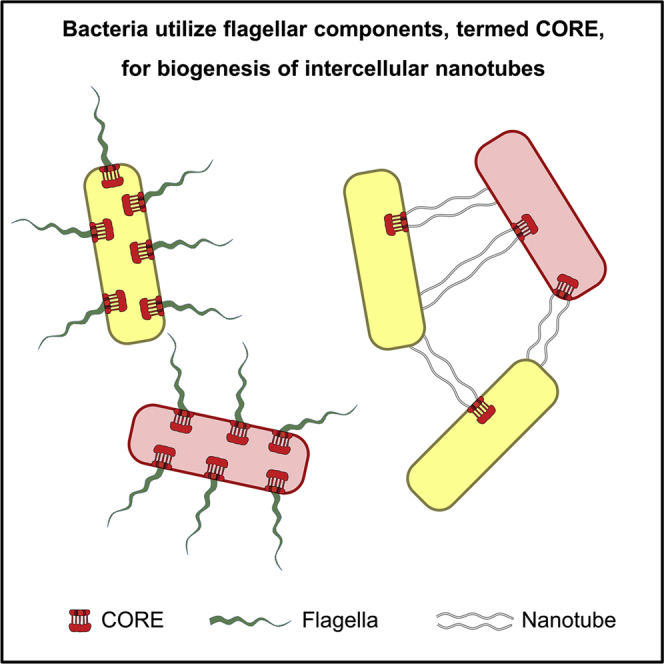
Summary
We have previously described the existence of membranous nanotubes, bridging adjacent bacteria, facilitating intercellular trafficking of nutrients, cytoplasmic proteins, and even plasmids, yet components enabling their biogenesis remain elusive. Here we reveal the identity of a molecular apparatus providing a platform for nanotube biogenesis. Using Bacillus subtilis (Bs), we demonstrate that conserved components of the flagellar export apparatus (FliO, FliP, FliQ, FliR, FlhB, and FlhA), designated CORE, dually serve for flagellum and nanotube assembly. Mutants lacking CORE genes, but not other flagellar components, are deficient in both nanotube production and the associated intercellular molecular trafficking. In accord, CORE components are located at sites of nanotube emergence. Deleting COREs of distinct species established that CORE-mediated nanotube formation is widespread. Furthermore, exogenous COREs from diverse species could restore nanotube generation and functionality in Bs lacking endogenous CORE. Our results demonstrate that the CORE-derived nanotube is a ubiquitous organelle that facilitates intercellular molecular trade across the bacterial kingdom.
Keywords: Bacillus subtilis, nanotubes, flagellar export apparatus, flagella type III secretion system, bacterial communication, contact-dependent molecular exchange, bacterial community
Graphical Abstract
Highlights
-
•
Conserved flagellar CORE components dually serve for flagella and nanotube assembly
-
•
CORE mutants are deficient in nanotube formation and intercellular molecular trade
-
•
CORE-dependent nanotube production is conserved among distinct bacterial species
-
•
The CORE-nanotube organelle can provide a common path for bacterial molecular trade
Bhattacharya et al. show that bacterial intercellular nanotubes, facilitating cytoplasmic molecular exchange among cells, emerge from conserved CORE components of the flagellar export apparatus. CORE-mediated nanotube formation is widespread among bacterial species. The results establish the CORE-derived nanotube as a ubiquitous organelle, facilitating intercellular molecular trafficking across the bacterial kingdom.
Introduction
Bacteria residing in natural communities maintain intricate molecular crosstalk with proximal prokaryotic and eukaryotic cells. Dedicated machineries such as type III, IV, and VI secretion systems are used by bacteria to conduct contact-dependent molecular delivery among species, as well as across kingdoms (Costa et al., 2015, Hayes et al., 2010). We have previously described a type of bacterial contact-dependent interaction mediated by membranous conduits, termed nanotubes, bridging neighboring cells of the bacterium Bacillus subtilis (Bs) (Dubey and Ben-Yehuda, 2011, Dubey et al., 2016). In fact, nanotube-like structures were shown to be produced by multiple bacterial species, mainly when grown on solid surfaces or in biofilm assemblies (e.g., McCaig et al., 2013, Pande et al., 2015, Wei et al., 2014); however, their function remains mostly elusive. Our previous analysis revealed that nanotubes serve as conduits for an intercellular exchange of cytoplasmic proteins and even plasmids (Baidya et al., 2018, Dubey and Ben-Yehuda, 2011, Dubey et al., 2016, Stempler et al., 2017). Furthermore, we and others reported that nanotubes facilitate interspecies molecular trafficking, including that of nutrients and toxins (Benomar et al., 2015, Pande et al., 2015, Stempler et al., 2017).
Inspired by these results, we recently discovered that enteropathogenic E. coli (EPEC) uses nanotubes to interact with infected epithelial cells (Pal et al., 2019). The formation of these nanotubes was dependent on five inner membrane proteins, composing the export apparatus of the type III secretion system (T3SS), embedded within the injectisome, the major virulence machinery of EPEC (Pal et al., 2019). The Gram-positive Bs lacks injectisome, which is restricted to Gram-negative pathogens, but its flagellum harbors integral T3SS, including an export apparatus (Abby and Rocha, 2012, Diepold and Armitage, 2015, Diepold and Wagner, 2014). We thus hypothesized that the Bs flagellar export apparatus, termed here CORE (CORE-FBs), is required not only for motility (Hueck, 1998) but also for nanotube generation. The CORE, situated at the basal body of the flagella, is composed of five channel-forming integral membrane proteins, FliP, FliQ, FliR, FlhB, and FlhA, exhibiting a stoichiometry of 5:4:1:1:9, respectively (Dietsche et al., 2016, Fukumura et al., 2017, Kuhlen et al., 2018) (Figure 1A). In addition, FliO, a non-structural component of the CORE, serves as a scaffold for the assembly of FliP multimeric ring, the presumed nucleation step of the CORE complex assembly (Fabiani et al., 2017, Fukumura et al., 2017).
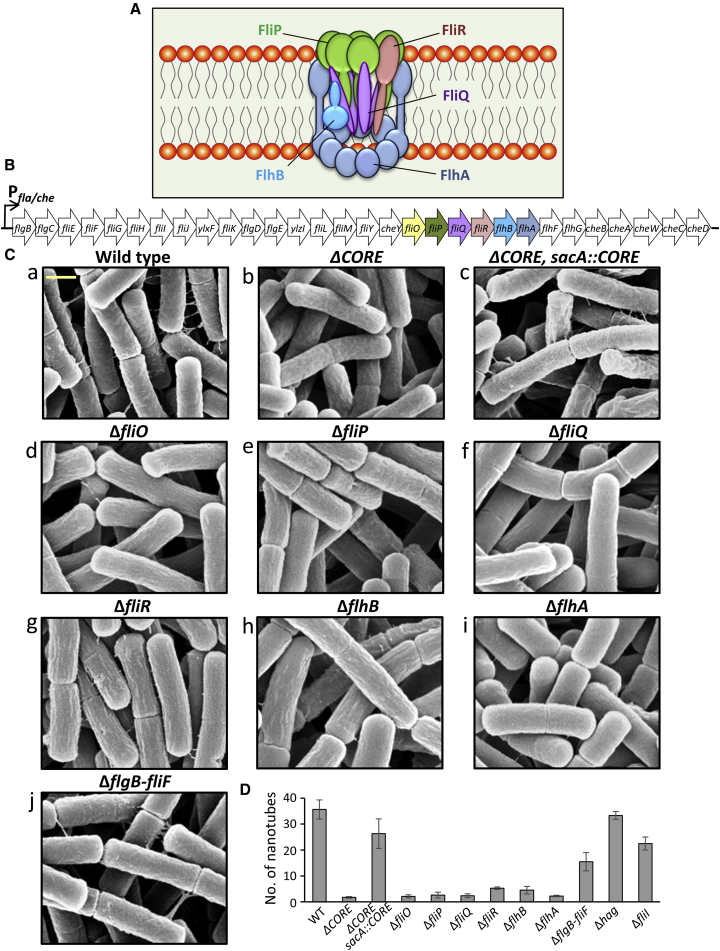
Figure 1.
Bs CORE Mutants Are Impaired in Nanotube Formation
(A) Schematic illustration of the flagellar CORE apparatus on the basis of Fukumura et al. (2017) and Kuhlen et al. (2018). The CORE consists of FliP, FliQ, FliR, FlhB, and FlhA (5:4:1:1:9) transmembrane proteins and the chaperone FliO (not shown), which only transiently associates with the CORE complex.
(B) A map depicting the fla/che operon of Bs, encoding the components required for flagellar basal body formation. Genes encoding the CORE proteins are highlighted with the same color code as in (A).
(C) The indicated Bs CORE mutant strains were visualized using XHR-SEM to monitor the formation of intercellular nanotubes. Strains were grown to the mid-logarithmic phase, spotted onto EM grids, incubated on LB agar plates for 4 h at 37°C, and visualized using XHR-SEM. Scale bar represents 500 nm.
(D) Quantification of the average number of nanotubes displayed per 50 cells by the indicated Bs mutant strains following XHR-SEM analysis described in (C). Shown are average values and SD of at least three independent experiments (n ≥ 200 for each strain).
See also Figure S1.
Here we describe that the CORE complex, the central constituent of the bacterial flagellum, has a dual utility, serving as a platform for both flagellum and nanotube biogenesis. We reveal that the CORE is positioned at sites of nanotube emergence, enabling their production along with the associated intercellular molecular exchange. Subsequently, by using bioinformatics and functional analyses, we show that the sequence and utility of CORE components are conserved among distinct bacterial species. Taken together, our results expose the existence of a bacterial organelle related to flagella and injectisome that is likely to provide a major and prevalent route for intercellular molecular exchange in bacteria.
Results
Flagellar CORE Proteins Are Required for Nanotube Formation
To investigate whether CORE-FBs is required for nanotube formation, we generated a non-polar deletion of a chromosomal fragment, containing the five CORE genes fliP, fliQ, fliR, flhB, and flhA, as well as fliO, encoding the CORE-associated chaperone (ΔCORE) (Figures 1A and 1B) (Fabiani et al., 2017, Fukumura et al., 2017). We then used extreme-high-resolution scanning electron microscopy (XHR-SEM) to examine the capacity of this mutant to form nanotubes on a solid surface, conditions restricting flagella formation and facilitating nanotube biogenesis (Dubey and Ben-Yehuda, 2011). Remarkably, the ΔCORE mutant exhibited severe deficiency in the formation of intercellular nanotubes (Figures 1Ca, 1Cb, and 1D). Consistently, the mutant was blocked in generating extending nanotubes (Figure S1A), shown to be produced at low cell density (Dubey et al., 2016). Ectopic complementation of the mutant with CORE genes restored nanotube formation to that of wild-type levels (Figures 1Cc and 1D). Importantly, nanotube structures were readily evident in bacteria deleted of genes encoding non-CORE key flagellar components. These include mutants lacking hag, encoding flagellin; flgB, flgC, fliE, and fliF (ΔflgB–fliF), encoding non-CORE basal body components; and fliI, encoding the flagellar ATPase, a key component of the CORE-associated sorting platform (Figures 1B, 1Cj, 1D, and S1B) (Minamino and Imada, 2015). These results indicate that the defect observed in nanotube production can be directly attributed to the function of the CORE, suggesting a dual function for this complex in biogenesis of either flagella or nanotubes.
To elucidate whether all six CORE genes are required for nanotube projection, we deleted each gene individually. Strains lacking fliO, fliP, fliQ, or flhA were severely deficient in nanotube construction, whereas flhB mutant could seldom form nanotubes, but these were very slim and frequently torn apart (Figures 1Cd–1Ci and 1D). Interestingly, fliR mutant generated very short projections, exhibiting a bristle-like shape (Figure 1Cg), suggesting that nanotube formation was initiated but their elongation process was halted. Reintroducing each CORE gene into the corresponding CORE mutant restored nanotube development (Figures S1C and S1D). Thus, all CORE components are required for production of proper nanotubes, though lack of each component affects the process with slight variations, highlighting further complexity layer in nanotube generation.
CORE-FBs Is Required for Intercellular Molecular Trade
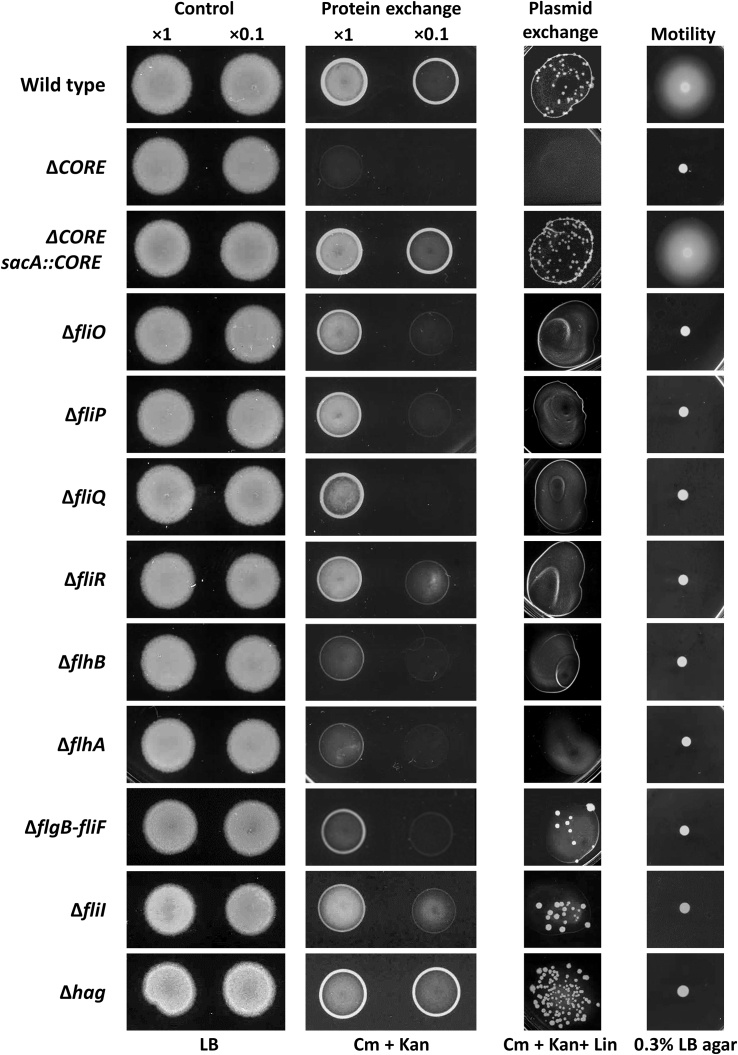
Having established that the CORE genes are required for nanotube development, we next sought to explore their influence on intercellular molecular exchange. Previously, we found that mixing two strains, each harboring a different antibiotic resistance gene, results in the exchange of antibiotic resistance enzymes in a nanotube-dependent path, yielding a population of cells transiently resistant to both antibiotics (protein exchange). Furthermore, in a similar manner, we detected the delivery of a non-conjugative plasmid from donor to recipient (plasmid exchange) (Dubey and Ben-Yehuda, 2011). Indeed, a Bs mutant lacking the CORE genes was severely deficient in both protein and plasmid exchange (Figure 2). Consistently, both defects were cured by reintroducing the CORE genes into the mutant strain (Figure 2). Analyzing the CORE single mutants revealed that they were all significantly impaired in protein exchange, with ΔflhA and ΔflhB showing the most severe phenotype, whereas fliR mutant, harboring the bristle-like nanotubes, was the least affected (Figures 1Cg and 2). Moreover, each and every single mutant was fully defective in plasmid exchange (Figure 2), indicating that such event exemplifies a more challenging task than protein trade. Importantly, none of the CORE mutants exhibited any measurable growth defect (Figures S1E and S1F). The capacity to exchange proteins and plasmids was regained by complementing the individual CORE mutants with the corresponding CORE gene (Figure S2A). Mutants lacking non-CORE flagellum genes (flgB–fliF, fliI, and hag) were capable of performing molecular exchange, while their motility was fully blocked (Figure 2). Notably, ΔflgB–fliF and ΔfliI strains exhibited minor deficiency in both protein and plasmid exchange that is consistent with the modestly reduced number of nanotube structures detected (Figures 1D and 2). Because the expression of CORE genes was not perturbed by these mutations (Figure S1G), this effect might imply some role of these proteins in stabilizing the CORE complex. Overall, our results strengthen the view that the CORE-FBs mediates intercellular molecular trafficking via nanotubes.
Figure 2.
Bs CORE Mutants Are Deficient in Molecular Exchange
Assessing molecular exchange in CORE mutants. For protein exchange assay, pairs of a donor (SB463: amyE::Phyper-spank-cat-spec) (CmR, SpecR) and a recipient (SB513: amyE::Phyper-spank-gfp-kan) (KanR) parental strains (wild-type) were used. The investigated mutants harbor the corresponding genotypes and carry the indicated null mutation in both donor and recipient strains. Donor and recipient strains were mixed in 1:1 ratio (at two concentrations, 1× and 0.1×) and incubated in LB supplemented with 1 mM IPTG for 4 h at 37°C with gentle shaking. Equal numbers of cells were then spotted onto LB agar (control) and LB agar containing chloramphenicol (Cm) and kanamycin (Kan) (protein exchange) and photographed after 18 h. For plasmid exchange assay, pairs of a donor (GD110: amyE::Phyper-spank-cat-spec, pHB201/cat, erm) (CmR, SpecR, MlsR) and a recipient (SB513: amyE::Phyper-spank-gfp-kan) (KanR) parental strains (wild-type) were used, with the investigated mutants harbor in addition the indicated null mutation in both donor and recipient strains. Cells were mixed in 1:1 ratio (concentration 1×), processed as described for protein exchange, and spotted onto LB agar containing Cm, Kan, and lincomycin (Lin) (plasmid exchange). Cells were incubated at 37°C, and colonies were photographed after 36 h of incubation. For motility assay, wild-type (PY79) and the indicated mutant strains were grown to the mid-logarithmic phase and spotted onto LB plates containing 0.3% agar and photographed after 7 h of incubation at 37°C (motility). See also Figures S1 and S2.
Next, we examined the impact of CORE on nanotube-dependent interspecies interactions. We have previously shown that Bs kills neighboring B. megaterium (Bm) cells by delivering the tRNase toxin WapA via nanotubes (Stempler et al., 2017). GFP-labeled Bs was mixed with Bm, and cells were visualized using time-lapse microscopy. Bs cells, lacking CORE, indeed failed to arrest Bm growth and division, inferring that the transfer of WapA toxin into Bm cells was impeded (Figure S2B). Consistently, ectopic expression of CORE genes restored this capability (Figure S2B). Thus, interspecies delivery of WapA via nanotubes is CORE dependent.
CORE Components Are Localized to the Site of Nanotube Emergence
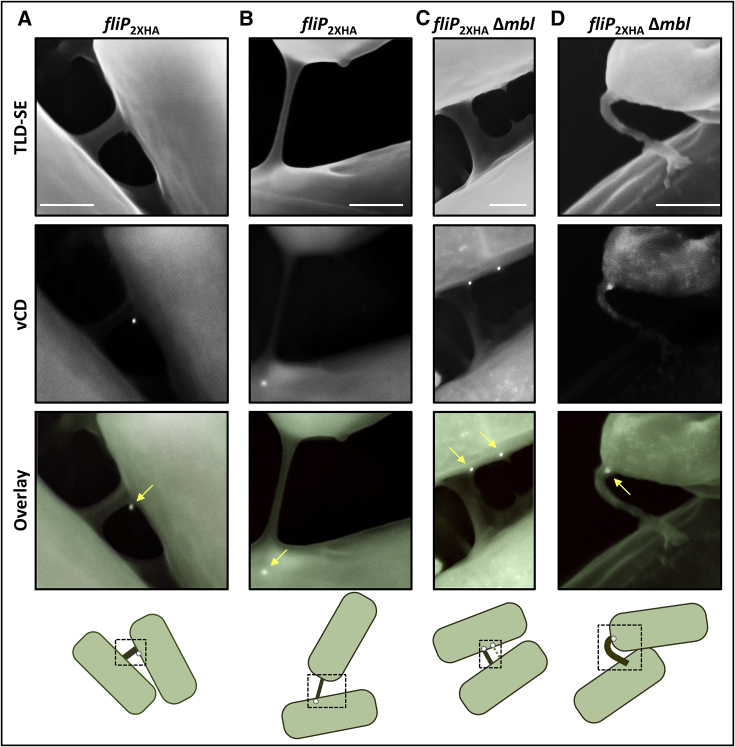
Since the CORE complex is a prime constituent of the flagellar basal body (Diepold and Wagner, 2014), we posited that in a similar fashion, the CORE forms a platform from which nanotubes emerge and extend. The major CORE component FlhA, harboring a large cytoplasmic domain (Figure 1A) (Morimoto et al., 2014), was fused to GFP. The fusion rendered the protein partially functional (Figures S3A–S3C); however, the few nanotubes produced by these cells emerged from sites adjacent to FlhA-GFP focal assemblies (Figures S3D and S3E), hinting that nanotubes are projected from CORE-containing complexes. To further establish this notion, we co-visualized nanotubes and CORE components at high resolution by using immuno-XHR-SEM. To this end, FliP was HA-tagged in its C terminus, predicted to be exposed at the cell surface (Fukumura et al., 2017) (Figure 1A), and introduced into the Bs genome ectopically. Next, cells were subjected to immuno-XHR-SEM, using primary anti-HA antibody followed by gold-labeled secondary antibodies. Strikingly, a gold particle was recurrently observed at the site of nanotube emanation, signifying that indeed the CORE serves as the basal body for nanotube construction (Figures 3A and 3B). Consistently, signal from FliP was also detected at the origin of extending nanotubes (Figure S4A). Tagging FliP at a site predicted to form a surface-exposed loop (Fukumura et al., 2017) showed a similar outcome (Figure S4B). Furthermore, use of a mutant (Δmbl) exhibiting a defect in cell wall assembly (Jones et al., 2001) to improve antibody accessibility to CORE complexes, or a strain harboring tagged fliP as a sole copy, yielded a similar gold labeling pattern (Figures 3C, 3D, and S4C). Significantly, in all viewed cases (25), the signal from FliP was restricted to only one end of a connecting nanotube, suggesting that the CORE is exploited for nanotube projection by the producing cell, while docking on a recipient cell is CORE independent. Of note, detection of only a single gold particle (18 nm) associated with the FliP multimeric ring (10 nm in diameter; Fukumura et al., 2017, Kuhlen et al., 2018) is most likely due to resolution limit or steric hindrance. As a control, no gold signal was obtained from cells lacking the HA tagged protein (Figure S4D) or from cells expressing a membrane protein tagged at a site facing the cytoplasm (Figures S4E and S4F) (Rudner et al., 2002). Furthermore, cells harboring a surface-exposed tag in an unrelated membrane protein yielded a scattered pattern, which did not coincide with nanotube emanation sites (Tzipilevich et al., 2017) (Figures S4G and S4H). Hence, we conclude that the CORE provides a platform for nanotube emergence.
Figure 3.
FliP Localizes to the Base of Nanotubes
Cells expressing HA-tagged FliP (SH93: amyE::Phyper-spank-fliP2xHA, sacA::Phyper-spank-ymdB, A and B, or SH110: amyE::Phyper-spank-fliP2xHA, sacA::Phyper-spank-ymdB, Δmbl, C and D) were spotted onto EM grids and subjected to immuno-gold XHR-SEM, using primary antibodies against HA and secondary gold-conjugated antibodies. Samples were not coated before observation. Examples of FliP2xHA localization (white dots) at the base of nanotubes are presented (indicated by arrows). Shown are XHR-SEM images that were acquired using TLD-SE (through-lens detector-secondary electron) for nanotubes visualization and vCD (low-kV high-contrast backscattered detector) for gold particle detection, as well as overlay of both images. Schematic below depicts the interpretive cell layout and highlights the nanotube region with gold signal (dashed box) captured by XHR-SEM. Scale bars represent 200 nm. See also Figures S3 and S4.
Flagellar CORE Complexes Are Used for Nanotube Formation by Diverse Species
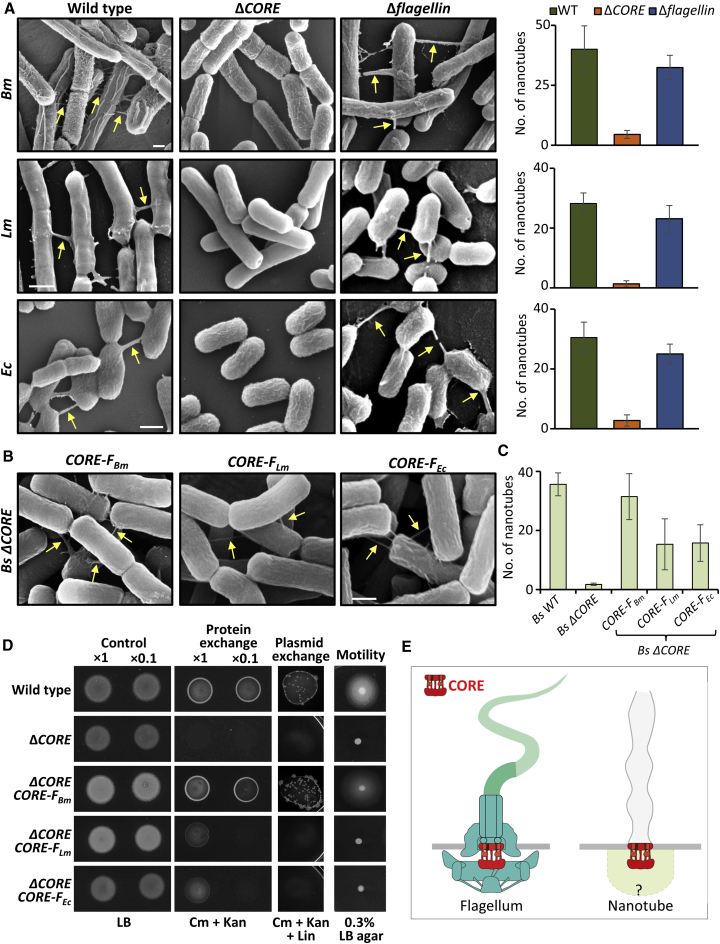
On the basis of our results, we asked whether the flagellar CORE of other bacterial species is bi-functional and similarly to CORE-FBs can give rise to both flagella and nanotubes. We thus deleted the CORE genes from the genomes of Bm, a close relative of Bs, the Gram-positive human pathogen Listeria monocytogenes (Lm), and the evolutionary distant Escherichia coli (Ec) (Table S1). Wild-type, ΔCORE, and mutants lacking the flagellum filament (Δflagellin) from all tested species were incubated on a solid surface to induce nanotube formation and subsequently examined using XHR-SEM. While nanotube appendages were plentiful in wild-type and in strains devoid of flagellum filaments, they were absent from the corresponding CORE mutants (Figure 4A), hence corroborating that the flagellar CORE is used by diverse species for nanotube production.
Figure 4.
CORE Is a Widespread Complex with Conserved Functions across Species
(A) Wild-type Bm (OS2), Lm (10403S), and Ec (MG1655) and their corresponding mutant strains lacking CORE (ΔCORE) or gene encoding flagellin (Δflagellin) were grown to the mid-logarithmic phase, spotted onto EM grids followed by incubation on LB agar plates for 4 h at 37°C, and visualized using XHR-SEM. Arrows indicate nanotubes. Scale bar represents 500 nm. Right: quantification of the average number of nanotubes displayed per 50 cells by the indicated strains following XHR-SEM analysis. Shown are average values and SD of at least three independent experiments (n ≥ 200 for each strain).
(B) Cells of Bs (ΔCORE) complemented with Bm flagellar CORE (CORE-FBm), Lm flagellar CORE (CORE-FLm), and Ec flagellar CORE (CORE-FEc) were processed as in (A) and visualized using XHR-SEM. Arrows indicate nanotubes. Scale bar represents 500 nm.
(C) Quantification of the average number of nanotubes displayed per 50 cells by the indicated strains following XHR-SEM analysis described in (B). Shown are average values and SD of at least three independent experiments (n ≥ 200 for each strain).
(D) Assessing molecular exchange in Bs ΔCORE strain complemented with exogenous CORE genes. For protein exchange assay, pairs of a donor (SB463: amyE::Phyper-spank-cat-spec) (CmR, SpecR) and a recipient (SB513: amyE::Phyper-spank-gfp-kan) (KanR) parental strains (wild-type) were used. The investigated strains ΔCORE and ΔCORE complemented with CORE-FBm, CORE-FLm, and CORE-FEc (controlled by Bs Pfla/che promoter) harbor the corresponding genotypes of donor and recipient. Donor and recipient strains were mixed in 1:1 ratio (at two concentrations, 1× and 0.1×) and incubated in LB supplemented with 1 mM IPTG for 4 h at 37°C with gentle shaking. Equal numbers of cells were then spotted onto LB agar (control) and LB agar containing chloramphenicol (Cm) and kanamycin (Kan) (protein exchange) and photographed after 18 h. For plasmid exchange assay, pairs of a donor (GD110: amyE::Phyper-spank-cat-spec, pHB201/cat, erm) (CmR, SpecR, MlsR) and a recipient (SB513: amyE::Phyper-spank-gfp-kan) (KanR) parental strains (wild-type) were used. The investigated strains complemented with exogenous COREs harbor the corresponding genotypes of donor and recipient strains. Cells were mixed in 1:1 ratio (concentration 1×), processed as described for protein exchange, and spotted onto LB agar containing Cm, Kan, and lincomycin (Lin) (plasmid exchange). Cells were incubated at 37°C, and colonies were photographed after 36 h of incubation. For motility assay, wild-type (PY79) and the indicated strains were grown to mid-logarithmic phase, spotted onto LB plates containing 0.3% agar, and photographed after 7 h of incubation at 37°C (motility).
(E) A schematic model depicting the modularity of CORE complexes in flagellum and nanotube. CORE-associated components of the nanotube basal body are missing. Flagellum structure was adapted from (Dietsche et al., 2016).
We then investigated the cross-species functional conservation of COREs by testing the capacity of foreign flagellar CORE complexes to complement nanotube formation by CORE deficient Bs. Introducing CORE-FBm genes into Bs ΔCORE mutant prompted the formation of intercellular nanotubular structures to a level close to that of wild-type Bs (Figures 4B and 4C). Compellingly, also CORE-FLm and CORE-FEc genes induced the formation of nanotubes in Bs ΔCORE, although of an apparent thin morphology (Figures 4B and 4C). Interestingly, however, motility was reestablished by CORE-FBm but could not be restored by CORE-FLm or CORE-FEc (Figure 4D). Consistent with the complete restoration of nanotubes by CORE-FBm, both protein and plasmid exchange levels were regained to levels comparable with that of wild-type Bs (Figure 4D). In line with the limited nanotube restitution (Figure 4C), CORE-FLm and CORE-FEc enabled partial protein exchange but could not support plasmid exchange (Figure 4D), indicating that additional non-CORE components that differ among species are involved in this process. Taken together, our results suggest that distinct bacterial species carrying flagellar CORE genes have the potential to form nanotubes.
CORE Complexes Are Universal and Phylogenetically Widespread
We next addressed the ubiquity of the CORE genes across the bacterial kingdom. Since previous such analyses included primarily genomes of Gram-negative bacteria (Abby and Rocha, 2012, Hu et al., 2017, Snyder et al., 2009), here we attempted to include representatives of the majority of the bacterial phyla. Using the STRING database homology data (Szklarczyk et al., 2017, von Mering et al., 2007), we analyzed 400 species belonging to 18 phyla. We found CORE genes to be widespread in all the examined bacterial phyla, typically as part of the flagellum or its homologous injectisome apparatus (Figure S5; Table S2). Interestingly, some bacteria lacking a substantial fraction of the gene cohort required for formation of these organelles still harbor the CORE genes that could function to nucleate exclusively nanotube biogenesis. These bacteria include Myxococci species (phylum Proteobacteria), Chloracidobacterium thermophilum (phylum Acidobacteria), and Succinatimonas hippie (phylum Proteobacteria) (Figure S5; Table S2) (Abby and Rocha, 2012, Hu et al., 2017). Overall, this examination emphasizes that a wide spectrum of species possesses the potential to produce nanotubes and use them for molecular exchange.
Discussion
We uncovered that the export apparatus of the flagella, designated CORE, is a dual-functional complex, communally serving as a foundation for both flagella and nanotube generation (Figure 4E). This complex is likely to promote flagella formation by planktonic bacteria but alternates to support nanotube formation by bacteria grown on solid surfaces. The functional conservation of CORE complexes from distinct bacterial species was revealed by their ability to support nanotube formation in Bs mutant lacking endogenous CORE genes. Our findings insinuate that CORE-dependent molecular trafficking among bacteria is extensive and broad and includes a large array of cytoplasmic molecules. As such, it fundamentally expands the bacterial metabolic flexibility, facilitating the acquisition of new features. Furthermore, the observation that even plasmid trafficking uses CORE-dependent nanotubes highlights the potential impact of CORE complexes on horizontal gene transfer in nature. In the accompanying study, we discovered a similar set of highly conserved injectisome CORE proteins in EPEC to be implicated in extraction of cytoplasmic molecules from infected human host cells via nanotubes (Pal et al., 2019). On the basis of our findings, we postulate that the CORE represents an evolutionary ancestral functional unit, which subsequently evolved by acquisition of additional components, into nanotube, flagellum, or injectisome.
Some bacteria, including Vibrio cholerae and Helicobacter pylori, form flagella encased by membranous protrusions, termed “sheathed flagella.” Intriguingly, these bacteria were reported to occasionally produce “empty sheaths,” with a structure highly resembling nanotubes (Allen and Baumann, 1971, Geis et al., 1993, McCarter, 2001). Furthermore, the existence of isolated orphan CORE components in various bacterial genomes, frequently referred to as truncated or incomplete systems (Pallen and Matzke, 2006, Ren et al., 2005), insinuates their potential nanotube designation.
The discovery that CORE serves as a platform for nanotube biogenesis could provide the foundation for elucidating central aspects of nanotube functionality, such as the selection of the delivered molecular cargo, the source of energy used for cargo transportation, the directionality of nanotube operation, and the fusion mechanism with recipient cell membrane. Further mechanistic insight into nanotube biogenesis and function may escort the development of innovative methodologies, enabling control of intercellular molecular trade among bacteria, to attain intelligent bacterial community design.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal anti-HA antibodies (Rabbit) | Thermo Fisher Scientific | Cat#:71-5500; RRID: AB_2533988 |
| Polyclonal anti-GFP antibodies (Rabbit) | Laboratory stock (Dubey and Ben-Yehuda, 2011) | NA |
| 18 nm gold-conjugated IgG secondary antibody Goat anti-Rabbit. | Jackson ImmunoResearch Laboratories | Cat#: 111-215-144; RRID: AB_2338017 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Chloramphenicol | Sigma-Aldrich | Cat#: C0378 |
| Tetracycline | Sigma-Aldrich | Cat#: 87128 |
| Kanamycin | US Biological | Cat#: K0010 |
| Lincomycin | Sigma-Aldrich | Cat#: 62143-5G |
| Erythromycin | Sigma-Aldrich | Cat#: E0774 |
| Spectinomycin | Sigma-Aldrich | Cat#: S4014-5G |
| Ampicillin | Sigma-Aldrich | Cat#: A9518-25G |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Sigma-Aldrich | Cat#: I6758-5G |
| D-Xylose | Sigma-Aldrich | Cat#: X1500-1KG |
| Sucrose | J.T.Baker | Cat#: 4072-05 |
| Polyethylene glycol 8000 | Promega | Cat#: V3011 |
| D-Sorbitol | Sigma-Aldrich | Cat#: S6021 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#: 15710 |
| Glutaraldehyde | Electron Microscopy Sciences | Cat#: 16020 |
| Sodium Cacodylate Buffer (pH 7.4) | Electron Microscopy Sciences | Cat#: 11650 |
| FM4-64 | Molecular probes/ Thermo Fisher Scientific | Cat#: T13320 |
| Critical Commercial Assays | ||
| Q5 High-Fidelity DNA Polymerase | NEW ENGLAND BioLabs | Cat#: M0491S |
| Gibson Assembly Master Mix | NEW ENGLAND BioLabs | Cat#: E2611L |
| Quick Ligation Kit | NEW ENGLAND BioLabs | Cat#: M2200S |
| RQ1 RNase-free DNase | Promega | Cat#: M6101 |
| qScript cDNA synthesis kit | Quanta Biosciences | Cat#: 95047-25 |
| PerfeCTa SYBR Green FastMix | Quanta Biosciences | Cat#: 95073-250 |
| Experimental Models: Organisms/Strains | ||
| PY79 (B. subtilis wild type) | Youngman et al., 1984 | NA |
| AR16 (amyE::PrrnE-gfp-spec) | Rosenberg et al., 2012 | NA |
| GD215 (Δhag::erm) | Dubey and Ben-Yehuda, 2011 | NA |
| GD110 (amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | Dubey and Ben-Yehuda, 2011 | NA |
| GD267 (Δhag::erm, amyE::Phyper-spank-cat-spec) | Laboratory stock | NA |
| GD268 (Δhag::erm, amyE::Phyper-spank-gfp-kan) | Laboratory stock | NA |
| SB463 (amyE::Phyper-spank-cat-spec) | Dubey and Ben-Yehuda, 2011 | NA |
| SB513 (amyE::Phyper-spank-gfp-kan) | Dubey and Ben-Yehuda, 2011 | NA |
| GB61 (ΔymdB::tet) | Dubey et al., 2016 | NA |
| GB168 (ΔymdB::tet, amyE::Phyper-spank-ymdB-spec, Δhag::erm) | Dubey et al., 2016 | NA |
| IB11 (Δmbl::erm) | Bejerano-Sagie et al., 2006 | NA |
| ET13 (amyE::Phyper-spank-yueB-yfp-spec) | Tzipilevich et al., 2017 | NA |
| BDR524 (amyE::PxylA-spoIVFB-gfp-cat) | Rudner et al., 2002 | NA |
| SH8 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD) | This study | NA |
| SH9 [ΔCORE (fliO-flhA)::tet, Pfla/che-flhF-cheD] | This study | NA |
| SH30 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE::PrrnE-gfp-spec) | This study | NA |
| SH31 (sacA::Pfla/che-fliO-flhA-spec) | This study | NA |
| SH33 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec) | This study | NA |
| SH47 (flhA-gfp-kan) | This study | NA |
| SH55 (flhA-gfp-kan, ΔymdB::tet, amyE::Phyper-spank-ymdB-spec, Δhag::erm) | This study | NA |
| SH79 (sacA::Phyper-spank-ymdB-kan) | This study | NA |
| SH86 (amyE::Phyper-spank-fliP2xHA-spec) | This study | NA |
| SH93 (amyE::Phyper-spank-fliP2xHA-spec, sacA::Phyper-spank-ymdB-kan) | This study | NA |
| SH103 (ΔfliO::tet, Pfla/che-fliP-cheD) | This study | NA |
| SH104 (ΔfliP::tet, Pfla/che-fliQ-cheD) | This study | NA |
| SH105 (ΔfliQ::tet, Pfla/che-fliR-cheD) | This study | NA |
| SH106 (ΔfliR::tet, Pfla/che-flhB-cheD) | This study | NA |
| SH107 (ΔflhB::tet, Pfla/che-flhA-cheD) | This study | NA |
| SH108 (ΔflhA::tet, Pfla/che-flhF-cheD) | This study | NA |
| SH110 (amyE::Phyper-spank-fliP2xHA-spec, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) | This study | NA |
| SH115 [sacA::Pfla/che-CORE-FEc (fliOPQRflhBA)-spec] | This study | NA |
| SH116 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FEc (fliOPQRflhBA)-spec] | This study | NA |
| SH150 (amyE::Phyper-spank-fliP2xHA-LOOP-spec) | This study | NA |
| SH151 (amyE::Phyper-spank-fliP2xHA-LOOP-spec, sacA::Phyper-spank-ymdB-kan) | This study | NA |
| SH161 (amyE::Phyper-spank-fliP2xHA-LOOP-spec, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) | This study | NA |
| SH169 [amyE::Phyper-spank-CORE-FEc (fliOPQRflhBA)-spec] | This study | NA |
| SH170 [ΔfliO-flhA::tet, amyE::Phyper-spank-CORE-FEc (fliOPQRflhBA)-spec] | This study | NA |
| SH177 (ΔfliI::tet, Pfla/che-fliJ-cheD) | This study | NA |
| SH203 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec) | This study | NA |
| SH204 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec) | This study | NA |
| SH205 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec) | This study | NA |
| SH206 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec) | This study | NA |
| SH207 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec) | This study | NA |
| SH208 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec) | This study | NA |
| SH209 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec] | This study | NA |
| SH210 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec] | This study | NA |
| SH244 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec) | This study | NA |
| SH245 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP2xHA-spec) | This study | NA |
| SH247 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP2xHA-spec, Δmbl::erm) | This study | NA |
| SH255 (amyE::Phyper-spank-yueB-yfp-spec, sacA::Phyper-spank-ymdB-kan) | This study | NA |
| SH256 (amyE::PxylA-spoIVFB-gfp-cat, sacA::Phyper-spank-ymdB-kan) | This study | NA |
| SH257 (amyE::Phyper-spank-yueB-yfp-spec, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) | This study | NA |
| SH258 (amyE::PxylA-spoIVFB-gfp-cat, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) | This study | NA |
| SH12 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH13 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH41 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH119 (ΔfliO::tet, Pfla/che-fliP-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH120 (ΔfliP::tet, Pfla/che-fliQ-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH121 (ΔfliQ::tet, Pfla/che-fliR-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH122 (ΔfliR::tet, Pfla/che-flhB-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH123 (ΔflhB::tet, Pfla/che-flhA-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH124 (ΔflhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH131 [ΔfliO-flhA::tet, sacA:: Pfla/che-CORE-FEc (fliOPQRflhBA)-spec, amyE::Phyper-spank-gfp-kan] | This study | NA |
| SH182 (ΔfliI::tet, Pfla/che-fliJ-cheD, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH213 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH214 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH215 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH216 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH217 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH218 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH219 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec, amyE::Phyper-spank-gfp-kan] | This study | NA |
| SH220 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec, amyE::Phyper-spank-gfp-kan] | This study | NA |
| SH248 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec, amyE::Phyper-spank-gfp-kan) | This study | NA |
| SH16 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD, amyE::Phyper-spank -cat-spec) | This study | NA |
| SH17 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank -cat-spec) | This study | NA |
| SH58 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH125 (ΔfliO::tet, Pfla/che-fliP-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH126 (ΔfliP::tet, Pfla/che-fliQ-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH127 (ΔfliQ::tet, Pfla/che-fliR-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH128 (ΔfliR::tet, Pfla/che-flhB-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH129 (ΔflhB::tet, Pfla/che-flhA-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH130 (ΔflhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH132 [ΔfliO-flhA::tet, sacA:: Pfla/che-CORE-FEc (fliOPQRflhBA)-spec, amyE::Phyper-spank-cat-spec] | This study | NA |
| SH183 (ΔfliI::tet, Pfla/che-fliJ-cheD, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH223 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH224 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH225 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH226 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH227 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH228 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH229 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec, amyE::Phyper-spank-cat-spec] | This study | NA |
| SH230 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec, amyE::Phyper-spank-cat-spec] | This study | NA |
| SH249 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec, amyE::Phyper-spank-cat-spec) | This study | NA |
| SH22 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH23(ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH62 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH134 (ΔfliO::tet, Pfla/che-fliP-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH135 (ΔfliP::tet, Pfla/che-fliQ-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH136 (ΔfliQ::tet, Pfla/che-fliR-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH137 (ΔfliR::tet, Pfla/che-flhB-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH138 (ΔflhB::tet, Pfla/che-flhA-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH139 (ΔflhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH181 (Δhag::erm, pHB201/cat, erm) | This study | NA |
| SH185 (ΔfliI::tet, Pfla/che-fliJ-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH233 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH234 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec, amyE::Phyper-spank-cat-spe, pHB201/cat, erm) | This study | NA |
| SH235 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec, amyE::Phyper-spank-cat-spe, pHB201/cat, erm) | This study | NA |
| SH236 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec, amyE::Phyper-spank-cat-spe, pHB201/cat, erm) | This study | NA |
| SH237 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH238 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| SH239 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm] | This study | NA |
| SH240 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm] | This study | NA |
| SH250 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) | This study | NA |
| OS2 (Bacillus megaterium wild type isolate) | Stempler et al., 2017 | NA |
| Bm ΔCORE (fliO-flhA) | This study | NA |
| Bm Δflagellin (hag) | This study | NA |
| Escherichia coli K-12 MG1655 | Laboratory stock | NA |
| XTL634 (Template for tet::sacB cassette) | Li et al., 2013 | NA |
| RP7802 [Escherichia coli K-12 MG1655 Δ fliOPQR] | This study | NA |
| RP7809 [Escherichia coli K-12 MG1655 ΔCORE (fliOPQRflhBA)] | This study | NA |
| Ec DY378 (1974) [W3110 λcI857, Δ(cro-bioA)] | Yu et al., 2000 | |
| SK2462 (Ec DY378 ΔfliC::kan) | This study | NA |
| SK2468 [Ec K-12 MG1655 Δflagellin (fliC::kan)] | This study | NA |
| Listeria monocytogenes 10403S | Kindly provided by Ran Nir-Paz (Hebrew U) | NA |
| Lm 10403S ΔCORE (fliP-flhA) | This study | NA |
| Lm 10403S Δ flagellin (flaA) | This study | NA |
| Oligonucleotides | ||
| Primers used in this study are listed in Table S3. | All primers were designed during this study, and synthesized by Integrated DNA Technologies (IDT). | NA |
| Recombinant DNA | ||
| pDR111 (amyE::Phyper-spank-spec) | Kindly provided by David Rudner (Harvard U) | NA |
| pHB201 (cat, erm) | Bron et al., 1998 | NA |
| pKL168 (gfp-kan) | Lemon and Grossman, 1998 | NA |
| pKD46 (λ RED genes, Ampr) | Datsenko and Wanner, 2000 | NA |
| pRP7358 [Ptac-CORE-FEc (fliOPQRflhBA)] | Pal et al., 2019 | NA |
| pSH13 (flhA-gfp-kan) | This study | NA |
| pSH17 (amyE::Phyper-spank-fliP2xHA-spec) | This study | NA |
| pSH19 (amyE::Phyper-spank-fliP2xHA-LOOP-spec) | This study | NA |
| pSH21 [amyE::Phyper-spank- CORE-FEc (fliOPQRflhBA)-spec] | This study | NA |
| pLR16- Phes | Kindly provided by Anat Herskovits (Tel Aviv U) (Argov et al., 2017) | NA |
| pLR16-AB1 | This study | NA |
| pSH22 (pLR16-Lm flaA int) | This study | NA |
| pSH23 (pDG1514-Bm hag int) | This study | NA |
| Other | ||
| Carbon film on 300 square mesh copper grids | Electron Microscopy Sciences | Cat#: CF300-Cu |
| Lysing Matrix B Bulk | MP Biomedicals | Cat#: 6540-428 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sigal Ben-Yehuda (sigalb@ekmd.huji.ac.il).
Experimental Model and Subject Details
Details on bacterial strain construction
Bs strains are derivatives of the wild-type strain PY79, Bm is a wild-type soil isolate (OS2), Ec strains are derivatives of the wild-type strain K-12 MG1655, Lm strains are derivatives of the wild-type strain 10403S. Bacterial strains and plasmids are listed in Key resources table, and primers are listed in Table S3.
For gene replacement strategy in Bs, indicated primer pairs (P1-P4; Table S3) were used to amplify the flanking genomic regions of the corresponding gene. The fla/che promoter region was amplified using the primers PflgB1-2. PCR products were used for Gibson assembly (NEB, USA), together with the respective antibiotic resistance gene (Guérout-Fleury et al., 1996) The resultant product was used to transform PY79 to obtain the mutant allele.
SH8 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers flgB-fliF KO P1-P4 and PflgB1-2. SH9 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers CORE KO P1-P4 and PflgB1-2. SH12 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH8 with genomic DNA (gDNA) from strain SB513. SH13 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH9 with gDNA from strain SB513. SH16 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD, amyE:: Phyper-spank-cat-spec) was constructed by transforming SH8 with gDNA from strain SB463. SH17 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE:: Phyper-spank -cat-spec) was constructed by transforming SH9 with gDNA from strain SB463. SH22 (ΔflgB-fliF::tet, Pfla/che-fliG-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH8 with pHB201. SH23 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH9 with pHB201. SH30 (ΔfliO-flhA::tet,Pfla/che-flhF-cheD, amyE::PrrnE-gfp-spec) was constructed by transforming gDNA SH9 with from strain AR16. SH31 (sacA::Pfla/che-fliO-flhA-spec) was constructed using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, Bs CORE comp1-2 and Comp PflgB1-2. SH33 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec) was constructed by transforming SH9 with gDNA from strain SH31. SH41 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH33 with gDNA from strain SB513. SH47 (flhA-gfp-kan) was constructed by transforming PY79 with pSH13. SH55 (flhA-gfp-kan, ΔymdB::tet, amyE::Phyper-spank-ymdB-spec, Δhag::erm) was constructed by transforming GB168 with gDNA from strain SH47. SH58 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH33 with gDNA from strain SB463. SH62 (ΔfliO-flhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-fliO-flhA-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH58 with pHB201. SH79 (sacA::Phyper-spank-ymdB-kan) was constructed using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4 and Comp PHS1-ymdB R. Phyper-spank-ymdB was amplified from GB168 gDNA using Comp PHS1-ymdB R primers. SH86 (amyE::Phyper-spank-fliP2xHA-spec) was constructed by transforming PY79 with pSH17. SH93 (amyE::Phyper-spank-fliP2xHA-spec, sacA::Phyper-spank-ymdB-kan) was constructed by transforming SH79 with gDNA from strain SH86. SH96 (flhA-gfp-kan, amyE::Phag-hagT209C-spec) was constructed by transforming DS1895 with gDNA from strain SH47. SH103 (ΔfliO::tet, Pfla/che-fliP-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers fliO KO P1-P4 and PflgB1-2. SH104 (ΔfliP::tet, Pfla/che-fliQ-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers fliP KO P1-P4 and PflgB1-2. SH105 (ΔfliQ::tet, Pfla/che-fliR-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers fliQ KO P1-P4 and PflgB1-2. SH106 (ΔfliR::tet, Pfla/che-flhB-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers fliR KO P1-P4 and PflgB1-2. SH107 (ΔflhB::tet, Pfla/che-flhA-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers flhB KO P1-P4 and PflgB1-2. SH108 (ΔflhA::tet, Pfla/che-flhF-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers flhA KO P1-P4 and PflgB1-2. SH110 (amyE::Phyper-spank-fliP2xHA-spec, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) was constructed by transforming SH93 with gDNA from strain IB11. SH115 [sacA::Pfla/che-CORE-FEc (fliOPQRflhBA)-spec] was constructed using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, Ec CORE comp 1-2 and Comp PflgB1-2a. CORE-FEc (fliOPQRflhBA) was amplified from pRP7358 using primers Ec CORE comp 1-2. SH116 [ΔfliO-flhA::tet, sacA: Pfla/che-CORE-FEc (fliOPQRflhBA)-spec] was constructed by transforming SH9 with gDNA from strain SH115. SH119 (ΔfliO::tet, Pfla/che-fliP-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH103 with gDNA from strain SB513. SH120 (ΔfliP::tet, Pfla/che-fliQ-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH104 with gDNA from strain SB513. SH121 (ΔfliQ::tet, Pfla/che-fliR-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH105 with gDNA from strain SB513. SH122 (ΔfliR::tet, Pfla/che-flhB-cheD, amyE::Phyper-spank-gfp-kan)was constructed by transforming SH106 with gDNA from strain SB513. SH123 (ΔflhB::tet, Pfla/che-flhA-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH107 with gDNA from strain SB513. SH124 (ΔflhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH108 with gDNA from strain SB513. SH125 (ΔfliO::tet, Pfla/che-fliP-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH103 with gDNA from strain SB463. SH126 (ΔfliP::tet, Pfla/che-fliQ-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH104 with gDNA from strain SB463. SH127 (ΔfliQ::tet, Pfla/che-fliR-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH105 with gDNA from strain SB463. SH128 (ΔfliR::tet, Pfla/che-flhB-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH106 with gDNA from strain SB463. SH129 (ΔflhB::tet, Pfla/che-flhA-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH107 with gDNA from strain SB463. SH130 (ΔflhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH108 with gDNA from strain SB463. SH131 [ΔfliO-flhA::tet, sacA:: Pfla/che-CORE-FEc (fliOPQRflhBA)-spec, amyE::Phyper-spank-gfp-kan] was constructed by transforming SH116 with gDNA from strain SB513. SH132 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FEc (fliOPQRflhBA)-spec, amyE::Phyper-spank-cat-spec] was constructed by transforming SH116 with gDNA from strain SB463. SH134 (ΔfliO::tet, Pfla/che-fliP-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH125 with pHB201. SH135 (ΔfliP::tet, Pfla/che-fliQ-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH126 with pHB201. SH136 (ΔfliQ::tet, Pfla/che-fliR-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH127 with pHB201. SH137 (ΔfliR::tet, Pfla/che-flhB-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH128 with pHB201. SH138 (ΔflhB::tet, Pfla/che-flhA-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH129 with pHB201. SH139 (ΔflhA::tet, Pfla/che-flhF-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH130 with pHB201. SH150 (amyE::Phyper-spank-fliP2xHA-LOOP-spec) was constructed by transforming PY79 with pSH19. SH151 (amyE::Phyper-spank-fliP2xHA-LOOP-spec, sacA::Phyper-spank-ymdB-kan) was constructed by transforming SH79 with gDNA from strain SH150. SH161 (amyE::Phyper-spank-fliP2xHA-LOOP-spec, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) was constructed by transforming SH150 with gDNA from strain IB11. SH169 [amyE::Phyper-spank-CORE-FEc (fliOPQRflhBA)-spec] was constructed by transforming PY79 with pSH21. SH170 [ΔfliO-flhA::tet, amyE::Phyper-spank-CORE-FEc (fliOPQRflhBA)-spec] was constructed by transforming SH9 with gDNA from strain SH169. SH177 (ΔfliI::tet, Pfla/che-fliJ-cheD) was constructed using Gibson assembly kit (NEB, USA) utilizing primers fliI KO P1-P4 and PflgB1-2. SH181 (Δhag::erm, pHB201/cat, erm) was constructed by transforming GD215 with pHB201. SH182 (ΔfliI::tet, Pfla/che-fliJ-cheD, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH177 with gDNA from strain SB513. SH183 (ΔfliI::tet, Pfla/che-fliJ-cheD, amyE::Phyper-spank-cat-spec) was constructed by transforming SH177 with gDNA from strain SB463. SH185 (ΔfliI::tet, Pfla/che-fliJ-cheD, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH183 with pHB201. SH203 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec) was constructed by transforming SH103 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, fliO comp1-2 and Comp PflgB1-R. SH204 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec) was constructed by transforming SH104 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, fliP comp1-2 and Comp PflgB1-R. SH205 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec) was constructed by transforming SH105 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, fliQ comp1-2 and Comp PflgB1-R. SH206 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec) was constructed by transforming SH106 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, fliR comp1-2 and Comp PflgB1-R. SH207 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec) was constructed by transforming SH107 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, flhB comp1-2 and Comp PflgB1-R. SH208 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec) was constructed by transforming SH108 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, flhA comp1-2 and Comp PflgB1-R. SH209 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec] was constructed by transforming SH9 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, Lm CORE comp1-2 and Comp PflgB1-R. CORE-FLm (fliPQRflhBA) was amplified from Lm (10403S) gDNA using primers Lm CORE comp 1-2. SH210 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec] was constructed by transforming SH9 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, Bm CORE comp1-2 and Comp PflgB1-R. CORE-FBm (fliOPQRflhBA) was amplified from Bm (OS2) gDNA using primers Bm CORE comp 1-2. SH213 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH203 with gDNA from strain SB513. SH214 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH204 with gDNA from strain SB513. SH215 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH205 with gDNA from strain SB513. SH216 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH206 with gDNA from strain SB513. SH217 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH207 with gDNA from strain SB513. SH218 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH208 with gDNA from strain SB513. SH219 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec, amyE::Phyper-spank-gfp-kan] was constructed by transforming SH209 with gDNA from strain SB513. SH220 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec, amyE::Phyper-spank-gfp-kan] was constructed by transforming SH210 with gDNA from strain SB513. SH223 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH203 with gDNA from strain SB463. SH224 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH204 with gDNA from strain SB463. SH225 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH205 with gDNA from strain SB463. SH226 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH206 with gDNA from strain SB463. SH227 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH207 with gDNA from strain SB463. SH228 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH208 with gDNA from strain SB463. SH229 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm(fliPQRflhBA)-spec, amyE::Phyper-spank-cat-spec] was constructed by transforming SH209 with gDNA from strain SB463. SH230 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec, amyE::Phyper-spank-cat-spec] was constructed by transforming SH210 with gDNA from strain SB463. SH233 (ΔfliO::tet, Pfla/che-fliP-cheD, sacA::Pfla/che-fliO-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH223 with pHB201. SH234 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliP-spec, amyE::Phyper-spank-cat-spe, pHB201/cat, erm) was constructed by transforming SH224 with pHB201. SH235 (ΔfliQ::tet, Pfla/che-fliR-cheD, sacA::Pfla/che-fliQ-spec, amyE::Phyper-spank-cat-spe, pHB201/cat, erm) was constructed by transforming SH225 with pHB201. SH236 (ΔfliR::tet, Pfla/che-flhB-cheD, sacA::Pfla/che-fliR-spec, amyE::Phyper-spank-cat-spe, pHB201/cat, erm) was constructed by transforming SH226 with pHB201. SH237 (ΔflhB::tet, Pfla/che-flhA-cheD, sacA::Pfla/che-flhB-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH227 with pHB201. SH238 (ΔflhA::tet, Pfla/che-flhF-cheD, sacA::Pfla/che-flhA-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH228 with pHB201. SH239 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FLm (fliPQRflhBA)-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm] was constructed by transforming SH229 with pHB201. SH240 [ΔfliO-flhA::tet, sacA::Pfla/che-CORE-FBm (fliOPQRflhBA)-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm] was constructed by transforming SH230 with pHB201. SH244 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec) was constructed by transforming SH104 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, fliO comp 1-fliP comp 2 and Comp PflgB1-R. SH245 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP2xHA-spec) was constructed by transforming SH104 with complementation construct generated using Gibson assembly kit (NEB, USA) utilizing primers sacA P1-P4, Comp PflgB1-R, and fliOP2xHA construct. fliOP was amplified using fliO comp 1 and fliP ORF R (2X HA), followed by a second round of PCR using primers fliP comp 1 and 2X HA R to finally obtain fliOP2xHA construct. SH247 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP2xHA-spec, Δmbl::erm) was constructed by transforming SH245 with gDNA from strain IB11. SH248 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec, amyE::Phyper-spank-gfp-kan) was constructed by transforming SH244 with gDNA from strain SB513. SH249 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec, amyE::Phyper-spank-cat-spec) was constructed by transforming SH244 with gDNA from strain SB463. SH250 (ΔfliP::tet, Pfla/che-fliQ-cheD, sacA::Pfla/che-fliO-fliP-spec, amyE::Phyper-spank-cat-spec, pHB201/cat, erm) was constructed by transforming SH249 with pHB201. SH255 (amyE::Phyper-spank-yueB-yfp-spec, sacA::Phyper-spank-ymdB-kan) was constructed by transforming ET13 with gDNA from strain SH79. SH256 (amyE::PxylA-spoIVFB-gfp-cat, sacA::Phyper-spank-ymdB-kan) was constructed by transforming BDR524 with gDNA from strain SH79. SH257 (amyE::Phyper-spank-yueB-yfp-spec, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) was constructed by transforming ET13 with gDNA from strain IB11. SH258 (amyE::PxylA-spoIVFB-gfp-cat, sacA::Phyper-spank-ymdB-kan, Δmbl::erm) was constructed by transforming BDR524 with gDNA from strain IB11.
Method Details
General growth conditions
All general methods for Bs were carried out as described previously (Harwood and Cutting, 1990). Bs cultures were inoculated at OD600 0.05 from an overnight culture and growth was carried out at 37°C in LB medium (Difco). For strains harboring genes under inducible promoters, 1 mM IPTG (Sigma-Aldrich) or 0.5% xylose (Sigma-Aldrich) was added to the medium. Antibiotics were used at the following concentrations: kanamycin (5 μg/ml, US Biological), chloramphenicol (6 μg/ml, Sigma-Aldrich), lincomycin (25 μg/ml, Sigma-Aldrich), erythromycin (1 μg/ml, Sigma-Aldrich), tetracycline (10 μg/ml, Sigma-Aldrich), spectinomycin (100 μg/ml, Sigma-Aldrich).
Transformation into Bm (OS2) cells was carried out as previously described (Moro et al., 1995). Bm cells were grown up to 1.0 OD600 (10 ml). Cells were then washed with electroporation buffer [25% PEG 8000 (Promega) and 0.1 M sorbitol (Sigma-Aldrich)] and resuspended in 1 mL of the same buffer. Electroporation was carried out with 0.1 mL of cells supplemented with 500 ng of linear DNA or plasmid DNA at 1500 V (Bio-Rad). Cells were then resuspended in 1 mL LB, incubated at 37°C for 1 hr, and plated on LB plates containing 5 μg/ml tetracycline.
Scarless deletions of E. coli were constructed using λ Red system and tet-sacB cassette as described (Datsenko and Wanner, 2000, Li et al., 2013). 50 bp of upstream and downstream sequences of fliOPQR ORFs were included in the forward primer 3907 and reverse primer 3908, respectively. These primers were used for PCR amplifying tet-sacB cassette from XTL634. Next, the cassette was inserted into MG1655 strain, containing pKD46 carrying λ Red genes (γ, β and exo) and a temperature sensitive origin. Around 1 Kb of recombination sequences upstream and downstream of fliOPQR were PCR amplified using primers 3909, 3912, 3950 and 3951. The upstream and downstream recombination sequences were ligated together by isothermal assembly (Gibson, 2011). The tet-sacB cassette was then replaced with the ligated DNA using similar λ Red system and selected on 6% sucrose (J.T.Baker). Next, the plasmid pKD46 was cured at 42°C to construct the strain RP7802 (ΔfliOPQR). Next, tet-sacB cassette was PCR amplified using primers 3914 and 3915. The cassette was inserted in RP7802 strain containing pKD46 using λ Red system. Around 1 Kb of recombination sequences upstream and downstream of flhBA were PCR amplified using primers 3952, 3953, 3954 and 3955. The upstream and downstream recombination sequences were ligated together by isothermal assembly and the tet-sacB cassette was then replaced with the ligated DNA using similar λ Red system. Next, the plasmid pKD46 was cured, and strain RP7809 (ΔCORE [fliOPQRflhBA]) constructed. ΔfliC::kan allele was PCR amplified with primers 311 and 312 using pKD4 as a template. Wild-type fliC of DY378 was disrupted by λ-red recombination of ΔfliC::kan allele and SK2462 strain was constructed (Datsenko and Wanner, 2000). Then ΔfliC::kan allele was transferred to wild-type MG1655 by P1 transduction with SK2462 lysate to construct SK2468 (Thomason et al., 2007).
Deletion mutant in Lm was constructed using pLR16-Phes as previously described (Argov et al., 2017). In brief, pLR16-AB1 was transformed into E. coli SM-10 strain and subsequently transferred to Lm 10403S by conjugation. Lm transconjugants were selected by plating on LB agar plates supplemented with 2% glucose and containing 7.5 μg/ml chloramphenicol and 100 μg/ml Streptomycin. Transconjugants were then passaged in LB supplemented with 2% glucose and plated to obtain single colonies. Colonies were tested for sensitivity to chloramphenicol. Chloramphenicol sensitive colonies were further verified to contain the desired mutation by PCR. For disruption of the flaA gene, pSH22 was transformed into E. coli SM-10 strain and transferred to Lm 10403S by conjugation. Lm transconjugants were selected by plating on LB agar plates supplemented with 2% glucose and containing 7.5 μg/ml chloramphenicol and 100 μg/ml Streptomycin. Transconjugants were then passaged in LB supplemented with 2% glucose and 10 μg/ml chloramphenicol at 41°C and plated to obtain flaA disruption mutants.
Details on plasmid construction
Plasmid constructions were performed in E. coli DH5α using standard methods.
pSH13 (flhA-GFP-kan) was constructed by amplifying the 3′ region of flhA using the gDNA of wild-type Bs strain PY79, using primers flhA CT-F-EcoRI and flhA CT-R-XhoI. The PCR-amplified DNA was digested with EcoRI and XhoI and was cloned into pKL168 digested with the same enzymes.
pSH17 (amyE::Phyper-spank-fliP2xHA-spec) was constructed by amplifying fliP using the gDNA of wild-type Bs strain PY79, using primers fliP ORF F (HindIII) and fliP ORF R (2xHA), followed by a second round of PCR using primers fliP ORF F (HindIII) and 2xHA R (SphI). The PCR-amplified DNA was digested with HindIII and SphI and was cloned into pDR111 digested with the same enzymes.
pSH19 (amyE::Phyper-spank-fliP2xHA-LOOP-spec) was constructed by amplifying the 5′ and 3′ regions of fliP using the gDNA of wild-type Bs strain PY79. 5′ region of fliP was amplified using primers fliP ORF F (pDR111) and fliP NT R (GSS/2xHA), and the 3′ region of fliP was amplified using primers fliP CT F (GSS/2xHA) and fliP CT R (pDR111). The PCR-amplified DNA was cloned into pDR111 digested with HindIII and SphI using Gibson assembly kit (NEB, USA).
pSH21 [amyE::Phyper-spank-CORE-FEc (fliOPQR flhBA)-spec] was constructed by amplifying the Ec CORE using the plasmid pRP7358, using primers Ec fliO-flhA F (pDR111) and Ec fliO-flhA R (pDR111). The PCR-amplified DNA was cloned into pDR111 digested with HindIII and SphI using Gibson assembly kit (NEB, USA).
pLR16-AB1 was constructed by amplifying approximately 1000bp fragments upstream of fliP and downstream of flhA using primers Lm CORE P1-P2 and Lm CORE P3-P4 respectively. The fragments were joined using Gibson assembly kit (NEB, USA) and further amplified by PCR using primers Lm CORE P1 and P4. The resultant PCR product was digested with XhoI and SalI and cloned into pLR16-Phes digested with the same enzymes.
pSH22 (pLR16-Lm flaA int) was constructed by amplifying approximately 450bp fragment of the Lm flaA gene, using primers Lm flaA int F and Lm flaA int R. The PCR-amplified DNA was cloned into pLR16-Phes digested with SalI using Gibson assembly kit (NEB, USA).
pSH23 (pDG1514-Bm hag int) was constructed by amplifying approximately 450bp fragment of the Bm hag gene, using primers Bm hag int F (SalI) and Bm hag int R (BamHI). The PCR-amplified DNA was digested with SalI and BamHI and was cloned into pDG1514 digested with the same enzymes.
Nanotube visualization by XHR-SEM
Nanotube visualization was carried out as previously described (Dubey et al., 2016). Accordingly, Bs, Bm, Ec and Lm cells grown to mid logarithmic phase were spotted onto EM grids (mesh copper grids, EMS) placed over LB agar plates and incubated for 4 hr at 37°C. Cells were then washed 3 times with PBS × 1, fixed with 2% paraformaldehyde (Electron Microscopy Sciences) and 0.01% glutaraldehyde (Electron Microscopy Sciences) in sodium cacodylate buffer (0.1 M, pH 7.2, Electron Microscopy Sciences) for 10 min at 25°C. Cells were left overnight for fixation in 2% glutaraldehyde in sodium cacodylate buffer (0.1 M, pH 7.2) at 4°C. For cell dehydration, EM grids underwent a series of washes in increasing concentrations of ethanol (25, 50, 75, and 96%) (J.T.Baker) and kept in vacuum till visualization. Samples were coated and observed using Through-Lens Detector operated at Secondary Electron (TLD-SE) mode by Magellan XHR SEM (FEI).
Immuno-XHR-SEM analysis
Immuno-XHR-SEM analysis was carried out as previously described (Dubey et al., 2016, Stempler et al., 2017). Bs cells were grown on EM grids (mesh copper grids, EMS), and grid-attached cells were washed three times with PBS × 1, fixed with 2% paraformaldehyde and 0.01% glutaraldehyde in sodium cacodylate buffer (0.1 M, pH 7.2) for 10 min at 25°C. Subsequently, grids were washed 3 times in PBS × 1, incubated in PBS × 1 containing 2% BSA (Amresco) and 0.1% Tween 20 (J.T.Baker) for 30 min at 25°C, and washed twice with PBS × 1. Next, grids were incubated for 2 hr at 25°C with rabbit anti-HA antibodies (Thermo Fisher Scientific, USA) or rabbit anti-GFP, diluted 1:1000 in PBS × 1 containing 1% BSA. Grids were then washed 3 times with PBS × 1 and incubated for 1 hr at 25°C with 18nm gold-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, USA), diluted 1:500 in PBS × 1. Grids were washed 3 times with PBS × 1 and fixed with 2.5% glutaraldehyde in sodium cacodylate buffer (0.1 M, pH 7.2) for 1 hr at 25°C. Grids were then washed gently with water, and cells were dehydrated by exposure to a graded series of ethanol washes (25, 50, 75, 95, and 100% ( × 2); 10 min each), samples were kept in vacuum till visualization. Specimens were imaged without coating by Magellan XHR SEM (FEI) using Through-Lens Detector operated at Secondary Electron (TLD-SE) and Low-voltage high-Contrast backscatter electron Detector (vCD).
Molecular exchange assay
For detecting molecular exchange, antibiotic transfer assay was carried out as described previously (Dubey and Ben-Yehuda, 2011) with some modifications. Donor and recipient strains used for the molecular exchange assays are listed in the Key resources table. Respective donor and recipient strains were grown to mid logarithmic phase, after which cells were mixed in 1:1 ratio (OD600 = 0.8 or 0.08) and incubated in LB supplemented with 1 mM IPTG for 4 hr at 37°C with gentle shaking. Equal numbers of cells were spotted onto either double selective LB plates containing chloramphenicol (6 μg/ml) and kanamycin (5 μg/ml) for detection of protein exchange, or triple selective LB plates containing chloramphenicol, kanamycin and lincomycin (25 μg/ml) for detection of plasmid exchange. As a control, cells were spotted onto LB plates lacking antibiotics. Plates were incubated at 37°C.
Motility assay
Motility assay was carried out as previously described (Kearns and Losick, 2003) with some modifications. Cells were grown to mid logarithmic phase in LB and concentrated 10 times to OD600 5.0. 5 μL of the cell suspension was spotted on freshly prepared LB plates containing 0.3% agar, incubated at 37°C for 7-9 hr, and imaged over time.
Fluorescence microscopy
For visualization of FlhA-GFP, SpoIVFB-GFP and YueB-YFP, exponentially growing cells were harvested at an OD600 0.5, washed with PBS × 1 and observed by fluorescence microscopy. For staining bacterial membrane and nanotubes, exponentially growing cells were harvested and resuspended in PBS × 1 containing 1 μg/ml FM4-64 (Molecular Probes, Thermo Fisher Scientific) and visualized by fluorescence microscopy. Bs-Bm competition assays were carried out as described previously (Stempler et al., 2017). Overnight cultures of Bs and Bm were diluted to OD600 0.1, mixed, and mounted onto a metal ring (A-7816, Invitrogen) filled with LB agarose (1.5%). Cells were incubated in a temperature-controlled chamber at 37°C, and followed by fluorescence microscopy. Cells were visualized by Eclipse Ti microscope (Nikon, Japan), equipped with CoolSnap HQII camera (Photometrics, Roper Scientific, USA). System control and image processing were performed with NIS Elements AR 4.3 (Nikon, Japan).
RNA isolation and qRT-PCR
RNA was extracted from Bs cells grown to the mid logarithmic phase by FastRNA Pro Blue kit (MP Biomedicals) according to the manufacturer protocol. RNA concentration was determined using NanoDrop 2000C (Thermo Scientific). 2 μg RNA from each sample was treated with RQ1 DNase (2 Units, Promega), and subjected to cDNA synthesis using qScript cDNA synthesis kit (Quanta Biosciences), according to the manufacturer protocol. qRT-PCR reactions were conducted using PerfeCTa SYBR Green FastMix (Quanta Biosciences), and fluorescence detection was performed using Applied Biosystems StepOnePlus system according to manufacturer instructions. RNA from 16S rRNA was used to normalize expression. Relative gene expression and melt curve analysis was done using the Applied Biosystems StepOnePlus software (v. 2.3). Each assay was performed in duplicates with at least two RNA templates prepared from independent biological repeats. qRT-PCR primers were designed using Primer3 software (v. 0.4.0, available online).
Phylogenetic analysis
In order to study the evolutionary conservation of the CORE complex we focused on seven proteins of Bs 168: the five CORE complex proteins and two exclusive proteins of the flagellar apparatus. In addition, we analyzed three proteins from E. coli O157:H7 str. EDL933 and one protein from C. trachomatis D/UW-3/CX, which are unique to the T3SS. These were searched in each of the core genomes in the STRING database, using STRING’s protein homology data (Szklarczyk et al., 2017) and analyzed by custom python scripts to determine the conservation of the query proteins (Table S2). Sequence similarity scores < 0.1 were discarded. The conservation vectors of the species in each phylum were clustered using hierarchical clustering (Figure S5). Additionally, all species from all phyla were clustered together, and species clustering along with M. fulvus were inspected further to assess whether they include only the CORE proteins without other T3SS or flagella proteins, in a similar manner to M. fulvus.
PSI-BLAST was used to search several flagella unique proteins from Bs, along with several T3SS unique proteins from both Enterohemorrhagic E. coli and C. trachomatis, against the proteomes of the suspected species from the NCBI Assembly database. Hits adhering to stringent threshold of E-value < 0.01, spanning longer than half the length of the query with identity > 0.2, were considered putative homologs. Furthermore, a literature search was carried out by searching PubMed for the relevant species names alongside “Type III secretion system” and “flagella.” Species that had putative hits for < 20% of the proteins in all categories and had no relevant results in the literature search were considered to harbor only the CORE proteins.
Quantification and statistical analysis
Unless stated otherwise, bar charts display a mean ± SD from at least 3 independent biological experiments. Quantification of nanotubes were done manually. MS Excel was used for all statistical analysis, data processing and presentation.
Acknowledgments
We thank E. Blayvas and A. Ben-Hur (Hebrew University) for help with XHR-SEM and R. Nir-Paz (Hadassah Hospital) and A. Herskovits (Tel Aviv University) for strains and plasmids. We are grateful to S. Wagner (Tubingen University) for help generating Figure 4E and valuable insight. We are indebted to A. Rouvinski (Hebrew University) and members of the Ben-Yehuda and Rosenshine laboratories for valuable discussions. This work was supported by a European Research Council Advance grant (339984) awarded to S.B.-Y.; a grant from the Israel Science Foundation (617/15) awarded to I.R.; a European Research Council Synergy grant (810186) awarded to S.B.-Y. and I.R.; and a grant from the Israel Science Foundation (876/17) awarded to H.M. S.B. is partially funded by the Lady Davis Fellowship Trust, and Y.E.G. is partially supported by the Hoffman Program.
Author Contributions
S.B., A.K.B., R.R.P., G.M., and Y.E.G. performed the experiments. S.B., A.K.B., R.R.P., Y.E.G., H.M., S.B.-Y., and I.R. conceived the experiments and analyzed the data. S.B., A.K.B., R.R.P., S.B.-Y., and I.R. wrote the manuscript. S.B.-Y. and I.R. managed the project.
Declaration of Interests
The authors declare no competing interests.
Published: March 28, 2019
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.celrep.2019.02.055.
Contributor Information
Ilan Rosenshine, Email: ilanr@ekmd.huji.ac.il.
Sigal Ben-Yehuda, Email: sigalb@ekmd.huji.ac.il.
Supplemental Information
Each row represents a single species from the STRING core genomes and each column represents one protein. The protein names are followed by the species of origin (Bacillus: B. subtilis proteins; EHEC: Enterohemorrhagic E. coli proteins; Chlamydia: C. trachomatis protein). The sequence similarity scores are based on the STRING database homology data. Species lacking any homolog were excluded from the analysis.
References
- Abby S.S., Rocha E.P. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R.D., Baumann P. Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. J. Bacteriol. 1971;107:295–302. doi: 10.1128/jb.107.1.295-302.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argov T., Rabinovich L., Sigal N., Herskovits A.A. An effective counterselection system for Listeria monocytogenes and its use to characterize the monocin genomic region of strain 10403S. Appl. Environ. Microbiol. 2017;83:e02927-16. doi: 10.1128/AEM.02927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidya A.K., Bhattacharya S., Dubey G.P., Mamou G., Ben-Yehuda S. Bacterial nanotubes: a conduit for intercellular molecular trade. Curr. Opin. Microbiol. 2018;42:1–6. doi: 10.1016/j.mib.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Bejerano-Sagie M., Oppenheimer-Shaanan Y., Berlatzky I., Rouvinski A., Meyerovich M., Ben-Yehuda S. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Benomar S., Ranava D., Cárdenas M.L., Trably E., Rafrafi Y., Ducret A., Hamelin J., Lojou E., Steyer J.P., Giudici-Orticoni M.T. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat. Commun. 2015;6:6283. doi: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- Bron S., Bolhuis A., Tjalsma H., Holsappel S., Venema G., van Dijl J.M. Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J. Biotechnol. 1998;64:3–13. doi: 10.1016/s0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- Costa T.R., Felisberto-Rodrigues C., Meir A., Prevost M.S., Redzej A., Trokter M., Waksman G. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Armitage J.P. Type III secretion systems: the bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20150020. doi: 10.1098/rstb.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Wagner S. Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 2014;38:802–822. doi: 10.1111/1574-6976.12061. [DOI] [PubMed] [Google Scholar]
- Dietsche T., Tesfazgi Mebrhatu M., Brunner M.J., Abrusci P., Yan J., Franz-Wachtel M., Schärfe C., Zilkenat S., Grin I., Galán J.E. Structural and functional characterization of the bacterial type III secretion export apparatus. PLoS Pathog. 2016;12:e1006071. doi: 10.1371/journal.ppat.1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey G.P., Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Dubey G.P., Malli Mohan G.B., Dubrovsky A., Amen T., Tsipshtein S., Rouvinski A., Rosenberg A., Kaganovich D., Sherman E., Medalia O., Ben-Yehuda S. Architecture and characteristics of bacterial nanotubes. Dev. Cell. 2016;36:453–461. doi: 10.1016/j.devcel.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Fabiani F.D., Renault T.T., Peters B., Dietsche T., Gálvez E.J.C., Guse A., Freier K., Charpentier E., Strowig T., Franz-Wachtel M. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol. 2017;15:e2002267. doi: 10.1371/journal.pbio.2002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura T., Makino F., Dietsche T., Kinoshita M., Kato T., Wagner S., Namba K., Imada K., Minamino T. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 2017;15:e2002281. doi: 10.1371/journal.pbio.2002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis G., Suerbaum S., Forsthoff B., Leying H., Opferkuch W. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J. Med. Microbiol. 1993;38:371–377. doi: 10.1099/00222615-38-5-371. [DOI] [PubMed] [Google Scholar]
- Gibson D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A.M., Frandsen N., Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Harwood C.R., Cutting S.M. Wiley; 1990. Molecular biological methods for Bacillus. [Google Scholar]
- Hayes C.S., Aoki S.K., Low D.A. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hu Y., Huang H., Cheng X., Shu X., White A.P., Stavrinides J., Köster W., Zhu G., Zhao Z., Wang Y. A global survey of bacterial type III secretion systems and their effectors. Environ. Microbiol. 2017;19:3879–3895. doi: 10.1111/1462-2920.13755. [DOI] [PubMed] [Google Scholar]
- Hueck C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L.J., Carballido-López R., Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kearns D.B., Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kuhlen L., Abrusci P., Johnson S., Gault J., Deme J., Caesar J., Dietsche T., Mebrhatu M.T., Ganief T., Macek B. Structure of the core of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 2018;25:583–590. doi: 10.1038/s41594-018-0086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon K.P., Grossman A.D. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- Li X.T., Thomason L.C., Sawitzke J.A., Costantino N., Court D.L. Positive and negative selection using the tetA-sacB cassette: recombineering and P1 transduction in Escherichia coli. Nucleic Acids Res. 2013;41:e204. doi: 10.1093/nar/gkt1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig W.D., Koller A., Thanassi D.G. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J. Bacteriol. 2013;195:1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L.L. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Imada K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015;23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Morimoto Y.V., Ito M., Hiraoka K.D., Che Y.S., Bai F., Kami-Ike N., Namba K., Minamino T. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol. Microbiol. 2014;91:1214–1226. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- Moro A., Sanchez J.C., Serguera C. Transformation of Bacillus megaterium by electroporation. Biotechnol. Tech. 1995;9:589–590. [Google Scholar]
- Pal R.R., Baidya A.K., Mamou G., Bhattacharya S., Socol Y., Kobi S., Katsowich N., Ben-Yehuda S., Rosenshine I. Pathogenic E. coli extracts nutrients from infected host cells utilizing injectisome components. Cell. 2019;177 doi: 10.1016/j.cell.2019.02.022. Published online March 28, 2019. [DOI] [PubMed] [Google Scholar]
- Pallen M.J., Matzke N.J. From The Origin of Species to the origin of bacterial flagella. Nat. Rev. Microbiol. 2006;4:784–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- Pande S., Shitut S., Freund L., Westermann M., Bertels F., Colesie C., Bischofs I.B., Kost C. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat. Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- Ren C.P., Beatson S.A., Parkhill J., Pallen M.J. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 2005;187:1430–1440. doi: 10.1128/JB.187.4.1430-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A., Sinai L., Smith Y., Ben-Yehuda S. Dynamic expression of the translational machinery during Bacillus subtilis life cycle at a single cell level. PLoS ONE. 2012;7:e41921. doi: 10.1371/journal.pone.0041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D.Z., Pan Q., Losick R.M. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. U S A. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L.A., Loman N.J., Fütterer K., Pallen M.J. Bacterial flagellar diversity and evolution: seek simplicity and distrust it? Trends Microbiol. 2009;17:1–5. doi: 10.1016/j.tim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Stempler O., Baidya A.K., Bhattacharya S., Malli Mohan G.B., Tzipilevich E., Sinai L., Mamou G., Ben-Yehuda S. Interspecies nutrient extraction and toxin delivery between bacteria. Nat. Commun. 2017;8:315. doi: 10.1038/s41467-017-00344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason L.C., Costantino N., Court D.L. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007;Chapter 1:Unit 1.17. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- Tzipilevich E., Habusha M., Ben-Yehuda S. Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell. 2017;168:186–199.e12. doi: 10.1016/j.cell.2016.12.003. [DOI] [PubMed] [Google Scholar]
- von Mering C., Jensen L.J., Kuhn M., Chaffron S., Doerks T., Krüger B., Snel B., Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Vassallo C.N., Pathak D.T., Wall D. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J. Bacteriol. 2014;196:1807–1814. doi: 10.1128/JB.00850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Perkins J.B., Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each row represents a single species from the STRING core genomes and each column represents one protein. The protein names are followed by the species of origin (Bacillus: B. subtilis proteins; EHEC: Enterohemorrhagic E. coli proteins; Chlamydia: C. trachomatis protein). The sequence similarity scores are based on the STRING database homology data. Species lacking any homolog were excluded from the analysis.