Abstract
The watermelon (Citrullus lanatus) is one of the most important horticultural crops for fruit production worldwide. However, the production of watermelon is seriously restricted by one kind of soilborne disease, Fusarium wilt, which is caused by Fusarium oxysporum f. sp. niveum (Fon). In this study, we identified an efficient PGPR strain B. velezensis F21, which could be used in watermelon production for Fon control. The results of biocontrol mechanisms showed that B. velezensis F21 could suppress the growth and spore germination of Fon in vitro. Moreover, B. velezensis F21 could also enhance plant basal immunity to Fon by increasing the expression of plant defense related genes and activities of some defense enzymes, such as CAT, POD, and SOD. To elucidate the detailed mechanisms regulating B. velezensis F21 biocontrol of Fusarium wilt in watermelon, a comparative transcriptome analysis using watermelon plant roots treated with B. velezensis F21 or sterile water alone and in combination with Fon inoculation was conducted. The transcriptome sequencing results revealed almost one thousand ripening-related differentially expressed genes (DEGs) in the process of B. velezensis F21 triggering ISR (induced systemic resistance) to Fon. In addition, the Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment indicated that numerous of transcription factors (TFs) and plant disease resistance genes were activated and validated by using quantitative real-time PCR (qRT-PCR), which showed significant differences in expression levels in the roots of watermelon with different treatments. In addition, genes involved in the MAPK signaling pathway and phytohormone signaling pathway were analyzed, and the results indicated that B. velezensis F21 could enhance plant disease resistance to Fon through the above related genes and phytohormone signal factors. Taken together, this study substantially expands transcriptome data resources and suggests a molecular framework for B. velezensis F21 inducing systemic resistance to Fon in watermelon. In addition, it also provides an effective strategy for the control of Fusarium wilt in watermelon.
Keywords: watermelon, Fusarium oxysporum f. sp. niveum (Fon), biological control, Bacillus velezensis, induced systemic resistance, transcriptome profiling
Introduction
The watermelon (Citrullus lanatus) is one of the most important horticultural crops for fruit production in the world and is known as the most popular fruit in the summer. In fact, according to research statistics, watermelon accounts for almost 2% of the world’s total area devoted to vegetable production. The cultivated area and yield of watermelon have increased yearly due to increasing consumption and its comparatively high economic value in China (Sun et al., 2017). However, the production of watermelon is seriously restricted by soilborne diseases, such as Fusarium wilt, anthracnose, and gummy stem blight and so on. Watermelon Fusarium wilt, caused by Fon, is a devastating disease affecting a large variety of economically important crops worldwide (Demers et al., 2015). The symptoms of Fusarium wilt disease are necrotic lesions, followed by foliar wilting as the pathogen invades the vascular system of plants, and eventually plant death symptoms can manifest in a few weeks (Janvier et al., 2007). The disease can result in 100% yield losses under favorable environmental conditions during the watermelon production. Due to the long-term survival and accumulation of pathogens in soil, there is currently no effective control strategy. In addition, as a result of the substantial loss incurred by the devastating pathogen, effective non-chemical, environmentally friendly strategies to prevent or reduce the damage caused by Fon in watermelon production are valuable and urgently needed.
Biological control of soilborne diseases has been studied for over 100 years, and it has been considered a viable alternative method to chemical control (Heydari and Pessarakli, 2010). It is one of the highest potential strategies for soilborne disease control due to its advantages of environmental friendliness, safety and lack of the induction of pesticide resistance occurring (Jiang et al., 2015). To date, studies on the biological control of watermelon Fusarium wilt are still in progress and an efficient strategy for Fon control is still in discovery. For instance, Paenibacillus polymyxa SQR-21 has been identified by Ling et al. (2011) as a potential agent for the biocontrol of Fusarium wilt in watermelon. They demonstrated that P. polymyxa SQR-21 could systemically affect root exudates of watermelon to decrease the conidial germination of Fon. In addition, the substantial potential of species of Streptomyces to suppress plant diseases caused by fungal pathogens has also been reported in recent years. For example, the strain S. goshikiensis YCXU was evaluated as an efficient biocontrol agent (BCA) that showed antifungal activity against Fon by producing CAT, chitinase, β-1,3-glucanase and urease enzymes (Faheem et al., 2015). Lu et al. (2016) reported that a new species of Streptomyces, S. rimosus M527, was isolated and identified as a BCA against F. oxysporum f. sp. cucumerinum. They demonstrated that M527 not only displayed broad-spectrum antifungal activity but also showed the strongest antagonistic activity against the spore germination of F. oxysporum f. sp. cucumerinum (Lu et al., 2016). Moreover, Bacillus spp. have been investigated for their potential ability to serve as a BCA against a variety of plant diseases (Zhao et al., 2014). The high potential of B. amyloliquefaciens SN16-1 to control F. oxysporum f. sp. lycopersici was recently reported (Wan et al., 2017). Yang et al. (2014) found that B. subtilis B006 could manage cucumber wilt disease through the suppression of Fusarium oxysporum f. sp. cucumerinum in rhizosphere. In addition to the biocontrol bacteria listed above, many other species are also being discovered that serve as BCAs for Fusarium wilt disease control in watermelon.
In recent years, the research on the biological control of F. oxysporum is has been devoted not only to the discovery of new BCAs but also to the biocontrol mechanisms of BCAs. So far this year, a few reports on the biocontrol mechanisms of BCAs against Fusarium wilt have been reported. The biocontrol mechanisms operating against Fusarium wilt include the production of diffusible and volatile antimicrobial compounds, changes in soil microbial diversity and ISR. Wang et al. (2013) reported that the antagonistic effect of B. amyloliquefaciens W19 is the main mechanism of banana Fusarium wilt biocontrol. Three kinds of antifungal lipopeptides (iturin, surfactin, and bacillomycin D) secreted by W19 were identified in their study. In addition, more than 18 volatile antifungal compounds with significant antagonistic effects against F. oxysporum were identified (Wang et al., 2013). Otherwise, the microbiome of plants playing a crucial role in both plant and ecosystem health, and the maintenance of microbial diversity in the soil can promote plant growth, prevent diseases, and increase stress resistance. Berg et al. (2017) reported that microbial diversity was a key factor in preventing diseases and could be implemented as a biomarker in plant protection strategies. Shen et al. (2014) reported that the application of BIO could suppress the bacterial community associated with banana Fusarium wilt disease. They found that the abundances of Gemmatimonadetes and Acidobacteria were high in BIO-treated soil, while that of Bacteroidetes was low (Shen et al., 2014). In addition, it is envisaged that in suppressive soils plant roots are associated with microbial communities that have an overall beneficial effect on plant health. This phenomenon is known as ISR triggered by plant beneficial microbes (Choudhary et al., 2007). Many effective biocontrol PGPR can elicit ISR and related research results have been reported in recent years. For example, the Pseudomonas fluorescens WCS417r-mediated ISR has been shown to function against various plant pathogens in Arabidopsis by activating the JA signaling pathway, there by affecting such pathogens as the bacterial leaf pathogen P. syringae pv. tomato and the fungal root pathogen Fusarium oxysporum f. sp. raphani (Pieterse et al., 1998). Niu et al. (2011) first demonstrated that the PGPR Bacillus cereus AR156 can induce systemic resistance in Arabidopsis thaliana by simultaneously activating SA-and JA/ETH-dependent signaling pathways. In subsequent studies, two TFs, WRKY11 and WRKY70, were identified as important regulators involved in the ISR triggered by B. cereus AR156 (Jiang et al., 2016). Current research shows that ISR is also one of the important mechanisms of Fusarium biological control. It has been reported that plant growth-promoting rhizobacteria (PGPR) B. fortis and B. subtilis have ISR against Fusarium wilt disease in controlled environments (Akram et al., 2013). Bekkar et al. (2018) found that Bacillus strains application could induce systemic resistance against Fusarium wilt in chickpeas. Therefore, in-depth analysis of ISR triggered by PGPR is of great significance to the biological control of Fusarium wilt.
In this study, we identified an efficient PGPR strain B. velezensis F21 which can be used for F. oxysporum f. sp. niveum control in watermelon. B. velezensis F21 can not only promote watermelon growth but also protect watermelon from Fon invasion. These results indicated that B. velezensis F21 could suppress the growth and spore germination of Fon in vitro. Moreover, the PGPR strain could enhance plant basal immunity to Fon by increasing the expression of plant defense related genes and activities of some defense enzymes. Otherwise, the transcriptome sequencing is a highly efficient, widely used, molecular biological research method used to obtain detailed transcriptome information. In this study, a total of 20,985 genes in watermelon were identified and the transcriptome analysis revealed almost 1,000 ripening-related DEGs in the process of B. velezensis F21 triggering ISR to Fon. These genes were subjected to scatter plot analysis, the GO classification and KEGG pathway enrichment. Furthermore, a series of TFs and plant disease resistance genes (PRGs) were identified and validated by using quantitative real-time PCR (qRT-PCR), which showed significantly different expression levels in the roots of watermelon with different treatments. Finally, we analyzed the genes involved in the MAPK signaling pathway and plant hormone signaling pathways and validated the gene expression level and hormone accumulation content by qRT-PCR and ELISA, respectively. Collectively, this study suggests a molecular framework for the induction of systemic resistance to Fon in watermelon by B. velezensis F21, and provides detailed transcriptome data resources for further molecular mechanistic research.
Materials and Methods
Plant Material, Bacteria, Fungi, Growth Condition, and Experimental Design
In this study, we used one kind of watermelon cultivar (Sumi #1) as a test plant. The variety was susceptible to F. oxysporum f. sp. niveum. It originated from the Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences, China.
All the PGPR that we used in this study came from our biocontrol bacteria library, which consisted of beneficial bacteria isolated by our lab from the field where Fusarium wilt previously occurred in different provinces of China. All the bacteria were grown on LB agar media and incubated at 28°C for 24 h. One single colony on the medium was picked and then inoculated into LB broth and incubated at 28°C for 24 h while shaking at 200 rpm. The bacterial fermented liquid was centrifuged at 6,000 rpm for 10 min, and the supernatant was discarded, suspended in sterile water and adjusted to a concentration of 1 × 108 CFU/mL for further experiments.
F. oxysporum f. sp. niveum, F. oxysporum f. sp. cubense, F. graminearum, Mycosphaerella melonis, and B. cinerea were used in antagonistic in vitro studies. They were cultured on potato dextrosa agar (PDA) medium at 25°C under dark conditions for 5 days. The microconidial suspension of F. oxysporum f. sp. niveum used in spore germination and greenhouse experiments was prepared as described by Meldrum et al. (2013).
Effects of B. velezensis F21 on Hydrolase Activity, Fungal Antagonism, and Spore Germination
To evaluate the biocontrol potential of B. velezensis F21, we evaluated the effect of some hydrolase activities and fungal antagonism. For the hydrolase activity evaluation, we detected the activities of protease, chitinase, ferric enzyme, glucanase, and cellulose as described by Jiang et al. (2015). For the fungal antagonism, we adopted a flat confrontation experiment on PDA medium in vitro. The Five-millimeter plugs from 5-day-old cultures of Fon were inoculated in the center of the PDA medium, and then B. velezensis F21 was inoculated on both sides of the culture dish by using filter paper. Sterile water was used as a control. Subsequently, all the plates were incubated at 25°C (Jiang et al., 2018), and the colony diameters were measured and recorded every 12 h after. To verify the broad-spectrum effect of B. velezensis F21, we also used other pathogenic fungi besides Fon, such as F. oxysporum f. sp. cubense, F. graminearum, Mycosphaerella melonis, and Botrytis cinerea.
Spore germination assays were performed as described by Mujoko et al. (2014) with minor modifications. The B. velezensis F21 culture in agar media was fragmented by a cork borer, and the top layer was sliced. Then, the top surface of the slice containing B. velezensis F21 was removed and assumed to contain metabolites produced by B. velezensis F21. The slice was transferred into deck glass for spore germination observation. The agar disk was inoculated with 10 μl Fon conidia suspension at a concentration of 1 × 105 conidia/ml, kept in the slide box and incubated for 5 h at 25°C. The germination of conidia was observed and photographed using an Olympus DP71 camera with an Olympus BX51-P microscope (Olympus America, Inc.).
Experimental Design in Greenhouse and Filed Trials
To evaluate the plant growth-promoting effect of B. velezensis and the biocontrol effect on watermelon Fusarium wilt, greenhouse and field experiments were carried out in this study. For the greenhouse experiment, the watermelon cultivar (Sumi #1) was used as the test plant. The seeds of watermelon were sown into sterile pots with one seedling per pot and then cultivated in a greenhouse with a 14 h day (200 μE m-2 s-1 at 28°C) and a 10 h night (at 20°C) cycle at 70% relative humidity. The seedlings were treated with B. velezensis F21 by sprinkling the root in combination with spraying the leaf when the seedlings grew to three leaves, and sterile water served as a control. Five days later, the watermelon seedlings in each treatment were challenged with Fon by inoculation with spore suspensions in 5 days, and the inoculation rate was 20 ml (1 × 105 conidia/ml) per seedling. The Fusarium wilt symptoms were recorded and photographed 15 days after inoculation. In addition, the plant materials at different time points were harvested for RNA sequencing, plant defense related gene expression and related enzyme activity analysis. The plant growth-promoting effect of B. velezensis F21 on watermelon was also evaluated in this study, and the results were recorded and photographed after 30 days of B. velezensis F21 treatment.
Moreover, the plant growth promotion effect of B. velezensis and the biocontrol effect on watermelon Fusarium wilt were also verified in the field. The field experiment was conducted in Guangzhou, China in watermelon growing seasons by using one kind of watermelon cultivar (Sumi #1). The trial experiment was carried out on private land: the field was located in Guangzhou Province, China, with GPS coordinates of N 23°23′28.68″, E113°26′7.14″ and the field studies did not involve endangered or protected species. Two treatments were established, and water was used as a mock control. Twenty milliliters of a 1 × 108 CFU/mL suspension of B. velezensis F21was poured onto the roots and both sides of the leaves were sprayed with the suspension when the watermelon seedlings grew to one real leaf status. Sixty days later, the growth-promoting effect on watermelon and disease occurrence were recorded.
RNA Extraction, Gene Expression, and Activities of Plant Defense-Related Enzyme Analysis
The roots of watermelon in each treatment for qRT-PCR analysis were harvested and soaked in liquid nitrogen. The total RNA of plants was extracted by using the TRIZOL reagent (Invitrogen, Cat#: 15596-026), following the manufacturer’s recommendations.
In this study, we analyzed the expression of DEGs and some genes related to the plant defense signaling pathway, such as the SA and JA signaling pathways. RT-PCR was developed with 0.5 mg total RNA, followed by DNase I treatment (gDNA Wiper from VazymeTM, Cat#: R133-01). Then reverse transcription was conducted using HiScriptTM Q Select RT SuperMix (VazymeTM, Cat#: R133-01). qRT-PCR was conducted on an ABI 7500 system (ABI, United States) by using the SYBR premix Ex-Taq mixture (Takara). The reaction was performed under the following conditions: 94°C for 5 min, followed by 45 cycles of 94°C for 10 s, 55°C for 20 s, and 72°C for 30 s, and end at 72°C for 5 min. The actin gene of watermelon (Cla008455) was employed as the internal standard. All the gene information and PCR primers used in our study are shown in Supplementary Table S1.
In addition, we also analyzed the activities of plant defense related enzymes (such as SOD, CAT, and POD) during the process of B. velezensis F21 triggering ISR to Fusarium wilt in watermelon. The activities of POD, SOD, and CAT in plant roots were measured by using the corresponding test kits as described by Jiang et al. (2015), who used a ‘POD detection kit,’ ‘SOD detection kit,’ and ‘CAT detection kit,’ respectively, (Nanjing Jiancheng Biological Engineering Institute, Nanjing, China) according to the manufacturer’s instructions.
Library Construction, RNA Sequencing, and de novo Assembly
In the present study, a total of 20 samples were used for library construction. Twelve of the samples were obtained from watermelon treated with B. velezensis F21 or sterile water alone at 0 dpt, 3 dpt, 5 dpt, 8 dpt, 11 dpt, and 13 dpt. Moreover, the remaining eight samples obtained from watermelon pretreated with BCA and then combined with Fon challenge at 0 dpt, 3 dpt, 5 dpt, and 8 dpt. All samples were harvested from at least five individual watermelon roots, and the mixture was quickly placed in liquid nitrogen. The total RNA of each sample was isolated according to the method described above.
The RNA sequencing library preparation was performed with 10 μg RNA sample by using a TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, United States) following the manufacturer’s instructions. Briefly, the mRNAs collected from the roots of watermelon with different treatments were purified, broken into short fragments and further used to synthesize first-strand cDNAs with hexamer and reverse transcriptase (Promega). Subsequently, the double-stranded cDNA was synthesized with DNA polymerase I and RNase H. Then the final cDNA library was generated using Phusion High-Fidelity DNA polymerase (Thermo ScientificTM, United States, CA#: F530L) after the end of the repairment process with a single adenine residue and adapter ligation. The quality of the library was evaluated by using bioanalyzer plots. Illumina sequencing was carried out in the Beijing Genomics Institute (BGI), China, and performed using the HiSeqTM 2500 platform according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). The sequencing data were saved as FASTQ files, and deposited to National Center for Biotechnology Information (NCBI) (BioProject accessions: PRJNA503589; BioSample accessions: SAMN10365249, SAMN10365250, SAMN10365251, SAMN10365252, SAMN10365253, SAMN10365254, SAMN10365255, SAMN10365256, SAMN10365257, SAMN10365258, SAMN10365259, SAMN10365260, SAMN10365261, SAMN10365262, SAMN10365263, SAMN10365264, SAMN10365265, and SAMN10365266).
Raw Read Processing, de novo Assembly, and Functional Annotation
To obtain the sequencing information, the raw reads were initially processed by Illumina Pipeline Software. Then, the reads were cleaned by removing adaptor sequences and low-quality reads (Q < 20) with more than 10% uncertain (N) bases using in-house Perl scripts (Zhang et al., 2015). All the following analyses were based on the high-quality clean data we obtained. De novo assembly of the transcriptome was performed by using the trinity platform1 (Grabherr et al., 2011). After that, the data were mapped back onto the corresponding contigs with all the clean reads. Then, all contigs were assembled to produce unigenes with no extension on either end (Chen Y. et al., 2017). The assembled transcripts with significant gene expression values were subjected to a similarity search against the NCBI non-redundant protein database using the BlastX (E-value ≤ 10-5) program (Altschul et al., 1997). Blast annotations (NCBI id) were mapped back to the UniProt protein database and GO terms (molecular function, biological process, and cellular component) were extracted from the UniProt database. To identify more putative functions, all assembled unigenes were searched against several publicly available databases such as the NCBI non-redundant (NR) protein database, the Swiss-Prot protein database, the KEGG pathway database, and the GO database (Wu et al., 2016).
Identification of Differentially Expressed Genes (DEGs), GO Enrichment, Pathway Analysis and Transcription Factor and PRG Identification
To quantify the identified gene expression, the number of fragments per kilobase of exon per million fragments (FPKM) mapped was used. The differential expression analysis of each sample was performed using the DESeq R package(1.10.1) and the DEGs were determined with a log-fold expression change (log FC) greater than 2 or less than -2 using a threshold of false discovery rates (FDR < 0.001) and a high statistically significant value (P < 0.05). GO and KEGG pathway enrichment analyses for all the DEGs were then carried out. The obtained GO annotation was enriched and refined using the GOseq R package with the “ELIM” method and Kolmogorov–Smirnov test. KEGG pathways were performed in KOBAS 2.0 plates and enriched by using in-house scripts according to Fisher’s exact test PRI. Enriched p-values were calculated and adjusted by using the Bonferroni correction. Then the corrected p-value of 0.05 was selected as the threshold to determine significant enrichment of the gene sets. Furthermore, to identify the TFs represented in all watermelon roots, all assembled unigenes were searched against the plant TF database (PlnTFDB2) by using a BLASTX search with the stringency of E-value 1e-06 (Moriya et al., 2007). Finally, to identify the plant resistance genes (PRGs) that function on B. velezensis F21 to trigger ISR to Fusarium wilt in watermelon, all the assembled unigenes were searched by using the PRG database (PRG-Wiki3).
Validation of DEGs by qRT-PCR
To confirm the transcriptome data, 38 DEGs were randomly selected and verified (Supplementary Table S1). The expression of validated genes was analyzed by qRT-PCR on ABI 7500 Real-Time PCR System (Applied Biosystems, United States). The qRT-PCR procedure was performed as described previously. All the information of selected genes and primers designed by Primer premier 6.0 are shown in Supplementary Table S1. Finally, to ensure the reliability and reproducibility of the validation results, four technical replicates of each biological replicate and three independent biological replicates for each sample were arranged.
Validation of Hormone Accumulation by Enzyme-Linked Immunosorbent Assay (ELISA)
In this study, the transcriptome results indicated that some plant hormone signaling pathways involved in B. velezensis F21 trigger ISR to Fusarium wilt in watermelon. In addition to validate the expression of genes involved in phytohormone synthesis and regulation, the accumulation of endogenous phytohormones, such as ABA, ETH, BR, JA, and SA, was also detected by ELISA with corresponding phytohormone ELISA test kits purchased from Nanjing Jin Yi Bai Biotechnology Co., Ltd.4 In this part, a total of 20 samples were used, and 12 of the samples were obtained from watermelon treated with B. velezensis F21 or sterile water alone at 0 dpt, 3 dpt, 5 dpt, 8 dpt, 11 dpt, and 13 dpt. Moreover, the remaining eight samples obtained from watermelon pretreated with a BCA and then combined with a Fon challenge at 0 dpt, 3 dpt, 5 dpt, and 8 dpt. All samples were harvested from at least three individual watermelon roots, and the mixture was quickly placed in liquid nitrogen. The detailed method for phytohormone extraction and quantification refer to the respective ELISA kit instructions. Moreover, for each treatment, four technical replicates of each biological replicate and three independent biological replicates for each sample were arranged to ensure the reliability and reproducibility of validation results.
Data Analysis
All bioassays and experiments were conducted three times with 36 seedlings per treatment. Significant differences between means were compared by using the LSD test (Fisher’s protected least-significant differences test) at P = 0.05. A P-value of <0.05 was considered statistically significant. All statistical analyses were performed by using analysis of variance in SPSS 24.0 (IBM SPSS Inc., United States).
Results
B. velezensis F21 Could Serve as a Potential Biocontrol Agent to Control Fusarium Wilt Disease in Watermelon
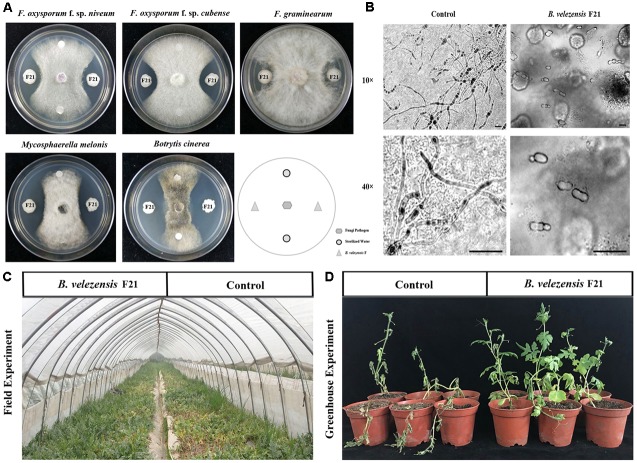
B. velezensis F21 was isolated by using the antagonistic and hydrolase activity examination assessment system previously established by our laboratory (Jiang et al., 2018). The results indicated that B. velezensis F21 had antagonistic activity against Fon in vitro (Figure 1A). Moreover, we found that the antagonism of B. velezensis F21 to fungal pathogens might be broad-spectrum. The strain also showed antagonistic effects against other fungal pathogens, such as F. oxysporum f. sp. cubense, F. graminearum, M. melonis, and B. cinerea (Figure 1A). In addition, it was also found that B. velezensis F21could inhibit the germination of F. oxysporum spores (Figure 1B). The results of hydrolase activity examination indicated that B. velezensis F21 had multiple hydrolase activities, such as protease, ferric enzyme, glucanase and cellulose, but no chitinase activity (Supplementary Figure S1). Based on the above results, we found that B. velezensis F21 had the potential to control watermelon Fusarium wilt. To verify this hypothesis, we conducted greenhouse experiments and field experiments and the results indicated that B. velezensis F21 could significantly control Fusarium wilt disease in the greenhouse and in the field. The symptoms of watermelon Fusarium wilt in the B. velezensis F21 treatment were significantly weaker in compared with the control treatment (Figure 1C,D) and the biocontrol efficiency of B. velezensis F21 in the greenhouse and in the field was 80.35% and 65.81%, respectively (Supplementary Table S2).
FIGURE 1.
B. velezensis F21 served as a potential biocontrol agent for control of Fusarium wilt disease in watermelon. B. velezensis F21 can significantly control watermelon Fusarium wilt disease. (A) Antagonistic effect of biocontrol bacteria on various pathogenic fungi, such as Fusarium oxysporum f. sp. niveum, Fusarium oxysporum f. sp. cubense, Fusarium graminearum, Mycosphaerella melonis, and Botrytis cinerea. (B) Effect of B. velezensis F21 on spore germination were observed by microscope, the bars equal 5 μm. (C) Biological control effect of B. velezensis F21 to Fusarium wilt disease on watermelon in the field. (D) The symptoms of Fusarium wilt disease development on watermelon seedlings 30 days post Fusarium oxysporum f. sp. niveum inoculation in each treatment in greenhouse experiment.
As is well known, an efficient BCA also plays a role in promoting plant growth. Therefore, in this study, we also evaluated the plant growth promoting effect of B. velezensis F21 on watermelon through greenhouse and field experiments. As shown in Supplementary Figure S2, the growth state of watermelon treated with B. velezensis F21 was significantly better than that of the control treatment, and the field experiment results still showed that the size and yield of watermelon were significantly higher than those of the control treatment (Supplementary Figure S2). Taken together, it could be concluded that B. velezensis F21 could serve as a potential BCA to control Fusarium wilt disease in watermelon.
B. velezensis F21 Treatment Triggers ISR to Fon by Inducing Defense-Related Gene Expression and Raising the Activities of Plant Defense-Related Enzymes in Watermelon
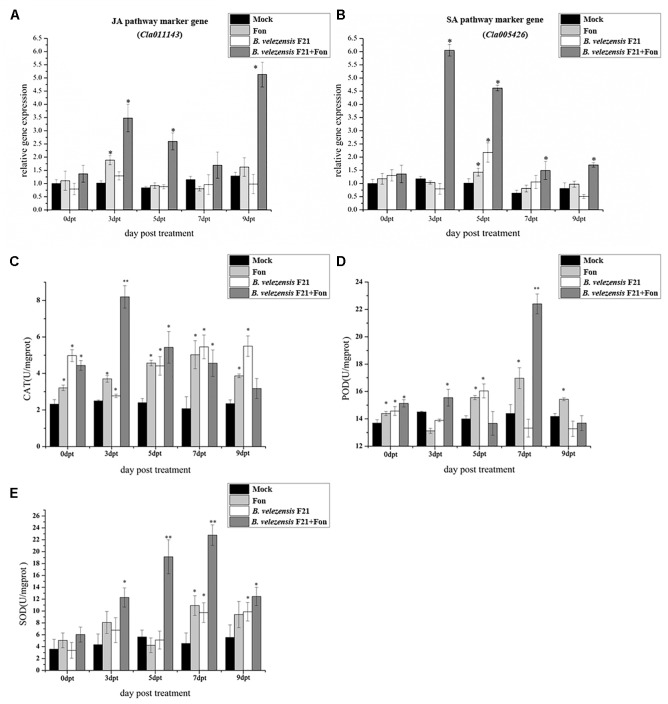
Biocontrol agent-mediated ISR is often combined with priming for the enhanced expression of defense-related genes and the activities of defense-related enzymes. To determine the mechanism of B. velezensis F21-mediated ISR in Fusarium wilt, the expression levels of defense related genes, such as Cla011143 (JA pathway marker gene) andCla005426 (SA pathway marker gene) were detected. Moreover, the activities of defense related enzymes (CAT, POD, and SOD) were also determined in the roots of watermelon only inoculated with Fon and in those pretreated with B. velezensis F21 before inoculation with Fon. As shown in Figure 2A,B, the transcripts of Cla01114 and Cla005426 accumulated in the roots of watermelon from 0 dpt to 9 dpt. The transcripts of two tested genes were both generally stronger in the roots of watermelon pretreated with B. velezensis F21 and then challenged with Fon from 3 dpt to 9 dpt and reached their maximum at 3 dpt in comparison with the control treatment. In addition, we also found that B. velezensis F21 could result in a quicker response of related genes (Figure 2A,B). These results indicated that B. velezensis F21 could prime the watermelon to enhance resistance to Fon by simultaneously activating the SA-and JA-mediated defense signaling pathways. Moreover, the activities of defense related enzymes (CAT, POD, and SOD) were also determined and the results in Figure 2 demonstrated that compared with the control treatment, B. velezensis F21 could increase the activities and advance those defense-related enzymes to maximum activity in advance in response to Fon invasion (Figure 2C–E). As shown in Figure 2, the activities of CAT, POD, and SOD in the roots of watermelon pretreated with B. velezensis F21 combined with the challenge with Fon were higher and showed a high peak at 3 dpt, 7 dpt, and 7 dpt, respectively (Figure 2C–E).
FIGURE 2.
Effect on the expression of defense related gene and activities of defense-related enzymes in the process of controlling watermelon Fusarium wilt by B. velezensis F21. (A,B) Expression of defense related gene in watermelon root in response to interaction between B. velezensis F21 and F. oxysporum f. sp. niveum. Time course of expression of JA signaling pathway marker gene Cla011143 (A) and SA signaling pathway marker gene Cla005426 (B) in the root of watermelon treated with B. velezensis F21and challenged with Fon. The expression values of the individual genes were normalized using Actin gene as an internal standard. (C–E) The activities of defense-related enzymes (CAT, POD, and SOD) in the root of watermelon pretreated with B. velezensis F21 and challenged with Fon. All data were presented as means of three replicates ± SD, and error bars represent SD for three replicates. Means with asterisk have significant differences (∗P < 0.05 and ∗∗P < 0.01; LSD test). All experiments were performed three times, and similar results were obtained.
Identification and Functional Annotation of Differentially Expressed Genes (DEGs) During the Process of B. velezensis F21 Biocontrol of Fusarium Wilt in Watermelon
To attempt to elucidate the detailed mechanisms of B. velezensis F21 in the biocontrol of Fusarium wilt in watermelon, a comparative transcriptome analysis using watermelon plant roots treated with B. velezensis F21 or sterile water alone and in combination with Fon inoculation was conducted. All libraries from different samples were sequenced by using a HiSeqTM 2500 platform. Approximately 21.61 Gb total clean reads were obtained after cleaning and quality checking. The Q20 percentage of each library was from 97 to 99.11%; the Q30 percentage of each library was from 89 to 95%; and the clean reads ratio percentage of each library was up to 99% (Supplementary Table S3). These data showed that the RNA-Seq quality was applicable for further analysis.
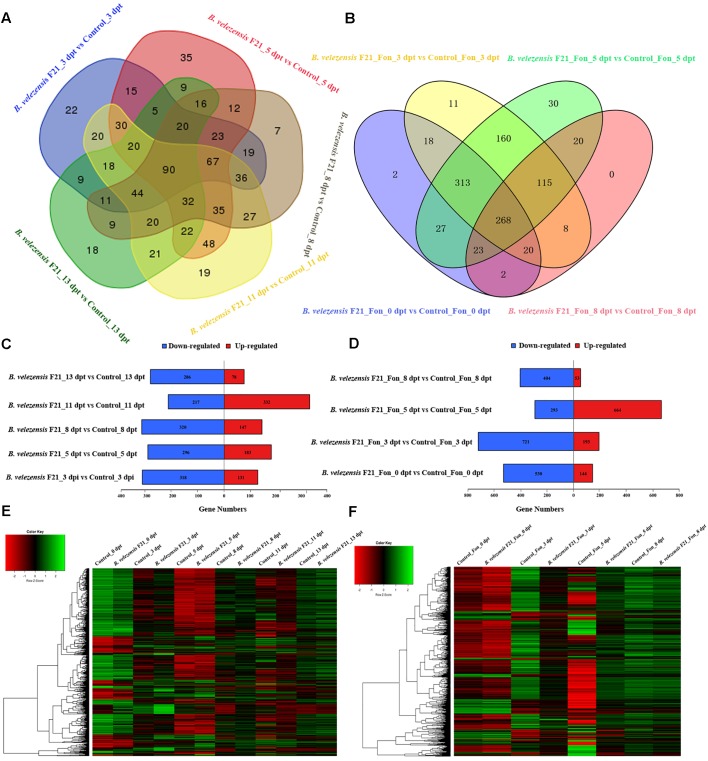
To delineate the mechanisms of B. velezensis F21 biocontrol Fusarium wilt in watermelon, two comparison groups were set with sequencing data from the 20 libraries. One group was the “pretreatment” group, using B. velezensis F21-treated samples alone compared with mock treatment samples at different time points and six sets of comparisons were made. The other group was the “ISR” group by using B. velezensis F21 pretreated samples for 5 days and then challenging the plants with Fon compared with control treatment samples at different time points and four sets of comparisons were made. It was observed that whether B. velezensis F21 was treated alone or pretreated before inoculating the pathogen, a large number of gene expression levels in watermelon could be changed (Supplementary Table S4 and Figure 3A,B). The comparative results revealed that 768 significant DEGs were identified in the samples treated with B. velezensis F21 alone and the gene transcription levels were most affected at 11 dpt, with 549 DEGs (217 down-regulated and 332 up-regulated) (Figure 3C,E). However, during the process of B. velezensis F21 ISR to Fon, over 1,000 significant DEGs were identified and the gene transcription levels were influenced most strongly at 3 dpt and 5 dpt. There were 894 DEGs (721 down-regulated and 193 up-regulated) and 957 DEGs (293 down-regulated and 664 up-regulated), respectively (Figure 3D,F). The results indicated that B. velezensis F21 could result in a quicker and stronger watermelon response to Fon invasion. In summary, the results showed that B. velezensis F21 could succeed in inducing the resistance of watermelon to Fusarium wilt by regulating the transcription of some related genes.
FIGURE 3.
Classification of differential expressed genes (DEGs) in the root of watermelon in response to B. velezensis F21 treatment or in combination with Fon inoculation. (A) Venn diagram of DEGs in the root of watermelon treated by B. velezensis F21 alone at different time points. (B) Venn diagram of DEGs in the root of watermelon pretreated with B. velezensis F21 and challenged with Fon at different time points. (C,D) Numbers of DEGs in response to B. velezensis F21 treatment or in combination with Fon inoculation. X-axis represents DEG numbers, Y-axis represents comparison method between each group. Red color represents up-regulated DEGs, blue color represents down-regulated DEGs. (E) Cluster analysis of DEGs in root of watermelon treated with B. velezensis F21 alone and in root of mock-treated watermelon seedlings based on the expression profiles measured by RNA-seq. (F) Cluster analysis of DEGs in root of watermelon pretreated with B. velezensis F21 and challenged with Fon based on the expression profiles measured by RNA-seq. The color scale in the heat map corresponds to log2 (FPKM) value of genes in each samples.
Otherwise, the global functional analysis of DEGs was carried out by using GO annotation to derive “biological process,” “cellular component,” and “molecular function” (Supplementary Figure S3 and Figure 4). As shown in Supplementary Figure S3 and Figure 4, in comparison with mock treatment or treatment inoculated with Fon alone, the most enriched terms for “biological process” were “metabolic process,” “cellular process,” and “single-organism process” in the libraries constructed from watermelon plants treated with B. velezensis F21 alone or pretreated with B. velezensis F21 and then inoculated with Fon. Moreover, the “cell,” “cell part,” and “membrane” were most enriched in the term “cellular component.” “The catalytic activity” term was the most frequent in the “molecular function,” followed by “transporter activity” and “binding” (Supplementary Figure S3 and Figure 4). The above results show that B. velezensis F21 could change the physiological and biochemical functions of watermelon during biological control in Fusarium wilt.
FIGURE 4.
GO classification of up-regulated and down-regulated genes. GO classification of DEGs in the comparisons of B. velezensis F21 pretreated or mock treated watermelon seedlings, both challenged with Fon at different time points. X-axis represents GO term. Y-axis represents the amount of up- and down-regulated genes.
Pathway Enrichment Analysis for DEGs Identified in the Process of B. velezensis F21 Biocontrol Fusarium Wilt Disease
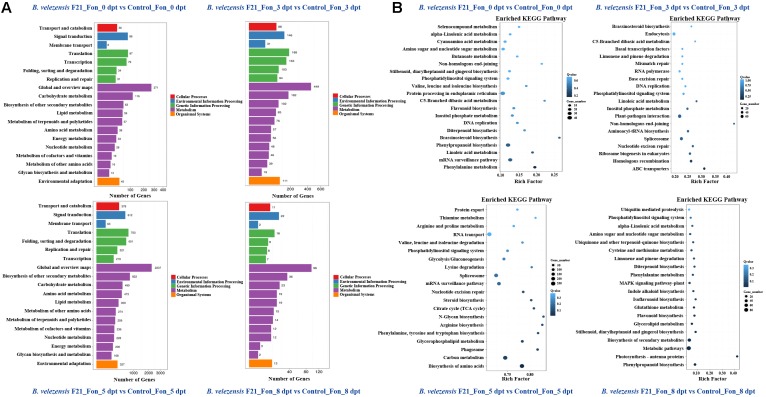
Pathway enrichment analysis with DEGs could provide guidance to identify significant metabolic pathways. To further investigate the biochemical pathways of these DEGs in the process of B. velezensis F21 ISR to Fon, we mapped all DEGs identified in the RNA sequencing to terms in the KEGG database. As shown in Supplementary Figure S4 and Figure 5A, the DEGs affected by B. velezensis F21 could be classified into five pathways, including cellular processes, environmental information processing, genetic information processing, metabolic and organism systems, whether the watermelon was treated by B. velezensis F21 alone or pretreated with B. velezensis and then challenged with Fon. Moreover, the metabolic pathway contained the largest number of DEGs, followed by genetic information processing (Supplementary Figure S4 and Figure 5A). In addition, we found that a 5-day treatment of B. velezensis F21 alone for 5 days had the most significant effect on the metabolic pathways in watermelon Supplementary Figure S4. While in the process of biological control of Fusarium wilt at 5 days post pathogen inoculation, the effects on plant metabolic pathways and the number of DEGs reached their maximum (Figure 5A). In addition, we also analyzed the enrichment in the KEGG pathway with DEGs to predict the biochemical pathway. The top 20 enriched KEGG pathways associated with DEGs affected by B. velezensis F21 at different timepoints are shown in Supplementary Figure S5. It was demonstrated that some KEGG pathways associated with plant immunity were most enriched, such as the MAPK signaling pathway, plant–pathogen interaction, and plant hormone signaling transduction (Supplementary Figure S5). The results indicated that B. velezensis F21 treatment could enhance plant basal immunity. Otherwise, we also analyzed the enrichment in the KEGG pathway with DEGs identified in the process of B. velezensis F21 ISR to Fon. Likewise, the top 20 enriched KEGG pathways associated with DEGs influenced by B. velezensis F21 at different timepoints are shown in Figure 5B. From the results, we found that KEGG pathways, such as plant–pathogen interaction, ABC transporter plant hormone signaling transduction, and MAPK signaling pathways play important roles in the process of B. velezensis F21 ISR to Fon (Figure 5B).
FIGURE 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification and functional enrichment of DEGs. (A) Pathway classification of DEGs in the comparisons of B. velezensis F21 pretreated or mock treated watermelon seedlings then both challenged with Fon at different time points, X-axis represents number of DEG. Y-axis represents functional classification of KEGG. There are seven branches for KEGG pathways: cellular processes, environmental information processing, genetic information processing, human disease (for animals only), metabolism, organismal systems and drug development. (B) Pathway functional enrichment of DEGs in the comparisons of B. velezensis F21 pretreated or mock treated watermelon seedlings, both challenged with Fon at different time points, X-axis represents enrichment factor. Y-axis represents pathway name. The color indicates the q-value (high: white, low: blue), the lower q-value indicates the more significant enrichment. Point size indicates DEG number (the bigger dots refer to larger amount). Rich Factor refers to the value of enrichment factor, which is the quotient of foreground value (the number of DEGs) and background value (total gene amount). The larger the value, the more significant enrichment.
Transcription Factors and Plant Disease Resistance Genes Prediction of DEGs Consistent With B. velezensis F21-Triggered Immunity to Fon
Transcription factors play important roles in plant growth and development, hormonal signaling, leaf senescence, and plant resistance to pathogens. To explore which TFs functioned on B. velezensis F21 triggered immunity to Fon, we analyzed the DEGs identified in each library. As shown in Supplementary Figure S6, B. velezensis F21 could affect the expression of TFs when treated with watermelon alone at different time points (Supplementary Figure S6A). Moreover, in the present study, a total of 36 kinds of TFs were identified when the watermelon was treated with B. velezensis F21 alone, including WRKY, MYB, bZIP, AP2, and NAC, which function in plant disease resistance. However, we also found that the types of TFs changed the most when watermelon was treated with B. velezensis F21 alone for 5 days (Supplementary Figure S6B). In addition, we also found that the expression of TFs was significantly influenced in the watermelons that were pretreated with B. velezensis F21 for 5 days and then challenged with Fon at different time points (Figure 6A). The numbers and types of TFs reached their maximum when challenged with Fon for 5 days, the number of types of TFs was 55. Similarly, it included some TFs related to plant disease resistance, such as MYB, WRKY, ERF, ARF, bZIP, AP2, and NAC (Figure 6B).
FIGURE 6.
Transcription factors (TFs) and plant disease resistance genes prediction of DEGs. (A) Expression heat map of TFs coding DEGs in the comparisons of B. velezensis F21 pretreated or mock treated watermelon seedlings then both challenged with Fon at different timepoints. (B) DEGs classification on TFs family in the comparisons of watermelon seedlings in each treatment. (C) DEGs classification on plant disease resistance genes in the comparisons of watermelon seedlings in each treatment.
In addition, the numbers and kinds of PRGs were analyzed with the DEGs in each sample in our study as well. As shown in Supplementary Figure S7, eleven kinds of PRGs could be affected by B. velezensis F21, including CN, CNL, Mlo-like, N, NL, RLK, RLK-GNK, RLP, RPW0-NL, T and TNL, respectively. In addition, we also found that the PRGs could be influenced the most strongly, and almost 148 PRGs were identified when B. velezensis F21 alone was treated for 5 days (Supplementary Figure S7). Moreover, a total of 595 PRGs of eleven species were identified in the process of B. velezensis F21-triggered immunity to Fon, and the effect of PRGs reached a maximum when challenged with pathogens for 5 days (Figure 6C). The above results correspond to the expression intensity of defense-related genes and defense-related enzymes at the same time points as previously explored, and taken together, we could conclude that B. velezensis F21 could trigger plant immunity to Fon via enhancing the expression of TFs and PRGs.
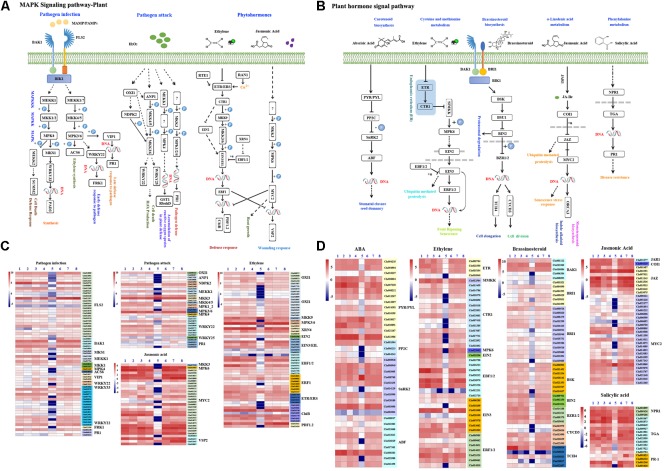
B. velezensis F21-Triggered Immunity to Fon Depended on the MAPK Signaling Pathway
Previous studies have demonstrated that the MAPK signaling pathway widely exists in eukaryotic organisms and participants in plant growth and responds to abiotic and biotic stress. As shown in Supplementary Figure S8A and Figure 7A, the MAPK signaling pathway response to plant pathogens could be divided into three parts, and a large number of genes were involved (Supplementary Figure S8A and Figure 7A). To investigate whether the MAPK signaling pathway was involved in B. velezensis F21-triggered immunity to Fon in watermelon, the above genes involved in the MAPK signaling pathway were analyzed with DEGs in each library. The results in Supplementary Figure S8B suggested that B. velezensis F21 could affect the expression of numerous of MAPK signaling pathway-related genes involved in plant responses to pathogen infection, pathogen attack and hormone. Moreover, we also found that the effect of B. velezensis F21 on the MAPK signaling pathway was the most significant when treated for 5 days (Supplementary Figure S8B). We also analyzed the expression levels of genes involved in the MAPK signaling pathway during the process of B. velezensis F21 triggering plant immunity to Fon. As shown in Figure 7C, numerous MAPK signaling pathway-related genes were influenced especially when the plants were pretreated with B. velezensis F21 and then challenged with Fon for 5 days (Figure 7C). The above results show that the MAPK signaling pathway plays a key role in B. velezensis F21 triggered plant immunity to Fon.
FIGURE 7.
Analysis of DEGs related to MAPK signaling pathway and plant hormone signaling pathway. Analysis of DEGs related to MAPK signaling pathway and plant hormone signaling pathway in the comparisons of B. velezensis F21 pretreated or mock treated watermelon seedlings then both challenged with Fon. (A) The model of MAPK signaling pathway in plant. (B) The model of plant hormone signaling pathway in plant. (C,D) The heat map for expression of DEGs involved in MAPK signaling pathway and plant hormone signaling pathway, respectively, in the comparisons of B. velezensis F21 pretreated or mock treated watermelon seedlings then both challenged with Fon at different time points. The number 1, 3, 5, 7 means the samples got from the watermelon which was pretreated with sterile water then challenged with Fon at 0 dpt, 3 dpt, 5 dpt, and 8 dpt, respectively; the number 2, 4, 6, 8 means the samples got from the watermelon which was pretreated with B. velezensis F21 then challenged with Fon at 0 dpt, 3 dpt, 5 dpt, and 8 dpt, respectively.
Serials of Phytohormone Signaling Pathways Were Involved in B. velezensis F21 Triggered Immunity to Fon in Watermelon
Phytohormone signaling has been shown to have broad and crucial roles in plant development and response to biotic and abiotic stress. Therefore, in our study, we also studied the effect of B. velezensis F21 on changes in the phytohormone signaling pathway during the process of B. velezensis F21 triggering plant immunity to Fon. As shown in Supplementary Figure S9A and Figure 7B, many phytohormones participated in the plant immunity, such as ABA, ETH, BR, JA, and SA, and a large number of genes were involved in the synthesis and regulation of these phytohormones (Supplementary Figure S9A and Figure 7B). To validate which genes were involved in B. velezensis F21-triggered plant immunity to Fon, the expression of these genes was analyzed with the DEGs identified from transcriptome sequencing data. As shown in Supplementary Figure S9B, the numbers of genes involved in phytohormone synthesis and regulation were significantly affected by B. velezensis F21 (Supplementary Figure S9B). Likewise, the results in Supplementary Figure S8B demonstrated that the expression of these genes was influenced during B. velezensis F21-triggered plant immunity to Fon (Figure 7D). In addition, we also detected the accumulation of these phytohormones including ABA, ETH, BR, JA, and SA by ELISA in the process of B. velezensis F21-triggered plant immunity to Fon. It was found that the accumulation of these phytohormones in watermelon changed to varying degrees at different time points during the process of B. velezensis F21-triggered plant immunity to Fon (Supplementary Figure S10). The above results indicated that B. velezensis F21 could trigger plant immunity to Fon via activating the phytohormone signaling pathways.
To confirm the expression patterns identified by the transcriptome sequencing data of genes that were involved in the MAPK signaling pathway and phytohormones, the transcript levels of 15 genes during treatment with B. velezensis and 21 genes in the process of B. velezensis F21-triggered plant immunity to Fon were examined by qRT-PCR. The detailed information of the selected genes and primer pairs used in this study is shown in Supplementary Table S1. All the genes selected in this study were successfully amplified and the patterns of gene expression detected by qRT-PCR were consistent with those from the transcriptome sequencing data (Supplementary Figures S11, S12). Therefore, it turns out that the DEGs and gene expression profiles from transcriptome sequencing data were reliable.
Discussion
Watermelon Fusarium wilt, caused by Fon, is a devastating disease affecting a large variety of economically important crops worldwide (Demers et al., 2015). The disease can result in 100% yield losses under favorable environmental conditions during watermelon production. At present, the control of the disease mainly depends on resistant varieties and chemical control, but the effect of the above control strategies is not ideal. PGPR were reported as an important functional group of beneficial bacteria used for plant growth promotion and the control of soilborne disease (Lemessa and Zeller, 2007). However, the exploitation of rhizobacteria as BCAs against this Fusarium wilt disease in watermelon has rarely been considered. In the present study, we found that B. velezensis F21 could serve as a potential BCA to control Fusarium wilt disease in watermelon. BCA not only promoted watermelon seedling growth (Supplementary Figure S2) but also significantly controlled Fusarium wilt disease in the greenhouse and filed (Figure 1C,D). However, the biocontrol of Fusarium wilt by Bacillus spp. has been reported in previously. For example, Rania et al. (2016) reported that B. amyloliquefaciens SP1 could significantly control F. oxysporum. However, Fusarium wilt disease in watermelon has rarely been considered. In the present study, we report for the first time that B. velezensis F21 could be used as a BCA for the control of Fusarium wilt disease. Moreover, we also found that B. velezensis F21showed remarkable production of many hydrolases (Supplementary Figure S1). Therefore, we hypothesized that this bacterium has strong and broad-spectrum bacteriostatic activity (Figure 1A).
In addition, as is well known, determining the mode of action is important and necessary for developing Bacillus sp. as a useful and successful BCA for the biocontrol of Fusarium wilt disease in watermelon (Jangir et al., 2018). In recent years, the focus was on the study of the antimicrobial activity of secondary metabolites. For example, Jangir et al. (2018) mentioned that one kind of Bacillus sp. B44, which was isolated from rhizospheric soil of tomato, could significantly protect against F. oxysporum f. sp. lycopersici, and study of the biocontrol mechanism indicated that the strain could produce numerous lytic enzymes and antifungal secondary metabolites (Jangir et al., 2018). Moreover, it was also reported that B. pumilus MSUA3 produced chitinolytic enzymes and utilized heat-stable antibiotic surfactin mediated antagonistic activity to control F. oxysporum (Agarwal et al., 2017). However, in our study, we demonstrated that B. velezensis F21 could not only directly suppress the growth of Fon by secreting some secondary metabolites with antimicrobial activity but also induce systemic resistance to Fon in watermelon. Many effective biocontrol PGPR have been reported to elicit ISR in recent years, including those that could elicit ISR to Fusarium. For instance, Duijff et al. (1998) investigated that P. fluorescens WCS417r could induce systemic resistance to suppress of Fusarium wilt of tomato. Otherwise, some non-pathogen fungi, such as Phytophthora cryptogea, were also reported to have the ability to induce systemic resistance to Fon in the host (Attitalla et al., 2010). In our study, as shown in Figure 2, 3, we found that the transcripts of two tested genes, Cla01114 and Cla005426, were both generally stronger in the roots of watermelon pretreated with B. velezensis F21 and then challenged with Fon from 3 dpt to 9 dpt and reached their maximums at 3 dpt compared with the control treatment. Moreover, the activities of defense-related enzymes (CAT, POD, and SOD) were also determined and the results indicated that compared with the control treatment, B. velezensis F21 could increase the activities and advance those defense-related enzymes to maximum activity in advance in response to Fon invasion. Taken together, we conclude that B. velezensis F21 could prime the watermelon to enhance resistance to Fon by simultaneously activating the SA-and JA-mediated defense signaling pathways. This is also the first report that B. velezensis can control Fusarium wilt disease by inducing systemic resistance to Fon in watermelon by simultaneously activating the SA-and JA-mediated defense signaling pathways.
Recent studies show that the application of transcriptomics can provide precise information on gene networks for identifying the functions of genes and unknown genes on regulation (Tohge et al., 2005). In this study, we identified thousands of genes that function on B. velezensis F21 ISR to Fon in watermelon. As shown in Supplementary Table S4 and Figure 3A,B, the expression of a large number of genes in watermelon could be changed or modified by B. velezensis F21 treated alone or pretreated before the inoculation of pathogens (Supplementary Table S4 and Figure 3A,B), indicating that B. velezensis F21 could induce systemic resistance to Fon via altering transcriptional levels of genes in watermelon. In addition, from the results, we hypothesize that there must be many genes involved in regulation in this process. TFs have been demonstrated to play essential roles in plants by controlling the expression of genes involved in various cellular processes (Han et al., 2014; Chen F. et al., 2017). It consists of many families, such as WRKY, AP2-ERF, MYB, and NAC. WRKY factors have been verified to fulfill essential regulatory functions to modulate pathogen-triggered cellular responses in a number of plant species, and most WRKY factors participate in the SA signaling pathway (Tsuda and Somssich, 2015; Chen F. et al., 2017). However, some ERF factors have been demonstrated, which participate in the JA signaling pathway are involved in the interaction between plants and insects of some fungal pathogens (Mizoi et al., 2012). In addition, NAC proteins are another kind of plant-specific TF that constitute a large family. At present, many functional studies have shown that NAC TFs could regulate immune responses in plants, and over 100 NAC genes have been identified in Arabidopsis (Puranik et al., 2012; Lee et al., 2017). In our study, by comparing the RNA sequencing data, we also identified numerous TFs involved in the process of B. velezensis F21inducing systemic resistance to Fon in watermelon, including WRKY, MYB, bZIP, AP2, and NAC (Supplementary Figure S6 and Figure 6A,B). Moreover, we found that two families were both identified in our sequencing data, WRKY and AP2-ERF. The results indicated that the two signaling pathways, both SA and JA, take part in the process of B. velezensis F21 inducing systemic resistance to Fon in watermelon. The above results coincide with the conclusion of a previous expression level of gene determination by qRT-PCR.
Plant MAPKs are involved in plant growth, development, responses to endogenous and environmental cues. Serials of studies have demonstrated that MAPKs could be activated by external sensors to cellular responses (Yu and Tang, 2004). Otherwise, some studies verified that MAPKs were involved in the interaction between plant and pathogens, and they played key roles in plant response to pathogen invasion. For instance, many effectors secreted via pathogens have been found to inhibit MAP kinase cascade (Bi and Zhou, 2017). In our study, we also analyzed the transcriptional levels of genes related to MAP kinase cascade, and as the results shown in Supplementary Figure S8, many genes were affected by B. velezensis F21, this indicated that the perception of B. velezensis F21 in watermelon also depends on MAPK pathway. From the result shown in Figure 7C, we could demonstrate that the pretreatment of B. velezensis F21 could speed up the recognition of pathogens in plants, make corresponding resistance response quickly, and enhance the immunity of plants. The above hypothesis is only based on the results of current bioinformatics analysis. In-depth analysis and verification work needs to be completed in future research.
Phytohormones are some substances biosynthesized in very low concentrations but able to regulate serials of cellular processes in plants. They play key roles and coordinate various signal transduction pathways during abiotic and abiotic-stress response (Voß et al., 2014). For instance, some phytohormones, such as ABA, JA, ETH, SA, etc., have been identified as stress hormones. They played critical roles in plant growth regulation, development, stomatal closure and mediating abiotic and biotic stress responses (Li et al., 2010). In this study, we also evaluated the effect of velezensis F21 on phytohormones synthesis and regulation. Therefore, we analyzed the expression level of some genes involved in some phytohormones and detected the accumulation of these phytohormones in watermelon at different treatment by ELISA, such as ABA, JA, ETH, SA, and so on. As the results shown in Figure 7D and Supplementary Figures S9, S10, number of genes involve in phytohormones synthesis and regulation was significantly affected by B. velezensis F21. Moreover, we also found that the accumulation of these phytohormones in watermelon changed in varying degrees at different time points during the process of B. velezensis F21 triggered plant immunity to Fon. These results suggested that phytohormones in watermelon played critical roles in B. velezensis F21 triggered plant immunity to Fon. However, it still requires us to invest more time in future work to figure out how the BCAs regulate phytohormone synthesis and which kind of phytohormones play the main roles in B. velezensis F21 triggered plant immunity to Fon.
Conclusion
In this study, we identified an efficiently PGPR strain B. velezensis F21, which could be used for control of Fon in watermelon. In-depth mechanism study indicated that B. velezensis F21 could suppress the growth and spore germination of Fon in vitro. Moreover, B. velezensis F21 could also enhance plant basal immunity to Fon by increasing the expression of plant defense related genes and activities of some defense enzymes, such as CAT, POD, and SOD. In addition, a comparative transcriptome analysis was employed to figure out more detail mechanisms and the results revealed almost thousands of DEGs in the process of B. velezensis F21 triggered ISR to Fon, among these identified DEGs, a large number of genes belong to some TFs, plant resistant genes, some genes involved in MAP kinase cascade and phytohormone biosynthesis and regulation. Taken together, this study substantially expands transcriptome data resources and suggests a molecular framework for B. velezensis F21 inducing systemic resistance to Fon in watermelon. It also provides a new idea for the mechanism study to understand how BCAs biocontrol Fusarium wilt disease in watermelon. Besides, it also provides an effective strategy for the control of Fusarium wilt in watermelon.
Author Contributions
C-HJ, X-FY, and J-HG designed the experiments. D-DM, Z-JL, and B-YY performed the experiments with assistance from YZ, C-HJ, Y-JQ, and J-HG analyzed and discussed the results. C-HJ wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Chen Y. (Zhejiang University, Hangzhou) for kindly providing us with pathogen Fusarium oxysporum f. sp. niveum.
Abbreviations
- ABA
abscisic acid
- BIO
bio-organic fertilizer
- BR
brassinosteroid
- CAT
catalase
- DEGs
differentially expressed genes
- ELISA
enzyme-linked immunosorbent assay
- ETH
ethylene
- Fon
Fusarium oxysporum f. sp. niveum
- GO
Gene Ontology
- ISR
induced systemic resistance
- JA
jasmonic acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LB medium
Luria-Bertani medium
- MAPK
mitogen-activated protein kinase
- PDA medium
potato dextrosa agar medium
- PGPR
plant growth promotion rhizobacteria
- POD
peroxidase
- SA
salicylic acid
- SOD
superoxide dismutase
- TFs
transcription factors
Funding. This study was supported by the Natural Science Foundation of Jiangsu Province (Young Scholars Program) (BK20170709), National Natural Science Foundation of China (Young Scholars Program) (31701829), National Natural Science Foundation of China (31672075), National Postdoctoral Program for Innovative Talents (BX201600074), and China Postdoctoral Science Foundation (2017M611839).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00652/full#supplementary-material
References
- Agarwal M., Dheeman S., Dubey R. C., Kumar P., Maheshwari D. K., Bajpai V. K. (2017). Differential antagonistic responses of Bacillus pumilus MSUA3 against Rhizoctoniasolani and Fusarium oxysporum causing fungal diseases in Fagopyrum esculentum moench. Microbiol. Res. 205 40–47. 10.1016/j.micres.2017.08.012 [DOI] [PubMed] [Google Scholar]
- Akram W., Anjum T., Ali B., Ahmad A. (2013). Screening of native Bacillus strains to induce systemic resistance in tomato plants against Fusarium Wilt in split root system and its field applications. Int. J. Agric. Biol. 15 1289–1294. [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids. Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attitalla I. H., Johnson P., Brishammar S., Quintanilla P. (2010). Systemic resistance to Fusarium wilt in tomato induced by Phytopthora cryptogea. J. Phytopathol. 149 373–380. 10.1111/j.1439-0434.2001.tb03865.x [DOI] [Google Scholar]
- Bekkar A. A., Zaim S., Belabid L. (2018). Induction of systemic resistance in chickpea against Fusarium wilt by Bacillus strains. Arch. Phytopathol. Plant Protect. 51 1–11. 10.1080/03235408.2018.1438819 [DOI] [Google Scholar]
- Berg G., Köberl M., Rybakova D., Müller H., Grosch R., Smalla K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93 fix050. 10.1093/femsec/fix050 [DOI] [PubMed] [Google Scholar]
- Bi G., Zhou J. M. (2017). Map kinase signaling pathways: a hub of plant-microbe interactions. Cell Host Microbe. 21:270. 10.1016/j.chom.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Chen F., Hu Y., Vannozzi A., Wu K., Cai H., Qin Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36 311–335. 10.1080/07352689.2018.1441103 22579867 [DOI] [Google Scholar]
- Chen Y., Shen Q., Lin R., Zhao Z., Shen C., Sun C. (2017). De novo transcriptome analysis in dendrobium and identification of critical genes associated with flowering. Plant Physiol. Biochem. 119 319–327. 10.1016/j.plaphy.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Choudhary D. K., Prakash A., Johri B. N. (2007). Induced systemic resistance (ISR) in plants: mechanism of action. Indian J. Microbiol. 47 289–297. 10.1007/s12088-007-0054-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers J. E., Gugino B. K., Jiménezgasco M. M. (2015). Highly diverse endophytic and soil Fusarium oxysporum populations associated with field-grown tomato plants. Appl. Environ. Microbiol. 81 81–90. 10.1128/AEM.02590-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijff B. J., Pouhair D., Olivain C., Alabouvette C., Lemanceau P. (1998). Implication of systemic induced resistance in the suppression of Fusarium wilt of tomato by Pseudomonas fluorescens WCS417r and by nonpathogenic Fusarium oxysporum Fo47. Eur. J. Plant Pathol. 104 903–910. 10.1023/a:1008626212305 [DOI] [Google Scholar]
- Faheem M., Raza W., Zhong W., Nan Z., Shen Q., Xu Y. (2015). Evaluation of the biocontrol potential of Streptomyces goshikiensis YCXU against Fusarium oxysporum f. sp. niveum. Biol. Control. 81 101–110. 10.1016/j.biocontrol.2014.11.012 [DOI] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Kumar D., Chen H., Wu S., Kim J. Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J. Exp. Bot. 65 1737–1749. 10.1093/jxb/ert422 [DOI] [PubMed] [Google Scholar]
- Heydari A., Pessarakli M. (2010). A review on biological control of fungal plant pathogens using microbial antagonists. J. Biol. Sci. 10 273–290. 10.3923/jbs.2010.273.290 [DOI] [Google Scholar]
- Jangir M., Pathak R., Sharma S., Sharma S. (2018). Biocontrol mechanisms of Bacillus sp. isolated from tomato rhizosphere against Fusarium oxysporum f. sp. lycopersici. Biol. Control. 123 60–70. 10.1016/j.biocontrol.2018.04.018 [DOI] [Google Scholar]
- Janvier C., Villeneuve F., Alabouvette C., Edel-Hermann V., Mateille T., Steinberg C. (2007). Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol. Biochem. 39 1–23. 10.1016/j.soilbio.2006.07.001 [DOI] [Google Scholar]
- Jiang C. H., Huang Z. Y., Xie P., Gu C., Li K., Wang D. C., et al. (2016). Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringaepv. tomato DC3000 in Arabidopsis. J. Exp. Bot. 67 157–174. 10.1017/S0079497X00011130 [DOI] [PubMed] [Google Scholar]
- Jiang C. H., Liao M. J., Wang H. K., Zheng M. Z., Xu J. J., Guo J. H. (2018). Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control. 126 147–157. 10.1016/j.biocontrol.2018.07.017 [DOI] [Google Scholar]
- Jiang C. H., Wu F., Yu Z. Y., Xie P., Ke H. J., Li H. W., et al. (2015). Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol. Res. 170 95–104. 10.1016/j.micres.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Lee M. H., Jeon H. S., Kim H. G., Park O. K. (2017). An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 214 343–360. 10.1111/nph.14371 [DOI] [PubMed] [Google Scholar]
- Lemessa F., Zeller W. (2007). Screening rhizobacteria for biological control of Ralstonia solanacearum in Ethiopia. Biol. Control. 42 336–344. 10.1016/j.biocontrol.2007.05.014 [DOI] [Google Scholar]
- Li S., Zhou X., Chen L., Huang W., Yu D. (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 29 475–483. 10.1007/s10059-010-0059-2 [DOI] [PubMed] [Google Scholar]
- Ling N., Huang Q., Guo S., Shen Q. (2011). Paenibacillus polymyxa SQR-21 systemically affects root exudates of watermelon to decrease the conidial germination of Fusarium oxysporum f.sp. niveum. Plant Soil 341 485–493. 10.1007/s11104-010-0660-3 [DOI] [Google Scholar]
- Lu D., Ma Z., Xu X., Yu X. (2016). Isolation and identification of biocontrol agent Streptomyces rimosus M527 against Fusarium oxysporum f. sp. cucumerinum. J. Basic Microb. 56 929–933. 10.1002/jobm.201500666 [DOI] [PubMed] [Google Scholar]
- Meldrum R. A., Daly A. M., Ltt T. N., Eab A. (2013). The effect of surface sterilants on spore germination of Fusarium oxysporum f. sp. cubense tropical race 4. Crop Prot. 54 194–198. 10.1016/j.cropro.2013.08.014 [DOI] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul. Mech. 1819 86–96. 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35 W182–W185. 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujoko T., Sastrahidayat I. R., Hadiastono T., Djauhari S. (2014). Antagonistic effect of Streptomyces spp. on spore germination and mycelial growth of Fusarium oxysporum f.sp. lycopersici. IJB 5 414–422. 10.12692/ijb/5.9.414-422 [DOI] [Google Scholar]
- Niu D. D., Liu H. X., Jiang C. H., Wang Y. P., Wang Q. Y., Jin H. L., et al. (2011). The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. MPMI 24 533–542. 10.1094/MPMI-09-10-0213 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M., van Wees S. C., van Pelt J. A., Knoester M., Laan R., Gerrits H., et al. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10 1571–1580. 10.1105/tpc.10.9.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Rania A. B. A., Jabnoun-Khiareddine H., Nefzi A., Mokni-Tlili S., Daami-Remadi M. (2016). Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biol. Control 164 811–824. 10.1111/jph.12501 [DOI] [Google Scholar]
- Shen Z., Wang D., Ruan Y., Xue C., Zhang J., Li R., et al. (2014). Deep 16s rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS One 9:e98420. 10.1371/journal.pone.0098420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang Y., Han L. R., Zhang X., Feng J. T. (2017). Antifungal activity and action mode of cuminic acid from the seeds of Cuminum cyminum L. against Fusarium oxysporum f. sp. niveum (FON) Causing Fusarium Wilt on Watermelon. Molecules 22:2053. 10.3390/molecules22122053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Nishiyama Y., Hirai M. Y., Yano M., Nakajima J. I., Awazuhara M., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. 10.1111/j.1365-313x.2005.02371.x [DOI] [PubMed] [Google Scholar]
- Tsuda K., Somssich I. E. (2015). Transcriptional networks in plant immunity. New Phytol. 206 932–947. 10.1111/nph.13286 [DOI] [PubMed] [Google Scholar]
- Voß U., Bishopp A., Farcot E., Bennett M. J. (2014). Modelling hormonal response and development. Trends Plant Sci. 19 311–319. 10.1016/j.tplants.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T., Zhao H., Wang W. (2017). Effect of the biocontrol agent Bacillus amyloliquefaciens SN16-1 on the rhizosphere bacterial community and growth of tomato. J. Phytopathol. 166 324–332. 10.1111/jph.12690 [DOI] [Google Scholar]
- Wang B. B., Yuan J., Zhang J., Shen Z. Z., Zhang M. X., Li R., et al. (2013). Effects of novel bioorganic fertilizer produced by Bacillus amyloliquefaciens W19 on antagonism of Fusarium wilt of banana. Biol. Fert. Soils 49 435–446. 10.1007/s00374-012-07395 [DOI] [Google Scholar]
- Wu Q., Wu J., Li S. S., Zhang H. J., Feng C. Y., Yin D. D., et al. (2016). Transcriptome sequencing and metabolite analysis for revealing the blue flower formation in waterlily. BMC Genomics 17:897. 10.1186/s12864-016-3226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. Y., Jia K., Geng W. Y., Guo R. J., Li S. D. (2014). Management of cucumber wilt disease by Bacillus subtilis B006 through suppression of Fusarium oxysporum f. sp. cucumerinum in rhizosphere. Plant Pathol. J. 13 160–166. 10.3923/ppj.2014.160.166 [DOI] [Google Scholar]
- Yu S. W., Tang K. X. (2004). MAP kinase cascades responding to environmental stress in plants. Acta Bot. Sin. 46 127–136. [Google Scholar]
- Zhang Y., Li W., Dou Y., Zhang J., Jiang G., Miao L., et al. (2015). Transcript quantification by RNA-Seq reveals differentially expressed genes in the Red and yellow fruits of Fragaria vesca. PLoS One 2015:10. 10.1371/journal.pone.0144356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P. C., Quan Y., Wang J., Fan S. (2014). Bacillus amyloliquefaciens Q-426 as a potential biocontrol agent against Fusarium oxysporum f. sp. spinaciae. J. Basic Microbiol. 54 448–456. 10.1002/jobm.201200414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.