Abstract
An ontology offers a human-readable and machine-computable representation of the concepts in a domain and the relationships among them. Mappings between ontologies enable the reuse and interoperability of biomedical knowledge. We sought to map concepts of the Radiology Gamuts Ontology (RGO), an ontology that links diseases and imaging findings to support differential diagnosis in radiology, to terms in three key vocabularies for clinical radiology: the International Classification of Diseases, version 10, Clinical Modification (ICD-10-CM), the Radiological Society of North America’s radiology lexicon (RadLex), and the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT). RGO (version 0.7; Jan 2018) incorporated 16,918 terms (classes) for diseases, interventions, and imaging observations linked by 1782 subsumption (class-subclass) relations and 55,569 causal (“may cause”) relations. RGO classes were mapped to RadLex (46,656 classes, version 3.15), SNOMED CT (347,358 classes, version 2018AA), and ICD-10-CM (94,645 classes, version 2018AA) using the National Center for Biomedical Ontology (NCBO) Annotator web service. We identified 1275 exact mappings from RGO to RadLex, 5302 to SNOMED CT, and 941 to ICD-10-CM. RGO terms mapped to one ontology (n = 3401), two ontologies (n = 1515), or all three ontologies (n = 198). The mapped ontologies provide additional terms to support data mining from textual information in the electronic health record. The current work builds on efforts to map RGO to ontologies of diseases and phenotypes. Mappings between ontologies can support automated knowledge discovery, diagnostic reasoning, and data mining.
Keywords: Ontology, Differential diagnosis, Radiology, Gamuts, ICD, RadLex, SNOMED
Introduction
An ontology offers a human-readable and machine-computable representation of the concepts in a domain and the relationships among them [1]. Mappings between ontologies enable the reuse and interoperability of biomedical knowledge. The Radiology Gamuts Ontology (RGO) is an ontology that provides a formal representation of differential diagnosis in radiology [2]. RGO comprises 16,918 classes that represent disorders (e.g., giardiasis; RGO:3525), interventions (e.g., oral medication; RGO:3968), and imaging findings (e.g., fused ribs; RGO:30094). RGO’s subsumption (is_a) relation defines subclasses: concepts that are more specific than their parent class; e.g., congenital splenomegaly is a subclass of splenomegaly. The ontology has a relatively flat hierarchical structure, in that it has only 1782 subclass relations; 90% of the ontology’s entities are subclasses of the top-level Entity class. The ontology had a maximum depth of 4 subclass relations [3].
In addition to subsumption, RGO defines a causal relation (may_cause, and its inverse, may_be_caused_by) to express the links between diagnoses and imaging observations. This relation is neither tautological nor exhaustive; that is, it does not express the absolute certainty of logical implication (“if A, then B”), nor does it require that all possible causes of a finding are defined. However, the relation does allow one to express the differential diagnosis of imaging findings, and the relation’s transitive property allows one to explore chains of inference [3].
A key application of RGO has been in named-entity recognition of diagnoses and imaging findings in narrative-text radiology reports. To further expand RGO’s applicability in “deep phenotyping” studies—which seek to identify all phenotypic findings in a set of subjects—it is important to expand the set of synonyms and coded terms that are connected to RGO. This investigation sought to map RGO concepts to terms in three key vocabularies used in clinical radiology.
The International Classification of Diseases, version 10, Clinical Modification (ICD-10-CM) is an adaptation of the World Health Organization disease coding scheme that is used in the USA as a source of diagnostic codes. ICD-10-CM codes typically are used to encode the indications for a radiology exam, and thus are incorporated into the patient’s clinical records and billing information [4–6]. RadLex is an ontology of radiology terms developed and published by the Radiological Society of North America (RSNA) [6–8]. It was developed to address gaps in general clinical vocabularies [9], and incorporates terms for anatomy, diseases, and imaging signs [10]. RadLex has been used to index radiological literature, learning materials, and radiological procedures. The Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) is the world’s most comprehensive clinical healthcare terminology; it enables consistent representation of clinical content in electronic health records and has been used widely in biomedical information systems [6, 11].
Methods
RGO (version 0.7; Jan 2018) incorporated 16,918 terms (classes) linked by 1782 subsumption (class-subclass) relations and 55,569 causal (“may cause”) relations. We defined “disorders” as entities that could cause other entities in the ontology, and “observations” as entities that could be caused by other entities. Whether or not an entity was considered an intervention was encoded as one of its properties. We tallied the numbers of disorders, interventions, and observations in RGO.
RGO terms were mapped to RadLex (46,656 classes, version 3.15), SNOMED CT (347,358 classes, version 2018AA), and ICD-10-CM (94,645 classes, version 2018AA) using the National Center for Biomedical Ontology (NCBO) Annotator web service [12, 13]. The ontologies were accessed through the National Center for Biomedical Ontology (NCBO) BioPortal web site (http://bioportal.bioontology.org/) and web services [14, 15]. An automated script was created to query the Annotator web service for each RGO term and to retrieve all partial and exact string matches. Preferred terms and their synonyms were considered in RGO and the target ontologies; only exact string matches were included for mapping. To confirm the mappings, we randomly selected 10% of all exact mappings for manual review and identified any errors.
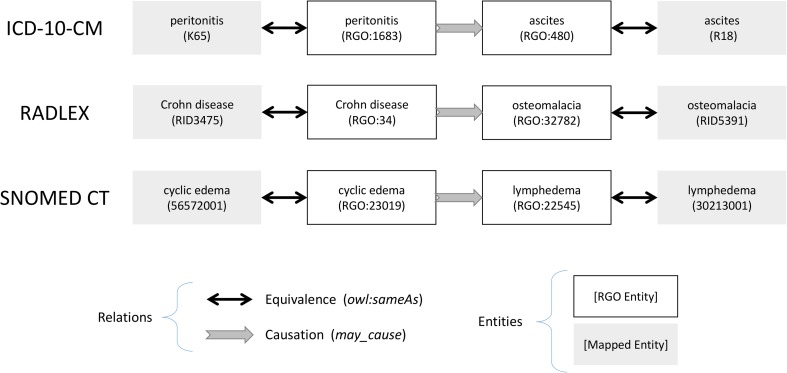
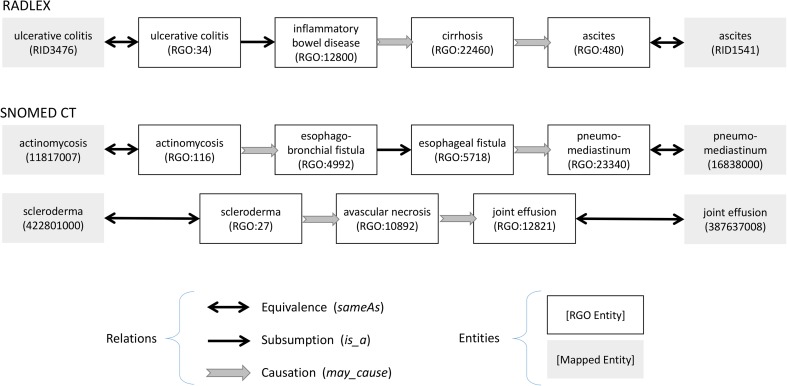
Within each target ontology, we tallied the number of causal relationships that could be expressed using the equivalence relationships to causally linked RGO terms. Examples are shown in Fig. 1. For the purposes of this study, we also defined an “indirect causal relationship” as an axiom between two terms in a target ontology linked by one or two causal relations and zero or more is_a relations in RGO, in any order of occurrence. Examples are shown in Fig. 2.
Fig. 1.
Causal relations between terms within each target ontology. For example, the RadLex term Crohn’s disease is causally related to osteomalacia through their equivalence relations to the corresponding RGO terms
Fig. 2.
“Indirect” causal relations as defined in this study are based on up to two causal relations and zero or more subclass (is_a) relations in RGO. For example, as shown graphically, actinomycosis may cause esophagobronchial fistula, which is an esophageal fistula, which may cause pneumomediastinum
Mappings were stored initially in a MySQL relational database, and then exported as a delimited text file containing the Gamuts entity name and the mapped term name. From this file, an OWL file was constructed for submission to NCBO so that Gamuts could be parsed and displayed; the OWL file included equivalence (sameAs) relationships as well as hierarchical (is_a) relationships. Equivalence relationships between Gamuts and ICD-10, RadLex, and SNOMED-CT were added one at a time using the BioPortal API (endpoint https://data.bioontology.org/mappings). Each post included a JSON object that documented the relationship and the entities that should be mapped; for example:
{
"creator": "rwfilice",
"relation": "http://www.w3.org/2002/07/owl#sameAs",
"classes": {
"http://www.gamuts.net/entity#hematoma_of_neck": "GAMUTS",
"http://purl.bioontology.org/ontology/SNOMEDCT/447220009": "SNOMEDCT"
}
}
Mapped relations were limited to equivalence (sameAs) relations because NCBO does not support hierarchical relationships between different ontologies. Mappings were added one at a time to allow for error checking to identify transactions that timed out or had other errors.
Results
RGO incorporated 12,878 disorders, i.e., entities that could cause other RGO entities; examples included multiple periapical condensing osteitis (RGO:9089), Stewart-Treves syndrome (RGO:23924), and mineralizing vasculopathy (RGO:7770). The ontology included 524 interventions, such as surgical repair of esophageal atresia (RGO:3550), body cast (RGO:4194), and beta-blocker (RGO:24937). RGO included 4662 imaging findings, i.e., entities that could be caused by other RGO entities, such as sloughed calcified renal papilla (RGO:20192), underdeveloped pubic rami (RGO:31943), septicemia (RGO:3317), and brachytelephalangy (RGO:34143). These three subsets were not disjoint.
We identified 51,049 potential matches from RGO, of which there were a total of 7518 exact matches: 941 to ICD-10-CM, 1275 to RadLex, and 5302 to SNOMED CT. In total, 5114 (30.2% of 16,918) RGO terms mapped to one or more ontology: 3401 RGO terms mapped to exactly one ontology, 1515 terms mapped to exactly two ontologies, and 198 terms mapped to all three ontologies. For example, esophageal web (RGO:68) was mapped as equivalent to RadLex term RID34693, SNOMED CT term 19216006, and ICD-10-CM code Q39.4. The numbers of mappings of disorders, interventions, and observations into each of the target ontologies is shown in Table 1. A randomly selected sample of 510 mapped terms was reviewed manually; no errors were identified.
Table 1.
The numbers of mapped Gamuts entities are shown by category (disorder, intervention, or observation) for each of the target ontologies. Note that the categories are not disjoint; hence, the total does equal the sum of cells to its left. Column totals are not provided; Gamuts entities may be mapped to more than one target ontology
| Number of mapped Gamuts entities | ||||
|---|---|---|---|---|
| Ontology | Disorders | Interventions | Observations | Total |
| ICD10CM | 798 | 1 | 340 | 941 |
| RADLEX | 1162 | 63 | 318 | 1275 |
| SNOMEDCT | 4153 | 346 | 1238 | 5302 |
RGO’s causal relations allowed one to express 10,958 direct causal relationships between concepts within the target ontologies: 696 in ICD-10-CM, 1160 in RadLex, and 9102 in SNOMED CT. We explored only causal relations between entities within the same ontology using their mappings to RGO terms. In RadLex, for example, scleroderma (RID34592) may cause achalasia (RID3458), based on the causal relation between the corresponding RGO terms. There were a total of 31,845 indirect causal relations (1849 for ICD-10-CM, 3596 for RadLex, and 26,400 for SNOMEDCT). Figure 2 shows examples of indirect causal relationships. RGO and its equivalence mappings have been published to NCBO BioPortal, where they are freely available.
Discussion
The mapped ontologies provide additional terms to support data mining from textual information in the electronic health record. Overall, 30% of RGO terms were mapped to concepts in at least one of the target ontologies; this value represents a quite good result, especially given that RGO terms are highly specific to radiology, and include complex terms such as sloughed calcified renal papilla (RGO:20192). For comparison, equivalent SNOMED CT concepts were identified for 30% of classes of the Human Phenotype Ontology, an ontology similar to RGO in size [16]. The current work builds on efforts to map RGO to other biomedical ontologies such as the Orphanet Rare Disease Ontology (ORDO), which enabled analysis of 12.4 million radiology reports to estimate the frequency of rare diseases in radiology reports [17, 18].
The current study has two principal limitations. First, it did not explore partial matches. Partial mappings can provide a “next-best approach” when equivalence mapping is limited due to differences in the ontologies’ focus and granularity [16]. However, the need for manual review of a large number of potential matches—here, 51,049—limits such an approach. Second, the current study relied upon an audit of 10% of exact matches rather than exhaustive review of all matches. Future work may include exploring partial mappings to SNOMED CT, which admits “pre-coordinated” concepts (such as chronic pain) and “post-coordinated” concepts that entail an expression made up of other concepts (e.g., pain, with clinical qualifier chronic) [19].
Recently, longitudinal electronic health record (EHR) information such as ICD billing codes have been coupled with genetic data to perform phenome-wide association scans (PheWAS) for disease-gene associations [20]. EHR systems also frequently incorporate SNOMED CT terms to encode clinical findings. Because a large fraction of EHR data exists as unstructured narrative text, ontologies such as SNOMED CT and RadLex can provide critical bases to guide named-entity recognition in textual data, such as clinic notes and radiology reports. Mappings between ontologies can support automated knowledge discovery, diagnostic reasoning, and data mining.
The current work helps link interface terminologies and reference terminologies. An “interface terminology” is a maintained set of unique, identified terms designed to be compatible with the natural language of the user. A “reference terminology” is one in which every concept has a formal, machine-usable definition that can support data aggregation and retrieval. A reference terminology is designed to provide common semantics for diverse implementations. Integration of these different types of terminologies is critical to promote semantic interoperability [21]. One goal of precision medicine is to more precisely classify patients in order to improve diagnosis and medical treatment [22]. Ontologies can support precision medicine through their systematic representations of knowledge that allow researchers to integrate and analyze large collections of heterogeneous data [23].
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bodenreider O: Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform 67–79, 2008 [PMC free article] [PubMed]
- 2.Budovec JJ, Lam CA, Kahn CE Jr: Radiology Gamuts Ontology: differential diagnosis for the Semantic Web. RadioGraphics 34:254–264, 2014 [DOI] [PubMed]
- 3.Kahn CE Jr: Transitive closure of subsumption and causal relations in a large ontology for radiology diagnosis. J Biomed Inform 61:27–33, 2016 [DOI] [PubMed]
- 4.Barta A, McNeill G, Meli P, Wall K, Zeisset A: ICD-10-CM primer. J AHIMA 79:64–66, 2008 [PubMed]
- 5.Jonassen K, Saboe R. The use of text encoding in the development of a terminology and knowledge system associated with the Norwegian version of the ICD-10. Medinfo 8 Pt. 1995;1:51–55. [PubMed] [Google Scholar]
- 6.Wang KC. Standard lexicons, coding systems and ontologies for interoperability and semantic computation in imaging. J Digit Imaging. 2018;31:353–360. doi: 10.1007/s10278-018-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langlotz CP. RadLex: a new method for indexing online educational materials. RadioGraphics. 2006;26:1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 8.Rubin DL. Creating and curating a terminology for radiology: ontology modeling and analysis. J Digit Imaging. 2008;21:355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langlotz CP, Caldwell SA. The completeness of existing lexicons for representing radiology report information. J Digit Imaging. 2002;15(Suppl 1):201–205. doi: 10.1007/s10278-002-5046-5. [DOI] [PubMed] [Google Scholar]
- 10.Shore MW, Rubin DL, Kahn CE Jr: Integration of imaging signs into RadLex. J Digit Imaging 25:50–55, 2012 [DOI] [PMC free article] [PubMed]
- 11.Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. J Am Med Inform Assoc. 2014;21:e11–e19. doi: 10.1136/amiajnl-2013-001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonquet C, Shah NH, Musen MA. The open biomedical annotator. Summit Transl Bioinform. 2009;2009:56–60. [PMC free article] [PubMed] [Google Scholar]
- 13.Shah NH, Bhatia N, Jonquet C, Rubin D, Chiang AP, Musen MA. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey MA, Chute CG, Musen MA. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhombres F, Bodenreider O. Interoperability between phenotypes in research and healthcare terminologies--investigating partial mappings between HPO and SNOMED CT. J Biomed Semantics. 2016;7:3. doi: 10.1186/s13326-016-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn CE Jr: Integrating ontologies of rare diseases and radiological diagnosis. J Am Med Informatics Assoc 22:1164–1168, 2015 [DOI] [PMC free article] [PubMed]
- 18.Kahn CE Jr: An ontology-based approach to estimate the frequency of rare diseases in narrative-text radiology reports. Stud Health Technol Inform 245:896–900, 2017 [PubMed]
- 19.Rector A, Iannone L. Lexically suggest, logically define: quality assurance of the use of qualifiers and expected results of post-coordination in SNOMED CT. J Biomed Inform. 2012;45:199–209. doi: 10.1016/j.jbi.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz S, Rodrigues JM, Rector A, Chute CG. Interface terminologies, reference terminologies and aggregation terminologies: a strategy for better integration. Stud Health Technol Inform. 2017;245:940–944. [PubMed] [Google Scholar]
- 22.National Research Council . Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 23.Haendel MA, Chute CG, Robinson PN. Classification, ontology, and precision medicine. N Engl J Med. 2018;379:1452–1462. doi: 10.1056/NEJMra1615014. [DOI] [PMC free article] [PubMed] [Google Scholar]