Abstract
Synthetic mRNA has emerged as a powerful tool for the transfer of genetic information, and it is being explored for a variety of therapeutic applications. Many of these applications require prolonged intracellular persistence of mRNA to improve bioavailability of the encoded protein. mRNA molecules are intrinsically unstable and their intracellular kinetics depend on the UTRs embracing the coding sequence, in particular the 3′ UTR elements. We describe here a novel and generally applicable cell-based selection process for the identification of 3′ UTRs that augment the expression of proteins encoded by synthetic mRNA. Moreover, we show, for two applications of mRNA therapeutics, namely, (1) the delivery of vaccine antigens in order to mount T cell immune responses and (2) the introduction of reprogramming factors into differentiated cells in order to induce pluripotency, that mRNAs tagged with the 3′ UTR elements discovered in this study outperform those with commonly used 3′ UTRs. This approach further leverages the utility of mRNA as a gene therapy drug format.
Keywords: RNA stability, mRNA therapeutics, 3’ UTR library, mRNA pharmacokinetics, cancer immunotherapy, cellular reprogramming, iPSCs
Orlandini von Niessen et al. describe a cell-based selection process for the identification of 3′ UTRs that augment the expression of proteins encoded by synthetic mRNA. They show in two examples of mRNA application that the discovered 3′ UTRs outperform commonly used regions. These findings leverage the utility of mRNA as a gene therapy drug format.
Introduction
Protein-encoding mRNA has emerged as an attractive new technology for gene delivery.1 Synthetic mRNA has the same structure as natural mRNA molecules and features a cap, 5′ and 3′ UTRs and a poly(A)-tail embracing the encoded gene of interest. In contrast to DNA, mRNA does not need to enter the nucleus, and, therefore, it can be expressed in non-dividing cells. mRNA does not integrate into the genome, is only transiently active, and is considered to be safer than DNA and viral vectors.1, 2 Manufacturing of synthetic mRNA by in vitro transcription is simple, fast, and cost efficient at any scale. As a consequence, the development of mRNA drugs has garnered broad interest, and it is being explored for delivery into various cell types and applications as diverse as cancer vaccination, cell reprogramming, and protein replacement therapies.
Delivery of cancer vaccine antigens into antigen-presenting cells, which represents the, to date, most advanced practical application of mRNA, is in late-stage clinical development. One approach is to load human dendritic cells (hDCs) with mRNA in cell culture and adoptively transfer these engineered cells. Another approach is to inject naked mRNA into lymph nodes (reviewed in Vallazza et al.3) for efficient macropinocytotic uptake by resident hDCs.4, 5 A third option, which is in clinical testing, uses intravenously administered nanoparticle formulations targeting mRNA specifically into lymphoid organs for body-wide expression of the antigen by antigen-presenting cells.6 Irrespective of the route, the objective is to achieve translation of mRNA-delivered tumor antigen in the cytoplasm of antigen-presenting cells (reviewed in Sahin et al.1) and presentation of immunogenic epitopes to T cells in the context of major histocompatibility complexes (MHCs) and costimulatory molecules. The potency of the antigen-specific immune response correlates positively with the persistence of the synthetic mRNA in the hDCs and the amount of translated protein.7, 8
Regenerative medicine is another field in which mRNA is used for the conversion of cell fate, including the generation of induced pluripotent stem cells (iPSCs) from easily accessible adult somatic cells (reviewed in Steinle et al.9). iPSCs resemble human embryonic stem cells (hESCs) but are not associated with the respective ethical concerns.10 Reprogramming of somatic cells into iPSCs requires the efficient, transient expression of exogenous pluripotency-associated transcription factors until the cell’s endogenous pluripotency-maintaining network is fully activated.11 For clinical translation of the iPSC approach, mRNA gene transfer appears more attractive than conventional retroviral delivery of the transcription factors, as it lacks the risk of genomic integration and oncogenic transcriptional reactivation in iPSC-derived differentiated cells.12, 13 We have recently reported a reprogramming protocol with a mixture of mRNAs. These mRNAs encode the established iPSC-promoting transcription factors OCT4, SOX2, KLF4, cMYC, NANOG, and LIN28 (OSKMNL) and vaccinia virus immune evasion proteins E3, K3, and B18R (EKB).14, 15, 16 The latter set of genes counteracts type I interferon-dependent cellular defense mechanisms activated by mRNA delivery, which inhibit mRNA translation and may compromise cell viability upon repetitive transfection.17, 18 Transfecting these two classes of factors along with double-stranded reprogramming-enhancing microRNAs (miRNAs) from the 302/367 cluster19, 20 on 4 consecutive days has been shown to improve the efficiency and speed of generating stable iPSC lines from human neonatal and adult fibroblasts and to enable, for the first time, iPSC generation from human blood-derived endothelial progenitor cells (EPCs).17
Regardless of the application, the bioavailability of the mRNA-encoded protein is crucial, and its improvement is continuously being explored by a variety of approaches.3 One of the key regulators of intracellular kinetics of an mRNA molecule is the 3′ UTR (reviewed in Guhaniyogi and Brewer21). The length of the 3′ UTR has an optimal length requirement, as mRNAs with longer 3′ UTRs have a shorter half-life22 whereas mRNAs with shorter 3′ UTRs are less efficiently translated.23 The commonly used 3′ UTRs in mRNA therapeutics are derived from α- and β-globins,24, 25 the most abundant proteins of erythrocytes, which as enucleated cells lack active mRNA synthesis.26 Having originally been validated by their ability to confer increased mRNA half-life in cultured erythroleukemic Friend cells27 and human reticulocytes,28 globin 3′ UTRs are now being broadly used for mRNA delivery into various cell types.
The scope of this study was to discover naturally occurring 3′ UTRs that augment total expression of the encoded proteins by increasing mRNA stability and are comparable or superior to the broadly used human β-globin (hBg) 3′ UTR. To this end, we adapted systematic evolution of ligands by exponential enrichment (SELEX)29, 30 to develop a versatile cell culture-based selection process. The 3′ UTRs discovered by this approach improve therapeutic mRNA-based gene transfer for various applications.
Results
Development of a Method for Systematic Enrichment of Naturally Occurring mRNA Motives with mRNA-Stabilizing Capability
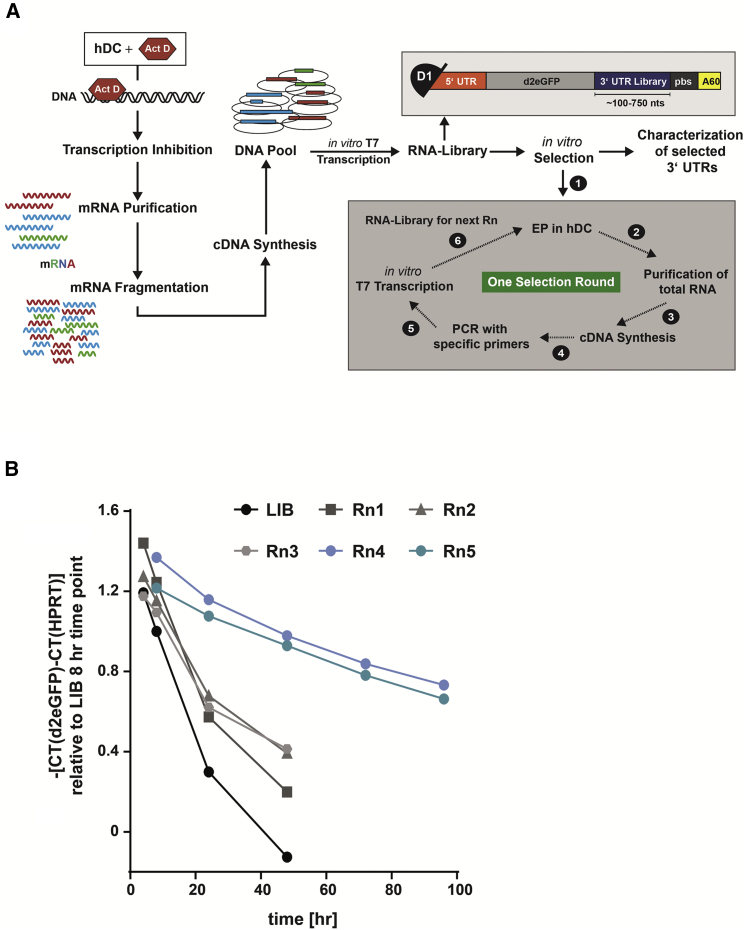
To identify naturally occurring 3′ UTR sequence elements that stabilize synthetic mRNA, we combined two previously described approaches31, 32 and developed a systematic evolution and cell culture-based selection process (Figure 1A).
Figure 1.
Development of a Systematic and Unbiased Method to Enrich 3′ UTR Sequence Motives with mRNA-Stabilizing Capacity
(A) Cell-based selection process. mRNA was purified from hDCs treated with transcription blocker Actinomycin D (ActD), fragmented, and reverse transcribed. The cDNA was fused as 3′ UTR to the reporter gene d2eGFP, and a 3′ UTR mRNA library was generated by in vitro transcription. Synthetic mRNAs were co-transcriptionally capped with the beta-S-ARCA (D1) cap analog.55 Six consecutive rounds (Rn) of selection were performed. In each round, the mRNA library was electroporated into host cells (1), incubated for increasing periods of time to increase stringency (Rn1, 24 hr; Rn2/3, 48 hr; and Rn4/5, 72 hr) (2), followed by total RNA harvest (3) and reverse transcription using oligo-d(T)-primer (4). The cDNA pool was amplified by PCR (5) and in vitro transcribed for initiation of the subsequent round by transfection in hDCs (6). EP, electroporation; pbs, primer-binding site. (B) Average stability of the RNA pool was assessed by qRT-PCR before (LIB) and after selection rounds 1–5 from hDCs harvested at the indicated time points. The transcript levels were determined via ΔΔCT calculation relative to the hypoxanthine-guanine phosphoribosyltransferase (HPRT) and are shown relative to the 8 hr time point of the starting library.
Immature hDCs were cultured in the presence of the mRNA synthesis-blocking compound Actinomycin D (ActD) to enrich the stable mRNA species in the surviving cells (Figures S1A and S1B).33 hDCs were chosen as the environment for evolution, as they survive in cell culture for an extended period of time without cell division, which would counteract the enrichment of mRNA species. Moreover, hDCs are the host cells for one of the mRNA applications in which we are interested, namely, vaccine antigen delivery. A library was engineered from the ActD-treated hDC mRNA pool. We randomly fragmented the purified mRNAs to a size range of 100–750 nt, aiming at the aforementioned optimal length of the 3′ UTR. cDNAs were generated from these mRNA templates by reverse transcription and fused as 3′ UTR ends in between destabilized EGFP (d2eGFP) and a poly(A)-tail of 60 adenosines. Complexity analysis of the starting library was performed via next-generation sequencing (NGS). Most of the alignments could be assigned to RNA transcripts (data not shown).
Six consecutive selection rounds were conducted. Each started with an electroporation of the mRNA pool into hDCs, incubation of the cells for increasing periods of time for highly stringent selection of stable mRNA species, purification, and amplification of the remaining mRNA pool to prepare the selected 3′ UTRs for the next round. Decay curves were generated by analyzing mRNA levels over time by qRT-PCR to assess the stability and half-life of the mRNAs (Figure 1B). The average stability of the RNA pool increased in each round and reached a plateau after round four, indicating an enrichment of stabilizing 3′ UTR sequences. Comparing the half-life of the round five library to the starting mRNA library, we found an about 6-fold increase (Table S1). In the sixth and final round, the stringency of skewing toward highly stable mRNAs was further increased by culturing the cells electroporated with the library recovered from round five for 96 hr, 1 week, or 2 weeks. A total of 402 individual clones from round five and all three time points of round six was harvested, and the regions downstream of the reporter gene were sequenced. Sequence alignment analysis of these individual clones revealed that the majority of them mapped, at least partially, to the 5′ or 3′ UTR of coding mRNAs (Table 1).
Table 1.
Most Frequent Motives Recovered by the Selection Process
| BLAST Result with Representative Sequence of Each Group | Abbreviation | MR | Motive Core Area (nt) | Number of Clones | Percentage of Clones | RNA Library Reads (%) × 10−3 |
|---|---|---|---|---|---|---|
| DnaJ (Hsp40) homolog, subfamily C, member 4, mRNA | DNAJC4 | 5′ UTR + CDS | 170 | 13 | 3.2 | 0.7 |
| Fc fragment of IgG, receptor, transporter, alpha, mRNA | FCGRT | 3′ UTR | 143 | 50 | 12.4 | 0.6 |
| MRS2 magnesium homeostasis factor homolog, mRNA | MRS2 | 5′ UTR + CDS | 142 | 11 | 2.7 | 0.6 |
| Lymphocyte-specific protein 1, mRNA | LSP1 | 3′ UTR | 149 | 22 | 5.5 | 4.6 |
| Chemokine (C-C motif) ligand 22, mRNA | CCL22 | 3′ UTR | 155 | 13 | 3.2 | 6.4 |
| Amino-terminal enhancer of split, mRNA | AES | 3′ UTR | 136 | 4 | 1.0 | 0.3 |
| Phospholipase D family, member 3, mRNA | PLD3 | CDS + 3′ UTR | 190 | 15 | 3.7 | 1.7 |
| Polymerase I and transcript release factor, mRNA | PTRF | 5′ UTR + CDS | 172 | 8 | 2.0 | 0.3 |
| Mitochondrially encoded 12S rRNA | mtRNR1 | ncRNA | 142 | 17 | 4.2 | 14.0 |
| Major histocompatibility complex, class II, DR beta 4, mRNA | HLA-DRB4 | 3′ UTR | 233 | 22 | 5.5 | 1.4 |
| Coiled-coil domain containing 124, mRNA | CCDC124 | CDS | 170 | 16 | 4.0 | 2.2 |
| Prothymosin, alpha, mRNA | PTMA | 5′ UTR | 142 | 8 | 2.0 | 6.4 |
| Myosin, heavy chain 9, non-muscle, mRNA | MYH9 | CDS | 167 | 5 | 1.2 | 0.0 |
| Chemokine (C-C motif) ligand 3, mRNA | CCL3 | 5′ UTR + CDS | 109 | 4 | 1.0 | 0.1 |
| Glutaminase, nuclear gene encoding mitochondrial protein, mRNA | GLS | 5′ UTR | 126 | 8 | 2.0 | 0.2 |
The mRNA origins of the most frequent motives as determined by BLAST are shown. The number of analyzed clones aligned to the respective motive and their percentage within the total 402 clones harvested in rounds 5 and 6, the region they map to in the respective genes of origin (MR), and the length of their core area defined as motive and used for further analysis in nucleotides (nt) are provided. Moreover, it is listed which fraction of reads in the NGS data of the starting mRNA library these motives represent. CDS, coding sequence; ncRNA, non-coding RNA.
More than half (n = 216) of the sequenced clones were derived from only 15 different endogenous transcripts, many of which were immune cell-specific mRNAs functionally relevant for hDCs, such as Fc fragment of immunoglobulin G (IgG) receptor and transporter (FCGRT), lymphocyte-specific protein 1 (LSP1), chemokine ligand 22 (CCL22), major MHC class II DR beta 4 (HLA-DRB4), and chemokine ligand 3 (CCL3). All transcripts except for one are abundantly expressed in hDCs (Table 1; Table S2). Of note, as many 5′ as 3′ UTRs were found in the most stable transcripts. Fragment sequences mapping to the same region of one of the 15 genes and represented by at least four individual clones were further analyzed (Table 1). Within each alignment, we identified a core area present in all clones (defined as motives) and flanked by up- and downstream areas of different length (Tables S2 and S3). Compared to the percentage of reads that could be aligned to these 15 core areas from NGS data of the starting library, all motives were highly enriched during the selection process (Table 1).
Characterization of 3′ UTR Motives Promoting High Protein Expression
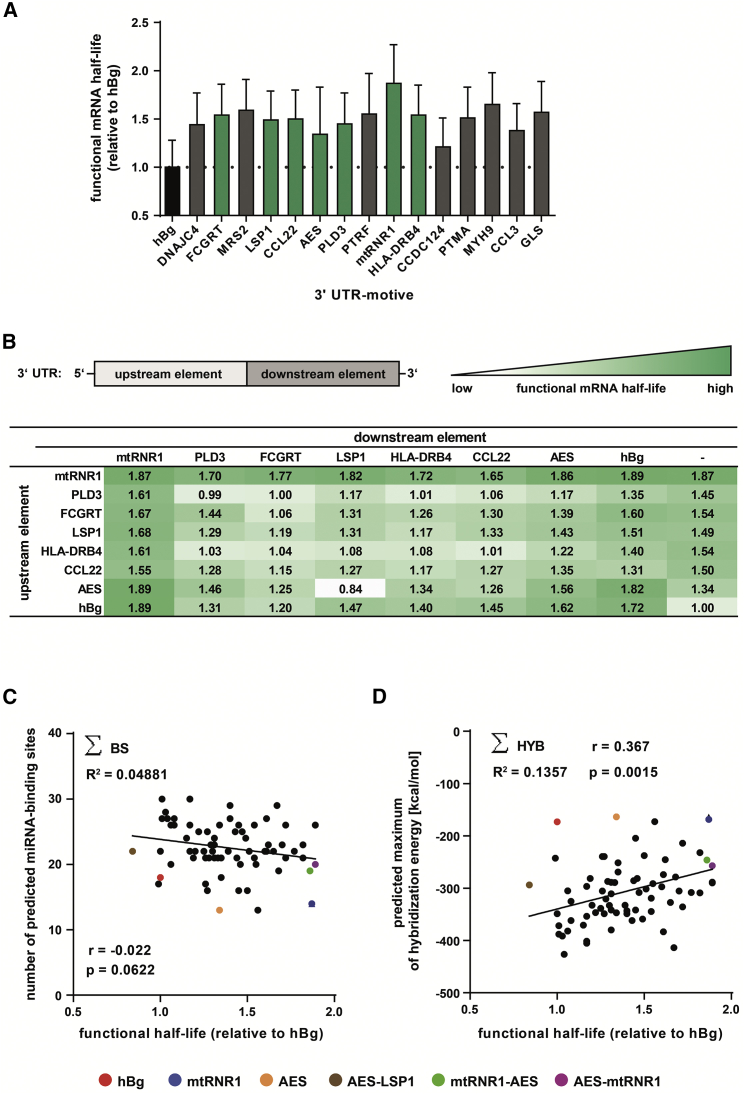
To assess the stabilizing capacity of these 15 core areas, we attached them as 3′ UTRs to a luciferase reporter gene variant (Luc2CP) with a degradation-promoting C-terminal PEST domain,34 and we fused them to a poly(A)-tail of 60 adenosines. Luc2CP has a short protein half-life of 2 hr, and together with the short poly(A)-tail, which has lower mRNA-stabilizing activity,7 the kinetics of the mRNA and of the encoded protein depend largely on the 3′ UTR. For the determination of the functional half-life of the reporter gene signal, which is the time needed for a 50% decay of the protein encoded by the synthetic mRNA, we employed the expression and decay kinetics of the luciferase reporter gene (Figure 2A). This metric is more informative than the physical half-life deduced directly from the mRNA transcript by qRT-PCR, as it additionally considers translational competence of an mRNA species. All tested 3′ UTRs were found to perform better than the commonly used standard hBg.
Figure 2.
Identification of 3′ UTR Motives Resulting in Higher Functional Half-Lives of mRNA and Correlation to Predicted miRNA-Binding Sites
(A and B) Sequences of interest were cloned as single (A) or as double (B) 3′ UTR of a destabilized luciferase reporter gene (Luc2CP). Equal amounts of respective mRNAs were electroporated into hDCs, luciferase activity was measured over 72 hr, and functional mRNA half-lives were calculated and compared to the standard hBg 3′ UTR. A summary of 11 experiments is shown. (C and D) Correlation of functional half-lives of mRNAs tagged with single or double 3′ UTR motives, respectively, with the sum of predicted miRNA-binding sites located on the indicated motive (∑BS) (C) and predicted maximum hybridization energy resulting from miRNA-binding sites located on the indicated motive (∑HYB) (D). Putative binding sites for miRNAs and their respective maximum hybridization energies in the sequences of interest were identified via IntaRNA using a miRNA set specific for hDCs. **p < 0.01; n.s., p > 0.05; r, Pearson correlation coefficient.
We had previously shown in hDCs that two head-to-tail-cloned hBg 3′ UTRs (2hBg) are superior to a single copy in improving mRNA stability.7 mRNA-encoded vaccine antigens equipped with 2hBg generate superior immune responses and survival benefit in mouse tumor models,35, 36, 37 and they are being tested in clinical trials (ClinicalTrials.gov: NCT01684241, NCT02035956, NCT02316457, NCT02410733, and NCT03418480).5
Therefore, we selected those motives mapping to 3′ UTRs (FCGRT, LSP1, CCL22, amino-terminal enhancer of split [AES], PLD3, and HLA-DRB4) (Table 1), the best performer (mtRNR1), and hBg pairwise to form double (d)UTRs. Functional half-lives of all 64 dUTR combinations were tested in the Luc2CP system against hBg. Almost all dUTRs outperformed hBg (Figure 2B; >1.00), and dUTRs carrying the mtRNR1 element, in particular in combination with AES and hBg motives, were moderately superior to 2hBg (Figure 2B). We mapped putative binding sites of miRNAs typically expressed in naive hDCs38, 39 to the single (s)UTRs and dUTRs. Functional half-lives of mRNAs correlated negatively with the number of predicted miRNA-binding sites in the 3′ UTRs with which they were equipped (Figure 2C), whereas they correlated positively with the total hybridization energies as a surrogate for impaired miRNA binding (Figure 2D). These results are consistent with the mechanism of miRNA action, which is to inhibit the translation of their target mRNA and to mediate its degradation.40 Of note, the mtRNR1 and AES motives were among the 3′ UTRs that had the lowest number of predicted binding sites for miRNAs and the highest total hybridization energies. Moreover, in line with published data, the length of the 3′ UTR elements correlated negatively with their stabilizing effect (Figure S2).22
We focused on the mtRNR1, AES, and hBg motives, as well as heterologous combinations thereof, for a more detailed characterization. Homologous dUTRs (fusion of two copies of the same element) were not analyzed, as they are prone to recombination and lead to undesired by-products,41 which is one of the drawbacks of the 2hBg 3′ UTR.
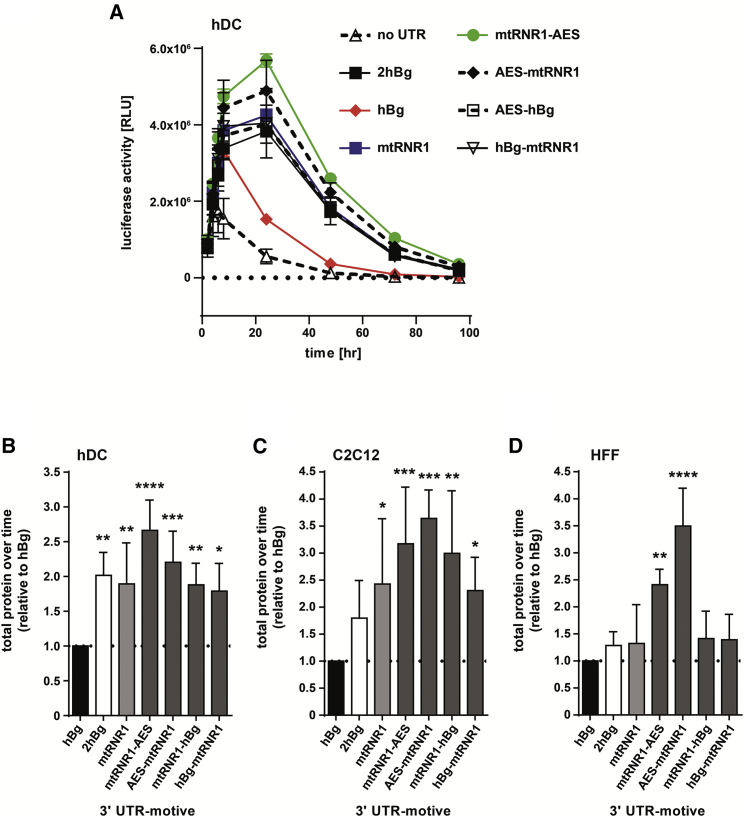
The mtRNR1 and/or AES motive-containing 3′ UTRs were cloned downstream of the standard luciferase (Luc) reporter gene and transfected into hDCs. Protein expression kinetics, which integrate all functionally relevant translational characteristics of mRNA, were measured.23, 42 The hBg 3′ UTR mediated the lowest protein expression, whereas the dUTRs combining mtRNR1 and AES motives’ protein expression was augmented and prolonged (Figure 3A). We analyzed the effects of the various UTRs on the individual parameters integrated into protein expression kinetics in comparison to hBg in more detail. Functional mRNA half-life and time to maximum protein expression, which are both impacted by mRNA stability, were significantly enhanced across all tested 3′ UTRs (Figures S3A and S3B). The maximum protein level and the initial ascending slope of the protein expression kinetics curve both characterize translational efficiency. Here the mtRNR1-AES 3′ UTR performed best by achieving significantly higher maximum protein levels (Figures S3C and S3D). As a consequence, the total amount of protein produced over time (measured as area under the protein decay curve) in hDCs by mRNAs with all tested 3′ UTRs was significantly higher in comparison to hBg and particularly pronounced for the mtRNR1-AES 3′ UTR (Figure 3B).
Figure 3.
mtRNR1- and AES-Containing 3′ UTRs Promote High Protein Expression
(A) 3′ UTR motives were cloned downstream of a standard luciferase reporter gene (Luc). After electroporation of the respective mRNAs with equal amounts into hDCs, luciferase activity was measured in relative light units (RLU) to obtain protein decay kinetics. (B) Based on these data, total protein expressed over time (the area under the curve) was calculated and is depictured relative to the hBg 3′ UTR-containing mRNA. One-way ANOVA, Dunnett’s post-test; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05 (three independently performed experiments). (C and D) mRNA coding for luciferase with the indicated 3′ UTR motives was electroporated into murine myoblasts (C2C12) (C) or human foreskin fibroblasts (HFFs) (D), and, based on protein decay kinetics, total protein expressed over time was calculated and compared to mRNA containing hBg 3′ UTR, as described above. One-way ANOVA, Dunnett’s post-test; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05 (three independently performed experiments).
We tested the effects of the 3′ UTR motives on total amount of protein produced over time in other cell types. In the murine myoblast cell line C2C12, all tested 3′ UTRs significantly outperformed hBg by 2.3- to 3.6-fold (Figure 3C). In human foreskin fibroblasts (HFFs), mtRnR1- and AES-element combinations were superior to hBg by 2.4- to 3.5-fold (Figure 3D).
Altogether these data motivated further assessment of mtRNR1 and AES combination 3′ UTRs in two typical mRNA therapeutics applications: delivery of a vaccine antigen into antigen-presenting cells for the induction of an immune response and delivery of factors for reprogramming of somatic cells into iPSCs.
Increased Translation and Induction of Strong Immune Responses in Mice upon the Application of mRNA Tagged with mtRNR1 and AES 3′ UTR Motives
Robust induction of an immune response by a cancer vaccine requires efficient expression of the vaccine antigen-encoding mRNA specifically in DCs to achieve a high density of epitope-loaded MHCs.1, 42 To assess the utility of the identified 3′ UTRs for this application, an intravenously (i.v.) administered vaccine was used, which is based on liposome-formulated antigen-encoding mRNA. This formulation delivers mRNA systemically into lymphoid compartments, and it is selectively taken up by antigen-presenting cells.6
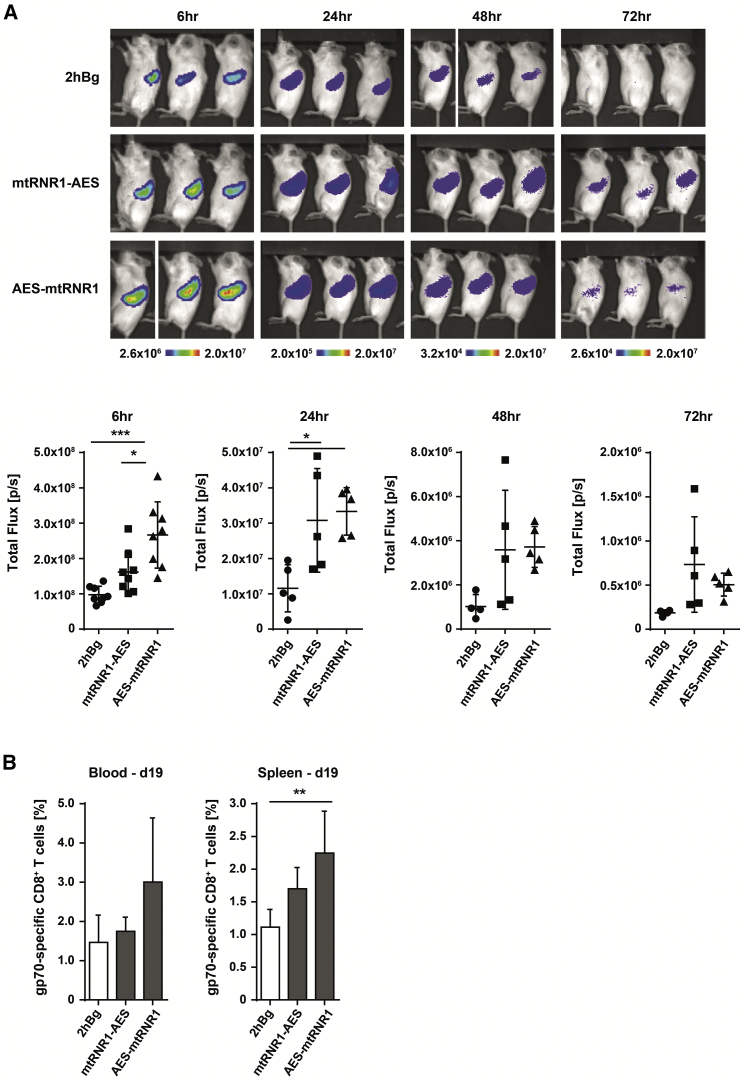
BALB/c mice were injected with liposome-formulated luciferase mRNA containing 2hBg, mtRNR1-AES, or AES-mtRNR1 3′ UTRs. Splenic luciferase expression in mice injected with mtRNR1-AES or AES-mtRNR1 3′ UTR-tagged mRNA was higher and more prolonged as compared to mice injected with mRNA containing the 2hBg 3′ UTR. This difference was pronounced and significant within the first 24 hr, with AES-mtRNR1-tagged reporter mRNA being particularly superior (Figure 4A). To assess the effect of the prolonged antigen prevalence in lymphatic compartments on the induction of immune responses, BALB/c mice were immunized three times with mRNA encoding glycoprotein 70 (gp70), the dominant antigen of the endogenous murine leukemia virus, tagged with the different 3′ UTRs. Mice immunized with AES-mtRNR1 3′ UTR-tagged gp70 generated a significantly higher frequency of gp70 antigen-specific CD8+ T cells in the spleen as compared to mice immunized with the 2hBg-tagged mRNA (Figure 4B).
Figure 4.
Increased Translation and Induction of Strong Immune Responses in Mice upon the Application of mRNA Tagged with mtRNR1 and AES 3′ UTR Motives
(A) In vivo translation of luciferase-encoding mRNAs containing 2hBg, mtRNR1-AES, or AES-mtRNR1 3′ UTRs in BALB/c mice after i.v. vaccination. Three representative mice per group and time point are shown. Quantification was done for all available mice per time point. One-way ANOVA, Tukey’s post-test; *p < 0.05, ***p < 0.001 (one experiment with four to eight mice as indicated). (B) Immune response in BALB/c mice induced by three vaccinations with gp70-encoding mRNAs tagged with the indicated 3′ UTRs. The fraction of gp70 tetramer-positive cells within the CD8+ T lymphocyte population was analyzed in the blood and spleen 5 days after the last vaccination (day 19). One-way ANOVA, Tukey’s post-test; **p < 0.01 (one experiment with five mice).
Improved Reprogramming of Human Fibroblasts into iPSCs by mRNAs Tagged with mtRNR1 and AES 3′ UTR Motives
Next we assessed the utility of 2hBg, AES-mtRNR1, or mtRNR1-AES 3′ UTRs for the induction of iPSCs from HFFs. To this end, we applied a previously reported mRNA-based reprogramming technology based on six reprogramming transcription factors (OSKMNL) and three vaccinia virus immune evasion proteins (EKB).17 In line with that protocol, fibroblasts were lipofected with these mRNA combinations together with double-stranded miRNAs 302a-d and 367 (Figure 5A).
Figure 5.
Improved Reprogramming with mRNAs Tagged with mtRNR1 and AES 3′ UTR Motives
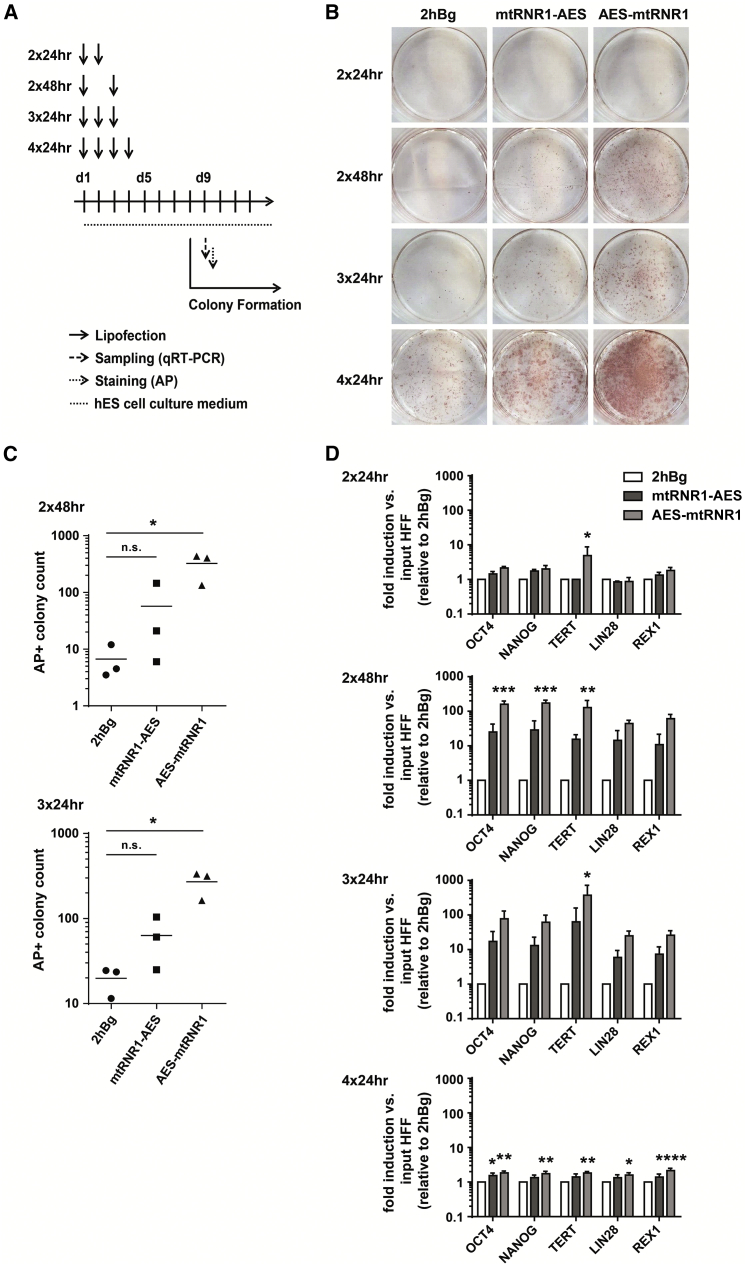
(A) Experimental setup for the reprogramming of human foreskin fibroblasts (HFFs) using synthetic mRNA coding for reprogramming transcription factors, immune evasion proteins, and pluripotency-promoting miRNAs.19, 20 HFFs were lipofected two (2× 24 hr, 2× 48 hr), three (3× 24 hr), or four (4× 24 hr) times. Analysis of colonies was performed on day 9. (B) Representative alkaline phosphatase (AP) staining of HFF-derived iPSCs. (C) Day 9 colony count of AP-positive iPSC colonies comparing two (day 1, day 3) versus three (day 1, day 2, day 3) cycles of lipofection. Colony counts with four lipofections could not be reliably analyzed due to overgrowth and are not shown. One-way ANOVA, Dunnett’s post-test; *p < 0.05; n.s., not significant (three independently performed experiments). (D) Expression of endogenous hESC marker in iPSC colonies was analyzed by qRT-PCR on day 9. Relative expression changes of each transcript in the probe of interest as compared to the day 1 starting control (input HFF) were quantified using the ΔΔCT method and normalized to the housekeeping gene HPRT. The fold inductions obtained by this calculation relative to the corresponding 2hBg benchmark are displayed as mean ± SEM. Two-way ANOVA, Tukey’s post-test; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, relative to 2hBg (three independently performed experiments).
Generation of iPSCs with gene delivery systems that do not integrate and are not self-replicating requires several cycles of transfection with exogenous reprogramming factors until endogenous transcription factors reach sufficiently high protein levels and take over. This interferes with cell viability, increases complexity, and is a hurdle for clinical translation.
Therefore, when evaluating the UTRs for mRNA-based reprogramming, we assessed cells not only after four cycles of transfection, as established in the original protocol, but also tested three and even two cycles, which are insufficient for 2hBg-tagged reprogramming factors (Figure 5A). mRNAs containing the 3′ UTRs discovered in this study performed better than 2hBg 3′ UTR-tagged mRNAs, with the AES-mtRNR1 3′ UTR variant being significantly superior (up to 60-fold higher colony-forming units as compared to 6.5-fold), as shown by alkaline phosphatase (AP)-staining of colonies on day 9 (Figures 5B and 5C). More importantly, even when cells were transfected only two times within 48 hr with mRNAs containing the UTRs discovered in this study, colony formation of iPSCs was robustly detectable from day 8 on. The colonies had distinct well-defined borders, consisted of tightly packed small cells with hESC morphology (Figure S4A), were highly active for AP (Figure S4B), and stained positive for the hESC surface marker TRA-1-81 (Figure S4B). Of note, the activity of the pluripotency-associated TRA-1-81 and AP markers in mtRNR1-AES- or AES-mtRNR1-derived iPSC colonies and on the cell level was comparable to that of 2hBg-derived colonies. The mRNAs tagged with the 3′ UTR discovered in this study (with AES-mtRNR1 being superior) induced a significantly higher level of endogenous hESC marker transcripts OCT4, NANOG, LIN28, TERT, and REX1 as compared to 2hBg 3′ UTR-containing mRNAs, even after two transfection rounds, i.e., under conditions where 2hBg-tagged reprogramming factor mRNAs do not show a robust effect (Figure 5D).
Discussion
Building on previously published approaches,31, 32 we developed a cell culture-based method for the systematic enrichment of naturally occurring RNA sequences that improve the kinetics of gene-encoding mRNA in vitro and in vivo when used as 3′ UTRs.
A general challenge of cell culture-based enrichment and selection methods is the delivery of the mRNA pool.43 Typically, only a small amount of mRNA pool resembling just a tiny fraction of the theoretical complexity of a randomized library can be delivered into the selection environment, which limits the discovery space substantially.44 As a consequence, with conventional synthetic libraries, relevant sequences may be lost during electroporation. We mitigated this risk by constructing a library of reduced complexity from an amplified starting pool, thus ensuring representation of the full repertoire of sequences in several copies.
Over 50% of the sequences enriched by several selection rounds were represented by 15 distinct motives defined by identical core areas but diverse 5′/3′ ends. Six of the motives mapped to mRNAs (AES, PLD3, PTRF, CCDC124, PTMA, and MYH9), which are known to have a high median mRNA half-life.22 The selection rounds enriched for motives mapping to the UTR of the respective mRNAs, but not to their on average much longer coding regions. Altogether, these findings indicate a directed and efficient evolution.
The AES-mtRNR1 and mtRNR1-AES combination motives that we determined functionally as the best performers have a far lower number of putative miRNA-binding sites as compared to the other dUTRs we had discovered and tested. This is in line with reports that mRNA translation decreases with a higher number of miRNAs binding to the 3′ UTR.45
mtRNR1- and AES-derived motives were previously not known to stabilize mRNA. mtRNR1 is the mitochondrially encoded non-coding 12S rRNA, which is involved in the translation of the 13 mtDNA protein-coding genes in mitochondria.46 Mutations in mtRNR1 are associated with deafness.47 AES is a distinct member of the Groucho/Transducin-like enhancer of split gene family, and it regulates androgen receptor transcriptional activity and Notch and Wnt signaling and acts as a tumor suppressor gene.48, 49, 50
We validated the identified 3′ UTR motives in two common applications for mRNA-based gene delivery. First, we studied their usefulness for vaccination with antigen-encoding mRNA. Vaccine antigen-encoding mRNA can be administered by different routes, e.g., subcutaneously, intradermally, into lymph nodes, or systemically. Each route aims to deliver the mRNA into professional antigen-presenting cells. As our lab has developed and clinically translated systemic targeting of lymphatic compartments by mRNA encapsulated in liposomal nanoparticles, we used this model. By doing so, we demonstrated that AES-mtRNR1 and mtRNR1-AES tagging improved expression of the mRNA in mouse lymphatic compartments as compared to the 2hBg 3′ UTR. The optimized expression translated into stronger induction of antigen-specific immune responses in the vaccinated mice. Future studies have to explore whether the discovered elements also improve expression and immune responses of mRNA delivered by routes used by others for cancer vaccination.
Second, we studied the efficiency and robustness of cellular reprogramming of human fibroblasts to iPSCs with mRNA-encoded transcription factors, which is considered to benefit from prolonged mRNA stability.17, 51 Our initial screening had enriched elements for augmented transcript stability in hDCs, which are the target cells of gene delivery for vaccination. Due to the cell type-specific expression of trans-acting factors, such as miRNAs and RNA-binding proteins, we had not expected that mtRNR1-AES and AES-mtRNR1 3′ UTRs would be profoundly superior in conveying transcript stability in human fibroblasts, which are broadly used target cells for iPSC generation-directed gene delivery.
Alphaviral self-replicating RNA is an alternative vector for fast reprogramming, which requires only one transfection.52, 53 It, however, comes with the disadvantage that clinical grade iPSCs have to be free of residual contaminations of self-replicating vectors. With mRNAs tagged with the AES-mtRNR1 3′ UTR, two transfection rounds were sufficient for the generation of iPSC colonies from human neonatal fibroblasts. One of the implications of this improvement is to make refractory cell lines or cell types like EPCs that require even more rounds of transfections amenable to iPSC generation. Optimized 3′ UTRs further increase the efficiency of a potent, simple, safe, and good manufacturing practice (GMP)-compatible gene delivery by mRNA for integration-free iPSC generation from easily accessible adult human somatic cells.
In summary, AES-mtRNR1- and mtRNR1-AES-based 3′ UTRs, discovered by the approach presented herein, enhance the applications of mRNA-based transfer of genetic information shown exemplarily for antigen-encoding mRNA vaccination and mRNA-based reprogramming of human fibroblasts, and they promise to contribute to clinical translation of mRNA gene delivery.
It was not the scope of this study to test the discovered elements across a broader range of cell types. In fact, as factors regulating mRNA stability are cell type specific, there may be cell types in which AES-mtRNR1-based elements are not superior over the conventionally used 3′ UTRs. The enrichment platform we present here can be adapted with regard to the cell types used for library generation and as a screening environment to find cell type-specific UTR elements for mRNA stabilization, and thus it offers a broadly applicable procedure for the improvement of mRNA therapeutics.
Materials and Methods
Animals
Age-matched (6–12 weeks) female BALB/c mice (Janvier Labs, Le Genest-Saint-Isle, France) were used throughout all experiments. Mouse studies were approved by the German regulatory authorities for animal welfare, and mice were kept in accordance with federal and state policies on animal research at BioNTech.
Reagents
All reagents, including chemicals, growth media, supplements, enzymes, growth factors, kits, and solutions, in this study were supplied by Thermo Fisher Scientific (Waltham, MA), unless stated otherwise.
Cell Culture
Peripheral blood mononuclear cells (PBMCs) were purified from buffy coat donors (Transfusion Center, University Medical Center, Mainz, Germany) by Ficoll-Paque Plus (GE Healthcare, Uppsala, Sweden) density gradient centrifugation. Human monocytes were enriched, differentiated into immature hDCs as described previously,7 and cultured in hDC medium (RPMI 1640 with 10% fetal calf serum (FCS), 1% non-essential amino acids (NEAAs), 0.5 U/mL penicillin, and 1 μg/mL streptomycin). Neonatal HFFs (SBI Biosciences, Palo Alto, CA) were cultivated in minimum essential medium (MEM) with 15% FCS and 1% NEAAs. C2C12 mouse myoblasts (ATCC, Manassas, VA) were cultivated in DMEM with 10% FCS. All aforementioned media in this study contained GlutaMAX and 1 mM sodium pyruvate. iPSCs and cells in reprogramming experiments were cultured in NutriStem XF/FF hESC culture medium (ReproCELL USA, Beltsville, MD) supplemented with 10 ng/mL basic fibroblast growth factor and 0.5 μM Thiazovivin (ReproCELL). All cells were cultivated at 37°C and a CO2 content of 5%.
Plasmid Construction and In Vitro Transcription
All plasmids used in this study were based on pST1-2hBgUTR-A120 (pST1) described previously.7 For library construction and the selection process, plasmids with destabilized versions of the reporter genes d2eGFP and Luc2CP were modified by removing the T7-promoter and the poly(A)-tail and introducing a specific cloning site for insertion of the library and a primer-binding site (pbs) for PCR amplification downstream of the reporter gene. For analysis of translational efficiency, Luc2CP was replaced by Luc using Cold Fusion technology (SBI). For in vivo vaccination experiments, pST1-plasmids with a poly(A)-tail of 60 adenosines instead of 120 were used. For cellular reprogramming, all genes were codon optimized and assembled by gene synthesis (Geneart, Regensburg, Germany), and they were inserted into pST1-plasmids containing 2hBg, mtRNR1-AES, or AES-mtRNR1 3′ UTR. For in vitro transcription, plasmids were linearized downstream of the poly(A)-tail or a PCR product was used. In vitro transcription reaction and purification of mRNA were performed as previously described.7, 54 The synthetic mRNA was co-transcriptionally capped with beta-S-ARCA (D1).55 Quality of purified mRNA was assessed by spectrophotometry (NanoDrop 2000c; VWR International, Darmstadt, Germany) and on-chip electrophoresis on the 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA).
Library Construction, Amplification, and Selection Process
For library construction, immature hDCs were seeded at a cell density of 0.5 × 106 cells/mL and treated with 3 μM ActD (Sigma-Aldrich, St. Louis, MO). After incubation for 3 hr, cells were harvested and total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s protocol. mRNA was purified from total RNA using the Poly(A)Purist Kit, according to the manufacturer’s instructions, and fragmented via Nuclease P1 (NP1; Sigma-Aldrich) digestion. NP1 was dissolved in 30 mM Tris-HCl (pH 7.9), and 0.075 U NP1/μg mRNA was used for 55 min at 20°C in 50 mM sodium acetate buffer (pH 5.5). First- and second-strand cDNA synthesis was done using the RevertAid Premium First Strand cDNA Synthesis Kit with random hexamer primers followed by proofreading via Pfu Polymerase (30 min at 72°C). Double-stranded cDNA was next treated with the Fast DNA End Repair Kit and cut with NotI-HF (New England Biolabs, Frankfurt, Germany) prior to ligation into linearized modified d2eGFP-plasmids upstream of the pbs region. Ligations were carried out with the T4 DNA ligase according to the manufacturer’s instructions. To produce the final mRNA library, the cDNA library ligation mixture was directly used as the template for amplification, with a forward primer (5′-CGCCTCGAGAATTAATACGACTCACTATAGGGCGAACTAGTACTCTTCTGGTCCCCACAGACTC-3′) binding the hAg-Kozak sequence and containing a T7 promoter and a reverse primer (5′-T60-CTCTTTGCCGTATCCCATCTTAG-3′) binding the pbs and containing a poly(A)-tail of 60 nt.
Amplification was done with Phusion Hot Start Polymerase in 3% DMSO following the manufacturer’s instructions, with 90 s at 98°C followed by 25 cycles of 20 s at 98°C, 30 s at 65°C, and 45 s at 72°C, followed by 5 min at 72°C. PCR products were purified via magnetic beads (Agencourt AMPure XP; Beckman Coulter, Pasadena, CA), according to the manufacturer’s instructions. The amplified cDNA library was then in vitro transcribed into mRNA library as described above.
The cell-based selection process comprised several rounds, starting with (1) the transcription of the library, (2) electroporation of the mRNA library into hDCs, and (3) extraction and amplification of stable sequences after defined time points. During the selection process, first-strand cDNA synthesis was done from total RNA isolations using the SuperScript II Kit and oligo(d)T-primers, according to manufacturer’s instructions and including an RNase H treatment for 20 min at 37°C. All other steps were done as described above.
Analysis of Selected 3′ UTRs
Selected 3′ UTRs were cloned into the Luc2CP-plasmid as described for the library construction above. PCR reactions were performed using 20 ng cDNA of selection rounds five and six each as the template with 90 s at 98°C followed by 10 cycles of 15 s at 98°C, 30 s at 65°C, and 25 s at 72°C, followed by 5 min at 72°C and using primers binding the 3′ end of the d2eGFP reporter gene and the pbs. Ligations were transferred into One Shot OmniMAX 2 T1R cells, and at least 100 clones from each selection round were picked, purified (NucleoSpin 8 Plasmid Kit; Macherey-Nagel, Düren, Germany), and sequenced (Eurofins, Luxembourg, Luxembourg). Sequencing results were analyzed via Clone Manager Software (Sci-Ed Software, Denver, CO), and the origin of the sequences and alignment region (5′ UTR, CDS, or 3′ UTR) was verified via BLAST (NCBI, Bethesda, MD). Moreover, a qualitative and quantitative comparison to naturally occurring mRNAs in hDCs was performed (NextBio; Illumina, San Diego, CA). For analysis of miRNA-binding sites, miRNA data from published data38, 39 considering hDC specificity determined by copy number and p value were combined to assemble a miRNA set typically found in hDCs. With this miRNA set, binding sites for miRNAs were analyzed via IntaRNA, a program for fast and accurate prediction of RNA-RNA interactions.56
Next Generation Sequencing
Sequence variability of the starting library was determined by NGS. Touch-Down-PCR was done to amplify the 3′ UTR library region using primers with a BpmI restriction site and 50 ng of starting cDNA library as template. The protocol of PCR was performed using Phusion Hot Start Polymerase in 6% DMSO, following the manufacturer’s instructions with 90 s at 98°C followed by 10 cycles of 20 s at 98°C, 30 s at 82°C–64°C, and 45 s at 72°C and 20 cycles of 90 s at 98°C, 30 s at 64°C, and 45 s at 72°C, followed by 5 min at 72°C. PCR products were digested with BpmI (New England Biolabs) to ensure that only the amplified 3′ UTR library region was left over for sequencing. Afterward they were prepared according to the TruSeq DNA Sample Preparation Kit (Illumina), omitting the fragmentation and size selection steps and sequenced using a HiSeq 2000 (Illumina). Analysis of sequence alignment of the reads was performed with STAR 2.3.057 using the human reference genome (hg19).
Electroporation of Cells
Electroporation of in vitro-transcribed mRNA was done as described previously,4, 7, 58 using a square-wave electroporation device (BTX ECM 830, Harvard Apparatus, Holliston, MA). Optimal electroporation parameters for each cell type were determined in preliminary experiments: hDCs (750 V/cm, one pulse of 12 ms); C2C12 (600 V/cm, five pulses of 5 ms within an interval of 400 ms); and HFFs (625 V/cm, one pulse of 24 ms). For the cell-based selection process, 10 μg mRNA library was used for 1 × 107 hDCs. For the analysis of mRNA expression kinetics, the following mRNA and cell amounts were used: hDCs (10 pmol, 1 × 106 cells); C2C12 (3 pmol Luc2CP and 2.6 pmol GFP as spike-in control, 1 × 106 cells); and HFFs (3 pmol Luc2CP and 2.6 pmol GFP as spike-in control, 1 × 106 cells).
Flow Cytometry Analysis
hDC purity and viability were analyzed using α-CD83-APC and α-DC-Sign antibodies (BD Biosciences, Franklin Lakes, NJ) as well as 7-Aminoactinomycin D (7-AAD; Beckman Coulter). Cells were fixed with 1% paraformaldehyde (Carl Roth, Karlsruhe, Germany) in PBS and were stained with the aforementioned antibodies in fluorescence-activated cell sorting (FACS) buffer (PBS, 5% FCS, and 5 mM EDTA). Flow cytometric data of stained or GFP-expressing cells were acquired on a FACS Canto II flow cytometer (BD Biosciences) and analyzed using FlowJo 7.6.5 software (Tree Star, Ashland, OR). For in vivo vaccination studies, peripheral blood was collected from the orbital sinus and spleen; single-cell suspensions were prepared in PBS by mashing tissue against the surface of a 70-μm cell strainer (Corning, Corning, NY), using the plunger of a 3-mL syringe (BD Biosciences). Erythrocytes were removed by hypotonic lysis. Quantification of antigen-specific CD8+ T cells recognizing gp70 AH1-A1 was performed using an H2-Ld/AH1-A1423–431 (SPSYVYHQF) tetramer kindly provided by the NIH tetramer core facility (Emory University Vaccine Center, Atlanta, GA) and co-staining with α-CD8 (BD Biosciences). Viability was determined using fixable viability dye. Flow cytometric data were acquired as described above.
Reporter Gene Assays
Luciferase expression in vitro was analyzed as described58 using the Bright-Glo Luciferase Assay System (Promega, Madison, WI), and stability and translational efficiency of different mRNA constructs were calculated as described before59 via R program (R Foundation for Statistical Computing, Vienna, Austria) using internal script codes. Half-lives of mRNAs in every experiment of Figure 2 were thereby determined to a corresponding internal 2hBg 3′ UTR-containing luciferase control and then calculated relative to the hBg 3′ UTR mean of all experiments. In all other experiments, half-lives of mRNAs were determined directly relative to hBg 3′ UTR. In vivo bioluminescence was evaluated using the Xenogen IVIS Spectrum Imaging System (Caliper Life Sciences, Waltham, MA). An aqueous solution (1.6 mg/250 μL) of D-luciferin (BD Biosciences) was injected intraperitoneally at the indicated time points after mRNA injection. Emitted photons of live animals were quantified 5 min later, with an exposure time of 1 min. Regions of interest (ROIs) were quantified as total flux (photons/s), represented by color bars using the complementary software IVIS Living Image 4.0. Quantification of d2eGFP RNA was performed by qRT-PCR analysis as described previously.59 Relative expression of d2eGFP RNA was quantified via ΔΔCT calculation. Half-lives of expression were calculated from fitted one-phase decay curves using GraphPad Prism 6.04.
In Vivo Vaccination Experiments
For the characterization of antigen translation and immunostimulatory capacity in vivo, BALB/c mice (n = 8) were immunized i.v. with 10 μg liposome-formulated Luc mRNA, as described previously.6 Translation of Luc mRNA was evaluated by in vivo bioluminescence imaging as described above. After 6 hr, only five mice could be followed up for the following time points, and one mouse was lost to follow-up after 24 hr. For immunogenicity studies, BALB/c mice (n = 5) were immunized three times i.v. (days 0, 7, and 14) with 5 μg formulated mRNA encoding the H-2Ld-restricted AH1-A5423–431 (SPSYAYHQF) epitope derived from Moloney Murine Leukemia Virus envelope gp70 with an amino acid substitution at position five (V/A) described previously.60 The frequency of antigen-specific CD8+ T cells of total CD8+ T cells was determined in the blood and spleen 5 days after the last vaccination by flow cytometry.
Reprogramming of Human Fibroblasts
Mature, double-stranded miRNAs 302a-d and 367 were mixed in equimolar ratio and stored at −80°C until use. Lipofections of synthetic mRNA were performed in 12-well plates previously coated with 0.1% gelatine solution (Merck Millipore, Burlington, MA) for 20 min at 37°C. 4 × 104 fibroblasts were seeded per well in 1.0 mL NutriStem medium. The first lipofection was performed after cells adhered, usually between 4 and 5 hr after plating. To prepare lipoplexes, 2.5 μL RNAiMAX was diluted in 104 μL Opti-MEM serum-free medium. 0.58 μg total RNA was diluted in 104 μL Opti-MEM and combined immediately with the diluted RNAiMAX-preparation. In more detail, the reprogramming mRNA mixtures were composed of 0.33 μg transcription factor mRNA encoding OSKMNL (3:1:1:1:1:1) supplemented with 0.083 μg of each EKB (1:1:1) (total mRNA = 0.58 μg) and 0.17 μg miRNAs 302a-d and 367 (0.4 μM each). After 15-min incubation, 200 μL lipoplex solution was added per well to the cells and incubated for 24 hr. To perform daily lipofections, culture supernatant containing lipoplexes of the previous lipofection was removed and fresh 1.0 mL NutriStem medium was added before lipofection was repeated. Medium was further exchanged on day 5 and day 8.
Characterization of Human iPSCs
AP activity of iPSCs was determined using the AP Staining Kit Red-Color (SBI Biosciences). Shortly, iPSCs grown were washed with 2 mL PBS and fixed for 1 min with 1 mL Fixation Solution. Thereafter cells were washed twice with 2 mL PBS and stained for active AP with 0.8 mL AP-Staining Solution (Solution A:Solution B:PBS, 1:1:4) in the dark. After 15 min, AP-Staining Solution was removed, and cells were washed twice with 2 mL PBS and analyzed by microscopy. The induction of endogenous hESC marker was performed by qRT-PCR exactly as described in detail previously.17 Relative expression changes of each transcript in the probe of interest as compared to the day 1 starting control were quantified using the ΔΔCT method and normalized to the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT). The fold inductions obtained by this calculation were presented relative to the corresponding 2hBg benchmark.
Statistics
Unless stated otherwise, results are expressed as mean ± SD. One- or two-way ANOVA was performed when more than two groups were compared, followed by multiple comparisons using Tukey’s or Dunnett’s post-test as indicated. Pearson two-tailed correlation analysis was performed to determine correlation coefficients. All statistical analysis was performed with GraphPad Prism 6.04. No statistical methods were applied to pre-determine sample size for animal experiments.
Author Contributions
Conceptualization, A.G.O.v.N., A.N.K., and U.S.; Methodology, A.G.O.v.N., A.N.K., V.B., and M.D.; Formal Analysis, A.G.O.v.N., M.A.P., L.M.K., and M.L.; Investigation, A.G.O.v.N., M.A.P., A.P., C.R., and L.M.K.; Resources, V.B., M.L., and U.S.; Writing – Original Draft, A.G.O.v.N. and M.A.P.; Writing – Review & Editing, M.A.P., L.M.K., A.N.K., Ö.T., and U.S.; Visualization, A.G.O.v.N., M.A.P., and L.M.K.; Supervision, A.N.K., V.B., B.V., T.B., S.F., M.D., M.L., and U.S.; Funding Acquisition, Ö.T. and U.S.
Conflicts of Interest
M.A.P., C.R., A.P., L.M.K., V.B., S.F., B.V., A.N.K., Ö.T., and U.S. are employees of BioNTech RNA Pharmaceuticals GmbH, a company that develops mRNA-based therapeutics. M.D. is a consultant for BioNTech RNA Pharmaceuticals GmbH. In addition, A.G.O.v.N., M.A.P., L.M.K., S.F., M.D., B.V., T.B., A.N.K., Ö.T., and U.S. are inventors on patent applications, which cover the optimization of RNA-based therapeutics and RNA-based reprogramming techniques. The remaining authors declare no competing interests.
Acknowledgments
We thank F. Wille, E. Böhm, T. Sticker, M. Drude, J. Beckerle, R. Roth, N. Krimmel, A. Breitkreuz, T. Hempel, M. Suchan, and L. Leppien for technical assistance; J. de Graaf and K. Walzer for technical support and advice; and S. Witzel and B. Tillmann for cloning of DNA plasmids. Furthermore, we like to acknowledge R. Rae for critical reading. The study was supported by the GO-Bio (grant ID 0315640) and Ci3 cluster of excellence funding of the Federal Ministry of Education and Research (BMBF), Germany.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, four figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.12.011.
Supplemental Information
References
- 1.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 2.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallazza B., Petri S., Poleganov M.A., Eberle F., Kuhn A.N., Sahin U. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. Wiley Interdiscip. Rev. RNA. 2015;6:471–499. doi: 10.1002/wrna.1288. [DOI] [PubMed] [Google Scholar]
- 4.Diken M., Kreiter S., Selmi A., Britten C.M., Huber C., Türeci Ö., Sahin U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18:702–708. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 5.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 6.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 7.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 8.Princiotta M.F., Finzi D., Qian S.B., Gibbs J., Schuchmann S., Buttgereit F., Bennink J.R., Yewdell J.W. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 9.Steinle H., Behring A., Schlensak C., Wendel H.P., Avci-Adali M. Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges. Stem Cells. 2017;35:68–79. doi: 10.1002/stem.2402. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-David U., Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 13.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 14.Davies M.V., Elroy-Stein O., Jagus R., Moss B., Kaufman R.J. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romano P.R., Zhang F., Tan S.L., Garcia-Barrio M.T., Katze M.G., Dever T.E., Hinnebusch A.G. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 1998;18:7304–7316. doi: 10.1128/mcb.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Symons J.A., Alcamí A., Smith G.L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 17.Poleganov M.A., Eminli S., Beissert T., Herz S., Moon J.I., Goldmann J., Beyer A., Heck R., Burkhart I., Barea Roldan D. Efficient Reprogramming of Human Fibroblasts and Blood-Derived Endothelial Progenitor Cells Using Nonmodified RNA for Reprogramming and Immune Evasion. Hum. Gene Ther. 2015;26:751–766. doi: 10.1089/hum.2015.045. [DOI] [PubMed] [Google Scholar]
- 18.Wilkins C., Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A., Morrisey E.E. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Wang G., Lu C., Guo X., Hong W., Kang J., Wang J. Synergetic cooperation of microRNAs with transcription factors in iPS cell generation. PLoS ONE. 2012;7:e40849. doi: 10.1371/journal.pone.0040849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guhaniyogi J., Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 22.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 23.Tanguay R.L., Gallie D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peixeiro I., Silva A.L., Romão L. Control of human beta-globin mRNA stability and its impact on beta-thalassemia phenotype. Haematologica. 2011;96:905–913. doi: 10.3324/haematol.2010.039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waggoner S.A., Liebhaber S.A. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. (Maywood) 2003;228:387–395. doi: 10.1177/153537020322800409. [DOI] [PubMed] [Google Scholar]
- 26.Russell J.E., Morales J., Liebhaber S.A. The role of mRNA stability in the control of globin gene expression. Prog. Nucleic Acid Res. Mol. Biol. 1997;57:249–287. doi: 10.1016/s0079-6603(08)60283-4. [DOI] [PubMed] [Google Scholar]
- 27.Aviv H., Voloch Z., Bastos R., Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell. 1976;8:495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- 28.Ross J., Sullivan T.D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- 29.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 30.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 31.Chrzanowska-Lightowlers Z., Lightowlers R.N. Fending off decay: a combinatorial approach in intact cells for identifying mRNA stability elements. RNA. 2001;7:435–444. doi: 10.1017/s1355838201001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuhashi K., Ohnishi S., Kodama T., Koike K., Kanamori H. In vitro selection of the 3′-untranslated regions of the human liver mRNA that bind to the HCV nonstructural protein 5B. Virology. 2014;450-451:13–23. doi: 10.1016/j.virol.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 33.Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E., Jr. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C.C., Kain S.R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 35.Diken M., Kreiter S., Vascotto F., Selmi A., Attig S., Diekmann J., Huber C., Türeci Ö., Sahin U. mTOR inhibition improves antitumor effects of vaccination with antigen-encoding RNA. Cancer Immunol. Res. 2013;1:386–392. doi: 10.1158/2326-6066.CIR-13-0046. [DOI] [PubMed] [Google Scholar]
- 36.Kreiter S., Diken M., Selmi A., Diekmann J., Attig S., Hüsemann Y., Koslowski M., Huber C., Türeci Ö., Sahin U. FLT3 ligand enhances the cancer therapeutic potency of naked RNA vaccines. Cancer Res. 2011;71:6132–6142. doi: 10.1158/0008-5472.CAN-11-0291. [DOI] [PubMed] [Google Scholar]
- 37.Van Nuffel A.M., Benteyn D., Wilgenhof S., Pierret L., Corthals J., Heirman C., van der Bruggen P., Coulie P.G., Neyns B., Thielemans K., Bonehill A. Dendritic cells loaded with mRNA encoding full-length tumor antigens prime CD4+ and CD8+ T cells in melanoma patients. Mol. Ther. 2012;20:1063–1074. doi: 10.1038/mt.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimi S.T., Fulcher J.A., Chang M.H., Gov L., Wang S., Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114:404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brümmer A., Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. BioEssays. 2014;36:617–626. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 41.Bradley R.D., Hillis D.M. Recombinant DNA sequences generated by PCR amplification. Mol. Biol. Evol. 1997;14:592–593. doi: 10.1093/oxfordjournals.molbev.a025797. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn A.N., Diken M., Kreiter S., Vallazza B., Türeci Ö., Sahin U. Determinants of intracellular RNA pharmacokinetics: Implications for RNA-based immunotherapeutics. RNA Biol. 2011;8:35–43. doi: 10.4161/rna.8.1.13767. [DOI] [PubMed] [Google Scholar]
- 43.Thiel W.H., Bair T., Wyatt Thiel K., Dassie J.P., Rockey W.M., Howell C.A., Liu X.Y., Dupuy A.J., Huang L., Owczarzy R. Nucleotide bias observed with a short SELEX RNA aptamer library. Nucleic Acid Ther. 2011;21:253–263. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javorovic M., Pohla H., Frankenberger B., Wölfel T., Schendel D.J. RNA transfer by electroporation into mature dendritic cells leading to reactivation of effector-memory cytotoxic T lymphocytes: a quantitative analysis. Mol. Ther. 2005;12:734–743. doi: 10.1016/j.ymthe.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 45.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chinnery P.F., Hudson G. Mitochondrial genetics. Br. Med. Bull. 2013;106:135–159. doi: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schon E.A., DiMauro S., Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa A.M., Pereira-Castro I., Ricardo E., Spencer F., Fisher S., da Costa L.T. GRG5/AES interacts with T-cell factor 4 (TCF4) and downregulates Wnt signaling in human cells and zebrafish embryos. PLoS ONE. 2013;8:e67694. doi: 10.1371/journal.pone.0067694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okada Y., Sonoshita M., Kakizaki F., Aoyama N., Itatani Y., Uegaki M., Sakamoto H., Kobayashi T., Inoue T., Kamba T. Amino-terminal enhancer of split gene AES encodes a tumor and metastasis suppressor of prostate cancer. Cancer Sci. 2017;108:744–752. doi: 10.1111/cas.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonoshita M., Aoki M., Fuwa H., Aoki K., Hosogi H., Sakai Y., Hashida H., Takabayashi A., Sasaki M., Robine S. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19:125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshioka N., Dowdy S.F. Enhanced generation of iPSCs from older adult human cells by a synthetic five-factor self-replicative RNA. PLoS ONE. 2017;12:e0182018. doi: 10.1371/journal.pone.0182018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshioka N., Gros E., Li H.R., Kumar S., Deacon D.C., Maron C., Muotri A.R., Chi N.C., Fu X.D., Yu B.D., Dowdy S.F. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13:246–254. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kreiter S., Konrad T., Sester M., Huber C., Türeci O., Sahin U. Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA. Cancer Immunol. Immunother. 2007;56:1577–1587. doi: 10.1007/s00262-007-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kowalska J., Lewdorowicz M., Zuberek J., Grudzien-Nogalska E., Bojarska E., Stepinski J., Rhoads R.E., Darzynkiewicz E., Davis R.E., Jemielity J. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA. 2008;14:1119–1131. doi: 10.1261/rna.990208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright P.R., Georg J., Mann M., Sorescu D.A., Richter A.S., Lott S., Kleinkauf R., Hess W.R., Backofen R. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–W123. doi: 10.1093/nar/gku359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koste L., Beissert T., Hoff H., Pretsch L., Türeci Ö., Sahin U. T-cell receptor transfer into human T cells with ecotropic retroviral vectors. Gene Ther. 2014;21:533–538. doi: 10.1038/gt.2014.25. [DOI] [PubMed] [Google Scholar]
- 59.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Türeci O., Sahin U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 60.Slansky J.E., Rattis F.M., Boyd L.F., Fahmy T., Jaffee E.M., Schneck J.P., Margulies D.H., Pardoll D.M. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529–538. doi: 10.1016/s1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.