethyl acetate-extracted C4-HSL was measured by using bioassays as described previously (17). Our positive control was an ethyl acetate extract of a P. aeruginosa PAO1 culture, and our negative control was an ethyl acetate extract of a P. aeruginosa PAO1 lasI, rhlI double-deletion mutant. We used a previously described method to measure HHQ and PQS (6).
Assessment of Extracellular Protease Production.
Production of extracellular protease was assessed by spotting bacteria grown in LB-Mops broth on casein agar plates and monitoring the size of the proteolytic zone forming around the bacterial colony after 48 h at 37 °C. Even protease-negative bacteria formed colonies on this agar, presumably by using LB-Mops carried with the inoculum and/or the low levels of peptides and amino acids present in the sodium caseinate.
Pyocyanin and Cyanide Assays.
We used a previously described method to measure pyocyanin levels in P. aeruginosa cultures (39). We used a cyanogenic test paper method described elsewhere (36) to detect cyanide. Briefly, cells grown overnight in LB-Mops broth were used to spot-inoculate peptone agar plates, and after 6–12 h at 37 °C, the plates were overlaid with test paper and incubated an additional 18 h at 37 °C. The paper turns blue when exposed to cyanide. To measure cyanide resistance, cells were diluted 100-fold from an overnight LB-Mops culture and grown to 37 °C until late-log phase (OD600, 0.8). Cells were then back-diluted to an OD600 of 0.02, treated for 2 h with the indicated concentration of potassium cyanide, and then serially diluted and plated on LB agar to determine the colony-forming units (cfu). Potassium cyanide was prepared in 35 mM potassium hydroxide.
Social Evolution Experiments.
The evolution experiments were in casein broth. Initial inocula were 50 μL of an overnight LB-Mops broth culture. Two to three days after the initial inoculation, when growth was evident, we transferred 50 μL to fresh casein broth (3 mL), after which, we transferred 50 μL to fresh casein broth daily. The relative abundance of protease-deficient cells was assessed by plating isolates on skim milk agar as described previously (14).
Growth Curves.
We used overnight cultures grown in LB-Mops as inoculum for growth-curve experiments. The starting density (OD600) was 0.02.
Competition Experiments.
For competition experiments, we used P. aeruginosa strains with or without a gentamicin-resistance marker, after first demonstrating that the gentamicin marker is neutral under these conditions (Fig. 5B). Fresh cultures (without antibiotics) of the P. aeruginosa E80 mutants or E80 wild type (OD600, 1) were used to inoculate fresh LB-Mops broth at a relative abundance of nine E80 wild-type cells: one competitor cell. The initial OD600 in competition experiments was 0.1. The relative abundances after 24 h of incubation were determined by plate counting with and without gentamicin. Competitive indexes were calculated as the ratio of mutant to wild type at 24 h divided by the ratio at the time of inoculation. For frequency-dependent competition experiments in casein broth (Fig. 5C), the E80 inoculum was prepared from a densely grown (48 h) casein culture, and the PqsR-mutant inoculum was prepared from log-phase LB-Mops culture (OD600, 0.8). The volume of E80 inoculum was held constant (50 μL in a 3-mL culture), while the volume of PqsR-mutant inoculum varied to create a range of starting ratios of E80 and the PqsR mutant (Fig. 5C). The relative abundance of PqsR-mutant cells was assessed at the time of inoculation and after 24 h by plating isolates on skim milk agar as described previously (14).
DNA Sequencing.
Whole-genome sequencing and assembly was done commercially by Total Genomics Solution Company, and PCR sequencing was used to confirm results of genomic sequencing. Genomic sequences of E80 and the E80 PQS mutants have been deposited in the GenBank database (accession nos. PRJNA475978 and PRJNA515291) (40, 41).
Supplementary Material
Acknowledgments
We thank Dr. Meizhen Wang for sharing a protocol for measurement of cyanide sensitivity ahead of publication. This research was supported by US Public Health Service Grants P30DK089507 and GM59026 and Innovative Team Program of Guangdong Province Grant 2013S034. QS research in E.D.’s laboratory is supported by Canadian Institutes of Health Research Operating Grant MOP-142466, and E.D. holds the Canada Research Chair in sociomicrobiology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. PRJNA475978 and PRJNA515291).
See Commentary on page 6525.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819801116/-/DCSupplemental.
References
- 1.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuster M, Greenberg EP. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 3.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson RM, Zimprich CA, Rust L. A second operator is involved in Pseudomonas aeruginosa elastase (lasB) activation. J Bacteriol. 1999;181:6264–6270. doi: 10.1128/jb.181.20.6264-6270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Déziel E, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao G, et al. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 8.Déziel E, et al. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: Multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 9.Diggle SP, et al. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 10.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 11.Park SJ, et al. Protease IV, a quorum sensing-dependent protease of Pseudomonas aeruginosa modulates insect innate immunity. Mol Microbiol. 2014;94:1298–1314. doi: 10.1111/mmi.12830. [DOI] [PubMed] [Google Scholar]
- 12.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 14.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci USA. 2015;112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang HB, et al. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68:4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman LR, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feltner JB, et al. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. MBio. 2016;7:e01513–e01516. doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjarnsholt T, et al. Scandinavian Cystic Fibrosis Study Consortium Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One. 2010;5:e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer-Hamblett N, et al. Pseudomonas aeruginosa phenotypes associated with eradication failure in children with cystic fibrosis. Clin Infect Dis. 2014;59:624–631. doi: 10.1093/cid/ciu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 24.Smith RS, Iglewski BH. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J Clin Invest. 2003;112:1460–1465. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Köhler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci USA. 2009;106:6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Argenio DA, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Loughlin CT, et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boursier ME, et al. Structure-function analyses of the N-butanoyl-L-homoserine lactone quorum-sensing signal define features critical to activity in RhlR. ACS Chem Biol. 2018;13:2655–2662. doi: 10.1021/acschembio.8b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekimpe V, Déziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 30.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee S, et al. The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2018;115:E9411–E9418. doi: 10.1073/pnas.1814023115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung JCS, et al. Genomic variation among contemporary Pseudomonas aeruginosa isolates from chronically infected cystic fibrosis patients. J Bacteriol. 2012;194:4857–4866. doi: 10.1128/JB.01050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostylev M, et al. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci USA. 2019;116:7027–7032. doi: 10.1073/pnas.1819796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshri RD, Zrihen KS, Shner I, Omer Bendori S, Eldar A. Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 2018;12:2458–2469. doi: 10.1038/s41396-018-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castric KF, Castric PA. Method for rapid detection of cyanogenic bacteria. Appl Environ Microbiol. 1983;45:701–702. doi: 10.1128/aem.45.2.701-702.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 38.Hmelo LR, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J, et al. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol. 2013;9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 40.Chen R, Greenberg EP, Schaefer AL. 2019 Pseudomonas aeruginosa E80 genome sequencing. NCBI Bioproject. Available at https://www.ncbi.nlm.nih.gov/bioproject/475978. Deposited February 22, 2019.
- 41.Chen R, Greenberg EP, Schaefer AL. 2019 Variant analysis of Pseudomonas aeruginosa E80-derived, protease-negative strains. NCBI Bioproject. Available at https://www.ncbi.nlm.nih.gov/bioproject/515291. Deposited January 15, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.