Abstract
Lifestyle factors, such as food choices and exposure to chemicals, can alter DNA methylation and lead to changes in gene activity. Two such exposures with pharmacologically active components are coffee and tea consumption. Both coffee and tea have been suggested to play an important role in modulating disease-risk in humans by suppressing tumour progression, decreasing inflammation and influencing estrogen metabolism. These mechanisms may be mediated by changes in DNA methylation. To investigate if DNA methylation in blood is associated with coffee and tea consumption, we performed a genome-wide DNA methylation study for coffee and tea consumption in four European cohorts (N = 3,096). DNA methylation was measured from whole blood at 421,695 CpG sites distributed throughout the genome and analysed in men and women both separately and together in each cohort. Meta-analyses of the results and additional regional-level analyses were performed. After adjusting for multiple testing, the meta-analysis revealed that two individual CpG-sites, mapping to DNAJC16 and TTC17, were differentially methylated in relation to tea consumption in women. No individual sites were associated with men or with the sex-combined analysis for tea or coffee. The regional analysis revealed that 28 regions were differentially methylated in relation to tea consumption in women. These regions contained genes known to interact with estradiol metabolism and cancer. No significant regions were found in the sex-combined and male-only analysis for either tea or coffee consumption.
Introduction
Both coffee and tea include hundreds of different compounds, which can be found and measured in blood (1–4) and that might be associated with different outcomes. Coffee consumption has received much attention in research, which has resulted in associations between coffee consumption and lower risk of several diseases, for example, Alzheimer’s disease, dementia (5), Parkinson’s disease (6) and type 2 diabetes (7). On the other hand, coffee consumption has also been associated with an increased risk of high blood pressure (8,9), different types of cancer (10,11) and myocardial infarction (12). Both the beneficial and negative health effects may spring from different compounds within coffee.
Even though most studies considered the impact of age, sex, body mass index (BMI), genetics and smoking, the health effects of coffee consumption might still be hard to separate from the effects of people’s lifestyles and previous health problems. Studies have shown that caffeine, the major pharmacologically active constituent of both coffee and tea, is the explanatory molecule for the reduced risk of Parkinson’s disease attributed to coffee consumption (13). Caffeine also reduces dopaminergic neurotoxicity in animal models of Parkinson’s disease (13,14). On the other hand, the roasting of coffee leads to production of polycyclic aromatic hydrocarbons, which are known to be carcinogenic (15). Tea contains substances, such as polyphenols, which are linked to lower risk of heart disease, cancer and diabetes (16–20). Polyphenols are antioxidants that latch onto and neutralize chemicals called oxidants. Oxidants have been linked with increased risk of cardiovascular disease (21,22).
Studies on smoke exposure and diet have shown that consumption of bioactive components can leave a persistent mark on the epigenetic information layer (23,24). Epigenetics is generally used to denote the regulation of genes that cannot be attributed to nucleotide variation in the DNA sequence. It includes a number of different mechanisms of which DNA methylation is among the most well studied. However, a systematic genome-wide analysis of the influence of coffee and tea on DNA methylation has not been performed previously.
One reason to study the influence of coffee and tea consumption on DNA methylation is that previous genetic studies have presented evidence that individual coffee consumption are related to genetic factors (25,26). The genetic background of tea consumption is less conclusive at present. However, it has previously been demonstrated that tea catechins inhibit DNA methylation in vitro and in cultured cancer cells (27,28), arguing that some of the health effects of tea may be mediated by epigenetics. In this study, we performed an Epigenome-Wide Association Study (EWAS) and a regional-level analysis to determine the association between coffee and tea consumption and changes in DNA methylation.
Results
Following exclusion of individuals lacking phenotype data, DNA methylation data or information on any of the covariates, a final set of 3,096 individuals from NSPHS (N = 723); PIVUS (N = 804), HWFS (N = 948) and EGM (N = 621) were used in the meta-analysis (Table 1). Coffee and tea consumption as well as mean age for all cohorts are included in Table 1. Mean age of all included cohorts was 56 years. Tea and coffee consumption differed between men and women, tea consumption in women was significantly higher in all cohorts except EGM and coffee consumption was higher in men in all cohorts except in EGM where coffee consumption was significantly higher in women (Table 1).
Table 1.
Subject characteristics for all included populations
| COFFE (cups/month) | TEA (cups/month) | |||
|---|---|---|---|---|
| N (men/women) | Age (mean) | Mean (men/women) | Mean (men/women) | |
| NSPHS | 723 | 14–94 years | 106.2 (122.5/92.5) | 10.95 (7.4/13.9) |
| (335/380) | (50.0) | (3.61 × 10−6) | (P = 0.00033) | |
| PIVUS | 804 | 69.8–70.7 years | 89.7 (95.6/83.7) | 21.7 (19.51/24.0) |
| (405/399) | (70.1) | (P = 0.007) | (P = 0.007) | |
| HWFS | 948 | 36.5–76.8 years | 107.02 (117.8/98.6) | 57.2 (45.5/66.8) |
| (428/520) | (58.4) | (P = 3.74 × 10−7) | (1.24 × 10−9) | |
| EGM | 621 | 29.6–74.9 years | 28.8 (31.7/27.3) | 9.7 (9.4/10.3) |
| (223/398) | (52.1) | (P = 0.020) | (P = 0.50) |
N is the number of individuals included in the analysis for each cohort. P is calculated with a student t-test to estimate if there is a significant difference in the amount of coffee and tea consumed between males and female.
EWAS
Sex-combined analysis
In the sex-combined analyses, no individual CpG-sites were significantly associated to either coffee or tea consumption, using a Bonferroni threshold correcting for 421,695 sites (P < 1.19×10−7).
Sex-stratified analysis
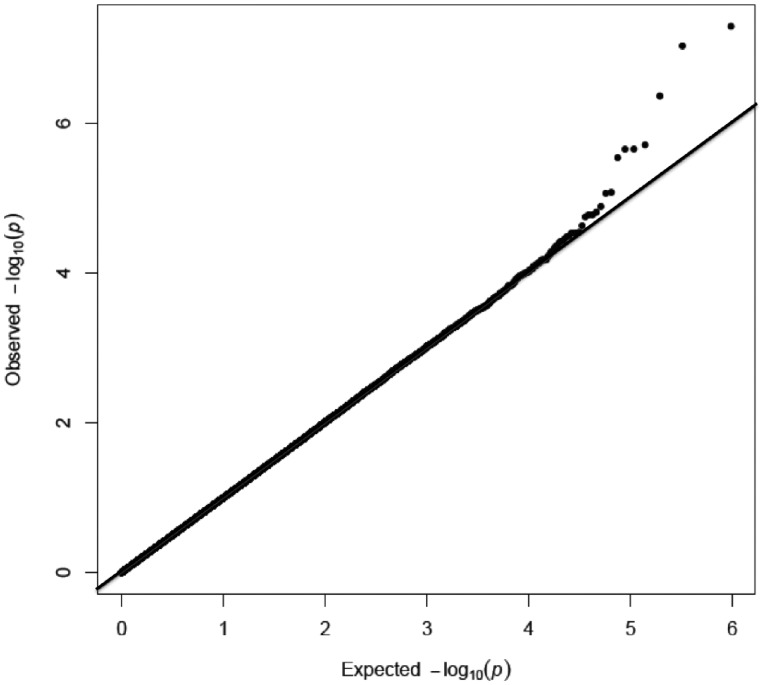
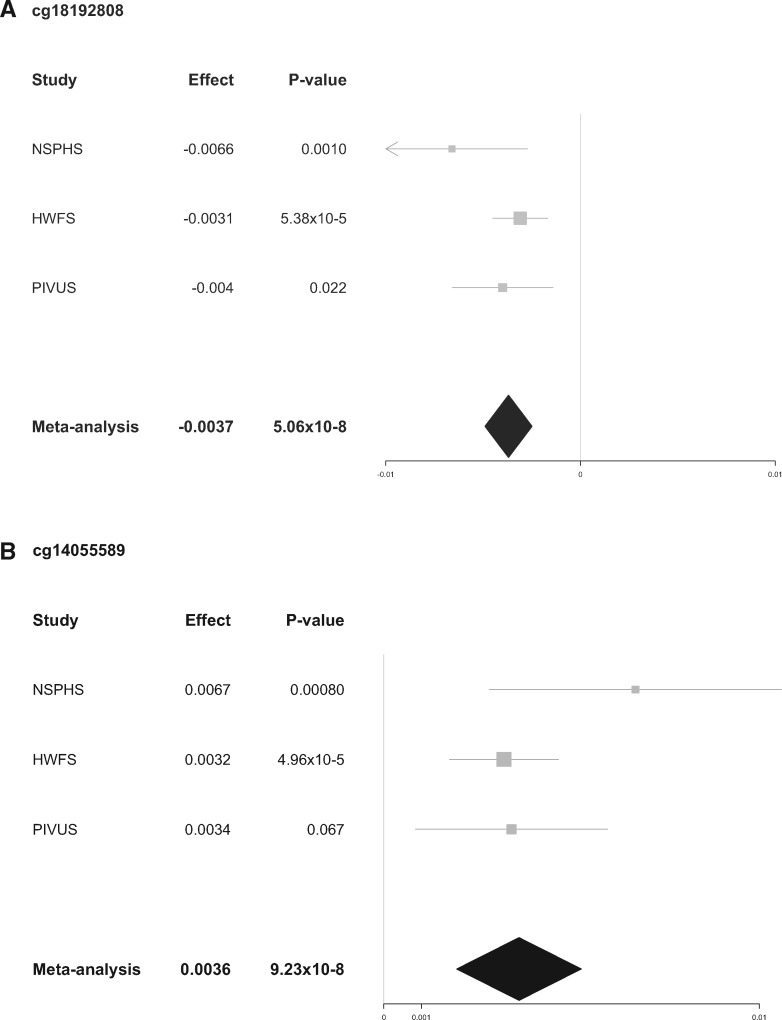
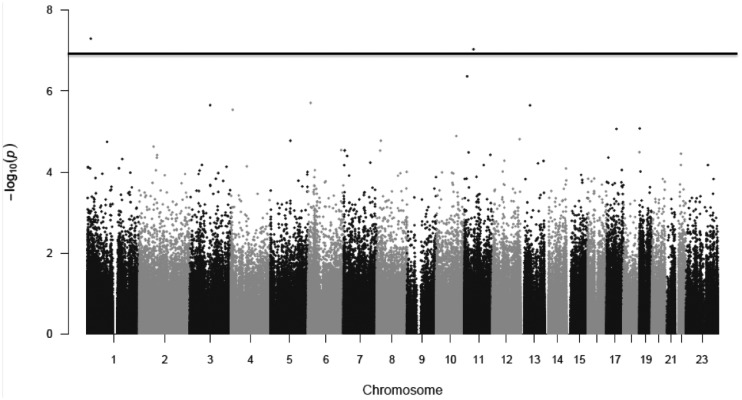
No CpG-sites were significantly associated with coffee or tea in men. Neither did we see an association between coffee and DNA-methylation in women. However, we did find an association between DNA-methylation and tea in women. After adjusting for inflation in each population (λNSPHS=0.95, λHWFS=1.14, λPIVUS=1.24, and λEGM=1.00), no inflation was seen in the meta-analysis (λ = 1.00) (Fig. 1). The lambda-adjusted analyses revealed two significant sites, CpG-site cg18192808 (5.06×10−8), mapped to DnaJ heat shock protein family (Hsp40) member C16 (DNAJC16) and cg14055589 (9.23×10−8,) mapped to Tetratricopeptide domain 17 (TTC17) (Figs 1 and 2). cg18192808 were nominally significant with the same direction of effect in all included populations separately (Table 2, Fig. 3A), and cg14055589 were nominally significant with the same direction of effect for all cohorts except PIVUS (P = 0.067) (Table 2, Fig. 3B). None of these two sites passed QC in the EGM cohort. In men, cg18192808 and cg14055589 were not significant in the meta-analysis and overall had the opposite direction of effect in most cohorts as compared to women (Table 2). None of the CpG-sites located on the sex chromosomes passed, for any of the phenotypes, even a chromosome-wide Bonferroni threshold (Px<4.45×10−6 and Py<1.2×10−4, respectively).
Figure 1.
QQ-plot for the meta-analysis for tea consumption in women.
Table 2.
Significant CpG-sites in the meta-analysis associated with tea consumption in women
| Meta-analysis |
Individual Cohorts |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NSPHS |
HWFS |
PIVUS |
EGM |
||||||||||
| CpG-site | I2 | P | Effect | P | Effect | P | Effect | P | Effect | P | Effect | Chr | Gene |
| cg18192808 | 30 | 5.06 × 10−8 | −0.0037 | 0.001 | −0.0066 | 5.38 × 10−5 | −0.0031 | 0.022 | −0.004 | na | na | 1 | DNAJC16 |
| (0.51) | (0.0004) | (0.71) | (-1×10−5) | (0.78) | (−0.0002) | (0.039) | (0.004) | ||||||
| cg14055589 | 25 | 9.23 × 10−8 | 0.0036 | 0.0008 | 0.0067 | 4.96 × 10−5 | 0.0032 | 0.087 | 0.0034 | na | na | 11 | TTC17 |
| (0.62) | (−0.005) | (0.24) | (−5×10−5) | (0.39) | (−0.001) | (0.64) | (0.0008) | ||||||
P-value and effect in men are presented below in italic within brackets. The values for each population is also included, i.e. NSPHS (N = 723), HWFS (N = 949), PIVUS (N = 804) and EGM (N = 621). The two significant CpG-sites were missing in the EGM cohort. N is the number of included populations and effect is the standard deviation of methylation change per cup tea consumed. I2 describes the percentage of the variability in effect estimates that is due to hetrogeneity rather than sampling error (chance).
Figure 3.
Forest plot of the association between DNA methylation and tea consumption in females. Shown are the effect (95% CI) and P-values for each individual cohort as well as the meta-analysis. (A) cg18192808 was nominally significant with the same direction of effect in all included cohorts, NSPHS, HWFS and PIVUS, and significant at Bonferroni adjusted threshold in the meta-analysis (P < 1.19 × 10−7). (B) cg14055589 was nominally significant with the same direction of effect in NSPHS and HWFS, but not significant in PIVUS.
Figure 2.
Manhattan plot for the meta-analysis for tea consumption in women. Bonferroni Significance threshold is marked with a only line (1.19 × 10−7).
The four cohorts mainly include adults and elderly people. Therefore, we cannot conclude that the effect would be the same in younger individuals. However, to investigate if the effect seems to be similar between younger and older individuals we stratified females from NSPHS into two groups. The first group included females younger than 50 years old (N = 215) and the second group included females at the age of 50 or older (N = 170). The association between tea consumption and DNA methylation for the two significant CpG-sites, cg18192808 and cg14055589, show that there are no significant difference between the effects for the two age groups for either of the two sites. For cg18192808, the effect was −5.43×10−5 (P = 0.042) and −7.08×10−5 (P = 0.042), respectively for the young and the older group, and for cg14055589, the effect was 6.95×10−05 (P = 0.069) and 0.00019 (P = 0.053), respectively for the young and the older group.
Regional-level analysis
Regional-level (locus) analysis was performed using the comb-p software (29) to identify differentially methylated regions of the genome associated with total coffee and tea consumption. Lambda adjusted P-values were used for this analysis. In the sex-combined analyses and when analyzing men separately, no significant regions were identified for tea and coffee. No significant regions were identified for coffee in women but when analyzing tea in women, comb-p identified 28 significantly associated regions, located on 13 different chromosomes, mapping to 17 different genes (Table 3).
Table 3.
Comb-p differentially methylated region analysis
| Region | N CpG-sites | P.adj* | Gene |
|---|---|---|---|
| chr11:2322286-2323459 | 23 | 1.07 × 10−6 | TSPAN32 |
| chr11:67383377-67384040 | 8 | 5.86 × 10−6 | |
| chr13:36871646-36872346 | 14 | 7.49 × 10−6 | SMAD9 |
| chr6:33244709-33246488 | 52 | 1.08 × 10−5 | VPS52/HCG25 |
| chr3:42977777-42978248 | 8 | 3.24 × 10−5 | |
| chr6:149805995-149806732 | 11 | 0.00030 | PCMT1 |
| chr6:169238138-169238443 | 5 | 0.00047 | THBS2 |
| chr2:84743142-84743743 | 12 | 0.00076 | DNAH6 |
| chr4:8412369-8412795 | 4 | 0.00101 | ACOX3 |
| chr4:1243849-1244086 | 7 | 0.00208 | CTBP1 |
| chr1:75198211-75199117 | 11 | 0.00225 | SLC44A5 |
| chr19:55549414-55549842 | 8 | 0.00267 | |
| chr1:211652549-211652741 | 3 | 0.00282 | |
| chrX:8751190-8751687 | 7 | 0.00333 | |
| chr17:37123638-37123949 | 9 | 0.00409 | ACACA |
| chr7:2653651-2653733 | 3 | 0.00561 | TTYH3 |
| chr6:31733619-31734580 | 20 | 0.00636 | CLIC1 |
| chr13:84660622-84660772 | 2 | 0.01012 | |
| chr17:27346732-27347260 | 6 | 0.01122 | |
| chr19:47287778-47288263 | 7 | 0.01148 | |
| chr6:72130209-72130799 | 12 | 0.02469 | RIMS1 |
| chr15:40268421-40268777 | 4 | 0.02623 | PAK6 |
| chr12:75785089-75785295 | 5 | 0.02746 | |
| chr6:31690580-31692375 | 38 | 0.03345 | ABHD16A |
| chrX:150151572-150151823 | 12 | 0.03486 | |
| chr12:53359155-53359506 | 3 | 0.03557 | |
| chr13:25670047-25670327 | 11 | 0.0473 | ATP8A2 |
| chr3:142666108-142666476 | 4 | 0.04955 | PLS1 |
Comb-p performs a Stouffer-Liptak correction on probes by assessing all CpG-sites within defined window, as weighted by their observed correlation. *Significant threshold is 0.05.
Discussion
In this study, we identified differentially methylated CpG-sites in whole blood that are associated with self-reported tea consumption in women only, but not in men. Tea consumptions appear to be associated with DNA methylation in women. Women drink higher amounts of tea compared to men, which may increase the power to find associations in women. Results from previous studies have shown that tea consumption reduces estrogen levels by catechins and theaflavines that inhibit aromatase, an enzyme which catalyses the conversion of androgens to estrogens. This highlights a potential difference between the biological response to tea in men and women (30–32). We did not find an association between DNA-methylation and coffee, in either women or men.
Age, sex and smoking are potential confounders on the association between tea or coffee consumption and DNA methylation. Therefore, all analyses were adjusted for these variables but did not stratify by reported smoking habits. In agreement with this, our two significant sites did not overlap with findings from a large meta-EWAS on smoking (33), making confounding by tobacco smoking an unlikely explanation of our findings. It is also important to note that tea and coffee findings were both corrected for each other, making it likely that a factor associated with tea, and not coffee, is driving the association.
The EWAS revealed two significant loci, cg18192808 and cg14055589. cg18192808 maps to DNAJC16, which is paralogue to DnaJ Heat Shock Protein Family (Hsp40) Member A3 (DNAJA3). DNAJA3 plays a critical role in tumor suppression through its interactions with oncogenic proteins (34). Interestingly, DNAJC16 has previously been shown to be up-regulated in HepG cells upon treatment of green tea (35). The second site, cg14055589 mapped to TTC17, which plays a role in ciliogenesis (36). Both these individual loci seem to mainly be driven by the HWFS cohort (Table 2 and Fig. 3A and B). This is probably due to the larger amount of tea consumed in HWFS, which provides a higher statistical power to detect associations within this cohort (Table 1). As an attempt to establish whether included studies are consistent; we present a statistical test of heterogeneity, I2, as presented in Table 2. I2 describes the percentage of the variability in effect estimates that is due to heterogeneity rather than by chance. The I2 values for our top CpG-sites describes a low to moderate heterozygosity (37).
Before adjusting for inflation, two additional individual sites were statistically significant for tea consumption in women, cg03155301 (unadjusted P = 7.47×10−9, adjusted P = 4.31×10−7) and cg24165638 (unadjusted P = 8.77×10−8, adjusted P = 8.36×10−6). cg03155301 maps to TEA domain transcription factor 1 (TEAD1) and showed a nominal significance in all included populations but with an opposite direction of effect in EGM. TEAD1 plays a key role in the Hippo signaling pathway, a pathway involved in organ size control and tumor suppression. Interestingly, studies have reported correlations between tea drinking and cancer (19,31,38). The second site, cg24165638, mapped to azurocidin 1 (AZU1), showed the same direction of effect in all populations, but was not nominally significant in NSPHS and EGM. AZU1 is an interesting gene since the protein it encodes binds heparin, which prevents blood coagulation (39). Diseases associated with AZU1 include heparin-induced thrombocytopenia and gastric cancer (40).
It has previously been shown that tea consumption is associated with a lower risk of developing cancer (19,31). In our regional analysis, five cancer-associated genes were differentially methylated with tea consumption in women: THBS2, which has been shown to function as a potent inhibitor of tumour growth (41) and angiogenesis (42); TSPAN32, which is located within an important tumour suppressor region (43); CTBP1, which is involved in promoting the carcinogenesis of human glioma (44); SLC44A5, a potential therapeutic target in hepatocellular carcinoma (45) and CLIC1 that is known to be overexpressed in malignant tumours (46,47) (Table 3).
As many as six genes known to interact with estradiol (THBS2, CTBP, ACACA, RIMS1, PLS1, CLIC1) were significantly associated with tea consumption in the regional analysis in women (41,42,48–51). Previous studies have reported tea consumption to lower estradiol levels, which is believed to protect against breast cancer (31,32). It is also biologically plausible that the interaction between tea consumption, estrogen levels and DNA methylation would be more apparent in women compared to men.
Limitations
When correcting for inflation, only two out of the four individual CpG-sites that were associated with tea consumption remained significant. Inflation is usually higher in EWAS studies compared to GWAS studies since we expect that the DNA methylation pattern will change in response to many different environmental factors. It is also reasonable to believe that part of the inflation is due to the large impact of tea metabolites on DNA methylation, since previous studies have shown that catechins affect DNA methylation (27,28). Adjusting for inflation may therefore decrease statistical power to find significant results and interesting findings may be revealed by the un-adjusted analysis. The only way to be sure that we correctly adjust for cofounding is to do controlled experiments.
Our hypothesis was that tea and coffee consumption affect DNA methylation. However, an EWAS does not allow for establishing causality of the observed effects. There is a possibility that confounding variables correlated with tea consumption are the truly causal factor behind the alterations in DNA methylation. The most obvious confounding factors that are associated with coffee and tea consumption are smoking and age. In order to eliminate confounding effects by smoking and age, these variables were including as covariates in all analyses. Reverse causation is also a possibility, where DNA methylation would, in itself, affect tea and coffee consumption. Mendelian randomisation (MR) approaches have been developed to answer this question, but these require large study sizes to perform, and although previous studies have shown growing evidence that individual coffee and caffeine consumption and response to caffeine are related to genetic factors (25,26) and show a high heritability (52,53) we don’t have the power to perform MR in this cohort. In contrast, the heritability and genetic background of tea consumption is less conclusive at present (30), making such MR analyses impractical to perform as of yet for tea to ascertain the directionality of the associations on which we report.
We did not have information about decaffeinated coffee consumption. Consumption of decaffeinated coffee is relatively uncommon, however, if caffeine is a factor associated with DNA methylation our study may be biased due to missing information on decaffeinated coffee consumption habits. Brewing technique may also influence DNA methylation. Cafestol and kahweol, known as coffee-specific anti-carcinogenic lipids (54) in coffee, are extracted by boiling water, but retained by coffee filters. Consequently, the lipid content of boiled un-filtered coffee may be as much as 60 times higher than the lipid content of filtered coffee (55). Since only the NSHDS cohort provided information on coffee brewing technique, for statistical power reasons, no sub-analysis according to brewing technique was performed.
The same is true for different types of teas that will consist of different important constituents such as catechins and teaflavins, e.g. black tea (fermented), and green tea (unfermented). We did not have information on which kind of tea that was consumed. In addition, the content of caffeine in tea also depends on the plant variety, processing of tealeaves, brewing time as well as brewing methods. Cup size is also different between countries, which lead to a bias in our measurement of the amounts of beverage consumed. However, this bias is unlikely to lead to false positives, but rather to decreased statistical power.
To perform the EWAS, we used whole blood samples. Blood has an inter-individual variability in the fractions of different DNA-containing blood cells. Previous studies have shown that differences in methylation can be the result of variability in cell composition when blood is used in DNA methylation studies (14). We therefore corrected for variability in cell composition (CD8T-, CD4T-, NK- and B-cells, monocytes and granulocytes) between individuals. However, we do not know if the association between DNA methylation and coffee or tea consumption is specific to only one cell type or present in all of the individual cell types in blood. Predicted cell proportions for the low-frequency cell types, such as T cells, may be less accurate than those of the higher frequency cell types. However, previous studies have also shown that cell composition does not have a significant effect and that it mainly depends on the exposure you are looking at (56,57). However, studies have shown that metabolites from coffee and tea consumption end up in the blood stream (1–4), making it a directly exposed tissue and thus relevant to study in this respect. We do not have other tissue specimens available to test if associations are also present in tissues more directly related to some of the health effects reported for tea consumption, such as reduced risk of breast cancer.
The two significant CpG-sites show nominal significance with the same direction of effect in all included cohorts, which strengthen our results. However, replication in an independent cohort would further confirm these associations and to clarify which components in tea that has an effect on gene regulation, we need to investigate the effect of tea on cell migration in vitro. It should also be noted that the results of the regional analysis should be interpreted with care since the consistency between the cohorts was unclear and comb-p has been reported to sometimes induce spurious results when the association is weak (58).
In conclusion, this study did not find an association between coffee consumption and DNA-methylation. In women, 6 genes interacting with estradiol were observed to be associated with tea consumption using region based analyses. Estradiol has previously shown to be decreased in blood due to tea consumption. We also found genes involved in cancer, previously shown to be associated with tea consumption.
Materials and Methods
Study populations
This study included participants from four European cohorts, including 3,096 subjects, further described below.
Northern Sweden Population Health Study (NSPHS)
The Northern Sweden Population Health Study (NSPHS) was initiated in 2006 to provide a health survey of the population in the parishes of Karesuando and Soppero, County of Norrbotten, and to study the medical consequences of lifestyle and genetics. This parish has about 3,000 inhabitants who meet the eligibility criteria in terms of age (> 14 years) of which 1,069 individuals participated in the study. The median age of these individuals was 50 years, ranging from 14 to 94 (Table 1). For each participant in the NSPHS, whole blood, plasma and serum samples were taken and immediately frozen and stored at −70°C. Genomic DNA for methylation analyses was extracted from previously frozen peripheral blood leukocytes using a phenol:chloroform protocol. The participants were asked to answer a questionnaire about diet and lifestyle factors including coffee and tea consumption. The NSPHS study was approved by the local ethics committee at the University of Uppsala (Regionala Etikprövningsnämnden, Uppsala Dnr 2005:325) in compliance with the Declaration of Helsinki. All participants gave their written informed consent to the study including the examination of environmental and genetic causes of disease. In case the participant was not of legal age, a legal guardian gave additional consent. The procedure used to obtain informed consent and the respective informed consent form have recently been discussed in the light of present ethical guidelines (59). More information about the NSPHS has been published previously (60).
Dutch Hunger Winter Families Study (HWFS)
The Dutch Hunger Winter Families Study has been described in detail elsewhere (61). In short: 2,417 singleton births with detailed birth records, born between 1 February 1945 and 31 March 1946 with mothers that were exposed to the Dutch famine of 1944-45 during or immediately preceding pregnancy were selected. Likewise 890 births from 1943 and 1947 were selected to include individuals whose mothers were not exposed to famine during this pregnancy. For 70% of the individuals, an address could be obtained and they were invited by mail to participate together with a same-sex sibling not exposed to the famine as a family-control. In total 1,075 interviews and 971 clinical examinations were performed between 2003 and 2005. The Institutional Review Board (IRB) of Columbia University Medical Center and the Medical Ethical Committee (MEC) of Leiden University Medical Center approved this study and participants provided verbal consent at the start of the telephone interview and written informed consent. Coffee and tea consumption was ascertained from a 140-item food frequency questionnaire that has been validated for the Dutch population (62).
EnivroGenoMarkers project (EGM)
The EnivroGenoMarkers project (www.envirogenomarkers.net) involved subjects from the European Prospective Investigation into Cancer and Nutrition study (EPIC-ITALY) and the Northern Sweden Health and Disease Study (NSHDS) (Table 1) (63–65). Both studies used population-based recruitment with standardized lifestyle (including smoking) and personal history questionnaires, anthropometric data and blood samples collected at recruitment (1993–1998 for EPIC-ITALY; 1990–2006 for NSHDS). The EnviroGenomarkers project and its associated studies and protocols were approved by the Regional Ethical Review Board of Umeå, as regards the Swedish cohort, and the Florence Health Unit Local Ethical Committee, as regards the Italian cohort, and all participants gave written informed consent. The studies were conducted in accordance with the approved guidelines. 659 buffy coat samples from both cohorts were thawed and DNA was isolated using the QIAamp Blood Mini Kit (QIAGEN), evaluating it spectrophotometrically and by agarose gel electrophoresis.
The prospective investigation of the Vasculature in Uppsala Seniors (PIVUS)
Eligible subjects were those aged 70 years living in the community of Uppsala, Sweden. The subjects were chosen from the register of community living and were invited in a randomized order. The subjects received an invitation by letter within 2 months of their 70th birthday. Of the 2,025 subjects invited, 1,016 subjects participated giving a participation rate of 50.1% (66). Median age of individuals in PIVUS was 70.1 years, ranging from 69.8 to 70.7 years (Table 1). The study was approved by the Ethics Committee of the University of Uppsala (Dnr:00-419) and the participants gave informed consent. All subjects were investigated in the morning after an overnight fast, where collection of blood for DNA preparation took place. No medication or smoking was allowed after midnight.
Determination of DNA methylation status
Genomic DNA for 743 samples from NSPHS, 659 samples from EGM, 1,016 samples from PIVUS and 971 from HWFS was bisulfite-converted using an EZ DNA methylation Kit (ZYMO research) according to the manufacturer's recommendations. The methylation status of the genomic DNA was then assessed using the Human Methylation450 BeadChip, (Illumina, San Diego, USA) according to the standard protocol. The Human Methylation450 BeadChip, (Illumina, San Diego, USA) has been validated in previous studies (67,68).
Analysis of the raw data in NSPHS was performed using minfi. Normalization was performed using Subset-quantile Within Array Normalisation (SWAN). A marker detection P-value ≤ 1.38×10−10 (Bonferroni adjusted P-value = 0.05, adjusted for the number of individuals * the number of CpG sites analyzed) was applied, a Probe Call rate of > 0.98, and an individual call rate of > 0.98 was used. DNA methylation data were obtained for 743 individuals with self-reported coffee and tea consumption available.
Analysis of the raw data in PIVUS was performed using GenomeStudio 2011.1 from Illumina Inc. After exclusion of replicates a total of 1,002 study participants had methylation data available for quality control procedures. Three samples were excluded based on poor bisulphite conversion efficiency, twelve samples due to low pass rate of CpG sites (<98.5% with a detection P-value >0.01), six samples based on low SNP genotype match (>1 SNP mismatches) between genotypes from the methylation array and Omni/Metabochip genotyping chips and 14 samples due to abnormal leukocyte cell counts (>10×109 cells/l). The signal intensities for the methylated and unmethylated state were then quantile-normalized for each probe type separately. DNA methylation data were obtained for 967 individuals, 804 with self-reported coffee and tea consumption available.
The preprocessing of methylation data from EGM has been described previously (65). All 659 samples passed the inbuilt Methylation450 BeadChip 450K quality control. Probes were excluded from the analysis when >20% of samples had detection P-values <0.05, which resulted in 53,053 probes being excluded. 432,524 probes were used in analysis and. DNA methylation data were obtained for 659 individuals, 621 with self-reported coffee and tea consumption available.
Raw 450k data processing of the HWFS described in detail before (69). In short: array QC was performed with MethylAid (70) and bisulfite conversion efficiency assessed using both the dedicated probes on the 450K array and Sanger Sequencing of a set of random samples. Sample swaps and admixtures were prevented by comparing the measurement of 21 single nucleotide polymorphisms (SNPs) by 450K array with data acquired by MASSARRAY and by confirming recorded sex with DNA methylation patterns of the X-chromosomal CpG sites. Normalization of the dataset was performed by Functional Normalization after applying NOOB background correction in the minfi package (71) using six principal components. All measurements with a detection P-value >0.01 or zero intensity value in one of the color channels was set as missing. The measurement success rate per sample was >99%. DNA methylation data were obtained for 950 individuals, 948 with self-reported coffee and tea consumption available.
Estimation of cell fractions from the methylation data
To ensure that the results were not influenced by variation in cell fraction between samples, we estimated the fraction of CD8T-, CD4T-, NK- and B-cells, monocytes and granulocytes in the study samples. This was done using the R package minfi (38) that allows for estimating cell fractions in Illumina 450K methylation data from whole blood. This method is based on the methylation data published for flow-sorted cells (72) and algorithms derived from the study by Houseman et al. (73).
Coffee and tea measurements
The phenotypes analysed were self-reported coffee and tea consumption converted to cups/month.
Statistical analysis
The level of methylation was reported as average beta values, array and batch (bisulfite conversion plate and scan batch). Association between coffee and tea consumption and the transformed methylation beta values were adjusted for sex and age. To test if smoking was correlated with coffee and tea consumption, we ran a linear regression adjusting for sex and age. We found both coffee and tea consumption to be correlated to tobacco smoking. Coffee was positively correlated (roh = 0.21, P = 1.97×10−9) and tea negatively (roh = −0.045, P = 0.030). This is an interesting result that is in line with previous studies showing that tea drinkers are overall more health conscious compared to coffee drinkers (74). Coffee consumption was further adjusted for tea consumption and tea was adjusted for coffee consumption. Analyses were performed in men and women, both separately and together.
Association between coffee and tea consumption and the transformed methylation beta values were analyzed in NSPHS, PIVUS and EGM using linear models. NSPHS is a population-based study including related individuals and special methods were therefore used to handle relatedness. All statistical analyses were performed using the stats library of R version 2.15.10 (75). A kinship matrix was used to adjust beta values for pedigree structure when analyzing the association between beverage consumption and methylation. Pairwise kinship matrices were calculated for genotyped SNPs (N = 180,212) using the ibs function implemented in GenABEL. This function computes a matrix of average IBS (identical by state) for a group of people. In addition to covariates included for the other cohorts, we also included year of collection (e.g. 2006 or 2009).
The HWFS contains same-sex sibling pairs and we therefore used generalized estimation equations (GEE), as implemented in the R geepack package, with a Gaussian link function to evaluate the association between DNA methylation beta values and coffee or tea consumption, controlling for sibships. In addition to covariates above, models were additionally adjusted for famine exposure status and the first three principal components calculated on the entire beta-value matrix to adjust for cell heterogeneity (given their almost perfect correlation with one or more cell fractions measures as imputed with the algorithm of Houseman et al. (73)).
Before performing the meta-analysis, all cross-hybridized CpG-sites where removed (N = 52,172), leaving 421,695 autosomal sites. 11,232 CpG-sites were located on the X chromosome and those were only analyzed in men and women separately; while 416 CpG-sites located on the Y chromosome were meta-analyzed only in men. Meta analyses were performed using an inverse variance weighted fixed-effects model implemented in PLINK (76) for men and women together and on the results from the analyses stratified by sex. Significant results were further adjusted for inflation by correcting the standard error and P-value for each included population using the inflation factor lambda (genomic control).
Regional-level analysis
To perform a regional (locus)-level analysis, we used the comb-p software (29). Briefly, it groups correlated CpG sites in a 200-bp sliding window. The program search for a single CpG-site with a Bonferroni-adjusted P-value less than 1×10−3, and providing it finds another CpG-site with an adjusted P-value less than 1×10−3 within 200 bases, a putative region will be defined. For this analysis, we used the lambda-adjusted meta-analysis. Significant regions were identified as those with at least two CpG-sites and a corrected P value < 0.05. Comb-p performs a Stouffer-Liptak correction on probes by assessing all CpG-sites within a defined window as weighted by their observed correlation.
Acknowledgements
W.E.E and Å.J planned the study and interpreted the data. W.E.E, Å.J, E.P, J.M.F, E.L and E.W.T analyzed the data. All authors wrote and critically reviewed the manuscript for important intellectual content. We are grateful to all the participants from the community for their interest and willingness to contribute to this study. Illumina genotyping, and DNA methylation analyses was performed by the SNP & SEQ Technology Platform in Uppsala, which is supported by Uppsala University, Uppsala University Hospital, Science for Life Laboratory (SciLifeLab) - Uppsala and the Swedish Research Council (Contracts 80576801 and 70374401). The computations were performed on resources provided by SNIC through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under projects b2011203, p2012153 and b2013110. We are grateful to the participants of the Dutch Hunger Winter Families study and the staff of TNO Quality of Life for contact tracing. At the Leiden University Medical Center we wish to acknowledge the staff of the department of Gerontology and Geriatrics Study Center for the physical examinations and the Central Clinical Chemical Laboratory for extracting DNA. The DNA Methylation study in NSPHS has been funded by the Swedish Medical Research Council (Project Number 2011-2354) and the Göran Gustafssons Foundation. The NSPHS study was funded by the Swedish Medical Research Council (Project Number K2007-66X-20270-01-3) and the Foundation for Strategic Research (SSF). NSPHS as part of EUROSPAN (European Special Populations Research Network) was also supported by European Commission FP6 STRP grant number 01947 (LSHG-CT-2006-01947). This work has also been supported by the Swedish Society for Medical Research (SSMF), the Kjell och Märta Beijers Foundation, The Marcus Borgström Foundation, the Åke Wiberg foundation and the Vleugels Foundation. JMF acknowledges funding from Breast Cancer Now, CRUK programme grant A13086, the Imperial College Experimental Cancer Medicine Centre (ECMC) and the National Institute for Health Research (NIHR) Biomedical Research Centre (BRC). The Hunger Winter Families Study was supported by the U.S. National Institutes of Health [AG042190 to LHL and BTH, HL067914 to LHL] and the European Union’s Seventh Framework Program IDEAL (259679) and EWT was supported by a NWO VENI grant (91617128). The EnviroGenomarkers project was partly funded by the European Union (Grant number 226756).
Conflict of Interest statement. Erik Ingelsson is a scientific advisor for precision Wellness, Cellink and Olink Proteomics for work unrelated to the present project.
Funding
Uppsala University, Uppsala University Hospital, Science for Life Laboratory (SciLifeLab) - Uppsala and the Swedish Research Council (Contracts 80576801 and 70374401), Swedish Medical Research Council (Project Number 2011-2354 and K2007-66X-20270-01-3) and the Göran Gustafssons Foundation, Foundation for Strategic Research (SSF), European Commission FP6 STRP grant number 01947 (LSHG-CT-2006-01947), Swedish Society for Medical Research (SSMF), Kjell och Märta Beijers Foundation, The Marcus Borgström Foundation, Åke Wiberg foundation and the Vleugels Foundation, Breast Cancer Now, CRUK programme grant A13086, the Imperial College Experimental Cancer Medicine Centre (ECMC), National Institute for Health Research (NIHR) Biomedical Research Centre (BRC), U.S. National Institutes of Health, European Union’s Seventh Framework Program IDEAL (259679) , NWO VENI grant (91617128), and European Union (Grant number 226756).
References
- 1. Neilson A.P., Green R.J., Wood K.V., Ferruzzi M.G. (2006) High-throughput analysis of catechins and theaflavins by high performance liquid chromatography with diode array detection. J. Chromatogr. A, 1132, 132–140. [DOI] [PubMed] [Google Scholar]
- 2. Lee M.J., Prabhu S., Meng X., Li C., Yang C.S. (2000) An improved method for the determination of green and black tea polyphenols in biomatrices by high-performance liquid chromatography with coulometric array detection. Anal. Biochem., 279, 164–169. [DOI] [PubMed] [Google Scholar]
- 3. Maiani G., Serafini M., Salucci M., Azzini E., Ferro-Luzzi A. (1997) Application of a new high-performance liquid chromatographic method for measuring selected polyphenols in human plasma. J. Chromatogr. B Biomed. Sci. Appl., 692, 311–317. [DOI] [PubMed] [Google Scholar]
- 4. Kuznicki J.T., Turner L.S. (1986) The effects of caffeine on caffeine users and non-users. Physiol. Behav., 37, 397–408. [DOI] [PubMed] [Google Scholar]
- 5. Eskelinen M.H., Kivipelto M. (2010) Caffeine as a protective factor in dementia and Alzheimer’s disease. J. Alzheimers. Dis., 20 Suppl 1, S167–S174. [DOI] [PubMed] [Google Scholar]
- 6. Ross G., Abbott R., Petrovitch H., Morens D., Grandinetti A., Tung K., Tanner C., Masaki K., Blanchette P., Curb J.. et al. (2000) Association of Coffee and Caffeine Intake With the Risk of Parkinson Disease. JAMA, 283, 2674–2679. [DOI] [PubMed] [Google Scholar]
- 7. Huxley R., Man C., Lee Y., Barzi F., Timmermeister L. (2009) Coffee, Decaffeinated Coffee, and Tea Consumption in Relation to Incident Type 2 Diabetes Mellitus. Am. Med. Asoc. J., 169, 2053–2063. [DOI] [PubMed] [Google Scholar]
- 8. Jee S.H., He J., Whelton P.K., Suh I., Klag M.J. (1999) The effect of chronic coffee drinking on blood pressure: a meta-analysis of controlled clinical trials. Hypertension, 33, 647–652. [DOI] [PubMed] [Google Scholar]
- 9. Nurminen M.L., Niittynen L., Korpela R., Vapaatalo H. (1999) Coffee, caffeine and blood pressure: a critical review. Eur. J. Clin. Nutr., 53, 831–839. [DOI] [PubMed] [Google Scholar]
- 10. Lueth N.A., Anderson K.E., Harnack L.J., Fulkerson J.A., Robien K. (2008) Coffee and caffeine intake and the risk of ovarian cancer: the Iowa Women’s Health Study. Cancer Causes Control, 19, 1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. MacMahon B., Yen S., Trichopoulos D., Warren K., Nardi G. (1981) Coffee and cancer of the pancreas. N. Engl. J. Med., 304, 630–633. [DOI] [PubMed] [Google Scholar]
- 12. El-Sohemy A., Cornelis M.C., Kabagambe E.K., Campos H. (2007) Coffee, CYP1A2 genotype and risk of myocardial infarction. In Genes and Nutrition, Vol. 2, p 155–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palacios N., Gao X., McCullough M.L., Schwarzschild M.A., Shah R., Gapstur S., Ascherio A. (2012) Caffeine and risk of Parkinson’s disease in a large cohort of men and women. Mov. Disord., 27, 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarzschild M. a., Chen J.-F., Ascherio A. (2002) Caffeinated clues and the promise of adenosine A(2A) antagonists in PD. Neurology, 58, 1154–1160. [DOI] [PubMed] [Google Scholar]
- 15. Orecchio S., Ciotti V.P., Culotta L. (2009) Polycyclic aromatic hydrocarbons (PAHs) in coffee brew samples: Analytical method by GC-MS, profile, levels and sources. Food Chem. Toxicol., 47, 819–826. [DOI] [PubMed] [Google Scholar]
- 16. Lambert J.D., Yang C.S. (2003) Mechanisms of cancer prevention by tea constituents. J. Nutr., 133, 3262S–3267S. [DOI] [PubMed] [Google Scholar]
- 17. Ju J., Lu G., Lambert J.D., Yang C.S. (2007) Inhibition of carcinogenesis by tea constituents. Semin. Cancer Biol., 17, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao Y., Cao R. (1999) Angiogenesis inhibited by drinking tea. Nature, 398, 381.. [DOI] [PubMed] [Google Scholar]
- 19. Jankun J., Selman S.H., Swiercz R., Skrzypczak-Jankun E. (1997) Why drinking green tea could prevent cancer. Nature, 387, 561.. [DOI] [PubMed] [Google Scholar]
- 20. Hodgson J.M. (2006) Effects of tea and tea flavonoids on endothelial function and blood pressure: A brief review. Clin. Exp. Pharmacol. Physiol., 33, 838–841. [DOI] [PubMed] [Google Scholar]
- 21. Mukhtar H., Ahmad N. (2000) Tea polyphenols: Prevention of cancer and optimizing health. In Am. J Clin. Nutr., 71, 1698S–1702S. [DOI] [PubMed] [Google Scholar]
- 22. Scalbert A., Johnson I., Saltmarsh M. (2005) Polyphenols: antioxidants and beyond. Am. Soc. Clin. Nutr., 81, 2155–2175. [DOI] [PubMed] [Google Scholar]
- 23. Choi S.-W., Friso S. (2010) Epigenetics: A New Bridge between Nutrition and Health. Adv. Nutr., 1, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Besingi W., Johansson Å. (2014) Smoke-related DNA methylation changes in the etiology of human disease. Hum. Mol. Genet., 23, 2290–2297. [DOI] [PubMed] [Google Scholar]
- 25. Yang A., Palmer A.A., De Wit H. (2010) Genetics of caffeine consumption and responses to caffeine. Psychopharmacology (Berl), 211, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amin N., Byrne E., Johnson J., Chenevix-Trench G., Walter S., Nolte I.M., Vink J.M., Rawal R., Mangino M., Teumer A.. et al. (2012) Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol. Psychiatry, 17, 1116–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee W.J., Shim J., Zhu B.T. (2005) Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol. Pharmacol., 68, 1018–1030. [DOI] [PubMed] [Google Scholar]
- 28. Zhu Fang M., Wang Y., Ai N., Hou Z., Sun Y., Lu H., Welsh W., Yang C.S. (2003) Tea polyphenol (-)-Epigallocatechin-3-Gallate Inhibits DNA Methyltransferase and Reactivates Methylation-Silenced Genes in Cancer Cell Lines. Cancer Res., 63, 7563–7570. [PubMed] [Google Scholar]
- 29. Pedersen B.S., Schwartz D.A., Yang I.V., Kechris K.J. (2012) Comb-p: Software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics, 28, 2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luciano M., Kirk K., Heath A., Martin N. (2005) The genetics of tea and coffee drinking and preference for source of caffeine in a large community sample of Australian twins. Addiction, 100, 1510–1517. [DOI] [PubMed] [Google Scholar]
- 31. Shrubsole M.J., Lu W., Chen Z., Shu X.O., Zheng Y., Dai Q., Cai Q., Gu K., Ruan Z.X., Gao Y.-T.. et al. (2009) Drinking green tea modestly reduces breast cancer risk. J. Nutr., 139, 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun C.-L., Yuan J.-M., Koh W.-P., Yu M.C. (2006) Green tea, black tea and breast cancer risk: a meta-analysis of epidemiological studies. Carcinogenesis, 27, 1310–1315. [DOI] [PubMed] [Google Scholar]
- 33. Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R., Guan W., Xu T., Elks C.E., Aslibekyan S.. et al. (2016) Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet., 9, 436 LP–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen C.Y., Chiou S.H., Huang C.Y., Jan C.I., Lin S.C., Hu W.Y., Chou S.H., Liu C.J., Lo J.F. (2009) Tid1 functions as a tumour suppressor in head and neck squamous cell carcinoma. J. Pathol., 219, 347–355. [DOI] [PubMed] [Google Scholar]
- 35. Seo E.-J., Wu C.-F., Ali Z., Wang Y.-H., Khan S.I., Walker L.A., Khan I.A., Efferth T. (2016) Both phenolic and non-phenolic green tea fractions inhibit migration of cancer cells. Front. Pharmacol., 7, 398.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bontems F., Fish R.J., Borlat I., Lembo F., Chocu S., Chalmel F., Borg J.P., Pineau C., Neerman-Arbez M., Bairoch A.. et al. (2014) C2orf62 and TTC17 are involved in actin organization and ciliogenesis in zebrafish and human. PLoS One, 9, e86476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Higgins J.P.T., Thompson S.G., Deeks J.J., Altman D.G. (2003) Measuring inconsistency in meta-analyses. BMJ Br. Med. J., 327, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakachi K., Suemasu K., Suga K., Takeo T., Imai K., Higashi Y. (1998) Influence of drinking green tea on breast cancer malignancy among Japanese patients. Jpn. J. Cancer Res., 89, 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Flodgaard H., Ostergaard E., Bayne S., Svendsen A., Thomsen J., Engels M., Wollmer A. (1991) Covalent structure of two novel neutrophile leucocyte-derived proteins of porcine and human origin. Neutrophile elastase homologues with strong monocyte and fibroblast chemotactic activities. Eur. J. Biochem., 197, 535–547. [DOI] [PubMed] [Google Scholar]
- 40. Ran X., Xu X., Yang Y., She S., Yang M., Li S., Peng H., Ding X., Hu H., Hu P.. et al. (2015) A quantitative proteomics study on olfactomedin 4 in the development of gastric cancer. Int. J. Oncol., 47, 1932–1944. [DOI] [PubMed] [Google Scholar]
- 41. Sun R., Wu J., Chen Y., Lu M., Zhang S., Lu D., Li Y. (2014) Down regulation of Thrombospondin2 predicts poor prognosis in patients with gastric cancer. Mol. Cancer, 13, 225.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fernandez-Madrid F., Tang N., Alansari H., Granda J.L., Tait L., Amirikia K.C., Moroianu M., Wang X., Karvonen R.L. (2004) Autoantibodies to annexin XI-A and other autoantigens in the diagnosis of breast cancer. Cancer Res., 64, 5089–5096. [DOI] [PubMed] [Google Scholar]
- 43. Koi M., Johnson L., Kalikin L., Little P., Nakamura Y., Feinberg A. (1993) Tumor cell growth arrest caused by subchromosomal transferable DNA fragments from chromosome 11. Science 260, 361–364. [DOI] [PubMed] [Google Scholar]
- 44. Liu B., Di G. (2016) C-Terminal Binding Protein is Involved in Promoting to the Carcinogenesis of Human Glioma. Mol. Neurobiol. [DOI] [PubMed] [Google Scholar]
- 45. Peng G., Ye Q., Wang R., Li M., Yang Z. (2016) No TitleKnockdown by shRNA identifies SLC44A5 as a potential therapeutic target in hepatocellular carcinoma. Mol. Med. Rep., 136, 4845–4852. [DOI] [PubMed] [Google Scholar]
- 46. Chen C.D., Wang C.S., Huang Y.H., Chien K.Y., Liang Y., Chen W.J., Lin K.H. (2007) Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics, 7, 155–167. [DOI] [PubMed] [Google Scholar]
- 47. Wang P., Zhang C., Yu P., Tang B., Liu T., Cui H., Xu J. (2012) Regulation of colon cancer cell migration and invasion by CLIC1-mediated RVD. Mol. Cell. Biochem., 365, 313–321. [DOI] [PubMed] [Google Scholar]
- 48. Li X., Liu R., Luo L., Yu L., Chen X., Sun L., Wang T., Hylemon P., Zhou H., Jiang Z.. et al. (2016) Role of AMP-activated protein kinase α1 in 17α-ethinylestradiol-induced cholestasis in rats. Arch. Toxicol., 91,481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ronis M., Gomez-Acevedo H., Blackburn M., Cleves M., Singhal R., Badger T. (2015) Uterine responses to feeding soy protein isolate and treatment with 17β-estradiol differ in ovariectomized female rats. Toxicol. Appl. Pharmacol., 297, 68–80. [DOI] [PubMed] [Google Scholar]
- 50. Lam H., Ho S., Chen J., Medvedovic M., Tam N. (2016) Bisphenol A disrupts HNF4α-regulated gene networks linking to prostate preneoplasia and immune disruption in noble rats. Endocrinology, 157, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jennen D., Magkoufopoulou C., Ketelslegers H., van Herwijnen M., Kleinjans J., van Delft J. (2010) Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci., 115, 66–79. [DOI] [PubMed] [Google Scholar]
- 52. Vink J.M., Staphorsius A.S., Boomsma D.I. (2009) A genetic analysis of coffee consumption in a sample of Dutch twins. Twin Res. Hum. Genet., 12, 127–131. [DOI] [PubMed] [Google Scholar]
- 53. Laitala V.S., Kaprio J., Silventoinen K. (2008) Genetics of coffee consumption and its stability. Addiction, 103, 2054–2061. [DOI] [PubMed] [Google Scholar]
- 54. Nkondjock A. (2009) Coffee consumption and the risk of cancer: An overview. Cancer Lett., 277, 121–125. [DOI] [PubMed] [Google Scholar]
- 55. Gross G., Jaccaud E., Huggett A.C. (1997) Analysis of the content of the diterpenes cafestol and kahweol in coffee brews. Food Chem. Toxicol., 35, 547–554. [DOI] [PubMed] [Google Scholar]
- 56. Flanagan J., Brook M., Orr N., Tomczyk K., Coulson P., Fletcher O., Jones M., Schoemaker M., Ashworth A., Swerdlow A.. et al. (2015) Temporal stability and determinants of white blood cell DNA methylation in the breakthrough generations study. Cancer Epidemiol. Biomarkers Prev., 24, 221–229. [DOI] [PubMed] [Google Scholar]
- 57. van Veldhoven K., Polidoro S., Baglietto L., Severi G., Sacerdote C., Panico S., Mattiello A., Palli D., Masala G., Krogh V.. et al. (2015) Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin. Epigenetics, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kolde R., Märtens K., Lokk K., Laur S., Vilo J. (2016) seqlm: an MDL based method for identifying differentially methylated regions in high density methylation array data. Bioinformatics, 10.1093/bioinformatics/btw304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mascalzoni D., Janssens A.C.J.W., Stewart A., Pramstaller P., Gyllensten U., Rudan I., van Duijn C.M., Wilson J.F., Campbell H., Quillan R.M.C. (2010) Comparison of participant information and informed consent forms of five European studies in genetic isolated populations. Eur. J. Hum. Genet., 18, 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johansson A., Marroni F., Hayward C., Franklin C.S., Kirichenko A.V., Jonasson I., Hicks A.A., Vitart V., Isaacs A., Axenovich T.. et al. (2009) Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet., 18, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lumey L., Stein A., Kahn H., Van der Pal-de Bruin K., Blauw G., Zybert P., Susser E. (2007) Cohort profile: the Dutch Hunger Winter families study. Int. J. Epidemiol., 36, 1196–1204. [DOI] [PubMed] [Google Scholar]
- 62. Grootenhuis P., Westenbrink P., Sie C., de Neeling J., Kok F., Bouter L. (1995) A semiquantitative food frequency questionnaire for use in epidemiologic research among the elderly: validation by comparison with dietary history. J. Clin. Epidemiol., 48, 859–868. [DOI] [PubMed] [Google Scholar]
- 63. Gonzalez C.A., Riboli E. (2010) Diet and cancer prevention: contributions from the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Eur. J. Cancer., 46, 2555–2562. [DOI] [PubMed] [Google Scholar]
- 64. Hallmans G., Agren A., Johansson G., Johansson A., Stegmayr B., Jansson J.-H., Lindahl B., Rolandsson O., Söderberg S., Nilsson M.. et al. (2003) Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand. J. Public Health. Suppl, 61, 18–24. [DOI] [PubMed] [Google Scholar]
- 65. Georgiadis P., Hebels D., Valavanis I., Liampa I., Bergdahl I., Johansson A., Palli D., Chadeau-Hyam M., Chatziioannou A., Jennen D.. et al. (2016) Omics for prediction of environmental health effects: Blood leukocyte-based cross-omic profiling reliably predicts diseases associated with tobacco smoking. Sci. Rep, 6, 20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lind L., Fors N., Hall J., Marttala K., Stenborg A. (2005) A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: The Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler. Thromb. Vasc. Biol., 25, 2368–2375. [DOI] [PubMed] [Google Scholar]
- 67. Roessler J., Ammerpohl O., Gutwein J., Hasemeier B., Anwar S., Kreipe H., Lehmann U. (2012) Quantitative cross-validation and content analysis of the 450k DNA methylation array from Illumina, Inc. BMC Res. Notes, 5, 210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sandoval J., Heyn H.A., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M. (2011) Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics, 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 69. Tobi E.W., Slieker R.C., Stein A.D., Suchiman H.E.D., Eline Slagboom P., Van Zwet E.W., Heijmans B.T., Lumey L.H. (2015) Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. Int. J. Epidemiol., 44, 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Van Iterson M., Tobi E.W., Slieker R.C., Den Hollander W., Luijk R., Slagboom P.E., Heijmans B.T. (2014) MethylAid: visual and interactive quality control of large Illumina 450k datasets. Bioinformatics, 30, 3435–3437. [DOI] [PubMed] [Google Scholar]
- 71. Hansen K., Aryee M. (2013) minfi: Analyze Illumina’s 450k methylation arrays, Vol. R package version 1.8.7.
- 72. Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.-E., Greco D., Söderhäll C., Scheynius A., Kere J. (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One, 7, e41361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schwarz B., Bischof H.P., Kunze M. (1994) Coffee, tea, and lifestyle. Prev. Med. (Baltim), 23, 377–384. [DOI] [PubMed] [Google Scholar]
- 75. R Development Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/. R Found. Stat. Comput. Vienna, Austria.
- 76. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J.. et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]