Abstract
Background:
Inhibition of Hsp90 is desirable due to potential downregulation of oncogenic clients. Early generation inhibitors bind to the N-terminal domain (NTD) but C-terminal domain (CTD) inhibitors are a promising class because they do not induce a heat shock response. Here we present a new structural class of CTD binding molecules with a unique allosteric inhibition mechanism.
Methods:
A hit molecule, NSC145366, and structurally similar probes were assessed for inhibition of Hsp90 activities. A ligand-binding model was proposed indicating a novel Hsp90 CTD binding site. Client protein downregulation was also determined.
Results:
NSC145366 interacts with the Hsp90 CTD and has anti-proliferative activity in tumor cell lines (GI50 = 0.2–1.9 μM). NSC145366 increases Hsp90 oligomerization resulting in allosteric inhibition of NTD ATPase activity (IC50 = 119 μM) but does not compete with NTD or CTD-ATP binding. Treatment of LNCaP prostate tumor cells resulted in selective client protein downregulation including AR and BRCA1 but without a heat shock response. Analogs had similar potencies in ATPase and chaperone activity assays and variable effects on oligomerization. In silico modeling predicted a binding site at the CTD dimer interface distinct from the nucleotide-binding site.
Conclusions:
A set of symmetrical scaffold molecules with bisphenol A cores induced allosteric inhibition of Hsp90. Experimental evidence and molecular modeling suggest that the binding site is independent of the CTD- ATP site and consistent with unique induction of allosteric effects.
General significance:
Allosteric inhibition of Hsp90 via a mechanism used by the NSC145366-based probes is a promising avenue for selective oncogenic client downregulation.
Keywords: Hsp90 inhibitor, C-terminal domain, Small molecule probes, Allostery, In silico modeling, Prostate cancer therapeutic
1. Introduction
Heat shock protein 90 (Hsp90) is a highly-conserved member of a multi-chaperone complex and one of the most abundant proteins in eukaryotic cells. Two cytosolic isoforms are present, constitutively active Hsp90β and the stress-inducible Hsp90α [1–3]. Hsp90 facilitates protein homeostasis by enhancement of protein stability through reduction of aggregation and misfolding [4,5] via its chaperone cycle. This conformationally-dynamic, multi-domain protein functions as an obligate dimer and has ATPase activity. ATP binding to the N-terminal (amino) domain (NTD) and hydrolysis by Hsp90 drive a conformational cycle necessary for chaperone function [6–8]. Binding of ATP to each monomer shifts Hsp90 to a “closed” formation that can bind, fold, and activate client proteins [1,9].
The C-terminal domain (CTD) of Hsp90 plays important roles in Hsp90 chaperone function. The CTD contains a secondary nucleotidebinding site [10–12]. This site, which only becomes available for binding after adenine occupation of the N-terminal binding site, lacks nucleotide-binding specificity and has low affinity for nucleotides [13]. The Hsp90 CTD alone has no independent ATPase activity but has a higher affinity for ATP than full-length Hsp90 providing further evidence for interdomain modulation of conformation [12–14]. The Hsp90 CTD exhibits chaperone activity in vitro, independent of the N- terminal ATPase activity, and is necessary for dimerization and oligomerization of Hsp90 [7,12,15].
Hsp90 is commonly overexpressed in cancer cells and stabilizes many clients known to promote oncogenesis, including transcription factors, signaling proteins and kinases [9,16]. Multiple studies have shown that certain tumor cells are more dependent on Hsp90 activity compared to non-malignant cells [17–19]. First-generation Hsp90 inhibitors radicicol and geldanamycin (GA) and their derivatives bind to the NTD through ATP structural mimicry and inhibit ATP hydrolysis, which leads to degradation of oncogenic client proteins [20–22]. Hsp90 inhibitors that target the ATP binding pocket have shown potent antiproliferative effects though they are ineffective over time due to the compensatory mechanism involving induction of a heat shock response. The expression of chaperones Hsp27, Hsp70, Hsp40, and Hsp90 increase [23–25] leading to undesirable chemoprotective effects even though client proteins are being degraded simultaneously [26–28]. Clinical resistance has been attributed to this chemoprotective effect, and dosage increases to overcome resistance are not a viable option due to toxicity. These results are motivating the pursuit of alternative strategies for modulating heat shock protein complexes [29–31].
An alternative molecular mechanism of inhibition is through interaction with the C-terminal ATP binding pocket. The aminocoumarin antibiotics novobiocin and coumermycin A1 (Cm A1) have been shown to bind weakly to the C-terminal ATP binding site and induce degradation of Hsp90 client proteins [7]. Mechanistically, C-terminal ATP site inhibitors are proposed to cause client protein release and degradation through induction of a conformational change that results in separation of the Hsp90 dimer [15]. In general, these types of inhibitors appear less potent than N-terminal inhibitors and none have reached the clinic, although novobiocin derivatives have demonstrated efficacy in prostate cancer cell lines [32,33]. A deguelin derivative was shown to disrupt ATP-binding to the CTD and have anti-proliferative activity in non-small cell lung cancer cell lines [34,35]. Recent evidence suggests that compounds binding to the CTD inhibit Hsp90 function and induce client protein degradation without a heat shock response [16,32,33,36,37], giving new promise to Hsp90 inhibition for cancer treatments. Additional compounds have been implicated in binding the CTD including a curcumin analog, [38] and peptide-based inhibitors that inhibit client protein interactions and dimerization [39].
The aim of this study was to elucidate the biochemical mechanism of action by which a novel chemical scaffold modulates Hsp90 function. Based upon our previous observations for NSC145366 binding to Hsp90 [40], a set of probe molecules related by a common symmetrical scaffold found in bisphenol A (BPA) have been evaluated in a series of biochemical assays. The evidence indicates that these probes bind to the Hsp90 CTD and induce allosteric effects that result in inhibition of the NTD ATPase activity. Testing of NSC145366 in prostate tumor cells provides evidence for Hsp90 client downregulation in the absence of a global heat shock induction. A molecular docking model for these new inhibitors implicates a novel site at the C-terminal dimer interface, which is independent of the CTD ATP-binding site or other proposed inhibitor binding sites.
2. Materials and methods
2.1. Recombinant protein purification
Proteins were purified as described previously [41]. GST-Hsp9α CTD (amino acids 626–732; Addgene #22483) and GST-Hsp90α NTD (amino acids 9–236; Addgene #22481) were expressed in E. coli BL21-DE3 cells. Briefly, BL21-DE3 expression strains were grown overnight and used to inoculate LB medium at 25 °C supplemented with 100 μg/mL ampicillin to an OD600 of 0.4–0.6 followed by O/N induction of protein expression with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 25 °C. After induction, cells were harvested by centrifugation at 4000 × g and lysed using B-Per bacterial protein extraction reagent (ThermoFisher). GST-tagged Hsp90 CTD And NTD proteins were affinity purified with glutathione agarose resin (Thermo Scientific) and eluted using elution buffer (50 mM Tris-HCl pH 8.0, 250 mM glutathione, 0.3 M NaCl). Protein aliquots were made and supplemented with 5% glycerol and stored at − 80 °C.
pET28a( + )-hHsp90β (530–724) was provided by Dr. Thomas Ratajczak (University of Western Australia). Plasmid was transformed into E. coli BL21-DE3 cells and expressed as previously described [42]. Briefly, expression strain was grown overnight and used to inoculate LB medium supplemented with 40 μg/mL Kanamycin to an OD600 of 0.4–0.6 followed by 3 h induction of protein expression with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37 °C. After induction, cells were harvested by centrifugation at 4000 × g and lysed using B- Per bacterial protein extraction reagent (ThermoFisher). The hexameric histidine (His6)-tagged Hsp90 CTD protein was affinity purified with Ni-NTA resin (Sigma Aldrich) and eluted using elution buffer (50 mM Tris-HCl (pH 8.0), 250 mM imidazole, 0.3 M NaCl). Proteins were aliquoted, supplemented with 5% glycerol and stored at − 80 °C.
2.2. Drug Affinity Response Target Stability (DARTs) assay using recombinant purified Hsp90β and Hsp90α
Recombinant Hsp90α protein purified from baculovirus culture was used (Stressmarq Biosciences Inc, SPR-102) for the DARTs assay as previously described [43]. Proteins were incubated with 200 μM compound (AUY922 or NSC145366), and binding buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 5% glycerol, 0.1% Triton X-100) to 20 μL final volume for 2 h at room temperature. After compound treatment, samples were digested with pronase (Roche) at varying concentrations for 10 min at RT. Protease activity was stopped by adding 5 μL of 5× SDS loading dye and boiling samples at 95 °C for 5 min. Samples were run in 8% SDS-PAGE gels at 150 V for 60 min followed by western blotting. For domain studies, the Hsp90 CTD and NTD were incubated with 200 μM compound and binding buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 5% glycerol, 0.1% TritonX-100) 20 μL final volume for 2 h at room temperature treated as previously described. Western blot images were acquired using the ChemiDoc (Biorad) and quantification was completed using ImageLab (Biorad). Briefly, following automated lane and band selection treated samples were compared to vehicle samples. Arbitrary units were converted to percentages and graphed using GraphPad Prism.
2.3. Microscale thermophoresis
Microscale thermophoresis (MST) experiments were performed on a Monolith NT.115 system (NanoTemper Technologies) using 20% LED and 60% IR-laser power. Laser-on and off-times were set at 30 s and 5 s, respectively. Conditions used were nearly identical to a manufacturer technical note that measured 17-DMAG binding to Hsp90 [44] and binding of peptidic inhibitors to Hsp90 [39]. Recombinant Hsp90β was labeled with NT647 (NanoTemper Technologies) and used at a final concentration of 40 nM. A twofold dilution series of compound (NSC145366 or 17-AAG) was prepared for binding experiments in binding buffer (20 nM NaPO4, 100 mM NaCl, 1 mM MgCl2, 0.1% BSA, 0.05% NP-40). Ten μL were transferred to 10 μL labeled Hsp90β, with the final compound concentration ranging from 3 nM to 100 μM. DMSO concentration was held at 2% across samples. Samples were loaded into hydrophilic capillaries for measurements. Each measurement was performed 3 times independently. Data fitting was performed with the Monolith software as per manufacturer instructions.
2.4. Dimerization and oligomerization assays
Hsp90 dimerization was assessed by chemical crosslinking using amine-reactive cross-linker bis(sulfosuccinimidyl) suberate (BS3) (Pierce) as previously described [15]. Protein was buffer exchanged into crosslinking buffer (50 mM NaPO4, 150 mM NaCl) using Zeba™ Spin Desalting Columns (Thermo Scientific). 2 μM protein was preincubated with compound for 1 h at room temperature to 25 μL final volume. BS3 was added to a final concentration of 66–200 μM of crosslinker depending on the desired amount of capture of dimer or oligomer. Samples were incubated for 1 h at room temperature and crosslinking was quenched by addition of 50 mM Tris-HCl pH 7.5 for 15 min. Following quenching, samples were mixed with 5X SDS loading dye and boiled at 95 °C for 5 min. Samples were run in 10% SDS-PAGE gels at 150 V for 60 min followed by western blotting. Blots were probed with anti-GST antibody (P1A12, Developmental Studies Hybridoma Bank, University of Iowa).
2.5. Anti-aggregation activity assays
Anti-aggregation function was measured using an alcohol dehydrogenase (ADH) aggregation assay adapted from previous methods [45]. Forty μL reaction mixtures were prepared containing 6.2 μM equine ADH (Sigma-Aldrich) in the presence and absence of 500 nM GST- Hsp90α CTD and compound. Aggregation was induced at 55 °C while measuring absorbance at 360 nm every minute using an Epoch 2 microplate reader (BioTek) for 90 min. Experiments were performed three times and the end point read at 90 min was used to compare efficiency of inhibition. Mean value for ADH aggregation alone was set to 100%. Another chaperone activity assay was used based on insulin aggregation: 200 μM insulin is pre-incubated with 10 μM Hsp90 CTD + /− 100 μM compound in reaction buffer (10 mM NaPO4 + 100 mM NaCl) at 30 °C for 1 h. Aggregation was induced using 20 mM DTT and monitored by absorbance at 650 nM using an Epoch 2 microplate reader (Biotek) for 40 min.
2.6. Luciferase refolding assay to assess in vitro chaperone function
Compound ability to inhibit Hsp90 chaperone activity was assessed using a luciferase refolding assay. QuantiLum Recombinant Luciferase (Promega) was diluted to 100 μg/mL in denaturation buffer (25 mM Tricine, pH 7.8, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA, 10%(v/v) glycerol, and 10 mg/mL bovine serum albumin) and denatured at 41 °C for 30 min. 0.5 μL denatured luciferase was added to 20 μL rabbit reticulocyte lysate (Promega) that was pre-incubated at 30 °C with compound. Two μL samples were removed every 15 min for 90 min and added to 48 μL Bright Glo Luciferase assay system and luminescence was read using a Synergy 4 microplate reader (Biotek). Percent luciferase refolding was determined using luminescence of DMSO at 90 min as 100% and comparing all samples to this value. Each experiment was completed in triplicate (N = 3).
2.7. Competitive displacement studies of Hsp90 domains binding to ATP
Displacement studies were performed using recombinant GST- Hsp90α CTD and NTD with ATP-agarose (Sigma) as previously described [46]. Briefly, 6 μM Hsp90 CTD or NTD was pre-incubated with 200 μM compound for 30 min. ATP-agarose slurry was added to the reaction and incubated at 30 °C for 30 min. After washing, Hsp90 domains were eluted with 5 mM ATP and protein levels were assessed via western blot.
2.8. Competitive displacement of FITC-GA by Hsp90 inhibitors
Displacement studies were performed with recombinant Hsp90β (Stressmarq Biosciences Inc, SPR-102) and appropriate compounds (GA, AUY922 and NSC145366). Reaction mix was created containing 60 nM Hsp90β, 5 nM FITC-GA (Enzo life sciences) and fluorescence polarization (FP) assay buffer (20 mM Hepes pH 7.3, 50 mM KCl, 0.01% NP-40), aliquoted into a 96-well plate and pre-incubated for 3 h at room temperature. Compounds were serially diluted in FP assay buffer and added to the reaction plate. DMSO concentration is normalized to 2% (v/v) in all reactions. 25 μL reaction mixtures were transferred to shallow 384 well, black microplates (ProxiPlate-384 F plus, Perkin Elmer) and incubated for 16 h at room temperature. Endpoint FP values in mP were recorded with a Synergy 4 hybrid microplate reader (BioTek). The measured FP values were plotted against competitor concentration.
2.9. Hsp90 ATPase assay
The Transcreener™ ADP kit (Bellbrook Labs) was used to assay for ATPase activity. Conditions for the Hsp90 ATPase reaction were 50 mM HEPES (pH 7.4), 20 mM KCl, 4 mM MgCl2, and 0.01% NP-40 in a 10-μL total assay volume in shallow 384 well, black microplates (ProxiPlate- 384 F plus, Perkin Elmer) as previously described [47]. Compounds were incubated in the presence of 250 μM ATP and 0.5 μM Hsp90β at 37 °C for 30 min. ATPase activity was stopped via addition of 1X stop & detect buffer (200 mM HEPES (pH 7.5), 400 mM EDTA, and 0.2% Brij-35) with 100 mg/mL ADP2 Antibody-IRDye® QC-1 and 8 nM ADP Alexa594 Tracer and incubated for 1 h at room temperature. Endpoint fluorescence intensity (RFU) was measured using a Synergy 4 hybrid microplate reader (BioTek). Experiments were performed three times and the measured RFU values were plotted against competitor concentration using GraphPad Prism and curves were fit to Sigmoidal, four parameter logistic, where X is log(concentration).
2.10. NCI-60 cancer cell line testing and chemoinformatics analysis
The NCI-60 tumor cell line screen was completed by the Developmental Therapeutics Program at National Cancer Institute (http://dtp.cancer.gov) as described previously [32,48]. NSC145366 tumor cell data was publically available on the PubChem website but was recently re-screened upon our request to confirm anti-proliferative effects of the compound. A 5-concentration dose response against the NCI 60 cell lines was determined and similar results were obtained. The concentration resulting in 50% reduction of net protein increase in control cells (measured via SRB staining) was calculated and presented as 50% growth inhibition (GI50).
The CellMiner web application was used to search the NCI-60 cellline database to identify compounds that had pharmacological responses similar to NSC145366. The log10(GI50) range value for NSC145366 of 1.12 was less than the 1.2 cutoff used to remove compounds from the CellMiner database. The GI50 data were converted to standardized values and represented as z-scores, and the resulting pattern was submitted to the CellMiner pattern-comparison tool, which returns other compounds, genes, and microRNAs based on the Pearson correlation between all patterns in the database. To compare Pearson correlation coefficients, the Fisher transformation was performed. These transformed values were used for the two-sample t-test, and the reported means were transformed back. CellMiner was then queried for the drug activities of the top ten hits for NSC145366 and twelve GA analogs and five other Hsp90 inhibitors in the CellMiner database. A custom R script was used to parse the drug activities from the resulting Excel files, formatted as a matrix of z-scores representing 28 drugs in the analysis. These data were visualized as a heat-map with z-scores binned by deciles using CellMiner Companion [49]. Hierarchical clustering was performed on the Euclidean distances between the z- scores using average linkage to order the columns of the heat-map.
2.11. Cell culture and western blot analysis
LNCaP cells were purchased from the American Tissue Culture Collection (ATCC; Manassas, VA) and maintained in media conditions identical to those recommended by ATCC. Cell lines were verified using cell line authentication testing from DDC Medical (Fairfield, OH) [50]. All experiments were completed within 20 passages of acquisition from ATCC. Cells were harvested with trypsin and cell pellets were washed in 1x PBS and then lysed in RIPA buffer (Rockland MB-030–0520) containing protease inhibitor cocktail (Sigma P8340), 1 mM PMSF, 0.1% Triton X-100, and 1 mM DTT. Protein concentrations were determined using Quick start Bradford (Biorad) before analyzing cell samples via western blot. A Biorad ChemiDoc was used for quantitation because it has a broad linear dynamic range allowing accurate quantitation of relative protein levels. Bands were selected using Image Lab software (Biorad) and the treated samples were compared to vehicle samples and the ratios of band intensities converted to percentages. Western blots were performed a minimum of three independent times and a representative image is shown.
2.12. Statistical analysis
Experimental data was subject to One-way ANOVA analysis using Graphpad Prism version 7.0. Statistical significance descriptors including error bars and number of replicates are noted in figure legends.
2.13. Synthesis of NSC145366 analogs
Analogs were synthesized based on the parent molecule NSC145366; 1-(4-(2-(4-(2-hydroxy-3-(2,4,4-trimethylpentan-2-ylami-no)propoxy)phenyl)propan-2-yl)phenoxy)-3-(2,4,4-trimethylpentan-2- ylamino)propan-2-ol hydrochloride. All analogs were synthesized from bisphenol A diglycidyl ether (BADGE) (Sigma-Aldrich). Badge (1 equivalent) was dissolved in methanol at room temperature. The corresponding amine was added (2.2 equivalents) and the reaction was stirred at room temperature. Progress was evaluated by thin layer chromatography. Upon completion, as judged by disappearance of the BADGE spot, the reaction mixture was concentrated at 37 °C. The resulting oil was purified by flash chromatography and HPLC (where appropriate). Fractions containing the desired products were finally pooled, and dried under vacuum overnight.
Compound identification: Summary of assigned NMR resonances, spectra and FPLC traces demonstrating purity are available in the supplementary information (Table S1). Synthesized analogs were verified as follows and were named A1, A2, A3 and A8.
A1: 3,3’-((propane-2,2-diylbis(4,1-phenylene))bis(oxy))bis(1-mor-pholinopropan-2-ol) 1H NMR (500 MHz, Chloroform-d) δ 7.26 (s, 0H), 7.15–7.09 (m, 4H), 6.85–6.78 (m, 4H), 5.30 (s, 2H), 4.09 (dq, J = 9.4, 4.9 Hz, 2H), 3.96 (d, J = 4.9 Hz, 4H), 3.78–3.67 (m, 9H), 3.43 (s, 2H), 2.66 (dt, J = 9.8, 4.5 Hz, 5H), 2.59–2.51 (m, 4H), 2.46 (dt, J = 9.9, 4.2 Hz, 5H); 13C NMR (500 MHz, Chloroform-d) δ 156.58, 143.58, 127.85, 113.95, 70.17, 67.13, 65.51, 61.19, 53.86, 41.82, 31.15; HRMS: (ESI) calculated for C29H43N2O6 [M + H] + : 515.3122, found: 515.3117
A2: 3,3’-((propane-2,2-diylbis(4,1-phenylene))bis(oxy))bis(1-(4- methylpiperazin-1-yl)propan-2-ol); 1H NMR (500 MHz, Chloroform-d (90%), Methanol-d (10%)) δ 7.08–7.03 (m, 4H), 6.73–6.67 (m, 4H), 3.89 (ddd, J = 37.9, 9.8, 5.1 Hz, 4H), 3.83–3.66 (m, 10H), 3.62 (d, J = 5.2 Hz, 8H), 3.31 (d, J = 6.3 Hz, 4H), 2.85 (s, 6H), 1.55 (s, 7H). 13C NMR (500 MHz, Chloroform-d (90%), Methanol-d (10%)) δ 155.81, 144.12, 127.89, 113.88, 69.48, 64.55, 59.81, 50.11, 43.05, 41.74, 30.83. HRMS: (ESI) calculated for C31H49N4O4 [M + H] + : 541.3754, found: 541.3745.
A3: 3,3’-((propane-2,2-diylbis(4,1-phenylene))bis(oxy))bis(1-((fur- an-2-ylmethyl)amino)propan-2-ol); 1H NMR (500 MHz, Chloroform-d (90%), Methanol-d (10%)) δ 7.42 (dd, J = 13.2, 1.9 Hz, 2H), 7.09–7.01 (m, 5H), 6.70 (td, J = 7.5, 1.6 Hz, 5H), 6.50 (d, J = 3.3 Hz, 1H), 6.35 (dd, J = 3.3, 1.9 Hz, 1H), 4.63–4.52 (m, 1H), 4.21 (s, 3H), 4.05–3.98 (m, 1H), 3.93 (s, 2H), 3.84 (tt, J = 9.6, 5.1 Hz, 3H), 3.39–3.29 (m, 3H), 3.14 (dd, J = 12.7, 2.9 Hz, 1H), 3.02 (ddd, J = 12.4, 9.9, 2.0 Hz, 1H), 1.55 (s, 6H). 13C NMR (500 MHz, Chloroform-d (90%), Methanol-d (10%)) δ 155.85, 144.49, 127.81, 113.87, 113.07, 111.18, 69.49,65.17, 43.30, 41.75, 30.92. HRMS: (ESI) calculated for C31H39N2O6 [M + H] + : 535.2809, found: 535.2799.
A8: 3,3’-((propane-2,2-diylbis(4,1-phenylene))bis(oxy))bis(1-(iso-butylamino)propan-2-ol); 1H NMR (500 MHz, Chloroform-d (90%), Methanol-d (10%)) δ 7.07–7.01 (m, 4H), 6.73–6.67 (m, 4H), 4.28 (dq, J = 9.0, 4.7, 3.5 Hz, 2H), 4.10 (s, 4H), 3.94 (dt, J = 10.8, 5.4 Hz, 2H), 3.86 (dd, J = 9.7, 5.8 Hz, 2H), 3.20 (dd, J = 12.6, 2.8 Hz, 2H), 3.07 (dd, J = 12.6, 10.0 Hz, 2H), 2.82 (d, J = 7.3 Hz, 4H), 2.01 (non, J = 6.8 Hz, 2H), 1.55 (s, 6H), 0.97 (dd, J = 6.7, 2.6 Hz, 12H). 13C NMR (500 MHz, Chloroform-d (90%), Methanol-d (10%)) δ 155.95, 143.93, 127.83, 113.84, 69.53, 65.10, 55.45, 51.07, 41.74, 30.91, 25.75, 19.85, 19.66. HRMS: (ESI) calculated for C29H47N2O4 [M + H] + : 487.3536, found: 487.3525.
2.14. In silico modeling of ligand-protein interactions
The Hsp90 crystal structure used for all computational modeling was derived from previously published data (PDB ID: 3Q6N) [51]. Chains C, D, E, and F were removed, leaving a middle domain and CTD dimer structure, which was most appropriate for docking studies with the CTD-binding analogs. All modeling efforts were carried out within the Schrodinger Maestro suite of programs. The ligands were first ionized and all possible tautomers generated at a pH 7.4 ± 2, using Epik [52,53]. The resulting structures were minimized using the OPLS- 2005 force field. The Hsp90 dimer was prepared via the Protein Preparation wizard, wherein all relevant hydrogen bonds (Asn, Gln, His) and residue orientations were optimized at pH 7.4 [54]. Water molecules with less than two hydrogen bonds to non-waters were removed and the whole protein structure was minimized to an RMSD of 0.3 Å (heavy atoms only). Flexible docking of the ligands was carried out via the Induced Fit docking utility [55–57]. First, a grid box was defined based on the binding “hot spots” identified using an FTMap unbiased search [58] of the complete surface of the dimer including the middle domain and CTD. Of the several identified hot spots, only one coincided with the CTD. This region is comprised of residues 672, 676, 677, 678, 681, and 688 (occurring symmetrically on both HSP90 monomers), and does not overlap with the putative ATP-binding site (residues 611, 615, 624, 627, 632, and 680). Using the induced fit utility [55–57], NSC145366 was flexibly-docked to the identified location, and the protein structure was iteratively refined within a 10 Å radius from each point on the ligand. Finally, the most favorable protein conformation was held rigid, while the ligand was flexibly redocked with the XP algorithm to yield the best possible docking pose. This was subsequently used as a starting point for docking new analogs, using the same settings. A small region in the 3Q6N structure has a set of residues that are unassigned (617–630) and this is in common for many but not all other Hsp90 structures. This unstructured gap was filled in with equivalent residues of the yeast Hsp90 structure (2CGE) as previously described [59] for visualizing the ATP-binding site.
3. Results and discussion
3.1. NSC145366 directly interacts with human Hsp90β/α via the CTD
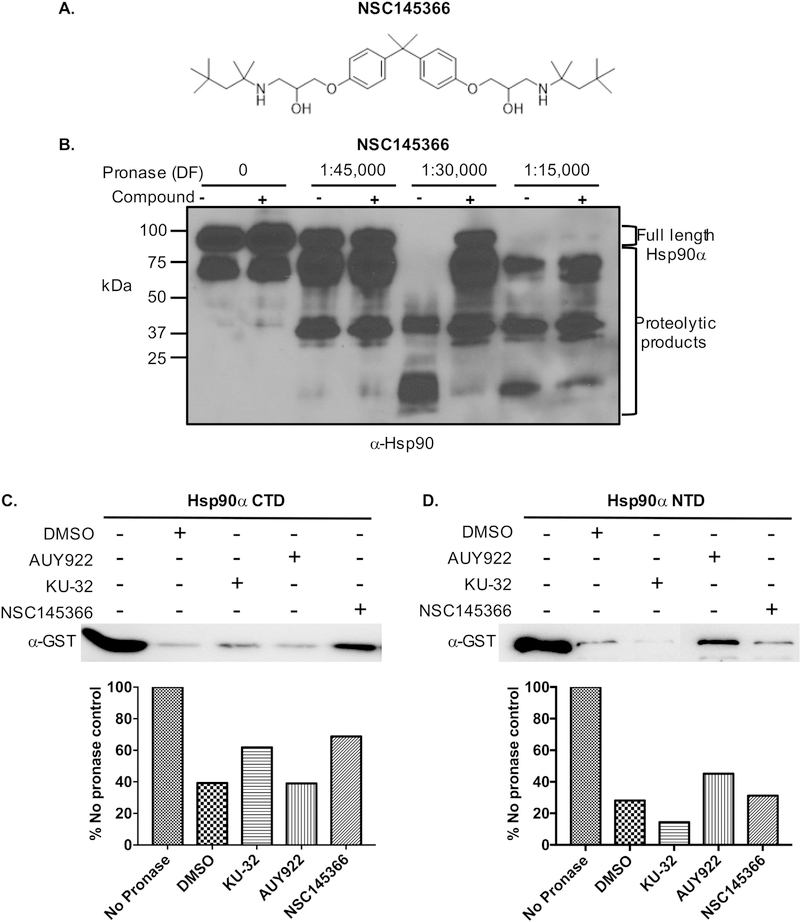
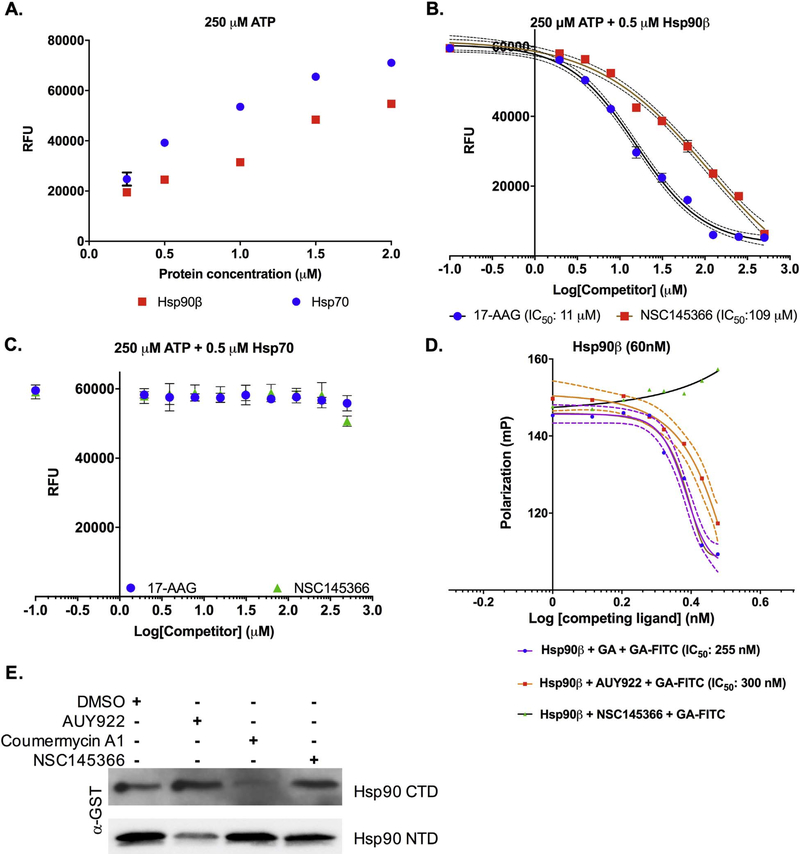
A liquid-culture based phenotypic screen employing Saccharomyces cerevisiae haploid deletion strains with differential sensitivities to Hsp90 inhibitors was used to identify small molecules with potential heat shock protein interactions [40]. NSC145366 emerged as a hit compound and possible Hsp90 inhibitor that affects CTD chaperone activity (Fig. 1A). We had used the DARTs assay to confirm the ability of the compound to directly bind Hsp90 in tumor cell lysates and also recombinant Hsp90β and Hsp90α [40]. Recombinant human Hsp90α was pre-incubated with 200 μM compound and exposed to proteolysis by pronase (Fig. 1). Protection from protease degradation indicates compound binding stabilizes a protein conformation more resistant to proteolysis [43]. NSC145366 protection of Hsp90α was clearly observed (Fig. 1B, lane with 1:30,000 dilution factor). A similar observation was made using the NTD inhibitor AUY922 (200 μM) although a protection pattern distinct from that seen with NSC145366 implicates a different mode of binding (Fig. S1). MST analysis was also used to determine the Kd of NSC145366 binding to Hsp90. To determine the compound binding affinity, Hsp90β was labeled and mixed with varying concentrations of compound. MST signals were plotted and indicated a value of approximately 3.8 μM (Fig. S2). The control compound 17-AAG yielded a Kd of 140 nM similar to the value published previously and a manufacturer technical note for a closely related analog, 17-DMAG [44,60].
Fig. 1.
NSC145366 interacts with human Hsp90α and specifically binds to the Hsp90 CTD. A) The chemical structure of NSC145366. B) The DARTs assay demonstrating NSC145366 binds to Hsp90α. Hsp90α was incubated with 200 μM of compound and pronase added at the indicated dilution factors (DF). Addition of NSC145366 results in protection of Hsp90α. C) The presence of NSC145366 protects GST-Hsp90α CTD (626–732) from proteolysis in the DARTs assay. KU-32 is a known CTD binder and AUY922 is a NTD binder. D) NSC145366 does not inhibit the proteolysis of GST-Hsp90α NTD (9–236) in the DARTs assay but AUY922 does inhibit proteolysis of the NTD.
DARTs assays with recombinant GST tagged Hsp90α CTD and NTD, consisting of amino acids 626–732 and 9–236 respectively, were pursued to further characterize NSC145366 interactions with Hsp90. The ligand binding in different Hsp90 domains appear to elicit different mechanisms of cellular action [7,32,35,61]. NSC145366 protects the Hsp90 CTD from pronase degradation at levels comparable to known CTD inhibitor KU-32 [62] and as expected, AUY922 does not (Fig. 1C). There is no protection observed when NSC145366 is pre-incubated with the Hsp90 NTD using identical experimental conditions, whereas AUY922 does protect the NTD from pronase degradation (Fig. 1D). These results demonstrate that protection of full-length Hsp90 is mediated by the interaction of NSC145366 with the CTD. The lack of compound protection of the NTD is consistent with iNSC145366 binding specifically in the CTD. We also demonstrated that NSC145366 protects Hsp90 in tumor cell lysates but does not protect actin from degradation under the same proteolytic conditions [40].
3.2. NSC145366 inhibits Hsp90 CTD anti-aggregation activity and FL Hsp90 chaperone activity
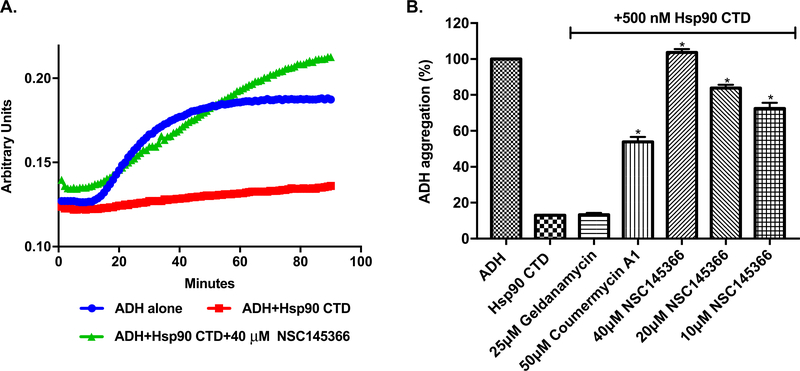
After establishing a direct interaction between NSC145366 and Hsp90 CTD, we assessed the effect of binding on Hsp90 functional activity. Previous studies have shown CTD nucleotide-binding site inhibitors, including novobiocin and coumarin derivatives, inhibit the anti-aggregation function of Hsp90 CTD through disruption of dimerization [15]. Hsp90 CTD anti-aggregation function can be measured using an ADH aggregation assay [45]. At elevated temperatures, ADH forms aggregates that can be detected using light absorption at 360 nm. This aggregation is greatly reduced in the presence of chaperone proteins including the Hsp90 CTD [12,15].
The ADH aggregation assay demonstrated that GST-Hsp90α-CTD can induce an ~84% reduction in ADH aggregation as previously documented [45] (Fig. 2A-B). Addition of NSC145366 increases the level of ADH aggregation in a dose dependent manner from 10 μM to 40 μM (Fig. 2B). At 40 μM, NSC145366 completely eliminates CTD chaperone activity while the positive control, Cm A1 (50 μM), is only able to increase ADH aggregation by ~ 40%. An independent chaperone assay based on DTT-induced aggregation of insulin also demonstrated that NSC145366 inhibits CTD chaperone activity (Fig. S3). In combination, these results effectively demonstrate that NSC145366 is a more potent in vitro inhibitor of CTD anti-aggregation function than Cm A1.
Fig. 2.
NSC145366 inhibits Hsp90 CTD anti-aggregation function. A) The ADH aggregation assay was used to monitor CTD chaperone activity. ADH aggregation was induced by incubation at 55 °C and the level of aggregation monitored by measuring absorbance. A representative set of curves are shown showing inhibition of chaperone activity with 40 μM of NSC145366 compared to lack of compound or the CTD. B) Graph showing quantitation of all absorbance curves. Mean values at 90 h (n = 3) are shown and the SEM error bars are displayed. *significant between untreated (Hsp90 C-terminus alone) and compound treated groups P ≤ 0.0001.
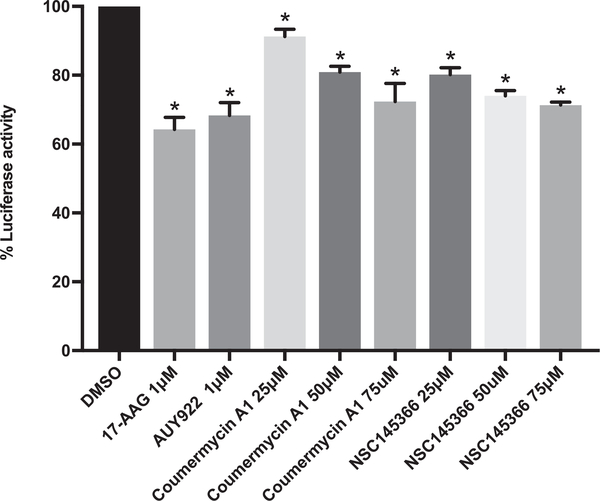
To demonstrate that NSC145366 inhibits Hsp90 chaperone function, we performed a luciferase refolding assay. Denatured luciferase was incubated with rabbit reticulocyte lysate as previously described [63]. This lysate-based assay demonstrates that Hsp90 inhibition occurs in the presence of the complex system of co-chaperones and other cellular components that are involved in normal protein folding. The known Hsp90 inhibitors 17-AAG, AUY922, and Cm A1 were all used as positive controls. Addition of NSC145366 to the lysate system showed that NSC145366 decreased luciferase activity as compared to vehicle treated lysates. The level of inhibition achieved by NSC145366 was greater than C-terminal inhibitor Cm A1 at 25 and 50 μM concentrations, and was comparable to that of N-terminal controls when used at 75 μM (Fig. 3). The complete time course data are available in Fig. S4.
Fig. 3.
NSC145366 inhibits Hsp90 chaperone function. Luciferase refolding in rabbit reticulocyte lysate following pre-incubation with Hsp90 inhibitors. Mean values at 90 min are shown and SEM error bars are displayed. *significance between the DMSO control and compound treated lysates P < 0.006.
3.3. NSC145366 induces oligomerization of Hsp90
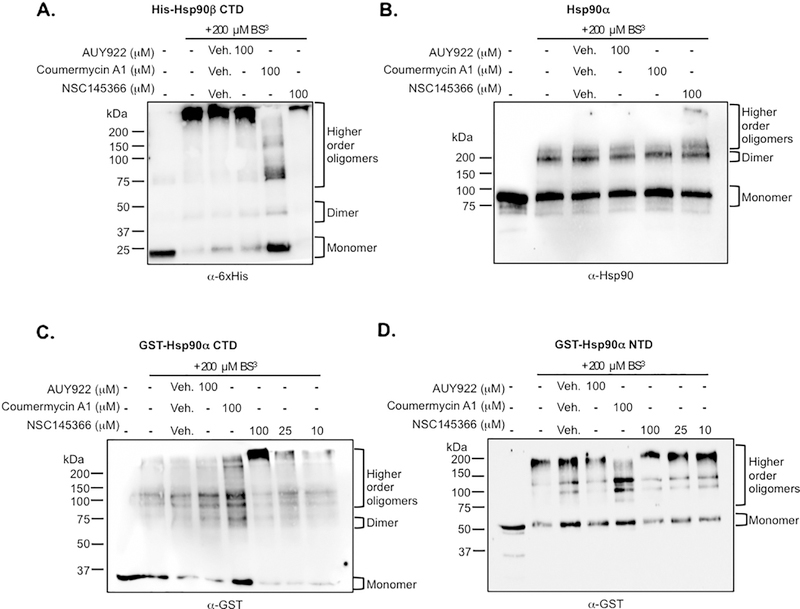
Since C-terminal-based inhibitors have been shown to disrupt Hsp90 dimerization [7,15,32] and eliminate N-terminal ATPase function, we investigated the compound effect on Hsp90 dimerization. Previous studies have shown that the CTD and Hsp90 dimerization can be assessed using crosslinking [15,33]. Dimerization status of His6-tagged Hsp90β CTD or full-length recombinant Hsp90α were evaluated. The CTD protein was pre-incubated with compound and exposed to amine reactive crosslinking as described [15,33]. The addition of crosslinker captured the dominant oligomerized state of the CTD as evidenced by the presence of a slower migrating species compared to the uncrosslinked monomeric species, as previously observed for the Hsp90β isoform [15](Fig. 4A). Surprisingly, the addition of NSC145366 resulted in a band representing a species with markedly decreased mobility suggesting an even higher order oligomerized species (Fig. 4A lane 6). Interestingly, Cm A1 increases the level of monomeric Hsp90 CTD and slightly increases the monomer level of full-length Hsp90α, while N-terminal inhibitor 17-AAG had no effect on dimerization or oligomerization as expected (Fig. 4A-B). The disruption of dimerization by Cm A1 has been previously described [15] and validates our crosslinking conditions. Aminocoumarins are widely reported to inhibit nucleotide binding [7,35,62,64,65] but it is not clear if this inhibition is a result of direct overlap in binding sites or if it involves an allosteric mechanism [59]. The compound-induced oligomerization of full-length Hsp90α (Fig. 4B lane 6) was less evident than the oligomerization level induced for the Hsp90β CTD. Hsp90α does not oligomerize as readily as Hsp90β and prefers the dimer state [66,67]. In addition, the CTD binding site may be more accessible when separated from the full-length protein.
Fig. 4.
NSC145366 increases the oligomerization state of the Hsp90 CTD. A) Crosslinking of the His6-Hsp90β CTD in solution followed by SDS-PAGE. Addition of the BS3 crosslinker captures CTD oligomerization compared to no crosslinking. Addition of NSC145366 increases the oligomerization state while decreasing monomer and dimer states. B) Crosslinking of full length Hsp90α demonstrates the presence of a higher-order oligomer when NSC145366 is added. C) Crosslinking of the GST-Hsp90α CTD. Presence of NSC145366 increases the oligomerization state whereas Cm A1 increases monomer levels. AUY922 has no effect on oligomerization level. D) Crosslinking of GST-Hsp90α NTD results in higher order oligomers. Note that GST can dimerize and is effectively substituting for the native Hsp90 CTD in this case. NSC145366 does not affect oligomerization of the NTD but Cm A1 decreases oligomerization despite being a CTD inhibitor. Veh = DMSO vehicle used to dissolve compounds. All proteins were used at 2 μM concentration.
We also investigated the effect of compound in crosslinking experiments comparing GST-Hsp90α CTD with GST-Hsp90α NTD. We confirmed that induced CTD oligomerization increases in a dose dependent manner from 10 μM to 100 μM (Fig. 4C) while NSC145366 had no effect on oligomerization of the NTD (Fig. 4D). The Cm A1 control compound increases the level of CTD monomers but also appears to decrease oligomerization of the NTD. To further ensure that induced oligomerization is specific to the Hsp90 CTD, we investigated the effect of NSC145366 on the oligomerization of BSA, which has a well- established oligomerization pattern [68–70]. NSC145366 does not induce oligomerization of BSA (Fig. S5). In total, these results show the induced oligomerization of Hsp90 is a novel and specific effect promoted by NSC145366 binding to the CTD in both Hsp90 isoforms.
3.4. NSC145366 allosterically modulates Hsp90 ATPase function
Previous Hsp90 inhibitors have been shown to inhibit N-terminal ATPase activity via two distinct mechanisms. NTD-based inhibitors compete with ATP-binding to prevent ATPase activity, while CTD-based inhibitors disrupt dimerization preventing the necessary conformational switch to a closed Hsp90 conformation and abrogate N-terminal ATPase function [7,15,47,71].
The considerable shift in Hsp90 oligomerization induced by NSC145366 suggests large conformational changes in the protein. To assess the effect of the compound on Hsp90 ATPase activity, we employed the Transcreener™ ADP assay for Hsp90 ATPase activity using an endpoint fluorescence readout [47]. Both Hsp90β and Hsp70 had readily detectable ATPase activity in a protein concentration dependent manner (Fig. 5A) in the Transcreener™ assay. A protein concentration with sufficient signal for inhibition activity assays was chosen to assess the effects of NSC145366 on ATPase function (0.5 μM Hsp90β and Hsp70). As expected, known NTD-based Hsp90 inhibitor 17-AAG inhibits Hsp90β ATPase function (IC50 ~11 μM) similar to previously published IC50 [19,72]. Interestingly, NSC145366 also inhibits Hsp90β ATPase function (IC50~109 μM) (Fig. 5B). The IC50 of ~109 μM corresponds to compound concentrations which induced oligomerization of the CTD in crosslinking experiments. To determine specificity of compound inhibition, NSC145366 was also measured against Hsp70 and was found to have no inhibitory activity up to 200 μM (Fig. 5C). As expected, the known Hsp90 inhibitor 17-AAG had no effect on Hsp70 ATPase function. These results confirm that NSC145366 inhibits Hsp90 N-terminal based ATPase function.
Fig. 5.
NSC145366 allosterically inhibits Hsp90 ATPase function. A) Concentration dependent assessment of Hsp90β and Hsp70 ATPase activity using the Transcreener™ assay. RFU = relative fluorescence units. 0.5 μM of protein generated sufficient signal for subsequent inhibitor activity experiments. B) Inhibition of Hsp90β ATPase activity by 17-AAG and NSC145366. N = 3 with SEM error bars. C) ATPase activity of Hsp70 is not affected by NSC145366 or 17-AAG. N = 3 with SEM error bars. D) Competitive displacement assay of FITC- GA. AUY922 and GA displace FITC-GA from Hsp90β but NSC145366 does not displace. Competitive inhibitor concentrations range from 3 nM to 3 μM and are presented as Log (competing ligand) (nM) SEM is represented by the dashed trendlines. E) ATP-agarose pulldown assay of the Hsp90 CTD and NTD. Addition of NSC145366 does not inhibit pulldown of either domain. AUY922 inhibits NTD pulldown and coumermycin A1 preferentially inhibits the CTD pulldown. Hsp90 CTD or NTD (6 μM) were pre-incubated with 200 μM compound before pulldown with ATP-agarose. ATP-agarose pulldowns were performed three times and similar results were observed.
To ensure this inhibition of ATPase function was not occurring through direct competition at the N-terminal ATP-binding site, ATP-site competitive fluorescence polarization, and ATP-agarose pulldown assays were executed. As expected, ATP competitive inhibitors, GA and AUY922, compete with fluorescently labeled GA, FITC-GA. NSC145366 does not compete with FITC-GA binding, confirming that the observed inhibition of ATPase activity is not occurring by binding to the ATP site (Fig. 5D). We also performed a direct ATP-binding assay in which 200 μM of each compound was pre-incubated with 6 μM of CTD or NTD and assessed the capacity to inhibit the pulldown of the protein domains by ATP-agarose. As expected, AUY922 competes with ATP binding to the NTD but not the CTD. Cm A1, a known CTD ATP-binding site competitor, inhibits CTD binding to ATP but not the NTD consistent with its known mode of interaction. Interestingly, NSC145366 does not disrupt NTD or CTD binding to ATP, indicating that NSC145366 is binding to a site within the CTD other than the nucleotide-binding site (Fig. 5E).
These results are consistent with NSC145366 behaving as an allosteric modulator of Hsp90 NTD ATPase activity through a novel CTD binding mechanism. The NSC145366 site does not compete with the nucleotide binding on the Hsp90 CTD but inhibits chaperone activity and NTD ATPase activity while inducing oligomerization of Hsp90. Previously characterized NTD inhibitors directly compete with ATP at the N-terminal site to inhibit ATPase activity while the CTD inhibitors used here bind to the CTD nucleotide-binding site and inhibit dimerization [7,35].
3.5. NSC145366 exhibits broad activity across the NCI-60 cancer cell panel and correlates with established Hsp90 inhibitor profiles
NSC145366 was previously screened against the NCI-60 panel of tumor cell lines and shown to have activity in nearly all cell lines (PubChem CID: 5351240). We further evaluated NSC145366 NCI-60 sensitivity profile using CellMiner [73] for the possible relationships of tumor-cell growth inhibition with over 20,000 drugs and chemicals from the DTP database. The DTP data indicated that NSC145366 had robust GI50 values ranging from Log10 − 6.7 to − 5.6 with an average GI50 corresponding to 1.12 μM (Table S3) across the tested NCI-60 cell lines. To query CellMiner, the GI50 values for NSC145366 were converted to a z-score and submitted as a vector to search for compounds with a similar pattern of response across the NCI-60 cell lines. Although NSC145366 is not included in CellMiner, a query returned compounds screened by the DTP with vectors of GI50 values that strongly correlated with NSC145366 GI50 values. More importantly, upon analysis with the CellMiner tool, the established Hsp90 inhibitors such as GA analogs, macbecin and herbimycin were found to return highly correlated GI50 vectors. The average Pearson correlation coefficient for these known inhibitors is 0.438, which is considerably higher than that for all other compounds (mean 0.141) in the CellMiner database (two-sample t test; t = 8.7477, df = 16.056, p-value = 1.662e-07; see methods). To further analyze the activity of NSC145366, we performed hierarchical clustering on the NCI-60 activities of NSC145366, the top ten compounds with the highest Pearson correlation coefficient to NSC145366, and known Hsp90 inhibitors and GA analogs in the CellMiner database. GI50 values were converted to z-scores using the mean and standard deviation of the 28 compounds in this analysis. Clustering on the Euclidean distance of the z-scores revealed both compound potency and cell line specificity. This analysis was performed using CellMiner Companion [49] and was visualized as a heatmap which demonstrated that macbecin I (NSC330499; r2 = 0.573), macbecin II (NSC33050; r2 = 0.519) and a geldanamycin analog (NSC330512; r2 = 0.626) were the most similar in cell-line specificity among the known Hsp90 inhibitors (Fig. S6). The robust GI50 values and correlation with established Hsp90 inhibitors support that NSC145366 is a Hsp90 inhibitor. Macbecin exhibits off target toxicities involving redox cycling and arylation of nucleophiles so it has been limited in clinical use [74] and it has been demonstrated to have several different binding features compared to geldanamycin [75]. Although NSC145366 had reasonable correlation with both NTD inhibitors, stronger correlation is not expected because of the different mode of binding to Hsp90. We also note that the NCI DTP had tested NSC145366 for preliminary assessment of toxicity and antitumor activity in mouse xenograft models. NSC145366 (3.12 mg/kg) appears to have very limited antitumor activity (68% treated/control) in a Lewis lung carcinoma mouse model and is toxic at 6.25 mg/kg or higher concentrations (http://dtp.cancer.gov).
To further verify the cancer cell line sensitivity profile, we requested the National Cancer Institute to rescreen against the NCI-60 tumor cell panel including prostate and breast cancer cell lines as described previously [32,48]. The re-screened results were very similar to the original screen and showed broad-spectrum cell growth inhibition activity across the NCI-60 tumor cell lines similar to KU174, a CTD inhibitor that disrupts Hsp90 dimerization [32] (Table S3). GI50 values ranged from 0.20 μM to 1.86 μM. Notably, NSC145366 seems to have the most activity across colon cancer cell lines, followed by leukemias and melanomas. In the prostate cancer cell lines, PC3 and DU-145, NSC145366 showed GI50 of 1.85 μM and 1.38 μM respectively. Based on previous prostate cancer studies using several CTD inhibitors [32,33,76], we focused on assessment of NSC145366 in LNCaP cells to further characterize its cellular effects.
3.6. NSC145366 reduces AR and BRCA1 expression while generating a non-traditional heat shock response
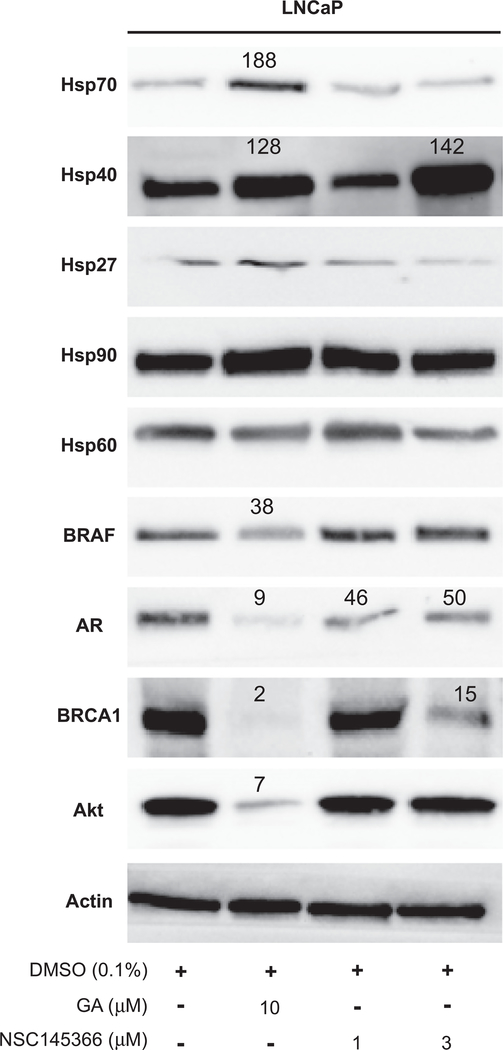
Decreased Hsp90 client expression is a hallmark of Hsp90 inhibition. To determine if NSC145366 inhibited Hsp90 function in LNCaP cells, levels of Hsp90 clients including the Androgen Receptor (AR), BRAF, Akt and BRCA1 were examined after 48 h cell treatment (Fig. 6). Following NSC145366 treatment (1 μM and 3 μM), a decrease in AR expression to a 50% level compared to DMSO was observed. Though this decrease is modest, it is consistent with recently published CTD inhibitor KU675 treatment of LNCaP cells [33]. BRAF and Akt do not show significant downregulation in the presence of NSC145366. BRCA1 expression is considerably decreased in the presence of both GA (9% level) and NSC145366 (15% level at 3 μM). Because this decrease in BRCA1 expression is more pronounced than the decrease in AR expression levels, and NSC145366 does not affect BRAF or Akt expression, NSC145366 may be exhibiting some selectivity with respect to client degradation. The selective client down-regulation exhibited by NSC145366 could lead to a therapeutic advantage but further investigation is needed to elucidate the selectivity mechanism.
Fig. 6.
NSC145366 mediates AR and BRCA1 degradation without inducing classical heat shock response. LNCaP cells were incubated with indicated treatments for 48 h. Cell lysates were prepared and Protein levels were quantified in the linear dynamic range of a digital imager. The percentage expression level compared to DMSO is displayed for notable signals. Western blots were performed on a minimum of three independent biological replicates and similar results were observed for all replicates.
Interestingly, the induction of a classical heat shock response (induction of Hsp70, Hsp27, and Hsp40) was not detectable compared to the NTD Hsp90 inhibitors, GA, which did induce the classical heat shock response (Fig. 6 lane 2), NSC145366 only appears to induce Hsp40 expression after 48 h cellular treatment at the highest concentration (3 μM) (Fig. 6 lane 4). It should be noted that classical heat shock response was also observed for GA after 24 h (Fig. S7) and all treatment concentrations did not induce cellular apoptosis (Fig. S8). Because Hsp40 induction was only observed at the highest concentration and 5 μM NSC145366 treatment is completely cytotoxic to LNCaP cells (data not shown), this Hsp40 response is attributed to a general heat shock response caused by overwhelming cellular stress induced by compound toxicity. The response would not represent the classical compensatory effect to Hsp90 inhibition as evidenced by a lack of induced Hsp70 expression. In addition, it should be noted that Hsp40 levels are not commonly assessed in most studies investigating heat shock responses [32,33,35].
3.7. The symmetrical core of NSC145366, Bisphenol A (BPA) di-(2-hydroxyethyl) ether, is an important molecular determinant to inhibit Hsp90 function
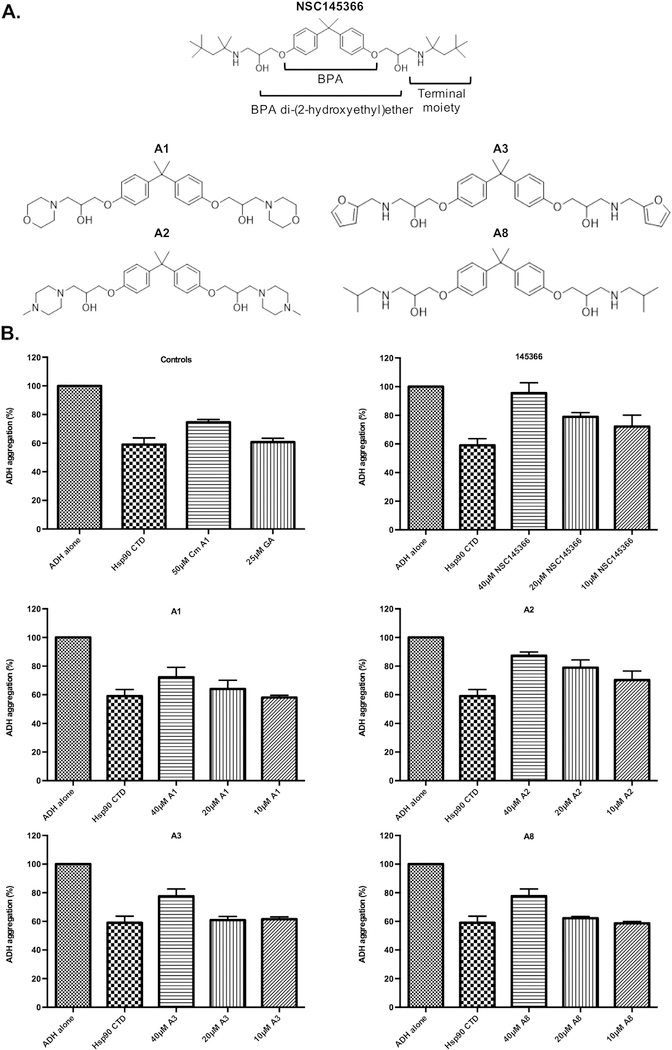
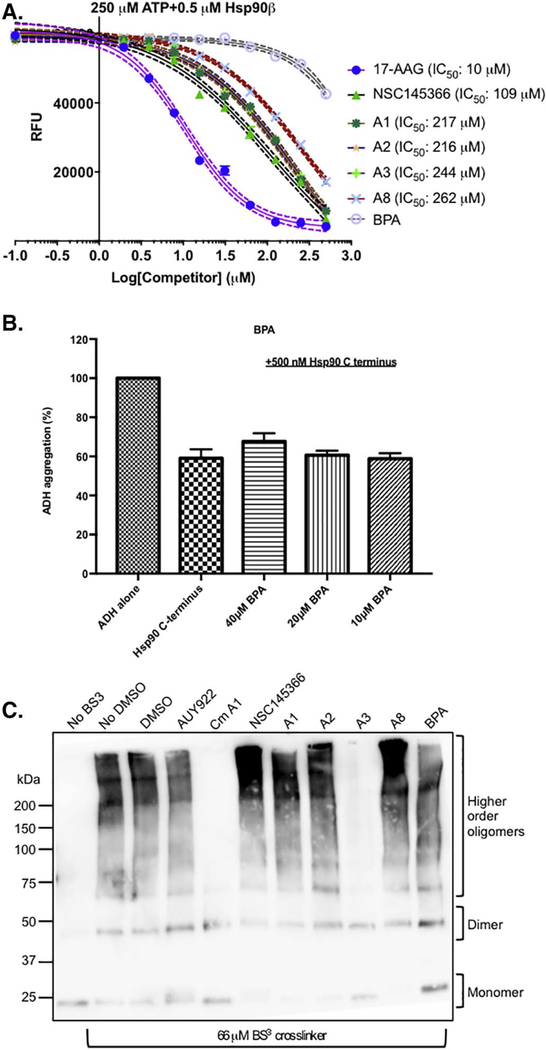
To further test the hypothesis for the existence of a CTD specific binding site, chemical structure variants of NSC145366 were evaluated in the biochemical assays. The initial assumption is that the symmetrical features of NSC145366 have some role in binding; this assumption was used to guide selection of a minimal set of compounds for analysis. Bisphenol A core and the diglycidyl ether are both symmetrical agents that contain the central core. Four additional symmetrical analogs were also synthesized (A1, A2, A3 and A8) to vary the cLogP (1.6–5.1), LogS ( − 4.1 to − 5.6) and basicity values (2–3 ionizable functional groups with pKa varying from 3.1 to 14) (Fig. 7A). Each of these analogs was screened for inhibitory properties in both the ADH chaperone activity assay and the ATPase assay. A1–3 and A8 inhibited Hsp90 CTD chaperone activity at 40 μM but were less efficient than NSC145366 at lower concentrations (Fig. 7B). A2 is the most potent of the analogs demonstrating inhibition to ~75% of NSC145366 levels. BPA did not exhibit inhibition and BADGE exhibited slight inhibition at 40 μM (Fig. S9). When the panel was tested using the Transcreener™ ADP assay, it was evident that analogs A1–3 and A8 were able to allosterically modulate N-terminal Hsp90 ATPase function similarly to NSC145366 (Fig. 8A). Consistent with the ADH assay, A2 was the most potent (IC50~216.3 μM) and A8 was the least effective (IC50~262.1 μM). Again, the BPA core was not able to exhibit considerable inhibition (Fig. 8B).
Fig. 7.
Inhibition of Hsp90 CTD chaperone activity is driven by BPA di-(2-hydroxyethyl)ether. A) Structure of NSC145366 and four symmetrical analogs (A1, A2, A3 and A8); all contain the same Bisphenol A (BPA) di-(2-hydroxyethyl) ether core but differ in their terminal moieties. B) Inhibitor activity of the analogs in the ADH chaperone assay. Cm A1 = coumermycin A1. All assays used identical conditions including 500 nM of Hsp90 CTD. *significant between untreated (Hsp90 C-terminus alone) and compound treated groups P ≤ 0.02 ‡significant between NSC145366 and analog groups P ≤ 0.003.
Fig. 8.
The BPA di-(2-hydroxyethyl) ether drives allosteric modulation of Hsp90 ATPase function in the context of side chains but BPA alone is not sufficient for inhibitory activity. A) Inhibition of ATPase activity by the analogs compared to NSC145366 and BPA. IC50 for each analog is shown. BPA does not have a measurable IC50. N = 3 with SEM error bars. B) ADH aggregation assay examining the ability of BPA to inhibit ADH aggregation. Minimal inhibition occurs at 40 μM. C) Crosslinking assay demonstrating the oligomerization of His6-Hsp90β CTD in the presence of all analogs and compounds. 66 μM of BS3 crosslinker was used in these samples. An identical experiment using 200 μM of BS3 (Fig. S10) enabled improved visualization of the A3-mediated dimer disruption.
To assess if all the analogs affected oligomerization of Hsp90 at similar levels to the parent compound, we also tested them in the crosslinking assay using the His6-tagged Hsp90β CTD under similar conditions as Fig. 4A. The CTD monomer band was eliminated in the lanes containing NSC145366, A1, A2 and A8 and increased signal was observed in bands corresponding to the higher order oligomeric species (Fig. 8C). However, a dramatic reversal in behavior was observed for A3 in which oligomer formation was completely inhibited and the compound actually behaves in a very similar manner to the Cm A1 control. A3 appears to disrupt dimer formation more efficiently than Cm A1 (Fig. S10). This result implicates a binding site capable of triggering disparate allosteric changes in Hsp90 depending on the composition of the terminal moieties of the analogs. The disruption of dimerization by A3 should result in inhibition of the NTD ATPase activity because CTD dimerization is required for trapping ATP at the NTD and for maximal ATP hydrolysis [77]. Further studies are needed to assess the roles of oligomer formation (or not) in the biochemical mechanisms of action.
While NSC145366 and all four active analogs share the BPA di-(2- hyroxyethyl) ether core, it does not appear to impart the inhibitory capacity of NSC145366 or the analogs tested to date. The comparison offered by this minimalistic structure-activity analysis implicates the variant regions in A1–3 and A8 as determinants for functional antagonism in both assays. If the BPA core alone is capable of binding to the CTD, it is not sufficient to induce inhibition of chaperone activity or the NTD ATPase activity. It is also noted that a relatively wide range of structural variants are tolerated and can alter the course for oligomerization vs CTD dimer disruption. This chemical scaffold warrants further investigation to delineate a binding site(s) and suitable SAR of NSC145366 to better inform a pharmacophore model with improved biological properties and assess if improved potency and selectivity can reduce toxicity and retain inhibitor-mediated client degradation.
3.8. In silico modeling of Iigand-Hsp90 interactions
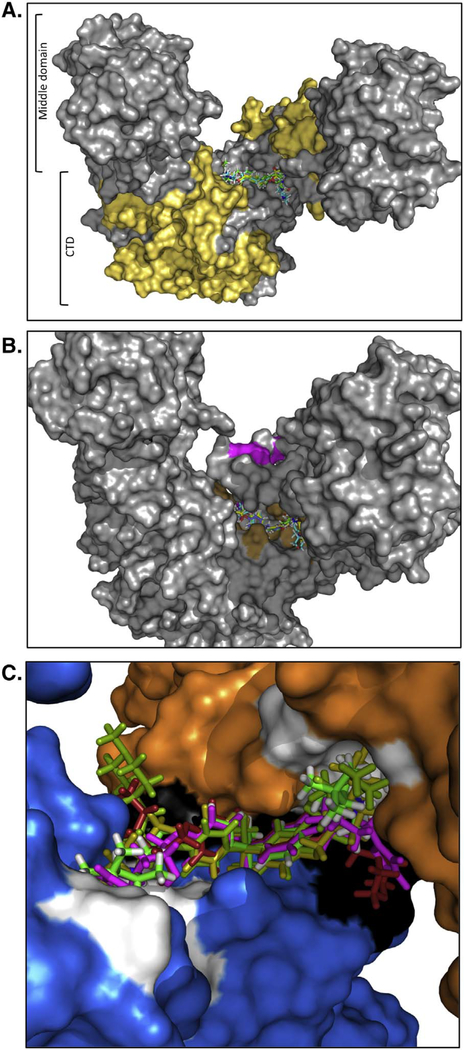
The biochemical results with NSC145366 and the additional symmetrical analogs provided a basis for identification of the Hsp90 binding site. The experimental data constrain the binding model to account for the symmetrical features of NSC145366, existing in a location in the CTD of Hsp90 distinct from the ATP and/or Cm A1 sites, and with potential to modulate the dimeric features. The Hsp90α hexameric crystal structure (PDB ID: 3Q6N) [51] was used after removal of chains to render the middle domain and CTD dimer structure. A search for binding “hot spots” was conducted using FTMap for an unbiased screen [58] of the complete surface of the middle domain and CTD dimer (Fig. S11). Of the binding sites identified, one hot spot site emerged as the main candidate due to its location at the tips of the CTD, where a dimer interface exists. This region displays inter-subunit distances that fall within the variable regions in NSC145366 and A1–3, A8. It is also comprised of symmetrically disposed areas on each HSP90 monomer. None of the other identified hot spots appears near the CTD and/or bear symmetry features, which can accommodate NSC145366 or its analogs. A further constraint on the search for the binding site is based upon the minimal protein construct used for our DARTs and crosslinking experiments, which encompassed residues 626–732 (GST-Hsp90α CTD) (Fig. 9A). The proposed NSC145366 binding site is proximal to, but distinct from, the proposed binding sites of other C-terminal binders [7,64,65,78] and the recently modeled putative nucleotide-binding site within the CTD [59]. The identified hot spot is comprised of residues 672, 676, 677, 678, 681, and 688, whereas the nucleotide-binding site consists of residues 611, 615, 624, 627, 632, and 680 [59] (Fig. 9B and S11). These relationships would suggest that NSC145366 does not compete with ATP for the same binding site, also consistent with the ATP-agarose pulldown results.
Fig. 9.
In silico model of ligand binding to the CTD. A) Structure of the middle domain and CTD of Hsp90α. Yellow represents residues (530–625) in the CTD that that are missing in the minimal CTD construct that can be bound by NSC145366 (GST-Hsp90α CTD 626–732). B) Structural view showing the residues predicted to contact the ligand (Brown) and the residues that are involved in nucleotide binding (Pink). An alternative view is depicted in Fig. S12. C) Close-up view of all ligands docked with near identical overlap between their respective BPA cores.
The four new analogs (A1–3, A8) docked readily in the same location as NSC145366, with near perfect overlap of their respective BPA cores and similar docking scores (− 10.923 ± 0.927). There was notable variation in side chain positions interacting with two distinct symmetrically disposed areas on each monomer (Fig. 9C). The surface areas for molecular contacts consist of residues A608, N609, E611, G675, and F676 (A), and Q501, L672, P681, and Q682 (B). However, the residues involved in hydrogen bond and salt bridge formation are shared among the five ligands and include E497, Q501, L672, G675, S677, and Q682. Previous docking simulations combined with experimental data suggest a partially unstructured region (617–630) that is stabilized by nucleotide binding [59] and may enhance dimerization. The unstructured region was filled in as previously described [59] and the ATP-binding site colored pink showing that it is adjacent to but distinct from the NSC145366 binding site (Fig. S12).
Based on the biochemical evaluation of the ligands, it is evident that the BPA scaffold alone is insufficient to induce N-terminal effects. The molecular fragments that engaged the A and B sub-pockets are implicated in important roles for eliciting the effects seen with NSC145366 and analogs. In addition, the models indicate that the A3 terminal moieties extend in a pose that is distinct from the other analogs and warrants further investigation to determine how the furan ring impedes dimerization. The current docking model also reveals important information about the BPA scaffold. While the BPA scaffold alone is insufficient for inhibition of Hsp90, it does interact with the dimer in a conserved manner and is instrumental in directing the proper orientation of the side chains. Further, it is important to note that the binding site is composed of identical and symmetrically disposed residues in both dimer subunits, which accounts for the importance of the symmetrical BPA core. Confirmation of the proposed binding site and additional mechanistic insights to reveal how these molecules modulate the Hsp90 oligomerization state and induce allosteric effects on the NTD ATPase will be the subject of a separate focused study requiring additional experimental approaches.
4. Conclusion
The inhibition of Hsp90 has been a long-sought therapeutic strategy and efforts have mainly focused on compounds that bind to the NTD; more recently, attention has refocused to other Hsp90 domains. In addition to the novobiocin-based derivatives that target the CTD [7,33,65], recent efforts have identified alternative modes of inhibition including the identification of compounds that bind to the Hsp90 middle domain [37,79] and the CTD [35,80,81]. Most CTD-based inhibitors compete with ATP-binding although the precise binding location and ability to directly bind to the nucleotide-binding site is unclear [59]. Several CTD inhibitors appear to disrupt dimer or oligomer formation while others have not been assessed for this property. We describe a unique mechanism of Hsp90 inhibition that increases the oligomerization state of Hsp90 and our biochemical binding studies in concert with in silico modeling suggest a noncompetitive binding site with respect to ATP at the dimer interface. The minimal domain construct used in this study that binds NSC145366 also contains the dimerization domain, nucleotide-binding site and exhibits chaperone-like anti-aggregation activity. In addition, we have evidence that the NSC145366 binding site is a particularly sensitive allosteric site because substitution to a furan moiety in the case of A3 results in disruption of CTD dimerization. Since in silico modeling cannot explain the oligomerization process, further investigation is required to probe the mechanistic details. Hsp90 appears to have multiple allosteric regulation points that are part of its conformational cycle; these may be modulated by interactions with co-chaperones [82,83] and mutations appear to affect oligomerization [83]. Although Hsp90 is a homodimer, it also exists in other oligomeric forms that are important for its biological activity including the ability to interact with clients and/or co-chaperones [66,84,85]. This ligand class exploits the natural oligomerization properties of Hsp90 resulting in a unique and promising approach to Hsp90 inhibition with selective client down- regulation. NSC145366 has a narrow NCI-60 selectivity GI50 range and very limited efficacy in the NCI xenograft models but due to its novel mechanism of action and chemical structure it offers insight into a unique approach to Hsp90 inhibition. This alternative site to modulating Hsp90 function could potentially improve the scope and utility of Hsp90 targeted probes and future experimental therapeutics.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Ratajczak (University of Western Australia) for the Hsp90 constructs and Dr. Brian Blagg (Kansas University) for the KU-32 compound. We thank Dr. Chang-Deng Hu and members of the VJD and TRH labs for project discussions.
Funding
This study was supported by the Purdue University Center for Cancer Research (PUCCR), NIH grant P30 CA023168, Phase 1 Concept grant from PUCCR and R21NS061667 from NIH (TH, VJD). KMG was supported by a SIRG fellowship from PUCCR.
Footnotes
Conflicts of interest
None to declare.
Transparency document
The http://dx.doi.org/10.1016/j.bbagen.2017.05.006 associated with this article can be found, in the online version.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2017.05.006.
References
- [1].Pearl LH, Prodromou C, Structure and mechanism of the Hsp90 molecular chaperone machinery, Annu. Rev. Biochem 75 (2006) 271–294. [DOI] [PubMed] [Google Scholar]
- [2].Voss AK, Thomas T, Gruss P, Mice lacking HSP90beta fail to develop a placental labyrinth, Development 127 (2000) 1–11. [DOI] [PubMed] [Google Scholar]
- [3].Wandinger SK, Richter K, Buchner J, The Hsp90 chaperone machinery, J. Biol. Chem 283 (2008) 18473–18477. [DOI] [PubMed] [Google Scholar]
- [4].Sato S, Fujita N, Tsuruo T, Modulation of Akt kinase activity by binding to Hsp90, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 10832–10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang T, Bisson WH, Maser P, Scapozza L, Picard D, Differences in conformational dynamics between Plasmodium falciparum and human Hsp90 orthologues enable the structure-based discovery of pathogen-selective inhibitors, J. Med. Chem. 57 (2014) 2524–2535. [DOI] [PubMed] [Google Scholar]
- [6].Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH, Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex, Nature 440 (2006) 1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Donnelly A, Blagg BS, Novobiocin and additional inhibitors of the Hsp90 C- terminal nucleotide-binding pocket, Curr. Med. Chem. 15 (2008) 2702–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Prodromou C, Pearl LH, Structure and functional relationships of Hsp90, Curr. Cancer Drug Targets 3 (2003) 301–323. [DOI] [PubMed] [Google Scholar]
- [9].Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ, Targeting HSP90 for cancer therapy, Br. J. Cancer 100 (2009) 1523–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marcu MG, Chadli A, Bouhouche I, Catelli M, Neckers LM, The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP- binding domain in the carboxyl terminus of the chaperone, J. Biol. Chem 275 (2000) 37181–37186. [DOI] [PubMed] [Google Scholar]
- [11].Marcu MG, Schulte TW, Neckers L, Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins, J. Natl. Cancer Inst 92 (2000) 242–248. [DOI] [PubMed] [Google Scholar]
- [12].Garnier C, Lafitte D, Tsvetkov PO, Barbier P, Leclerc-Devin J, Millot JM, Briand C, Makarov AA, Catelli MG, Peyrot V, Binding of ATP to heat shock protein 90: evidence for an ATP-binding site in the C-terminal domain, J. Biol. Chem 277 (2002) 12208–12214. [DOI] [PubMed] [Google Scholar]
- [13].Soti C, Vermes A, Haystead TA, Csermely P, Comparative analysis of the ATP- binding sites of Hsp90 by nucleotide affinity cleavage: a distinct nucleotide specificity of the C-terminal ATP-binding site, Eur. J. Biochem 270 (2003) 2421–2428. [DOI] [PubMed] [Google Scholar]
- [14].Scheibel T, Siegmund HI, Jaenicke R, Ganz P, Lilie H, Buchner J, The charged region of Hsp90 modulates the function of the N-terminal domain, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 1297–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Allan RK, Mok D, Ward BK, Ratajczak T, Modulation of chaperone function and cochaperone interaction by novobiocin in the C-terminal domain of Hsp90: evidence that coumarin antibiotics disrupt Hsp90 dimerization, J. Biol. Chem 281 (2006) 7161–7171. [DOI] [PubMed] [Google Scholar]
- [16].Moses MA, Henry EC, Ricke WA, Gasiewicz TA, The heat shock protein 90 inhibitor, (−)-epigallocatechin gallate, has anticancer activity in a novel human prostate cancer progression model, Cancer Prev. Res. (Phila.) 8 (2015) 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Trepel J, Mollapour M, Giaccone G, Neckers L, Targeting the dynamic HSP90 complex in cancer, Nat. Rev. Cancer 10 (2010) 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Workman P, Burrows F, Neckers L, Rosen N, Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress, Ann. N. Y. Acad. Sci 1113 (2007) 202–216. [DOI] [PubMed] [Google Scholar]
- [19].Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ, A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors, Nature 425 (2003) 407–410. [DOI] [PubMed] [Google Scholar]
- [20].Mimnaugh EG, Chavany C, Neckers L, Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin, J. Biol. Chem 271 (1996) 22796–22801. [DOI] [PubMed] [Google Scholar]
- [21].Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH, Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin, J. Med. Chem 42 (1999) 260–266. [DOI] [PubMed] [Google Scholar]
- [22].Sidera K, Patsavoudi E, HSP90 inhibitors: current development and potential in cancer therapy, Recent Pat. Anticancer Drug Discov 9 (2014) 1–20. [PubMed] [Google Scholar]
- [23].Clarke PA, Hostein I, Banerji U, Stefano FD, Maloney A, Walton M, Judson I, Workman P, Gene expression profiling of human colon cancer cells following inhibition of signal transduction by 17-allylamino-17-demethoxygeldanamycin, an inhibitor of the hsp90 molecular chaperone, Oncogene 19 (2000) 4125–4133. [DOI] [PubMed] [Google Scholar]
- [24].Erlichman C, Tanespimycin: the opportunities and challenges of targeting heat shock protein 90, Expert Opin. Investig. Drugs 18 (2009) 861–868. [DOI] [PubMed] [Google Scholar]
- [25].McCollum AK, Teneyck CJ, Sauer BM, Toft DO, Erlichman C, Up-regulation of heat shock protein 27 induces resistance to 17-allylamino-demethoxygeldanamycin through a glutathione-mediated mechanism, Cancer Res. 66 (2006) 10967–10975. [DOI] [PubMed] [Google Scholar]
- [26].Demidenko ZN, Vivo C, Halicka HD, Li CJ, Bhalla K, Broude EV, Blagosklonny MV, Pharmacological induction of Hsp70 protects apoptosis-prone cells from doxorubicin: comparison with caspase-inhibitor- and cycle-arrest- mediated cytoprotection, Cell Death Differ. 13 (2006) 1434–1441. [DOI] [PubMed] [Google Scholar]
- [27].Gabai VL, Budagova KR, Sherman MY, Increased expression of the major heat shock protein Hsp72 in human prostate carcinoma cells is dispensable for their viability but confers resistance to a variety of anticancer agents, Oncogene 24 (2005) 3328–3338. [DOI] [PubMed] [Google Scholar]
- [28].Pocaly M, Lagarde V, Etienne G, Ribeil JA, Claverol S, Bonneu M, Moreau- Gaudry F, Guyonnet-Duperat V, Hermine O, Melo JV, Dupouy M, Turcq B, Mahon FX, Pasquet JM, Overexpression of the heat-shock protein 70 is associated to imatinib resistance in chronic myeloid leukemia, Leukemia 21 (2007) 93–101. [DOI] [PubMed] [Google Scholar]
- [29].Wang RE, Targeting heat shock proteins 70/90 and proteasome for cancer therapy, Curr. Med. Chem 18 (2011) 4250–4264. [DOI] [PubMed] [Google Scholar]
- [30].Wang H, Tan MS, Lu RC, Yu JT, Tan L, Heat shock proteins at the crossroads between cancer and Alzheimer’s disease, Biomed. Res. Int 2014 (2014) 239164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Murphy ME, The HSP70 family and cancer, Carcinogenesis 34 (2013) 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eskew JD, Sadikot T, Morales P, Duren A, Dunwiddie I, Swink M, Zhang X, Hembruff S, Donnelly A, Rajewski RA, Blagg BS, Manjarrez JR, Matts RL, Holzbeierlein JM, Vielhauer GA, Development and characterization of a novel C- terminal inhibitor of Hsp90 in androgen dependent and independent prostate cancer cells, BMC Cancer 11 (2011) 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu W, Vielhauer GA, Holzbeierlein JM, Zhao H, Ghosh S, Brown D, Lee E, Blagg BS, KU675, a concomitant heat-shock protein inhibitor of Hsp90 and Hsc70 that manifests isoform selectivity for Hsp90αlpha in prostate cancer cells, Mol. Pharmacol. 88 (2015) 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nguyen MP, Lee D, Lee SH, Lee HE, Lee HY, Lee YM, Deguelin inhibits vasculogenic function of endothelial progenitor cells in tumor progression and metastasis via suppression of focal adhesion, Oncotarget 6 (2015) 16588–16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee SC, Min HY, Choi H, Kim HS, Kim KC, Park SJ, Seong MA, Seo JH, Park HJ, Suh YG, Kim KW, Hong HS, Kim H, Lee MY, Lee J, Lee HY, Synthesis and evaluation of a novel deguelin derivative, L80, which disrupts ATP binding to the C-terminal domain of heat shock protein 90, Mol. Pharmacol 88 (2015) 245–255. [DOI] [PubMed] [Google Scholar]
- [36].Burlison JA, Avila C, Vielhauer G, Lubbers DJ, Holzbeierlein J, Blagg BS, Development of novobiocin analogues that manifest anti-proliferative activity against several cancer cell lines, J. Org. Chem 73 (2008) 2130–2137. [DOI] [PubMed] [Google Scholar]
- [37].Armstrong HK, Koay YC, Irani S, Das R, Nassar ZD, Australian Prostate Cancer B, Selth LA, Centenera MM, McAlpine SR, Butler LM, A novel class of Hsp90 C-terminal modulators have pre-clinical efficacy in prostate tumor cells without induction of a heat shock response, Prostate 76 (2016) 1546–1559. [DOI] [PubMed] [Google Scholar]
- [38].Fan Y, Liu Y, Zhang L, Cai F, Zhu L, Xu J, C0818, a novel curcumin derivative, interacts with Hsp90 and inhibits Hsp90 ATPase activity, Acta Pharm. Sin. B 7 (2017) 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bopp B, Ciglia E, Ouald-Chaib A, Groth G, Gohlke H, Jose J, Design and biological testing of peptidic dimerization inhibitors of human Hsp90 that target the C-terminal domain, Biochim. Biophys. Acta 1860 (2016) 1043–1055. [DOI] [PubMed] [Google Scholar]
- [40].Thomas FM, Goode KM, Rajwa B, Bieberich AB, Avramova LV, Hazbun TR, Davisson VJ, A chemogenomic screening platform used to identify chemotypes perturbing HSP90 pathways, SLAS Discov. (2017), 10.1177/2472555216687525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ward BK, Allan RK, Mok D, Temple SE, Taylor P, Dornan J, Mark PJ, Shaw DJ, Kumar P, Walkinshaw MD, Ratajczak T, A structure-based mutational analysis of cyclophilin 40 identifies key residues in the core tetratricopeptide repeat domain that mediate binding to Hsp90, J. Biol. Chem 277 (2002) 40799–40809. [DOI] [PubMed] [Google Scholar]
- [42].Li Y, Zhang T, Jiang Y, Lee HF, Schwartz SJ, Sun D, (−)-Epigallocatechin-3- gallate inhibits Hsp90 function by impairing Hsp90 association with cochaperones in pancreatic cancer cell line Mia Paca-2, Mol. Pharm. 6 (2009) 1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lomenick B, Jung G, Wohlschlegel JA, Huang J, Target identification using drug BBA - General Subjects 1861 (2017) 1992–2006 affinity responsive target stability (DARTS), Curr. Protoc. Chem. Biol 3 (2011) 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McLaughlin SH, Binding of the geldanamycin derivative 17-DMAG to Hsp90 measured with fluorescence label and label-free, NanoTemper Technologies, 2011. [Google Scholar]
- [45].Clark JI, Huang QL, Modulation of the chaperone-like activity of bovine alpha- crystallin, Proc. Natl. Acad. Sci. U. S. A 93 (1996) 15185–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Csermely P, Kahn CR, The 90-kDa heat shock protein (hsp-90) possesses an ATP binding site and autophosphorylating activity, J. Biol. Chem 266 (1991) 4943–4950. [PubMed] [Google Scholar]
- [47].Rowlands M, McAndrew C, Prodromou C, Pearl L, Kalusa A, Jones K, Workman P, Aherne W, Detection of the ATPase activity of the molecular chaperones Hsp90 and Hsp72 using the TranscreenerTM ADP assay kit, J. Biomol. Screen 15 (2010) 279–286. [DOI] [PubMed] [Google Scholar]
- [48].Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A, et al. , Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines, J. Natl. Cancer Inst 83 (1991) 757–766. [DOI] [PubMed] [Google Scholar]
- [49].Wang S, Gribskov M, Hazbun TR, Pascuzzi PE, CellMiner Companion: an interactive web application to explore CellMiner NCI-60 data, Bioinformatics 32 (2016) 2399–2401. [DOI] [PubMed] [Google Scholar]
- [50].Vickman RE, Crist SA, Kerian K, Eberlin L, Cooks RG, Burcham GN, Buhman KK, Hu C-D, Mesecar AD, Cheng L, Ratliff TL, Cholesterol sulfonation enzyme, SULT2B1b, modulates AR and cell growth properties in prostate cancer, Mol. Cancer Res 14 (2016) 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lee CC, Lin TW, Ko TP, Wang AH, The hexameric structures of human heat shock protein 90, PLoS One 6 (2011) e19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Greenwood JR, Calkins D, Sullivan AP, Shelley JC, Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution, J. Comput. Aided Mol. Des 24 (2010) 591–604. [DOI] [PubMed] [Google Scholar]
- [53].Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M, Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules, J. Comput. Aided Mol. Des 21 (2007) 681–691. [DOI] [PubMed] [Google Scholar]
- [54].Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W, Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments, J. Comput. Aided Mol. Des 27 (2013) 221–234. [DOI] [PubMed] [Google Scholar]
- [55].Farid R, Day T, Friesner RA, Pearlstein RA, New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies, Bioorg. Med. Chem 14 (2006) 3160–3173. [DOI] [PubMed] [Google Scholar]
- [56].Sherman W, Day T, Jacobson MP, Friesner RA, Farid R, Novel procedure for modeling ligand/receptor induced fit effects, J. Med. Chem 49 (2006) 534–553. [DOI] [PubMed] [Google Scholar]
- [57].Sherman W, Beard HS, Farid R, Use of an induced fit receptor structure in virtual screening, Chem. Biol. Drug Des 67 (2006) 83–84. [DOI] [PubMed] [Google Scholar]
- [58].Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S, The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins, Nat. Protoc 10 (2015) 733–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Roy SS, Kapoor M, In silico identification and computational analysis of the nucleotide binding site in the C-terminal domain of Hsp90, J. Mol. Graph. Model 70 (2016) 253–274. [DOI] [PubMed] [Google Scholar]
- [60].Onuoha SC, Mukund SR, Coulstock ET, Sengerova B, Shaw J, McLaughlin SH, Jackson SE, Mechanistic studies on Hsp90 inhibition by ansamycin derivatives, J. Mol. Biol 372 (2007) 287–297. [DOI] [PubMed] [Google Scholar]
- [61].Neckers L, Schulte TW, Mimnaugh E, Geldanamycin as a potential anti-cancer agent: its molecular target and biochemical activity, Investig. New Drugs 17 (1999) 361–373. [DOI] [PubMed] [Google Scholar]
- [62].Kusuma BR, Zhang L, Sundstrom T, Peterson LB, Dobrowsky RT, Blagg BS, Synthesis and evaluation of novologues as C-terminal Hsp90 inhibitors with cytoprotective activity against sensory neuron glucotoxicity, J. Med. Chem. 55 (2012) 5797–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].McConnell JR, Alexander LA, McAlpine SR, A heat shock protein 90 inhibitor that modulates the immunophilins and regulates hormone receptors without inducing the heat shock response, Bioorg. Med. Chem. Lett 24 (2014) 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Matts RL, Dixit A, Peterson LB, Sun L, Voruganti S, Kalyanaraman P, Hartson SD, Verkhivker GM, Blagg BS, Elucidation of the Hsp90 C-terminal inhibitor binding site, ACS Chem. Biol 6 (2011) 800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Moroni E, Zhao H, Blagg BS, Colombo G, Exploiting conformational dynamics in drug discovery: design of C-terminal inhibitors of Hsp90 with improved activities, J. Chem. Inf. Model 54 (2014) 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nemoto T, Sato N, Oligomeric forms of the 90-kDa heat shock protein, Biochem. J 330 (Pt 2) (1998) 989–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nemoto T, Ohara-Nemoto Y, Ota M, Takagi T, Yokoyama K, Mechanism of dimer formation of the 90-kDa heat-shock protein, Eur. J. Biochem 233 (1995) 1–8. [DOI] [PubMed] [Google Scholar]
- [68].Babcock JJ, Brancaleon L, Bovine serum albumin oligomers in the E- and B-forms at low protein concentration and ionic strength, Int. J. Biol. Macromol 53 (2013) 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vaiana SM, Emanuele A, Palma-Vittorelli MB, Palma MU, Irreversible formation of intermediate BSA oligomers requires and induces conformational changes, Proteins 55 (2004) 1053–1062. [DOI] [PubMed] [Google Scholar]
- [70].Bhattacharya A, Prajapati R, Chatterjee S, Mukherjee TK, Concentration- dependent reversible self-oligomerization of serum albumins through intermolecular beta-sheet formation, Langmuir 30 (2014) 14894–14904. [DOI] [PubMed] [Google Scholar]
- [71].Sydor JR, Normant E, Pien CS, Porter JR, Ge J, Grenier L, Pak RH, Ali JA, Dembski MS, Hudak J, Patterson J, Penders C, Pink M, Read MA, Sang J, Woodward B, Zhang Y, Grayzel DS, Wright J, Barrett JA, Palombella VJ, Adams J, Tong JK, Development of 17-allylamino-17-demethoxygeldanamycin hydroquinone hydrochloride (IPI-504), an anti-cancer agent directed against Hsp90, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 17408–17413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Guo W, Reigan P, Siegel D, Zirrolli J, Gustafson D, Ross D, Formation of 17- allylamino-demethoxygeldanamycin (17-AAG) hydroquinone by NAD(P)H:quinone oxidoreductase 1: role of 17-AAG hydroquinone in heat shock protein 90 inhibition, Cancer Res. 65 (2005) 10006–10015. [DOI] [PubMed] [Google Scholar]
- [73].Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, Doroshow J, Pommier Y, CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set, Cancer Res. 72 (2012) 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chang CH, Drechsel DA, Kitson RR, Siegel D, You Q, Backos DS, Ju C, Moody CJ, Ross D, 19-Substituted benzoquinone ansamycin heat shock protein-90 inhibitors: biological activity and decreased off-target toxicity, Mol. Pharmacol 85 (2014) 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Martin CJ, Gaisser S, Challis IR, Carletti I, Wilkinson B, Gregory M, Prodromou C, Roe SM, Pearl LH, Boyd SM, Zhang MQ, Molecular characterization of macbecin as an Hsp90 inhibitor, J. Med. Chem 51 (2008) 2853–2857. [DOI] [PubMed] [Google Scholar]
- [76].Huggins C, Hodges CV, Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941, J. Urol 168 (2002) 9–12. [DOI] [PubMed] [Google Scholar]
- [77].Weikl T, Muschler P, Richter K, Veit T, Reinstein J, Buchner J, C-terminal regions of Hsp90 are important for trapping the nucleotide during the ATPase cycle, J. Mol. Biol 303 (2000) 583–592. [DOI] [PubMed] [Google Scholar]
- [78].Sgobba M, Forestiero R, Degliesposti G, Rastelli G, Exploring the binding site of C-terminal hsp90 inhibitors, J. Chem. Inf. Model 50 (2010) 1522–1528. [DOI] [PubMed] [Google Scholar]
- [79].Yim KH, Prince TL, Qu S, Bai F, Jennings PA, Onuchic JN, Theodorakis EA, Neckers L, Gambogic acid identifies an isoform-specific druggable pocket in the middle domain of Hsp90beta, Proc. Natl. Acad. Sci. U. S. A 113 (2016) E4801–E4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Teracciano S, Chini MG, Vaccaro MC, Strocchia M, Foglia A, Vassallo A, Saturnino C, Riccio R, Bifulco G, Bruno I, Identification of the key structural elements of a dihydropyrimidinone core driving toward more potent Hsp90 C- terminal inhibitors, Chem. Commun. (Camb.) 52 (2016) 12857–12860. [DOI] [PubMed] [Google Scholar]
- [81].Lee SC, Min HY, Choi H, Bae SY, Park KH, Hyun SY, Lee HJ, Moon J, Park SH, Kim JY, An H, Park SJ, Seo JH, Lee S, Kim YM, Park HJ, Lee SK, Lee J, Lee J, Kim KW, Suh YG, Lee HY, Deguelin analogue SH-1242 inhibits Hsp90 activity and exerts potent anticancer efficacy with limited neurotoxicity, Cancer Res. 76 (2016) 686–699. [DOI] [PubMed] [Google Scholar]
- [82].Vettoretti G, Moroni E, Sattin S, Tao J, Agard DA, Bernardi A, Colombo G, Molecular dynamics simulations reveal the mechanisms of allosteric activation of Hsp90 by designed ligands, Sci. Rep 6 (2016) 23830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rehn A, Moroni E, Zierer BK, Tippel F, Morra G, John C, Richter K, Colombo G, Buchner J, Allosteric regulation points control the conformational dynamics of the molecular chaperone Hsp90, J. Mol. Biol 428 (2016) 4559–4571. [DOI] [PubMed] [Google Scholar]
- [84].Lepvrier E, Moullintraffort L, Nigen M, Goude R, Allegro D, Barbier P, Peyrot V, Thomas B, Nazabal A, Garnier C, Hsp90 oligomers interacting with the Aha1 cochaperone: an outlook for the Hsp90 chaperone machineries, Anal. Chem 87 (2015) 7043–7051. [DOI] [PubMed] [Google Scholar]
- [85].Moullintraffort L, Bruneaux M, Nazabal A, Allegro D, Giudice E, Zal F, Peyrot V, Barbier P, Thomas D, Garnier C, Biochemical and biophysical characterization of the Mg2+-induced 90-kDa heat shock protein oligomers, J. Biol. Chem 285 (2010) 15100–15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.