Significance
Immune response to infection is accompanied by active demethylation of thousands of CpG sites. Yet, the causal relationship between changes in DNA methylation and gene expression during infection remains to be elucidated. Here, we investigated the role of DNA methylation in the regulation of innate immune responses to bacterial infections. We found that virtually all changes in gene expression in response to infection occur prior to detectable alterations in the methylome. We also found that the binding of most infection-induced transcription factors precedes loss of methylation. Collectively, our results show that changes in methylation are a downstream consequence of transcription factor binding, and not essential for the establishment of the core regulatory program engaged upon infection.
Keywords: DNA methylation, epigenetic, immune responses, tuberculosis, dendritic cells
Abstract
DNA methylation is considered to be a relatively stable epigenetic mark. However, a growing body of evidence indicates that DNA methylation levels can change rapidly; for example, in innate immune cells facing an infectious agent. Nevertheless, the causal relationship between changes in DNA methylation and gene expression during infection remains to be elucidated. Here, we generated time-course data on DNA methylation, gene expression, and chromatin accessibility patterns during infection of human dendritic cells with Mycobacterium tuberculosis. We found that the immune response to infection is accompanied by active demethylation of thousands of CpG sites overlapping distal enhancer elements. However, virtually all changes in gene expression in response to infection occur before detectable changes in DNA methylation, indicating that the observed losses in methylation are a downstream consequence of transcriptional activation. Footprinting analysis revealed that immune-related transcription factors (TFs), such as NF-κB/Rel, are recruited to enhancer elements before the observed losses in methylation, suggesting that DNA demethylation is mediated by TF binding to cis-acting elements. Collectively, our results show that DNA demethylation plays a limited role to the establishment of the core regulatory program engaged upon infection.
Innate immune cells, such as dendritic cells (DCs) and macrophages, are the first mediators recruited in response to an invading pathogen. Upon stimulation, these cells considerably shift their transcriptional program, activating hundreds of genes involved in immune-related processes in a rapid and highly choreographed fashion. This is achieved through the binding of signal-dependent transcription factors (TFs), including NF-κB/Rel, AP-1, and IFN regulatory factors (IRFs), to gene regulatory regions of the genome where recruitment of various coactivators is initiated (1, 2). Alterations to the epigenome, such as histone modifications and DNA methylation, are recognized as important permissive or suppressive factors that play an integral role in modulating access of TFs to cis-acting DNA regulatory elements via the regulation of chromatin dynamics.
Many studies have highlighted the importance of histone modifications in regulating complex gene expression programs underlying immune responses (3, 4). However, the exact role that DNA methylation plays in innate immune response regulation remains ambiguous. We have previously shown that infection of postmitotic DCs is associated with an active loss of methylation at enhancers and that such demethylation events are strongly predictive of changes in expression levels of nearby genes (5). Many other studies correlate these two processes (6–13), but it remains unclear whether altered methylation patterns directly invoke transcriptional modulation or whether such patterns are the downstream consequence of TF binding to regulatory regions. Thus, the causal relationship between changes in DNA methylation and gene expression during infection remains unresolved. To address this question, we characterized in parallel genome-wide patterns of DNA methylation, gene expression, and chromatin accessibility in noninfected and Mycobacterium tuberculosis (MTB)-infected DCs at multiple time points. Our results show that the loss of DNA methylation observed in response to infection is not required for the activation of most enhancer elements and that, instead, demethylation is a downstream consequence of TF binding.
Results
Bacterial Infection Induces Stable DNA Demethylation at Enhancers of Dendritic Cells.
We infected monocyte-derived DCs from four healthy individuals with a live virulent strain of M. tuberculosis (MTB) for 2, 18, 48, and 72 h. We chose to work with DCs because they play an essential, nonredundant role in protective immunity to TB (14). In the absence of DCs, CD4+ T cell responses are impaired and bacterial load is uncontrolled (15–17), arguing for an important role for DCs in resistance to mycobacteria infections (18). At each time-point, we obtained single base pair resolution DNA methylation levels for over 130,000 CpG sites using a customized capture-based bisulfite sequencing panel (SeqCap Epi, ref. 19; see Materials and Methods), in matched noninfected and MTB-infected DCs. Our customized SeqCap Epi panel interrogates 33,059 regions highly enriched among putative enhancer elements [58% are associated with the H3K4me1 enhancer mark (20); SI Appendix, Fig. S1A], which are the main targets of methylation changes in response to infection (5). In total, we generated ∼717 million single-end reads (mean = 17.5 million reads per sample; Dataset S1), resulting in an average coverage of ∼70× per CpG site (SI Appendix, Fig. S1B). Methylation values between samples were strongly correlated, attesting to the high quality of the data (SI Appendix, Fig. S1C; median r across all samples = 0.94).
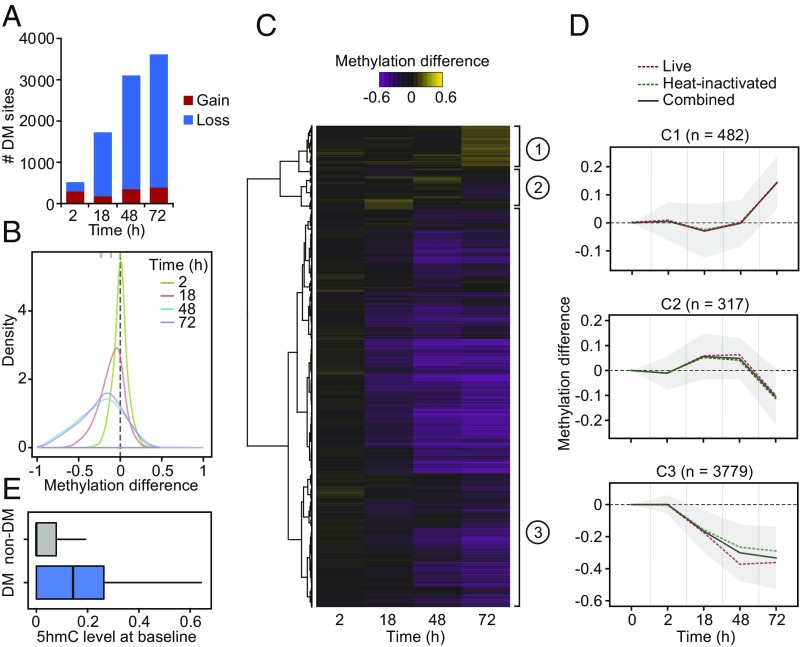
We next assessed temporal changes in methylation levels in response to infection using the DSS software (21). We defined differentially methylated (DM) CpG sites as those showing a significant difference of methylation between infected and noninfected samples at a False Discovery Rate (FDR) < 0.01 and an absolute mean methylation difference above 10%. Using these criteria, we identified 6,174 DM CpG sites across the time course of infection. Consistent with previous findings (5), the vast majority of changes in methylation (87%) were associated with the loss of DNA methylation in infected cells (SI Appendix, Fig. S1 A and B). To evaluate if the loses in methylation induced by MTB infection were specific to DCs, we collected additional methylation profiles on monocyte-derived macrophages from two additional individuals before and after infection with MTB for 20 h. Consistent with our findings in DCs, the vast majority of changes observed in macrophages (∼80%) were associated with the loss of DNA methylation in infected cells (SI Appendix, Fig. S2). Moreover, ∼30% of the CpG sites changing methylation in response to infection overlap CpG sites that are also differentially methylated in DCs, which is a 10-fold enrichment compared with random CpG sites from our targeted SeqCap-Epi assay (P = 8.12 × 10−25; SI Appendix, Fig. S2).
Next we tested if live bacteria were required to induce the observed changes in DNA methylation. Changes in methylation in response to live and heat-killed MTB were strikingly correlated, particularly at later time-points postinfection (r ≥ 0.84 at 18 h and above; Fig. 1 and SI Appendix, Fig. S3). These results show that DCs do not require exposure to a live pathogen to elicit the overall demethylation detected in response to infection. Indeed, the stimulation of DCs for 24 h with LPS, which activates Toll-like receptor (TLR) 4, and beta-glucan, a Dectin-1 ligand, and a classical molecule used for trained immunity experiments, is sufficient to induce demethylation at the same CpG sites altered upon MTB infection—albeit at a lower magnitude—suggesting that the activation of single innate immune receptors is sufficient for the induction of active changes in DNA methylation (SI Appendix, Fig. S4). Hierarchical clustering analysis of the DM sites observed when considering samples exposed to either live or heat-killed bacteria showed that >80% of the sites exhibited a gradual loss of methylation over the time course of infection until methylation marks were almost completely erased (DM Cluster 3; Fig. 1 C and D and Dataset S2).
Fig. 1.
(A) Barplots showing the number of differentially methylated (DM) CpG sites identified at a |methylation difference| > 10% and FDR < 0.01 (y-axis) at each time point after MTB infection (2, 18, 48, and 72 h [h]) (x-axis). (B) Distribution of differences in methylation between infected and noninfected cells at DM sites, at each time point. (C) Heatmap of differences in methylation constructed using unsupervised hierarchical clustering of the 4,578 DM sites (identified at any time point using live and heat-inactivated MTB-infected samples combined; y-axis) across four time points after infection, which shows three distinct patterns of changes in methylation. (D) Mean differences in methylation of CpG sites in each cluster across all time points; shading denotes ±1 SD. For visualization purposes, we also show the 0h time point, where we expect no changes in methylation. (E) Boxplots comparing the distribution of 5hmC levels in noninfected DCs between non-DM and DM sites (Cluster 3).
Monocyte-derived DCs do not proliferate in response to infection (5) and, therefore, any observed losses in methylation must occur through an active mechanism involving the ten-eleven translocation (TET) enzymes, a family of enzymes that converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (22). Thus, we used Tet-assisted bisulfite sequencing (TAB-seq) data collected from noninfected DCs (5) to assess if DM sites had significantly different levels of 5hmC compared with non-DM sites. We found that DM sites (Cluster 3) show high levels of 5hmC even before infection (Fig. 1E; 3.2-fold enrichment compared with non-DM sites; Wilcoxon test; P < 1 × 10−16), suggesting that DM sites are likely prebound by TET enzymes (likely TET2 [23, 24], the most expressed TET enzyme in DCs [SI Appendix, Fig. S5]) and that 5hmC may serve as a stable mark that acts to prime enhancers (25–27).
Up-Regulation of Inflammatory Genes Precedes DNA Demethylation.
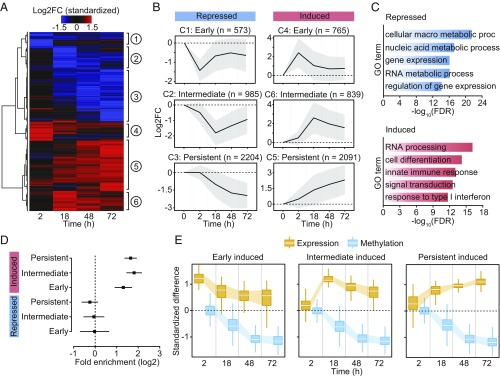
We collected RNA-seq data from matched noninfected and infected samples at each time point, for a total of 34 RNA-seq profiles across time-treatment combinations (28) (mean = 42.2 million reads per sample; Dataset S1). The first principal component of the resulting gene expression data accounted for 63% of the variance in our dataset and separated infected and noninfected DCs (SI Appendix, Fig. S6A). We found extensive differences in gene expression levels between infected and noninfected DCs: of the 13,956 genes analyzed, 1,987 (14%), 4,371 (31%), 4,591 (33%), and 5,189 (37%) were differentially expressed (DE) at 2, 18, 48, and 72 h postinfection, respectively (FDR < 0.01 and absolute log2[fold change] > 1; Dataset S3). We also collected RNA-seq data in samples stimulated with heat-inactivated MTB and found that, similar to changes in methylation, changes in gene expression in response to live and heat-inactivated MTB were strongly correlated (r ≥ 0.94; SI Appendix, Fig. S6B). We next grouped the set of DE genes across the time course (7,457 in total) into six distinct temporal expression clusters (Fig. 2 A and B and Dataset S3). These clusters cover a variety of differential expression patterns, including genes which show increasing up-regulation over time (DE Cluster 5: Persistent induced; n = 2,091) to genes in which the highest levels of expression occur at 2 or 18 h followed by a decrease toward basal levels (DE Cluster 4: Early induced [n = 765], and DE Cluster 6: Intermediate induced [n = 839], respectively) (Fig. 2B). Gene ontology (GO) enrichment analysis revealed that induced genes were strongly enriched among GO terms directly related to immune function, including defense response (FDR = 1.2 × 10−11) and response to cytokine (FDR = 8.2 × 10−12), whereas repressed genes were primarily enriched for gene sets associated with metabolic processes (Fig. 2C and Dataset S4).
Fig. 2.
(A) Heatmap of differences in expression (standardized logtwofold changes) constructed using unsupervised hierarchical clustering of the 7,457 differentially expressed genes (identified at any time point) across four time points after MTB infection. (B) Mean logtwofold expression changes of genes in each cluster across all time points; shading denotes ±1 SD. For visualization purposes, we also show the 0 h time point, where we expect no changes in expression. (C) Gene ontology enrichment analyses among genes that are repressed or induced in response to MTB infection. (D) Enrichment (in log2; x-axis) of differentially expressed genes associated with differentially methylated CpG sites (Cluster 3). Error bars show 95% confidence intervals for the enrichment estimates. (E) Boxplots showing the distribution of standardized differences in methylation of DM sites in Cluster 3 (blue) along with the corresponding standardized differences in expression of the associated genes (orange), across all time points.
We next tested whether genes located near DM sites—particularly focusing on those sites exhibiting a stable loss of methylation (i.e., Cluster 3 in Fig. 1 C and D)—were more likely to be differentially expressed upon MTB infection relative to all genes in the genome. We found that genes associated with one or more DM sites were strongly enriched among genes that were up-regulated in response to infection, regardless of the time point at which expression levels started to change: early (2.5-fold, P = 3.23 × 10−11), intermediate (3.5-fold, P = 3.59 × 10−25), and persistent (3.1-fold, P = 3.80 × 10−33) (Fig. 2 D and E).
If demethylation is required for the activation of enhancer elements and the subsequent up-regulation of their target genes, we would expect demethylation to occur prior to changes in gene expression; instead, we found the opposite pattern. Among up-regulated genes associated with DM sites (n = 593), 37% exhibited at least a twofold increase in gene expression levels at 2 h postinfection, although differential methylation did not begin to be detectable until 18 h postinfection (Fig. 2E). To better delineate the relationship between changes in DNA methylation and changes in gene expression, we collected data from three individuals at additional early/intermediate time points—4, 5, and 6 h postinfection. Again, we did not detect changes in DNA methylation until after 6 h postinfection (SI Appendix, Fig. S7). However, by 6 h, 5,110 genes are already differentially expressed at a stringent FDR of 1% and |log2FC| > 1. Among the set of genes associated with both changes in methylation and up-regulated upon infection at any time point, 83.1% show a change in gene expression before any detectable change in methylation. In contrast, only 1.3% (eight genes) show a change in DNA methylation that precedes a statistically detectable change in gene expression (SI Appendix, Figs. S7 and S8), suggesting that no definitive causal relationship between DNA demethylation and gene activation exists.
Given that our SeqCapEpi panel mostly interrogates enhancer elements, we cannot exclude the possibility that rapid changes in methylation nearby early response genes may occur in their promoter region. Thus, we performed whole-genome bisulfite sequencing (WGBS) on three of our samples in matched noninfected and MTB-infected DCs (2 h postinfection). In total, we generated ∼1.4 billion paired-end reads, resulting in an average coverage of ∼5× per CpG site. Using these data, we found no evidence that promoter regions of differentially expressed genes—regardless of their response dynamics—significantly changed methylation levels in response to infection compared with nondifferentially expressed genes (SI Appendix, Fig. S9). This finding recapitulates what we have previously reported at 18 h postinfection, where promoter regions appeared to be largely refractory to methylation status changes in response to infection (5).
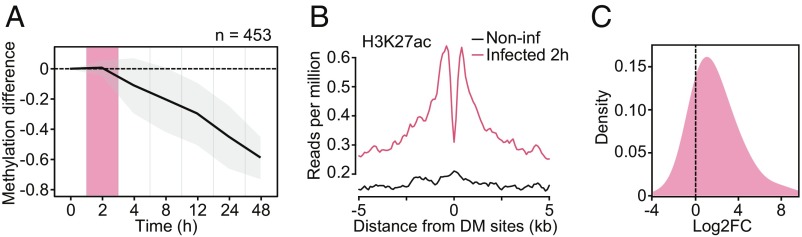
To confirm that our findings were generalizable to other innate immune cell types and pathogenic infections, we performed a separate time-course analysis of differential methylation in Salmonella-infected macrophages from one additional donor over six time points (Dataset S1). We discovered hundreds of CpG sites that exhibited a progressive loss of methylation over the time course of infection, corroborating our findings in MTB-infected DCs (Fig. 3A). To assess whether demethylation arises after the activation of associated enhancers, we collected ChIP-seq data for acetylation of histone 3 lysine 27 (H3K27ac) at 2 h postinfection, as changes in DNA methylation have yet to occur at this point (29). We found that the deposition of activating H3K27ac marks preceded demethylation at these CpG sites (Fig. 3B). Moreover, using previously published RNA-seq expression data from Salmonella-infected macrophages (30), we found that most genes associated with these sites were up-regulated at 2 h postinfection (Fig. 3C), before any changes in methylation. Collectively, these findings indicate that DNA demethylation is not required for the activation of most enhancer elements and that the vast majority of methylation changes induced by infection are a downstream consequence of transcriptional activation.
Fig. 3.
(A) Mean differences in methylation (y-axis) in CpG sites that show stable losses of methylation (similar to Cluster 3 DM sites in Fig. 1 C and D; n = 453) in Salmonella-infected macrophages, across six time points after infection (2, 4, 8, 12, 24, and 48 h [h]; x-axis). Shading denotes ±1 SD. For visualization purposes, we also show the 0 h time point, where we expect no changes in methylation. (B) Composite plots of patterns of H3K27ac ChIP-seq signals ±5 kb around the midpoints of hypomethylated sites (x-axis) in macrophages at 2 h postinfection with Salmonella. (C) Distribution of logtwofold expression changes (between noninfected and Salmonella-infected macrophages at 2 h) for genes associated with DM sites in A (n = 269).
The Binding of Most Infection-Induced TFs Does Not Require Active Demethylation.
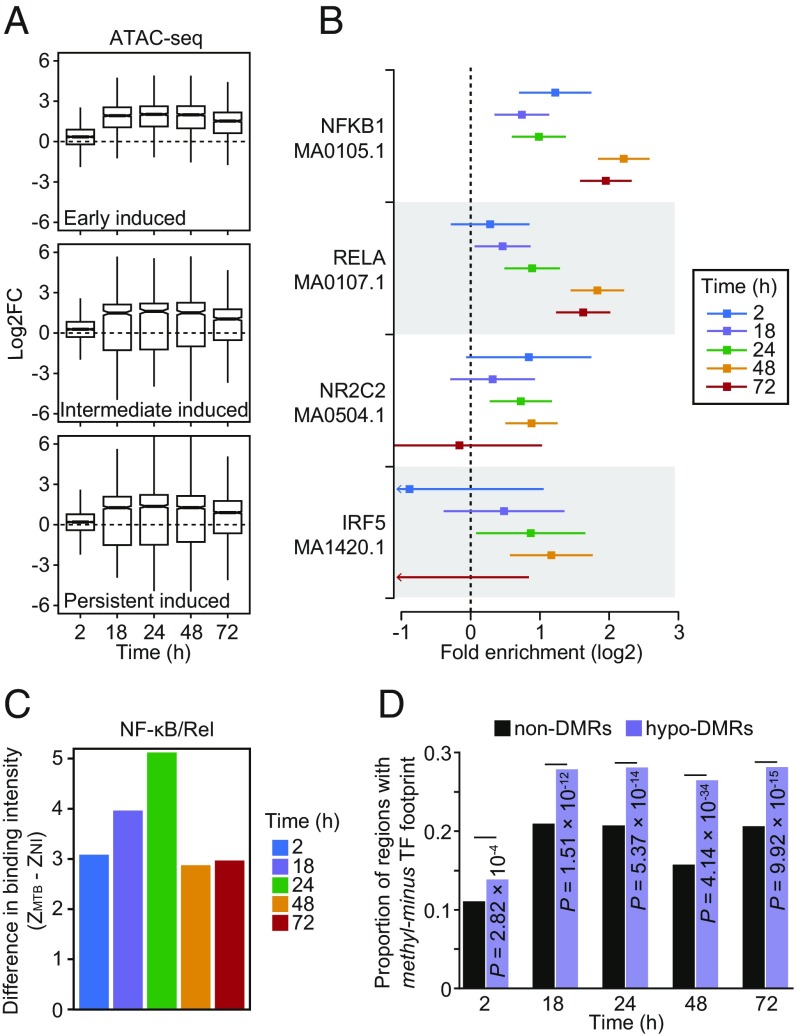
We next asked whether MTB-induced gene expression changes were associated with changes in chromatin accessibility. To do so, we profiled regions of open chromatin in noninfected and infected DCs at the same time points (plus one additional time point at 24 h) using ATAC-seq (31, 32). We found that the response to MTB infection was accompanied by an increase in chromatin accessibility across regulatory regions associated with genes up-regulated upon MTB infection, regardless of their expression profiles (Fig. 4A).
Fig. 4.
(A) Boxplots showing the distribution of logtwofold changes in chromatin accessibility between noninfected and MTB-infected DCs across the five time points of infection (2, 4, 18, 24, 48, and 72 h) for open chromatin regions associated with the three classes of induced genes described in Fig. 2 A and B. (B) TF binding motifs for which the number of well-supported footprints (posterior probability > 0.99) within hypomethylated regions (i.e., the combined set of DM sites for all four time-points) were enriched (FDR < 0.01) relative to non-DMRs (with 250 bp flanking the start and end) in MTB-infected DCs. The enrichment factors (x-axis) are shown in a log2 scale and error bars reflect the 95% confidence intervals. A complete list of all TF binding motifs for which footprints are enriched within hypomethylated regions can be found in Dataset S5. (C) Barplots showing significant differences in TF occupancy score predictions for NF-κB/Rel motifs between MTB-infected and noninfected DCs (ZMTB − ZNI; y-axis; see Materials and Methods) across all time points (x-axis). A positive Z-score difference indicates increased TF binding in hypomethylated regions after MTB infection. (D) Proportion of regions that overlap a methylation-sensitive (“methyl-minus”; reported in Yin et al. [33]) TF footprint (y-axis) observed among non-DMRs and hypomethylated regions (or hypo-DMRs; see Materials and Methods).
To investigate the relationship between DNA methylation and TF occupancy, we performed TF footprinting analysis on our target regions (i.e., the set of putative enhancers tested for dynamic DNA methylation). We classified target regions as “hypomethylated regions” (n = 1,877) or “non-differentially methylated regions” (non-DMRs) (n = 31,182) according to whether or not these regions overlap DM CpG sites (from differential methylation Cluster 3, specifically). We found that hypomethylated regions were significantly enriched for the binding of immune-related TFs relative to regions exhibiting consistent methylation levels. These immune-related TFs include several master regulators of the innate immune response, such as NF-κB/Rel family members NFKB1 (up to 4.6-fold enrichment across the time course [FDR = 4.78 × 10−29]) and RELA (up to 3.6-fold enrichment across the time course [FDR = 1.95 × 10−18]) Fig. 4B and Dataset S5).
We next used CentiDual (5) to test for differential binding of TFs between noninfected and infected samples, specifically focusing on the set of TF family members known to orchestrate innate immune responses to infection (i.e., NF-κB/Rel, AP-1, STATs, and IRFs). We found increased binding at NF-κB/Rel binding motifs starting at 2 h postinfection, despite the fact that no changes in methylation were observed at such early time points (P = 0.002; Fig. 4C and Dataset S5; see Materials and Methods). A similar pattern was observed for AP-1 (P = 0.01; SI Appendix, Fig. S10). These data show that, while demethylated regions overlap areas bound by immune-induced TFs, the binding of these TFs occurs before DNA demethylation.
Although demethylation does not appear to be required for the binding of key TFs involved in regulation of innate immune responses, it is plausible that the removal of methylation marks at DM sites might enable occupancy of methylation-sensitive factors at later time points (33–35). In support of this hypothesis, we found that, at later time points (18 h and above), there was a stronger enrichment for the binding of TFs that preferentially bind to unmethylated motifs (or “methyl-minus” as defined by Yin et al. [33]) within hypomethylated regions (up to 1.7-fold enrichment; χ2-test; P = 4.14 × 10−34; Fig. 4D; see Materials and Methods). Collectively, these results suggest that, although demethylation is likely not required for the engagement of the core regulatory program induced early after infection, it might play a role in fine-tuning the innate immune response by facilitating the binding of salient methyl-sensitive TFs that mediate later immune responses.
Discussion
Our results show that bacterial infection leads to marked remodeling of the methylome of phagocytic cells. Strikingly, virtually all changes in gene expression in response to infection occurred before detectable alterations in DNA methylation, suggesting that the observed demethylation is a downstream consequence of TF binding and transcriptional activation. This pattern holds true genome-wide as well as when focusing the analyses to genes known to be associated with immunity to TB (SI Appendix, Figs. S11 and S12). We note, however, two limitations of our bisulfite sequencing data. First, there might be subtle changes in methylation that occur at early time points that we cannot detect given our small sample sizes, or changes in methylation that occur in regions not covered by our targeted array. Second, our data do not allow us to distinguish between 5mC and 5hmC. Thus, it is possible that the gain of 5hmC in DM sites, which do not show a loss of 5mC at 2 h postinfection, precedes the activation of certain enhancers, as was recently suggested in T cells (8). In SI Appendix, Fig. S13, we provide a schematic representation of our proposed model that links changes in DNA methylation with changes in gene expression in the context of an innate immune response.
The observed changes in methylation most likely occur via TET2-mediated active demethylation, as previously shown (5, 23, 36). Consistent with this hypothesis, we found that CpG sites that lose methylation upon infection display high levels of 5hmC at baseline, suggesting that these regions are actively bound by TET2 even before infection. Moreover, TET2 is strongly up-regulated 2 h after infection (∼2.5 fold; SI Appendix, Fig. S14). 5hmC could be a stable intermediate that serves as an epigenetic priming mark, ensuring the rapid response of DCs against infection (25–27, 36–39). Interestingly, albeit not significant, we noticed a clear trend toward higher levels of 5hmC among early induced genes compared with later induced genes (P = 0.1, SI Appendix, Fig. S15), suggesting that 5hmC could be particularly important for the up-regulation of early response genes.
Using footprint analysis, we show that NF-κB/Rel, a master regulator of inflammation, is recruited to hypomethylated regions as soon as 2 h postinfection. This finding is consistent with ChIP-seq data collected from mouse macrophages stimulated with Kdo2-Lipid A (KLA), a highly specific TLR4 agonist, which shows that the NF-κB subunit p65 is rapidly recruited to enhancer elements within 1 h poststimulation (40). We hypothesize that the rapid binding of NF-κB, and of other immune-induced TFs, instigates chromatin opening which is then followed by the recruitment of histone acetyltransferase p300 and the subsequent deposition of activating H3K27ac marks in these regions (41). Interestingly, p300 can acetylate TET2, conferring enhanced enzyme activity (42), which might account for the eventual loss of DNA methylation in response to infection. Incorporating time-course ChIP-Seq data for NF-κB (or other immune-induced TFs) with methylation and gene expression data will be an important next step to validate the link between TF binding, gene activation, and losses in DNA methylation.
Our results indicate that most changes in gene expression that occur in response to infection are independent of DNA demethylation, further supporting a lack of repressive capacity of DNA methylation (43). Similar to previous findings (36, 44–49), our results further reinforce the idea that site-specific regulation of DNA demethylation is mediated by TFs that bind to cis-acting sequences. Interestingly, several recent reports have shown that other epigenetic modifications, such as the H3K4me1 enhancer mark, have a similar passive regulatory function (50–52). However, our results do not exclude the possibility that demethylation might be necessary for the binding of a second wave of TFs that only play a role at later stages of infection. In agreement with this hypothesis, we observed a significant enrichment of binding of TFs known to preferentially bind unmethylated CpGs in hypomethylated regions, primarily at later stages postinfection. Ultimately, this suggests that DNA demethylation is not a key regulatory mechanism of early innate immune responses but that it could still play a role in fine-tuning later innate immune responses by facilitating the binding of methylation-sensitive TFs at enhancers. This conclusion should be further validated on a cellular system where one can prevent demethylation to occur upon infection (e.g., by using TET2-deficient cells) to study the downstream impact of such changes in the overall immune response.
After an infection is cleared, TFs are expected to unbind, and gene expression as well as DNA methylation levels are anticipated to return to basal state. However, our 72-h time course study of DNA methylation shows that levels of methylation at DM sites gradually decrease with time postinfection and never revert back to higher levels. Thus, we speculate that demethylation in response to infection could have a specific biological role in innate immune memory (53–56), and that regions that stably lose methylation may act as primed enhancers, potentially allowing for a faster response to a secondary infection.
Materials and Methods
Details of the experimental and statistical procedures can be found in SI Appendix, SI Materials and Methods. Buffy coats from healthy donors were purchased from Indiana Blood Center and all participants signed a written consent. The ethics committee at the CHU Sainte-Justine approved the project (protocol #4023). Blood mononuclear cells from each donor were isolated by Ficoll-Paque centrifugation and blood monocytes were purified from peripheral blood mononuclear cells (PBMCs) by positive selection with magnetic CD14 MicroBeads (Miltenyi Biotec). Monocytes were then derived into DCs (5) or macrophages (30) and subsequently infected with MTB or Salmonella typhimurium. RNA-seq libraries were prepared using the TruSeq RNA Sample Prep Kit v2. ATAC-seq libraries were generated from 50,000 cells, as previously described (32). SeqCap Epi and whole-genome bisulfite sequencing libraries were generated using the KAPA Library Preparation Kit. Bisulfite sequencing reads were mapped to the human reference genome using Bismark (57), and MTB-induced differences in methylation were identified using the R package DSS (21), which implements the BSmooth smoothing method (58). We used ClueGO (59) to test for enrichment of functionally annotated gene sets among differentially expressed genes. TF footprinting analyses were performed using the Centidual algorithm (5) and JASPAR annotated human TF binding motifs (60).
Supplementary Material
Acknowledgments
We thank Calcul Quebec and Compute Canada for managing and providing access to the supercomputer Briaree from the University of Montreal. This study was funded by grants from the Canadian Institutes of Health Research (301538 and 232519), and the Canada Research Chairs Program (950-228993) (to L.B.B.). A.P. and F.M.-L. were supported by a fellowship from the Fonds de recherche du Québec – Santé (FRQS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE116406 (ATAC-seq), GSE116411 (ChIP-seq), GSE116405 (RNA-seq), and GSE116399 (SeqCap Epi).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814700116/-/DCSupplemental.
References
- 1.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 2.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 4.Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacis A, et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015;25:1801–1811. doi: 10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marr AK, et al. Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PLoS Pathog. 2014;10:e1004419. doi: 10.1371/journal.ppat.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 8.Ichiyama K, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murayama A, et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinclair SH, Yegnasubramanian S, Dumler JS. Global DNA methylation changes and differential gene expression in Anaplasma phagocytophilum-infected human neutrophils. Clin Epigenetics. 2015;7:77. doi: 10.1186/s13148-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiencke JK, et al. The DNA methylation profile of activated human natural killer cells. Epigenetics. 2016;11:363–380. doi: 10.1080/15592294.2016.1163454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, et al. DNA methylation dynamics during ex vivo differentiation and maturation of human dendritic cells. Epigenetics Chromatin. 2014;7:21. doi: 10.1186/1756-8935-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cizmeci D, et al. Mapping epigenetic changes to the host cell genome induced by Burkholderia pseudomallei reveals pathogen-specific and pathogen-generic signatures of infection. Sci Rep. 2016;6:30861. doi: 10.1038/srep30861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber HA, Sandor M. The role of dendritic cells in mycobacterium-induced granulomas. Immunol Lett. 2010;130:26–31. doi: 10.1016/j.imlet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao X, et al. Dendritic cells are host cells for mycobacteria in vivo that trigger innate and acquired immunity. J Immunol. 2002;168:1294–1301. doi: 10.4049/jimmunol.168.3.1294. [DOI] [PubMed] [Google Scholar]
- 16.Tian T, Woodworth J, Sköld M, Behar SM. In vivo depletion of CD11c+ cells delays the CD4+ T cell response to Mycobacterium tuberculosis and exacerbates the outcome of infection. J Immunol. 2005;175:3268–3272. doi: 10.4049/jimmunol.175.5.3268. [DOI] [PubMed] [Google Scholar]
- 17.Wolf AJ, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 18.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacis A, et al. 2018 SeqCap Epi data. NCBI Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116399. Deposited June 28, 2018.
- 20.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng H, Conneely KN, Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42:e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Zhang Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat Rev Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 23.Klug M, Schmidhofer S, Gebhard C, Andreesen R, Rehli M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013;14:R46. doi: 10.1186/gb-2013-14-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Álvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol. 2015;15:7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 25.Mahé EA, et al. Cytosine modifications modulate the chromatin architecture of transcriptional enhancers. Genome Res. 2017;27:947–958. doi: 10.1101/gr.211466.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calo E, Wysocka J. Modification of enhancer chromatin: What, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pacis A, et al. 2018 RNA-seq data. NCBI Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116405. Deposited June 28, 2018.
- 29.Pacis A, et al. 2018 ChIP-seq data. NCBI Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116411. Deposited June 28, 2018.
- 30.Nedelec Y, et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 31.Pacis A, et al. 2018 ATAC-seq data. NCBI Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116406. Deposited June 28, 2018.
- 32.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin Y, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domcke S, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 35.Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17:551–565. doi: 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vento-Tormo R, et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 2016;17:4. doi: 10.1186/s13059-015-0863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sérandour AA, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40:8255–8265. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hon GC, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatt D, Ghosh S. Regulation of the NF-κB-mediated transcription of inflammatory genes. Front Immunol. 2014;5:71. doi: 10.3389/fimmu.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YW, et al. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol Cell. 2017;65:323–335. doi: 10.1016/j.molcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ford EE, et al. 2017. Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. bioRxiv:10.1101/170506. Preprint, posted August 16, 2017.
- 44.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495, and correction (2012) 484:550. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 45.Han L, Lin IG, Hsieh CL. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol Cell Biol. 2001;21:3416–3424. doi: 10.1128/MCB.21.10.3416-3424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem Biophys Res Commun. 2006;344:845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 49.de la Rica L, et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rickels R, et al. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat Genet. 2017;49:1647–1653. doi: 10.1038/ng.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorighi KM, et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell. 2017;66:568–576 e4. doi: 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandenbon A, Kumagai Y, Lin M, Suzuki Y, Nakai K. 2017. Waves of chromatin modifications in mouse dendritic cells in response to LPS stimulation. bioRxiv:10.1101/066472. Priprint, posted April 16, 2018.
- 53.Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: Towards a better understanding of host defense mechanisms. Curr Opin Immunol. 2014;29:1–7. doi: 10.1016/j.coi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 56.Kaufmann E, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172:176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 57.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen KD, Langmead B, Irizarry RA. BSmooth: From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bindea G, et al. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan A, et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284. doi: 10.1093/nar/gkx1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.