Abstract
In the past decade, significant progress has been made in complex disease research across multiple omics layers from genome, transcriptome and proteome to metabolome. There is an increasing awareness of the importance of biological interconnections, and much success has been achieved using systems biology approaches. However, because of the typical focus on one single omics layer at a time, existing systems biology findings explain only a modest portion of complex disease. Recent advances in multi-omics data collection and sharing present us new opportunities for studying complex diseases in a more comprehensive fashion, and yet simultaneously create new challenges considering the unprecedented data dimensionality and diversity. Here, our goal is to review extant and emerging network approaches that can be applied across multiple biological layers to facilitate a more comprehensive and integrative multilayered omics analysis of complex diseases.
Keywords: integrative omics, network approaches, systems biology, computational methods
Introduction
Recent advances in multiple biological layers, such as the genome, transcriptome, proteome and metabolome, have significantly facilitated research into complex diseases. Traditional genome-wide association analysis (GWAS) has brought valuable insights into the genetic basis of human disease, such as PICALM for Alzheimer’s disease (AD) [1, 2] and SNCA for Parkinson’s disease [3]. The increasing awareness of biological interconnections and the wide application of systems biology approaches are yielding high-level insights into disease mechanisms, with a particular focus on networks and pathways. For example, using network analysis, Zhang et al. [4] found an immune/microglia subnetwork that was strongly related to the pathophysiology of late-onset AD, with TYROBP as a key regulator. In a separate in vitro study, this gene was also found to be directly involved with amyloid-beta turnover [4] and was recently reported to inhibit the expression of a well-known AD risk gene TREM2 in HeLa cells [5]. Despite these achievements, many existing studies still treat the genome, transcriptome, proteome and metabolome as isolated biological layers without fully acknowledging their interconnections. This shortcoming is largely because of the limited availability of multi-omics data collected on the same group of individuals, as well as the limited availability of sufficiently powerful tools for high-dimensional analysis. In view of the limited information carried by a single omics layer, there is the potential for multilayered analyses to be much more powerful in facilitating our understanding of disease complexity [6, 7], hence the necessity of an integrative approach to omics.
Recent efforts in collecting multi-omics data in the same group of individuals open numerous opportunities for more comprehensive analyses of complex diseases. Example projects include the Alzheimer’s Disease Neuroimaging Initiative (ADNI) [8], The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov/) and the International Cancer Genome Consortium (ICGC; http://icgc.org/). Instead of limiting their perspective to a single omics layer, these data collections create a molecular landscape spanning the genome, transcriptome, proteome and even metabolome [9]. By capturing the abnormalities across multiple molecular dimensions, these data sets are believed to hold great potential for revealing a multilayered molecular basis of complex diseases and are likely to provide insights for developing novel therapeutic interventions [10]. Although to date, there has been limited work in AD, integrative omics analysis has already been performed on the TCGA data and has helped to drive the progress of cancer research by revealing a large-scale integrative view of the molecular aberrations in various cancers [11–13].
Our goal is to perform a detailed review of network approaches across multiple biological layers to help future analyses of the emerging multi-omics data in complex disease studies. Networks constitute the foundation of biological systems, and substantial efforts have been dedicated to network analysis within each biological layer. For the genome, epistatic interactions have been evaluated that account for disease status or quantitative traits (QTs), and these gene–gene interactions can constitute one or more networks [14, 15]. For the transcriptome and proteome, network inference, pathway enrichment analysis and network module identification are three principal topics. Network inference aims to reconstruct the underlying dependency structure between entities [e.g. gene regulatory networks (GRNs)]; pathway enrichment analysis and network module identification help identify risk factors (e.g. perturbed pathways or network modules) by mapping candidate genes/proteins onto pathways or prior networks, such as protein–protein interaction (PPI) or gene co-expression networks. Protective effects can similarly be analyzed in a network framework. In the biomarker discovery field, these known networks can also serve as priors to help guide machine learning models, so that biologically meaningful biomarkers can be identified.
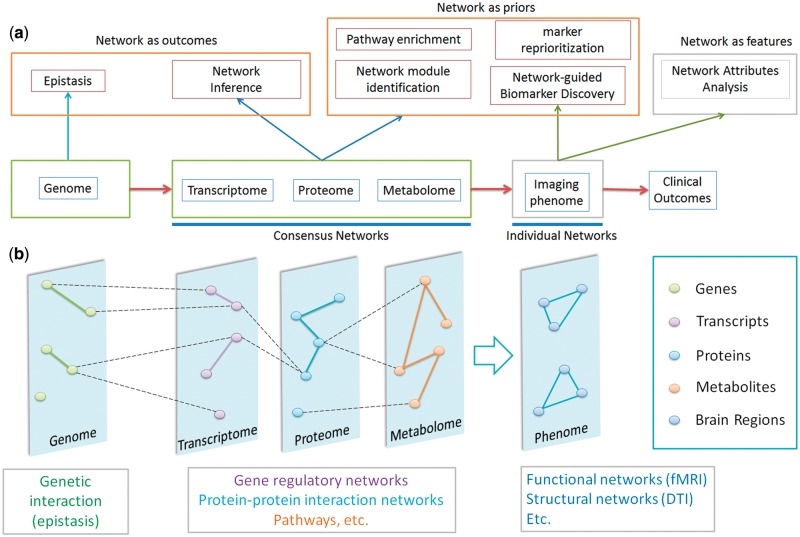
Based on the role of networks, analytic approaches can be divided into three groups. The first aims to explore the relationships between entities resulting in network generation; the second uses existing network(s) as prior knowledge to guide the analytic procedure; and the third analyzes the prior network(s) regarding their topology and attributes (both nodes and edges). This review is specifically focused on methods with wide applications in molecular omics layers from genome, transcriptome and proteome to metabolome. They can be further divided, based on the topic, into six subcategories as shown in Figure 1: (1) epistasis, (2) network inference, (3) pathway enrichment, (4) module identification, (5) marker reprioritization and (6) network-guided biomarker discovery. While the first two are mainly for exploration of disease mechanisms, the latter ones are expected to provide critical insights into perturbed pathways and to reveal potential diagnostic and therapeutic targets. As (3)–(6) are closely related topics, we will discuss them together in ‘Pathway and network analysis’ section.
Figure 1.
Networks and approaches in omics layers. (A) There are primarily three categories of network approaches across omics layers, where networks are generated as outcomes, used as priors or analyzed as features, respectively. (B) The ultimate goal of integrative omics is to illuminate the interplay across biological layers that potentially drives the progress of complex diseases.
In each section, we review state-of-the-art network approaches and associated tools, as well as their applications in disease studies. In addition, we summarize recent efforts on integrative analysis, as well as findings that take advantage of multi-omics data sets as a whole rather than one at a time. Our overarching goal is to provide a critical review and global map of network approaches spanning several of the most accessible data modalities in studies of complex diseases and to present a conceptual and methodological foundation for the emerging multi-omics research paradigm that is transforming medical research.
Epistasis
Though GWAS has dominated the genetic discovery of complex diseases over the past decade and identified hundreds of disease-associated single-nucleotide polymorphisms (SNPs), existing findings cannot fully explain either disease heritability or phenotypic variance [15–17]. This missing genetic variance remains largely unknown with some portion hypothesized to be because of the interactions between genetic loci (known as epistasis) [18]. Despite little progress in finding the missing heritability [19], epistasis analyses in model organisms have revealed a substantial portion of phenotypic variance explained by genetic interactions only [20]. The high dimensionality of genotype data makes the detection of epistasis effects computationally challenging; high-order epistasis involving more than two loci is even more complicated and rarely evaluated, given the daunting number of possible interactions [19]. Thus, we focus our review on those approaches for detection of pairwise interactions.
Regression-based models
Logistic regression and linear regression are two models widely applied to detect epistasis effects on disease case/control status and QTs, respectively. Usually, there will be two models: a saturated model and a reduced model. In the saturated model, it takes both genetic variants and their product as an input, which models the interaction effect, and estimates the total phenotypic variance explained by these predictors. In the reduced model, the multiplicative interaction component is excluded, and only the variance explained by the two genetic variants is estimated. Then, by directly comparing the variance explained in the saturated model relative to the reduced model, one can obtain the variance explained only by the interaction. In addition, by examining the weight and corresponding statistical value of the interaction term in saturated model, the statistical significance of the interaction effect can be derived and evaluated.
PLINK [21] and interSNP [22] are two widely used packages that implement these regression-based method. However, these analyses can be time-consuming, especially given the increasing genotyping resolution. Cordell et al. [23] reported that a pairwise interaction test of 89 294 SNPs would take over 300 h to complete in PLINK. Schupbach et al. [24] proposed a faster implementation through parallelization, which decreases the running time to around 24 h on a similar scale data set. Note that this implementation is only applicable to QTs, not case/control studies. BOOST is another high-performance tool that takes advantage of Boolean operation. BOOST was shown to be capable of evaluating all pairs of ∼360 000 SNPs within 60 h on a standard desktop computer and helped to identify a number of significant interactions in a study of diabetes [25]. SNPHarvester [26] improves the computation efficiency with an extra dimension reduction step. As removing nonsignificant SNPs is part of the dimension reduction strategy, this method has difficulties in identifying interaction pairs with weak or no main effects. Other tools based on regression methods include PIAM [27], epiGPU [28] and PLATO [29].
Contrast test approaches
For case/control studies, epistasis effects can be detected through examination of differences in the multi-locus association between cases and controls. For example, if disease risk is increased by the interaction of two SNPs, then the co-occurrence of the corresponding alleles should be enriched in the case group. Then, the significance of the interaction between two SNPs can be easily captured by contrasting the statistic measuring the association between each SNP pair between case and control groups. Such an approach will be computationally much more efficient, as degree of freedom reduces to one rather than four as in regression models mentioned earlier. Typical statistics used for the contrast test are chi-square, linkage disequilibrium [30], Pearson’s correlation [31] and odds ratio [21] metrics. One package implementing this strategy is SIXPAC [32], which coupled LD contrast test and Boolean operations for fast analysis. This package is capable of handling interactions between 450K SNPs within 8 h in a normal desktop computer [32]. Another tool, EPIQ [33], was proposed in a recent study to extend the contrast tests to QT analysis by coupling it with linear regression models.
Data mining/machine learning approaches
Multifactor dimensionality reduction (MDR) [14] is one of the earliest data mining strategies for exploring genetic interactions. For each genotypic class defined by the genotype of two SNPs, MDR classifies it as high-risk or low-risk based on the ratio of cases and controls in that class. In such a way, an n-dimensional problem is able to be transformed into a single-dimension problem, and the issues of sparse cells and the concerns of multiple parameters can be avoided [34]. To further extend the application of MDR to continuous QT, Lee et al. [35] later proposed an extension by introducing a traditional regression-based approach in the cell classification. Like all exhaustive search methods, MDR does not scale well to present data volumes, and thus, additional methods are built in as part of the software package to help filter SNPs before epistasis analysis [36]. A parallelized version of MDR with better scalability was later developed to facilitate the processing of large data sets [37].
Bayesian models are alternative approaches that do not require explicit modeling of all SNP pairs. Bayesian Epistasis Association Mapping (BEAM [38]) is specifically designed to capture the loci with either a main effect or an interaction effect. BEAM partitions all genotypes into three distinctive groups: one without any effect, one with independent main effect and one with joint effect on the outcomes based on the posterior probability. BEAM is capable of handling a data set with ∼100 000 SNPs in 50 cases and 50 controls [38]. However, considering that the scale of current genotyping platforms usually falls between 500K and 1M SNPs before imputation, a genome-wide interaction analysis remains a challenging task for the BEAM program, and additional data filtering techniques may be required.
Many other approaches have been explored for interaction analysis, such as information theory or entropy-based approaches [39, 40]. Detailed reviews are available in Cordell et al. [23], Wei et al. [41] and Upton et al. [42], which summarize the epistasis methods from different perspectives. A review by Koo et al. [43] focused on machine learning methods for epistasis analysis and detailed their application in various diseases.
Overall, despite its extremely challenging nature, genome-wide epistasis analysis has now become much more feasible. Exploration of genetic interactions that affect QTs has progressed by taking advantage of high-performance methodologies and tools. For example, Prabhu et al. [32] used the software SIXPAC to search for epistasis influencing bipolar disorder and identified a pair of interacting SNPs that had not been previously shown to have an effect in GWAS. Hemani et al. [44] used epiGPU to perform an exhaustive search and found multiple genetic interactions influencing gene expression in human peripheral blood. Despite the accumulating evidence indicating the importance of epistasis, many efforts to replicate these findings in independent cohorts have been unsuccessful [32, 45], and we are still far from understanding the underlying biological mechanisms. One possible solution is to perform extra downstream analyses for more insight into the underlying mechanisms, e.g. mapping variants to genes and then performing pathway enrichment analysis [46]. However, to have an influence on disease diagnosis and treatment, more work replicating and experimentally validating these epistasis findings in the future is needed.
Network inference
The emergence of high-throughput methods has revolutionized the study of diseases in the past decade and has greatly facilitated the exploration of interactions between biological entities. However, examining all of these experimentally still remains technically and financially infeasible. Reconstructing the underlying dependent relationships from observed data arises as an alternative strategy, which can also help potentially generate new biological hypotheses. These techniques have been widely explored in transcriptomics, where microarray gene expression data are used to infer GRNs. Other inferential networks of interest are transcript-binding network, PPI networks, gene co-expression networks and metabolic networks. As most methods are generalizable, we will review methods with a focus on transcriptome applications. Such data-driven network inference strategies have recently demonstrated great potential in discovering networks perturbed in disease, as discussed later in this section.
The simplest way to estimate the pairwise relevance is by correlation coefficients or mutual information (MI). On top of that, we can either define module networks, such as implemented in the weighted gene co-expression network analysis toolbox (WGCNA) [47], or generate a normal network by opting out those edges with relevance below a certain threshold. Networks generated in this way are undirected and known as correlation/relevance networks. Caution should be taken in direct application of these methods, as they are likely to generate numerous indirect connections as false positives. To overcome this limitation, context likelihood of relatedness (CLR) [48] derived a new score based on the distribution of MI to serve as the edge attribute, so that false-positive rate can be controlled. Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNE) [49] performed an extra filtering step in which the weakest edge in each triplet would be interpreted as indirect interaction and therefore be removed. This approach was found to be helpful in a GRN inference study in mammalian cells [49], but its computation complexity increases significantly with the network size. MRNet, which combines both criteria in CLR and ARACNE, was implemented in the R package MINET [50]. Such methods, though superior in simplicity, have limitations in identifying joint regulation effects, and their computational advantages disappear when extra filtering steps are involved to control the false-positive rate.
Network inference can also be formulated as a regression problem, such as in TIGRESS [51]. For example, the expression of one gene (response) is considered as a function of the expression of all other genes (predictors). In TIGRESS, lasso is applied to help yield sparse patterns, as links identified in this way are less likely to be indirect. The output of this strategy is also well known as an estimator of partial correlation relationships between genes. Owing to the predefined setting of ‘predictor’ and ‘response’, networks inferred in this way are both weighted and directional. GENIE3 [52] is a similar algorithm that implements random forest regression instead of lasso. This algorithm was recently extended in iRafNet [53] (available as an R package), where multiple data are integrated to significantly reduce the search space.
Probabilistic graph models, such as Bayesian network (BN) inference [54], could also be used to estimate direct influences. They compute the probability of the observed data given various a priori networks. Then, the one with the highest probability is selected as the most probable network. BN algorithms can capture linear, nonlinear, stochastic and combinatorial relationships between variables and are powerful for handling noisy data given their probabilistic nature. However, these BN algorithms are usually time-consuming. BNFinder [55] addresses this concern by taking advantage of multiple CPU cores, speeding up the algorithms linearly with increasing the number of cores. BNFinder has been recently used to identify genes with transcriptomic changes in smokers and to estimate the directional relationships between these changes [56]. In this work, the search space was further narrowed by performing BN inference only on those genes that were differentially expressed between groups.
BNs are directed acyclic graphs and do not allow feedback loops, which are important features in many biological networks. To overcome this limitation, dynamic Bayesian network (DBN) algorithms have been proposed. In these algorithms, DBNs are estimated as a function of time series observations, where each entity is unfolded into several nodes corresponding to the time points. Then, the algorithms construct priors, indicating the statistical dependence between variables at the initial time point, and a transition network, indicating the dependence between nodes at consecutive time points. Unlike BN algorithms, DBN algorithms have the power of detecting cyclic loops through the transition networks. Most tools, such as Banjo [57], SEBINI [58] and BNFinder [55], are designed for both static BN and DBN inference. An alternative approach for time series data is ordinary differential equations, which are specifically designed for inferring dynamic interactions between entities. In these models, the change in one measure, rather than the measure itself, is assumed to be the outcome of all other variables. Simple module methods and correlation/relevance network inference approaches are also applicable to time series data. TD-ARACNE [59] and the recently proposed algorithm, MIDER [60], are two example packages that allow time series data as input.
One natural question when facing many so methods to choose from is which of the methods is the best in a given circumstance. However, this question has not yet been conclusively answered. Even though many new methods claim themselves to be superior, it could also be argued that these methods are complementary rather than competitive to each other. All methods have their own advantages and limitations, especially when applied under different experimental settings. Marbach et al. [61] did an extensive comparison study on most of abovementioned methods. They evaluated those methods based on the area under precision–recall curve, which accounts for both false-positive rate and true-positive rate. Interestingly, they found that no method performed optimally across all experimental settings. In contrast, integration of multiple network inference methods shows the most robust and high performance across diverse data sets. This finding implies that different methods can only capture partial network structures individually, but fortunately can complement each other well. The same conclusion was also made in a previous work [62]. Inspired by this, efforts have been made to develop tools that combine networks inferred by multiple methods (e.g. NAIL [63]). However, most of these methods will become limited when the data volume is significantly increased. Relevance networks would perform better in high data volume situations, but unfortunately they are not directional and will be more likely to include many indirect edges with a high false-positive rate. A simple yet effective strategy to solve this problem is to narrow down the search space using prior knowledge as discussed in [53] and [56]. Though the majority of existing network inference methods are currently used for GRNs and co-expression networks, they are equally valuable for inference of other networks, such as metabolic networks [64]. Finally, most current studies are focused on network inference under various experimental conditions, but some have extended these methods to assess networks that are disturbed in diseases. For example, two recent studies [65, 66] have successfully used these methods to identify several key regulators of transcriptomic changes in AD.
Pathway and network analysis
Pathway and network analyses are two common procedures for exploration of high profile perturbations with candidate genes/proteins [67]. They allow us to benefit from knowledge of prior networks and pathways and gain extra insights from a higher-level perspective, which is of particular importance for large-scale analysis. In this section, we will cover four categories of pathway and network analysis approaches.
Pathway enrichment analysis
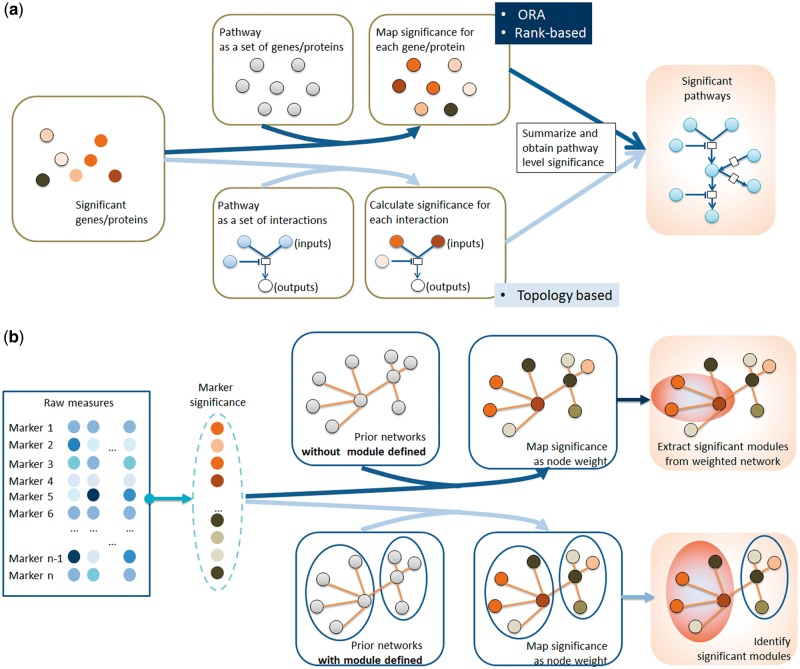
There are three dominant analytic techniques for pathway enrichment analysis (Figure 2A): (1) overrepresentation analysis (ORA), (2) rank-based approaches and (3) topology-based approaches. ORA is the first-generation approach for enrichment analysis and is still widely used because of its easy implementation. ORA assesses the significance of overrepresentation through hypergeometric distribution, chi-square or Fisher’s exact statistics. This method has been integrated into many tools including WebGestalt [68] and DAVID [69]. Notably, in ORA, the input candidates are assumed to be significant, and an arbitrary threshold is required for the prefiltering. In the enrichment step, all included markers are treated equally without further examination. This assumption leads to another key drawback of ORA, especially for large-scale analysis, where some risk markers may fall below the threshold and thus will be excluded. Also, significance scores in ORA also tend to vary considerably with small changes in overlap sizes [70]. In contrast, rank-based approaches account for the differences of all markers by taking their significance as an extra input. GSEA [71] is one typical example that first ranks all markers in the list and then generates the enrichment score through random walk using the weighted Kolmogorov–Smirnov-like statistic. Other similar tools include GenGen [72] and MAXMEAN [73]. Compared with ORA, the performance of rank-based approaches is not subject to an arbitrary threshold, but may be heavily affected by a few highly significant markers depending on the type of statistics applied [67].
Figure 2.
Common pipelines for pathway enrichment and network module identification. (A) Demonstration of three types of pathway enrichment analysis. In ORA and rank-based methods (following the dark blue line), the pathway-level significance is based on node counts (ORA) or the significance of individual nodes (rank-based); in topology-based methods (following the dark blue line), the topology of pathways, such as reactant and product in a single biochemical reaction, is taken into account. (B) Demonstration of two types of network analyses. The first one (following the dark blue line) identifies the module based on the weighted network after mapping gene/protein significance onto the node; the second one (following the light blue line) predefines the module based on the original network topology.
While the first two approaches purely treat pathways as simple sets of genes/proteins, third-generation approaches perform enrichment analysis in a more refined way by making the best use of topology information. Considering that pathway structure has long been known to be critical in biological function, topology-based approaches are believed to hold great promise for revealing more in-depth information. One example tool is EnrichNet [70], which generates an enrichment score for each pathway by estimating the distance of that pathway to all candidate genes in a network using a random walk with restart (RAR) algorithm. A recent method, named SAFE, also takes advantage of the random walk algorithm, but uses it for functional enrichment of the whole biological network [74]. SPIA [75] is another well-known topology-based enrichment analysis tool, which combines the evidence obtained from the classical ORA analysis with a novel measure of the actual perturbation on a given pathway. This family of algorithms has been recently extended in a tool called PhenoNet that accounts for topology information in both PPI networks and pathways via a two-step procedure [76]. The majority of existing topology-based tools are designed for transcriptomic microarray data. However, some of them only require differentially expressed gene list as the input, such as MetaCore (Thomson Reuters, http://www.thomsonreuters.com) and EnrichNet [70], and thus can be directed for other enrichment purposes. In addition, these enrichment methods are not only applicable to risk markers. By taking advantage of the databases characterizing functional relationship networks (e.g. STRING [77]), genetic interactions (e.g. BioGRID [78]) or physical interactions (HPRD [79]), the risk set can be easily extended by selectively incorporating their neighbors. This extension strategy will not only help to reveal the interaction of risk markers but also to discover other potential genes that might participate in disease because of interaction effects. By accounting for the topology information, the third-generation enrichment approaches have shown better performance than those only using the pathway memberships. However, it is worth noting that these approaches rely heavily on the network information documented in existing databases, which may be inaccurate, inconsistent, incomplete or not cell-type specific. One possible solution, as proposed recently by Ma et al. [80], is to first estimate a more reliable network by using existing networks as priors and then perform the enrichment analysis. However, whether this additional estimation step can provide a more accurate and more complete network remains to be seen, as the superior performance is only shown relative to classical ORA approaches.
Network module identification
Module-based approaches are an alternative to enrichment analyses, where the significance of each module is derived by aggregating the significance of all its member markers (Figure 2B). Usually, modules are predefined through analysis of gene co-expression networks [81] or PPI networks [82]. For example, both HotNet2 [83] and dmGWAS [84] map significance of single markers onto PPI nodes and extract a list of significant subnetworks by analyzing the topology of weighted network. In contrast, NIMMI [85] first constructs subnetworks of PPI based on the Google page rank algorithm and prioritizes them by the integrated statistics of their members.
Modules can be further examined by pathway enrichment analysis for broader findings. With the enrichment analysis of significant modules, Leiserson et al. [83] were able to illuminate the critical role of the combination of rare mutations in cancer and cross talks between cancer pathways. While these existing studies largely focus on statistical significance of modules, another potential extension will be to estimate the module-level polygenic risk of complex disease. Instead of a gene set obtained by thresholding GWAS results, densely connected genes within a module are more likely to function together in a collaborative way and exert polygenic effect toward QTs or disease status.
Marker prioritization
Another relevant topic that has been increasingly studied is prioritization of GWAS results. One dominant strategy is to rank genes based on their similarity to candidate disease genes in PPI network. The similarity can be measured by direct neighborhood relationship [86], shortest distance [87], random walk distance [88], etc. This strategy is capable of identifying novel biomarkers that may not be collected in a specific data set. But as a large part of disease genes remain unknown, candidate genes with close connections to them will inevitably be excluded if we purely rely on the similarity measures. Another recent strategy NetWAS by Greene et al. [89] proposes to reprioritize the genes based on their connections with GWAS findings without any prior knowledge of disease, where the whole network topology was used as the only input in a classification model. However, to be framed as a classification problem, NetWAS requires an arbitrary threshold of GWAS P-values, and its selection may affect the final reprioritization results. A regression model that can directly take the P-values or their log transformation may yield more stable results.
Prior-guided association analyses
Similar to third-generation enrichment analyses, another approach also uses the network topology information, but in the very first step, to generate a summary measure of each pathway/network module. These methods significantly reduce the dimensionality of the data, and derived scores allow for several types of further analysis, including disease classification, regression and survival prediction [90]. Pathifier [91], a tool designed for microarray data, generates a principle curve for each pathway and provides a pathway-level dysregulation score based on the projection in the principle curve. Pathologist, a Matlab-based tool, derives pathway-level measures by looking at each individual interaction and deriving two metrics, ‘activity’ and ‘consistency’ scores, to estimate each interaction’s possibility to occur [92]. This algorithm has been used in a cancer research for discovery of drug sensitivity predictors in cell lines. Glaab et al. [93] recently provided a thorough review of various dimension reduction algorithms used in existing pathway/network-based classification approaches and discussed their robustness, accuracy and biological interpretability. Compared with enrichment analysis, this approach is particularly desirable for capturing joint effects among markers that would be otherwise undetectable using methods that purely rely on the differential performance of individual markers.
In contrast to approaches separating pathway/network feature extraction and association analysis in two steps, some recent efforts have been made in advanced modeling to merge them into one step. Most of these approaches formulate the feature selection step as an optimization problem and incorporate the topology information of specific pathways/networks into a so-called ‘network-constrained penalty’ such that markers with joint effect can be detected. In a recent paper [94], a transcriptomic co-expression network of 15 amyloid genes was used as a prior to perform a bi-multivariate association analysis between amyloid imaging measures and genetic variants. A similar approach has also been used in Li et al. [95], where molecular interaction networks were incorporated using a graph Laplacian matrix. Both studies demonstrated better association performance when accounting for the topology information of prior networks than when simply using individual markers. With sparse models, a group of significant subnetworks can also be generated [95]. However, these methods cannot provide pathway/network-level significance toward phenotypes unless with further examination; only one network at a time is allowed as prior. Approaches serving this purpose are still evolving and much less explored than previously mentioned network approaches. Most of them are accessible as Matlab or R packages, such as HDBIG-SCCA [94], but for best usage, some computation background is required.
Multi-omics data and integrative analysis
In previous sections, we discussed state-of-the-art network analyses and approaches as well as their application to omics data. A list of the tools categorized by type is shown in Table 1. For each type of data or analytic method, network approaches have been developed and successfully adopted. However, the majority of work to date remains within single omics layer and does not address the connections between multi-omic layers. Even though pathway and network analyses use more than one omics data type to some extent, their findings still rely primarily on one single omics layer without effectively combining them together. However, science cannot truly progress, even with replicable significant findings, without combining together the multi-omics data into an integrated framework as none of the single dimensions can provide enough context or knowledge by themselves for full interpretation of a biological system [99]. Particularly for large GWAS, without integrative analysis, our knowledge of disease remains limited even if significant genetic variations are identified, as this analysis alone cannot tell the effects of these genetic alterations on downstream layers.
Table 1.
A list of network tools for omics studies
| Epistasis | Features | Reference |
|---|---|---|
| PLINK | Regression based | [21] |
| InterSNP | Regression based | [22] |
| Parallelized PLINK (FastEpistasis) | Regression based; not applicable to case control analysis | [24] |
| BOOST | Regression based; high-performance tool | [25] |
| SNPHarvester | Regression based; high-performance tool | [26] |
| SIXPAC | Contrast test based; only for case control analysis; high-performance tool | [32] |
| EPIQ | Contrast test based; QT analysis; high-performance tool | [33] |
| MDR | Data mining based; exhaustive search | [14] |
| BEAM | Bayesian based | [38] |
| Network inference | ||
| WGCNA | Correlation coefficient based; generating modular networks | [47] |
| CLR | MI based | [48] |
| ARACNE | MI based; pruning out indirect edges with extra filtering steps | [49] |
| MRNET | MI based | [50] |
| TIGRESS | Regression based | [51] |
| GENIE3 | Regression based; using random forest regression | [52] |
| iRafNet | Regression based; using prior biological knowledge to narrow the search space | [53] |
| BNFinder | BN inference | [55] |
| NAIL | An integration of many state-of-art network inference methods | [63] |
| Pathway and network analysis | ||
| WebGestalt | Overrepresentation-based enrichment tool | [68] |
| DAVID | Overrepresentation-based enrichment tool | [69] |
| GSEA | Rank-based gene set enrichment tool | [71] |
| GenGen | Rank-based enrichment tool | [72] |
| MAXMEAN | Rank-based enrichment tool | [73] |
| EnrichNet | Topology-based enrichment tool; combining rank-based enrichment score | [70] |
| PhenoNet | Topology-based enrichment tool; accounting for topology information in both PPI networks and pathways | [76] |
| SPIA | Topology-based enrichment tool; only focus on signaling pathway | [75] |
| HotNet2 | Network module identification | [83] |
| dmGWAS | Network module identification | [84] |
| NIMMI | Network module identification | [85] |
| GIANT | Reprioritization tool; no prior needed | [89] |
| HDBIG | Topology-guided association tool; biological network as prior; performing prediction tasks | [94] |
| Pathifier | Topology-guided association tool; pathway as prior; reducing the feature dimension by generating pathway-level measures | [91] |
| Pathologist | Topology-guided association tool; pathway as prior; examining each interaction in pathways and reducing the feature dimension by generating pathway-level measures | [92] |
| Integrative analysis | ||
| PARADIGM | Integrating copy number and gene expression data to estimate the activation status of each pathway for each sample | [96] |
| Lemon-Tree | Integrating multi-omics data for module network inference | [97] |
| ATHENA | Meta-dimensional multi-omics analysis package with both data filtering and interaction modeling | [98] |
Substantial attention has been devoted to integrative omics, as the TCGA data release for a comprehensive and thorough understanding of human health and disease. As a public funded project, the TCGA provides comprehensive genomic profiles for over 30 human tumors, e.g. genetic mutations, gene expression, microRNA (miRNA) sequencing, etc., to help advance the discovery of cancer-causing genomic alterations. A similar project is the ICGC, an international collaborative effort that also focuses on cancer data collection and distribution. Based on the TCGA data, plenty of efforts have been made to integrate the multidimensional genomic data for a comprehensive understanding of the underlying cancer biology. In [100], TCGA researchers performed a comprehensive analysis of cervical cancer using copy number, microRNA, mRNA and methylation data. In addition to examining each data type individually, they further integrated these multidimensional data and managed to identify three subgroups with distinct molecular characteristics. Similar approaches have also been applied to ovarian cancer [12], lung cancer [101], bladder cancer[11], etc. While these studies primarily aim to discover multiple types of genomic abnormalities associated with diseases, some other researchers take advantage of the multi-omics data to search for robust markers with evidence found in multiple omics layers [102]. In [103], Zhang et al. examined the association between gene expression, miRNA, DNA methylation and bone mineral density. Three genes and one miRNA were found to have consistent association evidence in both expression and methylation data, which suggests a consistent signal across layers. Although there is some debate about the precision oncology initiative given some initial trial failures [104, 105], findings from TCGA analyses have helped expand our current knowledge of cancer biology [12, 100], and we remain positive about the potentials of these comprehensive data sets to better understand human disease.
The ADNI is another example project that has led the efforts to collect multi-omics data in a human cohort with a primary focus on AD. Specifically, ADNI has collected genomic, transcriptomic and proteomic data for each individual in the study. In addition, they also have metabolomic data and a broad range of phenotypes available, including multimodal imaging measures and results from various clinical and cognitive assessments [9]. Further, as ADNI is a longitudinal study, this data set is still evolving and is advancing rapidly with more longitudinal profiles. Another AD-focused project, the Imaging and Genetic Biomarkers for AD (ImaGene), also provides a wide collection of omics data, such as GWAS, methylation and longitudinal blood gene expression data along with clinical, cognitive and imaging phenotypes [106]. To promote data sharing and analysis, the AMP-AD project (https://www.nia.nih.gov/alzheimers/amp-ad) was recently established, and multiple omics data sets are being incorporated into one knowledge portal to advance the integration of multi-omics studies in AD. These efforts have provided a global multi-omics landscape of individuals to allow studies to begin to fully characterize AD progression in multiple layers.
Another interesting topic of integrative omics is to understand the molecular interplays between different omic layers by taking advantage of computational network approaches. For example, Shin et al. [107] examined the effects of genetic variations on human blood metabolite networks by integrating GWAS with metabolite network data, inferred using Gaussian graphical models, and were able to generate nearly 100 new potential SNP-metabolite/disease correlations for further biomedical and pharmacogenetic assessment. Another work [108] explored a human blood metabolome/transcriptome interface where the between-level network was built using a thresholded correlation matrix and confirmed the cross talk between the biological layers at both a pathway level and a regulatory level.
Despite these progresses, computational network approaches in integrative omics are still notably under-explored and most studies use simple network approaches instead. While simple network approaches are easy to implement, caution should be taken when applying them, as their drawbacks may introduce certain bias especially with an escalating number of markers. As multi-omics data collection is much easier for simple model organisms, several leading groups have been working on more advanced computational approaches for integrative analysis in these systems. For example, Zhu et al. [99] estimated the causal network between genes and metabolites in yeast using a BN reconstruction algorithm in which genetic information, DNA–protein binding and PPI network were used as priors. These advanced approaches could be borrowed for multi-omics network analysis of human disease. With the significant progress of multi-omics data in human individuals, advanced network approaches to handle more layers are expected to be developed and implemented in future studies of human disease.
In addition to network approaches, integrative research tools are also in urgent need to extract the most salient knowledge out of the multi-omics data sets. Pathway Recognition Algorithm using Data Integration on Genomic Model (PARADIGM) is one of the earliest tools developed for integrative analysis [96]. By combining copy number and gene expression data, it takes into account different types of relationships within pathways using a probabilistic graphical model and is capable of providing a value for the activation status of each pathway for each sample. In the past few years, it has been extensively applied to TCGA analyses, which have revealed several key pathways associated with various cancers [12, 100]. Lemon-Tree [97], one recent effort in multi-omics network analysis, has recently been expanded to allow integration of multi-omics data for module network inference. Application of this tool identified several novel candidate driver genes in glioblastoma tumor. Another tool, ATHENA [98], is also dedicated to comprehensive analysis of multidimensional omics data by integrating data filtering and modeling together. In a recent breast cancer study based on copy number alteration, gene expression, protein expression and methylation data, ATHENA successfully identified complex nonlinear interactions across biological levels that contributed to cancer survival [109]. Despite the promising findings, these tools are far from enough to tackle the extensive multi-omics data necessary to fully understand human diseases. With the advance in individual multi-omics data collection and the intensive attention on the interplay between biological levels, integrative network approaches will be needed to enable the discovery of novel relationships that are causal for complex human diseases.
Conclusion
Network approaches have generated substantial interest based on their great potential for integrative omics analysis and are expected to facilitate a new era of precision understanding of complex diseases. However, this research field is still in its infancy, from concepts and approaches, to databases and enabling tools, with more expected in the near future. Biomedical research appears poised to take advantage of existing network approaches in single omics layers and soon will benefit from emerging multi-omics methods and tools.
Considering the dimensionality and heterogeneity of the contemporary biomedical data, a critical first step is data filtering, which will be a key enabler of initial multi-omics analyses. In this step, candidate markers from each omics layer can be extracted through traditional GWAS, statistical identification of differentially expressed genes, epistasis, etc. This method will significantly narrow the search space and allow further pathway enrichment, network module identification and direct application of the existing network inference approaches to explore the interplays between candidate markers, as many of existing approaches cannot yet manage large data scales. Reprioritization of initially identified candidate markers is an important follow-up stage, where new risk markers may be revealed by evaluating their distance to candidate markers in prior networks, e.g. PPI, gene regulatory and metabolic networks. It is also noteworthy that network-based prioritization can play an essential role in replication. Replication of findings in independent data sets is a convincing approach that has become a requisite step in single biological layers [110], but will be particularly challenging in multi-omics studies because of both limited data availability and the nature of network modules, which present challenges for the definition of replication. For example, is it sufficient for replication for the same network module to emerge with a particular gene or analyte? Network-based prioritization, which allows partial replication as long as markers identified in multiple data sets are closely related in prior networks, is increasingly being adopted in systems biology-level research. This strategy may be effective even in the absence of independent data sets with exactly same data modalities or measures for replication, as prioritization can help infer different modality markers if a prior network includes multimodal nodes, e.g. metabolic network with metabolites and genes. In addition to statistical analysis, informative integrative visualization of high-dimensional multi-omics networks is another challenging goal where accelerated progress is needed. Despite few extant multi-omics visualization tools, biomedical research would greatly benefit from more powerful network visualization tools.
Compared with traditional analysis with a focus on single biological layers, integrative multi-omics analysis is a new discipline with much higher dimensionality and much more complexity. It is ideally suited to the problems of complex diseases such as most forms of AD and cancer that are polygenic and multifactorial. Successful multi-omics research requires extensive collaborative efforts from a wide array of scientists, including disease experts, computer scientists, bioinformaticians, biologists and many others. While we can make progress with existing network approaches, novel methods, approaches and strategies are emerging that can be expected to bring a better understanding of important and interacting biological processes underlying complex human diseases.
Key Points
Understanding the complex relationships among multiple omics layers rather than individual genes/proteins is critical to enable a more complete view of complex disease.
Network approaches have generated substantial interest based on their great potential for integrative omics analysis and are expected to facilitate a new era of precision understanding of complex diseases.
From concepts and approaches, to databases and enabling tools, integrative omics is still in its infancy, with much more expected in the near future.
The ability to take advantage of existing network approaches in single omics layers is important to push the frontier, while new multi-omics methods and tools are in development.
Funding
The National Institutes of Health (grant numbers R01 EB022574, R01 LM011360, U01 AG024904, R01 AG19771, P30 AG10133, R01 CA129769, UL1 TR001108 and K01 AG049050); Department of Defense (grant numbers W81XWH-14-2-0151, W81XWH-13-1-0259 and W81XWH-12-2-0012); and National Collegiate Athletic Association (grant number 14132004), as well as by the Indiana University Network Science Institute (IUNI), the Alzheimer’s Association, the Indiana Clinical and Translational Science Institute and the Indiana University/IU Health Strategic Neuroscience Research Initiative (in part).
Biographies
Jingwen Yan is an assistant professor in the Department of BioHealth Informatics, School of Informatics and Computing, Indiana University Purdue University Indianapolis, USA.
Shannon L. Risacher is an assistant professor in the Department of Radiology and Imaging Sciences, Indiana University School of Medicine, USA.
Li Shen is an associate professor in the Department of Radiology and Imaging Sciences, Indiana University School of Medicine, USA.
Andrew J. Saykin is a professor in the Department of Radiology and Imaging Sciences, Indiana University School of Medicine, USA.
References
- 1. Shen L, Kim S, Risacher SL, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage 2010;53(3):1051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013;45(12):1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 2009;41(12):1308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 2013;153(3):707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pottier C, Ravenscroft TA, Brown PH, et al. TYROBP genetic variants in early-onset Alzheimer’s disease. Neurobiol Aging 2016;48:222.e9–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Civelek M, Lusis AJ.. Systems genetics approaches to understand complex traits. Nat Rev Genet 2014;15(1):34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature 2009;461(7261):218–23. [DOI] [PubMed] [Google Scholar]
- 8. Weiner MW, Aisen PS, Jack CR Jr. et al. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement 2010;6(3):202.e7–11.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saykin AJ, Shen L, Yao X, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimers Dement 2015;11(7):792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brubaker D, Difeo Chen AY, et al. Drug Intervention Response Predictions with PARADIGM (DIRPP) identifies drug resistant cancer cell lines and pathway mechanisms of resistance. Pac Symp Biocomput 2014:125–35. [PMC free article] [PubMed] [Google Scholar]
- 11. The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507(7492):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474(7353):609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163(2):506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 2001;69(1):138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moore JH, Williams SM.. Epistasis and its implications for personal genetics. Am J Hum Genet 2009;85(3):309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan J, Kim S, Nho SK, et al. Hippocampal transcriptome-guided genetic analysis of correlated episodic memory phenotypes in Alzheimer’s disease. Front Genet 2015;6:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tyler AL, McGarr TC, Beyer BJ, et al. A genetic interaction network model of a complex neurological disease. Genes Brain Behav 2014;13(8):831–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hemani G, Knott S, Haley C.. An evolutionary perspective on epistasis and the missing heritability. PLoS Genet 2013;9(2):e1003295.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sackton TB, Hartl DL.. Genotypic context and epistasis in individuals and populations. Cell 2016;166(2):279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kryazhimskiy S, Rice DP, Jerison ER, et al. Microbial evolution. global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 2014;344(6191):1519–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herold C, Steffens M, Brockschmidt FF, et al. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics 2009;25(24):3275–81. [DOI] [PubMed] [Google Scholar]
- 23. Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet 2009;10(6):392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schupbach T, Xenarios I, Bergmann S, et al. FastEpistasis: a high performance computing solution for quantitative trait epistasis. Bioinformatics 2010;26(11):1468–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wan X, Yang C, Yang Q, et al. BOOST: a fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am J Hum Genet 2010;87(3):325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang C, He Z, Wan X, et al. SNPHarvester: a filtering-based approach for detecting epistatic interactions in genome-wide association studies. Bioinformatics 2009;25(4):504–11. [DOI] [PubMed] [Google Scholar]
- 27. Liu Y, Xu H, Chen S, et al. Genome-wide interaction-based association analysis identified multiple new susceptibility Loci for common diseases. PLoS Genet 2011;7(3):e1001338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hemani G, Theocharidis A, Wei W, et al. EpiGPU: exhaustive pairwise epistasis scans parallelized on consumer level graphics cards. Bioinformatics 2011;27(11):1462–5. [DOI] [PubMed] [Google Scholar]
- 29. Grady BJ, Torstenson E, Dudek SM, et al. Finding unique filter sets in PLATO: a precursor to efficient interaction analysis in GWAS data. Pac Symp Biocomput 2010:315–26. [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao JY, Jin L, Xiong MM.. Test for interaction between two unlinked loci. Am J Hum Genet 2006;79(5):831–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kam-Thong T, Czamara D, Tsuda K, et al. EPIBLASTER-fast exhaustive two-locus epistasis detection strategy using graphical processing units. Eur J Hum Genet 2011;19(4):465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prabhu S, Pe’er I.. Ultrafast genome-wide scan for SNP-SNP interactions in common complex disease. Genome Res 2012;22(11):2230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arkin Y, Rahmani E, Kleber ME, et al. EPIQ-efficient detection of SNP-SNP epistatic interactions for quantitative traits. Bioinformatics 2014;30(12):i19–i25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Motsinger AA, Ritchie MD.. Multifactor dimensionality reduction: an analysis strategy for modelling and detecting gene-gene interactions in human genetics and pharmacogenomics studies. Hum Genomics 2006;2(5):318–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee SY, Chung Y, Elston RC, et al. Log-linear model-based multifactor dimensionality reduction method to detect gene gene interactions. Bioinformatics 2007;23(19):2589–95. [DOI] [PubMed] [Google Scholar]
- 36. Greene CS, Penrod NM, Kiralis J, et al. Spatially uniform reliefF (SURF) for computationally-efficient filtering of gene-gene interactions. BioData Min 2009;2(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bush WS, Dudek SM, Ritchie MD.. Parallel multifactor dimensionality reduction: a tool for the large-scale analysis of gene-gene interactions. Bioinformatics 2006;22(17):2173–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Liu JS.. Bayesian inference of epistatic interactions in case-control studies. Nat Genet 2007;39(9):1167–73. [DOI] [PubMed] [Google Scholar]
- 39. Moore JH, Gilbert JC, Tsai CT, et al. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol 2006;241(2):252–61. [DOI] [PubMed] [Google Scholar]
- 40. Chanda P, Zhang A, Brazeau D, et al. Information-theoretic metrics for visualizing gene-environment interactions. Am J Hum Genet 2007;81(5):939–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei WH, Hemani G, Haley CS.. Detecting epistasis in human complex traits. Nat Rev Genet 2014;15(11):722–33. [DOI] [PubMed] [Google Scholar]
- 42. Upton A, Trelles O, Cornejo-Garcia JA, et al. Review: high-performance computing to detect epistasis in genome scale data sets. Brief Bioinform 2016;17(3):368–79. [DOI] [PubMed] [Google Scholar]
- 43. Koo CL, Liew MJ, Mohamad MS, et al. A review for detecting gene-gene interactions using machine learning methods in genetic epidemiology. Biomed Res Int 2013;2013:432375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hemani G, Shakhbazov K, Westra HJ, et al. Detection and replication of epistasis influencing transcription in humans. Nature 2014;508(7495):249–53. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Chiesa A, Lia L, Han C, et al. Investigation of epistasis between DAOA and 5HTR1A variants on clinical outcomes in patients with Schizophrenia. Genet Test Mol Biomarkers 2013;17(6):504–7. [DOI] [PubMed] [Google Scholar]
- 46. Upton A, Trelles O, Perkins J.. Epistatic analysis of Clarkson disease. Procedia Comput Sci 2015;51:725–34. [Google Scholar]
- 47. Langfelder P, Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Faith JJ, Hayete B, Thaden JT, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 2007;5(1):54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 2006;7:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meyer PE, Lafitte F, Bontempi G.. minet: A R/Bioconductor package for inferring large transcriptional networks using mutual information. BMC Bioinformatics 2008;9:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haury AC, Mordelet Vera-Licona FP, et al. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst Biol 2012;6:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huynh-Thu VA, Irrthum A, Wehenkel L, et al. Inferring regulatory networks from expression data using tree-based methods. PLoS One 2010;5(9):e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petralia F, Wang P, Yang JL, et al. Integrative random forest for gene regulatory network inference. Bioinformatics 2015;31(12):197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Friedman N, Linial M, Nachman I, et al. Using Bayesian networks to analyze expression data. J Comput Biol 2000;7(3–4):601–20. [DOI] [PubMed] [Google Scholar]
- 55. Wilczynski B, Dojer N.. BNFinder: exact and efficient method for learning Bayesian networks. Bioinformatics 2009;25(2):286–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jennen DG, van Leeuwen DM, Hendrickx DM, et al. Bayesian network inference enables unbiased phenotypic anchoring of transcriptomic responses to cigarette smoke in humans. Chem Res Toxicol 2015;28(10):1936–48. [DOI] [PubMed] [Google Scholar]
- 57. Yu J, Smith VA, Wang PP, et al. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics 2004;20(18):3594–603. [DOI] [PubMed] [Google Scholar]
- 58. Taylor RC, Shah A, Treatman C, et al. SEBINI: Software Environment for BIological Network Inference. Bioinformatics 2006;22(21):2706–8. [DOI] [PubMed] [Google Scholar]
- 59. Zoppoli P, Morganella S, Ceccarelli M.. TimeDelay-ARACNE: reverse engineering of gene networks from time-course data by an information theoretic approach. BMC Bioinformatics 2010;11:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Villaverde AF, Ross J, Moran F, et al. MIDER: network inference with mutual information distance and entropy reduction. PLoS One 2014;9(5):e96732.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marbach D, Costello JC, Kuffner R, et al. Wisdom of crowds for robust gene network inference. Nat Methods 2012;9(8):796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. De Smet R, Marchal K.. Advantages and limitations of current network inference methods. Nat Rev Microbiol 2010;8(10):717–29. [DOI] [PubMed] [Google Scholar]
- 63. Hurley DG, Cursons J, Wang YK, et al. NAIL, a software toolset for inferring, analyzing and visualizing regulatory networks. Bioinformatics 2015;31(2):277–8. [DOI] [PubMed] [Google Scholar]
- 64. Peterson C, Vannucci M, Karakas C, et al. Inferring metabolic networks using the Bayesian adaptive graphical lasso with informative priors. Stat Interface 2013;6(4):547–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aubry S, Shin W, Crary JF, et al. Assembly and interrogation of Alzheimer’s disease genetic networks reveal novel regulators of progression. PLoS One 2015;10(3):e0120352.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rembach A, Stingo FC, Peterson C, et al. Bayesian graphical network analyses reveal complex biological interactions specific to Alzheimer’s disease. J Alzheimers Dis 2015;44(3):917–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ramanan VK, Shen L, Moore JH, et al. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet 2012;28(7):323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang J, Duncan D, Shi Z, et al. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41:W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huang da W, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Glaab E, Baudot A, Krasnogor N, et al. EnrichNet: network-based gene set enrichment analysis. Bioinformatics 2012;28(18):i451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34(3):267–73. [DOI] [PubMed] [Google Scholar]
- 72. Wang K, Li MY, Bucan M.. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 2007;81(6):1278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tintle NL, Borchers B, Brown M, et al. Comparing gene set analysis methods on single-nucleotide polymorphism data from genetic analysis workshop 16. BMC Proc 2009;3(Suppl 7):S96.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Baryshnikova A. Systematic functional annotation and visualization of biological networks. Cell Syst 2016;2(6):412–21. [DOI] [PubMed] [Google Scholar]
- 75. Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics 2009;25(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ben-Hamo R, Gidoni M, Efroni S.. PhenoNet: identification of key networks associated with disease phenotype. Bioinformatics 2014;30(17):2399–405. [DOI] [PubMed] [Google Scholar]
- 77. Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res 2015;43:D470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Keshava Prasad TS, Goel R, Kandasamy K, et al. Human protein reference database–2009 update. Nucleic Acids Res 2009;37:D767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ma J, Shojaie A, Michailidis G.. Network-based pathway enrichment analysis with incomplete network information. Bioinformatics 2016;32(20):3165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010;466(7309):973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Baranzini SE, Srinivasan R, Khankhanian P, et al. Genetic variation influences glutamate concentrations in brains of patients with multiple sclerosis. Brain 2010;133(9):2603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Leiserson MD, Vandin F, Wu HT, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 2015;47(2):106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jia P, Zheng S, Long J, et al. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics 2011;27(1):95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Akula N, Baranova A, Seto D, et al. A network-based approach to prioritize results from genome-wide association studies. PLoS One 2011;6(9):e24220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Oti M, Snel B, Huynen MA, et al. Predicting disease genes using protein-protein interactions. J Med Genet 2006;43(8):691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Franke L, van Bakel H, Fokkens L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet 2006;78(6):1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kohler S, Bauer S, Horn D, et al. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet 2008;82(4):949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Greene CS, Krishnan A, Wong AK, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet 2015;47(6):569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pyatnitskiy M, Mazo I, Shkrob M, et al. Clustering gene expression regulators: new approach to disease subtyping. PLoS One 2014;9(1):e84955.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Drier Y, Sheffer M, Domany E.. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci USA 2013;110(16):6388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Greenblum SI, Efroni S, Schaefer CF, et al. The PathOlogist: an automated tool for pathway-centric analysis. BMC Bioinformatics 2011;12:133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Glaab E. Using prior knowledge from cellular pathways and molecular networks for diagnostic specimen classification. Brief Bioinform 2015;17(3):440–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yan J, Du L, Kim S, et al. Transcriptome-guided amyloid imaging genetic analysis via a novel structured sparse learning algorithm. Bioinformatics 2014;30(17):i564–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li CY, Li HZ.. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 2008;24(9):1175–82. [DOI] [PubMed] [Google Scholar]
- 96. Vaske CJ, Benz SC, Sanborn JZ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 2010;26(12):i237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bonnet E, Calzone L, Michoel T.. Integrative multi-omics module network inference with lemon-tree. PLoS Comput Biol 2015;11(2):e1003983.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Holzinger ER, Dudek SM, Frase AT, et al. ATHENA: the analysis tool for heritable and environmental network associations. Bioinformatics 2014;30(5):698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhu J, Sova P, Xu QW, et al. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol 2012;10(4):e1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ramirez LM, Goukasian N, Porat S, et al. Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol Aging 2016;39:82–9. [DOI] [PubMed] [Google Scholar]
- 104. Prasad V. Perspective: the precision-oncology illusion. Nature 2016;537(7619):S63.. [DOI] [PubMed] [Google Scholar]
- 105. Abrahams E, Eck SL.. Molecular medicine: precision oncology is not an illusion. Nature 2016;539(7629):357. [DOI] [PubMed] [Google Scholar]
- 100. The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017;543(7645):378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489(7417):519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Whitaker JW, Boyle DL, Bartok B, et al. Integrative omics analysis of rheumatoid arthritis identifies non-obvious therapeutic targets. PLoS One 2015;10(4):e0124254.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang JG, Tan LJ, Xu C, et al. Integrative analysis of transcriptomic and epigenomic data to reveal regulation patterns for BMD variation. PLoS One 2015;10(9):e0138524.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet 2014;46(6):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bartel J, Krumsiek J, Schramm K, et al. The human blood metabolome-transcriptome interface. PLoS Genet 2015;11(6):e1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim D, Li R, Dudek SM, et al. Predicting censored survival data based on the interactions between meta-dimensional omics data in breast cancer. J Biomed Inform 2015;56:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ritchie MD, Holzinger ER, Li R, et al. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet 2015;16(2):85–97. [DOI] [PubMed] [Google Scholar]