Abstract
A collection of over 1,600 sequenced bacteriophages isolated on a single host strain, Mycobacterium smegmatis mc2155, can be grouped into over two dozen types that have little or no nucleotide sequence similarity to each other. One group, Cluster K, can be divided into several subclusters, and the well-characterized and much exploited phage TM4 lies in Subcluster K2. Many of the Cluster K phages have broad host ranges and infect both fast- and slow-growing mycobacterial strains. Here we describe phage ZoeJ, a new Subcluster K2 member, which infects a broad spectrum of mycobacterial hosts including M. smegmatis, Mycobacterium tuberculosis, and Mycobacterium avium. ZoeJ has extensive sequence similarity to TM4, and comparative analysis reveals the precise deletion conferring the lytic phenotype of TM4. The ZoeJ immunity repressor was identified as gene 45, which is prophage-expressed, is required for lysogeny, and is sufficient to confer superinfection immunity to ZoeJ. ZoeJ gp45 also confers immunity to Subcluster K2 phage Milly, and Subcluster K1 phages Adephagia and CrimD, but surprisingly not to TM4. RNAseq analysis reveals the temporal pattern of early and late gene expressions in ZoeJ lytic growth and suggests a role for the ESAS motifs for gene regulation.
Introduction
Mycobacteriophages – phages of mycobacterial hosts – span considerable genetic diversity, illustrated by the large numbers of different genomic types1. However, nearly all of these phages were isolated using Mycobacterium smegmatis mc2155 as the host, and relatively little is known about the host range of these phages, or if genomically distinct phages could be isolated using different mycobacterial strains for isolation2. A relatively small subset of the phages have been shown also to infect Mycobacterium tuberculosis H37Rv, or its avirulent derivatives3.
Approximately 10,000 individual phages have been isolated on M. smegmatis mc2155, of which 1,600 have sequenced genomes (http://phagesdb.org)4. Comparative analysis shows that there are over two dozen distinct genomic types, and the phages can be grouped in clusters of similarity according to these relationships. Currently, there are 29 clusters (Clusters A-Z, AA-AC) and five singletons, each of which lack a close relative. The initial grouping of phages was based on the sharing of nucleotide sequence similarity spanning over 50% of the genome lengths, but has been revised such that the sharing of 35% or more genes based on amino acid sequence similarity helps to resolve ambiguous assignments5. This reflects an underlying continuum of genetic diversity of the phage population, but with unequal representation of different types6. Such a continuum and the blurring of taxonomic boundaries is anticipated from the mosaic architecture of phage genomes and the pervasive swapping of genes by horizontal genetic exchange7–9.
Over 115 sequenced mycobacteriophages are grouped in Cluster K, which is divided into seven subclusters (K1-K7), based on average nucleotide identities (http://phagesdb.org)10. Several cluster K phages have been shown to infect M. tuberculosis H37Rv, including those in Subclusters K1, K2, K3, and K4, although one of the K1 phages tested (Angelica) and the one K5 phage tested (Larva) do not efficiently infect M. tuberculosis3. Little is known about the host range of the other Cluster K phages, although some of them replicate poorly at temperatures above 30° C (e.g. Anaya), conditions that are not c onducive for M. tuberculosis growth.
TM4 is a Subcluster K2 phage which has been used extensively as a tool for mycobacterial genetics 11, 12. Although most Cluster K phages are temperate, TM4 forms clear plaques and does not form stable lysogens10, 11. It efficiently infects M. tuberculosis and was one of the first phages used to construct shuttle phasmids11 which were adapted for transposon delivery13, 14, introducing allelic exchange substrates15, 16, and as a tool for rapid diagnosis and drug susceptibility test of tuberculosis clinical isolates17–19. Comparison of TM4 with the genomes of other Cluster K phages showed that it has lost a central portion of the genome that includes the repressor and the integrase genes10, although at that time no other Subcluster K2 phages had been isolated and thus the nature of the deletion and which genes had been lost was ill-defined10.
Cluster K phage genomes (average genome length, 60 kbp) contain several unusual features including start-associated sequences (SAS) and extended start-associated sequences (ESAS)10. The SAS motifs are 13 bp asymmetric sequences (consensus ‘5-GGGATAGGAGCCC) positioned 3–8bp upstream of the translation initiation codons of a subset of the phage genes, in the positions normally located by the ribosome binding sites (RBS). The SAS RNA sequences partially correspond to the 16S rRNA, but four of the sequence positions are highly conserved and do not pair with the 16S rRNA10. Only 11–19 genes have SAS motifs, predominantly in the right arms of the genomes containing non-structural genes, and it is unclear if or how the translation of these genes differs from other genes. A subset of the SAS-associated genes contains a second motif, the ESAS, composed of two 17 bp partially symmetric sequences in inverted orientation and spaced 4–13 bp apart10. These are typically located in small intergenic spaces and are likely also involved in gene expression or regulation, although their specific roles are not known.
Here, we describe mycobacteriophage ZoeJ, a Subcluster K2 phage that is closely related to TM4 and reveals the nature of the TM4 deletion and the genes that have been deleted. ZoeJ infects both fast- and slow-growing mycobacteria, is temperate, and we identify the phage repressor gene (45). We describe the transcriptomic profiles of the ZoeJ lysogen and ZoeJ lytic growth – the first for any of the Cluster K mycobacteriophages – and suggest a plausible role for the ESAS sequence motifs.
Materials and methods
Bacterial strains and media
M. smegmatis mc2155 and M. tuberculosis mc27000 (an avirulent derivative of M. tuberculosis H37Rv)20 were grown as described previously3. M. avium Va14 (O)21, M. bovis BCG, M. avium subsp. silvaticum ATCC 49884, M. simiae ATCC 25275, M. abscessus ATCC 19977, and M. interjectum ATCC 51457 were grown similarly.
Construction of ZoeJΔ45 and recombinant plasmid pKC01-P
The ZoeJΔ45 mutant was constructed using the BRED methodology as described previously22,23. In brief, a 400 bp gBlock (Integrated DNA Technologies) was synthesized containing sequences homologous to those flanking ZoeJ gene 45 and amplified by PCR using primers ZoeJgBlockFwd and ZoeJgBlockRev (Table S1); the product was purified and quantified. This BRED substrate together with ZoeJ genomic DNA was electroporated into recombineering-proficient M. smegmatis cells24. Primary plaques were screened by PCR for the presence of the wild-type or mutant alleles. Mixed plaques were identified, replated, and secondary plaques screened by PCR using the same primers as above. A mutant was identified, amplified, and sequenced as described previously25, verifying the deletion of gene 45 as predicted. The ZoeJΔ45 derivative also has four single base substitutions at A10712G, C46053T, G52462C, and C54263G. These mutations were most likely present in the parent phage strain used for engineering, rather than being introduced during recombineering.
To construct plasmid pKC01-P, ZoeJ gene 45 together with the 45-46 intergenic region was amplified using primers ZoeJRepFwd and ZoeJRepRev (Table S1) and inserted into the integration-proficient vector pMH9426 using Gibson assembly (New England Biolabs). Plasmid pKC01-P was verified by restriction digest and sequencing (Genewiz).
Host range determination
Phage lysates were serially diluted in phage buffer (10 mM Tris-HCl, pH 7.5; 10 mM MgSO4; 68.5 mM NaCl; 1 mM CaCl2) and spotted onto top agar lawns containing bacterial cultures. Plates were incubated at 37°C for 24–28 hours for M. smegmatis mc2155 and 6 days for M. tuberculosis mc27000, M. avium Va14 (O), M. bovis, M. interjectum, M. avium subsp. silvaticum, M. abscessus, and M. simiae.
RNAseq analysis
Total RNA was extracted from two separate logarithmically growing M. smegmatis mc2155(ZoeJ) or mc2155 cells infected with wild-type ZoeJ (MOI of 3) at 30, 90, 150 and 210 min after adsorption. DNA was removed using Turbo-DNase-Free kit (Ambion) following the manufacturer’s instructions. Removal of rRNA was completed using the Ribo-Zero kit (Illumina). Libraries were constructed using TruSeq Stranded RNAseq Kit (Illumina) and verified using a BioAnalyzer. Libraries were multiplexed on an Illumina MiSeq. Analysis of the data was as described previously22. The RNAseq data set is deposited in the Gene Expression Omnibus (GEO) with accession number GSE124840.
Results
Genome organization of ZoeJ and relationship to TM4
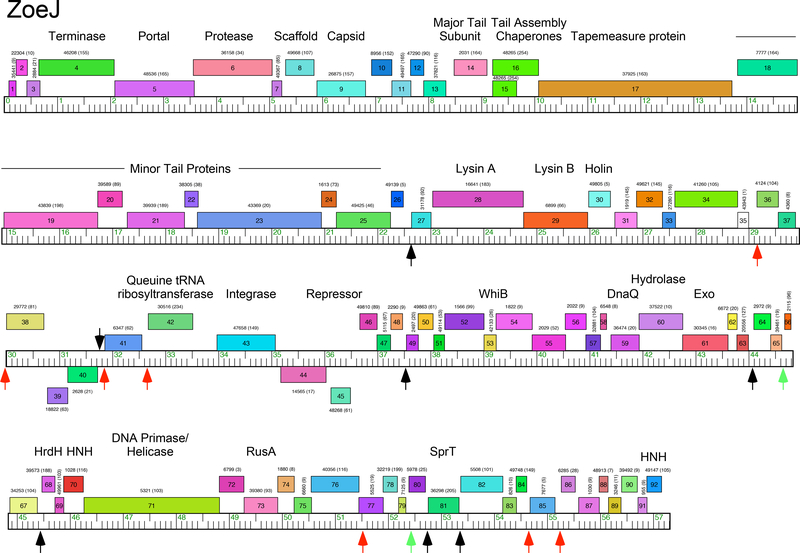
Phage ZoeJ was isolated from Providence, Rhode Island, using M. smegmatis as a host, and its genome sequence has been previously reported (Accession No. KJ510412)6. Annotation of the 57,315 bp genome identified 92 open reading frames, all but four of which are transcribed rightwards (Fig. 1). The left part of the genome (genes 1-25) contains the virion structure and assembly genes canonically arranged as for other phages with siphoviral morphologies, although no terminase small subunit gene was identified (Fig. 1). To the right of the structural genes is the lysis cassette including lysin A, lysin B, and putative holin genes (28, 29, 30 respectively). Candidate integrase and repressor genes (43 and 45, respectively) lie close to the genome center, and the right part of the genome (genes 46-92) has 47 orfs, of which ten can be assigned putative functions, including a WhiB-like regulator (53), exonucleases (59, 61) DNA primase/helicase (71), RusA (73), and two HNH genes (70, 92; Fig. 1). Overall, the ZoeJ genome organization is similar to that of other Cluster K phages10.
Figure 1. Genome map of mycobacteriophage ZoeJ.
The genome of ZoeJ is represented as a ruler with kbp markers with the predicted genes as colored boxes above or below the genome corresponding to rightwards- and leftwards-transcription, respectively. Gene names are shown within each box, and the corresponding phamily number shown above or below each gene with the number of phamily members in parentheses. The map was drawn using Phamerator 36 and the database ‘actinobacteriophages_draft’ as of July 2018. Black and red vertical arrows indicate the locations of Start Associated Sequences (SAS; see Table 1), with the red arrows indicating those that also include an Extended Start Associated Sequence (ESAS; see Table 2). Green vertical arrows indicate positions of ESAS site that lack a linked SAS. Putative gene functions are indicated.
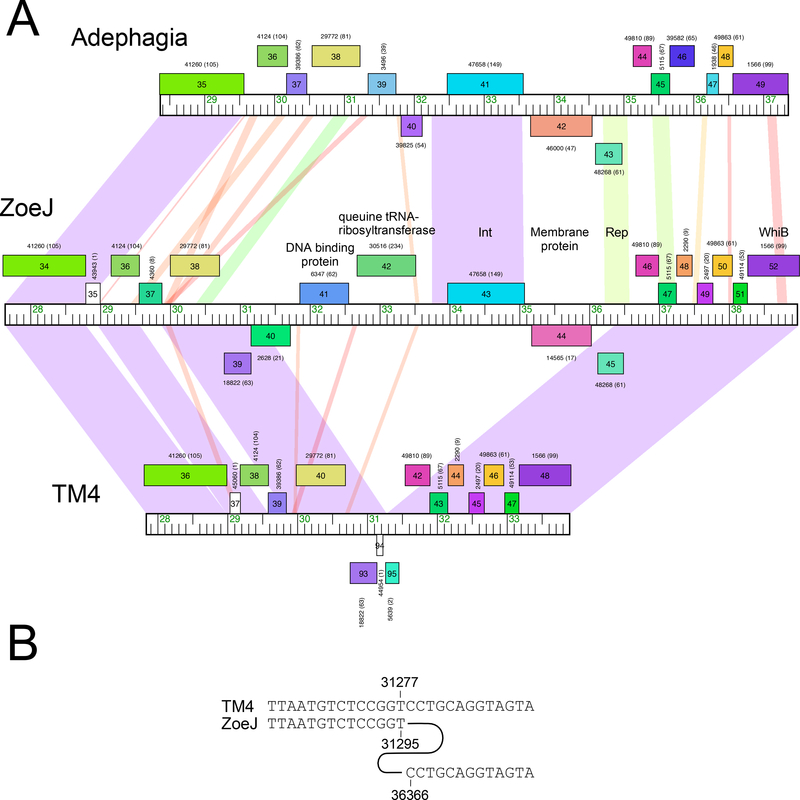
Sequence comparisons show that ZoeJ is a member of Subcluster K2, along with TM4. Alignment of the two genomes show that they are very closely related, with the primary differences being the presence of different lysin A genes, mosaic substitutions of ZoeJ genes 37 and 86 for TM4 genes 39 and 86, respectively (all of unknown function), the loss of TM4 genes 63 and 81 from ZoeJ, and several regions of nucleotide differences (Fig. S1). Relative to ZoeJ, TM4 has a 5,069 bp deletion that removes all or part of six genes (ZoeJ genes 40-45; Fig. 2A). The nucleotide sequences are sufficiently closely related to determine the nature of the deletion, which is precise removal of ZoeJ coordinates 31,296 to 36,366 (Fig. 2B). There is no evidence for even a short stretch of sequence homology, suggesting that the deletion occurred by a truly sequence-independent illegitimate recombination event (or more than one event), as has been postulated as a general process for the creation of mosaic genomes8. The deleted genes include the phage integrase, and part of ZoeJ gene 45, a candidate for the immunity repressor gene. It also removes ZoeJ gene 44 which may be co-transcribed with the repressor, and codes for a putative membrane protein with six predicted transmembrane domains. Most of ZoeJ gene 40 is deleted, leaving a fragment that was annotated as TM4 gene 94 but which is almost certainly non-functional. ZoeJ gp40 is an interesting gene product as it is predicted to be a lipoprotein, and has sequence similarity to MPT63 (Rv1926c), a major secreted antigen of M. tuberculosis27, 28. ZoeJ gene 42 codes for a putative Queuine tRNA ribosyltransferase, and gene 41 is predicted to code for a DNA-binding protein. The specific roles of these ZoeJ genes in the growth or adaptation of the phage is unknown, although we note that the genomic location (i.e. between the immunity and lysis functions) is similar to that of the prophage-mediated viral defense genes in the Cluster N mycobacteriophages22.
Figure 2. Mapping a deletion in the phage TM4 genome.
A. Genome maps of the central portions of phages Adephagia, ZoeJ, and TM4 are shown, with pairwise nucleotide sequence similarity shown as spectrum-colored shading between genomes; violet indicates closest similarity, and red the least similarity above a threshold E value of 10-4. Maps are annotated as in Fig. 1. B. Alignment of TM4 and ZoeJ shows that the deletion in TM4 occurred precisely between ZoeJ coordinates 31,295 and 36,366.
ZoeJ host range
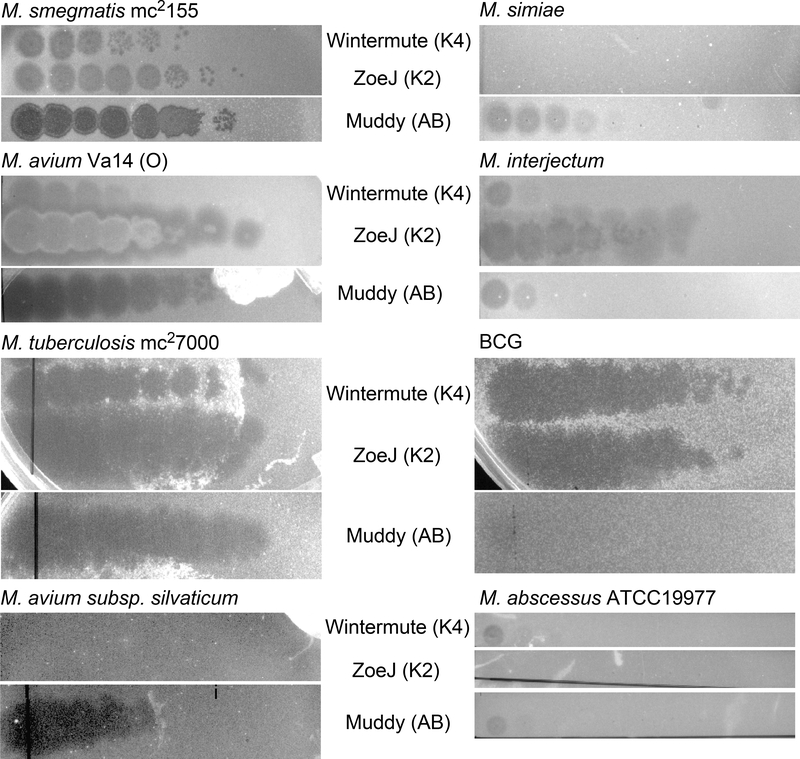
To evaluate the host range of ZoeJ, serial dilutions of a lysate prepared on M. smegmatis mc2155 were plated on a variety of different mycobacterium strains (Fig. 3). ZoeJ was found to have a broad host range and infects M. tuberculosis mc27000 [an avirulent derivative of H37Rv 20], M. avium Va14 (O), M. bovis BCG, and M. interjectum ATCC 51457 with a plating efficiency equivalent to that on M. smegmatis. No infection was observed on M. avium subsp. silvaticum, M. abscessus ATCC 19977, or M. simiae (Fig. 3), or on M. avium subsp. avium ATCC 25291, Mycobacterium nonchromogenicum ATCC 19530 or Mycobacterium terrae ATCC15755 (data not shown), even when plated at high titer (Fig. 3). We note that individual Cluster K phages plate differently on these strains, and Wintermute (Cluster K4) is an example that also infects M. tuberculosis mc27000, M avium Va14 (O), and BCG, but plates poorly on M. simiae, M. interjectum, M. abscessus ATCC 19977, and M. avium subsp. silvaticum (Fig. 3). The infection by ZoeJ of M. avium Va14 (O) and M. interjectum in addition to M. tuberculosis is of interest as both are also associated with opportunistic infections of the lung29, 30. However, there may be variation in infectivity of ZoeJ among different strains of these species, as it does not infect M. avium subsp. avium.
Figure 3. Host range of mycobacteriophage ZoeJ.
Lawns of mycobacterial strains were spotted with ten-fold serial dilutions (left to right) of Cluster K phages ZoeJ (Subcluster K2) and Wintermute (Subcluster K4) together with control phage Muddy (Cluster AB) and incubated. The highest titer phage spot corresponds to a 10−1 dilution of the stock lysate of each phage.
ZoeJ SAS and ESAS sequences
TM4 and other Cluster K phages contain conserved motifs closely-linked but upstream of the translation initiation site of known function, but which are presumably associated with gene expression and its regulation. A search for motifs related to the consensus SAS described for Cluster K phages (5’-GGGATAGGAGCCC) permitting two mismatches identified 14 ZoeJ SAS sites (Table 1). Eleven of the sites correspond to those previously reported for TM410, but three are located in the region deleted in TM4, upstream of the rightwards-transcribed genes 41 and 42, and the leftwards transcribed gene 40 (Fig. 1). The site upstream of gene 40 is positioned immediately adjacent to the translation initiation site, which is unusual but also observed for gene 27 and its homolog in TM4 (Table 1).
Table 1.
ZoeJ Start-Associated Sequences
| Gene2 | Sequence1 | Orientation2 | Coordinates3 |
|---|---|---|---|
| 27 | TGGATAGGAGCACCGTG | + | 22627–22639 |
| 36 | GGGATAGGAGCCCAAAATG | + | 29139–29151 |
| 38 | GGGATAGGAGCCCAAAATG | + | 29987–29999 |
| 40 | GGGATAGGAGACCCATG | - | 31723–31735 |
| 41 | GGGATAGGAGCCGACGACATG | + | 31839–31851 |
| 42 | GGGATAGGAGCCCACGACATG | + | 32658–32670 |
| 49 | GGGATAGGAGCCACTTGTTATG | + | 37526–37538 |
| 64 | GGGATAGGAGCGAAACAGCATG | + | 44078–44090 |
| 68 | GGGATAGGAGCCCCGAGAACATG | + | 45451–45463 |
| 77 | GGGATAGGAGCC CAC GAAATG | + | 51501–51513 |
| 81 | GGGATAGGAGCC CACGAGATG | + | 52738–52750 |
| 82 | GGGATAGGAGCCCCTGCAATG | + | 53351–53363 |
| 85 | GGGATAGGAGCCCAAAATG | + | 54661–54673 |
| 86 | GGGATAGGAGCCCAAAATG | + | 55250–55262 |
| consensus | GGGATAGGAGCCC |
Translation start codon is underlined
Rightwards- and leftwards-transcribed genes are denoted ‘+’ and ‘-‘ respectively.
Coordinates of the 13 bp SAS are shown.
The ESAS sequences in Cluster K phages are complex, being composed of two closely-related 17 bp motifs spaced 4–13bp apart, each of which has imperfect dyad symmetry10. A search for similar motifs in ZoeJ identified nine putative ESAS sequences (Table 2, Fig. 1), seven of which are associated with SAS motifs (Fig. 1). Two of these are within the region deleted in TM4, positioned upstream of the SAS motifs associated with genes 41 and 42. We note that homologues of ZoeJ gene 42 are prevalent in some other mycobacteriophages, notably in the Cluster B1 phages, but do not have conserved motifs analogous to the SAS or ESAS sites. Two of the ESAS motifs (upstream of genes 66 and 80) are not associated with SAS sites, although the site upstream of gene 66 is positioned similarly with respect to the downstream translation start site as with the other ESAS motifs, and a highly redundant SAS motifs can be envisaged. We note that the leftmost inverted repeats of six of the ESAS motifs have a canonical SigA-specific −35 hexamer (5-TTGACA) and a plausible −10 hexamer spaced from them by 16–18 bp (Table 2), as in other Cluster K phages10. This suggests that at least a subset of the ESAS motifs may have promoter activity using SigA-associated RNA polymerase.
Table 2.
ZoeJ Extended Start-Associated Sequences (ESAS)
| Gene | Sequence1 | Orientation |
|---|---|---|
| 36 | TGTTGACGCGCCAACAGG------TTTGCGCCTAAGCTGTTGGGTAGTCAACA | + |
| 38 | TGTTGACGGGTCAACAGC--------TCGCGGTGTACTGTTGAGGTATCAACA | + |
| 41 | TGTTGAGCAGTCAACAGT--------TTGTATGCTACTGTTGAGGCATCAACA | + |
| 42 | TGTTGAGGTGTCAACACG-----------TGTGCTACTGTTGAGGTGTCAACA | + |
| 66 | TGTTGAACGCTCAACACT--GAGCACTCGGCATGACCGTTATCGCTGAACACA | + |
| 77 | TGTTGACAGCTCAACAGATCGTCGGTAACGTCGCCGGTGTTGACCAATCAACA | + |
| 80 | TGTTGACAGCTCAACACC------GCGCATGCTTAACTGTTGACAGCTCAACA | + |
| 85 | TGTTGACACCTCAACACC--------CCGCGGTGTAGTGTTGAGGTATCAACA | + |
| 86 | TGTTGACACCTCAACACC--------CCGAGGTGTACTGTTGAGCTATCAACA | + |
| Consensus | TGTTGAcrnsTCAACAs-------------------sTGTTGaGswrTCAACA |
The ESAS sequences are shown, with inverted repeats shown in bold type, and ‘-‘ characters inserted to align them. Putative promoter −35 hexamers in the center repeat are underlined, and putative −10 hexamers in or near the center repeat are also underlined. For the consensus, nucleotides present in 5–6 motifs or in seven or more, are shown in lower and upper case letters respectively. Redundant codes show preferences in those positions: r = A or G, s= G or C, w = A or T, , n = A, C, G or T.
ZoeJ immunity functions
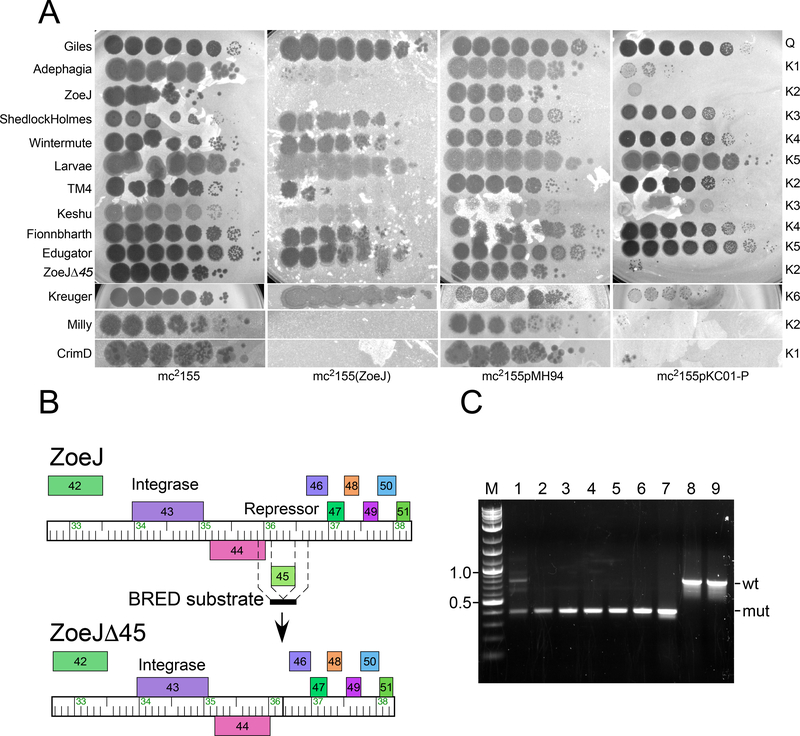
ZoeJ is temperate and forms turbid plaques from which stable lysogens can be recovered, which are immune to superinfection with ZoeJ (Fig. 4A). The ZoeJ lysogen is also immune to superinfection by Milly and CrimD (Subclusters K2 and K1, respectively) and TM4 infects but with a three orders of magnitude reduction in plating efficiency; Adephagia (K1) shows poor infection of the ZoeJ lysogen and forms very turbid spots (Fig. 4A). ZoeJ does not confer immunity to other Cluster phages including ShedlockHolmes, and Keshu (K3), Wintermute and Fionnbharth (K4), Larvae and Edugator (K5), or Kreuger (K6) (Fig. 4A).
Figure 4. ZoeJ gene 45 codes for repressor-mediated superinfection immunity.
A. Lawns of four strains – M. smegmatis mc2155, a ZoeJ lysogen [mc2155(ZoeJ)], a vector-containing strain (mc2155pMH94), and a strain expressing the ZoeJ repressor (mc2155pKC01-P) were spotted with ten-fold serial dilutions of phages as indicated at the left. The highest titer phage spot corresponds to a 10−1 dilution of the stock lysate of each phage. The cluster designation is shown at the right. B. Scheme for construction of the ZoeJΔ45 mutant. The position of the 400 bp BRED dsDNA substrate is shown with 200 bp of sequence identities flanking gene 45. Recombination between phage genomic DNA and the BRED substrate results in deletion of gene 45. C. Identification of a ZoeJΔ45 mutant by PCR. Following co-electroporation of ZoeJ genomic DNA and the BRED substrates, primary plaques were screened by PCR, a mixed primary plaque (lane 1) contains both wild-type (wt; 770 bp) and mutant (mut; 400 bp) products was identified. After replating, an isolated plaque containing only the mutant allele (lane 3) was identified, and after purification, four individual plaques (lanes 4–7) all have only the mutant allele. Wild-type ZoeJ phage lysate (lane 8) and ZoeJ phage DNA (lane 9) contain only the wild type allele. M: DNA Marker, sizes shown in kbp.
ZoeJ gene 45 is a strong candidate for encoding the immunity repressor, based on its genomic location (Fig. 1) and amino acid sequence relationship (72% identity) with the Adephagia repressor (gp45; Fig. 2 & S1)31; these putative repressors share helix-turn-helix (HTH) DNA binding motifs near their N-termini (Fig. S2). We thus constructed a strain expressing ZoeJ gene 45 (M. smegmatis mc2155pKC01-P) and tested it for immune specificity (Fig. 4A). In general, the immunity patterns mirror those of the ZoeJ lysogen, (Fig. 4A). with the notable exception that it fails to confer immunity to TM4. Thus, the reduction in plating of TM4 on the ZoeJ lysogen is not repressor-mediated, and presumably results from other prophage-expressed ZoeJ genes (Fig. 4A).
To confirm that the ZoeJ repressor is required for lysogeny, we constructed a derivative in which the repressor gene (45) is deleted, and which contains no non-ZoeJ sequences (Fig. 4B). A dsDNA substrate was constructed containing the terminal six codons of gene 45 and flanking sequences, which was co-electroporated with ZoeJ phage DNA. Plaques were recovered and screened by PCR, and a mixed plaque was identified that contained both wild-type and mutant alleles, as is typical for the BRED strategy23. This mixed primary plaque was re-plated and secondary plaques were picked and screened by PCR (Fig. 4C). A candidate mutant phage was identified, propagated, and sequenced. The sequence of the deletion junction was confirmed, and the mutant contains four additional single base substitutions (A10712G, C46053T, G52462C, C54263G) relative to the GenBank accession entry for ZoeJ, but which may have been in the precursor phage stock used for the mutagenesis. ZoeJΔ45 forms clear plaques on lawns of M. smegmatis and does not form stable lysogens (Fig. 4A). Furthermore, it remains subject to ZoeJ immunity, and does not efficiently infect either a ZoeJ lysogen, or a recombinant strain expressing ZoeJ gene 45 (Fig. 4A), although plaques likely corresponding to immunity escape mutants are observed at low frequency (Fig. 4A).
ZoeJ gene 43 codes for a tyrosine integrase that is closely related (84% amino acid identity) to the previously described CrimD integrase10. The region upstream contains the putative ZoeJ attP site, including a 23 bp common core (coordinates 33,860 – 33,882) that is identical to CrimD attP and to the putative M. smegmatis attB site, which overlaps the 3’ end of the tmRNA gene (Msmeg_2093) such that the tmRNA is predicted to be intact and functional following integration. The tmRNA gene and the attB site are well conserved and present in M. tuberculosis H37Rv (Rcnc0046). Interestingly the attP region corresponding to CrimD is constrained to the common core and 50 bp to its right that contains a putative transcriptional terminator 10. The regions flanking these are predicted to contain the arm-type integrase binding sites that are recognized by the N-terminal domain of integrase. These are not easy to predict in ZoeJ, although two direct repeats of the sequence 5’-TGTGGATnnnA located to the right of the core and one to the left of the core may correspond to arm-type binding sites. The use of the tmRNA-attB site was confirmed by PCR analysis of ZoeJ lysogens (data not shown).
RNAseq analysis of a ZoeJ lysogen shows that the repressor (45) is expressed as expected, together with the closely-linked downstream gene (44) (Fig. 5). The integrase gene (43) is expressed at a surprisingly high level as in lysogens of other mycobacteriophages it is typically expressed at only low levels if at all 22, 32. However, expression presumably extends from the host tmRNA gene, one of the most highly expressed M. smegmatis genes 33. We also observed some expression of genes 36-38 as well as genes at the right end of the genome including genes 77-86 and 89 (Fig. 5); we note that there is some variation in relative gene expression levels in independently grown lysogenic cultures, and the profile for a second culture is shown in Figure S3. The expression of these genes does not obviously reflect leaky lytic gene expression, as few RNA reads are detected for gene 67, which is the most highly expressed early lytic gene (see below). The functions of most of these lysogenically expressed genes is unknown, although we note that ZoeJ gp81 is a predicted SprT-like metalloprotease (Fig. 1). Interestingly, expression is observed from all six of the ESAS motifs that are predicted include SigA-like transcriptional promoters (Fig. 5).
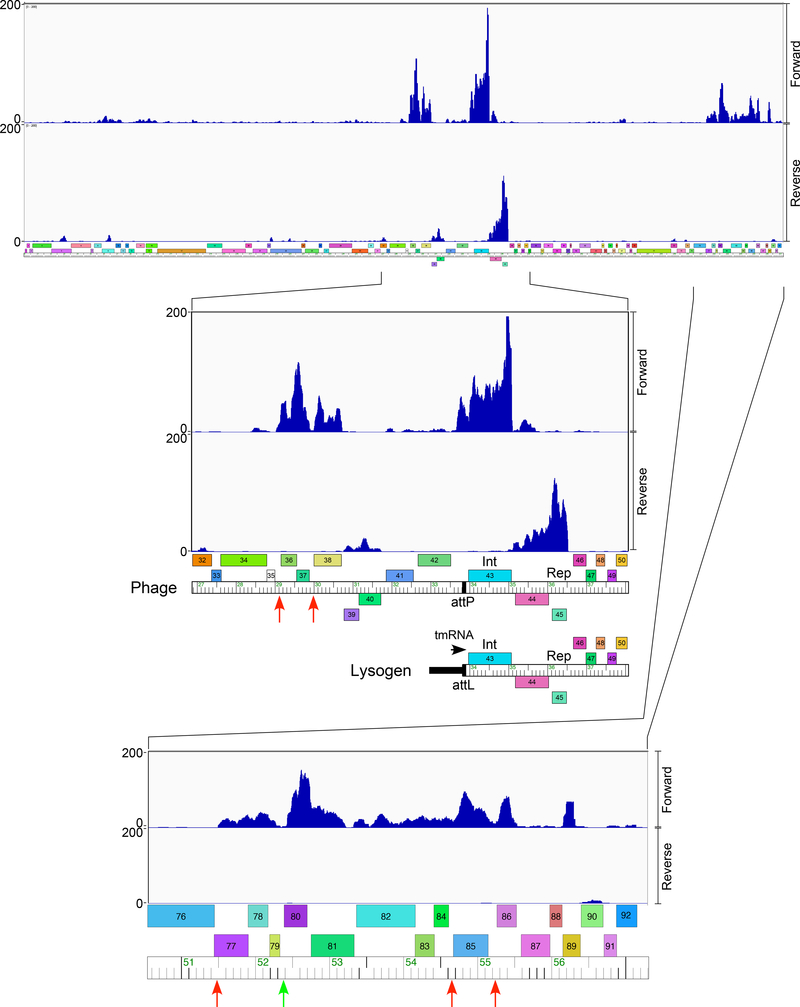
Figure 5. ZoeJ lysogenic transcription.
Lysogenic expression of ZoeJ prophage genes is shown at the top, with expanded views of the att-proximal region and the extreme genome right end are shown below. RNAseq reads are mapped to the ZoeJ prophage, but are represented on the linear viral form of the genome. The expanded view of the central genome part includes a map indicating the prophage orientation near the attL integration junction. The RNAseq reads are strand-specific and those mapping to forward and reverse DNA orientations are indicated. Locations of ESAS motifs associated with either an SAS motif or not (red and green respectively) are shown as vertical arrows. We note that although the overall expression patterns are similar in duplicate experiments with independent lysogenic cultures that there are subtle variations in relative gene expression. Data for an independently tested lysogen are shown in Figure S3.
ZoeJ lytic gene expression
To examine transcriptional patterns during ZoeJ lytic growth, RNA was isolated at 30’, 90’, 150’ and 210’ after adsorption of M. smegmatis, analyzed using RNAseq, and the sequence reads mapped to the ZoeJ and M. smegmatis genomes (Fig. 6). At 30’ after infection, ~10% of the sequence reads map to ZoeJ, and an ‘early’ pattern of gene expression is observed, including genes 39 and 40, genes 46-79, and genes 87-92 (Fig. 6). The expression level of most of the genes is quite low, as in other mycobacteriophages early lytic expression begins at a rightwards promoter in the intergenic region between the divergently transcribed repressor (ZoeJ 45) and a Cro-like protein (ZoeJ 46), and the genes at the beginning of the operon are most highly expressed22, 32, 34. Equally surprising is that gene 67 is highly expressed and is the most highly expressed early lytic gene (Fig. 6), although its function is unknown. We note that little expression of the repressor is observed, perhaps reflecting a low lysogenization frequency under these conditions, and low repressor expression relative to the lytic genes.
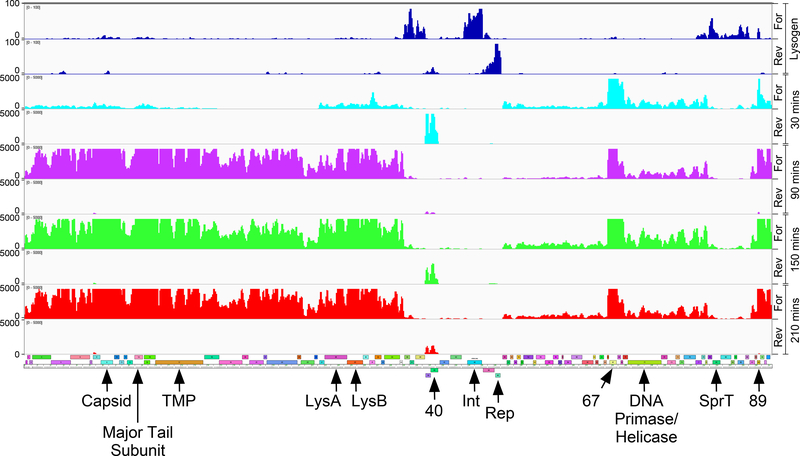
Figure 6. ZoeJ transcription in lytic growth.
RNA was isolated at 30, 90, 150, and 210 minutes (shown in aqua, purple, green, and red, respectively) after infection of M. smegmatis with ZoeJ; reads mapping to forward (For) and reverse (Rev) strands are shown as indicated. A map of the ZoeJ genome is shown below (see Fig. 1 for details). The lysogen RNAseq data is shown for comparison in blue at the top. The positions of several key genes are indicated (9, capsid subunit; 14, major tail subunit; 45, repressor; TMP, Tape Measure Protein).
At later times (90’ 120’ and 150’ after infection) the ZoeJ genome is actively expressed and ~50% of all sequence reads map to the ZoeJ genome (Fig. 6). Most of the early transcripts persist, although the 39-40 transcript diminishes somewhat at the later times (Fig. 6). A distinct ‘late’ pattern is observed with high levels of expression of the left arm genes (1-35) including the virion structural genes (1-25) and the lysis cassette (28-30) (Figs. 1, 6). The origins of these transcripts may be at the right genome end within gene 89, although we note that there are even greater abundant transcripts for the scaffold and capsid genes (8-9) as well as the major tail subunit and tail assembly chaperones (14-16) (Fig. 6), suggesting that there are additional expression signals upstream of these.
Five of the six of the ESAS sites containing putative SigA-like promoters that are active in the lysogen are down-regulated in lytic growth relative to other ZoeJ genes (Fig. 6); whether the sixth site, upstream of gene 77 is also down-regulated is unclear, as it may be transcribed with the upstream genes (Fig. 6). It therefore plausible that the ESAS motifs play a role in repressing expression of the associated genes in lytic growth, perhaps through binding of an early lytic gene product. Because most of the ESAS sites are associated with SAS-linked genes, it is also plausible that the SAS motifs are involved in regulation of lytic gene translation.
Discussion
We have described here mycobacteriophage ZoeJ, a member of Subcluster K2. It is closely related to the well-studied phage TM4 but is temperate and forms stable lysogens in M. smegmatis. Comparison with TM4 reveals the precise deletion in TM4 of 5.0 kbp relative to ZoeJ that removes the repressor and the integrase genes, and all or part of genes 40, 41, 42 and 44. ZoeJ gene 40 (and downstream gene 39) is expressed early in lytic growth but is evidently not required for lytic propagation. ZoeJ has a broad host range within the mycobacteria and can infect both fast- and slow-growing strains.
ZoeJ gene 45 codes for the immunity repressor, is expressed in lysogeny, confers superinfection immunity, and is required for lysogeny. The repressor presumably regulates an early rightwards-transcribing lytic promoter in the 45-46 intergenic region, although operator sites have not been identified and are not easily predicted bioinformatically. However, early lytic transcription is weak from this region, raising the question as to whether the repressor also regulates the highly expressed gene 67. The specificity of the repressor is of interest, in that it confers immunity to some other Subcluster K2 phages such as Milly, and also to some Subcluster K1 phages such as Adephagia and CrimD (Fig. 4). However, the ZoeJ repressor does not confer immunity to TM4, in spite of the genomic similarities between ZoeJ and TM4. Furthermore, a ZoeJ lysogen strongly inhibits TM4 infection, but with escape mutants arising at a frequency of approximately 10-4. Because this defense against TM4 infection is not repressor-mediated, it presumably involves an alternative prophage-expressed ZoeJ gene. A plausible candidate is ZoeJ gene 37, which is predicted to code for a membrane protein with three transmembrane helices. ZoeJ gp37 could thus confer homotypic exclusion, similar to that reported for HK97 gp1535.
The ZoeJ genome has a repertoire of Start Associated Sequences and Extended Start Associated Sequences that are well-conserved among all of the Cluster K phages10. The specific roles of these has remained elusive, but transcriptional analysis of ZoeJ suggests that at least some of the ESAS sites are used for promoting transcription in lysogeny and for repression of such expression during lytic growth. Although there is substantial sequence variation among the Cluster K phages, the overall genome organizations are similar, and it seems likely that the ESAS sites play similar roles throughout this phage group. Although the roles of the SAS motif is still unresolved, it is possible that they act similarly to the ESAS motifs but to downregulate translation at some time during lytic growth.
ZoeJ impressively subverts the transcriptional machinery towards its own genome such that during late lytic growth ~50% of all RNAseq reads map to the ZoeJ genome, even though it is only 1% the size of the host chromosome. ZoeJ does not code for its own RNA polymerase, so late lytic transcription must use a modified form of the host RNA polymerase. The regulatory mechanism is not known, but the RNAseq analysis suggests that late lytic transcription begins upstream of genes 89, 8, and 14. However, alignment of the putative sequences does not reveal regulatory signals that promote late transcription and further investigation is warranted.
Finally, the broad host range of ZoeJ suggests that it could have therapeutic potential for mycobacterial infections. It would be unsuitable as the wild-type virus, but the Δ45 derivative which is strictly lytic rather than temperate is a candidate. Its broad range is advantageous and facilitates the use of M. smegmatis for propagation of the phage which could be used against infections with slow-growing mycobacteria.
Supplementary Material
Genome maps of Adephagia, ZoeJ, and TM4 are shown with rulers indicating kbp markers. Pairwise nucleotide sequence similarity is shown as spectrum-colored shading between genomes; violet indicates closest similarity, and red the least similarity above a threshold E value of 10-4. The predicted genes are shown as colored boxes above or below the genome corresponding to rightwards- and leftwards-transcription, respectively. Gene names are shown within each box, and the corresponding phamily number is shown above or below each gene with the number of phamily members in parentheses. Genes in the same phamily are colored similarly, although we note that there are instances in which genes with high nucleotide sequence similarity are not necessarily grouped in the same phamily due to the pham-building methods using kclust6. The TM4 annotation reflects revisions reported previously10.
Proteins were aligned using ClustalX with absolutely conserved residues indicated with asterisks above the alignment. The position of the putative helix-turn-helix DNA recognition motif is indicated.
Data are presented as in Figure 5, except with an independently grown ZoeJ lysogen. Lysogenic expression of ZoeJ prophage genes is shown at the top, with expanded views of the att-proximal region and the extreme genome right end are shown below. RNAseq reads are mapped to the ZoeJ prophage, but are represented on the linear viral form of the genome. The expanded view of the central genome part includes a map indicating the prophage orientation near the attL integration junction. The RNAseq reads are strand-specific and those mapping to forward and reverse DNA orientations are indicated. Locations of ESAS motifs associated with either an SAS motif or not (red and green respectively) are shown as vertical arrows.
DNA substrates and oligonucleotides used in this study.
Acknowledgements
We thank William R. Jacobs and colleagues at the Albert Einstein College of Medicine for providing M. avium Va14 (O) and M. bovis BCG. We thank Welkin Pope for critical comments on the manuscript and Christian Gauthier for assistance with RNAseq analysis.
Footnotes
Research Data for this Article
The RNAseq data set is deposited in the Gene Expression Omnibus (GEO) with accession number GSE124840.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hatfull GF. Molecular Genetics of Mycobacteriophages. Microbiology Spectrum 2014;2:1–36. doi: doi: 10.1128/microbiolspec.MGM2-0032-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatfull GF. Mycobacteriophages: windows into tuberculosis. PLoS Pathog 2014;10:e1003953. doi: 10.1371/journal.ppat.1003953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing G, Evolutionary Science Sea-Phages P, Modlin RL, Hendrix RW, Hatfull GF. On the nature of mycobacteriophage diversity and host preference. Virology 2012;434:187–201. doi: 10.1016/j.virol.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell DA, Hatfull GF. PhagesDB: the actinobacteriophage database. Bioinformatics 2017;33:784–786. doi: 10.1093/bioinformatics/btw711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pope WH, Mavrich TN, Garlena RA, Guerrero-Bustamante CA, Jacobs-Sera D, Montgomery MT, Russell DA, Warner MH, Science Education Alliance-Phage Hunters Advancing G, Evolutionary S, Hatfull GF. Bacteriophages of Gordonia spp. Display a Spectrum of Diversity and Genetic Relationships. MBio 2017;8 doi: 10.1128/mBio.01069-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science, Phage Hunters Integrating Research and Education, Mycobacterial Genetics Course. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 2015;4:e06416. doi: 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A 1999;96:2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell 2003;113:171–182. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol 2002;184:4891–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pope WH, Ferreira CM, Jacobs-Sera D, Benjamin RC, Davis AJ, DeJong RJ, Elgin SCR, Guilfoile FR, Forsyth MH, Harris AD, Harvey SE, Hughes LE, Hynes PM, Jackson AS, Jalal MD, MacMurray EA, Manley CM, McDonough MJ, Mosier JL, Osterbann LJ, Rabinowitz HS, Rhyan CN, Russell DA, Saha MS, Shaffer CD, Simon SE, Sims EF, Tovar IG, Weisser EG, Wertz JT, Weston-Hafer KA, Williamson KE, Zhang B, Cresawn SG, Jain P, Piuri M, Jacobs WR Jr., Hendrix RW, Hatfull GF. Cluster K Mycobacteriophages: Insights into the Evolutionary Origins of Mycobacteriophage TM4. PLoS ONE 2011;6:e26750. doi: 10.1371/journal.pone.002675011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs WR Jr., Tuckman M, Bloom BR. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 1987;327:532–535. [DOI] [PubMed] [Google Scholar]
- 12.Ford ME, Stenstrom C, Hendrix RW, Hatfull GF. Mycobacteriophage TM4: genome structure and gene expression. Tuber Lung Dis 1998;79:63–73. doi: 10.1054/tuld.1998.0007 S0962-8479(98)90007-7 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, Bloom BR, Hatfull GF, Jacobs WR Jr. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 1997;94:10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A 2001;98:12712–12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardarov S, Bardarov Jr S Jr., Pavelka Jr MS Jr., Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs Jr WR Jr. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 2002;148:3007–3017. [DOI] [PubMed] [Google Scholar]
- 16.Tufariello JM, Malek AA, Vilcheze C, Cole LE, Ratner HK, Gonzalez PA, Jain P, Hatfull GF, Larsen MH, Jacobs WR Jr. Enhanced specialized transduction using recombineering in Mycobacterium tuberculosis. MBio 2014;5:e01179–01114. doi: 10.1128/mBio.01179-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain P, Weinrick BC, Kalivoda EJ, Yang H, Munsamy V, Vilcheze C, Weisbrod TR, Larsen MH, O’Donnell MR, Pym A, Jacobs WR Jr., Dual-Reporter Mycobacteriophages (Phi2DRMs) Reveal Preexisting Mycobacterium tuberculosis Persistent Cells in Human Sputum. MBio 2016;7 doi: 10.1128/mBio.01023-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain P, Hartman TE, Eisenberg N, O’Donnell MR, Kriakov J, Govender K, Makume M, Thaler DS, Hatfull GF, Sturm AW, Larsen MH, Moodley P, Jacobs WR Jr. phi(2)GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J Clin Microbiol 2012;50:1362–1369. doi: JCM.06192–11 [pii] 10.1128/JCM.06192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs WR Jr., Barletta RG, Udani R, Chan J, Kalkut G, Sosne G, Kieser T, Sarkis GJ, Hatfull GF, Bloom BR. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 1993;260:819–822. [DOI] [PubMed] [Google Scholar]
- 20.Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, Jacobs WR Jr., Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 2006;24:6309–6320. [DOI] [PubMed] [Google Scholar]
- 21.Falkinham JO 3rd. Factors influencing the chlorine susceptibility of Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum. Appl Environ Microbiol 2003;69:5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedrick RM, Jacobs-Sera D, Bustamante CA, Garlena RA, Mavrich TN, Pope WH, Reyes JC, Russell DA, Adair T, Alvey R, Bonilla JA, Bricker JS, Brown BR, Byrnes D, Cresawn SG, Davis WB, Dickson LA, Edgington NP, Findley AM, Golebiewska U, Grose JH, Hayes CF, Hughes LE, Hutchison KW, Isern S, Johnson AA, Kenna MA, Klyczek KK, Mageeney CM, Michael SF, Molloy SD, Montgomery MT, Neitzel J, Page ST, Pizzorno MC, Poxleitner MK, Rinehart CA, Robinson CJ, Rubin MR, Teyim JN, Vazquez E, Ware VC, Washington J, Hatfull GF. Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol 2017;2:16251. doi: 10.1038/nmicrobiol.2016.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE 2008;3:e3957. doi: 10.1371/journal.pone.0003957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nature Methods 2007;4:147–152. [DOI] [PubMed] [Google Scholar]
- 25.Russell DA. Sequencing, Assembling, and Finishing Complete Bacteriophage Genomes. Methods Mol Biol 2018;1681:109–125. doi: 10.1007/978-1-4939-7343-9_9 [DOI] [PubMed] [Google Scholar]
- 26.Lee MH, Pascopella L, Jacobs WR Jr., Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A 1991;88:3111–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goulding CW, Parseghian A, Sawaya MR, Cascio D, Apostol MI, Gennaro ML, Eisenberg D. Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at 1.5-A resolution. Protein Sci 2002;11:2887–2893. doi: 10.1110/ps.0219002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz MA, Lee BW, Dillon BJ, Harth G. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 1995;92:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sotello D, Hata DJ, Reza M, Satyanarayana R, Arunthari V, Bosch W. Disseminated Mycobacterium interjectum Infection with Bacteremia, Hepatic and Pulmonary Involvement Associated with a Long-Term Catheter Infection. Case Rep Infect Dis 2017;2017:6958204. doi: 10.1155/2017/6958204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daley CL. Mycobacterium avium Complex Disease. Microbiol Spectr 2017;5 doi: 10.1128/microbiolspec.TNMI7-0045-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrova ZO, Broussard GW, Hatfull GF. Mycobacteriophage-repressor-mediated immunity as a selectable genetic marker: Adephagia and BPs repressor selection. Microbiology 2015;161:1539–1551. doi: 10.1099/mic.0.000120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dedrick RM, Mavrich TN, Ng WL, Hatfull GF. Expression and evolutionary patterns of mycobacteriophage D29 and its temperate close relatives. BMC Microbiol 2017;17:225. doi: 10.1186/s12866-017-1131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape. Plos Genetics 2015;11 doi: 10.1371/journal.pgen.1005641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dedrick RM, Marinelli LJ, Newton GL, Pogliano K, Pogliano J, Hatfull GF. Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol Microbiol 2013;88:577–589. doi: 10.1111/mmi.12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cumby N, Edwards AM, Davidson AR, Maxwell KL. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol 2012;194:5012–5019. doi: 10.1128/JB.00843-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 2011;12:395. doi: 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome maps of Adephagia, ZoeJ, and TM4 are shown with rulers indicating kbp markers. Pairwise nucleotide sequence similarity is shown as spectrum-colored shading between genomes; violet indicates closest similarity, and red the least similarity above a threshold E value of 10-4. The predicted genes are shown as colored boxes above or below the genome corresponding to rightwards- and leftwards-transcription, respectively. Gene names are shown within each box, and the corresponding phamily number is shown above or below each gene with the number of phamily members in parentheses. Genes in the same phamily are colored similarly, although we note that there are instances in which genes with high nucleotide sequence similarity are not necessarily grouped in the same phamily due to the pham-building methods using kclust6. The TM4 annotation reflects revisions reported previously10.
Proteins were aligned using ClustalX with absolutely conserved residues indicated with asterisks above the alignment. The position of the putative helix-turn-helix DNA recognition motif is indicated.
Data are presented as in Figure 5, except with an independently grown ZoeJ lysogen. Lysogenic expression of ZoeJ prophage genes is shown at the top, with expanded views of the att-proximal region and the extreme genome right end are shown below. RNAseq reads are mapped to the ZoeJ prophage, but are represented on the linear viral form of the genome. The expanded view of the central genome part includes a map indicating the prophage orientation near the attL integration junction. The RNAseq reads are strand-specific and those mapping to forward and reverse DNA orientations are indicated. Locations of ESAS motifs associated with either an SAS motif or not (red and green respectively) are shown as vertical arrows.
DNA substrates and oligonucleotides used in this study.