Abstract
The ATPase family, AAA domain-containing protein 2 (ATAD2) has a C-terminal bromodomain, which functions as a chromatin reader domain recognizing acetylated lysine on the histone tails within the nucleosome. ATAD2 is overexpressed in many cancers and its expression is correlated with poor patient outcomes, making it an attractive therapeutic target and potential bio-marker. We solved the crystal structure of the ATAD2 bromodomain and found that it contains a disulfide bridge near the base of the acetyllysine binding pocket (Cys1057-Cys1079). Site-directed mutagenesis revealed that removal of a free C-terminal cysteine (C1101) residue greatly improved the solubility of the ATAD2 bromodomain in vitro. Isothermal titration calorimetry experiments in combination with the Ellman’s assay demonstrated that formation of an intramolecular disulfide bridge negatively impacts the ligand binding affinities and alters the thermodynamic parameters of the ATAD2 bromodomain interaction with a histone H4K5ac peptide as well as a small molecule bromodomain ligand. Molecular dynamics simulations indicate that the formation of the disulfide bridge in the ATAD2 bromodomain does not alter the structure of the folded state or flexibility of the acetyllysine binding pocket. However, consideration of this unique structural feature should be taken into account when examining ligand-binding affinity, or in the design of new bromodomain inhibitor compounds that interact with this acetyllysine reader module.
Keywords: acetyllysine, ATAD2, bromodomain inhibitor, chromatin reader domain, disulfide bridge, epigenetics, histone, post-translational modification
1 |. INTRODUCTION
Eukaryotic DNA is packaged for gene regulation and nuclear organization into nucleosomal units consisting of approximately 146 base pairs of DNA wrapped around an octamer of histones.1 Residues on the N-terminal tails protruding from the histone core are targeted by post-translational modifications, which contribute to gene regulation by supporting or inhibiting access to the genetic information. These heritable modifications are collectively called the “histone code,” and they can propagate alternative phenotypes without any changes to the DNA sequence.2,3 Aberrant changes in the epigenome are linked to the development of disease, and have been shown to be one of the major mechanisms contributing to oncogenesis.4–6
The ε-N-lysine acetylation motif is one of the most common post-translational modifications found in proteins.7 Addition of an acetyl group neutralizes the positive charge on the lysine side-chain, and interrupts the interaction of histone proteins with the negatively-charged DNA, resulting in opening of the chromatin structure to increase gene expression.7 Acetylation levels are tightly controlled by histone acetyl transferases (HATs; which create acetylation marks) and histone deacetylases (HDACs; which remove acetylation marks).8 Histone acetylation is generally associated with euchromatin and gene active transcription, and these marks can also be important for DNA repair and replication.9 Inappropriate histone acetylation has been linked to the abnormal expression of disease-promoting genes, particularly in cancer.10
Bromodomains (BRDs) are well-characterized “reader” modules that recognize ε-N-acetylated lysines found on the histone tails through a conserved asparagine residue in their hydrophobic binding pocket.11 Bromodomains are comprised of four α-helices (αZ, αA, αB, αC) that are connected by highly variable loop regions (ZA and BC loops), which form the rim of the histone binding pocket.11 In addition, bromodomain structures frequently contain three stabilizing hydro-phobic residues near the core of their helical bundle, as well as a tyro-sine in the short αAZ helix, and an aspartate capping the αA helix, which forms a hydrogen bond with a backbone amide.12 In the bromodomain and extra terminal domain (BET) subfamily of bromodomains a “WPF shelf” motif and a conserved gatekeeper residue were found to make hydrophobic contacts important for the selection of histone ligands13 As an evolutionarily conserved domain, bromodomains function to guide the bromodomain-containing protein and any associated subunits to the chromatin.14 Due to their presence in many transcriptional complexes, aberrant expression of bromodomain-containing proteins is increasingly recognized as a contributing factor in cancer cell proliferation and survival.15
One such bromodomain-containing protein is the ATPase family, AAA domain-containing protein 2 (ATAD2, UniProtKB Q6PL18, also called ANCCA/PRO 2000).16 ATAD2 has been shown to recognize his-tone H4 that is acetylated at lysine 5 (H4K5ac) and lysine 12 (H4K12ac),17,18 and has recently been proposed to function as a reader of newly synthesized H4K5acK12ac di-acetyllysine marks during DNA replication.19 Several studies suggest a role for ATAD2 in the pathogenesis of cancer where it functions as a co-regulator of oncogenic transcription factors including E2F,20,21 the estrogen receptor-α,16 the androgen receptor22, and MYC.23,24 Once initiated, ATAD2 expression leads to several positive feedback loops, dependent on tissue type, the products of which will further upregulate ATAD2,16 cell proliferation, and survival genes. ATAD2 overexpression is associated with a myriad of unrelated cancers, including breast,20,23,25–28 colorectal,29–31 endometrial32–34 gastric,31,35,36 hepatocellular carcinoma,37–40 lung,25,41,42 ovarian,43,44 and prostate.16,45,46 Up regulation of ATAD2 is often correlated with poor patient outcomes, and can be used as prognostic marker.25,26,30,35,43 This pattern across multiple cancers makes the ATAD2 bromodomain an attractive target for cancer therapy, and several inhibitors are currently in development.17,47–50
To further investigate structural features of the ATAD2 bromodomain that contribute to histone ligand and small molecule inhibitor recognition we optimized the expression and solubility of this protein. Crystallographic studies coupled with site-directed mutagenesis and isothermal titration calorimetry binding assays revealed that formation of a disulfide bridge near the base of the ATAD2 bromodomain binding pocket impacts protein solubility, histone ligand, and small molecule inhibitor binding. Molecular dynamics simulations provide evidence that the overall rigidity of the four-helix bundle and flexibility of the binding site loops are not affected by the disulfide bridge. This study highlights an additional structural element to be taken into consideration when examining ligand binding affinity and working to develop new therapeutics targeting the ATAD2 bromodomain.
2 |. MATERIALS AND METHODS
2.1 |. Plasmid construction
The human ATAD2 bromodomain plasmid was a gift from Dr. Nicola Burgess-Brown (Addgene plasmid # 38916). Residues 981–1108 encoding the 128 amino acid ATAD2 bromodomain were PCR amplified and recloned into the pDEST15 vector (Invitrogen) with an N-terminal GST (glutathione transferase) tag followed by a PreScission Protease site (GE Healthcare) using the Gateway Cloning technology (Invitrogen). The DNA sequence was verified and the plasmid was transformed into Escherichia coli Rosetta 2 (DE3) pLysS competent cells (Novagen), which supply tRNAs for rare codons to express the wild-type ATAD2 bromodomain (WT ATAD2). To further improve protein expression of the human ATAD2 bromodomain in E. coli the ATAD2 bromodomain gene sequence was codon-optimized by DAPCEL, synthesized (Bio Basic Inc.) and recloned into the pDEST15 vector containing an N-terminal GST tag and PreScission Protease site as described above. The codon optimized ATAD2 bromodomain DNA sequence was verified (sequence provided in Supporting Information figure S1), and then transformed into E. coli One Shot BL21 Star (DE3; Invitrogen) cells (ATAD2 CO). An ATAD2 bromodomain C1101A mutant protein was created using site-directed mutagenesis with the QuikChange II kit (Agilent). Primers were designed to introduce the C1101A mutation using standard PCR from the pDEST15 plasmid containing the codon optimized ATAD2 bromodomain as a template. The ATAD2 C1057A, C1079A, and C1101A triple cysteine mutant was made in the same way from the codon-optimized plasmid. The DNA sequences for all mutants were verified before transformation into E. coli One Shot BL21 Star (DE3; Invitrogen) cells (ATAD2 CO C1101A and ATAD2 CO C1057A/C1079A/C1101A).
2.2 |. ATAD2 bromodomain expression and purification
The wild-type and codon optimized mutant versions of the ATAD2 bromodomain protein were expressed from E. coli Rosetta 2 (DE3) pLysS or BL21 Star (DE3) cells grown in TB (Terrific Broth) at 37°C. Once the culture OD600 reached 1.2, the temperature was reduced to 20°C for 1 hour before induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubated for 16 hours at 20°C. The cells were harvested by centrifugation and soniciated in lysis buffer containing 50 mM Tris pH 7.5, 500 mM NaCl, 0.05% Nonidet P-40, 5% glycerol, and 1 mM EDTA. The ATAD2 bromodomain protein was purified using glutathione agarose resin (Thermo Scientific) in a 2.5 × 5 cm BioRad econo-column with wash buffer (50 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol and 1 mM EDTA). The GST tag was cleaved overnight at 4°C by addition of PreScission Protease (GE Healthcare), and the ATAD2 bromodomain was eluted with wash buffer, and the eluted fractions were pooled, concentrated and dialyzed into the appropriate buffers described below. The ATAD2 bromodomain total protein concentration was determined from its absorbance at 280 nm using the extinction coefficient of 7450 M−1 cm−1. Thermo Scientific Pierce Ellman’s Reagent (DTNB, 5,5’-dithio-bis-[2-nitrobenzoic acid]) was used to calculate the free sulfhydryl groups in the purified ATAD2 bromodomain proteins by measuring their absorbance at 405 nm on a Eppendorf BioPhotometer-plus and using the molar extinction coefficient of TNB (2-nitro-5-thiobenzoic acid) 14 050 M−1 cm−1 at 25°C in ITC buffer (20 mM NaH2PO4 pH 7.0, 150 mM NaCl).51 The absorbance of the measured TNB product was used to calculate the millimolar concentration of free sulfhydryl groups in each ATAD2 bromodomain sample according to the reagent instructions. The following formula was used to assess the percentage of ATAD2 bromodomain protein with a formed disulfide bridge. First, we multiplied the ATAD2 bromodomain total protein concentration by a factor of 2 or 3 for the ATAD2 CO C1101A bromodomain or the ATAD2 WT bromodomain, respectively, to obtain the potential concentration of free Cys residues found in each sample. Then the concentration of free cysteine’s measured in the Ellman’s assay was subtracted from the total Cys concentration possible ([Total Cys]-[Free Cys]) in order to detect the concentration of Cys residues that are bound in disulfide bonds, ([Bound Cys]). The concentration of bound Cys was divided by two to account for the two Cys residues in the ATAD2 bromodomain that can participate in the intra-molecular disulfide bridge. Then we used the ratio of [bound Cys] to [total protein] to determine the percentage of disulfide bridge formed. However, this is an approximation for the WT ATAD2 bromodomain sample as we expect the C-terminal C1101 residue (or even C1079/C1057) to form intermolecular disulfide bonds between other bromodomain proteins in solution.
To remove any bromodomains that did not have a fully formed disulfide bridge we further purified the ATAD2 CO C1101A bromodomain protein sample by incubating it with activated thiol sepharose 4B resin (GE Healthcare), which binds to any free cysteine residues, at 4°C for 2 hours. The suspension was applied to a 2.5 × 20 cm BioRad econo-column and the free bromodomain protein (containing the disulfide bridge) was eluted with wash buffer. The eluted ATAD2 CO C1101A bromodomain sample containing protein with a fully formed disulfide bridge was concentrated to 1 mL total volume and applied to a HiPrep 16/60 Sephacryl S-100 size exclusion column (GE Healthcare), equilibrated with wash buffer, to remove any free 2-thiopyridone from the sample. The ATAD2 CO C1101A bromodomain protein was concentrated to approximately 0.2 mM and dialyzed for 48 hours at 4°C into ITC Buffer (20 mM NaPO4 pH 7.0, 150 mM NaCl). The final percentage of disulfide bridges in the sample was confirmed using the Ellman’s reagent assay described above immediately before ITC experiments.
2.3 |. X-ray crystallography
The WT ATAD2 bromodomain protein was purified using the lysis and wash buffers as described above that were supplemented with 1 mM dithiothreitol (DTT). Once the protein was eluted from the GST column it was concentrated and further purified by gel filtration chromatography using a HiPrep 16/60 Sephacryl S-100 High Resolution column (GE Healthcare) equilibrated with crystallization buffer (25 mM HEPES-HCl pH 7.5, 150 mM NaCl, and 1 mM DTT). Eluted fractions corresponding to the WT ATAD2 bromodomain were pooled and concentrated, and the purity of the sample was checked on an SDS-PAGE gel. 0.517 mM of the purified 131-residue WT ATAD2 bromodomain protein (containing 128 residues from the bromodomain and -GPL from the N-terminal GST tag) was mixed with1.637 mM of the H2AK5ac peptide ligand (residues 1–12, SGRGKacQGGKARA) and incubated in a microcentrifuge tube for 1 hour prior to tray setup. Crystallization screens were performed using the sitting-drop method in 96-well CrystalEX plates with drops consisting of 0.8 μL of the protein-peptide mixture plus 0.8 μL reservoir solution and a reservoir volume of 50 μL.
Crystals of the ATAD2 bromodomain in complex with the H2AK5ac ligand grew at 4°C in condition number 16 of the Hampton Crystal Screen 1 (0.1 M HEPES sodium pH 7.5, 1.5 M Lithium sulfate monohydrate). Optimizations were performed using the hanging-drop method (1 μL protein solution plus 1 μL mother liquor) in 24-well VDX plates (Hampton Research) containing 500 μL mother liquor in the reservoir. After screening against pH and precipitant concentrations, the optimal crystallization condition for the ATAD2 bromo:H2AK5ac complex was found to be 0.1 M HEPES sodium pH 7.5 and 1.6 M lithium sulfate at 277 K. Crystals were cryoprotected by soaking for 60 seconds in a solution containing 2.0 M lithium sulfate and 0.1 M HEPES sodium pH 7.5 before flash-cooling in liquid nitrogen at 100 K. Crystal screening and data collection was carried out at the Center for X-ray Crystal-lography at the University of Vermont on a Bruker D8 Quest generator equipped with a PHOTON II detector at a wavelength of 1.5418 Å. The diffraction data were processed using the Proteum3 suite (Bruker). The structure was solved by molecular replacement using Phaser with the apo ATAD2B bromodomain structure (PDBID: 3DAI) as the starting model.52 The model was built using COOT.53 Notably, isomorphous difference maps did not support placement of the H2AK5ac peptide. Instead, the anomalous difference map indicated a HEPES molecule from the crystallization buffer bound into the ATAD2 bromodomain binding pocket (Supporting Information figure S2). PHENIX and COOT were used for iterative rounds of refinement, density modification and model building.53,54 The final structure at 1.93 Å resolution was validated using MolProbity and Polygon.55,56
2.4 |. Isothermal titration calorimetry
ITC measurements were performed using a MicroCal iTC200 (GE Healthcare) as described previously.57 The WT and the mutant bromodomain proteins were prepared in a 20 mM NaH2PO4 pH 7.0, 150 mM NaCl ITC buffer by dialysis for 48 hours. The histone H4K5ac peptide (residues 1–10, SGRGKacGGKGL) was synthesized by the Peptide Core Facility at the University of Colorado Denver. Both the histone peptide and the Compound 38 (C-38) ligand were prepared in the ITC dialysis buffer. Titration experiments with the histone H4K5ac peptide were set up for optimal heat of binding reactions at 5°C over 20 injections, with one preliminary injection of 0.5 μL, followed by 19 2.0 μL injections, using 200 μM ATAD2 bromodomain protein in the sample cell, and 5 mM of histone peptide in the injection syringe.
Reverse titration experiments were used to measure the nanomolar binding affinities of the C-38 ligand with the ATAD2 CO C1101A bromodomain protein. In this case 300 μM of ATAD2 CO C1101A bromodomain was loaded in the injection syringe, while 20 μM of the C-38 ligand was in the sample cell. ITC experiments with the C-38 ligand were carried out at 25°C over 20 injections, with one preliminary injection of 0.5 μL, followed by 19 2.0 μL injections. For all samples each injection was spaced apart by time intervals of 150 seconds after an initial delay of 60 seconds and the titration cell was continuously stirred at 750 RPM. The preliminary injection was excluded from integration and calculation of the KDs. The raw data were integrated, corrected for nonspecific heats of dilution, and analyzed according to a 1:1 binding model assuming a single set of identical binding sites to calculate the binding affinity, stoichiometry (N), and the thermodynamic values. Data were analyzed using the software ORIGIN 7.0 (OriginLab Corporation). All experiments where binding occurred were performed in triplicate, while nonbinding experiments were performed in duplicate and standard errors are reported as standard deviations.
2.5 |. Molecular dynamics simulations
Molecular dynamics simulations were used to investigate the structural flexibility, on the sub-μs time scale, of the ATAD2 bromodomain in the presence or absence of the disulfide bridge between residues C1057 and C1079. The starting coordinates of two independent runs were taken from the solved crystal structure (PDBID: 6CPS) and the two alternate rotameric states of C1079 were used to model the presence or absence of the disulfide bridge. The structures were prepared with the CAMPARI v3 software package (http://campari.sourceforge.net) as follows. We added hydrogens and missing side chains, removed the expression tags and capped the N- and C-terminus with acetyl and N-methylamide groups respectively. To reproduce neutral pH conditions we set the protonation states of the side chains of D/E (negatively charged), K/R (positively charged) and H (neutral, ε-N protonated). HEPES and buffer ions were removed. We solvated the system in a cubic box with 85 Å side length and added K+ and Cl− ions to neutralize the system and approximate an ionic strength of 150 mM. A short relaxation with molecular dynamics in torsional space58 was run to reduce major clashes in the structure. The equilibration and production simulations were run with GROMACS 201659 using the CHARMM36 force field60 with modified TIP3P water.61 An equilibration of 1 ns in the NPT ensemble (constant number of molecules, pressure and temperature) at 1 bar and 310 K was run to let the box volume adapt to the correct density of the system. The average box side length of84.04 Å calculated from this run was set for the following equilibration (0.5 ns) and production runs in the NVT ensemble (constant number of molecules, volume and temperature) at 310 K with velocity rescaling thermostat62 with a coupling time of 2 ps. The electrostatic interactions were evaluated with the generalized reaction field63 and we used a threshold of 12 Å for truncating all non-bonded interactions. All covalent bonds were constrained with LINCS64 and the integration time was set to 2 fs. The two production simulations (with and without disulfide bridge) were run for 300 ns with saving frequency of 10 ps.
3 |. RESULTS AND DISCUSSION
3.1 |. The ATAD2 bromodomain contains a disulfide bridge
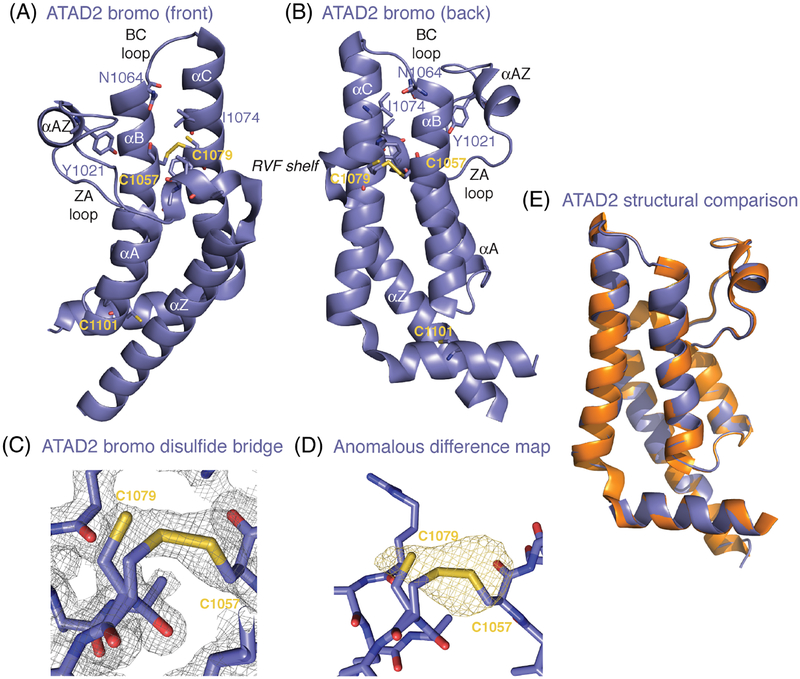
We determined the crystal structure of the wild-type human ATAD2 bromodomain with a HEPES molecule from the crystallization buffer located in the acetyllysine-binding pocket (PDBID: 6CPS). The crystal-lographic data collection details and the refinement statistics are summarized in Table 1. As expected, the ATAD2 bromodomain contains the canonical bromodomain protein-fold consisting of a left-handed four-helix bundle (αZ, αA, αB, and αC), with an extended C-terminal αC kinked helix (Figure 1A,B). The arrangement of the helical bundle creates a deep binding pocket that is surrounded by two variable loop regions that contribute to histone ligand binding specificity. Figure 1A, B shows the overall structure of the ATAD2 bromodomain from the front and back. Residues and structural features known to be important for coordination of H4K5ac18 are labeled. These include the conserved asparagine N1064 that is responsible for co-ordinating the acetylated lysine group, Y1021, which coordinates an ordered water in the binding pocket, the RVF shelf motif (res 1007–1009, corresponding to the WPF shelf in BET bromodomains) and the gatekeeper residue I1074, both of which make important hydrophobic contacts with the histone ligand. Labeled in yellow are the positions of three cysteine residues in the ATAD2 bromodomain. There is one free C-terminal cysteine located after the kink in the extended αC helical region at position C1101. There are also two cysteine residues in the αB and αC helices at positions C1057 and C1079, respectively, which are located on the opposite the RVF shelf motif in the histone binding pocket. One unique feature of our ATAD2 bromodomain structure is the clear presence of continuous electron density connecting the two cysteine residues in a disulfide bridge, with anomalous signal corresponding to the location of the sulfur atoms (Figure 1C,D, and Supporting Information figure S2). The C1079 residue adopts one alternate conformation; one rotamer participates in the disulfide bridge, and a second conformation is rotated out of the disulfide bond, and each conformer is refined to 50% occupancy. At this time there are over 30 structures of the ATAD2 bromodomain deposited in the Protein Data Bank representing the apo state12, as well as the ATAD2 bromodomain in complex with histone ligands,17,18 and numerous small molecules.49,65,66 However, this is currently the only deposited structure of the ATAD2 bromodomain with clear electron density supporting formation of a disulfide bridge near the base of the bromodomain binding pocket. Previous structural studies on the ATAD2 bromodomain used higher concentrations of DTT (10 mM, compared to our 1 mM) during the purification17,49,66 and/or storage steps,12,18 which likely abrogated formation of the disulfide bridge. As this is a unique structural feature of a bromodomain module we wanted to investigate if it has a role in histone ligand recognition, which could also potentially be important for current drug discovery efforts.
TABLE 1.
Summary of the data collection and refinement statistics for the ATAD2 bromodomain in complex with HEPES
| Collection on a single crystal PDB code | ATAD2 / HEPES 6CPS |
|---|---|
| Data collection | |
| Space group | P6522 |
| Cell dimensions | |
| a, b, c (Å) | 79.359, 79.359, 138.96 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 39.68–1.93 (1.99–1.93) |
| R-pim (%) | 2.6 (58.7) |
| CC1/2 | 0.999 (0.545) |
| Mean I/σ(I) | 20.0 (1.32) |
| Completeness (%) | 99.37 (97.0) |
| Redundancy | 16.4 (6.9) |
| Refinement | |
| Resolution (Å) | 38.68–1.93 |
| No. unique reflections | 36 485 |
| Rwork/Rfree | 18.33 / 21.46 |
| No. atoms: | |
| Protein | 1108 |
| Ligand/ion | 25 |
| Waters | 238 |
| Average B-factors (Å2) | |
| Protein | 25.35 |
| Water | 38.25 |
| Ligand (HEPES) | 37.74 |
| R.M.S deviations: | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.27 |
| Ramachandran plot (%) | |
| Favored | 100 |
| Allowed | 0 |
| Outliers | 0 |
FIGURE 1.
Disulfide bridge formation in the ATAD2 bromodomain. A and B, Three-dimensional structure of the human ATAD2 bromodomain (slate), showing the front and back (180° rotation) with secondary structural elements labeled. Residues known to be important for histone ligand coordination are shown as sticks, and include the conserved Asn1064, which coordinates the acetyllysine, the gatekeeper residue Ile1074, Tyr1021 that coordinates ordered water in the binding pocket, and “RVF shelf” motif residues Arg1007, Val1008 and Phe1009, which are important for ligand selectivity. The residues forming the disulfide bridge include Cys1079 and Cys1057, and link the αB-αC helices near the base of the bromodomain binding pocket. The location of the C-terminal Cys1101 is also labeled. C, The 2Fo-Fc electron density map contoured at 1 σ around the ATAD2 bromodomain disulfide bridge region. D, The anomalous difference map contoured at 3 σ around the ATAD2 bromodomain disulfide bridge region. E, Structural alignment comparing ATAD2 proteins with and without an intact disulfide bridge. The apo structure of the ATAD2 bromodomain with no disulfide bridge (3DAI, orange) was taken from the Protein Data Bank and aligned to our ATAD2 structure with a 50% formed disulfide bridge (6CPS, slate) in PyMOL.80 All figures were generated using PyMOL80
3.2 |. Free sulfhydryl groups contribute to ATAD2 bromodomain aggregation in vitro
We used Ellman’s reagent to estimate the number of free sulfhydryl groups available in the ATAD2 bromodomain samples by monitoring the formation of 2-nitro-5-thiobenzoic acid (TNB) by UV-Vis at 405 nm.51 We estimated that the WT ATAD2 bromodomain protein expressed from Rosetta 2 cells has on average, 56% disulfide bridge formation (Table 2). As the crystal structure of the ATAD2 bromodomain also indicates that the disulfide bridge is partially formed, we concluded that when each of the three sulfhydryl groups is free, they are likely forming intermolecular disulfide bridges. Free thiol groups within protein structures are known to be highly reactive and can interact with other proteins in the solution creating intermolecular disulfide bridges linking two or more proteins together, or cause biologically active disulfide bridges to become mixed. Both of these scenarios can result in protein mis-folding, aggregation of protein oligomers, and precipitation.67 In our experience, the WT ATAD2 bromodomain protein begins to become insoluble and precipitate out of solution when it reaches a concentration of approximately0.2–0.5 mM. Codon optimization of human proteins for heterologous expression in bacterial systems is well known to improve protein production,68,69 folding70–72 and stability73, and is a strategy that has been used to improve expression of other bromodomain-containing proteins.74 In order to enhance the expression and co-translational folding of the WT human ATAD2 bromodomain protein in E. coli BL21 cell strains we developed a codon-optimized version of the ATAD2 bromodomain gene sequence (WT/CO ATAD2).
TABLE 2.
Expression and solubility of ATAD2 bromodomain proteins. The ATAD2 bromodomain protein from the native wild-type ATAD2 gene sequence (ATAD2 WT), the codon optimized ATAD2 C1101A mutant gene sequence (ATAD2 CO C1101A), and the codon optimized ATAD2 triple cysteine mutant (ATAD2 CO C1057A/C1079A/C1101A) were expressed and purified from E. coli host cell strains to optimize the protein production and solubility. The status of disulfide bridge formation for each protein sample calculated using absorbance measurements at 405 nm with the Ellman’s reagent is also indicated
| ATAD2 construct | E. coli host expression strain | Total protein, mM (in ITC buffer) | A405 | Total Cys, mM Possiblea | Free Cys, mMb | Bound Cys, mM (total Cys-free Cys) | Bound Cys, mMc/protein, mM | % Disulfide bridge |
|---|---|---|---|---|---|---|---|---|
| ATAD2 WT | Rosetta2 (DE3) pLysS | 0.589 | 0.139 | 1.767 | 1.108 | 0.659 | 0.559 | 56% |
| ATAD2 CO C1101A | BL21 star (DE3) | 0.410 | 0.052 | 0.820 | 0.415 | 0.405 | 0.493 | 49% |
| ATAD2 CO C1101A, after thiol sepharose purification | BL21 star (DE3) | 0.197 | 0 | 0.394 | 0 | 0 | 0 | 100% |
| ATAD2 CO C1057A/C1079A/C1101A | BL21 star (DE3) | 0.443 | 0 | 0 | 0 | 0 | 0 | 0% |
Calculated by multiplying the protein concentration by the number of Cys residues (3 for ATAD2 WT and 2 for ATAD2 CO C1101A).
Measured using the Ellman’s assay.
The mM of bound Cys is divided by 2 to account for the number of Cys residues participating in disulfide bridge formation (then this number is divided by the total protein concentration).
Codon-optimization of the ATAD2 bromodomain improved its expression, but did not improve overall solubility. Thus, we hypothesized that as the ATAD2 bromodomain proteins come in close contact with each other in solution, the availability of free sulfhydryl groups at positions C1057 and C1079 and C1101 allow for the formation of disulfide bridges between adjacent ATAD2 bromodomains causing them to aggregate. We believe this phenomenon makes it difficult to obtain a highly concentrated solution of the ATAD2 bromodomain protein in vitro, as we are only routinely able to reach concentrations slightly between 0.5 and 0.6 mM with the WT ATAD2 bromodomain, even in the presence of reducing agents such as DTT or TCEP. To circumvent aggregation we designed a C1101A mutant of the codon optimized ATAD2 bromodomain construct (ATAD2 CO C1101A) and tested its histone binding activity after expression in BL21 Star (DE3) cells. The ATAD2 bromodomain has been reported to bind to histone H4 that is acetylated at lysine 5 (H4K5ac).17 As seen in Table 3, and Supporting Information figure S3A the purified WT ATAD2 bromodomain protein with 56% bridge formation binds to histone H4K5ac (residues 1–10) with a dissociation constant of 91.4 ± 6.1 μM using ITC. Comparably, the ATAD2 CO C1101A mutant bromodomain protein has 49% disulfide bridge formation, and ITC titrations demonstrated it binds to the histone H4K5ac ligand with a KD of 96.5 ± 2.1 μM (Tables 2 and 3 and Supporting Information figure S3C), in-line with what we observed for the WT ATAD2 bromodomain protein. However, the ATAD2 CO C1101A mutant bromodomain is significantly more soluble, as we were able to concentrate it to above 1.4 mM without any observable protein aggregation. Thus, removal of the free C-terminal cysteine greatly improved the solubility of this protein, likely by reducing the formation of unwanted intermolecular disulfide bridges.
TABLE 3.
Effect of disulfide bridge formation on ligand binding by the ATAD2 bromodomain. Solution binding constants, binding stoichiometry (N; ligand-to-bromodomain ratio), and thermodynamic parameters (enthalpy, ΔH and entropy, ΔS) of the interaction of the wild type (WT) and codon optimized (CO) mutant ATAD2 bromodomain proteins with the histone H4K5ac peptide (res 1–10) and GlaxoSmithKline inhibitor Compound 38 as measured by ITC
| Complex | % DSB formed | KD | N | ΔH cal/Mol | ΔS cal/Mol/deg |
|---|---|---|---|---|---|
| ATAD2 WT H4K5ac |
56% | 91.4 ± 6.1 μM | 0.93 ± 0.08 | −8297.3 ± 947.8 | −11.6 ± 3.6 |
| ATAD2 CO C1057A/C1079A/C1101A H4K5ac |
No DSB | 112.8 ± 14.7 μM | 0.84 ± 0.03 | −4203.7 ± 795.7 | 3.0 ± 3.2 |
| ATAD2 CO C1101A H4K5ac |
49% | 96.5 ± 2.1 μM | 1.22 ± 0.17 | −5839.3 ± 767.5 | −2.0 ± 2.42 |
| ATAD2 CO C1101A H4K5ac |
100% | 209.6 ± 21.0 μM | 1.26 ± 0.47 | −3569.3 ± 148.0 | 3.0 ± 1.26 |
| ATAD2 CO C1057A/C1079A/C1101A C-38 |
No DSB | 223.4 ± 33.2 nM | 1.14 ± 0.08 | −6894.3 ± 1609.8 | 7.31 ± 5.9 |
| ATAD2 CO C1101A C-38 |
49% | 249.2 ± 27.1 nM | 1.10 ± 0.02 | −8016.0 ± 351.4 | 3.3 ± 1.05 |
| ATAD2 CO C1101A C-38 |
100% | 875.1 ± 96.3 nM | 1.09 ± 0.84 | −11746.0 ± 6804.0 | −20.2 ± 11.0 |
3.3 |. Disulfide bridge formation impacts ligand binding affinity
The two cysteine residues that form a disulfide bridge in the ATAD2 bromodomain are located near the base of the bromodomain binding pocket, opposite from the RVF shelf motif. Disulfide bridges play many important roles in protein structure. Formation of disulfide bonds is known to increase protein rigidity, thermal stability and contribute to folding of protein modules.75 We postulated that disulfide bridge formation could contribute to the overall conformation of the bromodomain acetyllysine binding pocket, and play a role in the recognition of histone ligands.
As free sulfhydryl groups also negatively impact the ATAD2 bromodomain stability in solution we aimed to develop a method to isolate the ATAD2 bromodomain protein with 100% of the disulfide bridges formed. To do this, we expressed the ATAD2 CO C1101A mutant bromodomain protein in BL21 Star (DE3) cells, and added an additional purification step after elution from the glutathione agarose column. By applying the ATAD2 CO C1101A protein to an activated agarose 4B column that binds to any solutes containing free thiol groups, we were able to recover the fraction of the ATAD2 CO C1101A bromodomain sample that contains a fully formed disulfide bridge in the eluted protein. This protein was then further purified by size exclusion chromatography to desalt the sample prior to ITC experiments to test its histone-binding activity. The Ellman’s assay confirmed we were able to isolate the ATAD2 CO C1101A bromodomain with a 100% formed disulfide bridge (Table 2). Also, removal of any free cysteines improved the protein solubility significantly as we were able to concentrate it up to 1.9 mM without any signs of precipitation. Interestingly, the presence of the fully formed disulfide bridge reduces the binding affinity of the ATAD2 CO C1101A bromodomain for histone H4K5ac to 209.6 ± 21.0 μM, from the 96.5 ± 2.1 μM affinity observed with the ATAD2 CO C1101A bromodomain containing a 49% formed disulfide bridge (Table 3 and Supporting Information figure 3C–D). Similarly, when we tested the binding affinity of Compound 38 (C-38), a small molecule inhibitor of the ATAD2 bromodomain developed by GlaxoSmithKline49, we observed a binding affinity of 249.2 ± 27.1 nM when the protein has 49% disulfide bridge for mation, but this is reduced to 875.1 ± 96.3 nM when the disulfide bridge is fully formed (Tables 2 and 3. and Supporting Information figure 2F–G).
The presence or absence of an intact disulfide bridge may be a regulatory element for the ATAD2 bromodomain. For example, disulfide bridges are an important feature of the signal transducer and activator of transcription protein 3 (STAT3). Unphosphorylated STAT3 contains two disulfide bridges that are essential for the formation of STAT3 dimers that can shuttle into the nucleus and interact with DNA. Mutation of the four cysteine residues needed for the formation of intermolecular disulfide bridges in dimerization completely abolishes the DNA-binding activity of STAT3.76 Expression of the ATAD2 protein is tightly regulated, and it is predominantly found in the nucleus during S phase where it is associated with DNA replication sites.19 To investigate how complete removal of the disulfide bridge from the ATAD2 binding pocket would affect ligand binding we generated a triple cysteine mutant with residues C1057, C1079, and C1101 changed to alanine (ATAD2 CO C1057A/C1079A/C1101A). As seen in Table 3 (and Supporting Information figure S3) ITC titration experiments with the H4K5ac and C-38 inhibitor compound showed they bound to the ATAD2 triple cysteine mutant with an affinity of 112.8 ± 14.7 μM and 223.4 ± 32.2 nM, respectively. Together our binding data demonstrate that removal of the disulfide bridge does not have a significant impact on the binding activity of the ATAD2 bromodomain, but increasing the amount of disulfide bridge formed weakens ligand interactions. In support of this, a previously reported KD value for the ATAD2 bromodomain with the histone H4K5ac peptide ligand of 22 μM was lower than our KD for the WT protein (91 μM), likely due to strong reducing conditions with the addition of 10 mM DTT in the ITC experiment, and possibly from the use of a longer peptide spanning residues 1–20.17 Thus, in vitro it appears there is interplay between disulfide bridge formation in the ATAD2 bromodomain and its ligand binding affinity. The bromodomain alpha helical bundle is quite stable, and other parts of the bromodomain structure likely compensate for complete loss of the disulfide bridge, while higher percentages of disulfide bridge formation may limit ligand interactions. In the cell, redox regulation in the nucleus may be a mechanism used to influence the biological activity of a subset of bromodomain-containing proteins including ATAD2A. A similar scenario was recently observed for the plant homeodomain (PHD finger) of the protein partner of Sans-fille (PPS), where interaction with its H3K4me3 histone ligand is regulated by pH, and high pH values were shown to increase the ligand interaction.77
3.4 |. Effect of disulfide bridge formation on the thermodynamics of ATAD2 bromodomain ligand binding
Comparison of our ATAD2 bromodomain crystal structure with a 50% formed disulfide bridge (PDBID: 6CPS), to the previously deposited apo structure of the ATAD2 bromodomain with no disulfide bridge (PDBID: 3DAI) by structural alignment (RMSD = 0.209 Å over 109 Cα atoms), revealed that no major structural/conformational changes are induced by formation of the disulfide bridge (Figure 1E). The bridge forms in a region between the αB and αC helices, which is already structurally rigid. This is supported by the molecular dynamics simulations (see below).
Our ITC binding assays also indicate that the formation of the disulfide bridge does not lead to dramatic changes in the structure of the binding site. The data fits for the ligand binding to the ATAD2 bromodomain samples containing two fractions of the protein, with and without the formed disulfide bridge (ATAD2 WT and ATAD2 CO C1101A before thiol sepharose purification) indicate that only one type of binding sites are present in both the bridged and unbridged forms (Table 3, N ~ 1). The ITC measurements do show a two-fold to three-fold decrease in affinity for the histone H4K5ac ligand and C-38 small molecule inhibitor, respectively, when the ATAD2 bromodomain contains a 100% formed disulfide bridge.
Analysis of the thermodynamic parameters from the ITC titration experiments (Table 3) shows that all studied ATAD2 bromodomainligand interactions are enthalpy-driven. Enthalpy-dominated binding has been reported recently for small-molecule ligands of the BRPF1 bromodomain78, which belongs to the same subfamily IV as the ATAD2 bromodomain. When the disulfide bridge is 100% formed in the ATAD2 CO C1101A bromodomain, the binding affinity for the H4K5ac peptide is lower and there is a smaller change in enthalpy (−3569.3 ± 148.0 cal/mol) compared to the same ATAD2 bromodomains with 49% bridge formation (−5839.3 ± 767.5 cal/mol). The change in enthalpy observed upon C-38 binding with the ATAD2 CO C1101A bromodomain possessing a fully formed disulfide bridge is −11746.0 ± 6804.0 cal/mol, compared to −8016.0 ± 351.4 cal/mol with 49% bridge formation. When the disulfide bridge could not be formed in the ATAD2 bromodomain triple Cys mutant the enthalpy change was −4203.7 ± 795.7 cal/mol for the H4K5ac ligand and −6894.3 ± 1609.8 cal/mol for C-38. Overall, the largest change in enthalpy values were observed for the ATAD2 bromodomain in complex with the C-38 ligand.
The entropy changes observed with the disulfide bridge formation are contradictory. There is an increase in (ΔS) with more disulfide bridge formation for the ATAD2 CO C1101A bromodomains upon binding to the histone H4K5ac ligand. When the same ATAD2 bromodomain proteins bind to the C-38 ligand the entropy change induced by ligand binding becomes more negative with the percentage of disulfide bridge formed.
3.5 |. Atomistic simulations reveal similar flexibility of ATAD2 bromodomain with and without disulfide bridge
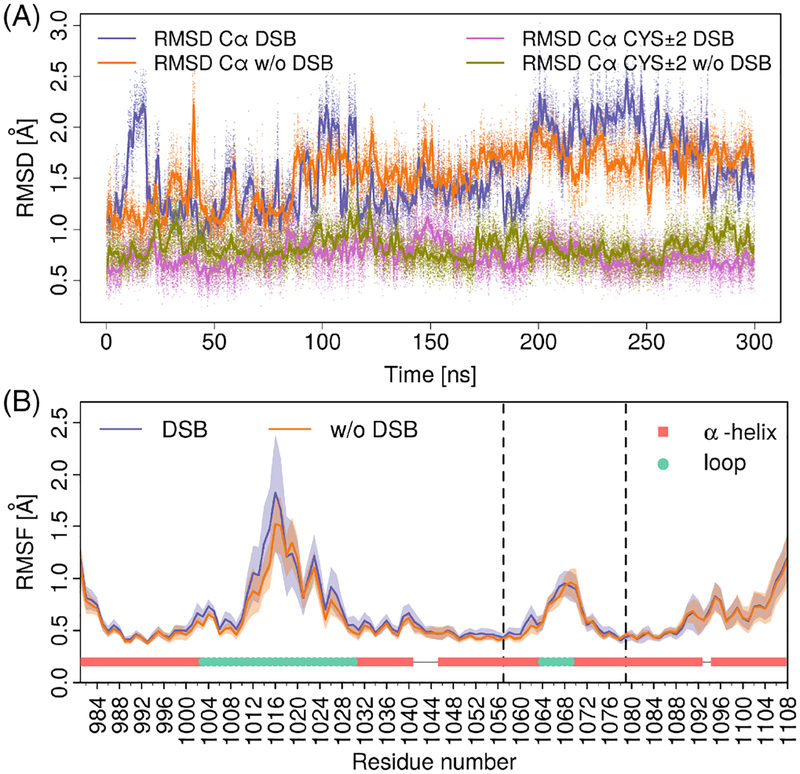
To examine how formation of the disulfide bridge might influence the flexibility of the bromodomain binding pocket and the ZA and BC loop regions we performed a molecular dynamics (MD) simulation on the ATAD2 bromodomain structure containing the disulfide bridge and compared it to a simulation of the structure with free cysteines C1057 and C1079. As starting coordinates of the runs we used the two conformations of C1079 that were fitted in the electron density and are present in our crystal structure (PDBID: 6CPS). We simulated both systems in the canonical ensemble, each for 300 ns (see Materials and Methods). The MD simulations show that the presence of the disulfide bridge does not influence the flexibility of the bromodomain. The time traces of the RMSD of the Cα atoms (Figure 2A, slate and orange solid line and dots) with respect to the crystal structure span a similar range, and the difference between the average values along the two trajectories is only 0.03 Å. For the Cα atoms of C1057 and C1079 and the two residues up- and downstream of them (10 atoms in total), the RMSD is almost always below 1 Å regardless of the presence of the disulfide bridge (Figure 2A, magenta and dark yellow line and dots). A possible explanation for the similar structural stability is the fact that the disulfide bond connects the middle of the αB and αC helices; hence it joins two segments of the protein that are already rigid. Moreover, in the run without the disulfide bridge, the average distance between the two sulfur atoms of C1057 and C1079 is 4.61 Å, which reflects an optimal van der Waals contact (Supporting Information figure S4). The root mean square fluctuation (RMSF) profile of the Cα atoms (Figure 2B), averaged over 2 ns intervals, confirms that the presence of the disulfide bridge does not alter the flexibility of the protein and particularly does not hinder the motion of the loops. This simulation result is not surprising as the two cysteines involved in the disulfide bridge are at a distance of more than 12 Å from the residues in the ZA and BC loops. In addition, the helical sections do not show any relevant motions at the backbone level with RMSF values around 0.5 Å for all but the C-terminal helix. The highest fluctuations are observed for the ZA loop in agreement with previous simulation studies.79
FIGURE 2.
Molecular dynamics analysis of ATAD2 bromodomain flexibility. A, Temporal evolution of the RMSD. We compared the RMSD of the system with (slate and magenta lines and dots) and without the disulfide bridge (orange and dark yellow lines and dots). Two different RMSD metrics were employed; the slate and orange colors represent the RMSD of all Cα except three terminal residues (122 atoms in total) after alignment on the same set. The magenta and dark yellow colors encode the RMSD of the Cα of C1057, C1079 and two residues up- and downstream of them (10 atoms in total) after alignment on the four α-helices forming the bromodomain bundle. For each case all data points (dots) and a moving average (with a window of 100 data points) are reported. B, Root mean square fluctuations of the Cα atoms as a function of residue for the trajectory interval between 100 and 300 ns, averaged over stretches of 2 ns. The RMSF values were calculated by first aligning the trajectory to the crystal structure by using all Cα atoms. Then, we divided the trajectory into intervals of 2 ns and calculated the average structure for each interval. Finally, for each interval and residue we calculated the standard deviation (SD) of the coordinates of the Cα atom. The solid lines are the average over all the 100 intervals and the shaded envelope is at one SD distance. The calculation was carried out separately for the run with (slate) and without (orange) disulfide bridge. The vertical dashed lines mark the position of C1057 and C1079. A structural annotation of helical and loop (ZA and BC) regions is added at the bottom of the plot
Differences in binding affinity could also originate from different rotameric states of the residues in the binding site. Thus, we investigated two residues in the ATAD2 bromodomain binding site that are involved in hydrophobic interactions with natural and synthetic ligands. The statistical weights of the rotameric states of the gate-keeper residue I1074 and “RVF” shelf residue V1008 are not affected by the presence of the disulfide bridge (Supporting Information figure S5). Overall, the very similar behavior observed in the molecular dynamics simulations with and without disulfide bridge is consistent with the relatively small difference in binding affinity as measured by ITC (less than 1 kcal/mol).
3.6 |. Disulfide bridges are found in a select number of human bromodomain-containing proteins
Our results indicate that formation of the disulfide bridge between residues C1057 and C1079 in the ATAD2 bromodomain negatively impacts its acetylated histone binding ability. We examined the structure based sequence alignment of the human bromodomain family published by Filippakopoulos et al.,12 to determine if any other bromodomain proteins have a cysteine pair similar to the ATAD2 bromodomain, with one Cys in each of the αB and αC helices. Based on previous phylogenetic analysis, the ATAD2 bromodomain is a member of the human bromodomain subfamily IV.12 Within this subfamily, the other subfamily IV bromodomain proteins contain an Ile, Val, or Cys residue at position 1057 in helix αB, while in the αC helix at position 1079, a Val or Lys residue is usually found. Only the ATAD2 and ATAD2B bromodomains possess a Cys residue at position 1079. Thus, ATAD2 and its paralog ATAD2B are the only two bromodomains within the subfamily IV bromodomain-proteins with the ability to form a disulfide bridge. Inspection of the sequences for all of the other bromodomain subfamilies reveals that many others do have a Cys residue in either the αB or αC helix, but Cys residues are not commonly found in both helices. Only the subfamily I bromodomain proteins were found to have Cys residues in both the αB and αC helices. For example, the GCN5 bromodomain has a Cys residue in helices αB and αC. In the X-ray crystal structure of the GCN5 bromodomain these two Cys residues are in close enough proximity to form a disulfide bridge, but a reduced state with no bridge formed is observed (PDBID: 3D7C). In the structures of the PCAF and BPTF bromodomains (PDBID: 3GG3 and 3UV2), the two cysteine residues are not positioned in the correct geometry to form a disulfide bridge, but with modest conformational changes to the side chains of these residues it may be possible for bridge formation to occur. The subfamily I bromodomain in the CECR2 protein, also contains two cysteine residues, but they are located too far apart from each other in the αB and αC helices to be able to form a disulfide bridge (PDBID: 3NXB). The only other two bromodomain proteins that contain a Cys residue in each of the αB and αC helices include the TRIM66 bromodomain in subfamily V, and the PRKCBP1 bromodomain in subfamily VII. The structure of the TRIM66 bromodomain has not been solved, but there is a structure available for the human PRKCBP1 bromodomain as a PHD-Bromo-PWWP cassette (PDBID: 4COS). However, in this structure the Cys residues in the αB and αC helices are on distal ends of the helices, and are not positioned close enough to form a disulfide bridge. Thus, formation of a disulfide bridge appears to be a unique structural feature of the ATAD2 and ATAD2B bromodomains, but it may also form in the GCN5, PCAF, and BPTF subfamily I bromodomains within the human bromodomain-containing proteins.
In summary, formation of a disulfide bridge near the ATAD2 binding pocket is a unique structural feature of this bromodomain that should be taken into consideration when examining ligand binding affinity, or in drug discovery efforts to develop new therapeutic compounds that interact with this module.
Supplementary Material
ACKNOWLEDGMENTS
We are especially thankful to Dr. Anton Komar at DapCel, Inc. for designing the codon-optimized ATAD2 bromodomain, and to Dr. Robert J. Hondal in the Biochemistry Department at the University of Vermont for helpful discussions on how to measure free sulfhydryl groups within a protein to study the disulfide bridge formation. AC and CL are also grateful to Dr. Ilian Jelezarov and Dr. Andreas Vitalis for interesting discussions. This study was supported by the National Institute of General Medical Sciences, National Institutes of Health under award number R15GM104865 to KCG, and a Swiss National Science Foundation grant to AC (grant number 31003A-169007). JTL was the recipient of an ACPHS graduate research assistantship from 2016–2017, and CME was the recipient of an ACPHS Student Summer Research Award in 2016 and an ACPHS graduate research assistantship from 2017–2018. Crystal growth, screening and data collection was carried out at the Center for X-ray Crystallography at the University of Vermont, which is supported by the National Cancer Institute of the National Institutes of Health under award numbers P01CA098993 and R01CA52040, to Sylvie Doublié. DNA sequencing was performed in the University of Vermont Cancer Center Advanced Genome Technologies Core and through Eurofins Genomics LLC. The structure of the ATAD2 bromodomain in complex with HEPES was deposited into the Protein Data Bank (PDBID: 6CPS).
Funding information
National Institute of General Medical Sciences, National Institutes of Health, Grant/Award Number: R15GM104865; Swiss National Science Foundation, Grant/Award Number: 31003A-169007; National Cancer Institute, National Institutes of Health, Grant/Award Numbers: R01CA52040, P01CA098993
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
FINANCIAL INTERESTS DISCLOSURE
None.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature. 1997;389(6648):251–260. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286(5439):481–486. [DOI] [PubMed] [Google Scholar]
- 5.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14(11):1008–1016. [DOI] [PubMed] [Google Scholar]
- 6.Heightman TD. Therapeutic prospects for epigenetic modulation. Expert Opin Ther Targets. 2011;15(6):729–740. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. [DOI] [PubMed] [Google Scholar]
- 8.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T Chromatin modifications and their function. Cell. 2007; 128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 10.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26(37): 5395–5407. [DOI] [PubMed] [Google Scholar]
- 11.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. [DOI] [PubMed] [Google Scholar]
- 12.Filippakopoulos P, Picaud S, Mangos M, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung CW, Coste H, White JH, et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem. 2011;54(11):3827–3838. [DOI] [PubMed] [Google Scholar]
- 14.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26(37):5521–5527. [DOI] [PubMed] [Google Scholar]
- 15.Jain AK, Barton MC. Bromodomain histone readers and cancer. J Mol Biol. 2017;429(13):2003–2010. [DOI] [PubMed] [Google Scholar]
- 16.Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci USA. 2007;104(46):18067–18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poncet-Montange G, Zhan Y, Bardenhagen JP, et al. Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2. Biochem J. 2015;466(2):337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morozumi Y, Boussouar F, Tan M, et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J Mol Cell Biol. 2016; 8(4):349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koo SJ, Fernandez-Montalvan AE, Badock V, et al. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget. 2016;7(43):70323–70335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsia EY, Kalashnikova EV, Revenko AS, Zou JX, Borowsky AD, Chen HW. Deregulated E2F and the AAA+ coregulator ANCCA drive proto-oncogene ACTR/AIB1 overexpression in breast cancer. Mol Cancer Res. 2010;8(2):183–193. [DOI] [PubMed] [Google Scholar]
- 21.Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol Cell Biol. 2010;30(22): 5260–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou JX, Guo L, Revenko AS, et al. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69(8):3339–3346. [DOI] [PubMed] [Google Scholar]
- 23.Ciro M, Prosperini E, Quarto M, et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69(21):8491–8498. [DOI] [PubMed] [Google Scholar]
- 24.Lin CY, Loven J, Rahl PB, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caron C, Lestrat C, Marsal S, et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010;29(37):5171–5181. [DOI] [PubMed] [Google Scholar]
- 26.Kalashnikova EV, Revenko AS, Gemo AT, et al. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010;70(22):9402–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez SV, Robertson FM, Pei J, et al. Inflammatory breast cancer (IBC): clues for targeted therapies. Breast Cancer Res Treat. 2013; 140(1):23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salhia B, Kiefer J, Ross JT, et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PLoS One. 2014;9(1): e85448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou M, Huang R, Song Y, Feng D, Jiang Y, Liu M. ATAD2 overexpression is associated with progression and prognosis in colorectal cancer. Jpn J Clin Oncol. 2016;46(3):222–227. [DOI] [PubMed] [Google Scholar]
- 30.Luo Y, Ye GY, Qin SL, Yu MH, Mu YF, Zhong M. ATAD2 overexpression identifies colorectal cancer patients with poor prognosis and drives proliferation of cancer cells. Gastroenterol Res Pract. 2015; 2015:936564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S, Bi M, Yan Z, Sun D, Ling L, Zhao C. Silencing of ATPase family AAA domain-containing protein 2 inhibits migration and invasion of colorectal cancer cells. Neoplasma. 2016;63(6):846–855. [DOI] [PubMed] [Google Scholar]
- 32.Krakstad C, Tangen IL, Hoivik EA, et al. ATAD2 overexpression links to enrichment of B-MYB-translational signatures and development of aggressive endometrial carcinoma. Oncotarget. 2015;6(29):28440–28452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raeder MB, Birkeland E, Trovik J, et al. Integrated genomic analysis of the 8q24 amplification in endometrial cancers identifies ATAD2 as essential to MYC-dependent cancers. PLoS One. 2013;8(2):e54873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang P, Meng F, Liu Y, Chen X. Overexpression of ANCCA/ATAD2 in endometrial carcinoma and its correlation with tumor progression and poor prognosis. Tumour Biol. 2015;36(6):4479–4485. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Zhang C, Du W, Yang X, Chen Z. ATAD2 is overexpressed in gastric cancer and serves as an independent poor prognostic bio-marker. Clin Translat Oncol. 2016;18(8):776–781. [DOI] [PubMed] [Google Scholar]
- 36.Murakami H, Ito S, Tanaka H, Kondo E, Kodera Y, Nakanishi H. Establishment of new intraperitoneal paclitaxel-resistant gastric cancer cell lines and comprehensive gene expression analysis. Anticancer Res. 2013;33(10):4299–4307. [PubMed] [Google Scholar]
- 37.Wu G, Liu H, He H, et al. miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer. 2014;14:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang HW, Ha SY, Bang H, Park CK. ATAD2 as a poor prognostic marker for hepatocellular carcinoma after curative resection. Cancer Res Treat. 2015;47(4):853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Q, Lin B, Liu H, et al. RNA-Seq analyses generate comprehensive transcriptomic landscape and reveal complex transcript patterns in hepatocellular carcinoma. PLoS One. 2011;6(10):e26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Huang J, Luo L, Chen Z, Guo Y, Guo L. Significance of PRO2000/ANCCA expression, a novel proliferation-associated protein in hepatocellular carcinoma. Cancer Cell Int. 2014;14:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouret R, Laffaire J, Hofman P, et al. A comparative and integrative approach identifies ATPase family, AAA domain containing 2 as a likely driver of cell proliferation in lung adenocarcinoma. Clin Cancer Res. 2012;18(20):5606–5616. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Sun Y, Li Y, et al. ANCCA protein expression is a novel independent poor prognostic marker in surgically resected lung adenocarcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S577–S582. [DOI] [PubMed] [Google Scholar]
- 43.Wan WN, Zhang YX, Wang XM, et al. ATAD2 is highly expressed in ovarian carcinomas and indicates poor prognosis. Asian Pac J Cancer Prev. 2014;15(6):2777–2783. [DOI] [PubMed] [Google Scholar]
- 44.Guan X, Zong ZH, Chen S, et al. The role of miR-372 in ovarian carcinoma cell proliferation. Gene. 2017;624:14–20. [DOI] [PubMed] [Google Scholar]
- 45.Duan Z, Zou JX, Yang P, et al. Developmental and androgenic regulation of chromatin regulators EZH2 and ANCCA/ATAD2 in the prostate via MLL histone methylase complex. Prostate. 2013;73(5): 455–466. [DOI] [PubMed] [Google Scholar]
- 46.Altintas DM, Shukla MS, Goutte-Gattat D, et al. Direct cooperation between androgen receptor and E2F1 reveals a common regulation mechanism for androgen-responsive genes in prostate cells. Mol Endocrinol. 2012;26(9):1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bamborough P, Chung CW, Demont EH, et al. A chemical probe for the ATAD2 Bromodomain. Angewandte Chemie. 2016;55(38):11382–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romero FA, Taylor AM, Crawford TD, Tsui V, Cote A, Magnuson S. Disrupting acetyl-lysine recognition: Progress in the development of Bromodomain inhibitors. J Med Chem. 2016;59(4):1271–1298. [DOI] [PubMed] [Google Scholar]
- 49.Bamborough P, Chung CW, Furze RC, et al. Structure-based optimization of Naphthyridones into potent ATAD2 Bromodomain inhibitors. J Med Chem. 2015;58(15):6151–6178. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Montalvan AE, Berger M, Kuropka B, et al. Isoform-selective ATAD2 chemical probe with novel chemical structure and unusual mode of action. ACS Chem Biol. 2017;12:2730–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eyer P, Worek F, Kiderlen D, et al. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal Biochem. 2003; 312(2):224–227. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4): 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server):W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urzhumtseva L, Afonine PV, Adams PD, Urzhumtsev A. Crystallo-graphic model quality at a glance. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 3):297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poplawski A, Hu K, Lee W, et al. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. J Mol Biol. 2014;426(8):1661–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitalis A, Pappu RV. A simple molecular mechanics integrator in mixed rigid body and dihedral angle space. J Chem Phys. 2014;141(3): 034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abraham MJ, Murtola T, Schulz R, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 60.Best RB, Zhu X, Shim J, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput. 2012;8(9):3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durell SR, Brooks BR, Ben-Naim A. Solvent-induced forces between two hydrophilic groups. J Phys Chem. 1994;98(8):2198–2202. [Google Scholar]
- 62.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. [DOI] [PubMed] [Google Scholar]
- 63.Tironi IG, Sperb R, Smith PE, Gunsteren WF. A generalized reaction field method for molecular dynamics simulations. J Chem Phys. 1995; 102(13):5451–5459. [Google Scholar]
- 64.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12): 1463–1472. [Google Scholar]
- 65.Harner MJ, Chauder BA, Phan J, Fesik SW. Fragment-based screening of the bromodomain of ATAD2. J Med Chem. 2014;57(22):9687–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Demont EH, Chung CW, Furze RC, et al. Fragment-based discovery of low-micromolar ATAD2 Bromodomain inhibitors. J Med Chem. 2015; 58(14):5649–5673. [DOI] [PubMed] [Google Scholar]
- 67.Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides on protein stability. Curr Protein Pept Sci. 2009;10(6):614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Angov E, Hillier CJ, Kincaid RL, Lyon JA. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS One. 2008;3(5):e2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komar AA, Guillemet E, Reiss C, Cullin C. Enhanced expression of the yeast Ure2 protein in Escherichia coli: the effect of synonymous codon substitutions at a selected place in the gene. Biol Chem. 1998;379(10): 1295–1300. [PubMed] [Google Scholar]
- 70.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462(3):387–391. [DOI] [PubMed] [Google Scholar]
- 71.Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, Nussinov R. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol. 2008;383(2): 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim SJ, Yoon JS, Shishido H, et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348(6233): 444–448. [DOI] [PubMed] [Google Scholar]
- 73.Hu S, Wang M, Cai G, He M. Genetic code-guided protein synthesis and folding in Escherichia coli. J Biol Chem. 2013;288(43):30855–30861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flynn EM, Huang OW, Poy F, et al. A subset of human Bromodomains recognizes Butyryllysine and Crotonyllysine histone peptide modifications. Structure. 2015;23(10):1801–1814. [DOI] [PubMed] [Google Scholar]
- 75.Betz SF. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2(10):1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butturini E, Gotte G, Dell’Orco D, et al. Intermolecular disulfide bond influences unphosphorylated STAT3 dimerization and function. Biochem J. 2016;473(19):3205–3219. [DOI] [PubMed] [Google Scholar]
- 77.Tencer AH, Gatchalian J, Klein BJ, et al. A unique pH-dependent recognition of methylated histone H3K4 by PPS and DIDO. Structure. 2017;25(10):1530–1539e1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu J, Zhou C, Caflisch A. Structure-based discovery of selective BRPF1 bromodomain inhibitors. Eur J Med Chem. 2018;155:337–352. [DOI] [PubMed] [Google Scholar]
- 79.Langini C, Caflisch A, Vitalis A. The ATAD2 bromodomain binds different acetylation marks on the histone H4 in similar fuzzy complexes. J Biol Chem. 2017;292(40):16734–16745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.