Abstract
Agarum clathratum, a brown macroalgae species, has recently become a serious environmental problem on the coasts of Korea. In an effort to solve this problem, fungal diversity associated with decaying A. clathratum was investigated and related β-glucosidase and endoglucanase activities were described. A total of 233 fungal strains were isolated from A. clathratum at 15 sites and identified 89 species based on morphology and a multigene analysis using the internal transcribed spacer region (ITS) and protein-coding genes including actin (act), β-tubulin (benA), calmodulin (CaM), and translation elongation factor (tef1). Acremonium, Corollospora, and Penicillium were the dominant genera, and Acremonium fuci and Corollospora gracilis were the dominant species. Fifty-one species exhibited cellulase activity, with A. fuci, Alfaria terrestris, Hypoxylon perforatum, P. madriti, and Pleosporales sp. Five showing the highest enzyme activities. Further enzyme quantification confirmed that these species had higher cellulase activity than P. crysogenum, a fungal species described in previous studies. This study lays the groundwork for bioremediation using fungi to remove decaying seaweed from populated areas and provides important background for potential industrial applications of environmentally friendly processes.
Keywords: β-glucosidase, cellulase activity, endoglucanase, fungal enzyme, seaweed
1. Introduction
Agarum clathratum, a brown macroalgal species, is generally found on rocks in the low intertidal to sub-tidal zones. It is widely distributed in Alaska and East Asia, including Hokkaido in Japan, the Kuril Islands, and northern area of Korea [1]. Brown algae, including A. clathratum, are composed of 30–50% carbohydrates as cellulose which is the primary component of the cell wall [2,3]. Interest in A. clathratum and other seaweed species has risen recently due to its immunomodulatory and antioxidant activities [4–6]. However, increasing masses of seaweed waste have been reported worldwide due to climate change and eutrophication by fertilizer run-off [7,8]. A large amount of seaweed wastes are deposited on the shores of Korea peninsula [9], and A. clathratum mass in particular has accumulated on the northeast coast of Korea causing serious environmental problems. Despite the severity of this problem, no effective method has been developed despite the severity of this problem.
Cellulose has previously been degraded using costly and energy demanding industrial methods such as chemical hydrolysis under high pressure and temperatures [10]. Hydrolysis of cellulose using microbial enzymes requires less energy and is less expensive and more environment-friendly process than industrial processes [11]. Microbial enzymes have been studied and applied in various industries such as bio-energy production from seaweed [12], mostly focused on bacterial cellulases. Numerous fungi have been reported from a wide range of brown seaweed, many with abundant cellulase, protease, and xylanase activities [13–15]. However, there is no study about fungal diversity and enzymes activity associated with A. clathratum.
Marine fungi interact with marine organisms as parasites, symbionts, or decomposers and are crucial for organic and inorganic nutrient cycling in marine ecosystems [13,16]. Approximately 1500 species of marine fungi have been reported worldwide [13], and are found in various substrates including seaweed, plants, sediments, and wood [16–18]. Marine fungi are categorized into two groups depending on their origin and salinity tolerance, obligate marine fungi from the marine origins and facultative fungi from the terrestrial environments [19]. The number of fungi associated with seaweed accounts for one-third of reported marine fungi [13].
Historically, morphological characters have been used to identify fungi; however, they often lead to misidentification because many intraspecific fungal characters are variable depending on environmental conditions [20]. Thus, current identification methods employ both morphology and molecular markers. Among these molecular markers, the internal transcribed spacer (ITS) region has been widely used as a fungal barcode marker [21], and increased resolution can be attained using a multi-locus approach. Specifically, the actin locus (act) has been used for Cladosporium, β-tubulin (benA) for Penicillium, calmodulin (CaM) for Aspergillus, and translation elongation factor (tef1) for Fusarium and Trichoderma [20,22].
In this study, A. clathratum samples were collected from fifteen different sites along the northeast coast of Korea and fungal diversity associated with A. clathratum degradation was investigated. In addition, extracellular enzymes associated with fungi, including β-glucosidase and endoglucanase, were evaluated in order to discover potential candidates for the treatment of seaweed waste.
2. Materials and methods
2.1. Sampling and fungal isolation
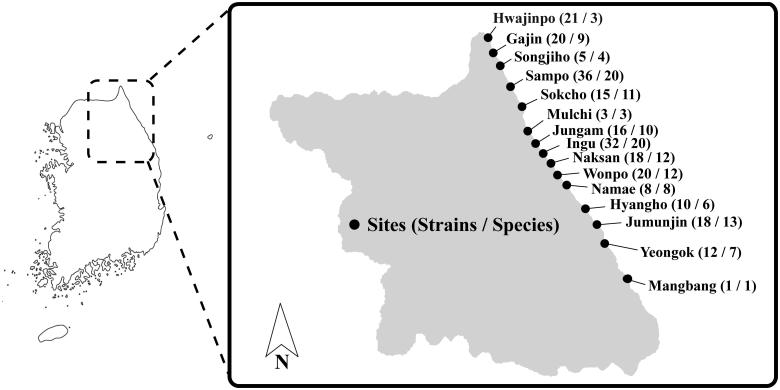
Sampling was conducted from accumulated A. clathratum from fifteen different sites along the eastern coast of Korea in August 2015 (Figure 1). Before fungal isolation, each sample was washed with sterilized Artificial Sea Water (ASW) [23] to remove any debris on the surface. Sections of each sample (5 mm in diameter) were placed on three different media plates containing artificial seawater: potato dextrose agar (PDA; BD-Difco, Sparks, MD) and glucose yeast extract agar (GYA; 1 g/L glucose, 0.1 g/L yeast extract, 0.5 g/L peptone, and 15 g/L agar), and dichloran rose bengal chloramphenicol agar (DRBC; BD-Difco). All plates were incubated at 25°C and isolates with distinguishable morphology were transferred to a new PDA plate. Each pure culture was stored in 20% glycerol at –80°C at the Seoul National University Fungus Collection (SFC) or Marine Bio-Resource Information System (MRS).
Figure 1.
Map showing the location of sampling sites for A. clathratum collected along eastern coastline of Korea. Numbers in the parentheses for each site indicate strains and species isolated from decaying A. clathratum.
2.2. Molecular identification procedures
Fungal isolates were grouped based on their morphological characteristics and several representative strains were chosen from each group for molecular identification. DNA was extracted using a cetyltrimethylammonium bromide (CTAB) protocol described by Rogers and Bendich [24].
PCR was conducted in two steps. First, the ITS region was amplified from all representative strains. Then, different protein-coding genes (act, benA, CaM, and tef1) were used to identify strains to the species level for the genera Cladosporium (act), Penicillium (benA), Aspergillus (CaM), Fusarium (tef1), and Trichoderma (tef1). Each PCR was performed using AccuPower® PCR PreMix (Bioneer, Daejeon, Korea) in a final volume of 20 μl, containing 10 pmol of each primer and 10 ng of gDNA. The PCR amplification of ITS, act, benA, CaM, and tef1 were performed using the primers ITS1F/ITS4 [25], ACT-512F/ACT-783R [26], Bt2a/Bt2b [27], CF1/CF4 [22], and EF1/EF2 [28], respectively, with C1000 Thermal Cycler (Bio-Rad, Hercules, CA) as described by Park et al. [29]. PCR products were purified using ExpinTM PCR Purification Kit (Geneall Biotechnol., Seoul, Korea) following the manufacturer's instruction. The purified amplicons were sequenced using corresponding PCR primers by Macrogen (Seoul, Korea) in both forward and reverse directions using ABI Prism 3730 genetic analyzer (Life Technol., Gaithersburg, MD).
Sequences were assembled, proofread, edited, and aligned using MEGA v.5 [30] and were deposited in GenBank (Table 1). For multiple sequence alignments, MAFFT v.7 [31] was used, and each sequence was checked and adjusted manually. After alignment, maximum likelihood (ML) phylogenetic trees were constructed. The phylogenetic trees were generated using RAxML 8.0.2 [32] and the GTR + GAMMA model of evolution with 1000 bootstrap replicates.
Table 1.
GenBank accession numbers and clear zones of isolated strains from Agarum clathratum.
| Representative strain | No. of strain | GenBank accession number |
Top Scoring BLAST Mach in GenBank | Clear zone (mm)
a
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | ACT | BenA | CaM | tef1 | GA | EA | |||
| SFC102468 | 34 | MH374541 | Acremonium fuci | 2.9 | 9 | ||||
| SFC102380 | 3 | MH374571 | Alfaria terrestris | 14 | 10 | ||||
| SFC102269 | 1 | MH374617 | Alternaria broccoli-italicae | – | – | ||||
| SFC102318 | 1 | MH374593 | Arthrinium malaysianum | – | – | ||||
| SFC102355 | 1 | MH374579 | MH367021 | Aspergillus chevalieri | – | – | |||
| SFC102407 | 4 | MH374559 | MH367023 | Aspergillus costaricaensis | 1 | 6 | |||
| SFC102289 | 3 | MH374606 | MH367020 | Aspergillus fumigatus | – | – | |||
| SFC102359 | 1 | MH374578 | MH367022 | Aspergillus insulicola | 5 | 5 | |||
| SFC102419 | 1 | MH374555 | MH367024 | Aspergillus terreus | 1.5 | 4.5 | |||
| SFC102281 | 1 | MH374611 | MH367019 | Aspergillus welwitschiae | – | – | |||
| SFC102362 | 3 | MH374575 | Asteromyces cruciatus | – | – | ||||
| SFC102361 | 1 | MH374576 | Chaetomium globosum | – | – | ||||
| SFC102453 | 1 | MH374547 | Chloridium sp. | 4.5 | 3.5 | ||||
| SFC102433 | 5 | MH374553 | MH367029 | Cladosporium cladosporioides | 1.5 | – | |||
| SFC102263 | 1 | MH374620 | MH367026 | Cladosporium grevilleae | – | 4 | |||
| SFC102255 | 1 | MH374622 | MH367025 | Cladosporium perangustum | – | – | |||
| SFC102395 | 3 | MH374563 | MH367028 | Cladosporium rectoides | 3 | 1 | |||
| SFC102352 | 1 | MH374581 | MH367027 | Cladosporium sphaerospermum | – | – | |||
| SFC102394 | 11 | MH374564 | Clonostachys miodochialis | – | – | ||||
| SFC102311 | 1 | MH374597 | Clonostachys rosea | 5 | 5 | ||||
| SFC102291 | 2 | MH374605 | Coniella quercicola | – | – | ||||
| SFC102400 | 26 | MH374560 | Corollospora gracilis | 4.5 | 5 | ||||
| SFC102442 | 2 | MH374552 | Corollospora maritima | 1 | – | ||||
| SFC102459 | 1 | MH374544 | Corollospora sp. | – | 4 | ||||
| SFC102272 | 1 | MH374616 | Diaporthe sp. | – | – | ||||
| SFC102458 | 1 | MH374545 | Didymella bellidis | 1 | 1 | ||||
| SFC102377 | 1 | MH374572 | Didymella pomorum | – | – | ||||
| SFC102337 | 2 | MH374586 | Didymella sp. | – | – | ||||
| SFC102399 | 1 | MH374561 | Discosia artocreas | 5 | 4 | ||||
| SFC102353 | 1 | MH374580 | Epicoccum sorghinum | 1 | – | ||||
| SFC102301 | 1 | MH374600 | Epicoccum sp. | 1 | – | ||||
| SFC102238 | 1 | MH374628 | Eutypella scoparia | 7 | 3 | ||||
| SFC102386 | 4 | MH374567 | MH367033 | Fusairum cf. equiseti | – | – | |||
| SFC102286 | 1 | MH374609 | MH367031 | Fusairum acuminatum | 1.5 | 1.5 | |||
| SFC102314 | 2 | MH374596 | MH367032 | Fusairum graminearum | – | – | |||
| SFC102248 | 1 | MH374626 | MH367030 | Fusarium sp. | – | – | |||
| SFC102330 | 1 | MH374590 | Galactomyces sp. | – | – | ||||
| SFC102393 | 1 | MH374565 | Hypocrealessp. | 2 | – | ||||
| SFC102449 | 4 | MH374551 | Hypoxylon perfoatum | 9 | 7.5 | ||||
| SFC102275 | 1 | MH374614 | Lophiostoma sp. | – | – | ||||
| SFC102294 | 1 | MH374602 | Myrmecridium schulzeri | – | – | ||||
| SFC102316 | 3 | MH374594 | Neopestalotiopsis clavispora | – | – | ||||
| SFC102383 | 1 | MH374569 | Nigrospora oryzae | – | – | ||||
| SFC102348 | 1 | MH374582 | Paraconiothyrium fuckelii | – | 2 | ||||
| SFC102409 | 3 | MH374558 | Paradendryphiella arenariae | – | 1.5 | ||||
| SFC102461 | 5 | MH374543 | Paraphaeosphaeria sp. | – | – | ||||
| SFC102388 | 3 | MH374566 | Paraphaeosphaeria sporulosa | – | – | ||||
| SFC102385 | 1 | MH374568 | MH367039 | Penicillium antarcticum | – | 2 | |||
| SFC102451 | 1 | MH374549 | MH367035 | Penicillium aurantiogriseum | – | 5 | |||
| SFC102254 | 1 | MH374623 | MH367045 | Penicillium bialowiezense | 8 | 5.5 | |||
| SFC102288 | 1 | MH374607 | MH367043 | Penicillium bilaiae | – | – | |||
| SFC102305 | 8 | MH374599 | MH367042 | Penicillium citrinum | 2 | 2.5 | |||
| SFC102287 | 1 | MH374608 | MH367044 | Penicillium cremeogriseum | – | – | |||
| SFC102320 | 2 | MH374592 | MH367041 | Penicillium daejeonium | 1 | 1 | |||
| SFC102452 | 5 | MH374548 | MH367034 | Penicillium guanacastense | 2 | 2 | |||
| SFC102420 | 2 | MH374554 | MH367037 | Penicillium madriti | 13.5 | 12.5 | |||
| SFC102415 | 1 | MH374557 | MH367038 | Penicillium oxalicum | 3 | 2.5 | |||
| SFC102343 | 3 | MH374583 | MH367040 | Penicillium roseomaculatum | – | – | |||
| SFC102243 | 2 | MH374627 | MH367046 | Penicillium spinolusum | – | – | |||
| SFC102450 | 3 | MH374550 | MH367036 | Penicillium virgatum | 2.5 | 3.5 | |||
| SFC102471 | 1 | MH374540 | Pestalotiopsis lespedezae | 3 | 1 | ||||
| SFC102282 | 4 | MH374610 | Pestalotiopsis sp. | – | 2 | ||||
| SFC102465 | 3 | MH374542 | Phaeosphaeria oryzae | – | 2 | ||||
| SFC102360 | 1 | MH374577 | Pleosporales sp. 1 | 8 | 6 | ||||
| SFC102369 | 5 | MH374573 | Pleosporales sp. 2 | 10 | 5 | ||||
| SFC102333 | 2 | MH374589 | Pleosporales sp. 3 | 7 | 1 | ||||
| SFC102334 | 1 | MH374588 | Pleosporales sp. 4 | 1 | 3 | ||||
| SFC102342 | 1 | MH374584 | Pleosporales sp. 5 | 11.5 | 6.5 | ||||
| SFC102260 | 1 | MH374621 | Pleosporales sp. 6 | – | – | ||||
| SFC102306 | 1 | MH374598 | Pleosporales sp. 7 | 1 | 3 | ||||
| SFC102397 | 4 | MH374562 | Pleosporales sp. 8 | – | – | ||||
| SFC102457 | 1 | MH374546 | Pleosporales sp. 9 | 2 | 2 | ||||
| SFC102382 | 1 | MH374570 | Pleosporales sp. 10 | – | – | ||||
| SFC102267 | 1 | MH374619 | Porostereum spadiceum | 1 | – | ||||
| SFC102329 | 2 | MH374591 | Roussoella sp. | 5 | 5 | ||||
| SFC102268 | 1 | MH374618 | Schizophylum commune | 1.5 | – | ||||
| SFC102315 | 1 | MH374595 | Septoriellahubertusii | 4 | 3 | ||||
| SFC102335 | 1 | MH374587 | Sesquicillium microsporum | 8.5 | 5.5 | ||||
| SFC102276 | 1 | MH374613 | Stagonosporopsis cucurbitacearum | – | – | ||||
| SFC102417 | 1 | MH374556 | Stemphylium solani | 7 | 5.5 | ||||
| SFC102277 | 3 | MH374612 | Stereum sp. | – | – | ||||
| SFC102341 | 1 | MH374585 | Teichospora sp. | – | – | ||||
| SFC102274 | 2 | MH374615 | Trametes hirsuta | – | – | ||||
| SFC102367 | 1 | MH374574 | MH367051 | Trichoderma atroviride | – | – | |||
| SFC102249 | 2 | MH374625 | MH367047 | Trichoderma guizhouense | – | – | |||
| SFC102293 | 1 | MH374603 | MH367050 | Trichoderma sp. 1 | – | – | |||
| SFC102292 | 1 | MH374604 | MH367049 | Trichoderma sp. 2 | – | – | |||
| SFC102252 | 6 | MH374624 | MH367048 | Trichoderma sp. 3 | – | – | |||
| SFC102299 | 1 | MH374601 | Zymoseptoria verkleyi | 1 | 2 | ||||
The number in the last column indicates the diameter of the clear zone for each media plate.
GA: β-glucosidase; EA: Endoglucanase.
2.3. Enzyme assays
The plate screening assays for cellulase activity were conducted for representative strains from each species (Table 1). Mandel’s medium was used supplemented with 0.5% D-cellobiose (CB; Sigma-Aldrich, St. Louis, MO) for β-glucosidase and 1% carboxymethylcellulose (CMC; Sigma-Aldrich) for endoglucanase [33]. The five species that showed the highest activity from each enzyme assay were chosen from the plate screening assays and then all strains of these five species were further screened for enzyme activity to select the strain with highest enzyme activity among them.
Fungal strains with the highest level of enzyme activity were incubated in a shaking bath at 25°C with minimal liquid media (0.3 g/L urea, 1.4 g/L KH2PO4, 2.0/L g of (NH4)2SO4, 0.3 g/L CaCl2, 0.3 g/L MgSO4, 0.25 g/L yeast extract, 0.75 g/L peptone, 5 mg/L FeSO4·7H2O, 36 mg/L COCl2·6H2O, 1.8 mg/L MnSO4·H2O, and 2.5 mg/L ZnSO4·7H2O) and ground A. clathratum as a carbon source. After a week of incubation, culture broths were collected by filtration and their cellulase activity was measured by micro-assay based on the dinitrosalicylic acid (DNS) method [34]. The cellulase activity was compared to that of P. crysogenum (FU42), which has shown high cellulase activity [33].
3. Results
3.1. Identification and diversity
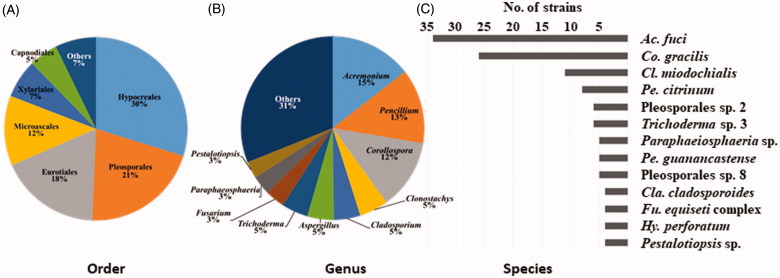
A total of 233 fungal strains were isolated from decaying A. clathratum at 15 sites, and 89 fungal taxa were determined by morphological characters and ITS analysis. Next, the strains were identified using additional genetic markers: act, CaM, benA, or tef1 (Table 1). The use of an additional locus (in conjunction with the ITS), significantly improved species identification. Sixty-two species were identified to the species level, while 27 remained unidentified species due to ambiguous phylogenetic relationships (Figure 2). The species were grouped into two phyla, five classes, 14 orders, 42 genera, and 89 species (Figure 2, Table 1). Ninety-seven percent of strains (226 strains of 85 species) were identified as Ascomycota and 3% (7 strains of 4 species) as Basidiomycota. At the order level, 30% of species (n = 70) belonged to the Hypocreales, 21% to the Pleosporales (n = 49), 18% to the Eurotiales (n = 42), 12% to the Microscales (n = 29), and 7% to the Xylariales (n = 14). At the genus level, over one-third of strains were represented by the Acremonium (15%), Penicillium (13%), and Corollospora (12%) (Figure 2). The genus Penicillium was represented by 13 species, while Aspergillus and Cladosporium were represented by 5 species. Acremonium fuci was the most dominant species (34 strains) and was found in most sites, followed by Corollospora gracilis (26 strains), Clonostachys miodochialis (11 strains), and P. citrinum (8 strains) (Figure 2). Fungal abundance and diversity varied depending on the sampling site. The highest number of strains was recovered at Sampo, while only one strain was found at Mangbang (Figure 1).
Figure 2.
Composition of the dominant fungi isolated from A. clathratum at the order level (A); at genus level (B); and species level (C).
3.2. Enzyme activity
Of 89 species, 49 exhibited cellulase activity: 41 species had β-glucosidase activity and 42 species had endoglucanase activity. The highest β-glucosidase activity was observed in five species: A. terrestris, P. madriti, Pleosporales sp. 5, H. perforatum, and Sesquicillium microsporum. The highest endoglucanase activity was observed in A. fuci, A. terrestris, H. perforatum, P. madriti, and Pleosporales sp. 5 (Table 1). Considering the isolation frequency and enzyme activity, five species were selected for further experiments; A. fuci, A. terrestris, H. perforatum, P. madriti, and Pleosporales sp. 5.
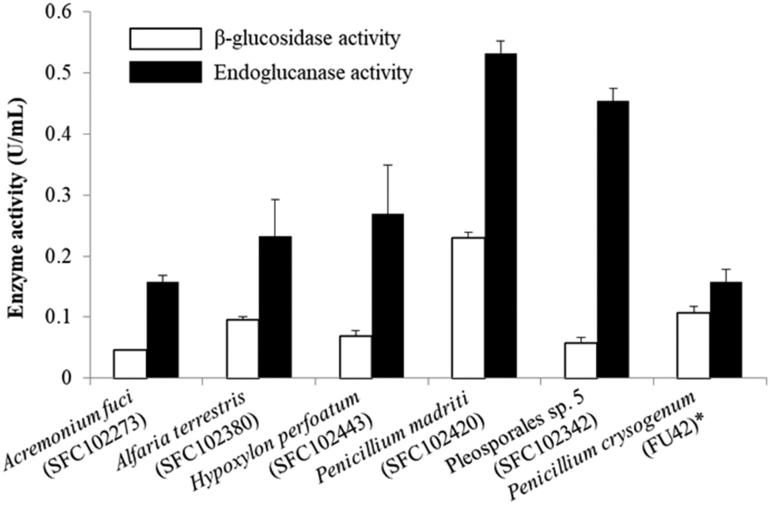
All strains were screened in order to choose strains with the highest enzyme activity. These strains were A. fuci (SFC102273; SFC20190110-M01; MRS002000115463), A. terrestris (SFC102380; SFC20190110-M02; MRS002000115464), H. perforatum (SFC102443; SFC20161014-M23; MRS002000066796), P. madriti (SFC102420; SFC20160317-M24; MRS002000066660) and Pleosporales sp. 5 (SFC102342; SCF20190110-M03; MRS002000115465) (Table S1). These selected fungal strains were then measured for cellulose enzyme activity with A. clathratum as a substrate. Endoglucanase activity from selected strains was higher than P. crysogenum (FU42). Penicillium madriti had the highest endoglucanase activity, which was approximately three times that of P. crysogenum. Most strains, however, had similar, or slightly less, β-glucosidase activity as P. crysogenum, with the exception of P. madriti (SFC102420), which had approximately twice as much higher β-glucosidase activity as P. crysogenum (Figure 3).
Figure 3.
The enzyme quantification comparison of selected species when A. clathratum was given. ‘*’ indicates positive control which showed good fungal enzyme activity in the previous study [33].
4. Discussion
Macroalgae is composed of large amounts of polysaccharides, primarily cellulose [35]. Marine fungi acquire nutrients from various organisms such as algae, sponges, and mangroves [13,16], and play a crucial role in ecosystem nutrient cycling by converting carbohydrate polymers (e.g., cellulose) to monosaccharides, which are easily metabolized by myriad organisms [13,16,36]. While macroalgae are a primary source of substrate for marine fungi [13,16], most studies have focused on their ecological role as symbionts within living algae, rather than as decomposers [37,38].
4.1. Diversity of fungi associated with A. clathratum
High fungal diversity (89 species) was detected from a single substrate, decaying A. clathratum, on the east coast of Korea. The recovery of such diversity is likely the result of using three different media used for isolation as well as a multi-locus molecular approach (ITS, ITS, act, benA, CaM, and tef1). The majority of species were classified into the phylum Ascomycota, which is consistent with a previous study [39]. However, four Basidiomycota wood decay fungi were also isolated. These fungi are commonly found in terrestrial environments, especially forests, thus, these species were likely opportunistic fungi of terrestrial origin. Similar results have been reported from intertidal zones and marine sediments [40].
The order Pleosporales is one of the most dominant fungal groups in marine environments, while species in the Hypocreales are rare in marine habitats, with the exception of the family Bionectriaceae, in which the Acremonium belong [13]. In our study, Hypocreales species, including A. fuci, were abundant. Within the order Pleosporales, several unidentified species were detected. In general, species in this order have similar morphological features and molecular markers for this order have not been developed, thus, many Pleosporales species remain unidentified [41,42].
A diverse array of species of Penicillium and Aspergillus were isolated from decaying A. clathratum. Although these species are also found in terrestrial environments, many species in these genera have been reported from marine environments such as macroalgae, coral, and sea sands [13,39]. In general, species diversity in these genera is much higher in brown algae than either red or green algae due to different cellulose components [13,36,38,43,44]. Many Penicillium species, in particular, have been reported in macroalgae including A. clathratum [15,44].
Acremonium fuci and C. gracilis, which are obligate marine fungi, accounted for most isolates of Acremonium and Corollospora (Figure 2). Acremonium separated into two main clades based on the ITS data: one clade likely had a terrestrial origin, while the other clade likely had a marine origin [45]. Specifically, A. fuci belongs to a marine-derived clade and is commonly isolated from brown seaweed [45]. Corollospora species are commonly found in sand, shell fragments, and algal thalli as obligate marine fungi [19,46,47], and species in this genus are produce antibacterial metabolites called corollosporine [48]. For example, C. gracilis has modified ascomycetous pores that can control the flow of seawater, a likely adaptation to aquatic habitats [13,47].
4.2. Fungal enzyme activity
Identifying fungi with high cellulolytic activity is the first step toward the application of these fungi to the removal of decaying seaweed in marine environments, in particular, populated shore areas and beaches. Some fungal spores of terrestrial origin that were isolated in this study may exhibit salt-resistance. Among these strains, five species were selected based on their high cellulase activity relative to other species. Species in the Pleosporales, Eurotiales, and Hypocreales degrade cellulose, and species in the genera Aspergillus and Penicillium are especially efficient at cellulose degradation [13].
When A. clathratum was chosen as a substrate, endoglucanase was higher than β-glucosidase activity and all species exhibited higher cellulase activity compared to P. crysogenum (Figure 3). Secretion of cellulolytic enzymes is likely influenced by the proportion of cellulose components but is also induced or repressed by other enzymes. For example, β-glucosidase is known to induce endoglucanase synthesis [49,50]. Thus, increased endoglucanase secretion of fungi associated with A. clathratum is expected compared to fungi provided with only a single carbon source. These cellulolytic enzymes are commonly found in other fungi from different substrates such as sediments and sponges, but they exhibited different enzyme activity depending on the substrate [51]. Both β-glucosidase and endoglucanase are used in the paper and detergent industries, however, both rely on massive amounts of water during cellulose hydrolysis and rely less on enzymatic catalysis of the natural hydrolytic process [52,53]. Thus, in addition to bioremediation using natural fungi, industrial applications of the fungi and their enzymes described here hold potential for more environmentally friendly manufacturing methods.
In this study, we found no significant relationship between fungal dominance and enzyme activity. This discordance in marine environments between microbial dominance and biological activity has previously been reported [54–56]. According to these previous studies, secondary metabolites and enzymes such as tannase were active in facultative fungi despite their lower abundance [54,55]. In nature, enzymatic activity of fungi is influenced by several factors including interactions with other microbial communities, the availability of organic matter, and other environmental factors [56]. Facultative fungi may produce increased metabolites and enzymes relative to obligate marine fungi in order to adapt to extreme environments [54,55].
Marine fungi have received increased attention in recent years, especially fungi associated with seaweed. These fungi play an important role in marine ecosystems as decomposers, and the use of fungal cellulases is potentially an important method with which combat environmental problems caused by seaweed waste. In this study, 89 fungal species were identified from 233 strains associated with the macroalgae, A. clathratum. Enzyme activity of ∼50% of the isolated strains exhibited β-glucosidase and endoglucanase activity. We expect that this study will provide critical, basic information regarding the fungi associated with A. clathratum decay in nature and that the enzymes produced by selected fungi have potential industrial applications.
Supplementary Material
Funding Statement
This research was supported by the Marine Biotechnology Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) [No. 20170431].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1. Kang JW. The geographical distribution of marine algae in Korea. Bull Pusan Fish Coll. 1966;7:1–25. [Google Scholar]
- 2. Usov AI, Smirnova GP, Klochkova NG. Polysaccharides of algae: polysaccharide composition of several brown algae from Kamchataka. Russ J Bioorganic Chem. 2001;27:395–399. [DOI] [PubMed] [Google Scholar]
- 3. Kim EJ, Fathoni A, Jeong G-T, et al. . Microbacterium oxydans: a novel alginate- and laminarin – degrading bacterium for the reutilization of brown seaweed waste. J Environ Manage. 2013;130:153–159. [DOI] [PubMed] [Google Scholar]
- 4. Jeon YE, Yin X, Lim SS. Antioxidant activities and acetylcholinesterase inhibitory activities from seaweed extracts. J Food Science Nutr. 2012;41:443–449. [Google Scholar]
- 5. Park SJ, Min KJ, Park TG. Nutritional characteristics and screening of biological activity of Agarum cribrosum . J Food Science Nutr. 2012;25:842–849. [Google Scholar]
- 6. Cho ML, Lee DJ, Kim JK, et al. . Molecular characterization and immunomodulatory activity of sulfated fucans from Agarum cribrosum . Carbohydr Polym. 2014;113:507–514. [DOI] [PubMed] [Google Scholar]
- 7. Lapointe BE, Bedford BJ. Drift rhodophyte blooms emerge in Lee County: Florida: USA: evidence of escalating coastal eutrophication. Harmful Algae. 2007;6:421–437. [Google Scholar]
- 8. Hu C, Li D, Chen C, et al. . On the recurrent Ulva prolifera blooms in the Yellow Sea and East China Sea. J Geophys Res Oceans. 2013;115:C05017. [Google Scholar]
- 9. Hwang EK, Lee SJ, Ha DS, et al. . Sargassum golden tides in the Shinan-gun and Jeju Island, Korea. Kor J Fish Aquat Sci. 2016;49:689–693. [Google Scholar]
- 10. McMillan JD. Pretreatment of lignocellulosic biomass In: Himmel ME, Baker JO, Overend RP, editors. Enzymatic conversion of biomass for fuels production. Washington: DC: American Chemical Society; 1994. p. 292–324. [Google Scholar]
- 11. Bhat MK. Cellulases and related enzymes in biotechnology. Biotechnol Adv. 2000;18:355–383. [DOI] [PubMed] [Google Scholar]
- 12. Lee SM, Choi IS, Kim SK, et al. . Production of bio-ethanol from brown algae by enzymic hydrolysis. Kor Sci Biotechnol Bioeng J. 2009;24:483–488. [Google Scholar]
- 13. Jones EBG, Pang KL. Marine fungi and fungal-like organisms. Berlin/Boston: Walter de Gruyter; 2012. [Google Scholar]
- 14. Hong JH, Jang S, Heo YM, et al. . Investigation of marine derived fungal diversity and their expliotiable biological activities. Marine Drugs. 2015;13:4137–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park MS, Lee S, Oh SY, et al. . Diversity and enzyme activity of Penicillium species associated with macroalgae in Jeju Island. J Microbiol. 2016;54:646–654. [DOI] [PubMed] [Google Scholar]
- 16. Bugni TS, Ireland CM. Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Nat Prod Rep. 2004;21:143–163. [DOI] [PubMed] [Google Scholar]
- 17. Schulz B, Draeger S, Rheinheimer J, et al. . Screening strategies for obtaining novel: biologically active: fungal secondary metabolites from marine habitats. Botanica Marina. 2008;51:219–234. [Google Scholar]
- 18. Godinho VM, Furbino LE, Santiago IF, et al. . Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. ISME J. 2013;7:1434–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohlmeyer J. Higher fungi as parasites and symbionts of algae. Veröff Inst Meeresforsch Bremerh. 1974;5:339–356. [Google Scholar]
- 20. Visagie CM, Houbraken J, Frisvad JC, et al. . Identification and nomenclature of the genus Penicillium . Stud Mycol. 2014;78:343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schoch CL, Seifert KA, Huhndorf S, et al. . Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker of fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peterson SW, Vega FE, Posada F, et al. . Penicillium coffeae: a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;1:659–666. [DOI] [PubMed] [Google Scholar]
- 23. Huang XL, Gao Y, Xue DQ, et al. . Streptomycindole: an indole alkaloid from a marine Streptomyces spp. DA22 associated with South China Sea sponge Craniella australiensis . Helv Chim Acta. 2011;94:1838–1842. [Google Scholar]
- 24. Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants: algae and fungi In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Netherlands: Springer; 1994. p. 183–190. [Google Scholar]
- 25. White TJ, Bruns T, Lee S, et al. . Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322. [Google Scholar]
- 26. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;1:553–556. [Google Scholar]
- 27. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O' Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 29. Park MS, Eom JE, Fong JJ, et al. . New record and enzyme activity of four species in Penicillium section Citrina from marine environments in Korea. J Microbiol. 2015;53:219–225. [DOI] [PubMed] [Google Scholar]
- 30. Tamura K, Stecher G, Peterson D, et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. [DOI] [PubMed] [Google Scholar]
- 33. Lee H, Lee YM, Heo YM, et al. . Halo-tolerance of marine-derived fungi and their enzymatic properties. BioResources. 2015;10:8450–8460. [Google Scholar]
- 34. Lee YM, Lee H, Kim GH, et al. . Miniaturized enzyme production and development of micro-assays for cellulolytic and xylanolytic enzymes. J Microbiol Methods. 2011;86:124–127. [DOI] [PubMed] [Google Scholar]
- 35. Mabeau S, Fleurence J. Seaweed in food products: biochemical and nutritional aspects. Trends Food Sci Technol. 1993;4:103–107. [Google Scholar]
- 36. Raghukumar S. Methods to study marine fungi: in fungi in coastal and oceanic marine ecosystems. Cham: Springer; 2017. [Google Scholar]
- 37. Flewelling AJ, Ellsworth KT, Sanford J, et al. . Macroalgal endophytes from the Atlantic Coast of Canada: a potential source of antibiotic natural products? Microorganisms. 2013;1:175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Furbino LE, Godinho VM, Santiago IF, et al. . Diversity patterns: ecology and biological activities of fungal communities associated with the endemic macroalgae across the Antarctic Peninsula. Microb Ecol. 2014;67:775–787. [DOI] [PubMed] [Google Scholar]
- 39. Jones EBG, Suetrong S, Sakayaroj J, et al. . Classification of marine Ascomycota: Basidiomycota: Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015;73:1–72. [Google Scholar]
- 40. Zhang T, Wang NF, Zhang YQ, et al. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden. Svalbard (High Arctic) Sci Rep. 2015;5:14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones EBG, Sakayaroj J, Suetrong S, et al. . Classification of marine Ascomycota: anamorphic taxa and Basidiomycota. Fungal Divers. 2009;35:1–187. [Google Scholar]
- 42. Suetrong S, Schoch CL, Spatafora JW, et al. . Molecular systematics of the marine Dothideomycetes. Stud Mycol. 2009;64:155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suryanarayanan TS, Venkatachalam A, Thirunavukkarasu N, et al. . Internal mycobiota of marine macroalgae from the Tamilnadu coast: distribution: diversity and biotechnological potential. Botanica Marina. 2010;53:457–468. [Google Scholar]
- 44. Park MS, Lee S, Lim YM. A new record of four Penicillium species isolated from Agarum clathratum in Korea. J Microbiol. 2017;55:237–246. [DOI] [PubMed] [Google Scholar]
- 45. Zuccaro A, Summerbell RC, Gams W, et al. . A new Acremonium species associated with Fucus spp and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud Mycol. 2004;50:283–297. [Google Scholar]
- 46. Zuccaro A, Schoch CL, Spatafora JW, et al. . Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus . Appl Environ Microbiol. 2008;74:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hsieh SY, Moss ST, Jones EBG. Ascoma development in the marine ascomycete Corollospora gracilis (Halosphaeriales: Hypocreomycetidae: Sordariomycetes). Botanica Marina. 2007;50:302–313. [Google Scholar]
- 48. Ohzeki T, Mori K. Synthesis of corollosporine, an antibacterial metabolite of the marine fungus Corollospora maritima . Biosci Biotechnol Biochem. 2001;65:172–175. [DOI] [PubMed] [Google Scholar]
- 49. Kubicek CP. Involvement of a conidial endoglucanase and a plasma-membrane-bound β-glucosidase in the induction of endoglucanase synthesis by cellulose in Trichoderma reesei . Microbiol. 1987;133:1481–1487. [DOI] [PubMed] [Google Scholar]
- 50. Wu B, Zhao Y, Gao PJ. Estimation of cellobiohydrolase I activity by numerical differentiation of dynamic ultraviolet spectroscopy. Acta Biochim Biophys Sin (Shanghai). 2006;38:372–378. [DOI] [PubMed] [Google Scholar]
- 51. Burtseva YV, Sova VV, Pivkin MV, et al. . Distribution of O-glycosylhydrolases in marine fungi of the Sea of Japan and the Sea of Okhotsk: characterization of exocellular N-acetyl-β-D-glucosaminidase of the marine fungus Penicillium canescens . Appl Biochem Microbiol. 2010;46:648–656. [PubMed] [Google Scholar]
- 52. Dinçer A, Telefoncu A. Improving the stability of cellulase by immobilization on modified polyvinyl alcohol coated chitosan beads. J Mol Catal B Enzym. 2007;45:10–14. [Google Scholar]
- 53. Tebeka IR, Silva AG, Petri DF. Hydrolytic activity of free and immobilized cellulase. Langmuir. 2009;25:1582–1587. [DOI] [PubMed] [Google Scholar]
- 54. Höller U, Wright AD, Matthee GF, et al. . Fungi from marine sponges: diversity: biological activity and secondary metabolites. Mycol Res. 2000;104:1354–1365. [Google Scholar]
- 55. Panno L, Bruno M, Voyron S, et al. . Diversity: ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanic. N Biotechnol. 2013;30:685–694. [DOI] [PubMed] [Google Scholar]
- 56. Arnosti C, Bell C, Moorhead DL, et al. . Extracellular enzymes in terrestrial: freshwater: and marine environments: perspectives on system variability and common research needs. Biogeochem. 2014;117:5–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.