Abstract
Enhancers and promoters both regulate gene expression by recruiting transcription factors (TFs); however, the degree to which enhancer vs. promoter activity is due to differences in their sequences or to genomic context is the subject of ongoing debate. We examined this question by analyzing the sequences of thousands of transcribed enhancers and promoters from hundreds of cellular contexts previously identified by cap analysis of gene expression. Support vector machine classifiers trained on counts of all possible 6-bp-long sequences (6-mers) were able to accurately distinguish promoters from enhancers and distinguish their breadth of activity across tissues. Classifiers trained to predict enhancer activity also performed well when applied to promoter prediction tasks, but promoter-trained classifiers performed poorly on enhancers. This suggests that the learned sequence patterns predictive of enhancer activity generalize to promoters, but not vice versa. Our classifiers also indicate that there are functionally relevant differences in enhancer and promoter GC content beyond the influence of CpG islands. Furthermore, sequences characteristic of broad promoter or broad enhancer activity matched different TFs, with predicted ETS- and RFX-binding sites indicative of promoters, and AP-1 sites indicative of enhancers. Finally, we evaluated the ability of our models to distinguish enhancers and promoters defined by histone modifications. Separating these classes was substantially more difficult, and this difference may contribute to ongoing debates about the similarity of enhancers and promoters. In summary, our results suggest that high-confidence transcribed enhancers and promoters can largely be distinguished based on biologically relevant sequence properties.
Keywords: enhancers, promoters, gene regulation, machine learning, sequence analysis
THE regulation of gene expression plays an important role in all biological processes. In multicellular organisms, the repertoire of expressed genes varies depending on cell type, developmental stage, and the presence of stimuli (Orozco et al. 2012; Bauer et al. 2013; Brown et al. 2013; Busche et al. 2015). Differential expression of genes is implicated in many diseases, and is often mediated by genetic variation in regulatory sequences (Fortini et al. 2014; Claussnitzer et al. 2015; GTEx Consortium 2015; Lupiáñez et al. 2015).
Promoters and enhancers are regulatory sequences that work in concert to control gene expression at the transcriptional level. Promoters are traditionally defined as sequences immediately upstream of a transcription start site (TSS) that are directly involved in recruiting general transcription factors (TFs) and RNA polymerase II to the gene, and directing transcription (Kwak et al. 2013). Enhancers are sequences that recruit proteins that interact with promoters to facilitate and modulate transcription of genes, but can be thousands of base pairs away from their targets (Levine 2010). Many different assays have been developed to identify regions with promoter and enhancer activity (Benton et al. 2018; Rickels and Shilatifard 2018). For example, cap analysis of gene expression (CAGE)-based approaches map the locations of capped 5′ ends of transcribed RNA to the genome to identify regions involved in the regulation of transcription [Andersson et al. 2014; FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. 2014]. Since many enhancers are transcribed, this enables the identification of enhancers as well as promoters. Another common identification approach is to profile different histone modifications that are characteristic of each type of region. Trimethylation at the fourth lysine of histone 3 proteins (H3K4me3) is enriched near promoters, while monomethylation of the same residue (H3K4me1) is common at enhancers (Heintzman et al. 2007). However, we note that histone mark patterns at promoters and enhancers are complex and incompletely understood, and are not necessarily the cause of the regulatory activity (Calo and Wysocka 2013; Kim and Shiekhattar 2015).
Given the challenges of mapping regulatory regions, and enhancers in particular, there has been extensive work on using genomic sequence characteristics to identify enhancers. Enhancer-finding algorithms based solely on sequence information have successfully predicted active enhancers in many tissues (Burzynski et al. 2012; Ghandi et al. 2014; Kleftogiannis et al. 2016). We have previously published a support vector machine (SVM) framework capable of identifying enhancers and segregating them by their activity across tissues (Colbran et al. 2017). Several studies have reported distinct TF-binding preferences between enhancers and promoters (Rada-Iglesias et al. 2011; Shen et al. 2012; Thurman et al. 2012; Core et al. 2014; Nguyen et al. 2016), and some of these differences may be due to differences in GC content (Andersson et al. 2014). However, sequence-based enhancer predictors often also mistakenly identify promoters (Herman-Izycka et al. 2017).
There are many similarities between promoters and enhancers. Both contain sequences to recruit and bind TFs, in some cases the same ones (Bienz and Pelham 1986). In particular, promoters that lack CpG islands (CGIs) have similar sequences, recruit similar TFs, and have similar chromatin structure to enhancers (Andersson 2015; Andersson et al. 2015). In addition, enhancers and promoters are both transcribed, at least in some contexts (Natoli and Andrau 2012). Furthermore, the same sequence can have both promoter and enhancer activity, contingent on the context and particular complement of TFs bound (Nguyen et al. 2016).
As a result of these similarities, recent work has proposed blurring the traditional distinction between promoters and enhancers (Raab and Kamakaka 2010; Andersson et al. 2015; Kim and Shiekhattar 2015). Indeed, classifiers trained to distinguish tissue-specific promoters from the genomic background can also be used to predict tissue-specific enhancers, indicating the presence of similarity in their sequences (Taher et al. 2013).
In this study, we directly compare transcribed enhancers and promoters identified by the Functional ANnotation Of the Mammalian genome (FANTOM) Consortium by evaluating the similarity of the DNA sequence patterns underlying their activity across multiple cell types and contexts. In spite of their known sequence similarities, we show that machine learning classifiers can be trained to distinguish promoters from enhancers and to predict their activity levels across cellular contexts using short DNA sequence patterns (6-mers). It is possible to distinguish enhancers and promoters even when stratifying regions by CGI overlap. Furthermore, sequence-based models trained to predict enhancers and their levels of activity can also identify promoters; however, models that predict promoters are far less accurate at identifying enhancers. Interpreting the patterns learned by our classifiers revealed substantial differences in the sequence content of enhancers and promoters. For example, the association of GC content with promoter activity is present beyond the influence of CGIs. We also identify several DNA sequence patterns that are associated with enhancer (or promoter) activity. Many of these sequences match binding motifs for different TFs (e.g., AP-1 for enhancers and ETS for promoters) and have been identified in previous Massively Parallel Reporter Assay (MPRA) studies. Their importance in our classifiers (which consider hundreds of cellular contexts) suggests their broader relevance beyond the few contexts considered in the MPRAs. Finally, we find that accurately distinguishing enhancers and promoters identified by the Roadmap Epigenomics Consortium based on histone modifications using 6-mer sequence patterns is significantly more challenging. This difference may contribute to the ongoing debate about the similarity of enhancers and promoters, and should be addressed in future work. Collectively, our results suggest that while sequences with enhancer and promoter activity have many similarities, there are consistent differences between them at the sequence level, and enhancers and promoters defined by different experimental assays have different sequence relationships.
Materials and Methods
Enhancer data
We analyzed enhancers identified by CAGE from the FANTOM Consortium across all 411 different tissues and cellular contexts they examined (Andersson et al. 2014). CAGE tags and isolates RNAs with a 5′ cap, which includes mRNAs and enhancer RNAs. With this method, active enhancers can be distinguished by bidirectional transcription, whereas promoters show a strong bias toward the sense direction. For enhancers, this approach explicitly excluded regions near known transcription start sites and exons of mRNAs (both protein-coding and noncoding), and long noncoding RNAs. We defined enhancers as the 600-bp regions flanking the midpoint between the paired bidirectional CAGE peaks. Given that the average distance between paired enhancer CAGE peaks is 180 bp, this results in consideration of ∼390 bp upstream and ∼210 bp downstream of each enhancer TSS. However, there is a wide amount of variability in the overall length of the original FANTOM enhancers, so these sequences likely have varying proportions of regulatory influence. Of the 38,538 robust enhancers, we defined the top 5% enhancers active in most contexts as the “broadly active” set; this corresponded to enhancers active in > 45 contexts. Altering this threshold did not significantly alter classifier performance (Supplemental Material, Figure S2). Correspondingly, we defined the lowest 5% as “narrowly active” or “context-specific” enhancers, which corresponded to enhancers active in a single context. The enhancers with the broadest activity were active in all 411 contexts.
We used shuffleBed (Quinlan and Hall 2010) to obtain random sets of length-matched nonenhancer regions for each enhancer set. We also generated negative regions matched on GC content and chromosome, as well as length. For this, we used a custom script that finds all regions on a chromosome of the same length with similar GC content and randomly selects a representative for each positive region. The negative region sets excluded all enhancers from the full permissive CAGE enhancer data set (43,011 total sequences), Encyclopedia of DNA Elements (ENCODE) blacklist regions, genome (hg19) assembly gaps, and experimentally verified VISTA enhancers (downloaded in March 2014) (Visel et al. 2007).
We also trained a direct promoter–enhancer classifier using regions defined based on histone marks from the Roadmap Epigenomics Project (Kundaje et al. 2015). Specifically, these consisted of H3K27ac and H3K4me1 chromatin immunoprecipitation sequencing (ChIP-seq) peaks in 98 primary tissues. We defined enhancers for each tissue as the intersection of the two marks, then merged all regions across tissues that overlapped by ≥1 bp. We considered the central 600 bp of the resulting regions to control for length. We also filtered for those regions that were never considered to be promoters by excluding all regions that overlapped with a promoter in any tissue.
Promoter data
We defined promoters based on CAGE peaks predicted to be TSSs by FANTOM (FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. 2014). In cases where these regions overlapped on the same strand, suggesting they were alternate start sites for the same gene, we combined the two regions and used the mean activity, which left us with 27,227 promoter regions. To match the sequences analyzed for the enhancers, we considered the central 600 bp of the resulting regions (expanding 100 bp in both directions on the 300-bp upstream and 100-bp downstream definition used by FANTOM). In most cases, this resulted in a promoter being 400-bp upstream and 200-bp downstream of the TSS. We defined the top and bottom 5% by activity to be our broadly and narrowly active promoters, which corresponded to promoters active in > 372 contexts and < 9 contexts, respectively. The promoters with the broadest activity were active in 382 of the 411 contexts. Because of the differences in their overall levels of activity and the number of contexts assayed, the thresholds for the broad and narrow promoter and enhancer classes corresponded to different numbers of contexts (Figure S14). As we did with enhancers, we generated both length- and GC-matched random background sets, excluding hg19 assembly gaps, ENCODE blacklist regions, and our full promoter set.
To match the Roadmap enhancer data, we defined promoters for each tissue as the H3K27ac peaks that did not overlap with H3K4me1 peaks, then merged all regions across tissues that overlapped by ≥ 1 bp. We considered the central 600 bp of the resulting regions to control for length. We also filtered for those regions that were never considered to be enhancers by excluding all regions that overlapped with an enhancer in any tissue.
Regulatory region prediction from DNA sequence patterns
For all classification tasks, we trained 6-mer spectrum kernel SVMs to distinguish between sets of genomic regions. The spectrum kernel is a string kernel that defines the similarity of two sequences based on the occurrence of all possible short-sequence patterns of length k within them, including reverse complements (Leslie et al. 2002). All SVM analyses were performed using the SHOGUN Machine Learning Toolbox v4.0.0 (Sonnenburg et al. 2010). We set the soft margin constant, C, based on the balance of positives and negatives in the training set (Ben-Hur and Weston 2010); since nearly all of our training sets were balanced, this resulted in a value of 1.0 for most analyses. Performance of the classifiers was evaluated using 10-fold cross-validation across the full data set. We partitioned the data into 10 nonoverlapping subsets with the same number of positives and negatives in each, trained a classifier on 90% of the data, and then evaluated the classifier’s performance on the remaining 10%. We repeated this procedure 10 times using each partition as the evaluation set once, and calculated receiver operator characteristic (ROC) and precision recall (PR) areas under the curve (AUCs) by averaging over the 10 cross-validation runs. All figures plot the average, maximum, and minimum curves obtained unless otherwise stated. Because we did not tune hyperparameters or select models based on performance on this data set, we did not use a separate validation set. We computed the average weights for each possible 6-mer in each SVM (Guyon et al. 2002), and when comparing to the genomic background, we averaged across runs vs. four independent random negative sets.
To evaluate the ability of models to directly distinguish FANTOM promoters from enhancers, we trained and evaluated on nine nonoverlapping random sets of 3000 regions from each class. To ensure that our results were not sensitive to the particular sets chosen, we trained an independent classifier as described above (including 10-fold cross-validation) for each pair of sets and reported the average performance of the classifiers over these subsets. We controlled for length differences by expanding or contracting enhancers and promoters in each set to be 600-bp long—approximately the maximum enhancer length—while maintaining their original centers. For the Roadmap replication sets, we trained and evaluated on 10 random subsets of 4000 regions from each class. For all classifiers, we compared feature weights using the mean weight of each 6-mer across the nonoverlapping subsets.
Training and evaluation of classifiers for distinguishing broadly active regions of each type vs. genomic background or context-specific regions proceeded similarly, using the regions generated as described above. When the positive and negative sets were of different size, we took a random subset of the larger set, so there would be a 50% chance of picking a positive or negative at random.
To assess the performance of classifiers trained on enhancers at predicting promoters, and vice versa, we calculated the relative ROC AUC (ROC AUCpred/ROC AUCtrain), where ROC AUCpred is obtained from evaluation of the classifiers trained on one type of region at predicting the other types of regions, and ROC AUCtrain is obtained by evaluating the classifiers on the same types of regions that they were trained on.
Principal component analysis
To summarize and visualize the sequence similarities of regulatory regions, we performed principal component analysis (PCA) on their sequences. We counted the occurrences of all possible 6-mers in each promoter and enhancer, then transformed the counts using log10(count+1) and standardized them before conducting PCA using prcomp in the R stats package with the default settings.
TF-binding motif and expression analysis
We obtained 248 human TF-binding motifs from the HOCOMOCOv9 database (Kulakovskiy et al. 2013), 402 from the HOCOMOCO v11 CORE database (Kulakovskiy et al. 2018), and 519 motifs from the JASPAR 2016 vertebrate database (Mathelier et al. 2016). We used tissue specificity scores (TSPS) for 332 TFs from the FANTOM Consortium (Ravasi et al. 2010). A TF with uniform expression across all 34 tissues considered is assigned a TSPS of zero, while a TF expressed in only a single tissue receives the highest TSPS (∼5). Following the original analysis of TSPS, we classified 157 TFs as “specific” (TSPS ≥ 1) and 175 as “broad” that are expressed in a wider range of contexts (TSPS < 1).
We counted the occurrences of all TF motifs in sequences of interest by first controlling for length by expanding or contracting all regions to be 600-bp long, focused on the center of each region. We used FIMO under default settings (Grant et al. 2011), then tallied the counts by specificity. We tested for enrichment of predicted binding sites by activity using a binomial test for the mean proportion of broad motifs in a given set of regulatory regions vs. the overall proportion of broad motifs in the database being used. We also compared the distribution of those proportions between regulatory region sets using a Wilcoxon rank sum test.
We used Tomtom version 4.10.1 to calculate the similarity between 6-mers and each TF-binding motif, with default parameters (Gupta et al. 2007). We compared the full distributions of P-values for the broad and specific TF groups using the Wilcoxon rank sum test. For matching TFs to specific 6-mers in the promoter vs. enhancer classifiers (Figure 3D and Figure S10), we used the HOCOMOCO v11 CORE database (Kulakovskiy et al. 2018).
Figure 3.
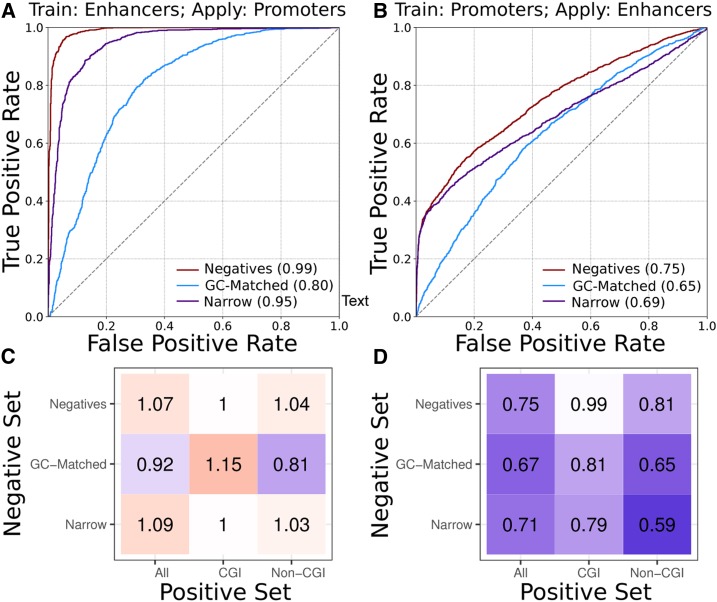
Classifiers trained on enhancers can accurately predict promoters and their breadth of activity, but not vice versa. (A) ROC curves for classifiers trained on enhancers and then used to classify promoters. (B) ROC curves for classifiers trained on promoters and then used to classify enhancers. Classifiers were trained using broadly active regions as positives and genomic background regions (red, “Negatives”), GC-matched background regions (blue, “GC-Matched”), or narrowly active regions (purple, “Narrow”) as negatives. PR curves are given in Figure S5. (C and D) Relative ROC AUCs (AUCpred/AUCtraining) for cross-region classifiers predicting promoters and enhancers reveal that enhancer-trained classifiers (C) generalize well to promoter prediction tasks, but the promoter-trained classifiers (D) do not. The superior generalization of the enhancer classifier held when regions with and without CpG islands were analyzed separately. ROC and PR curves for (C and D) are in Figure S6. AUC, area under the curve; CGI, CpG island; PR, precision recall; ROC, receiver operator characteristic.
Data availability
Data and scripts used in this study are available on GitHub (https://github.com/colbrall/enhancer_promoter_manuscript), or from the authors upon request.
Results
Promoters and enhancers have distinct DNA sequence patterns
Motivated by the similarities in the sequence, and functional architectures of enhancers and promoters, we investigated the ability of a machine learning algorithm to distinguish the two types of regions based on sequence characteristics. We considered 38,538 enhancers and 27,227 promoters from 411 cellular contexts identified using CAGE by the FANTOM Consortium [Andersson et al. 2014; FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. 2014]. We trained SVM classifiers to distinguish enhancers and promoters using the frequencies of all 4096 possible 6-bp-long sequences (6-mers) in a 600-bp window centered on the FANTOM-defined regions (Materials and Methods). To facilitate training and evaluation, we split the promoters and enhancers into nine random nonoverlapping subsets of 3000 promoters and enhancers, and performed 10-fold cross-validation within each subset to evaluate performance (Materials and Methods).
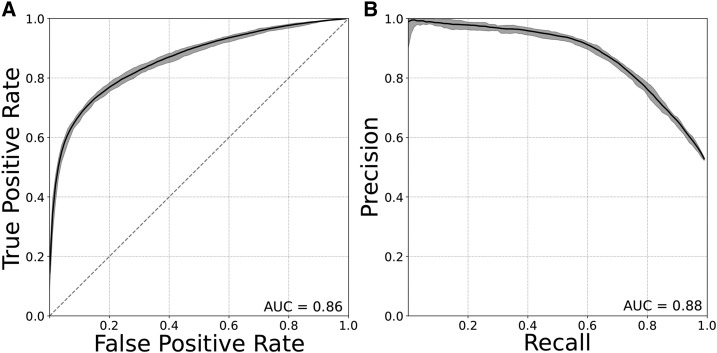
The classifiers were able to distinguish promoters from enhancers with high accuracy. They achieved average areas under ROC AUCs of 0.86 and areas under PR AUCs of 0.88 (Figure 1, A and B). The strong performance of the models was consistent across independent subsets of enhancer and promoters (ROC AUC range 0.85–0.91; PR AUC range 0.85–0.92).
Figure 1.
Promoters and enhancers can be distinguished from one another based on DNA sequence properties alone. (A) ROC curve and (B) PR curve evaluating SVM classifiers trained to distinguish FANTOM promoters from enhancers using patterns of short DNA sequences (6-mers) as features. Plots show the mean, maximum, and minimum curves obtained from classifiers, trained and evaluated on nine unique subsets of 3000 promoters and 3000 enhancers from the full data sets. AUC, area under the curve; PR, precision recall; ROC, receiver operator characteristic; SVM, support vector machine.
The accuracy of the classifiers was somewhat surprising given recent work that has found strong commonalities between promoter and enhancer sequences, especially between enhancers and non-CGI promoters [Andersson et al. 2014; FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. 2014]. Furthermore, machine learning models trained to accurately distinguish active from inactive promoters based on sequence characteristics (TF-binding site motifs, specifically) are also able to predict enhancers (Taher et al. 2013). However, this previous study focused on promoters and enhancers with tissue-specific activity, so it is possible that the similarity of promoters and enhancers varies based on breadth of activity. In other words, promoters and enhancers specific to a given context may be very similar in their sequence properties; meanwhile, consistent with our results, more broadly active regions may have less similarity. It is also possible that enhancers and promoters identified by different experimental techniques may have different levels of sequence similarity. We explore these possibilities over the next few sections by investigating the sequence patterns underlying breadth of activity in transcribed enhancers and promoters, characterizing their similarities and differences, and examining other enhancer and promoter sets.
Sequence-based classifiers are able to distinguish the breadth of activity of enhancers and promoters across tissues
Many enhancers are active in a small number of tissues, while most promoters are active in a large number of tissues (Figure S1). We previously demonstrated that SVM classifiers can predict enhancers and their breadth of activity using characteristics of their sequences (Colbran et al. 2017). Thus, we hypothesized that the ability to distinguish enhancers from promoters based on sequence patterns could be influenced by the differences in their activity levels. To test this, we identified sets of broadly and narrowly active regions of each type. For both enhancers and promoters, we defined the top and bottom 5% by activity as broadly and narrowly active regions (Materials and Methods).
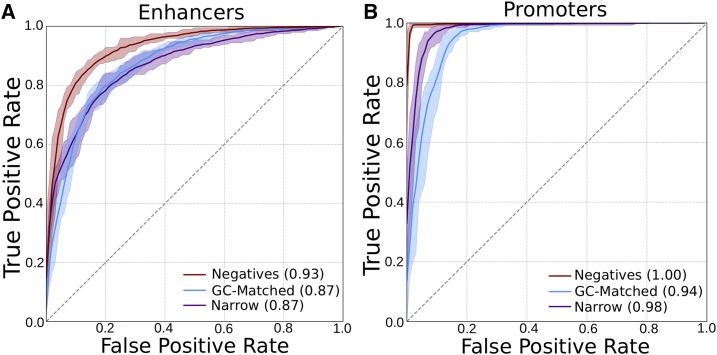
As expected from our previous work, SVM classifiers were able to distinguish enhancers with broad activity across tissues from those with narrow activity and from the genomic background based on sequence properties (Figure 2A, ROC AUCs of 0.87 and 0.93, respectively), and this ability is not contingent on the specific thresholds used to define broad and narrow activity (Figure S2). In contrast, the classifier was no better than random when attempting to distinguish enhancers and promoters when their labels were shuffled (Figure S3). Next, we evaluated the ability of 6-mer spectrum SVM classifiers to distinguish broadly active promoters from narrowly active promoters and negative control regions without promoter activity. The classifiers were able to distinguish broadly active promoters from context-specific promoters and from background regions with high accuracy (Figure 2B and Figure S4, ROC AUCs of 0.98 and ∼1.0, respectively). In fact, the promoter-trained classifiers were even more accurate at distinguishing broadly active promoters from the negative regions than the corresponding enhancer-trained classifiers across all classification tasks: ROC AUCs between 0.94 and ∼1.0 (Figure 2B and Figure S4). Because these classifiers were all trained on balanced data, it is important to note that the reported AUCs are not necessarily reflective of their performance over the whole genome.
Figure 2.
DNA sequence-based classifiers can accurately distinguish broadly active regulatory regions from the genomic background and narrowly active regions. Average ROC curves for 6-mer spectrum SVMs trained using broadly active (A) enhancers and (B) promoters as positives. The negatives were an equal number of random length-matched genomic background regions (red), length- and GC-matched background regions (blue), or narrowly active regulatory regions (purple). The shaded areas give the maximum and minimum curves observed over 10-fold cross-validation. PR curves also indicated strong performance (Figure S4, PR AUCs of 0.88–1.0). PR, precision recall; ROC, receiver operator characteristic; SVM, support vector machine.
Because of the known correlations between GC content and regulatory activity (Colbran et al. 2017), we also tested the ability to distinguish promoters and enhancers from background regions matched for GC content (Figure 2). For both region types, performance when predicting positives vs. GC-matched regions dropped compared to vs. non-GC-matched regions. However, the promoter classifier’s performance was nearly as good as vs. non-GC-matched regions; this suggests that it is “easier” to distinguish between broadly and narrowly active promoters than between broadly and narrowly active enhancers.
Classifiers trained on enhancers are generalizable to promoters, but not vice versa
Given that the breadth of activity of both enhancers and promoters could be accurately predicted from DNA sequence patterns (Figure 2), we next sought to test how well the sequence patterns informative about breadth of activity generalized between promoters and enhancers. We evaluated this by applying 6-mer sequence models trained to distinguish enhancer activity to predict the activity levels of promoters, and vice versa.
In general, classifiers trained on enhancers performed well on the corresponding promoter classification task: ROC AUCs between 0.80 and 0.99 (Figure 3A and Figure S5A). However, the reverse was often not true; promoter-trained classifiers applied to the prediction of enhancers had much lower ROC AUCs: between 0.6 and 0.8 (Figure 3B and Figure S5B).
To place the performance of the classifiers applied across regulatory region types in the context of their performance on their training data, we calculated a relative ROC AUC (AUCpred/AUCtraining). Enhancer-trained classifiers were generally able to predict promoters at least as well as they performed on the corresponding enhancer data, and in some cases, they performed better (Figure 3C). This suggests that sequence characteristics learned by the models to distinguish enhancers and their activity are also sufficient for distinguishing levels of promoter activity across cellular contexts. In contrast, classifiers trained on promoters always had worse performance when applied to enhancers (Figure 3D). This suggests that the enhancer classifiers have learned sequence patterns that influence enhancer activity levels and are not captured in promoter classifiers.
Overall, these results suggest that, while promoters and enhancers share key sequence features, broad enhancer activity may be influenced by additional characteristic sequence patterns that classifiers trained on promoters fail to learn.
CGI overlap does not account for all CpG content differences between enhancers and promoters
GC content and CGIs are known to be important for broad activity in promoters, and they are enriched in promoters compared to enhancers (Roider et al. 2009; Fenouil et al. 2012). To explore if our classifiers learned these distinctions and evaluate the importance of CGIs compared to more general CpG content, we conducted several analyses.
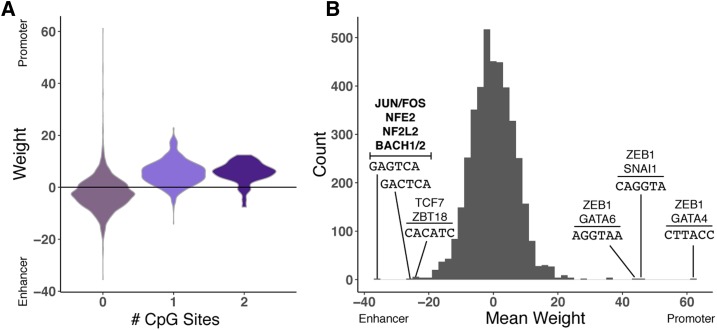
First, given the known importance of CGIs to promoter activity, we examined the importance of GC and CpG content to the classifiers. As expected, in the classifiers trained to directly distinguish promoters from enhancers, 6-mers assigned weights indicative of promoter activity were positively correlated with both GC content of the 6-mer (Spearman’s ρ = 0.38, P < 2.2E−16) and its CpG content (Figure 4A; Spearman’s ρ = 0.56, P < 2.2E−16). Results were similar for the breadth of activity classifiers; CpG content and overall GC content are higher in 6-mers that are predictive of broad activity for both promoters and enhancers (Figure S7).
Figure 4.
Interpreting the weights assigned to 6-mers by the promoter vs. enhancer classifier. (A) The distribution of weights assigned to 6-mers by the promoter vs. enhancer classifier (Figure 1), stratified by the number of CpG sites in the 6-mer. Each 6-mer is represented by its mean weight across classifiers trained on nine nonoverlapping subsets of the regions. Positive weights indicate that the 6-mer is predictive of promoter activity, and negative weights are indicative of enhancer activity. (B) The distribution of mean 6-mer weights. The 6-mers with the highest and lowest weights are labeled with their sequences and matches to transcription factor motifs from the HOCOMOCO v11 CORE database. Significant matches after multiple testing correction (false discovery ratio < 0.05) are shown in bold; for high-weight 6-mers without matches that meet this threshold, the top two nominally significant (P < 0.05) matches are listed. The top enhancer-associated 6-mers match motifs associated with components of AP-1 (JUN and FOS) and several other families. There were no significant matches for the promoter-associated 6-mers after multiple testing correction, but all contain the GGTA sequence, and nominally match ZEB1 and GATA factor motifs. CGI-stratified analyses are provided in Figure S10. CGI, CpG island.
To directly explore if the importance of GC and CpG content was driven by CGIs, we trained classifiers based on enhancers and promoters stratified based on CGI overlap. These classifiers were still able to accurately distinguish promoters from enhancers when both were in CGIs (ROC AUC = 0.80, PR AUC = 0.80) and when they did not overlap CGIs (Figure S8; ROC AUC = 0.76, PR AUC = 0.76). This was likely partially due to the fact that matching for CGI status does not necessarily match for GC content. Despite this, both classifiers performed worse than when not stratifying due to the lack of information provided by CGI status. Nonetheless, given the known motif-level similarities between transcribed enhancers and non-CGI promoters [Andersson et al. 2014; FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. 2014], the ability to distinguish them reasonably accurately suggests meaningful sequence differences.
Even with CGI stratification, GC content was significantly correlated with the weights assigned to 6-mers for both CGI (Spearman’s ρ = 0.13, P = 5.6E−16) and non-CGIs (Spearman’s ρ = 0.23, P < 2.2E−16) classifiers. We also evaluated CGI-stratified breadth of activity classification. In the breadth of activity classifiers that excluded CGIs from analysis, CpG content still explained a significant amount of the weight assigned to 6-mers predictive of broad promoter activity (R2 = 0.31; P < 2.2E−16). However, the association was much lower for non-CGI enhancers (Figure S9; R2 = 0.05; P < 2.2E−16), suggesting that, while CpG content is indicative of broad promoter activity even outside CGIs, the same is not true of enhancers. We used R2 for this analysis because we further investigated partial correlations (Supplemental Text). PCA on the 6-mer spectra of enhancers and promoters with varying activities also suggested that there are sequence differences between enhancers and promoters beyond the established difference in CGI prevalence (Figure S15 and Supplemental Text).
Finally, we stratified the cross-region classifiers by CGI status. The superior generalization of the enhancer classifiers compared to promoter classifiers held both in and outside CGIs (Figure 3, C and D). However, the non-CGI classifiers are of particular interest, as they were trained without the benefit of CGI presence. When the negative sets were matched for GC content, the enhancer classifier was still superior to the corresponding promoter classifier, but was noticeably worse at identifying promoters than the other enhancer-trained classifiers (relative ROC AUC of 0.81 vs. > 0.92 for the others). This suggests that, even after accounting for the greater CGI and GC content of promoters, enhancer classifiers learned sequence characteristics distinct from those of promoters.
The 6-mers most important to distinguish enhancers and promoters match different TF motifs
To explore how these sequence differences could affect function, we analyzed occurrence of binding motifs for TFs from the HOCOMOCO v11 CORE database (Kulakovskiy et al. 2018) among the highly weighted 6-mers. In particular, 6-mers most characteristic of enhancers significantly (false discovery ratio < 0.05) matched binding motifs of several JUN and FOS proteins (key components of the AP-1 complex), as well as motifs for NFE2, NF2L2, and BACH1/2 (Figure 4B). The top promoter-associated 6-mers in the direct classifier matched binding motifs for TFs such as ZEB1, GATA4, and SNAI1; however, these associations did not pass multiple testing correction.
We next analyzed how the motifs for the most highly weighted 6-mers changed in the CGI-stratified classifiers. For the classifier trained on CGI-containing regions, nearly all significant motif matches (for both enhancer and promoter 6-mers) were to ETS TF family members and its core GGAA sequence. ETV2 and ETV4 were associated with enhancer 6-mers, and ELK1, ELK4, ETV1, and ELF2 matched promoter 6-mers (Figure S10A). Nominally significant hits included RFX2 with promoter 6-mers, and ZEB1 to both enhancer and promoter 6-mers. For the non-CGI classifier, the TF motifs matching high-weight 6-mers were similar to those in the nonstratified classifier, with significant enrichment for AP-1 components among the enhancer-associated 6-mers (Figure S10B).
To further validate the relevance of these sequences to regulatory activity, we compared them to motif activity patterns estimated from three recent MPRA studies. First, we compared the 6-mers that most distinguished enhancer and promoter activity levels in our models to conclusions from a human MPRA study, which directly tested and compared enhancer and promoter sequence activity (Nguyen et al. 2016). Several of the TF motifs matched by the high-weight 6-mers from the promoter and enhancer activity classifiers were also observed in the MPRA analysis. For example, the RFX and ETS TF families were found to be intrinsically biased toward the generation of promoter activity, and binding motifs for TFs from these families matched 6-mers strongly associated with promoter activity in our analyses (Figure 4B and Figure S10). Similarly, 6-mers we identified as important to enhancer activity significantly matched the binding motifs for components of the AP-1 complex (Fos and Jun family members), agreeing with observations from the MPRA study that AP-1-binding sites generate strong enhancers with little promoter activity.
We also compared our results to two other recent MPRA studies of regulatory region activity in HepG2 and K562 cells (Ernst et al. 2016; Klein et al. 2018). Klein et al. (2018) focused on liver enhancers, and they also found enrichment for liver-expressed AP-1 complex member (FosL2 and JunD)-binding sites in active regions, further supporting these conclusions. Ernst et al. (2016) found ETS motifs to be among the strongest activating among both HepG2 and K562 cells. They also found sequences matching RFX motifs to have repressive activity in HepG2 cells. However, since these studies did not explicitly separate enhancers and promoters, it is difficult to directly relate their findings to our own. Nonetheless, the identification of many similar TF families supports the potential biological relevance of our findings. Furthermore, the fact that we observe these patterns in analyses of enhancers and promoters across hundreds of cellular contexts suggests that these sequences are broadly important to enhancer and promoter activity beyond the few cell types analyzed in the MPRAs.
Broadly active promoters and enhancers have more potential TF-binding sites
To explore how these sequence differences could affect breadth of activity across many cellular contexts, we hypothesized that regulatory regions with broad activity would have more predicted binding sites than their narrowly active counterparts after controlling for length. Indeed, both broadly active promoters and enhancers have more predicted TF-binding sites than their narrowly active counterparts (mean 165.3 vs. 67.8 and 107.9 vs. 61.4, respectively; P < 2.2E−16 for both, Wilcoxon rank sum test).
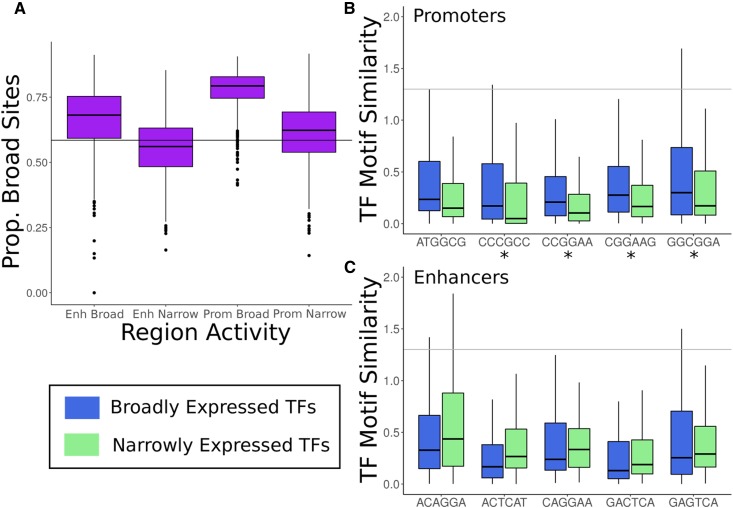
Next, we tested for differences in the breadth of expression across tissues of TFs with binding sites in broadly active and narrowly active regions. Promoters and enhancers on average have a significantly higher proportion of binding sites for broadly expressed TFs than would be expected by chance (Figure 5A; P < 2.2E−16 for all, binomial test). Furthermore, broadly active regions had significantly higher proportions of sites for broad TFs than narrowly active regions (promoters: 0.78 vs. 0.61, P = 3.5E−289; enhancers: 0.68 vs. 0.56, P = 4.3E−179, Wilcoxon rank sum test). These analyses were based on TF motifs from the HOCOMOCO database, and the results were similar when using motifs from the JASPAR database (Figure S12).
Figure 5.
Broadly active promoters and enhancers have more TF-predicted binding sites. (A) The distributions of the proportion of binding sites for broadly expressed TFs in each regulatory region (enhancers and promoters) contrasted between those with broad and narrow activity across tissues. The horizontal line indicates the proportion of broad TFs overall. While all regions except for narrow enhancers are enriched for broad predicted TF-binding sites (P < 2.2E−16 for all), broadly active regulatory regions are significantly more enriched than narrowly active ones (P = 3.5E−289 for promoters and P = 4.3E−179 for enhancers). (B) The most highly weighted DNA 6-mers by the SVMs for distinguishing broadly active promoters from narrowly active promoters match different sets of TFs. Sequence patterns predictive of broadly active promoters are enriched for similarity to broadly active TF motifs (blue), in particular ETS family members (CCGGAA, CGGAAG, and GGCGGA). (C) Same as (B), but for SVMs trained to distinguish broadly active enhancers from narrowly active enhancers. In contrast to promoters, patterns predictive of broadly active enhancers are not enriched for broadly expressed TFs and show a preference for AP-1 complex components. Similarity is quantified as the –log10 of the 6-mer–TF motif match P-value (gray line: P = 0.05). The box plots show the median and first/third quartiles, and outliers were not plotted. *P < 0.05, Wilcoxon rank sum test. TF-binding motifs were taken from HOCOMOCO; results were similar for motifs from the JASPAR database (Figures S11–S13). SVM, support vector machine; TF, transcription factor.
Next, we hypothesized that the 6-mers most predictive of broad activity would be more similar to broadly expressed TF-binding motifs than to context-specific TF motifs, and vice versa. This hypothesis is supported by the fact that broadly active enhancers are enriched for GC content, as are the binding motifs of broadly active TFs (Colbran et al. 2017). Supporting this hypothesis, the five highest-weighted 6-mers from the broadly active vs. narrowly active promoter classifier all are more similar to broadly active TF motifs than to context-specific TF motifs (Figure 5B). The most negatively weighted 6-mers follow the opposite trend and are more similar to context-specific TF motifs. Similarly, the highest- and lowest-weight 6-mers from the promoter vs. genomic background classifiers follow the same trends (Figure S12). In contrast, the high- and low-weight 6-mers from the corresponding enhancer classifiers did not exhibit a clear pattern. In fact, two of the high-weight 6-mers are more similar to context-specific TF motifs (Figure 5C and Figure S13). In summary, the motifs significantly similar to 6-mers predictive of broadly active promoters belong to broadly active TFs, while those for enhancers are a mix of broad and context-specific TFs.
Sequence differences between enhancers and promoters are inconsistent across regulatory region identification strategies
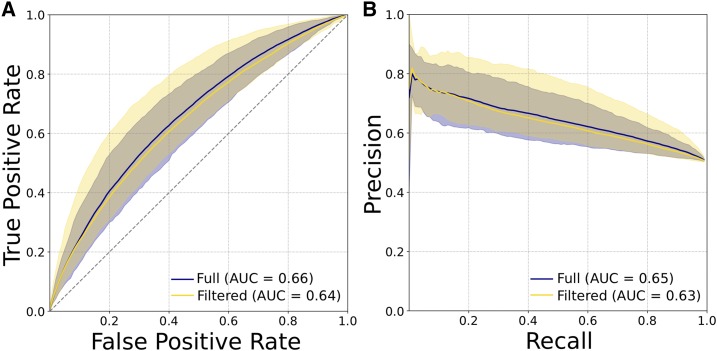
Many strategies have been developed to identify regulatory regions active in different cellular contexts, and there is increasing evidence that regulatory regions defined by different methodologies have different functional and evolutionary properties (Benton et al. 2018). Thus, we repeated our analyses on two additional enhancer and promoter data sets. First, we considered enhancers and promoters defined from histone modifications across 98 tissues from the Roadmap Epigenomics Project by intersecting H3K27ac ChIP-seq peaks with H3K4me1 peaks (Kundaje et al. 2015). The presence or absence of H3K4me1 was used as a proxy for enhancer or promoter activity, respectively. We found 388,416 enhancers and 201,090 promoters across all tissues, and ran analyses on nine random subsets of 4000 elements of each type. The 6-mer SVM was able to distinguish promoters from enhancers better than random based on the 600 bp centered on the annotated regulatory region, but the performance was substantially lower than for the transcribed regulatory regions identified by FANTOM (Figure 6; e.g., mean ROC AUC = 0.66 vs. 0.86).
Figure 6.
Enhancers and promoters defined by histone marks (H3K27ac with or without H3K4me1) from the Roadmap Epigenomics Project are less distinguishable by short sequence patterns than those defined by transcription patterns by FANTOM. (A) ROC curves and (B) PR curves evaluating SVM classifiers trained to distinguish promoters from enhancers using patterns of 6-mers as features. Plots show the mean, maximum, and minimum curves obtained from classifiers trained and evaluated on nine unique subsets of 4000 promoters and enhancers from the full data sets. The two curves on each plot represent an analysis of all enhancers and promoters, or a set with regions that have enhancer activity in some cellular contexts and promoter activity in others removed. Promoters and enhancers identified by bidirectional transcription via cap analysis of gene expression assays were much easier to distinguish from sequence patterns (Figure 1, A and B). AUC, area under the curve; PR, precision recall; ROC, receiver operator characteristic; SVM, support vector machine.
More than 25% of regions identified as Roadmap promoters were identified as enhancers in other tissues. While this potentially reflects the dynamic landscape of histone marks and regulatory regions (Wu and Sun 2006; Riccio 2010), we were concerned that the inclusion of these sequences with dual activity may have confounded the classifier. Thus, we repeated the training and evaluation, based on random subsets of 4000 elements each from a filtered set of nonoverlapping enhancers and promoters. Performance did not improve (Figure 6; mean ROC AUC = 0.64), which suggests that regulatory regions as defined by histone mark combinations are less distinct from one another at the sequence level than those defined by transcription patterns. However, similar to the results for FANTOM regions, the 6-mer weights most indicative of promoters were positively correlated with GC content (Spearman’s ρ = 0.25, P < 2.2E−16) and CpG content (Spearman’s ρ = 0.33, P < 2.2E−16). This suggests that there are consistent differences in sequence between enhancers and promoters, including overall GC content and the importance of CGIs, but the scale of the difference varies by the methodology used to identify the regions.
Discussion
We investigated similarities and differences in the sequence determinants of promoters, enhancers, and their activity levels. An SVM classifier trained on short DNA sequence patterns was able to distinguish between transcribed enhancers and promoters with high accuracy (ROC AUC = 0.86), indicating that there are substantial sequence differences between the two groups in aggregate. Nonetheless, there are similarities in the determinants of the breadth of promoter and enhancer activity; classifiers trained to recognize broadly active enhancers were able to identify broadly active promoters as accurately as classifiers trained on broadly active promoters. However, the reverse was not true; promoter-trained classifiers could not distinguish broadly active enhancers. This further suggests that broadly active enhancers exhibit different sequence patterns than promoters, and that they may have greater sequence complexity that includes promoter-like patterns. Indeed, some enhancers are known to have weak promoter activity (Kowalczyk et al. 2012). It is possible that this could have been influenced by the greater variability in length of the FANTOM enhancers, which could thereby alter the amount of regulatory relevance of our final regions. It is also possible that the promoters are easier for the SVMs to distinguish from the negative sets we considered, and this could also influence their generalization. However, the worse generalization of the promoter classifiers held in each analysis (using genomic background, GC-matched background, and tissue-specific activity regions as negatives). There are also differences in the TF-binding motifs associated with the highest-weighted sequence patterns in the enhancer and promoter classifiers. Sequences characteristic of broadly active promoters were more similar to binding motifs for broadly active TFs, in particular ETS family members, while sequences matching motifs for both broad and context-specific TFs, such as components of the AP-1 complex like JUN and FOS, contribute to broad enhancer activity. These TF motif patterns also reflect the more general sequence differences between promoters and enhancers.
Our findings are consistent with a model in which promoters often achieve broad activity by binding broadly expressed TFs, while enhancers obtain broad activity using combinations of both broad and context-specific TFs. The AP-1 family members associated with enhancers have many context-specific binding partners with similar binding motifs, and these TFs influence the enhancer motif similarity distributions toward context-specific TFs. AP-1 factors are broadly active in general, but the complex is often made up of context-specific components that are involved in more tissue-specific processes such as differentiation (Angel and Karin 1991; Karin et al. 1997). Furthermore, most members of the ETS TF family, which is enriched among high-weight promoter 6-mers, are involved in processes relevant to all cells like cell growth and apoptosis (Oikawa and Yamada 2003).
A recent MPRA-based study of the enhancer and promoter activity of short sequences bound by CREBBP in cortical neurons observed many similar patterns in sequences with enhancer vs. promoter activity (Nguyen et al. 2016). For example, they found that elevated CpG content is more strongly associated with promoter activity than enhancer activity. They also concluded that there are significant differences between enhancers and promoters driven by binding of specific TFs. There was considerable agreement in the TFs that they found to be characteristic of promoters (e.g., ELK and RFX family members) and enhancers (AP-1), with the sequence patterns given high weights by our machine learning models. Our findings also broadly agree with TFs identified as important to regulatory activity in several other MPRA studies that did not directly compare enhancers and promoters (Ernst et al. 2016; Klein et al. 2018). This argues that these are general patterns that apply across cellular contexts and tissues.
There are a few caveats to consider when interpreting our results. We defined enhancers and promoters based on CAGE assays; however, there are many other strategies for identifying regulatory regions, which can alter the scientific conclusions drawn (Benton et al. 2018; Klein et al. 2018; Halfon 2019). We attempted to replicate our results using histone mark-defined regulatory regions, and found that sequence-based differences between enhancers and promoters were much less profound for these regions. This is despite the fact that our findings are corroborated by recent MPRA studies of enhancers and promoters (Nguyen et al. 2016). Furthermore, in a previous study of CAGE-defined enhancers, we found similar sequence patterns to be predictive of activity in enhancers defined by histone marks and DNase hypersensitivity (Colbran et al. 2017). We did not consider DNase hypersensitivity here, so it is possible that this would increase the ability to distinguish histone mark-defined enhancers and promoters. Overall, this suggests differences in the classification and sequence properties of regulatory regions depending on how they are identified, and it emphasizes the need for more accurate models of gene regulatory elements and their architectures.
Another limitation of our study is that several analyses focus on the extremes of the enhancer and promoter activity distributions. This approach enabled us to detect stark sequence-level differences, and we found that our results are robust to the specific threshold used to define broad and narrow activity (Figure S2). However, more work will be needed to map sequence patterns and dynamics of regions with intermediate levels of activity.
Overall, promoters and enhancers share many similar characteristics. For example, the most broadly active enhancers often contain CGIs and have characteristic 6-mers similar to broadly active TFs. These enhancers resemble promoters at the sequence level, and it is possible that many of them also have promoter activity. However, most enhancers do not exhibit the strong relationship between CpG count and activity seen in promoters, and they generally contain binding sites for different sets of TFs. Thus, while some enhancers are very promoter-like, and vice versa, most display distinct sequence properties.
Acknowledgments
We thank Mary Lauren Benton, Alexandra Fish, Emily Hodges, and Corinne Simonti for helpful discussion and comments on the manuscript. L.L.C. was supported by the National Institutes of Health (NIH) (T32 GM-080178). J.A.C. was supported by the NIH [R01 GM-115836 and R35 GM-127087], a March of Dimes Innovation Catalyst award, the Burroughs Wellcome Fund, and institutional funds from Vanderbilt University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare they have no competing interests.
Footnotes
Communicating editor: C. Kaplan
Literature Cited
- Andersson R., 2015. Promoter or enhancer, what’s the difference? Deconstruction of established distinctions and presentation of a unifying model. Bioessays 37: 314–323. 10.1002/bies.201400162 [DOI] [PubMed] [Google Scholar]
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., et al. , 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Sandelin A., Danko C. G., 2015. A unified architecture of transcriptional regulatory elements. Trends Genet. 31: 426–433. 10.1016/j.tig.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Angel P., Karin M., 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072: 129–157. 10.1016/0304-419X(91)90011-9 [DOI] [PubMed] [Google Scholar]
- Bauer D. E., Kamran S. C., Lessard S., Xu J., Fujiwara Y., et al. , 2013. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342: 253–257. 10.1126/science.1242088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hur A., Weston J., 2010. A user’s guide to support vector machines, pp. 223–239 in Methods in Molecular Biology (Clifton, N.J.), edited by Carugo O., Eisenhaber F. Humana Press, Totowa, NJ: 10.1007/978-1-60327-241-4_13 [DOI] [PubMed] [Google Scholar]
- Benton M. L., Talipineni S. C., Kostka D., Capra J. A., 2018. Genome-wide enhancer maps differ significantly in genomic distribution, evolution, and function. bioRxiv. 10.1101/176610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M., Pelham H. R., 1986. Heat shock regulatory elements function as an inducible enhancer in the Xenopus hsp70 gene and when linked to a heterologous promoter. Cell 45: 753–760. 10.1016/0092-8674(86)90789-0 [DOI] [PubMed] [Google Scholar]
- Brown C. D., Mangravite L. M., Engelhardt B. E., 2013. Integrative modeling of eQTLs and Cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 9: e1003649 10.1371/journal.pgen.1003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzynski G. M., Reed X., Taher L., Stine Z. E., Matsui T., et al. , 2012. Systematic elucidation and in vivo validation of sequences enriched in hindbrain transcriptional control. Genome Res. 22: 2278–2289. 10.1101/gr.139717.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busche S., Shao X., Caron M., Kwan T., Allum F., et al. , 2015. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol. 16: 290 10.1186/s13059-015-0856-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Wysocka J., 2013. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49: 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M., Dankel S. N., Kim K.-H., Quon G., Meuleman W., et al. , 2015. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 373: 895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbran L. L., Chen L., Capra J. A., 2017. Short DNA sequence patterns accurately identify broadly active human enhancers. BMC Genomics 18: 536 10.1186/s12864-017-3934-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Martins A. L., Danko C. G., Waters C. T., Siepel A., et al. , 2014. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46: 1311–1320. 10.1038/ng.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Melnikov A., Zhang X., Wang L., Rogov P., et al. , 2016. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat. Biotechnol. 34: 1180–1190. 10.1038/nbt.3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest A. R., Kawaji H., Rehli M., Baillie J. K., et al. , 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouil R., Cauchy P., Koch F., Descostes N., Cabeza J. Z., et al. , 2012. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 22: 2399–2408. 10.1101/gr.138776.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini B. K., Tring S., Plummer S. J., Edlund C. K., Moreno V., et al. , 2014. Multiple functional risk variants in a SMAD7 enhancer implicate a colorectal cancer risk haplotype. PLoS One 9: e111914 10.1371/journal.pone.0111914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M., Lee D., Mohammad-Noori M., Beer M. A., 2014. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 10: e1003711 (erratum: PLoS Comput. Biol. 10: e1004035) 10.1371/journal.pcbi.1003711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L., Noble W. S., 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium , 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S., 2007. Quantifying similarity between motifs. Genome Biol. 8: R24 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I., Weston J., Barnhill S., Vapnik V., 2002. Gene selection for cancer classification using support vector machines. Mach. Learn. 46: 389–422. 10.1023/A:1012487302797 [DOI] [Google Scholar]
- Halfon M. S., 2019. Studying transcriptional enhancers: the founder fallacy, validation creep, and other biases. Trends Genet. 35: 93–103. 10.1016/j.tig.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., et al. , 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39: 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Herman-Izycka J., Wlasnowolski M., Wilczynski B., 2017. Taking promoters out of enhancers in sequence based predictions of tissue-specific mammalian enhancers. BMC Med. Genomics 10: 34 10.1186/s12920-017-0264-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Liu Z., Zandi E., 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9: 240–246. 10.1016/S0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- Kim T. K., Shiekhattar R., 2015. Architectural and functional commonalities between enhancers and promoters. Cell 162: 948–959. 10.1016/j.cell.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleftogiannis D., Kalnis P., Bajic V. B., 2016. Progress and challenges in bioinformatics approaches for enhancer identification. Brief. Bioinform. 17: 967–979. 10.1093/bib/bbv101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J. C., Keith A., Agarwal V., Durham T., Shendure J., 2018. Functional characterization of enhancer evolution in the primate lineage. Genome Biol. 19: 99 10.1186/s13059-018-1473-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M. S., Hughes J. R., Garrick D., Lynch M. D., Sharpe J. A., et al. , 2012. Intragenic enhancers act as alternative promoters. Mol. Cell 45: 447–458. 10.1016/j.molcel.2011.12.021 [DOI] [PubMed] [Google Scholar]
- Kulakovskiy I. V., Medvedeva Y. A., Schaefer U., Kasianov A. S., Vorontsov I. E., et al. , 2013. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 41: D195–D202. 10.1093/nar/gks1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy I. V., Vorontsov I. E., Yevshin I. S., Sharipov R. N., Fedorova A. D., et al. , 2018. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46: D252–D259. 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., et al. , 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., Fuda N. J., Core L. J., Lis J. T., 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie C., Eskin E., Noble W. S., 2002. The spectrum kernel: a string kernel for SVM protein classification. Pac. Symp. Biocomput. 7: 564–575. [PubMed] [Google Scholar]
- Levine M., 2010. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20: R754–R763. 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez D. G., Kraft K., Heinrich V., Krawitz P., Brancati F., et al. , 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A., Fornes O., Arenillas D. J., Chen C., Denay G., et al. , 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44: D110–D115. 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G., Andrau J.-C., 2012. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 46: 1–19. 10.1146/annurev-genet-110711-155459 [DOI] [PubMed] [Google Scholar]
- Nguyen T. A., Jones R. D., Snavely A. R., Pfenning A. R., Kirchner R., et al. , 2016. High-throughput functional comparison of promoter and enhancer activities. Genome Res. 26: 1023–1033. 10.1101/gr.204834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Yamada T., 2003. Molecular biology of the Ets family of transcription factors. Gene 303: 11–34. 10.1016/S0378-1119(02)01156-3 [DOI] [PubMed] [Google Scholar]
- Orozco L. D., Bennett B. J., Farber C. R., Ghazalpour A., Pan C., et al. , 2012. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell 151: 658–670. 10.1016/j.cell.2012.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., Kamakaka R. T., 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11: 439–446. 10.1038/nrg2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., et al. , 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Vi. Cannistraci C., Katayama S., Bajic V. B., et al. , 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140: 744–752. 10.1016/j.cell.2010.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A., 2010. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat. Neurosci. 13: 1330–1337. 10.1038/nn.2671 [DOI] [PubMed] [Google Scholar]
- Rickels R., Shilatifard A., 2018. Enhancer logic and mechanics in development and disease. Trends Cell Biol. 28: 608–630. 10.1016/j.tcb.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Roider H. G., Lenhard B., Kanhere A., Haas S. A., Vingron M., 2009. CpG-depleted promoters harbor tissue-specific transcription factor binding signals—implications for motif overrepresentation analyses. Nucleic Acids Res. 37: 6305–6315. 10.1093/nar/gkp682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., et al. , 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg S., Ratsch G., Henschel S., Widmer C., Behr J., et al. , 2010. The SHOGUN machine learning toolbox. J. Mach. Learn. Res. 11: 1799–1802. [Google Scholar]
- Taher L., Smith R. P., Kim M. J., Ahituv N., Ovcharenko I., 2013. Sequence signatures extracted from proximal promoters can be used to predict distal enhancers. Genome Biol. 14: R117 10.1186/gb-2013-14-10-r117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., et al. , 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Minovitsky S., Dubchak I., Pennacchio L. A., 2007. VISTA enhancer browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35: D88–D92. 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Sun Y. E., 2006. Epigenetic regulation of stem cell differentiation. Pediatr. Res. 59: 21R–25R. 10.1203/01.pdr.0000203565.76028.2a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and scripts used in this study are available on GitHub (https://github.com/colbrall/enhancer_promoter_manuscript), or from the authors upon request.