Abstract
Melanoma resistant to MAPK inhibitor(s) (MAPKi) displays loss-of-fitness upon experimental MAPKi withdrawal and, clinically, may be re-sensitized to MAPKi therapy after a drug holiday. Here, we uncovered and therapeutically exploited the mechanisms of MAPKi-addiction in MAPKi-resistant MUTBRAF or MUTNRAS melanoma. MAPKi-addiction phenotypes evident upon drug-withdrawal spanned transient cell-cycle slowdown to cell-death responses, the latter of which required a robust p-ERK rebound. Generally, drug withdrawal-induced p-ERK rebound up-regulated p38-FRA1-JUNB-CDKN1A and down-regulated proliferation, but only a robust p-ERK rebound resulted in DNA damage and parthanatos-related cell death. Importantly, pharmacologically impairing DNA damage repair during MAPKi withdrawal augmented MAPKi-addiction across-the-board by converting a cell-cycle deceleration to a caspase-dependent cell-death response or by furthering parthanatos-related cell death. Specifically in MEKi-resistant MUTNRAS or atypical MUTBRAF melanoma, treatment with a type I RAF inhibitor intensified p-ERK rebound elicited by MEKi-withdrawal, thereby promoting a cell-death predominant MAPKi-addiction phenotype. Thus, MAPKi discontinuation upon disease progression should be coupled with specific strategies that augment MAPKi-addiction.
INTRODUCTION
The combination of BRAF plus MEK inhibitors (BRAFi+MEKi) extends the survival benefits of BRAFi monotherapy in V600BRAF-mutant melanoma by counteracting MAPK-reactivating resistance mechanisms (1–9). However, acquired resistance to the BRAFi+MEKi is the norm rather than the exception. In cell line and patient-derived xenograft (PDX) models of V600BRAF mutant melanoma adapted to BRAFi monotherapy, loss-of-fitness due to BRAFi withdrawal in a process termed drug-addiction has been documented (2, 10). Moreover, the magnitude of drug-addiction increases with adaptation to BRAFi+MEKi (2). Regardless of the extent of drug-addiction, rebound p-ERK levels induced by MAPKi (BRAFi or BRAFi+MEKi) withdrawal seemed critical for this phenotype, since a low dose of ERKi was sufficient to block this p-ERK rebound and reversed drug-addiction (2). However, how p-ERK rebound mediates tumor cell-cycle deceleration and/or cell-death is unknown. Identifying the factor(s) that, together with p-ERK rebound, incite tumor cell-death or regression (rather than mere tumor stabilization or transient tumor cytostasis) may inform potential clinical strategies.
Anecdotal case series of patients with advanced V600BRAF-mutant melanoma suggest that rechallenge with a MAPKi, after previous evidence of disease progression and a brief drug holiday, can lead to clinical benefits, including objective tumor regression and enhanced life quality (11–16). More recently, a prospective clinical trial demonstrated that, following an interval of at least 12 weeks since disease progression and off MAPKi, rechallenge with BRAFi+MEKi led to 32% partial responses and 40% disease stabilization (17). Thus, an intentional drug holiday may select against MAPKi-resistant melanoma, leading to a re-sensitization phenomenon. Maximizing this counter-selection may lead to greater rates of re-sensitization and, if applied against microscopic resistance earlier during MAPKi therapy, could lead to longer durations of response or disease control.
Currently, MAPKi therapy is clinically approved only for patients with advanced V600BRAF-mutant melanoma. This is because type I RAF inhibitors (vemurafenib, dabrafenib) specifically inhibit monomeric V600BRAF mutants (18) but paradoxically activate the MAPK pathway in NRAS mutant and/or dimeric RAF-active melanoma (19–22). MEKi monotherapy has clear clinical activity against advanced V600BRAF mutant melanoma (23) but more limited activity against advanced MUTNRAS melanoma (24). Thus, strategies against MEKi-resistant melanoma, including MUTNRAS melanoma specifically and other subsets of melanoma with potential MAPKi sensitivity, may have clinical utility.
In this study, we sought to understand the basis of variable cellular responses (cell cycle versus death) to drug withdrawal in MAPKi-resistant melanoma. We also provided in vivo (PDX and murine melanoma in immune competent mice) proof-of-concepts that specific therapeutic approaches could potentially augment MAPKi-addiction by favoring tumor cell death over transient cell cycle suppression. Using three independent MEKi-resistant melanoma models with MUTNRAS or atypical BRAF mutations, we derived data suggesting that MAPKi-addiction is not exclusive to V600BRAF mutant melanoma and may be a hallmark of MAPKi-resistant melanoma.
RESULTS
Depth of drug-addiction in resistant melanoma is determined by slow-cycling or cell-death responses to MAPKi withdrawal
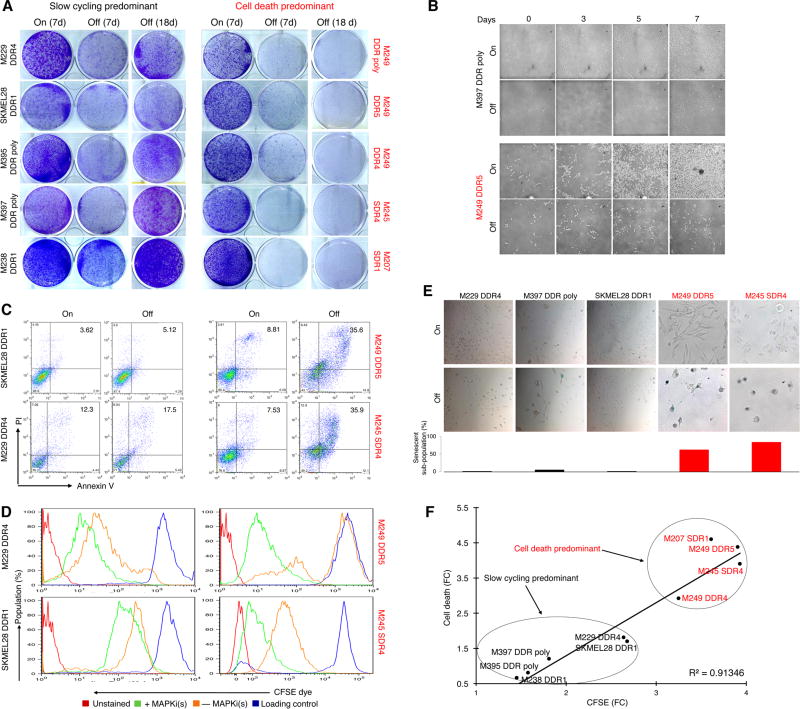
Previously we have shown that acquired MAPKi resistance mechanisms vary in their degrees of ERK reactivation (1, 3, 25) and that the levels of MAPKi-addiction correlate with the degrees of p-ERK rebound upon drug(s) withdrawal (2). To assess the spectrum of variations in the MAPKi-addiction phenotype, we analyzed the cellular responses of V600BRAF mutant, double-drug resistant (DDR) melanoma cell lines to BRAFi+MEKi withdrawal. We extended our analysis to Q61NRAS mutant melanoma sublines with acquired MEKi resistance (M207 and M245 single-drug resistant or SDR). All ten melanoma cell lines with diverse mechanisms (Supplementary Fig. S1A) of acquired MAPKi-resistance (R-lines) displayed MAPKi-addiction (Fig. 1A). This loss-of-fitness response (to MAPKi withdrawal) varied from transient in some R-lines to persistent in others, suggesting reversible slow-cycling and cell-death responses respectively (Fig. 1A). By vital imaging (Fig. 1B), we observed that a subset of R-lines responded to MAPKi-withdrawal by slowing down proliferation, whereas a distinct subset responded predominantly by cell-death (Fig. 1C). Consistently, cell-cycle slow-down predominant R-lines off-MAPKi displayed relatively lower CFSE dye dilution compared to on-MAPKi (Fig. 1D). On the other hand, residual cells that escaped a predominantly cell-death response to drug-withdrawal tended to retain dye strongly relative to the same R-lines on MAPKi and the slow-cycling R-lines off-MAPKi (Fig. 1D). Furthermore, greater fractions of persisting cells after MAPKi withdrawal displayed senescence-associated β-galactosidase staining in the cell-death predominant R-lines (Fig. 1E), consistent with a reduced proliferative potential. Thus, a cell-death predominant drug-addiction phenotype characterizes MAPKi-resistant melanoma cells that, upon drug withdrawal, undergo cell death in the majority subpopulation and a robust and persistent cell cycle deceleration in the minority, remainder subpopulation. On the other hand, a slow-cycling predominant drug-addiction phenotype characterizes MAPKi-resistant melanoma cells that, upon drug withdrawal, undergo a relatively weak and transient cell cycle deceleration (Fig. 1F).
Figure 1.
MAPKi-resistant melanoma display distinct drug-addiction phenotypes characterized by slow-cycling versus cell-death responses. (A) Clonogenic growth of double-drug resistant or DDR (MUTBRAF) or single-drug resistant or SDR (MUTNRAS) melanoma cell lines plated 24 hrs with BRAFi (vemurafenib)+MEKi (selumetinib) at 1 µM or MEKi (trametinib) at 0.1 µM followed by 7 d with (On) or 7 and 18 d without (Off) inhibitor withdrawal. (B) Temporal vital images of MAPKi-resistant or R-lines on or off BRAFi+MEKi. (C–E) Percentages of annexin V/PI-positive dead cells (C), CFSE dye dilution patterns (D), and levels of SA-βgal staining (E) in R-lines on or off MAPKi(s) for 6 d. Loading control (D) refers to the intensity of the CFSE dye initially loaded into the cells. (F) Correlation between fold-changes (FC) in CFSE dye dilution and % cell death off vs. on MAPKi(s).
Extent of ERK rebound induced by MAPKi withdrawal dictates tumor cell slow-cycling or death responses
We then assessed whether the extents of MAPKi withdrawal-induced p-ERK rebound (fold-change from baseline) are causally related to distinct phenotypic outcomes. Cell-death predominant R-lines displayed greater p-ERK rebound when compared to the slow-cycling predominant R-lines (Supplementary Fig. S1B, S1C). Moreover, a sub-optimal or low dose of an ERK inhibitor (ERKi) (Supplementary Fig. S1D), when added to cell-death predominant R-lines concomitant with MAPKi withdrawal, completely blocked cell death (Supplementary Fig. S1E) and, when added to both groups of R-lines during MAPKi withdrawal, reversed cell cycle deceleration (Supplementary Fig. S1F). Consistently, sub-optimal ERKi protected all R-lines from clonogenic growth suppression spurred by BRAFi+MEKi (in MUTBRAF R-lines) or MEKi (in MUTNRAS R-lines) withdrawal (Supplementary Fig. S1G). We then tested whether augmenting MAPKi withdrawal-induced p-ERK rebound in a slow-cycling predomiant R-line would enhance MAPKi-addiction. V600EBRAF amplification drives acquired MAPKi resistance via ERK-reactivation (6) and has been associated with BRAFi (10) or BRAFi+MEKi (2) addiction. We engineered exogenous V600EBRAF in the slow-cycling predominant R-lines SKMEL28 DDR1 (Supplementary Fig. S1H), which increased p-ERK levels on and especially off double-drug treatment. Functionally, V600EBRAF over-expression, upon double-drug withdrawal, enhanced cell-death and cell-cycle deceleration, and suppressed long- and short-term growth (Supplementary Fig. S1I–S1L). Hence, the extent of p-ERK rebound upon MAPKi withdrawal determines the addiction phenotype of MAPKi-resistant melanoma (Supplementary Fig. S1M).
Effectors of cell cycle or death responses to MAPKi withdrawal in resistant melanoma
To identify the effectors of distinct MAPKi-addiction phenotypes in drug-resistant melanoma, we generated RNA-seq profiles of cell-death predominant (M249 DDR5) and slow-cycling predominant (SKMEL28 DDR1) R-lines on (6 h) or off (6 h and 24 h) BRAFi+MEKi. From genes that were induced (≥ 2 fold) by double-drug withdrawal in both R-lines and at both time points (Supplementary Fig. S2A), we analyzed for transcription factor (TF)-binding motif enrichment of the differentially up-expressed genes (Supplementary Fig. S2B). Among the six TFs whose binding motifs were enriched, four (JUNB, FOSL1/FRA1, FOSL2, c-JUN) belonged to the AP-1 family. Among these, JUNB and FOSL1/FRA1 transcripts were induced by MAPKi withdrawal (Supplementary Fig. S2C). At the protein level, both total FRA1 (and p-FRA1) and total JUNB were induced by MAPKi withdrawal, together with induction of total c-FOS (and p-c-FOS), FOSB, and p-p38, the upstream kinase (Supplementary Fig. S2D and see below). In slow-cycling predominant R-lines, double-drug withdrawal induced levels of p-FRA1, p-p38 and the cell cycle inhibitor p21 (Supplementary Fig. S2E). Treatment with a p38 inhibitor (p38i) during MAPKi withdrawal reduced the phosphorylation of its substrate (p-HSP27) but not p21. However, treatment with p38i in combination with FRA-1 knockdown during MAPKi withdrawal abolished p21 induction, accelerated cell-cycling and recovered 40–60% of growth inhibition induced by MAPKi withdrawal in these slow-cycling predominant R-lines (Supplementary Fig. S2E–S2H). Furthermore, as JUNB was induced by MAPKi withdrawal (Supplementary Fig. S2C and S2I), we knocked down JUNB. Interestingly, joint JUNB and FRA1 knockdown abolished p21 induction by MAPKi-withdrawal and recovered the growth of slow-cycling predominant R-lines after double-drug withdrawal (Supplementary Fig. S2I and S2J). However, shFRA1 transduction together with p38i treatment in a cell-death predominant R-line, M249 DDR5, blunted MAPKi withdrawal-induced p21 but failed to alter the cell-cycling profile, likely because the major growth inhibition phenotype, i.e., cell death, was not impacted (Supplementary Fig. S2K–S2M). Consistently, M249 DDR5 was still strongly addicted to BRAFi+MEKi despite FRA1 knockdown and p38 inhibition (Supplementary Fig. S2N). Thus, p38-FRA1/JUNB signaling and p21 accumulation induced by MAPKi withdrawal are necessary for the slow-cycling but not the cell-death predominant phenotype of drug addiction.
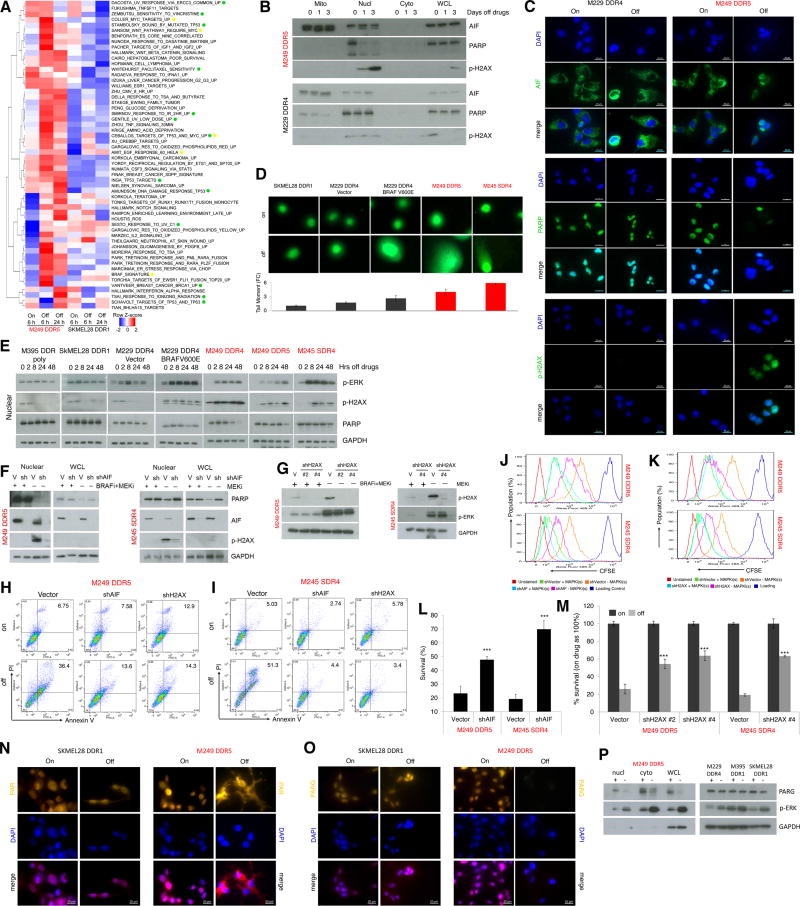
To identify processes that drive the cell-death predominant MAPKi-addiction phenotype, we analyzed the RNA-seq data for differential gene set enrichment between SKMEL28 DDR1 and M249 DDR (using the C2 (CGP subset), C6, and hallmark gene sets in MSigDB version 5.1) under each condition (on drug for 6 h, off drug for 6 h, off drug for 24 h). We selected gene sets which showed higher (median log2 difference of 1.25-fold or 25% up-expression across all genes in the set) gene set enrichment in one R-line compared to the other in either the 6 h or 24 h off drug condition, provided that this differential enrichment was higher (by 25%) than that observed in the baseline (on drug for 6 h) condition. The gene sets that were more strongly enriched in the cell-death predominant R-line (M249 DDR5) off MAPKi for 6 h or 24 h are shown in Figure 2A. As expected, gene sets induced by MAPK hyper-activation were preferentially enriched in the cell-death predominant R-line (M249 DDR5) off double-drugs. Several gene sets induced by DNA damage were also preferentially enriched in M249 DDR5 when withdrawn from MAPKi. Consistently, when we probed the levels of p-H2AX, a marker of DNA damage and mediator of repair, only the cell-death predominant R-line, M249 DDR5, displayed p-H2AX induction during MAPKi withdrawal (Fig. 2B, 2C).
Figure 2.
Excessive ERK activation induces DNA damage and AIF-mediated death in the cell-death predominant MAPKi-addiction phenotype. (A) Heatmap showing the gene set variance analysis scores of differentially enriched gene signatures (median log fold-change (FC) ≥ 1.25 in the off-drug condition compared to the on-drug condition in either cell line; additionally, differential enrichment between the cell-death predominant and slow-cycling predominant R-lines must also be higher (by at least 25%) in the off-drug condition than in the on-drug condition). (B–C) Levels of AIF, PARP, and p-H2AX measured by Western blots (WB) (B) or immunofluorescence (IF) (C) in R-lines on or off MAPKi(s) for 1 and 3 d or 3 d, respectively. For b, mitochondrial (Mito), nuclear (Nucl), cytoplasmic (Cyto) cellular fractions or whole cell lysates (WCL). For c, nuclei visualized by DAPI; ruler, 20 µm. (D) DNA strand break measurements by the comet assay of R-lines on or off MAPKi(s) for 3 d. Tail length/moment FCs were quantified (n=5; mean ± SDs). (E) Temporal levels of p-ERK, p-H2AX and PARP in the nuclear fraction of R-cell lysates measured by WBs at indicate hrs off MAPKi(s). (F–G) Levels of indicated proteins by WBs in the nuclear (F, G) or WCL (F) fractions of R-lines, on and off MAPKi(s), transduced with control (V) or AIF (F) or H2AX (G) shRNA lentiviruses. (H–M) Percentages of annexin V/PI-positive dead cells (H, I), CFSE dye dilution patterns (J, K), and viable cell counts (L, M) in R-lines on or off MAPKi(s) for 6 d, transduced with control (Vector) or AIF (J, L) or H2AX (K, M) shRNA lentiviruses. For l, M, n=6; mean ± SDs; ***p <0.001 based on ANOVA. (N–O) Sub-cellular localization of PAR (N) or levels of PARG (O) by IF in R-lines. Nuclei visualized by DAPI; ruler, 20 µm. (P) PARG and p-ERK levels by WBs in the indicated fractions of M249 DDR5 or nuclear fractions of indicated additional R-lines. For WB, GAPDH, loading control.
We hypothesized that a strong p-ERK rebound might induce mitochondrial dysfunction and ROS and thereby DNA damage. Indeed, MAPKi withdrawal induced pronounced levels of mitochondrial ROS, swelling, and depolarization only in the cell-death predominant R-lines (Supplementary Fig. S3). Pan-caspase inhibition did not rescue cell-death predominant R-lines from cell-death or long-term growth inhibition induced by MAPKi withdrawal (Supplementary Fig. S4A, S4B). Consistently, MAPKi withdrawal failed to induce cleaved caspase 3 by measuring its activity or staining (Supplementary Fig. S4C–S4D). We then tested whether AIF, which becomes cleaved and activated by mitochondrial dysfunction or depolarization and nuclear-localized to induce cell death via necrosis or parthanatos (26), might play a role in the cell-death predominant MAPKi-addiction phenotype. Consistently, cleaved AIF accumulated in the nuclear fraction of cell-death predominant R-line, M249 DDR5, but not in the slow-cycling predominant R-line, M229 DDR4, after MAPKi withdrawal, in conjunction with nuclear loss of PARP-1, a binding partner of AIF and p-H2AX induction (Fig. 2B, 2C). By using the comet assay, we corroborated increased DNA damage preferentially in the cell-death predominant phenotype (Fig. 2D). In fact, when a slow-cycling predominant R-line (M229 DDR4) was engineered with V600EBRAF over-expression, it then displayed increased DNA damage after MAPKi-withdrawal, consistent with transition to a cell-death predominant MAPKi-addiction phenotype (Fig. 2D). Furthermore, p-ERK rebound coincided temporally with p-H2AX induction in cell-death predominant R-lines (Fig. 2E). By manipulating the levels of p-ERK rebound pharmacologically, we showed that the extent of p-ERK rebound among cell-death predominant R-lines was strictly associated with the degree of DNA damage, as measured by the comet assay or levels of p-H2AX, or with the level of clonogenic growth (Supplementary Fig. S5A to S5E). In fact, p-ERK and p-H2AX levels, quantified by immunofluorescent detection, across all cell-death predominant lines and tumors (see below) were highly correlated (Supplementary Fig. S5F).
To assess the functional contributions of AIF or H2AX to the cell-death predominant drug-addiction phenotype, we engineered M249 DDR5 and M245 SDR4 to express shVector, shAIF or shH2AX (Fig. 2F, 2G). Importantly, AIF or H2AX knockdown strongly diminished p-H2AX accumulation induced by MAPKi withdrawal and abrogated cell death while reducing cell-cycle deceleration and loss of viable cells (Fig. 2F to 2M). On the other hand, AIF knockdown did not rescue the growth-inhibitory effect of MAPKi withdrawal in slow-cycling predominant R-lines (Supplementary Fig. S6). Consistent with PAR as a key signal underlying excessive DNA damage-PARP-AIF-mediated parthanatos (a recently characterized variant of programmed cell death (26)), we observed PAR cytoplasmic localization as well as loss of PARG, the major enzyme responsible for PAR catabolism, upon MAPKi withdrawal only in the cell-death predominant R-line, M249 DDR5, but not the slow-cycling predominant R-line, SKMEL28 DDR1 (Fig. 2N to 2P). Thus, non-apoptotic programmed cell death driven by excessive DNA damage underlies the cell-death predominant MAPKi-addiction phenotype.
Pharmacologic induction of DNA damage promotes death across all MAPKi-resistant cell lines but selectively during MAPKi withdrawal
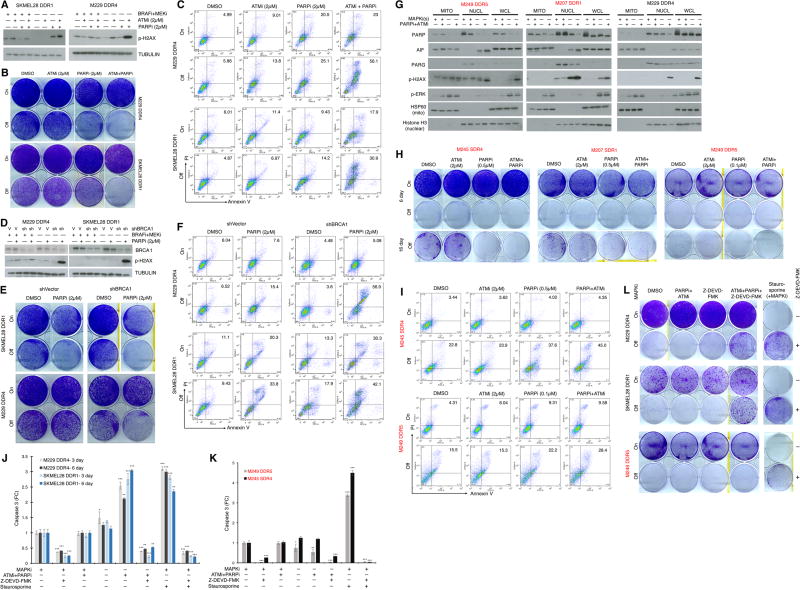
Since the prior results suggested a synthetic lethal relationship between excessive pERK level/rebound and DNA damage in the cell-death predominant drug-addiction phenotype, we tested whether enhancing DNA damage via inhibition of DNA damage repair in slow-cycling predominant R-lines would shift the drug-addiction phenotype toward cell death. We treated four slow-cycling predominant R-lines with ATMi, PARPi or both after MAPKi withdrawal. In SKMEL28 DDR1, M229 DDR4, M238 DDR1, and M395 DDR poly, ATMi+PARPi added upon BRAFi+MEKi withdrawal strongly promoted p-H2AX accumulation (Fig. 3A; Supplementary Fig. S7A), suppressed clonogenic growth or cell viability (Fig. 3B; Supplementary Fig. S7B, S7C), and induced cell death 6- to 10-fold (while only 2- to 4-fold on MAPKi) (Fig. 3C; Supplementary S7D). To confirm the functional importance of DNA damage or its impaired repair in determining the cellular fate (death vs. slow-cycling) of R-lines off MAPKi or during p-ERK rebound, we knocked down BRCA1 in the slow-cycling predominant R-lines (Supplementary Fig. S7E) and determined the p-H2AX levels induced by MAPKi withdrawal, with or without PARPi (Fig. 3D). We found that down-regulating DNA damage repair via BRCA1 knockdown and PARP1/2 inhibition, specifically after MAPKi withdrawal, strongly induced DNA damage (p-H2AX levels), suppressed clonogenic growth (Fig. 3E) or cell viability (Supplementary Fig. S7F), and induced cell death 4- to 10-fold (vs. 0- to 3-fold on MAPKi) (Fig. 3F).
Figure 3.
Impairing DNA damage repair augments MAPKi-addiction. (A) Western blot (WB) analysis of p-H2AX levels in slow-cycling predominant R-lines on or off MAPKi(s), with or without ATMi and/or PARPi treatment for 3 d. (B–C) Clonogenic growth (B) or percentages of annexin V/PI-positive dead cells (C) in slow-cycling predominant R-lines on or off MAPKi(s) for 5 d, with or without ATMi and/or PARPi treatment. (D) WB analysis of BRCA1 and p-H2AX levels in in slow-cycling predominant R-lines, transduced with control (V) or BRCA1-specific shRNA lentiviruses, on or off MAPKi(s), with or without PARPi treatment for 3 d. (E–F) R-line cells from D were subjected to the same assays as in B and C, respectively. For WBs, TUBULIN, loading control. (G) Levels of indicated proteins by WBs in mitochondrial (MITO), nuclear (NUCL) subcellular fractions or whole-cell lysates (WCL) of slow-cycling versus cell-death predominant R-lines, on and off MAPKi(s), with or without ATMi and PARPi treatment for 3 d. HSP60, mitochondrial fraction control; Histone H3, nuclear fraction control. (H–I) Clonogenic growth (H) or percentages of annexin V/PI-positive dead cells (I) in cell-death predominant R-lines on or off MAPKi(s) for 6 d and 16 (only in H), with or without ATMi and/or PARPi treatment. (J–L) Levels of caspase-3 activity (J, K) or clonogenic growth (L) with indicated MAPKi(s) treatment, with or without ATMi and PARPi, with or without caspase-3 inhibitor, Z-DEVD-FMK (20 µM), in slow-cycling predominant R-lines (J) for 3, 6 d or cell-death predominant R-lines (K) for 3 d. For J, K, n=5; mean ± SDs; ***p <0.001 based on ANOVA. Staurosporine (1 µM) used to induce caspase-3 activity and death.
We then assessed whether we could further enhance synthetic lethality in R-lines that were already cell-death predominant in their drug-addiction phenotype. In cell-death predominant R-lines (but not in a slow-cycling predominant R-line), PARPi+ATMi added upon MAPKi withdrawal further induced the nuclear levels of AIF (Fig. 3G). Further boosting the induction of DNA damage (p-H2AX levels) and nuclear AIF levels resulted in further clonogenic growth suppression that was evident after prolonged culture off MAPKi (Fig. 3H), consistent with the highest level of cell death detected (early during the course of culture) in cell-death predominant R-lines taken off MAPKi along with PARPi+ATMi co-treatment (Fig. 3I). As earlier data indicated that the cell-death predominant drug-addiction phenotype did not involve induction of caspase 3 activity and could not be reversed by a pan-caspase inhibitor (Supplementary Fig. S4A–S4D), we went further to test whether cell-death induced by MAPKi-withdrawal plus DNA damage repair inhibition in slow-cycling predominant R-lines would upregulate caspase 3 activity or function in a caspase 3-dependent manner (Fig. 3J; Supplementary Fig. S4E). As shown clearly, caspase 3 was induced in slow-cycling predominant R-lines specifically during MAPKi-withdrawal plus ATMi+PARPi co-treatment but not in a cell-death predominant R-line (Fig. 3K; Supplementary Fig. S4E). Accordingly, in slow-cycling predominant R-lines off MAPKi and on ATMi+PARPi, a caspase 3 inhibitor was able to rescue, at least partially, the drug-resistant melanoma cells from clonogenic suppression (Fig. 3L). In contrast, caspase 3 inhibition, consistent with pan-caspase inhibition (Supplementary Fig. S4A, S4B), failed to reverse the cell-death predominant phenotype (Fig. 3L).
Pharmacologically augmenting ERK rebound and DNA damage to induce tumor regression of MEKi-resistant MUTNRAS- or atypical MUTBRAF-melanoma
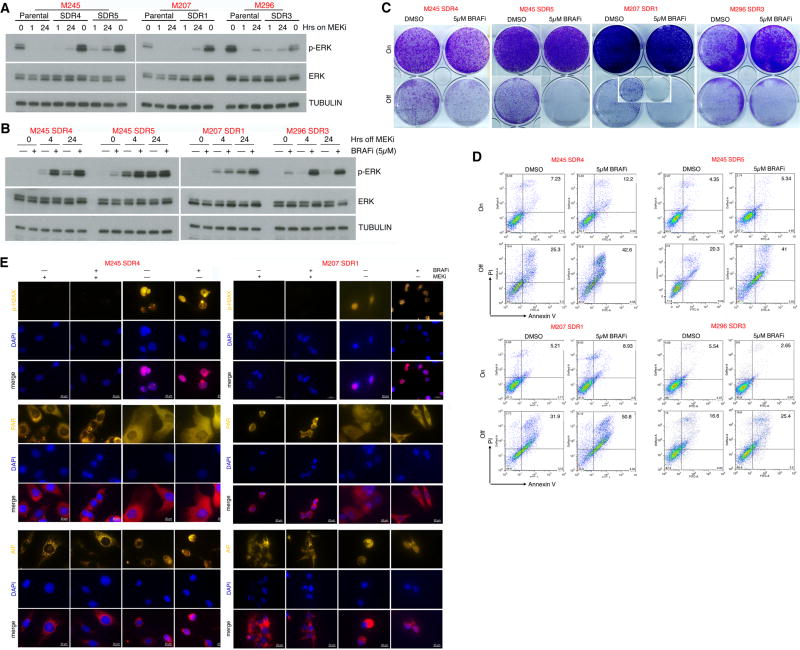
Although MEKi monotherapy has been shown recently to have clinical activity against MUTNRAS melanoma, resistance developed readily (27). Thus, we sought a strategy that would exploit the aforementioned synthetic lethality to augment MEKi-addiction in MEKi-resistant MUTNRAS melanoma. Since type I RAF inhibitors, including vemurafenib and dabrafenib, paradoxically activates ERK in the WTBRAF/MUTNRAS context (19–22), we hypothesized that vemurafenib may augment MEKi withdrawal-induced p-ERK rebound and therefore a cell-death predominant drug-addiction phenotype. To this end, we tested four MUTNRAS melanoma cell lines (from three genetic backgrounds) resistant to MEKi, which displayed variable degrees of ERK reactivation (on drug) or hyper-activation (off drug) (Fig. 4A). Importantly, initiating BRAFi treatment at the time of MEKi withdrawal in these MUTNRAS melanoma SDR clones resulted in a more rapid and robust p-ERK rebound, growth suppression, and cell death, compared with MEKi withdrawal alone (Fig. 4B to 4D). As was shown with cell-death predominant MUTBRAF DDR-lines, MEKi withdrawal with or without BRAFi treatment in these MUTNRAS SDR-lines induced p-H2AX accumulation and altered PAR and AIF localization (Fig. 4E).
Figure 4.
Paradoxical ERK activation by BRAFi potentiates drug addiction in MEKi-resistant MUTNRAS melanoma. (A–B) Western blot levels of p-ERK, ERK, and loading control TUBULIN in MUTNRAS parental and isogenic MEKi-resistant SDR-lines with indicated hrs on MEKi/trametinib (0.1 µM) treatment (A) or in SDR-lines with indicated hrs off MEKi/trametinib (0.1 µM) treatment, with or without BRAFi/vemurafenib (5 µM) treatment (B). (C–D) Clonogenic growth (C) and percentages of annexin V/PI-positive dead cells (D) in MUTNRAS SDR-lines on or off MEKi/trametinib (0.1 µM) for 6 d, with or without BRAFi/vemurafenib (5 µM) Inset for M207 SDR1 off MEKi for 16 d. (E) Levels and/or sub-cellular localization of p-H2AX, PAR, and AIF in cell-death predominant MEKi-addicted R-lines on or off MEKi for 3 d, with or without BRAFi (5 µM) treatment.
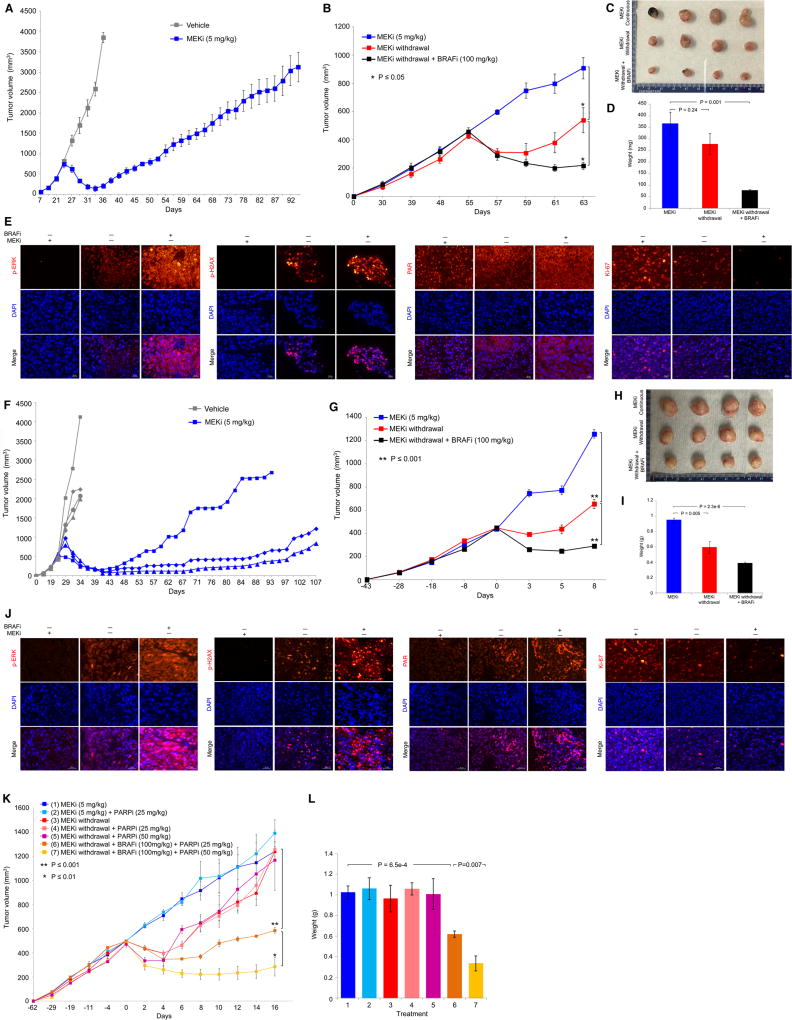
We also tested in vivo the synthetic lethal relationship (between supra-physiologic ERK activity and excessive DNA damage) underlying a cell-death predominant drug-addiction phenotype. We focused on a MEKi-resistant MUTNRAS patient-derived xenograft (PDX) model (Fig. 5A to 5E) and a MEKi-resistant non-BRAF V600 or atypical BRAF mutant (S365LBRAF) PDX model (Fig. 5F to 5J), because targeted therapies against these melanoma genotypes are currently lacking. To derive a MEKi-resistant MUTNRAS PDX model, F1 PDX fragments were transplanted in NSG mice, and mice with tumors of similar volumes were treated with trametinib at 5 mg/kg daily (day 25), which led to maximal tumor regression within 10 days (day 33) followed by acquired MEKi resistance (Fig. 5A). One resistant tumor, excised at day 85, was fragmented and serially transplanted into mice with continuous daily MEKi treatment until a cohort of mice with similar tumor volumes was assembled. This group of mice was divided into three sub-groups: continuous trametinib, trametinib withdrawal, and vemurafenib treatment with trametinib withdrawal (Fig. 5B). Importantly, withdrawing trametinib (half-life of 4 hrs) led to a transient tumor regression. However, vemurafenib treatment upon trametinib withdrawal led to a significantly more sustained tumor regression (Fig. 5B–5D). Analysis of these three groups of tumors revealed that, while trametinib withdrawal induced p-ERK, vemurafenib treatment on top of trametinib withdrawal further up-regulated p-ERK (Fig. 5E). Consistent with cell line findings (Fig. 4), trametinib withdrawal, especially with vemurafenib treatment, induced DNA damage or p-H2AX (Fig. 5E). Interestingly, trametinib withdrawal plus vemurafenib treatment led to redistribution of nuclear PAR to the cytoplasm (Fig. 5E). Consistent with tumor volumes/sizes/weights, tumors withdrawn from trametinib and then treated with vemurafenib displayed the lowest proliferation or Ki-67 level (Fig. 5E). To derive a MEKi-resistant atypical MUTBRAF PDX model, we treated F1 PDX S365LBRAF melanoma tumors in NSG mice with trametinib at 5 mg/kg daily (day 25), which led to maximal tumor regression within 20 days (day 45) followed by acquired MEKi resistance (Fig. 5F). The fastest growing MEKi-resistant tumor was selected for serial transplantation on continuous MEKi therapy and subjected to three experimental goups: continuous trametinib, trametinib withdrawal, and vemurafenib treatment with trametinib withdrawal. As with MEKi-resistant MUTNRAS PDX tumors, S365LBRAF PDX tumors resistant to MEKi responded to MEKi withdrawal but more robustly when a type I RAF inhibitor (vemurafenib) was added during MEKi withdrawal (Fig. 5G–5I). Consistent with the notion that non-V600E BRAF mutants operate as RAF dimers and acquired MAPKi resistance due to MAPK-reactivation can further drive RAF dimerization (18), we observed that BRAFi addition during MEKi withdrawal robustly up-regulated the p-ERK level (along with levels of p-H2AX, PAR) coincident with Ki-67 down-regulation (Fig. 5J and Supplementary Fig. S8A).
Figure 5.
Excessive ERK and DNA damage induce regression of MEKi-resistant PDX tumors. (A) Tumor volumes (mean ± SEM) of a MUTNRAS PDX transplanted in NSG mice in response to daily gavage with the vehicle (n = 3) or trametinib (5 mg/kg; n = 5). Asterisk indicates the resistant tumor, R1, which was serially transplanted for experiment in B. (B) R1 trametinib-resistant MUTNRAS PDX tumors were grown in NSG mice for 55 days with daily trametinib (5 mg/kg) gavage until initiating indicated daily treatments or regimens (n = 4 in each group). Tumor volumes are shown as means ± SEM. P values, Student’s t-test. MEKi, trametinib (5 mg/kg); BRAFi, vemurafenib (100 mg/kg). (C) Pictures of tumors from three experimental groups in B at day 63. (D) Tumor weights (means ± SEM; p value, unpaired t-test) of three experimental groups in B. (E) Levels of indicated proteins in representative tissue sections of tumors in C. Ruler, 20 µm. (F–J) As in A to E except experiments used a distinct PDX model harboring S365LBRAF and individual tumor growth curves were plotted separately in F. The MEKi-resistant tumor (R3) which arose first was fragmented, serially passaged and used in G. (K–L) R1 trametinib-resistant MUTNRAS PDX tumors were serially passaged in NSG mice with daily trametinib (5 mg/kg) gavage until segregation into seven groups (1, 2, 3, 5, n=3 per group; 4, 6, 7, n=5 per group). Tumor volumes and weights are shown as means ± SEM. P values, Student’s t-test.
We then tested whether we could effect deeper and more sustained tumor regression by triggering DNA damage during p-ERK rebound (Fig. 5K). We noticed in vitro (in MUTNRAS SDR lines) that PARPi treatment alone (without ATMi) could enhance to some extent clonogenic suppression and cell death induced by MEKi withdrawal (Fig. 3H, 3I). Similarly, in long-term clonogenic assays (Supplementary Fig. S8B), while BRAFi or PARPi each could further suppress the growth of MUTNRAS SDR lines withdrawn from MEKi, their combination led to the most profound suppression of growth. Importantly, in vivo, MEKi withdrawal combined with BRAFi+PARPi led to the most profound and sustained regression of MEKi-resistant MUTNRAS melanoma (Fig. 5K, 5L), which was associated with the strongest induction of p-ERK, p-H2AX, and PAR and suppression of Ki-67 (Supplementary Fig. S8C, S8D). These and additional results (Supplementary Fig. S8E, S8F) suggest that inhibition of PARP specifically (or DNA damage repair in general) may be a potential strategy to augment MAPKi-addiction.
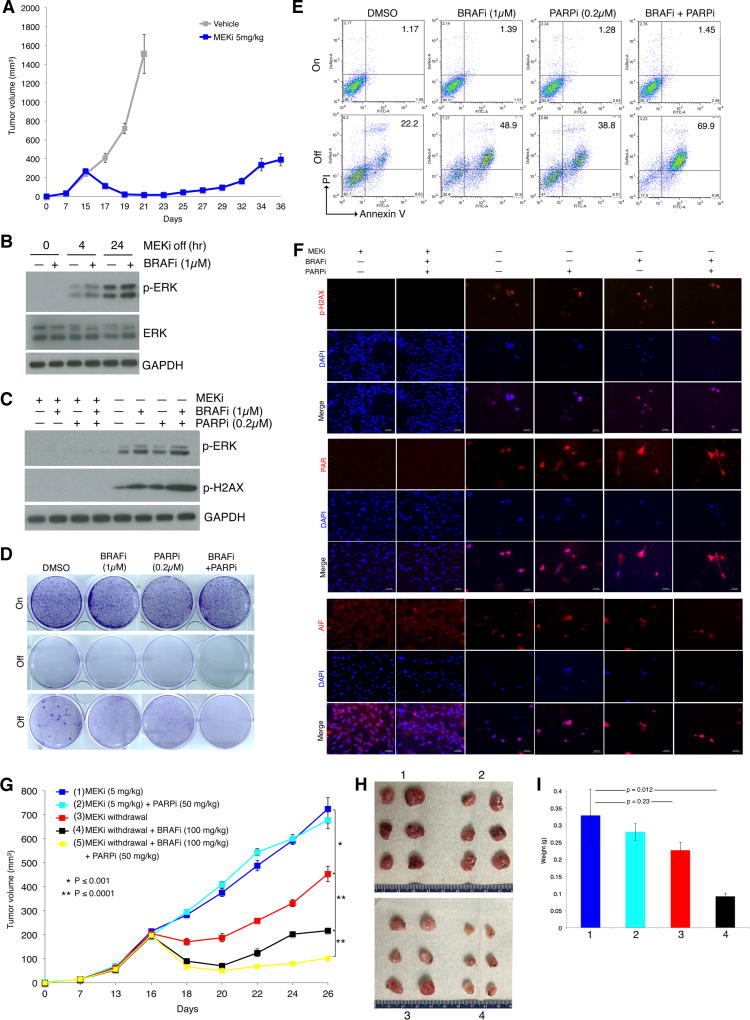
We also assessed whether the synthetic lethality (between supraphysiologic levels of activated ERK and DNA damage) observed across MAPKi-resistant human melanoma cell lines and PDX tumors could also be observed in an immune-competent context. To do this, we generated a syngeneic and transplantable model of MEKi-resistant MUTNras melanoma. First, we derived a NrasQ61R/Ink4a/Lkb1 murine melanoma cell line (TpLN61R for Tyrosinase p16 Lkb1 NrasQ61R) from a tumor of the Tyr-CRE-NRAS Q61R genetically engineered murine melanoma model (28) and adapted TpLN61R to efficient in vivo subcutaneous growth in C57BL/6 mice. These tumors were then chronically treated with trametinib at 5 mg/kg daily until MEKi-resistant tumors arose (Fig. 6A). We then dissociated a resistant tumor and cultured the tumor cells with trametinib in vitro (up to 0.1 µM over three weeks). In vitro, this MEKi-resistant cell line, termed NILR2R (Nras, Ink4a, Lkb1, Resistant mouse 2 Right flank) displayed robust time-dependent induction of p-ERK levels by MEKi withdrawal, which was accelerated by BRAFi treatment (Fig. 6B). Consistent with robust p-ERK rebound levels, p-H2AX was induced by MEKi withdrawal and further induced by BRAFi treatment during MEKi withdrawal (Fig. 6C). Importantly, PARPi treatment (at a low concentration effective in augmenting cell killing among cell-death predominant R-lines, Fig. 3) during MEKi withdrawal and BRAFi treatment resulted in even greater p-H2AX accumulation (Fig. 6C). Consistently, while BRAFi and/or PARPi had no appreciable effects on clonogenic growth or cell death levels of NILR2R in the presence of MEKi, BRAFi or PARPi treatment during MEKi withdrawal reduced clonogenic growth and increased the levels of cell death (Fig. 6D, 6E). Co-treatment of BRAFi and PARPi during MEKi withdrawal further reduced growth and increased death (Fig. 6D, 6E). Consistent with a cell-death predominant drug addiction phenotype observed among MAPKi-resistant human melanoma cell lines and PDX tumors, this MEKi-resistant murine melanoma cell line also displayed upregulation of PAR levels and nuclear localization of AIF during induction of the drug addiction phenotype (Fig. 6F). Finally, we re-implanted NILR2R back into C57BL/6 mice treated with trametinib (5 mg/kg via daily gavage) and identified five groups of mice with closely matched tumor volumes (Fig. 6G). We then tested the individual tumor-shrinkage effect of BRAFi or combined effect of BRAFi+PARPi treatments in vivo. As expected based on prior results, PARPi intraperitoneal treatments had no significant effect on the growth of NILR2R tumors on continuous MEKi oral treatments. MEKi discontinuation alone led to the most transient tumor-shrinkage. Importantly, BRAFi oral treatment beginning with MEKi discontinuation prolonged the tumor shrinkage effect of MEKi withdrawal (Fig. 6G to 6I). PARPi+BRAFi co-treatment during MEKi withdrawal led to the most sustained tumor regression (Fig. 6G). Thus, pharmacologically augmenting ERK rebound and DNA damage to maximize regression or shrinkage of MEKi-resistant MUTNras during MEKi withdrawal may also be feasible in an immune competent host.
Figure 6.
BRAF and PARP inhibitors augment MEKi addiction of MUTNras murine melanoma in an immune competent host. (A) Tumor volumes (mean ± SEM) of TpLN61R murine melanoma cells transplanted in C57BL/6 mice in response to daily gavage with the vehicle (n = 6) or trametinib (5 mg/kg; n = 6). One resistant tumor on day 36 was dissociated and cultured as a MEKi-resistant cell line (NILR2R). (B) Western blot levels of p-ERK, ERK, and the loading control GAPDH in the MEKi-resistant MUTNras SDR line, NILR2R, with indicated hrs off MEKi/trametinib (0.1 µM) treatment, with or without BRAFi/vemurafenib (1 µM) treatment. (C) Analysis of NILR2R protein lysates by Western blots of p-ERK and p-H2AX levels on or off MAPKi(s), with or without BRAFi (1µM) and/or PARPi (0.2µM) treatment for 3 d. (D–E) Clonogenic growth (D; 8 d) and percentages of Annexin V/PI-positive dead cells (E; 5 d) in NILR2R, on or off MEKi/trametinib (0.1 µM), with or without BRAFi/vemurafenib (1 µM) and/or PARPi (0.2µM) treatments. For D, cultures were seeded at 30K cells per well, except for the third row where cultures were seeded at 150K cells per well. (F) Levels and/or subcellular localization of p-H2AX, PAR, and AIF in NILR2R, on or off MEKi for 3 d, with or without BRAFi and/or PARPi treatment. Ruler, 20 µm. (G) Trametinib-resistant NILR2R cells were transplanted subcutaneously in C57BL/6 mice with daily trametinib (5 mg/kg) gavage until segregation into five groups (n=6 per group). Tumor volumes are shown as means ± SEM. P values, Student’s t-test. (H) Pictures of tumors from the first four experimental groups (mice sacrificed due to tumor ulceration) in G at day 26. (I) Tumor weights (means ± SEM; p value, unpaired two-way t-test) of the first four experimental groups in G.
DISCUSSION
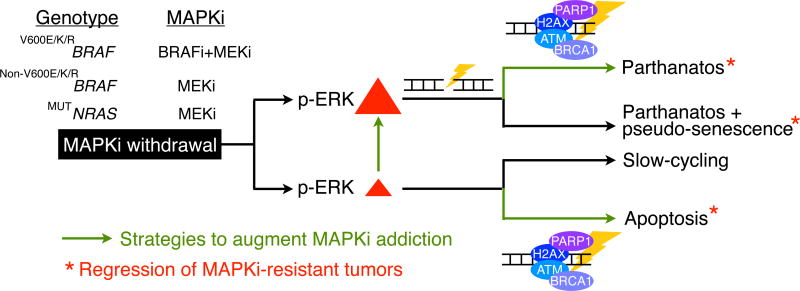
Understanding mechanisms underlying the cancer vulnerability of MAPKi-addiction availed us with potential therapeutic opportunities (Fig. 7). During a predominantly tumor cell-death rather than cell-cycle deceleration response upon drug withdrawal, MAPKi-resistant melanoma, regardless of the specific underlying driver(s) of resistance, displays a synthetic lethality between acute, supra-basal ERK hyper-activation and excessive DNA damage. Death by this synthetic lethality in the context of a robust p-ERK rebound is characterized by AIF-dependent but caspase 3-independent death. Furthermore, impairing DNA damage repair pharmacologically augmented DNA damage already induced by strong p-ERK rebounds and further boosted AIF-dependent death of MAPKi-resistant melanoma cells. On the other hand, a weak MAPKi withdrawal-induced p-ERK rebound was sufficient to elicit only a cell-cycle slow-down, leading to persisters that will resume rapid proliferation. However, upon MAPKi withdrawal, an innately weak p-ERK rebound coupled with impairment of the DNA damage repair machinery turned a predominantly slow-cycling response to a cell-death response that was caspase 3-dependent. Based on these findings, one would expect an even stronger synthetic lethality for drug-withdrawn, MAPKi-resistant melanoma harboring genetic loss of BRCA1/2 or displaying BRCAness (29) (either preexising or acquired during MAPKi therapy). Thus, after the emergence of MAPKi resistant clones, targeting the DNA damage repair pathways subsequent to cessation of MAPKi dosing, may specifically select against disease progression across the spectrum of MAPKi-addiction phenotypes.
Figure 7.
Strategies to select against MAPKi-resistant melanoma. Schematic showing MAPKi-addiction phenotypes being driven by p-ERK rebound levels and potential therapeutic strategies (enhancing p-ERK or impairing DNA damage repair) that promote tumor cell-death (apoptosis or parthanatos) or regression.
In the particular contexts of MEKi-resistant MUTNRAS melanoma, type I RAF inhibitors such as vemurafenib further augmented ERK rebound (on top of what was induced by MEKi withdrawal) and cell death. Counter to the dogma that type I RAF inhibitor is contraindicated for MUTNRAS melanoma, its ability to boost ERK levels in the context of MEKi-addiction suggest a novel context-dependent application. Furthermore, MEKi-resistant melanoma with atypical MUTBRAF mutations likely up-regulated RAF dimer levels (as a mechanism of resistance). This may explain the ability of vemurafenib to enhance p-ERK induction and tumor regression during MEKi withdrawal. Given the potential liability of prolonged BRAFi treatment in these genetic contexts, further studies are required to define an optimal BRAFi treatment duration in the context of augmenting drug-addiction.
This study highlights the importance of future studies capitalizing on MAPKi-addiction mechanisms to control MAPKi-resistant subclones (before the development of clinically evident disease progression). Our recent study identified melanoma tumors, even during MAPKi-induced regression, to be undergoing dynamic and stereotypic tumor cell and immune compartmental adaptations (30). It will also be important to characterize the immune infiltration/composition and identify specific adaptive immune resistance mechanisms associated with tumor regression resulting from a pharmacologically augmented drug addiction phenotype. Furthermore, MAPK inhibitors with ideal clinical pharmacokinetic properties (short half-lives, e.g., encorafenib, binimetinib) may be particularly useful for implementation of strategies based on MAPKi-addiction in human subjects. Finally, understanding in further detail how excessive ERK and DNA damage levels engage distinct programmed cell-death pathways promises to guide the development of rotational therapies that adapt to evolving cancer phenotypes and vulnerabilities.
METHODS
Cell Culture, Sub-line Derivation, Constructs and Inhibitors
All cell lines were routinely tested for Mycoplasma, and cell line and sub-line identities have been ensured by RNA-seq and the GenePrint 10 system (Promega) at routine intervals during the course of this study for banking and experimental studies. All cell lines were maintained in DMEM high glucose with 10% heat-inactivated FBS (Omega Scientific) and 2mM glutamine in humidified, 5% CO2 incubator. All M series cell lines were established from patient-derived tumors at UCLA. The TpLN61R cell line was derived at UNC and adapted to in vivo growth at UCLA in 2016. The NILR2R cell line was established in vitro from a MEKi-resistant tumor at UCLA in 2017. SKMEL28 was obtained from Dr. Alan Houghton (between 2008 and 2010). To derive resistant clones, parental melanoma cells seeded at low density were treated with BRAFi+MEKi (vemurafenib+selumetinib) (MUTBRAF) or MEKi (trametinib) (MUTNRAS) every 2–3 days for 6–12 weeks, and proliferative colonies were ring-isolated (except those designated “poly” for polyclonal where ring-cloning was not performed) and expanded. shH2AX, shFRA-1, shJUNB and vector (pLKO.1) were obtained commercially (ThermoFisher). shBRCA1 and vector (pGIPZ) were accessed through the Molecular Screening Shared Resource at UCLA. V600EBRAF was subcloned into the doxycycline-repressible lentiviral vector pLVX-Tight-Puro (ClontechLaboratories). Over-expression and knockdown constructs were packaged into lentiviral particles for infection. Inhibitors were obtained from the following: vemurafenib, in vitro (Plexxikon), vemurafenib, in vivo, selumetinib in vitro, trametinib in vitro and in vivo (LC Laboratories), ERKi/SCH772984 (Merck), p38i/SB203580, PARPi/Olaparib, ATMi/KU-55933, caspasei/Z-VAD-FMK (Selleckchem).
Protein Detection
Cells were lysed in RIPA buffer (Sigma) with protease inhibitor (Roche) and phosphatase inhibitor (Santa Cruz Biotechnology) cocktails for Western blotting. For immunohistochemistry (IHC), tissues were either fixed in 4% paraformaldehyde (PFA) and sucrose cryoprotected in OCT or in formalin and embedded in paraffin (FFPE). For FFPE tissues, after de-paraffinization and re-hydration, tissue sections were subjected heat for antigen retrieval. PFA/OCT sections were not subjected to antigen retrieval. Both IHC and immunocytochemistry of cell lines were performed with Alexa Fluor-conjugated secondary antibodies (Life Technologies) on 4% PFA-fixed cells. Nuclei were counterstained by DAPI. Fluorophore signals were captured with a Zeiss microscope (AXIO Imager A1) mounted with a charge-coupled device camera (Retiga EXi QImaging), and the images captured by Image-pro plus 6.0. Western blots and immunofluorescence assays were performed using the following antibodies: p-ERK1/2 (T202/Y204), p-c-FOS (S32), p-FRA1 (S265), p-p38 MAPK (T180/Y182), p-HSP27 (S82), p-Histone H2A.X (S139), total ERK1/2, c-FOS, FRA1, FOSB, p21, AIF, PARG, BRCA1, GAPDH (Cell Signaling Technology), TUBULIN (Sigma), PARP1, BRAF (Santa Cruz), PAR (Enzo), and Ki-67 (EMD Millipore). Western blot quantification was performed using NIH ImageJ.
Cell Line-based Assays
Clonogenic assays were performed by plating cells at single-cell density in six-well plates, and inhibitor/media replenished every 2 days for 7 days, unless noted otherwise. Colonies were fixed in 4% paraformaldehyde and stained with 0.05% crystal violet. Viable cell counts were performed in triplicate wells (in six-well plates). Cell death assays were performed by plating indicated cell lines with or without MAPKi(s) for 6 days (unless otherwise indicated), and cells were stained with Annexin V-FITC and PI for 15 minutes at room temperature before sample loading (LSR II Flow Cytometry, BD Bioscience). CFSE (Molecular Probes) dilution detected by flow cytometry was used to monitor cell division. Cell were loaded with 3 µM of CFSE and cultured for 6 days. Samples were collected and fixed in 2% PFA and analyzed with LSRII. Flow cytometry data were analyzed by FlowJo. Senescence was assessed by Senescence Associated β-Galactosidase staining kit (Cell Signaling Technology) with MAPKi withdrawn for 6 days. For vital imaging, cells were plated onto gridded dishes (Sigma) and imaged at indicated time-points at pre-designated areas. Comet assays (Cell Biolabs, Inc.) were performed by plating cell lines in six-well plates in the presence or absence of MAPKi for 3 days. Stranded breaks were detected by following the manufacturer’s recommendations, imaged with a Zeiss microscope (AXIO Imager A1) and distance of tails measured with Image-pro plus 6.0. Caspase-3 activity was assessed using Caspase-Glo 3/7 kit (Promega) by plating cells in white-walled, 96-well plates in the presence or absence of indicated inhibitors for 3 or 6 days.
RNA-seq Analysis
RNA-seq data from cell lines were generated using 2×100bp paired end sequencing using the Illumina HiSeq2000 platform. Paired end reads were mapped to the UCSC hg19 reference genome using Tophat2(31). Normalized expression levels of genes were expressed in FPKM values as generated by cuffquant and cuffnorm. Both programs were run with the option "--frag-bias-correct" and "--multi-read-correct" to improve sensitivity. For differential gene expression calls, a gene was defined as differentially expressed when its expression increased or decreased by at least two-fold. RNA-seq runs on multiple sequencing lanes were independently mapped, and the expression values of each gene (in FPKM) were averaged across multiple lanes. To overcome noise in differential expression values caused by extremely low FPKM levels, we added a pseudo-FPKM value of 0.1 to all expression values. RNA-seq data have been made available through Gene Expression Omnibus (GEO) at the accession number GSE87326.
Transcription Factor Enrichment Analysis
We collected transcription factor (TF) binding motifs in the form of position weight matrices (PWMs) from the JASPAR database (32). Instead of using a fixed PWM score cutoff(s) as done in other PWM matching programs, we estimated the significance of the PWM score in a gene’s promoter, which is defined as −1500 to +1500 bp from TSS, by comparing the score with a background distribution of the same PWM’s scores on non-promoter regions from randomly selected genes. Specifically, we collected 10,000 random 3 KB intragenic regions (excluding the genes’ promoter regions) and, for each sequence, computed the best score of a PWM. These scores defined an empirical background distribution of the PWM, and we defined a significant match of the PWM if and only if i) the PWM score was greater than or equal to the 95th percentile of the background PWM scores (i.e, P ≤ 0.05) and ii) a PWM score was at least 0.75. This approach avoided applying the same absolute PWM score cutoff on PWMs with differing lengths and complexities. To estimate enrichment of a TF’s PWM W in a set of co-regulated genes G, we compared the number of significant matches of W in the promoter regions in G (accounting for possible multiple TSS for each gene) and the number of matches against a set of randomly selected promoter regions of the same size. We repeated the latter step 100,000 times to estimate the empirical enrichment P-value of the PWM. Finally, we corrected the PWM enrichment P-values across all tested PWMs for multiple hypothesis testing using the Benjamini-Hochberg (FDR) method. A TF’s PWM was defined as enriched in a set of genes when its adjusted enrichment P-value was ≤ 0.05.
Gene Set Enrichment Analysis
Paired gene set enrichment analyses between off- versus on-drug conditions or between two different cell lines were performed as described previously(1). We computed differential gene set enrichments of the gene sets in the C2 CGP, C6 and Hallmark subsets from the Molecular Signature Database of the Broad Institute using the following steps: i) Calculating log2 fold changes (log2 FC) of mRNA expression of each gene in M249 DDR5 compared to SKMEL28 DDR1 at all three treatment conditions, i.e., on-drug 6 h, off-drug 6 and 24 h, ii) Based on the log2 FC values, we computed the differential enrichment of each gene set between M249 DDR5 and SKMEL28 DDR1 in all three treatment conditions (cutoff for differential enrichment, Wilcoxon rank-sum test between genes within the gene set and the rest of the genes; p ≤ 0.05; median of up-expression across all genes in the gene set ≥ 25%, i.e, median log2 FC ≥ 0.322), iii) To exclude differential enrichment already present between on-drug condition, for each of the gene sets meeting the cutoffs in step 2, we required that the difference between the median log2 FC in either off-drug condition to be higher than the median log2 FC in the on-drug condition by at least 0.322 (1.25 fold higher), and iv) For visualization, we computed the single sample enrichment GSVA scores (33) of the selected gene sets from step 3 using log2 CPM values as input.
PDX Models and Murine Melanoma
Mouse experiments were approved by the Animal Research Committee. Tumor fragments derived from a Q61RNRAS and a S365LBRAF metastatic melanoma (F0), which were obtained from two distinct patients with approval by the local Institutional Review Board, were transplanted subcutaneously in sex-matched NSG mice (6–8 week old). Tumors were measured with a calliper every 2 days, and tumor volumes were calculated using the formula (length × width2)/2. Trametinib-resistant tumors were cut into fragments, which were serially transplanted. NILR2R cells were injected at 1 million cells per flank in C57BL/6 mice. Mice were treated with vehicle (0.5%HPMC-0.2%Tween80, pH8), trametinib (5mg/kg), or vemurafenib (100 mg/kg) by oral gavage daily or olaparib (25 or 50 mg/kg) by daily IP injections.
Supplementary Material
SIGNIFICANCE.
Discontinuing targeted therapy may select against drug-resistant tumor clones, but drug-addiction mechanisms are ill-defined. Using melanoma resistant to but withdrawn from MAPKi, we defined a synthetic lethality between supra-physiologic levels of p-ERK and DNA damage. Actively promoting this synthetic lethality could rationalize sequential/rotational regimens that address evolving vulnerabilities.
Acknowledgments
Grant Support
This work has been funded by Burroughs Wellcome Fund (to R.S. Lo), the National Institutes of Health (1R01CA176111 and P01CA168585 to R.S. Lo), the Ressler Family Foundation (to R.S. Lo), the Melanoma Research Alliance (to R.S. Lo and W. Hugo), the Ian Copeland Melanoma Fund (to R.S. Lo), the SWOG/Hope Foundation (to R.S. Lo), Steven C. Gordon Family Foundation (to R.S. Lo), the Melanoma Research Foundation (to R.S. Lo), American Skin Association (to W. Hugo), American Association for Cancer Research-Amgen, Inc. Fellowship in Clinical/Translational Cancer Research (16-40-11-HUGO to W. Hugo), Department of Defense Horizon Award (to A. Hong), and Dermatology Foundation (to G. Moriceau).
We thank G. Bollag (Plexxikon Inc.) for providing PLX4032 and S. Hu-Lieskovan (UCLA), J. Zaretsky (UCLA), P. J. Kaplan-Lefko (UCLA) and A. Ribas (UCLA) for assisting with animal work.
Footnotes
Authors declare no conflicts of interest.
Supplementary Material
Access the most recent supplemental material at: http://cancerdiscovery.aacrjournals.org/content/suppl/2017/09/18/2159-8290.CD-17-0682.DC1
References
- 1.Hugo W, Shi H, Sun L, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–85. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moriceau G, Hugo W, Hong A, et al. Tunable-Combinatorial Mechanisms of Acquired Resistance Limit the Efficacy of BRAF/MEK Cotargeting but Result in Melanoma Drug Addiction. Cancer Cell. 2015;27:240–56. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi H, Hugo W, Kong X, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, Moriceau G, Kong X, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer Discov. 2012;2:414–24. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, Moriceau G, Kong X, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Allen EM, Wagle N, Sucker A, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N, Van Allen EM, Treacy DJ, et al. MAP Kinase Pathway Alterations in BRAF-Mutant Melanoma Patients with Acquired Resistance to Combined RAF/MEK Inhibition. Cancer Discov. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das Thakur M, Salangsang F, Landman AS, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amann VC, Hoffmann D, Mangana J, Dummer R, Goldinger SM. Successful retreatment with combined BRAF/MEK inhibition in metastatic BRAFV600-mutated melanoma. J Eur Acad Dermatol Venereol. 2017 doi: 10.1111/jdv.14268. [DOI] [PubMed] [Google Scholar]
- 12.Rogiers A, Wolter P, Bechter O. Dabrafenib plus trametinib rechallenge in four melanoma patients who previously progressed on this combination. Melanoma Res. 2017;27:164–7. doi: 10.1097/CMR.0000000000000320. [DOI] [PubMed] [Google Scholar]
- 13.Romano E, Pradervand S, Paillusson A, et al. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin Cancer Res. 2013;19:5749–57. doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]
- 14.Roux J, Pages C, Malouf D, et al. BRAF inhibitor rechallenge in patients with advanced BRAF V600-mutant melanoma. Melanoma Res. 2015;25:559–63. doi: 10.1097/CMR.0000000000000196. [DOI] [PubMed] [Google Scholar]
- 15.Seghers AC, Wilgenhof S, Lebbe C, Neyns B. Successful rechallenge in two patients with BRAF-V600-mutant melanoma who experienced previous progression during treatment with a selective BRAF inhibitor. Melanoma Res. 2012;22:466–72. doi: 10.1097/CMR.0b013e3283541541. [DOI] [PubMed] [Google Scholar]
- 16.Seifert H, Fisher R, Martin-Liberal J, et al. Prognostic markers and tumour growth kinetics in melanoma patients progressing on vemurafenib. Melanoma Res. 2016;26:138–44. doi: 10.1097/CMR.0000000000000218. [DOI] [PubMed] [Google Scholar]
- 17.Schreuer M, Jansen Y, Planken S, et al. Combination of dabrafenib plus trametinib for BRAF and MEK inhibitor pretreated patients with advanced BRAFV600-mutant melanoma: an open-label, single arm, dual-centre, phase 2 clinical trial. Lancet Oncol. 2017;18:464–72. doi: 10.1016/S1470-2045(17)30171-7. [DOI] [PubMed] [Google Scholar]
- 18.Yao Z, Torres NM, Tao A, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell. 2015;28:370–83. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010 doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 20.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010 doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su F, Viros A, Milagre C, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–15. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 24.Dummer R, Schadendorf D, Ascierto P, et al. Results of NEMO: A phase III trial of binimetinib (BINI) vs dacarbazine (DTIC) in NRAS-mutant cutaneous melanoma. J Clin Oncol. 2016;34 abstr 9500. [Google Scholar]
- 25.Villanueva J, Infante JR, Krepler C, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–9. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad M, Angeli JP, Vandenabeele P, Stockwell BR. Regulated necrosis: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2016;15:348–66. doi: 10.1038/nrd.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dummer R, Schadendorf D, Ascierto PA, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18:435–45. doi: 10.1016/S1470-2045(17)30180-8. [DOI] [PubMed] [Google Scholar]
- 28.Burd CE, Liu W, Huynh MV, et al. Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 2014;4:1418–29. doi: 10.1158/2159-8290.CD-14-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–25. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song C, Piva M, Sun L, et al. Recurrent tumor cell-intrinsic and -extrinsic alterations during MAPKi-induced melanoma regression and early adaptation. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathelier A, Fornes O, Arenillas DJ, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–5. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.