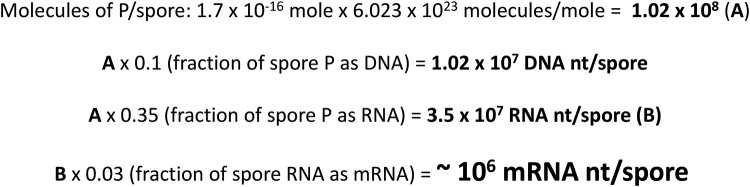Previous work indicates that dormant Bacillus subtilis spores have many hundreds of mRNAs, some of which are suggested to play roles in spores’ “return to life” or revival. The present work finds only ∼46 mRNAs at ≥1 molecule spore, with others in only fractions of spores in populations, often very small fractions. Less-abundant spore mRNAs are not contaminants in spore preparations, but how spores accumulate them is not clear. Almost all abundant spore mRNAs are synthesized in the developing spore late in its development, most encode proteins in spores, and abundant mRNAs in spores are relatively stable at 4°C. These findings will have a major impact on thinking about the roles that spore mRNAs may play in spore revival.
KEYWORDS: Bacillus subtilis, spore mRNAs, spores
ABSTRACT
Large-scale shotgun sequencing (RNA-seq) analysis of mRNAs in dormant Bacillus subtilis spores prepared on plates or in liquid generally found the same ∼46 abundant mRNA species, with >250 mRNAs detected at much lower abundances. Knowledge of the amount of phosphate in a single B. subtilis spore allowed calculation of the amount of mRNA in an individual spore as ∼106 nucleotides (nt). Given the levels of abundant spore mRNAs compared to those of other mRNAs, it was calculated that the great majority of low-abundance mRNAs are present in only small fractions of spores in populations. Almost all of the most abundant spore mRNAs are encoded by genes expressed late in sporulation in the developing spore under the control of the forespore-specific RNA polymerase sigma factor, σG, and most of the encoded proteins are in spores. Levels of the most abundant spore mRNAs were also relatively stable for a week at 4°C after spore harvest. RNA-seq analysis of mRNAs in highly purified and less-well-purified spores made in liquid, as well as from spores that were chemically decoated to remove possible contaminating mRNA, indicated that low-abundance mRNAs in spores were not contaminants in purified spore preparations, and several sources of low-abundance mRNAs in spores are suggested. The function of at least the great majority of spore mRNAs seems most likely to be the generation of ribonucleotides for new RNA synthesis by their degradation early in spore revival.
IMPORTANCE Previous work indicates that dormant Bacillus subtilis spores have many hundreds of mRNAs, some of which are suggested to play roles in spores’ “return to life” or revival. The present work finds only ∼46 mRNAs at ≥1 molecule spore, with others in only fractions of spores in populations, often very small fractions. Less-abundant spore mRNAs are not contaminants in spore preparations, but how spores accumulate them is not clear. Almost all abundant spore mRNAs are synthesized in the developing spore late in its development, most encode proteins in spores, and abundant mRNAs in spores are relatively stable at 4°C. These findings will have a major impact on thinking about the roles that spore mRNAs may play in spore revival.
INTRODUCTION
Spores of various Firmicutes species are formed in sporulation and are metabolically dormant and extremely resistant to a large variety of harsh treatments (1, 2). While these spores can survive for long periods, they can rapidly return to life in germination and then outgrowth if given the proper stimulus (3, 4). Germination itself is of significant interest, as in only a few minutes, a dormant spore is converted into a metabolically active germinated spore, and this can happen in the absence of exogenous nutrients. One of the first germination events is the release of the spore core’s large depot, ∼25% of the core dry weight, of a 1:1 chelate of pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]) with Ca2+ (CaDPA) and replacement of the CaDPA in the core by water. In Bacillus subtilis spores, this event raises the core water content from 35% of the wet weight to ∼45%. CaDPA release can take place in 1 to 2 min for an individual spore and most likely does not require ATP (4–6). The second major germination event follows CaDPA release, the hydrolysis of the large peptidoglycan cortex, which is outside the spore core and is thought to be important in at least attaining the low water content in the spore core (1). Cortex hydrolysis then allows the spore core to expand and take up more water, reaching the 80% of the core wet weight in growing cells. It also appears likely that this second major germination event requires minimal, if any, ATP (4–6). However, completion of the latter process has been suggested to require at least some spore core protein synthesis, but this requirement may not be absolute (7, 8).
Spores do not contain significant levels of common high-energy small molecules such as ATP, although abundant AMP is present, and ATP does not accumulate significantly until spore germination is approximately complete (4, 6, 9). For many years, it was thought that spores did not contain functional mRNA, as when spores were germinated in the presence of transcription inhibitors, there was minimal if any protein synthesis (10–13). However, there were several reports ∼50 years ago that spores contain low-molecular-weight RNA in addition to tRNA, and there was evidence that this was mRNA (14–16). Notably, at least some of the low-molecular-weight RNA is degraded early after germination is initiated, thus providing ribonucleotides for RNA synthesis early in outgrowth (10). This degradation is essential for RNA synthesis in this period of development, as dormant spores lack multiple enzymes of nucleotide biosynthesis, and these enzymes are synthesized only at defined times in spore outgrowth (3, 10). Only then can outgrowing spores carry out new RNA synthesis upon germination in a minimal medium.
While the results noted above are consistent with dormant spores having no functional mRNAs, nevertheless, beginning in ∼2007, there have been multiple reports that spores of a number of Bacillales and Clostridiales species have a significant store of specific mRNAs (17–25). These spore mRNAs were initially detected by microarray analysis, reverse transcriptase PCR, and, more recently, large-scale shotgun sequencing (RNA-seq) (23). Surprisingly, the number of spore mRNAs detected in those studies has ranged from a low of ∼20 abundant transcripts (21) to a high of several thousand (23). Generally, many of the most abundant spore mRNAs have been from genes expressed late in gene expression in the developing spore, with this transcription carried out by RNA polymerase holoenzyme with the RNA polymerase promoter specificity factor σG active only in the developing spore, although the transcription of a substantial number of these genes is also directed by the earlier-acting forespore-specific factor σF (26). Surprisingly, almost always, studies have found that some of the less abundant spore mRNAs are encoded by genes not known to be expressed in the developing spore, with some thought to be expressed only in the mother cell within which the spore develops. While suggestions have been made as to how mRNAs made in the mother cell get into the developing forespore (24), the exact mechanism for this and the significance of the plethora of spore mRNAs found in some studies are not clear. Consequently, we have used RNA-seq to determine levels and identities of mRNAs in spores of B. subtilis and to investigate several possible explanations for the presence of so many mRNAs in spores, including some mother cell-specific transcripts, and the significance of these many spore mRNAs.
RESULTS
mRNAs in highly purified dormant spores.
RNA-seq analysis was carried out on rRNA-depleted total RNA from dormant B. subtilis spores prepared on plates and highly purified as described in Materials and Methods. Illumina sequencing was carried out on RNAs from 3 independent and highly purified spore preparations, yielding ∼14 million reads per RNA sample (see Table S1 in the supplemental material). When all mRNAs that were detected at reads per kilobase of transcript per million mapped reads (RPKM) values of ≥5 for a given gene in at least one sample were summed, a total of 302 dormant spore mRNAs were detected (Tables 1 and 2). There was also a large spread in the RPKM values for the different mRNAs detected (Fig. S1A). The 29 most abundant mRNA species were each, on average, ∼4,000-fold more abundant than each of the 224 least abundant mRNAs (Table 2, sixth column). Notably, 2 of the 46 most abundant spore mRNAs, yhcO and sscA, and some of the ∼260 lower-abundance mRNAs detected in this work, for example, cotD, cotV, cotG, and lytH (see the GEO accession number in Materials and Methods), are thought to be generated only in the mother cell compartment of the sporulating cell, while other low-abundance mRNAs, for example, serS, speD, and ansR, are thought to be generated in growing cells (26). In contrast, of the 46 most abundant mRNAs detected, at least 41 are encoded by genes expressed only late in the developing forespore under σG control, although 19 of these same genes are also expressed under the control of σF, which acts just prior to σG in the developing forespore (Table 1). Notably, the proteins encoded by at least 42 of the 46 most abundant mRNAs have been identified in spores, although the function of only a few of these proteins is known (Table 1; also see Discussion). For the 36 most abundant genes, the coverage of the reads for these genes was relatively uniform across the full-length annotated mRNA (Fig. S2A to D and data not shown), while the coverage for at least some of the less abundant mRNAs was less uniform (Fig. S2E and data not shown).
TABLE 1.
Characteristics of the 46 most abundant dormant B. subtilis spore mRNAsa
| Gene | Length (nt)b | No. of reads | RPKM valuec | Sigma factor(s) |
|---|---|---|---|---|
| RPKM > 7,000 | ||||
| sspA | 210 | 2,479 | 10,848 | F and G |
| sspE | 255 | 159,687 | 577,971 | F and G |
| sspF | 186 | 26,976 | 131,260 | G |
| sspJ | 141 | 20,095 | 133,439 | G |
| sspK | 153 | 3,947 | 23,696 | G |
| sspM | 105 | 2,878 | 25,626 | G |
| sspN* | 147 | 32,488 | 201,551 | F and G |
| tlp* | 252 | 56,102 | 202,343 | F and G |
| sspO† | 147 | 16,078 | 102,766 | F and G |
| sspP† | 147 | 13,486 | 86,361 | F and G |
| yfhD | 192 | 36,871 | 177,547 | F and G |
| yhcN | 570 | 4,689 | 7,464 | F and G |
| yhcQ | 654 | 9,699 | 13,540 | G |
| yhcV | 423 | 74,504 | 161,470 | F and G |
| yhdB | 243 | 22,294 | 82,876 | G |
| yizC | 198 | 16,362 | 75,142 | G |
| ykzE‡ | 177 | 51,891 | 271,743 | G |
| ykzP‡ | 156 | 81,390 | 480,746 | G |
| ymfJ | 258 | 24,076 | 85,309 | F and G |
| yozQ | 294 | 3,085 | 9,528 | G |
| ypzF | 147 | 133,054 | 827,125 | G |
| ypzG | 153 | 110,878 | 652,271 | G |
| yqfX | 390 | 16,853 | 39,843 | G |
| yrzQ§ | 132 | 11,482 | 81,387 | F and G |
| yrzR§ | 192 | 3,663 | 17,332 | F and G |
| ytzC | 273 | 80,936 | 269,462 | F and G |
| ytzL | 159 | 26,849 | 157,337 | F and G |
| yuzA | 237 | 2,512 | 9,616 | G |
| yxeD | 354 | 2,697 | 7,175 | G |
| RPKM = 2,000–7,000 | ||||
| sspB | 204 | 809 | 2,972 | F and G |
| sspH | 180 | 593 | 2,415 | G |
| yhcM | 456 | 1,457 | 4,044 | F and G |
| yraE¶ | 198 | 54 | 3,249 | G |
| yrrD | 525 | 1,853 | 2,674 | G |
| yusG | 237 | 628 | 2,087 | UK |
| yusN | 333 | 942 | 2,082 | F and G |
| RPKM = 567–2,000 | ||||
| pdhA | 1,116 | 823 | 683 | A |
| sscA | 87 | 124 | 1,767 | K |
| sspI | 216 | 148 | 843 | F and G |
| yhcO | 969 | 1,502 | 1,429 | E and K |
| ykzD | 138 | 108 | 714 | G |
| yoyE | 126 | 137 | 1,462 | G |
| ypzI | 132 | 119 | 850 | F and G |
| yraD¶ | 300 | 284 | 843 | G |
| yrzB | 282 | 208 | 567 | UK |
| yutC | 633 | 516 | 636 | G |
Data are averages from RNA-seq analyses on RNA from 3 independent highly purified spore preparations prepared on plates, all as described in Materials and Methods. Genes in boldface type are those for which the protein product has been identified in dormant spores (33, 34). RNA polymerase σ factors that direct the transcription of individual genes were identified using information in subtiwiki and the literature (26), including information on gene expression in sporulation and correlation of a gene’s expression with that from a gene definitively established as being under the control of a specific sigma factor. For the yusG and yrzB genes, their σ factor dependence is unknown (UK). Two genes with the same symbol are almost certainly cotranscribed, as seen by their appropriate genomic arrangement and the complete coverage of both genes by RNA-seq reads, with no gaps in intergenic spaces (data not shown; see also Fig. S2F in the supplemental material).
Note that the mRNAs for coding genes are invariably longer than the coding sequences shown; this is ∼50 nt for these 46 most abundant mRNAs (data not shown).
The average standard deviations for the average RPKM values were 8% for the 29 most abundant mRNAs, 21% for the 7 next most abundant mRNAs, and 17% for the 10 next most abundant mRNAs.
TABLE 2.
Average numbers of molecules per spore for different-abundance mRNAs in highly purified B. subtilis sporesa
| Avg RPKM group | No. of genes (no. of σG-dependent genes)b | Length (nt)c | RPKM value | No. of reads/gened | RelmRNAs/genee | No. of mRNA nt/sporef | No. of mRNA nt/spore (Σ = 106)g | No. of mRNAs/sporeh |
|---|---|---|---|---|---|---|---|---|
| >7,000 | 29 (29) | 339 | 1.7 × 105 | 57,630 | 4,433 | 4.4 × 107 | 9.87 × 105 | 100 |
| 2,000–7,000 | 7 (6) | 404 | 2,789 | 1,127 | 87 | 2.5 × 105 | 5.6 × 103 | 2 |
| 567–2,000 | 10 (6) | 500 | 979 | 490 | 38 | 1.9 × 105 | 4.3 × 103 | 0.9 |
| 111–566 | 24 (8) | 565 | 221 | 125 | 10 | 1.4 × 105 | 3.1 × 103 | 0.2 |
| 2–110 | 232 (24) | 472 | 28 | 13 | 1 | 1.1 × 105 | 2.5 × 103 | 0.02 |
| Total | 302 | |||||||
RPKM values are averages of data from RNA-seq analyses carried out on 3 independent RNA preparations from highly purified dormant B. subtilis spores prepared on plates for 2 days at 37°C, as described in Materials and Methods. Values in the third through sixth and ninth columns are averages for all mRNAs in each group. Values in the second, seventh, and eighth columns are totals for all genes or mRNAs in this group.
Values in parentheses are numbers of σG-dependent genes. There may be more σG-dependent genes in the two lowest groups.
The numbers of nucleotides in mRNAs in the original RNA-seq data include the coding sequences plus the stop codons. Consequently, the values in this column have been increased by 50 nt to take into account the 3′ and 5′ untranslated regions in mRNAs.
Calculated as [(value in fourth column) × (value in third column)]/1,000 to correct for the fact that RPKM values are based on reads per kilobase of transcript.
RelmRNAs/gene are relative levels of spore mRNA that were calculated as (value in fifth column)/13, the average number of reads per gene for the mRNAs of the group with the lowest average RPKM value.
Values were calculated as (value in sixth column) × (value in second column) × (value in third column) for each group.
Calculated as (value in seventh column) × 0.0224 to obtain a total of 106 nt for all groups.
These values were calculated as (value in eighth column)/(value in second column) × (value in third column) and are the average numbers of each individual mRNA/spore in each group.
In at least some respects, the data described above are generally similar to what has been obtained when RNAs from spores of other species, from both Bacillales and Clostridiales, have been analyzed (17–25). In these studies, mRNAs from σG-dependent genes were among the most abundant dormant spore mRNAs, and in most studies, multiple additional mRNAs were also detected, some of which are thought to be expressed only in the mother cell compartment of the sporulating cell and under the control of mother cell-specific σ factors such as σK or σE (26). A major question, then, concerns the significance of the additional mRNAs not under σG control, especially those present in rather low abundance. To answer the latter question, we took advantage of knowledge of the amount of P in a single B. subtilis spore (1.7 × 10−16 mol) and the percentages of this P in RNA (35%) and DNA (10%) (27) to calculate the molecules of RNA and DNA nucleotides in spores, as described in Materials and Methods (Fig. 1). The close agreement between the calculated amount of DNA nucleotides in individual B. subtilis spores, which are monogenomic (28), and the size of the B. subtilis genome indicates that calculations of the number of RNA nucleotides per spore from the number of moles of P per spore would be relatively accurate. The remaining value needed to complete the calculation of mRNA nucleotides in spores is the percentage of spore RNA as mRNA. This value has been measured as 3% in growing B. subtilis cells (29, 30), and that this value is not an unreasonably low estimate for the percentage of spore RNA as mRNA is supported by (i) analysis of total reads from rRNA and mRNA in an ∼100-million-read RNA-seq experiment on highly purified spore RNA that was not ribodepleted, which gave ∼9.5 × 107 reads from rRNA and 1.7 × 105 reads from all 46 abundant spore mRNAs, or mRNA as 0.2% of total RNA (data not shown), and (ii) Agilent RNA ScreenTape analysis of total RNA from vegetative cells and highly purified spores (Fig. S3). This analysis showed that the level of RNA in the size region of the 46 most abundant spore mRNAs (340 to 500 nucleotides [nt]) (Table 2) relative to the 16S rRNA level was ∼40% higher in spore RNA than in vegetative cell RNA, which could be due to a higher percentage of spore RNA as mRNA than in vegetative cells. Thus, mRNA levels in spores likely lie somewhere between 0.2 and 4.2% of total RNA. Using a value of 3% for the percentage of spore RNA as mRNA, the value in growing cells, we calculate that B. subtilis spores contain an average of 106 mRNA nt. Notably, even a 2-fold increase in the level of total spore mRNA nucleotides would not appreciably increase the numbers of mRNAs present at ≥1 molecule/spore (Table 2), as this would require spore mRNA percentages that are 20-fold higher than those in growing cells.
FIG 1.
Calculation of mRNA nucleotides per B. subtilis spore. The values of moles of P per spore and the fractions of spore P as DNA and RNA were reported previously (27), and the calculations of molecules of DNA nucleotides per spore, RNA nucleotides per spore, and mRNA nucleotides per spore are described in Materials and Methods.
Assuming that 1 molecule of each of the 224 least abundant mRNAs is present per spore, and knowing the relative abundance of various groups of spore mRNAs (Table 2, sixth column), the number of genes in each group, and their average mRNA nucleotide length (Table 2, second and third columns), allows calculation of the total mRNA nucleotides in a spore as ∼4.4 × 107 (Table 2, sixth column), a value ∼44-fold higher than the number of mRNA nucleotides expected in each spore. Notably, the total numbers of nucleotides in the 29 most abundant spore mRNAs are ∼50-fold higher than the number of nucleotides in all other spore mRNAs combined (Table 2, sixth column). Given that the 29 most abundant mRNAs identified by RNA-seq account for ∼98% of the 106 mRNA nt per spore (Table 2, eighth column), these 29 mRNAs are present at an average of ∼100 molecules/spore, and the two groups of dormant spore mRNAs present in the next highest abundance are at ∼2 and 0.9 molecules/spore, respectively (Table 2, ninth column). However, all other 256 mRNAs detected are at well below 1 molecule/spore, with the lowest-abundance mRNAs being detected by RNA-seq in only ∼2% of the spores in the population examined (Table 2, ninth column). Notably, while 89% of the 46 most abundant spore mRNAs detected were under σG control, the percentages of σG-dependent genes coding for the less abundant spore mRNAs were lower (Table 2, second column, values in parentheses).
The analysis described above of numbers of various mRNAs per spore was also carried out using previously reported RNA-seq data generated by Nagler et al. using dormant spore mRNAs from another wild-type B. subtilis strain (23); again, it was assumed that spore mRNA was 3% of spore RNA and that there are ∼106 mRNA nt/spore (Table 3; Table S2). This calculation found that 29 of the many mRNAs identified in this work were each present at an average of ∼86 molecules/spore, with a further 49 each being present at ∼1 molecule/spore. Notably, of the 46 most abundant mRNAs in these spores, 36 (78%) were under σG control, including all but 1 of the 29 most abundant mRNAs identified in the present work (Table S2), close to the value, 89%, for the 46 most abundant mRNAs found in the present work. In addition, of the 46 most abundant mRNAs found in the present work, all but 8 were among the 78 most abundant mRNAs reported by Nagler et al. (23). Nonetheless, a few abundant spore mRNAs detected in the previously reported work were not observed in the present study, with coxA mRNA being a notable example. The reasons for these small numbers of differences are not known but could be due to differences in culture conditions in the different laboratories or the different B. subtilis strains used, with possible sequence differences between the strain used and the reference genome preventing, for example, coxA mRNA identification in our analyses.
TABLE 3.
Average levels of molecules per spore for different-abundance mRNAs in dormant B. subtilis spores calculated from data in previously reported worka
| Abundance level | No. of genes | Length (nt)b | RPKM value | No. of reads/genec | RelmRNAs/gened | No. of mRNA nt/sporee | No. of mRNA nt/spore (Σ = 106)f | No. of mRNA/sporeg |
|---|---|---|---|---|---|---|---|---|
| Top | 29 | 272 | 353,000 | 96,016 | 842 | 6.6 × 106 | 6.8 × 105 | 86 |
| Next lowest | 17 | 323 | 4,970 | 1,605 | 14 | 7.7 × 104 | 7.9 × 103 | 1.4 |
| Next lowest | 32 | 487 | 2,740 | 1,334 | 12 | 1.9 × 105 | 2 × 104 | 1.3 |
| Lowest | 2,711 | 1,039 | 110 | 114 | 1 | 2.8 × 106 | 2.9 × 105 | 0.1 |
RNA-seq data on relative abundances of mRNAs detected in B. subtilis spores prepared in liquid were reported previously (23). Values in in the third through sixth and ninth columns are averages for the mRNAs in each group of genes. Values in the second, seventh, and eighth columns are totals for all genes or mRNAs in each group.
The numbers of nucleotides in mRNAs in original RNA-seq data include the coding sequence plus the stop codon. Consequently, the values in this column have been increased by 50 nt to take into account the 3′ and 5′ untranslated regions in mRNAs.
Calculated as [(value in the fourth column) × (value in the third column)]/1,000 to correct for the fact that RPKM values are based on reads per kilobase of transcript.
RelmRNAs/gene are relative levels of spore mRNA that were calculated as (value in the fifth column)/114, the average number of reads/gene for the lowest-abundance group.
Values were calculated as (value in the sixth column) × (value in the second column) × (value in the third column) for each group.
Calculated as (value in the seventh column) × 0.103 to obtain a total of 106 nt for all groups.
These values were calculated as (value in the eighth column)/(value in the second column) × (value in the third column) and are the average numbers of each individual mRNA per spore in each group.
mRNAs in less-highly-purified dormant spores.
A major question raised by the results given above concerns the source of the low-abundance mRNAs detected in dormant spores. One obvious possibility is that spores used for RNA extraction are contaminated with nucleic acids, including mRNAs, from outside the spore core. To test this possibility, we prepared spores in liquid, as were almost all Bacillus spores used previously for analysis of dormant spore mRNAs, and spores were either (i) highly purified over 7 days or (ii) less well purified over 7 days, all as described in Materials and Methods, RNA was then isolated, and RNA-seq was used to identify spore mRNAs (Tables 4 and 5; Fig. S1). Strikingly, relative levels of most of the 46 most abundant mRNAs in the highly purified and less-well-purified spores prepared in liquid were almost identical, as determined by relative RPKM values (Table 4; Fig. S1B and C). This was also true when RPKM values for mRNAs of even lower abundances chosen to represent mRNAs from genes under the control of σF/G, σE/K, or σA were compared (Table 5). The rosters of almost all abundant mRNAs in spores prepared in liquid medium are largely identical to those in spores prepared on plates (compare data in Tables 1 and 4). However, the relative levels of ∼50% of both the highest-abundance mRNA group and the 26 very-low-abundance mRNAs selected were ≥2-fold higher in highly purified spores prepared on plates than the levels in highly purified spores prepared in liquid, although there were some mRNAs that were more abundant in spores prepared in liquid (Tables 4 and 5). Importantly, analysis of the absolute numbers of various mRNAs/spore in less-well-purified spores prepared in liquid gave values very similar to those obtained from spores prepared on plates and highly purified (compare data in Table S3 and Table 2).
TABLE 4.
Levels of the most abundant mRNAs in B. subtilis spores prepared and purified in different waysa
| Gene | RPKM value with spore prepn |
|||||
|---|---|---|---|---|---|---|
| Plates, 7 days, HPb | Plates, 7 days, HP, DCc | Plates, 1 day, DCc | Liquid, 7 days, HPd | Liquid, 7 days, LPd | Liquid, 7 days, HP, DCe | |
| Most abundant | ||||||
| sspA | 10,848 | 7,101 | 2,558 | 82,252 | 74,270 | 7,964 |
| sspE | 577,971 | 195,633 | 307,920 | 1,680,099 | 1,682,101 | 207,023 |
| sspF | 131,260 | 256,602 | 141,093 | 73,546 | 76,555 | 128,480 |
| sspJ | 133,439 | 68,934 | 125,571 | 162,439 | 146,117 | 102,419 |
| sspK | 23,696 | 28,553 | 45,106 | 9,312 | 10,937 | 35,108 |
| sspM | 25,626 | 58,428 | 42,904 | 6,428 | 6,833 | 55,389 |
| sspN | 201,551 | 101,257 | 231,754 | 70,065 | 69,888 | 227,852 |
| sspO | 102,766 | 94,637 | 113,429 | 19,089 | 18,530 | 90,525 |
| sspP | 86,361 | 63,492 | 74,849 | 13,805 | 13,860 | 52,905 |
| tlp | 202,343 | 176,853 | 186,473 | 67,845 | 56,567 | 204,990 |
| yfhD | 177,547 | 176,800 | 272,126 | 124,322 | 119,052 | 216,922 |
| yhcN | 7,464 | 7,715 | 13,405 | 3,651 | 3,007 | 6,249 |
| yhcQ | 13,540 | 9,189 | 20,905 | 11,083 | 1,130 | 9,728 |
| yhcV | 161,470 | 102,408 | 111,900 | 325,933 | 347,985 | 79,119 |
| yhdB | 82,876 | 111,283 | 141,329 | 32,185 | 42,392 | 108,510 |
| yizC | 75,142 | 31,984 | 55,033 | 64,672 | 68,169 | 43,288 |
| ykzE | 271,743 | 232,006 | 241,481 | 83,154 | 82,219 | 230,510 |
| ykzP | 480,746 | 460,080 | 436,876 | 169,218 | 168,247 | 534,888 |
| ymfJ | 85,309 | 74,482 | 136,945 | 56,091 | 60,290 | 95,673 |
| yozQ | 9,528 | 11,683 | 18,217 | 1,622 | 1,480 | 9,246 |
| ypzF | 827,125 | 847,377 | 95,592 | 360,382 | 370,698 | 1,071,770 |
| ypzG | 465,198 | 577,580 | 593,031 | 412,715 | 412,193 | 717,523 |
| yqfX | 39,843 | 67,657 | 78,389 | 59,215 | 57,190 | 44,988 |
| yrzQ | 81,387 | 129,290 | 95,592 | 41,102 | 42,065 | 101,654 |
| yrzR | 17,332 | 17,844 | 23,238 | 8,397 | 8,505 | 20,738 |
| ytzC | 269,462 | 199,868 | 182,090 | 137,144 | 132,765 | 218,097 |
| ytzL | 157,337 | 216,142 | 187,257 | 48,820 | 49,262 | 174,765 |
| yuzA | 9,616 | 8,584 | 7,101 | 6,148 | 5,671 | 6,864 |
| yxeD | 7,175 | 5,323 | 10,280 | 4,581 | 4,867 | 6,115 |
| Less abundant | ||||||
| sspB | 2,972 | 4,257 | 2,201 | 38,450 | 35,951 | 3,706 |
| sspH | 2,415 | 4,473 | 8,782 | 1,608 | 1,443 | 6,035 |
| yhcM | 4,044 | 4,010 | 9,478 | 2,982 | 2,850 | 4,903 |
| yraE | 3,249 | 6,239 | 12,225 | 553 | 368 | 7,292 |
| yrrD | 2,674 | 2,254 | 2,218 | 4,748 | 4,193 | 2,074 |
| yusG | 2,087 | 2,098 | 1,919 | 1,366 | 1,320 | 1,513 |
| yusN | 2,082 | 2,095 | 4,330 | 859 | 820 | 2,336 |
| Much less abundant | ||||||
| pdhA | 683 | 811 | 881 | 55 | 52 | 807 |
| sscA | 1,767 | 117 | 139 | 127 | 148 | 40 |
| sspI | 843 | 2,187 | 2,462 | 341 | 297 | 2,117 |
| yhcO | 1,429 | 1,578 | 2,475 | 1,269 | 1,429 | 1,179 |
| ykzD | 714 | 1,889 | 2,251 | 416 | 395 | 2,341 |
| yoyE | 1,462 | 4,147 | 6,214 | 1,371 | 1,442 | 5,416 |
| ypzI | 850 | 1,264 | 3,695 | 596 | 498 | 1,372 |
| yraD | 843 | 1,391 | 3,446 | 150 | 139 | 1,342 |
| yrzB | 567 | 521 | 1,662 | 291 | 234 | 667 |
| yutC | 636 | 671 | 955 | 698 | 681 | 553 |
Values shown are from RNA-seq, as described in Materials and Methods, on RNA from (i) three spore preparations harvested after 2 days that were highly purified (HP) over ∼7 days to give three RNA preparations, after which RNA-seq data for all three were averaged; (ii) one spore preparation prepared on plates and harvested after 1 day and then either highly purified over 7 days and then chemically decoated (DC) or purified for only ∼4 h and then chemically decoated; (iii) one spore preparation made in liquid, harvested after 2 days, and divided into three equal aliquots, one of which was highly purified over 7 days and two of which were less well purified (LP) over 7 days, after which RNA-seq data for the RNA from the two less-well-purified spore preparations were averaged; and (iv) one spore preparation prepared in liquid, highly purified over 7 days, and then chemically decoated, all as described in Materials and Methods. The order of mRNAs is alphabetical within each group.
Spores were from one preparation that was divided into three equal samples that were purified separately to give three RNA samples.
Spores were prepared on the same batch of sporulation plates and harvested at the same time but purified for only 1 day or 7 days prior to decoating and RNA isolation.
Spores were from one batch of liquid medium and harvested at the same time but purified differently.
Spores were prepared separately from those described in footnote d.
TABLE 5.
Levels of very-low-abundance mRNAs in B. subtilis spores prepared and purified differentlya
| Gene | RPKM value with spore prepn |
|||||
|---|---|---|---|---|---|---|
| Plates, 7 days, HPb | Plates, 7 days, HP, DCc | Plates, 1 day, DCc | Liquid, 7 days, HPd | Liquid, 7 days, LPd | Liquid, 7 days, HP, DCe | |
| Genes under the control of σF/G | ||||||
| ygzC | 229 | 212 | 416 | 172 | 194 | 195 |
| yjzC | 329 | 616 | 2,098 | 440 | 469 | 742 |
| ykuS | 130 | 225 | 612 | 470 | 383 | 193 |
| sspL | 441 | 976 | 1,693 | 349 | 391 | 976 |
| yphA | 83 | 122 | 206 | 13 | 10 | 147 |
| spoIVB | 82 | 138 | 211 | 93 | 95 | 146 |
| yraF | 167 | 258 | 818 | 37 | 49 | 341 |
| yraG | 414 | 581 | 1,767 | 60 | 68 | 811 |
| adhB | 179 | 417 | 934 | 29 | 26 | 408 |
| spoIIQ | 37 | 89 | 128 | 62 | 62 | 72 |
| Genes under the control of σE/K | ||||||
| cotD | 161 | 83 | 71 | 74 | 62 | 65 |
| cotV | 174 | 39 | 27 | 4 | 20 | 26 |
| spoVID | 99 | 67 | 38 | 1 | 1 | 41 |
| gerE | 10 | 175 | 7 | 3 | 28 | 89 |
| lytH | 41 | 198 | 71 | 92 | 93 | 70 |
| cotG | 74 | 66 | 1,059 | 12 | 106 | 56 |
| Genes under the control of σΑ | ||||||
| ansA | 37 | 86 | 151 | 3 | 5 | 69 |
| ansB | 40 | 105 | 174 | 6 | 7 | 78 |
| ansR | 183 | 253 | 177 | 174 | 171 | 111 |
| ptsG | 25 | 62 | 19 | 7 | 6 | 33 |
| speD | 292 | 251 | 274 | 135 | 102 | 123 |
| tcyL | 99 | 77 | 41 | 2 | 32 | 91 |
| sufB | 36 | 52 | 76 | 51 | 62 | 54 |
| serS | 41 | 86 | 39 | 13 | 17 | 53 |
| Genes under the control of an unknown σ factor | ||||||
| yqxK | 151 | 199 | 85 | 154 | 162 | 106 |
| ytjP | 16 | 66 | 13 | 3 | 3 | 38 |
Values shown are from RNA-seq, as described in Materials and Methods, on RNA from (i) three spore preparations harvested after 2 days that were highly purified over ∼7 days to give three RNA preparations, after which RNA-seq data for all three were averaged; (ii) one spore preparation prepared on plates and harvested after 1 day and then either highly purified (HP) over 7 days and then chemically decoated (DC) or purified for only ∼4 h and then chemically decoated; (iii) one spore preparation that was made in liquid, harvested after 2 days, and divided into three equal aliquots, one of which was highly purified over 7 days and two of which were less well purified (LP) over 7 days, after which RNA-seq data with the RNA from the two less-well-purified spore preparations were averaged; and (iv) one spore preparation prepared in liquid, highly purified over 7 days, and then chemically decoated, all as described in Materials and Methods.
Spores were from one preparation that was divided into three equal samples that were purified separately to give three RNA samples.
Spores were prepared on the same batch of sporulation plates and harvested at the same time but purified for only 1 day or 7 days prior to decoating and RNA isolation.
Spores were from one batch of liquid medium and harvested at the same time but purified differently.
Spores were prepared separately from those described in footnote d.
As an additional check on the contribution of mRNAs outside the spore core to the mRNAs detected by RNA-seq in spores, samples of spores prepared in liquid or on plates were highly purified over 7 days as described in Materials and Methods. The final purified spores were then chemically decoated prior to RNA extraction to remove mRNA trapped in spores’ outer layers. RNA-seq analysis of mRNAs from chemically decoated spores and comparison of RPKM values between spores that were and those that were not decoated (Tables 4 and 5; Fig. S1D and E) found almost no mRNAs that were at ≥2.5-fold-lower levels in the decoated spores. Exceptions were sspE mRNA, which was at a ∼3-fold-lower level in the decoated spores prepared on plates but is a known forespore-specific gene, and sscA mRNA, which was at a ∼15-fold-lower level in the decoated spores and was also at a low level in spores prepared on plates and decoated ∼6 days earlier than spores highly purified for 7 days. Levels of almost all of the 26 much less abundant mRNAs were also not significantly reduced in decoated spores (Table 5). Taken together, the results reported above rule out the possibility that the low levels of the great majority of mRNAs found in spores are due to contaminants in spore preparations.
In contrast to the few mRNAs whose levels were reduced in decoated spores, levels of a large number of mRNAs were >2.5-fold higher in decoated spores made in liquid and highly purified, with 21/46 of the most abundant mRNAs and 9/26 of the much less abundant mRNAs being detected at higher levels in the decoated spores than in intact spores (Tables 4 and 5). However, this was not seen with spores prepared on plates.
Stability of dormant spore mRNAs during spore purification.
As noted above, spores were normally purified over ∼7 days at 4°C, and it is possible that spore mRNA pools could change during this purification period. Consequently, we also prepared spores on plates and harvested them after 1 day when spores were released from ≥95% of sporangia, as determined by phase-contrast microscopy. Immediately after the spores were scraped from these plates, the preparation was divided in half. One half was purified by centrifugation, washing, and sonication for ∼ 4 h at 4°C immediately after harvest; chemically decoated; and then frozen overnight. Note that the decoating treatment will remove germinated spores and growing cells as well as sporulating cell debris. The next day, the frozen decoated spores were thawed on ice and disrupted, and RNA was isolated and frozen. Spores from the second half of the preparation were highly purified over 7 days at 4°C and then chemically decoated, and RNA was extracted. RNA-seq data from decoated spores purified either for <24 h or over 7 days (Tables 4 and 5; Fig. S1E and F) revealed that the most abundant mRNAs from these spores were the same as those seen in spores prepared either on plates or in liquid and then highly purified. However, the levels of 8 of the most abundant mRNAs (Table 4) and 6 mRNAs of the low-abundance group (Table 5) were ≥2-fold higher in spores from which RNA was isolated soon after spore harvest than the levels in spores held for 7 days at 4°C and then decoated prior to RNA isolation, but there were also some mRNAs that were at higher levels in the 7-day spores (Tables 4 and 5). Overall, these results suggest that most spore mRNAs, but perhaps not all, are relatively stable over 7 days at 4°C. Indeed, the fact that spore decoating for 30 min at 70°C and at pH 13 to 14 had no major effects on the levels of spore mRNAs prepared in liquid (Tables 4 and 5) further indicates that these mRNAs are relatively stable in the novel environment of the spore core.
DISCUSSION
Probably the most important conclusion from this work is that only ∼46 mRNAs detected by RNA-seq in B. subtilis spores are present at ≥1 molecule/spore. Presumably, this is also the case in spores of other Bacillus species and perhaps Clostridiales species as well. However, RNA-seq has not been carried out on all these spores’ mRNAs, nor have average levels of mRNA nucleotides per spore been determined. Given that only the ∼46 most abundant mRNAs are present at ≥1 copy/B. subtilis spore, these mRNAs are then the ones that could give rise to proteins during spore revival and might influence the behavior of a spore population. What is known about the group of genes that encode these mRNAs? First, the great majority are transcribed only in the developing spore late in sporulation under the control of σG, which directs the synthesis of the last group of mRNAs made in the developing spore, although the transcription of some of these genes is also directed by σF acting earlier in the forespore than σG (26). However, not all known σG-dependent mRNAs are found among the most abundant spore mRNAs, as the latter group does not include (i) genes encoding spores’ nutrient germinant receptors; (ii) genes encoding the proteins composing the SpoVA channel by which CaDPA moves across spores’ inner membrane (IM); (iii) the gene encoding at least one cortex-lytic enzyme, SleB, involved in spore cortex hydrolysis in spore germination; or (iv) many other genes, often of unknown function (26). Notably, the expression levels of the latter genes are generally much lower than those of the genes encoding some of the most abundant spore mRNAs, in particular the sspA, sspB, and sspE genes, which, when transcribed, encode ∼40% of all protein synthesized in the developing spore (31). The spoVT gene is transcribed under the control of both σF and σG, and SpoVT activates and represses the transcription of different σG-dependent genes (26). Consequently, it was possible that the expression of the most abundant spore mRNAs is activated by SpoVT. However, the expression of only ∼33% of the 46 most abundant spore mRNAs is activated by SpoVT, while the expression of ∼41% is repressed (26). A second possibility is that the σG-dependent mRNAs found in spores are much more stable than other mRNAs. This certainly remains a possibility, but at least the B. subtilis sspA and sspB mRNAs have been reported to exhibit a stability in the developing forespore that is similar to that of bulk mRNA (32). Two additional features of the abundant mRNAs found in spores are that (i) these mRNAs are rather small, averaging 339 to 505 nt, compared to the ∼1-kb average size for protein-coding genes, and (ii) the proteins encoded by almost all of these abundant mRNAs have been detected in proteomic analyses on spores (33, 34). In dormant spores prepared on plates and highly purified, 42 of the 46 most abundant spore mRNAs encode proteins detected in spores, while 3 of the remaining 4 mRNAs encode small 29- to 46-amino-acid (aa) polypeptides (Table 1) that might easily be missed in proteomic analyses.
Certainly, a major question about the abundant mRNAs in dormant spores is whether these gene products play a role in spore properties such as resistance, stability, germination, and outgrowth. As far as is known, the genes encoding the ∼46 most abundant spore mRNAs are not essential for cell growth, but as is evident from the large percentage that are “y” genes, the exact function of many of the proteins encoded by these genes is not known. However, for many of the ssp genes, all of which encode proteins present in spores, their deletion has no significant effects on spore germination or outgrowth (3, 35–37). In contrast, some of the most abundant spore mRNAs, most notably from the sspA, sspB, and sspE genes, encode proteins crucial for spore resistance properties (SspA/SspB) and as sources of amino acids for protein synthesis early in spore outgrowth (SspA, SspB, and SspE) (1, 2, 11). Consequently, it is difficult to imagine any benefit to spores in translating these proteins’ mRNAs in spore revival. Indeed, at least the presence of SspA/SspB-type proteins has drastic negative effects on macromolecular synthesis in outgrowing spores or growing cells due to the interaction of these proteins with DNA (38, 39). The latter effects are presumably why the degradation of SspA and SspB takes place soon after germination is initiated (3, 11), not only to generate free amino acids but also to destroy these potentially deadly proteins. However, one important function that these abundant spore mRNAs could fulfill when spores return to life is to be degraded rapidly to serve as a source of preformed nucleotides for the synthesis of new RNAs. This is crucial for a spore which has germinated in an environment lacking preformed nucleic acid bases, as enzymes for the synthesis of ribonucleotides from amino acids and other precursors are lost in sporulation and are synthesized only at defined times in spore outgrowth (3, 10). Thus, the abundant spore mRNAs can serve as a reservoir of RNA precursors, and degradation of spore RNA, including mRNA, has been observed early after germination is initiated (19, 21). Consequently, the most likely function for the great majority of spore mRNAs is to serve as a ribonucleotide reserve. It is, however, certainly possible that a few spore mRNAs direct the synthesis of some important proteins early in spore outgrowth, and this is a topic for further study.
Overall, what is known about the expression of genes encoding the most abundant spore mRNAs and the presence of most of the encoded proteins in spores suggest that the great majority of the most abundant dormant spore mRNAs are translated in the developing forespore late in its development. However, soon after these mRNAs’ translation takes place, the developing B. subtilis spore core undergoes (i) a decrease in pH from 7.8 to 6.5, (ii) a decrease in the percentage of wet weight as water from 80% to ∼45%, and (iii) an accumulation of ∼25% of the dry weight as CaDPA, which further reduces the core water content to 35% of the wet weight (1). Presumably, the completion of these events in effect “freezes” the spore cytoplasm, such that the mRNAs left are now quite stable (40, 41). Indeed, the mobility of a soluble protein in the dormant spore core falls ≥4 orders of magnitude during sporulation (41). These abundant mRNAs then await spore germination and outgrowth when they are degraded to allow synthesis of new mRNAs.
An obvious question about the abundant mRNAs in spores is whether they are stable or are degraded as spores age. Our initial experiment to test this suggests that there may be degradation of some, but by no means all, mRNAs in spores held at 4°C for a week but not when incubated for 30 min at 70°C at pH 13 to 14. However, it will be interesting to see what happens when spores are incubated at higher temperatures (37°C to 70°C) for extended times; indeed, rRNA is known to be fragmented significantly in spores held for long periods at these temperatures (7, 24). Some spore mRNAs have also been shown to be degraded during spore storage at elevated temperatures (24). Additional questions about the abundant spore mRNAs include (i) what might be the function of the products of many of the “y” genes, and in particular, do some of the encoded proteins have redundant functions, as do at least some ssp genes (1), and (ii) might a few of these mRNAs be translated early in spore germination/outgrowth and serve a crucial function in this period of spore development, although this seems unlikely for most of them, as noted above? Clearly, there are still some important things to be learned about the genes encoding abundant spore mRNAs and the mRNAs themselves.
A final major question is why there are so many mRNAs in only small percentages of spore populations, especially since some are thought to be expressed in the mother cell compartment of the sporulating cell. Our results rule out that this is due to contamination of spore preparations with mother cell constituents, including trapping of mRNA in spores’ outer layers. This is consistent with previous work (24) but is now shown directly in the present work. We envisage at least three possible explanations for the presence of low-abundance mRNAs in spores, as follows. First, there is a connection between the mother cell and the developing spore, which has been termed a “feeding tube” (42–45) and is thought to be in essence an umbilical cord through which low-molecular-weight nutrients such as amino acids, ATP, and other ribonucleoside triphosphates are made available to the forespore; note that forespores have lost enzymes for amino acid and nucleotide biosynthesis as well as the tricarboxylic acid cycle (3). Perhaps some mRNAs can be delivered to the developing spore through this feeding tube, as was suggested several years ago (24). This would certainly explain why mRNAs from mother cell-expressed genes would be in spores. This feeding tube could also move developing spore mRNAs into the mother cell. While the expression of such developing spore genes in the mother cell is clearly minimal, quite low levels would probably either be not detected or passed off as contamination. A second possibility is that the transcription apparatus in the developing spore is not 100% selective and, in addition to transcribing large numbers of the abundant spore mRNAs, occasionally carries out low levels of inappropriate transcription, hence the presence of low levels of large numbers of mRNAs not under σG control in spores. Indeed, the σ factor directing the transcription of housekeeping genes, σA, has been detected in significant levels in dormant B. subtilis spores (33). However, the mother cell-specific σ factors σE and σK have not been detected in dormant spores (26, 33). Finally, a third possibility is that some of the mRNAs that are found in the spore yet thought to be transcribed only in the mother cell compartment of the sporulating cell, such as the yhcO and sscA mRNAs, in fact have a promoter recognized in the forespore, perhaps even one recognized by σA or at an extremely low level by σF or σG. It is not clear which of the possible explanations for the high level of low-abundance mRNAs in dormant spores mentioned above are correct, but again, this seems to be an important topic for future study.
MATERIALS AND METHODS
Spore preparation and purification.
The B. subtilis strain used in this work was PS533 (46), a laboratory strain 168 derivative that contains plasmid pUB110, which provides resistance to kanamycin (10 μg/ml). Spores of this strain were routinely produced at 37°C on double-strength Schaeffer’s glucose (2×SG) medium agar plates for ∼2 days and harvested (47, 48). Purification was routinely performed over ∼7 days by daily multiple washes with 4°C water and intermittent sonication for 3 min on ice, and obvious contaminating material was washed off surfaces of spore pellets with a gentle stream of water, followed by centrifugation through 50% HistoDenz (2); these spores are termed highly purified (see reference 2 for a bright-field microscopic image of spores purified in this manner). Note that upon centrifugation through 50% HistoDenz, dormant spores pellet because of their high core wet density, while impurities will float. Sporulation plates were also incubated for only 24 h, and spores were harvested and either highly purified over ∼7 days as described above or washed only 3 times by centrifugation with 4°C water, with sonication between centrifugations. The final pellets of the latter two spore preparations were then chemically decoated for 30 min at 70°C in a solution containing 1 M dithiothreitol, 4 M NaCl, 1 M NaOH, and 20% sodium dodecyl sulfate and washed six times at 4°C with 0.15 M NaCl and then with water as described previously (24, 49). Spores were also prepared at 37°C in liquid 2×SG medium for 2 days, which is the minimum time to obtain release from the sporangium of >90% of the spores from sporulating cells. These spores were then either (i) highly purified over 7 days as described above, (ii) purified over 7 days as described above but with sonication, removal of surface material on spore pellets, and centrifugation through 50% HistoDenz being omitted, giving what were termed less-well-purified spores, or (iii) highly purified, followed by chemical decoating, as described above. Total RNA was extracted from spores prepared in various ways and purified as described previously (7). Quantity and purity ratios were determined for each sample using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Purified RNAs were sent to the UConn Center for Genome Innovation facility for quality control, rRNA depletion, library construction, and RNA-seq analyses (see below).
Illumina transcriptome library preparation, sequencing, and analysis.
To further assess RNA quality, total RNA was analyzed on an TapeStation 4200 system (Agilent Technologies, Santa Clara, CA) using the RNA high-sensitivity assay. Ribosomal integrity number (RINe) values were recorded for each sample. Only samples with RINe values above 7.0 were considered for library preparation. Total RNA samples, 300 ng of Qubit (Thermo Fisher Scientific)-quantified total RNA input, were prepared for prokaryotic RNA-seq by first ribodepleting bacterial rRNA using the Ribo-Zero rRNA removal kit (Bacteria) from Illumina (San Diego, CA). Libraries were prepared from ribodepleted RNA using the TruSeq stranded mRNA sample preparation kit according to the manufacturer’s protocol for purified mRNA as the input (Illumina). Libraries were validated for length and adapter dimer removal using the Agilent TapeStation 4200 D1000 high-sensitivity assay and then quantified and normalized using the double-stranded DNA (dsDNA) high-sensitivity assay for Qubit 3.0 (Life Technologies, Carlsbad, CA). Sample libraries were prepared for Illumina MiSeq sequencing using v3 chemistry (paired-end 2- by 75-bp read length). Sequencing read depth was targeted at 10 million to 15 million total paired-end reads/sample.
Analysis of RNA-seq data and assignment of coding genes to regulons.
Analysis of the RNA-seq (fastq) data was performed using the Assembler module in MacVector (v16.0.9) (MacVector, Inc., Apex, NC), using default settings. Raw paired-end read data from each library were aligned to the B. subtilis strain 168 reference genome (GenBank accession number AL009126.3) using Bowtie. The resulting read count (i.e., total number of mapped reads per reference gene), reads per kilobase of transcript per million mapped reads (RPKM), and transcripts per million mapped reads (TPM) values for each library were exported as text for comparison. Mapped reads were visualized over the reference genome using the Map option in MacVector, which graphically displays read coverage across individual genes or putative operons. Summaries of RNA-seq results for each RNA preparation analyzed can be found in Table S1 in the supplemental material. Assignment of the RNA polymerase σ factors directing the transcription of various mRNAs was based on information in the subtiwiki database (http://subtiwiki.uni-goettingen.de) (50) and the literature (26), including correlations between an individual mRNA’s expression under different conditions with those of genes whose expression has been well documented to be under the control of specific σ factors, in particular the forespore-specific σ factor σG (3, 35–37).
Calculations of levels of various mRNAs in spores.
In order to calculate average levels of various mRNA species in dormant B. subtilis spores (Fig. 1), we took advantage of the previously reported values for the levels of phosphorus (P) molecules in a dormant B. subtilis spore, 1.7 × 10−16 mol/spore (27), which, when multiplied by Avogadro’s number, gives ∼108 P molecules/spore. The percentage of spore P in DNA determined in the previous study was 10% (27), and the value for DNA nucleotides in spores now calculated from these data is ∼107 nt/spore, close to the value of 8.4 × 106 nt for the double-stranded B. subtilis genome, and B. subtilis spores are monogenomic (28). With confidence in the relative accuracy of the values for numbers of P molecules per spore, we then calculated that an individual spore, in which 35% of spore P is in RNA (27), contains ∼3.5 × 107 RNA nt. If 3% of spore RNA is assumed to be mRNA, as has been found to be the case in B. subtilis vegetative cells (29, 30), this means that there are only ∼106 mRNA nt/spore.
Data availability.
All RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) (51) and are accessible through GEO series accession number GSE125530.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an award from the Army Research Office to P.S. under contract number W911NF-09-1-0286. R.M. was supported by a grant from the German Aerospace Center (DLR-FuE-Projekt ISS LIFE, Programm RF-FuW, Teilprogramm 475). M.J.C. was supported by the NIH/NIAD under award numbers R01AI029735, R21AI128379, R21AI126146, and R21AI139940.
We are grateful to B. Reese of the UConn Center for Genome Innovation facility for advice and assistance with RNA-seq.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00007-19.
REFERENCES
- 1.Setlow P. 2016. Spore resistance properties, p 201–215. In Driks A, Eichenberger P (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC. [Google Scholar]
- 2.Setlow P. 2019. Observations on research with spores of Bacillales and Clostridiales spores. J Appl Microbiol 126:348–358. doi: 10.1111/jam.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paidhungat M, Setlow P. 2002. Spore germination and outgrowth, p 537–548. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and its relatives: from genes to cells. ASM Press, Washington, DC. [Google Scholar]
- 4.Setlow P, Wang S, Li Y-Q. 2017. Germination of spores of the orders Bacillales and Clostridiales. Annu Rev Microbiol 71:459–477. doi: 10.1146/annurev-micro-090816-093558. [DOI] [PubMed] [Google Scholar]
- 5.Dills SS, Vary JC. 1978. An evaluation of respiration chain-associated functions during initiation of germination of Bacillus megaterium spores. Biochim Biophys Acta 541:301–311. doi: 10.1016/0304-4165(78)90190-3. [DOI] [PubMed] [Google Scholar]
- 6.Setlow P, Kornberg A. 1970. Biochemical studies of bacterial sporulation and germination. XXII. Energy metabolism during early stages of germination of spores of Bacillus megaterium. J Biol Chem 245:3637–3644. [PubMed] [Google Scholar]
- 7.Korza G, Setlow B, Rao L, Li Q, Setlow P. 2016. Changes in Bacillus spore small molecules, rRNA, germination, and outgrowth after extended sublethal exposure to various temperatures: evidence that protein synthesis is not essential for spore germination. J Bacteriol 198:3254–3264. doi: 10.1128/JB.00583-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinai L, Rosenberg A, Smith Y, Segev E, Ben-Yehuda S. 2015. The molecular timeline of a reviving bacterial spore. Mol Cell 57:695–707. doi: 10.1016/j.molcel.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Korza G, Maciejewski M, Setlow P. 2015. Analysis of metabolism in dormant spores of Bacillus species by 31P nuclear magnetic resonance analysis of low-molecular-weight compounds. J Bacteriol 197:992–1001. doi: 10.1128/JB.02520-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setlow P, Kornberg A. 1970. Biochemical studies of bacterial sporulation and germination. XXIII. Nucleotide metabolism during spore germination. J Biol Chem 245:3645–3652. [PubMed] [Google Scholar]
- 11.Setlow P, Primus G. 1975. Protein metabolism during germination of Bacillus megaterium spores. J Biol Chem 250:623–630. [PubMed] [Google Scholar]
- 12.Steinberg W, Halvorson HO, Keynan A, Weinberg E. 1965. Timing of protein synthesis during germination and outgrowth of spores of Bacillus cereus strain T. Nature 208:710–711. doi: 10.1038/208710a0. [DOI] [Google Scholar]
- 13.Torriani A, Levinthal C. 1967. Ordered synthesis of proteins during outgrowth of spores of Bacillus cereus. J Bacteriol 94:176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambon P, Deutscher MP, Kornberg A. 1968. Biochemical studies of bacterial sporulation and germination. XX. Ribosomes and nucleic acids of vegetative cells and spores of Bacillus megaterium. J Biol Chem 243:5110–5116. [PubMed] [Google Scholar]
- 15.Deutscher MP, Chambon P, Kornberg A. 1968. Biochemical studies of bacterial sporulation and germination. XXI. Protein-synthesizing systems from vegetative cells and spores of Bacillus megaterium. J Biol Chem 243:5117–5125. [PubMed] [Google Scholar]
- 16.Jeng YH, Doi RH. 1974. Messenger ribonucleic acid of dormant spores of Bacillus subtilis. J Bacteriol 119:514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassi D, Cappa F, Cocconcelli PS. 2013. Array-based transcriptional analysis of Clostridium sporogenes UC9000 during germination, cell outgrowth and vegetative life. Food Microbiol 33:11–23. doi: 10.1016/j.fm.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Bassi D, Colla F, Gazzola S, Puglisi E, Delledonne M, Cocconcelli PS. 2016. Transcriptome analysis of Bacillus thuringiensis spore life, germination and cell outgrowth in a vegetable-based food model. Food Microbiol 55:73–85. doi: 10.1016/j.fm.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Bettegowda C, Huang X, Lin J, Cheong I, Kohli M, Szabo SA, Zhang X, Diaz LA Jr, Velculescu VE, Parmigiani G, Kinzler KW, Vogelstein B, Zhou S. 2006. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat Biotechnol 12:1573–1580. doi: 10.1038/nbt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dembek M, Stabler RA, Witney AA, Wren BW, Fairweather NF. 2013. Transcriptional analysis of temporal gene expression in germinating Clostridium difficile 630 endospores. PLoS One 8:e64011. doi: 10.1371/journal.pone.0064011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keijser BJF, Ter Beek A, Rauwerda H, Schuren F, Montijn R, van der Spek H, Brul S. 2007. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J Bacteriol 189:3624–3634. doi: 10.1128/JB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson WL, Moeller R, PROTECT Team, Horneck G. 2012. Transcriptomic responses of germinating Bacillus subtilis spores exposed to 1.5 years of space and simulated Martian conditions on the EXPOSE-E experiment PROTECT. Astrobiology 12:469–487. doi: 10.1089/ast.2011.0748. [DOI] [PubMed] [Google Scholar]
- 23.Nagler K, Krawczyk AO, De Jong A, Madela K, Hoffmann T, Laue M, Kuipers OP, Bremer E, Moeller R. 2016. Identification of differentially expressed genes during Bacillus subtilis spore outgrowth in high-salinity environments using RNA sequencing. Front Microbiol 7:1564. doi: 10.3389/fmicb.2016.01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segev E, Smith Y, Ben-Yehuda S. 2012. RNA dynamics in aging bacterial spores. Cell 148:139–149. doi: 10.1016/j.cell.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 25.Van Melis CC, Nierop Groot MN, Tempelaars MH, Moezelaar R, Abee T. 2011. Characterization of germination and outgrowth of sorbic acid-stressed Bacillus cereus ATCC 14579 spores: phenotype and transcriptome analysis. Food Microbiol 28:275–283. doi: 10.1016/j.fm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu T, Greenfield A, Shuster B, Barry SN, Gallitto M, Liu B, Kacmarczyk T, Santoriello F, Chen J, Rodrigues CD, Sato T, Rudner DZ, Driks A, Bonneau R, Eichenberger P. 2015. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol 11:839. doi: 10.15252/msb.20156236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson DL, Kornberg A. 1970. Biochemical studies of bacterial sporulation and germination. XIX. Phosphate metabolism during sporulation. J Biol Chem 245:1137–1145. [PubMed] [Google Scholar]
- 28.Hauser PM, Karamata D. 1992. A method for determination of bacterial spore DNA content based on isotopic labelling, spore germination and diphenylamine assay; ploidy of spores of several Bacillus species. Biochimie 74:723–733. doi: 10.1016/0300-9084(92)90145-5. [DOI] [PubMed] [Google Scholar]
- 29.Brown S, Coleman G. 1975. Messenger ribonucleic acid content of Bacillus amyloliquefaciens throughout its growth cycle compared with Bacillus subtilis 168. J Mol Biol 96:345–352. doi: 10.1016/0022-2836(75)90353-8. [DOI] [PubMed] [Google Scholar]
- 30.Midgley JE. 1969. The messenger ribonucleic acid content of Bacillus subtilis. Biochem J 115:171–181. doi: 10.1042/bj1150171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dignam SS, Setlow P. 1980. In vivo and in vitro synthesis of the spore specific proteins A and C of Bacillus megaterium. J Biol Chem 255:8417–8423. [PubMed] [Google Scholar]
- 32.Leventhal JM, Chambliss GH. 1982. Synthesis of acid-soluble spore proteins by Bacillus subtilis. J Bacteriol 152:1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swarge BN, Roseboom W, Zheng L, Abhyankar WR, Brul S, de Koster CG, de Koning LJ. 2018. “One-pot” sample processing method for proteome-wide analysis of microbial cells and spores. Proteomics Clin Appl 12:e1700169. doi: 10.1002/prca.201700169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng L, Abhyankar W, Ouwerling N, Dekker HL, van Veen H, van der Wel NN, Roseboom W, de Koning LJ, Brul S, de Koster CG. 2016. Bacillus subtilis spore inner membrane proteome. J Proteome Res 15:585–594. doi: 10.1021/acs.jproteome.5b00976. [DOI] [PubMed] [Google Scholar]
- 35.Bagyan I, Setlow B, Setlow P. 1998. New small, acid-soluble proteins unique to spores of Bacillus subtilis: identification of the coding genes and regulation and function of two of these genes. J Bacteriol 180:6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabrera-Hernandez A, Sanchez-Salas JL, Paidhungat M, Setlow P. 1999. Analysis of the regulation of four genes encoding small, acid-soluble spore proteins in Bacillus subtilis. Gene 232:1–10. doi: 10.1016/S0378-1119(99)00124-9. [DOI] [PubMed] [Google Scholar]
- 37.Cabrera-Hernandez A, Setlow P. 2000. Analysis of the regulation and function of five genes encoding small, acid-soluble spore proteins of Bacillus subtilis. Gene 248:169–181. doi: 10.1016/S0378-1119(00)00125-6. [DOI] [PubMed] [Google Scholar]
- 38.Hayes CS, Setlow P. 2001. An α/β-type small, acid-soluble spore protein which has a very high affinity for DNA prevents outgrowth of Bacillus subtilis spores. J Bacteriol 183:2662–2666. doi: 10.1128/JB.183.8.2662-2666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setlow JK, Randesi M, Adams JG, Setlow B, Setlow P. 1992. Mutation and killing of Escherichia coli expressing a cloned Bacillus subtilis gene whose product alters DNA conformation. J Bacteriol 174:2943–2950. doi: 10.1128/jb.174.9.2943-2950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaieda S, Setlow B, Setlow P, Halle B. 2013. Mobility of core water in Bacillus subtilis spores by 2H NMR. Biophys J 105:2016–2023. doi: 10.1016/j.bpj.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowan AE, Koppel DE, Setlow B, Setlow P. 2003. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc Natl Acad Sci U S A 100:4209–4214. doi: 10.1073/pnas.0636762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camp AH, Losick R. 2008. A novel pathway of intercellular signaling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camp AH, Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meisner J, Wang X, Serrano M, Henriques AO, Moran CP Jr.. 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci U S A 105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crawshaw AD, Serrano M, Stanley WA, Henriques AO, Salgado PS. 2014. A mother cell-to-forespore channel: current understanding and future challenges. FEMS Microbiol Lett 358:129–136. doi: 10.1111/1574-6968.12554. [DOI] [PubMed] [Google Scholar]
- 46.Setlow B, Setlow P. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J Bacteriol 178:3486–3495. doi: 10.1128/jb.178.12.3486-3495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J Bacteriol 182:5505–5512. doi: 10.1128/JB.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson WL, Setlow P. 1990. Sporulation, germination and outgrowth, p 391–450. In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 49.Bagyan I, Noback M, Bron S, Paidhungat M, Setlow P. 1998. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 212:179–188. doi: 10.1016/S0378-1119(98)00172-3. [DOI] [PubMed] [Google Scholar]
- 50.Zhu B, Stülke J. 2018. SubtiWiki in 2018: from genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res 46:D743–D748. doi: 10.1093/nar/gkx908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) (51) and are accessible through GEO series accession number GSE125530.