Bacteria alter their metabolism in response to nutrient availability, growth conditions, and environmental stresses using a number of different mechanisms. One is lysine acetylation, a posttranslational modification known to target many metabolic enzymes. However, little is known about this regulatory mode. We investigated the factors inducing changes in lysine acetylation by comparing growth on glucose and xylose. We found that the specific sugar used for growth did not alter the pattern of acetylation; rather, the amount of sugar did, with more sugar causing more acetylation. These results imply that lysine acetylation is a global regulatory mechanism that is responsive not to the specific carbon source per se but rather to the accumulation of downstream metabolites.
KEYWORDS: acetyl phosphate, acetylation, acetylome, Escherichia coli, fermentation, mass spectrometry
ABSTRACT
Lysine acetylation is thought to provide a mechanism for regulating metabolism in diverse bacteria. Indeed, many studies have shown that the majority of enzymes involved in central metabolism are acetylated and that acetylation can alter enzyme activity. However, the details regarding this regulatory mechanism are still unclear, specifically with regard to the signals that induce lysine acetylation. To better understand this global regulatory mechanism, we profiled changes in lysine acetylation during growth of Escherichia coli on the hexose glucose or the pentose xylose at both high and low sugar concentrations using label-free mass spectrometry. The goal was to see whether lysine acetylation differed during growth on these two different sugars. No significant differences, however, were observed. Rather, the initial sugar concentration was the principal factor governing changes in lysine acetylation, with higher sugar concentrations causing more acetylation. These results suggest that acetylation does not target specific metabolic pathways but rather simply targets accessible lysines, which may or may not alter enzyme activity. They further suggest that lysine acetylation principally results from conditions that favor accumulation of acetyl phosphate, the principal acetate donor in E. coli.
IMPORTANCE Bacteria alter their metabolism in response to nutrient availability, growth conditions, and environmental stresses using a number of different mechanisms. One is lysine acetylation, a posttranslational modification known to target many metabolic enzymes. However, little is known about this regulatory mode. We investigated the factors inducing changes in lysine acetylation by comparing growth on glucose and xylose. We found that the specific sugar used for growth did not alter the pattern of acetylation; rather, the amount of sugar did, with more sugar causing more acetylation. These results imply that lysine acetylation is a global regulatory mechanism that is responsive not to the specific carbon source per se but rather to the accumulation of downstream metabolites.
INTRODUCTION
An abundant posttranslational modification in many bacteria is Nε-lysine acetylation (1, 2). Multiple studies have shown that lysine acetylation predominantly targets the enzymes involved in central metabolism (2–5). Because some of these lysines are catalytically active, their acetylation may regulate metabolism in bacteria. This hypothesis is supported by several in vitro studies showing that lysine acetylation indeed alters the activity of some enzymes involved in central metabolism (4, 6–8). However, it is still not clear what role lysine acetylation plays in regulating metabolism. One compelling model is that lysine acetylation provides a global mechanism by which cells regulate metabolism in response to their energy status. The response to energy status occurs through the availability of the acetyl group donors acetyl coenzyme A (acetyl-CoA) and acetyl phosphate, two metabolic intermediates that are related through a single reaction. According to this model (2), lysine acetylation reduces carbon flux through central metabolism when these metabolic intermediates accumulate in the cells. This occurs when the carbon flux exceeds the capacity of the tricarboxylic acid (TCA) cycle, for example, by acetate overflow metabolism (9).
In the present work, we sought to determine whether the carbon source also affects lysine acetylation patterns in Escherichia coli. We tested this by profiling changes in lysine acetylation during growth of E. coli on hexose d-glucose, which is metabolized principally through the Embden-Meyerhof-Parnas pathway, or pentose d-xylose, which is metabolized through the nonoxidative pentose phosphate pathway. This design enabled us to distinguish between two mutually exclusive hypotheses, that either the carbon source and its associated metabolic pathway matter or they do not. We chose these two sugars because (i) E. coli grows faster on glucose than on xylose and thus may have different lysine acetylation patterns (10) and (ii) their metabolism involves a number of different enzymes that may also be differentially acetylated. In addition, these are the two most abundant sugars in plant biomass (11). E. coli has been genetically engineered to produce a wide variety of valuable chemicals and fuels from these sugars, with the goal of replacing petroleum-based feedstocks with renewable plant-based ones (12, 13). These designs require that carbon flux be redirected toward producing these compounds. Thus, any knowledge regarding the regulation of carbon flux during growth on these two sugars will aid in these engineering efforts.
RESULTS AND DISCUSSION
Effects of carbon source identity and concentration on protein expression.
To measure changes in lysine acetylation, cells from three biological replicates were harvested after 12 h of growth in M9 minimal medium (Fig. 1; see Fig. S1 in the supplemental material) and then subjected to label-free mass spectrometry (data-independent acquisition [DIA]) (14, 15). Protein expression was similar under the four different growth conditions, with only 61 proteins identified exhibiting significant changes in expression (Fig. S2). Many differentially expressed proteins were less abundant during growth at high sugar concentrations. Reduced expression likely results from catabolite repression due to high sugar concentrations (Table S1), as many of these proteins are expressed from cyclic AMP receptor protein (CRP)-dependent promoters (16). Fewer proteins exhibited increased expression during growth at high sugar concentrations. Four proteins (AmtB, RutA, GlnK, and Nac) are members of the NtrC (NRI) regulon, which is induced when the carbon-to-nitrogen ratio is high (17). Two proteins (ProV and ProX) are components of the glycine betaine/proline ABC transporter, which is induced in response to osmotic stress (18). These results are consistent with high sugar concentrations inducing osmotic stress and being sensed by the cell as nitrogen limitation.
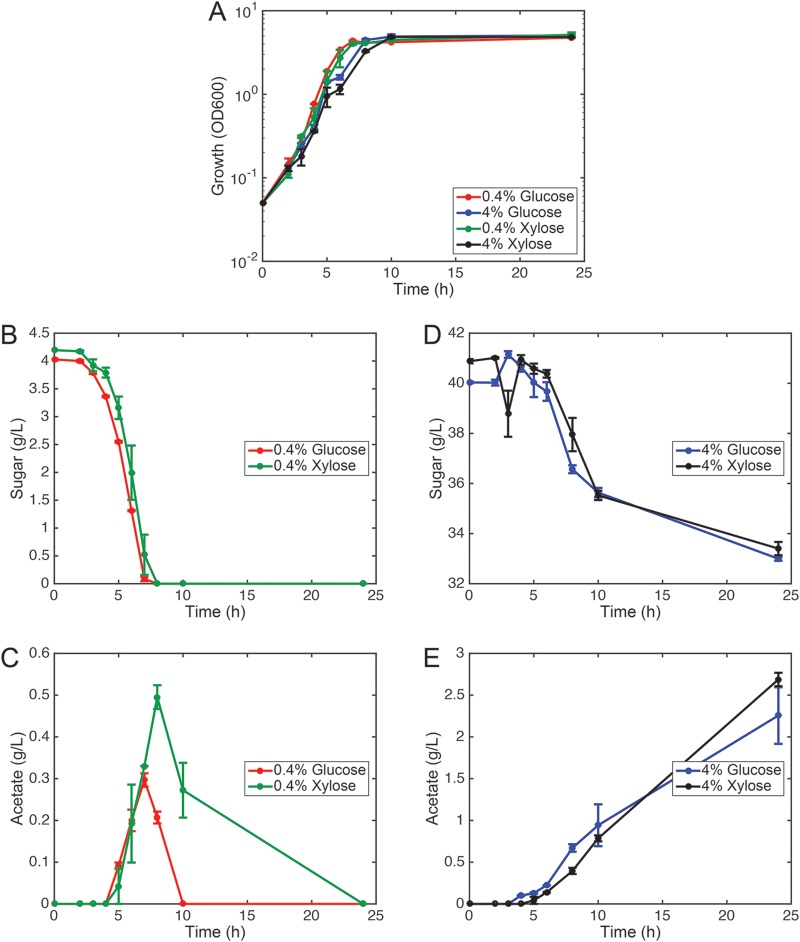
FIG 1.
Growth of E. coli on M9 minimal medium containing 0.4% glucose, 4% glucose, 0.4% xylose, or 4% xylose as the sole carbon source. Error bars denote the standard deviation for the results from three biological replicates. (A) Cell growth as determined by optical absorbance. (B) Sugar consumption during growth on 0.4% glucose or xylose. (C) Acetate production during growth on 0.4% glucose or xylose. (D) Sugar consumption during growth on 4% glucose or xylose. (E) Acetate production during growth on 4% glucose or xylose. During growth on higher concentrations of sugar, consumption is incomplete due to acidification of the growth medium.
Effects of carbon source identity and concentration on protein acetylation.
We next used label-free mass spectrometry to identify the acetylated lysines. Label-free mass spectrometry identified 3,840 unique acetyllysine sites on 978 proteins (Tables S2 and S3). Ninety-five percent of the sites identified during growth on 4% glucose or 4% xylose were identical (Table S3).
For more in-depth quantification of all sites and conditions, we also acquired all immunoenriched samples in DIA or SWATH acquisition mode. Our specific DIA acquisition mode allows for comprehensive quantification even in replicates where very low-level signals are observed, as long as robust peak area groups (peptide signals) could be integrated in other biological replicates (19, 20). Of all observed acetylation sites, 278 lysines on 157 unique proteins exhibited significant changes in acetylation, showing fold changes of >2 (and with P values of <0.05) (Fig. 2A and Table S4).
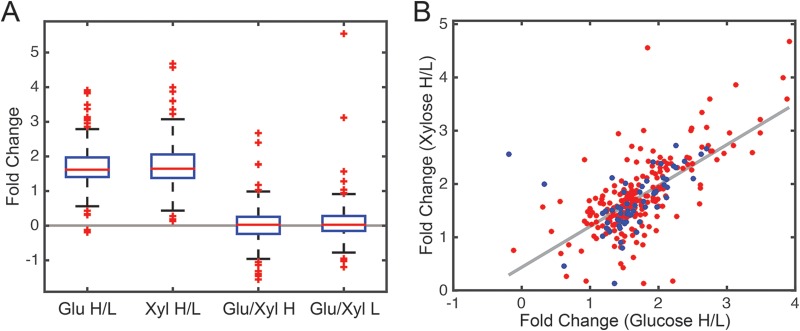
FIG 2.
Relative changes in lysine acetylation under the four growth conditions. (A) Box plot showing relative change in acetylation for the four different growth conditions. (B) Comparison of differentially acetylated lysines during growth on xylose versus glucose. The blue dots denote lysines on the metabolic enzymes depicted in Fig. 3. Glu H/L, 4% glucose versus 0.4% glucose; Xyl H/L, 4% xylose versus 0.4% xylose; Glu/Xyl H, 4% glucose versus 4% xylose; and Glu/Xyl L, 0.4% glucose versus 0.4% xylose.
The relative degree of acetylation increased at least 2-fold for 260 sites on 149 proteins during growth on 4% versus 0.4% glucose. No sites were found to exhibit a 2-fold decrease in the relative degree of acetylation at the higher sugar concentration. Similarly, the relative abundance of acetylation increased at least 2-fold for 256 sites on 147 proteins during growth on 4% versus 0.4% xylose. Once again, no sites exhibited a 2-fold decrease in the relative abundance of acetylation at the higher sugar concentration.
We explored whether observed increases in the relative abundance of acetylation at higher sugar concentrations were correlated during growth on glucose versus xylose. As shown in Fig. 2B, changes in acetylation are moderately correlated during growth on the two sugars (R2 = 0.45). In other words, many of those lysines where acetylation increased during growth on high glucose concentrations often increased during growth on high xylose concentrations. We separately confirmed these results using antiacetyllysine Western blotting, which again showed that global acetylation is correlated with the concentration but not the identity of the carbon sources (Fig. S3).
These results suggest that many differentially acetylated lysines are sensitive not to the specific growth sugar but rather to the amount of sugar available. They also support the hypothesis that most acetylation is a result of acetate overflow metabolism and is independent of the specific route for catabolism. Indeed, cells grown on higher concentrations of sugar produced significantly more acetate, which would explain the increase in acetylation (Fig. 1).
We note that cells grown on 0.4% xylose produced more acetate than those grown on 0.4% glucose. However, acetylation was not increased during growth on 0.4% xylose compared to growth on 0.4% glucose (Fig. 2A). Compared to growth on 4% sugar, the amount of acetate produced during growth on 0.4% sugar is significantly less, and any differences between glucose and xylose are likely insufficient to cause significant changes in acetylation. In addition, we note that cells grown on 4% sugar could not reassimilate the acetate, causing it to accumulate in the growth medium. This inability to reassimilate the acetate likely results from acidification of the growth medium and the inability of E. coli to consume acetate at low pH (21).
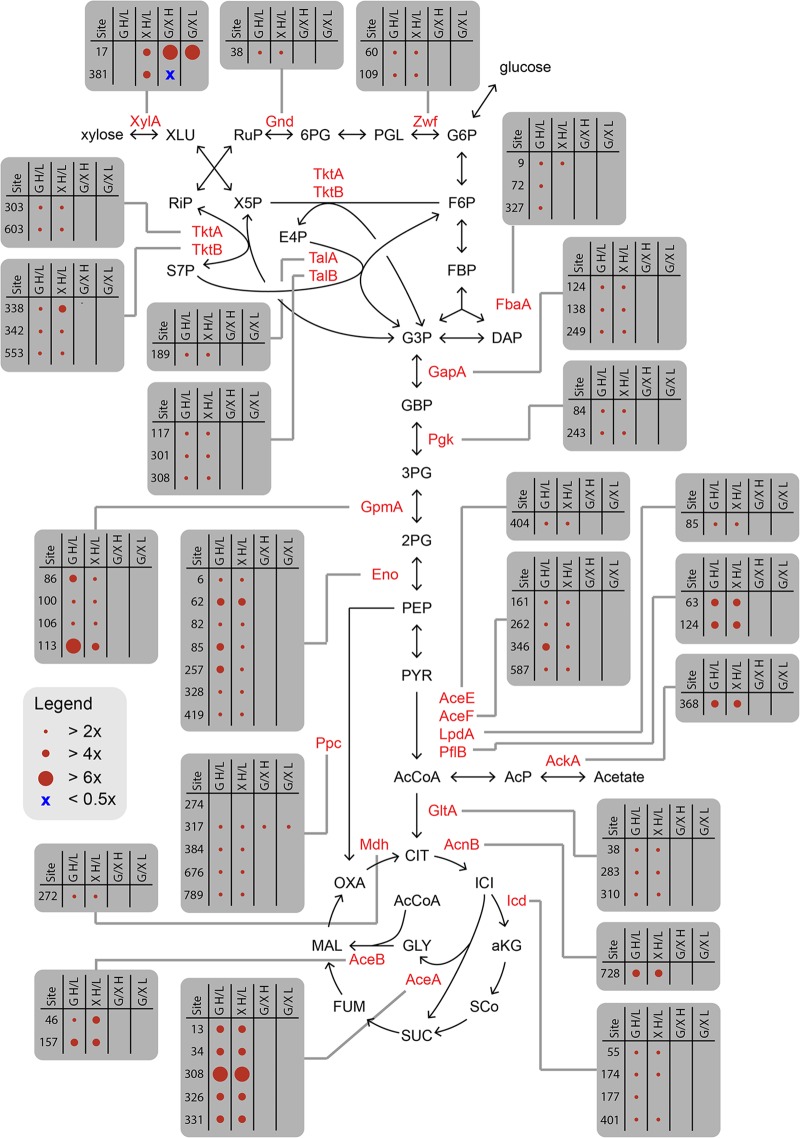
We further examined those acetylated proteins exhibiting significant changes in their relative acetylation abundance, focusing on those involved in central metabolism (Fig. 3). Only two of these enzymes exhibited significant (2-fold) changes among the four growth conditions, xylose isomerase (XylA) and phosphoenolpyruvate carboxylase (Ppc). In fact, they were the only two central metabolic enzymes where the relative degree of acetylation was sensitive to the choice of growth sugar. XylA catalyzes the first step in xylose metabolism and is differentially acetylated at two sites. Interestingly, the relative abundance of acetylation for one lysine (XylAK17) increased, while the other (XylAK381) decreased during growth on 4% glucose versus 4% xylose; in fact, it is the only lysine on a metabolic enzyme to exhibit a decrease in the relative abundance of acetylation. The acetylation of lysine 17 also increased during growth on 4% versus 0.4% xylose, suggesting that it is sensitive to the energy state of the cells. These results imply that acetylation of these lysines may alter enzymatic activity. However, both lysines are on the surface of the enzyme and not near the active site. Nonetheless, we replaced these lysines with glutamines or arginines, which mimic acetylated lysine or unacetylated lysine, respectively (22–24). We observed no effect on during growth on xylose (Fig. S4). While these experiments may not accurately model lysine acetylation, they nonetheless suggest that acetylation does not regulate XylA activity.
FIG 3.
Enzymes in central metabolism exhibiting changes in lysine acetylation under the four growth conditions. Specific lysines are shown in gray boxes. Data are also available in Table S3. XLU, xylulose; RuP, ribulose 5-phosphate; 6PG, gluconate 6-phosphate; PGL, phosphogluconolactone; G6P, glucose 6-phopshate; RiP, ribulose 5-phosphate; X5P, xylulose 5-phosphate; F6P, fructose 6-phosphate; S7P, sedoheptulose 7-phosphate; E4P, erythrose 5-phosphate; FBP, fructose 1,6-bisphosphate; G3P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; 3PG; GBP, 1,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl-CoA, AcP, acetyl-phosphate; CIT, citrate; ICI, isocitrate; aKG, α-ketoglutarate; SCo, succinyl-CoA; SUC, succinate; FUM, fumarate; MAL, malate; OXA, oxaloacetate; Glu H/L, 4% glucose versus 0.4% glucose; Xyl H/L, 4% xylose versus 0.4% xylose; Glu/Xyl H, 4% glucose versus 4% xylose; Glu/Xyl L, 0.4% glucose versus 0.4% xylose.
Ppc catalyzes the carboxylation of phosphoenolpyruvate to form oxaloacetate and is involved in replenishing metabolites in the TCA cycle. It is acetylated at five sites, and none are proximal to the active site or known to be involved in catalysis. The relative abundance of one lysine (Ppc317) increased during growth on glucose versus xylose at both high (4%) and low (0.4%) sugar concentrations, indicating that this lysine is sensitive to the growth sugar. However, this lysine is located on the surface of the enzyme and away from the dimerization interfaces, suggesting that it also has no effect on enzyme activity.
These results further indicate that acetylation does not target specific metabolic pathways but rather targets accessible lysines. Of the two central-metabolic enzymes showing different patterns of acetylation during growth on glucose versus xylose, acetylation does not appear to have any regulatory role, indicating that this posttranslational modification does not always have a regulatory role. To date, only a few enzymes have been shown to be sensitive to lysine acetylation (4, 6–8), and the vast majority of acetylated lysines likely have no effect on protein function or activity. That being said, it is also clear that some enzymes are sensitive to acetylation and, in these regards, have coopted this promiscuous modification for regulatory roles. Further work is needed to identify which acetylated lysines have regulatory roles.
Conclusion.
We profiled protein acetylation in E. coli during growth on glucose and xylose at both high and low sugar concentrations. Our goal was to see whether lysine acetylation differed during growth on these two different sugars. To our initial surprise, we did not observe major differences among the lysines acetylated during respective growth on these two sugars. Rather, the observed changes in lysine acetylation were principally correlated with the initial sugar concentration, with higher sugar concentrations causing more acetylation. These results further support the hypothesis that lysine acetylation results from the buildup of metabolic intermediates, principally acetyl phosphate, under conditions that favor acetate production (Fig. 1). In other words, whenever cells produce acetate, they will also acetylate their proteins. These results also indicate that acetylation is agnostic to the metabolic route and simply targets accessible lysines, which may or may not alter enzyme activity. In these regards, acetylation is a general regulatory mechanism and not one tailored for specific pathways or sugars. Finally, these results arguably change the outstanding question from “how is acetylation regulated?” to “how does the cell cope with, or perhaps utilize for its advantage, acetylation that it cannot avoid?”
MATERIALS AND METHODS
Chemicals and media.
High-performance liquid chromatography (HPLC) grade acetonitrile and water were obtained from Burdick & Jackson (Muskegon, MI). Reagents for protein chemistry, including iodoacetamide, dithiothreitol (DTT), ammonium bicarbonate, formic acid (FA), and urea, were purchased from Sigma-Aldrich (St. Louis, MO). Sequencing grade trypsin was purchased from Promega (Madison, WI). Hydrophilic-lipophilic-balanced (HLB) Oasis solid-phase extraction (SPE) cartridges were purchased from Waters (Milford, MA). M9 minimal medium (11.3 g/liter M9 salts [5×], 0.1 mM CaCl2, 1 mM MgSO4, 2.45 μM ferric citrate, and 0.03 mM thiamine) supplemented with 0.4% glucose, 0.4% xylose, 4% glucose, or 4% xylose was used for all growth experiments. M9 salts were purchased from BD Difco (Franklin Lakes, NJ); all other chemicals were purchased from Sigma-Aldrich.
Strains and growth conditions.
All strains are derivatives of Escherichia coli BW25113 [F− λ− Δ(araD-araB)567 Δ(rhaD-rhaB)568 ΔlacZ4787 rrnB3 rph-1 hsdR514] (25). Strains encoding the acetylation-mimicking mutants of XylA were generated using the flexible recombineering using integration of thyA (FRUIT) technique (26). Briefly, the xylA allele of a ΔthyA mutant (BW25113 ΔthyA) was replaced with a PCR product containing the variant allele using lambda Red recombination. Then, the thyA deletion was replaced with a PCR product containing the wild-type thyA gene via lambda Red recombination. Finally, the plasmid pKD46 was cured by growing the cells at 42°C. The resulting strains were verified by sequencing the xylA gene. Bacteria were grown in 250-ml shake flasks at 37°C in an incubated shaker at 225 rpm and flask-to-medium ratio of 10:1.
Measurement of cell growth and sugar concentrations.
Cell growth was measured by optical density at 600 nm (OD600). Glucose and xylose concentrations were measured using a Shimadzu high-performance liquid chromatography system equipped with a RID-10A refractive index detector, an Aminex HPX-87H carbohydrate analysis column (Bio-Rad Laboratories, Hercules, CA), and a Micro-Guard cation H cartridge (Bio-Rad Laboratories). The column and guard cartridge were kept at 65°C, and 5 mM H2SO4 was used as the mobile phase at a constant flow rate of 0.6 ml/min. Prior to analysis, culture samples were first pelleted, and then the supernatant was passed through a 0.22-µm polyethersulfone syringe filter. Peaks were identified and quantified by retention time comparison to authentic standards.
Cell lysis, proteolytic digestion of protein lysates, and affinity enrichment of acetylated peptides.
For mass spectrometric analysis, wild-type E. coli (BW25113) was grown overnight in M9 minimal medium supplemented with 0.4% glucose or 0.4% xylose. Overnight cultures grown in 0.4% glucose were diluted to an OD600 of 0.05 into M9 medium supplemented with 0.4% or 4% glucose. Overnight cultures grown in 0.4% xylose were diluted to an OD600 of 0.05 into M9 medium supplemented with 0.4% or 4% xylose. After 12 h of aeration, cells were harvested by centrifugation at 1,877 × g for 20 min at 4°C. Cell pellets were suspended in 6 ml of phosphate-buffered saline (PBS) and centrifuged at 4°C and 1,877 × g for 20 min. The pellets were then frozen at −80°C. Isolated frozen bacterial pellets from each group (3 biological replicates each) were suspended in 6 ml of PBS and centrifuged at 4°C and 15,000 × g for 20 min. The firm cell pellet was collected and resuspended and denatured in a final solution of 6 M urea, 100 mM Tris, 75 mM NaCl, and the deacetylase inhibitors trichostatin A (1 mM) and nicotinamide (3 mM). Samples were sonicated on ice (5 times each for 15 s), cellular debris was removed, and the supernatants were collected for proteolytic digestion. Protein lysate containing 1.5 mg of protein was reduced with 20 mM DTT (37°C for 1 h) and subsequently alkylated with 40 mM iodoacetamide (30 min at room temperature [RT] in the dark). Samples were diluted 10-fold with 100 mM Tris (pH 8.0) and incubated overnight at 37°C with sequencing grade trypsin (Promega) added at a 1:50 (wt/wt) enzyme/substrate ratio. Subsequently, samples were acidified with formic acid and desalted using HLB Oasis SPE cartridges (Waters) (27). Proteolytic peptides were eluted, concentrated to near dryness by vacuum centrifugation, and resuspended in NET buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA). An aliquot of each protein digestion (∼500 μg) was saved for quantitative analysis of protein expression changes. The remaining proteolytic peptide samples were used for affinity purification of acetylated peptides (Kac). Acetylated peptides were enriched using 1/4 of a tube containing an antiacetylysine antibody-bead conjugate, which is part of the PTMScan acetyllysine motif [Ac-K] kit (Cell Signaling Technologies) for each of the 1-mg protein lysate samples, according to the manufacturer’s instructions (15, 28). Prior to mass spectrometric analysis, the acetylated peptide enrichment samples were concentrated and desalted using C18 ZipTips (Millipore, Billerica, MA).
Mass spectrometric analysis.
Samples were analyzed by reverse-phase HPLC-electrospray ionization-tandem mass spectrometry (HPLC-ESI-MS/MS) using an Eksigent NanoLC Ultra Plus 2D HPLC system (Dublin, CA) with a cHiPLC system (Eksigent) which was directly connected to a quadrupole time of flight (QqTOF) TripleTOF 6600 mass spectrometer (Sciex, Concord, Ontario, Canada) (15, 28). After injection, peptide mixtures were loaded onto a C18 precolumn chip (200-µm by 0.4-mm ChromXP C18-CL chip, 3 µm, 120 Å; Sciex) and washed at 2 µl/min for 10 min with the loading solvent (H2O–0.1% formic acid) for desalting. Subsequently, peptides were transferred to the 75-µm by 15-cm ChromXP C18-CL chip (3 µm, 120 Å; Sciex) and eluted at a flow rate of 300 nl/min with a 3-h gradient using aqueous and acetonitrile solvent buffers (15, 28).
(i) Data-dependent acquisitions.
To build a spectral library for protein-level quantification, the mass spectrometer was operated in data-dependent acquisition (DDA) mode, where the 30 most abundant precursor ions from the survey MS1 scan (250 ms) were isolated at 1 m/z resolution for collision-induced dissociation–tandem mass spectrometry (CID-MS/MS; 100 ms per MS/MS, “high-sensitivity” product ion scan mode) using the Analyst 1.7 (build 96) software with a total cycle time of 3.3 s, as previously described (14, 15).
(ii) Data-independent acquisitions.
For quantification, all peptide samples were analyzed by data-independent acquisition (DIA; e.g., SWATH) (19), using 64 variable-width isolation windows (29). The SWATH cycle time of 3.2 s included a 250-ms precursor ion scan, followed by 45-ms accumulation time for each of the 64 variable SWATH segments.
Mass spectrometric data processing and bioinformatics.
Mass spectrometric DDA were analyzed using the database search engine ProteinPilot (30) (Sciex Beta 4.5, revision 1656) using the Paragon Algorithm (4.5.0.0,1654). The following sample parameters were used: trypsin digestion, cysteine alkylation set to iodoacetamide, urea denaturation, acetylation emphasis, and species E. coli. All data files were searched using the Swiss-Prot 2014_05 database, with a total of 8,870 E. coli protein sequences. A cutoff peptide confidence value of 99 was chosen. The ProteinPilot false-discovery rate (FDR) analysis tool (PSPEP) algorithm (30) provided a global FDR of 1% and a local FDR of 1% in all cases.
Mass spectral data sets were also analyzed and searched using an in-house Mascot server. For Mascot searches (31), peak lists were generated using the Sciex mgf data converter version 1.3, and the data subsequently were searched using a Mascot server version 2.3.02 with the Swiss-Prot 2014_05 protein database (see above). Search parameters were as follows: trypsin digestion with four missed cleavages to account for the inability of trypsin to cleave at acetylated lysine residues. Variable modifications included lysine acetylation, protein N-terminal acetylation, methionine oxidation, and conversion of glutamine to pyroglutamic acid. Carbamidomethyl cysteine was set as a fixed modification, and precursor ion and fragment ion mass tolerances were set to 50 ppm and 0.3 Da, respectively. For all Mascot searches, automatic decoy searches were performed, and peptide expectation values (E values) lower than 0.01 were chosen, with all FDR rates below 0.01. Table S2 in the supplemental material shows an overview of all identified acetylation sites and acetylated proteins.
Relative quantification of acetylation site changes and protein expression changes.
Using the database search engine results generated and discussed above, MS/MS spectral libraries are generated in Skyline daily version 4.1.1 (32), an open-source data processing workspace for quantitative proteomics. DIA raw data files are imported into Skyline, and both MS1 precursor ion scans and MS2 fragment ion scans are extracted for all acetylated peptides present in the spectral libraries. In Skyline, typically, 6 to 10 MS2 fragment ions are extracted per acetylated peptide based on ranking from the corresponding MS/MS spectra in the spectral libraries, and fragment peak areas are summed per peptide. Relative quantification of acetylation levels comparing the different growth media, specifically, M9 supplements (4% Glc versus 0.4% Glc, 4% Xyl versus 0.4% Xyl, 4% Glc versus 4% Xyl, and 0.4% Glc versus 0.4% Xyl) can be performed directly in Skyline using integrated statistical algorithms. Relative quantification of acetylation levels was called significant with the following values for significance cutoffs: condition A versus B ratios with changes of >2-fold or <−2-fold at a P value of <0.05. For relative quantification details and acetylation site changes, see Table S4. Protein expression changes were assessed using the SWATH Acquisition MicroApp 2.0 in the PeakView (Sciex) (33).
Antiacetyllysine Western blotting.
Wild-type E. coli (BW25113) was grown exactly as described above for mass spectrometry sample acquisition. Bacteria were harvested by centrifugation and lysed using the BugBuster protein extraction reagent (Novagen, Merck Millipore, Billerica, MA). The amount of cell lysate loaded on the gel was normalized to the total protein concentration, as determined by the bicinchoninic acid (BCA) assay (Thermo Scientific Pierce, Waltham, MA). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and normalization was verified by Coomassie staining. Protein acetylation was determined using a rabbit polyclonal antiacetyllysine antibody (Cell Signaling, Danvers, MA) at a dilution of 1:1,000, as described previously (14, 15).
Growth assay of XylA acetylation mimic variants.
To ensure growth of strains encoding the xylA acetylation mimics, all strains were first grown overnight in M9 medium supplemented with 0.4% g/liter glucose. A volume of culture to yield an OD600 of 0.05 was harvested and pelleted. The cell pellet was washed three times in PBS to remove residual carbon. The cell pellet was resuspended in either M9 medium with 0.4% glucose or M9 medium with 0.4% xylose. Growth was monitored over 8 h and again at 24 h.
Data availability.
All mass spectrometry data files were uploaded to public repositories (MassIVE ID number MSV000082949 [ftp://massive.ucsd.edu/MSV000082949] and ProteomeXchange PXD011098). The repository also contains additional supplemental tables, such as (i) tables with all search engine results for acetylation sites, (ii) additional statistical details for acetylation site changes, and (iii) tables with relative quantification for protein expression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NCRR shared instrumentation grant for the TripleTOF 6600 (grant 1S10 OD016281, Buck Institute) and by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under award DE-SC0012443.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00768-18.
REFERENCES
- 1.Hentchel KL, Escalante-Semerena JC. 2015. Complex regulation of the sirtuin-dependent reversible lysine acetylation system of Salmonella enterica. Microb Cell 2:451–453. doi: 10.15698/mic2015.11.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe AJ. 2016. Bacterial protein acetylation: new discoveries unanswered questions. Curr Genet 62:335–341. doi: 10.1007/s00294-015-0552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler CA, Veith PD, Nieto MF, Dashper SG, Reynolds EC. 2015. Lysine acetylation is a common post-translational modification of key metabolic pathway enzymes of the anaerobe Porphyromonas gingivalis. J Proteomics 128:352–364. doi: 10.1016/j.jprot.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Nakayasu ES, Burnet MC, Walukiewicz HE, Wilkins CS, Shukla AK, Brooks S, Plutz MJ, Lee BD, Schilling B, Wolfe AJ, Muller S, Kirby JR, Rao CV, Cort JR, Payne SH. 2017. Ancient regulatory role of lysine acetylation in central metabolism. mBio 8:e01894-17. doi: 10.1128/mBio.01894-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouidir T, Kentache T, Hardouin J. 2016. Protein lysine acetylation in bacteria: current state of the art. Proteomics 16:301–309. doi: 10.1002/pmic.201500258. [DOI] [PubMed] [Google Scholar]
- 6.Venkat S, Gregory C, Sturges J, Gan Q, Fan C. 2017. Studying the lysine acetylation of malate dehydrogenase. J Mol Biol 429:1396–1405. doi: 10.1016/j.jmb.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkat S, Chen H, Stahman A, Hudson D, McGuire P, Gan Q, Fan C. 2018. Characterizing lysine acetylation of isocitrate dehydrogenase in Escherichia coli. J Mol Biol 430:1901–1911. doi: 10.1016/j.jmb.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunk E, Chang RL, Xia J, Hefzi H, Yurkovich JT, Kim D, Buckmiller E, Wang HH, Cho BK, Yang C, Palsson BO, Church GM, Lewis NE. 2018. Characterizing posttranslational modifications in prokaryotic metabolism using a multiscale workflow. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1811971115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez A. 2017. Overflow metabolism: from yeast to marathon runners. Academic Press, Cambridge, MA. [Google Scholar]
- 10.Aidelberg G, Towbin BD, Rothschild D, Dekel E, Bren A, Alon U. 2014. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol 8:133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll A, Somerville C. 2009. Cellulosic biofuels. Annu Rev Plant Biol 60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen J, Keasling JD. 2016. Engineering cellular metabolism. Cell 164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Jagtap SS, Rao CV. 2018. Microbial conversion of xylose into useful bioproducts. Appl Microbiol Biotechnol 102:9015–9036. doi: 10.1007/s00253-018-9294-9. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2014. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, Sorensen DJ, Zemaitaitis B, Gibson BW, Wolfe AJ. 2015. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol 98:847–863. doi: 10.1111/mmi.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng D, Constantinidou C, Hobman JL, Minchin SD. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res 32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol 57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 18.Dattananda CS, Gowrishankar J. 1989. Osmoregulation in Escherichia coli: complementation analysis and gene-protein relationships in the proU locus. J Bacteriol 171:1915–1922. doi: 10.1128/jb.171.4.1915-1922.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. 2012. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins BC, Hunter CL, Liu Y, Schilling B, Rosenberger G, Bader SL, Chan DW, Gibson BW, Gingras AC, Held JM, Hirayama-Kurogi M, Hou G, Krisp C, Larsen B, Lin L, Liu S, Molloy MP, Moritz RL, Ohtsuki S, Schlapbach R, Selevsek N, Thomas SN, Tzeng SC, Zhang H, Aebersold R. 2017. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat Commun 8:291. doi: 10.1038/s41467-017-00249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orr JS, Christensen DG, Wolfe AJ, Rao CV. 2019. Extracellular acidic pH inhibits acetate consumption by decreasing gene transcription of the tricarboxylic acid cycle and the glyoxylate shunt. J Bacteriol 201:e00410-18. doi: 10.1128/JB.00410-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki W, Tachiwana H, Kawaguchi K, Shibata T, Kagawa W, Kurumizaka H. 2011. Comprehensive structural analysis of mutant nucleosomes containing lysine to glutamine (KQ) substitutions in the H3 and H4 histone-fold domains. Biochemistry 50:7822–7832. doi: 10.1021/bi201021h. [DOI] [PubMed] [Google Scholar]
- 23.Lima BP, Thanh Huyen TT, Basell K, Becher D, Antelmann H, Wolfe AJ. 2012. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J Biol Chem 287:32147–32160. doi: 10.1074/jbc.M112.365502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Basell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ. 2013. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol 195:4174–4186. doi: 10.1128/JB.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stringer AM, Singh N, Yermakova A, Petrone BL, Amarasinghe JJ, Reyes-Diaz L, Mantis NJ, Wade JT. 2012. FRUIT, a scar-free system for targeted chromosomal mutagenesis, epitope tagging, and promoter replacement in Escherichia coli and Salmonella enterica. PLoS One 7:e44841. doi: 10.1371/journal.pone.0044841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. 2007. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics 6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basisty N, Meyer JG, Wei L, Gibson BW, Schilling B. 2018. Simultaneous quantification of the acetylome and succinylome by ‘one-pot’ affinity enrichment. Proteomics 18:1800123. doi: 10.1002/pmic.201800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schilling B, Gibson BW, Hunter CL. 2017. Generation of high-quality SWATH acquisition data for label-free quantitative proteomics studies using TripleTOF mass spectrometers. Methods Mol Biol 1550:223–233. doi: 10.1007/978-1-4939-6747-6_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 32.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert JP, Ivosev G, Couzens AL, Larsen B, Taipale M, Lin ZY, Zhong Q, Lindquist S, Vidal M, Aebersold R, Pawson T, Bonner R, Tate S, Gingras AC. 2013. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat Methods 10:1239–1245. doi: 10.1038/nmeth.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry data files were uploaded to public repositories (MassIVE ID number MSV000082949 [ftp://massive.ucsd.edu/MSV000082949] and ProteomeXchange PXD011098). The repository also contains additional supplemental tables, such as (i) tables with all search engine results for acetylation sites, (ii) additional statistical details for acetylation site changes, and (iii) tables with relative quantification for protein expression.