Significance
Based on the Sanger COSMIC (Catalogue of Somatic Mutations in Cancer) database, >30% of missense mutations of the PTEN gene result in disruption of arginine residues. We identified arginine methylation of PTEN using mass-spectrometry analysis and found that PRMT6 methylates PTEN on R159, resulting in suppression of the PI3K–AKT cascade. PTEN R159 is highly conserved, and R159 mutations have been found in cancer-susceptibility syndromes and in sporadic cancers. Disruption of PRMT6 or PTEN methylation leads to activation of PI3K–AKT signaling. Moreover, transcriptome analysis revealed that PTEN methylation influences mature mRNA transcripts through modulation of pre-mRNA alternative splicing, which may influence multiple cellular processes.
Keywords: PTEN, arginine methylation, PI3K–AKT cascade, pre-mRNA splicing, PRMT6
Abstract
Arginine methylation is a ubiquitous posttranslational modification that regulates critical cellular processes including signal transduction and pre-mRNA splicing. Here, we report that the tumor-suppressor PTEN is methylated by protein arginine methyltransferase 6 (PRMT6). Mass-spectrometry analysis reveals that PTEN is dimethylated at arginine 159 (R159). We found that PTEN is mutated at R159 in cancers, and the PTEN mutant R159K loses its capability to inhibit the PI3K–AKT cascade. Furthermore, PRMT6 is physically associated with PTEN, promotes asymmetrical dimethylation of PTEN, and regulates the PI3K–AKT cascade through PTEN R159 methylation. In addition, using transcriptome analyses, we found that PTEN R159 methylation is involved in modulation of pre-mRNA alternative splicing. Our results demonstrate that PTEN is functionally regulated by arginine methylation. We propose that PTEN arginine methylation modulates pre-mRNA alternative splicing and influences diverse physiologic processes.
In human cancers, the PTEN tumor-suppressor gene is frequently mutated (1, 2). PTEN germline mutations result in inherited cancer susceptibility syndromes, including Cowden syndrome and Bannayan–Riley–Ruvalcaba syndrome, that are collectively referred to as PTEN hamartoma tumor syndrome (3–5). Although PTEN is primarily considered to be a tumor suppressor, it is well established that PTEN loss also leads to pathologic phenotypes, including developmental defects (6, 7), neural dysfunction (8–10), heart disease (11, 12), dysregulation of the immune system (13–17), and metabolic disorders (18–20). PTEN dephosphorylates phosphatidylinositol-3,4,5-trisphosphate (PIP3) and thus serves as a pivotal cellular antagonist of the PI3K–AKT pathway (21–23). PTEN also functions in multiple cellular processes independent of PI3K–AKT signaling to maintain physiologic homeostasis (24). Posttranslational modifications are critical for regulation of PTEN function. PTEN protein is modified by phosphorylation (25–30), oxidation (31–33), acetylation (34–36), ubiquitination (37–39), SUMOylation (40, 41), and PARylation (42), and these modifications regulate PTEN–AKT signaling or AKT-independent PTEN functions. However, arginine methylation of PTEN has not been reported.
Protein arginine methylation is a common posttranslational modification involved in cellular signaling transduction, mRNA splicing, cell-fate decision, DNA replication, DNA-repair epigenetic regulation, and gene transcription (43–45). Arginine methylation that involves the addition of methyl groups to nitrogen atoms is a modification as common as phosphorylation and ubiquitination. Three forms of methylated arginine have been identified in eukaryotes, based on whether one or both nitrogens in arginine are methylated and whether each is methylated once or one twice and the other not at all. Thus, there are three forms of arginine methylation, including monomethylated arginine (MMA), symmetrical dimethylated arginine (sDMA), and asymmetrical dimethylated arginine (aDMA) (44). Protein arginine methyltransferases (PRMTs) are divided into three types based on catalytic activity. Type I consists of PRMT1, PRMT2, PRMT3, PRMT4/CARM1, PRTM6, and PRMT8 that catalyze MMA and aDMA. Type II includes PRMT5 and PRMT9 that catalyze MMA and sDMA (44, 46). PRMT7 is categorized as type III and functions only as a MMA methyltransferase for histones (47). PRMT6 is a type I PRMT primarily recognized as an oncogene that inhibits p53, p21, and p16 functions involved in regulation of cell-cycle arrest and apoptosis (48–52). Previous observations suggest that PRMT6 is overexpressed in breast (53), prostate (54), bladder, and lung cancer (55). PRMT6 depletion in PTEN-null prostate cancer cell lines represses the PI3K–AKT cascade (54). In the present study, we found that PRMT6 promotes asymmetrical dimethylation at PTEN R159, which is essential for activation of PTEN to oppose the PI3K–AKT cascade. In cancers, higher levels of PRMT6 predict better prognoses. Based on these findings, we propose that PRMT6 is an important cellular regulator of the PTEN–AKT axis and likely functions as a tumor-suppressor gene under certain conditions.
Recent evidence suggests that PI3K–AKT signaling is involved in modulation of pre-mRNA alternative splicing (56). Moreover, PTEN interacts with hnRNPK to regulate G6PD pre-mRNA splicing, which is independent of the PI3K–AKT signaling (57). In view of these observations, we employed transcriptome analysis to study the relationship of PTEN methylation and pre-mRNA splicing. Our data suggest that PTEN modulates pre-mRNA splicing in a methylation-related manner as well as in a methylation-unrelated manner, providing insight into the multifaceted roles of PTEN in diverse biologic processes.
Results
Dimethylation of PTEN R159 Within a Non-RG-Rich Motif Is Critical for Suppression of the PI3K–AKT Cascade.
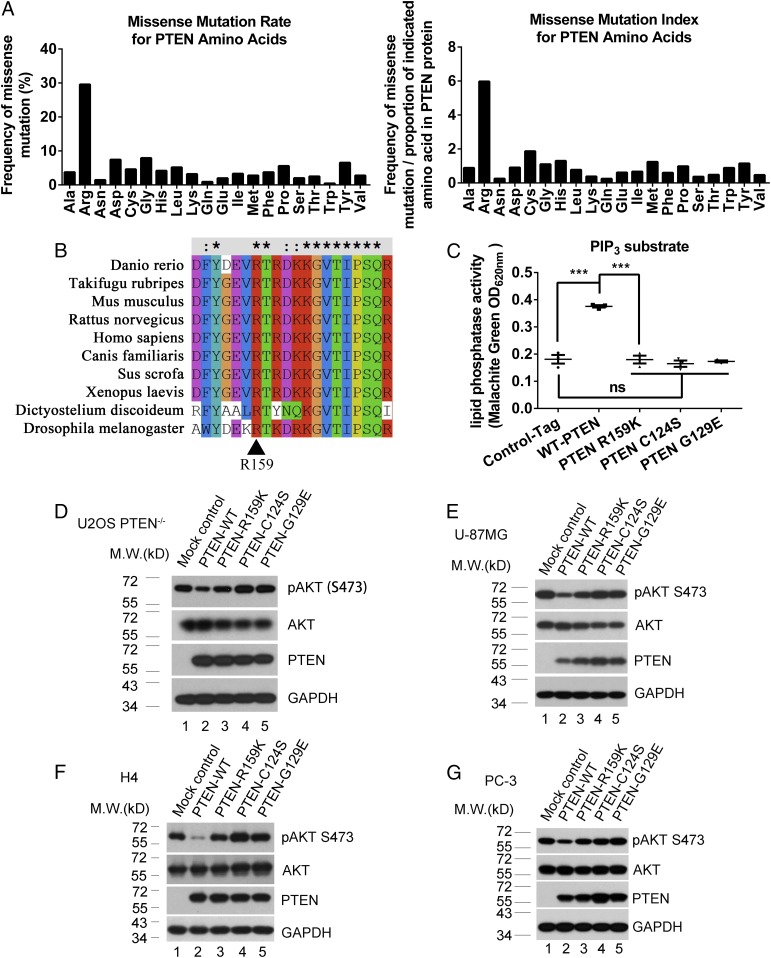
The Sanger COSMIC (Catalogue of Somatic Mutations in Cancer) database includes PTEN gene-sequencing data derived from >65,000 sporadic human cancer samples. In surveying somatic missense mutations of the PTEN gene, we found that arginine residues are the most frequently mutated PTEN residues (Fig. 1A; analysis of raw data from Sanger COSMIC database), suggesting that modification of arginine residues is particularly important for PTEN function. PTEN does not contain the canonical RG-rich motif that was previously considered to be a characteristic feature of protein arginine methylation (58). However, a recent proteomic study has shown that arginine methylation is as common as phosphorylation, and PRMTs target RG-rich and non-RG-rich motifs with similar frequency (59). This prompted us to determine whether PTEN arginine residues are modified by methylation. Ectopically expressed FLAG-tag-PTEN protein was purified from an extract of HEK293FT cells by using anti-FLAG affinity gel. Purified PTEN protein was subjected to mass-spectrometry analysis, which showed dimethylation of PTEN residue R159 (SI Appendix, Fig. S1A). PTEN R159 shows a high degree of evolutionary conservation and is found in invertebrates and vertebrates (Fig. 1B). Consistent with this degree of evolutionary conservation, PTEN germline mutations R159T and R159G have been found in patients with PTEN hamartoma tumor syndromes (60), suggesting that the R159 residue is important for PTEN anticancer activity. Arginine-to-lysine mutation is often used as a nonmethylated mimic to analyze the function of protein arginine methylation (46, 61). The methylation-defective PTEN R159K mutant has been found in somatic human cancers, including glioma, melanoma, and thyroid cancer (SI Appendix, Fig. S1B; from the COSMIC database). This supports use of the R159K mutant as a model for study of the role of PTEN R159 methylation in tumor suppression. R159 is located in the N-terminal phosphatase domain of PTEN (SI Appendix, Fig. S1C). This raises the question as to whether the methylation-defective PTEN R159K mutant loses its phosphatase activity. PTEN mutations C124S and G129E are the most famous pathogenic mutations that cause spontaneous tumorigenesis in mice (62, 63). The PTEN G129E mutant only loses lipid phosphatase activity (3), while the PTEN-C124S mutant loses both protein and lipid phosphatase activity (64). We thus used these PTEN mutants as controls in some of our analyses. PTEN−/− U2OS cells were infected with lentivirus expressing control S-tag, wild-type PTEN, PTEN R159K, PTEN C124S, or PTEN G129E and were then lysed for purification of wild-type or mutant PTEN proteins. In vitro evaluation of phosphatase activity showed that PTEN R159K failed to dephosphorylate PIP3, which is comparable to the PTEN C124S and G129E mutants (Fig. 1C). PTEN R159K protein purified from SF9 insect cells failed to dephosphorylate PIP3 (SI Appendix, Fig. S1D) or phospho-GluTyr (4:1) peptides (SI Appendix, Fig. S1E), indicating that both its lipid- and protein-phosphatase activities were lost. In PTEN−/− U2OS cells and PTEN-null cancer cells, including U-87 MG, H4, and PC-3, evaluation of phospho-AKT (Ser-473) levels showed that PTEN R159K, C124S, and G129E mutants failed to efficiently repress phospho-AKT (Ser-473), compared with wild-type PTEN (Fig. 1 D–G, lanes 3–5 vs. lane 2). These results argue that methylation of PTEN R159 is critical for PTEN phosphatase activity and is thus critical for opposing the PI3K–AKT cascade.
Fig. 1.
Dimethylation of PTEN R159 is required for repression of PI3K–AKT signaling. (A) Raw data for missense mutations of the PTEN gene in sporadic cancer samples were derived from the COSMIC database. There were a total of 2,271 clinical samples with missense mutations of the PTEN gene (updated on August 22, 2018), and the mutation rate of each type of amino acid was analyzed (A, Left). (A, Right) The analysis was also normalized to the amino acid constitution of PTEN protein. (B) Evolutionary conservation of the PTEN R159 residue was analyzed with ClustalX. Sequence data of PTEN from different species were derived from the UniProt database. (C) S-tagged PTEN proteins were precipitated from cell lysates with S-protein beads and subjected to PTEN enzyme reactions (PIP3 substrate). OD620nm reflects the quantity of PO4 released in the reaction. Data were analyzed by using the paired two-tailed Student’s t test. Error bars indicate SEM (n = 3). ns, no significant difference; ***P < 0.001. (D–G) pAKT (S473) levels in PTEN−/− U2OS (D), PTEN-null U-87 MG (E), H4 (F), and PC-3 (G) cells expressing S-tag control, wild-type PTEN (PTEN-WT), PTEN R159K, PTEN C124S, or PTEN G129E were analyzed by using immunoblotting. Levels of pAKT (S473) and total AKT expression were analyzed. GAPDH served as a loading control.
PRMT6 Suppresses PI3K–AKT Signaling.
Previous studies have demonstrated the impact of PRMTs on the PI3K–AKT pathway. PRMT1 methylates ERα on R260, and methylation of ERα promotes AKT activation, resulting in cell proliferation and migration in a subset of breast cancers (65). It has been reported that PRMT5 represses PTEN expression at the transcription level and thus activates the PI3K–AKT pathway (66). Conversely, PRMT1 also methylates downstream targets of AKT, such as FOXO1 (67) and BAD (68), and therefore represses AKT signaling.
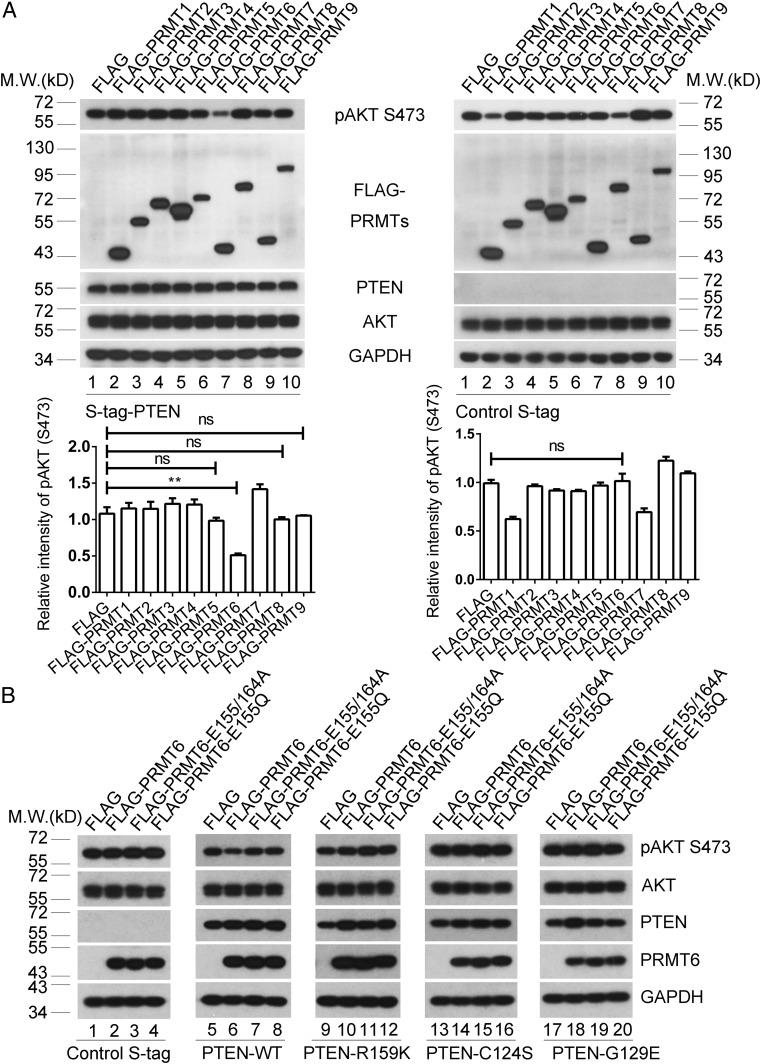
Because PTEN R159 methylation is critical for repression of the PI3K–AKT cascade, we evaluated the individual effect of each PRMT in this cascade to determine which PRMT acts on PTEN R159. In PTEN−/− U2OS cells, PRMT6, but not other PRMT family members, strongly repressed the level of phospho-AKT (S473) in the presence of ectopically expressed S-tagged PTEN (Fig. 2 A, Left, lane 7). For controls, each PRMT protein was individually overexpressed in PTEN−/− U2OS cells expressing S-tag, and phospho-AKT (S473) levels were determined. PRMT6 did not influence the level of phospho-AKT (S473) on this PTEN-deficient background (Fig. 2 A, Right, lane 7), indicating that regulation of the PI3K–AKT pathway by PRMT6 is PTEN-dependent. In PTEN−/− U2OS cells, PRMT6 repressed the level of phospho-AKT (S473) in the presence of ectopically expressed wild-type PTEN (Fig. 2 B, lane 6 vs. lane 5), but not in the presence of control-tag, PTEN R159K, C124S, or G129E mutants (Fig. 2 B, lane 2 vs. lane 1; lane 10 vs. lane 9; lane 14 vs. lane 13; and lane 18 vs. lane 17). However, the methyltransferase dead mutant PRMT6 E155/164A (52) and the cancer-derived mutant PRMT6 E155Q [bladder urothelial carcinoma (The Cancer Genome Atlas; TCGA) from the International Cancer Genome Consortium] both lost the capacity to suppress PI3K–AKT signaling (Fig. 2 B, lanes 7 and 8 vs. lane 6), indicating that PRMT6 function in the PTEN–AKT axis is dependent on its methyltransferase activity. The CRISPR-Cas9 system (69) was utilized to induce PRMT6 deficiency in PTEN-proficient U2OS and SF763 cells (SI Appendix, Figs. S2 A–C). PRMT6 deficiency resulted in activation of PI3K–AKT signaling (SI Appendix, Fig. S2D, lane 2 vs. lane 1; and lane 4 vs. lane 3). These data suggest that PRMT6, by virtue of its methyltransferase activity, acts as a regulator of PTEN–AKT signaling.
Fig. 2.
PRMT6 is involved in repression of the PI3K–AKT cascades. (A) Exogenous methyltransferases PRMT1–9 were individually introduced into PTEN−/− U2OS cells reexpressing wild-type PTEN (Left) or control S-tag (Right). pAKT (S473) and total AKT levels were analyzed. GAPDH served as a loading control. pAKT (S473) quantification relative to total AKT is shown below the blots. Data were analyzed with the unpaired two-tailed Student’s t test. Error bars indicate SEM (n = 3). ns, no significant difference. **P < 0.01. (B) PTEN−/− U2OS cells with reexpression of control S-tag, wild-type PTEN (PTEN-WT), PTEN R159K, PTEN C124S, or PTEN G129E were introduced with Flag-tag control, wild-type PRMT6, methyltransferase-dead PRMT6 E155/164A mutant, or the cancer-derived PRMT6 E155Q mutant. pAKT (S473) and total AKT levels were analyzed. GAPDH served as a loading control.
PRMT6 Promotes PTEN Arginine Methylation.
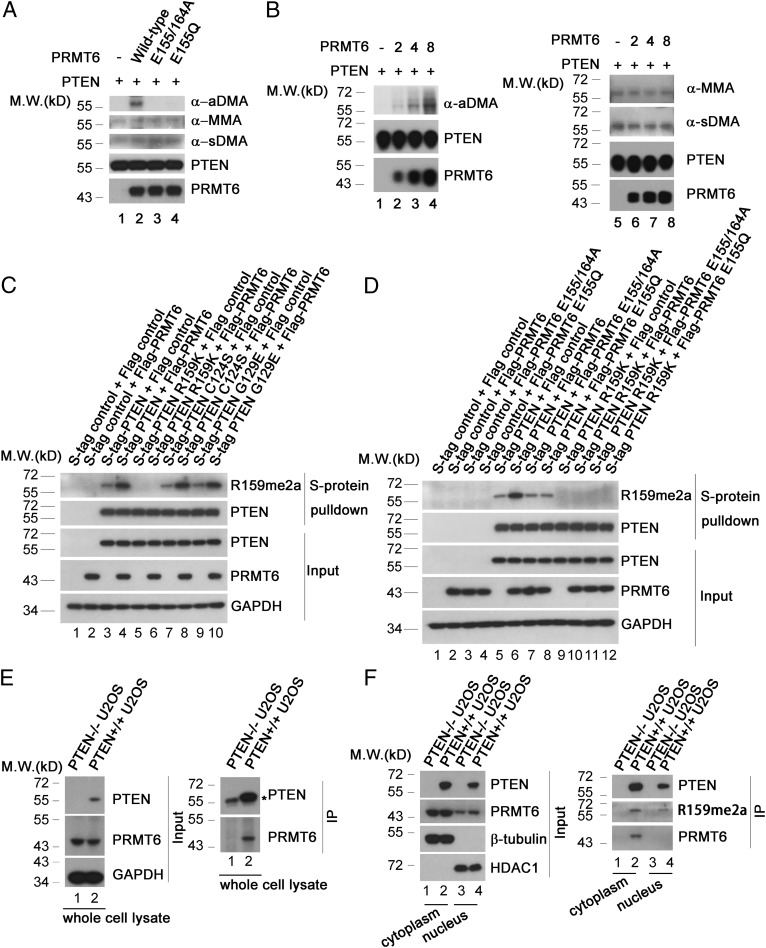
PRMT6 did not reduce the level of phospho-AKT (S473) in the presence of ectopic expression of the methylation-defective PTEN R159K mutant (Fig. 2 B, lane 10 vs. lane 9), raising the possibility that PRMT6 acts as an antagonist of the PI3K–AKT pathway through methylation of PTEN R159. PRMT6 promotes monomethylation and asymmetrical dimethylation (70), and mass-spectrometry analysis revealed dimethylation of PTEN R159 (SI Appendix, Fig. S1A). This strongly suggests that PRMT6 specifically asymmetrically dimethylates PTEN R159 (designated PTEN R159me2a). To confirm the mechanism of this methylation, in vitro methylation assays were carried out. Wild-type PRMT6, but not the methyltransferase dead mutant PRMT6 E155/164A or the cancer-derived mutant PRMT6 E155Q, promoted asymmetrical dimethylation of PTEN (Fig. 3 A, lane 2 vs. lanes 1, 3, and 4). PRMT6 promoted PTEN asymmetrical dimethylation in a dose-dependent manner (Fig. 3 B, lanes 2–4 vs. lane 1), but did not influence symmetrical dimethylation or monomethylation (Fig. 3 A and B, lanes 6–8 vs. lane 5). We generated a polyclonal antibody specific for PTEN R159me2a and found that overexpression of PRMT6 increased the level of PTEN R159me2a in wild-type PTEN and the PTEN C124S and G129E mutants (Fig. 3 C, lane 4 vs. lane 3; lane 8 vs. lane 7; and lane 10 vs. lane 9; Fig. 3 D, lane 6 vs. lane 5). However, this modification was disrupted in the PTEN R159K mutant (Fig. 3 C, lanes 5 and 6; Fig. 3 D, lanes 9–12). In addition, PRMT6 mutants failed to increase levels of PTEN R159me2a (Fig. 3 D, lanes 7 and 8 vs. lane 6). Physical interaction of PTEN with PRMT6 was shown in PTEN-proficient U2OS cells by coimmunoprecipitation (Fig. 3 E, Right, lane 2 vs. lane 1). Consistent with the role of PRMT6 in suppression of PI3K–AKT cascade, PRMT6 physically interacted with PTEN in cytoplasm but not in nucleus (Fig. 3F, Right, lane 2 vs. lane 4). We therefore conclude that PRMT6 asymmetrically dimethylates PTEN.
Fig. 3.
PRMT6 methylates PTEN in vitro and in cells. (A) PRMT6 promotes PTEN dimethylation in vitro. Four micrograms of purified His-PTEN was incubated with 8 μg of purified His-tagged wild-type PRMT6, PRMT6 E155/164A, or PRMT6 E155Q mutant in 50 μL of HMT buffer for 2 h at 37 °C. Methylation of PTEN protein was analyzed by immunoblotting with antibodies including anti-MMA motif (α-MMA), anti-sDMA motif (α-sDMA), and anti-aDMA motif (α-aDMA). Loading of purified His-PTEN and -PRMT6 proteins is shown. (B) PRMT6 asymmetrically dimethylates PTEN in vitro in a dose-dependent manner. Two micrograms of purified His-PTEN was incubated with different amounts of purified His-PRMT6 (0, 2, 4, or 8 μg) in 25 μL of HMT buffer for 2 h at 37 °C. Methylation of PTEN protein was analyzed with immunoblotting with antibodies including anti-MMA motif (α-MMA), anti-sDMA motif (α-sDMA), and anti-aDMA motif (α-aDMA). Loading of purified His-PTEN and -PRMT6 proteins is shown. (C) S-tagged PTEN, PTEN R159K, PTEN C124S, or PTEN G129E was coexpressed with FLAG-tagged PRMT6 in PTEN−/− U2OS cells and purified with S-protein beads. Purified proteins were analyzed by Western blotting with PTEN R159me2a and PTEN antibodies. Levels of exogenous PRMT6 are shown, and GAPDH served as a loading control. (D) S-tagged PTEN or PTEN R159K was coexpressed with FLAG-tagged wild-type PRMT6 or PRMT6 mutants as indicated in PTEN−/− U2OS cells and purified with S-protein beads. Purified proteins were analyzed by Western blotting with PTEN R159me2a and PTEN antibodies. Levels of indicated exogenous PRMT6 proteins are shown, and GAPDH served as a loading control. (E) Lysates from PTEN+/+ and PTEN−/− U2OS cells were immunoprecipitated with a PTEN mouse monoclonal antibody (sc-7974) and subjected to Western blotting with a PRMT6 antibody. * indicates IgG heavy chain. In E, Left, the expression levels of PTEN and PRMT6 were analyzed, and GAPDH served as a loading control. (F) Cytoplasmic and nuclear extracts from PTEN+/+ and PTEN−/− U2OS cells were immunoprecipitated with anti-PTEN agarose resin. The immunoprecipitates were eluted by glycine–HCl (pH 3.5), neutralized with Tris⋅HCl (pH 8.8), and subjected to Western blotting using anti-PTEN, anti-PTEN R159me2a, and anti-PRMT6 antibodies. In F, Left, the expression levels of PTEN and PRMT6 were analyzed. β-tubulin served as a loading control in the cytoplasmic extracts, and HDAC1 served as a loading control in the nuclear extracts.
Loss of PTEN R159 Methylation Impairs PTEN-Mediated Tumor Suppression.
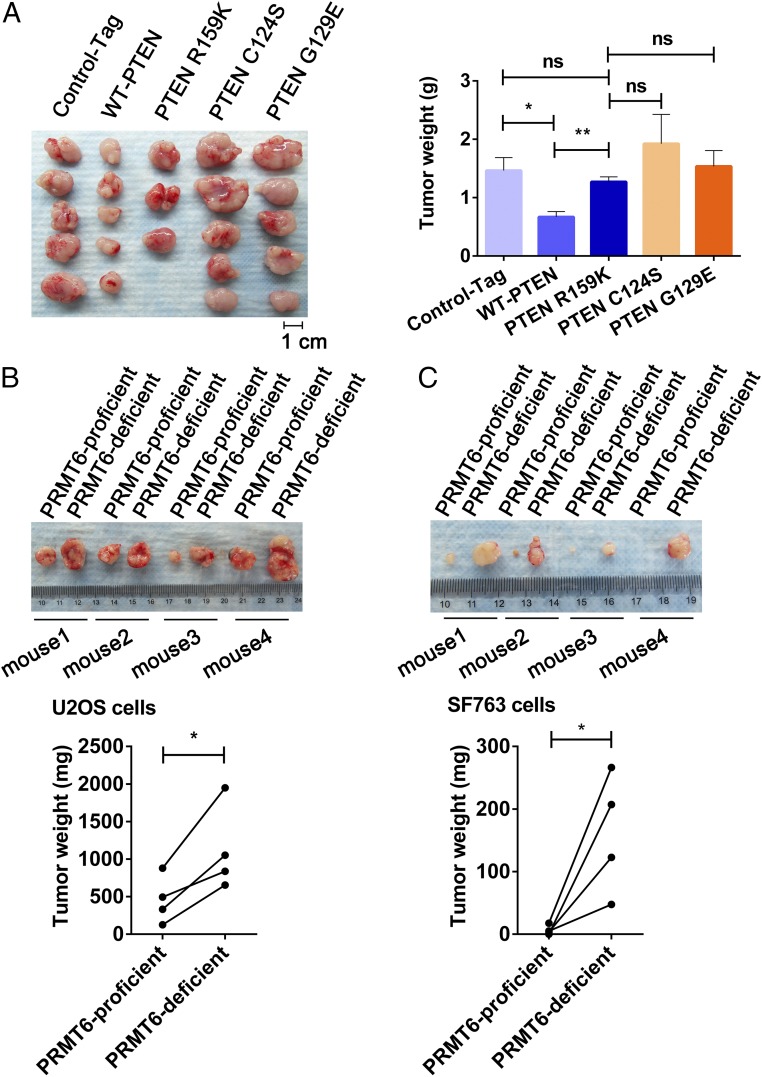
To better demonstrate the biologic importance of PTEN R159me2a as an antagonist of tumorigenesis, PTEN-null U87-MG cells with reexpression of S-tag control, wild-type PTEN, PTEN R159K, PTEN C124S, or PTEN G129E were assayed by using mouse xenograft models. Xenograft-derived tumors were weighed 15 d after s.c. injection. Cells expressing the PTEN R159K mutant yielded significantly larger tumors than those expressing wild-type PTEN, with a growth enhancement comparable to those expressing control tag, PTEN C124S, or G129E mutants (Fig. 4A). Consistent with the promotion of PTEN R159me2 by PRMT6, PRMT6-deficient U2OS and SF763 cells gave rise to larger tumors in xenograft models (Fig. 4 B and C). These findings indicate that PTEN R159me2a is required for suppression of tumorigenesis.
Fig. 4.
PTEN R159 methylation is important for PTEN tumor-suppressive function. (A) PTEN-null U-87 MG cells expressing S-tag control (Control-Tag), wild-type PTEN (WT-PTEN), PTEN R159K, PTEN C124S, or PTEN G129E yielded tumors (Left). Two nude mice bearing tumors expressing PTEN R159K died significantly earlier than the other nude mice, and their tumors were excluded from statistical analysis. (A, Right) Tumors shown in A, Left were weighed and analyzed. Data were analyzed with the unpaired two-tailed Student’s t test. Error bars indicate SEM. *P < 0.05; **P < 0.01. ns, no significant difference. (B and C) PRMT6-proficient and -deficient U2OS (B) or SF763 (C) cells were injected into symmetrical sites on the back in the same nude mouse. Data were analyzed with the paired two-tailed Student’s t test. Error bars indicate SEM. *P < 0.05.
PTEN Modulates Methylation-Related and -Unrelated Pre-mRNA Splicing.
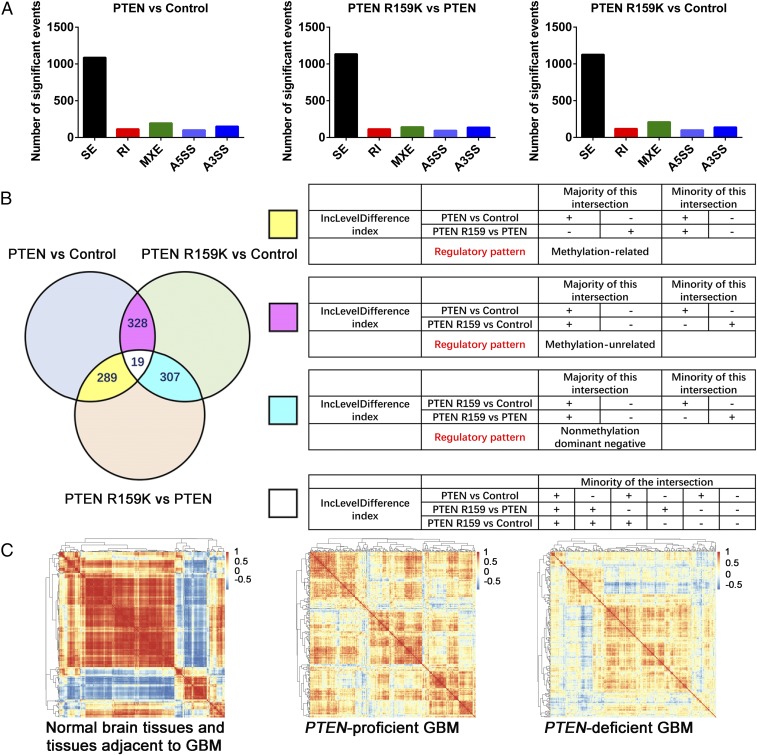
PTEN methylation was found in cytoplasm and nucleus (Fig. 3 F, Right, lanes 2 and 4), suggesting that this modification may be involved in both cytoplasmic and nuclear functions of PTEN. There is increasing evidence that PI3K–AKT signaling plays a critical role in the regulation of pre-mRNA splicing (56). In addition, nuclear PTEN modulates G6PD pre-mRNA splicing in an AKT-independent manner (57). In view of these observations, we hypothesized that PTEN R159 methylation is involved in modulation of pre-mRNA splicing. To test this hypothesis, we utilized deep RNA-sequencing (RNA-seq) to profile the alternative mRNA splicing events in PTEN-null H4 glioma cells expressing control tag, wild-type PTEN, or the methylation-defective PTEN R159K mutant, as this mutant is found most frequently in glioma. For replicate multivariate analysis of transcript splicing, we obtained RNA-seq data from three biological replicates in each group. Three pairwise comparisons (PTEN vs. control; PTEN R159K vs. PTEN; and PTEN R159K vs. control) were performed among these three groups, showing differentially expressed alternative splicing events (mainly skipped exons; Fig. 5A and Datasets S1–S3). We evaluated the regulatory pattern of these differentially expressed alternative splicing events by intersection analysis and found that PTEN modulates pre-mRNA splicing in multiple ways. These mainly include methylation-related, methylation-unrelated, and nonmethylation dominant negative manners (Fig. 5B). Functional annotation of the gene products affected by the PTEN methylation status indicates that PTEN methylation regulates important cellular processes such as DNA repair and cell-cycle regulation through modulation of pre-mRNA splicing (SI Appendix, Fig. S3 and Dataset S4). PTEN may also influence cardiac muscle development, neuron development, antigen presentation, metabolic homeostasis, signaling transduction, and cellular transportation in methylation-unrelated and nonmethylation dominant negative manners (SI Appendix, Figs. S4 and S5 and Datasets S5 and S6).
Fig. 5.
PTEN R159 methylation is involved in regulation of pre-mRNA splicing. (A) Quantification of differentially expressed alternative splicing events in three pairwise comparisons (PTEN vs. control, PTEN R159K vs. PTEN, and PTEN R159K vs. control). A5SS or A3SS, alternative 5′ or 3′ splice site; MXE, mutually exclusive exons; RI, retained introns; SE, skipped exons. Significant events are listed in Datasets S1–S3. (B) Intersections of alternative splicing events among three pairwise comparisons (Left) containing 18 combinatorial modes of change, as listed in Right. Alternative splicing events in 6 of the 18 combinatorial modes account for about 96% of all intersected events and can be categorized into three regulatory modes, including those which are regulated by PTEN in a methylation-related manner (majority in yellow), a methylation-unrelated manner (majority in pink), or a nonmethylation dominant negative manner (majority in cyan). The IncLevelDifference index is important for understanding pairwise comparison, intersection analysis, and the related datasets. When the expression level of the inclusive form of a splicing event in X group is higher or lower than in Y group, then the IncLevelDifference index is, respectively, >0 (+) or <0 (−) in pairwise comparison of X vs. Y. For example, the regulatory pattern of methylation-related includes two conditions: (i) The expression level of the inclusive form of a splicing event is higher in the PTEN group, compared with the control group or PTEN R159K group. In this condition, the splicing event does not show a significant difference in pairwise comparison of PTEN R159K vs. control, the IncLevelDifference index is “+” in pairwise comparison of PTEN vs. control, and is “−” in pairwise comparison of PTEN R159K vs. PTEN. (ii) The expression level of the inclusive form of a splicing event is lower in the PTEN group, compared with the control or PTEN R159K group. In this condition, the splicing event does not show a significant difference in pairwise comparison of PTEN R159K vs. control, the IncLevelDifference index is “−” in pairwise comparison of PTEN vs. control and is “+” in pairwise comparison of PTEN R159K vs. PTEN. (C) Heatmap showing pairwise gene coexpression matrices for 220 known or putative splicing factors in normal brain tissues and tissues adjacent to GBM (Left), PTEN-proficient GBM (Center), and PTEN-deficient GBM (Right).
We validated some interesting alternative spliced gene products that are regulated by PTEN in a methylation-related manner, such as alternatively spliced RYR3 and MDM4. RYR3 is an intracellular calcium release channel, located on the endo/sarcoplasmic reticuli, widely expressed in many tissues including brain (71), while MDM4 is a p53 regulator involved in suppression of cell-cycle arrest and apoptosis (72). The PTEN R159K mutant lost the capacity to regulate the alternatively spliced RYR3 and MDM4, compared with wild-type PTEN (SI Appendix, Fig. S6 A and B, lane 2 vs. lane 3), which is comparable to the control group and the catalytically dead mutants PTEN C124S and G129E (SI Appendix, Fig. S6 A and B, lane 2 vs. lane 1, 4, or 5). The use of PI3K inhibitor LY294002 mimicked regulation by wild-type PTEN (SI Appendix, Fig. S6 A and B, lane 2 vs. lanes 6–10), further indicating that the regulation of alternatively spliced RYR3 and MDM4 by PTEN relies on suppression of the PI3K–AKT cascade. Since the catalytically dead mutants PTEN C124S and G129E can be modified by R159me2a (Fig. 3 C, lanes 7–10), this regulation of the alternatively spliced RYR3 and MDM4 is related to but not dependent on PTEN methylation. Thus, PTEN methylation can modulate pre-mRNA splicing through suppression of PI3K–AKT signaling.
Splicing factors are coordinated to establish a series of complexes to regulate alternative splicing (73, 74). Through coexpression analyses with the TCGA dataset, we found that the expression levels of splicing factors are exquisitely coordinated in normal brain tissues and tissues adjacent to glioblastoma multiforme (GBM) (Fig. 5 C, Left). In PTEN-proficient GBM samples, this coordination is not as exacting as that in normal tissues and tissues adjacent to GBM (Fig. 5 C, Center vs. Left), but is nonetheless impressive compared with that in PTEN-deficient GBM samples (Fig. 5 C, Center vs. Right). Together with RNA-seq analyses, these observations indicate a significant role of PTEN in modulation of pre-mRNA splicing, which opens a window for understanding PTEN biology.
Discussion
Multiple types of PTEN posttranslational modification have been identified (24). PTEN arginine methylation can now be added to this list. Methylated arginine residues were previously considered to be located in RG-rich motifs (58), which are absent in PTEN protein. Recent mass-spectrometry analysis showed that 69% of methylation sites reside outside of RG-rich motifs (59). Our data reveal an important methylation site located in a non-RG-rich motif in PTEN. We demonstrate that PTEN is asymmetrically dimethylated on R159, which exhibits a high degree of evolutionary conservation. Moreover, the methylation-dead mutant PTEN R159K has been identified in multiple types of cancer (75–78). The R159K mutant loses both lipid- and protein-phosphatase activity, and its ability to oppose the PI3K–AKT cascade is thus abolished. This mutant consequently fails to suppress tumorigenesis, arguing that methylation at this site is required for the tumor-suppression function of PTEN.
Arginine methylation by PRMT6 plays multiple roles in cellular activities. It has been reported that PRMT6 methylates p21 at R156, facilitating cytoplasmic localization of p21 and dysregulation of p21 function (51). In addition, PRMT6 inactivates the p21 promoter independent of H3R2 methylation (H3R2me2a) (49, 53). Similarly, PRMT6 inactivates p16 at the transcriptional level (49) and methylates p16 protein to disrupt p16 function (50). Mouse embryonic fibroblasts derived from PRMT6 knockout mice exhibit p53 transactivation, with resultant growth defects and cellular senescence (48). However, analysis of TCGA datasets shows that high expression of PRMT6 is associated with improved survival in patients with breast, colorectal, renal, or ovarian cancer (79) (SI Appendix, Fig. S7; data from the Human Protein Atlas is available from www.proteinatlas.org/), consistent with the concept that PRMT6 functions as a tumor suppressor in these cancers. In the presence of PTEN, we found that PRMT6 critically represses the PI3K–AKT cascade, while other PRMTs are not involved in repression of this cascade, and that PRMT6 deficiency results in activation of the PI3K–AKT cascade, promoting tumorigenesis. This contrasts with, but does not contradict, previous observations that PRMT6 depletion in PTEN-null prostate cancer cell lines represses the PI3K–AKT cascade (54), providing insight into the mechanism by which PRMT6 represses tumorigenesis. PRMT6 conventional knockout mice do not show any particular characteristic phenotypic features (48). In the future, the role of PRMT6 in tumorigenesis and in other PTEN-related diseases can be more specifically evaluated by using PRMT6 knockout mice with additional genetic alterations or with conditional knockout and inducible knockout mouse models.
Pre-mRNA splicing is a complicated multistep cellular process that must be accurately regulated by an exquisite network to maintain physiologic homeostasis (80). Recent proteomic studies have shown that pre-mRNA splicing regulating factors are a major subset of proteins modified by arginine methylation (59, 81). Here, we show that PTEN R159 methylation is important for modulation of alternative mRNA splicing. The intersection analysis reveals that PTEN regulates pre-mRNA splicing in methylation-related, -unrelated, and nonmethylation dominant negative manners. In addition, the coexpression analysis unveils that the coordinated expression of splicing factors is influenced by PTEN status. These findings suggest an important role of PTEN in modulation of pre-mRNA splicing. Functional annotation of the regulated gene products indicates that PTEN exerts influence on multiple cellular and physiologic processes through modulation of pre-mRNA splicing, thus providing insight into the complexity of PTEN function.
Emerging evidence highlights the potential of targeting pre-mRNA splicing to provide therapeutic benefit in cancer (82). For example, several independent studies have shown that PRMT5, a master regulator of pre-mRNA splicing, is a promising therapeutic target for certain types of cancer (83–85). Our study suggests the possibility that targeting pre-mRNA splicing might be a therapeutic strategy for PTEN-deficient cancers. To this end, further mechanistic details of how PTEN modulates pre-mRNA splicing should be unraveled in future studies.
PRMT6 interacts with PTEN in cytoplasm, consistent with the role of PRMT6 in suppression of PI3K–AKT signaling. However, subcellular fractionation showed that PTEN R159K mutation does not affect the subcellular localization of the PTEN protein (SI Appendix, Fig. S8). In addition, PTEN R159me2a was found in cytoplasm as well as in nucleus. These findings suggest that PTEN is methylated by PRMT6 in cytoplasm, and methylated PTEN can shuttle from cytoplasm to nucleus. Thus, methylated PTEN can exert its influence in the cytoplasm and nucleus, which might be not limited to suppression of PI3K–AKT signaling and modulation of pre-mRNA splicing.
In summary, our study reveals a type of PTEN modification by protein arginine methylation and provides perspective for understanding the mechanism underlying the complexity of PTEN regulation and function.
Experimental Procedures
Materials and methods used for mass-spectrometry analysis, cell culture, antibodies, plasmids, S-protein pulldown, immunoblotting, immunoprecipitation, PTEN enzyme reactions, in vitro methylation assay, CRISPR-based gene editing, mouse xenograft, RNA-seq library preparation, RNA-seq and analysis of alternative splicing events, Gene Ontology term analysis, and coexpression analysis were performed as described in detail in SI Appendix, SI Experimental Procedures. Animal procedures followed the Peking University Guidelines for “Using Animals in Intramural Research” and were approved by the Animal Care and Use Committee of Peking University.
Supplementary Material
Acknowledgments
We thank L. Liang, Z. Dai, Y. Li, W. Wang, and other members of the Y.Y. laboratory for technical assistance and critical discussion. This study was supported by National Key Research and Development Program of China Grant 2016YFA0500302 (to Y.Y.); National Natural Science Foundation of China Grants 81430056, 31420103905, 81372491, and 81321003 (to Y.Y.); National Natural Science Foundation of China Grant 31600618 (to J.F.); Beijing Natural Science Foundation Key Grant 7161007 (to Y.Y.); Peking University Health Science Center Grant BMU20150503 (to J.F.); and the Lam Chung Nin Foundation for Systems Biomedicine at Peking University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811028116/-/DCSupplemental.
References
- 1.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 4.Hobert JA, Eng C. PTEN hamartoma tumor syndrome: An overview. Genet Med. 2009;11:687–694. doi: 10.1097/GIM.0b013e3181ac9aea. [DOI] [PubMed] [Google Scholar]
- 5.Tan MH, et al. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18:400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki A, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 8.Backman SA, et al. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- 9.Kwon CH, et al. Pten regulates neuronal soma size: A mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 10.Marsh DJ, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 11.Crackower MA, et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110:737–749. doi: 10.1016/s0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 12.Oudit GY, et al. Loss of PTEN attenuates the development of pathological hypertrophy and heart failure in response to biomechanical stress. Cardiovasc Res. 2008;78:505–514. doi: 10.1093/cvr/cvn041. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14:523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 14.Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. 2003;4:287–294. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 15.Heit B, et al. PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat Immunol. 2008;9:743–752. doi: 10.1038/ni.1623. [DOI] [PubMed] [Google Scholar]
- 16.Shrestha S, et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol. 2015;16:178–187. doi: 10.1038/ni.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol. 2016;17:241–249. doi: 10.1038/ni.3311. [DOI] [PubMed] [Google Scholar]
- 18.Stiles B, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected] Proc Natl Acad Sci USA. 2004;101:2082–2087, and erratum (2004) 101:5180. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiles BL, et al. Selective deletion of Pten in pancreatic beta cells leads to increased islet mass and resistance to STZ-induced diabetes. Mol Cell Biol. 2006;26:2772–2781. doi: 10.1128/MCB.26.7.2772-2781.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z, et al. PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes. 2007;56:2449–2456. doi: 10.2337/db06-1731. [DOI] [PubMed] [Google Scholar]
- 21.Sulis ML, Parsons R. PTEN: From pathology to biology. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 22.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: The long and the short of it. Trends Biochem Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worby CA, Dixon JE. Pten. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 25.Adey NB, et al. Threonine phosphorylation of the MMAC1/PTEN PDZ binding domain both inhibits and stimulates PDZ binding. Cancer Res. 2000;60:35–37. [PubMed] [Google Scholar]
- 26.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez F, et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J Biol Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 28.Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 29.Maccario H, Perera NM, Davidson L, Downes CP, Leslie NR. PTEN is destabilized by phosphorylation on Thr366. Biochem J. 2007;405:439–444. doi: 10.1042/BJ20061837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yim EK, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon J, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SR, et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 33.Leslie NR, et al. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okumura K, et al. PCAF modulates PTEN activity. J Biol Chem. 2006;281:26562–26568. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- 35.Ikenoue T, Inoki K, Zhao B, Guan KL. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2008;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- 36.Meng Z, Jia LF, Gan YH. PTEN activation through K163 acetylation by inhibiting HDAC6 contributes to tumour inhibition. Oncogene. 2016;35:2333–2344. doi: 10.1038/onc.2015.293. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trotman LC, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun. 2012;3:911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 41.Bassi C, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341:395–399. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 2015;29:157–170. doi: 10.1101/gad.251785.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem. 2013;288:37010–37025. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012;40:9513–9521. doi: 10.1093/nar/gks764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein C, Riedl S, Rüthnick D, Nötzold RR, Bauer UM. The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 2012;40:9522–9533. doi: 10.1093/nar/gks767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, et al. Suppression of PRMT6-mediated arginine methylation of p16 protein potentiates its ability to arrest A549 cell proliferation. Int J Biochem Cell Biol. 2012;44:2333–2341. doi: 10.1016/j.biocel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Nakakido M, et al. PRMT6 increases cytoplasmic localization of p21CDKN1A in cancer cells through arginine methylation and makes more resistant to cytotoxic agents. Oncotarget. 2015;6:30957–30967. doi: 10.18632/oncotarget.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu H, et al. Structural basis of arginine asymmetrical dimethylation by PRMT6. Biochem J. 2016;473:3049–3063. doi: 10.1042/BCJ20160537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phalke S, et al. p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40:9534–9542. doi: 10.1093/nar/gks858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almeida-Rios D, et al. Histone methyltransferase PRMT6 plays an oncogenic role of in prostate cancer. Oncotarget. 2016;7:53018–53028. doi: 10.18632/oncotarget.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshimatsu M, et al. Dysregulation of PRMT1 and PRMT6, type I arginine methyltransferases, is involved in various types of human cancers. Int J Cancer. 2011;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 56.Lee G, Blenis J. Akt-ivation of RNA splicing. Mol Cell. 2014;53:519–520. doi: 10.1016/j.molcel.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Hong X, et al. PTEN antagonises Tcl1/hnRNPK-mediated G6PD pre-mRNA splicing which contributes to hepatocarcinogenesis. Gut. 2014;63:1635–1647. doi: 10.1136/gutjnl-2013-305302. [DOI] [PubMed] [Google Scholar]
- 58.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 59.Larsen SC, et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 60.Tan MH, et al. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jansson M, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, et al. Allele-specific tumor spectrum in pten knockin mice. Proc Natl Acad Sci USA. 2010;107:5142–5147. doi: 10.1073/pnas.0912524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papa A, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelen MR, et al. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 65.Le Romancer M, et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 66.Banasavadi-Siddegowda YK, et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene. 2017;36:263–274. doi: 10.1038/onc.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 68.Sakamaki J, et al. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci USA. 2011;108:6085–6090. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frankel A, et al. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 71.Kushnir A, Wajsberg B, Marks AR. Ryanodine receptor dysfunction in human disorders. Biochim Biophys Acta Mol Cell Res. July 21, 2018 doi: 10.1016/j.bbamcr.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 72.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papasaikas P, Tejedor JR, Vigevani L, Valcárcel J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol Cell. 2015;57:7–22. doi: 10.1016/j.molcel.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 75.Hou P, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13:1161–1170. doi: 10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- 76.Cecener G, et al. Investigation of MMAC/PTEN gene mutations and protein expression in low grade gliomas. Cell Mol Neurobiol. 2009;29:733–738. doi: 10.1007/s10571-009-9397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dahlman KB, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012;2:791–797. doi: 10.1158/2159-8290.CD-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cryan JB, et al. Clinical multiplexed exome sequencing distinguishes adult oligodendroglial neoplasms from astrocytic and mixed lineage gliomas. Oncotarget. 2014;5:8083–8092. doi: 10.18632/oncotarget.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uhlen M, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 80.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shishkova E, et al. Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat Commun. 2017;8:15571. doi: 10.1038/ncomms15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 84.Kryukov GV, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marjon K, et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.